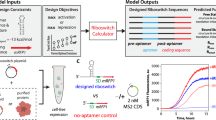
Abstract
Re-engineering natural riboswitches into orthogonal RNA switches by making them functional in response to exogenous ligands but unresponsive to endogenous cognate ligands is a promising yet less explored strategy for developing gene regulatory tools. Herein, we rationally engineer the aptamer domain of one of the largest and biotechnologically relevant flavin mononucleotide (FMN) riboswitch class, which specifically binds to synthetic ligands with a high binding affinity (KD = ~ 54-75 nM) and regulates gene expression in vitro, in prokaryotic, and eukaryotic system, while being unresponsive to FMN. To develop the orthogonal aptamers, we rationally alter key tertiary interactions, such as A/G minor motifs and base triples located in the periphery of the FMN binding pocket. The biophysical and structural probing analysis of the orthogonal aptamer and synthetic ligand complex shows binding mediated by favorable enthalpic and unfavorable entropic contributions. Our rational design approach, coupled with the adaptability to FMN aptamers derived from diverse bacterial strains, suggests the broad applicability of this strategy to numerous FMN riboswitches, each possessing a unique expression platform. This will greatly expand the current repertoire of synthetic riboswitches available for biomedical applications.
Similar content being viewed by others
Introduction
Synthetic riboswitches are regulatory RNA elements engineered to control and fine-tune gene expression in both prokaryotes and eukaryotes and are becoming increasingly significant for biomedical applications1,2,3,4,5,6. Synthetic riboswitches are constructed with a modular design comprising an aptamer domain and an expression platform. Like their natural counterparts, the aptamer domain remains an indispensable component of synthetic riboswitches. When a ligand binds to the aptamer domain, it induces structural changes in the adjacent expression platform, thereby regulating downstream gene expression through mechanisms like transcription termination, inhibition of translation initiation, or self-cleavage7,8,9. Synthetic riboswitches offer exciting potential for regulating genes conditionally for diverse applications due to their inherent characteristics, such as reliance on small molecules for cis-regulation without the need for proteins and their non-immunogenic regulation, making them straightforward for use across different species1,2,3,4,5,6,7.
Drawing inspiration from these characteristics, researchers have developed numerous synthetic riboswitches capable of reprogramming bacterial and mammalian cells for applications in biotechnology, medicine, and bioremediation1,10,11. The majority of aptamer domains used in creating synthetic riboswitches are chosen through a method called Systematic Evolution of Ligands by EXponential enrichment (SELEX)12,13. SELEX represents an iterative procedure where RNA fragments that strongly bind to a specific molecule are chosen from a large RNA library. While SELEX yields aptamers with high binding affinity to their target molecules, only a few synthetic aptamers have been effectively integrated into functional synthetic riboswitches, such as those recognizing theophylline, tetracycline, and neomycin14,15,16,17. This limitation arises because achieving not only high binding affinity but also coupling the binding event with conformational changes in the regulatory domain is critical for synthetic riboswitch success5,18,19,20,21.
An alternative approach for creating synthetic riboswitches involves utilizing naturally occurring riboswitches18,19,20,21. Presently, there are over 55 different classes of natural riboswitches, with thousands more predicted to exist22. The natural riboswitches have evolved with built-in switching sequences (expression platform) and an aptamer domain exhibiting remarkable specificity and high affinity for a wide range of chemically distinct ligands22. Moreover, a few mutations in the aptamer domain can alter the ligand binding specificity and riboswitch function19,20,21,23, making them excellent candidates for initiating the development of synthetic riboswitches24,25,26. However, the regulatory cognate ligand for natural riboswitches is present within cells and can interfere with the intended artificial regulation27. To address this challenge, attention has shifted towards modifying natural riboswitch aptamers to become orthogonal so that they selectively recognize synthetic ligands rather than their native cognate ligand19,20,21. In this context, the add A-riboswitch19 and PreQ1 riboswitch20 have been re-engineered to develop orthogonal riboswitches by using saturation mutagenesis and alteration of hydrogen-bonding interactions, respectively. Although promising, to date, only a few natural riboswitches with small structures have been engineered to recognize synthetic ligands19,20. The complex secondary and tertiary structural architectures of the natural riboswitches, particularly in larger riboswitches with >200 nucleotides, such as flavin mononucleotide (FMN)28,29 and cobalamin30, may explain why only a few attempts have been made to re-engineer natural riboswitches. Indeed, the scarcity of suitable aptamer domains highlights the necessity for engineering more classes of natural riboswitches to broaden the number and diversity of orthogonal aptamer-ligand pairs for conditional gene regulation.
Here, we have used a rational mutagenesis approach to re-engineer the FMN riboswitch (RFN element) aptamer domain derived from biotechnologically relevant bacterial strain Lactobacillus plantarum31, into an orthogonal riboswitch that selectively functions in response to a synthetic flavin analog. We chose the FMN riboswitch for this study owing to its widespread occurrence in biotechnologically relevant bacterial strains32,33, the plasticity and large opening of the FMN-binding pocket29,34, the well-characterized structure of the aptamer domain in the FMN-free (apo)34 as well as in FMN-bound (holo) states29,34, and nanomolar range binding affinities35. We have introduced mutations in various tertiary interactions, such as A/G minor motifs and base triples, which are found to facilitate FMN recognition by framing the ligand-binding pocket in such a manner that the resultant aptamers can no longer recognize FMN and instead bind to synthetic ligands with high affinities. Using this approach, a functional orthogonal riboswitch-ligand combination was developed that effectively regulated reporter gene expression in vitro, in bacterial and mammalian cells. These findings encourage the applicability of our approach in the development of orthogonal riboswitches by re-engineering the numerous natural riboswitches.
Results
Design, synthesis of uncharged flavin ligands, and rational mutagenesis of FMN riboswitch aptamer domain
The secondary structure of FMN riboswitch aptamer consists of six stems and five loops (Fig. 1a) that form a flexible pocket for binding FMN (riboflavin-5′-phosphate) ligand (Fig. 1b) via the formation of various tertiary interactions (Fig. 1c–f). The FMN ligand mainly binds to the aptamer domain through the stacking of its isoalloxazine ring between A48 and A85, coupled with specific Watson-Crick-like hydrogen bonding between the uracil-like face of the isoalloxazine ring and highly conserved A99 (Fig. 1c)29. The FMN ribityl moiety forms hydrogen bonds with only one of its four oxygens and its phosphate oxygens create additional bonds with the Watson-Crick edges of several conserved guanines (Fig. 1c). In addition, the FMN phosphate group oxygen interacts with N7 position of G33 through Mg2+ mediated interactions along with forming several water-mediated contacts with neighboring nucleotides (Fig. 1c)29. These structural informations suggest that the contribution of the ribityl chain hydroxyl groups in FMN recognition is minimal compared to the isoalloxazine ring interactions. Thus, the FMN ribityl chain could be modified without altering the π-π stacking and hydrogen bond interactions of the isoalloxazine ring with the aptamer. We intended to design and synthesize flavin analogs that do not contain the phosphate group (uncharged ligands), as they offer the advantage of not relying on proteins for their transport and phosphorylation for biological activity36,37,38. Consequently, we systematically modified the ribityl chain by removing the phosphate and hydroxyl groups (Supplementary Synthesis Scheme 1 and 2) to yield three potential ligands: Di-hydroxy ethyl flavin (DHEF), Mono-hydroxy ethyl flavin (MHEF) and Formyl methyl flavin (FMF) as depicted in Fig. 2a. In addition to above synthetic ligands, we included two natural analogs of FMN; lumichrome and alloxazine in our study to gain insights into the effect of ribityl chain on ligand binding. Lumichrome, which is 7,8-dimethylalloxazine, differs from alloxazine by the presence of two methyl groups at the 7 and 8 positions. However, a major challenge in developing uncharged ligands (with no terminal phosphate) is the drastic decrease in the binding affinity (by ~1000-fold), as seen in the case of riboflavin (a dephosphorylated precursor of FMN)29. Therefore, to break the interactions of natural aptamer with cognate ligands and to achieve orthogonal properties, we re-engineered the natural FMN riboswitch aptamer domain derived from Lactobacillus plantarum (Fig. 2c) in such a way that it does not rely on the phosphate group for ligand recognition and function and designed the synthetic analogs with no terminal phosphate to complement the introduced modifications in the aptamer.
a Schematic of the conserved secondary structure of FMN aptamer with key long-range tertiary interactions involved in the formation of FMN binding pocket. The different stems (P1-P6) and loops (L2-L6) are depicted in different colors. b Chemical structure of flavin mononucleotide (FMN). c Interactions of the FMN ligand with FMN riboswitch aptamer derived from Fusobacterium nucleatum (PDB ID: 3F2Q). The hydrogen bond contacts of the ligands with the nucleotides in the aptamer domains are shown with the dashed line. d–f Distinct alignment of nucleotides in G-minor motif G11-(G84-C31), base triple G12-(G93-C30), and aptamer-FMN interaction mediated by Mg2+ shown as green ball forming G62-(C83-G32) base triple, respectively.
a Chemical structures of synthetic and natural analogs of FMN used in this study, wherein the black part represents the preserved moiety and the red part denotes the modified ribityl chain. b Sequence alignment of aptamers from Lactobacillus plantarum (LP), Pseudomonas fluorescens (PF), and Fusobacterium nucleatum (FN), the color code is given as per the structure of FMN aptamer in Fig. 1a. c The predicted secondary structure of aptamer LP. d, e Relative fluorescence intensity of FMN and DHEF after the addition of Aptamer LP (0-5 μM) and Aptamer M15 (0-5 μM), respectively (n = 3 replicates). f The predicted secondary structure of aptamer PF. The nucleotides involved in stacking interactions are highlighted in blue, and the position of mutated nucleotides is shown in red (Fig. 2c, f). g, h Relative fluorescence intensity of FMN and DHEF after the addition of Aptamer PF (0-3 μM) and Aptamer MPF (0-10 μM), respectively (n = 3 replicates) in a buffer containing 50 mM MOPS (pH-7.5), 100 mM NaCl, 2 mM MgCl2, 0.05% Tween-20, and 3% DMSO. For all relevant panels, data are shown as mean ± standard deviation (SD). Source Data are provided as a Source Data file.
