Abstract
Adenosine 5′-triphosphate (ATP) is the currency of energy in cells; it plays essential roles in virtually all cellular processes, ranging from basic metabolism to signaling in development and disease. The opportunistic bacterial pathogen Legionella pneumophila utilizes the Dot/Icm type IV secretion system to deliver over 300 effectors into host cells, some of which utilize ATP to perform biochemical reactions catalyzed by their unique enzymatic activities. However, whether L. pneumophila directly regulates ATP level in host cells is unknown. Here, we discover that the Dot/Icm substrate Ceg14 (Lpg0437, a.k.a. SidL) is an ATP/dATPase, which after being activated by the host protein actin, efficiently converts ATP and dATP into adenosine and deoxyadenosine monophosphate, respectively by a mechanism that requires its S-HxxxE (x, any amino acid) motif. The activity of Ceg14 is regulated by its metaeffector AnkJ (Lpg0436, a.k.a. LegA11), which inhibits its ATPase activity via direct protein-protein interactions. Ceg14 and AnkJ impose temporal regulation of ATP levels in L. pneumophila-infected cells. Our results demonstrate that L. pneumophila modulates the energy level of host cells to create an environment permissive for its growth.
Similar content being viewed by others
Introduction
The Gram-negative bacterium Legionella pneumophila is the causative agent of Legionnaires’ disease1. This intracellular pathogen primarily is associated with freshwater amoebae in natural environments2. Inhalation of aerosolized bacteria introduces the pathogen into the lung where it is phagocytosed by alveolar macrophages. Instead of being digested, L. pneumophila survives and replicates within these immune cells by creating a phagosome called Legionella-containing vacuole (LCV). Biogenesis of the LCV requires the Dot/Icm type IV system that delivers at least 330 effectors into infected cells3,4. These effectors employ diverse biochemical activities to modulate a cohort of cellular processes including vesicle transport, lipid metabolism, autophagy, protein translation, and cytoskeleton dynamics to support the development of the LCV5,6. The function of the majority of these effectors remains elusive due to their limited similarity with proteins of known activity7,8. Additionally, study of these effectors is complicated by the fact that deletion of one or more effector genes typically does not impact intracellular replication of the bacterium in current cell-based virulence assays5,9,10,11, which poses a challenge to discern their exact role in L. pneumophila infection.
Bacterial effectors remodel host cell environment to evade host immunity often by functioning as enzymes that target specific host proteins important for signaling by post-translational modification (PTM)5,12. These virulence factors have been shown to catalyze various PTMs, and such examples include phosphorylation13, acetylation14, ubiquitination15, glycosylation16, AMPylation17, ADP-ribosylation18,19, fatty acylation20 and protein cleavage21,22. A number of effectors have been shown to eliminate or alter their abundancy of small molecules essential for metabolism or signaling in host cells, including lipids23 and nicotinamide adenine dinucleotide (NAD)24.
All biological systems utilize ATP as the energy currency to power molecular events critical for cellular functions25. ATP also plays an important role in signal transduction in cells in which it is utilized by enzymes such as kinases to modify both protein and non-protein substrates in response to specific stimuli26,27. Moreover, ATP is the substrate for RNA synthesis or, after being converted to dATP, for DNA synthesis. Because of these important roles, ATP and its derivative are common targets in the tug of war between host and pathogen. For example, viruses directly utilize ATP or dATP from the host cell as materials for their own replication28,29. Some bacteria limit phage replication by depleting ATP/dATP through the defensive two-component system (Detocs)30. Limiting the availability of ATP or its analogs is also an effective defense strategy for higher organisms, including humans. For instance, SAMHD1 limits human immunodeficiency virus-1 replication by hydrolyzing dNTP, thus restricting reverse transcription31.
A number of L. pneumophila Dot/Icm effectors have been shown to directly use ATP for their activities in infected cells. At least eight Dot/Icm effectors are kinases that modify proteins or phosphoinositides (PIs)32. Glutamylation of members of the SidE family effectors by SidJ involves AMPylation-mediated activation of the target glutamate residue using ATP33,34,35; this nucleotide is also consumed by SidM-induced AMPylation that functions to lock the small GTPase Rab1 in its GTP-loaded active form36. Recent studies found that LnaB converts phosphoribosyl ubiquitin (PR-Ub) into ADP-ribosylated ubiquitin (ADPR-Ub) via an AMPylation reactions using ATP as the donor for the AMP moiety, thus maintaining ubiquitin homeostasis in L. pneumophila-infected cells37,38.
Ceg14 (Lpg0437, a.k.a. SidL) is a 666-residue protein originally identified as a Dot/Icm substrate of L. pneumophila by virtue of its co-regulation with components of the Dot/Icm machinery39, and was reidentified by a machine learning method40. Ceg14 is toxic to yeast and such toxicity can be effectively suppressed by overexpressing yeast profilin41. Furthermore, recombinant Ceg14 inhibits actin polymerization in biochemcial assays41. These results led to a conclusion that Ceg14 functions to interfere with the structure and/or regulation of the actin cytoskeleton of host cells41. In addition, recombinant Ceg14 had been shown to inhibit protein translation in a cell-free assay using lysates of rabbit reticulocytes42. One weakness of these studies is the lack of a precise biochemical activity for Ceg14, which prevents detailed mechanistic analysis of its impact on these activities. In this study, we show that Ceg14 is an actin-dependent ATPase which efficiently converts ATP and dATP to AMP and dAMP, respectively and that hydrolysis of ATP by Ceg14 downregulates ATP levels in infected cells. We also show that the activity of Ceg14 is inhibited by its metaeffector AnkJ (Lpg0436, a.k.a. LegA11), which blocks its ATPase activity by direct binding.
Results
The S-HxxxE motif is critical for yeast toxicity of Ceg14
To determine the biochemical activity of Ceg14, we performed careful bioinformatics analysis using PSI-BLAST (Position-Specific Iterative Basic Local Alignment Search Tool) searches43 at the NCBI against the non-redundant protein database. These efforts led to the identification of a putative S-HxxxE motif associated with over a hundred toxins from multiple bacterial species38. Ceg14 is one of the two proteins with a predicted S-HxxxE motif in the genome of L. pneumophila strain Philadelphia 144. The three residues that form the S-HxxxE motif in Ceg14 are S527, H571, and E575 (Fig. 1a). The other S-HxxxE protein LnaB has recent been shown to function as an AMPylase that, upon being activated by the host protein actin, converts PR-Ub to ADPR-Ub using ATP37,38.
a Sequence alignment of the S-HxxxE motif in proteins from a diverse array of bacterial species. Proteins were identified by PSI-BLAST using Ceg14 as an entry. Information about the bacterial species, accession number of the proteins was shown in the left of the alignment. Highly conserved amino acids were highlighted in red background and residues with similar property were in red. The bacterial species included are: Lp Legionella pneumophila, Rb Rickettsiaceae bacterium, Lb Lentisphaerota bacterium, Vs Vibrio sinus, Pl Photorhabdus luminescens. b The predicted S-HxxxE motif is crucial for Ceg14-induced yeast toxicity. Yeast cells expressing Ceg14 or mutants defective in the S-HxxxE motif from the galactose-inducible promoter were serially diluted and spotted on the specified medium. Images were captured after 2 d incubation at 30 °C (left panel). The expression of Ceg14 and its mutants in yeast was probed by immunoblotting, the metabolic enzyme 3-phosphoglycerate kinase (PGK) was probed as a loading control (right panel). 1, uninduced samples (glucose); 2, induced samples (galactose). Similar results were obtained in three independent experiments. Source data are provided as a Source Data file.
To investigate whether the S-HxxxE motif in Ceg14 is important for its activity, we first determined how it impacts yeast toxicity of the effector. To this end, we constructed substitution mutants each with one of the three residues being replaced by alanine. All three mutants can be stably expressed in yeast, but none of them retained toxicity (Fig. 1b). These results indicate that the S-HxxxE motif is critical for the activity of Ceg14.
Ceg14 is an actin-dependent ATPase that hydrolyzes ATP into AMP and pyrophosphate
Actin is required for self-AMPylation by several members of the S-HxxxE family37,38. We thus examined whether Ceg14 possesses actin-dependent AMPylation activity. Since Ceg14 has been shown to potentially bind actin41, we first reexamined the interactions between these two proteins. Wild-type Ceg14 cannot be expressed in mammalian cells at levels for experiments such as immunoprecipitation (IP) due to its toxicity (Fig. S1a), we thus used the Ceg14S/A mutant for IP assays. GFP-Ceg14S/A effectively co-precipitated both Flag-actin and endogenous actin. Similar results were obtained in reciprocal experiments (Fig. 2a). Interactions between Ceg14 and actin were further analyzed by pulldown experiments with recombinant proteins, which showed that Ceg14 and Ceg14S/A retained actin indistinguishably (Fig. 2b). To investigate which regions of Ceg14 are important for binding to actin, we created truncations of Ceg14S/A by removing 100, 200, or 300 amino acids from either the N-terminus or the C-terminus and used IP experiments to detect binding between endogenous actin and these Ceg14 truncation mutants. These results showed that the mutant lacking 100 amino acids from the N-terminus (Ceg14ΔN100) retained the ability to bind actin and further deletion abolished such interactions. In contrast, truncation of 100 amino acids from the C-terminus (Ceg14ΔC100) abolished its ability to bind actin (Fig. S1b). Together, these results indicate that a large portion of the C-terminus of Ceg14 is important for actin binding.
