Abstract
Enzymatic depolymerization of polyethylene terephthalate (PET) towards monomer recycling offers a green route to a circular plastic economy, with scale-up currently underway. Yet, inconsistent assessment methods hinder clear comparisons between various PET hydrolases. This Perspective aims to identify critical gaps in this dynamic research field and outline key principles for selecting and tailoring novel enzymes, such as using uniform PET samples and standardizing reaction settings that mimic industrial conditions. Applying these guidelines will improve enzyme screening efficiency, increase data reproducibility, deepen the understanding of interfacial biocatalysis, and ultimately accelerate the development of more robust and cost-effective bio-based PET recycling methods.
Similar content being viewed by others
Introduction
Polyethylene terephthalate (PET) is extensively utilized in beverage bottles, textiles, and food packaging, making it one of the most prevalent plastics in global waste streams. Consequently, its significant contribution to plastic pollution has raised urgent demand for innovative recycling technologies from both scientific and industrial sectors. In many countries, PET, especially single-use bottles, may achieve up to 90% recycling rates, making it the most recycled among all mass-produced fossil-fuel-derived plastics1,2. PET in bottles and food packaging is rarely blended with other polymers and typically lacks or contains only trace amounts of chemical additives3. This high purity and homogeneity of its chemical composition render PET the most broadly studied and preferred target for recycling advancements, encompassing mechanical, chemical, and biological approaches4,5,6.
PET has a polyester backbone, which is notably susceptible to hydrolytic cleavage. Since the 1970s, scientists have been inspired to investigate the broad variety of natural ester hydrolases for their potential in depolymerizing synthetic polyesters, including PET. In 2005, Müller et al. reported the first widely recognized enzymatic depolymerization of amorphized waste PET bottles7. The last two decades have witnessed the identification, characterization, and engineering of numerous PET hydrolases6,8, as well as the successful demonstration of biological recycling of waste PET on an industrial scale9. This achievement has been recognized as one of the ten emerging chemical technologies in 2023 by IUPAC10.
Recent techno-economic analysis (TEA)11 and life cycle assessment (LCA)12 have indicated that although PET bio-recycling holds promise, substantial challenges persist regarding energy consumption, economic feasibility, and overall environmental impact, particularly due to mechanical processing required to amorphize waste PET and pH regulation during the depolymerization reaction. While innovations addressing these process-related bottlenecks are infrequently reported in recent scientific literature13, an increasing number of reports claiming superior enzymes based on limited experimental evidence have emerged. Moreover, redundant studies often appear that merely replicate findings derived from a set of thoroughly examined benchmark enzymes, particularly informed by two seminal studies: the 2016 Science article by Yoshida et al., which identified IsPETase from Ideonella sakaiensis14, and the 2020 Nature publication by Tournier et al., which described several engineered variants of the leaf-branch compost cutinase (LCC) allowing for rapid PET waste depolymerization on an industrial scale9. By contrast, although researchers in recent years have discovered numerous novel PET hydrolases from diverse habitats with potentially distinct characteristics, most are abandoned after initial publications. Another unfavorable scenario in this research domain is that many studies used polymer substrates from unknown origins or performed plastic depolymerization under conditions unrelated to industrial settings. Consequently, it is imperative to standardize and disclose a full array of these essential parameters in future research to prevent overoptimistic assessments of performance, erroneous data interpretations and the formulation of non-reproducible or invalid hypotheses, in alignment with previous expert viewpoints5,11,12. A recent publication by Arnal et al.15 has experimentally highlighted this issue by providing a reliable assessment of four important PET hydrolases under industry-mimicking recycling conditions. Following the identification of significant inconsistencies in depolymerization performance compared to the original studies, the necessity of establishing standards for future enzyme activity evaluations and enabling cross-study comparisons is increasingly evident.
This Perspective aims to provide a brief overview of the historical context and current advancements in this research area, focusing on methodologies for the discovery, screening, and characterization of novel PET hydrolases, as well as assessing their viability for industrial-scale plastic recycling, to complement recent expert reviews with a broader scope6,8,16,17. For the former research objective, we intend to emphasize research gaps in method selection and further development concerning potential final applications of PET hydrolases with distinct features. To achieve the latter research goal of developing more effective industrial biocatalysts for plastic recycling, we recommend prioritizing the use of standardized substrates that are most similar to industrial waste PET feedstocks, as well as selecting reaction conditions that resemble industrial environments in lab-scale or pilot-scale reactor experiments. We anticipate the emergence of an improved multi-pronged future research, including (i) the advancement of scientific understanding of interfacial biocatalysis and enzymatic depolymerization mechanisms, (ii) the validation of screening assays that can be readily implemented in most laboratories without the need for specialized equipment, and (iii) the design of novel PET hydrolases that are less related to a limited number of thoroughly investigated benchmark enzymes.
Approaches for uncovering the expanding diversity of PET hydrolases
Early efforts to discover polyester hydrolases strongly relied on classical microbiological techniques, including the isolation, cultivation, and enrichment of microbial strains capable of hydrolyzing natural polyesters. This led to the identification of fungal and bacterial cutinases, such as those from Fusarium18 and Thermobifida species19, which were later proven to depolymerize synthetic polyesters like PET7,20. Despite over half a century of research advancements, this method remains valuable for identifying novel plastic-degrading strains and enzymes from plastic-contaminated environments. These include the thoroughly studied PET-metabolizing bacterium I. sakaiensis, which secretes the mesophilic IsPETase14, and likewise other microorganisms with similar characteristics isolated from marine sediments21 or wastewater22.
Metagenomics has emerged as an efficient approach to identifying novel PET hydrolases since 2012. The earliest and most extensively studied example is LCC, identified from a plant compost metagenome using a fosmid library and functional screening against tributyrin, an esterase model substrate23. However, functional metagenomics has contributed minimally to further significant PET hydrolase discoveries. Instead, most recent PET hydrolases have been identified through sequence-based enzyme mining, regardless of whether metagenomic DNA was physically extracted24,25 or sourced from online databases26,27,28,29. These sequence-based methods have become central, with gene sources extending to annotated yet uncharacterized sequences found in public repositories like NCBI and UniProt16,29,30,31. However, these bioinformatics-based searches have often been biased by employing established enzymes such as LCC, IsPETase, and those from Thermobifida species as templates, limiting exploration to a narrow phylogeny and sequence space27. Interestingly, despite their sequence similarities, many of these newly discovered PET hydrolases were found to stem from geographically and ecologically distinct habitats, including extreme environments such as deep-sea hydrothermal vents28, glaciers32, and even human saliva metagenomes33, calling into question the hypothesis that PET hydrolases evolved naturally in response to environmental plastic pollution, which has only been present for less than 70 years. It is not surprising that many natural carboxylesterases, found in nearly all organisms, are anticipated to exhibit at least minimal hydrolytic activity on the ester bonds in PET.
The restricted structural diversity of reported PET hydrolases, attributable to their close phylogenetic relationships, creates an “echo chamber” effect that overrepresents structure-based mechanistic studies toward particular benchmark enzymes. This is evident in the Protein Data Bank (PDB), where out of more than 130 PET hydrolase-associated structures, over one-third represent IsPETase and its variants. Similar redundancy is seen with other benchmark enzymes9,24,34,35,36, with numerous nearly identical apo structures deposited. Instead, co-structures complexed with substrates or analogues may yield significant insights for semi-rational and artificial intelligence (AI)-driven enzyme engineering; nonetheless, they are limited, with less than 16% of PET hydrolase structures in the PDB solved in a substrate (analogue)-bound state16. This highlights the ongoing challenge of obtaining wet-lab data on substrate interactions, essential for advancing functional studies and developing more efficient PET hydrolases.
