Abstract
Real-time monitoring of plant stress signaling molecules is crucial for early disease diagnosis and prevention. However, existing methods are often invasive and lack sensitivity, rendering them inadequate for continuous monitoring of subtle plant stress responses. In this study, we develop a non-destructive near-infrared-II (NIR-II) fluorescent nanosensor for real-time detection of stress-related H2O2 signaling in living plants. This nanosensor effectively avoids interference from plant autofluorescence and specifically responds to trace amounts of endogenous H2O2, thereby providing a reliable means to real-time report stress information. We validate that it is a species-independent nanosensor by effectively monitoring the stress responses of different plant species. Additionally, with the aid of a machine learning model, we demonstrate that the nanosensor can accurately differentiate between four types of stress with an accuracy of more than 96.67%. Our study enhances the understanding of plant stress signaling mechanisms and offers reliable optical tools for precision agriculture.
Similar content being viewed by others
Introduction
Abiotic stresses, such as extreme temperatures, drought, salinity, and flooding, along with biotic stresses caused by pathogens and pests, have significantly reduced crop productivity and pose substantial challenges to food security1,2,3. It is essential to obtain stress-related information by monitoring stress-induced physiological signals and understanding the mechanisms underlying the stress responses of plants4,5,6.
Detecting stress-induced signals in living plants presents significant challenges due to their low concentrations and the frequent coexistence of various other active substances7,8,9,10. Current methods for sensing stress-induced signals primarily rely on histochemical reagents following the isolation and purification of plant extracts11,12,13. However, these methods are typically destructive and do not permit real-time tracking of the endogenous dynamic signals. Furthermore, genetically encoded molecular sensors for stress signaling, as a non-destructive approach, predominantly focus on the stress-specific signal transduction pathways triggered in the model plant Arabidopsis thaliana, making them time-consuming for phenotypic expression and screening14,15,16,17. Given that most crops are non-model plants, studying the stress signaling patterns of these crops places greater demands on the existing detection techniques. Nanotechnology-based sensors are species-independent and can detect stress responses in various wild-type plants by monitoring plant health in real time through electronic devices, eliminating the need for genetic engineering18,19,20,21,22,23,24,25. The latest plant advancements in wearable sensors and electrochemical sensors enable non-destructive and continuous acquisition of phenotypic data. Wearable electronic devices monitor plant conditions by capturing physical signals such as growth, surface temperature, and humidity26,27, as well as chemical signals like volatile organic compounds21, and bioelectrical potential signals28. From flexible sensors wrapped around the stem to the epidermal electronics that can be seamlessly attached to leaves, most efforts have focused on acquiring physiological information from the plant’s surface. Direct detection of trace molecules in the cytosol and intercellular spaces can provide deeper insights into the signaling network involved in plant stress responses. In contrast to lagging physiological indicators (such as strain, moisture, or leaf temperature), stress-induced real-time signaling molecules (including reactive oxygen species, phytohormones, and inorganic ions) reflect early stress responses before the appearance of visible stress symptoms.
In this work, we present a non-destructive sensor that employs NIR-II fluorescence imaging technology for the real-time detection of stress-induced H2O2 signals in living plants. H2O2 is a crucial signaling molecule that plays a key role in sensing multiple stresses and rapidly activating the plant’s stress response network29,30,31,32,33,34,35,36. Over the past decade, fluorescence imaging technology in the second near-infrared (NIR-II, 1000–1700 nm) region has enabled high-contrast and long-term in vivo plant imaging and sensing by significantly reducing interference from background signals originating from the chlorophyll autofluorescence and increasing the depth of penetration22,37. The aggregation-induced emission (AIE) fluorophore, recognized for its stable luminescent properties38,39,40, serves as the signal reporter and is co-assembled with polymetallic oxomolybdates (POMs) as a fluorescence quencher to create NIR-II fluorescence “turn-on” nanosensor. Under stress conditions, H2O2-selective POMs undergo oxidation, which diminishes their quenching effect and activates a bright NIR-II fluorescence signal from the AIE fluorophore. In contrast to “always-on” sensors, activatable “turn-on” sensors provide a visual representation of plant stress information while effectively suppressing non-target background signals41,42,43,44,45. The sensor’s sensitivity of 0.43 μM and response time of 1 min surpass those of existing NIR-II sensors, enabling rapid, real-time monitoring of trace levels of H2O2. We utilized an NIR-II microscopy system and a macroscopic whole-plant imaging system to elucidate the stress-induced H2O2 signals. The versatility of the nanosensor enables the monitoring of H2O2 signaling across various plant species, including Arabidopsis, lettuce, spinach, pepper, and tobacco. Finally, by employing a machine learning (ML) model that incorporates the dataset collected by the nanosensor, we demonstrate that fluorescence signals obtained from the NIR-II imaging system can be utilized to classify plant responses to stress with high accuracy.
Results
Design, synthesis, and characterization of NIR-II nanosensor
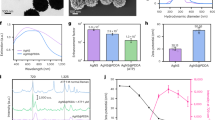
The nanosensor consists of NIR-II fluorophore and POMs as components. The NIR-II dye exhibiting AIE properties was selected as a fluorescence reporter. Simultaneously, a series of fluorescent quenchers, known as POMs, with varying NIR absorption characteristics, were co-assembled with AIE nanoparticles (AIENPs). This assembly led to a significant quenching of the NIR-II fluorescence signal, achieving a “turn-off” state (Fig. 1a). The inherent oxygen vacancies in POMs confer unique H2O2-responsive properties46,47,48. Upon interaction with the H2O2 signaling molecule, the NIR absorbance of POMs decreased dramatically, resulting in the recovery of the bright NIR-II signal of AIENPs (Fig. 1b).
a Schematic of the fabrication process for AIENPs@POM. b Schematic illustrating the responsive mechanism of the nanosensors toward H2O2. c Overlap of the absorbance spectrum of POMs and the fluorescence emission spectrum of AIE1035NPs. d Plots for the absorption intensity of Mo-POM, Mo/Fe-POM, and Mo/Cu-POM at different concentrations of H2O2. Data are presented as mean ± s.d. from three independent experiments. e Absorbance response of Mo/Cu-POM to H2O2 and other molecules commonly found in plant systems. Data are presented as mean ± s.d. from three independent experiments. Each dot represents an individual data point. A0 and A represent the absorption peak values of Mo/Cu-POM in water and interferences, respectively. f Normalized fluorescence intensity of AIE1035NPs@Mo/POM at 1035 nm with varying POM/AIE1035 ratios from 0 to 25. g, h Transmission electron microscopy image (g) and elemental mapping image (h) of AIE1035NPs@Mo/Cu-POM. Scale bar: 100 nm. Insert: size distribution and particle distribution index of the synthesized AIE1035NPs@Mo/Cu-POM. i X-ray photoelectron spectroscopy with deconvoluted 3d peaks of Mo. j Zeta potentials of AIE1035NPs (left), Mo/Cu-POM (middle), and AIE1035NPs@Mo/Cu-POM (right). Data are presented as mean ± s.d. from three independent measurements. k NIR images of a series of AIE1035NPs@Mo/Cu-POM hybrid nanosensors. l Heat map summary of the NIR signal of nanosensors doped with different amounts of AIE1035 and Mo/Cu-POM. Laser excitation: 808 nm, 15 mW. Filter: 900 nm long-pass filter. m Normalized fluorescence intensity of AIE1035NPs@Mo/Cu-POM nanosensor as a function of Cu-based POM/AIE1035 ratio (left) and H2O2 concentration (right), extracted from (I) and (Supplementary Fig. 18). Left insert: NIR-II images of sensor doped with (+) or without (−) Mo/Cu-POM. Right insert: NIR-II images of sensor reacting with (+) or without (−) H2O2.
The NIR-II fluorophore with AIE properties was chosen as a signal reporter due to its enhanced fluorescence efficiency in aggregates and photostability (Supplementary Fig. 1)49,50. This design features a strong donor-acceptor-donor (D-A-D) molecular structure, incorporating rotating units and π-conjugated bridges that introduce specific spatial hindrance. The strong electron-withdrawing group benzo[1,2-c:4,5-c′]bis[1,2,5]thiadiazole (BBTD) serves as the acceptor unit with a quinoidal structure, which facilitates greater electron delocalization and consequently reduces the bandgap51. The donor units trimethylamine (TPA), act as molecular rotors in AIE1035, ensuring the intramolecular rotation. TPA’s strong electron-donating capability and larger molecular structure inhibit intermolecular interactions in the aggregated state. The planar thiophene ring acts both as a secondary donor group and a π-conjugated unit, facilitating the intramolecular charge transfer (ICT) from TPA to BBTD. The branched carbon alkyl chains on the thiophene provide tunable steric hindrance, preventing excessive aggregation52. Subsequently, the NIR-II AIE dye was encapsulated into polystyrene (PS) nanospheres using the organic solvent swelling method (Supplementary Fig. 2).
