Abstract
The Escherichia coli helicase, YoaA, and DNA polymerase III subunit, χ, form a complex (YoaA-χ) that promotes tolerance to the DNA chain-terminator 3ˈ-azidothymidine (AZT). Single-stranded DNA binding protein (SSB), which accumulates at stalled replication forks, also contributes to AZT tolerance through interactions with χ. Here we show that in vitro, χ mediates interactions between YoaA and SSB that modulate helicase activity in a substrate-specific manner with little effect on overhang DNA but inhibiting unwinding of forked DNA. SSB similarly affects the activity of the YoaA paralog, DinG. Single-molecule experiments show that SSB translocates with YoaA-χ, increasing both the lifetime and frequency of SSB binding events. Mutational analyses show that χ binds at the back of YoaA relative to the direction of translocation supporting a model in which YoaA-χ pulls SSB along DNA as it translocates. To our knowledge, this is the first demonstration of a mechanoenzyme pulling SSB along ssDNA.
Similar content being viewed by others
Introduction
XPD/Rad3-like helicases are critical enzymes for preserving genomic integrity across various organisms1,2,3,4,5. These superfamily 2 helicases translocate 5ˈ to 3ˈ along ssDNA and unwind DNA/DNA and DNA/RNA duplexes in an ATP-dependent manner2,6,7,8,9,10,11. The two Escherichia coli XPD/Rad3-like helicases are DinG and YoaA. In vitro, YoaA binds χ, a subunit of the DNA polymerase III holoenzyme (pol III HE), to form the active helicase complex YoaA-χ, while DinG functions on its own11,12. The expression of DinG and YoaA is DNA damage-inducible, indicating DinG and YoaA-χ function in DNA damage repair13,14,15,16. DinG has been implicated in resolving replication and transcription conflicts and unwinds R-loops, D-loops, and G-quadruplexes in vitro9,17,18,19, whereas YoaA is less well-characterized. YoaA and χ are both required in E. coli for promoting tolerance to azidothymidine (AZT), a replication chain-terminating agent, and for resisting methyl methanesulfonate (MMS) damage in cells lacking AP endonuclease activity20,21.
To better understand how YoaA-χ is involved in DNA damage repair, the interactions between YoaA-χ and single-stranded DNA-binding protein (SSB) were investigated. SSB binds ssDNA within the cell to protect it from degradation and prevent the formation of secondary structures (reviewed in refs. 22,23). SSB also interacts with and coordinates other genomic maintenance proteins, including helicases (reviewed in refs. 23,24). While direct interactions between YoaA and SSB have not been identified, the interactions between SSB and χ are well-studied. Chi, though catalytically inactive, plays a crucial role in replication by mediating interactions between pol III HE and SSB25,26. This promotes stability of the pol III HE during replication27,28,29. Chi is bound to pol III HE via ψ. The last four C-terminal residues of the SSB tail have been crystallized with χψ, identifying SSB F177 and χ R128 as critical residues for SSB-χ binding30. The importance of χ R128 for binding SSB has also been shown by biochemical studies, with the Kd between χ R128A and SSB being too weak to measure compared to wild-type (wt) χψ binding SSB with a Kd of approximately 9 μM30,31.
The functional significance of YoaA-χ-SSB interactions are of interest because AZT-treated cells form single-stranded (ss) gaps and SSB foci32. Importantly, χ-SSB interactions are required for χ to promote AZT tolerance, as the χ R128A mutation, which disrupts SSB binding, diminishes AZT tolerance20. While not the focus of this study, it should be noted that current research has not yet established if the χ-SSB interaction required for AZT tolerance is that of χ complexed with the pol III HE or with YoaA.
We present here that χ and SSB interact functionally when χ is in complex with YoaA. YoaA-χ-SSB interactions are compared with DinG-SSB interactions due to the previously established interaction between DinG and SSB33. Both helicases are inhibited by SSB on forked DNA but are indifferent to the presence of SSB on overhang DNA. We also discovered via single-molecule experiments that SSB translocates along ssDNA with YoaA-χ and present evidence of a model where YoaA-χ pulls SSB along DNA like a caboose on a train.
Results
SSB has a substrate-specific effect on the DNA unwinding activity of YoaA-χ
To determine if SSB affects the DNA unwinding of YoaA-χ, helicase activity was measured in the presence and absence of SSB using a FRET-based assay11. Briefly, a 20-bp duplexed DNA substrate with a 5ˈ 65-nucleotide (nt) ssDNA overhang was labeled with a Cy3 donor and Cy5 acceptor on the 3ˈ and 5ˈ strands of the blunt end, respectively (Supplementary Table 3, O1). The increase in Cy3 fluorescence with time correlates to the amount of DNA unwound. The sequence of the 5ˈ ssDNA overhang consisted of dTs (T65) to reduce the possibility of DNA secondary structure in the absence of SSB. SSB (75 nM) was added to DNA (50 nM) before the addition of the YoaA-χ helicase (100 nM). SSB gave a small but reproducible increase in the rate of unwinding on this overhang substrate (Fig. 1A, G, green striped bar). A noncomplementary 10-nt 3ˈss DNA overhang was added to this substrate to create a fork substrate (forked DNA) with both 5ˈ and 3ˈ overhangs (Supplementary Table 3, F1). As shown previously, YoaA-χ unwound the forked substrate (F1) faster than the overhang substrate (O1) in the absence of SSB (Fig. 1A, B, black traces)11. In contrast to the 5ˈ overhang substrate, addition of SSB to the forked substrate inhibited unwinding by YoaA-χ (Fig. 1B, G, solid green bar).
Cy5 (red circle) and Cy3 (yellow circle) were attached to the end of a 20-bp duplex DNA containing a 65-nt 5ˈ overhang with and without a 10-nt 3ˈ overhang to monitor DNA unwinding. A Time courses of 5ˈ overhang DNA duplex (50 nM, O1) unwound by YoaA-χ (100 nM) without SSB present (black) and with SSB (75 nM) pre-bound to the DNA (green). B Time courses of forked DNA duplex (50 nM, F1) with a 3ˈ overhang unwound by YoaA-χ (100 nM) without SSB present (black) and with SSB (75 nM) pre-bound to the DNA (green). C Time courses of 5ˈ overhang DNA duplex (50 nM, O1) unwound by DinG (5 nM) without SSB present (black) and with SSB (75 nM) pre-bound to the DNA (green). D Time courses of forked DNA duplex (50 nM, F1) with a 3ˈ overhang unwound by DinG (5 nM) without SSB present (black) and with SSB (75 nM) pre-bound to the DNA (green). E Time courses of 5ˈ overhang DNA duplex (O2) unwound by YoaA-χ (100 nM) so as to form an in situ forked DNA structure, without SSB present (black) and with SSB (75 nM) pre-bound to the DNA (green). F Time courses of pre-formed forked DNA duplex (50 nM, F2) matching the substrate sequence in panel E with a 3ˈ overhang unwound by YoaA-χ (5 nM) without SSB present (black) and with SSB (75 nM) pre-bound to the DNA (green). G Quantification of observed rates for each condition shown in panels (A–F) obtained by fitting each unwinding curve with an exponential model, showing averages of three reactions with standard deviations. Reactions with and without SSB conditions were performed side-by-side in the same experiment. Three replicates of unwinding reactions as a function of helicase concentration are shown in Supplementary Figs. 1 and 2. Source data are provided as a source data file.
To determine if SSB affects the activity of both E. coli Rad3/XPD helicases in the same way, these FRET-based helicase experiments were performed with DinG. Because DinG unwinds these substrates faster than YoaA-χ, a lower concentration of DinG (5 nM) was used. As with YoaA-χ, SSB did not have a large effect on the unwinding rate of DinG on the overhang substrate (O1) (Fig. 1C, G, striped green bar). On the forked substrate (F1), DinG was also inhibited by the addition of SSB (Fig. 1D, G, solid green bar). Due to the faster unwinding rates of DinG even at low concentrations, the error in measured reaction rates was greater compared to YoaA-χ. A range of concentrations were tested for YoaA-χ (50, 20, and 10 nM) and DinG (10, 5, and 2 nM) on the overhang and forked substrates with and without SSB and showed the same phenomena (Supplementary Figs. 1 and 2).
To further define the effects of SSB on unwinding activity, YoaA-χ unwinding was measured on an overhang substrate with a 30-bp duplex (Supplementary Table 3, O2). The rationale is that this substrate will form a 20-bp duplex with a 10-nt 3ˈ ssDNA overhang in situ when partially unwound, similar to the substrate used in Fig. 1B and D. SSB stimulated unwinding activity on this overhang substrate by a factor of approximately 2 (Fig. 1E, G, striped green bar). Because the sequence of the duplex was different from the initial DNA substrates, DNA unwinding was measured on a pre-formed fork DNA substrate with a 20-bp duplex of the same sequence (Supplementary Table 3, F2) as in Fig. 1E. As before, unwinding by YoaA-χ was inhibited by SSB on the pre-formed forked substrate (Fig. 1F, G, solid green bar).
Effects of SSB on helicase activity are dependent on the lengths of the 3′ and 5′ ssDNA overhangs
To determine why SSB inhibits DNA unwinding by YoaA-χ on DNA substrates with pre-formed forks, the lengths of both the 5ˈ and 3ˈ ssDNA overhangs were varied. On DNA substrates with a 20-bp duplex and 5ˈ ssDNA overhangs of 20−, 35−, and 65−nt, identified as T20, T35, and T65, respectively (Supplementary Table 3, O1, O3, and O4), rates of DNA unwinding by YoaA-χ were similar when no SSB was present (Fig. 2A, black traces in upper panels and 2B, striped black bars). When a 3ˈ ssDNA overhang of 10-nt was added (Supplementary Table 3, F1, F3, and F4), rates of unwinding for these forked substrates were faster than for the overhang substrates in all cases (Fig. 2A, black traces and 2B, solid black bars). When SSB was added to the overhang substrates, there was little to no effect on unwinding rates for the T20 and T35 overhang substrates. The unwinding rate for T65 increased approximately 2-fold (Fig. 2A, green traces in upper panels and 2B, striped green bars). In contrast, the addition of SSB inhibited unwinding on all three forked substrates with a larger effect on the shorter T20 (8.6-fold) and T35 (11.9-fold) substrates than T65 (3.9-fold) (Fig. 2A, green traces in lower panels and 2B, solid green bars).
