Abstract
Viruses from the Flaviviridae family contain human relevant pathogens that generate subgenomic noncoding RNAs during infection using structured exoribonuclease resistant RNAs (xrRNAs). These xrRNAs block progression of host cell’s 5′ to 3′ exoribonucleases. The structures of several xrRNAs from mosquito-borne and insect-specific flaviviruses reveal a conserved fold in which a ring-like motif encircles the 5′ end of the xrRNA. However, the xrRNAs found in tick-borne and no known vector flaviviruses have distinct characteristics, and their 3-D fold was unsolved. Here, we verify the presence of xrRNAs in the encephalitis-causing tick-borne Powassan Virus. We characterize their secondary structure and obtain a mid-resolution map of one of these xrRNAs using cryo-EM, revealing a unique double-loop ring element. Integrating these results with covariation analysis, biochemical data, and existing high-resolution structural information yields a model in which the core of the fold matches the previously solved xrRNA fold, but the expanded double loop ring is remodeled upon encountering the exoribonuclease. These results are representative of a broad class of xrRNAs and reveal a conserved strategy of structure-based exoribonuclease resistance achieved through a unique topology across a viral family of importance to global health.
Similar content being viewed by others
Introduction
RNA viruses present a substantial global health threat, with urbanization and climate change exposing a growing portion of the population to infection1,2,3. Among these, the Flaviviridae family contains many arthropod-borne viruses such as Zika virus, Dengue virus, yellow fever virus, West Nile virus, Tick-Borne Encephalitis virus, and Powassan virus, many of which are major human health threats4,5,6,7,8. The genus Flavivirus contains positive sense single stranded RNA (+ssRNA) viruses consisting of a single genomic RNA4,9,10, with one open reading frame flanked by 5’ and 3’ untranslated regions (UTR) that contain RNA elements essential for regulating and facilitating the viral replication cycle11,12,13,14.
In addition to replication of the genomic RNA, infection by flaviviruses results in smaller noncoding viral RNAs called subgenomic flaviviral RNAs (sfRNAs)15. These sfRNAs result from incomplete degradation of the viral genome by host 5′ to 3′ exoribonucleases (primarily Xrn1) (Fig. 1A)16,17. Specifically, structured RNA elements in the 3′ UTR, called exoribonuclease resistant RNAs (xrRNAs), halt the 5′ to 3′ progression of Xrn1 at discrete locations18,19,20. Importantly, known xrRNAs are specific for blocking the progression of 5′ to 3′ (not 3′ to 5′) exoribonucleases and they function independently of bound protein factors20,21,22,23,24. That is, the structure of the xrRNA that the 5′ to 3′ exoribonuclease encounters as it approaches from the 5′ side is necessary and sufficient to specifically halt the processive enzyme.
A Cartoon representation of sfRNA production and function through Xrn1-mediated degradation of the viral genome. B Overview of the phylogenetic relationship within Flaviviridae based on the sequence of viral protein NS5. The tree files, alignments, and analysis were reanalyzed from data published by Bamford et al.84 and Mifsud et al.85 C Structural features of Flaviviridae xrRNAs are shown on the cartoon representations of the secondary structures of each class or subclass. Known long-range tertiary interactions are indicated with colored lines and boxes in class 1. Analogous predicted interactions in class 2 are indicated with dashed colored lines and boxes. The blue letters indicate conserved junction nucleotides, blue no symbol in subclass 1b denotes the absence of a nucleotide in this position. Source data is provided as a Source Data file.
Often, flaviviruses contain multiple tandem xrRNAs in their 3′ UTR leading to sfRNAs of different sizes, though whether different sfRNAs are redundant or have independent functions is unknown25,26,27. The sfRNAs have been implicated in several cytopathic and pathological effects such as disrupting host mRNA turnover, inhibiting RNAi, altering the host interferon response, and facilitating host switching16,17,18,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44. No analogous xrRNA structures have yet been identified in cellular RNAs; this and the ubiquitous presence of xrRNAs in Flaviviridae make them potential targets for anti-flaviviral therapeutics.
Current understanding of the structure-based mechanism of sfRNA formation comes from high-resolution crystal structures of xrRNAs from several mosquito-borne flaviviruses (MBFV). These include Zika Virus (ZIKV), Murray Valley Encephalitis Virus (MVEV), and the distantly related Tamana Bat Virus (TABV)21,23,24,45. These xrRNAs all form a compact structure based on a three-way helical junction. The defining structural characteristic is a unique ring-like feature that encircles the 5′ end of the xrRNA. Versions of this ring-like feature have been observed in all xrRNA structures solved to date, including those from plant-infecting viruses unrelated to flaviviruses46,47,48. The presence of this ring-like feature, combined with functional and biophysical studies, show that this ring acts as a molecular brace against the surface of the exoribonuclease22,45,49,50,51,52. This feature appears to create a mechanical block that specifically prevents processive 5′ to 3′ exoribonucleases from progressing past a defined point. Tertiary interactions that stabilize flavivirus xrRNAs’ unique fold include pseudoknots, base triples, non-Watson-Crick base pairing, base stacking, and metal ion coordination21,24,45,53. Evidence that this specific structure and stabilizing interactions are conserved among flaviviral clades comes from comparative sequence alignments of xrRNAs found throughout Flaviviridae20,21,22,24,25,45,47,48,54,55,56.
xrRNAs are ubiquitous in Flaviviridae where evolution gave rise to variation in sequence and structure17,23,25,54,56,56. Examination of covariance models of all Flaviviridae xrRNAs and the crystal structures mentioned above reveals differences that allow xrRNAs to be classified into two major classes: class 1 (subdivided into subclass 1a and subclass 1b) and class 2 (Fig. 1B)23,54. These classes are based on differences in the size, secondary structural elements, and tertiary interactions that stabilize the 3-D fold. The division is also supported bioinformatically with comparative sequence alignments54. Subclass 1a contains xrRNAs from MBFV, while subclass 1b xrRNAs are present in insect-specific flaviviruses (ISFV), TABV, and members of Pegivirus, Pestivirus, and Hepacivirus genera54. Although subclasses 1a and 1b have many similarities, differentiating structural features are the presence or absence of an unpaired nucleotide between P2 and P3 and fundamental differences in the tertiary interactions that close the ring feature24 (Fig. 1C).
The class 2 xrRNAs lack 3-D structural characterization compared to class 1 and have thus far only been found in members of the tick-borne flaviviruses (TBFV) and no known vector flaviviruses (NKVFV)22,23,34,38,57,58 (Fig. 1B). The secondary structures of the class 2 xrRNAs have been depicted several ways, but recent analyses led to versions showing features reminiscent of the class 1 with some key differences23,55 (Fig. 1C). Specifically, within the helical junction, an unpaired junction nucleotide is conserved as an A in all known sequences of class 2, while it is always a C in the subclass 1a and is always absent in the subclass 1b. Additionally, an important pseudoknot (PK1) in class 2 appears to be significantly longer (6-10 base pairs) than in class 1 (1-3 base pairs)23,55 (Fig. 1C).
Based on its similarities to class 1, it is hypothesized that the class 2 xrRNAs also contain a ring-like feature important for blocking Xrn1, but there is no 3-D structural information to verify this. Likewise, it is not clear how a ring-like structure could be accommodated given the larger proposed PK1. Also, the location where the exoribonuclease halts on class 2 xrRNAs is within the putative expanded PK1 element rather than in unpaired RNA upstream of the xrRNA ring structure22. If class 2 xrRNAs contain a ring-like feature, it likely has substantial deviations from class 1, given the differences in the proposed secondary structures. Finally, bioinformatic searches for new xrRNAs would be enhanced by structural insight, as current knowledge of class 2 comes mostly from biochemical studies.
Although we have successfully crystallized several class 1 xrRNAs, attempts to obtain diffraction-quality crystals of representative class 2 xrRNAs were unsuccessful. However, single particle cryo-electron microscopy (cryo-EM) has emerged as an invaluable method for determining the structure of biomolecules previously inaccessible by traditional structural approaches. Cryo-EM was recently applied to several RNAs to determine tertiary structure and dynamics, and we and others recently presented a scaffold-based approach to facilitate the structural determination of diverse RNAs by cryo-EM59,60,61,62,63,64,65. We chose to apply a scaffold-based approach to the structure of a class 2 xrRNA from a lineage II Powassan virus (POWV) isolate. POWV is an emerging tick-borne human pathogen found in both North America and north-east Asia that can cause severe encephalitis66,67,68. The presence of xrRNAs in the POWV genomic RNA and its ability to form infection-critical sfRNAs had not been verified, and the structures of any putative class 2 xrRNAs in POWV were unknown.
Here we report the results of combined virological, biochemical, bioinformatic, and structural methods applied to the xrRNAs from POWV. We verified the presence of two xrRNAs in POWV 3’UTR that result in sfRNA formation. We used cryo-EM to obtain mid-resolution maps of a POWV class 2 xrRNA that were sufficient to observe the overall topology of the RNA. This architecture contains a core fold that is similar to the subclass 1b xrRNAs, but a ring-like feature that is more extensive than those previously observed. Specifically, the ring forms a double loop around the 5′ end of the xrRNA that may interact with Xrn1 in unique ways compared to the ring in class 1. Integrating biochemical, functional, bioinformatic, and existing high-resolution data allowed us to construct a model suggesting key tertiary interactions akin to those in other xrRNA classes. Structure-informed bioinformatic analysis then helped us catalog a more complete set of class 2 xrRNAs and define key features of the class. Overall, our findings fill a key gap in understanding the diversity of 3-D structures in Flaviviridae xrRNAs, and further highlight the essential role of conserved but variable ring-like folds in xrRNAs.
