Abstract
The ephemeral, supraglacial meltwater ponds of the McMurdo Ice Shelf’s undulating ice serve as analogues for refugia where eukaryotic organisms could have thrived during the Cryogenian period. The seafloor sediment and debris lined ponds support the growth of a diverse array of cyanobacterial mat communities and provide habitats for a variety of protists and meiofauna. Here, we show that these eukaryotic assemblages, assessed by steroid biomarker and 18S rRNA gene analyses, inform long-standing questions regarding the diversity of, and controls on, community composition in these environments. Sixteen photosynthetically active microbial mats from meltwater ponds, a 700-year-old relict microbial mat, and a microbial mat from the Bratina Lagoon were analysed for their sterol compositions. These sterols were subjected to simulated diagenesis via catalytic hydrogenation/hydrogenolysis affording their sterane hydrocarbon counterparts, facilitating comparisons with ancient settings. Pond salinity appeared to be a factor influencing the sterol distributions observed. Analyses of 18S rRNA gene sequences conducted on the modern mats independently confirm that the ponds host diverse eukaryotes, including many types of microalgae, protists, and an array of unclassifiable organisms. Our findings support the hypothesis that supraglacial meltwater ponds like those of the McMurdo ice are strong candidates for refugia that sheltered complex life during Snowball Earth episodes.
Similar content being viewed by others
Introduction
The global prevalence of late Neoproterozoic glacial deposits at equatorial latitudes, together with a range of accompanying features such as the distinctive cap dolostones and unusual iron formations, led to the articulation of the Snowball Earth hypothesis1,2,3. Following extensive research in high precision geochronology, stratigraphy, palaeontology, geochemistry, and modelling, the Cryogenian period was established; the 720 to 635 million year ago interval is marked by two long-lived Sturtian and Marinoan glacial epochs with a warmer interglacial4,5,6. The extreme environmental transformation that took place during the Cryogenian has been cited as an evolutionary driver for the Ediacaran expansion of multicellular life7,8. The idea of a hard Snowball, a scenario in which the entire planet was encapsulated in ice so thick that sub-ice photosynthesis in the marine realm was prevented, has faced numerous critiques, with alternative proposals such as a soft Snowball, slushball9, or thin ice scenarios10 that would have been less disruptive to the marine carbon cycle. Whatever the case, it is instructive to consider the viability of eukaryotic refugia under these scenarios, specifically supraglacial settings, where aquatic photosynthesis could have been productive in support of ecosystems where complex life persisted and evolved11,12.
Supraglacial meltwater ponds, analogous to those presently scattered atop the McMurdo Ice Shelf, would have been prevalent along continental margins and supported microbial communities during the Cryogenian11,12,13. In turn, these communities would have sheltered antecedents for the succeeding Ediacaran Period’s diversification and proliferation of complex eukaryotic life, including the first animals5,14. Still, outstanding questions remain regarding the persistence and distribution of such eukaryotic refugia, as well as of the types of organisms that may have existed within them. The scarcity of fossil records of Cryogenian ecosystems, coupled with the low likelihood that evidence of life from supraglacial habitats could be preserved in the geologic record, highlights the value of detailed characterisations of contemporary analogues. While acknowledging that Cryogenian microbiota would have been different from those of today, it has long been recognised that the ecological and metabolic structuring of modern microbial mats serve as useful models for those of Earth’s past15.
Detailed characterisations of the eukaryotic communities of Antarctic supraglacial ecosystems are limited and, until recently, were primarily constructed through microscopy13,16,17,18,19. Such visual approaches necessitate the identification of distinct morphological characters—a prospect complicated by morphotype similarities for many eukaryotic microbial groups, including metazoa and SAR19. Early, microscopy-based identifications of diatoms, chlorophytes, chrysophytes, and metazoa have been confirmed and expanded upon via contemporary applications of environmental 18S rRNA gene sequencing techniques19,20,21. However, methodological challenges, such as the optimisation of primers and amplification techniques for psychrophiles, still limit the understanding of microbial eukaryotic diversity in Antarctica22. Some of these limitations may be addressed by the identification of sterols, the membrane-stabilising and cell-signalling lipid biomarkers of eukaryotes, which can be closely associated with distinct eukaryotic groups23,24. The detection of sterols and their chemically reduced counterparts, the stanols and steranes, in ancient and modern sedimentary settings enables an independent reconstruction of the eukaryotic community. Though sterol biosynthetic pathways may be shared across many eukaryotic taxa, broad classifications may be assigned, and steroid biomarker compositions may be compared across different environmental settings and time.
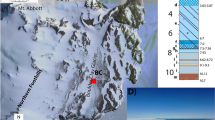
The McMurdo Ice Shelf’s undulating ice affords a readily accessible Cryogenian analogue environment with the opportunity to examine microbial eukaryotic distributions across a variety of supraglacial settings, including physicochemically-diverse meltwater ponds (Fig. 1 and Table 1) supporting photosynthetically active microbial communities (Fig. 2A), former pond basins that are now elevated and have desiccated (Fig. 2B), and an active meltwater lagoon system. This environment also hosts an array of marine debris on the ice surface (Fig. 2C, D). To characterise signatures of eukaryotic life across the undulating ice, steroid biomarker distributions were examined in 16 microbial mats collected from the landscape and compared with 18S rRNA gene characterisations of the mat communities. Sterols derived from the microbial mats were further subjected to simulated diagenesis via catalytic hydrogenation/hydrogenolysis to generate sterane assemblages that can be compared to those preserved in the fossil record. The results of these analyses support the notion that diverse communities of protists and meiofauna persist within supraglacial meltwater ponds, and that the meltwater refugia on the McMurdo Ice Shelf are plausible candidates for settings which harboured complex ecosystems during the Cryogenian Period.
A The geographical location of the study vicinity and Bratina Island. B Detailed depiction of the meltwater ponds off the coast of Bratina Island based on satellite imagery. Sites included in this study are outlined in black and coloured in blue. Not shown: Bratina Lagoon, located approximately 1 km west of the area depicted.
Results
Sterol TMS ethers
Diverse assemblages of C26 to C30 sterols were identified across the ponds, Bratina Lagoon, and within the relict microbial mat. Representative chromatograms for ponds Skua, Duet, and Brack are shown in Fig. 3; systematic names corresponding to peak numbers are denoted in Table 2. A list of common names corresponding to systematic names is available in Supplementary Table 1. Twenty-nine individual steroids were identified across the samples not including the internal standard, cholest-5-en-3β-ol (2H7). In some cases, sterols clearly co-eluted with another compound, as in peaks 16, 17, 21, 25, and 26. For peaks 21 and 26, the sterols co-eluted with non-steroidal molecules, such as α-tocopherol in the latter. 24-Ethylcholesta-5,24(28)Z-dien-3β-ol, peak 25, co-elutes with (24R)-ethyl-5α-cholestan-3β-ol on a standard DB-5MS column as shown in the supplementary material (Supplementary Fig. 1). Nine unknown steroids were identified for which standards and reference spectra were not available; their mass spectra and observed characteristics are reported in the supplementary material (Supplementary Figs. 2–10).
Sterols identified are numbered; their corresponding peaks are described in Table 2. Peaks labelled A represent carboxylic acids, peaks labelled S represent siloxanes, and the peak labelled U represents an unidentified non-sterol, non-acid compound.
A sole C26 sterol, 22-trans-24-norcholesta-5,22E-dien-3β-ol, was detected in the Brack Pond microbial mat. Most samples contained a range of C27 sterols and stanols, including 5α-cholest-22E-en-3β-ol, 5β-cholestan-3β-ol, 27-nor-(24S)-cholesta-5,22E-dien-3β-ol, cholesta-5,22E-dien-3β-ol, cholest-5-en-3β-ol, 5α-cholestan-3β-ol, and 5α-cholest-7-en-3β-ol. In nearly all microbial mat samples, cholest-5-en-3β-ol had the greatest percent abundance of all sterols except in the case of ponds Skua, Fogghorne, Conophyton, Orange, Legin, Brack, and in the relict microbial mat. In those ponds except Brack and Salt, a C29 sterol, (24R)-ethylcholest-5-en-3β-ol, was the dominant sterol. Brack and Salt Pond’s sterol assemblages were dominated by 24-methylcholesta-5,22E-dien-3β-ol, a C28 sterol. The C28 sterols 24-methylcholesta-5,22E-dien-3β-ol and (24R)-methylcholest-5-en-3β-ol were present in all microbial mats examined, while 24-methylcholesta-5,24(28)-dien-3β-ol was detected in all ponds except Skua, Roger, AMS, and the Bratina Lagoon. 24-Methyl-5α-cholest-7-en-3β-ol was detected in most of the mat samples except in those from ponds Roger, AMS, Salt, the Bratina Lagoon, and the relict microbial mat.