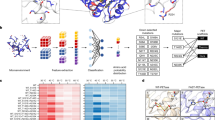
According to the ESTHER database on α/β-hydrolase fold proteins37, most PET hydrolases with known structures belong to the polyesterase-lipase-cutinase (PLC) family sourced from bacteria27,29. These enzymes can be further classified into three types (I, IIa, IIb) based on amino acid adoption at specific surface subsites38. Type I includes thermophilic enzymes like LCC23 and those from Thermobifida species20, while Type IIa is represented by the polyester hydrolase (PE-H) from Pseudomonas aestusnigri39 and Type IIb by IsPETase14. Figure 1 shows that, despite their classification, these enzymes have a highly conserved core fold with only minor variations from the structure of LCC, such as an extra α-helical turn due to additional residues. Archaeal PET hydrolase from the feruloyl esterase family has key structural similarities with bacterial PLC-family PET hydrolases, but with additional α-helices flanking the active site40. This partially covered active site, similar to certain lipase family polyester hydrolases41, is expected to limit its access to polymeric substrates. Eliminating these lid-like structures has demonstrated an improvement in PET hydrolyzing ability41. Fungal cutinase-like PET hydrolases possess the most unique structures compared to LCC, with the lowest root-mean-square deviation (RMSD) value42. Although fungal cutinases were among the first recognized effective PET hydrolases43, they remained largely neglected in this research domain until recently when several studies with significant polymer degradation performance were published44,45. PET hydrolases with alternative structural folds and distinct active site geometries but considerably low depolymerizing activity were recently discovered using bioinformatics and machine-learning-guided enzyme mining in multiple metagenomic libraries27. This study validated the hypothesis that high depolymerization efficiency is likely confined to bacterial PLC-like and fungal cutinase-like PET hydrolases. A more recent study significantly expanded the range of target PET hydrolase sequences by incorporating various blocks from the ESTHER database, although it remains confined to enzymes with an α/β-hydrolase fold. Landscape profiling by sampling a sequence cluster framework revealed a collection of highly active enzymes, including MiPa-P and KuBu-P, the latter of which can even catalyze PET glycolysis in the presence of highly concentrated ethylene glycol (EG)29. Still, the top-performing enzymes, KuBu-P (PDB code 8YTW) and MiPa-P (PDB code 8YTU), have relatively low RMSD values of 0.94 Å and 0.98 Å to LCC, respectively. These findings are complicating the potential for further engineering of distinct scaffolds for industrial applications.
Illustrated are selected bacterial polyesterase-lipase-cutinase (PLC)-like27,34,36,136 and fungal cutinase-like PET hydrolases42,137,138, alongside phylogenetically distinct enzymes from archaea40 and bacteria27,139 with minimal depolymerization activity but structural uniqueness. These PET hydrolases are grouped based on their Cα root-mean-square deviation (RMSD) relative to LCC as a reference. Ribbon plots are colored with a gradient from low RMSD (blue) to 4 Å and higher (white). The catalytic triad (serine, histidine, and asparagine in all structures) is shown as yellow sticks. PDB codes, RMSD values, and residues included in the calculations are shown below each structure.
Alternatively, emerging AI-driven techniques, such as de novo enzyme design46, have the potential to generate unique PET hydrolases. In a recent study47, novel, mostly α-helical hydrolase scaffolds were obtained using a diffusion model coupled with an ensemble generation method focused on the active site preorganization. Interestingly, new active serine hydrolase scaffolds have been generated, even specifically designed to catalyze PET hydrolysis. However, disclosing only kinetic data against an atypical small-molecule substrate is insufficient to assess its realistic PET depolymerization activity. Another study generated a terminally truncated PET hydrolase using LCC as a template, with a similar catalytic efficiency but markedly lower stability than its parental scaffold46. The next option for creating novel PET hydrolases is to incorporate the classical hydrolase catalytic triad into structurally stable, non-catalytic proteins48. Nonetheless, the resulting PET depolymerization activity is significantly constrained by the inherent protein architecture, which may hinder effective substrate access necessary for interfacial catalysis17,49. Although these studies demonstrated the feasibility of the computational methodology, it is yet to be determined whether AI-assisted design can generate novel PET hydrolase scaffolds with overwhelming activity that surpasses the naturally evolved α/β hydrolase folds.
PET properties and industrial bio-recycling implications
PET is a semi-crystalline thermoplastic composed of crystalline fractions with well-ordered, tightly packed polymer chains and surrounding amorphous fractions with randomly oriented, intertwined molecules50. The crystalline microstructures exhibit significant resistance to the diffusion of water and enzymes, which are essential reactants and catalysts for biocatalytic depolymerization, respectively8,51. In contrast, the amorphous PET fractions are significantly more susceptible to enzymatic hydrolysis, particularly when environmental temperatures reach or exceed the glass transition temperature (Tg). Shorter segments of amorphous PET undergo conformational changes during the glass transition, which is thought to improve enzyme accessibility and subsequent ester bond hydrolysis significantly8,50,52. The Tg of PET under anhydrous conditions has been determined to range between 65 and 81 °C, depending on the analytical methods used8,53. PET substrates are completely immersed in water during biocatalytic depolymerization, and the resulting water plasticization effect can lower the bulk Tg of PET by up to 16 °C8,53. Considering the enhanced mobility of surface-located polymer chains54, the Tg of the PET surface layer (surface Tg) may be as low as ~40 °C53, which explains why significant hydrolysis of highly amorphous PET can be observed with mesophilic bacteria like I. sakaiensis under ambient conditions14.
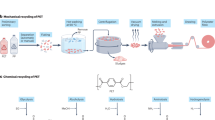
Melt-extrusion-based amorphization9,15,36,55,56,57,58,59 is commonly used as the first step before enzymatic PET decomposition due to the high crystallinity of over 30% in real-world waste PET streams (Fig. 2). Rapidly quenching the hot polymer extrudates in cold water or other solvents is required to achieve a low-crystallinity of ideally less than 15%, allowing for rapid biocatalytic depolymerization50,60,61. Thereafter, the amorphized PET typically undergoes a mechanical size-reduction process, either via cryogenic grinding or ambient temperature crushing9,15,36,57,58,59. The increased surface-to-volume ratios are considered advantageous for interfacial enzyme accessibility, thereby improving polymer conversion efficiency. Nevertheless, this hypothesis has not consistently held true in several recent studies. The smallest particles with the highest specific surface area (SSA) extracted from the same micronization batch of low-crystallinity (or amorphized) PET typically exhibited slightly or moderately enhanced initial hydrolysis rates, particularly at lower enzyme loading, but significantly reduced maximum achievable depolymerization extent compared to the larger ones with lower SSA55,62,63. The latter phenomenon has been ascribed to the increased crystallinity, which typically exhibits an inverse relationship with particle sizes, caused by strain-induced crystallization during micronization36,55,62,64. Larger ground particles and precursor amorphous PET materials prior to micronization (either granules from melt-quenching or commercial PET films) can be enzymatically depolymerized to comparably high extents after an adequate reaction duration. Given the high energy costs and negative environmental effects associated with the micronization process11,12, the trade-offs between fast hydrolysis kinetics using cryomilled plastics and prolonged depolymerization with non-ground plastics necessitate reassessment in industrial settings. A patent for PET pre-treatment aimed at enzymatic recycling suggests that using an underwater pelletizer (Fig. 2) to produce small pellets under 1 mm could replace the contentious and energy-intensive grinding process65.
To improve its economic viability, the cryogrinding step is considered optional and omitted here. The figure also displays key material-related parameters and reaction conditions that affect depolymerization performance. The crystallinity, molecular mass, and specific surface area of a particular PET sample are crucial interrelated parameters that influence the achievable degree of depolymerization. A simplified schematic surface plot illustrating the threshold values for optimal depolymerization performance emphasizes the interdependence of these parameters. Additional parameters influencing decomposition performance, such as varying Tg of the surface layer and interior bulk polymers, as well as the effect of water plasticization and polymer recrystallization under biocatalytic conditions, should be considered. Bioreactor parameters for reaction conditions must be consistently documented and standardized to ensure comparable high-level depolymerization results.