Three fluorescence quenchers (Mo-POM, Mo/Fe-POM, and Mo/Cu-POM) with different NIR absorption properties were synthesized to modulate the NIR-II emission of AIENPs and to suppress the fluorescence (Supplementary Figs. 3–7). The absorption spectra indicated that these POMs exhibited absorption peaks ranging from ultraviolet (UV) to NIR-II with a characteristic absorption peak at around 750 nm (Fig. 1c). X-ray photoelectron spectroscopy (XPS) analysis revealed that Mo in the POMs existed in a mixed valence state of Mo5+ and Mo6+ (Supplementary Figs. 8–11). According to the XPS peak area of Mo 3d, the average oxidation states of Mo in Mo-POM, Mo/Fe-POM, and Mo/Cu-POM were determined to be 5.38, 5.49, and 5.40, respectively (Supplementary Table 1), indicating a mixed-valence state resulting from the presence of oxygen vacancies53. The strong NIR absorption band of POMs was attributed to the localized surface plasmon resonance (LSPR) effect associated with the charge-transfer transition between Mo5+ and Mo6+ via oxygen vacancies54. Upon reaction with varying concentrations of H2O2, Mo5+ in the POMs was oxidized to Mo6+, leading to a decrease in the intervalence charge transfer (IVCT) between the mixed-valence Mo centers48, which is directly correlated with the reduction in the NIR absorbance (Fig. 1b). Oxygen vacancies introduce localized defect states that facilitate the adsorption and activation of H2O2 molecules on the surface of POMs and promote more efficient electron transfer55,56,57, thus expanding the redox transformation from Mo5+ to Mo6+. Notably, the absorption profiles of Mo/Cu-POM mixed with H2O2 solution exhibited significant changes, with rapid decay effects observed at NIR-I (750 nm) and NIR-II (1100 nm), indicating a more sensitive response to H2O2 property compared to Mo-POM and Mo/Fe-POM (Fig. 1d and Supplementary Fig. 8). Subsequently, the three POMs were evaluated for their responses to various endogenous molecules commonly found in plant tissues (Fig. 1e and Supplementary Fig. 12) and for their stability across a range of pH levels and under laser irradiation (Supplementary Fig. 13). These findings suggest that the POMs are highly selective for H2O2 and are suitable for long-term optical imaging applications. Co-assembling the AIE1035NPs with three POMs, we evaluated the quenching properties of POMs. The assembly of Mo/Cu-POM exhibited a stronger fluorescence “turn-off” effect, which was attributed to its enhanced NIR-II absorption properties and stronger electrostatic interactions (Fig. 1f and Supplementary Fig. 14).
A variety of characterization techniques were employed to confirm the successful synthesis of the nanosensor. TEM images and elemental mapping showed that Mo/Cu-POM was uniformly assembled on the surface of AIE1035NPs (Fig. 1g, h). The resulting hybrid nanosensors displayed good dispersion, with a particle distribution index (PDI) of 0.078 and an approximate diameter of 230 nm. The successful preparation of AIE1035NPs@Mo/Cu-POM was further confirmed by X-ray photoelectron spectroscopy (XPS) and zeta potential measurements (Fig. 1i, j and Supplementary Fig. 15). Mo/Cu-POM and AIE1035NPs were co-assembled to construct a series of nanosensors with mass ratios ranging from 0 to 100, allowing for flexible modulation of NIR-II fluorescence performance (Fig. 1k–m). The NIR-II signals of the nanosensors were gradually enhanced with increasing AIE dye doping. As the mass ratio of Mo/Cu-POM increased, the fluorescence emission of the nanosensors gradually decreased, indicating that Mo/Cu-POM effectively modulated the fluorescence of the AIE molecule. The results of dynamic light scattering (DLS) measurements lasting for 3 weeks indicated that the assembly of Mo/Cu-POM did not significantly affect the hydrodynamic diameter and PDI of the nanoparticles (Supplementary Fig. 16), thereby demonstrating the long-term stability of the nanosensor. Next, the responsiveness of AIE1035NPs@Mo/Cu-POM to H2O2 was evaluated. Upon the addition of H2O2, NIR-II fluorescence emission rapidly recovered within 1 min (Supplementary Fig. 17), ultimately achieving a 7.7-fold enhancement, which provided a limit of detection of 0.43 μM, better or comparable to previously reported NIR-II sensors (Fig. 1m, Supplementary Fig. 18, and Supplementary Table 2). Additionally, we investigated the sensing selectivity of the designed sensor against interfering substances such as phytohormones, reducing agents, metal ions, and other active compounds. The presence of these substances had a negligible influence on fluorescence, confirming the high specificity of AIE1035NPs@Mo/Cu-POM (Supplementary Fig. 19).
Superior NIR-II nanosensor improves imaging performance
To investigate the imaging performance of fluorescent molecules with different emission bands in plants, we co-assembled five AIE luminophores with varying excitation and emission wavelengths with Mo/Cu-POM (Fig. 2a–c). There was insufficient overlap between the absorption spectrum of Mo/Cu-POM and the ultraviolet-visible absorption and fluorescence emission spectra of AIE475 and AIE520 with a tetrastyrene structure (Supplementary Fig. 20). In contrast, the spectra of AIE712 showed partial overlap with the characteristic absorption peak of Mo/Cu-POM. Notably, the spectra of the NIR-II fluorescent AIE955 and AIE1035 nearly coincided with the broad absorption peak of Mo/Cu-POM. Due to these varying degrees of spectral overlap, the extent of fluorescence quenching differed significantly among the AIENPs (Fig. 2d). We employed the spectral overlap integral and the fluorescence quenching constant (K) to evaluate the ability of Mo/Cu-POM to modulate the fluorescence emission of the nanoparticles (Supplementary Figs. 20, 21). The results revealed that both the spectral overlap integral and the quenching constant of Mo/Cu-POM were greater for NIR-II FL nanoparticles compared to those that are visible and NIR-I fluorescent. These findings suggest that the fluorescence quenching effect of Mo/Cu-POM on AIENPs is critically determined by the spectral overlap and can be further optimized by adjusting the concentration of Mo/Cu-POM.
a Chemical structures of five AIE molecules and their optical responses. b, c Normalized absorption (b) and normalized fluorescence spectra (c) of the five molecules encapsulated in polystyrene nanospheres. d Fluorescence spectra of five AIENPs before and after assembly with Mo/Cu-POM and their response to 100 μM H2O2. e Schematic illustration of the investigation of deep penetration in pepper leaves. Created in BioRender. Hu, H. (2025) https://BioRender.com/6lm4x2yf Fluorescence images of AIENPs-filled 96-well plates through stacked pepper leaves in different detection windows. Leaf penetration experiments for AIE475NPs, AIE520NPs, and AIE712NPs were conducted using an IVIS Spectrum platform, while experiments for AIE955NPs and AIE1035NPs were performed on a home-built NIR imaging system. g Corresponding normalized fluorescence intensity of the five AIENPs with stacked pepper leaves in (f). h Photostability evaluation of the AIE1035NPs dispersion and ICG solutions under constant laser beam exposure for 60 min. Laser excitation: 808 nm, 100 mW. i NIR images of AIE1035NPs and ICG in MES buffer at various concentrations, using an 808 nm laser as the excitation source at 15 mW. Filters: 850 nm long-pass filter (left) and 1000 nm long-pass filter (right). j Quantitative analysis of NIR fluorescence intensity for AIE1035NPs and ICG at different concentrations as shown in (i). Data are presented as mean ± s.d. from three independent measurements.
Considering the strong autofluorescence background interference from chlorophyll in plant leaves and the weakening effect of chloroplastic tissues on the fluorescence signal, we evaluated the imaging performance of the AIENPs in vivo. In contrast to the other detection windows, NIR-II images of the leaves were acquired using long-pass filters and a laser excitation (at 808 nm), which is off-resonance with photosynthetic pigments, resulting in no observable autofluorescence signal from the leaves (Supplementary Fig. 22). Both AIE520NPs and AIE712NPs injections produced significantly lower fluorescence signals in the infiltrated region compared to the original nanoparticles in the solution (Supplementary Fig. 23a, b). In contrast, NIR-II AIE1035NPs showed excellent imaging performance by avoiding the spectral band where chloroplast autofluorescence is concentrated, and they exhibited good penetration and low scattering loss despite the presence of chloroplastic tissue barriers (Fig. 2e–g and Supplementary Fig. 23c), suggesting that the NIR-II nanosensor is well-suited for in vivo deep plant tissue imaging. Furthermore, under constant laser beam irradiation, the signal intensity of AIE1035NPs exhibited remarkable stability without significant attenuation, unlike indocyanine green (ICG), which is highly advantageous for the continuous monitoring of biological information in plants (Fig. 2h). The NIR signals of AIE1035NPs were significantly stronger than those of aggregation-induced quenching ICG under the same laser excitation and were notably enhanced with increasing concentrations in the 2-Morpholinoethanesulphonic (MES) acid buffer (Fig. 2i, j). These results indicate the potential of AIE1035NPs for in vivo imaging or monitoring that surpasses that of the conventional NIR dye ICG.