a, b Interactions between Ceg14 and actin. Lysates of HEK293T cells transfected to express the testing proteins were used for immunoprecipitation. Copurified proteins were detected by immunoblotting (a). His6-Ceg14 or His6-Ceg14S/A was mixed with actin and the potential protein complex was captured (b). 5% of mixed samples were loaded as input in each experiment. c, d. Actin-dependent ATP hydrolysis by Ceg14. ATP was incubated with the indicated reactants at 37 °C for 1 h. 90% of reaction products were analyzed by HPLC. Compounds were identified by their retention time comparing to that of standard AMP and ADP (c). 10% of the reactions were used for pyrophosphate detection, equal amounts of ATP, ADP, AMP, or PPi standards were used as controls (d). e The activity of Ceg14 does not produce ADP as a reaction intermediate. AMP-PNP, an ATP analog that cannot be cleaved at the nitrogen atom between the β-γ-phosphate was used as a substrate and the production AMP was detected. f, g Actin does not contribute to ATP hydrolysis induced by Ceg14. Reactions containing the indicated reactants were incubated at 37 °C for 2 h. 90% of reaction products were analyzed by HPLC (f). 10% of the reactions were used for pyrophosphate detection (g). h The S-HxxxE motif is required for the activity of Ceg14. The standard ATP, products of the indicated reactions were analyzed by HPLC. All reactions were allowed to proceed at 37 °C for 1 h. i Enzyme kinetics analysis. His6-LnaB, His6-Ceg14 or mutant Ceg14S/A each was incubated with actin and ATP for 15 min. The reduction in ATP was quantitated using HPLC. The values for Km and Kcat were calculated by fitting the Michaelis-Menten model using Prism 10. j Ceg14 is more effective than LnaB in ATP hydrolysis. The standard ATP and the products of the indicated reactions were analyzed by HPLC. All reactions were allowed to proceed for 1 h at 37 °C. Data shown in panels (a–c, e-f and h–j) each was a representative of three independent experiments with similar results, and quantitative results (mean ± s.e.) shown in panels (d and g) were from three independent experiments each done in triplicate. Source data are provided as a Source Data file.
We next attempted to identify cellular substrates of Ceg14 by proximity labeling using the TurboID method45. Flag-BirA*-Ceg14S/A was expressed in HEK293T cells in the presence of exogenous biotin. After enrichment with streptavidin beads, proteins were identified by mass spectrometry analysis. These efforts led to identification of several proteins involved in cytoskeleton structure, including ZYX46, PPP1R12A47, SH3PXD2B48 and SCRIB49. We then purified Flag-tagged versions of these proteins from mammalian cells after transient transfection and examined their modification using 32P-α-ATP. Despite extensive efforts, we were unable to detect Ceg14-induced modification of these proteins (Fig. S1c). Furthermore, self-AMPylation of Ceg14 was not detectable even after extended exposure (Fig. S1c). Similarly, self-AMPylation was not detected in reactions using biotin-labeled ATP (Fig. S1d). Finally, although overexpression of yeast Profillin (PFY) strongly suppressed the yeast toxicity of Ceg1441, we did not detect AMPylation of PFY by Ceg14 (Fig. S1e). In contrast, under the same experimental conditions, AMPylation of PR-Ub by LnaB occurred robustly and self-AMPylation of LnaB was readily detectable (Fig. S1d, e). Furthermore, mass spectrometric analysis did not detect any change in the molecular weight of PFY co-expressed with Ceg14 in yeast.
The AMPylation reaction is initiated by the attack of the α-phosphate in ATP by enzymes, leading to the transfer the AMP moiety from ATP to the substrate and the release of pyrophosphate50. We therefore, considered the hypothesis that Ceg14 functions to hydrolyze ATP. Although at low rates, incubation of Ceg14 with ATP led to production of ADP. In contrast, ATP hydrolysis and the production of AMP and pyrophosphate occurred efficiently when actin was added to the reactions. In reactions with actin, the production of ADP still occurred at rates similar to those of actin-free reactions (Fig. 2c, d). To determine whether ATP hydrolysis occurred via producing ADP as a reaction intermediate, we established reactions using adenylyl imidodiphosphate (AMP-PNP) harboring a cleavage-resistant nitrogen atom between the β and γ phosphate. AMP was robustly produced by Ceg14 and actin when this ATP analog was used as the substrate (Fig. 2e), indicating that the reaction does not involve the production of ADP as a reaction intermediate. Finally, activated Ceg14 is incapable of hydrolyzing ADP into AMP, which suggests that Ceg14 specifically targets ATP and hydrolyzes it into AMP and pyrophosphate (Fig. S2a).
Because actin has intrinsic ATPase activity51, we thus examined its potential contribution to ATP hydrolysis in our reactions. Incubation of ATP with actin alone did not lead to detectable ATP hydrolysis (Fig. 2c). In agreement with this observation, the mutant actinQ137A/D154A/H161A, which has completely lost the ATPase activity51 can still stimulate Ceg14 to hydrolyze ATP at rates indistinguishable to those of wild-type actin (Fig. 2f, g and Fig. S2b).
Next we determined whether the S-HxxxE motif is required for ATP hydrolysis by Ceg14. Recombinant protein of each of the three mutants, Ceg14S/A, Ceg14H/A, or Ceg14E/A was incubated with ATP and actin. Whereas robust AMP production occurred in reactions receiving Ceg14, no activity was detected in reactions containing any of these mutants (Fig. 2h). Notably, whereas the Ceg14S/A mutant retained the ability to produce ADP from ATP at levels comparable to wild-type protein, this activity had been compromised for both Ceg14H/A and Ceg14E/A mutants (Fig. 2h). Kinetic analysis indicated that Ceg14 has a robust ATP hydrolysis activity with a Km value of ~0.28 mM. As a comparison, the Km value for Ceg14S/A is about 57.2 mM, and for LnaB, it is about 16.1 mM (Fig. 2i). We further compared the ATP hydrolysis activity between Ceg14 and LnaB by establishing reactions containing equal amounts of enzymes and actin (500 nM). We observed that nearly 100% of ATP was converted into AMP by Ceg14 in 2 h whereas only 17.3% ATP was consumed by LnaB to produce ADP in the same reaction duration (Fig. 2j). These results establish Ceg14 as a novel actin-dependent ATPase that cleaves ATP into AMP and pyrophosphate.
To explore the mechanism of action of Ceg14, we docked ATP into the S-HxxxE domain of the highest-ranking model of Ceg14 structure predicted by AlphaFold52. These efforts revealed that N412, R443, S527, G528, H529, Y565 and H571 of Ceg14 are potentially involved in binding ATP by forming hydrogen bonds (Fig. S3a). We next validated the importance of these residues by constructing mutants in which each of these sites was substituted by alanine. In addition to S527 and H571 in the S-HxxxE motif, mutation of N412 or R443 to alanine also completely abolished the ability of Ceg14 to hydrolyze ATP. In addition, mutating G528, H529 or Y565 to alanine also significantly impaired the ability of Ceg14 to hydrolyze ATP (Fig. S3b).
We also investigated whether Ceg14 hydrolyzes other nucleotides, and found that dATP can be converted into dAMP by Ceg14. Again, this activity required actin and the S-HxxxE motif (Fig. S4a, b). Ceg14-induced hydrolysis of dGTP into dGDP was also detectable but this activity required the S-HxxxE motif but not actin (Figs. S4a and c). Finally, Ceg14 did not detectably hydrolyze DNA (Fig. S4d). Thus, Ceg14 is actin-activated enzyme capable of hydrolyzing ATP and dATP to AMP and dAMP, respectively.
Ceg14 inhibits cell growth by depleting cellular ATP
The finding that Ceg14 has ATP/dATPase activity prompted us to examine its role in the inhibition of mammalian cell growth. 24 h after transfection, the density of HEK293T cells transfected with the construct expressing wild-type Ceg14 was significantly lower than those transfected with the vector or plasmids expressing mutants defective in the S-HxxxE motif (Fig. 3a, b). Thus, Ceg14 inhibits the growth of mammalian cells in a manner that requires the S-HxxxE motif-dependent ATP hydrolysis activity. To further probe the mechanism of such inhibition, we determined membrane integrity of cells expressing the different alleles of Ceg14 by measuring the release of lactate dehydrogenase (LDH). Although Ceg14 severely inhibited cell proliferation, it did not damage cell membranes as LDH release did not occur in transfected cells (Fig. 3c). This observation suggests that the low cell numbers in samples expressing Ceg14 was caused by lower cell proliferation rates, primarily caused by ATP depletion.
a Expression of Ceg14 inhibits the growth of HEK293T cells. HEK293T cells of ~50% confluence were transfected with the empty vector (EV) or plasmid that expressed Flag-tagged Ceg14, Ceg14S/A, Ceg14H/A, or Ceg14E/A for 24 h prior to image acquisition (left panels). Note the low cell confluency of the sample expressing Ceg14. The expression of Ceg14 and its mutants in an equal number of transfected HEK293T cells was detected by immunoblotting using the Flag antibody. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was probed as a loading control (right panels). Scale bar, 20 μm. b, c HEK293T cells transfected to express the indicated proteins for 24 h were assessed for cell viability using the CCK-8 method (b) or for LDH release (c). Results (mean ± s.e.) shown were from three independent experiments each done in triplicate. d Ceg14 depleted cellular ATP. Total ATP was measured in HEK293T cells transfected to express Ceg14 or LnaB. Cells transfected with the empty vector were used as controls. All samples contained an equal number of cells. Results (mean ± s.e.) shown were from three independent experiments (upper). The expression of Flag-Ceg14 and its mutants was detected with Flag antibody and GAPDH was probed as a loading control (lower panels). e HEK293T cells were transfected with either an empty plasmid or a Flag-Ceg14 expression plasmid for 4 h, followed by treatment with different doses of cytochalasin D for an additional 20 h. Equal amounts of cells were then collected to measure ATP levels. Data shown in (a, d) (lower panels) are representative from three independent experiments. Data shown in b-e were mean ± s.e. from three replicates. Unpaired two-tailed Student’s t-tests were performed. Source data are provided as a Source Data file.
