Abstract
SERPINB9, an endogenous inhibitor of granzyme B (GzmB), has emerged as a critical factor in the resistance to immunotherapy by protecting cancer cells from GzmB-induced cytotoxicity. However, its role in chemosensitivity remains unknown. In this study, we show that gemcitabine (GEM) treatment upregulates SERPINB9 through transcription factor ATF-3. Interestingly, GEM also induces the expression of GzmB and knockout or knockdown of SERPINB9 results in enhanced response of tumor cells to GEM, suggesting a role of GzmB/SERPINB9 axis in regulating chemosensitivity. To facilitate the therapeutic translation of these findings, we engineer POEM nanocarrier (consisting of lipid-derivatized polylysine (PEG-PLL-Oleic acid, PPO), and GEM-conjugated polylysine (PEG-PLL-OA-GEM, PPOGEM), PPO/PPOGEM (POEM)) that is highly effective in codelivery of built-in GEM and loaded SERPINB9 short interfering RNA (siSPB9). GEM conjugation introduces an additional mechanism of carrier/siRNA interaction in addition to charge-mediated interaction and enables efficient i.v. delivery at lower N/P ratios. Here, we show that co-delivery of GEM and siSPB9 significantly improves antitumor efficacy and remodels the tumor immune microenvironment in pancreatic cancer models, supporting a promising therapeutic strategy.
Similar content being viewed by others
Introduction
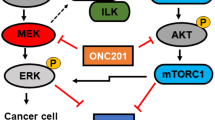
Immunotherapy, such as immune checkpoint blockade (ICB), represents a promising approach for various type of cancers1. However, only a small number of cancer patients benefit from ICB2,3. Strategies aimed at enhancing ICB effectiveness include facilitating the infiltration of cytotoxic T lymphocytes (CTLs) and boosting CTL activation at the different stages of immune response4. Despite these advances, relatively few therapeutic strategies have been explored that target the mechanisms of resistance after CTLs and NK cells are recruited and activated. Granzyme B (GzmB), a potent cytotoxic molecule secreted by activated CTLs or NK cells, triggers tumor cell apoptosis by cleaving and activating caspases-3 and -85. The gene expression as well as the activity of GzmB is regulated by various mechanisms at transcriptional and post-transcriptional levels6. Overexpression of SERPINB9 (SPB9), a potent endogenous inhibitor of GzmB, represents another important mechanism of cancer resistance to immunotherapy7.
SPB9, first defined in 19958, is a member of the largest and most widely distributed superfamily of proteinase inhibitors known as serpins, which specifically inhibit serine proteases9,10. SPB9 offers cytoprotective benefits against self-inflicted damage caused by GzmB in various immune cells, including cytotoxic T cells11, dendritic cells12,13, and neutrophils14. This protection allows immune cells to deploy GzmB to eliminate target cells without falling prey to their own cytolytic mechanisms. However, it has been established that SPB9 also protects tumor cells from GzmB mediated destruction, regardless of whether GzmB originates from cytotoxic lymphocytes or is self-produced by tumor cells15,16. In addition, SPB9 has been observed to shield immunosuppressive cells and tumor-associated fibroblasts (TAF) from GzmB induced destruction16. Several studies have reported that SPB9 expression is markedly elevated in several types of cancers, suggesting that its upregulation could be a crucial mechanism of resistance to immunotherapies, including ICB7,17,18,19. Therefore, selective inhibition of SPB9 activity in tumor cells and immunosuppressive immune cells while preserving its function in CTLs shall represent an attractive and effective strategy to boost ICB as well as other immunotherapies. So far, the specific role of SPB9 in modulating the response of cancer cells to chemotherapeutic agents remains poorly understood.
Gemcitabine (GEM) is one of the front-line chemotherapeutic agents for pancreatic cancer (PCa). Recent studies show that GEM is capable of boosting antitumor immune response through various mechanisms20. However, several immune suppressive mechanisms are also induced following GEM treatments, which limit the overall therapeutic outcome21.
In this work, we show that GEM also induces SPB9 expression in tumor cells or tissues in vitro and in vivo, suggesting another feedback mechanism that negatively regulates the antitumor immune response. We study the mechanism by which GEM induces the SPB9 gene expression at transcriptional level. We further evaluate the role of GzmB/SPB9 axis in regulating the chemosensitivity of PCa cells to GEM. Finally, POEM (PPO/PPOGEM) nanocarrier is developed and evaluated for codelivery of GEM and siSPB9 to overcome both chemotherapy and immune resistance.
Results
Gemcitabine-mediated induction of SERPINB9 in murine and human PDAC cell lines
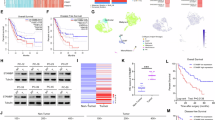
GEM is a front-line treatment for PCa but shows limited efficacy. To further understand the mechanisms of resistance in PCa, RNA sequencing was conducted to analyze changes in the gene expression profiles of murine wild type (WT, KPC-C2 (clone KPC 6694c2)22 and GEM resistant (KPC C2GEMR) PDAC cell lines (Fig. 1a). We established cutoff values for both log2(fold change) and log10(p value) at 2. Twenty-two (22) genes were shown to be upregulated in GEMR cells, including 3 genes (Atf3, Gadd45a, and Asns) that were previously shown to be involved in gemcitabine resistance23. The expression of Serpinb9b gene was most significantly induced among the 22 upregulated genes (Fig. 1b and Supplementary Fig. 1a–c). Upregulation of Serpinb9b was further confirmed by qRT-PCR in 3 murine GEMR cell lines (KPC-C2, KPC-C5 (clone KPC 6914c5)22, and Panc02) (Fig. 1b and Supplementary Fig. 1a–c). Serpinb9b is a murine orthologue of human SERPINB9 (SPB9), although humans do not have the SERPINB9 subtype SERPINB9b24. Several bioinformatic and experimental studies suggest that SPB9 might be implicated in immunotherapy resistance in both animal models and in cancer patients16,17,25, but its role in chemoresistance has not yet been explored. Therefore, we selected SPB9 as a primary target to elucidate its potential involvement in chemoresistance. To bridge our findings with clinical applications, we assessed the expression of Serpinb9 in these murine GEMR cell lines via qRT-PCR. We also observed the upregulation of this orthologue in these murine GEMR cell lines (Fig. 1c).
a Volcano plot for the RNA-seq analysis of KPC-C2 gemcitabine resistant (GEMR) cells versus KPC-C2 wide type (WT) cells. Statistical analysis of RNA-seq was performed by two-tailed Student’s t-test for comparison using Cuffdiff in the Cufflinks package. b qRT-PCR analysis of the fold changes of 8 genes selected from RNA-seq (a) in KPC-C2 GEMR, KPC-C5 GEMR, and Panc02 GEMR versus their WT cells. c qRT-PCR analysis of Serpinb9 expression in KPC-C2 GEMR, KPC-C5 GEMR, and Panc02 GEMR versus their WT cells. n = 6 independent experiments. d SERPINB9 expression in pancreatic adenocarcinoma (PAAD) patients’ tumor tissues (n = 179) and normal tissues (n = 171) from TCGA database. Center lines represent median expressions, the box limit indicates the lower quantile and upper quantile, and whiskers represent the minimal and maximal expressions. e Survival rate of PAAD patients with high levels of SERPINB9 versus those with low levels of SERPINB9 from TCGA database. Serpinb9 mRNA expression in KPC-C2 (f), KPC-C5 (g), or Panc02 (h) cells following treatment with various concentrations of gemcitabine. n = 3 independent samples. i Serpinb9 protein expression in KPC-C2, KPC-C5 or Panc02 cells following treatment with various concentrations of gemcitabine. j SERPINB9 mRNA expression in PANC-1, MIA PaCa-2, Panc 10.05, and Panc 02.03 cells following treatment with different concentrations of gemcitabine. n = 3 independent samples. Representative Western blot of SERPINB9 levels (k) in PANC-1, MIA PaCa-2, Panc 10.05 and Panc 02.03 cells after treatment with different concentrations of gemcitabine, and densitometry analysis of the protein bands in the Western blot (l). n = 3 independent samples and data were quantified by densitometry. Data are presented as mean ± s.e.m. in (c, f, g, h, j). Statistical analysis was performed by two-tailed Student’s t-test for comparison in (c), log rank test for comparison in (e) and one-way ANOVA with Tukey’s post hoc test for comparison in (f, g, h, j, l). Source data are provided as a Source Data file.
To further explore the clinical implications of SPB9, we analyzed its expression across various cancer types in the TCGA database (Fig. 1d). SPB9 overexpression was not only seen in pancreatic ductal adenocarcinoma (PDAC) (Fig. 1d) but also found in diffuse large B-cell lymphoma (DLBC), kidney renal clear cell carcinoma (KIRC), skin cutaneous melanoma (SKCM), and stomach adenocarcinoma (STAD) (Supplementary Fig. 1d). Importantly, elevated levels of SPB9 expression were negatively correlated with the clinical prognosis of cancer patients across all cancer types (PANCAN) (Supplementary Fig. 1e) including PDAC patients (Fig. 1e).
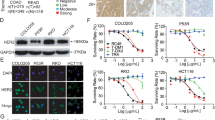
Figure 1f–h shows that GEM treatment induced the expression of SPB9 mRNA in a dose-dependent manner in KPC-C2, KPC-C5, and Panc02 cell lines. Upregulation of SPB9 mRNA by GEM was also observed in human PCa cell lines (PANC-1, MIA PaCa-2, Panc 10.05 and Panc 02.03) in both time- and dose-dependent manners (Fig. 1j). Induction of SPB9 by GEM was further confirmed at the protein level in several murine (C2, C5 and Panc02) (Fig. 1i and Supplementary Fig. 1f) and human pancreatic cancer cell lines (PANC-1, MIA PaCa-2, Panc 10.05 and Panc 02.03) (Fig. 1k–l). In addition to GEM, doxorubicin (DOX) and cisplatin (CDDP) induced SPB9 mRNA expression in several murine PCa cell lines tested (C2, C5, and Panc02) (Supplementary Fig. 1g–i).
Role of ATF3 in GEM-mediated induction of SERPINB9 at transcriptional level
The above data suggests that chemotherapy drugs induce SPB9 gene expression likely at transcriptional level. We then reanalyzed our earlier RNA-seq data to identify candidate transcription factors whose expression levels were also significantly induced. Similar to the cutoff value established earlier, we set cutoff values for both log2(fold change) and log10(p-value) at 2. ATF3, a transcription factor critical to stress responses, was identified as the most significantly upregulated gene (Supplementary Fig. 2a). The gene set ATF3_Q6 includes those genes with at least one ATF3 binding motif, CBCTGACGTCANCS, within a 4 kb region centered on their transcription starting sites (https://www.gsea-msigdb.org/gsea/msigdb/cards/ATF3_Q6). This gene set was also significantly upregulated (Supplementary Fig. 2b). These findings were validated by qRT-PCR in GEMR PCa cell lines (Supplementary Fig. 2c). A transient treatment with GEM for 24 or 48 h induced the expression of ATF3 at both mRNA and protein levels (Supplementary Fig. 2d–f). The positive correlation of the expression levels of SPB9 and ATF3 was further shown in tumor samples of PAAD patients from TCGA database (Supplementary Fig. 3a, b) and a referenced study26. A similar correlation was also seen in tumors from a cohort of Pan-Cancer patients from the TARGET program and TCGA database (Supplementary Fig. 3c, d). Supplementary Fig. 3e shows that siATF3 could achieve over 60% knockdown efficiency. Knockdown of ATF3 via siRNA significantly attenuated the GEM-mediated induction of SPB9 in both murine (KPC-C5) and human (MIA PaCa-2) PDAC cells at both mRNA and protein levels (Supplementary Fig. 3f–h). Interestingly, ATF3 KD had no impact on GEM-induced expression of murine orthologue Serpinb9b (Supplementary Fig. 3f).
The above data suggest that ATF3 plays a role in GEM-mediated induction of SPB9 gene expression. Three (3) putative ATF3-binding sites (-303-311 (ATF3-1), -390-397 (ATF3-2), and -1503-1510 (ATF3-3)) upstream of human SPB9 gene were identified based on an ATF3 chromatin immunoprecipitation sequencing (ChIP-Seq) and the UCSC Genome Browser27,28,29. A DNA fragment spanning all 3 putative binding sites was generated by PCR and cloned into the pGL3 vector (Supplementary Fig. 3i). In addition, 3 other reporters were constructed by cloning a 5-times repeat of each of the 3 putative ATF3 binding sequences, respectively. The transcriptional activity of each cloned sequence was examined by luciferase assay using MIA PaCa-2, PANC-1, and Panc 02.03 cell lines. As shown in Supplementary Fig. 3j, GEM induced the expression of luciferase reporter in a dose-dependent manner in cells transfected with the plasmid containing all 3 putative binding sequences, as well as the one with 5x repeat of ATF3-3 sequence. In contrast, minimal transcriptional activity was shown for ATF3-1 and ATF3-2 sequences (Supplementary Fig. 3j). In addition, deletion of ATF3-3 from the full-length sequence completely abolished its transcriptional activity (Supplementary Fig. 3k). Taking together our data suggests that ATF3 is a key transcriptional factor that drives the GEM-induced SPB9 gene expression through binding to ATF3-3 in SPB9 gene promoter (Supplementary Fig. 3l).
