Abstract
In patients with hepatocellular carcinoma (HCC) meeting the Milan criteria, liver transplantation (LT) is an effective therapy. This study aims to define the survival-related molecular biological features helping precisely identifying the patients with HCC beyond the Milan criteria who have acceptable outcomes. In the derivation cohort, integrated analyses of tumor tissues are conducted using RNA sequencing (RNA-seq), proteomic landscape, and transposase-accessible chromatin sequencing (ATAC-seq). Based on transcriptomics, three subgroups that significantly differ in overall survival were identified in the derivation cohort, and these findings are validated in an independent cohort. In-depth bioinformatics analysis using RNA-seq and proteomics reveals that the promotion of cancer stemness by cancer-associated fibroblasts (CAFs) can be responsible for the negative biological characteristics observed in high-risk HCC patients. The ATAC-seq identifies key factors regulating transcription, which may bridge CAF infiltration and stemness. Finally, we demonstrate that the CAF-derived CXCL12 sustains the stemness of HCC cells by promoting XRCC5 through CXCR4.
Similar content being viewed by others
Introduction
Liver transplantation (LT) is a radical method for treating hepatocellular carcinoma (HCC), especially for candidates within the Milan criteria (single tumor with a diameter of ≤5 cm, or up to 3 tumors each with a diameter of ≤3 cm; no extra-hepatic metastases; no extra-hepatic metastases; no major vessel involvement)1,2. LT in patients with HCC within the Milan criteria generally yields a 5-year overall survival (OS) of 70%. Expansion of the criteria for LT has been proposed for several years. In the last two decades, modified and expanded selection criteria have been developed2,3. Tumor morphology, including tumor size and number, was mainly increased in the expanded criteria. A high alpha fetal protein (AFP) value before LT has also been associated with a higher risk of HCC recurrence4,5,6.
Recent advances in systems biology techniques, such as genomic, transcriptomic, and proteomic analysis, have led to the discovery of molecular mechanisms that participate in the initiation and evolution of HCC7. Integrative multi-omic analyses have revealed several molecular subtypes of HCC associated with specific molecular characteristics and clinical outcomes8,9,10,11,12. For recipients with HCC beyond the Milan criteria, a gene set of progenitor cell markers can be used to predict poorer survival after LT13. The tumor immune microenvironment (TIME), characterized by various immune and stromal cells, is a driving factor facilitating tumor metastasis14. Specific cellular components, such as neutrophils15 and cancer-associated fibroblasts (CAFs)16, are closely associated with an immunosuppressive TIME, which may promote the metastasis of HCC into the bloodstream before LT.
In China, HCC accounts for more than 40% of all adult LT recipients, and most HCC cases are hepatitis B virus (HBV)-related17,18,19,20. However, few studies have explored the survival-related molecular biological features in liver transplant recipients with HBV-related HCC. In the present study, we conducted a multi-omic analysis of HBV-related HCC beyond the Milan criteria in a Chinese LT cohort. Integrated analyses of tumor tissues involved the assay for RNA sequencing (RNA-seq), proteomic landscapes and transposase-accessible chromatin sequencing (ATAC-seq) of patients at more advanced states. A transcriptomic subgroup model was established for LT candidate selection and was validated in an independent cohort. In addition, deconvolution analysis was performed to track the composition of cell types, and we identified CAF-stemness features associated with poor survival of patients after LT. The ATAC-seq revealed key factors bridging CAF infiltration and cancer stemness.
Results
Patient characteristics
To obtain a comprehensive molecular profile of HCC patients with LT, tumor samples from 122 HCC patients were obtained from liver explants for RNA-seq, ATAC-seq, and proteomic analyses based on stringent criteria (Supplementary Fig. 1). The samples from 100 HCC patients were suitable for RNA-seq (Supplementary Fig. 2), those from 92 patients were suitable for ATAC-seq, and those from 94 patients were suitable for proteomics (Supplementary Fig. 3A).
The clinicopathological data and outcomes of the patients are presented in Supplementary Table 1. Ninety-one percent of patients were male, and all patients had HBV infection. The median tumor size was 6.05 cm (range 0.6–10 cm). Tumors exceeded the Milan criteria due to a single tumor larger than 5 cm (18.0%), 2 to 3 tumors with the largest tumor larger than 3 cm (44.0%), or more than three tumors within the liver (38.0%). The median AFP level was 134.7 mg/dL (range 12.1–1207.5 mg/dL). The median model for end-stage liver disease score was 19 (range 10-34). The Child‒Pugh grade was Class A in 26.0% of patients, Class B in 37.0% of patients, and Class C in 37.0% of patients. At the end of follow-up, 45 patients had died, and the 5-year OS rate was 45.0%. 52 patients had HCC recurrence, and the 5-year recurrence rate was 52.0% (Supplementary Fig. 3B).
Transcriptome analysis defines clusters of HCC associated with survival after LT
To realize the goal of precisely identifying appropriate LT patients with HCC beyond the Milan criteria, we used transcriptomic information to establish models and cluster HCC patients into subgroups. Transcriptomic data can reflect the expression of genes and are the most mature and stable among various omics methods under current conditions. Through transcriptomic data analysis, 57 positive regulatory pathways and 1 negative regulatory pathway were screened (Supplementary Fig. 4). We then performed unsupervised clustering based on the transcriptomic activity of these pathways and identified three subgroups, namely, the low-risk group (n = 39), medium-risk group (n = 30) and high-risk group (n = 31), among the 100 HCC patients (Supplementary Fig. 5).
The significantly enriched pathways of the transcriptomic subgroups were shown in Fig. 1A (high-risk vs. low-risk, likelihood ratio test, p < 0.01) and the subgroups significantly differed in OS (p = 0.0003, Fig. 2B) and recurrence (p = 0.0012, Fig. 1C) and were determined to be independent prognostic indicators of LT for HCC beyond the Milan criteria based on multivariate analysis (medium-risk vs. low-risk, hazard ratio (HR): 2.780, p = 0.016; high-risk vs. low-risk, HR: 4.527, p < 0.001, Supplementary Table 2). The low-risk group achieved a 5-year OS rate of 69.5% (Fig. 1B). In addition, tumor size, tumor number and AFP level were not independent prognostic indicators (p > 0.05, Fig. 1D), demonstrating that traditional morphological parameters and tumor biomarkers cannot be used to accurately select suitable patients with HCC beyond the Milan criteria for LT. The cohort mentioned above was considered the derivation cohort in the current study.