The structural studies have shown that the FMN riboswitch aptamer is globally pre-organized in the absence of FMN and undergoes minimum structural rearrangements upon FMN binding29,34. In the FMN-bound state, the junctional regions are found to be stapled together by P2-P6 and P3-P5 peripheral domains (Supplementary Fig. 1). These peripheral domains, in turn, are stabilized by two tertiary interactions involving loop-loop (L2-L6 and L3-L5) and loop-helix (L6-P2 and L3-P5) interactions (Fig. 1a). The FMN is recognized by its aptamer primarily through Mg2+-mediated recognition of the FMN’s phosphate group with the formation of G62-(G32-C83) (Fig. 1c, f), followed by intercalation of its isoalloxazine ring by nucleotides A48, A85, and A9929. While the global structure remains unchanged, the region showing FMN-induced conformational changes is the region J1-2. The J1-2, along with J6-1, serves as an opening to the binding pocket. In addition to linking the P2 and P6 domains, the G-minor motif G11-(G84-C31) and base triple G12- (G93-C30) are actively involved in the recognition of the FMN’s phosphate group (Fig. 1d, e). It has been shown that to prevent a steric clash, G12 is moved out of the triple plane in a non-planar orientation, which results in the weakening of both G11-(G84-C31) and G12- (G93-C30). This weakening has been predicted to play an important role in regulating the mobility of the J1-2 junction29.
Together, these observations led us to hypothesize that if we introduce mutations at the nucleotide position G62 in J4-5, which is involved in Mg2+-mediated recognition of the FMN’s phosphate group by forming G62-(C83-G32) base triple, the resultant aptamer might not be able to interact with the FMN’s phosphate and hence FMN, a prerequisite to achieving orthogonality. Additionally, we aimed to convert the base triples G11-(G84-C31) and G12- (G93-C30) to A11-(G84-C31) and A12-(G93-C30), respectively. We envisioned that these mutations might hamper the flexibility of the junctional regions such that opening to the binding pocket can no longer allow the larger FMN to enter, but the rationally designed synthetic flavin analogs might compensate for the resultant loss of mobility. Thus, we selected nucleotide residues G8, G9, G74, and G96 in L. plantarum (corresponding to residues G11, G12, G62, and G84 in F. nucleatum, Fig. 2b) for mutagenesis and developed a library of 16 mutant aptamers. In aptamer M16, we have mutated the nucleotides A58, A97, and A111 (corresponding to residues A48, A85, and A99 in F. nucleatum) participating in FMN stacking interactions, along with mutations in aptamer M15 to completely disrupt the ligand recognition. The complete sequence and predicted secondary structure of aptamer LP and all its mutant aptamers are shown in Fig. 2b, c, and Supplementary Tables 2 and 3, and Supplementary Figs. 11–26, respectively.
Fluorescence-based screening to identify orthogonal mutants
To begin with, we screened the aptamer LP and its mutants (M1-16) for their ability to discriminate between FMN and its non-phosphorylated derivative DHEF in a fluorescence titration assay that utilizes the intrinsic fluorescence of the isoalloxazine ring35. The decrease in fluorescence intensity upon aptamer addition indicates binding between the aptamer and ligand35. The representative binding curves obtained for the aptamer LP and its orthogonal mutant M15, with FMN and the synthetic ligand DHEF, are shown in Fig. 2d, e. In the case of the LP aptamer, the aptamer concentration-dependent decrease in FMN fluorescence intensity suggests it’s bound. On the other hand, there is no significant change in DHEF fluorescence intensity with increasing aptamer concentration, indicating no interaction with aptamer LP (Fig. 2d). On the contrary, we observed the aptamer concentration-dependent decrease in DHEF fluorescence intensity and no significant change in that of FMN’s upon the addition of aptamer M15, which implies binding and nonbinding, respectively (Fig. 2e). Subsequently, the binding curves were fitted with a 1:1 binding model equation detailed in the methods section. Table 1 shows the equilibrium dissociation constant (KD) values of the natural and the mutant LP aptamers (M1-16) for FMN and DHEF; the fitted binding curves are shown in Supplementary Fig. 2a–p and Supplementary Fig. 3a–p. The KD of the aptamer LP for FMN was found to be 0.21 ± 0.06 × 10−7 L mol−1, which is consistent with the binding affinities observed for other natural FMN aptamers35. The aptamer LP did not show any interaction with the synthetic ligand DHEF. Meanwhile, the KD values of the mutant aptamers for DHEF ranged from 1.58 × 10−7 L mol−1 to 19.3 × 10−7 L mol−1 ( ~ 13-fold difference).
Based on the obtained results, the mutants were categorized into two distinct groups: those that bind to DHEF (M1, M2, M3, M5, M6, M8, M9, M10, M11, M12, M13, M14, and M15) ranging from 1.58 × 10−7 L mol−1 to 19.3 × 10−7 L mol−1, and those that do not bind to DHEF at all (M4, M7) as shown in Table 1. Among the mutants that bind to DHEF, except for M11, M13, and M15, all others retained significant binding affinity for FMN and thus were not considered suitable as orthogonal aptamers. The mutant M15 was observed to bind to DHEF with the highest binding affinity, followed by M11 with a KD value of 1.58 × 10−7 L mol−1 and 2.72 × 10−7 L mol−1, respectively (Table 1) and therefore, the mutant M15 was identified as the best candidate with orthogonal properties.
It was interesting to note that even a single mutation at the selected nucleotide positions increased the mutants’ sensitivity towards DHEF compared to the natural aptamer, except for M4 and M7 (Table 1). Increasing the number of mutations resulted in the increased sensitivity of the mutants to DHEF. Conversely, the sensitivity towards FMN decreased with the increasing number of mutations. These results can be attributed to the fact that mutations at nucleotide positions G8, G9, G74, and G96 might have rendered the aptamer to lose contact with the phosphate group of the FMN and also hamper the mobility of the junctional region J1-2 required for FMN to enter the binding pocket. As a result, the dephosphorylated ligand DHEF with no phosphate and a shorter ribityl chain could easily fit into the altered binding pocket. Importantly, the disruptive mutant M16 did not interact with FMN and DHEF, stating the importance of the stacking interactions for ligand recognition irrespective of the modified ribityl chain. In this view, the aptamer-ligand pair M15-DHEF was considered the best candidate for developing orthogonal riboswitches and was thus prioritized for further studies.
Next, to check the adaptability of our approach for diverse FMN riboswitch aptamers derived from other bacterial strains, we selected a natural 165-nucleotide-long FMN aptamer derived from Pseudomonas fluorescens (PF), a biotechnologically significant strain39. The complete FMN aptamer sequence and predicted secondary structure of the aptamer PF are shown in Fig. 2b, f, respectively. To convert the natural aptamer PF into an orthogonal aptamer, we introduced similar mutations (G10A, G11A, G114A, and G136A corresponding to the G11, G12, G62, and G84 of F. nucleatum) in the binding pocket of the aptamer PF and checked the binding preferences of the wild-type PF and its mutant between FMN and DHEF using a fluorescence titration assay. As a result, we observed that the wild-type PF displayed its natural characteristics and bound to FMN with a binding affinity of 0.22 × 10−7 L mol−1, and was completely nonresponsive to the synthetic ligand DHEF (Fig. 2g). On the other hand, the mutant PF did not interact with FMN and bound to DHEF with a high binding affinity of 2.30 × 10−7 L mol−1 (Fig. 2h). This implies that our approach of rational altering of the ligand-binding pocket to design orthogonal riboswitches can be successfully applied to the other members of the FMN riboswitch class.