Polymer chain scission can occur at each stage of the presently performed thermomechanical pre-treatment of PET waste (Fig. 2). Despite being an undesirable consequence when a similar melt-extrusion process is used for PET mechanical recycling66, the decreased molecular weights were found to facilitate the subsequent enzymatic depolymerization36,67. Once PET feedstocks have a number-averaged molecular mass (Mn) of less than 10,000 g mol−1, their rapid and complete enzymatic depolymerization becomes less dependent on the bulk crystallinity, even if it surpasses the previously recommended degradable threshold value of 20%57. By harsher pre-treatment involving microwaves, PET’s Mn can be reduced below the entanglement length of 3500 g mol−1 and readily hydrolyzed using an IsPETase variant at ambient temperature67. Within this Mn range, conventional indicators of high crystalline PET, such as the increased ratio of trans to gauche conformation, become irrelevant for describing its enzymatic degradability.
These findings highlight the importance of comprehensively documenting all of these polymer properties, including molecular mass, SSA (or particle size), and bulk crystallinity (preferably also the crystallinity of the surface polymer layer), throughout the depolymerization process in future research. This can assist in unambiguously defining standard PET feedstocks (Table 1) and building a standardized amorphization technique that balances optimal material qualities related to degradability and economic viability, reducing the cost of this most expensive step11,12. Currently, research groups lacking comprehensive polymer characterization equipment are advised to utilize commercially available amorphous PET film, such as that provided by Goodfellow Ltd., as a preliminary test material for PET hydrolases in initial laboratory-scale scientific studies. Several studies36,68 have recently published detailed polymer parameters for this material, ranging beyond mere crystallinity, establishing its optimal utility over waste PET from unknown sources with undefined properties.
The distinct polymer properties of PET also define the appropriate characteristics of the biocatalysts and the reaction conditions necessary for its fast depolymerization (Table 1). The flexibility of water-plasticized amorphous PET polymer chains enhances as reaction temperatures rise between 40-70 °C, facilitating their accessibility to enzymatic hydrolysis69. Higher temperatures, however, lead to faster polymer recrystallization into non-degradable crystalline microstructures than enzymatic depolymerization can occur9,15,50,60. Consequently, recent studies indicate a preference for reaction temperatures between 65 and 70 °C and the use of thermotolerant enzymes, such as LCC-ICCG9,15, TurboPETase57 and PES-H115,24,36,61,70,71, in pilot-scale processes that resemble industrial settings9,57,71. Accordingly, thermostabilization has been implemented as a primary strategy to enhance the depolymerization capability of mesophilic PET hydrolases, including Kubu-P29 and IsPETase14. Nonetheless, regarding IsPETase, despite substantial research having raised its melting point (Tm) by over 37 °C72,73,74, the most advanced variants remain inferior to LCC-ICCG in large-scale PET depolymerization tests15. This raises the question of whether a high Tm (above 85 °C) can serve as the sole criterion, as commonly employed in previous studies6,8, to determine if a specific enzyme inherently exhibits the necessary long-term stability and activity over several hours for the rapid PET decomposition at elevated temperatures. Instead, future research should evaluate the kinetic stability of an enzyme under operational conditions to provide a broader assessment of its thermostability.
The continuous release of terephthalate (TPA) monomer as a result of enzymatic PET hydrolysis lowers the pH value of the reaction supernatant, which may influence enzyme activity and stability, as well as the compositions of the product mixture15,75. Increased concentrations of unwanted oligomeric ester intermediates can hinder subsequent product purification and diminish the yield of pure TPA. Therefore, recently demonstrated depolymerization processes are frequently pH-controlled in the range of 7–8, which is appropriate for most known PET hydrolases9,43,57. Nonetheless, low-concentrated phosphate or Tris buffers are commonly used in lab-scale bioreactors with pH control9,57. This should be avoided in large industrial settings to reduce costs and ensure reactor durability. pH control is considered a concern regarding the economic and environmental impacts of industrial PET recycling12. For future lab-scale studies, these impacts can be mitigated by utilizing ammonia in place of NaOH for pH regulation12,58 or implementing small-scale bioreactors (<50 mL), such as the modified Chi.Bio systems instead of other larger commercial bioreactors76. Nonetheless, because small reactors can only accommodate less than 10% solid loading, it is recommended to validate the process in medium-sized reactors (in litre scales) that allow standard agitation with up to 30% solid loading before final industrial implementation at cubic meter scale (Box 1). A more ambitious long-term vision to address this pH-related issue may involve designing a PET hydrolase that is less sensitive to pH fluctuations or that is optimized for a low pH range to facilitate rapid accumulation of exclusively pure TPA.
Alongside the utilization of standardized PET feedstocks, establishing standardized bioreactor conditions by defining optimal solid (substrate) loading, enzyme loading, temperature, pH, agitation, and other parameters can ensure consistent, efficient, and scalable operations while maximizing monomer yield across diverse scales, from research laboratories to industrial environments. Nevertheless, as indicated in Tables 1 and 2, substantial discrepancies in specific key parameters reported to date, coupled with the sporadic reporting of others, highlight the necessity and challenges of conducting further comprehensive studies for standardization.
Alternative recycling scenarios using thermolabile PET hydrolases
Numerous recently discovered PET hydrolases are intrinsically thermolabile, rendering them less suitable candidates for free enzyme-based PET recycling without extensive and expensive thermostabilization engineering. Alternatively, they can be recombinantly expressed in (mostly mesophilic) microbial cell factories that can utilize at least one PET monomer for growth71,77,78,79. This concept of polymer degradation and monomer catabolism by a single microbe was recently coined PETtrophy80,81, analogous to consolidated bioprocessing in cellulose conversion82. However, achieving high enzymatic plastic depolymerization activities that align with the microbe’s growth conditions, including pH, temperature, and salt concentration, is challenging. Molecular engineering tools are frequently absent for modifying microbial thermophiles83, leading to the infrequent demonstration of thermotolerant whole-cell catalysts for PET decomposition at its optimal temperature. The only example to date is engineered Clostridium thermocellum expressing LCC and its variants, though lacking monomer metabolism capability63,84. Other PET-metabolizing strains are constructed based on trivial microbial chassis that grow at temperatures below 40 °C85. Consequently, mesophilic PET hydrolases must be expressed at sufficiently high levels and tailored to enable the rapid release of growth-essential monomers into the medium, outpacing their translocation into microbial cells and subsequent metabolism. The efficacy of PET depolymerization under these conditions is expected to be significantly lower than that at elevated temperatures exceeding 65 °C. Thus, an upcycling strategy utilizing these strains to produce value-added chemicals should always be considered to realize potential economic viability. In addition, focusing on “easier” substrates that can be depolymerized faster at ambient temperatures may enhance the utility of these PET-trophic strains for stakeholders. These include PET oligomers that have been rigorously pretreated or pre-degraded67,86, as well as alternative polyesters with similar chemical compositions, such as the commercially available polybutylene adipate terephthalate (PBAT)80. The time and cost savings from bypassing polymer and monomer separation may compensate the typically low depolymerization efficiency of whole-cell catalysts, providing significant benefits compared to free enzyme-based depolymerization.
Another extreme example is the in situ remediation of wastewater contaminated with synthetic microfibers, which, according to a current study, are primarily oligomeric PET87, for example, in effluents in textile production regions88 or general communal wastewater treatment plants. PET hydrolases derived from marine environments exhibiting significant salt tolerance28 can ultimately be utilized to construct environmentally resistant whole-cell catalysts when integrated into a comparably robust microbial chassis89,90. Other mesophilic wild-type enzymes may exhibit insufficient activity and stability under these harsh conditions, such as high salinity, extreme pH levels, or the presence of laundry detergents, necessitating further extensive enzyme optimization efforts91. Nonetheless, this may spark a heated debate about releasing genetically modified microbes (GMM)92, which is strictly prohibited in the EU (and other countries). In academic literature, this is referenced so far as a theoretical alternative in regions lacking other solutions93. However, as a result of intensive ongoing research activities, challenges associated with technical implementations and governmental regulations may be addressed shortly to maximize the benefits of GMMs in combating plastic pollution while mitigating risks of environmental release92.
Screening strategies for customizing superior PET hydrolases
Numerous high-throughput screening (HTS) assays have been developed in recent years for the discovery and engineering of PET hydrolases16,94. However, their specific limitations in areas such as technical availability, ease of handling, and reproducibility necessitate re-evaluation before a standard guideline can be established.