(24R)-Methyl-5α-cholestan-3β-ol minorly contributed to the total sterol abundances and was only detectable in ponds Skua, Duet, Fresh, Conophyton, Castenholz, Legin, Seventy, and Brack. The C29 sterols (24S)-ethylcholesta-5,22E-dien-3β-ol and (24R)-ethylcholest-5-en-3β-ol were present in all samples and together represented the largest proportion of sterols except in pond New, where they were present in equal proportion to cholest-5-en-3β-ol, and in ponds Salt and AMS. 24-ethylcholesta-5,24(28)Z-dien-3β-ol was not quantified due to its co-elution with (24R)-ethyl-5α-cholestan-3β-ol but its fragment ions, including 484, 386, 296, 281, 257, and 129 Da, were present within the wide C29 peaks across samples. The C29 phytosterol (24S)-ethyl-5α-cholesta-7,22E-dien-3β-ol was present in all environments except those of ponds Roger, AMS, Salt, the Bratina Lagoon, and within the relict microbial mat. 23,24-Dimethyl-5α-cholest-22E-en-3β-ol was detected, though in minor proportions across ponds Skua, Duet, Conophyton, Legin, and Pond Seventy. 4α,23,24-Trimethylcholest-22E-en-3β-ol was the sole C30 sterol detected within the sample set, in Skua Pond, and it co-eluted with a compound containing a strong 502 Da fragment, as well as 237, 221, and 73 Da fragments, which was tentatively identified to be α-tocopherol.
Sterane hydrocarbons
The simulated diagenesis of microbial mat sterols yielded an assortment of steranes with varying proportions across the sample set; the abundances of 4-desmethylsteranes are described in Table 3. C27 through C29 steranes dominated the total sterane distributions; minor proportions of C26 steranes, C30 4-desmethylsteranes, and C30 4-methylsteranes were also present. Mat sterols from all ponds generated the C26 norsteranes 24-norcholestane and 27-norcholestane in varying proportions, while the C30 4-desmethylsteranes 24-n-propylcholestane and 24-isopropylcholestane were recovered from ponds Skua, Fresh, Conophyton, Castenholz, Legin, Pond Seventy, Brack, Salt and from the Bratina Lagoon and were present in varying proportions. C30 4-methylsteranes were detected at either trace or quantifiable abundances in all the ponds and were not employed in abundance calculations. A putative C31 sterane (Supplementary Fig. 11) was detected after hydrogenation of the Pond Seventy sterols and comprised 0.02% of the total 4-desmethylsteranes; this compound was not detected elsewhere.
C27 to C29 sterane distributions varied across the mat samples. When plotted on a C27-C28-C29 ternary diagram, the mat-derived steranes cluster centrally, though steranes derived from mats from ponds with conductivities >4000 µS/cm have higher proportions of C28 steranes (Fig. 4). Steranes derived from the relict microbial plotted closely with those derived from Pond Seventy, Legin Pond, Duet Pond, and Castenholz Pond. Steranes analysed from the hydrocarbon fraction of the relict mats, which was not subjected to hydrogenation, plotted closely with its hydrogenated sterol counterpart.
Hydrogenated relict microbial mat steranes, noted by 1, are included as well as relict microbial mat steranes without hydrogenation, noted by F. Values were calculated using the relative intensities of ααα20R isomers. Ponds with conductivities >4000 µS/cm are noted with orange squares; ponds with conductivities <4000 µS/cm are noted with blue circles; ponds without conductivity information available are noted with white squares.
Catalytic hydrogenation/hydrogenolysis of the microbial mat sterol fractions yielded ααα20R and βαα20R sterane epimers; representative MRM chromatograms and peak identities from Fresh Pond are shown in Fig. 5. A total of ten 4-desmethylsteranes were detected, with the 5α epimer present in greater abundance than the 5β epimer in all cases. The isomer ratios are compiled for the steranes across the mats for C27 through C29 steranes in Supplementary Fig. 12. Across the sample set, the 5α:5β ratio ranged from close to 1:1 in the case of the relict mat and the mat from the Skua Pond margin, while steranes generated from the Bratina Lagoon mat exceeded a 7:1 ratio. Steranes generated from photosynthetically active microbial mats in meltwater ponds contained ratios intermediate to these endmembers. No clear trend was observed between carbon number and 5α:5β ratios.
In addition to the detection of C26 norsteranes across the sample set, a series of C27 to C29 norsteranes were present as shown in Fig. 5; they eluted slightly after the primary ααα20R and βαα20R cholestane, 24-methylcholestane, and 24-ethylcholestane peaks and appear to be the 27-nor series. In some cases, the C27 to C29 norsteranes were present in abundances too low for integration, such as in the case of Orange Pond, while in the relict microbial mat hydrogenated sterols, norsteranes comprised nearly 9% of the total steranes (Table 3). The presence of these norsteranes in the hydrocarbon fraction of the relict microbial mat, which was not subjected to any chemical treatment, confirm the natural occurrence of the C27 to C29 norsterane series detected (Supplementary Fig. 13).
18S rRNA gene analysis
The 18S rRNA gene sequencing of microbial eukaryotes in the microbial mat fragments revealed the presence of a variety of eukaryotic groups, including SAR, Opisthokonta, and Archaeplastida. These groups had the largest relative abundances (Fig. 6) across all ponds studied, followed by Amoebozoa and Excavata. Centrohelida comprised a small relative abundance in all ponds except Salt, where it was absent, while Cryptophyceae were minor constituents of ponds New, Fogghorne, Roger, Conophyton, Orange, Castenholz, Legin, Salt, and in the Bratina Lagoon. Haptophyta had low relative abundances in ponds Duet, Fresh, Conophyton, Orange, Castenholz, Seventy, and Brack. In some ponds, including Duet, Fresh, and AMS, the relative abundance of uncategorisable eukaryotes exceeded 50%; in other cases, some of the 18S rRNA gene sequences were grouped into incertae sedis.
Further evaluation revealed that certain subgroups dominated within the mats (Fig. 6 and Supplementary Fig. 14). Among the 18S rRNA genes assigned to Stramenopiles, Ochrophyta had the highest relative abundance, followed by minor contributions from Labyrinthulomycetes, Peronosporomycetes, and Bicosoecida. A small proportion of Stramenopiles were classified as incertae sedis and comprised less than 0.2% relative abundance in ponds Brack and New. Among the categorizable Alveolata, Ciliophora had the highest relative abundance, followed by Protalveolata and Dinoflagellata. Apicomplexa were detected in Fogghorne and Conophyton Ponds, corresponding to less than 0.1% relative abundance. All Rhizaria detected belonged to Cercozoa. Nearly all Archaeplastida sequences detected across all ponds grouped within Chloroplastida and mostly consisted of 18S rRNA gene sequences from the green alga Chlorophyta, followed by Charophyta. Among the ponds, Salt exclusively contained 18S rRNA gene sequences grouping into Chlorophyta, while it comprised the majority of the Archaeplastida in all ponds except AMS, where Charophyta was in the majority. The SAR group, comprising Stramenopiles, Alveolata, and Rhizaria, had the highest relative abundances in Salt Pond and in the Bratina Lagoon.
Within the Opisthokonta (Supplementary Fig. 15), Holozoa, including Metazoa and Choanoflagellida, comprised the largest relative abundance of 18S rRNA gene sequences in all ponds. Among the Nucletmycea, the Fungi and the Nucleariidae and Fonticula groups were detected and comprised at least 20% of the Opisthokonta relative abundance in ponds Brack, Orange, Duet, and Fresh.
Among the categorizable Amoebozoa (Supplementary Fig. 16), Tubulinea and Discosea had the largest relative abundances, followed by smaller contributions from Schizoplasmodiida. Protosteliida were only detected Roger Pond and comprised less than 0.2% of the relative abundance. Brack Pond, which had the highest relative abundance of Amoebozoa among all the ponds, also had the highest relative abundance of unclassified Amoebozoa among the microbial mats studied. Salt Pond had the largest relative abundance of Leptomyxida, while Fogghorne Pond had the highest relative abundance of Euamoebida; these groups were a minor constituent or absent in all other ponds. Centrohelida (Supplementary Fig. 17) were detected in all ponds except Salt; in these ponds, the group comprised less than 1% of the eukaryotic community.
