Abstract
Haptophytes are unicellular algae that produce 30 to 50% of biomass in oceans. Among haptophytes, a subset named coccolithophores is characterized by calcified scales. Despite the importance of coccolithophores in global carbon fixation and CaCO3 production, their energy conversion system is still poorly known. Here we report a cryo-electron microscopic structure of photosystem II (PSII)-fucoxanthin chlorophyll c-binding protein (FCPII) supercomplex from Chyrostila roscoffensis, a representative of coccolithophores. This complex has two sets of six dimeric and monomeric FCPIIs, with distinct orientations. Interfaces of both FCPII/FCPII and FCPII/core differ from previously reported. We also determine the sequence of Psb36, a subunit previously found in diatoms and red algae. The principal excitation energy transfer (EET) pathways involve mainly 5 FCPIIs, where one FCPII monomer mediates EET to CP47. Our findings provide a solid structural basis for EET and energy dissipation pathways occurring in coccolithophores.
Similar content being viewed by others
Introduction
Photosynthesis converts light energy into chemical energy and split water to generate molecular oxygen; both products are indispensable for sustaining almost all life forms on the earth. This reaction occurs in the thylakoid membranes of cyanobacteria, eukaryotic algae and higher plant cells, and involves several protein complexes and co-factors, including most importantly the photosystem I and photosystem II (PSII) machineries. PSII carries one of the early steps of photosynthesis by splitting two water molecules into one dioxygen and four protons1,2. To perform this reaction, a couple of PSII chlorophyll (Chl) a pigments (P680) receive light from light-harvesting complexes (LHCII) that surround the PSII core. LHCII subunits bind various pigments, which gather light energy and transmit it to the PSII core3,4,5. Although the structures of LHCIIs are similar among most plants, their organization around the PSII core, as well as their number and the pigments they bind, largely depend on the species. In recent years, the structure of PSII-LHCII supercomplexes from many species have been elucidated by cryo-electron microscopy (cryo-EM). Thus we now have a good understanding of how light energy is transferred from the LHCII subunits to the PSII core in plants, cyanobacteria and green algae6,7,8. Moreover, the structures of PSII-LHCIIs from diatoms (containing fucoxanthin chlorophyll c-binding proteins, FCPIIs) and cryptophytes (containing alloxanthin chlorophyll a/c-binding proteins, ACPIIs) have been solved in the last few years9,10,11,12,13,14,15, which showed the association of the PSII core with a different number of LHCII (FCPII or ACPII) subunits in different organisms, and different energy transfer pathways from the LHCII subunits to the PSII core. However, no structures of PSII-LHCII supercomplex from other species such as haptophytes have been reported yet. Such structures are needed to improve our knowledge of energy conversion and evolutionary processes occurring in photosynthetic organisms. Here, we focus on an important group of unicellular algae haptophytes.
Haptophytes are an important group of marine unicellular algae, and produce 30 to 50% of the biomass in oceans. Haptophytes originate from endosymbiosis of a red alga in the red lineage photosynthetic organisms, thus their plastid membranes have four layers16. They are characterized by a specific appendix called haptonema for the movement of the cells, which differs from the flagellum in the arrangement of microtubules and in its usage17. Among haptophytes, a subgroup named coccolithophores are particularly of interest, as they are the most calcifying organisms on the earth18. They are characterized by an exoskeleton of calcareous plates called coccoliths, which eventually fall into the deep ocean and constitute an abundant source of microfossils19,20. Coccolithophores fix carbon at ocean surfaces through photosynthesis, and release coccoliths in depths. This phenomenon also influences both ocean acidity at the surface, as well as the air/water CO2 exchange18. Overall, coccolithophores are responsible for 10% of global carbon fixation, and up to 50% of CaCO3 production in the ocean21,22. Thus, they play a major role in the marine carbon cycle, and constitute species of interest for research on climate issues. Despite the importance of coccolithophores, there is almost no description of light capturing systems carried out by photosystems in this group of haptophytes yet.
Here we solved the structure of a PSII-FCPII supercomplex purified from Chrysotila roscoffensis, a representative of coccolithophores, by cryo-EM at a resolution of 2.2 Å. Our results revealed an antenna organization of six antennas surrounding one monomeric PSII core that are exclusively composed of monomeric and dimeric FCPII subunits. Five of these subunits constitute the main energy harvesting center that is associated with the CP47 subunit. In this center, one of the monomeric FCPIIs constitutes the hub of energy transfer leading to the core, which locates at the interface between PsbH, CP47 and a newly sequenced PSII core subunit Psb36. Our findings reveal the unique pigment arrangement in this supercomplex, which provides a detailed molecular view of the energy transfer pathways in haptophytes and may help to decipher the light energy transfer and dissipation mechanisms in PSII of coccolithophores.
Results and discussion
Characterization and overall architecture of PSII-FCPII
The PSII-FCPII supercomplex was extracted from C. roscoffensis cells. Before harvesting the cells, light was turned off to increase the number of bounded light-harvesting complexes to the photosystems (see Methods). Thylakoid membranes were solubilized with 1.5% n-dodecyl-α-D-maltoside (α-DDM) and PSII-FCPII was purified by a sucrose density gradient centrifugation (Fig. 1a). SDS-PAGE showed a migration pattern typical of PSII, with bands corresponding to the main core subunits and additional antenna bands which were identified by N-terminal sequencing (Fig. 1b). Clear native (CN)-PAGE analysis showed two major pigment-containing complexes (Fig. 1c), which were identified to correspond to PSII dimers with 12 FCPIIs and 6 FCPIIs, respectively. The absorption spectrum showed one peak at 679 nm in the Qy absorption region (Fig. 1d). As typical PSII core has an absorption peak at around 674 nm, the red-shifted absorption wavelength reflects the association of antenna subunits. The absorption at the 450–550 region is due to the presence of carotenoids. The low-temperature fluorescence emission spectrum shows a peak at 680 nm (Fig. 1e), identical to the PSII-FCPII from diatoms described previously13. Notably, the fluorescence emission after 700 nm is negligible, indicating the absence of PSI contamination in the sample.
a Sucrose density gradient profile, with the lowest brown band corresponding to the PSII-FCPII supercomplex. b SDS-PAGE analysis of PSII-FCPII. Bands were identified by comparison with known PSII core complexes, and the FCPII bands were identified by N-terminal amino acid sequencing. This SDS-PAGE pattern was reproduced more than 3 times with independent samples. c Native PAGE profile of PSII-FCPII, with two main bands corresponding to PSII-12 FCPII and PSII-6 FCPII supercomplexes, respectively, which co-exist in the sample. After migration, the gel was stained with Coomassie blue to reveal the molecular markers. This migration pattern was obtained with 3 independent purifications. d Absorption spectrum of PSII-FCPII. e Low-temperature (77 K) fluorescence emission spectrum of PSII-FCPII. Uncropped photos and raw data of all these experiments are provided as a Source Data file.
The pigment composition of the purified PSII-FCPII was analyzed by HPLC, which showed the presence of Chl a, fucoxanthin (Fx), diadinoxanthin (Ddx) and Chl c (Supplementary Fig. 1). The images of this PSII-FCPII supercomplex were collected by cryo-EM, and analyzed using the cryoSPARC software (Supplementary Fig. 2). After a few rounds of 2D classification and particle selection, the results showed that the images contained two types of particles, with 52.8% of the particles corresponding to a PSII-12 FCPII supercomplex and 28.9% to a PSII-6 FCPII supercomplex. Further refinement resulted in two maps with 2.2 Å and 2.9 Å resolutions, respectively (Supplementary Figs. 2 and 3 and Supplementary Table 1). To build the structure of the PSII core, the genome of C. roscoffensis was sequenced using a RNAseq approach. Then, we built a local BLAST+ database from the sequenced fragments and the ORFs detected23. Using tblastn and blastp algorithms, all sequences of the PSII core subunits present in the density map were identified. For the FCPII sequences, we relied on both the accuracy of the electron density map and blast results of high similarity using cryptophyte and diatom FCPII proteins as queries13,14.