Localization of nanosensor in plant leaves
As stress-induced signaling molecules are transiently generated, precise control of the nanosensors’ location is essential for detecting plant signaling molecules18. To demonstrate the applicability of AIE1035NPs@Mo/Cu-POM in living plants, we infiltrated the nanosensors into the leaves of lettuce, spinach, pepper, and tobacco (Fig. 3a and Supplementary Figs. 24, 25). When AIE520 dye-swelled PS nanoparticles of 200 nm in size were introduced into the plant leaves, the fluorescent signals were observed around the stomatal pores and in the intercellular spaces of the epidermis using confocal microscopy (Fig. 3b and Supplementary Fig. 26). We employed a home-built NIR-II microscope to further confirm the localization of AIE1035NPs (Fig. 3c and Supplementary Fig. 27), which aligns with the model of nanoparticle uptake in plant systems, where particles with diameters larger than 100 nm are typically found around the stomatal pores and in the intercellular space58,59. Furthermore, we tracked the movement of AIE1035NPs within the lettuce leaf over a period of 120 min (Fig. 3d). The nanoparticles exhibited slight diffusion within the leaf tissue during the first 60 min post-infiltration but did not migrate to other regions due to the barrier posed by the leaf veins. Macroscopic imaging of the movement of fluorescent particles in leaves over 6 h was consistent with observations made through microscopy. Although the nanosensors in leaf tissues displayed small-scale, slow diffusion over time at the microscopic level, this resulted in negligible signal changes during the measurements (Fig. 3e, f and Supplementary Fig. 28).
a Schematic of the infiltration process for AIE1035NPs@Mo/Cu-POM. This schematic illustrates the infiltration of AIE1035NPs@Mo/Cu-POM through stomatal pores and its distribution within the plant tissues. Created in BioRender. Hu, H. (2025) https://BioRender.com/c2p1p6j. b Confocal fluorescence micrographs of AIE520 dye-swelled PS nanoparticles (200 nm in diameter, 0.1 mg mL–1) in the lettuce leaf tissue. The nanoparticles (green) were localized alongside the cell walls, intercellular spaces, and stomatal pores. Chlorophyll autofluorescence is displayed in red. Confocal fluorescence micrographs were obtained from three independent biological replicates and were presented as representative images. c NIR fluorescence micrographs and brightfield merged images of a lettuce leaf infiltrated with AIE1035NPs (0.1 mg mL–1) at ×20 and ×50 magnifications. All the NIR fluorescence micrographs were obtained with an 808 nm laser at 2 mW. NIR images are representative of at least three independent experiments. d NIR fluorescence micrographs and three-dimensional surface plots of the AIE1035NPs-embedded lettuce leaf over 120 min. Filter: 900 nm long-pass filter. Scale bar: 200 μm. e NIR fluorescence localization of AIE1035NPs in a living plant for 6 h. NIR fluorescence images were obtained with an 808 nm laser at 15 mW using a shutter. Scale bar: 5 mm. f NIR fluorescence intensity profiles along the white dashed line in (e) over 6 h and normalized signal intensity change of AIE1035NPs in the lettuce leaf (red dots).
When AIE1035NPs@Mo/Cu-POM is utilized for plant applications, it is crucial to ensure the biocompatibility of the nanosensor. AIE1035NPs@Mo/Cu-POM and its two components were infiltrated into plant leaves, and no significant changes in FV/FM (a critical indicator of a plant’s photosynthetic performance60,61,62) were observed, suggesting that the nanosensor exhibited no toxicity to the plants (Supplementary Figs. 29, 30). Furthermore, the propidium iodide assay indicated that the nanosensor did not cause appreciable cell death or negative effects on the cells (Supplementary Figs. 31, 32). Considering that various environmental factors, such as soil moisture and light conditions, can influence plant physiology, the compatibility of the nanosensor under different stress conditions was further evaluated (Supplementary Figs. 30, 32). In long-term assessments, nanomaterial treatments did not adversely affect the overall growth and chlorophyll content of Arabidopsis and peppers, nor did they impact the physical and chemical properties of the soil (Supplementary Fig. 33 and Supplementary Table 3).
Visualization of exogenous and endogenous H2O2
Motivated by the in vitro sensing performance and high stability of the nanosensor, we further investigated its capability to detect H2O2 in living plants. H2O2 solutions with concentrations ranging from 0 to 20 μM, along with mechanically induced damage, were utilized as sources of exogenous H2O2 and stress-induced endogenous H2O2, respectively (Fig. 4a, b). The addition of exogenous H2O2 resulted in a continuous recovery of NIR-II signal intensity, as observed through NIR-II microscopy, allowing for the detection of H2O2 concentrations as low as 5 μM (Fig. 4c–e and Supplementary Fig. 34). This observation was consistent with the trend of fluorescence changes in the nanosensor due to exogenous H2O2, as seen from a macroscopic perspective (Supplementary Fig. 35). The detection range of the proposed nanosensor, which includes physiologically relevant H2O2 concentrations ranging from nanomolar to micromolar levels32,63, demonstrates the capability of AIE1035NPs@Mo/Cu-POM to identify abnormal fluctuations of H2O2 in living plants.
a Schematic of exogenous and endogenous H2O2 generation in plants. b, c Experimental protocol (b) and illustration of NIR microscope settings (c) for exogenous (1) and endogenous (2) H2O2 visualization. The illustrations of (a–c) were created in BioRender. Hu, H. (2025) https://BioRender.com/0g60kqh. d NIR fluorescence micrographs and surface plots of lettuce leaves treated with the nanosensor and different concentrations of H2O2. e NIR-II signal intensity of the lettuce leaves in (d) was illustrated using a boxplot (five independent plants). f NIR fluorescence micrographs, surface plots, and integrated fluorescence intensity profiles along the white arrow of the lettuce leaves with various treatments as indicated. g NIR-II signal intensity of the lettuce leaves in (f) was illustrated by a boxplot (four independent plants). The bars of the boxplot in (e) and (g) represent the minimum, the lower (25th) quartile, the median, the upper (75th) quartile, and the maximum values from bottom to top. Asterisks indicate values that are significantly different between the two groups being compared (ns > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001,****P < 0.0001). Statistically significant differences were determined using a one-way ANOVA test with Tukey’s HSD test. h NIR fluorescence images of AIE1035NPs@Mo/Cu-POM nanosensor response towards H2O2 induced by wounding in WT Arabidopsis thaliana and rbohD mutant of Arabidopsis thaliana. Time denotes time points after wounding. The yellow dashed line and red dashed line represent the site of wounding and sensor infiltration, respectively. i Time profiles of normalized fluorescence intensity for AIE1035NPs@Mo/Cu-POM throughout the experiment. The black arrow indicates the time of wounding. The red and black line plots represent the WT and mutant of Arabidopsis thaliana, with the center line denoting the mean value and the shaded regions indicating the standard deviation from three independent measurements.
By inflicting mechanical damage to the leaf to simulate abiotic stress, the fluorescence changes induced by endogenous H2O2 in the model were observed. After incubating with AIE1035NPs@Mo/Cu-POM for 1 h, the damage was inflicted on an intact lettuce leaf, resulting in an activated NIR-II signal that was approximately 4.35-fold greater than that of the MES group (Fig. 4f, g). Following pretreatment with catalase as a reactive oxygen species (ROS) scavenger, the NIR-II fluorescence signals in plant tissues exhibited negligible changes compared to the MES group. This finding indicates the effectiveness of the fluorescence “turn-on” strategy in visualizing endogenous H2O2. In addition, the nanosensors were utilized to image the wound-induced H2O2 wave signals in both Arabidopsis wild-type (WT) and mutant defective in H2O2 generation. Previous studies have shown that the ROS signaling pathway during abiotic and biotic stress in Arabidopsis primarily relies on the activation of the respiratory burst oxidase homolog D (RbohD), a nicotinamide adenine dinucleotide phosphate (NADPH) oxidase located on the plasma membrane16. We compared the fluorescence intensity between the WT (Col-0) and rbohD mutants after the infiltration of the nanosensors. Following injury, WT plants exhibited an approximately 1.5-fold increase in the normalized fluorescence intensity of the nanosensor, while the rbohD mutants displayed a negligible sensor response (Fig. 4h, i). These results indicate that the nanosensor can effectively visualize both exogenously and endogenously produced H2O2 in real-time through a fluorescence “turn-on” mechanism.