Results
Powassan virus produces subgenomic flaviviruses during infection
We chose to use the putative class 2 xrRNAs from a lineage II Powassan virus (Spooner isolate) as a model system66. POWV is an emerging human pathogen found in North America for which no targeted therapies exist, and the sequence of its 3′ UTR suggested the presence of two class 2 xrRNAs (Fig. 2A)22,23. Additionally, POWV is not a select agent (as defined by the US National Institutes of Health), and can be cultured in the lab. To verify that POWV infection results in the production of sfRNAs, we infected Vero cells at various multiplicities of infection (MOI) and assessed sfRNA generation by northern blot (Fig. 2B). Blots revealed two discrete bands at sizes consistent with sfRNA 1 and sfRNA 2 species starting at the 5′ end of the first and the second putative xrRNA structures, respectively. Thus, POWV makes sfRNAs as expected.
A Cartoon representation of POWV 3’ UTR. Abbreviations: DB – dumbbell, 3′ SL – 3′ stem loop. B Northern blot of POWV-infected Vero cells at the indicated MOI and time post infection. Probes were specific for the 3′ UTR of POWV or the U6 RNA loading control. Three independent experiments were performed, results of one is shown. C In vitro degradation by Xrn1 of in vitro transcribed RNA of the POWV Spooner isolate 3′ UTR. The partially degraded but stable RNAs appear as bands in the lane with Xrn1, demonstrating the presence of xrRNAs. The gel is representative of experiments performed in triplicate. D, E Stop site analysis of POWV xrRNA1 and xrRNA2. RNA treated with Xrn1 was analyzed using reverse transcription primer extension. The intensity of the resultant bands is graphed as a function of nucleotide position. F Secondary structure diagram of the POWV xrRNA1 in the context of the full 3′ UTR with the results of 1M7 chemical probing. G In vitro RNA resistance assay demonstrating the functional importance of both PK1 and PK2. A diagram of the mutations is found in Fig. S3. The gel is representative of experiments performed in triplicate. Source data is provided as a Source Data file.
To verify that these sfRNAs were due to authentic xrRNAs, we used an established in vitro Xrn1 resistance assay. Briefly, in vitro transcribed RNA comprising the entire POWV 3′ UTR was challenged with pure recombinant Xrn1 then resolved by denaturing polyacrylamide gel electrophoresis (Fig. 2C). We observed two bands of the expected sizes that would result if the enzyme began degrading the input RNA but then halted at one of the two putative xrRNAs. The pattern matches that observed during infection as assayed by northern blot (Fig. 2A–C). These results confirm the production of sfRNAs by the xrRNAs in POWV during infection and demonstrate these structures function without protein cofactors, a hallmark of true xrRNAs. Overall, the results presented above establish these POWV xrRNAs as a model for class 2 xrRNAs for both cell culture-based experiments and in vitro biochemical assays.
The exonuclease halt site on POWV class 2 xrRNAs differs from the class 1
In class 1 xrRNAs, the Xrn1 halt site resides a few nucleotides upstream of the P1 helix and pseudoknot 1 (PK1), in unpaired 5′ nucleotides that emerge from the center of the ring-like structure20. In class 1, the 1-3 base pairs that comprise PK1 lie within the ring and are essentially inaccessible to the approaching enzyme. This configuration supports current models that the ring is preformed essentially as observed in the crystal structures, and the surface of the exoribonuclease braces against it. This prevents unwinding of base pairs that are within or behind the ring. However, in contrast to class 1, the proposed PK1 in class 2 is much longer (Fig. 1C)23,54,55. Indeed, the halt point on other class 2 xrRNAs has been mapped to a region that would lie within this longer base-paired pseudoknot (termed P1.1/P1.2 in some previous depictions)22.
We determined the exoribonuclease halt site on POWV xrRNAs. Briefly, we treated in vitro transcribed POWV 3′UTR RNA with Xrn1, recovered the xrRNA-protected products, and mapped the resulting 5′ ends by reverse transcription (Fig. 2D, E). In contrast to the class 1 xrRNAs – where the halt site exists primarily at a single position four nucleotides upstream of PK1 – in POWV the halt sites are located within PK1 for both the first and second POWV xrRNA (Fig. 2F). This is meaningful because the active site within Xrn1 only accommodates non-base-paired RNA, and ~5 nucleotides are needed to span the distance from the surface of the enzyme to the active site69,70. Thus, to reach the halt point, the enzyme must unwind parts of PK1. This contrasts with the class 1 flavivirus xrRNAs but may share some similarity with some xrRNAs found in plant viruses46,48.
The pattern of halt sites on the POWV xrRNAs also revealed several stop sites within the xrRNA rather than a well-defined single location (Fig. 2D–F). Specifically, on the upstream xrRNA1, the halt sites were spread across ~8 nucleotides in PK1, with the three most prominent. In the downstream xrRNA2, the halt sites were spread among a shorter stretch of ~3 nts in PK1. Overall, both xrRNA halt sites differ from the precision of the halt site of class 1. These results suggest differences in how Xrn1 interacts with class 1 vs. class 2 xrRNAs and perhaps some variability within different class 2 xrRNAs. Overall, the location and pattern of halt sites leads to the hypothesis that the configuration of the ring-like structure and extended PK1 in class 2 differs from class 1, and this may affect the way Xrn1 interacts with the xrRNA structure.
The POWV xrRNAs adopt a conserved secondary structure
Existing models of xrRNA allow robust predictions regarding the secondary structures of the POWV xrRNAs, but there may be features not obvious in those analyses. Also, the fact that the enzyme must partially unwind the xrRNA to reach the halt points raised the question of whether the xrRNA secondary structure could change after encountering the enzyme. We therefore experimentally queried the secondary structure of the POWV class 2 xrRNA in different contexts using chemical probing, specifically by DMS-MaP71 (Dimethyl sulfate (DMS; modifies Watson crick face of A and C)72) and SHAPE-MaP (selective 2′-hydroxyl acylation analyzed by primer extension (1-Methyl-7-Nitroisatoic Anhydride (1m7; modifies 2’OH in flexible backbone regions)73,74. To recapitulate the viral genomic context of the POWV xrRNA, we probed constructs consisting of the (1) full 3′ UTR, (2) sfRNA 1, and (2) sfRNA 2. The two sfRNA species were produced by in vitro treatment with Xrn1 followed by probing.
Overall, the measured reactivity profiles of the xrRNAs from the POWV 3′ UTR, sfRNA 1, and sfRNA 2 were all similar, indicating the secondary structure does not change in these different contexts (Fig. 2F, S1, S2). Nucleotide reactivities from both 1m7 and DMS were in agreement with features present in the proposed class 2 xrRNA secondary structure model23. The three-way helical junction is present in both xrRNAs, demonstrated by the overall high reactivities in the L2 and L3 loops that are not involved in tertiary interactions. The P1, P2, and P3 helices generally show low reactivity except for some of the nucleotides at the terminal end of the helix; this is expected in terminal base pairs. The large unpaired region after the proposed PK1 is accessible for modification in all constructs supporting the same secondary structure in all contexts.
Importantly, the presence of both pseudoknots was supported by both 1m7 and DMS probing. To further test their presence and importance, we made substitution mutations that disrupted either PK1 or PK2 and tested them for Xrn1 resistance in vitro (Fig. 2G, S3). Both mutants lost the ability to block the exoribonuclease, and resistance was rescued with compensatory mutations (Fig. 2G, S3). This matches results performed on xrRNAs from other viruses22,55. However, elevated reactivities within the 5′ portion of PK1 suggest that while the pseudoknot forms, there are conformational dynamics present at the 5′ end. We note that these more dynamic base pairs correspond to the location just upstream of the mapped stop sites in the xrRNAs, which is interesting given that the enzyme must unwind these nucleotides to reach the halt sites (Fig. 2F).
Cryo-EM yields an intermediate resolution map of POWV xrRNA structure
To better interpret the functional and biochemical data and understand the structural basis of exonuclease resistance in class 2 xrRNAs, we used cryo-EM to interrogate the structure of POWV xrRNA1. We chose xrRNA1 as it was well behaved biochemically, it has substantial sequence similarity and secondary structure redundancy with xrRNA2, and the more pronounced difference in the halting pattern suggested it might best reveal key differences with class 1 xrRNAs.
To perform cryo-EM, we employed our previously described scaffolding approach65. Briefly, the xrRNA was appended to a modified version of the Tetrahymena ribozyme, creating a large and structurally recognizable chimeric RNA amenable to cryo-EM. The connection point was through the xrRNA’s L2 loop. The L2 loop is not conserved in sequence, does not make any known long-range interactions, and analogous loops are known to be dispensable in other xrRNAs20,21 (Fig. S4A). The xrRNA’s L2 loop was connected to the ribozyme’s P6b element. This connection has been shown not to disrupt the folding of the scaffold RNA or the appended domain65.
To ensure appending of the xrRNA to the scaffold did not perturb xrRNA fold or function, we performed an Xrn1 resistance assay on this scaffolded version of the POWV xrRNA. The xrRNA maintained resistance to Xrn1 in vitro within the context of the chimeric RNA, and disruption of PK2 eliminated Xrn1 resistance in the chimeric RNA as it does in the isolated xrRNA (Fig. S4B, C). Likewise, the intron scaffold itself is not Xrn1 resistant. Because xrRNA function is necessarily dependent on the proper 3-D fold and formation of an intricate set of tertiary contacts, these results strongly suggest that the xrRNA adopts the correct functional fold within the cryo-EM construct.