Cryptophyceae (Supplementary Fig. 18) were present in low relative abundances in most ponds and were absent in ponds Seventy, Brack, Duet, Fresh, and AMS. The majority of the 18S rRNA gene sequences were assigned to Cryptomonadales, specifically Cryptophyta. The vast majority of Excavata detected (Supplementary Fig. 19) were assigned to Discoba and were present in all the microbial mats studied. Few 18S rRNA gene sequences from Haptophyta were detected (Supplementary Fig. 20); of the ponds that contained their sequences, Pond Seventy contained the largest relative abundance of 18S rRNA gene sequences assigned to Haptophyta, all of which were members of the Prymnesiophyceae class. Other ponds, such as Brack, Castenholz, Orange, Duet, and Fresh, contained 18S rRNA gene sequences from Pavlovophyceae. Haptophyte 18S rRNA gene sequences across the ponds belonged to either Prymnesiophyceae or Pavlovophyceae—no ponds in this work contained 18S rRNA gene sequences from both subgroups simultaneously. Small proportions of 18S rRNA gene sequences from some ponds were broadly categorised as incertae sedis (Supplementary Fig. 21), including Apusomonadidae and Amastigomonas in ponds Roger, Legin, and Seventy, as well as the Bratina Lagoon, as Breviata in ponds New, Duet, Roger, Conophyton, Orange, AMS, and Seventy, and as Telonema in Salt Pond and Legin Pond.
Principal component analysis
Principal component analysis (PCA) revealed relationships between the meltwater ponds and accompanying sterane and 18S rRNA gene data (Fig. 7). PCA biplots were comprised of loadings, which describe how strongly variables such as sterane or 18S rRNA gene categories influenced each principal component, and the principal component scores for each pond examined. For sterane distributions, the analysis recovered five principal components (PCs), while ten components were recovered for 18S rRNA gene distributions. PCs with the largest eigenvalues that explained at least 80% of the total variance were selected; this resulted in the retention of 2 PCs for each analysis. Environmental parameters such as pH, temperature, and conductivity were overlaid on individual pond PC scores to examine their potential impacts on the distribution of the data; no trends were observed for pH and temperature, which only varied slightly across ponds. Ponds Salt and Brack, which had the highest conductivities, had similar PC scores and clustered in both analyses, unlike ponds Duet and Fresh, which had the lowest conductivities but dissimilar scores across PCs.
Principal component scores for individual ponds are coloured according to conductivity, while loadings, representing the distributions of A steranes and B 18S rRNA genes, are coloured in dark purple. Certain 18S rRNA gene variables plotted very closely together near the origin and their names are abbreviated. Cr is short for Cryptophyceae, Ce is short for Centrohelida, and Ha is short for Haptophyta.
Within the sterane PCA analysis, minor sterane constituents exerted weak influence on the principal components, while the C27, C28, and C29 steranes strongly influenced PC1. The C27 and C29 steranes influenced PC2 while the C28 steranes did not. Ponds Brack and Salt were most influenced by C28 steranes, while ponds with lower conductivities were more influenced by C27 and C29 steranes. Duet Pond, which had the lowest conductivity, was most influenced by C29 steranes. In the 18S rRNA gene distribution PCA analysis, only Ophisthokonta, SAR, Archaeplastida, and Uncategorisable Eukaryota had strong influences on the PCs; Cryptophyceae, Centrohelida, Haptophyta, Excavata, and incertae sedis were largely uninfluential. Amoebozoa were slightly correlated with SAR. Ponds AMS and Fresh were influenced by Uncategorisable Eukaryota, while Conophyton was most influenced by Archaeplastida. Ponds Legin and Roger were most influenced by Opisthokonta. Ponds which plotted near the origin, such Castenholz, Seventy, Brack, and Salt, which had higher salinities than ponds with greater PC scores, were largely uninfluenced by any 18S rRNA gene distribution variables.
Discussion
The techniques applied in this work extend previous characterisations of microbial life from the Bratina meltwater pond landscape20,21,25,26,27, which mostly focused on the dominant bacterial diversity of the microbial mat communities. To characterise the eukaryotic communities living in the supraglacial meltwater pond landscape, sterol biomarkers were extracted, analysed, and converted to their fossil analogues via simulated diagenesis. This approach generates not only a would-be fossil assemblage for a type of organosedimentary system unlikely to be preserved in the fossil record12, but also a point-of-view from which low-abundance and/or potentially co-eluting sterols can be more readily detected and interpreted. Significant eukaryotic diversity remains under characterised in this Cryogenian analogue environment via modern genetic sequencing, underscoring the continued utility of steroid biomarker analyses of potential eukaryotic refugia.
18S rRNA gene eukaryotic communities in meltwater pond microbial mats
The eukaryotic 18S rRNA gene sequences analysed from the meltwater pond and Bratina Lagoon microbial mats were broadly classified into the major eukaryotic groups Amoebozoa, Archaeplastida, Centrohelida, Cryptophyceae, Excavata, Haptophyta, Opisthokonta, SAR, incertae sedis, and uncategorisable Eukaryota. Opisthokonta, along with SAR and Archaeplastida, comprised the greatest relative abundances of categorizable Eukaryota across the meltwater ponds. The high proportions of Opisthokonta detected likely reflect the presence of meiofauna and microfauna in the microbial mats, including nematodes, tardigrades, and rotifers. Due to the size difference between these meiofauna, microfauna, and protist and fungi cells, it is possible that the metazoa may bias the 18S rRNA gene survey results. It is for this reason that metazoa are commonly excluded from 18S rRNA gene surveys in polar environments20. The prevalence of 18S rRNA gene sequences which were not categorizable past Eukaryota and the broadest classification levels may be due to the use of short sequence reads, as well as the limited representation of polar microorganisms in reference databases22. It is also possible that these ponds may support cryptic or dark eukaryotic diversity that has yet evaded growing annotated environmental databases, as is currently the case with diverse, polyphyletic groups such as algae28 and SAR29.
The eukaryotic compositions of the meltwater ponds and lagoon varied greatly despite the close proximity and physicochemistries of the individual sites, a finding similarly observed during a previous 18S rRNA gene analysis of a small subset of these ponds19. The implication of this variation is that there is no archetypal microbial mat eukaryotic composition in these environments, though it is generally possible to expect the consistent presence of SAR, Opisthokonta, and Archaeplastida as the most abundant categorizable constituents. For example, both ponds Conophyton and Salt contain these three eukaryotic groups, but Conophyton Pond is dominated by Archaeplastida while Salt Pond is SAR-dominated (Fig. 6).
Potential controls, including environmental variables, have not been previously shown to majorly impact eukaryotic community composition20. However, the graphical representation of the PCA conducted in this work (Fig. 7b) revealed that salinity may play a more prominent role than previously determined. Ponds with more extreme principal component scores tended to have relatively low conductivities, while ponds closer to the origin, whose PC scores did not appear to be significantly influenced by the variables examined, were measured to have higher conductivities except for Castenholz. This potential subtle salinity control is more apparent in the sterane ternary (Fig. 4) and sterane PCA analysis (Fig. 7a) and is discussed further below.
Sterols
Sterol assemblages detected across the mats, although varied in their individual components and their distributions, broadly correspond to those produced by microalgae, protists, and potentially, metazoan meiofauna (Table 2).
Previous studies of Antarctic diatoms have recovered sterol distributions which closely overlap with those observed within the meltwater pond microbial mats, the components of which include 22-trans-24-norcholesta-5,22E-dien-3β-ol, cholesta-5,22E-dien-3β-ol, cholest-5-en-3β-ol, 24-methylcholesta-5,22E-dien-3β-ol, 24-methylcholesta-5,24(28)-dien-3β-ol, 24-methylcholest-5-en-3β-ol, 24-ethylcholesta-5,22E-dien-3β-ol, 24-ethylcholest-5-en-3β-ol, and 4α,23,24-trimethylcholest-22E-en-3β-ol30,31. Many of these compounds have been detected in other protists, such as dinoflagellates, and microalgae, including marine and freshwater eustigmatophytes, haptophytes, cryptophytes, chrysophytes, and prasinophytes23,32,33 Rampen et al.’s34 detailed classification of sterols from a variety of diatoms demonstrated that the most common sterols in this group were comprised of 24-methylcholesta-5,24(28)-dien-3β-ol, cholest-5-en-3β-ol, 24-methylcholest-5-en-3β-ol, and 24-ethylcholest-5-en-3β-ol, all of which are present in the majority of the microbial mats examined in this work34. Some sterols detected, such as 5α-cholest-22E-en-3β-ol and 27-nor-(24S)-methylcholesta-5,22E-dien-3β-ol, may derive from dinoflagellates35,36,37, while others, including the 22-trans-24-norcholesta-5,22E-dien-3β-ol detected in Brack Pond, and 24-methylcholesta-5,22E-dien-3β-ol, have been shown to be produced by both diatoms and dinoflagellates34,38,39. Certain Rhizaria, such as Cercozoa, have also been shown to produce 24-methylcholesta-5,22E-dien-3β-ol and (24S)-ethylcholesta-5,22E-dien-3β-ol32. Other sterols detected, including (24R)-methyl-5α-cholest-7-en-3β-ol, (24R)-ethyl-5α-cholestan-3β-ol and (24S)-ethyl-5α-cholest-7,22E-dien-3β-ol, have been detected in microalgae as well23,40,41. These potential overlapping sterol sources, including diatoms, dinoflagellates, and microalgae (Fig. 6) were all detected during the18S rRNA gene surveys of microbial mats from these ponds.