Real-time monitoring of H2O2 production in plants under stress
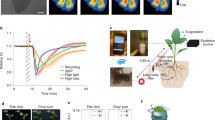
The applicability of the NIR-II fluorescence “turn-on” mode-based nanosensor was demonstrated for detecting mechanical damage responses in living plants (Fig. 5a–c). AIE1035NPs and AIE1035NPs@Mo/Cu-POM were infiltrated into two adjacent areas of a lettuce leaf, separated by the midrib. No signal response was triggered by either touching the leaf or by inserting the sensor into the leaf. The fluorescence intensity of AIE1035NPs@Mo/Cu-POM increased rapidly within minutes after injury, reaching a steady state within 20–30 min (Fig. 5d, e and Supplementary Fig. 36). In contrast, the fluorescence intensity of AIE1035NPs remained unaffected and constant throughout the experiment, serving as a reference sensor. When mechanical damage was inflicted multiple times at different locations on the leaf, the nanosensor exhibited characteristic time responses after each treatment, demonstrating the feasibility of the fluorescent probe for practical applications (Fig. 5f, g). In addition, the nanosensor can effectively monitor the stress status of plants under varying air humidity, soil moisture, and fertilization conditions, demonstrating its applicability to diverse greenhouse management strategies (Supplementary Figs. 37, 38).
a Schematic of in vivo visualization of stress-induced H2O2 by AIE1035NPs@Mo/Cu-POM nanosensor. b Schematic of the response of the nanosensor to various stresses in vivo. c Schematic of the experimental setup for in vivo detection. The illustrations of (a–c) were created in BioRender. Hu, H. (2025) https://BioRender.com/h4u7ond. d Brightfield image and false-colored NIR images of lettuce leaf in the intact plant infiltrated with AIE1035NPs and AIE1035NPs@Mo/Cu-POM. White arrows and the red dashed line indicate the location of AIE1035NPs and AIE1035NPs@Cu-POM, respectively. The yellow dashed line represents the location of touching and wounding. Time denotes the time points after wounding. Laser excitation: 808 nm, 15 mW. Filter: 900 nm long-pass filter. e Average time response of normalized AIE1035NPs@Mo/Cu-POM fluorescence intensity in the lettuce leaf. The black arrow represents the time point (10 min) of touching and wounding. Shaded regions represent standard error across five independent measurements. f Brightfield image and NIR images of lettuce leaf infiltrated with AIE1035NPs@Mo/Cu-POM after multiple woundings. The yellow dashed lines represent wounding locations, and the numbers represent the order of the wounding process. The red dashed lines indicate the location of AIE1035NPs@Mo/Cu-POM. Time denotes the time points after the first wounding. Damage caused every 30 min. g Average time response of normalized AIE1035NPs@Mo/Cu-POM fluorescence intensity during multiple woundings. Black, red, and blue arrows represent the time points of the first (10 min), second (40 min), and third (70 min) injuries, respectively. Shaded regions represent standard error across three independent measurements. h NIR fluorescence intensity changes of AIE1035NPs@Mo/Cu-POM in the leaf upon the stress of pathogen-associated peptide stress (flg22). Inset: NIR fluorescence images of the leaf from the representative period. Red and blue dots indicate the flg22-treated and no-stress groups, respectively, with the center dotted line representing the mean value and the shaded regions indicating the standard deviation of five independent measurements.
Next, we demonstrated that a nanosensor based on NIR-II fluorescence “turn-on” mode can also be utilized to monitor changes in endogenous H2O2 levels in plants subjected to various stresses. Compared to the non-stress control, the NIR emission intensity of AIE1035NPs@Mo/Cu-POM in the leaves significantly increased under high heat stress and the flg22 treatment (a model for studying microbial pathogen-induced responses64,65), exhibiting distinct H2O2 waveforms compared to those caused by mechanical damage (Fig. 5h and Supplementary Fig. 39). Three parameters, amplitude, lag time, and average velocity were defined to characterize the response of the nanosensor (Supplementary Fig. 40). To verify whether the wound-induced H2O2 signaling pathway in lettuce depends on the activation of RbohD and its relationship with the Ca2+ signaling pathway, lettuce leaves were treated with four different inhibitors and scavengers (Supplementary Fig. 41). The application of the NADPH oxidase inhibitor diphenyleneiodonium (DPI) and LaCl3 (a Ca2+ channel blocker) reduced the amplitude and average wave speed of the sensor response while increasing the lag time. These results indicate that the propagation of wound-induced H2O2 signals in lettuce requires the activation of NADPH oxidase and Ca2+ signaling, consistent with previous findings in Arabidopsis16,66.
Considering the species-independent design of the proposed nanosensor, we validated its applicability for stress detection in diverse plant species. In addition to lettuce and Arabidopsis, we demonstrated the utility of the nanosensor in monitoring wound-induced H2O2 signaling profiles in three other plants: spinach, pepper, and tobacco. Within minutes of wounding, the nanosensor in each species activated the “turn-on” mode, reflecting changes in NIR-II fluorescence intensity due to stress (Supplementary Fig. 42). The amount of H2O2 produced as a result of mechanical damage also exhibited different trends across the four species (Supplementary Fig. 43). The heterogeneity of the three previously defined parameters reflects the distinct conduction of H2O2 waves in different species, which may be related to their physiological structures or ecological adaptations. Plant immune signaling networks have evolved in response to their growth environments, explaining the differences in signal transmission mechanisms among species67,68.
Machine learning for stress identification and species differentiation
To evaluate the effectiveness of AIE1035NPs@Mo/Cu-POM in monitoring stress responses, controlled experiments were conducted on lettuce plants using four experimental conditions: no stress, flg22 treatment, heat stress, and mechanical wounding. Additionally, mechanical damage treatment was applied to different plants, including tobacco, lettuce, spinach, and pepper. The site of probe injection in each plant served as a fixed location for signal extraction, and the NIR-II signal was continuously recorded and automatically extracted for 1 h to generate the raw time series data. All signals were calibrated and normalized to ensure that the features extracted after data preprocessing were applicable to the overall dataset (Fig. 6a). Cellular homeostasis is characterized by a baseline level of ROS, which can be disrupted by various biotic and abiotic stresses, leading to ROS accumulation in different cellular compartments32. For each stress condition, the NIR-II signals of the nanosensors exhibited significant variation in response to each stressor (Fig. 6b). In the control experiment without stress, the fluorescence signal of the nanosensor remained relatively stable over the course of 1 h, as H2O2 was present at a lower concentration and was scavenged by a range of enzymatic and non-enzymatic antioxidants69. During the wounding experiments, the disruption of leaf tissue triggered a rapid accumulation of H2O2, leading to pronounced changes in the NIR-II fluorescence signal. Under heat stress, when membrane complexes involved in various electron transfer chains are compromised, H2O2 is produced in mitochondria and chloroplasts, subsequently accumulating in the cytoplasm and nucleus70. The signal changes observed in the nanosensor were consistent with immediate physiological responses of the plants. In the flg22-simulated pathogen stress experiments, H2O2 is primarily produced in the apoplast due to the activation of specific oxidases such as RBOHs, as well as in chloroplasts due to the disruption and imbalance of metabolic pathways71,72, which leads to an increase in the signal of the NIR-II nanosensor. During the wounding experiments across different plant species, each species showed distinct characteristic H2O2 signaling responses, which aligns with the heterogeneity of the characteristic parameters of the H2O2 signaling waves observed in the four species previously studied (Supplementary Fig. 42), indicating the diversity of the signaling networks utilized by these plant species. These results indicate that the proposed NIR-II fluorescent nanosensor can effectively monitor stress-induced H2O2 signaling in real time.
a Schematic of the ML architecture for signal preprocessing, feature extraction, supervised learning, stress identification, and species differentiation. b Heat maps of fluorescence changes in nanosensors monitoring different stress types and plant species over an hour. The intensity levels represent the changes in F/Fo of nanosensors in lettuce leaves under different stress conditions, including no stress, wound, heat, and flg22 treatment (top panel), as well as in response to mechanical damage across different plant species, including tobacco, lettuce, spinach, and pepper leaves (bottom panel). a Created or b partially created in BioRender. Hu, H. (2025) https://BioRender.com/jtldpq7. c, d t-distributed stochastic neighbor embedding plots for the no stress and stress datasets (c) and different species stress datasets (d). Visual clustering results showed the feature separation in two-dimensional space. e F1 scores (a metric of accuracy combining precision and recall) of different ML models for stress classification. f F1 scores of different ML models across an increasing number of ranges categorized by stress identification. The ranges of stress classification include 2 ranges (no stress/stress), 3 ranges (no stress/biotic stress/abiotic stress), and 4 ranges (no stress/flg22/wound/heat). g Confusion matrices of an XGBoost model displaying the classification accuracy for predicting each type of stress and different plant species.