We used the scaffolded xrRNA to prepare grids and collect cryo-EM data that was analyzed using a similar processing pipeline as previous work65. Following multiple rounds of 2D classification, classes were present in which the appended xrRNA domain is clearly observed (Fig. S5). The resulting 493,118 particles were then used for initial map generation. The maps had clear features corresponding to the xrRNA, revealing its global architecture. However, 3D variability analysis revealed a degree of structural heterogeneity among this set of particles. To recover the most complete map possible, we used 3-D classification to partition these states (Fig. S5). The resultant maps yielded several volumes corresponding to what appeared to be various levels of ‘melting’ of the distal regions of the structure. Interestingly, this supports the idea that these regions are conformationally dynamic as indicated in the chemical probing data and consistent with the fact that the enzyme must unwind them.
We proceeded with the most complete and well resolved map, corresponding to what we interpret as the fully base-paired extended PK1. Non-uniform refinement of these 55,485 particles yielded a map with a global resolution of 4.1 Å in the scaffold domain with a local resolution of the xrRNA of 5.1 Å (Fig. 3A, S5–S7). Within the intermediate resolution xrRNA portion of the map, clear structural features were observable. Importantly, both major and minor grooves were apparent and phosphate bumps were clear along the backbone for most of the map, allowing the backbone to be traced (Fig. 3B). Also, there is no observed interaction between the scaffold intron RNA and the xrRNA, further supporting the assertion that the xrRNA fold is not altered when attached to the intron.
A Local resolution map of the scaffolded POWV xrRNA. Resolution is colored as denoted in the inset key B. Cryo-EM map of the POWV xrRNA, the locations of secondary structure features are indicated. C Secondary structure diagram of the POWV xrRNA, colored to match D and E. D Structural model of the POWV xrRNA colored to match C. The yellow dashed backbone represents 9 nucleotides that were not visible in the map and hence not built. Structural features are labeled. E Comparison of POWV and TABV ring topologies colored corresponding to C.
Structural model of a POWV xrRNA reveals features reminiscent of other xrRNAs
Although the map could not reveal high-resolution details between bases ab initio, we integrated the map with existing structural information from other xrRNAs, sequence conservation, the known secondary structure, and the data presented here to construct a full model of POWV xrRNA1 (Fig. 3C, D, S6–8). While we could not make atomic resolution conclusions, the backbone could be readily traced, the overall topology was clear, and all nucleotides were placed. This model thus allowed us to compare the architecture of this class 2 xrRNA with the class 1.
The POWV xrRNA architecture has several features that match the class 1 xrRNAs, indicative of function dependent on a specific shared 3-D structure (Fig. 3C, D). First, the expected paired helical elements P1, P2, and P3 are present, as is pseudoknot 2 (PK2) formed through pairing between the sequence at the 3′ end with part of L3. These stems adopt an overall configuration similar to class 1 xrRNAs in which P1 stacks with P2, while P3 stacks on PK221,24,45. Interestingly, the overall angle between the helices and the trace of the backbone through the core of the POWV xrRNA structure is much more like the subclass 1b xrRNA from TABV than the subclass 1a from ZIKV (Fig. 3E, S9). Stated another way, the class 2 and class 1b xrRNA seem to share a common core fold within their three-way junctions. As with the class 1 xrRNAs, the POWV class 2 core structure gives rise to a ring-like motif, although it differs substantially from previous structures, further described and discussed in later sections of this manuscript.
Bioinformatic analyses reveals conserved determinants of exonuclease resistance
To better interpret the cryoEM and refine the structural model, we used our initial model to guide a bioinformatic analysis to (1) identify additional examples of class 2 xrRNAs, (2) develop a more robust understanding of the class 2 sequence and structural constraints, and (3) determine if the structural model of the POWV xrRNA1 is representative of class 2 xrRNAs in general. This type of approach has proven successful in identifying additional variants of structured RNAs as well as supporting proposed folds through statistically significant covariation75,77,78,79,79.
We used a comparative sequence alignment from 28 previously identified class 2 xrRNA sequences22,23 with the program Infernal80 to query a database containing all deposited viral genome sequences (retrieved 01/06/2023). This search identified 38 putative class 2 xrRNAs, 35 from TBFVs and 3 from NKVFVs (Fig. S10 Supplementary Data 1). Of the 13 recognized TBFV species, 10 were identified in this search, including both lineages of POWV as well as multiple subtypes of tick-borne encephalitis virus.
We calculated a consensus sequence and secondary structure from the identified sequences using CaCoFold visualized with R2R76,79,81. The resulting covariation patterns supported the secondary structure elements observed in the cryo-EM structure of the POWV xrRNA, recovering all stems and pseudoknots present in this class (Fig. 4A). The patterns also showed that the helical organization and base pairing schemes present in the POWV xrRNA are general to all class 2 xrRNAs. These results largely agree with previous work bioinformatically characterizing class 2 xrRNAs22,55. However, we were now able to integrate our structural model with this covariation analysis to provide evidence for several structural features critical for function. The covariation and conservation patterns also agreed with the 3-D structural model, further validating it.
A Consensus secondary structure model of class 2 xrRNAs calculated by R-Scape76,79. B Diagram denoting the location of three perfectly conserved A bases that were mutated to test their functional significance. C. Structure of the POWV xrRNA with the three conserved A bases indicated in B and C. D. In vitro RNA resistance assay demonstrating the functional importance of the three perfectly conserved (A) nucleotides in class 2 xrRNAs. The gel is representative of experiments performed in triplicate. Source data are provided as a Source Data file.
The most striking feature is the extended PK1 and double loop, supported by the chemical probing, bioinformatic, and cryoEM data. Several additional covariation-supported base pairs were identified, representing a more extensive base pairing context between the 5′ end of the structure and the seemingly unpaired sequence between P3 and P1. Indeed, this length ranges from 7 to 15 base pairs. To accommodate this extended interaction, the linker region between PK1 and P1 (S3) is sufficiently long in all cases to allow for the formation of PK1 (Fig. S10B, C).
The other pseudoknot present in all flavivirus xrRNA classes is PK2, which is variable in length in different xrRNAs. In class 2, the bioinformatic analysis confirms that PK2 is always present and is 4 base pairs in length. It likely stabilizes the ring-like motif through long-range base pairs and continuous stacking with P3.
Based on the alignment, the P1 helix consists of three Watson-Crick base pairs and a terminal non-Watson-Crick pair, consisting in most cases of an A•G pair. In some cases, this is replaced by a CC dinucleotide. Although the resolution of the maps precludes an analysis of detailed hydrogen bonding schemes, this CC dinucleotide is similar to that found in class 1b, and thus a similar pairing scheme is possible (Fig. S11)23,24,54. Higher resolution data will be needed to confirm the details of this putative interaction. If present, P2 ranges between 1 and 7 base pairs with an average of 5. The four-base pair P3 helix length is conserved, with ~92% of the aligned sequences containing a CGGG-CCCG pattern and the remaining 8% of sequences containing UGGG-CCCA or UGGG-CCUA (Fig. 4A, S10, S12).
A functionally important interaction that is found in all class 1 xrRNAs is a base triple formed between a nucleotide at the 5′ end of the xrRNA and a Watson-Crick base pair in stem L3. In class 1, this triple can be C+•G-C (class 1b) or U•A-U (class 1a), and mutational experiments show it to be essential for exoribonuclease halting activity20,21,22,24. The covariation analysis of class 2 presented here provides strong evidence for the presence of an analogous C+•G-Y (Y denotes a C or U) base triple in class 2, as the three corresponding nucleotides are universally conserved. The backbone trace provided by the map is consistent with the formation of this triple. Consistent with this, the mutation of C16 to a U to disrupt the putative triple resulted in loss of Xrn1 resistance (Fig. S13). However, the additional mutation of the G-C pair to a A-U pair to restore the putative triple did not restore resistance (Fig. S13). While this may indicate that the triple does not form, an equally valid interpretation is that the C+•G-Y triple, rather than the U•A-U triple, is specifically required to avoid alternate secondary structures from forming. This latter interpretation is consistent with the conservation analysis. We assert that the bulk of the evidence supports a base triple in this position. If true, a base triple appears to be a key interaction in all classes of flavivirus xrRNAs, forming between the 5′ end of the xrRNA and the three-way junction to position the 5′ end to pass through the center of the ring (Fig. S14). Final confirmation will require higher-resolution structural data.
The POWV xrRNA1 secondary structure contains a single unpaired nucleotide, A36, between P2 and P3. In the class 1a xrRNAs, a highly conserved and essential C nucleotide is in this position, where it is a defining characteristic, making interactions with the P1 helix. In our model of the POWV xrRNA1, the unpaired A23 occupies this same position. Interestingly, while both the class 2 and subclass 1a contain a nucleotide in this position, it is distinctly absent in the subclass 1b23,54. While the POWV xrRNA1 core is more class 1b-like, this specific nucleotide is likely more class 1a-like. Interestingly, Xrn1 resistance experiments testing substitutions, deletions, and insertions at this position show that each class of xrRNA is highly sensitive to the identity and presence or absence of the correct nucleotide in this position20,21,24,45,54. Analysis of the structures of different xrRNAs suggests this strict requirement may correlate to the size or nature of the ring-like structure and specific stabilizing interactions in different xrRNA classes.
Perfectly conserved A bases are necessary for function
Several perfectly conserved A bases are present that are not base-paired in the phylogenetically supported secondary structure or in our 3-D structural model. As noted previously, in class 2 xrRNAs, a perfectly conserved A base is present between P2 and P3. Two others are located adjacent to P1. The cryoEM map is not of sufficient resolution to allow us to determine the details of any interactions they are making (Fig. S15). However, their location and the fact that in class 1 xrRNAs, analogously-placed bases form non-Watson-Crick interactions that are essential to form the resistant motif suggest that they are also critical for class 2 xrRNA function21,24,45 (Fig. 4B, C).