Role of GzmB in GEM-mediated tumor killing
The GzmB-SPB9 axis is critically involved in the escape of cancer cells from immune surveillance as well as poor response to immunotherapy30. However, its role in chemotherapy resistance remains unexplored. We first generated KPC-C2 SPB9 KO cell line using the CRISPR/Cas9 system. Western blot analysis confirmed the disappearance of SPB9 protein, both the monomeric form and the SPB9-GzmB complex (Fig. 2a and Supplementary Fig. 4a). Notably, the levels of monomeric GzmB were increased in SPB9 KO cells (Fig. 2a and Supplementary Fig. 4a), demonstrating that GzmB is dissociated from its complex due to the depletion of SPB9. SPB9 KO cells showed a cell viability that was similar to that of WT cells as evaluated by MTT assay (Fig. 2a). However, SPB9 KO cells showed significant retardation in tumor formation in mice with tumor volumes and weights about 1/6 of those of WT tumors (Fig. 2b). The proportion of Annexin V+ tumor cells was also significantly increased in the KO group (Fig. 2c).
a Expression of SERPINB9-GranzymeB complex, SERPINB9 and granzymeB in KPC-C2 SERPINB9 KO (KO) cells and KPC-C2 control vector (CV) cells. Cell proliferation of KPC-C2 KO cells versus KPC-C2 CV cells. n = 3 independent experiments. b In vivo tumor growth curves and tumor weights of KPC-C2 KO cells compared to KPC-C2 CV cells. n = 5 mice. c, d Abundance of Annexin V+ tumor cells (c) and immune cells (d, CD45+, CD8+, CD8+IFNγ, CD4+IFNγ, dendritic cells, myeloid-derived suppressor cells (MDSCs), M2 macrophages) in KPC-C2 KO tumors and KPC-C2 CV tumors. n = 4 independent samples. e Cell viability of KPC-C2 KO or KPC-C2 gemcitabine resistant (GEMR) KO cells versus their CV cells following treatments with different concentrations of gemcitabine. n = 6 independent experiments. The impact of SPB9 knockdown on the cytotoxicity of GEM in KPC-C5 WT or KPC-C5 GEMR (f) or human pancreatic cancer cells (g, Panc 02.03, MIA PaCa-2). n = 6 independent experiments. h Tumor growth curves of KPC-C2 KO and KPC-C2 CV tumors following treatment of PBS or PPOGEM polymer. n = 5 mice. i Protein expression of granzymeB (GzmB) and SPB-GzmB complex following treatment with different concentrations of gemcitabine in KPC-C2 cells. j Abundance of GzmB+ KPC-C5 cells, KPC-C2 SPB9 KO cells or PANC-1 cells following treatment with different concentrations of gemcitabine. n = 3 independent samples. k The impact of granzymeB knockdown on the cytotoxicity of GEM in KPC-C2 SPB9 KO cells. n = 6 independent experiments. l The proportion of Annexin V+ KPC-C2 SPB9 KO cells following treatment with PBS, GEM, or GEM+granzymeB inhibitor (GzmBi). n = 3 independent samples. Data are presented as mean ± s.e.m. in (a–g, h, j, k, l). Statistical analysis was performed by two-tailed Student’s t-test for comparison in (b, c, d), one-way ANOVA with Tukey’s post hoc test for comparison in (j, l), and two-way ANOVA with Tukey’s post hoc test for comparison in (b, h). Source data are provided as a Source Data file.
KO of SPB9 significantly improved the tumor immune microenvironment as evident from increases in the infiltration of CD45+ immune cells (Fig. 2d). The numbers of both total CD8+ T cells and IFNγ+ T cells (both CD4+ and CD8+) were also increased in the SPB9-deficient tumors (Fig. 2d). In addition, there was a notable increase in the recruitment of dendritic cells in SPB9-deficient tumors (Fig. 2d). These changes were accompanied by a substantial reduction in myeloid-derived suppressor cells (MDSCs) (Fig. 2d). SPB9 KO further facilitated a reversal from the predominance of M2-like macrophages, suggesting a reprogramming of the tumor-associated macrophage phenotype (Fig. 2d).
Interestingly, SPB9 KO drastically increased the responsiveness of both WT and GEMR KPC-C2 cells to GEM (Fig. 2e). Similarly, SPB9 KD with siRNA (siSPB9) also enhanced the sensitivity of both WT and GEMR murine PCa cell lines (KPC-C2, KPC-C5 and Panc02) to GEM treatment (Fig. 2f and Supplementary Fig. 4b). Similar results were seen in human PCa cell lines (Panc 02.03 and MIA PaCa-2) (Fig. 2g). The proportion of Annexin V+ cells was significantly increased when GEM was combined with siSPB9 or SPB9 inhibitor 3034, suggesting a strong synergistic effect between GEM treatment and SPB9 KD or inhibition (Supplementary Fig. 4c, d). In line with in vitro results, GEM-based nanoparticles (NPs) effectively halted the tumor progression in all five SPB9 KO tumors (Fig. 2h).
To gain mechanistic insights, we examined the expression of GzmB following GEM treatment at both transcriptional and translational levels through PCR, Western blot, and flow cytometry. Both the expression levels of GzmB (Fig. 2i and Supplementary Fig. 5a–g) and the abundance of GzmB+ cells (Fig. 2j and Supplementary Fig. 5h) were increased by GEM treatment across several cell lines (KPC-C2 WT, KPC-C2 SPB9 KO, KPC-C5, Panc02, PANC-1, Panc 02.03, Panc 10.05, and MIA PaCa-2). Specifically, GEM induced the expression of GzmB in SPB9 KO cells at concentrations as low as 2 ng/mL (Fig. 2j). In addition, the activity of GzmB was also induced in murine KPC C2 cells and human PANC-1 cells (Supplementary Fig. 5i). Intriguingly, KD of GzmB in SPB9 KO cells using siRNA significantly attenuated the effects of GEM treatment (Fig. 2k). A similar effect was observed with the pharmacological inhibition of GzmB: treatment with the GzmB inhibitor 368050 led to decreased apoptosis (Fig. 2l). Interestingly, development of GEM resistance was associated with marked increases in SPB9 expression levels along with decreases in GzmB expression (Supplementary Fig. 6). The broad implications of these findings in cancer chemotherapy require more studies in the future. Nonetheless, our data suggests that SPB9 KD or inhibition in tumor cells may benefit increased sensitivity to GEM-based chemotherapy in addition to its benefits in improving antitumor immune response.
Development and characterization of POEM nanocarrier for codelivery of siSPB9 and GEM
Our data so far, along with data from other groups, suggests that SPB9 represents a promising therapeutic target for cancer, particularly in combination with other treatments such as chemotherapy. As discussed later, we chose to develop a therapy based on combination of GEM with siSPB9 instead of a small molecule inhibitor of SPB9. Various strategies for siRNA delivery, ranging from synthetic NPs to extracellular vesicles31,32,33, have been studied. Current lipid nanoparticle (LNP) systems have shown substantial efficacy in siRNA delivery to hepatocytes through i.v. administration34. However, these systems have shown limited success in targeting to extrahepatic organs or tissues, including solid tumors35. Polymer-based delivery systems have been explored as an alternative to LNPs due to their potential to enhance tumor targeting through various surface modifications and functionalization strategies35,36. Nevertheless, these polymeric systems often fall short in achieving the desired amounts of accumulation in tumors and/or knockdown efficiency37. We have recently reported a PMBOP-CP-based polymeric nanocarrier that is highly effective in selective codelivery of siRNA and lipophilic/hydrophobic drugs to tumors38. Here, we developed a simple and small-sized (~40 nm) platform that is highly effective in selective codelivery of siRNA and various hydrophilic nucleoside-based drugs as demonstrated by the “POEM” nanocarrier in codelivery of siSBP9 and GEM. This nanocarrier consists of lipid-derivatized polylysine (PEG-PLL-Oleic acid, PPO, positively charged) and lipid-derivatized, GEM-conjugated polylysine (PEG-PLL-OA-GEM, PPOGEM, nearly neutral charge). These components form compact nanoassembly PPO/PPOGEM (POEM) that efficiently loads siRNA into NPs (Fig. 3a). Design of this nanocarrier is based on the following rationales: (1) conjugation of GEM to a polymer enhances its delivery and protects it from degradation by deaminase39; (2) lipid-derivatization of both polymers aids in forming compact micelles and facilitate endosomal release40; (3) the electrostatic interactions between PPO and siRNA initiate the formation of POEM/siRNA complexes; (4) POEM/siRNA complexes may be further stabilized by π-π interactions and hydrogen bonding between the built-in GEM and siRNA41 (Fig. 3a). These multiple modes of interaction reduce the nitrogen/phosphate (N/P) ratio needed to form stable siRNA NPs, which shall decrease toxicity. In addition, this helps to reduce the surface positive charges, which shall decrease the non-selective uptake by lung and liver42; (5) The synthesis of PPO and PPOGEM is simple and can be readily scaled up. In addition, POEM/siRNA NPs can be prepared via a one-step of mixing followed by removal of solvents via dialysis. Systematic in vitro characterizations were conducted to optimize the ratio of the 3 components (PPO, PPOGEM, and siRNA) with a goal to identify top candidates that are small in sizes (<100 nm) and close to neutral in surface charges. The in vivo tumor targeting efficiency of the top candidates was then evaluated with near infrared fluorescence imaging using both subcutaneous and orthotopic tumor models.
a A schematic illustration of PPO/PPOGEM/siRNA (POEM/siRNA) nanocarrier delivery system. b Tissue distribution (H heart, S spleen, Lu lung, Li liver, K kidney, T tumor, T/Li tumor/liver, T/K tumor/kidney, T/Ti tumor/tissues) of POEM/siRNA with different PPO polymers. c Gel-retardation assay of PPO/siRNA complexes at various N/P ratios. d Gel-retardation, size, and zeta potentials (ZP) of POEM/siRNA complexes at various PPO/siRNA N/P ratios and PPOGEM/siRNA mass ratios. n = 3 independent samples. e Size and ZP of PPOGEM/PPO/siRNA (POEM/siRNA) at a ratio of 50/5/1, and Psugar/PPO/siRNA at a ratio of 40/5/1. n = 3 independent samples. f Upper left: Conformational change of POEM/siRNA system before and after MD simulation. Upper right: Radius of Gyration (RoG) of PPOGEM and Psugar during the final 150 ns MD simulation. Bottom: The interaction between siRNA and POEM. siRNA interacts with PPOGEM through both T-shaped π-π interactions (right dash line) and hydrogen bonding (left dash line). g Critical micelle concentration (CMC) of POEM/siRNA and Psugar/PPO/siRNA nanoparticles. h Spherical morphology of POEM/siRNA NPs by cryo-EM. Scale bar, 50 nm. i Cumulative release of gemcitabine at 72 h or 12 h from POEM/siRNA NPs under different conditions. n = 3 independent samples. j Gel-retardation assay of free siRNA or POEM/siRNA after incubation with RNAse. Data are presented as mean ± s.e.m. in (d, e, i). Statistical analysis was performed by one-way ANOVA with Tukey’s post hoc test for comparison in (i). Source data are provided as a Source Data file.
We first synthesized a small library of PPO cationic polymers that vary in the ratios of PEG/OA (y/z) and the amounts of cationic amino acids (histidine, lysine, and arginine) introduced (Fig. 3b). Polymers with unfavorable biophysical properties (low solubility and/or size over 400 nm) were triaged for further evaluations. Candidate PPO polymers selected from the initial screening were then mixed with PPOGEM and siRNA at an “empirical” ratio of 100/20/1 (w/w for PPOGEM/siRNA (100/1) and N/P for PPO/siRNA (20/1)) to form POEM/siRNA. Ex vivo near-infrared fluorescence imaging (NIRFI) was used to evaluate the Cy5.5-siRNA NPs with respect to their tumor-targeting efficiency. As depicted in Fig. 3b, a PPO with a PEG/OA ratio of 5:2 and without further amino acid modification formed the most effective siRNA NPs with PPOGEM in terms of the amounts of Cy5.5 signals in the tumor tissues, as well as the ratios of the signals in tumors over those in other normal tissues/organs. This PPO was selected for further optimization of POEM/siRNA via extensive characterizations of the biophysical properties and in vivo distribution of 18 NPs that vary in PPOGEM/PPO/siRNA ratio.