Certain sterols detected, including cholest-5-en-3β-ol, are commonly associated with animals, though they are widely produced by algae and other organisms24. It is likely that within the microbial mats, both algae and meiofauna contribute to the cholest-5-en-3β-ol abundances demonstrated, which is consistent with the 18S rRNA recovery of sequences from both groups. Seafloor metazoa and meiofauna may also contribute to the steroid assemblages as their remnants are distributed across the undulating ice shelf surface and line the base of each meltwater pond, and their steroids preserve for longer time periods in the environment than DNA (Fig. 2).
A mixture of 5α and 5β stanols were recovered in the microbial mats, possibly reflecting the natural reduction of sterols in a reducing setting42 as opposed to fecal inputs that might be expected in settings nearer to sites of human habitation24. Aged microbial mats, including the mat collected from the Skua Pond margin and the relict microbial mat, contained measurable 5β-cholestan-3β-ol, while microbial mats sampled from pond waters did not, suggesting that this molecule may be generated from the hydrogenation and epimerization of cholest-5-en-3β-ol, processes active particularly in dried, or decaying mats. A similar natural diagenetic process may have led to the production of the (24R)-ethyl-5β-cholestan-3β-ol detected in the Skua Pond margin microbial mat, which is derived from the reduction and subsequent epimerization of (24S)-ethylcholesta-5,22E-dien-3β-ol and (24R)-ethylcholest-5-en-3β-ol.
Steranes
The most abundant steranes generated by the simulated diagenesis of the microbial mat sterols corresponded to the C27, C28, and C29 precursor sterols discussed in Sterols. For example, during catalytic hydrogenation/hydrogenolysis, the C29 sterols (24S)-ethylcholesta-5,22E-dien-3β-ol, 24R-ethylcholest-5-en-3β-ol, and (24R)-ethyl-5α-cholestan-3β-ol all convert to 24-ethylcholestane as their double bonds are reduced and oxygens lost; these modifications mimic common chemical transformations that occur during early diagenesis. However, small amounts of other steranes, including norsteranes and C30 4-desmethylsteranes were detected, likely reflecting contributions from the meltwater communities themselves as well as contributions from seafloor communities which have been transported to the ice shelf surface over time (Fig. 2)12.
Norsteranes have been detected in a variety of oils and their source rocks, including the C26 21-, 24- and 27-norcholestanes43 and the C27 norsterane 27-nor-24-methylcholestane44. These molecules are thought to represent contributions from diatoms and dinoflagellates36,38,43,44,45,46, though some norsteranes have been hypothesised to derive from sponges47,48. Hydrogenation of the microbial mat sterols in this work resulted in the production of the C26 norsteranes, 24-norcholestane and 27-norcholestane, while 21-norcholestane, a possible product of thermal degradation, was notably absent43. In addition, a series of compounds that eluted closely following cholestane, 24-methylcholestane, and 24-ethylcholestane were observed, likely representing small quantities of isomers of analogous 27-norsteranes for C27, C28, and C29 steranes. To eliminate the possibility that these additional steranes were generated as a byproduct of the simulated diagenesis conducted on the microbial mats, the hydrocarbon fraction of the 700-year-old relict microbial mat was also analysed for its sterane contents. The same norsterane series evident in the hydrogenated 700-year-old microbial mat was also present in its free hydrocarbon fraction (Supplementary Fig. 13), which had not been subjected to hydrogenation after total lipid extraction. Further, hydrogenation experiments of pure sterol standards have been previously shown to not generate 27-norsteranes49, further suggesting that 27-norsterol precursors are indeed present in the microbial mats. Given the abundance of microalgal sterols detected in the microbial mats in this landscape, two of which are precursors molecules to 27-norsteranes, it is possible that these norsteranes are indicative of small microalgal or diatom inputs which are undetectable against more abundant sterols and stanols, though it is possible that some of the 27-norsterol derived from seafloor life as seafloor sediments line the bases of the meltwater ponds and are scattered across the undulating ice. These findings are in line with Moldowan et al.’s43 inference that unlike the primary cholestanes, 24-methylcholestane and 24-ethylcholestane, which result from a wide variety of precursor molecules, norsteranes and other less common steranes may be more informative for analyses that aim to link fossil molecules with their biological precursor producers43.
In addition to the norsteranes, the C30 4-desmethylsteranes 24-n-propylcholestane and 24-isopropylcholestane as well as 4-methylsteranes were detected in varying abundances across the mats. Precursor molecules to 24-n-propylcholestane, 24-n-propylidene-cholesterol and 24-n-propylcholesterol, have been detected in a variety of algae, including chrysophytes and pelagophytes32,50,51 Volkman et al.52 generated 24-n-propylcholestane from the hydrogenation of lipids extracted from a prasinophyte algae, though its C30 sterol precursor was not detected during the sterol composition analysis of that organism in that study52. Since other works have shown the detection of its precursor molecules in other algae32, it is possible that the precursor sterol was present in the original algal culture examined but in quantities undetectable by conventional full scan GC-MS methods. Mild catalytic hydrogenation methods like the ones employed by Volkman et al. in 1994 and in this study have not been demonstrated to selectively methylate their steroid reactants. In addition to algal sources, foraminifera may also contribute 24-n-propylcholestane precursors53.
Sterol precursors of the hydrocarbon 24-isopropylcholestane are commonly attributed to demosponges54,55 and feasibly represent inputs from demosponges transported to the ice shelf surface. Sponge remains were ubiquitous on the undulating ice at the time of sampling and have been identified on the McMurdo Ice Shelf surface for over a century56 (Fig. 2), having been transported from the seafloor via the conveyor belt mechanism referenced by Hawes et al. in 201812, and with many species of Antarctic demosponges having been previously characterised57. Recent studies have proposed alternative origins for 24-isopropylcholestane precursors, including bacterial sponge symbionts, pelagophyte algae, and Rhizaria49,58,59. Others have suggested generation of 24-isopropylcholestane via the thermally-driven methylation of C29 algal sterols60. Such a process is excluded at the near-freezing to below-freezing temperatures on the surface of the ice shelf and in the absence of geothermal heating. The ratios of 24-isopropylcholestane to 24-n-propylcholestane in the dataset vary by 2 orders of magnitude across the samples from which both molecules were detected after hydrogenation, and in some cases, 24-n-propylcholestane was absent where 24-isopropylcholestane was present (Table 3), potentially ruling out an algal source for 24-isopropylcholestane in the meltwater environments as both molecules would be expected from algal sources. Further, 26-methylstigmastane was not detected in any hydrogenated microbial mat lipid extract, eliminating Cercozoa from contention as producers of the 24-isopropylcholestane precursors. Cercozoa have previously been described to produce both 24-isopropylcholestane and 26-methylstigmastane49. The application of long-read sequencing for 18S rRNA gene metabarcoding, the characterisation of environmental genomes of polar aquatic communities, and even ancient DNA analyses may greatly assist the definitive source assignment of the C30 4-desmethylsteranes in this environment, though the simultaneous presence of algae, sponge fragments, and Rhizaria in the environment may be always confounding.
Further, the clear identification of precursor molecules for these compounds in sterol fractions may ultimately aid in source-assignment. However, the complex nature of environmental samples, particularly those with such a diverse array of C27 to C29 steroids, makes the detection of low-abundance steroids exceedingly difficult via traditional full-scan gas chromatography-mass spectrometric methods using nonpolar GC capillary columns. It is possible that the precursor molecules for the norsteranes as well as the C30 steranes detected, which must be present in the microbial mat sterol fractions, co-elute with other more abundant compounds or are present at levels below the instrumental detection limits during a full scan. However, once hydrogenated, they are readily detectable via dynamic multiple reaction monitoring, a technique which affords significant sensitivity and selectivity advantages compared to full scan mass spectrometry. Such is the case with the 4-methylsteranes, which were detected at measurable or trace abundances in all ponds studied via the 414 → 231 Da transition during multiple reaction monitoring (as in Fig. 5). Their precursor molecules are produced by dinoflagellates in the case of 4α,23,24-trimethylcholestanes and by prymnesiophyte algae in the case of 24-ethyl-4α-methylcholestanes54,61,62. However, these precursor molecules were similarly not detected within the sterol fractions analysed prior to hydrogenation except in the case of Skua Pond (Table 2), though a previous study of microbial mats in this environment successfully detected 4α,23,24-trimethylcholest-22E-en-3β-ol and its associated stanol26.