A Heatmap representing significantly enriched pathways (KEGG and WikiPathway databases) in three RNA-seq subgroups. B, C Kaplan‒Meier curves for OS or HCC recurrence based on RNA-seq subgroups (log-rank test). D Boxplots of AFP levels and tumor size, and column plot of tumor nodules in three RNA-seq subgroups (Low-risk n = 39, Medium-risk n = 30, High-risk n = 31). P values are derived from the Kruskal‒Wallis rank sum test, ANOVA F test and Pearson’s chi-squared test. P values are shown on the graphs. Boxplot shows the median (center line), 25th, and 75th percentile (lower and upper boundary). The upper whisker extends from the upper boundary to the maxima value no further than 1.5 × IQR from the boundary. The lower whisker extends from the lower boundary to the minima value at most 1.5 × IQR of the boundary. Data beyond the end of the whiskers are outlying points and are plotted individually. E The NTP method was used to divide the validation cohort (from the Second Affiliated Hospital of Dalian Medical University and the Affiliated Hospital of Qingdao University) samples into three subtypes. The heatmap illustrates the RNA-seq expression profiles of characteristic genes in samples with subtype prediction (FDR < 0.01). F, G Kaplan‒Meier curves for OS or HCC recurrence based on RNA-seq subgroups (log-rank test) in the validation cohort. HCC hepatocellular carcinoma, LT liver transplantation, OS overall survival, AFP alpha fetal protein, NES normalized enrichment score, HR hazard rate, IQR interquartile range, NTP Nearest Template Prediction, FDR false discovery rate; In OS status, 1 dead, 0 alive.
The volcano map (A) and enriched pathways (B) of significantly differentially expressed genes (FDR < 0.01, Two-tailed limma moderated t-test) between high-risk and low-risk RNA-seq subgroups. log2(fc) means log2 fold change between high-risk and low-risk groups. C Heatmap and quantitative analysis of differentially expressed genes in the CSC pathway (Two-tailed limma moderated t-test). The color of each cell represents the relative abundance of the gene. The length and color of the bar plot on the right side represents –log10 (adj.P. Val) of differentially expressed genes. Dashed line represented Benjamini-Hochberg method p value = 0.01. D Heatmap of differentially expressed proteins in three proteomic subgroups. E Kaplan‒Meier curves for OS based on proteomic subgroups (log-rank test) in the present HCC LT cohort. F Heatmap of CSC-related proteins in three proteomic subgroups. G OS Kaplan‒Meier curves (log-rank test) of certain CSC markers in the present cohort, depending on the 50% cut-off value of mass spectrometry-based protein quantification. OS overall survival, AFP alpha fetal protein, FDR false discovery rate, LT liver transplantation; HCC hepatocellular carcinoma, CSC cancer stemness cell, In OS status, 1 dead, 0 alive.
To validate the transcriptomic stratification, according to the same enrolling rules (Supplementary Fig. 1), we further collected fresh-frozen tissue samples of 80 HCC patients who received LT from two LT centers of China (Supplementary Table 3). Feature genes of subtypes were detected based on transcriptome profiling by limma moderated t-statistics. The Nearest Template Prediction (NTP) method was used to divide the validation cohort samples into three subtypes. The feature gene expression profiles in the validation cohort shown a significant association with the predicted subtypes (NTP, false discovery rate (FDR) < 0.01) (Fig. 1E, Supplementary Data 1). Similar OS and recurrence rate was found in low-risk group and high-risk group (Fig. 1F, G), and multivariate analysis also demonstrated the transcriptomic subgroups were the independent prognostic indicators (high-risk vs. low-risk, HR: 2.963, p < 0.05, Supplementary Table 4). We also applied our transcriptomic stratification to The Cancer Genome Atlas (TCGA) database. However, no differences were observed in OS and HCC recurrence (p > 0.5, Supplementary Fig. 6A, B) in patients receiving surgical resection, supporting the specificity and significance of our stratification system in patient selection and prognosis prediction of LT for HCC.
Comparison of our transcriptomic subgroup model with University of California San Francisco (UCSF) criteria and Up-to-7 criteria in the present cohort
We further chose two classical expanded criteria, UCSF21 and Up-to-722, and compared the efficiency of both criteria with our transcriptomic subgroup model in LT selection for HCC patients. A total of 180 patients from the derivation cohort and the validation cohort were included. No significant differences were found in the log-rank test for OS when UCSF and Up-to-7 criteria were used for grouping in our present cohort (p > 0.05, Supplementary Fig. 7A, B). The area under the curve value of the transcriptomic subgroup model at each time point was higher than those of the UCSF criteria and Up-to-7 criteria (Supplementary Fig. 7C, Supplementary Table 5). The value of the C-index of the transcriptomic subgroup model (0.667; 95% CI: 0.637–0.697) was superior to that of the UCSF criteria (0.509; 95% CI: 0.481–0.536; P < 0.01) and Up-to-7 criteria (0.516; 95% CI: 0.489–0.543; P < 0.01) (Supplementary Table 6). Thus, the transcriptomic subgroup model classified OS more appropriately than the other two classical expanded criteria in the present cohort.
A cancer stemness signature dominates poor survival of patients with HCC after LT
Comparing with the low-risk group, 952 differentially expressed genes were identified in the high-risk group of the derivation cohort, namely, 721 upregulated genes (fold change > 2) and 231 downregulated genes (fold change <0.5) (Fig. 2A). Pathway enrichment analysis revealed several significantly changed pathways (Fig. 2B). Among these, notably, the cancer stem cell (CSC) pathway was significantly upregulated in the high-risk group (high-risk vs. low-risk, Fisher’s exact test, p < 0.001), which may account for the poor survival of these patients. CSC is widely believed to drive cancer cell spread. Therefore, according to a commercial applicated CSC gene panel (QIAGEN, PAHS-176ZA), we analyzed the expression of 27 cancer stemness-related genes that were significantly changed in the three subgroups (Fig. 2C) and also part of cancer stemness-related genes in the validation cohort (Supplementary Fig. 8). The heatmap clearly shows that these genes were upregulated (high-risk vs. low-risk, fold change > 2, T-test p-value < 0.01) in the high-risk group.