Cy5 (red circle) and Cy3 (yellow circle) were attached to the end of a 20-bp duplex DNA containing variable lengths of 5ˈ overhangs with and without a 10-nt 3ˈ overhang to monitor DNA unwinding. A Time courses of DNA substrates (50 nM) with varying 5ˈ overhang lengths both with (F1, F3, and F4) and without (O1, O3, O4) a 10-nt 3ˈ overhang unwound by YoaA-χ (100 nM) both with SSB (green) and without SSB (black). B Quantification of observed rates for each of the 5ˈ overhang substrates shown in panel (A) obtained by fitting each unwinding curve with an exponential model, showing averages of three reactions with standard deviations. C Fluorescein located 6-nt back from the junction of a DNA substrate with varying 5ˈ overhang lengths and without a 3ˈ overhang (50 nM, O5, O6, and O7) is quenched upon addition of SSB, normalized to unquenched spectra shown in black. D Fluorescein located 6-nt back from the junction of a DNA substrate with varying 5ˈ overhang lengths and with a 10-nt 3ˈ overhang (50 nM, F5, F6, and F7) is quenched upon addition of SSB, normalized to unquenched spectra shown in black. E Quantification of quenches observed in panels (C, D) showing average values of three experiments with standard deviations. F Using the same DNA unwinding assay as in panel (A), DNA with a 65-nt 5ˈ overhang and varying 3ˈ overhang lengths (50 nM, O1, F1, F8, and F9) was unwound by YoaA-χ (100 nM) without SSB. G Using the same DNA unwinding assay as in panel (A), DNA with 65-nt 5ˈ overhang and varying 3ˈ overhang lengths (50 nM, O1, F1, F8, and F9) was unwound by YoaA-χ (100 nM) with SSB (75 nM). H Quantification of observed unwinding rates for each of the 3ˈ overhang substrates in panels (F) (without SSB, black) and (G). (with SSB, green), obtained by fitting each unwinding curve with an exponential model, showing averages of three experiments with standard deviations. Source data are provided as a source data file.
SSB-DNA binding is well-characterized, and SSB binds the T65 substrates stoichiometrically34,35. SSB binding to the shorter T20 and T35 substrates was measured to verify that inhibition of DNA unwinding was due to SSB binding forked DNA. An established fluorescence quench assay was used to measure SSB binding to DNA, in which physical interactions with SSB quench the intensity of fluorescein36. A fluorescein was placed within the 20-bp duplex 6-nt from the ds/ss DNA junction. The fluorescence was quenched when 75 nM SSB was added to 50 nM DNA for the forked substrates (Supplementary Table 3, F5, F6, and F7) with T20 and T35 overhangs, indicating that SSB binds these substrates (Fig. 2D). The quench in fluorescence was not as pronounced for the overhang substrates (Supplementary Table 3, O5, O6, and O7) of the same length (Fig. 2C), suggesting that SSB may bind these short substrates with higher affinity when a fork is present (Fig. 2E).
Forked DNA substrates with 3ˈ ssDNA overhangs of 3-, 7-, or 10-nt along with a 5ˈ ssDNA overhang of 65 nt (Supplementary Table 3, F1, F8, and F9) were used to determine how the length of the 3ˈ ssDNA overhang affects helicase activity. In the absence of SSB, unwinding rates for these forked substrates were similar to one another and were faster than for the overhang substrate (Fig. 2F). In reactions with SSB, unwinding rates decreased by a factor of 6.0 and 14.1 for the 7- and 10-nt 3ˈ arms, respectively, relative to the overhang substrate, whereas there was a smaller decrease (a factor of 1.4) for the 3-nt overhang (Fig. 2G, H, green bars). Together, results from varying the lengths of both the 5ˈ and 3ˈ ssDNA overhangs suggest that SSB may show a preference for binding adjacent to pre-formed forks compared with 3ˈ recessed ends to hinder unwinding forks more than overhangs.
A physical interaction between YoaA-χ and SSB is necessary for YoaA-χ to unwind SSB-bound DNA
By using their motor activity to translocate along DNA, DNA helicases can move other proteins on DNA such as SSB37,38, however, inhibition on pre-formed fork substrates suggests that YoaA-χ has difficulty moving SSB. To determine whether χ-SSB interactions are important for efficient unwinding activity by YoaA-χ on DNA bound by SSB, amino acid residues that participate in protein-protein interactions were mutated, and helicase activity was measured on the overhang substrate (O1). Arg-128 in χ was substituted with Ala (YoaA-χ R128A), and the C-terminal Phe residue (F177) was deleted from SSB (SSB ΔC1) in separate experiments30,39. Unwinding activity by YoaA-χ R128A alone was comparable to wt YoaA-χ in the absence of SSB, indicating the mutation in χ did not disrupt the helicase activity (Fig. 3A). In helicase assays with SSB, both the χ-R128A substitution (Fig. 3B) and the F177 deletion (Fig. 3C) decreased DNA unwinding rates to barely detectable levels. DinG was still able to unwind DNA in reactions with SSB ΔC1, but at a slower rate (Fig. 3D). Interestingly, in reactions with DinG, an increase in Cy3 fluorescence due to unwinding was followed by a slower decrease in fluorescence, suggesting DNA was slowly reannealing. Native gel electrophoresis confirmed the overhang substrate (Supplementary Table 3, O8) was slowly reannealed in reactions with DinG and SSB ΔC1 (Supplementary Fig. 3). The SSB binding site on DinG has not yet been identified, so the reciprocal experiment was not done.
Representative time courses for DNA unwinding of 65-nt 5ˈ overhang DNA (50 nM, O1) are shown for reactions with. A no SSB and 2, 5, 10, 20, and 50 nM of YoaA-χ R128A. B wt SSB (75 nM) prebound to DNA and 2, 5, 10, 20, and 50 nM of YoaA-χ R128A. C SSB ΔC1 (75 nM) prebound to DNA and 2, 5, 10, 20, and 50 nM of YoaA-χ. D SSB ΔC1 (75 nM) prebound to DNA and 2, 5, 10, 20, and 50 nM of DinG. All experiments were performed in triplicate. Darkness in blue corresponds to increasing concentrations of helicase. On the DNA schematic, the red circle indicates Cy5, the yellow circle indicates Cy3, YoaA-χ is denoted in blue, SSB in green, DinG in orange, and pink stars represent protein mutations. Source data are provided as a source data file.
SSB-χ interactions support concurrent binding of YoaA-χ and SSB to ssDNA
Physical interactions between χ and SSB are needed for efficient DNA unwinding by YoaA-χ (Fig. 3). To determine whether this is due to protein-protein interactions allowing both the helicase and SSB to bind DNA at the same time, or because χ-SSB interactions enable YoaA-χ to displace SSB from DNA, protein-DNA binding was measured in electrophoretic mobility shift assays (EMSAs). A Cy5-labeled 65-nt single-stranded dT DNA substrate (ssT65) was used to match the 5ˈssDNA overhang of the FRET substrates (Supplementary Table 2, S21). The nonhydrolysable ATP analog, ATPγS, was used to promote helicase-DNA binding but block translocation. When increasing concentrations of YoaA-χ (50 nM to 400 nM) were added to ssT65 (50 nM), helicase binding retarded migration of DNA and produced multiple bands that migrated different distances, indicating that more than one helicase molecule can bind ssT65 DNA (Fig. 4A). The total percentage of bound DNA as well as the percentage of DNA bound by multiple helicases increased with YoaA-χ concentration. A parallel experiment with SSB bound to DNA showed that SSB caused DNA to migrate more slowly and addition of YoaA-χ caused a supershift with a different banding pattern than YoaA-χ alone, indicating that both SSB and YoaA-χ bound DNA (Fig. 4A). The percentage of DNA bound by YoaA-χ, regardless of the number of helicases, with and without SSB was quantified and plotted (Fig. 4A). Similar results were obtained for DinG with multiple DinG molecules binding ssT65 in the absence of SSB, and supershift indicating that one DinG molecule likely bound in the presence of SSB (Fig. 4B). These results indicate that both SSB and the helicases can bind DNA with a 65-nt ssDNA overhang at the same time.
A EMSA of Cy5-labeled dT65 oligo (50 nM, S21) titrated with YoaA-χ (50, 100, 200, and 400 nM) with (right panel) and without (left panel) SSB (75 nM) pre-bound to the DNA. Quantification of percent of DNA bound by YoaA-χ in the absence (black) and presence (green) of SSB versus YoaA-χ concentration. B EMSA of Cy5-labeled dT65 oligo (50 nM, S21) titrated with DinG (50, 100, 200, and 400 nM) with (right panel) and without (left panel) SSB (75 nM) pre-bound to the DNA. Quantification of percent of DNA bound by DinG in in the absence (black) and presence (green) of SSB versus DinG concentration. C EMSA of Cy5-labeled dT65 oligo (50 nM, S21) titrated with YoaA-χ (50, 100, 200, and 400 nM) with (right panel) and without (left panel) SSB ΔC1 (75 nM) pre-bound to the DNA. Quantification of percent of DNA bound by YoaA-χ in the absence (black) and presence (pink) of SSB ΔC1 versus YoaA-χ concentration. D EMSA of Cy5-labeled dT65 oligo (50 nM, S21) titrated with YoaA-χ R128A (50, 100, 200, and 400 nM) with (right panel) and without (left panel) SSB (75 nM) pre-bound to the DNA. Quantification of percent of DNA bound by YoaA-χ R128A in the absence (black) and presence (green) of SSB versus YoaA-χ R128A concentration. E EMSA of Cy5-labeled dT65 oligo (50 nM, S21) titrated with DinG (50, 100, 200, and 400 nM) with (right panel) and without (left panel) SSB ΔC1 (75 nM) pre-bound to the DNA. Quantification of percent of DNA bound by DinG in the absence (black) and presence (green) of SSB ΔC1 versus DinG concentration. The (-) lanes contain zero helicase in the reaction. The gels shown are representative of three experiments. Dots represent individual experiments, dashed lines represent averages of these three experiments, and error bars indicate standard deviation. Source data are provided as a source data file.