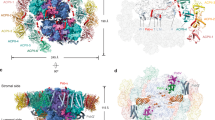
The molecular weight of the PSII-12 FCPII supercomplex identified by cryo-EM is 746 kDa, and its dimensions are 264 × 187 × 91 Å3. For the PSII-6 FCPII complex, these measurements are 614 kDa and 180 × 187 × 91 Å3. The size of PSII-6 FCPII only differ by length, but not by depth or width. In both complexes, the PSII core includes 14 of the canonical PSII subunits (including PsbA, PsbB, PsbC, PsbD, PsbE, PsbF, PsbH, PsbI, PsbK, PsbL, PsbM, PsbT, PsbW, and PsbX) (Fig. 2). In addition, we found a subunit previously named as PsbW in a red algal PSII24, or various names in several diatoms (Psb34 in a centric diatom12, Unknown0 in Chaetoceros gracilis (neogracilis)10, and Unknown1 in Thalassiosira pseudonana13), all with unknown sequences. This subunit is located at the interface of PSII monomer and FCPII proteins. Using cryo-EM density map and RNA sequencing results, we obtained its sequence and named it Psb36. Besides, PSII-FCPII is lacking all its canonical soluble subunits as well as the Mn4CaO5 cluster. This observation is somewhat contradictory to the results of SDS-PAGE gel which showed the presence of a band at around 33 kDa that corresponds to the PsbO subunit, suggesting that PsbO is present in the purified PSII-FCPII but lost upon grid preparation.
One set of PSII monomer–6 FCPII is shown in colors, whereas the other one is represented in gray. PSII-12 FCPII is made of two sets of 6 antennas surrounding both sides of PSII core. These include two peripheral FCPII heterodimers (FCPII-1a/FCPII-4 and FCPII-1b/FCPII-5, circled with dashed lines) and two inner FCPII monomers (FCPII-2 and FCPII-3). Phytol chains of Chls are hidden. Except for D1, CP47, CP43, and D2, all the Psb subunits are designated as a one letter identifier. The newly sequenced subunit Psb36 stands at the interface between the two monomers, close to the FCPII-2 subunit. The interface between the two PSII monomers is shown as a dashed line.
The 12 FCPII subunits are organized as two symmetric sets, with each consisting of 6 FCPII surrounding each PSII monomer. They are named FCPII-1 to FCPII-5, with FCPII-1 having two subunits FCPII-1a and FCPII-1b due to their similar structures and failure of distinguishing them from the sequences (Fig. 2). Among them, 5 FCPII are associated at the CP47/PsbH/Psb36 side, whereas 1 FCPII (FCPII-3) is associated in the opposite, CP43 side, suggesting that the CP47 side constitutes the main interface with FCPII subunits in this species. FCPII-2 and FCPII-3 are associated with the PSII core subunits as monomers, whereas FCPII-1a and FCPII-4, FCPII-1b and FCPII-5 form two heterodimers and are associated at the out periphery of FCPII-2 as well as FCPII-3 of the adjacent monomer. These heterodimers thus do not interact with the PSII core directly. This antenna organization is different from the diatom and cryptophyte PSII-LHCII supercomplexes described previously (Supplementary Fig. 4)9,10,11,12,13,14. Notably, FCPIIs in PSII-FCPII of C. roscoffensis are not arranged in an antenna belt nor in FCPII trimer or tetramer, which are seen in other species. FCPII-1a, FCPII-2, FCPII-3 and FCPII-5 are located in an area similar to the diatom C. neogracilis FCPIIs9,10, but their interaction interfaces are totally different (Supplementary Figs. 4 and 5). Furthermore, FCPII-2 and FCPII-3 are also facing the PSII core with non-canonical interfaces (Supplementary Fig. 5) and FCPII-4 is located in a region where no corresponding antenna is found in the structures of diatom and cryptophyte PSII (Supplementary Fig. 4). The main interaction surfaces involving FCPII subunits and PSII core within the PSII-FCPII supercomplex were analyzed using the PISA software25 (Supplementary Table 2). Apart from the dimeric interactions, the analysis revealed that the widest interfaces involve FCPII-2 with PsbH, Psb36 and CP47, and FCPII-3 with CP43. FCPII-2 also connects with both FCPII-1a and FCPII-5. In addition, the PISA analysis predicted interfaces between heterodimers, which involve FCPII-1a and FCPII-1b.
In the case of PSII-6 FCPII, one set of 6 FCPII is lacking from one PSII monomer (Supplementary Figs. 2 and 3). Such organization may suggest that the 12 FCPII and 6 FCPII forms are adapted to different light conditions: under strong light conditions PSII may lose half of its antennas, as they are not required for light harvesting, whereas under weak light conditions, an additional set of 6 FCPII subunits became associated with the PSII core to increase the light harvesting efficiency. However, the cells of C. roscoffensis used in this study were grown under rather weak light intensity, so the exact reason for the loss of one set of the antenna is unclear at present. This second set of 6 FCPII may bind weakly to the PSII core, although their organization is largely the same as the first set of the antennas. The symmetry between the two sets of 6 FCPII indicates that the excitation energy transfer (EET) pathways are likely to be similar in both 12 and 6 FCPII complexes, with similar regions of the PSII core involved. Moreover, both sets of 6 FCPII interact with the core through an axis that is not parallel to the thylakoid membrane, in a similar way to the PSII-FCPII structure from the diatom reported previously9,10,11,12,13.
A subunit Psb36 shared with red algae and diatoms
For the identification of Psb36, we first looked at the cryo-EM map and picked up several recognizable densities of sidechains, which include F10, Y13, I15, I17, and I18, constituting the sequence motif (FXXYXIXII) (Fig. 3a). We then searched this motif in our sequence database and found a 36 amino acid sequence that includes this motif (Fig. 3b). The entire density map of the chain was then fitted using the sequence that we identified. However, we could not see the N-terminal region in the cryo-EM density map, probably because of its high flexibility. We performed a BLAST analysis with the Psb36 sequence, which showed no similar proteins within the NCBI non-redundant protein sequence database. Nevertheless, 3 hypothetical proteins were found from the genome data of 2 haptophytes (Prymnesium Parvum and Emiliana Huxleyi) and 1 ochrophyta (Pelagophyceae sp. CCMP2097) (Fig. 3b). These 3 sequences are 91%, 83%, and 70% similar to the Psb36 sequence from C. roscoffensis, respectively.
a Map fits of the FXXYXIXII motif and the structure of the whole Psb36 chain. b Alignment between the Psb36 sequence from C. roscoffensis and the BLAST results obtained from the NCBI non-redundant protein sequence database. c Interactions between Psb36 with surrounding proteins, from its N-terminal part (upper panel) to its C-terminal part (lower panel). d Comparison of the Psb36 subunit secondary structure with previously published structures, using Matchmaker from UCSF ChimeraX.
In our structure, Psb36 consists of an N-terminal loop and one main helix at the center of the FCPII-2/PSII core interface. This N-terminal loop is located at the lumenal side and includes two hydrophobic residues involved in interactions with CP4 (Fig. 3c), as predicted by the PISA analysis (Supplementary Table 2). They are F10 which interacts with CP47-F162, and A7 which interacts with CP47-G89 at the PSII monomer interface. In addition, G11 is proximal to FCPII-2 (L222), with which it may interact weakly. Following this loop, the main helix of Psb36 contains a high amount of leucine and isoleucine residues (I15, I17, I18, L20, L22, I23, I27, L29, and L30), among which I23 and I27 interact with the phytol chain of Chl a from CP47 (a616), while L20 interacts with the phytol chain of CP47-a607 (Fig. 3c). The other side of the helix (T14 and L22) faces two Chl a from FCPII-2 (a303 and a304). Thus, in all directions, the side chains of the Psb36 helix are facing pigments either from the PSII core or FCPII-2. Following this helix, residue D35 of the C-terminal tail of Psb36 interacts with PsbH-R4. Thus, Psb36 may be important during PSII-core assembly by helping to position pigments at the PSII monomer interface, and it may also play a role in the positioning of CP47, PsbH and the FCPII-2 loops. However, its presence in this purified supercomplex and close interactions with Chls from FCPII-2 and CP47 indicate that its role may not be limited to the assembly, but also to facilitate energy transfer between FCPII-2 and CP47.