Several assays employ self-synthesized small-molecule surrogate substrates that resemble PET segments or degradation intermediates, such as mono- or bis(2-hydroxyethyl) terephthalate (MHET or BHET)95,96,97. While these proxy substrates offer practical benefits, including simplified dosing and the compatibility with established HTS platforms, they may result in a skewed identification of short ester hydrolyzing activity without PET depolymerization capability98,99,100, particularly in environmental samples or functional metagenomics. On the other hand, screening of mutant libraries derived from known PET hydrolases can benefit from this assay category96,101, as increased activity on MHET or BHET is frequently linked to the mitigation of their inhibitory effect, thereby improving overall PET decomposition performance. To minimize false positives, it is still highly advisable to consistently prioritize the direct use of PET polymers in HTS experimental setups. Optimal PET substrates must demonstrate uniform polymer properties, such as low-crystallinity and high specific surface area, facilitating rapid decomposition that can be observed within a constrained reaction volume, such as microplate wells or microdroplets, and a short timeframe, ideally under one hour. PET nanoparticles (NP), produced through solvent displacement processes102,103,104, are a polymeric substrate option characterized by a uniform size range of 100 ± 50 nm and lower molecular weights compared to their precursor materials (Table 1). These material properties promote their rapid enzymatic decomposition, even at ambient temperature36,48,102,103,104,105. PET NP can be used to detect microbial depolymerization activities by clear-zone assay when immobilized in agar plates. Similarly, the turbidity variations of homogenously suspended PET NP immobilized in other hydrogels can facilitate the determination of PET hydrolysis kinetics36,70,102. However, PET NP tend to agglomerate in buffered aqueous solutions, complicating their handling in small reaction volumes such as microplate wells or microdroplets. Therefore, when activity is quantified solely through the formation and detection of released monomers104,105, PET NP have no significant advantage over other amorphous PET materials, whether tailored from commercially available products or casted from polymer solution106,107,108. The casting method for preparing polymeric substrates can also incorporate fluorescent dyes, which can be detected in the supernatant during PET decomposition, providing an alternative to quantifying its monomers108. However, the use of toxic PET soluble organic solvents for HTS setups is considered suboptimal due to challenges in handling, high costs, environmental concerns, and laboratory safety issues.
TPA, MHET, and BHET are the main hydrolysis products of PET that can be detected at 240 nm. Aside from the standard quantification method of high-performance liquid chromatography (HPLC), the total amount of their formation can be easily estimated by simply measuring the total absorbance at 240 nm for HTS purposes101,106,109. When TPA needs to be precisely quantified as a final monomer, it can be converted to a hydroxylated form and detected by fluorimetric monitoring with a high detection sensitivity96,104,107,110. The quantification of the other monomer, EG, has been overlooked for a considerable period until recently. EG can now be accurately monitored by different fluorimetric assays based on enzymatic conversion111 or a biosensor112. In conjunction with other recently developed techniques for monitoring TPA105,113,114,115, living-cell based biosensors offer various options for assessing PET hydrolyzing activity, from precise endpoint determination of individual monomers to functional screening of metagenomic libraries. Microfluidics-based HTS also involves living cells, facilitating enzyme expression, biocatalysis, and product detection within a single microdroplet or microbead116. These methods may not effectively identify thermotolerant enzyme variants because the optimal temperatures for cell growth typically do not exceed 40 °C, while apparent PET depolymerization activity is generally detectable at temperatures above 50 °C. The same limitation may also pertain to other cell growth-based screening assays, such as enzyme surface display117,118. While these HTS assays primarily evaluate mesophilic PET hydrolases which typically necessitate costly and risky thermostabilization engineering prior to validating their potential for industrial PET recycling, these enzymes should be better suited to the alternative application scenarios at lower temperatures discussed above.
Consequently, an automated liquid handling system that incorporates: (i) enzyme expression and cell lysis at optimal cell-growth temperatures, (ii) PET depolymerization at elevated temperatures, and (iii) endpoint assessment of monomer release under optimal assay conditions should serve as an ideal standard workflow for screening thermotolerant PET hydrolases needed for industrial PET recycling (Table 2 and Fig. 3). A few recent studies demonstrated variations of this standard workflow, particularly characterized by the simultaneous detection of hydrolysis products and quantification of soluble recombinant enzymes in cell lysates through various optical techniques101,108. The latter can replace the labor-intensive and costly high-throughput protein purification step while maintaining high data accuracy of specific enzyme activities, which is crucial for machine-learning-guided enzyme engineering. To optimize these assays, proxy substrates like BHET and its derivatives96,101 can be replaced with, for example, cryomilled commercially available amorphous PET with defined characteristics62,68, particularly for HTS using microplate readers. This may enable solid loading of >10% in microplate wells to more accurately resemble standardized industrial conditions (Table 2).
The function-based HTS method is an essential component of the workflow. Given that agar-plate or microfluidics-based screening involves living cells, which prevents the detection of PET hydrolysis at elevated temperatures, microplate-based HTS (detailed in the lower part) may have the potential to be prioritized. Colorimetric or fluorometric methods, sensitive at different wavelengths, are optimal for the distinct quantification of functional enzyme concentrations in cell lysates and the monomers released from PET depolymerization. This approach enables rapid screening of large mutant libraries for efficient PET hydrolases while also producing high-quality data at a high throughput that can be used for machine-learning-guided enzyme engineering116.
Future methodological advancement to close knowledge gaps
Currently used endpoint measurements for assessing depolymerization efficacy allow for basic comparisons between engineered enzymes and their references, but they lack time-resolved data throughout the degradation process. As a result, molecular insights required to understand the mechanisms underlying interfacial biocatalysis reactions are frequently missing. Computational modeling based on recently resolved complex structures of various PET hydrolases with substrate analogues16 may elucidate the catalytic cycle in atomic detail. Still, it fails to account for other important aspects such as enzyme adsorption/desorption behavior and PET physical properties. Much evidence suggests that these latter aspects, which reflect non-covalent interactions, serve as key bottlenecks for the overall reaction. The most explicit example of this is the resilience of crystalline PET to enzymatic hydrolysis. However, small, soluble PET fragments, similar to those used in computational analyses, can be hydrolyzed very quickly119, which also implies that the main activation barrier for the overall process lies outside catalytic bond breakage. The classical way to single out the importance of different steps in a complex reaction is comparative kinetics, and this approach has two key requirements. Firstly, a simple yet realistic microkinetic reaction scheme, which introduces rate constants for the interconversion of substrate and intermediates, and secondly experimental methods that provide sufficient data to support kinetic modeling. At the current stage, none of these areas are well-developed for PET hydrolases, but some progress has been made. For example, limitations associated with an undefined concentration of the insoluble substrate have been addressed by introducing substrate area60,120, or attack site density119 as proxies for molar concentration. It has also proven useful to supplement the conventional conditions of substrate excess for steady-state kinetic analysis with measurements, where the enzyme is in excess concerning the substrate102,121. This latter experimental condition is the opposite of the conventional kinetic framework, and it has been termed inverse Michaelis–Menten kinetics122. A steady-state approximation under inverse conditions never occurs in homogenous reactions but may be relevant in two-phase systems as new reaction sites on the surface are replenished continuously123. Parameters from inverse kinetic analyses, particularly the maximal rate at high enzyme load, are of interest because they reflect both catalytic efficacy and the density of reaction or attack sites on the solid surface. Specifically, a combination of data obtained with either enzyme- or substrate-excess provides information on the maximal turnover (kcat in s−1), KM (Michaelis constant in M), and the density of hydrolysable sites on the PET surface (in molsites gPET−1)122. If experiments are only conducted under conditions of substrate excess, KM values will be in mass-based units, typically g L−1. This parameter can be useful for comparing different enzymes acting on the same substrate, but as mass-based KM values depend strongly on the SSA of the substrate, it is generally not meaningful to compare values obtained with different types of PET. Work along these lines has identified some general trends in PET hydrolase kinetics, which are listed in Table 2. For a number of the most proficient enzymes acting on insoluble PET at moderate temperature (30–50 °C), kcat was 0.5–2 s−1, and this was 1–2 orders of magnitude slower than their hydrolysis of the same bond in soluble PET fragments119,121,124. The same enzymes had KM values in the hundredths of nM range, and taken together, this implies strong substrate interactions but slow turnover compared to other hydrolases acting on native, soluble substrates125. A molecular description of these characteristics appears to be a promising avenue for further understanding of structure-function relationships for PET hydrolases. To achieve this, we will need better methods to quantify the accumulation and decay of intermediates; both enzyme-substrate complexes and PET fragments. Progress curves for these intermediates will elucidate the complex pathway from insoluble polymer to the monomers TPA and EG, and it will also provide the necessary experimental input for further kinetic modeling. Advanced knowledge in this respect may help to accelerate the late stage of the industrial recycling process, which is currently thought to be hampered by these hydrolysis intermediates. Finally, we encourage more experimental and computational research into the relationship between polymer conformation and enzyme activity. Some reports have pointed out a strong effect on catalysis of the trans-gauche isomerism in the PET chain52,67, and it has been proposed that constraints on isomerization may lessen gradually as PET chains are shortened in the enzymatic process126. Information on this would be important both for the fundamental understanding of the interfacial enzyme reaction and as a guide for enzyme engineering.