C27-C28-C29 sterane ternary analysis of the steranes generated by the simulated diagenesis of the microbial mat sterols (Fig. 4) reveals a distribution largely consistent with open marine and estuarine/bay ecosystems63. This distribution of steranes synthesised from Bratina microbial mat sterols contrasts with sterane distributions of Neoproterozoic and early Cambrian marine samples from around the globe, which cluster near the C27 or C29 endmembers during ternary analysis47,64,65,66. However, there is noticeable similarity between the central placement of the steranes generated from meltwater pond microbial mats and the distribution of steranes recovered from Phanerozoic sediments and oils64. The dominance of C27 or C29 steranes observed in most of the mats aligns closely with sterane distributions previously reported for Quaternary-aged sediments collected from the western Ross Sea67, though in two ponds, Brack and Salt, the major sterane contained 28 carbon atoms. While most of the mats were collected from ponds with conductivities which correspond to freshwater or mildly saline environments, ponds Brack and Salt had conductivities of 6690 and 28,400 µS/cm, respectively (Table 1). While these values are still lower than those measured in seawaters, they may be associated with a greater abundance of C28 sterol producers as demonstrated by sterane PCA analysis (Fig. 7). The sterane distributions align with previous research which suggested salinity as an abiotic factor influencing community composition in these environments, although that relationship was only demonstrable for prokaryotes20,21. Given the association observed between C28 steroid abundances and conductivity, it may be possible to make inferences about the conductivities of now-dry ponds from the hydrocarbon distributions of their relict microbial mats. The decaying microbial mat from Skua Pond, which was collected from the dry pond margin and therefore may represent a precursor relict mat, plotted among the other ponds with low conductivities (Fig. 4). The conductivity of the pond from which it derived was 1920 µS/cm, a number considerably lower than those measured in the ponds with higher proportions of C28 steranes. The 700-year-old relict microbial mat similarly plotted among mats with low conductivities, suggesting that it may have once grown within a freshwater or slightly saline pond.
Supraglacial meltwater oases as Cryogenian refugia
The detailed characterisation of the eukaryotic assemblages of supraglacial meltwater ponds is a prerequisite for their continued candidacy as possible Cryogenian eukaryotic refugia. In order to explain the ensuing expansion of eukaryotic and multicellular life in the Ediacaran as evidenced by the fossil record and molecular clock analyses8,68, Cryogenian refugia would have needed to support sufficient eukaryotic diversity; this is consistent with an emerging consensus that global glaciations did not impose an evolutionary bottleneck on eukaryotic life11,12.
The complementary application of 18S rRNA gene and sterol biomarker analyses revealed that microbial mats in supraglacial meltwater ponds are capable of supporting diverse eukaryotic assemblages. While 18S rRNA gene analysis provided information about living eukaryotic communities, sterol and sterane analysis enabled the assessment of the activities of current and former eukaryotic communities as well as of marine eukaryotic communities beneath the ice shelf whose remnants have accumulated atop the McMurdo Ice Shelf. A considerable proportion of the eukaryotic communities in the meltwater ponds was categorised as Archaeplastida and SAR through 18S rRNA gene surveys, consistent with the recovery of a diverse assortment of sterols commonly attributed to members of these groups. However, the abundance of uncategorisable eukaryotes, either due to methodological limitations or the lack of characterised reference genomes, potentially complicates analyses which attempt to examine physicochemical controls on 18S rRNA gene derived eukaryotic community compositions. Sterol and sterane assemblages can broadly describe eukaryotic communities in the absence of genetic data and revealed that eukaryotic assemblages in this environment vary with pond conductivity.
Many of the eukaryotic groups present in the microbial mats may have emerged prior to the onset of Cryogenian glaciation events according to molecular clock and fossil evidence. Fossil-calibrated molecular clocks indicate that major eukaryotic clades, including SAR, Excavata, Amoebozoa, and Opisthokonta diverged before 1000 Ma68. Archaeplastida, a constituent of all the microbial mats examined by 18S rRNA gene analysis, diversified significantly following the termination of the Sturtian glaciation, as evidenced by the Neoproterozoic sterane record64. Sterane biomarkers prior to the Sturtian are primarily comprised of cholestane attributed to red algae, while C29 steranes, more commonly associated with green algae, appeared before the initiation of the Marinoan glaciation64, though it is possible that additional extinct Neoproterozoic clades may have also contributed to the steranes recovered. The SAR group eukaryotes present in the pond microbial mats, including diatoms, dinoflagellates, and Cercozoa, diversified in the Mesozoic according to fossil and biomarker evidence69, and therefore would have been unlikely to have been major constituents of supraglacial Cryogenian eukaryotic assemblages. While the modern eukaryotic assemblages in the microbial mats in this study do not closely resemble pre-Cryogenian eukaryotic communities, their constituents support the hypothesis that these types of environments would have been capable of supporting diverse eukaryotic communities during the Cryogenian.
Methods
Study site
During the austral summer, cyanobacterial mats perennially grow in the meltwater ponds of the debris-covered ‘Dirty Ice’ located between Antarctica’s Bratina Island (78°00'27“S, 165°33'10“E) and the north tip of Brown Peninsula (78°02'01“S, 165°31'46“E) (Fig. 1). Known as the undulating ice, the landscape is shaped by the slow compression of ice against Bratina Island and is part of the McMurdo Ice Shelf’s ablation zone16,70,71. Dark-coloured moraine and seafloor debris cover most of the ice surface; previous measurements have estimated sediment heights from the surface of the ice to range from upwards of 10 cm to 30 cm in some places16,71,72. For a few weeks each year, meltwater ponds with various physicochemistries unfreeze atop the ice shelf, their floors lined with multi-annual cyanobacteria-dominated benthic microbial mat communities with stable community compositions (Fig. 2a)16,18,72,73,74,75,76.
Atop the ‘Dirty Ice,’ together with the photosynthetically active microbial mats, relict microbial mats from dried ponds are scattered throughout the region (Fig. 2b)27,77, as are marine debris, which are transported to the ice shelf surface from the seafloor via a combination of basal freezing and surface ablation, i.e., the conveyor belt mechanism detailed and supported by Hawes et al. in 2018 (Fig. 2c, d) and first formally suggested by Frank Debenham nearly a century earlier56. In addition to the marine debris pictured in Fig. 2, corals, shells, sponge spicules, frozen fish, and fragments of various marine invertebrates have been observed on the ice since the site’s first characterisations in the early twentieth century, beginning with those made during the Discovery expedition56,70,78,79. Located southwest of Bratina Island is the Bratina Lagoon (78°00 ‘56“S, 165°29’ 51“E). The tidal lagoon sits at the margins of the McMurdo Ice Shelf and Bratina Island and Brown Peninsula landmass, and it is flushed once-daily through a tide crack12,16,80. Hawes et al.80 measured a low conductivity of the tidal waters, indicating a meltwater, rather than seawater origin. Microbial mats line the lagoon shoreline as well80.
Sample collection
Sixteen microbial mat samples were collected from the study area in January 2018 using clean stainless steel spatulas21,27. Portions of the samples were frozen and stored in sterile tubes for rRNA analysis and in pre-combusted glass jars for lipid biomarker analysis21,27. Thirteen of the mats considered for this work were collected from active meltwater ponds, and two decaying microbial mat samples were collected from the margins of Skua Pond and the Bratina Lagoon. One desiccated relict mat was collected from a mound of dried mats located between Fresh and Brack ponds; its age was determined to be ca. 700 years old via radiocarbon dating at the National Ocean Sciences Accelerator Mass Spectrometry Facility as reported by Drozd et al. in 202327. At the time of collection, pH, temperature, and conductivity measurements were taken for some pond environments21. A summary of the samples and data collected are detailed in Table 1.
Sterol analysis
Sterol analysis was conducted on the same lipid extracts analysed by Evans et al.21; four additional samples were added to the sample set in this work, including the relict microbial mat, the lagoon microbial mat, as well as those from ponds Roger and AMS. Total lipids were manually extracted from the four samples as well as from microbial mat fragments from the original set of ponds via ultrasonication ahead of catalytic hydrogenation to convert mat sterols to their sterane hydrocarbon counterparts. Due to a lack of remaining biomass, total lipid extracts were not collected from ponds New or Fogghorne and so they are not included in the sterane analysis. Homogenised mat powders were mixed with dichloromethane and methanol (9:1 vol/vol) in ashed glass vials and sonicated for 20 min; extracted lipids were pipetted into a clean glass vial and the procedure was repeated a total of three times. A fourth extraction was conducted with acetone and methanol (7:2 vol/vol) to collect any remaining polar lipids from the microbial mat samples. The collected lipid extracts were concentrated under a gentle stream of N2.