Psb36 has been identified in several structures of diatoms (named Psb3412, Unknown010 or Unknown113) and a red alga (named PsbW24) previously, although its sequence was not identified previously. Among all these structures, the helix and N-terminal loop are in similar positions (Fig. 3d). However, the helix of haptophyte is shifted to become closer to the core. The C-terminal loop of Psb36 has two conformations. One is shared by centric diatoms C. meneghinianus12 and T. pseudonana13, a red alga24, and the haptophyte reported in the present study. In this conformation, the loop is interacting with PsbH, and faces the antenna side. The other conformation is found only in the diatom C. neogracilis structure10,11, in which the loop is directed to the other PSII monomer. Thus, the role of the C-terminal loop of Psb36 may be different depending on the species.
FCPII subunit structures and pigment organization
To assess the homology of FCPII subunits among them, we performed a multiple sequence alignment analysis and visualized it with ESpript26,27 (Supplementary Fig. 6 and Supplementary Table 3). The results show two groups of FCPII: one consists of FCPII-1a/FCPII-1b/FCPII-4/FCPII-5, and the other one consists of FCPII-2/FCPII-3. As expected, these two groups are consistent with the FCPII positions in the structure. Four antennas are distant to the PSII core (FCPII-1a, FCPII-1b, FCPII-4 and FCPII-5), which belong to the first group, whereas the other two are closer and interact with the PSII core directly (FCPII-2 and FCPII-3), which belong to the second group. The first group are found exclusively in a dimeric state, whereas the latter group is made by two monomeric FCPIIs. In the first group of the antennas, the three sequences are highly similar. We found 93% identity between FCPII-4/FCPII-5 as well as between FCPII-1a/b/FCPII-4, and 90% between FCPII-1a/b/FCPII-5. In contrast, the antennas of the second group (FCPII-2 and FCPII-3) have only 46% identity between them, even though they share a C-terminal helix of high similarity. The identities between the first and second groups are even lower. However, unlike FCPII-3, FCPII-2 features a unique loop extension made of 16 residues (S167-L183) that contains two beta-strands (Y166-F169 and V175-V178) (Supplementary Fig. 7b, red arrows).
All FCPII subunits are made of three main transmembrane helices, A, B, and C (Fig. 4), similar to major LHCIIs from other organisms, with helices A and C crossed and interacting with each other, whereas helix B is shorter and more distant28. All these helices are connected by loops of various lengths with a conserved distance between helices. In the FCPII-1a/FCPII-4 heterodimer, the C-terminal loop of FCPII-1a is well defined in the density map, which indicates that its position is stabilized by interactions, especially with pigments. FCPII-1a contains 10 Chls a, 3 Fxs, 1 Ddx, and 2 Chls c (Supplementary Table 4). Several of them (a301, a302, a303, a304, a308) surround helix A, and thereby are embedded in the core of FCPII-1a (Fig. 4a). Among the three carotenoids close to helix A (Ddx311 and Fx313, Fx314), Ddx311 is parallel to it and connects with Chls a301, a302, and a308. On the other hand, Fx313 and Fx314 bridge inner pigments with outer ones, with Fx314 connecting Chls a304 to a307 through Chl a309, whereas Fx313 is located at the intersection of Chls a303, a304, and a306, and close to Chl c315 in its other extremity. Other pigments that are associated with helix C of FCPII-1a (Chls a305, a307, a309, Fx312, and Chl c316) face the FCPII-2 subunit, and are orientated toward the PSII core. The B helix constitutes the dimerization interface of FCPII-1a, and includes a pigment (Chl a303) at the center of the dimer. On the other hand, the FCPII-4 subunit is located far from the PSII core, and close to the FCPII-1b antenna from the other FCPII heterodimer. The secondary structures of FCPII-4 are almost identical to FCPII-1a, except for the N and C-terminal loops that we could not see clearly due to poor local resolution. FCPII-4 features 5 Chls a, 1 Fx and 4 Chls c (Supplementary Table 4), which is remarkably less than FCPII-1a. Notably, Chls a301 and a310 of FCPII-1a are lacking in FCPII-4. The presence of these pigments in FCPII-1a may be important for energy exchanges at the PSII core side. Alternatively, these 2 pigments may be detached from FCPII-4 during the purification process due to their more accessible location to the solvent. Considering its position in PSII-FCPII, the FCPII-1a/FCPII-4 heterodimer probably transfers energy to other antennas such as FCPII-2, and it is unlikely that FCPII-1a/FCPII-4 constitutes a direct EET pathway to the core.
Chl a, Chl c, Fx and Ddx are shown in green, yellow, red, and blue, respectively. a FCPII-1a/4 heterodimer (FCPII-1a in dark yellow, FCPII-4 in light blue). b FCPII-1b/5 heterodimer (FCPII-1b in dark pink, FCPII-5 in dark orange). c FCPII-2 monomer (dark blue). d FCPII-3 monomer (light green). Phytol chains of Chls are omitted.
The second heterodimer FCPII-1b/FCPII-5 is located at the very outside of the complex, where FCPII-1b faces FCPII-4 of the FCPII-1a/FCPII-4 heterodimer described above (Fig. 2). FCPII-1b and FCPII-1a have an overall very similar structure, as both proteins are translated from the same gene, except for the loop between helices A and B (Supplementary Fig. 7a). In the FCPII-1b structure, this loop occupies a density filled by a Chl a molecule found in FCPII-1a (a305); this Chl is therefore absent in FCPII-1b. The Fx associated with Chl a306 in FCPII-1a (Fx313) is also lacking in FCPII-1b (Fig. 4b). This could reflect a better propensity of FCPII-1a over FCPII-1b to gather light energy and/or mediating energy transfer between different subunits. All other pigments have the same locations and similar orientations as in FCPII-1a. As a result, FCPII-1b contains 8 Chls a, 2 Fxs, 1 Ddx, and 3 Chls c (Supplementary Table 4). FCPII-5, the other component of the FCPII-1b/FCPII-5 heterodimer, is close to the FCPII-2 monomer. FCPII-5 includes 9 Chls a, 2 Fxs, and 3 Chls c but no Ddx (Supplementary Table 4), which makes the pigment content in the FCPII-1b/FCPII-5 heterodimer more homogeneous than in the FCPII-1a/FCPII-4 heterodimer. Similar to the FCPII-1a/FCPII-4 heterodimer, the location of FCPII-1b/FCPII-5 suggests that it does not transfer energy directly to the PSII core. Instead, it may gather light energy and transmit it to the proximal antennas FCPII-1a and FCPII-2.