Gel retardation assay revealed that siRNA could be effectively loaded into PPO polymer at a N/P ratio as low as 5/1 (Fig. 3c). However, PPO alone cannot form stable complexes with siRNA. Interestingly, incorporation of PPOGEM polymer into the PPO/siRNA system resulted in NPs with a single-peaked size distribution (Fig. 3d), indicating that PPOGEM helps stabilize the system. Judging from gel retardation assay, siRNA was well loaded into POEM NPs at an N/P ratio of 5/1, 10/1 and 20/1 respectively. Increasing the amounts of PPOGEM led to a gradual decrease of zeta potential likely due to the shielding effect from PPOGEM. Notably, at an N/P ratio of 5/1 and a PPOGEM/siRNA mass ratio of 50/1, the POEM/siRNA system demonstrated the most desirable biophysical properties with a size of ~45.6 nm and a zeta potential of around 9.3 mV (NPs with zeta potential in a range of −10 to +10 are considered close to neutral).
As an initial step to understand the interaction of siRNA with POEM carrier, we synthesized a control polymer with only sugar ring conjugated to the PLL backbone (Psugar) to elucidate a role of GEM aromatic ring in stabilizing the siRNA NPs (Supplementary Fig. 10a). Figure 3e shows that replacement of PPOGEM with Psugar led to formation of much larger-sized NPs (~160 nm compared to ~40 nm for PPOGEM) with higher zeta potential (~18 mV compared to ~9 mV for PPOGEM). Moreover, increasing the amounts of Psugar led to further increases in the sizes of the resulting NPs that eventually form precipitates due to poor stability (Supplementary Fig. 11). In addition, the zeta potential of the NPs could not be reduced to below 10 mV at all PPO/Psugar ratios examined. These data highlight the significant role of GEM in formulating the POEM NP system.
To gain further insight into the interactions between POEM and siRNA and the behavior of siRNA within the POEM carrier, we conducted all-atom molecular dynamics (MD) simulations. Considering that the system is very dynamic, we used the radius of gyration (RoG) to monitor the conformational variation of POEM/siRNA. The RoG measures how far the components of an object are spread out from its center of mass. A smaller RoG value implies that the mass of the object is more tightly clustered around the center, indicating a denser and more compact structure. The RoG of POEM system was compared to that of the Psugar system, as depicted in Fig. 3f. During the simulation, after the systems reached a plateau, it is obvious that the POEM system (blue curve, ~54.87 Å) exhibits lower RoG values than the control polymer (red curve, ~55.87 Å). This can be further explained by the representative conformation of POEM/siRNA, displayed at the bottom right of the panel, where siRNA effectively interacts with PPOGEM through both T-shaped π-π and hydrogen bonding interactions. It is emphasized that during the simulation, the whole NP system was constructed based on a molar ratio of PPOGEM (or Psugar), PPO, and siRNA of 33: 9: 1, thus, the simulation accurately represents the composition of the NPs. However, in the real-world scenario, NPs are composed of numerous copies of the simulation system, which can be considered as a unit. While the RoG difference between the POEM and Psugar systems is only around 1 Å in the simulation, this small difference at the unit level can lead to a significant variation in the overall size of the assembled NPs. Consequently, the POEM system (~40 nm) and the Psugar system (~160 nm) exhibit a substantial difference in their final dimensions.
POEM/siRNA exhibited a lower critical micelle concentration of 12 ng/mL compared to the Psugar system (Fig. 3g). The morphology of POEM/siRNA at a ratio of 50/5/1 was assessed using cryo-electron microscopy (cryo-EM), revealing spherical particles with a relatively uniform diameter (Fig. 3h).
The GEM release profile of POEM/siRNA NPs was evaluated using dialysis method. As shown in Fig. 3i, minimal GEM release was observed from the NPs in PBS (pH 7.4) with or without 10% of FBS or murine serum. However, a rapid release of GEM (over 30%) occurred at 12 h in the presence of 10 mM GSH (glutathione) due to the cleavage of built-in disulfide bond (-SS-), a concentration that is commonly found in tumor cells43 (Fig. 3i). In addition, POEM demonstrated a sustained release of GEM over a period of 72 h followed. These data suggests that GEM is well protected prior to delivery to tumor tissues and achieves sustained release upon intracellular delivery to tumor cells and exposure to high concentrations of GSH. Figure 3j shows that siRNA formulated in the POEM NPs was well protected from RNase-mediated degradation.
The in vitro gene knockdown efficiency of POEM NPs was evaluated using KPC-C2-Luc and Panc02-Luc cell lines (Supplementary Fig. 12). POEM NPs were more effective than the commercial siRNA transfection reagent RNAiMAX in mediating the siLuc transfection and silencing luciferase transgene expression in both cell lines.
POEM NPs showed efficient tumor targeting and favorable pharmacokinetic profile
The in vivo distribution of POEM NPs was first evaluated by IVIS imaging 24 h after i.v. injection into KPC-C2 tumor (subcutaneous)-bearing mice. These NPs were prepared with an N/P ratio of 5:1, which represents the minimum ratio required to effectively load siRNA, as demonstrated in Fig. 3c. Additionally, we investigated the biodistribution of the POEM system at different amounts of PPOGEM to examine how changes in size and zeta potential affect their in vivo behavior. Cy5.5 signals were predominantly accumulated in the kidney and were barely detectable in tumors following injection of free siRNA-cy5.5 (Fig. 4a). Positively charged PPO/siRNA-cy5.5 complexes were primarily found in lungs and kidneys, suggesting the initial aggregation in blood followed by gradual release of siRNA from the complexes. Incorporating PPOGEM at a ratio of 25/5/1 led to improved accumulation at the tumor site, with significant amounts of signals also observed in the liver and lungs, likely due to size reduction (~50 nm) but still insufficient shieling of surface positive charges (~20 mV) by PPOGEM. Increasing the PPOGEM ratio from 25/5/1 to 50/5/1 resulted in a gradual increase of the Cy5.5 signal in tumors and a decrease of signal in liver due to improvements in both size reduction and shieling of surface positive charge (~50 nm, ~10 mV) (Fig. 4a). Further increase in the amount of PPOGEM led to reduced tumor signals due to the larger size of the NPs (~86.5 nm). Subsequent studies were conducted with POEM/siRNA prepared at a PPOGEM/PPO/siRNA ratio of 50/5/1. Supplementary Fig. 13a shows similar results in Panc02 subcutaneous model. Effective tumor targeting of POEM/siRNA NPs was also demonstrated in PCa KPC-C2 and Panc02 orthotopic (o.t.) models (Fig. 4b and Supplementary Fig. 13b). However, more signals were seen in liver in o.t. models compared to subcutaneous models. PCa o.t. model is known to have a high propensity of developing liver metastasis44. More studies will be conducted in the future to further investigate if the increased liver uptake of NPs in o.t. model is attributed to the micrometastasis in liver.
a Ex vivo imaging and quantified radiant efficiency of KPC-C2 subcutaneous tumor and major organs (H heart, Li liver, S spleen, Lu lung, K kidney, T tumor) at 24 h following intravenous administration of free siSPB9-cy5.5 or various POEM/siSPB9-cy5.5 NPs. n = 3 independent samples. b Ex vivo imaging and quantified radiant efficiency of KPC-C2 orthotopic pancreatic tumor and major organs at 24 h following intravenous administration of free siSPB9-cy5.5 or POEM/siSPB9-cy5.5 NPs at a ratio of 50/5/1. n = 3 independent samples. c Percentage of injected dose and tissue concentration of siSPB9 in different organs following i.v. injection of free siSPB9 or POEM/siSPB9 at a dose of 1 mg/kg for siRNA. n = 3 independent samples. d Plasma concentrations of siSPB9 over time after i.v. injection of siSPB9 or POEM/siSPB9 at a dose of 1 mg/kg for siRNA. n = 3 independent samples. e Pharmacokinetic parameters of siSPB9 were analyzed by a one-compartmental model. t1/2 half-life, AUC∞ the area under the plasma concentration versus time curve, AUMC∞ the area under the plasma concentration versus the first moment curve (AUMC), CL clearance, Vd volume of distribution. Data are presented as mean ± s.e.m. in (a–d). Statistical analysis was performed by two-way ANOVA with Tukey’s post hoc test for comparison in (a–c). Source data are provided as a Source Data file.
We further quantitatively analyzed the tissue distribution and pharmacokinetics of siSPB9 via qRT-PCR. Free siRNA was found in all organs, with a notably higher concentration observed in the spleen (Fig. 4c). In contrast, siSPB9 was largely concentrated in tumors, with ~6% of the injected dose localized in tumors at 24 h following i.v. injection of POEM/siRNA (Fig. 4c). The concentration of siSPB9 in tumors was significantly higher than that in the liver (~3-fold) (Fig. 4c). It is important to note that there are some discrepancies between the imaging and qRT-PCR data, likely due to the limited sensitivity of whole-body and ex vivo imaging as reported in literature45. In addition, qRT-PCR detects intact siRNA while imaging detects all fluorescence signals. Figure 4d shows the kinetic changes in the concentrations of siSPB9 in blood after injection of free siSPB9 or POEM siSPB9 NPs into KPC-C2 tumor-bearing mice. It is apparent that POEM siSPB9 persisted in the circulation for a significantly longer time compared with free siSPB9. Encapsulation of siSPB9 within POEM substantially enhanced its half-life (t1/2) and area under the curve (AUC), while significantly reducing its volume of distribution (Vd) and clearance (CL) compared to the free form of siSPB9 (Fig. 4e). The extended circulation time of POEM/siSPB9 NPs was also confirmed by fluorescence measurement of serum samples (Supplementary Fig. 13c). The long circulation time of POEM/siRNA is likely attributed to the excellent stability of the NPs in blood, which contributes to the effective tumor targeting.
The effective tumor accumulation of POEM NPs is likely to benefit from a passive targeting through enhanced permeation and retention (EPR) effect due to their relatively small sizes (40–50 nm). To further elucidate a potential role of active transport in tumor targeting by POEM NPs, we conducted proteomics study to investigate the protein corona formed around POEM NPs, which is regarded as a key factor in influencing their in vivo behaviors, including tissue distribution46. Preliminary data indicated that fibronectin, which can contribute to NPs’ tumor targeting and penetration47, is involved in the formation of the NPs protein corona (Supplementary Fig. 14a). Our transcytosis study revealed that blocking fibronectin receptor ITGA5 with its antibody significantly inhibited the transcytosis of the NPs (Supplementary Fig. 14b). In line with this, cellular uptake experiments confirmed that both anti-ITGA5 and chlorpromazine were able to block the uptake of POEM NPs (Supplementary Fig. 14c).
Co-delivery of siSPB9 and GEM led to enhanced anti-tumor efficacy
We first evaluated the knockdown efficiency of siRNA delivered by the POEM system. A luciferase-expressing tumor cell line (KPC-C2-Luc) was used to establish a subcutaneous tumor model, and the gene knockdown efficiency was examined by following changes in luminescent intensity following treatment with POEM NPs loaded with siLuc. A non-targeting siRNA was included as a control (siCT). Figure 5a shows that repeated injections of siLuc NPs led to a gradual reduction in luciferase activity in KPC-C2-Luc tumor-bearing mice as assessed by whole-body bioluminescence imaging. Figure 5b shows that POEM/siSPB9 NPs effectively suppressed both basal and GEM-induced SPB9 mRNA levels in vivo.
a Whole body imaging and quantification of luminous intensity of tumors of KPC-C2-Luc tumor-bearing C57BL/6 mice receiving intravenous administration of POEM/siLuc NPs once every 3 days. siCT refers to non-targeting siRNA as a control. n = 5 mice. b The mRNA expression levels of SPB9 in KPC-C2 tumors following various treatments three times. n = 5 mice. Changes of average tumor volumes (c) and individual tumor volume (d) in KPC-C2 WT tumor (subcutaneous)-bearing mice receiving various treatments. n = 5 mice. e The scheme for the establishment of pancreatic orthotopic model and treatments. Created in BioRender. Mu, Y. (2025) https://BioRender.com/wu7dxg8. Whole-body imaging (f) and luminous intensity of tumors (g) of KPC-C2-Luc tumor (orthotopic)-bearing mice following different treatments. n = 6 mice. h Survival rate of KPC-C2-Luc tumor (orthotopic)-bearing mice receiving various treatments. n = 6 mice. Data are presented as mean ± s.e.m. in (a–c, f, g). Statistical analysis was performed by one-way ANOVA with Tukey’s post hoc test for comparison in (b, d), two-way ANOVA with Tukey’s post hoc test for comparison in (a, c, g), and log rank test for comparison in (h). Source data are provided as a Source Data file.