We next quantitated the amounts of ATP in cells transfected to express Ceg14 or its mutants. Whereas expression of the enzymatically inactive mutants did not significantly affect the abundancy of cellular ATP comparing to cells transfected with the empty vector, expression of wild-type Ceg14 led to ~90% reduction. As an additional control, similarly expressed LnaB also led to detectable decrease of cellular ATP, but the reduction rate was <25% (Fig. 3d), such effect clearly attributes to its use of ATP to add AMP to phosphorylated proteins37 and its low ATP hydrolysis activity (Fig. 2i, j). When cells were treated with cytochalasin D, Ceg14 further lowered the ATP levels (Fig. 3e), suggesting that Ceg14-induced reduction in intracellular ATP levels occurs independent of actin-mediated nutrient uptake.
Ceg14 has been shown to interfere with actin polymerization in biochemical assays41. Because ATP provides the energy for actin polymerization, the effect of Ceg14 in these assays may be indirect via depletion of ATP in the reactions. Indeed, inclusion of His6-Ceg14S/A defective in hydrolyzing ATP but retaining the actin-binding ability did not inhibit actin polymerization (Fig. S5a–c).
Overexpression of yeast PFY effectively suppressed Ceg14-mediated yeast toxicity41. In light of the observation that Ceg14 did not detectably modify PFY (Fig. S1e) and the fact that PFY is an actin-binding protein53, we considered the hypothesis that the toxicity suppression phenotype of PFY resulted from its competition for yeast actin (ACT1) with Ceg14. We first analyzed the binding among these three proteins by expressing GFP-PFY at different levels in HEK293T cells that expressed Flag-ACT1 and GFP-Ceg14. IP experiments with Flag beads showed that even in samples in which the expression level of GFP-PFY was lower than that of GFP-Ceg14, the amount of GFP-PFY co-purified with Flag-ACT1 was considerably higher than that of GFP-Ceg14 (Fig. S5d, lanes 3-5). Correspondingly, as the expression level of PFY increased, the amount of Ceg14 co-immunoprecipitated by Flag-ACT1 decreased (Fig. S5d). These results suggest that PFY can outcompete Ceg14 for binding ACT1, thus inhibiting its ATPase activity.
To further determine the effect of PFY on the actin-activated ATPase activity of Ceg14, we established a series of reactions containing equal amounts of ATP, Ceg14, ACT1, and increasing concentrations of PFY. After 3 h of incubation, ATP was almost completely depleted in reactions without PFY. In reactions receiving PFY, the ATPase activity of Ceg14 was inhibited in a dose-dependent manner. When the molar ratio between PFY and ACT1 was 7:1, ~37% of ATP remained unhydrolyzed in the reaction system and intact ATP was 70% in reactions in which such ratio was 63:1 (Fig. S5e). Importantly, the PFYK67A mutant, which has lost the ability to bind ACT154, failed to inhibit Ceg14-mediated ATP hydrolysis (Fig. S5e). These results indicate that PFY can block the activity of Ceg14 by titrating ACT1.
The metaeffector AnkJ inhibits the activity of Ceg14 by direct protein-protein interaction
A systematic analysis using yeast genetics suggests that AnkJ is a metaeffector of Ceg14 which functions to modulate its activity55. On the chromosome of L. pneumophila strain Philadelphia 1, AnkJ (Lpg0346) is a protein of 269 amino acids coded for by a gene adjacent to ceg1444.
Whereas several metaeffectors have been shown to function via posttranslational modification on the cellular target of their cognate effectors or the effectors themselves, a few have been shown to inhibit the activity of their cognate effectors by direct binding34,56,57. Similar to results from the earlier study55, coexpression of AnkJ with Ceg14 effectively suppressed its toxicity to yeast (Fig. 4a).
a AnkJ suppresses the yeast toxicity of Ceg14. Serially diluted cells of the indicated yeast strains were spotted on the indicated media. Images were captured after incubating at 30 °C for 2 d (left panels). Relevant proteins was detected using specific antibodies against Ceg14 or AnkJ. PGK was probed as a loading control (right panels). 1, uninduced; 2, galactose-induced. Ceg14 was driven by the galactose-inducible promoter. b Interactions between Ceg14 and AnkJ. Lysates from HEK293T cells transfected to express the indicated proteins were divided into three identical samples, which were subjected to immunoprecipitation with the indicated antibodies. Coprecipitated proteins were detected by immunoblotting. c Interactions between Ceg14 and AnkJ determined by GST pulldown. The indicated proteins were mixed at 25 °C for 30 min and GST beads were used to capture potential protein complexes. Interactions were assessed by Coomassie brilliant blue (CBB) staining. d AnkJ and Ceg14 interacts in L. pneumophila. Flag-Ceg14 was expressed in wild-type or the ∆ankJ mutant. Binding between Flag-Ceg14 and AnkJ was determined by immunoprecipitation and copurified proteins were detected by immunoblotting. Data shown in (a–d) each was a representative of three independent experiments with similar results. e AnkJ inhibits Ceg14-catalyzed ATP hydrolysis. Serially diluted AnkJ was preincubated with Ceg14 and actin and the mixtures were then added to reactions containing ATP, the reaction was allowed to proceed for 2 h at 37 °C prior to HPLC analysis. The quantity of AMP was determined using peak areas from standard AMP samples. The quantity of AMP was normalized to samples from reactions without AnkJ. f AnkJ stopped ongoing ATP hydrolysis by Ceg14. Ceg14 and actin were added to 10 identical ATP samples. At 10, 20, 30 min, a pair of samples were processed,one by rapidly freezing and the second by adding AnkJ and were allowed to proceed for an additional 1.5 h at 37 °C before HPLC analysis. AMP levels in each group were normalized to the 0 min control samples incubated at 37 °C for 2 h without AnkJ. For (e, f) data shown were mean ± s.e. from three replicates. Unpaired two-tailed Student’s t-tests were performed. Source data are provided as a Source Data file.
We next probed the mechanism of the suppression by examining interactions between Ceg14 and AnkJ55. IP with antibodies specific for GFP in lysates of HEK293T cells transfected to express GFP-Ceg14 and HA-AnkJ led to co-purification of the latter. Similar results were obtained in reciprocal IP experiments using HA beads (Fig. 4b). Moreover, pulldown experiments using GST beads allowed co-purification of His6-Ceg14 mixed with GST-AnkJ in protein solutions (Fig. 4c). Finally, IP with Flag beads in wild-type L. pneumophila expressing Flag-Ceg14 captured AnkJ, indicating that these two proteins form a complex in bacterial cells (Fig. 4d).
To determine the effect of AnkJ binding on the activity of Ceg14, we established a series of reactions that contained these two proteins at different ratios and the production of AMP was measured after adding ATP. Our results revealed a proportional inhibition of the ATPase activity of Ceg14 by AnkJ. A 1:5 molar ratio between AnkJ and Ceg14 led to 80% reduction in ATP hydrolysis and a complete inhibition was observed when the ratio was 1:1 (Fig. 4e). We also determined the inhibitory effect under conditions in which ATP hydrolysis had started. AnkJ was added to reactions (AnkJ:Ceg14 = 5:1) in which ATP hydrolysis had been initiated for 10, 20, and 30 min, respectively. At the time AnkJ was added, an identical reaction was terminated by flash freezing in liquid nitrogen to determine the level of hydrolysis. In each case, after adding AnkJ, the reactions were allowed to proceed for an additional 1.5 h to ensure maximal hydrolysis. Results from these experiments revealed that AnkJ effectively prevented further ATP hydrolysis once it was added to the reactions (Fig. 4f).
To determine the region of Ceg14 important for interacting with AnkJ, we constructed a series of deletion mutants lacking 100, 200, 300, 400, 500 or 600 residues from the carboxyl terminus and tested their binding to AnkJ. Removal of as few as 100 residues from the carboxyl end (Ceg14∆C100) abolished the interactions with AnkJ. When similarly constructed amino terminal deletion mutants were analyzed, we found that a mutant lacking 100 residues from this end (Ceg14∆N100) still robustly interacted with AnkJ (Fig. S6a). Thus, AnkJ binds a region of Ceg14 localized to its carboxyl terminal portion. Because the S-HxxxE motif is localized to the carboxyl end region of Ceg14 (S527, H571, and E575), these results suggest that AnkJ recognizes the Ceg14 domain harboring its catalytic center, which explains its strong inhibitory effects.
The region of Ceg14 that binds AnkJ appears to overlap with the region that binds actin (Fig. S1b and S6a). Thus, another possible mechanism for the inhibition of Ceg14 by AnkJ is that the binding of AnkJ to Ceg14 disrupts its interaction with actin. To test this hypothesis, we first purified Flag-actin and the Flag-actin-Ceg14 complex from HEK293T cells transfected to express these proteins using anti-Flag beads. The beads with bound proteins were then incubated for 1 h with AnkJ purified from E. coli. Our results showed that AnkJ can be captured by the actin-Ceg14 complex bound to beads, but not by beads coated by actin alone, and that the addition of AnkJ did not reduce the proportion of Ceg14 associated with the beads (Fig. S6b). These results indicate that the inhibitory effect of AnkJ on Ceg14 is not due to competition for the actin binding site, suggesting that these three proteins form a three-component complex. Interestingly, instead of inhibiting the interaction between Ceg14 and actin, inclusion of AnkJ in IP and biochemical pull-down assays led to a detectable increase in binding between actin and Ceg14, suggesting that this metaeffector facilitates interactions between these two proteins (Fig. 4b and Fig. S6c).