Isothermal titration calorimetry analysis of the aptamer-ligand binding
Next, we performed isothermal titration calorimetry (ITC) to determine the thermodynamic parameters for the formation of aptamer-ligand complexes. Representative ITC thermograms are shown in Fig. 3, and the equilibrium dissociation constant (KD), changes in enthalpy (ΔH°bind), entropy (ΔS°bind), and free energy (ΔG°bind) during the binding reaction at 25 °C are listed in Table 2. The ITC data indicate that the aptamer LP binds with FMN with a KD value of 0.051 ± 0.007 μM (Fig. 3a) and is enthalpy-driven, consistent with previous results observed with other natural FMN aptamers29,40. The favorable −ΔH°bind is associated with forming hydrogen bonds between the flavin pyrimidine ring with a uracil face and adenosine of aptamer-LP and Mg2+ ion-mediated interactions with FMN phosphate oxygens. The small unfavorable −TΔS°bind contribution indicates that only subtle conformational changes occur in the pre-organized aptamer binding pocket during FMN binding between the free and bound states. The aptamer LP and DHEF showed no interaction (Fig. 3b), consistent with our fluorescence titration studies.
Thermograms showing the interaction of wild-type LP aptamer binding to a FMN, b DHEF, c MHEF, and d FMF and M15 aptamer binding to e FMN, f DHEF, g MHEF, and h FMF. On top is the titration data showing the heat resulting from each injection of the ligand from the syringe to the aptamer solution into the cell. On the bottom are integrated heats after correcting for the heat of dilution. The binding experiments were performed at 25 °C in a buffer containing 50 mM MOPS (pH 7.5), 100 mM NaCl, 2 mM MgCl2, and 1% DMSO with an aptamer concentration of 7 μM and a ligand concentration of 100 μM. The equilibrium dissociation constants (KD) were calculated using the one-site binding (1:1) model provided by MicroCal PEAQ-ITC Analysis Software (n = 2 replicates). Source Data are provided as a Source Data file.
In contrast, the re-engineered aptamer M15 exhibited a remarkably strong binding affinity for DHEF with a KD value of 0.075 ± 0.01 μM and did not recognize FMN, as evidenced by the obtained ITC data (Fig. 3e, f). These results underline the excellent orthogonal selectivity of the M15 aptamer towards DHEF over FMN. Notably, for both pairs, aptamer LP with FMN and aptamer M15 with DHEF, we observed a large favorable −ΔH (Table 2). Compared to aptamer LP-FMN binding, a slight increase in the −TΔS in the case of aptamer M15-DHEF binding indicates relatively more conformational rearrangements in the aptamer M15 over aptamer LP. We further performed ITC of the aptamer LP and M15 with the other two ligands, MHEF and FMF. The aptamer LP did not show any interaction (Fig. 3c, d), while the aptamer M15 bound to the MHEF and FMF with a dissociation constant of 0.054 ± 0.005 μM and 0.068 ± 0.004 μM, respectively (Fig. 3g, h). Despite showing a similar binding interaction as that of DHEF, MHEF and FMF could not be used further in this study due to the lower solubility of MHEF and the highly reactive aldehyde group of FMF that may react with intracellular biomolecules if used for gene regulation, respectively41. The interactions disrupted aptamer M16 did not show interaction with FMN and DHEF (Supplementary Fig. 4a, b).
To further validate our hypothesis, we performed ITC experiments of aptamer LP and M15 with lumichrome and alloxazine. The lumichrome or alloxazine does not show interaction with natural LP riboswitch aptamers (Supplementary Fig. 5a, b). However, the aptamer M15 was observed to bind to lumichrome with a KD value of 0.035 ± 0.02 μM. On the other hand, the binding affinity for alloxazine was significantly low, with a KD value of 3.17 ± 0.58 μM (Supplementary Fig. 5c, d). The ~ 90-fold difference in the binding affinities of lumichrome and alloxazine towards aptamer M15 suggests that an intact isoalloxazine ring is indispensable for the synthetic ligands to be recognized by the re-engineered aptamers and, therefore, needs to be preserved while designing new flavin analogs.
Structural probing of the ligand-induced conformational changes
We carried out an in-line probing assay to trace the ligand-induced structural changes in the aptamers. The in-line probing assay helps to deduce the ligand-mediated conformational changes in the RNA structure by monitoring the product of spontaneous RNA cleavage in the absence and presence of the ligand42,43. Representative in-line probing data with the wild-type aptamer LP and the mutant aptamer M15 display different modulations in the intensity of the 3’ Cy3-labeled RNA in response to FMN and DHEF (Fig. 4). As shown previously, the FMN aptamer generally exists in a pre-organized state without FMN34,44. Therefore, the major conformational switching upon ligand binding is restricted to the junctional regions responsible for framing the ligand binding pocket29. The interaction of FMN with the FMN aptamer is marked by the Mg2+-mediated primary recognition of the phosphate moiety of FMN by its RNA, followed by intercalation of the isoalloxazine ring system. These interactions are essential to stabilize the J1-2 segment (G10–G12), followed by the J6-1 segment (A99–G104). Due to these FMN-induced structural rearrangements, the nucleotides clustered around the phosphate, and the isoalloxazine ring gets protected.
a PAGE analysis of in-line probing assays with 3’ Cy3-labeled Aptamer-LP and Aptamer-M15 RNA in the absence or presence of FMN and DHEF at a concentration of 10 μM. OH and T1 designate RNAs that have been partially digested with alkali or RNase T1, respectively (left panel). A schematic representation of a T-loop-like motif (right panel). b, c In-line probing gel images with lanes loaded with aptamer RNAs subjected to various concentrations of ligands with Aptamer LP and Aptamer M15, respectively, no reaction (NR). d, e The plot of the fraction of RNA cleaved versus the logarithm of the molar concentration of ligand of Aptamer LP with FMN and Aptamer M15 with DHEF, respectively. Fraction-cleaved values were estimated based on the extent of band intensity changes at site 1 in (b, c). Data are shown as mean ± standard deviation (SD) (n = 3 replicates). Source Data are provided as a Source Data file.
A similar pattern was observed with the aptamer LP, wherein the FMN binding showed a decrease in the spontaneous cleavage at various junctional regions (marked by black arrows), indicative of strong binding with FMN. On the other hand, the cleavage becomes more profound in the presence of DHEF (Fig. 4a left panel). This suggests that aptamer LP strongly discriminates between the two ligands and binds less tightly to DHEF due to the lack of terminal phosphate, a natural characteristic of the FMN aptamer. In contrast, the aptamer M15 showed increased scissions at different regions in the presence of FMN, while significant protection of the junctional regions was observed upon the addition of DHEF (Fig. 4a left panel). In light of this, the introduced mutations were rationalized to switch the specificity of the aptamer M15 from FMN to synthetic ligand DHEF. We observed interesting structural modulation resulting from aptamer LP-FMN and the mutant M15-DHEF binding event in the loop-5 region (highlighted in the dashed box in Fig. 4a), which forms the T-loop-like motif. The T-loop-like motif is known to be an RNA folding motif made up of five nucleotides UGAAA, from nucleotide 69 to 73 in F. nucleatum (corresponding to UGCAA from nucleotide 81 to 85 in L. plantarum, according to the predicted secondary structure of the aptamer LP in Fig. 2c), and is involved in various tertiary interactions that compact the RNA tertiary structure by bringing the distant nucleotides nearby45. In the absence of FMN, the nucleotides involved in T-loop formation remain free and are likely to be more accessible for spontaneous cleavage, while in the presence of FMN, a large gap between the fourth (A72) and fifth (A73) nucleotide enables intercalation of A38 to staple the loops L3 and L5 together (Fig. 4a, right panel) and thus supposed to inhibit the cleavage. Our in-line probing data of wild-type aptamer LP with FMN indicates FMN-induced local conformational rearrangements, which result in the protection of the nucleotides in the T-Loop region (Fig. 4a left panel). A similar cleavage pattern was observed in the T-loop region in the case of aptamer M15 in the presence of DHEF. This could only be possible if the aptamer M15 had retained the key structural features of its parent aptamer LP. Therefore, it is apparent from our probing analysis that the introduced mutations in the aptamer M15 do not perturb the global architecture of the aptamer and exert local structural changes to form the DHEF-responsive binding pocket, as must be the case with the wild-type aptamer LP-FMN complex.
Titration of aptamers LP and M15 with FMN and DHEF, respectively, showed that the modulation of band intensities was dose-dependent (Fig. 4b–e and Supplementary Fig. 6a–f). The results of our in-line probing assay, ITC, and fluorescence-titration assay are consistent with each other and thus mark the development of an orthogonal aptamer-ligand pair. These observations reinforce our rationale that the natural aptamer’s binding pocket can be rationally modified to occupy new ligands without breaking the direct contact of the aptamer with its ligand.