When χ-SSB interactions were weakened by protein mutations, YoaA-χ unwinding activity was inhibited on SSB-bound substrates. To determine whether the mutations hinder simultaneous binding of the helicase and SSB to DNA, EMSAs were performed with SSB ΔC1 and χ R128A. Binding of wt YoaA-χ to ssT65 was reduced when SSB ΔC1 was present compared to naked DNA (Fig. 4C). YoaA-χ R128A bound DNA similarly to wt YoaA-χ in the absence of SSB (black points in Fig. 4C, D). However, in the presence of SSB, YoaA-χ R128A binding was reduced (Fig. 4D). SSB ΔC1 binding to ssT65 also reduced DinG binding to DNA (Fig. 4E). These results indicate that inhibition of helicase activity when protein-protein interactions are weakened is due at least in part to reduced helicase binding to SSB-bound DNA. It is interesting to note that two bands were present in the no helicase lanes (-) that only contained SSB ΔC1 and ssT65 (Fig. 4C, E), indicating that deleting Phe-177 altered SSB ΔC1-DNA binding compared to wt SSB-DNA.
YoaA-χ binds DNA substrates used in helicase assays when SSB is bound
EMSAs showed that both SSB and YoaA-χ could bind to a 65-nt ssDNA. To determine whether YoaA-χ can bind overhang and forked helicase substrates when SSB is present, experiments were performed that leverage the distant-dependent quench that iron-sulfur (Fe-S) clusters in XPD/Rad3 helicases have on fluorophores11,40,41,42. A 30-nt DNA strand was annealed to a 95-nt strand to create 30-bp duplex with a 65-nt 5ˈ ss dT overhang. First, we verified that both YoaA-χ (1 μM) and DinG (1 μM) quenched fluorescein located at 4-, 7-, 11-, 16-, or 20-nt from the ds/ss junction in a distant-dependent manner (Fig. 5A). ATPγS was used in place of ATP to promote helicase binding but prevent translocation and unwinding.
A Relative intensity of fluorescein (glowing green circle) located either 4-, 7-, 11-, 16-, or 20-nt away from ds/ss junction on an overhang substrate (50 nM, O9) in the presence of either 1 μM YoaA-χ (blue) or DinG (orange). Relative intensity of fluorescein located 7-nt away from the ds/ss junction on an overhang substrate (50 nM, O10) is shown when either (B) YoaA-χ (50 nM, 100 nM, 250 nM, 500 nM, 1 μM, or 1.5 μM) or (C) DinG (50 nM, 100 nM, 250 nM, 500 nM, 1 μM, or 1.5 μM) is added. Relative intensity of fluorescein located 7-nt away from the ds/ss junction on a forked substrate (50 nM, F10) is shown when either (D) YoaA-χ (50 nM, 100 nM, 250 nM, 500 nM, 1 μM, or 1.5 μM) or (E) DinG (50 nM, 100 nM, 250 nM, 500 nM, 1 μM, or 1.5 μM) is added. For panels (B–E) black indicates naked DNA, green indicates SSB (75nM) prebound to DNA, and pink indicates SSB ΔC1 (75 nM) prebound to DNA. Dots represent individual experiments, dashed lines represent averages of these three experiments, and error bars indicate standard deviation. Source data are provided as a source data file.
The overhang substrate with fluorescein 7-nt from the ds/ss DNA junction (Supplementary Table 3, O10) was used for binding experiments because there is a large (> 50%) quench when the helicases bind DNA. The contribution of SSB (or SSB ΔC1) alone on fluorescence quenching is small at the concentration used (Supplementary Fig. 4A)36. Increasing concentrations of YoaA-χ (50 nM – 1.5 μM) were added to the overhang substrate (50 nM, O10) in the absence and presence of SSB (75 nM) (Fig. 5B and Supplementary Table 4). YoaA-χ quenched the intensity of fluorescein in a concentration-dependent manner, with higher concentrations of YoaA-χ correlating with lower fluorescence intensities both with and without SSB (Fig. 5B and Supplementary Table 4). DinG similarly quenched fluorescence when added to the overhang substrate with and without SSB (Fig. 5C and Supplementary Table 4). In both cases, the helicase concentration-dependent quench shows that the helicases bind the DNA substrates similarly, whether SSB is there or not. This experiment was also done with SSB ΔC1 that weakly interacts with the helicases. In contrast to experiments with wt SSB, there was a smaller quench in fluorescence for both helicases when SSB ΔC1 was present, consistent with EMSA results showing that SSB ΔC1 competes with the helicases for binding DNA (Fig. 5B, C and Supplementary Table 4).
One explanation for why SSB inhibits both helicases on the forked DNA substrate could be that SSB competes with the helicase to reduce DNA binding, as SSB ΔC1 does. The fluorescein quench assay was used to test this possibility. The sequence of the 3ˈ overhang on the fluorescein-labeled forked substrate (Supplementary Table 3, F10) was the same as for the FRET substrates (Figs. 1 and 2) to negate sequence-dependent effects. SSB alone did not quench fluorescence on the forked substrate (Supplementary Fig. 4B). Addition of increasing concentrations of YoaA-χ to the forked substrate (F10) quenched fluorescence in a concentration dependent manner to a similar extent with and without SSB (Fig. 5D). Addition of DinG to the forked substrate also quenched fluorescence to a similar extent with and without SSB (Fig. 5E). These results indicate that SSB does not prevent either helicase from binding the forked DNA substrate to inhibit helicase activity. Instead, SSB may bind more frequently adjacent to ss/ds DNA junctions on forked DNA compared with overhang DNA to hinder unwinding more on forked DNA.
Chi binds the C-terminal side of YoaA at the rear of the helicase relative to the direction of translocation
In vivo studies have shown that the last 18 amino acids of YoaA are needed for AZT tolerance16. Within this region, substitution of Arg-619 or Thr-620 with Ala increases AZT sensitivity in vivo and weakens YoaA-χ interactions in yeast two-hybrid assays16. To further support the presence of the χ binding site in this region of YoaA, three separate amino acid substitutions were made to YoaA, Arg-619, Thr-620, and Arg-621 to Ala. These YoaA mutants were co-expressed with χ and compared against wt YoaA-χ. In each case, YoaA had an N-terminal 6X histidine tag (His-tag) to bind an immobilized metal ion affinity column (IMAC) (Fig. 6A). Because χ lacks a His-tag, χ is only retained on the IMAC via binding to His-tagged YoaA. In the purification with wt YoaA, both YoaA and χ were retained on the IMAC and co-eluted. For the R619A and T620A mutants, YoaA was present in the soluble lysate fraction and retained on the IMAC, while χ was not retained on the IMAC despite also being highly expressed in the soluble fraction (Fig. 6A and Supplementary Fig. 5A, B). The YoaA R621A mutant was less soluble and found mostly in the cell pellet when expressed (Supplementary Fig. 5C). We also note that χ enhances the solubility of YoaA11 and both the R619A and T620A proteins precipitated while standing for 24 h at 4 °C whereas wt YoaA-χ did not. Together, these results support previous findings and show that both Arg-619 and Thr-620 are important for protein-protein interactions with χ.
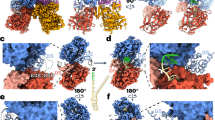
A SDS-PAGE gel showing lysate (L) and elution (E) fractions from IMAC purification of wt YoaA-χ and mutants. Compared to wt YoaA-χ, purification of YoaA R619A, T620A, and R621A mutants show a reduction of associated χ in the elution despite it being present in their respective lysates. The image shown is representative of three independent experiments. B AlphaFold 3 prediction of YoaA (pink is helicase domain I, blue is helicase domain II, light green is arch domain, and tan is Fe-S domain) binding a dT11 oligo (stick model) and χ (yellow)43. AlphaFold 3 predicts χ to bind the C-terminal end of YoaA. Residues with volume on YoaA indicate R619 and T620, and on χ indicate F64. Arg-128, responsible for binding SSB, is shown on χ opposite of YoaA. Source data are provided as a source data file.
A structural model of YoaA-χ bound to ssDNA (dT11) was generated with AlphaFold 3, and model parameters indicate a high confidence in the prediction (Supplementary Fig. 6A, B)43. The model is colored with the scheme used for the high-resolution structure of DinG44 with helicase domain 1 (HD1) in pink, helicase domain 2 (HD2) in light blue, the arch domain in green and FeS domain in tan (Fig. 6B). The overall fold agrees with that of DinG with ssDNA bound in the same orientation - the 5ˈ end at HD2 and 3ˈ end at HD1. The model also agrees with results from amino acid substitutions used to map the YoaA-χ interface. YoaA residues Arg-619 and Thr-620 are located at the interface of YoaA and χ. Similarly, χ residue Phe-64, which has been shown to be important for protein-protein interactions is also located at the interface12. The binding site on χ for SSB, indicated by Arg-128 shown as spheres, is on the opposite side of χ from the binding site for YoaA. Given that YoaA and other helicases in the Rad3/XPD family translocate in a 5ˈ to 3ˈ direction on DNA, this would place HD1, the arch, and FeS domains at the front of the moving helicase and HD2 and χ at the back of the helicase relative to the direction of travel.