Conclusions and outlook
The increasing demand for sustainable plastic recycling solutions, integral to the bio-based circular economy and climate crisis mitigation, positions the biotechnological valorization of PET as a dynamic research field in the foreseeable future.
The successful implementation of several extensively customized PET hydrolases, which allow for nearly complete polymer-to-monomer conversion in less than 12 h on pilot or larger industrial scales9,29,57, suggests that fast kinetics is unlikely to be the primary constraint in this research domain. However, to maintain competitiveness, PET hydrolases, similar to other industrial enzymes, must undergo ongoing performance improvements, including enhanced enzyme kinetics and long-term stability, tailored pH optima, etc. Consequently, standard guidelines derived from research endeavors so far are discussed above and summarized in Box 1 and Tables 1 and 2. Other related research avenues, which have received considerably less attention yet appear to hold significant promise, should be explored with greater focus in the future. For instance, enzyme immobilization may enhance stability and facilitate multiple applications127,128. Also, the development of synergistic enzyme cocktails may enable reduced dosages or address mixed plastic waste129, a more complex challenge within the plastic pollution dilemma. For the latter, a moderate temperature range of 40–50 °C may be considered when extended operation time is acceptable, particularly when not all applied enzymes exhibit adequate half-lives at higher temperatures.
Simultaneously, understanding and optimizing process conditions should be afforded equal attention and effort as enzyme engineering in future research. This may involve optimizing melt-extrusion parameters56, investigating the feasibility of omitting energy-intensive micronization stages, or implementing alternative pre-treatment techniques130, all of which could reduce the costs of the current process11,12. A thorough understanding of the material properties associated with degradability is urgently required to assess the feasibility of current research aimed at developing highly efficient enzymes specifically for crystalline PET. These forthcoming research endeavors may address our knowledge gaps concerning the absent information in the standardized parameter set (Tables 1 and 2) that collectively impacts the future development of industrial PET recycling.
At the enzyme discovery stage, researchers should move beyond the trivial homology-based database mining and increase the use of novel bioinformatics- or AI-based tools to discover or design new PET hydrolase scaffolds29,47. Moreover, the previously underutilized classical function-based metagenomics or microbiological techniques can still be remarkably beneficial for discovering novel sequence or structural diversity of depolymerizing enzymes that have not yet been included in the digital repository. We propose a more efficient data-sharing approach by characterizing novel enzymes and all known mutants of benchmark enzymes, summarizing their biocatalytic properties in the standardized format shown in Table 2, and making them available online for free access. These will ultimately contribute significantly to the advancement of AI-based PET hydrolase engineering or design, as well as the development of specific AI models tailored for interfacial biocatalysis.
Motivated by the successful industrialization of enzymatic PET recycling, other hydrolysable waste plastics, such as polyurethanes, polylactides, and polyamides, are logically emerging as the next wave of substrates for potential biocatalytic depolymerization13,131,132,133. This ambition necessitates considering the lessons and experiences learned from PET, such as using polymer feedstocks with precisely defined properties and the need for standardized conditions for evaluating novel depolymerizing enzymes. Ultimately, we expect this article will inspire researchers to collaborate and develop similar guidelines to standardize the future bio-based plastic recycling industry.
References
Xin, J. et al. Progress in the catalytic glycolysis of polyethylene terephthalate. J. Environ. Manag. 296, 113267 (2021).
Aryan, Y., Yadav, P. & Samadder, S. R. Life cycle assessment of the existing and proposed plastic waste management options in India: a case study. J. Clean. Prod. 211, 1268–1283 (2019).
Tsochatzis, E. D., Lopes, J. A. & Corredig, M. Chemical testing of mechanically recycled polyethylene terephthalate for food packaging in the European Union. Resour. Conserv. Recycl. 179, 106096 (2022).
Ragaert, K., Delva, L. & Van Geem, K. Mechanical and chemical recycling of solid plastic waste. Waste Manag. 69, 24–58 (2017).
Ellis, L. D. et al. Chemical and biological catalysis for plastics recycling and upcycling. Nat. Catal. 4, 539–556 (2021).
Tournier, V. et al. Enzymes’ power for plastics degradation. Chem. Rev. 123, 5612–5701 (2023).
Müller, R.-J., Schrader, H., Profe, J., Dresler, K. & Deckwer, W.-D. Enzymatic degradation of poly(ethylene terephthalate): rapid hydrolyse using a hydrolase from T. fusca. Macromol. Rapid Commun. 26, 1400–1405 (2005).
Wei, R. et al. Mechanism-based design of efficient PET hydrolases. ACS Catal. 12, 3382–3396 (2022).
Tournier, V. et al. An engineered PET depolymerase to break down and recycle plastic bottles. Nature 580, 216–219 (2020).
Gomollón-Bel, F. IUPAC’s 2023 top ten emerging technologies in chemistry. Chem. Int. 45, 14–22 (2023).
Singh, A. et al. Techno-economic, life-cycle, and socioeconomic impact analysis of enzymatic recycling of poly(ethylene terephthalate). Joule 5, 2479–2503 (2021).
Uekert, T. et al. Life cycle assessment of enzymatic poly(ethylene terephthalate) recycling. Green. Chem. 24, 6531–6543 (2022).
Wei, R., Weber, G., Blank, L. M. & Bornscheuer, U. T. Process insights for harnessing biotechnology for plastic depolymerization. Nat. Chem. Eng. 2, 110–117 (2025).
Yoshida, S. et al. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 351, 1196–1199 (2016).
Arnal, G. et al. Assessment of four engineered PET degrading enzymes considering large-scale industrial applications. ACS Catal. 13, 13156–13166 (2023).
Mican, J. et al. Exploring new galaxies: perspectives on the discovery of novel PET-degrading enzymes. Appl. Catal. B-Environ. 342, 123404 (2024).
Zhou, J., Cui, Z., Wei, R., Dong, W. & Jiang, M. Interfacial catalysis in enzymatic PET plastic depolymerization. Trends Chem. 7, 175–185 (2025).
Purdy, R. E. & Kolattukudy, P. E. Depolymerization of a hydroxy fatty acid biopolymer, cutin, by an extracellular enzyme from Fusarium solani f. pisi: Isolation and some properties of the enzyme. Arch. Biochem. Biophys. 159, 61–69 (1973).
Fett, W. F., Wijey, C., Moreau, R. A. & Osman, S. F. Production of cutinase by Thermomonospora fusca ATCC 27730. J. Appl. Microbiol. 86, 561–568 (1999).
Herrero Acero, E. et al. Enzymatic surface hydrolysis of PET: effect of structural diversity on kinetic properties of cutinases from Thermobifida. Macromolecules 44, 4632–4640 (2011).
Liu, R. et al. Polyethylene terephthalate (PET)-degrading bacteria in the pelagic deep-sea sediments of the Pacific Ocean. Environ. Pollut. 352, 124131 (2024).