Design and in vitro validation of orthogonal riboswitch functioning
To investigate the ability of the developed orthogonal aptamer-ligand pairs to be transformed into functional riboswitches, we designed artificial riboswitches based on the wild-type aptamer (Aptamer-LP), the orthogonal aptamer (Aptamer-M15), and the binding-disrupted aptamer (Aptamer-M16). The aptamers were fused to an expression platform designed in a way that the ribosome-binding site (RBS) would be freely accessible in a single-stranded region for the ribosome to bind in the absence of the ligand and the gene expression would remain ‘ON,’ whereas, in the presence of the ligand the RBS would be sequestered to form a stem-loop and will no longer be able to bind to ribosome resulting in turning ‘OFF’ the gene expression (Fig. 5a). The resulting riboswitches were positioned upstream of the Renilla luciferase gene under the control of the T7 promoter. The sequence and design of the artificial riboswitches are shown in Supplementary Table 4 and Supplementary Figs. 27–29. The level of gene regulation was evaluated by measuring the decrease in luminescence intensity using an in vitro coupled transcription-translation system, and the EC50 values were estimated by plotting the relative luciferase activity against the ligand concentration detailed in the Methods section. We observed that the aptamer LP-based riboswitch repressed the translation of the luciferase gene, indicated by a progressive decrease in the luminescence intensity with higher concentrations of FMN, with an EC50 value of 0.75 ± 0.04 μM. On the other hand, the gene expression decreased negligibly in the presence of DHEF (Fig. 5b). Conversely, the luciferase activity of the aptamer M15-based riboswitch was unaffected upon FMN addition and was repressed remarkably in a dose-dependent manner with an EC50 value of 6.29 ± 1.71 μM upon the addition of DHEF (Fig. 5c). Meanwhile, the luciferase gene expression remained unaffected under the control of M16-based riboswitch in the presence of FMN and DHEF (Supplementary Fig. 7a) which further confirms that the observed reduction in gene expression is due to the riboswitch-ligand binding-mediated inhibition of translation initiation. Collectively, the results of our reporter gene assay prove that the mutations G8A, G9A, G74A, and G96A in aptamer M15 not only alter the specificity of the re-engineered aptamer from natural ligand FMN to synthetic ligand DHEF but can also be successfully incorporated into artificial riboswitches to modulate gene expression.
a Schematic illustrating 5’ UTR orthogonal OFF riboswitch-mediated translational regulation via ribosome binding site (RBS) masking in the presence of the DHEF ligand. GOI denotes the gene of interest. The ribosome icon in a was created in BioRender. Rode (2025) https://BioRender.com/01y1hjp. In vitro coupled transcription-translation of synthetic riboswitches based on b Aptamer LP in the presence of FMN and DHEF (n = 2 replicates), and c Aptamer M15 in the presence of FMN and DHEF (n = 3 replicates), over a range of ligand concentrations (0 μM to 10 μM). The luciferase activity at 0 μM ligand concentration was defined as 100 %. d Relative mRFP1 expression for riboswitch constructs M15-RS A through E in the absence and presence of 2.5 mM DHEF (n = 3 biological replicates); the statistical significance was determined by two-way analysis of variance (ANOVA), with Sidak’s multiple comparisons test. e Dose-dependent suppression of mRFP1 expression under the control of the M15-RS-A riboswitch with varying DHEF concentrations (0-2.5 mM) (n = 3 biological replicates). f RT-qPCR analysis of relative mRFP mRNA levels in E. coli under untreated and DHEF-treated conditions (n = 3 biological replicates); the statistical significance was determined by the two-tailed t-test. g Representative Western blot showing mRFP1 detection in E. coli transformed with the M15-RS-A riboswitch under untreated and DHEF-treated conditions. GAPDH served as a loading control (left panel). The graph (right panel) shows Western blot quantification normalized to GAPDH (n = 3 biological replicates); the statistical significance was determined by the two-tailed t-test. For all the relevant panels, data are shown as mean ± standard deviation (SD). Source Data are provided as a Source Data file.
Orthogonal riboswitch-mediated gene regulation in bacteria
We further tested the capability of our developed orthogonal riboswitches for conditional gene regulation in bacteria. As the expression platform plays a crucial role in the gene regulation efficiency of the synthetic riboswitches, we designed five OFF-type synthetic riboswitches M15-RS-A to E with different pre- and post-aptameric sequences of varying lengths and sequence compositions (Supplementary Table 5), and inserted them in the 5’ UTR of the open reading frame of an mRFP1 reporter gene. The mechanism of riboswitch functioning was similar to that used for the in vitro coupled transcription-translation assay (Fig. 5a). All five riboswitch constructs were tested for their ability to inhibit reporter gene expression in the presence of DHEF. Based on the obtained results, the riboswitches could decrease the mRFP1 expression in the range of 38-81%. These results suggest that the efficiency of the riboswitch function can be modulated by the rational design of its expression platform for different purposes. Among them, the riboswitch M15-RS-A showed the highest reduction of up to 80.96% in the mRFP1 gene expression (Fig. 5d) at the DHEF concentration of 2.5 mM and was, therefore, selected for further validation. The E. coli culture harboring the M15-RS-A-based riboswitch was grown at various concentrations of DHEF (0–2.5 mM) and showed a reduction in the fluorescence intensity of the mRFP1 with the increasing ligand concentration (Fig. 5e). The M15-DHEF riboswitch-ligand combination exhibited dose-dependent gene repression with an EC50 value of 622 ± 61.1 μM. Next, we performed an RT-qPCR assay and observed no significant change in the mRFP mRNA levels of E. coli cultures with and without DHEF. The result indicates that the developed orthogonal riboswitch regulates gene expression at the translational level (Fig. 5f).
The Western blot analysis with an 87.6% decrease in the mRFP1 protein level in the presence of DHEF compared to no ligand (Fig. 5g) further validates the conditional gene regulation capability of the developed orthogonal riboswitches in bacteria.
Orthogonal aptamer-based aptazyme-mediated gene regulation in mammalian cells
Mammalian synthetic biology constantly demands advanced regulatory tools to manipulate gene expression for different purposes46. Ribozyme switches have been used to regulate gene expression in eukaryotic systems by modulating mRNA stability47. To demonstrate orthogonal riboswitch regulation in eukaryotic cells, we designed and constructed aptazyme-based switches that conditionally cleave the Poly(A) tail in 3’ UTR, triggering mRNA degradation and leading to a reduction in gene expression. The switch was designed using the wild-type aptamer (Aptamer-LP), the orthogonal aptamer (Aptamer-M15), and the disruptive mutant aptamer (Aptamer-M16). To construct the aptazyme switches, we replaced the stem-loop I of the hammerhead ribozyme with the respective aptamer domains (Supplementary Figs. 30–32). While integrating the aptamer domain with the ribozyme stem-loop, three base pairs were removed from the stem P1 region of the aptamer. This design was intended to disrupt the loop–loop tertiary interactions between stem I and stem II, which are critical for the ribozyme’s active conformation. Consequently, this disruption inhibits ribozyme-mediated self-cleavage of the transcript. The loop–loop tertiary interactions are restored upon ligand binding, enabling the aptazyme switch to self-cleave the transcript (Fig. 6a).
a Schematic illustrating 3’-UTR aptazyme-mediated conditional regulation of mRNA stability. DHEF ligand binding induces an active conformation in the ribozyme, leading to its activation and subsequent mRNA destabilization. GOI and (A)n denote the gene of interest and the poly-A tail, respectively. b Fluorescence microscopy images of HEK293T cells transiently transfected with the aptazyme-regulated mCherry-BFP dual reporter. Cells were treated with the indicated DHEF concentrations and incubated at 37 °C for 48 h before imaging, scale bar: 50 μm. c Relative mCherry/BFP fluorescence intensity ratios at the indicated DHEF concentrations (0 to 200 µM) (n = 3 biological replicates); the statistical significance was determined by one-way analysis of variance (ANOVA), followed by Dunnett’s multiple comparisons test. d RT-qPCR analysis of relative mCherry mRNA levels in HEK293T cells under untreated and DHEF-treated conditions (n = 3 biological replicates); the statistical significance was determined by the two-tailed t-test. e Immunoblot analysis of mCherry protein levels in HEK293T cells with and without DHEF treatment. β-Actin served as a loading control (left panel). The graph (right panel) shows Western blot quantification normalized to β-Actin (n = 3 biological replicates); the statistical significance was determined by the two-tailed t-test. For all the relevant panels, data are shown as mean ± standard deviation (SD). Source Data are provided as a Source Data file.
We cloned the designed constructs into the 3′ UTR of the mCherry gene within the mCherry-BFP dual reporter expression cassette PCS4092. The resulting constructs were evaluated for cleavage activity using an in vitro co-transcriptional RNA cleavage assay in the presence and absence of the ligand (see Supplementary Table 6 for aptazyme sequences). The relative levels of cleaved RNA transcripts in the presence and absence of the ligands were analyzed through polyacrylamide gel electrophoresis (PAGE). Consequently, the aptazyme designs, which showed increased cleavage in the presence of the desired ligands compared to the absence of ligands, were selected for cellular assays (Supplementary Fig. 8a). The selected M15 aptazyme showed increased RNA cleavage by ~ 50% only with DHEF and not with FMN when compared to the control in which no ligand was added, while the LP aptazyme showed ~70% increase in the RNA cleavage in the presence of the FMN only and not DHEF. The M16 construct showed no change in cleavage either with FMN or DHEF (Supplementary Fig. 8a). The results of our in vitro experiments on RNA cleavage during transcription are consistent with our earlier findings from biophysical studies and bacterial reporter gene regulation data. These constructs were further tested in HEK293T cells for gene regulation.