Our findings so far have shown that targeting SPB9 can directly sensitize PCa to GEM-based treatment in addition to enhancing antitumor immune response. In addition, we have developed a POEM nanocarrier capable of codelivery of GEM and siSPB9. Figures 5c–d show the therapeutic efficacy of POEM/siSPB9 NPs in a KPC-C2 subcutaneous tumor model. A pharmacologically “inert” control nanocarrier, POC (PPO/PCytidine), was also developed as a control NP to load siSPB9 via replacing gemcitabine in PPOGEM with cytidine. POC shows similar size and zeta potential compared to POEM. In addition, POC and POEM show similar interactions with siRNA as demonstrated by comparable radiuses in molecular dynamic simulation (Supplementary Fig. 15). Tumor-bearing mice received various treatments when the tumors reached ~50 mm3. Both free GEM and POC/siSPB9 showed modest antitumor activity, while POEM/siCT was more effective than free GEM. Combination of POC/siSPB9 with GEM led to a significantly improved antitumor activity (Fig. 5c), suggesting that POC/siSPB9, as a simple and stand-alone siRNA therapy, can be readily combined with current standard of care such as gemcitabine treatment. It is also apparent that POEM loaded with siSPB9 showed a further improvement in overall antitumor activity compared with GEM+POC/siSPB9, including stabilization of tumor growth in 2 mice (Fig. 5d), clearly demonstrating the therapeutic benefit of codelivery of gemcitabine and siSPB9 using POEM NPs.
There were no significant changes in body weights following the different treatments (Supplementary Fig. 19a, b). In addition, these treatments did not adversely affect liver and kidney functions, as indicated by minimal alterations in serum levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), and creatinine (Supplementary Fig. 19d–f). Furthermore, histological examination revealed no notable changes in major organs including heart, liver, spleen, lung, and kidney in all groups (Supplementary Fig. 19g). These findings underscore the excellent safety profile of co-delivery of siSPB9 and GEM using POEM NPs, at dosages that have shown significant therapeutic efficacy.
We further evaluated the therapeutic potential of POEM/siSPB9 in KPC-C2 o.t. model. Luciferase-expressing KPC-C2 cells (KPC-C2-luc) were orthotopically transplanted into the pancreas of mice48. Tumor-bearing mice received different treatments starting on day 5 post-inoculation of tumor cells (Fig. 5e). Figure 5f, g display the whole-body luminescence imaging of mice receiving different treatments and the changes in luminescence intensities of tumor areas over time, prior to any mortality in each group, respectively. POC/siSPB9 and POEM/siCT were both effective in controlling tumor growth, with the POEM/siSPB9 group demonstrating superior anti-tumor effects (Fig. 5f, g). The enhanced inhibition of tumor growth by POEM/siSPB9 was translated into a significant prolongation of survival time (Fig. 5h). Among all treatments evaluated, POEM/siSPB9 led to the longest survival.
GEM resistance presents a significant challenge in clinical treatment49. To further assess the effectiveness of POEM/siSPB9 NPs in combating GEM-resistant tumors, we evaluated their therapeutic efficacy in KPC-C2 GEMR model. Similar to the study in wild-type model, mice received various treatments 5 days after tumor inoculation (Fig. 6a, b). POEM/siCT showed a level of antitumor activity that is comparable to POC/siSPB9, supporting the notion that SPB9 is a viable therapeutic target in the treatment of GEM-resistant pancreatic cancer. However, GEM and siSPB9 combination was less effective in controlling the resistant tumor compared to what was shown earlier in WT tumor model (Figs. 6a, b). These results prompted us to further investigate the tumor immune microenvironment in the GEMR model as detailed below.
a Changes of average tumor volumes and individual tumor volume in mice bearing KPC-C2 gemcitabine resistant (GEMR) subcutaneous tumor receiving various treatments. n = 5 mice. b Tumor weights and images of KPC-C2 GEMR tumor-bearing mice receiving different treatments. n = 5 mice. Single-cell suspensions were prepared from KPC-C2 GEMR tumor samples and subjected to flow cytometry analysis of CD45+ cells (c), CD80+/CD163+ macrophages ratio (d), CD8+ T cells (e), CD8+GzmB+ T cells (f), CD4+IFNγ+ (g), CD8+IFNγ+ (h), CD4+Treg (i), Ki67+ tumor cells (j), Annexin V+ tumor cells (k), CD4+PD1+ (l) and CD8+PD1+ (m). n = 10 independent samples. n Changes of average tumor volumes and individual tumor volume in KPC-C2-GEMR tumor (subcutaneous)-bearing mice receiving PBS, anti-PD1, POEM/siSPB9, and combination treatment. n = 5 mice. Data are presented as mean ± s.e.m. in (a–n). Statistical analysis was performed by one-way ANOVA with Tukey’s post hoc test for comparison in (b–m) and two-way ANOVA with Tukey’s post hoc test for comparison in (a, n). Source data are provided as a Source Data file.
POEM NPs enhanced antitumor immunity and sensitized PDAC to anti-PD-1 therapy
As depicted in Fig. 6c, all the treatments resulted in increased infiltration of CD45+ immune cells. Figure 6d shows that formulations loaded with siSPB9, particularly POEM/siSPB9, led to the polarization of M1-like macrophages (CD80+) and an increase in the M1/M2 ratio (CD80+/CD163+). In addition, treatments with POEM/siCT and POEM/siSPB9 significantly increased the numbers of CD8+ T cells, along with GzmB+ CD8+ T cells, especially in the POEM/siSPB9 group (Fig. 6e, f). Activated CD8+ T cells (CD69+) were also increased (Supplementary Fig. 20). Moreover, there were significant increases of IFNγ-secreted CD4+ and CD8+ T cells, which are indicative of an enhanced immune response against the tumor (Fig. 6g, h). Meanwhile, regulatory T cells, which typically suppress immune responses and promote immune tolerance, are downregulated within the TME (Fig. 6i). Overall, the combination of GEM and siSPB9 using POEM NPs fostered an immunoactive tumor microenvironment, contributing to the inhibition of proliferation and killing of tumor cells as evident from decreased numbers of Ki67+ and increased numbers of Annexin V+ tumor cells (Fig. 6j, k).
Despite the favorable changes in various immune cell subsets as described above, treatment with GEM or siSPB9 significantly upregulated PD-1 expression in CD4+ and CD8+ T cells (Fig. 6l, m). PD-1 is a protein that inhibits the immune system’s ability to attack cancer cells50. This observation led us to explore the potential of combining anti-PD-1 antibody with our current treatment to enhance the overall therapeutic efficacy. As demonstrated in Fig. 6n, monotherapy with anti-PD-1 exhibited limited antitumor efficacy. However, the combination of anti-PD-1 with POEM/siSPB9 significantly enhanced the therapeutic outcome. Tumor growth was effectively suppressed following the 1st treatment, and by day 20, four out of five tumors were completely regressed upon gross examination. Moreover, all treatments were well tolerated, as indicated by minimal changes in body weights (Supplementary Fig. 19c). These results suggest that the combination of POEM/siSPB9 with anti-PD-1 treatment represents a promising regimen towards GEM resistant pancreatic cancer.
Discussion
As an endogenous inhibitor of GzmB, SPB9 has been shown to be critically involved in immune homeostasis, including viral infection, autoimmune diseases, and tumor immune escape. Inhibition of SPB9 in tumor cells has been proposed as a strategy for improving cancer immunotherapy. In this study, we extended our understanding of SPB9 by showing that GzmB/SPB9 axis is also involved in regulating the sensitivity of PCa cells to GEM through a non-immunological mechanism. Our data showed that GEM induces the gene expression of both SPB9 and GzmB (Figs. 1 and 2). SPB9 KO or KD led to sensitization of PCa cells to GEM in a GzmB-dependent manner. Interestingly, GEM resistance is associated with increased expression levels of SPB9 and decreased expression of GzmB (Supplementary Fig. 6). Our data suggests that KD of SPB9 can directly sensitize PCa to GEM treatment in addition to improving the antitumor immune response. It remains to be studied if GzmB/SPB9 axis also affects the sensitivity of PCa as well as other cancer types to other chemotherapeutic drugs.
POEM represents an effective nanocarrier in codelivery of GEM and siRNA (Fig. 3). The pyrimidine structure is critical in stabilizing the siRNA NPs through π-π stacking and hydrogen bonding. It is possible that the hydroxyl groups in the sugar ring are also important in promoting the interaction with siRNA, albeit less important compared to pyrimidine motif, which warrants more studies in the future. The use of two individual polymers (PPO and PPOGEM) each with cationic (-NH2) and GEM motif, respectively, will provide the flexibility of adjusting their ratio in formulation optimization. In addition, the very similar structure of the two polymers will facilitate their mixing to form compact and stable micelles. Although this study focuses on evaluating combination of GEM and siRNA, this strategy can be applied to codelivery of siRNA with other nucleoside analogue-based drugs such as AZA. In addition, other drugs such as cisplatin can be readily incorporated into POEM NPs following lipid-derivatization. Therefore, our system may be tailor-designed to develop precision medicines to suit different combination therapies.
Recently, protein corona (PC), a layer of proteins absorbed on the surface of NPs upon exposure to biological fluids, has been increasingly recognized to be crucial in defining the in vivo behaviors of the NPs, especially the tissue tropism. Much of this understanding has been derived from studies on lipid nanoparticles (LNPs) used for targeted delivery to hepatocytes34. We have recently reported that fibronectin was preferentially enriched in ultrasmall-sized PAZA NPs (~15 nm) but not the large-sized counterpart without AZA motif (~150 nm), which contributes significantly to both tumor accumulation and deep penetration of PAZA NPs47. It is interesting to note that fibronectin was also one of the proteins that were enriched in the protein corona of POEM NPs. The fact that fibronectin is involved in facilitating cellular uptake and transcytosis of POEM NPs by tumor cells (Supplementary Fig. 14) suggests that this protein, and possibly other proteins as well, may also play a role in enhancing the accumulation and penetration of POEM NPs. EPR shall also contribute to the tumor targeting by POEM NPs as well, especially considering their compact sizes (40–50 nm). The combination of EPR-mediated passive targeting and fibronectin-mediated active targeting contributes significantly to the effective tumor accumulation of POEM NPs in both subcutaneous and orthotopic models: around 6% of injected siRNA formulated in POEM NPs was found in the tumor tissues at 24 h, and the siRNA concentrations in tumors were 3–4 folds higher than those in liver. More mechanistic studies on tumor targeting may lead to further improved tumor delivery systems in the future.
Codelivery of siSPB9 with GEM led to significant antitumor activity in both WT and GEMR models, along with improved tumor immune microenvironment (Figs. 5 and 6). The enhanced antitumor activity with this combination therapy likely stems from multiple synergistic mechanisms. Firstly, GEM contributes directly to tumor cell death and also induces the production of GzmB in the tumor cells. Concurrently, SPB9 KD sensitizes tumor cells to both the direct pharmacological effect of GEM and the apoptosis induced by immune cells. Additionally, this combination therapy leads to a more robust antitumor immune response, creating a hostile environment for cancer progression. Importantly, combination of anti-PD1 antibody with this regimen leads to further improvement in the overall therapeutic effect (Fig. 6).
We are aware of the efforts in developing small molecule inhibitors of SPB9, such as 3034, due to the ease in clinical application. 3034 has been shown to inhibit tumor growth in several murine tumor models16. Codelivery of 3034 with DOX via NPs has also been shown to improve the overall therapeutic efficacy in a murine breast cancer model51. One concern with the small molecule inhibitor is the potential detrimental effect on T cells and NK cells via blocking the activity of SPB9 inside these cells. Despite the likely improved delivery of a SPB9 inhibitor to tumors using NPs, it is likely that T cells will also be exposed to the small molecule compound released inside tumor tissues. Systemic use of a SPB9 inhibitor may have more concerns on its impact on the immune homeostasis in normal organs and tissues. On the other hand, T cells are notoriously known to be hard to transfect52. Thus, uptake of siSPB9 NPs by T cells may cause minimal impact on the SPB9 levels inside these immune cells. Interestingly, POEM NPs exhibited minimal uptake by T cells compared to tumor cells, both in vitro and in vivo following systemic administration (Supplementary Fig. 17). This low uptake of POEM NPs by T cells is likely due to their limited interactions with T cells, in contrast to other immune cell types such as macrphages53. The efficient cellular uptake by TAFs (Supplementary Fig. 17c) indicates that our POEM NPs may serve as a powerful tool to deliver therapies to these cells directly and disrupt the TAFs-mediated pro-tumor processes. It is also interesting to note that POEM NPs were effectively taken by tumor endothelial cells as well (Supplementary Fig. 17c). We and others have recently reported that transcytosis across endothelial cells contributes significantly to the accumulation of various types of NPs54. A role of the active trans-endothelial transport in tumor targeting by POEM NPs requires more study in the future. In a preliminary study we showed that codelivery of siSPB9 and GEM using POEM NPs was more effective in inhibiting the tumor growth than POEM NPs-mediated codelivery of 3034 and GEM (Supplementary Fig. 18). More studies on the differential effect of different formulations and/or drug combinations on tumor cells and different immune cell subpopulations may lead to the development of an improved therapy targeting the GzmB/SPB9 axis.