The 2 FCPII monomers, FCPII-2 and FCPII-3, are the closest antennas to the PSII core within the supercomplex (Fig. 2). FCPII-2 is located near CP47, PsbH and Psb36, and is surrounded by the two FCPII-1b/FCPII-5 and FCPII-1a/FCPII-4 heterodimers at its outside. Thus FCPII-2 may serve as the main hub of energy transfer by connecting the entire antenna system to the PSII core. Its secondary structure is the most distinguishable one of the FCPII subunits (Supplementary Fig. 7b, red arrows), as it features an extended N-terminal part that interacts with FCPII-1a. Furthermore, its BC loop is longer than other FCPIIs (46 residues long), and includes two beta-strands (Y166-F169 and V175-V178) that cover a pigment pocket between FCPII-1a, PsbH and CP47. The end of this loop is facing residues from helix A, thereby creating an intern interface that may stabilize the loop region (V69, I73 and P149). FCPII-2 binds 6 Chls a, 4 Chl c and especially 6 Fxs, which are more than any other FCPII subunits (Fig. 4c). The 3 Fxs that were not found in FCPII-1a (Fx308, Fx309 and Fx310) are found at the peripheral part of FCPII-2, facing PsbH, CP47, and FCPII-3 of the adjacent monomer. As carotenoids are able to dissipate energy29, FCPII-2 may endorse its role in either energy harvesting or energy dissipation in the PSII-FCPII supercomplex.
The FCPII-3 monomer occupies a characteristic position in the supercomplex, as it is associated with the PSII core at the opposite side of FCPII-2, and is the only antenna connected to the other PSII monomer. The BC loop of FCPII-3 tends to be orientated towards the inner part of its structure, which interacts with a Chl molecule located at the outer part of other FCPII subunits (Supplementary Fig. 7c). Notably, this Chl is embedded only in peripheral FCPIIs (FCPII-1a, FCPII-1b, and FCPII-5). FCPII-3 is the antenna that binds the least pigments among FCPII subunits (5 Chls a and 2 Chls c) (Fig. 4d). One explanation of this is the absence of any close antenna partner, which prevents antenna-antenna energy transfer. Interestingly, FCPII-3 has a unique orientation among the previously published structures from diatoms9,10,11,12,13 and cryptophyte14,15 (Supplementary Figs. 4 and 5b), in which the B helix of FCPII-3 is orientated towards CP43 (Fig. 2), whereas in other structures this helix usually faces the other antennas to form a tetramer or a trimer. This particular orientation prevents canonical interactions between FCPII-3 and a putative missing antenna subunit in the structure. Moreover, considering the low light conditions that we used during the growth (see Methods), it is unlikely that other FCPII subunits may have been lost during purification, suggesting that no additional FCPII subunits may bind to the CP43 side. Therefore, FCPII-3 may play roles in harvesting the light energy and transfer it to CP43 as well as to dissipate excess energy from the PSII core, thereby regulating the energy distribution within the PSII-FCPII supercomplex.
FCPII-FCPII interactions
Within the PSII-FCPII supercomplex, every FCPII subunit interacts with other FCPIIs, except for FCPII-3. Such contacts may be important for both complex stability and EET pathways. The FCPII-1a/FCPII-4 heterodimer relies on two sorts of interactions to connect with both FCPII monomers (Fig. 5a). First, the basis of the helix B of FCPII-1a (W122, L123, I126) interacts with helix B of FCPII-4 (W87, F88, V91). Second, the other half of both B helices (D133, Y134, W135 of FCPII-1a, and D98, Y99, W100 of FCPII-4) interacts with Chls a (a303 of FCPII-1a and a304 of FCPII-4), which probably stabilizes the dimerization interface. On the most external part of the PSII-FCPII supercomplex, the FCPII-1b/FCPII-5 heterodimer has a similar dimeric interface (Fig. 5b), where both helices B at the lumenal side (W122, L123, I126 of FCPII-1b and W87, F88, V91 of FCPII-5) are connected. Moreover, the top of B helices (D133, Y134, W135 of FCPII-1b and D98, Y99, W100 of FCPII-5) interposes two sides of two Chls (a306 of FCPII-1b and c311 of FCPII-5), thereby stabilizing the binding of these two Chls.
Global view of interactions is shown in the top-left corner. FCPII-1a, FCPII-2, FCPII-3, FCPII-4, FCPII-1b and FCPII-5 are shown in dark yellow, light blue, green, dark blue, dark pink and dark orange, respectively. a Dimeric interface between FCPII-1a and FCPII-4. b Dimeric interface between FCPII-1b and FCPII-5. c First interface between loops of FCPII-1a and FCPII-1b. d Second interface between loops of FCPII-1b and FCPII-4. e Pigment located below the FCPII-1a and FCPII-2 interface. f Interaction between a304 of FCPII-5 and both FCPII-5 and FCPII-2 antennas.
In addition to the dimeric interfaces, both heterodimers face each other through an extended interface (Fig. 5c, d). The FCPII-1a/FCPII-4 heterodimer connects to amino acid residues from FCPII-1b and FCPII-2, respectively. A loop of FCPII-1b (L54 and N59) may interact with the same loop of FCPII-1a (L54 and N59) (Fig. 5c). In these loops, several amino acid residues are facing a putative partner from the other FCPII subunit. The same FCPII-1b loop (P47 and F51) is close to another loop from FCPII-4 (F102 and F106) (Fig. 5d). This interface could stabilize proximal pigments (FCPII-1b/Chl a300 and FCPII-1b/Chl c313) and hence enhance the energy transfer between the two heterodimers.
As predicted by the PISA analysis (Supplementary Table 2), FCPII-2 is connected to both heterodimers via interfaces involving FCPII-1a and FCPII-5 (Fig. 5e, f). FCPII-2 interacts with FCPII-1a through its N-terminal region (FCPII-2-R11 and FCPII-1a-E46) (Fig. 5e). In the C helix of FCPII-1a, D151 is also bridging an N-terminal residue from FCPII-2 (K16). Both of these interactions allow FCPII-2 to cover 3 Chls a (a305, a309 of FCPII-1a and a306 of FCPII-2), which possibly help energy transfer between these pigments and pigments in the CP47 subunit. On the other hand, FCPII-5 does not interact with other FCPII subunits directly, but stabilizes one common Chl (FCPII-5-a304) together with FCPII-2 (Fig. 5f), where L103 and F106 of FCPII-5 and L131 and V134 of FCPII-2 are connected to FCPII-5-a304. This region may therefore be important in the energy transfer involving the FCPII-1b/FCPII-5 heterodimer and the FCPII-2 subunit.
FCPII-PSII core interactions
According to the PISA analysis (Supplementary Table 2), only FCPII-2 and FCPII-3 interact with the PSII core complex directly within the PSII-FCPII supercomplex. This highlights the importance of these two subunits and of the amino acid residues involved at the interfaces.
The N-terminal extension of FCPII-2 includes a long region (21 residues, from A20 to Y40) that interacts directly with PsbH (15 residues, from E9 to P23) (Fig. 6a-i), among which FCPII-2-A20 and I23 interact with PsbH-P23. Following this, the aromatic residue of FCPII-2-Y30 connects with the side chain of K20 from PsbH and near these two residues, CP47-R127 is bridged with FCPII-2-Y30, forming a FCPII-2/PsbH/CP47 interface (Fig. 6a-ii). Finally, some residues of FCPII-2 (P47, P48, L49) constitute an interface between PsbH (I10) and Psb36 (L29 and L30) (Fig. 6a-iii). This whole interaction involving the N-terminal part of FCPII-2 is located above several Chls and Fxs (a303, a304, c316, Fx308, Fx310, and Fx311 from FCPII-2), and may be an anchor point at the stromal side to bring pigments of FCPII-2 closer to the core, thereby facilitating energy transfer. On the lumenal side, a similar interaction exists between FCPII-2 and Psb36, where P221 and L222 of FCPII-2 are proximal to G11 and T14 residues of Psb36 (Fig. 6a-iv). Thus, it is likely that pigments located in between the lumenal and the stromal sides (a303, a304, c316, Fx308, Fx310, and Fx311 of FCPII-2) may become easier to release when one of the two anchors are open. Such a mechanism may function in the regulation process occurring under various light conditions.
a Global view of the FCPII-2–core interface, which involves PsbH, CP47 and Psb36. The FCPII-2/PsbH-CP47-Psb36 interface is detailed from i to iii, including interactions with CP47 in ii and Psb36 in iii. A last interface with Psb36 is shown in iv, which occurs at the lumenal side. FCPII-2, PsbH, CP47, and Psb36 are represented in light blue, purple, green and dark pink, respectively. b A global view of the FCPII-3–CP43 interface, where two helices tightly interact with each other (shown in i). CP43 is shown in grey, and FCPII-3 is shown in green.