For cellular assays, the constructed cassettes were transiently transfected into HEK293T cells. The BFP reporter, expressed under an independent promoter, was used as a transfection control. Consequently, the cells containing the M15-aptazyme, when treated with DHEF, displayed a reduction in the mCherry gene expression due to the DHEF-mediated cleavage of the aptazyme in the 3’UTR of the mCherry gene as observed in the fluorescence imaging of the cells (Fig. 6b). Next, to quantify the decrease in the gene expression, we treated the cells with different concentrations of DHEF and performed cell lysis for fluorescence measurement. In the presence of 200 μM DHEF, the mCherry expression was suppressed up to 76.1% for the M15 aptazymes relative to the absence of ligands, confirmed by the reduction in normalized mCherry/BFP fluorescence (Fig. 6c), while the mCherry expression remained unaffected in the presence of FMN (Supplementary Fig. 8b). The observed suppression in mCherry expression was also consistent with results from the RT-qPCR and the Western blot assay, which showed up to 81.6% decrease in the mCherry expression (Fig. 6d, e, respectively). These results further support our approach to developing modular, versatile, and conditional gene regulatory tools that can be used in eukaryotic systems. Meanwhile, the mCherry gene expression decreased non-significantly in the cells harboring the LP and M16-based aptazymes when the growth media was supplemented with the DHEF (Supplementary Fig. 8c, d), indicating that DHEF binds specifically to the M15 aptazyme to exert its effect in the regulation of the gene expression.
Exploring orthogonal mutations in the natural pool of FMN aptamers
The mutations G8A, G9A, G74A, and G96A (corresponding to residues G11, G12, G62, and G84 in F. nucleatum) conferring orthogonal selectivity to the aptamer M15 prompted us to identify the natural FMN aptamers having mutations at these locations. Consequently, we selected five representative natural FMN aptamers having single and combined mutations at the mentioned nucleotide positions. The selection criteria for the natural FMN aptamers are mentioned in the Methods section. Out of which, four aptamers were found to possess a single mutation, namely Aptamer HO at nucleotide positions A9 (residue G12 in F. nucleatum) and AS, CB, and MS at A94, A80, and A116 (residue G84 in F. nucleatum), respectively. The fifth aptamer (Csp) showed triple mutations at A11, A12, and A62 nucleotide positions (residues G11, G12, and G62 in F. nucleatum). The complete sequence and predicted secondary structure of the aptamers are shown in Supplementary Table 2 and Supplementary Figs. 33–37. We performed a fluorescence titration assay to test how these variations affect the ligand preferences of these aptamers for FMN and DHEF. The binding curves obtained are shown in Supplementary Figs. 9, 10, and the estimated binding constant (KD) of the aptamers for FMN and DHEF are mentioned in the Table 3. It was observed that the aptamers HO (G12A) and Csp (G11A, G12A, and G62A) did not interact with FMN. Meanwhile, the aptamers AS, CB, and MS carrying G84A mutations bind with the FMN with a binding affinity of 6.27 × 10−8 L mol−1, 10.6 × 10−8 L mol−1, and 3.66 × 10−8 L mol−1, respectively which is comparable to the binding affinity observed for the mutant aptamer M4 (having similar mutation) and is also consistent with the other natural FMN aptamers wherein the G84 is conserved. In stark contrast, no aptamer was found to interact with DHEF. To rationalize these observations, we compared the aptamer sequences of these five aptamers with aptamer M11 and M15 (Fig. 7). The G12A mutation in aptamer HO was similar to M2 but could not recognize FMN. The sequence and structure analysis showed that the nucleotide G47, participating in G-G non-canonical base pairing with G10, was mutated in aptamer HO, which might have resulted in the aptamer being non-responsive to FMN. For the G84A mutation, similar results were observed in natural variants and M4, as the aptamers AS, CB, and MS bind to FMN but do not recognize DHEF. Despite having mutations similar to mutant M11, the aptamer Csp did not bind to DHEF. Notably, the aptamer Csp showed significant deviations from the consensus FMN sequence. In addition to the mutations at G11, G12, and G62 (similar to the mutations in aptamer M11), the nucleotides participating in G-G non-canonical base pairing involving nucleotides G10 and G47 and the T-loop-like motif (UGAAA) at position G70 were also found to be mutated in aptamer Csp, rendering the aptamer non-responsive to FMN and DHEF. This suggests that in addition to the mutations G11A, G12A, G62A, and G84A, the evolutionary preservance of other key interactions synergistically affects the aptamers’ ligand recognition ability and binding affinity.
Discussion
Given the significance of gene regulation in various synthetic biology endeavors, novel strategies are needed to devise and construct effective genetic regulatory tools. Within this context, synthetic riboswitches activated by external, non-natural compounds are emerging as potential tools for modulating gene expression. However, a limited pool of suitable combinations of aptamers (molecular recognition elements) and ligands (molecules binding to aptamers) can be incorporated into functional synthetic riboswitches. To address this limitation and broaden the range of appropriate aptamer-ligand pairs, the re-engineering of natural riboswitches into orthogonal variants shows promise. In our study, we have developed an orthogonal riboswitch based on flavin mononucleotide (FMN) that responds selectively to a synthetic flavin analog while remaining unresponsive to FMN itself.
Tertiary interactions are pivotal in shaping the active ligand-binding site for any riboswitch. In the case of FMN riboswitches, where the aptamer assumes a pre-organized state in its unbound form, random mutations may disrupt the overall folding of the riboswitch. Thus, a delicate balance of modifications is necessary to alter the ligand preference while preserving structural integrity. Our detailed examination of FMN and aptamer interaction helped us identify tertiary interactions essential for forming the active binding pocket. We focused on modifying key interactions such as the G-minor motif G11-(G84-C31) and base triples G12-(G93-C30) and G62-(G32-C83), which regulate the flexibility of the J1-2 segment and participate in Mg2+-mediated recognition of FMN’s phosphate group. These alterations might play a crucial role in reducing the flexibility of the J1-2 junction, hindering FMN from entering the binding pocket and disrupting Mg2+-mediated phosphate recognition, rendering the aptamer incapable of accommodating FMN.
Concurrently, we designed and synthesized synthetic flavin analogs lacking a terminal phosphate (uncharged ligands) to lessen their dependence on multiple proteins for activation and transport. These analogs also featured modified ribityl chains tailored to fit the altered binding pocket efficiently. Our binding studies demonstrate that the mutant aptamer M15 exhibits a high affinity for the synthetic ligand DHEF ( ~ 54-75 nM), while it does not recognize FMN.
Although lumichrome exhibited significant binding affinity towards aptamer M15, it seems incapable of interfering with M15-based riboswitch-mediated gene regulation for several reasons. First, lumichrome is predominantly secreted by plants and certain bacteria in minute nanomolar concentrations48, whereas the M15-based riboswitch requires ligand concentration within the millimolar range for its activation. Consequently, such low intracellular levels are unlikely to influence gene regulation. Second, certain bacterial species possess effective detoxification mechanisms49 to eliminate lumichrome promptly upon its generation, making it challenging to maintain the necessary concentration to trigger riboswitch function within cells. Thus, despite demonstrating active interaction in vitro, lumichrome may not exhibit in vivo gene regulation. Research indicates that lumichrome plays a pivotal role in plant growth and development. A low nanomolar concentration (5 nM) correlates with enhanced growth rate and increased biomass accumulation, while concentrations up to 50 nM impede root development and overall growth. Thus, other than gene regulation, the orthogonal aptamer M15, developed in our study, could be a valuable component for constructing riboswitch-based biosensors to detect lumichrome.
The structural probing data indicates that the mutations introduced in the aptamer LP to transform it into an orthogonal aptamer do not impact the overall global folding of the aptamer. The newly created synthetic riboswitch exhibited precise and dosage-dependent control over gene expression in vitro, in bacterial, and in mammalian systems when exposed to DHEF. Our rational design approach, coupled with the adaptability to FMN aptamers derived from diverse bacterial strains such as Lactobacillus plantarum (LP) and Pseudomonas fluorescens (PF), suggests the broad applicability of our strategy to numerous FMN riboswitches each possessing unique expression platform. This will greatly expand the current repertoire of synthetic riboswitches available for biomedical applications. Our approach might help lay the groundwork for re-engineering natural riboswitch classes that bind to negatively charged ligands with a unique binding mode. The design and synthesis of non-phosphorylated ligands with the complimentary modifications in the phosphate-binding domain of the aptamer can lead to the development of the orthogonal riboswitch-ligand system. The introduced orthogonal riboswitch holds promise for regulating genes involved in biosynthesis pathways, facilitating gene function analysis, and other biotechnological purposes requiring simultaneous control over multiple genes.