From the protein expression level, Isobaric tandem mass tag-based global proteomics identified an average of 4022 proteins per sample. We then performed unsupervised clustering based on proteins. The low-risk group was characterized by metabolism-related pathways, such as fatty acid degradation and pyruvate metabolism. In the high-risk group, pathways related to stemness, such as cancer stem cells and hematopoietic cell lineages, were enriched and effectively validated the transcriptomic subgroup model (Fig. 2D). The proteomic subgroups were significantly positive correlated with transcriptomic subgroups (Pearson chi-square test, p < 0.001, Fig. 2D) and also differed in survival (p < 0.001, Fig. 2E). To further explore the relationship between stemness-associated proteins and the prognosis of LT recipients with HCC, we performed clustering analysis of stem-associated proteins among patients with different survival risks (Fig. 2F). Several stemness biomarkers, such as CD44, ENG, MCAM and THY1, were selected as grouping proteins, whose higher expression predicted poor OS in the present cohort (p < 0.05, Fig. 2G). On the whole, the results of proteomics and RNA-seq revealed a cancer stemness signature dominates poor survival of patients with HCC after LT.
Accumulated CAFs are associated with high stemness in HCC
To determine the differences in the composition and proportion of cell subpopulations among the three subgroups, we performed deconvolution analysis based on transcriptomic data of the derivation cohort and the validation cohort (Fig. 3A; Supplementary Fig. 9). In all cell subpopulations with differences, we found that the different features of hematopoietic stem cells and CAFs (high-risk vs. low-risk, Kruskal-Wallis Rank Sum test, p < 0.01) among the three subgroups were consistent with their corresponding prognoses. In the derivation cohort, the scores of cell subpopulations with differences are presented, and among them, CAFs, the major components of the tumor microenvironment, were significantly enriched in the high-risk group (high-risk vs. low-risk, Kruskal-Wallis Rank Sum test, p < 0.01 Supplementary Fig. 9).
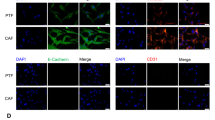
A Heatmap of xCell-derived immune, microenvironment, neutrophil and stroma scores across three RNA-seq subgroups. The dashed line represented the Kruskal-Wallis Rank Sum test FDR = 0.01. B Detection of CAFs by mIF, which were determined by α-SMA and FAP-α staining. C Boxplots of xCell-derived CAF scores in patients with high or low CAF scores from mIF (CAFs High n = 28, CAFs Low n = 49) by two-tailed Wilcoxon Rank Sum Tests. Boxplot shows the median (center line), 25th, and 75th percentile (lower and upper boundary). The upper whisker extends from the upper boundary to the maxima value no further than 1.5 × IQR from the boundary. The lower whisker extends from the lower boundary to the minima value at most 1.5×IQR of the boundary. Data beyond the end of the whiskers are outlying points and are plotted individually. D Heatmap of DEGs in the cancer stem cells pathway in patients with high or low CAF scores from mIF. Kaplan‒Meier curves for OS (E) and HCC recurrence (F) in patients with high or low CAF scores from mIF. G Heatmap of DEGs in the CSC pathway in patients with high or low CAF infiltration from an scRNA-seq data set of 10 independent HCC cohorts. HCC hepatocellular carcinoma, LT liver transplantation, CAF cancer-associated fibroblast, FDR false discovery rate, scRNA-seq single-cell RNA sequencing, OS overall survival, mIF multiplex immunofluorescence, IQR interquartile range, DEGs differentially expressed genes.
We next analyzed the infiltration of CAFs in HCC samples using multiplex immunofluorescence (mIF) staining (Fig. 3B) and found that the CAFs-high group has a 4.5 fold higher CAF score according to XCELL compared to the CAFs-low group. (Wilcoxon Rank Sum test, p < 0.001, Fig. 3C). To further explore the relationship between CAFs and HCC stemness, we analyzed the expression of stemness-related genes in the two groups with high and low CAF infiltration, and the results showed that the expression level of stemness-related genes was higher (fold change > 2, T-test p-value < 0.01) in the high CAF infiltration group (Fig. 3D). In addition, we analyzed the effect of CAF infiltration on prognosis, which showed that HCC patients with high CAF infiltration had a worse prognosis after LT (p < 0.05, Fig. 3E, F). Moreover, by analyzing a set of single-cell RNA sequencing (scRNA-seq) data from 10 HCC tissues in another published study23, we found that cancer stemness-related genes were highly expressed in HCC tissues with higher CAF infiltration compared to those with lower CAF infiltration (fold change > 2, T-test p-value < 0.01, Fig. 3G). The above results revealed that accumulated CAFs are associated with high stemness in HCC in the present cohort.
ATAC-seq reveals the transcription factors associated with stemness-related genes
CAFs can promote cancer cell survival by secreting cytokines and stimulating certain signaling pathways and transcription factors (TFs)24,25. We performed ATAC-seq to determine the key TFs driving stemness in HCC in the high-risk group. According to prognostic groupings based on RNA-seq data, the results of hierarchical clustering analysis of all peaks for each replicate are shown in Fig. 4A. Next, we analyzed the TFs with the greatest difference in transcription activity between the high-risk and low-risk groups (Fisher’s exact test, p < 0.05). Several DNA-binding proteins or TFs, such as X-ray repair cross-complementing group 5 (XRCC5), MYCN, TFAP2C, TFAP2A, NFYA, and NANOG, were identified as being activated in HCC of the high-risk group (adjusted p-value < 0.05, Fig. 4B). Among them, MYCN26 and NANOG27 were TFs widely believed closed with stemness in HCC. To further verify the significant role of cancer stem cells in LT patients with HCC, the network relationship of downstream stemness-related genes regulated by six TFs was analyzed by using the Transcription Regulatory Relationships Unraveled Sentence-based Text mining database28 (Fig. 4C). In addition, we analyzed the correlation between the activity of TFAP2C and XRCC5 and stemness-related gene expression in tumor tissues (Fig. 4D). The expression levels of CSC genes were determined according to the RNA-seq results. In summary, the results of ATAC-seq primarily revealed the positive correlation between several TFs and CSC gene expression in HCC in the present cohort.