Methods
Ethics statement
All animals were housed under pathogen free conditions according to AAALAC (Association for Assessment and Accreditation of Laboratory Animal Care) guidelines. All animal-related experiments were performed in full compliance with institutional guidelines and approved by the Animal Use and Care Administrative Advisory Committee at the University of Pittsburgh under protocol number 21099779. The maximum tumor volume permitted by the IACUC guidelines was 2000 mm3. Throughout the study, tumor size and animal health were monitored daily, and the maximum allowed tumor burden was not exceeded.
Reagents
Gemcitabine was purchased from LC Laboratories (MA, USA). Poly-ε-L-lysine HCl (MW 3500–4500 Da) was purchased from BIOSYNTH (KY, USA). Oleic acid, poly(ethylene glycol) methacrylate (average Mn = 950, PEG950), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), Dulbecco’s Modified Eagle’s Medium (DMEM), trypsin-EDTA solution, D-Luciferin were purchased from Sigma-Aldrich (MO, USA). RPMI-1640 medium, fetal bovine serum (FBS), and penicillin-streptomycin solution were purchased from Invitrogen (NY, USA). GlutaMAX (100×) was purchased from Thermo Fisher Scientific (MA, USA). Antibodies for Western blot and flow cytometry were listed in Supplementary Table 1.
Cell lines and animals
All cell lines were purchased from certified vendors (ATCC) and used at low passage. Authentication was performed by the suppliers using standard STR profiling. Routine mycoplasma testing was conducted using PCR-based assays to confirm the absence of contamination throughout the study. Panc02, PANC-1, MIA PaCa-2, Panc 02.03, and Panc 10.05 cell lines were obtained from ATCC (Manassas, VA). KPC-C2 and KPC-C5 cell lines were kindly gifted by Dr. Wen Xie (University of Pittsburgh). KPC-C2, KPC-C5 and Panc02 gemcitabine resistant cell lines were generated according to published protocols55. SERPINB9 KO KPC-C2, SERPINB9 KO KPC-C2 GEMR, Panc02-Luc, and KPC-C2-Luc cells were produced by following lentiviral/retroviral infection protocol as detailed in lentiviral infection section below. Panc02 and PANC-1 were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (100 U/ml). KPC-C2, SERPINB9 KO KPC-C2, and KPC-C5 were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), 1% GlutaMAX (100×) and penicillin/streptomycin (100 U/ml). MIA PaCa-2 was cultured in DMEM supplemented with 10% fetal bovine serum (FBS), 2.5% horse serum, and penicillin/streptomycin (100 U/ml). Panc 02.03 and Panc 10.05 were cultured in RPMI-1640 supplemented with 15% fetal bovine serum (FBS), 10 Units/ml human recombinant insulin, and penicillin/streptomycin (100 U/ml). Gemcitabine resistant cell lines were cultured in the full medium with gemcitabine (260 ng/mL). All cells were cultured at 37 °C in a humidified atmosphere with 5% CO2.
Female C57BL/6 mice aged between 4–6 and 8–10 weeks were purchased from The Jackson Laboratory (ME, USA). Mice were housed at an ambient temperature of 22 °C (range: 22–24 °C) and a humidity of 45%, with a 14/10 day/night cycle (lights on at 6:00, off at 20:00), and allowed access to food ad libitum.
Vector, RNA interference, and lentiviral infection
Edit-R Mouse Serpinb9 mCMV-EGFP All-in-one Lentiviral sgRNA was purchased from Horizon Discovery Ltd. (Cambridge, UK). psPAX2 and pMD2.G were kindly provided by Dr. Da Yang (University of Pittsburgh). pLentipuro3/TO/V5-GW/EGFP-Firefly Luciferase Plasmid was purchased from Addgene (MA, USA).
Murine and human SERPINB9 siRNA (siSPB9), murine and human ATF3 siRNA (siATF3), and control siRNA (siCT) were purchased from Sigma-Aldrich (MO, USA). Murine granzyme B siRNA (siGzmB) was purchased from Thermo Fisher Scientific (MA, USA). SiSPB9-cy5.5 was synthesized by Sigma-Aldrich. Cy5.5 was introduced to siSPB9 via phosphate linkage using phosphoramidite chemistry. The sequences of siSPB9, siSPB9-cy5.5, siATF3, and siCT are shown in Supplementary Table 2.
SERPINB9 KO cell lines were generated by using CRISPR technology. Cells were infected with the lentivirus packaged by Serpinb9-All-in-one lentiviral sgRNA-CRISPR-Cas9 plasmid encoding EGFP. The successfully knocked out cells were selected by cell sorting of EGFP+ population, followed by single clone culture and further confirmed through Western blot analysis for the lack of SERPINB9 proteins.
SERPINB9 control vector cell line was generated by using a control lentiviral vector with Cas9 coding sequence but without the specific guiding sequences (Lenti CRISPR plasmid without sgRNA sequence).
The luciferase-expressing cell lines were generated according to a published protocol56. Cells were infected with the lentivirus packaged from pLentipuro3/TO/V5-GW/EGFP-Firefly Luciferase plasmid encoding EGFP. The luciferase-expressing cells were selected by cell sorting of the EGFP+ population, followed by single clone culture, and further confirmed through flow cytometry analysis for EGFP expression.
RNA sequencing analysis
KPC-C2 gemcitabine resistant cells and wide type cells were harvested for RNA sequencing (RNA-seq), which was performed by the Health Sciences Sequencing Core at Children’s Hospital, University of Pittsburgh. Raw sequence data was analyzed by HISAT-Stringtie workflow as described in previously published protocol57,58,59 to generate transcript level gene expression. The Gene Set Enrichment Analysis (GSEA)60 was further performed based on the gene lists ranked by the log2FC between two groups.
qRT-PCR
To examine the mRNA level of different genes from RNA-seq results, murine pancreatic cancer cell lines (Panc02, KPC-C2, and KPC-C5; WT vs GEMR) were collected and subjected to qRT-PCR as detailed below.
To examine the effect of GEM on the mRNA level of GranzymeB, SERPINB9, and ATF3, different pancreatic cancer cells were treated with various concentrations of GEM. Cells were collected 24 or 48 h later and subjected to qRT-PCR as detailed below.
cDNA was generated from the purified RNA extracted from the indicated cultured cells using High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, MA, USA) according to the manufacturer’s instructions. qRT-PCR was performed using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific, MA, USA) on a 7900HT Fast Realtime PCR System. Relative target mRNA levels were analyzed using delta-delta-Ct calculations. The primers are shown in Supplementary Table 3.
Western blot assay
The total protein was extracted from the indicated cells by using radioimmunoprecipitation assay (RIPA) lysis buffer (Thermo Fisher Scientific, MA, USA) through gently shaking on ice for 30 min. After centrifugation at 12,500 × g for 10 min, the supernatants were collected, and the concentrations of proteins were determined using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, MA, USA). The protein samples were mixed with 5× SDS loading buffer, denatured at 98 °C for 10 min, loaded onto 10% SDS-polyacrylamide gel electrophoresis (PAGE) gel for electrophoresis, followed by transferring to PVDF membrane. The membranes were then blocked in 3% BSA dissolved in phosphate buffer containing 0.05% Tween-20 (PBST) for 1 h at room temperature. Afterwards, the membranes were incubated with primary antibody in diluted buffer (3% BSA in PBST) with gentle agitation overnight at 4 °C. After washing with PBST for three times, the membranes were subsequently incubated with horseradish peroxidase-conjugated secondary antibody at room temperature for 1 h. The membranes were then washed three times with PBST before being incubated with Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific, MA, USA). Signal was visualized by films with the AX700LE film processor (Alphatek) or iBright™ FL1500 Imaging System.
Granzyme B activity assay
Granzyme B activity was measured using the GranToxiLux PLUS kit. Cells subjected to different treatments were incubated with the Granzyme B substrate for 1 h at 37 °C. The cleaved Granzyme B substrate (GranToxiLux) exhibited an excitation peak at 488 nm and an emission peak at 520 nm. GranToxiLux fluorescence was measured by flow cytometry, and cell populations positive for GranToxiLux were considered to have active Granzyme B.
Construction of luciferase reporter assay plasmid
The UCSC genome browser identified a 1601 bp promoter region of human SERPINB9 gene, revealing 3 putative ATF3-binding motifs. The specific sequence of the SERPINB9 promoter region was shown in Supplementary Table 4. Three plasmids, each with a 5× repeated sequences of one of the three motifs (Supplementary Table 4) were constructed, respectively by using pGL3-Basic (Promega, CA, USA) as the backbone. The three DNA fragments with different 5x repeated sequences were synthesized by AZENTA (MA, USA) with added restriction enzyme sites. NheI (Thermo Fisher Scientific, MA, USA) and HindIII (Thermo Fisher Scientific, MA, USA) restriction enzymes were used to digest the pGL3-Basic plasmid and the synthesized fragments. The 3 DNA fragments were individually ligated with the backbone by using T4 DNA ligase (Invitrogen, NY, USA). Single colonies were picked and expanded in the DH5α competent cells (Thermo Fisher Scientific, MA, USA). After plasmid extraction, the concentration was determined by NanoDrop, and the sequence was confirmed by Sanger sequence.
Similarly, a full length promoter sequence containing all three binding motifs was synthesized by Twist Bioscience (CA, USA) and cloned into pGL3-Basic. The mutated plasmid was constructed through site-directed mutagenesis. In brief, a pair of primers for mutagenesis were designed and synthesized (IDT, Coralville, IA, USA). This pair of primers was designed to include the front and back sequence of each binding motif but without the motif sequence. Then the PCR reaction with this pair of primers was performed using the previously generated full-length plasmid as a template. The PCR product was treated with Dpn1 enzyme to remove the template. Then, the PCR product was used for the transformation in DH5α competent cells. After plasmid extraction, the concentration was determined by NanoDrop. The Sanger sequencing confirmed the sequence.
Luciferase reporter assay
PANC-1, MIA PaCa-2, and Panc 02.03 cells were transfected with each of the plasmids described above using Lipofectamine 3000 (Invitrogen, MA, USA) for 48 h. Subsequently, the cells were treated with gemcitabine for an additional 48 h. Cell lysis was performed using the lysis buffer from the Pierce Firefly Luciferase Glow Assay Kit (Thermo Fisher Scientific, MA, USA). D-Luciferin was added to the cell lysate, and bioluminescence was detected using a luminometer, according to the manufacturer’s instructions.
In vitro cytotoxicity assay
Cytotoxicity was evaluated by MTT assay with indicated cell lines. Cells were seeded in 96-well plates (attached 96 well tissue culture plate (CELLTREAT, MA)) at a density of 2 × 103 cells/well with 100 μL of culture medium. Cells received various treatments including GEM alone, GEM+siCT, or GEM+siSPB9 combination at various GEM and siRNA concentrations for 48 h. For the combination treatment, cells were transfected with siRNA using Lipofectamine RNAiMAX (Invitrogen, MA, USA) for 48 h followed by drug treatment for another 48 h. MTT assay was then performed on the cultured cancer cells. The absorbances of each well were measured at 590 nm. The cell viability was determined via the following formula: (ODtreated- ODblank)/(ODcontrol-ODblank) × 100 %.
Cell apoptosis study
Cells were digested following different treatments for 24 h and stained with Zombie NIR and BV421 anti-mouse Annexin V or FITC anti-mouse Annexin V for flow cytometry analysis. The Zombie NIR−, Annexin V+ population was considered apoptotic cells.
Synthesis and characterization of PEG1K-COOH
A solution of PEG1K (5 g, 5 mmol) in chloroform (15 mL) was added to a dry flask containing succinic anhydride (2.5 g, 25 mmol). Subsequently, a solution of DMAP (3.05 g, 25 mmol) in 10 mL of chloroform was introduced into the reaction mixture. The mixture was then refluxed at 60 °C for 24 h. After the reaction, the solvent was reduced to about half of the original volume using rotary evaporation. The product was recrystallized from cold ether, filtered, and the precipitate was collected. The synthesis route and NMR data for PEG1K-COOH are presented in Supplementary Fig. 7.
Synthesis and characterization of PPO polymer library
To a stirred solution of PEG1K-COOH and oleic acid (OA) in 30 mL DMSO, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) and N-hydroxysuccinimide (NHS) were added. The mixture was stirred for 2 h, after which Poly-ε-L-lysine HCl (PLL) and triethylamine (TEA) were added. Twelve hours later, the reaction mixture was purified by dialysis using a dialysis membrane (MWCO 3500 Da) in a DMSO/H2O (4:1) mixture for 24 h, followed by water for an additional 24 h. The product (PEG-PLL-Oleic acid, PPO) was lyophilized and stored at 4 °C. The synthesis route and NMR data are demonstrated in Supplementary Fig. 8.