YoaA-χ pulls SSB while it translocates on DNA
None of the experiments performed so far reveals the fate of SSB when YoaA-χ translocates on DNA. EMSAs showed SSB is bound to DNA when YoaA-χ binds, but once YoaA-χ translocates, it is unknown if SSB remains or is pushed off the ssDNA. In single-molecule experiments, SSB was site-specifically labeled at residue 12245,46 with Alexa Fluor 647 (SSB-AF647) and was visualized on ssDNA using confocal fluorescence in the absence and presence of unlabeled YoaA-χ. These experiments were performed by tethering a 20,452-nt ssDNA biotinylated at both ends between two streptavidin-coated polystyrene beads (Fig. 7). An experimentally determined concentration of SSB-AF647 (20 pM) that does not saturate the DNA was added, binding as expected to the ssDNA while held at a constant force of 10 pN (Fig. 7A)47,48. Short binding events, most of which had a lifetime of 2 s or less, were measured (number of binding events (n) = 442 on 22 DNA molecules) (Fig. 7D). The longest binding events lasted for 20 s. To accurately obtain rates of SSB movement, only tracks longer than 4 s were included in calculations to minimize noise from shorter tracks. Only 8 tracks were longer than 4 s, with an average slope of 0.010 ± 0.004 μm/s. These tracks appeared to exhibit inconsistent, diffusive motion and did not move systematically in any direction. This agrees with previous literature, where it is well-documented SSB freely diffuses on ssDNA in single-molecule experiments46.
All kymographs were collected with biotinylated ssDNA (20,452-nt) tethered between two streptavidin beads at constant force of 10 pN. A Representative kymograph of ssDNA in a channel containing AF647-labeled SSB (red, 20 pM) and ATP (4 mM). B A kymograph is shown for the same DNA strand that was used in panel (A) and moved to a different channel containing AF647-SSB (red, 20 pM), YoaA-χ (100 nM), and ATP (4 mM). C Representative kymograph of ssDNA in a channel with AF647-labeled SSB (red, 20 pM), ATP (4 mM), and YoaA-χ R128A (100 nM). D Binding events of each experimental condition were binned into intervals of 2 s and shown in histograms. The SSB negative control (top, green) contained 442 total tracks measured on 22 different DNA molecules. Wild-type YoaA-χ (middle, gray) contained 745 total tracks measured on 15 different DNA molecules. YoaA-χ R128A (bottom, purple) contained 490 total tracks measured on 35 different DNA molecules. Source data are provided as a source data file.
To determine if YoaA-χ removes SSB from DNA when it translocates, 100 nM YoaA-χ was added to ssDNA with 20 pM SSB-AF647 (Fig. 7B). In the presence of YoaA-χ, SSB binding events increased in frequency (n = 745 on 15 DNA molecules) (Fig. 7D). SSB was not stripped from the DNA and, surprisingly, was retained on DNA for longer time periods, as shown by longer lifetime binding events. With wt YoaA-χ, SSB lifetimes on DNA were as long as 67 s (Fig. 7D). Unlike in the case of SSB binding DNA alone, many longer SSB tracks in the presence of YoaA-χ were visibly sloped over their duration. There were 115 tracks longer than 4 s, with an average slope of 0.023 ± 0.001 μm/s. Critically, all these tracks sloped in the same direction within each kymograph, which matches what is expected of the 5ˈ to 3ˈ unidirectional movement of YoaA-χ. This suggests that SSB does not fall off the ssDNA while YoaA-χ translocates, but instead actually moves with the helicase.
To determine if the movement of SSB with YoaA-χ is due to the physical interaction between χ and SSB, the same experiment was performed with YoaA-χ R128A (Fig. 7C). With YoaA-χ R128A, SSB exhibited shorter lifetimes on DNA and few SSB molecules moved with the helicase (Fig. 7D). This condition had the least number of SSB binding events per DNA molecule (n = 490 on 35 DNA molecules), and kymographs were similar to SSB-only kymographs. The longest track measured was 31 s, and only 9 tracks were longer than 4 s, with an average slope of 0.024 ± 0.004 μm/s. These longer tracks and their slopes closely resembled those of SSB with wt YoaA-χ. The decrease in the number of long binding events with unidirectional motion indicates that these events are predominantly due to protein-protein interactions between χ and SSB. The few events observed with YoaA-χ R128A could result from weak binding of this mutant to SSB or from the helicase pushing SSB along DNA with its motor activity.
Discussion
Genetic studies have shown that χ-SSB interactions are important for DNA damage tolerance and repair20 and the goal of this work was to explore biochemical functions of χ-SSB interactions in the context of the YoaA-χ helicase. Chi-SSB interactions have been well-characterized structurally and biochemically30,31,49 as χ also plays an important role in mediating interactions with SSB at the replication fork in the context of DNA polymerase III holoenzyme27,28. Chi residue Arg-128 is important for binding SSB, and substitution of Arg-128 with Ala negatively affected both the DNA binding and helicase activities of YoaA-χ when SSB was present (Figs. 3 and 4). This shows that YoaA-χ interacts with SSB predominantly through χ rather than through YoaA-SSB binding. In contrast, the YoaA paralog DinG interacts directly with SSB, although the specific SSB binding site on DinG has not yet been identified33.
SSB affects the DNA unwinding activity of YoaA-χ and DinG in a substrate-dependent manner. On a DNA substrate with a duplex and 5ˈ ssDNA overhang, SSB modestly stimulated or had no effect on the unwinding activities of the helicases. This result is important because SSB binds the ssDNA overhang and could have blocked the helicases from binding to DNA and accessing the ds/ss DNA junction, but it did not. While SSB did not affect DinG helicase activity in our assays, SSB stimulated the helicase activity of DinG in another study33. This difference may be due to the use of different DNA substrates, as M13mp18 circular ssDNA with a 55-nt primer was used in the other work, whereas our study used a 65-nt ssDNA overhang. It is possible the large ssDNA region of the M13mp18 DNA, coated by SSB, directed DinG to the ds/ss junction more effectively than the shorter overhang in our study. Additional studies must be conducted to resolve this difference.
In contrast to the 5ˈ overhang substrate, both helicases were inhibited on forked DNA that contained a 10-nt 3ˈ ssDNA overhang in addition to the 5ˈ ssDNA overhang. This result is intriguing because an overhang structure becomes forked as it is unwound (Fig. 1E). However, SSB inhibition of YoaA-χ requires a pre-formed forked substrate (Fig. 1F). To determine how SSB inhibits YoaA-χ on a forked substrate, DNA unwinding activity of YoaA-χ was measured on DNA substrates where the length of the 3ˈ and 5ˈ arms of the fork were varied. The rationale was that these changes in the substrates would affect the manner in which SSB binds DNA to tease out why SSB inhibits YoaA-χ on pre-formed forks. SSB is a tetramer, and each of the four oligonucleotide/oligosaccharide binding (OB) folds can bind ssDNA. Depending on how many OB-folds bind ssDNA at a time, different DNA binding modes are possible. High-affinity binding modes occlude 35, 56, or 65 nt of DNA, with the 65-nt mode being fully wrapped such that all four OB-folds bind DNA (reviewed in ref. 22). SSB can also bind DNA with a single OB-fold but with lower affinity34,50,51. These binding modes are not static, and SSB can rapidly convert between binding modes as well as slide along DNA (reviewed in ref. 22).
SSB binding to forked and overhang DNA substrates with 5ˈ arms of 20-, 35-, and 65-nt was measured using a fluorescence quench assay with the same concentration of SSB used in the helicase assays. As expected for both substrates, quenching increased as the 5ˈ ssDNA length decreased because SSB is forced to bind closer on average to the duplex region where the fluorophore is located. Quenching was greater when the forked substrate was truncated compared to the overhang substrate, indicating that a greater fraction of forked DNA is bound by SSB, SSB is bound closer to the duplex where the fluorophore is located, or both. On the short 20-nt overhang substrate, SSB is likely stabilized by binding to both arms of the fork.
Consistent with these differences in SSB binding, YoaA-χ unwinding activity was strongly inhibited on forked substrates with 20- and 35-nt 5ˈ arms in comparison with the overhang substrates of the same length. When the 5ˈ arm of the forked DNA was kept at a constant length (65 nt) and the 3ˈ arm of the fork was truncated, similar unwinding activities were measured for 3ˈ arms 3-, 7-, and 10-nt in length in the absence of SSB. However, when SSB was added, unwinding was inhibited for 3ˈ arms of 7- or 10-nt in length, but not 3-nt. This is interesting in light of recent results showing that SSB can bind DNA in a mode where 8 nt of ssDNA are occluded48. Thus, when the 3ˈ arm of the fork is at least 7-nt in length, SSB likely binds both strands of the fork and hinders access of YoaA-χ to inhibit unwinding. As a whole, these results show that SSB has a tendency to bind at pre-formed forks, interfering with DNA unwinding by YoaA-χ (Fig. 8). The physiological implications of these findings are intriguing. When SSB is present at a fork, it may act to hinder YoaA-χ activity, preventing the enzyme from functioning at inappropriate times. A role for SSBs in directing enzyme activities at replication forks has been documented. For example, the human SSB, RPA, directs the activity of SMARCAL1 to promote DNA fork regression when obstacles arise during replication52.
SSB preferentially binds near the junction of pre-formed forked DNA substrates. Once bound at the ss/ds DNA junction, SSB interacts with both 5ˈ and 3ˈ DNA overhangs and hinders YoaA-χ from accessing the duplex, preventing unwinding. In contrast, SSB bound on a 5ˈ overhang without a 3ˈ overhang does not block YoaA-χ from accessing the duplex, resulting in unimpeded unwinding. On ssDNA, YoaA-χ is capable of pulling SSB behind it during translocation due to YoaA and SSB binding on opposite interfaces of χ Created in BioRender. Padgett-Pagliai, K. (2025) https://BioRender.com/w15ivvp.