A Heatmap of differentially expressed open chromatin regions detected by ATAC-seq and grouped by RNA-seq-based subgroups. B Bubble plots represent TFs significantly enriched for genes with differentially expressed ATAC-seq peaks (Fisher’s exact test). C Network of significantly enriched TFs with target genes, whose expression levels were determined according to RNA-seq results. D Boxplots presenting gene expression in patients with transcriptional activity of TFPA2C (Positive n = 20, Negative n = 48) and XRCC5 (Positive n = 34, Negative n = 34) by two-tailed Student’s t-test. Pearson correlations between gene expression and transcriptional activity greater than 0.3 were present. Transcriptional activity was represented by the normalized enrichment score from the single-sample gene set enrichment analysis method. Boxplot shows the median (center line), 25th, and 75th percentiles (lower and upper boundaries). The upper whisker extends from the upper boundary to the maxima value no further than 1.5 × IQR from the boundary. The lower whisker extends from the lower boundary to the minima value at most 1.5×IQR of the boundary. Data beyond the end of the whiskers are outlying points and are plotted individually. ATAC-seq, assay for transposase-accessible chromatin sequencing, OS overall survival; AFP alpha fetal protein, IQR interquartile range.
XRCC5, upregulated by CAFs, promotes cancer stemness features in HCC
In order to ascertain the link between CAFs and cancer stemness in the current study, we chose XRCC5, a DNA repair protein, which is rarely reported to be involved in fostering cancer stemness. XRCC5 levels were increased in HCC cells cultured in medium conditioned by CAFs (Fig. 5A) or when co-inoculated with CAFs into nude mice. The xenografts of HuH-7 cells co-inoculated with CAFs grew significantly faster than simple HuH-7 cell xenografts (on day 20, average tumor volume, HuH-7 cells + CAFs vs HuH-7 cells, 257 mm3 vs 27 mm3, p < 0.05 Fig. 5B, C). We then used a lentiviral-based approach to establish stable XRCC5 knockdown in HCC cells (Fig. 5A). XRCC5 knockdown resulted in nearly 70% less hepatosphere formation (Fig. 5D) and led to about an 80% reduction in migration ability, or a restoration to the migration ability induced by CAFs (p < 0.05, Fig. 5E, F). A reduction in XRCC5 suppressed mRNA expression of the CSC markers or abolished the CAF-induced upregulation of the CSC markers, such as CD24, CD44, CD133, and EPCAM (p < 0.05, Fig. 5G, H). Moreover, we also examined the tumorigenicity of HCC cells with XRCC5 knockdown in vivo by injecting shNC or shXRCC5 HuH-7 cells subcutaneously into NOD/SCID mice at three dilutions (2 × 106, 2 × 105 and 2 × 104) and allowing them to grow for four weeks (Fig. 5I). The confidence intervals (CIs) for 1/(stem cell frequency) using extreme limiting dilution were calculated29. The stem cell frequency (1/CI) in XRCC5 knockdown group was 1/1868283, compared with 1/184087 in NC HuH-7 cells (p = 0.00184). Similarly, overexpression of XRCC5 also significantly promoted the phenotypes of cancer stemness in HCC cells (p < 0.05, Fig. 5J, K, L and M). Finally, to preliminarily confirm the clinical roles of XRCC5, we performed IHC staining of XRCC5 in 82 HCC samples from the derivation cohort (Fig. 5N). The heatmap of certain CSC-related genes is shown (high-risk vs. low-risk, T-test, p < 0.05, Fig. 5O), and high XRCC5 expression predicted much poorer OS in the cohort (p < 0.001, Fig. 5P). High XRCC5 expression was determined to be an independent prognostic indicator of LT for HCC beyond the Milan criteria based on multivariate analysis (HR: 4.082, p = 0.002, Supplementary Table 7).
A HuH-7 cells and Hep 3B cells with XRCC5 knockdown (shXRCC5-1 and shXRCC5-2) or not were cultured CAF-conditioned medium (CAF+) or not and the expression of XRCC5 was detected using western blotting. Band intensities were semi-quantified using IMAGE LAB software and normalized with β-actin. Values were presented as the means under the bands. B The volume curves of the tumor nodules in groups of HuH-7 cells and HuH-7 cells co-inoculated with CAF in nude mice (n = 7 per group). C Tumor nodules were resected after 20 days and XRCC5 expression were detected by IHC staining. The effect of XRCC5 knockdown or not on stemness of HCC cells were shown by in vitro self-renewal capacity (D), cell migration (E and F), mRNA expression levels of stemness-related genes (G, H) and limiting dilution xenograft formation in NOD/SCID mice (I, shXRCC5 represents XRCC5 knockdown, n = 5 per group), and stem cell frequency was calculated. J In HuH-7 cells and Hep 3B cells with XRCC5 overexpression (OE) or not (vector), XRCC5 was detected using western blotting. The stemness of above cells were shown according to the in vitro self-renewal capacity (K), mRNA expression levels of stemness-related genes (L) and cell migration (M). N IHC staining of XRCC5 in patients with HCC undergoing LT. O Heatmap of differentially expressed genes in the CSC pathway in patients with high or low XRCC5 expression from IHC. P Kaplan‒Meier curves for OS in patients with high or low XRCC5 expression from IHC. The data of cell functional assays and RT-qPCR analysis were presented as mean ± SD of three individual experiments, and the data from animal experiments were presented as mean ± SEM. Two-tailed Student’s t-test was used for comparisons, and P values are shown on the graphs. CAFs cancer-associated fibroblasts, HCC hepatocellular carcinoma; LT liver transplantation, IHC immunohistochemistry, OS overall survival, NC negative control, CI confidence interval. Source data are provided in the Source Data file.