FCPII-3 is associated with CP43 at the opposite side of the PSII monomer (Figs. 2 and 6b), and forms a helix-helix interface with its B helix and a helix from CP43, involving amino acid residues from the stromal side to the lumenal side. Many pairs of hydrophobic residues interact between FCPII-3 and CP43 (FCPII-3-S147 with CP43-T226, FCPII-3-I143 with CP43-W222 and CP43-I221, FCPII-3-I143, and A139 with CP43-V218, FCPII-3-A139 with CP43-F214, FCPII-3-A135, and A132 with CP43-W211) (Fig. 6b-i). These interactions may enhance the binding of FCPII-3 to the PSII core. However, as FCPII-3 only features a few pigments (5 Chls a and 2 Chls c) with no direct interactions with other FCPII subunits either in the same monomer or from the adjacent monomer in a PSII dimer, the role of light-harvesting and energy transfer from FCPII-3 to the PSII core is limited. As mentioned before, FCPII-3 may block potential interactions involving CP43 with other antennas through this region. In the FCPII-1a/FCPII-4 and FCPII-1b/FCPII-5 heterodimers, the B helix constitutes the main dimerization interface. The interactions of FCPII-3 with CP43 are similar to the interactions found between each FCPII monomer within the dimers. This argument supports the hypothesis that FCPII-3 is no longer able to interact with other putative missing antennas in the complex. Thus, it may bind to CP43 to limit the number of antenna complexes associated with the CP43 side, thereby preventing excessive light harvesting.
Energy transfer pathways
In total, the 12 FCPII subunits feature 86 Chls a, 28 Fxs, 36 Chls c, and 4 Ddxs. Within a set of 6 FCPIIs, the PSII core is able to gather energy from pigments of FCPII-2 and FCPII-3. The pigments from the other four peripheral antennas (FCPII-1a, FCPII-1b, FCPII-4, and FCPII-5) absorb light and transfer the energy to the PSII core mainly through FCPII-2 (Fig. 7).
The PSII core complex is colored in grey, whereas FCPII-1a, FCPII-2, FCPII-3, FCPII-4, FCPII-1b and FCPII-5 are shown as dark yellow, light blue, green, dark blue, dark pink and dark orange, respectively. Chls a and c are shown in green and yellow, respectively. Magenta arrows indicate stromal EET pathways, whereas red arrows stand for lumenal EET pathways. Every EET interface is detailed for the left set of 6-FCPII subunits, which also occurs in the symmetric 6-FCPII set in the right part. EET pathways are shown between FCPII-5 and FCPII-2 (a), FCPII-2 and CP47 (b), FCPII-1b and FCPII-4 (c), the FCPII-1a/FCPII-2/CP47 interface (d), FCPII-5 and FCPII-3 (e), and FCPII-3 and CP43 (f). Phytol chains of Chls are omitted.
FCPII-5 may transfer energy to FCPII-2 directly. At the FCPII-5/FCPII-2 interface, Chls a304, a306, and c313 of FCPII-5 are close to Chls a302 and c313 of FCPII-2 at both lumenal and stromal sides (Fig. 7a). This pathway may also be efficient for the energy collected via FCPII-1b within the FCPII-1b/FCPII-5 dimer. After collecting energy from FCPII-5, FCPII-2 may transfer energy to the pigments of CP47 at both lumenal and stromal sides through interactions between FCPII-2-Chl a303/a304 and CP47-Chl a616/a607 (Fig. 7b). Moreover, one intermediate pigment (Chl c316) located close to the stromal pigments (Chls a301 and a303) of FCPII-2 is close to CP47-Chl a607, suggesting that it may also mediate energy transfer from FCPII-2 to the PSII core. This is particularly probable if we consider the high energy transfer efficiency from Chl c to Chl a compared to the energy transfer efficiency from Chl a to Chl c30. Thus, FCPII-2 is involved in EET pathways leading to CP47 at both the stromal and lumenal sides.
Another way for FCPII-2 to gather light energy is to receive it from the FCPII-1a/FCPII-4 heterodimer. Within this heterodimer, one interface allows energy transfer between the two heterodimers (Fig. 7c). FCPII-1b features Chls a302, c313, and c315 that face FCPII-4 pigments (a302 and c306) at the stromal side, and Chls a305, a308, and a309 of FCPII-1b are close to Chls a300 and c309 pigments of FCPII-4 at the lumenal side. Eventually, the transferred energy can reach a last interface that involves FCPII-1a, CP47, and the unique BC loop and N-terminal extension of FCPII-2 (Fig. 7d). In this area, 3 pigments of FCPII-1a (Chls a305, a307, and a309) could form an EET pathway with Chls a602, a603, and a609 of CP47. One pigment of FCPII-2 (Chl a306) stands between FCPII-1a-Chl a305 and CP47-Chl a609 at the stromal side, with a close distance but an orientation facing the lumen. This particular pigment could redirect energy from the stromal side to the lumenal side before transferring it to the core.
One last possible EET pathway may occur through FCPII-3, because of its proximity with CP43. Considering the negligible energy transfers from Chl a to Chl c compared to the energy transfers from Chl c to Chl a30, we can hypothesize on the direction of the energy transfer in this region. The FCPII-1b/FCPII-5 heterodimer in one PSII monomer could receive energy from FCPII-3 (Chl c305) of the adjacent PSII monomer through Chl a305 of FCPII-5 at the lumenal side (Fig. 7e). Due to their relatively close distances, these two Chls may constitute the most probable EET pathway between FCPII-5 and FCPII-3, whereas two other pairs of stromal Chls (between Chls a307, c312 of FCPII-5 and Chls c306, a300 of FCPII-3) are at a longer distance and the energy transfer between them will have a lower efficiency. FCPII-3 may transfer light energy directly to CP43-Chl a507 via its Chl a302 pigment at the stromal side (Fig. 7f), as a close distance is found between these two Chls. In conclusion, FCPII-3 may transfer light energy to CP43 at the same monomer, and also redirect energy to FCPII-5 of the adjacent PSII monomer at the lumenal side efficiently. Alternatively, it could use two long-distance EET pathways at the stromal side (involving Chls c306 and a300) to equilibrate EET between CP47 and CP43. In any cases, considering the few pathways available and the absence of any direct interaction with proximal antennas, the FCPII-3/CP43 interface seems to be a less efficient EET pathway for light harvesting than the CP47 side. However, receiving the energy from the adjacent PSII monomer through FCPII-3 may provide some advantages for the formation of a PSII dimer.
In order to confirm these observations, we calculated the Förster resonance energy transfer (FRET) rate constants between adjacent Chls a (Supplementary Fig. 8 and Supplementary Table 5) according to a previously published calculation method31. These results offer a partial view of the EET pathways, as PSII-FCPII contains a lot of Chls c (36) and there is no calculation method available for the FRET rates between Chls a and c so far. Moreover, this calculation gives estimations of the FRET rates, as it does not take into account the contribution of electrostatic environment of Chls32 nor the generalized Föster and Redfield theories33. In spite of this, the calculations confirmed that the main FRET events occur through Chls a of antennas located at the CP47 side (FCPII-1a/4, FCPII-2 and FCPII-1b/5), whereas Chls a from FCPII-3 are isolated from other Chls a of FCPIIs. Notably, the calculation results highlight the close connection between Chls a of FCPII-2 (Chls a305, a307 and a309) with Chls a of CP47 (a602, a603, and a609) mentioned above, thereby showing a dense network at the FCPII-2/CP47 interface (Supplementary Fig. 8, Supplementary Table 5).