To test the importance of interactions formed by these A’s in the POWV class 2 xrRNA, we prepared RNA in which each A was individually mutated to a G and challenged these RNAs with Xrn1 in vitro. Point mutations of any one of these bases resulted in the loss of resistance (Fig. 4D). All three As are universally conserved in class 2 xrRNAs, which suggests they are critical features for forming the proper resistant motif. Comparisons with class 1 xrRNAs, our structural model, and our consensus secondary structure support the hypothesis that A1 is involved in structuring the base of P1 and mediates the transition between PK1, PK2, and P1. A2 is important in defining the ring like feature like is seen in class 1 xrRNAs21,24,45. Based on its positioning in our map and the similarities to a long-range interaction in class 1 xrRNAs21,23,24,45,54, A3 is likely involved in a long-range base pair with a G in L3. For each of these, a G is likely not tolerated as it cannot form the specific hydrogen bonding interactions needed, but higher resolution information is needed to verify this and observe the specific tertiary interactions formed by these universally conserved and functionally critical As.
The POWV class 2 xrRNA contains a unique double-loop structure
While many features of the POWV class 2 xrRNA match or are analogous to class 1, a dramatic difference is in the size of PK1. The resultant expansion of the ring-like feature creates a form that we refer to as a double loop (Fig. 5A). In class 1 xrRNAs, the 2-3 base pair long PK1 forms stacking interactions with P2 and positions the 5′ end of the xrRNA to pass through the center of the fold21,24,45. The 13-base pair long PK1 in the POWV xrRNA1 also positions RNA to pass through the middle of the fold but forms a stem that extends away from the core of the structure (Figs. 2E, 5A). In fact, the expansion of PK1 makes it the longest base paired element in the structure and the junction within class 2 flavivirus xrRNAs is technically a four-way junction between P1, P2, P3, and PK1. The 3′ end of PK1 is linked to the 5′ end of P3 by a stretch of nine unpaired nucleotides (S3) that are not visible in our cryo-EM maps and hence could not be modeled. This is likely due to a high degree of conformational flexibility in these unpaired nucleotides, consistent with the chemical probing (Fig. 2F).
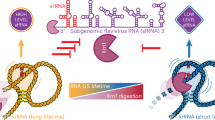
A The ring-like structure (magenta) encircling the 5′ end (blue) of the xrRNA from ZIKV (PDB: 5TPY)21, Murray Valley Encephalitis virus (MVE) (PDB: 4PQV)45, TABV (PDB: 7K16)24, POWV (this study), and Sweet Clover Necrotic Mosaic Virus (SCNMV) (PDB: 6D3P)48. The open circle denotes the start of the ring structure, the boxed 1 indicates the completion of one loop. In POWV, the RNA loops around the 5′ end a second time, indicated with green arrows and a boxed 2. B Proposed mechanistic model of Xrn1 encountering a class 2 xrRNA based on the three-dimensional structure and biochemical experiments. Depicted is the double loop consisting of the primary ring-like motif found in all solved xrRNAs to date (1) and the second ring resulting from the extended PK1 interaction in class 2 xrRNAs (2).
The expansion of PK1 and the resultant extended stem is due to a large insertion into the stretch of nucleotides that make up the ring structure (Fig. 3E). In the class 1 xrRNA, the length of the ring is conserved at ~15 nucleotides, forming a continuous and tight loop around the 5′ end of the xrRNA21,24,45. In contrast, in the POWV class 2 the large insertion between P2 and P3 creates an apparent break in the ring at the point of the insertion (Fig. 3D, E). However, closer examination reveals that the ring is still formed but is now expanded such that it wraps around the 5′ end twice (Fig. 5A). Thus, whereas all other xrRNA structures obtained to date show only a single loop around the 5′ end, the POWV class 2 forms a remarkable double loop ring with a full helical turn of base-paired RNA embedded within it.
Discussion
xrRNAs are ubiquitous in several genera of the viral family Flaviviridae, and are responsible for the production of non-coding viral sfRNAs. Understanding of the RNA structure-based mechanism of exoribonuclease resistance has come from solved structures of flavivirus class 1 xrRNAs, but this understanding lacked information on the 3-D structure of the class 2 xrRNAs. Here, we demonstrate the production of sfRNAs in POWV due to the presence of authentic xrRNAs and verify their secondary structures. We then integrated bioinformatic, biochemical, functional, existing high-resolution structural data, and intermediate-resolution cryo-EM maps to develop a 3-D structural model of a POWV class 2 xrRNA. The structural model, while unable to directly inform on detailed intramolecular interactions, reveals critical and unique features of the class 2 xrRNAs and furthers our understanding of the structural diversity of flavivirus xrRNAs.
One of the most distinctive features of the class 2 xrRNAs compared to previously solved xrRNAs is the extended PK1 and the associated expanded double loop ring structure (Fig. 5A). In other solved flavivirus xrRNA structures, the ring wraps around the 5′ end of the RNA only once, using a loop ~15 nucleotides in length21,24,45. This is also true in xrRNAs from some plant-infecting viruses46,47,48. The purpose of this expansion in the class 2 is not clear, however it is possible that the folding pathway is influenced by the extended PK1; that is, it helps prevent misfolding. However, xrRNAs with a smaller PK1 and ring structure can fold correctly. One other possibility is that the extended structure has a function beyond exoribonuclease resistance, such as binding a specific protein needed for other functions of the 3′UTR such as is seen in Zika virus82,83. Finally, there may be some advantage to a double loop ring around the 5′ end of the xrRNA that may change the mechanism or tune the efficiency of exonuclease resistance. Determining which of these or other possibilities will be an important focus of future studies.
In subclass 1a xrRNAs, such as those found in Zika virus and Murray Valley Encephalitis virus, PK2 is critical in forming the proper resistant fold and stacks with P321,45. Variation in the number of base-pairs in PK2 may change the stability of the xrRNA and has been shown to alter the dynamics of the RNA51. Adding base pairs may increase Xrn1 resistance in the presence of destabilizing mutations51, emphasizing its importance in folding and resistance. In class 2 xrRNAs, PK2 is always present and 4 base pairs long. Like in subclass 1a xrRNAs, class 2 PK2 stacks on P3, likely also participating in the stabilization of the overall fold and resistance.
The shared structural features between the class 1 and class 2 xrRNAs suggest a shared mechanism where the xrRNA braces against the surface of the enzyme, presenting a topological block that the enzyme cannot resolve. This in turn suggests that, similar to the class 1, class 2 xrRNAs can resist other 5’ to 3’ exoribonucleases such a yeast Dxo1 and bacterial RNase J22, but additional experiments are needed to verify this.
The presence of the double-loop ring structure in the class 2 xrRNAs raises questions of how this affects the details of exoribonuclease resistance compared to other xrRNAs. Unlike the class 1 xrRNAs, where the exoribonuclease halts on an unpaired region of RNA outside of the ring and pseudoknot, in the class 2 the enzyme halts at locations within PK1 in a region that pairs with nucleotides in the ring structure. This location of the enzyme halt site within PK1, knowledge of the structure and behavior of xrRNA, and the POWV xrRNA 3-D structure now allows a model to be proposed. Specifically, as the enzyme degrades in a 5′ to 3′ direction, it must unwind part of the PK1 stem. In fact, because ~5 nts of single-stranded RNA are needed to span the distance between Xrn1’s surface and its catalytic center, almost the entire PK1 stem will be unwound when the enzyme reaches its 3′-most halt point. In this state, the full stretch of RNA between P3 and P1, ~20 nts, will be unpaired but trapped between the enzyme and the xrRNA core, essentially wrapped twice around the 5′ end of the xrRNA. Thus, in the class 2 xrRNAs, the ring itself may be remodeled by the enzyme (Fig. 5B).
We predict that the induced steric clashes produced by this remodeling add resistance to the enzyme’s continued movement. Furthermore, the increasing steric clash as the RNA ring is pressed between the enzyme and the xrRNA core could progressively increase resistance, explaining the ‘stuttering effect’ observed when the halt sites were mapped (Fig. 2D, E). This differs from the class 1 in which there is no evidence that parts of the structure are unwound by Xrn1 or that the ring is remodeled, and the halt site is more precise20,21,24,45. This less-precise halting mechanism could also introduce variability in how subtly different class 2 xrRNAs behave, for example, the different halting patterns of POWV xrRNA1 and xrRNA2. While direct biophysical evidence for this process is not available, this model presents a testable model for future exploration.
The structure of a class 2 xrRNA presented here, combined with previous subclass 1a and subclass 1b structures, provides a picture of xrRNA diversity in the known flavivirus xrRNAs. All use a similar architecture comprising a central helical junction and two interwoven pseudoknots that create a compact fold. The ring-like structure is also ubiquitous, although the nature of the ring differs between the classes 1 and 2, suggesting that different interactions between the ring and the exoribonuclease provide resistance.
Despite the overall similarities in the architecture and the core fold, there are conserved differences that show how each class has evolved a precise and specific set of intramolecular tertiary contacts that cannot be interchanged easily. For example, while all form a ring-like element, the interactions between the base of the P1 stem and L3 that close the ring are different in each class. Likewise, the identity and presence of a nucleotide between P2 and P3, and the overall compactness of the ring element, are different across the classes and are not readily interchanged. Hence, evolution has crafted a set of related RNA folds using different sets of interactions to create similar 3-D outcomes, demonstrating the remarkable structural plasticity and adaptability of RNA structure.
Methods
Phylogenetic tree construction
The phylogenetic tree for Flaviviridae was constructed using the published maximum likelihood data using the viral NS5, the viral RNA dependent RNA polymerase, described in Bamford et al.84 and Mifsud et al.85 The tree was rendered in Figtree v1.4.4 (https://github.com/rambaut/figtree/releases) and adjusted in Adobe Illustrator.