CAF-derived CXCL12 sustains the stemness of HCC cells by promoting XRCC5 through CXCR4
Ultimately, we were delving into the mechanisms that connect CAFs and XRCC5 in driving stemness. The chemokine receptor CXCR4 was high expressed in HCC tissues of the high-risk group (Fig. 2C), and its ligand, cytokine CXCL12, could be secreted by CAFs30. We confirmed a concentration of CXCL12 in the medium conditioned by the CAFs we used that was more than 3 times higher than that in the human hepatic stellate cell lines, LX-2 (Fig. 6A). Treatment using exogenously supplied CXCL12 resulted in the elevation of XRCC5 levels in HCC cells (Fig. 6B, C), which could be abrogated by knockdown of CXCR4 (Fig. 6D–F). In accordance with the XRCC5 variation, the phenotypes of cancer stemness could also be abrogated by CXCR4 knockdown (Figs. 6G–I) or CXCR4 inhibitor AMD3100 (Fig. 6J). Co-cultures of HCC cells and CAFs with direct contact(GFP+ HuH-7 cells, Supplementary Fig. 10, Supplementary Fig. 11A) or in direct contact (Fig. 6K) were conducted, showing similar trends in CSC gene expressions compared to the conditioned medium of CAFs (Fig. 6L, M, Supplementary Fig. 11B, C). This suggests that the direct interaction between HCC cells and CAFs may not impact the cancer stemness driven by CXCL12/CXCR4. We also performed IHC staining for CXCL12 and CXCR4 on the same HCC samples used for XRCC5 staining (Fig. 5N), revealing positive associations between XRCC5 and CXCL12, and XRCC5 and CXCR4 (Fig. 6N, O). In summary, our Integrated multi-omic analysis reveals that CAF-derived CXCL12 sustains the stemness of HCC cells by promoting XRCC5 through CXCR4 (Fig. 6P).
A CXCL12 concentration in CAF conditioned medium (CAF-CM) and LX-2 conditioned medium (LX-2-CM) was detected using ELISA. B, C XRCC5 expression was detected by western blotting in HCC cells treated with 0 nM, 10 nM, 100 nM, and 1 μM recombinant human CXCL12 protein for 48 h. Band intensities were semi-quantified using IMAGE LAB software and normalized with β-actin. Values were presented as the means under the bands. D Effect of siCXCR4 was detected by western blotting. E, F XRCC5 and CXCR4 expression was detected by western blotting in HCC cells treated with recombinant human CXCL12 protein (1 μM) and siCXCR4 or not. The effect of CXCL12 stimulation (1 μM) with CXCR4 knockdown or not, as well as CXCR4 inhibitor administration, AMD3100 (1 μM), or not on the stemness of HCC cells were shown by mRNA expression levels of stemness-related genes (G, H), cell migration (I), in vitro self-renewal capacity (J). K Schematic of the transwell co-culture model of HCC cells and CAFs. Created with BioRender.com. L, M The effect of co-culture with CXCR4 knockdown or not as well as AMD3100 (1 μM) administration or not on stemness of HCC cells were shown by mRNA expression levels of stemness-related genes. N IHC staining of CXCR4 and CXCL12 in patients with HCC undergoing liver transplantation. O Positive correlation between XRCC5 with CXCR4 and CXCL12 in 79 HCC samples. P Graphical summary of the mechanisms based on the combination of integrated multi-omics and experimental evidence. The data of ELISA (four individual experiments), cell functional assays and RT-qPCR analysis (three individual experiments) were presented as mean ± SD. Two-tailed Student’s t-test was used for comparisons, P values are shown on the graphs. ELISA, enzyme linked immunosorbent assay; HCC, hepatocellular carcinoma; IHC immunohistochemistry, TFs transcriptional factors, NC negative control, CAF cancer-associated fibroblast, ATAC-seq assay for transposase-accessible chromatin sequencing, CSC cancer stem cell. Source data are provided in the Source Data file.
Overall, using integrated multi-omic data, we revealed the key role of CAF-stemness in governing the poor outcomes of HCC patients undergoing LT (Fig. 7).
A total of 180 patients transplanted from 2015–2019 were included (100 patients as derivation cohort, 80 patients as validation cohort). Samples from the derivation cohort were subjected to RNA-seq, ATAC-seq, and proteomic analyses. In vivo, in vitro experiments, and formalin-fixed paraffin-embedded HCC samples were used to validate the mechanisms. HCC hepatocellular carcinoma, ATAC-seq assay for transposase-accessible chromatin sequencing, IHC immunohistochemistry, mIF multiplex immunofluorescence. The figure was created with BioRender.com.
Discussion
For selecting patients with HCC for LT, the Milan criteria are still the most acceptable criteria worldwide. Extended LT criteria have achieved significant progress and are mostly based on tumor size, tumor number and AFP value3,31,32. With the increasing understanding of tumor biological mechanisms, multiple studies have reported the correlation between genetic profiles and outcomes after hepatic resection in HCC patients7. A previous study based on a cohort of LT patients with HCC beyond the Milan criteria from the United States found that patients without gene signatures of progenitor markers had survival rates similar to those within the Milan criteria13. Another study analyzed several biomarkers at the RNA level and revealed a 4-gene signature for helping select HCC patients for LT in a cohort from Europe33. For HBV-related HCC, previous studies mainly included early-stage patients undergoing hepatic resection10,11. However, thus far, in HBV-related HCC, the use of biological markers for predicting the outcome of patients undergoing LT has been limited. Importantly, molecular signaling can be used to mechanistically determine the metastasis or recurrence of HCC and may help precisely extend LT criteria.