To synthesize PPO polymers conjugated with various amino acids, tert-butyloxycarbonyl (BOC)-protected amino acids (histidine, lysine, or arginine) were dissolved in DMSO. The activation of the amino acid carboxyl groups was achieved by adding 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) and N-hydroxysuccinimide (NHS). The reaction mixture was stirred for 2 h to promote conjugation. Next, the non-amino acid-decorated PPO polymer and triethylamine (TEA) were introduced, and the mixture was stirred for an additional 12 h. The reaction product was purified by dialysis using a dialysis membrane (MWCO 3500 Da). Dialysis was performed in a DMSO/H2O (4:1) mixture for 24 h, followed by 24 h in pure water. The final product was lyophilized and stored at 4 °C.
Synthesis and characterization of PPOGEM polymer
4,4′-Dithiodibutyric acid (2.5 g, 10 mmol) and 10 mL of acyl chloride were combined in a 50 mL flask and refluxed at 65 °C for 4 h. After completion, the excess acyl chloride was evaporated, and the inner anhydride was obtained without further purification. Subsequently, PPO (500 mg), TEA (0.9 mL, 6.42 mmol), and 30 mL DMSO were added to the flask. The mixture was stirred and reacted at 50 °C for 24 h. Purification was conducted by dialysis using a dialysis membrane (MWCO 3500 Da) with a DMSO/H2O mixture (4:1) for 24 h, followed by water for another 24 h. The compound PLL(COOH)-OA-PEG was then lyophilized and stored at 4 °C. The NMR results of PLL(COOH)-OA-PEG are illustrated in Supplementary Fig. 9.
For the subsequent reaction, PLL(COOH)-OA-PEG (300 mg), hydroxybenzotriazole (HOBt) (60 mg, 0.44 mmol), EDC (150 mg, 0.98 mmol), gemcitabine (560 mg, 2.1 mmol), and TEA (0.45 mL, 3.2 mmol) were dissolved in 30 mL of DMSO. The mixture was then reacted at 50 °C for 72 h. Following the reaction, the product underwent purification by dialysis using a dialysis membrane (MWCO 3500 Da) with a DMSO/H2O mixture (4:1) for 24 h and then water for another 24 h. The final product was lyophilized and stored at 4 °C. The synthesis routes are illustrated in Supplementary Fig. 9. The content of gemcitabine in the polymer was determined by UV absorbance.
Synthesis and characterization of Psugar and PCytidine polymers
The control polymers Psugar and PCytidine were synthesized using a method similar to that for the PPOGEM polymer. PLL(COOH)-OA-PEG (300 mg), hydroxybenzotriazole (HOBt) (60 mg, 0.44 mmol), EDC (150 mg, 0.98 mmol), D-Glucosamine sulfate (600 mg, 2.1 mmol), and TEA (0.45 mL, 3.2 mmol) were dissolved in 30 mL of DMSO to synthesize Psugar polymer. PLL(COOH)-OA-PEG (300 mg), hydroxybenzotriazole (HOBt) (60 mg, 0.44 mmol), EDC (150 mg, 0.98 mmol), cytidine (520 mg, 2.1 mmol), and TEA (0.45 mL, 3.2 mmol) were dissolved in 30 mL of DMSO to obtain PCytidine polymer. Both reactions were stirred at 50 °C for 72 h. The subsequent dialysis and lyophilization processes followed the methods described above. The final compound structures are demonstrated in Supplementary Fig. 10. The conjugation rates of cytidine and glucosamine were determined by UV absorbance.
Fabrication and physicochemical characterization of nanoparticles
To prepare the POEM/siRNA or Psugar/PPO/siRNA NPs, Psugar, PPOGEM, and PPO were dissolved in DMSO, separately (40 mg/mL). SiRNA was dissolved in deionized water at a concentration of 25 μg/mL. The DMSO solutions of PPO with PPOGEM or Psugar were prepared at different ratios, followed by rapid addition of siRNA (10 μg) aqueous solution. The mixture was centrifuged using an Amicon Ultra centrifugal filter device at 15,000 × g for 15 min. Deionized water was added, and the centrifugation was repeated at 15,000 g for 15 min to remove residual DMSO. The final nanoparticles in the remaining liquid in the tube were collected for subsequent studies.
The formation of stable nanoparticle complexes was confirmed using gel-retardation assay. The particle size was measured via Zetasizer (Malvern Panalytical, UK) from three batches of formulation. The critical micelle concentration (CMC) of POEM/siRNA and Psugar/PPO/siRNA were determined through measuring the light scattering intensity61.
To assess the resistance of POEM/siRNA NPs against nuclease-mediated degradation, the NPs were incubated with RNase (50 U/mL) (NEB, MA, USA) at 37 °C for 1 h. Subsequently, the NPs were disrupted by 0.05% SDS, and the integrity of siRNA was evaluated by electrophoresis. Free siRNA served as a control.
Molecular simulation systems
The chemical structures of PPO, PPOGEM, Psugar, and PCytidine are all composed of different numbers of three residue types, A, B, and C as illustrated in Supplementary Fig. 16. For PPOGEM, Psugar and PCytidine, the residue composition is x = 24 for A, y = 3 for B, z = 3 for C, while for PPO, the residue numbers of A, B, and C are present in a ratio of 23:5:2. The residue topologies of all residues were generated using the Antechamber software62. Specifically, even though the numbers of different residues in the polymer have been determined, the precise arrangement of these residues within the polymeric chain remains unknown. To maximize the separation of the same type of residues, the 1D sequence of PPOGEM, Psugar, and PCytidine is depicted as the follows: A-A-A-A-A-A-A-A-B-A-A-A-A-A-A-A-A-B-A-A-A-A-A-A-A-A-B-C-C-C, while the sequence of PPO is: A-A-A-A-A-B-A-A-A-A-A-B-A-A-A-A-B-A-A-A-A-A-B-A-A-A-A-B-C-C. Next, we utilized the NAB program in the Amber Tools to generate the siRNA structure based on the human siRNA sequence listed in Supplementary Table 2. Finally, POEM nanoparticle system and the control nanoparticle (Psugar/PPO/siRNA, PCytidine/PPO/siRNA) were constructed with PPOGEM (Psugar or PCytidine), PPO, and siRNA in a ratio of 33:9:1. To construct the topology of the simulation system, the GAFF2 force field63,64 was utilized to model lipids, while the OL3 and OL15 force fields were used to model RNA and DNA, respectively.
Molecular dynamics simulations
The micelle-like structure of POEM loaded with siRNA was formed through a series of molecular dynamics (MD) simulations using the PMEMD.cuda program in the AMBER 22 software package65,66. Initial system relaxation was achieved by a 1000-step conjugated-gradient energy minimization on the entire POEM/siRNA structure. Next, constant-pressure MD simulation was performed for ~3.5 nanoseconds (ns), facilitating system shrinking and stabilization, with an objective of achieving a density close to 1. Subsequently, a two-stage MD simulation protocol was applied to the shrunk system, with a 600 ns MD simulation to continue to stabilize the system in stage one. Upon the completion of stage one, the last snapshot was solvated in a rectangular water box filled in with TIP3P water and 0.15 M Na+ and Cl− ions. The whole system was neutralized and the simulation box has a dimension of 220 × 220 × 220 Å. Then a 300 ns MD simulation was conducted with period boundary condition enforced in the second stage. During the whole simulation phase, position restraints were applied to the siRNA with a force constant of 100 kcal/mol/Å2 to prevent its distortion, but the other atoms were allowed to move freely. The snapshot with the smallest root-mean-square deviation (RMSD) from the average structure of snapshots collected after the system reached equilibrium (the last 150 ns) was selected as the representative conformation. A similar protocol was applied for the PCytidine/siRNA and Psugar/siRNA system.
CryoEM methods
Samples were initially examined using negative-stain electron microscopy. A 3 μL aliquot was applied to a freshly glow-discharged continuous carbon copper grid and stained with 1% uranyl acetate solution. The grids were then inserted into a Tecnai TF20 electron microscope (Thermo Fisher Scientific, MA, USA), equipped with a field emission gun, and images captured using an XF416 CMOS camera (TVIPS GmbH, Gilching, Germany) to assess nanoparticle uniformity and concentration.
For cryo-electron microscopy, 3 μL of the sample was pipetted onto a Protochips C-flat CF-2/1-3CU-T grid (Protochips, NC, USA) that had been glow discharged at 25 mA for 30 s using an Emitech KX100 glow discharger. The grids were then processed in a Thermo Fisher Vitrobot Mk 4, set to a relative humidity of 95%. They were blotted for 3 s with a force setting of 4 and subsequently plunged into a 40/60 mixture of liquid ethane/propane that was precooled in a liquid nitrogen bath. The grids were then placed onto a Gatan 910 3-grid cryoholder (Gatan, Inc, CA, USA) and inserted into the TF20 microscope, maintaining a temperature of no higher than −175 °C throughout. The microscope was operated at 200 kV, with contrast enhanced by a 100 μm objective aperture. Cryo-electron micrographs were collected at a nominal magnification of 150,000× on the TVIPS XF416 CMOS camera, with a pixel size of 0.74 Å at the sample. Low dose methods were employed to minimize electron beam damage, and images were acquired using TVIPS EMplified software in movie mode to correct for drift.
Release kinetics of gemcitabine
The kinetics of gemcitabine release from POEM nanoparticles under different conditions was performed by dialysis method. In brief, 2 mL of POEM nanoparticles (PPOGEM/PPO/siSPB9; 5 mg/0.5 mg/0.1 mg) PBS solution under different conditions (10% FBS, 10% murine serum, 10 mM GSH and 100 mM GSH) were placed in a dialysis bag (MWCO 3.5 kDa) and immersed in 40 mL of 0.1 M PBS solution containing 0.5% (w/v) Tween 80. The experiment was performed in an incubation shaker at 37 °C with gentle shaking. At selected time intervals, 10 μL solution in the dialysis bag and 1 mL medium outside the dialysis bag were withdrawn, while same amount of fresh dialysis solution was added for replenishment. The concentration of gemcitabine was measured by HPLC-UV according to our previous method67.
In vitro knock down efficiency
To test the in vitro knockdown efficiency of POEM NPs, KPC-C2-Luc and Panc02-Luc cells were treated with different formulations of POEM NPs with varying ratios of PPOGEM and PPO. After 48 h treatment, cell lysates were collected and analyzed by measuring the bioluminescent intensity. Commercial Lipofectamine RNAiMAX was used as a control.
Cellular uptake
For the cellular uptake study, KPC C2 WT cells were pretreated with chlorpromazine, anti-ITGA5, or IgG for 1 h, in medium depleted of FBS. The cells were then incubated with POEM/siRNA/DiD dye NPs for 1 h. After incubation, the cells were digested, and the fluorescence intensity of DiD dye was measured.
Transcytosis study
KPC C2 cells were seeded into the upper chamber of a transwell and cultured for 3 days until a confluent monolayer was formed. Prior to adding POEM/siRNA/DiD NPs to the upper chamber, the cells were pretreated with anti-ITGA5 or IgG for 1 h. After 8 h of incubation, the medium in the lower chamber was collected, followed by measurement of its fluorescence intensity.
Murine tumor models
To establish the subcutaneous tumor models, 5 × 105 Panc02, KPC-C2, KPC-C2 SERPINB9 KO, KPC-C2 control vector cells, or KPC-C2 GEMR cells were inoculated into the right flank of C57BL/6 mice (4-6 weeks). The Panc02, KPC-C2, or KPC-C2 GEMR tumor-bearing mice received different treatments when the tumor size reached the indicated volume. Tumor growth in KPC-C2 SERPINB9 KO or KPC-C2 control vector tumor-bearing mice was monitored for 20 days, followed by analysis of tumor-infiltrating immune cells as detailed in “Analysis of tumor-infiltrating immune cells” section.
The pancreatic orthotopic tumor models were established according to a previous publication48. Briefly, anesthetized C57BL/6 mice (8–10 weeks) were shaved, and the surgical area on the left flank was disinfected. The pancreas was exposed through a small incision (~1 cm) on the left flank. Fifty microliters Panc02-Luc or KPC-C2-Luc single cell suspension (1 × 105 per mouse) was injected cell into the tail of the pancreas using 28-gauge hypodermic needles. The needle was slowly removed after the injection of cell suspension. After injection of tumor cells, the mice were maintained at the heating pad and observed until complete recovery. The development of pancreatic cancer was monitored by bioluminescence radiance intensity after the mice were injected with D-Luciferin (GoldBio, 150 mg/kg) intraperitoneally.