Physical interactions between SSB and SSB interacting proteins (SIPs) are important for SIPs to function on DNA that is bound by SSB (reviewed in refs. 24,53). SIPs bind the C-terminal acidic tip of SSB, with the last Phe residue playing a key role in binding. Protein-protein interactions between SIPs and SSB allow SIPs to rearrange SSB on DNA and bind. The E. coli helicases RecQ and PriA bind SSB-DNA by promoting the SSB 35-nt binding mode over the 65-nt binding mode via direct helicase-SSB interactions54,55. Because the 35-nt binding mode of SSB wraps less ssDNA than the 65-nt mode, RecQ and PriA induce the mode switch to expose more ssDNA for the helicases to bind. This may also be the case for YoaA-χ and DinG because their helicase activities are not hindered on overhang substrates by wt SSB binding but are inhibited by SSB ΔC1. Inhibition of helicase activity by SSB ΔC1 stems at least in part from inhibition of DNA binding. In the absence of protein-protein interactions, SSB competes with the helicases for binding DNA because both helicases bound DNA along with wt SSB, but SSB ΔC1 reduced helicase binding of DNA. Interestingly, EMSA binding experiments showed that SSB ΔC1 gave a different banding pattern than wt SSB, indicating a second molecule of SSB ΔC1 could bind to ssDNA at the concentrations used in our experiment. This result suggests that the deletion of the C-terminal Phe could influence the binding modes of SSB, which could contribute to the effects of this mutation on helicase activities. When the reciprocal χ-R128A mutation was made in YoaA-χ to reduce protein-protein interactions with SSB, helicase, and DNA binding activities of YoaA-χ on DNA bound by wt SSB were inhibited to a similar extent as by SSB ΔC1. Because the χ-R128A mutation does not affect YoaA-χ activity in the absence of SSB, this inhibition stems from loss of protein-protein interactions.
Our studies show that χ mediates interactions between YoaA and SSB, but we lack high-resolution structural data to define the protein-protein binding sites in YoaA-χ. Mutagenesis of χ showed that Phe-64 is important for binding YoaA and for AZT tolerance in vivo12. This would place the YoaA binding site on χ on the opposite side of the protein as the binding site for SSB (Figs. 6B and 8). The χ binding site on YoaA was mapped to the last 18 amino acids using a mutagenesis strategy16. Within this region, Arg-619 and Thr-620 were needed for AZT tolerance and protein-protein interactions measured in a yeast two-hybrid assay16. In this work, when Arg-619 or Thr-620 was substituted with Ala, neither mutant YoaA protein co-purified with χ, unlike wt YoaA, showing that these residues are important for high-affinity binding. These mutagenesis experiments place the χ binding site on YoaA at the C-terminal end of YoaA in HD2. It is interesting to note that when the structure of χ in complex with the ψ subunit of the DNA polymerase III holoenzyme was solved, the authors noted that χ had a fold similar to a helicase domain of a DExx-box helicase, PcrA56. YoaA, like PcrA, is a superfamily 2 helicase, and χ has a similar fold to YoaA HD2. It is not clear what purpose, if any, this structural similarity serves.
A predicted structure of YoaA-χ bound to a dT11 oligo was generated with AlphaFold 343. The predicted YoaA-χ-dT11 structure matches the expected orientation of ssDNA bound to an XPD/Rad3-like helicase, where HD1, the arch, and the Fe-S domains bind closer to the 3ˈ end of DNA and HD2 is at the 5ˈ end40,57. In accordance with this arrangement, the five models generated by AlphaFold 3 for YoaA-χ-dT11 also all place a dT11 oligo within YoaA such that the N-terminal HD1 binds the 3ˈ end of ssDNA, and the C-terminal HD2 binds the 5ˈ end of ssDNA43. This predicted structure of YoaA-χ-dT11 is similar to the orientation of the solved structure of DinG-dT12, with DinG’s HD1 binding the 3ˈ end of DNA and HD2 binding closer to the 5ˈ end44. The predicted YoaA-χ-dT11 structure also places YoaA and χ in an orientation that agrees with mutational analyses of protein-protein interactions. YoaA Arg-619 and Thr-620, as well as χ Phe-64, are critical residues in YoaA-χ interactions, and the AlphaFold 3 structure situates χ and the HD2 of YoaA such that these three residues are within the predicted binding interface (Fig. 6B)12,16. Because XPD/Rad3-family helicases translocate in a 5ˈ to 3ˈ direction, HD1 is the front of the helicase relative to the direction of motion, and HD2 and χ are at the back. Given that SSB binds χ near Arg-128 on the opposite side of the protein as YoaA, SSB would be at the very back of the complex like a caboose on a train (Figs. 6B and 8).
Our single-molecule experiments were performed with ssDNA held at a constant force of 10 pN, at which SSB binds approximately 17-nt of DNA47,48. This weaker wrapping mode of SSB explains the short-lived and sparse SSB-DNA binding events that occurred with only SSB in the reaction. Motor proteins are known to remove SSB from DNA, as seen with the HELQ helicase, which strips RPA from ssDNA in single-molecule experiments58. Given this weaker SSB binding mode and the fact that YoaA-χ is a motor protein, it seems plausible that YoaA-χ could push SSB off the ssDNA. However, YoaA-χ did not remove SSB from ssDNA during translocation. In fact, SSB-DNA binding events per DNA molecule increased in the presence of YoaA-χ, indicating YoaA-χ may even help recruit SSB to ssDNA. Kymographs showed that SSB moved unidirectionally along DNA when wt YoaA-χ was present, indicating that SSB was co-translocating with the moving helicase. When YoaA-χ R128A was present, there were far fewer long tracks of SSB movement, indicating that the majority of SSB tracks observed with wt YoaA-χ resulted from direct protein-protein interactions between χ and SSB. When these results are interpreted in the context of the structural model for YoaA-χ, the simplest conclusion is that YoaA-χ is pulling SSB like the caboose on a train. If YoaA-χ were instead pushing SSB ahead of the helicase using its motor activity, we would expect comparable numbers of long, sloped tracks with both wt YoaA-χ and YoaA-χ R128A. Since YoaA-χ R128A is as active as wt YoaA-χ in the absence of SSB, the decrease in tracks is not attributable to a reduction in motor activity. The few sloped tracks observed with YoaA-χ R128A may be due to weak SSB binding to the χ mutant or occasional pushing by the helicase. Although sloped, the longer SSB tracks measured with YoaA-χ R128A were generally shorter in duration than those observed with wt YoaA-χ, highlighting the importance of these protein-protein interactions in stabilizing the longer lifetime events. There were also fewer SSB-DNA binding events per molecule when YoaA-χ R128A was present than for SSB alone or when wt YoaA-χ was present. This suggests that when YoaA-χ no longer binds SSB with high affinity, it may displace SSB from DNA.
Because YoaA-χ appears to pull SSB, its potential to push SSB along DNA does not seem to be the primary mechanism for clearing SSB-bound DNA. Some helicases, such as Pif1, are capable of pushing SSB, with Pif1 pushing both E. coli SSB and human RPA along ssDNA37,38. YoaA-χ may initially access SSB-bound DNA by passive rearrangement between χ and SSB, which is not without precedent. When in complex with the DNA polymerase III clamp loader complex, χ remodels SSB on DNA passively without the use of motor activity59. However, in the case of YoaA-χ, once the helicase complex gains access to DNA, YoaA-χ actively pulls SSB via its motor activity. In our experiments, ssDNA was maintained at 10 pN, likely causing SSB to adopt its 17-nt binding mode. This binding mode may make SSB easier to pull along the DNA. If SSB were to adopt one of its higher-affinity binding modes through the use of different DNA tensions and occlude more DNA, it may prove less amenable to translocation with YoaA-χ, although this idea needs further investigation. It is similarly unknown if the pulling interaction between YoaA and χ persists during active DNA unwinding. Our single-molecule experiments only examined SSB movement in the context of ssDNA translocation, and additional studies are needed to determine whether this interaction continues during unwinding of duplex DNA.
The cellular function of YoaA-χ pulling SSB along on DNA is unknown. In other systems, helicases and SSB proteins coordinate, such as FANCJ and RPA working together to resolve G-quadruplexes60,61,62. A similar phenomenon may occur with YoaA-χ and SSB, where SSB travels with YoaA-χ to prevent newly unwound secondary structures from reannealing. If YoaA-χ is repairing a base-stalling lesion on the leading strand, it would be pulling SSB away from the parental duplex, possibly allowing other proteins to bind and access the DNA to repair the lesion. YoaA-χ and SSB may also have a role in fork regression, with SSB being pulled out of the way of other proteins. To our knowledge, this is the first report of a mechanoenzyme pulling SSB along DNA, taking the field of helicase and SSB interactions in a new and intriguing direction.
Methods
Buffers
Assay buffer A is 50 mM Tris HCl pH 7.5, 125 mM NaCl, 10 mM MgCl2, 50 μg/mL BSA, and 2 mM DTT at final concentration within the reaction. For single-molecule experiments, buffer B is LUMICKS 10x running buffer (PBS, 50 mM sodium azide, and 5 mM EDTA) diluted 1:10 in water, and buffer C is buffer B diluted 1:10 in water.
SDM of holC R128A, yoaA R619A, yoaA T620A, and yoaA R621A
A Q5 Site-directed Mutagenesis Kit (New England Biolabs) was used per the manufacturer’s instructions with the primers in Supplementary Table 1 and a pET15b-holC plasmid to mutate arginine 128 to alanine in the HolC protein. The same Q5 Site-directed Mutagenesis Kit (New England Biolabs) was used per the manufacturer’s instructions with the primers in Supplementary Table 1 and a pCOLA-his-yoaA plasmid to mutate arginine 619 to alanine, threonine 620 to alanine, and arginine 621 to alanine in the YoaA protein. DNA sequencing was performed to confirm the desired single mutations in each protein were made and other mutations were not present.