Taken together, these observations suggest that FCPII-2 likely serves as the major light-harvesting point for EET pathways, as it is involved in two energy transfer interfaces with CP47. It can receive energy from both FCPII heterodimers via both stromal and lumenal interfaces. It may also function as a main gate for energy dissipation, as it includes 6 Fx pigments (Supplementary Fig. 9) and these pigments may facilitate dissipation of excess energy when growing under strong light intensities.
In conclusion, a 2.2 Å resolution structure of a PSII-FCPII supercomplex from Chyrostila roscoffensis, a representative of coccolithophores among haptophytes, was revealed by cryo-EM in this study. The structure shows that PSII forms a dimer with each monomer containing 15 PSII core subunits and 6 FCPII subunits. We determined the first sequence of Psb36, a core subunit previously identified and modeled as a poly-A peptide in diatoms and red algal structures. This subunit is located at the interface between both PSII monomers (CP47 and PsbH) and antennas (FCPII-2), and may play a role in both stabilizing the PSII-FCPII and pigment organization as well as PSII core biogenesis. In contrast to the structures of diatoms and cryotophytes reported previously, FCPIIs in the C. roscoffensis PSII-FCPII supercomplex do not form tetramer or trimer, but instead forming two heterodimers (FCPII-1a/FCPII-4 and FCPII-1b/FCPII-5) in each PSII monomer, with the remaining FCPII subunits organized as monomers (FCPII-2 and FCPII-3). Notably, all FCPIIs subunits have orientations and positions different from other PSII-LHCII structures reported so far. Consequently, interaction interfaces among the FCPII subunits and between FCPIIs and the core subunits different from the previous studies are found. The pigment composition and positions revealed in the present study give rise to putative EET pathways leading to the core which were not found previously. The main EET pathways involve FCPII-2, which could receive energy gathered by proximal antennas of both heterodimers (FCPII-1a/FCPII-4 and FCPII-1b/FCPII-5) and transfer it to CP47. FCPII-3, the other FCPII monomer that closely interact with CP43 through a dimeric-like interface, contains only a limited number of pigments and has no apparent interactions with other FCPII subunits. Its interaction with CP43 may prevent binding of other FCPIIs in this region of PSII and limit its light harvesting capacity through the CP43 side, but FCPII-3 itself is an efficient energy transfer partner to CP43. These results provide a solid structural basis for the organization of PSII-LHCII supercomplexes and light harvesting mechanisms in the coccolithophores and haptophytes.
Methods
Cell culture and PSII-FCPII purification
Cells of C. roscoffensis were cultured in a F/2 medium made in sea water. The cultures were bubbled with air containing 3% CO2 at 20 °C, under continuous weak light at 4.60 μmol photons m−2s−1 using a B3-450W Treviewer for 20 days. Before harvesting, the light was turned off for the last 12 h of growth. After the cells reach an OD of 0.3–0.4 at 730 nm, they were collected by centrifugation (12,000 × g for 15 min at 4 °C). From here, each step was performed under minimal light and the sample was covered by aluminum. An aliquot of 4 g cells was resuspended in 50 mL of 20 mM HEPES (pH 7.0), 10 mM NaCl using a brush, and stored at −80 °C until further utilization.
Cells were thawed and disrupted with a French press at 900 psi (3 times for 50 mL), and centrifuged at 3000 × g for 10 min at 4 °C to remove unbroken cells. Thylakoids in the supernatants were collected by centrifuging at 90,000 × g for 40 min at 4 °C, and resuspended in 2–3 mL of a 10 mM HEPES (pH 7.5) buffer supplemented with 10 mM trypsin inhibitor, 10 mM PMSF and 10 mM leupeptin. The concentration of Chl a was determined in a 90% acetone solution as described34. The thylakoids were solubilized by 1.5% α-DDM at 0.5 mg Chl a per mL under gentle mixing by a vortex for a few seconds, and the non-solubilized membranes were removed by centrifugation at 40,000 × g for 15 min at 4 °C. The supernatants were subsequently loaded onto centrifugation tubes containing a 0.3 M–0.73 M (10%–25%) sucrose density gradient solution in 10 mM HEPES (pH 7.5) and 0.05% α-DDM, which were centrifuged at 230,000 × g for 17 h at 4 °C. The resulting tubes showed three major bands (Fig. 1a), with one brown band at 20–25% sucrose concentration containing the PSII-FCPII supercomplex. This band was collected and concentrated using a polyether sulfone membrane concentrator (Pierce) with a 100 kDa molecular weight cutoff, with centrifugation cycles performed at 5000 × g for 3 min at 4 °C. After few rounds of concentration, sucrose was diluted with 10 mM HEPES (pH 7.5), 50 mM NaCl, 0.05% α-DDM and removed by further cycles of centrifugation. After reaching the volume limit of the concentrator (i.e., 50 μL), Chl a concentration was measured, which amounts to 1.53 mg/mL. Small aliquots of the sample were then frozen under liquid nitrogen, and stored at −80 °C until use.
SDS-PAGE and native PAGE experiments
PSII-FCPII was analyzed by an SDS-PAGE gel containing 5% acrylamide for the stacking gel and 16% acrylamide for the resolving gel. Sample was diluted to 15 μL using a 10 mM HEPES (pH 7.5), 50 mM NaCl, 0.05% α-DDM buffer, supplemented with 5 μL of denaturation buffer containing 2% lithium lauryl sulfate, 60 mM dithiothreitol, and 60 mM Tris-HCl (pH 8.5), and heated for 10 min at 60 °C. Electrophoresis was performed under a constant current (30 mA) for 90 min. For native PAGE experiments, a 3%–12% acrylamide gradient gel was made using a gradient mixer and the PSII-FCPII samples were diluted to a final volume of 10 μL with 10 mM HEPES (pH 7.5), 50 mM NaCl, 0.05% α-DDM, and supplemented with 2 μL of an 80% glycerol solution. Electrophoresis was conducted without any dye in any buffers (clear-native PAGE, CN-PAGE), with a constant voltage (100 V) and a maximum current of 15 mA35,36. After the samples entered the gel, electrophoresis was performed at a constant current (15 mA) and a maximum voltage of 400 V. After electrophoresis, the CN-PAGE gel was stained with Coomassie blue R-250 to visualize the molecular markers.
N-terminal sequencing of SDS-PAGE bands
The SDS-PAGE bands corresponding to 32 pmol of CP47, FCPII-1a/b, FCPII-2, FCPII-4 or FCPII-5 were transferred from a non-stained SDS-PAGE gel to a PVDF membrane, cut and identified by Edman sequencing using a Shimadzu Protein Sequencer PPSQ 31 A following the instructions provided in the user manual.
Absorbance and low-temperature fluorescence measurements
Absorption spectrum of the purified PSII-FCPII was recorded from 400 nm to 750 nm in 10 mM HEPES (pH 7.5), 50 mM NaCl, 0.05% α-DDM at 15 μg/mL. Fluorescence emission spectrum was recorded at 77 K with a fluorescence spectrophotometer (F-7000 from Hitachi Co.). Excitation wavelength was set at 435 nm, and the emission spectrum was recorded between 550 and 750 nm in liquid nitrogen. The sample was diluted to a final concentration of 5 μg Chl/mL with a buffer containing 10 mM HEPES (pH 7.5), 50 mM NaCl, 0.05% α-DDM.