Whole-body NIRF imaging and ex vivo imaging
For tissue biodistribution study, mice bearing subcutaneous (~300 mm3) or orthotopic (~10 days after inoculation) pancreatic tumors were intravenously injected with free siSPB9-cy5.5 and POEM/siSPB9-cy5.5 NPs with different PPOGEM/siRNA ratios, respectively. The mice were sacrificed at 24 h after injection. Tumor and major organs, including heart, liver, spleen, lung, and kidney were collected and imaged by IVIS 200 system (Perkin Elmer, MA, USA) at a constant 1 s exposure time with excitation at 679 nm and emission at 702 nm. For the study of PK in blood, blood was collected in EDTA-containing tubes at 10 min, 0.5 h, 1 h, 2 h, 4 h, 8 h, 12 h, 48 h and 72 h timepoints and plasma samples were prepared by centrifugation at 21,000 × g for 10 min and imaged by the IVIS 200 system.
Blood pharmacokinetics and biodistribution of siSPB9
Groups of 3 C57BL/6 mice (4–6 weeks) bearing subcutaneous KPC-C2 tumors (~300 mm3) received tail vein injection of POEM/siSPB9-cy5.5 NPs or free siSPB9-cy5.5 at a dose of 1 mg/kg for siRNA. At different timepoints (10 min, 0.5 h, 1 h, 4 h, 24 h, and 48 h) post injection, blood was collected in the tubes containing EDTA. The amount of siRNA in the plasma was quantified by qRT-PCR as detailed below. The PK parameters were obtained by fitting the blood siRNA concentration versus time using a one-compartment model. Hearts, livers, spleens, lungs, kidneys, and tumors were collected at 24 h and homogenized. SiRNA in the tissues was quantified through qRT-PCR.
SiSPB9 quantification by qRT-PCR
The siSPB9 was measured by qRT-PCR as previously published38. Briefly, the plasma, major organs and tumors collected were homogenized in 1000 μL TRIzol. Chloroform (200 μL) was added to the homogenized tissues. After 5 min of incubation, the samples were centrifuged at 21,000 × g for 15 min. The upper aqueous phase was used for qRT-PCR. A standard curve was generated by spiking a known amount of siSPB9-cy5.5 (0–20 μg/mL) in the tissues obtained from non-treated control animals and used to calculate the amount of siRNA in the samples. A SuperScript III reverse transcription kit (Invitrogen, NY, USA) was used to convert siRNA into cDNA. For reverse transcription, 6 μL of RT Master Mix (2 μL of water, 2 μL of 5× buffer, 0.5 μL of 0.1 M dithiothreitol, 0.5 μL of 10 mM dNTPs (Invitrogen, NY, USA), 0.5 μL of RNAse OUT (Invitrogen, NY, USA) and 0.5 μL of SuperScript III enzyme) were combined with 2 μL of 0.5 μM GS primer and 2 μL of template in a 96-well plate. GS primer and template were premixed, heated at 85 °C for 2 min, snap-chilled on ice, and RT premix was added. The 10 μL RT mixture was incubated at 50 °C for 30 min, 85 °C for 5 min, cooled to r.t., and diluted 10-fold with 90 μL of water. Following reverse transcription, quadruplicate measurements of 2 μL of cDNA were made in 10 μL final reaction volumes by qPCR in a 384-well optical PCR plate using a 7900 HT PCR instrument (Applied Biosystems, MA, USA). SYBR green PCR mix contained 5 μL of 2× SYBR green PCR Master Mix (Applied Biosystems, MA, USA), 1.4 μL of water, 0.8 μL of 10 μM universal primer, 0.8 μL of 10 μM LNA-R primer, and 2 μL of sample. The primer sequences are shown in Supplementary Table 5.
In vitro T cell uptake
Spleen and KPC-C2 tumor tissues were harvested from C57BL/6 mice, followed by the preparation of single-cell suspensions. CD4+ and CD8+ T cells were isolated using CD4/CD8 MicroBeads (Miltenyi Biotec, North Rhine-Westphalia, Germany). Cells were cultured in RPMI-1640 medium and stimulated by anti-CD3 (Invitrogen, NY, USA) and anti-CD28 (Invitrogen, NY, USA). Subsequently, the cells were treated with PBS, free siSPB9-cy5.5, POEM/siSPB9-cy5.5, or Lipofectamine RNAiMAX/siSPB9-cy5.5 as a control for 4 h. Cellular uptake of siRNA was examined by flow cytometry.
Analysis of in vivo cellular uptake
KPC-C2 tumor-bearing mice were treated with POEM/siSPB9-cy5.5 NPs intravenously. Tumors were collected and prepared into single cell suspensions, which were then stained for antibody (PE-Cyanine7 anti-mouse CD45, BUV737 anti-mouse CD4, BV480 anti-mouse CD8, eF405 anti-mouse CD31, FITC anti-mouse CD140a, BV711 anti-mouse Gr1, and PE anti-mouse CD11b; dilution: 1/200 for antibody) for flow cytometry analysis68.
In vivo gene knockdown
POEM NPs loaded with luciferase siRNA (siLuc) or control siRNA (siCT) were intravenously injected into KPC-C2-Luc tumor-bearing mice at a dose of 1 mg/kg. The efficiency of gene knockdown was measured three times by whole body bioluminescence imaging on the 2nd day following the 1st, 2nd, and 3rd injection of the NPs once every 3 days, respectively. The exposure time was set at 60 s for every experiment. Mice were anesthetized according to protocol prior to imaging.
In vivo SPB9 knockdown
When the tumor volume reached ~50 mm3, KPC-C2 tumor-bearing mice were randomly assigned to different groups (n = 5). They were intravenously administered with PBS, PPO/PCytidine/siSPB9 (POC/siSPB9) NPs, POEM/siCT NPs, or POEM/siSPB9 NPs three times at an interval of 5 days (on day 5, 10, and 15). The doses for PPOGEM and siRNA were 50 mg/kg and 1 mg/kg, respectively. Twenty-four hours after the final injection, tumors were collected and subjected to qRT-PCR of SPB9 expression as detailed above.
Therapeutic treatment
Murine pancreatic cancer (KPC-C2 and KPC-C2 GEMR) models were established for in vivo antitumor efficacy study. When the tumor volume reached ~50 mm3, mice were treated with PBS, POC/siSPB9, POEM/siCT, and POEM/siSPB9 respectively (n = 5), every 3 days for a total of 5 times (PCytidine: 50 mg/kg; PPOGEM: 50 mg/kg; siSPB9: 1 mg/kg) through intravenous injection. Tumor volumes and mouse body weights were monitored every 3 days following the initiation of the treatment. The tumor volumes (V) were calculated by the formula: (Length × Width2)/2. After completing the in vivo experiment, tumor tissues and major organs were harvested for histochemical staining. In addition, blood sample was collected, and plasma was isolated after centrifugation at 21,000 × g for 10 min. Alanine aminotransferase (ALT), aspartate aminotransferase (AST), and creatinine levels in plasma were measured as indicators of hepatic and renal function.
For orthotopic pancreatic cancer model, the KPC-C2-Luc tumor-bearing mice were treated intravenously with PBS, POC/siSPB9, POEM/siCT, and POEM/siSPB9, respectively, at dosage described above. The treatments were conducted every 3 days for a total of five times. The whole-body tumor burden was monitored and quantified by measuring the luminous intensity using Living Imaging 4.1.0 software.
To test the therapeutic effect of the combinational therapy of POEM/siSPB9 NPs with anti-PD-1, the treatment was started when the KPC-C2 GEMR tumors reached ~50 mm3 in size. Mice were treated with anti-PD-1 (clone RMP1-14, Bio X Cell) alone, POEM/siSPB9 NPs alone, or the combination once every 5 days, three times. Anti-PD-1 was administered at 10 mg/kg, while POEM/siSPB9 NPs were given intravenously at dosage described above. Mice were followed until death or were killed if the tumor size reached 2000 mm3, the maximal tumor size permitted by the Animal Use and Care Administrative Advisory Committee at the University of Pittsburgh.
Histopathological analysis
Tumors and major organs, including heart, liver, spleen, lung, and kidney, were excised and fixed in 10% formalin at the end of the in vivo therapy study, followed by embedment in paraffin. The paraffin embedded samples were sectioned into slices at 4 μm using an HM 340E Rotary Microtome (Epredia). The tissue slices were then processed for hematoxylin and eosin staining (H&E) for histological evaluation according to published protocols69. The images were observed under a BZ-X710 Fluorescence Microscope (Keyence, Itasca, IL, USA).
Analysis of tumor-infiltrating immune cells
Flow cytometry was performed with LSRII (BD Biosciences) and Aurora (Cytek Biosciences) instruments and analyzed by FlowJo (BD Biosciences). Spleens and tumors were collected 1 day after the last treatment. Single-cell suspensions were prepared as previously described38. Briefly, tumors were dissected and transferred into RPMI-1640 medium. Tumors were disrupted mechanically using scissors, digested with a mixture of deoxyribonuclease I (0.3 mg/mL, Sigma-Aldrich) and TL Liberase (0.25 mg/mL, Roche) in serum-free RPMI-1640 at 37 °C for 30 min, and dispersed through a 40 μm cell strainer (BD Biosciences). After red blood cell lysis, live/dead cell discrimination was performed using a Zombie NIR Fixable Viability Kit (BioLegend, dilution: 1/1000) at 4 °C for 30 min in PBS. Surface staining was performed at 4 °C for 30 min in FACS staining buffer (1× PBS/5% FBS/0.5% sodium azide) containing designated antibody cocktails (PerCP anti-mouse CD45 antibody, Brilliant Violet 737 anti-mouse CD4 antibody, Brilliant Violet 480 anti-mouse CD8 antibody, Brilliant Violet 615 anti-mouse PD-1 antibody, APC anti-mouse CD11b antibody, Brilliant Violet 510 anti-mouse Gr-1 antibody, APC/Cyanine7 anti-mouse F4/80 antibody, Pacific Blue anti-mouse MHC II antibody, Brilliant Violet 737 anti-mouse CD80 antibody, PE anti-mouse CD163 antibody, FITC anti-mouse CD69; dilution: 1/200 for all antibodies). Cells were fixed and permeabilized using the BD Cytofix/Cytoperm kit, following the manufacturer’s instructions. For intracellular cytokine staining (PE-Cyanine7 anti-mouse IFN-γ antibody and eF450 anti-mouse GzmB antibody; dilution: 1/200 for antibody), cells were stimulated with phorbol 12-myristate-13-acetate (100 ng/mL) and ionomycin (500 ng/mL) for 6 h in the presence of Monensin. Cells were fixed/permeabilized using the BD Cytofix/Cytoperm kit before cell staining.
Statistics and reproducibility
Statistical analysis was performed with two-tailed Student’s t-test for comparison between two groups, one-way analysis of variance (ANOVA) for comparison between multiple groups, and log-rank (Mantel–Cox) test for survival analysis as indicated in figure legend. Results were considered statistically significant if P < 0.05. Prism 10.1.0 (GraphPad Software) was used for data analysis and graph plotting. All experiments were repeated at least three times with similar results. No data were excluded from the analyses.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data from the TCGA can be accessed through the GDC data portal (https://portal.gdc.cancer.gov/). The bulk messenger RNA-seq data mapped to the mouse genome (GRCm38: https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.20/) are available in the NCBI Gene Expression Omnibus (GEO) under accession number GSE292903. NMR data are provided in Supplementary Fig. 7-10. All data generated or analyzed during this study are included in this Article, Supplementary Information or Source Data file. Source data are provided with this paper.
References
Hallqvist, A., Rohlin, A. & Raghavan, S. Immune checkpoint blockade and biomarkers of clinical response in non-small cell lung cancer. Scand. J. Immunol. 92, e12980 (2020).
Roper, N. et al. Notch signaling and efficacy of PD-1/PD-L1 blockade in relapsed small cell lung cancer. Nat. Commun. 12, 3880 (2021).
Lauss, M. et al. Molecular patterns of resistance to immune checkpoint blockade in melanoma. Nat. Commun. 15, 3075 (2024).
Zhou, X. et al. Mechanisms of tumor resistance to immune checkpoint blockade and combination strategies to overcome resistance. Front. Immunol. 13, 915094 (2022).
Afonina, I. S., Cullen, S. P. & Martin, S. J. Cytotoxic and non-cytotoxic roles of the CTL/NK protease granzyme B. Immunol. Rev. 235, 105–116 (2010).
Boivin, W. A., Cooper, D. M., Hiebert, P. R. & Granville, D. J. Intracellular versus extracellular granzyme B in immunity and disease: challenging the dogma. Lab. Investig. 89, 1195–1220 (2009).
Ibanez-Molero, S. et al. SERPINB9 is commonly amplified and high expression in cancer cells correlates with poor immune checkpoint blockade response. Oncoimmunology 11, 2139074 (2022).
Sprecher, C. A. et al. Molecular cloning, expression, and partial characterization of two novel members of the ovalbumin family of serine proteinase inhibitors. J. Biol. Chem. 270, 29854–29861 (1995).