We next used AlphaFold v2.3.152 to predict the structure of the Ceg14-AnkJ complex and the model with the highest confidence out of 5 candidates was used for subsequent analysis of the interactions between these two proteins. Two regions of AnkJ directly contact Ceg14 through extensive polar and hydrophobic interactions. In interface 1, residue R17 of AnkJ forms hydrogen bonds with D547 and D549 of Ceg14. Additional hydrogen bonds in this interface include I2(A, AnkJ):K164(C, Ceg14), R6(A):E543(C), D51(A):K381(C), H85(A):R396(C), K86(A):D393(C). In interface 2, K61 of AnkJ contacts residues E356 and E366 of Ceg14 with hydrogen bonds, N96 and N123 of AnkJ engage in hydrogen-bonding interactions with K357 of Ceg14. Other hydrogen bonds in this interface include interactions between K128(A):D362(C), K168(A):E124(C), Q170(A):Q122(C), R174(A):E360(C), K180(A):R121(C) (Fig. S6d). Yet, single mutations of amino acids of AnkJ in the predicted interaction interface in the complex did not detectably disrupt the binding between Ceg14 and AnkJ (Fig. S6d, e). By truncating 43 amino acids from both ends of AnkJ, we found that the amino-terminal deletion mutant (AnkJ∆N43) weakened its interaction with Ceg14 while retaining partial inhibitory effect against its ATP hydrolysis activity (Fig. S6f, g). The lack of severe defects in binding in these mutants suggests that multiple residues are involved in the interactions. Alternatively, the interface predicted by the AlphaFold algorithm was not sufficiently accurate to for identification of residues important for the interactions. In contrast, the carboxyl-terminal deletion mutant (AnkJ∆C43) has completely abolished the binding of AnkJ to Ceg14, thus has entirely lost its inhibitory effect on the ATPase (Fig. S6f, g). These observations suggest that the carboxyl terminal portion of AnkJ is crucial for its binding to Ceg14. Thus, AnkJ is a metaeffector of Ceg14 that functions to inhibit its ATPase activity by binding to a region that harbors the S-HxxxE motif.
In a few cases, regulation by metaeffector is known to be important for intracellular replication of L. pneumophila. For example, deletion of mesI, the metaeffector of sidI led to a defect in intracellular growth58. Similarly, sidJ, the metaeffector of the SidE effector family is required for optimal intracellular bacterial growth59,60. We thus generated a ∆ankJ mutant and assessed its intracellular replication in both mouse bone marrow-derived macrophages (BMDMs) and Dictyostelium discoideum. The growth of the ∆ankJ mutant was similar to that of the wild-type stain in both hosts (Fig. S7a, b). Similarly, deletion of ceg14 did not detectably affect intracellular replication of L. pneumophila in these host cells (Fig. S7c, d). Since MesI can inhibit the translocation of SidI into host cells61. We further investigated the effect of AnkJ on Dot/Icm-mediated translocation of Ceg14 and found that overexpression of AnkJ significantly inhibited translocation of the TEM1-Ceg14 fusion by the Dot/Icm system (Fig. S8a, b). These results suggest that in addition to directly inhibiting its activity, AnkJ also blocks the translocation of Ceg14 into host cells by the bacterium.
Ceg14 and AnkJ regulate the ATP levels in L. pneumophila-infected cells
We next examined the effects of AnkJ on the cell toxicity of Ceg14. Co-expression of AnkJ abolished the growth inhibition of HEK293T cells caused by Ceg14 (Fig. 5a, left). Notably, the presence of AnkJ allowed Ceg14 to be expressed at levels comparable to that of the enzymatically inactive Ceg14H/A mutant (Fig. 5a, right). This suggests that the extremely low expression level of wild-type Ceg14 is likely a result of its depletion of host ATP/dATP, leading to a global inhibition of gene expression and cell growth arrest. As expected, co-expression of AnkJ restored the viability of cells expressing Ceg14 (Fig. 5b). Consistent with the normal cell growth phenotype, co-expression of AnkJ restored ATP in cells expressing Ceg14 to a level that is comparable to that of cells expressing the Ceg14H/A mutant (Fig. 5c).
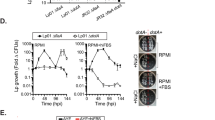
a AnkJ rescues Ceg14-induced cell growth arrest. HEK293T cells of 50% confluence were transfected to express the indicated proteins. Images were acquired 24 h post-transfection (left panels). The expression of the relevant proteins was probed with antibody specific for Flag or HA. GAPDH was probed as a loading control (right panels). V, empty vector. Data shown were a representative of three independent experiments with similar results. Scale bar, 20 μm. b AnkJ counteracted cell viability reduction induced by Ceg14. Samples expressed combinations of proteins were used to determined cell viability. c AnkJ rescued ATP depletion induced by Ceg14. HEK293T cells were transfected to express the indicated proteins for 24 h, and lysates of equivalent number of cells were used to measure ATP. The level of ATP in cells transfected with the empty vector was used as the baseline for comparison. Data shown (mean ± s.e.) in (b, c) were from three replicates. Unpaired two-tailed Student’s t-tests were performed. d, e Modulation of cellular ATP levels in Raw264.7 (d) and D. discoideum (e), by Ceg14 and AnkJ during L. pneumophila infection. ATP levels were determined in cells infected with the indicated bacterial strains at the indicated time points. The ATP level of uninfected cells at the 5 min time point was used as a control. Results (mean ± s.e.) shown were from three independent experiments. Ordinary one-way ANOVA with multiple comparisons was performed for data analysis. Source data are provided as a Source Data file.
We next investigated whether the Ceg14/AnkJ pair modulates ATP levels during L. pneumophila infection by measuring ATP levels in infected cells. The ATP levels in Raw264.7 cells began to decrease 4 h post-infection in a manner that requires the Dot/Icm system. At 8 and 12 h post-infection, the ATP levels in cells infected with the ∆ceg14 mutant were significantly higher compared to those in cells infected with wild-type bacteria (Fig. 5d). Importantly, complementation of the ∆ceg14 strain with ceg14 but not with the ceg14H/A mutant restored the mutant’s ability to reduce cellular ATP levels during infection (Fig. 5d). This difference was not due to a discrepancy in translocation efficiency, as Ceg14 and the Ceg14S/A or Ceg14H/A mutants exhibited comparable translocation efficiencies by the Dot/Icm system (Fig. S9a, b). Although there was no significant difference in statistical analysis, we also found that at 8 h post-infection, the ATP level in cells infected with the ∆ankJ mutant was lower than that of cells infected with wild-type L. pneumophila. Complementation of strain ∆ankJ with a plasmid expressing AnkJ abolished this difference (Fig. 5d). In infections using Raw264.7 cells, the transcription of ceg14 displayed a gradual decrease from 4 to 12 h which was accompanied by a slight increase for ankJ (Fig. S10a, b), suggesting a temporal regulation of ceg14 and ankJ expression in the intracellular life cycle of L. pneumophila.
Similar results were obtained in infection experiments using D. discoideum. At 8 h post-infection, cells infected with the ∆ankJ mutant exhibited a significant decrease in intracellular ATP comparing to that of cells infected with the wild-type strain (Fig. 5e). Again, complementation of the ∆ceg14 strain with ceg14, but not with the ceg14S/A mutant, restored the ability of the mutant to cause a reduction in cellular ATP levels (Fig. 5e). Thus, under our experimental conditions, the regulation of cellular ATP level by Ceg14 started to become apparent 8 h after infection, which coincided with the time phagocytosed L. pneumophila has begun to replicate62. Furthermore, deletion of ankJ led to further reduction in ATP levels in infected cells. These results indicate that Ceg14 functions to down regulate the ATP level of infected cells and that AnkJ serves as a brake for such inhibition.
Discussion
Here we demonstrate that Ceg14 and its metaeffector AnkJ work together to modulate cellular ATP levels, further extending the spectrum of host targets attacked by L. pneumophila. Several lines of evidence support the conclusion that Ceg14 directly hydrolyzes ATP and dATP. First, after being activated by actin, recombinant Ceg14 robustly hydrolyzed ATP with a Km value of 0.28 mM. Second, ectopic expression of Ceg14 drastically reduced cellular ATP concentrations, which is in contrast to other ATP-consuming effectors such as LnaB (Figs. 2–3). Third, consistent with its inhibition of Ceg14 activity in biochemical assays, AnkJ modulates ATP concentrations in cells infected with L. pneumophila (Fig. 5). Fourth, despite extensive efforts, we were unable to detect self-AMPylation or modification of host proteins by Ceg14 (Fig. S1). Finally, although enzymatically inactive mutants of Ceg14 interacts with actin indistinguishably to wild-type protein, they have lost the ability to inhibit actin polymerization (Fig. S5), supporting the notion that the inhibitory effects seen in earlier experiments were caused by ATP depletion. Several Dot/Icm effectors have been shown to impact energy metabolism in mitochondria. For example, Ceg3 and Larg1 regulate ADP/ATP translocation by catalyzing reversible ADP-ribosylation on translocases (ANTs)18,63. LpSpl (a.k.a LegS2) affects mitochondrial membrane potential (Δψm) by disrupting sphingolipid biosynthesis64. Because of the critical role of mitochondrion and ATP in immunity65,66, lowering the cellular ATP levels by the activities of these effectors will clearly benefit the bacterium. In addition, sensing a decrease in cellular ATP levels by AMPK activates various signaling pathways, leading to impaired cell signaling, hindered cell motility and transport, as well as cell apoptosis67, these changes may also benefit the bacterium.