Methods
Aptamer synthesis
DNA templates for RNA synthesis were prepared by primer extension PCR (Kapa, Taq PCR kit, KK1015) using oligonucleotides (Supplementary Tables 2, 3) purchased from Macrogen (Korea). All RNAs were transcribed from the DNA templates using MegaScript T7 Transcription Kit (Invitrogen, AMB1334-5) and were purified by denaturing polyacrylamide gel electrophoresis. Concentrations of the RNAs were determined by measuring the absorbance at 260 nm using a Jasco V-750 Spectrophotometer.
Calculation of equilibrium dissociation constant (K D)
Various concentrations (0–10 μM) of RNA aptamers were mixed with 100 nM flavin mononucleotide (FMN) or synthetic flavin analogs in a buffer containing 50 mM MOPS (pH 7.5), 100 mM NaCl, 2 mM MgCl2, 0.05% Tween-20, and 3% DMSO. The reaction mixtures were incubated at 70 °C for 10 min and cooled to 25 °C at 1 °C min−1. Aliquots of the reaction mixtures were transferred to wells of a 384-well black plate. The fluorescence signals of FMN and analogs were measured at 25 °C using I3x Spectramax (Molecular Devices) with 450 nm excitation and 530 nm emission. The binding between aptamers and FMN occurs in a simple 1:1 stoichiometry, as revealed by the crystal structure29. KD values at 25 °C were estimated from Kobs that were obtained by fitting the changes in ligand fluorescence as a function of aptamer concentration based on Eq. 1:
Where F is the fluorescence intensity of ligands at each concentration of the aptamer, Finitial is the initial fluorescence intensity of ligands without the aptamer, Ffinal is the fluorescence intensity when all of the ligand is bound to the aptamer, [ligand] is the total ligand concentration, and [aptamer] is the concentration of the aptamers.
Isothermal titration calorimetry (ITC)
For the ITC experiments, RNA folding and buffer compositions were similar to fluorescence-titration assays. ITC experiments were carried out with a MicroCal PEAQ-ITC (Malvern Instruments) with the sample cell (200 μL) containing 7 μM RNA and 60-100 μM ligand solution prepared in the same buffer in the injector syringe (40 μL). After thermal equilibration at 25 °C, an initial 60 s delay and one initial 0.4 μL injection were carried out. Then, 19 serial injections of 2.0 μL at intervals of 150 s at a stirring speed of 750 rpm were performed. Raw data were recorded as power (cal s−1) over time (min). The heat associated with each titration peak was integrated and plotted against the corresponding molar ratio of ligand and aptamer RNA. The dissociation constant (KD) was evaluated from a curve fit of the corrected data using the one-site binding (1:1) model provided by MicroCal PEAQ-ITC Analysis Software. Measurements were repeated at least twice.
Fluorescent cyanine 3 (Cy3) labeling of Aptamer
Aptamers (300 pmol) were ligated with 12 μM pCp-Cy3 (Jena Bioscience, NU-1706-CY3) in a buffer supplied by the manufacturer (New England Biolabs, M0204S) containing 1 mM ATP, 10% (v/v) DMSO, and 10 U μL-1 T4 RNA ligase 1 at 16 °C for 15 h. The labeled RNA product was purified using a QIAGEN RNeasy Mini Kit (74104). Briefly, the reaction mixture (50 μL) was mixed with RLT buffer (175 μL) and 100% ethanol (475 μL). The sample was then applied to the RNeasy Mini spin column, and the RNA sample was entrapped in the column by centrifugation. The column was placed in a new tube and washed with RPE buffer (500 μL), followed by 80% ethanol (500 μL). The RNA was eluted into nuclease-free H2O.
In-line probing
In-line probing experiments were performed with the aptamer LP and M1542,43. Briefly, 5 pmol of pCp-Cy3-labeled RNA aptamers were mixed with a concentration range of ligands (0-1 mM) in a buffer containing 50 mM Tris-HCl (pH 8.3), 25 mM MgCl2, 100 mM KCl, and 3% DMSO in a reaction volume of 10 µL. The in-line probing reactions were then refolded at 70 °C for 5 min, followed by incubation at 37 °C for 48 h. After the incubation was completed, a T1 ladder was prepared by mixing 5 pmol of labeled RNA in a buffer containing 0.25 M sodium citrate (pH 5.0), 5 M urea, 1 mM EDTA, and 1 unit RNase T1 (Sigma, R1003-100KU). The reaction was incubated at 55 °C for 15 min, after which the reaction was quenched by the addition of an equal volume of 2X loading buffer (10 M urea and 1.5 mM EDTA). The OH ladder was prepared by mixing 5 pmol of labeled RNA with a buffer containing 50 mM NaHCO3/Na2CO3 buffer (pH 9.5) and 1 mM EDTA. The reaction was incubated at 90 °C for 5 min, followed by adding an equal volume of 2X loading buffer. The in-line probing reactions were also quenched by adding an equal volume of 2X loading buffer. All the in-line probing reaction products and the ladders were then separated on 14% denaturing PAGE. The gel was imaged, and the band intensities were quantified using an Amersham Typhoon (Cytiva).
Construction of plasmids used for the in vitro coupled transcription-translation assay
DNA fragments coding different riboswitches were prepared by PCR (Supplementary Table 4). The PCR products were cloned into the Nhe I and BstB I sites of the previously constructed plasmid50. The resultant plasmids were transformed into E. coli DH5α with 100 μg/mL Ampicillin as a marker. Using the manufacturer’s protocol, the plasmids were isolated and purified using a Qiagen miniprep plasmid isolation kit (27104). The purified recombinant plasmid DNAs were sequenced, and after confirming the riboswitch insert, the plasmids were used to perform the experiments.
In vitro coupled transcription-translation Luciferase assay
The coupled transcription-translation reactions were performed in vitro. Reporter plasmids (1 ng μL−1) containing different riboswitches were mixed with the reaction mixture of E. coli T7 S30 Extract System for Circular DNA (Promega, L1130) in the presence of varying concentrations of ligands and incubated at 37 °C for 30 min. The reaction was stopped by adding RNase A (1.6 mg/μL) and puromycin (18 μM), followed by further incubation at 37 °C for 10 min. Luciferase activity was determined after the addition of substrate (Renilla-Glo Luciferase Assay system, Promega, E2710) for Renilla luciferase, coelenterazine, using a microplate reader. Relative gene expression was evaluated by normalization to the luciferase activity of the translated product in the absence of ligands. EC50 values were calculated by fitting the plot of the relative luciferase activity versus ligand concentration with Eq. 2:
Y is the relative luciferase activity at a ligand concentration of X, and LI and LS are the initial and saturated luciferase activities, respectively.
Construction of plasmids used for in vivo assessment of the riboswitch functionality in bacteria
DNA fragments coding different riboswitches were prepared by PCR (Supplementary Table 5). The PCR products were digested by XbaI and SacI restriction enzymes and cloned into the same restriction enzyme sites of the vector plasmid AEB-Theo45 (AEB-Theo45 was a gift from Howard Salis (Addgene plasmid # 63850; http://n2t.net/addgene:63850; RRID: Addgene_63850)) to make reporter gene constructs consisting of riboswitches upstream of the mRFP1 gene. The resultant plasmids were transformed into E. coli DH10B with 50 μg/mL Chloramphenicol as a marker. The purified recombinant plasmid DNAs were sequenced, and after confirming the riboswitch inserts, the plasmids were used to perform the experiments.
mRFP1 fluorescence measurement
All in vivo fluorescence measurements were carried out using the strain E. coli DH10B maintained in the exponential growth phase. Cells harboring the cloned plasmids expressing the mRFP1 reporter under the control of different riboswitches were grown overnight in 1 mL M9 media, pH 7.5 and 50 μg/mL chloramphenicol antibiotic at 37 °C and 180 rpm orbital shaking, overnight. The overnight-grown cultures were then diluted to an OD600 of 0.5 and added into 2 mL M9 media in a 1000-fold dilution with 50 μg/mL chloramphenicol and pre-defined concentrations of different ligands. The cultures were incubated at 37 °C and 220 rpm orbital shaking for 18 h. After incubation, the cells were collected and washed with 1X PBS to remove the traces of ligands and then resuspended in fresh 1X PBS. OD600 and fluorescence values were recorded by I3x Spectramax (Molecular Devices). The fluorescence of mRFP1 was measured in a black-well plate with an excitation wavelength of 555 nm and an emission wavelength of 608 nm. The obtained fluorescence was normalized with the OD600 to evaluate the change in the expression of the mRFP1 gene with the different ligands.
Western blotting for the bacterial system
The different cultures for western blot analysis were grown similarly, as mentioned in the fluorescence measurement section. After incubation, the bacterial cells were collected, washed, and resuspended in lysis buffer. The suspension was disrupted using a sonicator 20 s on and 40 s off cycle thrice at 50% amplitude and 0.5 cycles. The suspension was centrifuged at 13709 x g for 10 min at 4 °C, and the supernatant (cytoplasmic fraction) was transferred to a fresh tube. Protein estimation was performed using a standard BCA assay (Pierce BCA Protein Assy Kit, 23227). Protein samples (20 μg) were separated on a 12% polyacrylamide gel electrophoresis before being transferred onto a PVDF membrane (BIO-RAD, 1620177). The membrane was developed using rabbit-anti-RFP antibodies (GENETEX, GTX127897) used in 1:5000 dilution, rabbit-anti-GAPDH (BIOBHARTI LIFE SCIENCES, BB-AB0060) used in 1:2000 dilution as primary antibodies, and HRP-conjugated anti-rabbit as secondary antibodies (BIOBHARTI LIFE SCIENCES, BB-SAB01B) used in 1:10000 dilution using a Image Quant LAS4000 (GE). Protein expression was measured by densitometry using Image Quant.