The amount and rate of H2O2 production in plants vary under different stress conditions, yet data derived from manual processing often contain inaccuracies. Employing ML methods may offer a solution to this limitation, enabling more accurate monitoring of plant stress responses73,74. We developed a supervised ML model for NIR-II nanosensors to extract features and interpret H2O2 signaling in relation to stress type and plant species, thereby accelerating plant phenotyping. While ML techniques have been previously used for electrochemical75 and multimodal wearable sensors21 to monitor plant physiological information, there has yet to be a modeling approach that supports NIR-II fluorescent sensor for real-time monitoring of stress signals in plants. The stress data collected by the NIR-II imaging system are complex, consisting of continuously captured NIR-II images, which render traditional statistical methods inadequate. However, ML algorithms excel in such situations. They can model complex nonlinear relationships and interactions among variables, thereby enhancing the identification and understanding of potential associations between H2O2 signals and stress responses or plant species. Initially, the multidimensional feature space was projected into a two-dimensional space using the t-distributed stochastic neighbor embedding (t-SNE) visualization method, resulting in a scatter plot of the data features (Fig. 6c and Supplementary Fig. 44). t-SNE maps the high-dimensional data into a low-dimensional space while preserving the local similarities between data points, which can reveal the clustering structure of the data and facilitate the understanding of natural groupings within the dataset76,77. The data of unstressed and stressed conditions naturally formed different clusters, demonstrating the discriminative power of the features. Data from different species under stress formed partially overlapping clusters, suggesting that they may exhibit similar physiological response mechanisms when facing stress (Fig. 6d).
The eXtreme Gradient Boosting (XGBoost) algorithm was employed to develop three independent ML models: no stress/stress (2 ranges) model, categorization of biotic stress and abiotic stress (3 ranges) model, and identification of specific stress species (4 ranges) model. We trained and tested the models on lettuce plants across four experimental conditions: no stress, flg22, heat, and wound. The different ML models were evaluated using the F1 score, a metric of model accuracy that combines precision and recall, where a score of “1” represents perfect classification. The trained XGBoost model outperformed traditional ML models, including linear and radial basis function support vector machines (SVMs), logistic and ridge regression, and conventional decision trees (Fig. 6e, f and Supplementary Fig. 45a). Our XGBoost ML model achieves high accuracy, with an accuracy of 98.89% for stress detection, an accuracy more than 96.67% for categorization of biotic stress and abiotic stress, and an accuracy more than 96.67% for identification of specific stress species (Fig. 6g and Supplementary Figs. 45b, 46). We also collected a dataset of fluorescence signals from different plant species, including tobacco, lettuce, spinach, and pepper, subjected to mechanical damage, which was used for machine learning to differentiate H2O2 signal waves across the various species. The XGBoost ML model achieved over 93.33% recognition accuracy among these plant species (Fig. 6g). This ML analysis facilitates the rapid processing of real-time sensor data, is well-suited for large-scale screening efforts, and does not depend on subjective reporting, thereby providing a robust assessment tool for detecting plant health.
Discussion
This study represents an exploratory attempt to integrate an NIR-II fluorescent plant sensor with ML, yielding promising preliminary results in sensing performance and stress identification. We developed a NIR-II nanosensor based on the fluorescence “turn-on” mode for real-time, non-destructive monitoring of an important endogenous plant signaling molecule, H2O2, and demonstrated its application in both model and non-model plants. Compared to visible and NIR-I nanosensors, the designed NIR-II nanosensor effectively bypasses the strong fluorescent background signals emitted by plant leaves. This advancement allows for the direct monitoring of micromolar endogenous H2O2 in plant tissues from both microscopic and macroscopic perspectives without interference from background signals. The utility of this method was initially validated in leaves of Arabidopsis (Col-0) and the rbohD mutant. Subsequent studies were conducted on non-model plants, including lettuce, spinach, and pepper, as well as model plants such as tobacco, to demonstrate the universality of the fluorescence “turn-on” mode nanosensor. We showed that the NIR-II signal from the nanosensor encodes unique information regarding the intensity and velocity of plant signal waves. This information can be utilized to elucidate inter-species differences in signal wave propagation, thereby facilitating physiological studies of plant species that were previously inaccessible through transgenic approaches.
In addition, an XGBoost-based ML analysis framework was developed for stress detection and classification of multiple stressors. In recent years, the integration of ML with biosensors for plant disease detection, stress phenotyping, and predictive analysis has yielded significant results, owing to its remarkable advantages in data analysis and noise reduction. In this study, we employed three independent ML analyses: stress detection, categorization of biotic stress and abiotic stress, and identification of specific stressors. We demonstrated that the key physiological signals of plants in response to stress can be recognized through NIR-II imaging combined with ML models, achieving a classification accuracy of more than 96.67%. In addition, due to the variations in stress responses among species, we can also achieve efficient species classification through ML-powered NIR-II fluorescent sensing. This sensor platform, which does not rely on subjective reports, offers a highly robust sensing tool for various applications related to plant health monitoring and crop loss prevention.
In addition to the AIE fluorophores discussed in this study, several other remarkable luminophores, including lanthanide-doped nanocrystals57,78, near-infrared quantum dots79, and semiconducting polymers80,81, could also serve as potential candidates for in vivo fluorescence imaging of plants in the NIR-II window. To develop more advanced plant nanobionic sensors, it is essential to design components that are sensitive to plant signaling molecules. Creating hybrid nanosensors that respond to various endogenous signaling molecules in plants (e.g., phytohormones, reactive oxygen species, reactive sulfur species, inorganic ions, etc.) may be an effective strategy for understanding how plants respond to adverse environmental conditions. In addition to blade-based sensing, microneedle patches designed for stem applications provide direct access to vascular signals82,83, while nanosensors capable of root uptake facilitate stress detection in both hydroponic and soil environments84,85. These complementary approaches broaden the scope of plant stress sensing beyond leaf tissues, offering an integrated perspective on stress responses across different plant organs (Supplementary Table 4).
Considering that the NIR-II fluorescent nanosensor combined with the ML model enables rapid and accurate identification of plant stress, it presents new opportunities for agricultural and environmental monitoring. ML-based automated analysis methods can rapidly extract signals, analyze trends, and classify stress from large volumes of continuously captured images (tens of thousands), which far exceeds the capabilities of manual processing. Plants embedded with nanosensors serve as sentinels, facilitating the early, asymptomatic detection of biotic and abiotic stressors within a short timeframe (a few hours), thereby promoting targeted pest and disease management, as well as early stress response strategies to minimize crop losses. Furthermore, integrating nanosensor technology with environmental monitoring systems may facilitate the assessment of ecosystem health and the early detection of environmental degradation. For practical field applications, it is advisable to utilize portable and cost-effective electronic devices, such as Raspberry Pi, connected to small infrared cameras for imaging. However, ML models trained on datasets obtained under laboratory conditions may not be directly applicable to fluorescence signals collected by portable imaging devices, which typically exhibit lower sensitivity. Therefore, it is essential to train separate models on datasets sampled outdoor to enhance accuracy. Updated ML models must incorporate robust noise-filtering techniques to effectively distinguish stress signals from environmental noise. To facilitate field deployment, a conceptual workflow has been outlined to illustrate the transition from laboratory-based validation to real-world application (Supplementary Fig. 47). The relatively small number of experiments in the dataset for this study limits the scalability of the ML models and increases the potential for distortion due to outliers. Future research should focus on expanding the training dataset, extending sensor monitoring to encompass various stress conditions, and improving its applicability across a broader range of plant species. Given the potential feasibility of the approach demonstrated in this work, we envision that such ML-powered fluorescence “turn-on” mode nanosensors could be utilized to elucidate the complex signaling networks of plants under environmental stress and contribute to sustainable agriculture by enhancing plant phenotyping and improving management practices.
Methods
Synthesis of Mo-POM, Mo/Fe-POM, and Mo/Cu-POM
Mo-POM was prepared through a one-pot oxidation reaction. 0.2 g of Mo2C was dispersed in 1.5 mL of ultrapure water. Subsequently, 400 μL of H2O2 (30 wt.%) was added dropwise under vigorous stirring, and the reaction was allowed to proceed overnight. The Mo2C residue was removed by centrifugation at 2800×g. The supernatant was lyophilized to obtain Mo-POM powder. For the preparation of Mo/Fe-POM, 0.8032 g of (NH4)6Mo7O24·4H2O was dissolved in 10 mL of ultrapure water. Subsequently, 10 μL of FeCl3 (17.5 mg mL−1) was added under continuous stirring, while 2 mL of L-ascorbic acid solution (100 mg mL−1) was added into the above solution under vigorous stirring and the reaction was continued for 2 h. The reactants were centrifuged at 2800×g for 10 min to remove the precipitate. Mo/Fe-POM clusters were precipitated by adding 20 mL of ethanol, followed by collection through centrifugation. The clusters were washed three times with water and ethanol, and then dried in a lyophilizer. For the Mo/Cu-POM, 1.2 g of (NH4)6Mo7O24·4H2O and 0.05 g of CuCl2·2H2O were dissolved in 20 mL of ultrapure water. Under continuous stirring, 2 mL of L-ascorbic acid solution (100 mg mL−1) was added, stirring at room temperature for 2 h. The Mo/Cu-POM clusters were precipitated by adding 20 mL of ethanol, collected by centrifugation, and washed three times with water and ethanol. Finally, the samples were dried in a lyophilizer.