Wilkes, R. A. et al. Mechanisms of polyethylene terephthalate pellet fragmentation into nanoplastics and assimilable carbons by wastewater comamonas. Environ. Sci. Technol. 58, 19338–19352 (2024).
Sulaiman, S. et al. Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Appl. Environ. Microbiol. 78, 1556–1562 (2012).
Sonnendecker, C. et al. Low carbon footprint recycling of post-consumer PET plastic with a metagenomic polyester hydrolase. ChemSusChem 15, e202101062 (2022).
Meyer Cifuentes, I. E. et al. Molecular and biochemical differences of the tandem and cold-adapted PET hydrolases Ple628 and Ple629, isolated from a marine microbial consortium. Front. Bioeng. Biotechnol. 10, 930140 (2022).
Danso, D. et al. New insights into the function and global distribution of polyethylene terephthalate (PET)-degrading bacteria and enzymes in marine and terrestrial metagenomes. Appl. Environ. Microbiol. 84, e02773–02717 (2018).
Erickson, E. et al. Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity. Nat. Commun. 13, 7850 (2022).
Chen, J. et al. Global marine microbial diversity and its potential in bioprospecting. Nature 633, 371–379 (2024).
Seo, H. et al. Landscape profiling of PET depolymerases using a natural sequence cluster framework. Science 387, eadp5637 (2025).
Xi, X. et al. Secretory expression in Bacillus subtilis and biochemical characterization of a highly thermostable polyethylene terephthalate hydrolase from bacterium HR29. Enzym. Microb. Technol. 143, 109715 (2021).
Hong, H. et al. Discovery and rational engineering of PET hydrolase with both mesophilic and thermophilic PET hydrolase properties. Nat. Commun. 14, 4556 (2023).
Qi, X., Ji, M., Yin, C.-F., Zhou, N.-Y. & Liu, Y. Glacier as a source of novel polyethylene terephthalate hydrolases. Environ. Microbiol. 25, 2822–2833 (2023).
Eiamthong, B. et al. Discovery and genetic code expansion of a polyethylene terephthalate (PET) hydrolase from the human saliva metagenome for the degradation and bio-functionalization of PET. Angew. Chem. Int. Ed. 61, e202203061 (2022).
Sulaiman, S., You, D.-J., Kanaya, E., Koga, Y. & Kanaya, S. Crystal structure and thermodynamic and kinetic stability of metagenome-derived LC-cutinase. Biochemistry 53, 1858–1869 (2014).
Zeng, W. et al. Substrate-binding mode of a thermophilic PET hydrolase and engineering the enzyme to enhance the hydrolytic efficacy. ACS Catal. 12, 3033–3040 (2022).
Pfaff, L. et al. Multiple substrate binding mode-guided engineering of a thermophilic PET hydrolase. ACS Catal. 12, 9790–9800 (2022).
Chatonnet, A., Perochon, M., Velluet, E. & Marchot, P. The ESTHER database on alpha/beta hydrolase fold proteins—an overview of recent developments. Chem. Biol. Interact. 383, 110671 (2023).
Joo, S. et al. Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation. Nat. Commun. 9, 382 (2018).
Bollinger, A. et al. A novel polyester hydrolase from the marine bacterium Pseudomonas aestusnigri —structural and functional insights. Front. Microbiol. 11, 114 (2020).
Perez-Garcia, P. et al. An archaeal lid-containing feruloyl esterase degrades polyethylene terephthalate. Commun. Chem. 6, 193 (2023).
Biundo, A. et al. Polyphenol oxidases exhibit promiscuous proteolytic activity. Commun. Chem. 3, 62 (2020).
Dimarogona, M. et al. Structural and functional studies of a Fusarium oxysporum cutinase with polyethylene terephthalate modification potential. Biochim. Biophys. Acta Gen. Subj. 1850, 2308–2317 (2015).
Ronkvist, ÅM., Xie, W., Lu, W. & Gross, R. A. Cutinase-catalyzed hydrolysis of poly(ethylene terephthalate). Macromolecules 42, 5128–5138 (2009).
Lee, S. H. et al. Characterization and engineering of a fungal poly(ethylene terephthalate) hydrolyzing enzyme from Aspergillus fumigatiaffinis. ACS Catal. 14, 4108–4116 (2024).
Brinch-Pedersen, W. et al. Discovery and surface charge engineering of fungal cutinases for enhanced activity on poly(ethylene terephthalate). ACS Sustain. Chem. Eng. 12, 7329–7337 (2024).
Ding, Y., Zhang, S., Kong, X., Hess, H. & Zhang, Y. Replicating PET hydrolytic activity by positioning active sites with smaller synthetic protein scaffolds. Adv. Sci. 12, 2500859 (2025).
Lauko, A. et al. Computational design of serine hydrolases. Science 388, adu2454 (2025).
Robles-Martín, A. et al. Sub-micro- and nano-sized polyethylene terephthalate deconstruction with engineered protein nanopores. Nat. Catal. 6, 1174–1185 (2023).
Wei, R. & Bornscheuer, U. T. Designer catalytic nanopores meet PET nanoparticles. Nat. Catal. 6, 1105–1106 (2023).
Thomsen, T. B., Almdal, K. & Meyer, A. S. Significance of poly(ethylene terephthalate) (PET) substrate crystallinity on enzymatic degradation. N. Biotechnol. 78, 162–172 (2023).
Woodard, L. N. & Grunlan, M. A. Hydrolytic degradation and erosion of polyester biomaterials. ACS Macro Lett. 7, 976–982 (2018).
Wei, R. et al. Conformational fitting of a flexible oligomeric substrate does not explain the enzymatic PET degradation. Nat. Commun. 10, 5581 (2019).
Tarazona, N. A. et al. Rapid depolymerization of poly(ethylene terephthalate) thin films by a dual-enzyme system and its impact on material properties. Chem. Catal. 2, 3573–3589 (2022).
Shinotsuka, K., Bliznyuk, V. N. & Assender, H. E. Near-surface crystallization of PET. Polymer 53, 5554–5559 (2012).
Chang, A. C. et al. Understanding consequences and tradeoffs of melt processing as a pretreatment for enzymatic depolymerization of poly(ethylene terephthalate). Macromol. Rapid Commun. 43, 2100929 (2022).
Patel, A. et al. Melt processing pretreatment effects on enzymatic depolymerization of poly(ethylene terephthalate). ACS Sustain. Chem. Eng. 10, 13619–13628 (2022).
Cui, Y. et al. Computational redesign of a hydrolase for nearly complete PET depolymerization at industrially relevant high-solids loading. Nat. Commun. 15, 1417 (2024).
Xue, R. et al. Enzymatic upcycling of PET waste to calcium terephthalate for battery anodes. Angew. Chem. Int. Ed. 63, e202313633 (2024).
Cheng, Y., Cheng, Y., Zhou, S., Ruzha, Y. & Yang, Y. Closed-loop recycling of PET fabric and bottle waste by tandem pre-amorphization and enzymatic hydrolysis. Resour. Conserv. Recycl. 208, 107706 (2024).
Wei, R. et al. Biocatalytic degradation efficiency of postconsumer polyethylene terephthalate packaging determined by their polymer microstructures. Adv. Sci. 6, 1900491 (2019).
Thomsen, T. B. et al. Rate response of poly(ethylene terephthalate)-hydrolases to substrate crystallinity: basis for understanding the lag phase. ChemSusChem 16, e202300291 (2023).
Brizendine, R. K. et al. Particle size reduction of poly(ethylene terephthalate) increases the rate of enzymatic depolymerization but does not increase the overall conversion extent. ACS Sustain. Chem. Eng. 10, 9131–9140 (2022).
Liu, Y.-J. et al. Optimized whole-cell depolymerization of polyethylene terephthalate to monomers using engineered Clostridium thermocellum. J. Hazard. Mater. 488, 137441 (2025).
Jabarin, S. A. Strain-induced crystallization of poly(ethylene terephthalate). Polym. Eng. Sci. 32, 1341–1349 (1992).