RT-qPCR analysis of orthogonal riboswitch in bacteria
The different cultures for RT-qPCR analysis were grown similarly, as mentioned in the fluorescence measurement section. After incubation, the bacterial cells were collected, washed, and resuspended in Tri-Xtract (G-Biosciences, 786-652) for total RNA isolation. Reverse transcription was performed for 500 ng of total RNA using the RevertAid First Strand cDNA Synthesis kit (Thermo Scientific, K1622). The cDNAs generated and the gene-specific primers were used to perform RT-qPCR. The primers used for the RT-qPCR reaction are mentioned in Supplementary Table 7. All RT-qPCR experiments were performed on a Quantstudio 7 FLEX Real-Time PCR system (Life Technologies) using iTaq Universal SYBR Green Supermix (BIO-RAD, 1725121).
Construction of plasmids used for assessment of in vivo gene expression in mammalian cells
DNA fragments coding different aptazymes were prepared by PCR (Supplementary Table 6). The PCR products were digested by Bam HI and KpnI restriction enzymes and cloned into the same restriction enzyme sites of the vector plasmid pCS4092 (pCS4092 was a gift from Christina Smolke (Addgene plasmid # 131741; http://n2t.net/addegene:131741; RRID: Addgene_131741) to make reporter gene constructs consisting of different aptazymes in the 3’ end of the mCherry gene. The resultant plasmids were transformed into E. coli DH5α with 100 μg/mL Ampicillin as a marker. The purified recombinant plasmid DNAs were sequenced, and after confirming the aptazyme inserts, the plasmids were used to perform the experiments.
Cell culture
Human embryonic kidney 293 T (HEK293T) (ATCC-CRL-3216) cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM, Sigma Aldrich, D5648) at 37 °C in a 5% CO2-humidified incubator. All growth media were supplemented with 100 U/mL penicillin, 100 µg/mL streptomycin (Himedia, A002A), and 10% FBS (Himedia, RM10409).
In vitro characterization of the cleavage activity of the aptazymes
The DNA templates encoding the aptazyme were PCR amplified from the plasmid harboring the LP, M15, and M16-based aptazyme using primers mentioned in Supplementary Table 7. The RNAs were transcribed from the purified DNA templates (50 nM) in the presence and absence of the ligands using T7 RNA polymerase (Invitrogen) in a buffer containing 50 mM Tris-Cl (pH 7.5), 100 mM KCl, 20 mM MgCl2. The reaction mixtures were incubated at 37 °C for 2 h. After incubation, the reactions were treated with DNase and further incubated at 37 °C for 30 min. The reaction was stopped by adding an equal volume of 10 M urea, and the cleavage products were analyzed using a 9% denaturing PAGE. The gel was imaged using Amersham Typhoon (Cytiva). Additionally, an in vitro RNA cleavage assay was performed with the purified RNA in the presence and absence of the ligands.
Fluorescence microscopy
HEK293T cells were seeded in 96-well plates at a density of 1.0 × 104 cells per well. Transfection was performed using the Turbofect transfection reagent (Thermo Fisher Scientific, R0533) according to the manufacturer’s instructions, and 100 ng of plasmid was used in each transfection. Six hours post-transfection, the ligand at a concentration of 200 μM was added to the wells. After forty-eight hours, the media were removed, and the cells were washed with cold 1X PBS. The images were taken on a fluorescence microscope (Nikon) at a magnification of ×20.
mCherry fluorescence measurements
HEK293T cells were seeded at 10,000 cells per well of a 96-well plate 24 h before transfection. Transfection was performed using the Turbofect transfection reagent (Thermo Fisher Scientific, R0533) according to the manufacturer’s instructions, and 100 ng of plasmid was used in each transfection. Six hours post-transfection, different concentrations of the ligands were added to the wells. After forty-eight hours, the cells were washed with cold 1X PBS and lysed with RIPA (TCI, R0246) buffer according to the manufacturer’s instructions. The lysates were diluted 4 times in 1X PBS for fluorescence measurements. The fluorescence of blue fluorescent protein (BFP) and mCherry was measured in a black-well plate with an excitation/emission wavelength of 402/457 nm for BFP and 568/620 nm for mCherry. The obtained fluorescence of mCherry was normalized with the fluorescence of BFP (used as a transfection control) to evaluate the fold change in the expression of the mCherry gene in the different concentrations of the ligands.
RT-qPCR of orthogonal aptazyme in HEK293T cells
HEK293T cells were seeded at 50,000 cells per well of a 24-well plate 24 h before transfection. Transfection was performed using the Turbofect transfection reagent (Thermo Fisher Scientific, R0533) according to the manufacturer’s instructions, and 500 ng of plasmid was used in each transfection. Six hours post-transfection, the ligand at a concentration of 200 μM was added to the wells. After forty-eight hours, the cells were trypsinized, and total RNA was extracted using an RNeasy mini kit (Qiagen, 74104) according to the manufacturer’s instructions. Reverse transcription was performed for 500 ng of total RNA using the RevertAid First Strand cDNA Synthesis kit (Thermo Scientific, K1622). The cDNAs generated and the gene-specific primers were used to perform RT-qPCR. The primers used for the RT-qPCR reaction are mentioned in Supplementary Table 7. All RT-qPCR experiments were performed on a QuantStudio 7 FLEX Real-Time PCR system (Life Technologies) using iTaq Universal SYBR Green Supermix (BIO-RAD, 1725121).
Western blotting for the mammalian system
HEK293T cells were seeded at 50,000 cells per well of a 24-well plate 24 h before transfection. Transfection was performed using the Turbofect transfection reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions, and 500 ng of plasmid was used in each transfection. Six hours post-transfection, the ligand at a concentration of 200 μM was added to the wells. After forty-eight hours, the cells were collected, washed, and resuspended in a lysis buffer. The suspension was disrupted using a sonicator 20 s on and 40 s off cycle thrice at 50% amplitude and 0.5 cycles. The suspension was centrifuged at 13709 x g for 10 min at 4 °C, and the supernatant (cytoplasmic fraction) was transferred to a fresh tube. Protein estimation was performed using a standard BCA assay. Protein samples (20 μg) were separated on a 12% polyacrylamide gel electrophoresis before being transferred onto a PVDF membrane (BIO-RAD, 1620177). The membrane was developed using rabbit-anti-mCherry antibodies (GENETEX, GTX128508) used in 1:5000 dilution, mouse-anti-β-actin (CELL SIGNALLING TECHNOLOGY, 3700 T) used in 1:5000 dilution as primary antibodies, and HRP-conjugated anti-rabbit secondary antibodies (BIOBHARTI LIFE SCIENCES, BB-SAB01B) for anti-mCherry antibodies used in 1:10000 dilution and an anti-mouse secondary antibody (BIOLEGEND, 405306) used in 1:10000 dilution for anti-β-actin antibodies using a Image Quant LAS4000 (GE). Protein expression was measured by densitometry using Image Quant.
Sequence alignment and FMN aptamer selection
Rfam seed alignment for the FMN riboswitch was downloaded51. The sequences were then aligned using Clustal Omega. The nucleotide positions were identified and highlighted manually to select aptamer sequences with variations at the said nucleotide positions.
Statistical analysis
Statistical analyses were performed using one-way ANOVA, followed by Dunnett’s multiple comparisons test and two-way ANOVA, with Sidak’s multiple comparisons test and the two-tailed t-tests where appropriate. All analyses were performed using GraphPad Prism (version 10.4.1;www.graphpad.com). The numbers of biological replicates are indicated in the figure legends. Data are shown as the mean ± standard deviations. Differences were considered significant when α = 0.05.
Data availability
Data supporting the findings of this work are available within this paper and its Supplementary Information files. Source data are provided with this paper.
References
Luo, L., Jea, J. D., Wang, Y., Chao, P. W. & Yen, L. Control of mammalian gene expression by modulation of polyA signal cleavage at 5’ UTR. Nat. Biotechnol. 42, 1454–1466 (2024).
Etzel, M. & Morl, M. Synthetic riboswitches: from plug and pray toward plug and play. Biochemistry 56, 1181–1198 (2017).
Manna, S., Kellenberger, C. A., Hallberg, Z. F. & Hammond, M. C. Live cell imaging using riboswitch-spinach tRNA fusions as metabolite-sensing fluorescent biosensors. Methods Mol. Biol. 2323, 121–140 (2021).
Truong, J., Hsieh, Y. F., Truong, L., Jia, G. & Hammond, M. C. Designing fluorescent biosensors using circular permutations of riboswitches. Methods 143, 102–109 (2018).