Overexpression of proteins
YoaA-χ and YoaA-χ R128A were overexpressed using a previously published protocol11 in which BL21 (DE3) E. coli cells were transformed with pCOLADuet-his-YoaA and pET15b-HolC. Cells were grown at 37 °C, with shaking at 250 RPM, in Terrific Broth with kanamycin (50 μg/ml) and carbenicillin (100 μg/ml) until an OD600 of approximately 0.6. IPTG (1 mM) was added to induce protein expression along with iron supplements, iron (II) sulfate (0.1 mg/ml) and ammonium ferric citrate (0.1 mg/ml). Cells were grown for 4 h more at 25 °C with shaking at 250 RPM. The cells were pelleted by centrifugation at 6370 × g for 30 min, the media was removed, and the cell pellets stored at − 80 °C. Both preparations of wt and mutant YoaA-χ contain an N-terminal His-tag on YoaA.
For DinG, BL21 (DE3) E. coli cells were transformed with pET30-his-DinG vector expressing DinG with the N-terminal 6X His tag8. Cells were grown as for YoaA-χ except that only kanamycin (50 μg/mL) was used in the media.
The pGN62-SSB (a gift from Dr. Michael O’Donnell) and pET21a-SSBΔC1 (a gift from Dr. James Keck) were transformed into E. coli BL21 (DE3) pLysS cells for the overexpression of SSB and SSB ΔC1 respectively39. SSB ΔC1 denotes the last C-terminal residue, F177, is deleted. The cells were grown in Terrific Broth with ampicillin (100 μg/mL) and chloramphenicol (30 μg/mL) at 37 °C shaking at 250 RPM until an OD600 of approximately 0.6. Protein expression was induced with ITPG (1 mM). The cells were grown for 3 more hours at 37 °C with shaking at 250 RPM. The cells were pelleted by centrifugation at 6370 × g for 30 min, the media was removed, and pellets stored at − 80 °C.
Purification of proteins
YoaA-χ was purified by previously published methods. Cell pellets were resuspended in low imidazole buffer (20 mM sodium phosphate pH 7.8, 40 mM imidazole, 500 mM NaCl, 10% glycerol, Sigma FAST Protease Inhibitor Cocktail Tablet EDTA-free, and 2 mM DTT). After lysis by French press and centrifugation, the cleared lysate was loaded onto a HisTrap FF column (Cytiva) and washed with low imidazole buffer. Proteins were eluted with a linear gradient from 40 mM to 500 mM imidazole. Fractions from the HisTrap elution were dialyzed overnight in 25 mM Tris–HCl pH 7.8, 500 mM NaCl, 10% glycerol, and 2 mM DTT and then dialyzed for 6 h in 25 mM Tris–HCl pH 7.8, 250 mM NaCl, 10% glycerol, and 2 mM DTT. The samples were loaded onto a HiTrap Heparin HP column (Cytiva) and washed with low salt buffer (25 mM Tris–HCl pH 7.8, 250 mM NaCl, 10% glycerol, and 2 mM DTT). Proteins were eluted with a linear gradient of 250 mM to 1 M NaCl. Fractions containing YoaA-χ were dialyzed overnight into 25 mM Tris–HCl pH 7.8, 250 mM NaCl, 30% glycerol, and 2 mM DTT and stored at − 80 °C. Protein concentration was measured using a Bradford assay and measuring the absorbance of pure YoaA-χ at 595 nm. YoaA-χ R128A was purified using the same protocol11. Two different preparations of purified YoaA-χ R128A were used for all experiments that contained YoaA-χ R128A to confirm the effects of the R128A mutation.
For DinG, E. coli cell pellets were resuspended in low imidazole buffer (25 mM Tris HCl pH 8, 25 mM imidazole, 1 M NaCl, Sigma FAST Protease Inhibitor Cocktail Tablet EDTA-free, and 0.5 mM TCEP). After lysis by French press and centrifugation, the cleared lysate was loaded onto a HisTrap FF column (Cytiva) and washed with low imidazole buffer. DinG was eluted with a linear gradient from 25 mM to 500 mM imidazole, and fractions containing DinG were dialyzed overnight in 25 mM Tris HCl pH 8, 1 M NaCl, 0.5 mM TCEP, and then dialyzed for 6 h in 25 mM Tris HCl pH 8, 300 mM NaCl, 0.5 mM TCEP. This was loaded onto a HiTrap Heparin HP column (Cytiva). The column was washed with low salt buffer (25 mM Tris HCl pH 8, 300 mM NaCl, and 0.5 mM TCEP) and DinG was eluted with a linear gradient from 300 mM to 1 M NaCl. Fractions containing DinG were dialyzed overnight into 25 mM Tris HCl pH 8, 300 mM NaCl, 30% glycerol, and 0.5 mM TCEP and stored at − 80 °C. Protein concentration was measured using an A280 absorbance of DinG and molar absorptivity of 78,840 M−1 cm−1.
SSB and SSB ΔC1 were purified by previously published methods63 in which cell pellets were resuspended in lysis buffer (50 mM Tris HCl pH 7.5, 200 mM NaCl, 10 mM spermidine, 10% sucrose, 0.5 mM EDTA). After lysis by French press and centrifugation, the cleared lysate was treated with 0.24 g/ml ammonium sulfate. The pellet was repeatedly resuspended and pelleted in 100 ml of resuspension buffer (200 mM NaCl, 50 mM Tris HCl pH 7.5, 10% glycerol, 0.5 mM EDTA, 0.18 g/mL ammonium sulfate) six times and then twice in 100 mL of the same resuspension buffer with 0.13 g/mL ammonium sulfate. The pellet was resuspended in resuspension buffer again and dialyzed against the same buffer overnight. A HiTrap Heparin HP column (Cytiva) and washed with low salt buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 10% glycerol, 0.5 mM EDTA, and 2 mM DTT), and SSB was eluted with a linear gradient from 150 mM to 500 mM NaCl. Fractions with SSB were pooled and dialyzed overnight (200 mM NaCl, 20 mM Tris HCl pH 7.5, 10% glycerol, 0.5 mM EDTA, and 2 mM DTT). The pooled fractions were loaded onto a Mono Q column (Cytiva) and eluted with a linear gradient from 150 mM to 500 mM NaCl. Fractions with SSB were pooled and dialyzed overnight into 200 mM NaCl, 20 mM Tris-HCl, 10% glycerol, 0.5 mM EDTA, and 2 mM DTT and then stored at − 80 °C. Protein concentration was determined by measuring the absorbance at 280 nm under denaturing conditions and using an extinction coefficient of 1.5 mL mg−1 cm−1 for both SSB and SSB ΔC1.
Labeling SSB with Alexa Fluor 647
SSB A122C45,46 was labeled with Alexa Fluor 647 maleimide (ThermoFisher) per the manufacturer’s standard maleimide reaction protocol. The labeling reaction was designed such that, on average, all four monomers of the SSB tetramer were labeled. Labeled protein was purified from excess fluorophore using a BioGel P-6DG desalting column, followed by ion exchange chromatography using a HiTrap Q HP column64.
DNA annealing
Oligonucleotides were purchased from and purified by Integrated DNA Technologies (IDT) using HPLC for fluorophore-labeled oligonucleotides and PAGE for unlabeled oligonucleotides. Single-stranded DNA substrates were mixed at equal concentrations in 20 mM Tris HCl pH 7.5 and 50 mM NaCl, heated to 80 °C for 5 min, and cooled to room temperature over at least 4 h to anneal. Supplementary Table 2 contains the DNA sequences of oligonucleotides that were annealed to make the substrates in Supplementary Table 3.
FRET-based helicase assay
FRET-based helicase assays were used to measure DNA duplex unwinding11. DNA substrates (O1, O2, O3, O4, F1, F2, F3, F4, F8, and F9, Supplementary Table 3) were labeled at the blunt end with a Cy3-Cy5 FRET pair. The annealed DNA (50 nM) was mixed with ATP (2 mM) in assay buffer A. Cy3 fluorescence (565 nm, Iobs) was monitored continuously as a function of time while DNA, SSB (75 nM when present), and helicase were added to the cuvette sequentially. The data was analyzed using the previously published protocol11. The Cy3 signal for ds Cy3-Cy5 DNA without YoaA-χ was averaged to determine the intensity of ds DNA (Ids). The Cy3 signal for ss DNA, without YoaA-χ, was measured and averaged to determine the intensity of ss DNA (Iss). The fluorescent signals obtained in DNA unwinding reactions were converted to the concentration of DNA unwound by YoaA-χ using Eq. 1.
Observed reaction rates were calculated by fitting reaction time courses to exponential decays (Eq. 2) using KaleidaGraph Software.
Three technical repeats were performed as a function of helicase concentration in Suplementary Figs. 1 and 2.
SSB quenching assay
Reactions contained annealed DNA with a fluorescein located 6 bp into the duplex (50 nM, O5, O6, O7, F5, F6, and F7, Supplementary Table 3) and ATP (2 mM) in assay buffer A. A DNA-only emission spectrum from 505 to 575 nm was taken with a 490 nm excitation and 3-nm bandwidth prior to adding SSB (75 nM). A time-based scan was taken at 490 nm excitation and 515 nm emission to ensure full sample equilibration. An emission spectrum was then measured for the SSB-DNA complex after the signal plateaued at the same conditions as the DNA-only emission. The helicase-DNA emission spectrum was divided by the DNA-only emission spectrum at 516 nm for each trial to determine the relative quench in fluorescence. To correct for dilution, the relative helicase-DNA emission spectrum was divided by the relative DNA-only emission spectrum at 516 nm. Three technical repeats were performed for all SSB quenching assays.