Desrousseaux, M.-L. et al. A process for degrading plastic products. Patent WO2017198786A1. France (2017).
Schyns, Z. O. G. & Shaver, M. P. Mechanical recycling of packaging plastics: a review. Macromol. Rapid Commun. 42, 2000415 (2021).
Guo, B. et al. Conformational selection in biocatalytic plastic degradation by PETase. ACS Catal. 12, 3397–3409 (2022).
Cuthbertson, A. A. et al. Characterization of polymer properties and identification of additives in commercially available research plastics. Green. Chem. 26, 7067–7090 (2024).
Akram, E. et al. On the temperature dependence of enzymatic degradation of poly(ethylene terephthalate). Chin. J. Catal. 60, 284–293 (2024).
Jäckering, A. et al. From bulk to binding: decoding the entry of PET into hydrolase binding pockets. JACS Au 4, 4000–4012 (2024).
Welsing, G. et al. Two-step biocatalytic conversion of post-consumer polyethylene terephthalate into value-added products facilitated by genetic and bioprocess engineering. Bioresour. Technol. 417, 131837 (2025).
Cui, Y. et al. Computational redesign of a PETase for plastic biodegradation under ambient condition by the GRAPE strategy. ACS Catal. 11, 1340–1350 (2021).
Lu, H. et al. Machine learning-aided engineering of hydrolases for PET depolymerization. Nature 604, 662–667 (2022).
Bell, E. L. et al. Directed evolution of an efficient and thermostable PET depolymerase. Nat. Catal. 5, 673–681 (2022).
Świderek, K. et al. Mechanistic studies of a lipase unveil effect of pH on hydrolysis products of small PET modules. Nat. Commun. 14, 3556 (2023).
Denton, M. C. R. et al. Integration of pH control into Chi.Bio reactors and demonstration with small-scale enzymatic poly(ethylene terephthalate) hydrolysis. Biochemistry 63, 1599–1607 (2024).
Mückschel, B. et al. Ethylene glycol metabolism by Pseudomonas putida. Appl. Environ. Microbiol. 78, 8531–8539 (2012).
Tiso, T. et al. Towards bio-upcycling of polyethylene terephthalate. Metab. Eng. 66, 167–178 (2021).
Narancic, T. et al. Genome analysis of the metabolically versatile Pseudomonas umsongensis GO16: the genetic basis for PET monomer upcycling into polyhydroxyalkanoates. Microb. Biotechnol. 14, 2463–2480 (2021).
Brandenberg, O. F., Schubert, O. T. & Kruglyak, L. Towards synthetic PETtrophy: engineering Pseudomonas putida for concurrent polyethylene terephthalate (PET) monomer metabolism and PET hydrolase expression. Microb. Cell Fact. 21, 119 (2022).
Han, W. et al. Biodegradation of poly(ethylene terephthalate) through PETase surface-display: from function to structure. J. Hazard. Mater. 461, 132632 (2024).
Li, Z., Waghmare, P. R., Dijkhuizen, L., Meng, X. & Liu, W. Research advances on the consolidated bioprocessing of lignocellulosic biomass. Eng. Microbiol. 4, 100139 (2024).
Crosby, J. R. et al. Extreme thermophiles as emerging metabolic engineering platforms. Curr. Opin. Biotechnol. 59, 55–64 (2019).
Yan, F., Wei, R., Cui, Q., Bornscheuer, U. T. & Liu, Y.-J. Thermophilic whole-cell degradation of polyethylene terephthalate using engineered Clostridium thermocellum. Microb. Biotechnol. 14, 374–385 (2021).
Tiso, T. et al. The metabolic potential of plastics as biotechnological carbon sources—review and targets for the future. Metab. Eng. 71, 77–98 (2022).
Giraldo-Narcizo, S., Guenani, N., Sánchez-Pérez, A. M. & Guerrero, A. Accelerated polyethylene terephthalate (PET) enzymatic degradation by room temperature alkali pre-treatment for reduced polymer crystallinity. ChemBioChem 24, e202200503 (2023).
Yang, T., Xu, Y., Liu, G. & Nowack, B. Oligomers are a major fraction of the submicrometre particles released during washing of polyester textiles. Nat. Water 2, 151–160 (2024).
Deng, H. et al. Microplastic pollution in water and sediment in a textile industrial area. Environ. Pollut. 258, 113658 (2020).
Li, T., Menegatti, S. & Crook, N. Breakdown of polyethylene therepthalate microplastics under saltwater conditions using engineered Vibrio natriegens. AIChE J. 69, e18228 (2023).
Zhou, Y. et al. Development of a novel “4E” polyethylene terephthalate bio-recycling process with the potential for industrial application: efficient, economical, energy-saving, and eco-friendly. Bioresour. Technol. 391, 129913 (2024).
Zurier, H. S. & Goddard, J. M. PETase engineering for enhanced degradation of microplastic fibers in simulated wastewater sludge processing conditions. ACS EST Water 3, 2210–2218 (2023).
Chemla, Y., Sweeney, C. J., Wozniak, C. A. & Voigt, C. A. Design and regulation of engineered bacteria for environmental release. Nat. Microbiol. 10, 281–300 (2025).
de Lorenzo, V. Environmental galenics: large-scale fortification of extant microbiomes with engineered bioremediation agents. Philos. Trans. R. Soc. B-Biol. Sci. 377, 20210395 (2022).
Pirillo, V., Pollegioni, L. & Molla, G. Analytical methods for the investigation of enzyme-catalyzed degradation of polyethylene terephthalate. FEBS J. 288, 4730–4745 (2021).
Qiao, Y. et al. Fluorescence-activated droplet sorting of PET degrading microorganisms. J. Hazard. Mater. 424, 127417 (2022).
Shi, L. et al. Complete depolymerization of PET wastes by an evolved PET hydrolase from directed evolution. Angew. Chem. Int. Ed. 62, e202218390 (2023).
Taxeidis, G. et al. New labeled PET analogues enable the functional screening and characterization of PET-degrading enzymes. ACS Sustain. Chem. Eng. 12, 5943–5952 (2024).
Belisário-Ferrari, M. R., Wei, R., Schneider, T., Honak, A. & Zimmermann, W. Fast turbidimetric assay for analyzing the enzymatic hydrolysis of polyethylene terephthalate model substrates. Biotechnol. J. 14, 1800272 (2019).
von Haugwitz, G. et al. Structural insights into (tere)phthalate-ester hydrolysis by a carboxylesterase and its role in promoting PET depolymerization. ACS Catal. 12, 15259–15270 (2022).
Li, A. et al. Discovery and mechanism-guided engineering of BHET hydrolases for improved PET recycling and upcycling. Nat. Commun. 14, 4169 (2023).
Groseclose, T. M. et al. A high-throughput screening platform for engineering poly(ethylene terephthalate) hydrolases. ACS Catal. 14, 14622–14638 (2024).
Wei, R. et al. Turbidimetric analysis of the enzymatic hydrolysis of polyethylene terephthalate nanoparticles. J. Mol. Catal. B-Enzym 103, 72–78 (2014).
Vogel, K. et al. Enzymatic degradation of polyethylene terephthalate nanoplastics analyzed in real time by isothermal titration calorimetry. Sci. Total Environ. 773, 145111 (2021).
Pfaff, L., Breite, D., Badenhorst, C. P. S., Bornscheuer, U. T. & Wei, R. Fluorimetric high-throughput screening method for polyester hydrolase activity using polyethylene terephthalate nanoparticles. In Methods in Enzymology (eds Weber, G. et al.) 648, 253–270 (Academic Press, 2021).
Fu, X. et al. Single distal mutation enhances activity of known PETases via stabilisation of PET-binding. Preprint at https://www.biorxiv.org/content/10.1101/2024.09.11.612432v1 (2024).
Thomsen, T. B., Schubert, S. W., Hunt, C. J., Westh, P. & Meyer, A. S. A new continuous assay for quantitative assessment of enzymatic degradation of poly(ethylene terephthalate) (PET). Enzym. Microb. Technol. 162, 110142 (2023).