Our earlier works suggest that Ceg14 inhibits actin polymerization and protein synthesis41,42. The discovery of its ATPase activity suggests that its effects on these biochemical or cell-free assays are indirect because ATP is essential for the reactions occurred in both assays68. Consistent with this notion, Ceg14 mutants defective in the ATPase activity have lost the ability to inhibit actin polymerization (Fig. S5a–c). In addition, overexpression of PFY had been shown to effectively suppress the yeast toxicity of Ceg1441. Overexpression of actin is toxic to yeast69, which prevented us from testing the restoration of Ceg14 toxicity through simultaneous actin overexpression. Nevertheless, our results support a model in which the suppression effects likely arise from the titration of cellular actin by elevated PFY, leading to reduced Ceg14 activity and enhanced survival of yeast cells. This conclusion is supported by at least three lines of evidence. First, the yeast actin preferentially bond PFY (Fig. S5d). Second, PFY inhibited the actin-activated ATPase activity of Ceg14 in a dose-dependent manner. Third, the mutant PFYK67A defective in binding actin had lost the ability to inhibit Ceg14-induced ATP hydrolysis (Fig. S5e). Notably, a relatively large amount of PFY was required to effectively inhibit the ATPase activity of Ceg14 (Fig. S5), which likely is due to the fact that only a fraction of recombinant PFY is active. Although the activity of Ceg14 accounted for the majority of the such reduction, it is conceivable that binding to actin by Ceg14 and other actin-interacting proteins from L. pneumophila may interfere with nutrient uptake by affecting processes such as micropinocytosis, leading to lower cellular ATP level (Fig. 3e). Taken together, these results indicate that Ceg14 functions to reduce cellular ATP levels during L. pneumophila infection.
LnaB is another Dot/Icm effectors that utilizes an S-HxxxE motif for catalysis. This enzyme functions to convert PR-Ub produced by reversing ubiquitination induced by members of the SidE family effectors70,71 into ADPR-Ub37,38 and to impair phosphosignaling in L. pneumophila-infected cells by transferring the AMP moiety to phosphorylated proteins37. Akin to Ceg14, LnaB requires actin as a co-factor38. Albeit at lower rates, LnaB is also capable of hydrolyzing ATP (Fig. 3), this activity provides an explanation for the need of actin as the co-factor for its activity as a constitutive ATPase activity for effectors intended for host cells would be disastrous for the pathogen. Intriguingly, unlike Ceg14, cleavage of ATP by LnaB and actin produces ADP, which is in line with the fact that this enzyme utilizes this nucleotide as the donor of AMP to target PR-Ub and phosphorylated proteins37,38. In agreement with this notion, ADP allowed LnaB and actin to convert PR-Ub into ADPR-Ub37,38. The S-HxxxE motif is associated with a large number of bacterial toxins37,38. Intriguingly, self-AMPylation of several of these proteins appeared to be actin-dependent38. Whether other members of this toxin family require actin or other host proteins as a co-factor awaits further study.
Similar to most of Dot/Icm effector genes5,11, deletion of ceg14 or ankJ did not cause detectable defects in intracellular replication of L. pneumophila in commonly used hosts, including D. discoideum (Fig. S7). Furthermore, deletion of ceg14 did not completely restore ATP levels in L. pneumophila-infected cells to those seen in cells infected by mutants defective in the Dot/Icm system (Fig. 5), suggesting that additional effectors contribute to regulate cellular ATP during infection. The cohort of effectors with kinase activity32 certainly will consume ATP, so will the AMPylase SidM36 and the pseudo-kinase SidJ35,60.
The activity of a number of L. pneumophila Dot/Icm effectors is regulated by their respective metaeffector56. The relationship between AnkJ and Ceg14 is akin to that between MseI and SidI, in which the metaeffector MseI inhibits the activity of SidI by direct binding in both host and bacterial cells, and the binding in bacterial cells inhibits SidI translocation by the Dot/Icm system61,72. Deletion of mseI caused a sidI-dependent defect in intracellular bacterial growth, probably caused by unregulated activity of SidI61,72. An earlier study using strain AA100 showed that AnkJ is required for optimal intracellular growth73. In contrast, we did not detect such defects for the ∆ankJ mutant in BMDMs or D. discoideum (Fig. S7). Such discrepancies may result from the use of different bacterial strains, the host cells or a combination of both.
Intriguingly, binding of AnkJ to Ceg14 increased its ability to bind actin (Fig. 4b and Fig. S6c). We speculate that binding of actin and Ceg14 causes a conformational change in the latter and makes it more accessible to actin. Given the effective inhibition of Ceg14 activity by AnkJ (Fig. 4e, f), such increase in actin binding clearly does not affect the inhibitory effect of AnkJ. Future structural analysis of the binary complex of Ceg14-actin and of Ceg14-AnkJ, and potentially the Ceg14-actin-AnkJ tripartite complex will allow better elucidation of the mechanism of AnkJ-induced binding between Ceg14 and actin. Finally, the sequestration of actin by Ceg14 may also attribute to the modulation of host cytoskeleton by L. pneumophila, which is an important aspect its virulence5.
Although the exact biological significance remains largely unknown, regulation of energy metabolism in host cells has also been observed in other pathogens. Among these, Salmonella enterica infection results in a decrease in the concentrations both ATP and NAD, which is accompanied by lysosomal degradation of the Sirt1/LKB1/AMPK complex in macrophages74. These events occur in a manner that requires a functional pathogenicity island 2, suggesting the involvement of effectors translocated by this secretion system74. BtpA and BtpB act as NADase to deplete NAD in cells infected by Brucella species, which significantly diminishes the availability of intracellular ATP24.
Hydrolysis of ATP into AMP and pyrophosphate by enzymes is not unprecedented, it has been reported that nucleotide pyrophosphatases such as Enpp1 negatively regulate bone mineralization by hydrolyzing extracellular nucleotide triphosphates (NTPs) into NMP and PPi75. Given the central role of ATP in cellular processes, Ceg14 likely impacts multiple aspects of signaling. It will be of great interest to determine whether other effectors specifically function to regulate ATP levels in L. pneumophila-infected cells. Mutants lacking ceg14 and additional effector genes dedicated to ATP depletion may display defects in intracellular growth, such phenotypes will provide opportunities to answer the question of how modulation of ATP levels in host cells contributes to successful bacterial infection.
Methods
Mice
SPF-grade A/J mice (Cavens Laboratory Animal Co., Ltd, China) were housed in isolator cages under specific pathogen-free conditions in accordance with the Guide for the Care and Use of Laboratory Animals. Environmental parameters were strictly maintained at 22 ± 2 °C ambient temperature with 55 ± 5% relative humidity, accompanied by automated 12 h photoperiod regulation (07:00-19:00 light phase). Sterilized pelleted feed (autoclaved at 121 °C for 20 min) and reverse osmosis-purified water were provided ad libitum through IVC systems. All experimental protocols underwent rigorous review and received approval (Approval No. 20200669) by the Institutional Animal Care and Use Committee of the First Hospital of Jilin University.
Plasmid constructions, bacterial strains and media
Plasmids, bacterial strains and the sources of key reagents used in this study were listed in Supplementary Data 1 and Supplementary Data 2, respectively. E. coli strains DH5α, DH5αλπ were used for molecular cloning, strain BL21(DE3) was used for recombinant protein expression. E. coli was grown in LB medium at 37 °C. The antibiotic concentrations used for E. coli are as follows: ampicillin at 100 μg/mL and kanamycin at 30 μg/mL. The sequences of primers used for molecular cloning are listed in Supplementary Data 3.
All L. pneumophila strains used in this study were derived from the Philadelphia 1 strain Lp0276, and the dotA- mutant Lp0376 was used as a control. L. pneumophila was cultured in ACES-buffered yeast extract (AYE) liquid broth or on charcoal yeast extract (CYE) agar plates following a previously described procedure76. Deletion mutants were constructed followed an established method77. When needed, antibiotics in AYE or CYE were used at the following concentrations: kanamycin, 20 μg/mL; streptomycin, 100 μg/ml.
Bacterial infection and intracellular bacterial growth
To assess bacterial intracellular growth, BMDMs were differentiated from bone marrows of 6–8 weeks old A/J mice using an established procedure77. D. discoideum was grown as described earlier78. Cells seeded in 24-well plates were challenged with relevant L. pneumophila strains grown to the post-exponential phase at an MOI of 0.05. Bacterial intracellular growth was quantified at specific time points using colony counting through plate plating as described earlier77.
Yeast toxicity assays
The S. cerevisiae strain W30379 were used for toxicity assays. Yeast strains were cultured in YPD media or appropriate SD dropout media for selection of transformed plasmids. Yeast transformation was performed according to a standard procedure were incubated at 30 °C. Yeast transformation was performed according to a previously described procedure80.
To examine the yeast toxicity of Ceg14, wild type ceg14 and its mutants were inserted into pSB157m which contains galactose-inducible promoter for inducible expression in yeast81. Yeast cells grown in suitable liquid dropout media with glucose were washed and serially diluted in sterile water. Subsequently, 5 μL of each dilution was spotted onto selective plates containing either 2% glucose or 2% galactose. Images of the plates were captured following a 2 day incubation at 30 °C.
To determine the ability of AnkJ to suppress the yeast toxicity of Ceg14, AnkJ was cloned into p425GPD82, and the resulting construct was transformed into strain W303(pSB157::Ceg14). Yeast cells were spotted onto selection medium containing glucose or galactose following a procedure similar to the one described above.
Protein expression and purification
To produce His6- or GST-tagged recombinant protein, saturated E. coli cultures were diluted at a ratio of 1:50 into the specified volume of LB broth and grown at 37 °C until OD600 reached 0.6–0.8. IPTG was added at a final concentration of 0.25 mM to induce protein expression for 16–20 h at 18 °C on a shaker (180 rpm).