Fabrication of AIENPs@POM
AIENPs were obtained by embedding AIE dyes into the PS nanoparticles using the swelling method. A solution containing 2 mg of AIE475/AIE520/AIE712/AIE955/AIE1035, 50 mg of PS nanoparticles, and 15 mL of 1-methoxy-2-propanol was added to 5 mL of ultrapure water. The mixture was stirred at room temperature for 2 h and then washed three times with ultrapure water. Following the electrostatic adsorption method, the positively charged polyethyleneimine branched (PEI) polymer layers and the negatively charged POM were sequentially adsorbed onto the surface of the negatively charged AIENPs. 50 mg of AIENPs were dispersed in 10 mL of PEI aqueous solution (2 mg mL−1) under sonication for 30 min. The resulting product was washed three times with ultrapure water to remove uncoated PEI. Subsequently, 1 mg of PEI-coated AIENPs was mixed with 10 mL of Mo-POM, Mo/Fe-POM or Mo/Cu-POM solution (10 mg mL−1) for electrostatic coating for 12 h. Finally, the obtained AIENPs@POM solution was washed three times with ultrapure water and stored at 4 °C for later use.
Performance of AIE1035NPs@Mo/Cu-POM in response to H2O2
100 µL of AIE1035NPs@Mo/Cu-POM (0.5 mg mL−1) was added to 900 µL of different concentrations of H2O2 and incubated for 10 min. A four-sided quartz cuvette containing 1 mL of the reaction solution was placed in a holder and excited at a wavelength of 808 nm, with the laser power focused on the sample set at 15 mW. NIR fluorescence spectra were collected in the wavelength range of 850–1500 nm.
Selectivity of AIE1035NPs@Mo/Cu-POM in response to plant hormones and reactive oxygen species (ROS)
For the selectivity test of the nanosensor against the plant hormone analytes and molecules commonly found in plants, a 1 mM solution of each analyte was prepared in MES buffer (10 mM MES, 10 mM MgCl2, pH 5.7). 100 µL of AIE1035NPs@Mo/Cu-POM (0.5 mg mL−1) was reacted with 900 µL of the following solutions: methyl jasmonate (100 µM), salicylic acid (100 µM), glucose (100 µM), Ca2+ (100 µM), tannic acid (100 µM), adenosine 5′-triphosphate (100 µM), ascorbic acid (100 µM), and H2O2 (100 µM). After reacting for 10 min, the NIR fluorescence spectra of the mixture were recorded. For ROS screening, the same experimental setup was used. Hydroxyl radicals (•OH) were generated in situ by adding a 100 µM FeCl2 solution to 100 µM H2O2 through the Fenton reaction. Singlet oxygen (1O2) was generated in situ by the addition of 100 µM CuCl2 solution into 100 µM H2O2. Superoxide anions (O2•−) were produced by dissolving solid KO2 in dimethyl sulfoxide (DMSO). ROS solution was infiltrated into a lettuce leaf approximately 1 cm away from the nanosensor for the in vivo selectivity test.
Plant growth conditions
Arabidopsis thaliana, spinach, pepper, and tobacco seeds were cultivated in a 65 × 65 mm seeding tray. Once the first two leaves of the seedlings fully expanded, they were transplanted into pots containing the same soil mix. Lettuce seedlings were grown hydroponically using a 25% Hoagland nutrient solution. The plants were maintained in an artificial climate chamber under controlled conditions: a light intensity of 200 μmol m−2 s−1, a 12 h-light/12 h-dark photoperiod, temperature cycles of 22/18 °C (day/night), and 60% relative humidity.
Introduction of nanoparticles to plant leaves
Nanoparticles were diluted to a concentration of 0.1 mg mL−1 in MES buffer and infiltrated into plant leaves using a needleless 1 mL syringe. Gentle pressure was applied to the leaf during infiltration to prevent any mechanical damage. Infiltration continued until the periphery of the site was thoroughly saturated with the nanoparticle solution. Subsequently, any remaining nanoparticles on the surface of the leaf blade were washed away with water. The infiltrated leaves were then allowed to stand for 1 h before conducting in vivo measurements.
NIR-II fluorescence micro-imaging in vivo
For the localization of nanoparticles in plant leaves, AIE1035NPs were diluted in MES buffer and infiltrated into lettuce leaves. NIR measurements were conducted using an 808 nm laser to minimize the background fluorescence signal from chlorophyll. The data acquisition was performed using ×20 and ×50 objectives, with a laser power of 2 mW incident on the sample and an exposure time of 500 ms per point. A simple schematic diagram of the micro-imaging system is depicted in Fig. 4c.
The migration and diffusion of nanoparticles within live plant leaves were observed from a wider angle of view using a ×5 objective. AIE1035NPs were diluted in MES buffer and subsequently infiltrated into the lettuce leaf. Following infiltration, the lettuce was placed in a conical flask containing 300 mL of Hoagland nutrient solution, and the localization of nanoparticles in the plant was monitored for 6 h using a home-built NIR microscope.
For the visualization of exogenous H2O2, AIE1035NPs@ Mo/Cu-POM and H2O2 solutions with final concentrations of 5 μM, 10 μM, and 20 μM were diluted in MES buffer and infiltrated into the lettuce leaf. The leaf surface was subsequently washed with ultrapure water to remove any excess nanoparticles. Following a 30-min incubation, the leaves were gently washed with ultrapure water and then observed using an NIR microscope.
To visualize endogenous H2O2, AIE1035NPs@Mo/Cu-POM were diluted in MES buffer and infiltrated into a healthy lettuce leaf. The plant was then placed in a growth chamber for 30 min. Subsequently, the leaf was wounded 1 cm away from the infiltration point near the leaf tip. The production of endogenous H2O2 in the leaf was immediately observed using an NIR microscope. In another group of leaves, catalase (200 units mL−1) was pre-treated for 1 h before the wound treatment to inhibit the production of endogenous H2O2 in the cells.
NIR-II fluorescence macro imaging in vivo
After infiltration, the leaf was subjected to in vivo NIR-II fluorescence imaging under the excitation of an 808 nm laser. The NIR-II signals emitted by AIE1035NPs@Mo/Cu-POM within the leaf were collected through a NIR antireflection fixed-focus lens equipped with optical filters (FEL0900) and then subsequently detected by a liquid nitrogen-cooled 2D InGaAs camera (640 × 512 pixels). Fluorescence images were acquired using LightField imaging software and quantitatively analyzed with Image J software. Data analysis was conducted using OriginPro 2018 and GraphPad Prism 9.5.
Real-time monitoring of wound-induced H2O2 in plants
A plant infiltrated with AIE1035NPs@Mo/Cu-POM was placed 0.5 m in front of the camera. A 900 nm long-pass optical filter was positioned in front of the InGaAs detector. Entire living plants were selected for monitoring wound-induced H2O2 signals, with one plant used for each wounding experiment. NIR-II fluorescence signals from the nanosensors were monitored under 808 nm laser excitation (15 mW) with an exposure time of 500 ms. A mechanical wound was created on the leaf tip, 1 cm away from the nanosensor, using tweezers. To conduct multiple wounding experiments, wounds were made above, below, and to the left of the sensor point, as illustrated in Fig. 5f. Each wounding process was completed within 5 s.
Detection of H2O2 in plants under heat stress and elicitor application
Heat stress was induced by placing metal rods preheated in a water bath at 60 °C in contact with the tips of the lettuce leaves for 30 s. To simulate microbial pathogen stress, the flg22 peptide was used. 100 μL of flg22 peptide solution (10 μM) was applied to the surface of the lettuce leaves. The solution was allowed to diffuse into the leaves by creating small holes in the AIE1035NPs@Mo/Cu-POM penetration area using an ultrafine needle tip.
Machine learning-powered plant stress status assessment
For the signal preprocessing and feature extraction, the monitoring and data collection process was completed within 1 h. However, plants under stress conditions produce H2O2 rapidly, resulting in significant changes in the NIR-II signal. Images were continuously acquired using the NIR-II imaging system at a sampling frequency of T = 20 s. The site of probe injection in the plant served as the fixed signal extraction location, and the NIR-II signal was extracted for 1 h to create the raw time series data. Feature extraction was validated before ML analysis by projecting the multidimensional feature space into 2D space by means of t-distributed stochastic neighbor embedding (t-SNE). To obtain the time series segments, all raw data were averaged in batches and summarized for input into the plant stress identification model. The features of each sample were normalized prior to the machine learning (ML) pipeline to enhance the model’s generalizability. After data collection and analysis, the training and testing datasets were shuffled and divided in a 7:3 ratio, respectively.