Gel-based helicase assay
To confirm DinG was annealing DNA in the presence of SSB ΔC1 on the overhang substrate, DNA products were measured using a gel helicase assay. Reactions contained Cy5-labeled duplex DNA (50 nM, O8, Supplementary Table 3), ATP (2 mM), assay buffer A, and SSB ΔC1 (75 nM). The reaction started with the addition of DinG (20 or 50 nM). Samples were removed from the reaction at 1, 5, 10, 15, 25, and 35 min and quenched with 1.5% SDS, 15 mM EDTA, and 37.5% glycerol. In the positive control (ssDNA), DinG was not added to the reaction mix, and the sample was heated at 95 °C for 5 min and put directly on ice. For the negative control (dsDNA), DinG was not added to the reaction mix, and the sample was not denatured. Substrates were separated from products on a 10% native gel (10% acrylamide:bis solution, 19:1), which was run at 0.02 amps in a 4 °C room for 20 min. The gel was imaged with an Amersham Typhoon (Cytiva) and quantified using ImageQuantTL. Three technical repeats were performed for the gel helicase assays.
Fluorescein quench assay
Reactions contained DNA with a fluorescein located within the duplex (50 nM, O9, O10, and F10, Supplementary Table 3), ATPγS (0.5 mM), and assay buffer A. If the reaction contained wt SSB or SSB ΔC1, SSB (75 nM) was preincubated with the DNA before the addition of the helicase. A DNA-only emission spectrum from 505 to 625 nm was taken with a 495 nm excitation and 2-nm bandwidth prior to adding helicase. Then, either YoaA-χ or DinG was added to the reaction, and a time-based scan was taken at 495 nm excitation and 525 nm emission. An emission spectrum was measured for the helicase-DNA complex after the signal plateaued at the same conditions as the DNA-only emission. The helicase-DNA emission spectrum was divided by the DNA-only emission spectrum at 516 nm for each trial to determine the relative quench in fluorescence. To correct for dilution, the relative helicase-DNA emission spectrum was divided by the relative DNA-only emission spectrum at 516 nm. Three technical repeats were performed for all fluorescein quench assays. As a control to show that wt SSB and SSB ΔC1 did not affect fluorescein fluorescence, the experiment was performed as above, except SSB (50, 75, and 100 nM) was titrated onto the overhang DNA (50 nM, O10) or forked DNA (50 nM, F10) without a helicase present.
Electrophoretic mobility shift assay (EMSA)
EMSAs were performed with dT65 ssDNA labeled with a Cy5 fluorophore (Supplementary Table 2, S21) (50 nM), ATPγS (0.5 mM), glycerol (6.75%), and assay buffer A. Reactions containing SSB or SSB ΔC1 were pre-incubated with 75 nM SSB before the helicase was added. The helicase was titrated in concentrations ranging from 50 to 400 nM. Reactions were incubated at room temperature for 20 min, were loaded onto a 5% native gel (5% acrylamide:bis solution, 37.5:1) and run at 0.02 amps in a 4 °C room. The gel was scanned using an Amersham Typhoon (Cytiva). EMSAs were replicated with three technical repeats.
Protein interactions between YoaA and χ
YoaA wt and mutants (R619A, T620A, R621A) with χ were expressed and purified as above with slight modifications11. Pellets were suspended using 3.5 mL g−1 cell lysis buffer11. The cell lysate was bound to a 5 mL HisTrap FF column (Cytiva) and washed with cell lysis buffer, with 1.8 mL fractions collected until absorbance reached baseline. Total wash volume was 60 mL for each sample. Protein was eluted using 8 mL of high imidazole buffer and collected into 0.2 mL fractions11. This experiment was performed using biological replicates and technical triplicates to confirm the effects of each mutation.
Single-molecule assay
Single-molecule experiments were performed on a LUMICKS C-Trap DYMO. Flow-cell passivation was performed by flowing 300 μL of the appropriate buffers in each channel, flushing each channel with 500 μL of LUMICKS passivation buffer Pluronic (diluted 1:10 in water), flushing each channel with 500 μL of LUMICKS passivation buffer BSA (diluted 1:10 in water), and then flowing 300 μL of the appropriate buffer for each channel at 1.6 bar. The microfluidic flow cell was set up with channel 1 containing 4.34 μm streptavidin-coated polystyrene beads (LUMICKS) in buffer B, channel 2 with biotinylated 20,452-nt ssDNA in buffer B (LUMICKS), channel 3 with buffer C, channel 4 with AF647-labeled SSB (20 pM), ATP (4 mM), Trolox (1 mM), GODCAT (glucose oxidase 0.54 mg/mL and catalase 0.048 mg/mL), and glucose (0.65%) in assay buffer A. If YoaA-χ or YoaA-χ R128A were used, they were also included in channel 4 at 100 nM final concentration. The overall power of the trapping laser was set to 20%. The 638 nm excitation laser was used at 1.6 μW to visualize the AF647-labeled SSB. Kymographs were collected at a constant force of 10 pN with no flow, using a pixel size of 50 nm and a pixel dwell time of 0.1 ms.
Kymographs were analyzed using Lakeview Pro with minimum photon counts set at 2, positional search range at 0.07 μm, maximum gap at 10 scan lines, minimum length at 10 points, expected spot size at 0.50 μm, and expected velocity at 0 μm/s. Manual refinements were performed when necessary during data analysis in Lakeview Pro to ensure track measurements were accurate. Binding events were binned into 2 s intervals. Data was exported from Lakeview Pro, and the slopes of SSB movement were manually calculated.
Structure Prediction
A model of YoaA-χ bound to ssDNA (dT11) was generated with AlphaFold 343. All five models were similar in terms of their statistics, ipTM and pTM, and the top model (model.0) is shown. A structure colored by pLDDT and a predicted aligned error plot is given in Supplementary Fig. 6.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data are provided in this paper.
References
Coin, F. et al. Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nat. Genet. 20, 184–188 (1998).
Cantor, S. et al. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc. Natl. Acad. Sci. USA 101, 2357–2362 (2004).
Lelij, P. vander et al. Warsaw breakage syndrome, a cohesinopathy associated with mutations in the XPD helicase family member DDX11/ChlR1. Am. J. Hum. Genet. 86, 262–266 (2010).
Barber, L. J. et al. SPAR1/RTEL1 Maintains genomic stability by suppressing homologous recombination. Cell 135, 261–271 (2008).
Ding, H. et al. Regulation of murine telomere length by Rtel an essential gene encoding a helicase-like protein. Cell 117, 873–886 (2004).
Sung, P. et al. Human xeroderma pigmentosum group D gene encodes a DNA helicase. Nature 365, 852–855 (1993).
Hirota, Y. & Lahti, J. M. Characterization of the enzymatic activity of hChlR1, a novel human DNA helicase. Nucleic Acids Res. 28, 917–924 (2000).
Voloshin, O. N., Vanevski, F., Khil, P. P. & Camerini-Otero, R. D. Characterization of the DNA damage-inducible helicase DinG from Escherichia coli. J. Biol. Chem. 278, 28284–28293 (2003).
Voloshin, O. N. & Camerini-Otero, R. D. The DinG protein from Escherichia coli is a structure-specific helicase. J. Biol. Chem. 282, 18437–18447 (2007).
Uringa, E. J., Youds, J. L., Lisaingo, K., Lansdorp, P. M. & Boulton, S. J. RTEL1: An essential helicase for telomere maintenance and the regulation of homologous recombination. Nucleic Acids Res. 39, 1647–1655 (2011).
Weeks-Pollenz, S. J. et al. Characterization of the Escherichia coli XPD/Rad3 iron-sulfur helicase YoaA in complex with the DNA polymerase III clamp loader subunit chi (χ). J. Biol. Chem. 299, 102786 (2023).
Sutera, V. A. et al. Alternative complexes formed by the Escherichia coli clamp loader accessory protein HolC (χ) with replication protein HolD (ψ) and repair protein YoaA. DNA Repair 100, 103006 (2021).
Lewis, L. K., Jenkins, M. E. & Mount, D. W. Isolation of DNA damage-inducible promoters in Escherichia coli: regulation of polB (dinA), dinG, and dinH by LexA repressor. J. Bacteriol. 174, 3377–3385 (1992).
Lewis, L. K. & Mount, D. W. Interaction of LexA repressor with the asymmetric dinG operator and complete nucleotide sequence of the gene. J. Bacteriol. 174, 5110–5116 (1992).
Courcelle, J., Khodursky, A., Peter, B., Brown, P. O. & Hanawalt, P. C. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158, 41–64 (2001).
Sutera, V. A. et al. Genetic analysis of DinG-family helicase YoaA and its interaction with replication clamp-loader protein HolC in Escherichia coli. J. Bacteriol. 203, https://doi.org/10.1128/jb.00228-21 (2021).
Bharti, S. K. et al. Specialization among Iron-Sulfur cluster helicases to resolve G-quadruplex DNA structures that threaten genomic stability*. J. Biol. Chem. 288, 28217–28229 (2013).
Piante, E. D. et al. Exploring the G-quadruplex binding and unwinding activity of the bacterial FeS helicase DinG. Sci. Rep. 13, 12610 (2023).
Boubakri, H., Septenville, A. L. D., Viguera, E. & Michel, B. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J. 29, 145–157 (2010).
Brown, L. T. et al. Connecting replication and repair: YoaA, a helicase-related protein, promotes azidothymidine tolerance through association with Chi, an accessory clamp loader protein. PLoS Genet. 11, e1005651 (2015).
Watanabe, K., Tominaga, K., Kitamura, M. & Kato, J. I. Systematic identification of synthetic lethal mutations with reduced-genome Escherichia coli: Synthetic genetic interactions among yoaA, xthA and holC related to survival from MMS exposure. Genes Genet. Syst. 91, 183–188 (2016).
Antony, E. & Lohman, T. M. Dynamics of E. coli single stranded DNA binding (SSB) protein-DNA complexes. Semin. Cell Dev. Biol. 86, 102–111 (2019).
Caldwell, C. C. & Spies, M. Dynamic elements of replication protein A at the crossroads of DNA replication, recombination, and repair. Crit. Rev. Biochem. Mol. 55, 482–507 (2020).