For His6-tagged proteins, cells harvested by centrifugation were suspended in a lysis buffer (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 10 mM imidazole), lysed using a low temperature ultra-high pressure continuous flow cell crusher. Cleared lysates obtained by centrifugation twice at a speed of 12,000 g for 10 min were mixed with Ni2+-NTA beads (QIAGEN) at 4 °C for 1 h on a rotary shaker. The resin was loaded into a column and unbound proteins were removed by washing with 5 bed volume of a washing buffer (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 20 mM imidazole). Target protein was eluted with 5 mL of elution buffer (20 mM Tris-HCl, pH 7.5; 150 mM NaCl; 300 mM imidazole). Purified protein was dialyzed in a dialysis buffer (20 mM Tris-HCl, pH 7.5; 150 mM NaCl; 10% (v/v) glycerol) at 4 °C for 14 h.
For GST-tagged proteins, cleared lysates prepared in a lysis buffer (50 mM Tris-HCl, pH 7.5; 150 mM NaCl) by a similar procedure were mixed with glutathione agarose beads (Pierce). Beads were washed with a buffer (50 mM Tris-HCl, pH 7.5; 150 mM NaCl) prior to elution with a buffer (20 mM Tris-HCl, pH 7.5; 150 mM NaCl; 10 mM reduced glutathione).
His6-Profilin-b was purified with a procedure similar to that used for protein from E. coli. His6-Profilin-y was purified from a yeast strain derived from W303. Briefly, the sequence coding for 6 His residues was added to the 5’ end of the coding region of yeast Profilin, and the fusion gene was into p425GPD82 and the resulting plasmid was transformed into W303. 20 mL exponentially grown cells were transferred to 500-mL SD-Leu medium containing 2% glucose, the new culture was grown at 30 °C for 18 h. His6-Profilin was obtained through denaturation and renaturation purification. The collected cells were resuspended and lysed in lysis buffer (50 mM Tris-HCl; 150 mM NaCl; 10 mM imidazole; 8 M urea), The lysate was centrifuged at 15,000 g for 30 min at 4 °C. After centrifugation, incubate the supernatant with Ni2+-NTA beads at 4 °C for 1 h. Wash the beads with wash buffer (50 mM Tris-HCl; 150 mM NaCl; 20 mM imidazole; 8.0 M urea), beads containing recombinant proteins were followed by elution with elution buffer (50 mM Tris-HCl; 150 mM NaCl; 300 mM imidazole; 8 M urea). The protein was dialyzed sequentially in a buffer (20 mM Tris-HCl, pH 7.5; 150 mM NaCl; 10% (v/v) glycerol) containing urea at 6.0 M, 4.0 M, 2.0 M, 1.0 M, 0.5 M, respectively, each for 2 h. The final dialysis was performed overnight in the same buffer without urea.
His6-PR-Ub was purified from E. coli strain BL21(DE3) expressing both GST-SdeA and His6-Ub38. Briefly, His6-PR-Ub was first purified with Ni2+-NTA beads from BL21(DE3) harboring both pGEX6P-1-SdeA and pET28A-Ub. The protein was further purified through size-exclusion chromatography using a Superdex 200 Increase column (GE Healthcare). The column was equilibrated with a buffer containing 20 mM Tris-HCl, pH 7.5, 150 mM NaCl.
Purification of potential protein substrates (ZYX, PPP1R12A, SH3PXD2B, SCRIB) of Ceg14. 12-μg plasmids expressing the Flag-tagged target protein were transfected into HEK293T cells in 10-cm cell culture dishes. 24 h after transfection, the cells were lysed on ice for 10 min with a cell lysis buffer (PBS, 1% Triton). Subsequently, the cells were centrifuged at 13,000 g for 10 min. Anti-Flag beads were added to the supernatant and co-incubated at 4 °C for 8 h. Then, the beads were washed 3 times with lysis buffer. 100 μl of 3xFlag peptide elution buffer (Beyotime, cat# P9801) was used to incubate with the beads by rotation at 4 °C for 4 h to collect the eluted target proteins.
Biochemical AMPylation assays
One microgram of His6-Ceg14 or its mutants, 1-μg actin (Cytoskeleton, cat# APHL99) and 5 μg His6-Profilin were incubated in a 20 μL reaction system containing 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 1 mM DTT and 100 μM biotin-17-ATP for 2 h at 37 °C. To verify that LnaB is capable of utilizing Bio-17-ATP to modify PR-Ub, 1 μg His6-LnaB, 1 μg actin, 2 μg PR-Ub were incubated in a 20 μL reaction system containing 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 1 mM DTT and 100 μM biotin-17-ATP for 2 h at 37 °C.
For the AMPylation assay of potential protein substrates of Ceg14, 1 μg His6-Ceg14 or His6-Ceg14S/A, 1 μg actin (Cytoskeleton, cat# APHL99) and 10 μL eluted target proteins were incubated in a 20 μL reaction system containing 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2 and 5 μCi ATP-α-32P (Perkin Elmer, cat# BLU003H250UC), and the reaction was allowed to proceed for 1 h at 37 °C. Samples were resolved by SDS-PAGE and gels were stained with Coomassie brilliant blue. Gels were then dried and the signals were detected with x-ray films.
Transfection and immunoprecipitation
HEK293T cells grown to 80% confluency were transfected with the appropriate plasmids using Lipofectamine 3000 (Invitrogen, cat# L3000150). 24 h after transfection, cells were collected and lysed with a cell lysis buffer (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 1% Triton X-100) by incubating on ice for 10 min. For immunoprecipitation, cell lysates were incubated with beads coated with antibodies specific for GFP (Proteintech cat# 50430-2-AP), Flag(Sigma cat#F1804) or HA (Sigma cat#A2095) at 4 °C for 16 h. Beads were washed 3x with pre-chilled cell lysis buffer, and were resuspend in an appropriate volume of 1x SDS protein loading buffer. After boiling for 5 min, samples were resolved by SDS-PAGE and followed by immunoblotting analysis with the specific antibodies.
Protein pulldown assays
GST pull-down assays were used to analyze interactions between Ceg14 and AnkJ. 50 μg GST or 50 μg GST-AnkJ was co-incubated with 50 μg His6-Ceg14 in pre-cooled PBS at 4 °C for 30 min. An equal amount of GST Beads was added to each sample, and the samples were incubated at 4 °C for an additional 1 h on a rotatory shaker. The beads were washed 4x with cold PBS, and an equal amount of 1xSDS loading buffer was added to each sample, after boiling for 5 min, proteins separated by SDS-PAGE were detected by Coomassie Brilliant Blue staining. To determine the interaction between actin and Ceg14 or its Ceg14S/A mutant, 30 μg actin was incubated with 30 μg His6-Ceg14 or His6-Ceg14S/A in a binding buffer containing 150 mM NaCl, 20 mM Tris pH 8.0, 20 mM imidazole, and 0.02% Triton X-100 at 4 °C for 1 h. 20 μL Ni2+-agarose beads were added to the samples and the reaction was allowed to proceed for 1 h. Beads were washed 3x with the binding buffer, samples were resolved by SDS-PAGE prior to detection by staining.
ATP hydrolysis and HPLC analysis
Two micrograms of His6-Ceg14 or its mutants and 1 μg actin were added to a 50 μL reaction containing 10 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 100 mM NaCl, and 1 mM ATP. The reactions were allowed to proceed at 37 °C for 2 h, before being terminated by chloroform extraction. HPLC measurements were performed with a SHIMADZU HPLC system equipped with a C18 column. The samples were injected onto the HPLC and run for 25 min with a mobile phase buffer containing 50 mM KH2PO4, 50 mM K2HPO4, 10 mM TPAB, and 20% (v/v) acetonitrile at a flow rate of 1 mL/min. AMP, ADP, and ATP were detected by UV light at 254 nm and quantified (by peak area) using LabSolutions when necessary. The same method was used to detect all the substrates.
To assess the inhibition of ongoing ATP hydrolysis catalyzed by Ceg14 by AnkJ, a reaction system containing 1 μg Ceg14, 1 μg actin, 1 mM ATP, 1 mM DTT, 5 mM MgCl2, 20 mM Tris-HCl (pH 7.5), and 100 mM NaCl was incubated at 37 °C for the indicated time and 2 μg AnkJ or an equivalent volume of dialysis buffer was then added, and the incubation was continued at 37 °C for 2 h. ATP and the products of the reaction were measured by HPLC.
Pyrophosphate detection
Pyrophosphate levels were determined by Fluorimetric Pyrophosphate Assay Kit (AAT Bioquest, Cat# 21611). 1 μg actin, 2 μg Ceg14 or both 1 μg actin and 2 μg Ceg14 were added to a 50-μL reaction system containing 10 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 100 mM NaCl, and 1 mM ATP. The reactions were allowed to proceed at 37 °C for 1 h. 10% of each reaction sample was taken and diluted 200-fold with ddH2O, and 50 µL of each diluted sample was measured according to the protocol provided by the kit. 50 µL of 5 µM ATP, ADP, AMP, or PPi standards were used as controls.
DNase activity assay
Genomic DNA of bacterial or mammalian cells was isolated with a TIANamp Genomic DNA kit (TIANGEN, Cat# DP304-02) and TIANamp Bacteria DNA kit (TIANGEN, Cat# DP302-02), respectively. 1μg DNA was incubated with 2 μg His6-Ceg14 or its mutants and 1 μg actin in 20 μL reaction buffer containing 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 5 mM MgCl2 and 1 mM DTT for 1 h at 37 °C. The samples were analyzed by 1% agarose gel electrophoresis and ethidium bromide staining.