Cell Culture and POWV growth
Vero (African green monkey kidney fibroblast; ATCC CCL-81; passage 15) were grown in 1x MEM (Corning) supplemented with 1% MEM non-essential amino acids (Gibco), 1% sodium pyruvate (Gibco), 1x pen/strep (Gibco), 1 mM HEPES (Gibco), and 10% heat inactivated FBS (Corning). BHK-21 (baby hamster kidney fibroblast cells; ATCC ccl-10) were maintained in DMEM (Cytiva) supplemented with 1 mM sodium pyruvate (Gibco), 27 mM sodium bicarbonate (Gibco), 1x Pen/strep (Gibco), and 1 µM amphotericin B (Gibco), and 8% heat inactivated FBS (Corning).
POWV stock generation and quantification
All viral culture was performed in a BSL-3 facility. One aliquot of POWV of unknown titer was blindly passaged onto an 80% confluent T-175 flask of Vero cells, and incubated for 10 days at 37 °C with intermittent rocking. Once 50–60% cell clearance was observed, the supernatant was collected and clarified by centrifugation at 300 x g for 20 minutes at 4 °C. From the clarified supernatant, concentrated viral stocks were generated via ultracentrifugation. Briefly, 20 mL of clarified supernatant was gently layered on top of 10 mL of 20% sucrose dissolved in PBS (4 °C, sterile filtered) in an ultra-clear 1 × 3.5′ centrifuge tube (Beckman 344058). The samples were ultracentrifuged at 140,000 x g in a SW28 rotor (Beckman) for 3.5 hours at 4 °C. The supernatant was aspirated, and the resultant viral pellet was resuspended in complete1x DMEM (1 mM sodium pyruvate, 27 mM sodium bicarbonate, 1x Pen/strep, and 1 µM amphotericin B) supplemented with 10% heat-inactivated FBS. Concentrated viral stocks were quantified by a plaque-forming unit assay with BHK-21 cells.
In vitro POWV infection and viral RNA extraction
Vero cells were grown in 6-well culture plate and inoculated with 0.5 mL of POWV stock or mock inoculum at a MOI of 1 or 5 in serum-free 1x complete MEM (1% MEM non-essential amino acids, 1% sodium pyruvate, 1x pen/strep, and 1 mM HEPES). After a one-hour incubation at 37 °C, the inoculum was then aspirated from the cells, washed with 1x PBS, and overlayed with 1x complete MEM. The cells were incubated at 37 °C for 24 or 48 hours. After incubation, approximately 1.0 × 106 Vero cells per condition were washed 3x in in cold PBS, scraped in buffer TLK (Omega Bio-tek), and homogenized in the Qiashredder (Qiagen) spin column disrupter. Total RNA was purified from the cellular lysate with E.Z.N.A Total RNA Kit (Omega Bio-tek) following the manufacturer’s instructions.
Northern Blot analysis of POWV sfRNA
1.0 µg of total RNA from the POWV or mock-infected cells was resuspended in 2x formamide loading buffer (Thermo) and run on a precast 6% Invitrogen Novex TBE gel at 160 V for one hour. The gels were stained with 0.5 µg/mL ethidium bromide and imaged on a UV imager. Next, the RNA was electroblotted to a sheet of HyBond-N+ nylon membrane (GE) using the Genie Electrophoretic Transfer Apparatus (Idea Scientific) at 1.6 V for 2 hours, following the manufacturer's instructions. UV crosslinking of the RNA to the nylon membrane was performed with a Stratalinker (Stratagene) using the optimal crosslink setting. Next, the membrane was cut into two separate pieces, separating the viral sfRNA from the U6 snRNA loading control. Each blot was transferred into individual glass vials and blocked with ULTRAhyb Oligo Hybridization Buffer (Ambion) at 42 °C for two hours with rotation. Source blots are included I the Source Data file.
During the blocking step, the radiolabeled DNA probes were prepared. 100 pmol of POWV_Probe and U6_probe DNA probes were radioactively labeled with 5 mCi of [γ32-P] ATP using 20 units of T4 polynucleotide kinase (NEB) for 2 hours at 37 °C. The radiolabeled probes were purified with P-30 spin columns (BioRad). The probes were heated to 90 °C for 2 minutes and quickly resuspended in 10 mL of 42 °C ULTRAhyb Oligo Hybridization Buffer (Ambion).
Next, the blocking ULTRAhyb Oligo Hybridization Buffer (Ambion) was removed from the membranes and replaced with the 10 mL buffer containing the radiolabeled probe and incubated overnight with rotation at 42 °C. The blots were then washed 4 times in 2x saline-sodium citrate (SSC buffer; 300 mM NaCl, 30 mM Na3C6H5O7, 0.5% SDS, pH 7.0) for 15 minutes at 42 °C. The blots were imaged on the Typhoon 9400 scanner (GE Life Sciences) and aligned with the ethidium bromide-stained UV image to get the appropriate sizing of the bands.
Class 2 xrRNA bioinformatic searches and covariance model analysis
An initial class 2 xrRNA alignment was created from a total of 28 sequences of proposed class 2 xrRNA from TBFV 3′ UTR. The sequences were manually aligned with RALEE v0.886 using previous TBFV class 2 secondary structure alignments as a reference. Using Infernal v1.1.480 a database of all available +ssRNA viral sequences downloaded from the National Center for Biotechnology Information (NCBI; last retrieved on 1/06/2023) was searched using default parameters. Hits from the Infernal searches were manually added to the comparative sequence alignment if they met all the following criteria: Infernal E value < 0.05, >15% nucleotide variation within each sequence, and location within the 3′ UTR. Sequences with higher E values were also inspected and added to the list if they met the last three requirements. For the final proposed covariance model of 36 class 2 xrRNA sequences, statistical validation was performed using R-scape v2.0.0.j76,79 and rendering the covariance model with R2R v1.0.581.
In vitro RNA transcription
DNA templates were ordered as gBlock DNA fragments (IDT) and cloned into pUC19. 200 µL PCR reactions using primers containing an upstream T7 promoter were used to generate dsDNA templates for transcription. Typical PCR conditions: 100 ng plasmid DNA, 0.5 µM forward and reverse DNA primers (Supplementary Data 2), 500 µM dNTPs, 25 mM TAPS-HCl (pH 9.3), 50 mM KCl, 2 mM MgCl2, 1 mM β-mercaptoethanol, and Phusion DNA polymerase (New England BioLabs). Templates for RNA used in aminoacylation assays were amplified using reverse primers containing two 5′-terminal 2′-O-methyl modified bases to ensure the correct 3′ end of the RNA. dsDNA amplification was confirmed by 1.5% agarose gel electrophoresis. Transcriptions were performed in 1 mL volume using 200 µL of PCR product (~0.1 µM template DNA) and 10 mM NTPs, 75 mM MgCl2, 30 mM Tris-HCl (pH 8.0), 10 mM DTT, 0.1% spermidine, 0.1% Triton X-100, and T7 RNA polymerase. Reactions were incubated at 37 °C overnight. After transcription, insoluble inorganic pyrophosphate was removed by centrifugation at 5000 x g for 5 minutes, then the RNA-containing supernatant was ethanol precipitated with 3 volumes of 100% ethanol at −80 °C for a minimum of 1 hour and then centrifuged at 21,000 x g for 30 minutes at 4 °C to pellet the RNA, and the ethanolic fraction was decanted. The RNA was resuspended in 9 M urea loading buffer then purified by denaturing 10% PAGE. Bands were visualized by UV shadowing then excised. Bands were then crush-soaked in diethylpyrocarbonate-treated (DEPC) milli-Q water at 4 °C overnight. The RNA-containing supernatant was then concentrated using spin concentrators (Amicon) to the appropriate concentration in DEPC-treated water. RNAs were stored at −80 °C with working stocks stored at −20 °C. Normal levels of batch-specific variations in RNA purity were observed between preparations of RNA.
Expression of Kluyveromyces lactis Xrn1∆C
The DNA sequence encoding the Xrn1 enzyme from K. lactis (residues 1-1245) containing an in-frame C-terminal hexahistidine affinity tag was a kind gift from Dr. Liang Tong at Columbia University. The protein was recombinantly expressed in BL21 (DE3) cells. Cells were grown in LB to an OD600 of 0.6, then protein expression was induced using 500 µM isopropyl β-D-1-thiogalactopyranoside (IPTG) overnight at 18 °C. Pelleted cells were resuspended in lysis buffer containing 20 mM Tris-HCl (pH 7.0), 500 mM NaCl, 2 mM β-mercaptoethanol (BME), 10% (v/v) glycerol, and cOmplete EDTA-free protease inhibitor cocktail tablets (Roche). Cell lysate was then sonicated on ice for 2 minutes of: 20 seconds on, 40 seconds off at 75 W. Cell lysate was clarified by centrifugation at 30,000xg for 30 minutes at 4 °C. The soluble fraction was purified by nickel affinity chromatography in a buffer containing 150 mM NaCl, 20 mM Tris-HCl (pH 7.0), 200 mM imidazole, 10% glycerol, and 2 mM β-mercaptoethanol. The protein was exchanged into a storage buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 2 mM DTT, and 50% glycerol using a spin concentrator (Amicon) and stored at 1.0 mg mL-1 at −80 °C, with working stocks stored at −20 °C.
Expression of Bdellovibrio bacteriovorus RppH
A plasmid containing BdRppH (Uniprot ID Q6MPX4_BDEBA, Q6MPX4) in frame with a hexahistidine tag was a kind gift of J. Belasco at New York University. BdRppH was purified in a manner identical to that of Xrn1∆C as described above. The protein was concentrated to 12 mg mL−1 in a buffer containing 150 mM NaCl, 25 mM Tris (pH 7.5), 1 mM DTT, and 50% (v/v) glycerol and then stored at −80 °C.