Vezeau, G. E., Gadila, L. R. & Salis, H. M. Automated design of protein-binding riboswitches for sensing human biomarkers in a cell-free expression system. Nat. Commun. 14, 2416 (2023).
Zhou, L. B. & Zeng, A. P. Engineering a lysine-ON riboswitch for metabolic control of lysine production in Corynebacterium glutamicum. ACS Synth. Biol. 4, 1335–1340 (2015).
Breaker, R. R. Riboswitches and translation control. Cold Spring Harb. Perspect. Biol. 10, a032797 (2018).
Winkler, W. C., Nahvi, A., Roth, A., Collins, J. A. & Breaker, R. R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428, 281–286 (2004).
Wachter, A. Riboswitch-mediated control of gene expression in eukaryotes. RNA Biol. 7, 67–76 (2010).
Ausländer, S. & Fussenegger, M. Synthetic RNA-based switches for mammalian gene expression control. Curr. Opin. Biotechnol. 48, 54–60 (2017).
Hallberg, Z. F., Su, Y., Kitto, R. Z. & Hammond, M. C. Engineering and in vivo applications of riboswitches. Annu. Rev. Biochem. 86, 515–539 (2017).
Ellington, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Shanidze, N., Lenkeit, F., Hartig, J. S. & Funck, D. A theophylline-responsive riboswitch regulates expression of nuclear-encoded genes. Plant Physiol. 182, 123–135 (2020).
Wrist, A., Sun, W. & Summers, R. M. The theophylline aptamer: 25 years as an important tool in cellular engineering research. ACS Synth. Biol. 9, 682–697 (2020).
Wunnicke, D. et al. Ligand-induced conformational capture of a synthetic tetracycline riboswitch revealed by pulse EPR. RNA 17, 182–188 (2011).
Overbeck, J. H. et al. Multi-site conformational exchange in the synthetic neomycin-sensing riboswitch studied by (19) F NMR. Angew. Chem. Int. Ed. Engl. 62, e202218064 (2023).
Wieland, M., Benz, A., Klauser, B. & Hartig, J. S. Artificial ribozyme switches containing natural riboswitch aptamer domains. Angew. Chem. Int. Ed. Engl. 48, 2715–2718 (2009).
Dixon, N. et al. Reengineering orthogonally selective riboswitches. Proc. Natl. Acad. Sci. USA 107, 2830–2835 (2010).
Wu, M. C. et al. Rational re-engineering of a transcriptional silencing PreQ1 riboswitch. J. Am. Chem. Soc. 137, 9015–9021 (2015).
Dixon, N. et al. Orthogonal riboswitches for tuneable coexpression in bacteria. Angew. Chem. Int. Ed. Engl. 51, 3620–3624 (2012).
Kavita, K. & Breaker, R. R. Discovering riboswitches: the past and the future. Trends Biochem. Sci. 48, 119–141 (2023).
Polaski, J. T., Webster, S. M., Johnson, J. E. Jr. & Batey, R. T. Cobalamin riboswitches exhibit a broad range of ability to discriminate between methylcobalamin and adenosylcobalamin. J. Biol. Chem. 292, 11650–11658 (2017).
Mandal, M. & Breaker, R. R. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 5, 451–463 (2004).
Nudler, E. & Mironov, A. S. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 29, 11–17 (2004).
Serganov, A. & Nudler, E. A decade of riboswitches. Cell 152, 17–24 (2013).
Nshogozabahizi, J. C., Aubrey, K. L., Ross, J. A. & Thakor, N. Applications and limitations of regulatory RNA elements in synthetic biology and biotechnology. J. Appl. Microbiol. 127, 968–984 (2019).
Winkler, W. C., Cohen-Chalamish, S. & Breaker, R. R. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. USA 99, 15908–15913 (2002).
Serganov, A., Huang, L. & Patel, D. J. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature 458, 233–237 (2009).
Ding, J. et al. Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM. Nucleic Acids Res. 51, 9952–9960 (2023).
Burgess, C. M., Smid, E. J., Rutten, G. & van Sinderen, D. A general method for selection of riboflavin-overproducing food grade micro-organisms. Micro. Cell Fact. 5, 24 (2006).
McCown, P. J., Corbino, K. A., Stav, S., Sherlock, M. E. & Breaker, R. R. Riboswitch diversity and distribution. RNA 23, 995–1011 (2017).
Burgess, C., O’Connell-Motherway, M., Sybesma, W., Hugenholtz, J. & van Sinderen, D. Riboflavin production in Lactococcus lactis: potential for in situ production of vitamin-enriched foods. Appl. Environ. Microbiol. 70, 5769–5777 (2004).
Vicens, Q., Mondragon, E. & Batey, R. T. Molecular sensing by the aptamer domain of the FMN riboswitch: a general model for ligand binding by conformational selection. Nucleic Acids Res. 39, 8586–8598 (2011).
Rode, A. B., Endoh, T. & Sugimoto, N. Tuning riboswitch-mediated gene regulation by rational control of aptamer ligand binding properties. Angew. Chem. Int. Ed. Engl. 54, 905–909 (2015).
Gastaldi, G. et al. Riboflavin phosphorylation is the crucial event in riboflavin transport by isolated rat enterocytes. J. Nutr. 130, 2556–2561 (2000).
Cecchini, G., Perl, M., Lipsick, J., Singer, T. P. & Kearney, E. B. Transport and binding of riboflavin by Bacillus subtilis. J. Biol. Chem. 254, 7295–7301 (1979).
Blount, K. F. et al. Novel riboswitch-binding flavin analog that protects mice against Clostridium difficile infection without inhibiting cecal flora. Antimicrob. Agents Chemother. 59, 5736–5746 (2015).
Retallack, D. M., Jin, H. & Chew, L. Reliable protein production in a Pseudomonas fluorescens expression system. Protein Expr. Purif. 81, 157–165 (2012).
Baird, N. J. & Ferre-D’Amare, A. R. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA 16, 598–609 (2010).
Fritz, K. S. & Petersen, D. R. An overview of the chemistry and biology of reactive aldehydes. Free Radic. Biol. Med. 59, 85–91 (2013).
Regulski, E. E. & Breaker, R. R. In-line probing analysis of riboswitches. Methods Mol. Biol. 419, 53–67 (2008).
Soukup, G. A. & Breaker, R. R. Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5, 1308–1325 (1999).
Wilt, H. M., Yu, P., Tan, K., Wang, Y. X. & Stagno, J. R. FMN riboswitch aptamer symmetry facilitates conformational switching through mutually exclusive coaxial stacking configurations. J. Struct. Biol. X 4, 100035 (2020).
Appasamy, S. D., Ramlan, E. I. & Firdaus-Raih, M. Comparative sequence and structure analysis reveals the conservation and diversity of nucleotide positions and their associated tertiary interactions in the riboswitches. PLoS ONE 8, e73984 (2013).
Lienert, F., Lohmueller, J. J., Garg, A. & Silver, P. A. Synthetic biology in mammalian cells: next generation research tools and therapeutics. Nat. Rev. Mol. Cell Biol. 15, 95–107 (2014).
Meaux, S. & Van Hoof, A. Yeast transcripts cleaved by an internal ribozyme provide new insight into the role of the cap and poly(A) tail in translation and mRNA decay. RNA 12, 1323–1337 (2006).
Dakora, F. D., Matiru, V. N. & Kanu, A. S. Rhizosphere ecology of lumichrome and riboflavin, two bacterial signal molecules eliciting developmental changes in plants. Front. Plant Sci. 6, 700 (2015).
Atilho, R. M., Perkins, K. R. & Breaker, R. R. Rare variants of the FMN riboswitch class in Clostridium difficile and other bacteria exhibit altered ligand specificity. RNA 25, 23–34 (2019).
Gupta, P., Ojha, D., Nadimetla, D. N., Bhosale, S. V. & Rode, A. B. Tetraphenylethene derivatives modulate the RNA Hairpin-G-quadruplex conformational equilibria in proto-oncogenes. Chembiochem 23, e202200131 (2022).
Ontiveros-Palacios, N. et al. Rfam 15: RNA families database in 2025. Nucleic Acids Res. 53, D258–D267 (2025).
Acknowledgements
We acknowledge the funding from the DBT Ramalingaswami re-entry fellowship (BT/RLF/Re-entry/19/2016 to A.R.), DBT Govt of India (BT/PR45158/DRUG/134/119/2022 to A.R.), and the SERB-CRG grant (CRG/2022/004570 to A.R.).
Author information
Authors and Affiliations
Contributions
A.R. conceived the project and obtained the funding, D.O. and A.R. performed experiments, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ojha, D., Rode, A.B. Rational development of FMN-based orthogonal riboswitch that functions in response to specific non-cognate ligand. Nat Commun 16, 5751 (2025). https://doi.org/10.1038/s41467-025-60722-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-60722-4
This article is cited by
-
Recent advances in metabolic engineering of Escherichia coli for riboflavin biosynthesis
World Journal of Microbiology and Biotechnology (2025)