Pigment extraction and HPLC analysis
An isolated PSII-FCPII sample equivalent to 30 μg of Chl a was dissolved in a 50:50 methanol/acetone and centrifuged. The supernatant was dried under nitrogen gas and resuspended in 500 μL of 60:40 acetone/ethyl acetate. After addition of 400 μL of H2O, the sample was centrifuged at 17,000 × g for 5 min and the upper ethyl acetate layer was recovered and dried under nitrogen, and the resultant sample was resuspended in 100% methanol. Before HPLC analysis, the sample was filtered through a 0.45 μm PTFE filter (Minisart RC 4, Millipore). Carotenoids and Chls were analyzed using a Shimadzu HPLC system equipped with a Wakosil-II 5C18-100 column (particle size: 5 μm; 250 mm × 4.6 mm, Wako) and a Wakosil-II 5C18-100 guard column (5 μm, 10 mm × 4.6 mm, Wako), with 100% methanol as the eluent at a flow rate of 2 mL/min. Elution profiles and absorption spectra were monitored using an SPD-M20A photodiode array detector (Shimadzu, Japan).
RNAseq analysis
C. roscoffensis cells were harvested and suspended in RNAlaterTM (Thermofisher Scientific), then the sample was immediately frozen in liquid nitrogen and stored at −80 °C. Total RNA extraction and RNA sequencing were conducted by Bioengineering Lab Co., Ltd.
Cryo-EM data collection
An aliquot of 4 μL of the C. roscoffensis PSII-FCPII supercomplex (1.53 mg of Chl mL−1) in a 10 mM HEPES (pH 7.5), 50 mM NaCl buffer containing 0.05% α-DDM was applied to a Quantifoil R1.2/1.3 Cu 300 mesh grid in the chamber of FEI Vitrobot Mark IV (Thermo Fisher Scientific). Then, the grid was blotted with a filter paper for 5 s at 8 °C under 100% humidity and plunged into liquid ethane cooled by liquid nitrogen. The frozen grid was transferred into a Titan Krios G4 electron microscope (Thermofisher Scientific) equipped with an E-CFEG electron source operated at 300 kV. The images were recorded with a post-column mounted Selectris X energy filter and a Falcon 4i camera operating in the electron event representation (EER) mode. The particle images were acquired at a pixel size of 0.727 Å/pixel (165 000 × magnification) and a total dose of 50 e−/Å2 in 1044 EER frames in 3.40 s with a target defocus range of −0.8 to −1.8 µm, with increment steps of 0.2 µm. In total, 20,887 micrographs were collected.
Data processing
The 20,887 micrographs collected were analyzed using cryoSPARC 4.5.137,38,39,40,41,42. The initial movies were processed using Patch Motion Correction and Patch CTF Estimation40,41. In a first instance, an initial subset of images (852 micrographs) was used to define the picking parameters. A first step of separated manual picking for side views and top views was performed using Manual Picker, which was then used for template search using Template Picker. From this step, 7122 particles with side views and 8339 particles with top views were classified using 2D classification. The best 2D classes were selected, and duplicates between side views and top views were removed. This initial subset of 2462 particles was used for ab-initio reconstruction with 2 classes, which allowed us to identify the PSII-12 FCPII supercomplex (1428 particles). Then 2D classes were improved from this last subset, and used as a template for the picking of the whole dataset (20,887 micrographs). We extracted 300,825 and 418,205 particles for side views and top views, respectively, removed duplicates, and performed 2D classification. After selecting the best 2D classes, 82,495 particles were used for an ab-initio reconstruction using 4 classes. Two classes corresponded to artefacts (6.45% and 11.85%), whereas the other two classes corresponded to PSII-12 FCPII (52.81%) and PSII-6 FCPII (28.90%) respectively. For PSII-6 FCPII, we performed homogeneous refinement, global and local CTF refinements, and non-uniform refinement successively. As a result, a 2.88 Å resolution map was obtained. For the PSII-12 FCPII complex, we further performed a first step of homogeneous refinement with C2 symmetry, followed by reference-based motion correction on the subset of the PSII-12 FCPII particles. This resulted in a reduced subset (38,582 particles) which was subsequently used for global and local CTF refinement, and a final non-uniform refinement step, all using a C2 symmetry. Eventually, the final map reached a resolution of 2.22 Å, which was used for model reconstruction and refinement. The resolution was estimated using the FSC validation from cryoSPARC, and the local resolution map presented in the Supplementary Fig. 10 was calculated with Local Resolution Estimation.
Model building and refinement
Reconstruction of the PSII-12 FCPII supercomplex based on the 2.22 Å density map was achieved by fitting the previously published PSII-ACPII structure from cryptophyte (PDB entry 8XR6)14 into the map using UCSF ChimeraX43. In parallel, we built a local database from RNAseq raw sequence data using BLAST+23, and used blastp and tblastn to find every PSII core subunit from C. roscoffensis that corresponds to the cryptophyte subunits that were fitted into the map. Thereby, the sequences of each core subunit of the PSII-12 FCPII complex from C. roscoffensis (PsbA, PsbB, PsbC, PsbD, PsbE, PsbF, PsbH, PsbI, PsbK, PsbL, PsbM, PsbT, PsbW, and PsbX) were identified, and they were put into the cryo-EM map. For the Psb36 subunit, we relied on a motif that was identified in the map (FXXYXIXII) and searched for this sequence in our BLAST+ database. This results in one sequence which was fitted into the map, which matched every density of the chain. For the FCPII subunits, we first fitted the ACPII-1 structure of PSII-ACPII from a cryptophyte (PDB entry 8XR6)14 into every antenna density. Then, we used BLAST+ with the antenna sequences of both cryptophyte and diatom (PDB entries 8J5K, 6J40, and 8IWH) as queries to find homologous sequences in the C. roscoffensis RNAseq BLAST+ database, which identified the sequences of each FCPII subunit in the supercomplex. Every PSII core and ACPII sequences in the initial model were replaced with the PSII core and FCPII sequences from C. roscoffensis by the Align and Mutate function in Coot44.
Following this, all misplaced pigments were removed and each chain was checked manually in Coot. For refinement, we used the real-space refinement of PHENIX45,46 to perform a first iteration with rigid body fit enabled. From here, successive rounds of manual check on Coot and real-space-refinement in PHENIX were conducted. Ramachandran and rotamer constraints were used in the earlier steps of refinement, and to perform a final round of refinement only the non-bonded weight factor was set to 500 with other options set to default. Chls a and c were distinguished according to the presence or absence of the phytol chain, the spaces that can accommodate the phytol chain surrounding the pigment and the flatness of the Chl ring. The statistics of the final model was calculated with MolProbity47,48 and are shown in Supplementary Table 1, and the list of all pigments in the structure is shown in Supplementary Table 4. The maps for representative subunits and ligands are shown in Supplementary Figs. 11 and 12, and the angular distribution of particles used for the PSII-12 FCPII and PSII-6 FCPII cryo-EM density map constructions are shown in Supplementary Fig. 13.
FRET rate calculation between chlorophylls a and interface analysis with PISA
Coordinates of all Chls a within the PSII-FCPII supercomplex were extracted from the PDB file. These coordinates were used as an input for FRET calculation using a previously published calculation method31. The output file was filtered and only the Chls a couples with a FRET rate higher than 0.2 were selected. Each couple was connected using a dotted line in UCSF ChimeraX, in order to represent the most efficient energy transfer between Chls a within the supercomplex. Full couples of Chls a with FRET rate values higher than 0.2 is shown in Supplementary Table 5.
The PISA25 interface analysis was conducted on the PSII-FCPII supercomplex with automatic processing mode, including all the ligands (lipids and pigments) in the structure. The interfaces between the PSII core and FCPII subunits identified by this analysis are shown in Supplementary Table 2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The cryo-EM maps of the haptophyte PSII-12 FCPII and PSII-6 FCPII supercomplexes have been deposited in the Electron Microscopy Data Bank with accession codes EMD-62499 and EMD-62790, respectively. The PSII-12 FCPII structure has been deposited in the Protein Data Bank under accession code 9KQB. Source data are provided with this paper.
References
Shen, J.-R. The structure of photosystem II and the mechanism of water oxidation in photosynthesis. Annu. Rev. Plant Biol. 66, 23–48 (2015).