In our current study, we first explored tumor gene expression in 100 patients with HCC beyond the Milan criteria undergoing LT. All the cases were HBV-related. A transcriptome-based molecular classification, also called transcriptomic subgroup model, was established, in which the low-risk group accounted for 39% of cases and was associated with an acceptable 5-year survival of nearly 70%. For patients with HCC beyond the Milan criteria, the median risk and poor risk groups had a 5-year survival of less than 31%, which was associated with the cancer stemness gene signature. Given that the diseased liver is removed intact after LT, it is speculated that extrahepatic residual tumor cells result in HCC recurrence. Cancer stemness, reflecting the capacity for tumorigenicity, self-renewal, metastasis, and chemotherapeutic drug resistance34,35, is widely believed to drive HCC spread36,37. Thus, it is reasonable that the cancer stemness signature dominates the poor survival of patients with HCC after LT. Importantly, we validated our transcriptomic stratification in an independent HCC LT cohort, in which 91.25% of the patients were HBV-related, supporting the efficiency of our transcriptomic stratification in HBV-related HCC patients undergoing LT. Moreover, to support the transcriptomic subgroup model and the cancer stemness feature, we performed proteomics in the same cohort. The proteomic clustering was significantly correlated with transcriptomic subgroups, in which the molecular features in the high-risk group were also included, cancer stem cells and hematopoietic cell lineages, supporting the reliable transcriptomic subgrouping procedure in our study. Interestingly, the low-risk group was characterized by metabolism-related pathways, which was consistent with a previous work with larger HCC samples10.
Moreover, depending on bulk RNA data, cellular component analysis further highlighted the hematopoietic stem cell enrichment in tumors of poor survival groups, which further supported stemness as the main adverse biological features in HCC patients undergoing LT beyond the Milan criteria. Interestingly, accumulated CAFs were found in tumors of the poor-survival group. CAFs are the main cellular components in the TIME. An increasing number of studies have reported the induction of cancer stemness by CAFs38,39,40. In HCC, CAFs play an important role in the induction of drug resistance41, immune evasion16, and stemness24. We further demonstrated the close positive association between CAF infiltration and the stemness gene signature. Thus, in the transcriptomic subgroup model, CAF-induced stemness may dominate the adverse biological features.
To support the transcriptomic subgroup model and preliminary explore the mechanism of CAFs induced stemness in HCC, we performed ATAC-seq in the same cohort. The ATAC-seq analysis revealed 6 TFs or DNA-binding proteins in the high-stemness group. Among these proteins, MYCN26 and NANOG27 are commonly believed to promote the stemness of HCC. Significantly, the ATAC-seq data emphasize the role of cancer stemness in our classification. XRCC5, a DNA repair protein, ranked as the first protein with high transcriptional regulatory capacity in our high-risk HCC samples. In HCC, XRCC5 could interact with the TF, NRF2, to regulate target gene expression and drives tumor progression42, while rare studies reported the relationship between XRCC5 and cancer stemness. Our primary analysis, incorporating ATAC-seq and transcriptome data revealed that high transcriptional activity of XRCC5 may trigger cancer stemness gene expression. We further demonstrated that CAFs could trigger XRCC5 upregulation, which potently promotes HCC stemness. In the high-risk group, CXCR4, the receptor of CXCL12, exhibited a notable increasedin expression. The CXCL12/CXCR4 axis could potentially facilitate the communication between CAFs and tumor cells43. Thus, to deeply link the CAFs and XRCC5, we demonstrated that the CAF-derived CXCL12 sustains the stemness of HCC cells by promoting XRCC5 through CXCR4. The activation of CXCL12/CXCR4/XRCC5 axis partially explained the CAF accumulation and high stemness in the high-risk group. The precise regulation of XRCC5 and other TFs we found are worthy to be deeply explored in in HCC patients undergoing LT. Clinically, XRCC5 was a potent biomarker for predicting poor survival in HCC patients after LT. The CXCL12/CXCR4/XRCC5 axis might serve as a significant indicator in predicting the outcome of HCC patients undergoing LT and could present as potential targets for pre-transplant downstaging therapy.
For clinical application, our molecular classification is not applicable to HCC patients undergoing tumor resection, which further supports the novelty and specificity of the classification in HCC patients undergoing LT. We also primarily demonstrated that the transcriptomic subgroup model was better than UCSF and Up-to-7 criteria in selecting candidates of LT, which may further extend its clinical application. However, the shortcomings are clear. The genomic heterogeneity of HCC and limited tissues may hinder accurate analysis of the genomic landscape.
In summary, our observations provide the first multi-omic landscape of patients with HBV-related HCC beyond the Milan criteria undergoing LT. According to our molecular classification, patients with low risk had a 5-year survival of nearly 70%. Our study provides a novel genetic tool to expand the selection criteria for LT. It may act as an effective supplement for previously reported extended criteria. Importantly, we reveal the key role of CAF-stemness in governing the poor outcomes of HCC patients undergoing LT. More laboratory study is needed to deeply explore the CAF-stemness-related molecular mechanisms in HCC.
Methods
Patient cohorts and tumor tissue samples
This study is approved by Institutional Review Board of The First Affiliated Hospital, School of Medicine, Zhejiang University (Reference number: 20191421), Shulan (Hangzhou) Hospital (Reference number: KY2021014), the Second Affiliated Hospital of Dalian Medical University (Reference number: 2022166), and the Affiliated Hospital of Qingdao University (Reference number: QYFY WZLL 29314). Informed written consent was obtained from all participants. A total of 699 HCC patients received LT between 2015 and 2019 at the First Affiliated Hospital, School of Medicine, Zhejiang University, and Shulan (Hangzhou) Hospital. We selected 100 of these patients (n = 39 and 61, respectively) with HCC beyond the Milan criteria based on pathological assessment. Liver explants were used for sample collection. Fresh-frozen tissue samples were used for RNA-seq and ATAC-seq. Formalin-fixed paraffin-embedded (FFPE) tumor tissue samples were used for proteomics. 82 FFPE tissue samples were used for immunohistochemistry (IHC) detection. RNA-seq data from fresh-frozen tissue samples of 80 HCC patients who received LT between 2015 and 2020 at two LT centers, the Second Affiliated Hospital of Dalian Medical University and the Affiliated Hospital of Qingdao University, were utilized as the validation cohort in this study. Sex and/or gender were determined based on self-reporting.
RNA-seq, ATAC-seq and proteomics by mass spectrometry
Details are provided in the Supplementary Methods.
Real-time quantitative polymerase chain reaction (RT-qPCR) analysis, western blot analysis, IHC, and mIF detection
Details are provided in the Supplementary Methods.