In vitro RNA degradation assays
Assays were performed using 2 µg of RNA in a 20 µL volume. RNA was refolded as described above, then divided into two 10 µL volumes. To both, 1 µL of BdRppH was added and to one 1 µL of KlXrn1∆C was added. To the other volume 1 µL of KlXrn1∆C storage buffer (150 mM NaCl, 50 mM Tris, 2 mM DTT, 50% (v/v) glycerol, pH 8.0) was added. The reactions were then incubated at 37 °C for 2 hours. The reactions were quenched with the addition of 1 volume of 9 M urea loading buffer and resolved on a 10% denaturing PAGE gel, then the RNA was visualized by methylene blue staining.
Stop site analysis
Full-length in vitro transcribed POWV 3′UTR was degraded with Xrn1 following the in vitro Xrn1 degradation assay outlined above. A FAM-labeled primer specific to the 3′ terminal end was annealed to the RNA and then immobilized on oligo-dT25 beads (Invitrogen). The RNA was reverse transcribed on the oligo-dT25 beads with 2.5 µL of reverse transcription mixture consisting of 1x first strand buffer (Invitrogen), 25 mM 1,4-Dithiothreitol (DTT), 4 mM dNTPs, and 20 units of Superscript III reverse transcriptase (Invitrogen). The mixture was incubated at 52 °C for 50 min and the RNA was subsequently hydrolyzed with 0.4 M NaOH at 65 °C for 20 min. The samples were cooled on ice for 2 min and then neutralized with 5 µL of an acid quench mixture consisting of 1.4 M NaCl, 0.6 N HCl, and 1.3 M NaOAc. The beads were washed 3x with 70% EtOH, dried, and the cDNA was eluted in 11 µL of a ROX-formamide mixture composed of 2.75 µL of ROX-500 ladder (Applied Biosystems) diluted in 1.2 mL of HiDi-formamide (Applied Biosystems) at room temperature for 15 min. The cDNA mixture was transferred to an optical plate and analyzed with an ABI 3500 Genetic Analyzer Capillary Electrophoresis machine (GE).
In parallel, a Sanger sequencing ladder was constructed through reverse transcription of unmodified POWV 3′UTR (Data S2). In brief, 2.5 µL of an RNA mixture was made consisting of .2 µL of 0.25 µM FAM primer, 1.5 µL of oligio-dT25 beads (Invitrogen), 0.5 µL of 2.4 µM RNA, and 0.25 µL of 5 M betaine. To this, 2.5 µL of a ladder reverse transcription mixture 2.5 µL of reverse transcription mixture consisting of 1x first strand buffer (Invitrogen), 25 mM 1,4-Dithiothreitol (DTT), 4 mM dNTPs, and 20 units of Superscript III reverse transcriptase (Invitrogen). The cDNA was eluted off the beads in 11 µL of a ROX-formamide outlined above. The 11 µL cDNA mixture was transferred to an optical plate and analyzed with an ABI 3500 Genetic Analyzer Capillary Electrophoresis machine (GE). After running the Xrn1 degraded 3′UTR and the Sanger Sequencing ladder on the ABI 3500 Genetic analyzer Capillary Electrophoresis machine, the HiTRACE MATLAB suite87 (https://ribokit.github.io/HiTRACE/) with MATLAB (v8.3.0.532) was used to analyze the stop site data.
In vitro probing of POWV 3’UTR, sfRNA 1, and sfRNA 2
1m7 and DMS probing of the POWV 3’UTR
1.0 µg of in vitro transcribed full-length POWV 3′UTR was heat denatured in 12 µL DEPC-treated H2O at 95 °C for two minutes and then immediately transferred to ice. To this 8.0 µL of concentrated RNA folding buffer was added with a final concentration of 100 mM NaCl, 50 mM Tris pH 7.5, 4 mM MgCl2 in 20 µL. The RNA was folded for 20 minutes at 55 °C and slowly cooled to room temperature for 10 minutes. After cooling, the reaction was split into two equal 0.5ug RNA aliquots to be modified with their respective chemical probe and control. One 0.5ug aliquot was labeled with 10 mM 1m7 (Sigma) or DMSO (Sigma) at 37 °C for 75 seconds (five 1m7 hydrolysis half-lives). In parallel, the other 0.5 µg of folded RNA was modified with 0.5% DMS (Sigma) or EtOH for 5 minutes at 37 °C, and then quenched with 2-Mercaptoethanol to a final concentration of 37.5% (Sigma). All conditions were purified and concentrated using the RNeasy Micro Kit (Qiagen).
1m7 and DMS probing of the POWV sfRNA 1 and sfRNA 2
2 µg of folded in vitro transcribed POWV 3′UTR was degraded with Xrn1 following the in vitro Xrn1 degradation assay outlined above. Once the reaction was complete, the RNA was modified with 0.5% DMS, 10 mM 1m7, and their appropriate controls. The reactions were cleaned up using the RNeasy Micro Kit (Qiagen), and the sfRNA 1 and sfRNA 2 bands were separated on a 6% denaturing PAGE gel. sfRNA bands were visualized by UV shadowing, cut from the gel, and eluted at 4 °C overnight in ~5 mL of elution buffer consisting of 50 mM Sodium cacodylate, pH 6.5, 0.1 mM EDTA, and 50 mM NaCl. The RNA was concentrated with a 30 K molecular weight cut-off Amicon Ultra spin Concentrators (Millipore-Sigma) and buffer exchanged into DEPC water.
Reverse transcription and second-strand synthesis
The following reverse transcription and second-strand synthesis method was applied to the full-length 3′UTR, sfRNA 1, and sfRNA 2 RNA species. Approximately 400 ng of modified or control RNA was incubated with 20 ng/µL random primer 9 (NEB; S1254S) and 1.0 µM of POWV_SHAPEMAP_RT_Primer (Supplemental Data S2) in a final volume of 11 µL. The RNA was incubated at 65 °C for 5 minutes and immediately cooled on ice. To each of the reactions 8.0 µL of concentrated MAP buffer (125 mM Tris pH 8.0, 187.5 mM KCl, 15 mM MnCl2, 25 mM DTT, 1.25 mM equimolar dNTP mix) was added to the reaction followed by 200 units of Superscript II reverse transcriptase (Invitrogen) for a total reaction volume of 20uL. The mixture was incubated in a thermocycler at 25 °C for 10 minutes, 42 °C for 3 hours, 70 °C for 15 minutes, with a final cooling step to 4 °C. The RNA/cDNA hybrids were then purified with G25 spin column (Cyteva) following the manufacturer's protocol. Second-strand synthesis was carried out using the NEBNext Ultra II Non-Directional RNA Second Strand Synthesis Module (NEB; E6111) following the manufacturer’s instructions. The resultant cDNA was purified using the DNA Clean & Concentrator-5 kit (Zymo) eluting in 10 µL of DEPC-treated H2O and quantified fluorometrically with the Qubit (Thermo) using the dsDNA HS Assay Kit (Thermo).
Library prep and sequencing
Next generation sequencing library prep was performed with the Nextera XT DNA library preparation kit (Illumina) using the Nextera XT Index Kit v2 Set B (Illumina) following the manufactures instructions. Samples were normalized and pooled together using a combination of the Qubit (Thermo) and the 4200 TapeStation System (Agilent Technologies) with the High Sensitivity D1000 Screen Tape (Agilent Technologies). Multiplexed libraries were sequenced on the NovaSeq 6000 platform (Illumina).
Data analysis
Raw sequencing reads were first assessed with FastQC and sequencing adapters were trimmed using Trimmomatic v0.4088. The paired end processed reads were then analyzed with Shapemapper289 using the default parameters with a –min-depth 5000. The normalized reactivity data was analyzed with a custom in house pipeline consisting of Python v3.9 and R v4.2.2 scripts. Data was visualized on the secondary structure using Varna 3.9390.
In Vitro RNA transcription and purification for Cryo-Electron Microscopy
DNA templates were ordered as gBlock DNA fragments (IDT) and cloned into pUC19 or as overlapping primers. 1 mL PCR reactions using primers containing an upstream T7 promoter were used to generate dsDNA templates for transcription. Typical PCR conditions: 1 ng/µL plasmid DNA, 0.5 µM forward and reverse DNA primers, 500 µM dNTPs, 25 mM TAPS-HCl (pH 9.3), 50 mM KCl, 2 mM MgCl2, 1 mM β-mercaptoethanol, and Phusion DNA polymerase (New England BioLabs). dsDNA amplification was confirmed by 1.5% agarose gel electrophoresis. Transcriptions were performed in 5 mL volume using 1 mL of PCR product ( ~ 0.1 µM template DNA) and 10 mM NTPs, 75 mM MgCl2, 30 mM Tris-HCl (pH 8.0), 10 mM DTT, 0.1% spermidine, 0.1% Triton X-100, and T7 RNA polymerase. Reactions were incubated at 37 °C for 3 hours. After transcription, insoluble inorganic pyrophosphate was removed by centrifugation at 5000 x g for 5 minutes, then the RNA-containing supernatant was ethanol precipitated with 3 volumes of 100% ethanol at −80 °C for a minimum of 1 hour and then centrifuged at 21000xg for 30 minutes at 4 °C to pellet the RNA, and the ethanolic fraction was decanted. The RNA was resuspended in 9 M urea loading buffer then purified by denaturing 6% PAGE. Bands were visualized by UV shadowing then excised. Bands were then crush-soaked in diethylpyrocarbonate-treated (DEPC) milli-Q water at 4 °C overnight. The RNA-containing supernatant was then concentrated using spin concentrators (Amicon) to the appropriate concentration in DEPC-treated water. RNAs were stored at −80 °C with working stocks stored at −20 °C.
POWV RNA refolding
RNA was refolded at the described concentrations as follows. To the RNA solution (usually 18 µL of 3 mg/mL), 1/20th the volume of 1 M Tris pH 7.5 was added. The solution was heated to 90 °C for 3 minutes, then allowed to cool to room temperature for 10 minutes. Subsequently, 1/20th the volume of 200 mM MgCl2 was added. The solution was then heated to 50 °C for 30 minutes then allowed to cool to room temperature for 10 minutes. Once cooled, the folded RNA solution was stored on ice until use.