The total lipid extracts from the 14 samples were reacted with 0.5 M HCl in methanol at 60 °C for 16 h; following dilution in 18.2 MΩ·cm H2O, organic compounds were extracted with hexane and dichloromethane (4:1 vol/vol) until the organic layer appeared colourless. The extracted organic compounds were concentrated under a gentle stream of N2 before column separation into three fractions. An aliquot of the acid hydrolysed TLE was loaded onto a short column containing deactivated silica gel (2% H2O total weight) and its fractions were eluted with 2 dead volumes (DV) of hexane, 3 DV hexane and dichloromethane (1:1 vol/vol), 3 DV dichloromethane and methanol (4:1 vol/vol), and 2 DV washes with pure methanol to elute any remaining polar columns from the gel. The sterols eluted in the dichloromethane and methanol fraction; the resulting polar fraction was concentrated under a gentle stream of N2.
An aliquot of the sterol-containing fractions from both sample sets was reacted with pyridine and N,O-Bis(trimethylsilyl)trifluoroacetamide (1:1 vol/vol) for 30 min at 70 °C to produce sterol trimethylsilyl ethers after the addition of a cholest-5-en-3β-ol(2H7) standard. The sterol fractions of Evans et al.21 were spiked with 1000 ng of a 10 ng/µL cholest-5-en-3β-ol(2H7) standard solution, corresponding to 10 ng/µL injected, while the sterol fractions of the additional four samples were spiked with cholest-5-en-3β-ol(2H7) to correspond to 2.5 ng/µL injected.
The resulting sterol trimethylsilyl ethers were analysed on an Agilent 7890A GC System equipped with an Agilent J&W DB-5MS fused silica capillary column (60 m × 250 μm × 0.25 μm) coupled to an Agilent 5975C Inert XL MSD with Triple-Axis Detector. Samples were run in full-scan mode (scanning masses 50 to 700 Da) using the following parameters: inject splitless on a split/splitless inlet at 300 °C and hold for 2 min, after another 2 min ramp oven from 60 °C to 315 °C at 3.5 °C/min and hold for 30.8 min. The flow rate of helium through the column was 1 mL/min and constant flow was achieved throughout the run. The 5975C Inert EI 350 source was held at 250 °C and its electron energy was set to 70 eV; the quadrupole was held at 150 °C. Masses were scanned from 50 to 700 Da and results were analysed using Agilent MassHunter Qualitative Analysis Navigator B.08.00. Relative response factors for detected compounds were calculated against the known quantity of cholest-5-en-3β-ol(2H7) standard within each sample, and blanks were run in between samples to monitor instrumental background and baseline conditions. Whenever possible, samples were run against authentic sterol standards for retention time and mass spectral matches. For the sterols with no commercially available standards for comparison, mass spectra were analysed and primarily matched against those described by Gillan in 198181, against spectra compiled by NIST, and against the reference spectra database on William H. Christie’s TheLipidWeb.
Sterane analysis
Aliquots of the sterol-containing fractions from the 14 microbial mats extracted in this work were subjected to simulated diagenesis to yield their sterane hydrocarbon counterparts. The conversion of sterols and stanols to steranes by catalytic hydrogenation/hydrogenolysis bypasses natural sterol diagenesis pathways which can result in the formation of sterenes, diasterenes, diasteranes. Hydrogenation/hydrogenolysis was conducted on the sterol-containing fraction using Adams’ catalyst (PtO2). Following addition of the catalyst, the lipid fraction was suspended in hexane; hydrogen gas was gently bubbled into the solvent-sample mixture while it was stirred with a magnetic stir bar for a total of 3 hours. Post-hydrogenation, the solvent-sample mixture was filtered over activated silica gel and separated into fractions. The nonpolar, sterane hydrocarbon-containing fraction was eluted using 2 DV hexane, while the polar fraction was eluted using 2 DV of hexane and dichloromethane (1:1 vol/vol) and 2 DV dichloromethane and methanol (4:1 vol/vol). Both fractions were concentrated under a gentle stream of N2. The nonpolar, sterane-containing fraction was resuspended in hexane for analysis.
The resulting steranes were analysed on an Agilent 7890B GC System equipped with an Agilent J&W DB5-MS fused silica capillary column (59.9 m × 250 μm × 0.25 μm) coupled to an Agilent 7010 GC/MS Triple Quad. Samples were run in dynamic multiple reaction monitoring (dMRM) mode using the following parameters: inject splitless in a multimode inlet at 60 °C and hold for 0.01 min before ramping to 340 °C at 700 °C/min. The oven was ramped from 60 °C to 220 °C at 8 °C/min before being ramped to 325 °C at 2 °C/min and held for 30.5 min. The flow rate of helium through the column was 1.2 mL/min and constant flow was achieved throughout the run. The 7010 High Efficiency source was held at 270 °C and its electron energy was set to 70 eV; quadrupoles 1 and 2 were held at 150 °C. Transitions were viewed and analysed using Agilent MassHunter Qualitative Analysis Navigator B.08.00. The following transitions were targeted for sterane analysis: 358.3 → 217.2 for C26-steranes, 372.3 → 217.2 for C27-steranes, 386.3 → 217.2 for C28-steranes, 400.4 → 217.2 for C29-steranes, 414.4 → 217.2 for C30-steranes, 414.4 → 231.2 for methylated C30-steranes, and 428.4 → 217.2 for C31-steranes. Samples were run alongside aliquots of the Baghewala-A oil from the late Neoproterozoic-early Cambrian Marwar Supergroup82 and of KG-8 from the Miocene-age Monterey formation83 to aid in peak identification for C26 steranes and C30-steranes, as well as blanks to monitor instrument background and baseline conditions.
Select samples were also run on the same instrument in product ion mode to capture the principal fragment ions of specific sterane peaks to differentiate 4-methylsteranes from 24-n-propylcholestane and 24-isopropylcholestane. Scan segment times were determined based on elution times of corresponding dMRM runs. Chromatographic conditions were identical to those in the dMRM run; the 7010 High Efficiency source was held at 270 °C and its electron energy was set to 70 eV; the quadrupoles 1 and 2 were held at 150 °C. Masses were scanned from 55 to 600 Da; the collision energy for the scan segments was set to 10 eV. Results were analysed using Agilent MassHunter Qualitative Analysis Navigator B.08.00.
18S rRNA gene analysis
18S rRNA gene sequencing data and sequence analysis was conducted on subsample fragments of the microbial mats as previously detailed in Evans et al.21. In brief, DNA was extracted from the mats using a MO BIO Laboratories PowerBiofilm DNA Isolation Kit and amplified using polymerase chain reaction. The V9 hypervariable regions of the 18S rRNA genes were amplified using the universal forward primer 1391F (5′-GTACACACCGCCCGTC-3′) and the eukaryotic reverse primer EukBR (5′-TGATCCTTCTGCAGGTTCACCTAC-3′) containing Illumina MiSeq sequencing adaptors84,85,86. Following purification with Axygen AxyPrep MAG PCR Clean-up Kits, samples were sequenced on an Illumina MiSeq system at the Natural History Museum’s sequencing facility in the United Kingdom. The resulting 18S rRNA gene amplicon libraries from the mats were deposited with accession PRJNA758785 in the Sequence Read Archive. The raw 18S rRNA gene sequences from the mats were processed with the QIIME 2 microbiome bioinformatics platform (version 2019.4)87. Reads were quality filtered and merged, and the DADA2 pipeline was used to denoise and produce amplicon sequence variants (ASVs) using the default settings88. Taxonomy was assigned to ASVs with the sklearn-based taxonomy classifier using the SILVA 132 SSURef_NR99 database89,90. To enable comparisons across different ponds, datasets for all samples were rarefied to 25,000 reads using default QIMME 2 settings which randomly subsample sequences from each sample. The rarefied data for the ponds are included in the Source Data file.
Principal component analysis
To identify potential patterns or correlations within the data, principal component analysis was conducted. C26-C30 sterane and 18S rRNA gene data were applied as variables, and pond measurements were overlaid on individual pond principal component scores to determine whether the measured environmental parameters exerted any controls on the distributions of principal component scores. Samples with missing pond measurements, sterane, or 18S rRNA gene data were excluded from the analysis. Analyses were conducted in GraphPad Prism 10.4.1.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The 18S rRNA gene amplicon libraries from the mats have been deposited in the Sequence Read Archive under accession code PRJNA758785. Source data for the 18S rRNA analyses are provided in the Source Data file. The sterol and sterane biomarker data from the mats have been deposited in the NASA Astrobiology Habitable Environment Database Data Repository under accession code 5m63-qq45. Source data are provided with this paper.
References
Harland, W. B. Critical evidence for a great infra-Cambrian glaciation. Geol. Rundsch. 54, 45–61 (1964).