Actin polymerization assays
G-actin stock was prepared by adding actin to ice cold G-buffer (0.2 mM ATP, 5 mM Tris-HCl pH 8.0 and 0.2 mM CaCl2) to a final concentration of 4 μM, and solution was incubated on ice for 1 h prior to centrifugation at 14,000 g for 30 min to obtain soluble G-actin. actin precipitation and polymerization kinetic analysis were performed as previously described41. Briefly, various proteins to be tested were added to the G-actin stock containing 10% pyrene-labeled actin, and incubate at 25 °C for 30 min, and 10x actin polymerization buffer (APB) (500 mM KCl, 10 mM MgCl2, 10 mM EGTA) was added to induce actin polymerization. A Bio-Tek Synergy H1 microplate reader was used to detect the polymerization of actin throughout the process. The excitation and emission wavelengths was set at 365 nm and 410 nm, respectively, and detection was performed every 60 s throughout the indicated time duration. For precipitation assay, samples were centrifuged at 100,000 g for 1 h and the pellets were resuspended in volumes of deionized water equal to those of the supernatant. Samples were analyzed by SDS-PAGE and Coomassie brilliant blue staining.
Cell viability assays
HEK293T cells were transfected with plasmids expressing Flag-Ceg14 or its mutants when the cells reached about 90% confluence. 24 h after transfection, samples were assessed using the Cell Counting Kit-8 (Beyotime Biotechnology), and the absorbance at 450 nm was measured with a Bio-Tek Synergy H1 microplate reader to determine cell viability.
LDH release assays
HEK293T cells seeded in 96-well plates were grown to a density of ~90%. Cells were transfected with the relevant plasmids for 24 h. LDH in culture supernatant was measured using the LDH Release Assay Kit (Beyotime Biotechnology) following instructions by the manufacturer. A Bio-Tek Synergy H1 microplate reader was used to quantitate the enzyme by measuring the absorbance at 490 nm.
Measurement of cellular ATP concentrations
Standard curves of ATP were established following the protocol provided in the assay kit (Beyotime Biotechnology). HEK293T cells grown to 90% confluence in 24-well plates were transfected with the relevant plasmids. 24 h after transfection, ATP content of the samples was measured as follows. After removing the culture medium, 3 × 105 cells were collected and lysed on ice for 5 min using pre-cooled lysis buffer. Cleared lysates were obtained by centrifugation at 13,000 g for 5 min, a fraction of the supernatant was added to the assay wells containing the detection reagents, and the relative light units (RLU) were measured using a luminometer. To detect ATP levels in RAW264.7 and D. discoideum cells under infection conditions, cells were infected with relevant L. pneumophila strains at a multiplicity of infection (MOI) of 10 for 2 h prior to removing extracellular bacteria by 3x washing with warm PBS. At the indicated times, cells were lysed on ice for 10 min using pre-cooled TBS containing 0.02% saponin. Luminescence was similarly measured.
AlphaFold analysis and ATP docking
AlphaFold v2.3.152 was employed for structure prediction. The model with the highest confidence was used for analysis. All figures of structure models were generated using ChimeraX83. For ATP docking, the ATP structure file was retrieved from the PubChem website (https://pubchem.ncbi.nlm.nih.gov/) and converted from SDF to PDB format using Open Babel 2.3.2 software. Using AutoDockTools software, the highest scoring structure predicted by AlphaFold was modified by adding hydrogen atoms and balancing charges. The receptor protein 2 and ligand ATP structure files were then converted to pdbqt format. Molecular docking of the receptor protein with the ligand molecule was performed using AutoDock Vina 1.1.2, and the highest scoring result was saved as the final PDB file for subsequent analysis.
β-lactamase translocation assay
To test the efficiency of Ceg14, Ceg14S/A and Ceg14H/A translocation into host cells, we cloned ceg14, ceg14S/A and ceg14H/A into the pZLQ-TEM161 to express β-lactamase-Ceg14 fusion proteins. The plasmids were then introduced into wild-type and dotA- mutant L. pneumophila strains. The resulting strains were induced with 0.2 mM IPTG for 6 h before infecting Raw264.7 cells at an MOI of 20. After 2 h of infection, CCF4-AM substrates (Invitrogen, Carlsbad, CA) were added to the cell culture medium and incubated in the dark at room temperature for 1.5 h. Images were then captured using an Olympus IX-83 fluorescence microscope. The translocation efficiency was assessed by calculating the proportion of cells exhibiting blue fluorescence. To test the effect of AnkJ on the translocation efficiency of Ceg14, we introduced the pZLQ-TEM1-ceg14 plasmid into the following strains: WT(pEV), dotA-(pEV), WT(pAnkJ), and the resulting L. pneumophila strains were used to determine the Dot/Icm-dependent translocation efficiency of Ceg14 following the same procedure.
Bacterial RNA isolation and reverse transcription quantitative PCR
RAW264.7 cells seeded in six-well plates were infected with wild-type L. pneumophila strains for 5 min, 4 h, 8 h, 12 h and 16 h at an MOI of 10. Infected cells were washed three times with PBS to remove extracellular bacteria. Total RNA was then extracted using the MolPure® Bacterial RNA Kit (Yeasen Biotechnology, Cat# 19301ES50). RNA was reverse-transcribed into cDNA using the PrimeScript™ RT Reagent Kit (Takara, Cat# RR037A) according to the manufacturer’s instructions. Transcription levels were quantified using the BlasTaq™ 2X qPCR Master Mix (Applied Biological Materials, Cat# G891) on a Bio-Rad CFX Connect real-time PCR system, following the manufacturer’s protocol. The dotA gene was used as an internal control, and relative gene expression was calculated using the 2−ΔΔCT method. The primer sequences used for qRT-PCR analysis are listed in Supplementary Data 3.
Data quantification and statistical analysis
Student’s t-test was employed to compare the mean levels between two groups, each comprising at least three independent samples. Ordinary one-way ANOVA was performed for multiple comparisons. The western blot results presented are representative of three independent experiments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data of this paper are either included in the Figures, Tables and supplementary Information or source data. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD056059. Source data are provided with this paper.
References
Fraser, D. W. et al. Legionnaires’ disease: description of an epidemic of pneumonia. N. Engl. J. Med. 297, 1189–1197 (1977).
Fields, B. S., Benson, R. F. & Besser, R. E. Legionella and Legionnaires’ disease: 25 years of investigation. Clin. Microbiol. Rev. 15, 506–526 (2002).
Isberg, R. R., O’Connor, T. J. & Heidtman, M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7, 13–24 (2009).
Ge, J. & Shao, F. Manipulation of host vesicular trafficking and innate immune defence by Legionella Dot/Icm effectors. Cell Microbiol. 13, 1870–1880 (2011).
Qiu, J. & Luo, Z. Q. Legionella and Coxiella effectors: strength in diversity and activity. Nat. Rev. Microbiol. 15, 591–605 (2017).
Hubber, A. & Roy, C. R. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev. Cell Dev. Biol. 26, 261–283 (2010).
Burstein, D. et al. Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat. Genet 48, 167–175 (2016).
de Felipe, K. S. et al. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J. Bacteriol. 187, 7716–7726 (2005).
Xu, L. & Luo, Z. Q. Cell biology of infection by Legionella pneumophila. Microbes Infect. 15, 157–167 (2013).
O’Connor, T. J., Adepoju, Y., Boyd, D. & Isberg, R. R. Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc. Natl. Acad. Sci. USA 108, 14733–14740 (2011).
Romanov, K. A. & O’Connor, T. J. Legionella pneumophila, a Rosetta stone to understanding bacterial pathogenesis. J. Bacteriol. 206, e0032424 (2024).
Salomon, D. & Orth, K. What pathogens have taught us about posttranslational modifications. Cell Host Microbe 14, 269–279 (2013).
Moss, S. M. et al. A legionella pneumophila kinase phosphorylates the Hsp70 chaperone family to inhibit Eukaryotic protein synthesis. Cell Host Microbe 25, 454–462.e456 (2019).
Mukherjee, S. et al. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science 312, 1211–1214 (2006).
Zhou, Y. & Zhu, Y. Diversity of bacterial manipulation of the host ubiquitin pathways. Cell Microbiol. 17, 26–34 (2015).
Pearson, J. S. et al. A type III effector antagonizes death receptor signalling during bacterial gut infection. Nature 501, 247–251 (2013).
Yarbrough, M. L. et al. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science 323, 269–272 (2009).
Fu, J. et al. Legionella pneumophila modulates host energy metabolism by ADP-ribosylation of ADP/ATP translocases. Elife 11, e73611 (2022).
Xu, Y. et al. A bacterial effector reveals the V-ATPase-ATG16L1 axis that initiates xenophagy. Cell 178, 552–566.e520 (2019).
Zhou, Y. et al. N(epsilon)-Fatty acylation of Rho GTPases by a MARTX toxin effector. Science 358, 528–531 (2017).
Shao, F., Merritt, P. M., Bao, Z., Innes, R. W. & Dixon, J. E. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell 109, 575–588 (2002).
Liu, Y. et al. A legionella effector disrupts host cytoskeletal structure by cleaving actin. PLoS Pathog. 13, e1006186 (2017).
van der Meer-Janssen, Y. P., van Galen, J., Batenburg, J. J. & Helms, J. B. Lipids in host-pathogen interactions: pathogens exploit the complexity of the host cell lipidome. Prog. Lipid Res. 49, 1–26 (2010).
Coronas-Serna, J. M. et al. The TIR-domain containing effectors BtpA and BtpB from Brucella abortus impact NAD metabolism. PLoS Pathog. 16, e1007979 (2020).