Law, R. H. et al. An overview of the serpin superfamily. Genome Biol. 7, 216 (2006).
Huang, H., Mu, Y. & Li, S. The biological function of Serpinb9 and Serpinb9-based therapy. Front. Immunol. 15, 1422113 (2024).
Zhang, M. et al. Serine protease inhibitor 6 protects cytotoxic T cells from self-inflicted injury by ensuring the integrity of cytotoxic granules. Immunity 24, 451–461 (2006).
Hirst, C. E. et al. The intracellular granzyme B inhibitor, proteinase inhibitor 9, is up-regulated during accessory cell maturation and effector cell degranulation, and its overexpression enhances CTL potency. J. Immunol. 170, 805–815 (2003).
Bladergroen, B. A. et al. The granzyme B inhibitor, protease inhibitor 9, is mainly expressed by dendritic cells and at immune-privileged sites. J. Immunol. 166, 3218–3225 (2001).
Zhang, M. et al. Serine protease inhibitor 6-deficient mice have increased neutrophil immunity to Pseudomonas aeruginosa. J. Immunol. 179, 4390–4396 (2007).
Medema, J. P. et al. Blockade of the granzyme B/perforin pathway through overexpression of the serine protease inhibitor PI-9/SPI-6 constitutes a mechanism for immune escape by tumors. Proc. Natl. Acad. Sci. USA 98, 11515–11520 (2001).
Jiang, L. et al. Direct tumor killing and immunotherapy through anti-SerpinB9 therapy. Cell 183, 1219–1233.e1218 (2020).
Jiang, P. et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 24, 1550–1558 (2018).
Bozorgui, B., Kong, E. K., Luna, A. & Korkut, A. Mapping the functional interactions at the tumor-immune checkpoint interface. Commun. Biol. 6, 462 (2023).
van Houdt, I. S. et al. Expression of the apoptosis inhibitor protease inhibitor 9 predicts clinical outcome in vaccinated patients with stage III and IV melanoma. Clin. Cancer Res. 11, 6400–6407 (2005).
Zhang, X. et al. Low-dose gemcitabine treatment enhances immunogenicity and natural killer cell-driven tumor immunity in lung cancer. Front. Immunol. 11, 331 (2020).
Rohila, D. et al. Syk inhibition reprograms tumor-associated macrophages and overcomes gemcitabine-induced immunosuppression in pancreatic ductal adenocarcinoma. Cancer Res. 83, 2675–2689 (2023).
Li, J. et al. Tumor cell-intrinsic factors underlie heterogeneity of immune cell infiltration and response to immunotherapy. Immunity 49, 178–193.e177 (2018).
Palam, L. R., Gore, J., Craven, K. E., Wilson, J. L. & Korc, M. Integrated stress response is critical for gemcitabine resistance in pancreatic ductal adenocarcinoma. Cell Death Dis. 6, e1913 (2015).
Kaiserman, D. et al. Comparison of human chromosome 6p25 with mouse chromosome 13 reveals a greatly expanded ov-serpin gene repertoire in the mouse. Genomics 79, 349–362 (2002).
Dervovic, D. et al. In vivo CRISPR screens reveal Serpinb9 and Adam2 as regulators of immune therapy response in lung cancer. Nat. Commun. 14, 3150 (2023).
Balagurunathan, Y. et al. Gene expression profiling-based identification of cell-surface targets for developing multimeric ligands in pancreatic cancer. Mol. Cancer Ther. 7, 3071–3080 (2008).
Zhao, J., Li, X., Guo, M., Yu, J. & Yan, C. The common stress responsive transcription factor ATF3 binds genomic sites enriched with p300 and H3K27ac for transcriptional regulation. BMC Genomics 17, 335 (2016).
Ku, H. C. & Cheng, C. F. Master regulator activating transcription factor 3 (ATF3) in metabolic homeostasis and cancer. Front. Endocrinol. 11, 556 (2020).
Li, X. et al. ATF3 promotes the serine synthesis pathway and tumor growth under dietary serine restriction. Cell Rep. 36, 109706 (2021).
Bladergroen, B. A. et al. Expression of the granzyme B inhibitor, protease inhibitor 9, by tumor cells in patients with non-Hodgkin and Hodgkin lymphoma: a novel protective mechanism for tumor cells to circumvent the immune system? Blood 99, 232–237 (2002).
Li, Y. et al. Multifunctional oncolytic nanoparticles deliver self-replicating IL-12 RNA to eliminate established tumors and prime systemic immunity. Nat. Cancer 1, 882–893 (2020).
Rong, Y. et al. Engineered extracellular vesicles for delivery of siRNA promoting targeted repair of traumatic spinal cord injury. Bioact. Mater. 23, 328–342 (2023).
Luo, Z., Chen, C. Y. & Li, S. Improving tumor targeting and penetration for nanoparticle-mediated cancer therapy. Small Methods e2401860 (2025). https://doi.org/10.1002/smtd.202401860. Online ahead of print.
Akinc, A. et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 14, 1084–1087 (2019).
Jadhav, V., Vaishnaw, A., Fitzgerald, K. & Maier, M. A. RNA interference in the era of nucleic acid therapeutics. Nat. Biotechnol. 42, 394–405 (2024).
Dilliard, S. A. & Siegwart, D. J. Passive, active and endogenous organ-targeted lipid and polymer nanoparticles for delivery of genetic drugs. Nat. Rev. Mater. 8, 282–300 (2023).
Beach, M. A. et al. Polymeric nanoparticles for drug delivery. Chem. Rev. 124, 5505–5616 (2024).
Chen, Y. et al. Targeting Xkr8 via nanoparticle-mediated in situ co-delivery of siRNA and chemotherapy drugs for cancer immunochemotherapy. Nat. Nanotechnol. 18, 193–204 (2023).
Wan, Z. et al. Overcoming pancreatic cancer immune resistance by codelivery of CCR2 antagonist using a STING-activating gemcitabine-based nanocarrier. Mater. Today 62, 33–50 (2023).
Gujrati, M. et al. Multifunctional cationic lipid-based nanoparticles facilitate endosomal escape and reduction-triggered cytosolic siRNA release. Mol. Pharm. 11, 2734–2744 (2014).
Xu, J. et al. Creatine based polymer for codelivery of bioengineered MicroRNA and chemodrugs against breast cancer lung metastasis. Biomaterials 210, 25–40 (2019).
Xiao, K. et al. The effect of surface charge on in vivo biodistribution of PEG-oligocholic acid based micellar nanoparticles. Biomaterials 32, 3435–3446 (2011).
Ding, Y., Dai, Y., Wu, M. & Li, L. Glutathione-mediated nanomedicines for cancer diagnosis and therapy. Chem. Eng. J. 426, 128880 (2021).
Zhang, G. & Du, Y. N. Orthotopic pancreatic tumor mouse models of liver metastasis. Methods Mol. Biol. 1882, 309–320 (2019).
Wang, H. & Guo, P. Radiolabeled RNA nanoparticles for highly specific targeting and efficient tumor accumulation with favorable in vivo biodistribution. Mol. Pharm. 18, 2924–2934 (2021).
Sun, Y., Zhou, Y., Rehman, M., Wang, Y. F. & Guo, S. Protein corona of nanoparticles: isolation and analysis. Chem. Bio. Eng. 1, 757–772 (2024).
Luo, Z. et al. In situ formation of fibronectin-enriched protein corona on epigenetic nanocarrier for enhanced synthetic lethal therapy. Adv. Sci. 11, e2307940 (2024).
Cai, X. et al. Inhibition of the SLC35B2-TPST2 axis of tyrosine sulfation attenuates the growth and metastasis of pancreatic ductal adenocarcinom. Cell Mol. Gastroenterol. Hepatol. 16, 473–495 (2023).
Zhou, X. et al. Persister cell phenotypes contribute to poor patient outcomes after neoadjuvant chemotherapy in PDAC. Nat. Cancer 4, 1362–1381 (2023).
Parvez, A. et al. PD-1 and PD-L1: architects of immune symphony and immunotherapy breakthroughs in cancer treatment. Front. Immunol. 14, 1296341 (2023).
Huang, T. et al. Immunogenic cell death effects induced by doxorubicin improved chemo-immunotherapy via restoration of granzyme B activity. Nano Res. 16, 13250–13258 (2023).
Olden, B. R., Cheng, E., Cheng, Y. & Pun, S. H. Identifying key barriers in cationic polymer gene delivery to human T cells. Biomater. Sci. 7, 789–797 (2019).
Est-Witte, S. E., Livingston, N. K., Omotoso, M. O., Green, J. J. & Schneck, J. P. Nanoparticles for generating antigen-specific T cells for immunotherapy. Semin Immunol. 56, 101541 (2021).
Li, X. et al. Transvascular transport of nanocarriers for tumor delivery. Nat. Commun. 15, 8172 (2024).
Shah, A. N. et al. Development and characterization of gemcitabine-resistant pancreatic tumor cells. Ann. Surg. Oncol. 14, 3629–3637 (2007).
Brennan, T. V., Lin, L., Huang, X. & Yang, Y. Generation of luciferase-expressing tumor cell lines. Bio. Protoc. 8, e2817 (2018).
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667 (2016).
Wang, Y. et al. Systematic identification of non-coding pharmacogenomic landscape in cancer. Nat. Commun. 9, 1–15 (2018).
Wan, Z. et al. Targeting metabotropic glutamate receptor 4 for cancer immunotherapy. Sci. Adv. 7, eabj4226 (2021).
Yuan, F. et al. Mirovascular permeability and interstitial penetration of sterically stabilized (stealth) liposomes in a human tumor xenograft. Cancer Res. 54, 3352–3356 (1994).
Luo, Z. et al. Inhibition of iRhom1 by CD44-targeting nanocarrier for improved cancer immunochemotherapy. Nat. Commun. 15, 255 (2024).
Wang, J., Wang, W., Kollman, P. A. & Case, D. A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph Model. 25, 247–260 (2006).
Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A. & Case, D. A. Development and testing of a general amber force field. J. Comput Chem. 25, 1157–1174 (2004).
He, X., Man, V. H., Yang, W., Lee, T. S. & Wang, J. A fast and high-quality charge model for the next generation general AMBER force field. J. Chem. Phys. 153, 114502 (2020).
Salomon-Ferrer, R., Gotz, A. W., Poole, D., Le Grand, S. & Walker, R. C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 9, 3878–3888 (2013).
Gotz, A. W. et al. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. J. Chem. Theory Comput 8, 1542–1555 (2012).
Sun, J. et al. High loading of hydrophobic and hydrophilic agents via small immunostimulatory carrier for enhanced tumor penetration and combinational therapy. Theranostics 10, 1136–1150 (2020).
Chen, Y. et al. An immunostimulatory dual-functional nanocarrier that improves cancer immunochemotherapy. Nat. Commun. 7, 13443 (2016).
Huang, H. et al. A novel immunochemotherapy based on targeting of cyclooxygenase and induction of immunogenic cell death. Biomaterials 270, 120708 (2021).
Acknowledgements
This work was supported by the fund from National Institute of Health grants R01CA219399, R01CA223788, RO1CA278608, RO1CA270623, RO1CA295774 (to S.L.), R01GM147673 (to J.W.) and The David and Betty Brenneman Scholar Fund (to S.L.). This research used the Olympus FV3000 Confocal Microscope, which is supported by 1S10OD030254-01A1 (to RBG). The Pittsburgh Center for CryoEM (RRID:SCR_025216) used for data collection in this project was supported, in part, by the University of Pittsburgh, the School of Medicine, and the Department of Structural Biology.
Author information
Authors and Affiliations
Contributions
Conceptualization: H.H., J.S., and S.L., Methodology: H.H., Y.M., Y.H., B.J., Y.W., C.Y.C., Y.C., Z.L., S.H.L., Z.Z., L.W., J.F.C., and D.Y., Investigation: H.H., Y.M., Y.H., and B.J., Visualization: H.H., Y.M., Y.H., B.J., Y.W., C.Y.C., Y.C., Z.L., S.H.L., Z.Z., L.W., J.F.C., and D.Y., Funding acquisition: J.W., and S.L., Project administration: H.H., J.W., J.S., and S.L., Supervision: J.W., J.S., and S.L., Writing – original draft: H.H., Y.M., B.J., Y.W., S.H.L., J.W., J.S., and S.L., Writing—review and editing: H.H., Y.M., B.J., S.H.L., J.W., J.S., and S.L.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Philippe Soubeyran, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Huang, H., Mu, Y., Huang, Y. et al. Rational development of gemcitabine-based nanoplatform for targeting SERPINB9/Granzyme B axis to overcome chemo-immune-resistance. Nat Commun 16, 4176 (2025). https://doi.org/10.1038/s41467-025-59490-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-59490-y