Hoffman, P. F. & Schrag, D. P. The snowball Earth hypothesis: testing the limits of global change. Terra Nova 14, 129–155 (2002).
Kirschvink, J. L. Late Proterozoic low-latitude global glaciation: the snowball Earth. In The Proterozoic Biosphere (eds Schopf, J.W. & Klein, C.) 51–52 (Cambridge University Press, 1992).
Macdonald, F. A. et al. Calibrating the Cryogenian. Science 327, 1241–1243 (2010).
Hoffman, P. F. et al. Snowball Earth climate dynamics and Cryogenian geology-geobiology. Sci. Adv. 3, e1600983 (2017).
Rooney, A. D., Strauss, J. V., Brandon, A. D. & Macdonald, F. A. A Cryogenian chronology: two long-lasting synchronous Neoproterozoic glaciations. Geology 43, 459–462 (2015).
Crockett, W. W., Shaw, J. O., Simpson, C. & Kempes, C. P. Physical constraints during Snowball Earth drive the evolution of multicellularity. Proc. R. Soc. B Biol. Sci. 291, 20232767 (2024).
Tang, Q. et al. Quantifying the global biodiversity of Proterozoic eukaryotes. Science 386, eadm9137 (2024).
Allen, P. A. & Etienne, J. L. Sedimentary challenge to Snowball Earth. Nat. Geosci. 1, 817–825 (2008).
Pollard, D. & Kasting, J. F. Snowball Earth: A Thin-ice solution with flowing sea glaciers. J. Geophys. Res. Oceans 110, 2004JC002525 (2005).
Hoffman, P. F. Cryoconite pans on Snowball Earth: supraglacial oases for Cryogenian eukaryotes?. Geobiology 14, 531–542 (2016).
Hawes, I., Jungblut, A. D., Matys, E. D. & Summons, R. E. The “Dirty Ice” of the McMurdo Ice Shelf: analogues for biological oases during the Cryogenian. Geobiology 16, 369–377 (2018).
Vincent, W. F. et al. Ice shelf microbial ecosystems in the high arctic and implications for life on snowball earth. Naturwissenschaften 87, 137–141 (2000).
Fedonkin, M. A. Cold water cradle of animal life. Paleontol. J. 30, 669–673 (1996).
Gutiérrez-Preciado, A. et al. Functional shifts in microbial mats recapitulate early Earth metabolic transitions. Nat. Ecol. Evol. 2, 1700–1708 (2018).
Howard-Williams, C., Pridmore, R. D., Broady, P. A. & Vincent, W. F. Environmental and biological variability in the McMurdo Ice Shelf ecosystem. In Antarctic Ecosystems (eds Kerry, K. R. & Hempel, G.) 23–31 (Springer, 1990).
Suren, A. Microfauna associated with algal mats in melt ponds of the Ross Ice Shelf. Polar Biol. 10, 329–335 (1990).
Sutherland, D. L. Microbial mat communities in response to recent changes in the physiochemical environment of the meltwater ponds on the McMurdo Ice Shelf, Antarctica. Polar Biol. 32, 1023–1032 (2009).
Jungblut, A. D., Vincent, W. F. & Lovejoy, C. Eukaryotes in Arctic and Antarctic cyanobacterial mats. FEMS Microbiol. Ecol. 82, 416–428 (2012).
Jackson, E. E., Hawes, I. & Jungblut, A. D. 16S rRNA gene and 18S rRNA gene diversity in microbial mat communities in meltwater ponds on the McMurdo Ice Shelf, Antarctica. Polar Biol. 44, 823–836 (2021).
Evans, T. W. et al. Lipid biomarkers from microbial mats on the McMurdo Ice Shelf, Antarctica: signatures for life in the cryosphere. Front. Microbiol. 13, 903621 (2022).
Bowman, J. S. Identification of microbial dark matter in Antarctic environments. Front. Microbiol. 9, 3165 (2018).
Volkman, J. K. Sterols in microorganisms. Appl. Microbiol. Biotechnol. 60, 495–506 (2003).
Peters, K. E., Walters, C. C. & Moldowan, J. M. The Biomarker Guide, Volume 1: Biomarkers and Isotopes in the Environment and Human History (Cambridge University Press, 2005).
Jungblut, A. D. Characterisation of Microbial Mat Communities in Meltwater Ponds of The Mcmurdo Ice Shelf, Antarctica (UNSW Sydney, 2007).
Jungblut, A. D., Allen, M. A., Burns, B. P. & Neilan, B. A. Lipid biomarker analysis of cyanobacteria-dominated microbial mats in meltwater ponds on the McMurdo Ice Shelf, Antarctica. Org. Geochem. 40, 258–269 (2009).
Drozd, J. K., Evans, T. W. & Summons, R. E. Lipid biomarker comparison of relict and active microbial mats from the McMurdo Ice Shelf, Antarctica. Org. Geochem. 179, 104591 (2023).
Hanschen, E. R. & Starkenburg, S. R. The state of algal genome quality and diversity. Algal Res 50, 101968 (2020).
Grattepanche, J. D. et al. Microbial diversity in the Eukaryotic SAR clade: illuminating the darkness between morphology and molecular data. BioEssays 40, 1–12 (2018).
Gillan, F. T., Mcfadden, G. I., Wetherbee, R. & Johns, R. B. Sterols and fatty acids of an antarctic sea ice diatom, Stauroneis amphioxys. Phytochemistry 20, 1935–1937 (1981).
Nichols, P. D., Palmisano, A. C., Rayner, M. S., Smith, G. A. & White, D. C. Occurrence of novel C30 sterols in Antarctic sea-ice diatom communities during a spring bloom. Org. Geochem. 15, 503–508 (1990).
Volkman, J. K. Sterols in microalgae. In The Physiology of Microalgae (eds. Borowitzka, M. A., Beardall, J. & Raven, J. A.) 485–505 (Springer International Publishing, 2016).
Giner, J. L. & Wikfors, G. H. “Dinoflagellate Sterols” in marine diatoms. Phytochemistry 72, 1896–1901 (2011).
Rampen, S. W., Abbas, B. A., Schouten, S. & Sinninghe Damste, J. S. A comprehensive study of sterols in marine diatoms (Bacillariophyta): implications for their use as tracers for diatom productivity. Limnol. Oceanogr. 55, 91–105 (2010).
Kokke, W. C. M. C., Fenical, W. & Djerassi, C. Sterols of the cultured dinoflagellate pyrocystis lunula. Steroids 40, 307–318 (1982).
Giner, J. L. Biosynthesis of marine sterol side chains. Chem. Rev. 93, 1735–1752 (1993).
Giner, J. L., Zhao, H. & Tomas, C. Sterols and fatty acids of three harmful algae previously assigned as Chattonella. Phytochemistry 69, 2167–2171 (2008).
Rampen, S. W. et al. On the origin of 24-norcholestanes and their use as age-diagnostic biomarkers. Geology 35, 419 (2007).
Volkman, J. K., Barrett, S. M., Dunstan, G. A. & Jeffrey, S. W. Geochemical significance of the occurrence of dinosterol and other 4-methyl sterols in a marine diatom. Org. Geochem. 20, 7–15 (1993).
Patterson, G. W. The distribution of sterols in algae. Lipids 6, 120–127 (1971).
Volkman, J. K. Lipids of geochemical interest in microalgae. in Hydrocarbons, Oils and Lipids: Diversity, Origin, Chemistry and Fate (ed. Wilkes, H.) 159–191 (Springer International Publishing, 2020).
Gaskell, S. J. & Eglinton, G. Rapid hydrogenation of sterols in a contemporary lacustrine sediment. Nature 254, 209–211 (1975).
Moldowan, J. M. et al. Analysis and occurrence of C26-steranes in petroleum and source rocks. Geochim. Cosmochim. Acta 55, 1065–1081 (1991).
Schouten, S., Sinninghe Damsté, J. S., Schoell, M. & de Leew, J. W. A novel sterane, 27-nor-24-methyl-5αcholestane, in sediments. Geochim. Cosmochim. Acta 58, 3741–3745 (1994).
Holba, A. G. et al. 24-norcholestanes as age-sensitive molecular fossils. Geology 26, 783 (1998).
Holba, A. G. et al. Application of 24-norcholestanes for constraining source age of petroleum. Org. Geochem. 29, 1269–1283 (1998).
Dutta, S., Bhattacharya, S. & Raju, S. V. Biomarker signatures from Neoproterozoic–early Cambrian oil, western India. Org. Geochem. 56, 68–80 (2013).
Silva, C. J. & Djerassi, C. Biosynthetic studies of marine lipids 36. The origin of common sterol side chains in eleven sponges using [3-3H]-Squalene. Comp. Biochem. Physiol. Part B Comp. Biochem. 101, 255–268 (1992).