Cell culture, plasmids, lentivirus infection, transfection, and Isolation of CAFs
Human HCC cell line, HuH-,7 and CAFs were cultured in Dulbecco’s modified Eagle medium (Pricella) or Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (Pricella) with 10% fetal bovine serum (Wisent), 100 mg/ mL penicillin G and 100 μg /mL streptomycin (Gibco) at 37 °C in a humidified atmosphere containing 5% CO2. The details about HCC cell line, lentivirus vectors for shRNA, XRCC5 delivery, transfection, and isolation of CAFs are provided in the Supplementary Methods.
Cancer cell stemness detection
Cell migration, self-renewal ability (cell sphere formation and extreme limiting dilution in non-obese diabetic/severe combined immunodeficiency (NOD/SCID) mice), stemness gene expression detection was established to detect the stemness of HCC cells. The details are contained in the Supplementary Methods.
Bioinformatics analysis
Details are provided in the Supplementary Methods.
Statistics and reproducibility
Prism 8 (GraphPad Software, CA, USA) and R (version 4.1) software were used for statistical analysis. Significance of differences between groups was performed using Student’s t-test. Values of p < 0.05 were considered significantly different. Correlation analyses were performed using χ² test for quantitative data. For survival analysis, survival was estimated using the Kaplan-Meier method. Details are provided in the Supplementary Methods. All experiments were repeated independently at least three times with similar results. The number of experiments was stated in the corresponding Supplementary Methods.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNA-seq data generated in this study have been deposited in the Genome Sequence Archive (GSA) database under accession code HRA010539 and HRA010528. The proteomics data generated in this study have been deposited in the iProX database under accession code PXD061119 (https://www.iprox.cn/page/project.html?id=IPX0011178000). The ATAC-seq data generated in this study have been deposited in the GSA database under accession code HRA010529. Clinical data for the patients included in this study are not publicly available per policy to protect patient privacy. Clinical data access can be made available from the corresponding authors for qualified researchers. Queries for data access will be answered within a time frame required to ensure high quality assessment and coordination of the proposed collaborative work, and a first response can be provided within 8 weeks. The remaining data generated in this study are provided in the Supplementary Information and Source Data file. The RNA-seq data from TCGA for HCC used in this study are available in the TCGA data portal (https://portal.gdc.cancer.gov/). The previously published scRNA-seq data of 10 HCC tissues used in this study are available in the Gene Expression Omnibus database under accession code GSE14961423,44. Source data are provided with this paper.
References
Mazzaferro, V. et al. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N. Engl. J. Med. 334, 693–699 (1996).
Chen, J. et al. Clinical practice guideline on liver transplantation for hepatocellular carcinoma in China (2021 edition). Chin. Med J. 135, 2911–2913 (2022).
Reig, M. et al. BCLC strategy for prognosis prediction and treatment recommendation: The 2022 update. J. Hepatol. 76, 681–693 (2022).
Duvoux, C. et al. Liver transplantation for hepatocellular carcinoma: a model including alpha-fetoprotein improves the performance of Milan criteria. Gastroenterology 143, 986–994 e983 (2012). quiz e914-985.
Mehta, N., Dodge, J. L., Roberts, J. P., Hirose, R. & Yao, F. Y. Alpha-Fetoprotein decrease from > 1,000 to <500 ng/mL in patients with hepatocellular carcinoma leads to improved posttransplant outcomes. Hepatology 69, 1193–1205 (2019).
Li, J. H. et al. The AGH score is a predictor of disease-free survival and targeted therapy efficacy after liver transplantation in patients with hepatocellular carcinoma. Hepatobil. Pancreat. Dis. Int 22, 245–252 (2023).
Wu, Y., Liu, Z. & Xu, X. Molecular subtyping of hepatocellular carcinoma: A step toward precision medicine. Cancer Commun. 40, 681–693 (2020).
Hoshida, Y. et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 69, 7385–7392 (2009).
Lupberger, J. et al. Combined analysis of metabolomes, proteomes, and transcriptomes of Hepatitis C virus-infected cells and liver to identify pathways associated with disease development. Gastroenterology 157, 537–551 e539 (2019).
Gao, Q. et al. Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma. Cell 179, 561–577 e522 (2019).
Jiang, Y. et al. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature 567, 257–261 (2019).
Li, B. et al. Multiomics identifies metabolic subtypes based on fatty acid degradation allocating personalized treatment in hepatocellular carcinoma. Hepatology https://doi.org/10.1097/HEP.0000000000000553 (2023).
Miltiadous, O. et al. Progenitor cell markers predict outcome of patients with hepatocellular carcinoma beyond Milan criteria undergoing liver transplantation. J. Hepatol. 63, 1368–1377 (2015).
Liu, M., Yang, J., Xu, B. & Zhang, X. Tumor metastasis: Mechanistic insights and therapeutic interventions. MedComm (2020) 2, 587–617 (2021).
Xue, R. et al. Liver tumour immune microenvironment subtypes and neutrophil heterogeneity. Nature 612, 141–147 (2022).
Zhu, G. Q. et al. CD36(+) cancer-associated fibroblasts provide immunosuppressive microenvironment for hepatocellular carcinoma via secretion of macrophage migration inhibitory factor. Cell Discov. 9, 25 (2023).
Ling, S. et al. Liver transplantation in patients with liver failure: Twenty years of experience from China. Liver Int 42, 2110–2116 (2022).
Xu, X. State of the art and perspectives in liver transplantation. Hepatobil. Pancreat. Dis. Int 22, 1–3 (2023).
Li, Q. et al. Burden of liver cancer: From epidemiology to prevention. Chin. J. Cancer Res 34, 554–566 (2022).
Miao, L. L. et al. Hypomethylation of glycine dehydrogenase promoter in peripheral blood mononuclear cells is a new diagnostic marker of hepatitis B virus-associated hepatocellular carcinoma. Hepatobil. Pancreat. Dis. Int 23, 35–42 (2024).
Yao, F. Y. et al. Liver transplantation for hepatocellular carcinoma: expansion of the tumor size limits does not adversely impact survival. Hepatology 33, 1394–1403 (2001).