Cox, N., Pantazis, D. A. & Lubitz, W. Current understanding of the mechanism of water oxidation in photosystem II and its relation to XFEL data. Annu. Rev. Biochem. 89, 795–820 (2020).
Liguori, N., Periole, X., Marrink, S. J. & Croce, R. From light-harvesting to photoprotection: structural basis of the dynamic switch of the major antenna complex of plants (LHCII). Sci. Rep. 5, 15661 (2015).
Wood, W. H. J. & Johnson, M. P. Modeling the role of LHCII-LHCII, PSII-LHCII, and PSI-LHCII interactions in state transitions. Biophys. J. 119, 287–299 (2020).
Croce, R. & van Amerongen, H. Light harvesting in oxygenic photosynthesis: structural biology meets spectroscopy. Science 369, eaay2058 (2020).
Gao, J., Wang, H., Yuan, Q. & Feng, Y. Structure and function of the photosystem supercomplexes. Front. Plant Sci. 9, 357 (2018).
Sheng, X., Liu, Z., Kim, E. & Minagawa, J. Plant and algal PSII-LHCII supercomplexes: structure, evolution and energy transfer. Plant Cell Physiol. 62, 1108–1120 (2021).
Wang, W. & Shen, J.-R. Structure, organization and function of light-harvesting complexes associated with photosystem II. In Photosynthesis: Molecular Approaches to Solar Energy Conversion (eds Shen, J.-R., Satoh, K. & Allakhverdiev, A. I.) 163–194 (Springer International Publishing, 2021).
Pi, X. et al. The pigment-protein network of a diatom photosystem II-light-harvesting antenna supercomplex. Science 365, eaax4406 (2019).
Nagao, R. et al. Structural basis for energy harvesting and dissipation in a diatom PSII-FCPII supercomplex. Nat. Plants 5, 890–901 (2019).
Nagao, R. et al. Structural basis for different types of hetero-tetrameric light-harvesting complexes in a diatom PSII-FCPII supercomplex. Nat. Commun. 13, 1764 (2022).
Zhao, S. et al. Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana. Nat. Commun. 14, 8164 (2023).
Feng, Y. et al. Structure of a diatom photosystem II supercomplex containing a member of Lhcx family and dimeric FCPII. Sci. Adv. 9, eadi8446 (2023).
Zhang, Y.-Z. et al. Structure of cryptophyte photosystem II-light-harvesting antennae supercomplex. Nat. Commun. 15, 4999 (2024).
Mao, Z. et al. Structure and distinct supramolecular organization of a PSII-ACPII dimer from a cryptophyte alga Chroomonas placoidea. Nat. Commun. 15, 4535 (2024).
Reyes-Prieto, A., Yoon, H. S. & Bhattacharya, D. Marine algal genomics and evolution. In Encyclopedia of Ocean Sciences (ed Steele, J. H.) 552–559 (Elsevier, 2009).
Tsuji, Y. & Yoshida, M. Biology of haptophytes: Complicated cellular processes driving the global carbon cycle. In Advances in Botanical Research (ed Hirakawa, Y.) Vol. 84, 219–261 (Elsevier, 2017).
Rost, B. & Riebesell, U. Coccolithophores and the biological pump: responses to environmental changes. In Coccolithophores: from molecular processes to global impact. (eds Thierstein, H. R. & Young, J. R.) 99–125 (Springer Berlin Heidelberg, 2004).
Meng, R. et al. Genome sequence of chrysotila roscoffensis, a coccolithphore contributed to global biogeochemical cycles. Genes 13, 40 (2021).
Arundhathy, M. et al. Coccolithophores: an environmentally significant and understudied phytoplankton group in the Indian Ocean. Environ. Monit. Assess. 193, 144 (2021).
Bach, L. T., Riebesell, U., Gutowska, M. A., Federwisch, L. & Schulz, K. G. A unifying concept of coccolithophore sensitivity to changing carbonate chemistry embedded in an ecological framework. Prog. Oceanogr. 135, 125–138 (2015).
Poulton, A. J., Adey, T. R., Balch, W. M. & Holligan, P. M. Relating coccolithophore calcification rates to phytoplankton community dynamics: Regional differences and implications for carbon export. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 54, 538–557 (2007).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009).
Ago, H. et al. Novel features of eukaryotic photosystem II revealed by its crystal structure analysis from a red alga. J. Biol. Chem. 291, 5676–5687 (2016).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673 (1994).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Wang, W. et al. Structural basis for blue-green light harvesting and energy dissipation in diatoms. Science 363, eaav0365 (2019).
Kirilovsky, D. Photosynthesis: dissipating energy by carotenoids: photosynthesis. Nat. Chem. Biol. 11, 242–243 (2015).
Frackowiak, D. & Wróbel, D. Energy transfer between chlorophyll C and chlorophyll A. Biophys. Chem. 1, 125–129 (1973).
Kim, E. Computational Analysis of Förster Resonance Energy Transfer in Photosynthetic Proteins. https://doi.org/10.5281/ZENODO.3250622 (Zenodo, 2019).
Madjet, M. E., Abdurahman, A. & Renger, T. Intermolecular coulomb couplings from ab initio electrostatic potentials: application to optical transitions of strongly coupled pigments in photosynthetic antennae and reaction centers. J. Phys. Chem. B 110, 17268–17281 (2006).
Do, T. N. et al. Ultrafast excitation energy transfer dynamics in the LHCII-CP29-CP24 subdomain of plant photosystem II. J. Phys. Chem. Lett. 13, 4263–4271 (2022).
Jeffrey, S. W. & Humphrey, G. F. New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochem. Physiol. Pflanz. 167, 191–194 (1975).
Kawakami, K., Iwai, M., Ikeuchi, M., Kamiya, N. & Shen, J.-R. Location of PsbY in oxygen-evolving photosystem II revealed by mutagenesis and X-ray crystallography. FEBS Lett. 581, 4983–4987 (2007).
Wittig, I. & Schägger, H. Features and applications of blue-native and clear-native electrophoresis. Proteomics 8, 3974–3990 (2008).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Rubinstein, J. L. & Brubaker, M. A. Alignment of cryo-EM movies of individual particles by optimization of image translations. J. Struct. Biol. 192, 188–195 (2015).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Zivanov, J., Nakane, T. & Scheres, S. H. W. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267 (2020).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Acknowledgements
Chrysotila roscoffensis (NIES-8) cells were provided by NIES through NBRP of MEXT, Japan. We thank Adrien Gregorj for his help for the FRET rate calculations between Chls a. This work was supported partially by the Core-Facility Portal (CFPOU) at Okayama University (RIIS-n01), and we thank the International Research Center for Structural Biology of Okayama University for helping collection of the cryo-EM data. This research was also supported by a JSPS KAKENHI grant No. JP22H04916 (J.-R.S.).
Author information
Authors and Affiliations
Contributions
J.-R.S. conceived the project and acquired the funding. R.L.R. and F.A. cultured and harvested the C. roscoffensis cells. R.L.R. conceived the methodology and performed the purification of the PSII-FCPII supercomplex with the help of P.C.T., R.L.R., and K.K. collected the cryo-EM data and solved the structure of the PSII-FCPII supercomplex. R.L.R. and Y.N. conducted the low-temperature fluorescence and pigment analysis experiments. R.L.R. wrote the original draft, and J.-R.S. revised and edited the final manuscript. All authors contributed to the discussion of the results and improvement of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Roman Kouril and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
La Rocca, R., Kato, K., Tsai, PC. et al. Structure of a photosystem II-FCPII supercomplex from a haptophyte reveals a distinct antenna organization. Nat Commun 16, 4175 (2025). https://doi.org/10.1038/s41467-025-59512-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-59512-9
This article is cited by
-
Cryo-EM structure of photosystem II supercomplex from a green microalga with extreme phototolerance
Nature Communications (2026)