Model selection for stress classification
All training models were constructed using Python (v.3.9) and were based on data collected from 120 lettuce samples subjected to four different stress treatments, as well as 120 samples of scratched tobacco, lettuce, spinach, and pepper, with 240 sets of 3600 s of NIR-II signal recordings. Several ML models were evaluated using F1 scores and confusion matrices, including linear and radial basis function support vector machines (SVMs), logistic regression, ridge regression, conventional decision trees, and trained boosting decision tree model, Extreme Gradient Boosting (XGBoost). The trained XGBoost model outperformed traditional ML models in both stress detection and stress type classification.
Statistics and reproducibility
The experiments performed in this study were independently repeated at least three times, yielding consistent results. Statistical analyses were performed using GraphPad Prism 9.5, and the data were presented as the mean ± s.d. (standard deviation). Microscope images and NIR-II images were obtained from three or more independent biological replicates and were presented as representative images in this work. Differences between the two groups were analyzed using an unpaired two-sided Student’s t-test. Multiple comparisons were conducted using a one-way analysis of variance (ANOVA) followed by Tukey’s test for post hoc analysis. No statistical method was used to predetermine sample size. No data were excluded from the analyses. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the findings of this study are available within the article and its supplementary files. Any additional requests for information can be directed to and will be fulfilled by the corresponding authors. Source data are provided with this paper.
References
Bailey-Serres, J., Parker, J. E., Ainsworth, E. A., Oldroyd, G. E. D. & Schroeder, J. I. Genetic strategies for improving crop yields. Nature 575, 109–118 (2019).
Lesk, C. et al. Stronger temperature-moisture couplings exacerbate the impact of climate warming on global crop yields. Nat. Food 2, 683–691 (2021).
Rezaei, E. E. et al. Climate change impacts on crop yields. Nat. Rev. Earth Environ. 4, 831–846 (2023).
Coatsworth, P., Gonzalez-Macia, L., Collins, A. S. P., Bozkurt, T. & Guder, F. Continuous monitoring of chemical signals in plants under stress. Nat. Rev. Chem. 7, 7–25 (2023).
Jiang, Z. et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 572, 341–346 (2019).
Munne-Bosch, S. & Villadangos, S. Cheap, cost-effective, and quick stress biomarkers for drought stress detection and monitoring in plants. Trends Plant Sci. 28, 527–536 (2023).
Sharma, P., Jha, A. B., Dubey, R. S. & Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 1–26 (2012).
Tian, Y. et al. Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the BRASSINAZOLE-RESISTANT1 transcription factor. Nat. Commun. 9, 1063 (2018).
Cerny, M., Habanova, H., Berka, M., Luklova, M. & Brzobohaty, B. Hydrogen peroxide: its role in plant biology and crosstalk with signalling networks. Int. J. Mol. Sci. 19, 2812 (2018).
Waszczak, C., Carmody, M. & Kangasjarvi, J. Reactive oxygen species in plant signaling. Annu. Rev. Plant. Biol. 69, 209–236 (2018).
Romero-Puertas, M. C. et al. Cadmium-induced subcellular accumulation of O2·− and H2O2 in pea leaves. Plant Cell Environ. 27, 1122–1134 (2004).
Orozco-Cardenas, M. & Ryan, C. A. Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc. Natl. Acad. Sci. USA 96, 6553–6557 (1999).
Mullineaux, P. M., Karpinski, S. & Baker, N. R. Spatial dependence for hydrogen peroxide-directed signaling in light-stressed plants. Plant. Physiol. 141, 346–350 (2006).
Nietzel, T. et al. The fluorescent protein sensor roGFP2-Orp1 monitors in vivo H2O2 and thiol redox integration and elucidates intracellular H2O2 dynamics during elicitor-induced oxidative burst in Arabidopsis. N. Phytol. 221, 1649–1664 (2019).
Exposito-Rodriguez, M., Laissue, P. P., Yvon-Durocher, G., Smirnoff, N. & Mullineaux, P. M. Photosynthesis-dependent H2O2 transfer from chloroplasts to nuclei provides a high-light signalling mechanism. Nat. Commun. 8, 49 (2017).
Miller, G. et al. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2, ra45 (2009).
Exposito-Rodriguez, M., Laissue, P. P., Littlejohn, G. R., Smirnoff, N. & Mullineaux, P. M. The use of HyPer to examine spatial and temporal changes in H2O2 in high light-exposed plants. Methods Enzymol. 527, 185–201 (2013).
Son, W. K. et al. In vivo surface-enhanced Raman scattering nanosensor for the real-time monitoring of multiple stress signalling molecules in plants. Nat. Nanotechnol. 18, 205–216 (2022).
Ang, M. C. et al. Decoding early stress signaling waves in living plants using nanosensor multiplexing. Nat. Commun. 15, 2943 (2024).
Lew, T. T. S. et al. Real-time detection of wound-induced H2O2 signalling waves in plants with optical nanosensors. Nat. Plants 6, 404–415 (2020).
Lee, G. et al. Abaxial leaf surface-mounted multimodal wearable sensor for continuous plant physiology monitoring. Sci. Adv. 9, eade2232 (2023).
Giraldo, J. P., Wu, H., Newkirk, G. M. & Kruss, S. Nanobiotechnology approaches for engineering smart plant sensors. Nat. Nanotechnol. 14, 541–553 (2019).
Zhang, M. et al. Nanomaterials: recent advances in plant disease diagnosis and treatment. Nano Today 57, 102326 (2024).
Wong, M. H. et al. Nitroaromatic detection and infrared communication from wild-type plants using plant nanobionics. Nat. Mater. 16, 264–272 (2017).
Lew, T. T. S., Park, M., Cui, J. & Strano, M. S. Plant nanobionic sensors for arsenic detection. Adv. Mater. 33, 2005683 (2021).
Yang, Y. et al. All-organic transparent plant e-skin for noninvasive phenotyping. Sci. Adv. 10, eadk7488 (2024).
Wang, S. et al. Sunflower-like self-sustainable plant-wearable sensing probe. Sci. Adv. 10, eads1136 (2024).
Armada-Moreira, A. et al. Plant electrophysiology with conformable organic electronics: deciphering the propagation of Venus flytrap action potentials. Sci. Adv. 9, eadh4443 (2023).
Castro, B. et al. Stress-induced reactive oxygen species compartmentalization, perception and signalling. Nat. Plants 7, 403–412 (2021).
Fichman, Y., Miller, G. & Mittler, R. Whole-plant live imaging of reactive oxygen species. Mol. Plant 12, 1203–1210 (2019).
Kim, M. J., Ciani, S. & Schachtman, D. P. A peroxidase contributes to ROS production during Arabidopsis root response to potassium deficiency. Mol. Plant 3, 420–427 (2010).
Mittler, R., Zandalinas, S. I., Fichman, Y. & Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell. Biol. 23, 663–679 (2022).
Rodrigues, O. & Shan, L. Stomata in a state of emergency: H2O2 is the target locked. Trends Plant Sci. 27, 274–286 (2022).
Van Breusegem, F., Bailey-Serres, J. & Mittler, R. Unraveling the tapestry of networks involving reactive oxygen species in plants. Plant. Physiol. 147, 978–984 (2008).
Zou, J. J. et al. Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. Plant Cell 27, 1445–1460 (2015).
Saxena, I., Srikanth, S. & Chen, Z. Cross talk between H2O2 and interacting signal molecules under plant stress response. Front. Plant Sci. 7, 570 (2016).
Qiu, Q. et al. Second near-infrared fluorescent metal-organic framework sensors for in vivo extracellular adenosine triphosphate monitoring. Biosens. Bioelectron. 251, 116114 (2024).
Mei, J. et al. Aggregation-induced emission: the whole is more brilliant than the parts. Adv. Mater. 26, 5429–5479 (2014).
Mei, J., Leung, N. L., Kwok, R. T., Lam, J. W. & Tang, B. Z. Aggregation-induced emission: together we shine, united we soar! Chem. Rev. 115, 11718–11940 (2015).
Kwok, R. T., Leung, C. W., Lam, J. W. & Tang, B. Z. Biosensing by luminogens with aggregation-induced emission characteristics. Chem. Soc. Rev. 44, 4228–4238 (2015).
Li, H. et al. Activity-based NIR enzyme fluorescent probes for the diagnosis of tumors and image-guided surgery. Angew. Chem. Int. Ed. 60, 17268–17289 (2021).
Lucero, M. Y. et al. Development of NIR-II photoacoustic probes tailored for deep-tissue sensing of nitric oxide. J. Am. Chem. Soc. 143, 7196–7202 (2021).