Cryo-EM sample preparation and data acquisition
RNA sample was applied to a glow discharged C-flat (1.2/1.3, 400 mesh) holey carbon grid. Samples were vitrified in liquid ethane using a Vitrobot Mark IV (5 s wait time, 5.5 s blot, −5 blot force, 100% humidity, 4 °C). Samples were imaged on a Titan Krios equipped with a Gatan K3 camera and Bioquantum energy filter. Movies were collected in fringe-free mode, with a physical pixel size of 0.788 Å, total dose of 50 e-/Å2, and a defocus range of −0.8 to −2.0 µm. SerialEM91 v4.1 was used for all data collection. Additional information on data collection can be found in Supplementary Table 1.
Cryo-EM data processing
The general data processing workflow was approximately equivalent for all datasets using cryoSPARC 4.0. Briefly, all data were patch motion corrected, and the contrast transfer function was locally fit. An initial set of particles was picked using blob picker from the first 1000 micrographs. Particles were extracted and 2D classified. Good classes for template picking were manually chosen. Template-based picking was performed using template picker on the entire dataset. Particles were then extracted and subjected to several rounds of 2D classification to sort out junk particles. Good 2D classes and the associated particles were manually curated after each round. The pruned particles were then used for initial map generation using the Ab Initio reconstruction job requesting 2 volumes. The resulting 2 volumes were used for heterogeneous refinement, where one class was the good class and the other a junk class to remove poor quality particles. Several rounds of heterogeneous refinement were performed, discarding the junk particles after each round until the map quality stopped improving. Subsequently, the good volume and particles were subjected to non-uniform refinement. Local motion correction, local CTF refinement, and global CTF refinement were then carried out on the data and the particles were subjected to further rounds of non-uniform refinement to yield the final maps. The final workflow, an abbreviated version of all processes attempted, and additional information on data processing for the map can be found in Supplementary Fig. 5.
Structure modeling
To build the atomic model, the Cryo-EM structure of the tetrahymena ribozyme was placed into the maps using ridged body fitting (PDB: 8TJX)65 followed by the solved crystal structure of the Tamana bat virus xrRNA (PDB:7K16)24. The model was then manually built using the TABV xrRNA as an initial reference and joined in COOT92 v0.8.9.1. The final model was refined against the map by way of several rounds of real-space refinement in PHENIX93 v1.21.1 and manual adjustments in COOT. The statistics of model refinement and validation are listed in Supplementary Table 1.
Data availability
Structure coordinates and map file have been deposited in the Protein Data Bank (PDB) and Electron Microscopy Data Bank (EMDB) under accession numbers 9DP9 and 47099, respectively. Raw chemical probing data is available at the NCBI’s Gene Expression Omnibus archive under accession GSE270001. The xrRNA class 2 alignment is available in Stockholm format as online supplementary material. Source data are provided with this paper (https://doi.org/10.6084/m9.figshare.27317046). Source data are provided with this paper.
References
Mackenzie, J. S., Gubler, D. J. & Petersen, L. R. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 10, S98–S109 (2004).
Garcia-Blanco, M. A., Ooi, E. E. & Sessions, O. M. RNA viruses, pandemics and anticipatory preparedness. Viruses 14, 2176 (2022).
ME, J. W., Adair, K. & Brierley, L. RNA Viruses: A case study of the biology of emerging infectious diseases. Microbiol. Spectr. 1, OH-0001-2012 (2013).
Pierson, T. C. & Diamond, M. S. The continued threat of emerging flaviviruses. Nat. Microbiol 5, 796–812 (2020).
Valderrama, A., Diaz, Y. & Lopez-Verges, S. Interaction of Flavivirus with their mosquito vectors and their impact on the human health in the Americas. Biochem. Biophys. Res Commun. 492, 541–547 (2017).
Guarner, J. & Hale, G. L. Four human diseases with significant public health impact caused by mosquito-borne flaviviruses: West Nile, Zika, dengue and yellow fever. Semin. Diagn. Pathol. 36, 170–176 (2019).
Daep, C. A., Munoz-Jordan, J. L. & Eugenin, E. A. Flaviviruses, an expanding threat in public health: focus on dengue, West Nile, and Japanese encephalitis virus. J. Neurovirol. 20, 539–560 (2014).
Calisher, C. H. & Gould, E. A. Taxonomy of the virus family Flaviviridae. Adv. Virus Res. 59, 1–19 (2003).
Simmonds, P. et al. ICTV Virus Taxonomy Profile: Flaviviridae. J. Gen. Virol. 98, 2–3 (2017).
Fields, B. N., Knipe, D. M. & Howley, P. M. Fields virology, (Wolters Kluwer/ Lippincott Williams & Wilkins Health, Philadelphia, PA, USA, 2013).
Brinton, M. A. Replication cycle and molecular biology of the West Nile virus. Viruses 6, 13–53 (2013).
Ng, W. C., Soto-Acosta, R., Bradrick, S. S., Garcia-Blanco, M. A. & Ooi, E. E. The 5’ and 3’ Untranslated Regions of the Flaviviral Genome. Viruses 9, 137 (2017).
Selisko, B., Wang, C., Harris, E. & Canard, B. Regulation of Flavivirus RNA synthesis and replication. Curr. Opin. Virol. 9, 74–83 (2014).
Brinton, M. A. & Basu, M. Functions of the 3’ and 5’ genome RNA regions of members of the genus Flavivirus. Virus Res. 206, 108–119 (2015).
Slonchak, A. & Khromykh, A. A. Subgenomic flaviviral RNAs: What do we know after the first decade of research. Antivir. Res. 159, 13–25 (2018).
Roby, J. A., Pijlman, G. P., Wilusz, J. & Khromykh, A. A. Noncoding subgenomic flavivirus RNA: multiple functions in West Nile virus pathogenesis and modulation of host responses. Viruses 6, 404–427 (2014).
Pijlman, G. P. et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 4, 579–591 (2008).
Funk, A. et al. RNA structures required for production of subgenomic flavivirus RNA. J. Virol. 84, 11407–11417 (2010).
Silva, P. A., Pereira, C. F., Dalebout, T. J., Spaan, W. J. & Bredenbeek, P. J. An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1. J. Virol. 84, 11395–11406 (2010).
Chapman, E. G., Moon, S. L., Wilusz, J. & Kieft, J. S. RNA structures that resist degradation by Xrn1 produce a pathogenic Dengue virus RNA. Elife 3, e01892 (2014).
Akiyama, B. M. et al. Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science 354, 1148–1152 (2016).
MacFadden, A. et al. Mechanism and structural diversity of exoribonuclease-resistant RNA structures in flaviviral RNAs. Nat. Commun. 9, 119 (2018).
Vicens, Q. & Kieft, J. S. Shared properties and singularities of exoribonuclease-resistant RNAs in viruses. Comput. Struct. Biotechnol. J. 19, 4373–4380 (2021).
Jones, R. A. et al. Different tertiary interactions create the same important 3D features in a distinct flavivirus xrRNA. RNA 27, 54–65 (2021).
Slonchak, A. et al. Structural analysis of 3’UTRs in insect flaviviruses reveals novel determinants of sfRNA biogenesis and provides new insights into flavivirus evolution. Nat. Commun. 13, 1279 (2022).
de Borba, L. et al. RNA structure duplication in the Dengue Virus 3’ UTR: Redundancy or host specificity? mBio 10, e02506–18 (2019).
Villordo, S. M., Carballeda, J. M., Filomatori, C. V. & Gamarnik, A. V. RNA structure duplications and flavivirus host adaptation. Trends Microbiol 24, 270–283 (2016).
Chang, R. Y. et al. Japanese encephalitis virus non-coding RNA inhibits activation of interferon by blocking nuclear translocation of interferon regulatory factor 3. Vet. Microbiol 166, 11–21 (2013).
Donald, C. L. et al. Full genome sequence and sfRNA Interferon antagonist activity of Zika Virus from Recife, Brazil. PLoS Negl. Trop. Dis. 10, e0005048 (2016).
Filomatori, C. V. et al. Dengue virus genomic variation associated with mosquito adaptation defines the pattern of viral non-coding RNAs and fitness in human cells. PLoS Pathog. 13, e1006265 (2017).
Schuessler, A. et al. West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response. J. Virol. 86, 5708–5718 (2012).
Manokaran, G. et al. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 350, 217–221 (2015).
Bidet, K., Dadlani, D. & Garcia-Blanco, M. A. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog. 10, e1004242 (2014).
Clarke, B. D., Roby, J. A., Slonchak, A. & Khromykh, A. A. Functional non-coding RNAs derived from the flavivirus 3’ untranslated region. Virus Res. 206, 53–61 (2015).
Goertz, G. P. et al. Mosquito small RNA responses to West Nile and insect-specific virus infections in Aedes and Culex mosquito cells. Viruses 11, 271 (2019).
Moon, S. L. et al. Flavivirus sfRNA suppresses antiviral RNA interference in cultured cells and mosquitoes and directly interacts with the RNAi machinery. Virology 485, 322–329 (2015).
Schnettler, E. et al. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and Mammalian cells. J. Virol. 86, 13486–13500 (2012).
Schnettler, E. et al. Induction and suppression of tick cell antiviral RNAi responses by tick-borne flaviviruses. Nucleic Acids Res. 42, 9436–9446 (2014).
Yeh, S. C. & Pompon, J. Flaviviruses produce a subgenomic Flaviviral RNA that enhances mosquito transmission. DNA Cell Biol. 37, 154–159 (2018).