Pinna, S. et al. A prebiotic basis for ATP as the universal energy currency. PLoS Biol. 20, e3001437 (2022).
Sieber, S. A., Cappello, S. & Kielkowski, P. From Young to Old: AMPylation Hits the Brain. Cell Chem. Biol. 27, 773–779 (2020).
Saravia, J., Raynor, J. L., Chapman, N. M., Lim, S. A. & Chi, H. Signaling networks in immunometabolism. Cell Res. 30, 328–342 (2020).
Dion, M. B., Oechslin, F. & Moineau, S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 18, 125–138 (2020).
DeLong, J. P. et al. Towards an integrative view of virus phenotypes. Nat. Rev. Microbiol. 20, 83–94 (2022).
Rousset, F. et al. A conserved family of immune effectors cleaves cellular ATP upon viral infection. Cell 186, 3619–3631.e3613 (2023).
Goldstone, D. C. et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 (2011).
Krysińska, M., Baranowski, B., Deszcz, B., Pawłowski, K. & Gradowski, M. Pan-kinome of Legionella expanded by a bioinformatics survey. Sci. Rep. 12, 21782 (2022).
Bhogaraju, S. et al. Inhibition of bacterial ubiquitin ligases by SidJ-calmodulin catalysed glutamylation. Nature 572, 382–386 (2019).
Gan, N. et al. Legionella pneumophila regulates the activity of UBE2N by deamidase-mediated deubiquitination. EMBO J. 39, e102806 (2020).
Black, M. H. et al. Bacterial pseudokinase catalyzes protein polyglutamylation to inhibit the SidE-family ubiquitin ligases. Science 364, 787–792 (2019).
Müller, M. P. et al. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329, 946–949 (2010).
Wang, T. et al. Legionella effector LnaB is a phosphoryl AMPylase that impairs phosphosignalling. Nature 631, 393–401 (2024).
Fu, J. et al. Legionella maintains host cell ubiquitin homeostasis by effectors with unique catalytic mechanisms. Nat. Commun. 15, 5953 (2024).
Zusman, T. et al. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol. Microbiol 63, 1508–1523 (2007).
Burstein, D. et al. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 5, e1000508 (2009).
Guo, Z., Stephenson, R., Qiu, J., Zheng, S. & Luo, Z. Q. A Legionella effector modulates host cytoskeletal structure by inhibiting actin polymerization. Microbes Infect. 16, 225–236 (2014).
Fontana, M. F. et al. Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS Pathog. 7, e1001289 (2011).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Chien, M. et al. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305, 1966–1968 (2004).
Branon, T. C. et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887 (2018).
Yoshigi, M., Hoffman, L. M., Jensen, C. C., Yost, H. J. & Beckerle, M. C. Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. J. Cell Biol. 171, 209–215 (2005).
Joo, E. E. & Yamada, K. M. MYPT1 regulates contractility and microtubule acetylation to modulate integrin adhesions and matrix assembly. Nat. Commun. 5, 3510 (2014).
Lányi, Á. et al. The homolog of the five SH3-domain protein (HOFI/SH3PXD2B) regulates lamellipodia formation and cell spreading. PLoS ONE 6, e23653 (2011).
Boëda, B. & Etienne-Manneville, S. Spectrin binding motifs regulate Scribble cortical dynamics and polarity function. Elife 4, e04726 (2015).
Casey, A. K. & Orth, K. Enzymes Involved in AMPylation and deAMPylation. Chem. Rev. 118, 1199–1215 (2018).
Funk, J. et al. Profilin and formin constitute a pacemaker system for robust actin filament growth. Elife 8, e50963 (2019).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Babich, M., Foti, L. R., Sykaluk, L. L. & Clark, C. R. Profilin forms tetramers that bind to G-actin. Biochem. Biophys. Res. Commun. 218, 125–131 (1996).
Mockrin, S. C. & Korn, E. D. Acanthamoeba profilin interacts with G-actin to increase the rate of exchange of actin-bound adenosine 5’-triphosphate. Biochemistry 19, 5359–5362 (1980).
Urbanus, M. L. et al. Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila. Mol. Syst. Biol. 12, 893 (2016).
Joseph, A. M. & Shames, S. R. Affecting the effectors: regulation of Legionella pneumophila effector function by metaeffectors. Pathogens 10, 108 (2021).
Valleau, D. et al. Discovery of ubiquitin deamidases in the pathogenic arsenal of Legionella pneumophila. Cell Rep. 23, 568–583 (2018).
Shames, S. R. et al. Multiple Legionella pneumophila effector virulence phenotypes revealed through high-throughput analysis of targeted mutant libraries. Proc. Natl. Acad. Sci. USA 114, E10446–e10454 (2017).
Liu, Y. & Luo, Z. Q. The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect. Immun. 75, 592–603 (2007).
Gan, N. et al. Regulation of phosphoribosyl ubiquitination by a calmodulin-dependent glutamylase. Nature 572, 387–391 (2019).
McCloskey, A., Perri, K., Chen, T., Han, A. & Luo, Z. Q. The metaeffector MesI regulates the activity of the Legionella effector SidI through direct protein-protein interactions. Microbes Infect. 23, 104794 (2021).
Tilney, L. G., Harb, O. S., Connelly, P. S., Robinson, C. G. & Roy, C. R. How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: implications for conversion of plasma membrane to the ER membrane. J. Cell Sci. 114, 4637–4650 (2001).
Fu, J. et al. Legionella pneumophila temporally regulates the activity of ADP/ATP translocases by reversible ADP-ribosylation. mLife 1, 51–65 (2022).
Escoll, P. et al. Reverting the mode of action of the mitochondrial F(O)F(1)-ATPase by Legionella pneumophila preserves its replication niche. Elife 10, e71978 (2021).
Ganeshan, K. & Chawla, A. Metabolic regulation of immune responses. Annu Rev. Immunol. 32, 609–634 (2014).
Mills, E. L., Kelly, B. & O’Neill, L. A. J. Mitochondria are the powerhouses of immunity. Nat. Immunol. 18, 488–498 (2017).
Mihaylova, M. M. & Shaw, R. J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 13, 1016–1023 (2011).
Carlier, M. F. Actin polymerization and ATP hydrolysis. Adv. Biophys. 26, 51–73 (1990).
Liu, H., Krizek, J. & Bretscher, A. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics 132, 665–673 (1992).
Shin, D. et al. Regulation of phosphoribosyl-linked serine ubiquitination by deubiquitinases DupA and DupB. Mol. Cell 77, 164–179.e166 (2020).
Wan, M. et al. Deubiquitination of phosphoribosyl-ubiquitin conjugates by phosphodiesterase-domain-containing Legionella effectors. Proc. Natl. Acad. Sci. USA 116, 23518–23526 (2019).
Chauhan, D., Joseph, A. M. & Shames, S. R. Intrabacterial regulation of a cytotoxic effector by its cognate metaeffector promotes legionella pneumophila virulence. mSphere 8, e0055222 (2023).
Habyarimana, F., Price, C. T., Santic, M., Al-Khodor, S. & Kwaik, Y. A. Molecular characterization of the Dot/Icm-translocated AnkH and AnkJ eukaryotic-like effectors of Legionella pneumophila. Infect. Immun. 78, 1123–1134 (2010).
Ganesan, R. et al. Salmonella typhimurium disrupts Sirt1/AMPK checkpoint control of mTOR to impair autophagy. PLoS Pathog. 13, e1006227 (2017).
Kato, K. et al. Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling. Proc. Natl. Acad. Sci. USA 109, 16876–16881 (2012).
Berger, K. H., Merriam, J. J. & Isberg, R. R. Altered intracellular targeting properties associated with mutations in the Legionella pneumophila dotA gene. Mol. Microbiol. 14, 809–822 (1994).
Luo, Z. Q. & Isberg, R. R. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. USA 101, 841–846 (2004).
Solomon, J. M., Rupper, A., Cardelli, J. A. & Isberg, R. R. Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect. Immun. 68, 2939–2947 (2000).
Thomas, B. J. & Rothstein, R. Elevated recombination rates in transcriptionally active DNA. Cell 56, 619–630 (1989).
Gietz, R. D., Schiestl, R. H., Willems, A. R. & Woods, R. A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, 355–360 (1995).
Li, G., Liu, H., Luo, Z. Q. & Qiu, J. Modulation of phagosome phosphoinositide dynamics by a Legionella phosphoinositide 3-kinase. EMBO Rep. 22, e51163 (2021).
Mumberg, D., Müller, R. & Funk, M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122 (1995).
Pettersen, E. F. et al. UCSF chimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China grants 32470179 (LS), 32370185 (JF), 32270185 (LS), 82472330 (XC), 32100137(SL), 32200141(YZ), 82225028 and 82172287 (SO) the National Key Research and Development Program of China 2021YFC2301403 (SO). Mass spectrometric analysis was performed in the core facility of the First Hospital of Jilin University.
Author information
Authors and Affiliations
Contributions
C.H., L.S., and Z.Q.L. conceived the ideas for this work. C.H. performed the biochemical experiments, infection experiments and the yeast experiments. J.F. and C.X.L.L. performed mass spectrometric analyses. L.S., T.T.C., and S.O. performed the molecular docking and analyzed protein binding using biophysical tools. C.L., Y.L., X.C., S.L., L.W., and Y.Z. contributed to the experimental design and data analysis. C.H. wrote the first draft and Z.Q.L. revised the paper.
Corresponding authors
Ethics declarations
Competing interests
The author declares no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
He, C., Li, C., Liu, Y. et al. Modulation of host ATP levels by secreted bacterial effectors. Nat Commun 16, 4675 (2025). https://doi.org/10.1038/s41467-025-60046-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-60046-3