Jiang, Y. & Pu, K. Molecular probes for autofluorescence-free optical imaging. Chem. Rev. 121, 13086–13131 (2021).
Zhang, J. et al. Stimuli-responsive AIEgens. Adv. Mater. 33, 2008071 (2021).
Zhao, M. et al. A tumor-microenvironment-responsive lanthanide-cyanine FRET sensor for NIR-II luminescence-lifetime in situ imaging of hepatocellular carcinoma. Adv. Mater. 32, 2001172 (2020).
Mei, J. et al. Biofilm microenvironment-responsive self-assembly nanoreactors for all-stage biofilm associated infection through bacterial cuproptosis-like death and macrophage re-rousing. Adv. Mater. 35, 2303432 (2023).
Shi, Y. et al. Fe-doped polyoxometalate as acid-aggregated nanoplatform for NIR-II photothermal-enhanced chemodynamic therapy. Adv. Healthc. Mater. 9, e2000005 (2020).
Zhao, H. et al. A polyoxometalate-based pathologically activated assay for efficient bioorthogonal catalytic selective therapy. Angew. Chem. Int. Ed. 62, e202303989 (2023).
Xu, W., Wang, D. & Tang, B. Z. NIR-II AIEgens: a win-win integration towards bioapplications. Angew. Chem. Int. Ed. 60, 7476–7487 (2021).
Ni, J. S. et al. Red/NIR-emissive benzo[d]imidazole-cored AIEgens: facile molecular design for wavelength extending and in vivo tumor metabolic imaging. Adv. Mater. 30, 1805220 (2018).
Liu, S. et al. Molecular motion in aggregates: manipulating TICT for boosting photothermal theranostics. J. Am. Chem. Soc. 141, 5359–5368 (2019).
Li, Y. et al. Design of AIEgens for near-infrared IIb imaging through structural modulation at molecular and morphological levels. Nat. Commun. 11, 1255 (2020).
Cheng, H., Kamegawa, T., Mori, K. & Yamashita, H. Surfactant-free nonaqueous synthesis of plasmonic molybdenum oxide nanosheets with enhanced catalytic activity for hydrogen generation from ammonia borane under visible light. Angew. Chem. Int. Ed. 53, 2910–2914 (2014).
Zhang, C. et al. A polyoxometalate cluster paradigm with self-adaptive electronic structure for acidity/reducibility-specific photothermal conversion. J. Am. Chem. Soc. 138, 8156–8164 (2016).
Gumerova, N. I. & Rompel, A. Synthesis, structures and applications of electron-rich polyoxometalates. Nat. Rev. Chem. 2, 0112 (2018).
Weinstock, I. A., Schreiber, R. E. & Neumann, R. Dioxygen in polyoxometalate mediated reactions. Chem. Rev. 118, 2680–2717 (2017).
Zhan, Y. et al. Activatable NIR-II lanthanides-polymetallic oxomolybdate hybrid nanosensors for monitoring chemotherapy induced enteritis. Adv. Funct. Mater. 33, 2301683 (2023).
Kwak, S. Y. et al. A nanobionic light-emitting plant. Nano Lett. 17, 7951–7961 (2017).
Lew, T. T. S. et al. Rational design principles for the transport and subcellular distribution of nanomaterials into plant protoplasts. Small 14, 1802086 (2018).
Gorbe, E. & Calatayud, A. Applications of chlorophyll fluorescence imaging technique in horticultural research: a review. Sci. Hortic. 138, 24–35 (2012).
Rolfe, S. A. & Scholes, J. D. Chlorophyll fluorescence imaging of plant-pathogen interactions. Protoplasma 247, 163–175 (2010).
Yan, Y. et al. Metal-organic framework-based biosensor for detecting hydrogen peroxide in plants through color-to-thermal signal conversion. ACS Nano 16, 15175–15187 (2022).
Allan, A. C. & Fluhr, R. Two distinct sources of elicited reactive oxygen species in tobacco epidermal cells. Plant Cell 9, 1559–1572 (1997).
Albert, M. Peptides as triggers of plant defence. J. Exp. Bot. 64, 5269–5279 (2013).
Felix, G., Duran, J. D., Volko, S. & Boller, T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276 (1999).
Monshausen, G. B., Bibikova, T. N., Weisenseel, M. H. & Gilroy, S. Ca2+ regulates reactive oxygen species production and pH during mechanosensing in Arabidopsis roots. Plant Cell 21, 2341–2356 (2009).
Thaler, J. S., Humphrey, P. T. & Whiteman, N. K. Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17, 260–270 (2012).
Akpinar, B. A., Avsar, B., Lucas, S. J. & Budak, H. Plant abiotic stress signaling. Plant Signal. Behav. 7, 1450–1455 (2012).
Sies, H. & Jones, D. P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 21, 363–383 (2020).
Schwarzländer, M., Fricker, M. D. & Sweetlove, L. J. Monitoring the in vivo redox state of plant mitochondria: effect of respiratory inhibitors, abiotic stress and assessment of recovery from oxidative challenge. Biochim. Biophys. Acta Bioenerg. 1787, 468–475 (2009).
Kámán-Tóth, E. et al. Contribution of cell wall peroxidase- and NADPH oxidase-derived reactive oxygen species to Alternaria brassicicola-induced oxidative burst in Arabidopsis. Mol. Plant Pathol. 20, 485–499 (2019).
Torres, M. A., Dangl, J. L. & Jones, J. D. G. Arabidopsis gp91 phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99, 517–522 (2001).
Singh, A., Ganapathysubramanian, B., Singh, A. K. & Sarkar, S. Machine learning for high-throughput stress phenotyping in plants. Trends Plant Sci. 21, 110–124 (2016).
Khait, I. et al. Sounds emitted by plants under stress are airborne and informative. Cell 186, 1328–1336.e1310 (2023).
Coatsworth, P. et al. Time-resolved chemical monitoring of whole plant roots with printed electrochemical sensors and machine learning. Sci. Adv. 10, eadj6315 (2024).
Liu, C. et al. A self-powered dual ratchet angle sensing system for digital twins and smart healthcare. Adv. Funct. Mater. 34, 2405104 (2024).
Maaten, L. V. D. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Yang, Y., Jiang, Q. & Zhang, F. Nanocrystals for deep-tissue in vivo luminescence imaging in the near-infrared region. Chem. Rev. 124, 554–628 (2024).
Ding, C., Huang, Y., Shen, Z. & Chen, X. Synthesis and bioapplications of Ag2S quantum dots with near-infrared fluorescence. Adv. Mater. 33, 2007768 (2021).
Li, J. & Pu, K. Development of organic semiconducting materials for deep-tissue optical imaging, phototherapy and photoactivation. Chem. Soc. Rev. 48, 38–71 (2019).
Zhang, S. et al. Self-illuminating NIR-II chemiluminescence nanosensor for in vivo tracking H2O2 fluctuation. Adv. Sci. 10, e2207651 (2023).
Wang, S. et al. Chromatic covalent organic frameworks enabling in-vivo chemical tomography. Nat. Commun. 15, 2943 (2024).
Wu, X. et al. Rapid and in-field sensing of hydrogen peroxide in plant by hydrogel microneedle patch. Small 20, 2402024 (2024).
Zeng, C., Long, Y., Tan, Y., Zeng, F. & Wu, S. Water-dispersible activatable nanoprobe for detecting cadmium-ion-induced oxidative stress in edible crops via near-infrared second-window fluorescence imaging. Anal. Chem. 94, 14021–14028 (2022).
Long, Y., Chen, J., Zeng, F. & Wu, S. An activatable NIR-II fluorescent probe for tracking heavy-metal ion and high-level salt-induced oxidative stress in plant sprouts. Aggregate 4, e288 (2022).
Acknowledgements
This research was supported by the National Natural Science Foundation of China (No. 32022059) awarded to Y.W., National Natural Science Foundation of China (No. 323B2053) awarded to H.H. and the Natural Science Foundation of Zhejiang Province (No. LZ23C130005) awarded to Y.W.
Author information
Authors and Affiliations
Contributions
H.H., Y.Y. and Y.W. designed the project and experiments. H.H., H.Y. and Z.W. synthesized and characterized the nanoparticles. H.H., H.Y. and S.S. fabricated the devices. H.H. and H.Y. carried out the measurements and analyzed the data. J.F., S.S. and Y.L. assisted with the experiments. H.H. wrote the original manuscript. H.H. and Y.W. wrote and reviewed the final version of the text. All the authors reviewed and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Haitao Sun and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hu, H., Yuan, H., Sun, S. et al. Machine learning-powered activatable NIR-II fluorescent nanosensor for in vivo monitoring of plant stress responses. Nat Commun 16, 5114 (2025). https://doi.org/10.1038/s41467-025-60182-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-60182-w