Hogg, J. R. Viral evasion and manipulation of Host RNA quality control pathways. J. Virol. 90, 7010–7018 (2016).
Moon, S. L. et al. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA 18, 2029–2040 (2012).
Goertz, G. P. et al. Subgenomic flavivirus RNA binds the mosquito DEAD/H-box helicase ME31B and determines Zika virus transmission by Aedes aegypti. Proc. Natl Acad. Sci. USA 116, 19136–19144 (2019).
Goertz, G. P. et al. Noncoding Subgenomic Flavivirus RNA Is Processed by the Mosquito RNA Interference Machinery and Determines West Nile Virus Transmission by Culex pipiens Mosquitoes. J. Virol. 90, 10145–10159 (2016).
Villordo, S. M., Filomatori, C. V., Sanchez-Vargas, I., Blair, C. D. & Gamarnik, A. V. Dengue virus RNA structure specialization facilitates host adaptation. PLoS Pathog. 11, e1004604 (2015).
Chapman, E. G. et al. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science 344, 307–310 (2014).
Steckelberg, A. L., Vicens, Q., Costantino, D. A., Nix, J. C. & Kieft, J. S. The crystal structure of a Polerovirus exoribonuclease-resistant RNA shows how diverse sequences are integrated into a conserved fold. RNA 26, 1767–1776 (2020).
Steckelberg, A. L., Vicens, Q. & Kieft, J. S. Exoribonuclease-Resistant RNAs Exist within both Coding and Noncoding Subgenomic RNAs. mBio 9, e02461–18 (2018).
Steckelberg, A. L. et al. A folded viral noncoding RNA blocks host cell exoribonucleases through a conformationally dynamic RNA structure. Proc. Natl Acad. Sci. USA 115, 6404–6409 (2018).
Zhao, M. & Woodside, M. T. Mechanical strength of RNA knot in Zika virus protects against cellular defenses. Nat. Chem. Biol. 17, 975–981 (2021).
Niu, X. et al. Molecular mechanisms underlying the extreme mechanical anisotropy of the flaviviral exoribonuclease-resistant RNAs (xrRNAs). Nat. Commun. 11, 5496 (2020).
Niu, X. et al. Pseudoknot length modulates the folding, conformational dynamics, and robustness of Xrn1 resistance of flaviviral xrRNAs. Nat. Commun. 12, 6417 (2021).
Becchi, M., Chiarantoni, P., Suma, A. & Micheletti, C. RNA pore translocation with static and periodic forces: effect of secondary and tertiary elements on process activation and duration. J. Phys. Chem. B 125, 1098–1106 (2021).
Mainan, A., Kundu, R., Singh, R. K. & Roy, S. Magnesium regulates RNA ring dynamics and folding in subgenomic Flaviviral RNA. J. Phys. Chem. B 128, 9680–9691 (2024).
Szucs, M. J., Nichols, P. J., Jones, R. A., Vicens, Q. & Kieft, J. S. A new subclass of Exoribonuclease-resistant RNA found in multiple genera of Flaviviridae. mBio 11, e02352–20 (2020).
Dilweg, I. W. et al. Xrn1-resistant RNA structures are well-conserved within the genus flavivirus. RNA Biol. 18, 709–717 (2021).
Dilweg, I. W. et al. All genera of Flaviviridae host a conserved Xrn1-resistant RNA motif. RNA Biol. 18, 2321–2329 (2021).
Ochsenreiter, R., Hofacker, I. L. & Wolfinger, M. T. Functional RNA structures in the 3’UTR of tick-borne, insect-specific and no-known-vector Flaviviruses. Viruses 11, 298 (2019).
Proutski, V., Gould, E. A. & Holmes, E. C. Secondary structure of the 3’ untranslated region of flaviviruses: similarities and differences. Nucleic Acids Res. 25, 1194–1202 (1997).
Kappel, K. et al. Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures. Nat. Methods 17, 699–707 (2020).
Ma, H. et al. Auto-DRRAFTER: automated RNA modeling based on Cryo-EM density. Methods Mol. Biol. 2568, 193–211 (2023).
Zhang, K. et al. Cryo-EM and antisense targeting of the 28-kDa frameshift stimulation element from the SARS-CoV-2 RNA genome. Nat. Struct. Mol. Biol. 28, 747–754 (2021).
Su, Z. et al. Cryo-EM structures of full-length Tetrahymena ribozyme at 3.1 A resolution. Nature 596, 603–607 (2021).
Kappel, K. et al. De novo computational RNA modeling into cryo-EM maps of large ribonucleoprotein complexes. Nat. Methods 15, 947–954 (2018).
Liu, D., Thelot, F. A., Piccirilli, J. A., Liao, M. & Yin, P. Sub-3-A cryo-EM structure of RNA enabled by engineered homomeric self-assembly. Nat. Methods 19, 576–585 (2022).
Langeberg, C. J. & Kieft, J. S. A generalizable scaffold-based approach for structure determination of RNAs by cryo-EM. Nucleic Acids Res. 51, e100 (2023).
Pesko, K. N., Torres-Perez, F., Hjelle, B. L. & Ebel, G. D. Molecular epidemiology of Powassan virus in North America. J. Gen. Virol. 91, 2698–2705 (2010).
Piantadosi, A. & Solomon, I. H. Powassan Virus Encephalitis. Infect. Dis. Clin. North Am. 36, 671–688 (2022).
Brackney, D. E. & Vogels, C. B. F. The known unknowns of Powassan virus ecology. J. Med Entomol. 60, 1142–1148 (2023).
Jinek, M., Coyle, S. M. & Doudna, J. A. Coupled 5’ nucleotide recognition and processivity in Xrn1-mediated mRNA decay. Mol. Cell 41, 600–608 (2011).
Langeberg, C. J. et al. Biochemical characterization of Yeast Xrn1. Biochemistry 59, 1493–1507 (2020).
Olson, S. W. et al. Discovery of a large-scale, cell-state-responsive allosteric switch in the 7SK RNA using DANCE-MaP. Mol. Cell 82, 1708–1723.e10 (2022).
Zubradt, M. et al. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat. Methods 14, 75–82 (2017).
Lee, B. et al. Comparison of SHAPE reagents for mapping RNA structures inside living cells. RNA 23, 169–174 (2017).
Saha, K. & Ghosh, G. Chemical probing of RNA structure in vivo using SHAPE-MaP and DMS-MaP. Methods Mol. Biol. 2666, 81–93 (2023).
Gao, W., Jones, T. A. & Rivas, E. Discovery of 17 conserved structural RNAs in fungi. Nucleic Acids Res. 49, 6128–6143 (2021).
Rivas, E., Clements, J. & Eddy, S. R. Estimating the power of sequence covariation for detecting conserved RNA structure. Bioinformatics 36, 3072–3076 (2020).
Rivas, E. Evolutionary conservation of RNA sequence and structure. Wiley Interdiscip. Rev. RNA 12, e1649 (2021).
Kalvari, I. et al. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 46, D335–D342 (2018).
Rivas, E., Clements, J. & Eddy, S. R. A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat. Methods 14, 45–48 (2017).
Nawrocki, E. P. & Eddy, S. R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935 (2013).
Weinberg, Z. & Breaker, R. R. R2R-software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinforma. 12, 3 (2011).
Slonchak, A. et al. Zika virus noncoding RNA cooperates with the viral protein NS5 to inhibit STAT1 phosphorylation and facilitate viral pathogenesis. Sci. Adv. 8, eadd8095 (2022).
Michalski, D. et al. Zika virus noncoding sfRNAs sequester multiple host-derived RNA-binding proteins and modulate mRNA decay and splicing during infection. J. Biol. Chem. 294, 16282–16296 (2019).
Bamford, C. G. G., de Souza, W. M., Parry, R. & Gifford, R. J. Comparative analysis of genome-encoded viral sequences reveals the evolutionary history of flavivirids (family Flaviviridae). Virus Evol. 8, veac085 (2022).
Mifsud, J. C. O. et al. Transcriptome mining extends the host range of the Flaviviridae to non-bilaterians. Virus Evol. 9, veac124 (2023).
Griffiths-Jones, S. RALEE-RNA ALignment editor in Emacs. Bioinformatics 21, 257–259 (2005).
Yoon, S. et al. HiTRACE: high-throughput robust analysis for capillary electrophoresis. Bioinformatics 27, 1798–1805 (2011).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Busan, S. & Weeks, K. M. Accurate detection of chemical modifications in RNA by mutational profiling (MaP) with ShapeMapper 2. RNA 24, 143–148 (2018).
Darty, K., Denise, A. & Ponty, Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 25, 1974–1975 (2009).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Acknowledgements
The authors thank current and past members of the Kieft Lab for critical reading of the manuscript and insightful comments. Theo Humphries (PNCC) assisted with microscope operations. David Beckham (UT Southwestern) provided helpful discussions for the BSL3 work performed herein. POWV Spooner isolate was generously provided by Dr. Aaron Brault (CDC, Division of Vector-Borne Diseases). This work was supported by R01AI133348 to J.S.K. A portion of this research was supported by NIH grant U24GM129547 and performed at the PNCC at OHSU and accessed through EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research.
Author information
Authors and Affiliations
Contributions
M.J.S., C.J.L, Q.V., and J.S.K conceived and planned the overall study. M.J.S, C.J.L., and M.E.S. conducted and analyzed the virological, biochemical and bioinformatic experiments. C.J.L. conducted the cryo-EM studies and built the structural model. C.J.L. and M.J.S. prepared the initial manuscript draft, which was edited and approved by all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors have no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Langeberg, C.J., Szucs, M.J., Sherlock, M.E. et al. Tick-borne flavivirus exoribonuclease-resistant RNAs contain a double loop structure. Nat Commun 16, 4515 (2025). https://doi.org/10.1038/s41467-025-59657-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-59657-7