Mazzaferro, V. et al. Predicting survival after liver transplantation in patients with hepatocellular carcinoma beyond the Milan criteria: a retrospective, exploratory analysis. Lancet Oncol. 10, 35–43 (2009).
Lu, Y. et al. A single-cell atlas of the multicellular ecosystem of primary and metastatic hepatocellular carcinoma. Nat. Commun. 13, 4594 (2022).
Lam, K. H. & Ma, S. Noncellular components in the liver cancer stem cell niche: Biology and potential clinical implications. Hepatology https://doi.org/10.1002/hep.32629 (2022).
Xiao, C., Li, C. & He, J. Novel contributor to chemotherapy resistance: an interferon-dependent subtype of cancer-associated fibroblast. Mol. Biomed. 4, 12 (2023).
Qin, X. Y., Su, T., Yu, W. & Kojima, S. Lipid desaturation-associated endoplasmic reticulum stress regulates MYCN gene expression in hepatocellular carcinoma cells. Cell Death Dis. 11, 66 (2020).
Chen, J. et al. ABI2-mediated MEOX2/KLF4-NANOG axis promotes liver cancer stem cell and drives tumour recurrence. Liver Int. 42, 2562–2576 (2022).
Han, H. et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 46, D380–D386 (2018).
Hu, Y. & Smyth, G. K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009).
Zhang, Z. et al. Cancer-associated fibroblasts-derived CXCL12 enhances immune escape of bladder cancer through inhibiting P62-mediated autophagic degradation of PDL1. J. Exp. Clin. Cancer Res. 42, 316 (2023).
Mehta, N. et al. Liver transplantation for hepatocellular carcinoma. Working Group Report from the ILTS Transplant Oncology Consensus Conference. Transplantation 104, 1136–1142 (2020).
Wang, J. et al. The predictive value of the modified AFP model for liver transplantation outcomes in multinodular hepatocellular carcinoma patients. World J. Surg. Oncol. 21, 104 (2023).
Pinto-Marques, H. et al. A gene expression signature to select hepatocellular carcinoma patients for liver transplantation. Ann. Surg. 276, 868–874 (2022).
Manni, W. & Min, W. Signaling pathways in the regulation of cancer stem cells and associated targeted therapy. MedComm (2020) 3, e176 (2022).
Xia, R., Xu, M., Yang, J. & Ma, X. The role of Hedgehog and Notch signaling pathway in cancer. Mol. Biomed. 3, 44 (2022).
Ling, S. et al. USP22 promotes hypoxia-induced hepatocellular carcinoma stemness by a HIF1alpha/USP22 positive feedback loop upon TP53 inactivation. Gut 69, 1322–1334 (2020).
Liang, N. et al. Mechanism of cancer stemness maintenance in human liver cancer. Cell Death Dis. 13, 394 (2022).
Yu, S. et al. A CD10-OGP membrane peptolytic signaling axis in fibroblasts regulates lipid metabolism of cancer stem cells via SCD1. Adv. Sci. 8, e2101848 (2021).
Ma, Z. et al. Interferon-dependent SLC14A1(+) cancer-associated fibroblasts promote cancer stemness via WNT5A in bladder cancer. Cancer Cell 40, 1550–1565 e1557 (2022).
Zhuang, J. et al. Cancer-associated fibroblast-derived miR-146a-5p generates a niche that promotes bladder cancer stemness and chemoresistance. Cancer Res. https://doi.org/10.1158/0008-5472.CAN-22-2213 (2023).
Eun, J. W. et al. Cancer-associated fibroblast-derived secreted phosphoprotein 1 contributes to resistance of hepatocellular carcinoma to sorafenib and lenvatinib. Cancer Commun. 43, 455–479 (2023).
Liu, T. et al. The NRF2-dependent transcriptional axis, XRCC5/hTERT drives tumor progression and 5-Fu insensitivity in hepatocellular carcinoma. Mol. Ther. Oncolytics 24, 249–261 (2022).
Sulaiman, R. et al. A landscape of patient-derived cancer-associated fibroblast signals in endometrial cancers. Am. J. Cancer Res 14, 467–489 (2024).
Yoon, S. H., Choi, S. W., Nam, S. W., Lee, K. B. & Nam, J. W. Preoperative immune landscape predisposes adverse outcomes in hepatocellular carcinoma patients with liver transplantation. NPJ Precis. Oncol. 5, 27 (2021).
Acknowledgements
This work was supported by grants from the National Key Research and Development Program of China [2022YFA1106800 (S.L.) and 2021YFA1100500 (X.X.)], the National Natural Science Foundation of China [92159202 (X.X.) and 82203070 (Q.S.)], the National Science and Technology Major Project of China [2017ZX10203205 (X.X.)], the Joint Funds of the Zhejiang Provincial Natural Science Foundation of China under Grant No. LHDMZ25H180001 (Q.S.) and the Innovative Teams Project in Key Areas of Dalian [2021RT01 (L.W.)].
Author information
Authors and Affiliations
Contributions
X.X., S.Z., and S.L. designed the research. J.Y., S.W., Q.S., H.X., J.C., L.W., and M.G. obtained HCC samples and clinical data. Q.Z., J.Y., Y.W., L.Z., W.Z., Q.Q., J.H., H.L., J.W., S.C., J.X., and S.X. performed experiments and acquisition of data. S.L., J.Y., Q.Z., P.L., X.P., N.W., and W.W. analyzed and interpreted data. S.L., J.Y., Q.Z., J.L., and X.X. wrote the paper and critically reviewed the manuscript. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval statement
The study protocol was approved by the Review Boards of the NHC Key Laboratory of Combined Multi-organ Transplantation. The Ethical review approval number: 2018-768.
Peer review
Peer review information
Nature Communications thanks Ricardo D’Oliveira Albanus and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ling, S., Yu, J., Zhan, Q. et al. Multi-omic analysis reveals a CAF-stemness-governed classification in HCC liver transplant recipients beyond the Milan criteria. Nat Commun 16, 4392 (2025). https://doi.org/10.1038/s41467-025-59745-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-59745-8