Nettersheim, B. J. et al. Putative sponge biomarkers in unicellular Rhizaria question an early rise of animals. Nat. Ecol. Evol. 3, 577–581 (2019).
Moldowan, J. M. et al. Sedimentary 12-n-propylcholestanes, molecular fossils diagnostic of marine algae. Science 247, 309–312 (1990).
Giner, J. L., Zhao, H., Boyer, G. L., Satchwell, M. F. & Andersen, R. A. Sterol chemotaxonomy of marine pelagophyte algae. Chem. Biodivers. 6, 1111–1130 (2009).
Volkman, J. K., Barrett, S. M., Dunstan, G. A. & Jeffrey, S. W. Sterol biomarkers for microalgae from the green algal class Prasinophyceae. Org. Geochem. 21, 1211–1218 (1994).
Grabenstatter, J. et al. Identification of 24-n-propylidenecholesterol in a member of the Foraminifera. Org. Geochem. 63, 145–151 (2013).
Peters, K. E., Walters, C. C. & Moldowan, J. M. The Biomarker Guide, Volume 2: Biomarkers and Isotopes in Petroleum Exploration and Earth History (Cambridge University Press, 2005).
Love, G. D. et al. Fossil steroids record the appearance of Demospongiae during the Cryogenian period. Nature 457, 718–721 (2009).
Debenham, F. A new mode of transportation by ice. Q. J. Geol. Soc. Lond. 75, 51–76 (1920).
Sarà, M., Balduzzi, A., Barbieri, M., Bavestrello, G. & Burlando, B. Biogeographic traits and checklist of Antarctic demosponges. Polar Biol. 12, 559–585 (1992).
Gold, D. A. et al. Sterol and genomic analyses validate the sponge biomarker hypothesis. Proc. Natl. Acad. Sci. USA 113, 2684–2689 (2016).
Brown, M. O., Olagunju, B. O., Giner, J.-L. & Welander, P. V. Sterol methyltransferases in uncultured bacteria complicate eukaryotic biomarker interpretations. Nat. Commun. 14, 1859 (2023).
Bobrovskiy, I. et al. Algal origin of sponge sterane biomarkers negates the oldest evidence for animals in the rock record. Nat. Ecol. Evol. 5, 165–168 (2021).
Summons, R. E., Volkman, J. K. & Boreham, C. J. Dinosterane and other steroidal hydrocarbons of dinoflagellate origin in sediments and petroleum. Geochim. Cosmochim. Acta 51, 3075–3082 (1987).
Volkman, J. K., Kearney, P. & Jeffrey, S. W. A new source of 4-methyl sterols and 5α(H)-stanols in sediments: prymnesiophyte microalgae of the genus Pavlova. Org. Geochem. 15, 489–497 (1990).
Huang, W. & Meinschein, W. G. Sterols as ecological indicators. Geochim. Cosmochim. Acta 43, 739–745 (1979).
Brocks, J. J. et al. The rise of algae in Cryogenian oceans and the emergence of animals. Nature 548, 578–581 (2017).
Grosjean, E., Love, G. D., Stalvies, C., Fike, D. A. & Summons, R. E. Origin of petroleum in the Neoproterozoic-Cambrian South Oman Salt Basin. Org. Geochem. 40, 87–110 (2009).
Hoshino, Y. et al. Cryogenian evolution of stigmasteroid biosynthesis. Sci. Adv. 3, e1700887 (2017).
Kvenvolden, K. A. & Rapp, J. B. Multiple sources of aikanes in Quaternary oceanic sediment of Antarctica. Org. Geochem. 11, 291–302 (1987).
Parfrey, L. W., Lahr, D. J. G., Knoll, A. H. & Katz, L. A. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc. Natl. Acad. Sci.USA 108, 13624–13629 (2011).
Knoll, A. H., Summons, R. E., Waldbauer, J. R. & Zumberge, J. E. The geological succession of primary producers in the oceans. Evol. Prim. Prod. Sea 133–163 https://doi.org/10.1016/B978-012370518-1/50009-6 (2007).
Swithinbank, C. W. M. Ice Movement in the McMurdo Sounds Area of Antarctica. In International Symposium on Antarctic Glaciological Exploration (eds Gow, A.J., Keeler, C., Langway, C.C. & Weeks, W.F.) 472–487 (International Association of the Science of Hydrology, 1970).
Glasser, N., Goodsell, B., Copland, L. & Lawson, W. Debris characteristics and ice-shelf dynamics in the ablation region of the McMurdo Ice Shelf, Antarctica. J. Glaciol. 52, 223–234 (2006).
Hawes, I., Howard-Williams, C. & Pridmore, R. D. Environmental control of microbial biomass in the ponds of the McMurdo Ice Shelf, Antarctica. Arch. Für Hydrobiol. 127, 271–287 (1993).
Vincent, W. F., Downes, M. T., Castenholz, R. W. & Howard-Williams, C. Community structure and pigment organisation of cyanobacteria-dominated microbial mats in antarctica. Eur. J. Phycol. 28, 213–221 (1993).
de Mora, S. J. & Whitehead, R. F. The chemical composition of glacial melt water ponds and streams on the McMurdo Ice Shelf, Antarctica. Antarct. Sci. 6, 17–27 (1994).
Archer, S. D. J., McDonald, I. R., Herbold, C. W., Lee, C. K. & Cary, C. S. Benthic microbial communities of coastal terrestrial and ice shelf Antarctic meltwater ponds. Front. Microbiol. 6, 485 (2015).
Jungblut, A. D. & Hawes, I. Using Captain Scott’s Discovery specimens to unlock the past: has Antarctic cyanobacterial diversity changed over the last 100 years?. Proc. R. Soc. B Biol. Sci. 284, 20170833 (2017).
Lezcano, M. Á et al. Comprehensive metabolic and taxonomic reconstruction of an ancient microbial mat from the McMurdo Ice Shelf (Antarctica) by integrating genetic, metaproteomic and lipid biomarker analyses. Front. Microbiol. 13, 799360 (2022).
Scott, R. F. Finding winter quarters: a fatal accident. In The Voyage of the Discovery 205–253 (Smith, Elder, & Co., 1905).
Swithinbank, C. W. M., Darby, D. G. & Wohlschlag, D. E. Faunal remains on an Antarctic Ice Shelf. Science 133, 764–766 (1961).
Hawes, I., Howard-Williams, C., Schwarz, A. M. & Downes, M. T. Environment and microbial communities In a tidal lagoon at Bratina Island, McMurdo Ice Shelf, Antarctica. In Antarctic Communities: Species, Structure and Survival 170–177 (1997).
Gillan, F. T. Lipids of Aquatic Ecosystems (University of Melbourne, 1981).
Bhattacharya, S., Dutta, S. & Summons, R. E. A distinctive biomarker assemblage in an Infracambrian oil and source rock from western India: molecular signatures of eukaryotic sterols and prokaryotic carotenoids. Precambrian Res. 290, 101–112 (2017).
Isaacs, C. M. & Rullkötter, J. The Monterey Formation: From Rocks to Molecules (Columbia University Press, 2001).
Amaral-Zettler, L. A., McCliment, E. A., Ducklow, H. W. & Huse, S. M. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS ONE 4, e6372 (2009).
Stoeck, T. et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 19, 21–31 (2010).
Caporaso, J. G. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2012).
Yilmaz, P. et al. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–D648 (2014).
Acknowledgements
The authors thank Marc Schallenberg for assistance with sampling and Antarctica New Zealand for enabling the expedition to collect the samples examined in this study. The authors additionally thank Benjamin Uveges for helpful discussions regarding statistical analysis. F.H. was supported by the MIT Presidential Fellowship, the MIT EAPS John H. Carlson Fellowship, and the MIT Hugh Hampton Young Memorial Fellowship for the duration of this research. T.W.E. was supported by the Alexander von Humboldt Foundation Feodor Lynnen Research Fellowship for the duration of this research. This work was funded by the NASA Exobiology program (grant numbers: 80NSSC19K0465 and 80NSSC23K1361, awarded to R.E.S.).
Author information
Authors and Affiliations
Contributions
R.E.S., A.D.J., and F.H. designed the study. R.E.S. and I.H. collected the samples. F.H., T.W.E., J.L.M., and A.D.J. performed the research. F.H. wrote the manuscript with contributions and approval from all co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Simon C. Brassell, who co-reviewed with Kelsey Doiron; Cristina Cid, who co-reviewed with Victor Muñoz-Hisado; and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. [A peer review file is available].
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Husain, F., Millar, J.L., Jungblut, A.D. et al. Biosignatures of diverse eukaryotic life from a Snowball Earth analogue environment in Antarctica. Nat Commun 16, 5315 (2025). https://doi.org/10.1038/s41467-025-60713-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-60713-5