Shereda, R. D., Kozlov, A. G., Lohman, T. M., Cox, M. M. & Keck, J. L. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 43, 289–318 (2008).
Xiao, H., Dong, Z. & O’Donnell, M. DNA Polymerase III accessory proteins: IV. characterization of Chi and Psi. J. Biol. Chem. 268, 11779–11784 (1993).
Olson, M. W., Dallmann, H. G. & McHenry, C. S. DnaX complex of Escherichia coli DNA polymerase III holoenzyme. J. Biol. Chem. 270, 29570–29577 (1995).
Kelman, Z., Yuzhakov, A., Andjelkovic, J. & O’Donnell, M. Devoted to the lagging strand—the χ subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. EMBO J. 17, 2436–2449 (1998).
Glover, B. P. & McHenry, C. S. The χψ subunits of DNA polymerase III holoenzyme bind to single-stranded DNA-binding protein (SSB) and facilitate replication of an SSB-coated template. J. Biol. Chem. 273, 23476–23484 (1998).
Witte, G., Urbanke, C. & Curth, U. DNA polymerase III χ subunit ties single-stranded DNA binding protein to the bacterial replication machinery. Nucleic Acids Res. 31, 4434–4440 (2003).
Marceau, A. H. et al. Structure of the SSB–DNA polymerase III interface and its role in DNA replication. EMBO J. 30, 4236–4247 (2011).
Naue, N., Fedorov, R., Pich, A., Manstein, D. J. & Curth, U. Site-directed mutagenesis of the χ subunit of DNA polymerase III and single-stranded DNA-binding protein of E. coli reveals key residues for their interaction. Nucleic Acids Res. 39, 1398–1407 (2011).
Cooper, D. L. & Lovett, S. T. Toxicity and tolerance mechanisms for azidothymidine, a replication gap-promoting agent, in Escherichia coli. DNA Repair 10, 260–270 (2011).
Cheng, Z., Caillet, A., Ren, B. & Ding, H. Stimulation of Escherichia coli DNA damage inducible DNA helicase DinG by the single-stranded DNA binding protein SSB. FEBS Lett. 586, 3825–3830 (2012).
Bujalowski, W. & Lohman, T. M. Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions II. Salt, temperature and oligonucleotide length effects. J. Mol. Biol. 207, 269–288 (1989).
Kozlov, A. G. & Lohman, T. M. Calorimetric studies of E. coli SSB protein-single-stranded DNA interactions. Effects of monovalent salts on binding enthalpy. J. Mol. Biol. 278, 999–1014 (1998).
Kozlov, A. G. & Lohman, T. M. Kinetic mechanism of direct transfer of Escherichia coli SSB tetramers between single-stranded DNA molecules. Biochemistry 41, 11611–11627 (2002).
Sokoloski, J. E., Kozlov, A. G., Galletto, R. & Lohman, T. M. Chemo-mechanical pushing of proteins along single-stranded DNA. Proc. Natl. Acad. Sci. USA 113, 6194–6199 (2016).
Mersch, K. N., Sokoloski, J. E., Nguyen, B., Galletto, R. & Lohman, T. M. Helicase” activity promoted through dynamic interactions between a ssDNA translocase and a diffusing SSB protein. Proc. Natl Acad. Sci. USA 120, e2216777120 (2023).
Shereda, R. D., Reiter, N. J., Butcher, S. E. & Keck, J. L. Identification of the SSB Binding Site on E. coli RecQ Reveals a Conserved Surface for Binding SSB’s C Terminus. J. Mol. Biol. 386, 612–625 (2009).
Pugh, R. A. et al. The iron-containing domain is essential in Rad3 helicases for coupling of ATP hydrolysis to DNA translocation and for targeting the helicase to the single-stranded DNA-double-stranded DNA junction. J. Biol. Chem. 283, 1732–1743 (2008).
Honda, M., Park, J., Pugh, R. A., Ha, T. & Spies, M. Single-molecule analysis reveals differential effect of ssDNA-binding proteins on DNA translocation by XPD helicase. Mol. Cell 35, 694–703 (2009).
Pugh, R. A., Honda, M. & Spies, M. Ensemble and single-molecule fluorescence-based assays to monitor DNA binding, translocation, and unwinding by iron-sulfur cluster containing helicases. Methods 51, 313–321 (2010).
Abramson, J., et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Cheng, K. & Wigley, D. B. DNA translocation mechanism of an XPD family helicase. ELife 7, 8412 (2018).
Roy, R., Kozlov, A. G., Lohman, T. M. & Ha, T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature 461, 1092–1097 (2009).
Zhou, R. et al. SSB functions as a sliding platform that migrates on DNA via reptation. Cell 146, 222–232 (2011).
Suksombat, S., Khafizov, R., Kozlov, A. G., Lohman, T. M. & Chemla, Y. R. Structural dynamics of E. coli single-stranded DNA binding protein reveal DNA wrapping and unwrapping pathways. ELife 4, e08193 (2015).
Naufer, M. N. et al. Multiprotein E. coli SSB–ssDNA complex shows both stable binding and rapid dissociation due to interprotein interactions. Nucleic Acids Res. 49, 1532–1549 (2021).
Shinn, M. K., Kozlov, A. G., Nguyen, B., Bujalowski, W. M. & Lohman, T. M. Are the intrinsically disordered linkers involved in SSB binding to accessory proteins?. Nucleic Acids Res. 47, 8581–8594 (2019).
Krauss, G., Sindermann, H., Schomburg, U. & Maass, G. Escherichia coli single-strand deoxyribonucleic acid binding protein: stability, specificity, and kinetics of complexes with oligonucleotides and deoxyribonucleic acid. Biochemistry 20, 5346–5352 (1981).
Bujalowski, W. & Lohman, T. M. Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. I. Evidence and a quantitative model. J. Mol. Biol. 207, 249–268 (1989).
Bétous, R., Glick, G. G., Zhao, R. & Cortez, D. Identification and characterization of SMARCAL1 protein complexes. PloS ONE 8, e63149 (2013).
Bonde, N. J., Kozlov, A. G., Cox, M. M., Lohman, T. M. & Keck, J. L. Molecular insights into the prototypical single-stranded DNA-binding protein from E. coli. Crit. Rev. Biochem. Mol. Biol. 59, 99–127 (2024).
Bhattacharyya, B. et al. Structural mechanisms of PriA-mediated DNA replication restart. Proc. Natl. Acad. Sci. USA 111, 1373–1378 (2014).
Mills, M. et al. RecQ helicase triggers a binding mode change in the SSB–DNA complex to efficiently initiate DNA unwinding. Nucleic Acids Res. 45, 11878–11890 (2017).
Gulbis, J. M. et al. Crystal structure of the chi:psi sub-assembly of the Escherichia coli DNA polymerase clamp-loader complex. Eur. J. Biochem. 271, 439–449 (2004).
Pugh, R. A., Wu, C. G. & Spies, M. Regulation of translocation polarity by helicase domain 1 in SF2B helicases. EMBO J. 31, 503–514 (2012).
Anand, R. et al. HELQ is a dual-function DSB repair enzyme modulated by RPA and RAD51. Nature 601, 268–273 (2022).
Newcomb, E. S. P., Douma, L. G., Morris, L. A. & Bloom, L. B. The Escherichia coli clamp loader rapidly remodels SSB on DNA to load clamps. Nucleic Acids Res. 50, 12872–12884 (2022).
Wu, Y., Shin-ya, K. & Brosh, R. M. FANCJ Helicase Defective in Fanconia Anemia and Breast Cancer Unwinds G-Quadruplex DNA To Defend Genomic Stability. Mol. Cell. Biol. 28, 4116–4128 (2008).
Lee, W. T. C. et al. Single-molecule imaging reveals replication fork coupled formation of G-quadruplex structures hinders local replication stress signaling. Nat. Commun. 12, 2525 (2021).
Brosh, R. M. & Wu, Y. An emerging picture of FANCJ’s role in G4 resolution to facilitate DNA replication. NAR Cancer 3, https://doi.org/10.1093/narcan/zcab034 (2021).
Yao, N., Hurwitz, J. & O’Donnell, M. Dynamics of β and proliferating cell nuclear antigen sliding clamps in traversing DNA secondary structure*. J. Biol. Chem. 275, 1421–1432 (2000).
Paschall, C. O. et al. The Escherichia coli clamp loader can actively pry open the β-sliding clamp*. J. Biol. Chem. 286, 42704–42714 (2011).
Acknowledgements
We thank Dr. Rafael Camerini-Otero (DinG), Dr. Michael O’Donnell (SSB), and Dr. James Keck (SSBΔC1) for providing protein expression plasmids. We also thank Elijah Newcomb and Andrea Murciano for labeling SSB with Alexa Fluor 647. We thank Alyssa Goodyear for her contribution to the fluorescein quench assays. Diagrams were generated with BioRender. This work was supported by the National Science Foundation [MCB 1817869 to L.B.B.] and the National Institutes of Health [GM 140166 to L.B.B.] and [NIEHS F31ES034652 to S.W.P.]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
S.J.W.P., M.J.P., K.A.P.P., and L.B.B. conceived and designed the analysis. S.J.W.P., M.J.P., K.A.P.P., R.D., and K.K.H. collected the data. S.J.W.P., M.J.P., K.A.P.P., R.D., K.K.H., and L.B.B. performed the analysis. S.J.W.P. and M.J.P. wrote the original draft. S.J.W.P., M.J.P., K.A.P.P., R.D., K.K.H., and L.B.B. reviewed and edited the draft. L.B.B. supervised and acquired funding for the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Borja Ibarra and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Weeks-Pollenz, S.J., Petrides, M.J., Padgett-Pagliai, K.A. et al. The YoaA-χ helicase modulates the dynamics of single-stranded DNA binding protein on DNA. Nat Commun 16, 4993 (2025). https://doi.org/10.1038/s41467-025-60215-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-60215-4