Weigert, S., Gagsteiger, A., Menzel, T. & Höcker, B. A versatile assay platform for enzymatic poly(ethylene-terephthalate) degradation. Protein Eng. Des. Sel. 34, gzab022 (2021).
Liu, K. et al. A dual fluorescence assay enables high-throughput screening for poly(ethylene terephthalate) hydrolases. ChemSusChem 16, e202202019 (2023).
Zhong-Johnson, E. Z. L., Voigt, C. A. & Sinskey, A. J. An absorbance method for analysis of enzymatic degradation kinetics of poly(ethylene terephthalate) films. Sci. Rep. 11, 928 (2021).
Wei, R., Oeser, T., Billig, S. & Zimmermann, W. A high-throughput assay for enzymatic polyester hydrolysis activity by fluorimetric detection. Biotechnol. J. 7, 1517–1521 (2012).
Gimeno-Pérez, M. et al. A coupled ketoreductase-diaphorase assay for the detection of polyethylene terephthalate-hydrolyzing activity. ChemSusChem 15, e202102750 (2022).
Li, M. et al. Customization of ethylene glycol (EG)-induced BMOR-based biosensor for the directed evolution of PET degrading enzymes. Adv. Sci. 12, 2413205 (2025).
Bayer, T. et al. Biosensor and chemo-enzymatic one-pot cascade applications to detect and transform PET-derived terephthalic acid in living cells. iScience 25, 104326 (2022).
Dierkes, R. F. et al. An ultra-sensitive Comamonas thiooxidans biosensor for the rapid detection of enzymatic polyethylene terephthalate (PET) degradation. Appl. Environ. Microbiol. 89, e01603–e01622 (2023).
Hernández-Sancho, J. M. et al. A versatile microbial platform as a tunable whole-cell chemical sensor. Nat. Commun. 15, 8316 (2024).
Gantz, M., Mathis, S. V., Nintzel, F. E. H., Lio, P. & Hollfelder, F. On synergy between ultrahigh throughput screening and machine learning in biocatalyst engineering. Faraday Discuss. 252, 89–114 (2024).
Heyde, S. A. H., Arnling Bååth, J., Westh, P., Nørholm, M. H. H. & Jensen, K. Surface display as a functional screening platform for detecting enzymes active on PET. Microb. Cell Fact. 20, 93 (2021).
Cribari, M. A. et al. Ultrahigh-throughput directed evolution of polymer-degrading enzymes using yeast display. J. Am. Chem. Soc. 145, 27380–27389 (2023).
Baath, J. A., Borch, K., Jensen, K., Brask, J. & Westh, P. Comparative biochemistry of four polyester (PET) hydrolases. ChemBioChem 22, 1627–1637 (2021).
Scandola, M., Focarete, M. L. & Frisoni, G. Simple kinetic model for the heterogeneous enzymatic hydrolysis of natural poly(3-hydroxybutyrate). Macromolecules 31, 3846–3851 (1998).
Erickson, E. et al. Comparative performance of PETase as a function of reaction conditions, substrate properties, and product accumulation. ChemSusChem 15, e202101932 (2022).
Kari, J., Andersen, M., Borch, K. & Westh, P. An inverse Michaelis Menten approach for general description of interfacial enzyme kinetics. ACS Catal. 7, 4904–4914 (2017).
Andersen, M., Kari, J., Borch, K. & Westh, P. Michaelis–Menten equation for degradation of insoluble substrate. Math. Biosci. 296, 93–97 (2018).
Arnling Bååth, J., Jensen, K., Borch, K., Westh, P. & Kari, J. Sabatier principle for rationalizing enzymatic hydrolysis of a synthetic polyester. JACS Au 2, 1223–1231 (2022).
Sousa, S. F., Ramos, M. J., Lim, C. & Fernandes, P. A. Relationship between enzyme/substrate properties and enzyme efficiency in hydrolases. ACS Catal. 5, 5877–5887 (2015).
Schubert, S. W. et al. Relationships of crystallinity and reaction rates for enzymatic degradation of poly (ethylene terephthalate), PET. ChemSusChem 17, e202301752 (2024).
Schwaminger, S. P. et al. Immobilization of PETase enzymes on magnetic iron oxide nanoparticles for the decomposition of microplastic PET. Nanoscale Adv. 3, 4395–4399 (2021).
Zurier, H. S. & Goddard, J. M. Directed immobilization of PETase on mesoporous silica enables sustained depolymerase activity in synthetic wastewater conditions. ACS Appl. Bio Mat. 5, 4981–4992 (2022).
Branson, Y. et al. One-pot depolymerization of mixed plastics using a dual enzyme system. ChemSusChem 18, e202402416 (2025).
Yan, Z.-F. et al. Design and construction of chemical-biological module clusters for degradation and assimilation of poly(ethylene terephthalate) waste. J. Environ. Manag. 361, 121258 (2024).
Bayer, T. et al. Structural elucidation of a metagenomic urethanase and its engineering towards enhanced hydrolysis profiles. Angew. Chem. Int. Ed. 63, e202404492 (2024).
Bell, E. L. et al. Natural diversity screening, assay development, and characterization of nylon-6 enzymatic depolymerization. Nat. Commun. 15, 1217 (2024).
Li, Z. et al. Structure-guided engineering of a versatile urethanase improves its polyurethane depolymerization activity. Adv. Sci. 12, 2416019 (2025).
Then, J. et al. Ca2+ and Mg2+ binding site engineering increases the degradation of polyethylene terephthalate films by polyester hydrolases from Thermobifida fusca. Biotechnol. J. 10, 592–598 (2015).
Schmidt, J. et al. Effect of Tris, MOPS, and phosphate buffers on the hydrolysis of polyethylene terephthalate films by polyester hydrolases. FEBS Open Bio 6, 919–927 (2016).
Chen, C.-C. et al. General features to enhance enzymatic activity of poly(ethylene terephthalate) hydrolysis. Nat. Catal. 4, 425–430 (2021).
Suzuki, K., Watanabe, T. & Kitamoto, H. A biodegradable plastic-degrading cutinase-like enzyme from the phyllosphere yeast Pseudozyma antarctica. PDB database entry https://doi.org/10.2210/pdb7CC4/pdb (2020).
Suzuki, K. & Koitabashi, M. Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9. PDB database entry https://doi.org/10.2210/pdb7CY3/pdb (2020).
Hwang, J. et al. Structural and biochemical insights into bis(2-hydroxyethyl) terephthalate degrading carboxylesterase isolated from psychrotrophic bacterium Exiguobacterium antarcticum. Int. J. Mol. Sci. 24, 12022 (2023).
Acknowledgements
The authors R.W., L.M.B., and U.T.B. acknowledge funding from the European Union’s Horizon 2020 research and innovation program under grant agreement no. 870294 for the MIX-UP project and no. 953214 for the upPE-T project. L.M.B. acknowledges the support of the Werner Siemens Foundation in the frame of the WSS Research Centre “catalaix”. P.W. acknowledges support from Innovation Fund Denmark (Grant 5150-00020A) and the Novo Nordisk Foundation (Grants NNF15OC0016606 and NNFSA170028392). G.W. acknowledges funding from the “Helmholtz Sustainability Challenge” project FINEST and the satellite project PUreValue, funded by the Investment and Networking Fund of the Helmholtz Association under grant agreement no. KA2-HSC-10 and KA-HSC-13, respectively.
Author information
Authors and Affiliations
Contributions
R.W., P.W., G.W., L.M.B., and U.T.B. collaboratively developed the concept of this article and wrote the manuscript, with R.W. taking the lead.
Corresponding author
Ethics declarations
Competing interests
U.T.B. is a member of the Scientific Advisory Board of Carbios. The remaining authors (R.W., P.W., G.W., and L.M.B.) declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Sierin Lim and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wei, R., Westh, P., Weber, G. et al. Standardization guidelines and future trends for PET hydrolase research. Nat Commun 16, 4684 (2025). https://doi.org/10.1038/s41467-025-60016-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-60016-9
This article is cited by
-
Recent advances in enzyme engineering for improved deconstruction of poly(ethylene terephthalate) (PET) plastics
Communications Materials (2025)