Abstract
Photoperiodic flowering in plants is orchestrated by the dynamic interaction between light signals and the endogenous circadian clock, but how light signals integrate into the clock remains to be fully elucidated. Here, we identify ELD1, a CCHC-type zinc finger protein that is essential for rice embryo survival. Notably, partial loss of ELD1 function results in early flowering under long-day conditions. Further investigations demonstrate that ELD1 physically interacts with OsNKAP, an orthologue of mammal NF-κB activating protein, as well as core splicing factors to regulate the splicing profile of OsCCA1, a core oscillator of the circadian clock. Molecular and genetic evidence indicate that OsCCA1 is the primary target of ELD1 in controlling flowering time. Additionally, ELD1 interacts with photoactivated phyB, mediating red-light-regulated alternative splicing of OsCCA1. Collectively, our findings establish a molecular connection between light signaling and the circadian clock, with ELD1 modulating OsCCA1 alternative splicing to control photoperiodic flowering.
Similar content being viewed by others
Introduction
Heading date (or flowering time) is a pivotal agronomic trait in crop cultivation, as it determines seasonal and regional adaptability. Photoperiod constitutes a critical environmental cue that regulates flowering time. Rice, a prototypical short-day (SD) plant, exhibits delayed flowering under long-day (LD) conditions and accelerated flowering under SD conditions. Previous studies have identified two major signaling pathways regulating flowering in rice. The first is the evolutionarily conserved photoperiodic flowering pathway, OsGI-Hd1-Hd3a/RFT, which is analogous to the Arabidopsis GI-CO-FT pathway. Heading date 1 (Hd1), the ortholog of Arabidopsis CONSTANS (CO), functions as a central regulator governing photoperiodic flowering in rice1. Hd1 generally promotes flowering under SD conditions by activating florigen genes Heading date 3a (Hd3a) and RICE FLOWERING LOCUS T 1 (RFT1), but inhibits flowering under LD conditions1,2. In addition to this conserved pathway, rice possesses another monocot-specific pathway, in which Ehd1 acts as a central hub and regulated by multiple flowering genes, including Ghd7, MADS50, Ehd2, Ehd3, Ehd43,4.
The plant circadian clock exerts a profound influence on plant growth and physiology, not only orchestrating daily events but also governing crucial developmental transitions throughout the plant’s life cycle. The coincidence between a circadian clock-controlled internal signal and a periodic external signal can initiate the flowering process. Therefore, the conserved components of circadian clock genes in rice and Arabidopsis are integrated into the photoperiodic flowering pathway5,6. For example, Hd1 is regulated by Rice CIRCADIAN CLOCK-ASSOCIATED 1 (OsCCA1), the core component of the circadian clock7. OsCCA1 exhibits peak expression at dawn and represses the transcriptional activity of OsGIGANTEA (OsGI). At dusk, OsCCA1 is inhibited by other clock components, leading to the accumulation of OsGI mRNA and the subsequent activation of Hd1 expression7,8,9,10.
In plant developmental processes, the circadian clock’s period and phase can be reset by exogenous signals, such as light and temperature11, allowing the clock to synchronize with the surrounding environment. Phytochromes, which are red and far-red light photoreceptors in plants, play a crucial role in this process by enabling plants sense light and transducing light signals to the circadian clock. Previous studies have shown that phytochromes regulate the circadian clock primarily through transcriptional control and protein degradation12. In Arabidopsis, LIGHT-REGULATED WD1 (LWD1) interacts with TCP20/22 at the TCP-binding site in the CCA1 promoter to activate its expression at dawn13. FAR-RED ELONGATED HYPOCOTYL3 (FHY3) and its paralog FAR-RED IMPAIRED RESPONSE1 (FAR1) physically interact with phytochrome-interacting factor 5 (PIF5) to regulate the light-induced CCA1 transcription14. Moreover, clock components, CCA1, GI, TIMING OF CAB 1 (TOC1), and EARLY FLOWERING 3 (ELF3) have also been found directly interact with phytochrome B (phyB)15.
Alternative splicing (AS) is widespread in eukaryotes, enhancing the diversity of the transcriptome and proteome. The process of pre-mRNA splicing is executed by a large dynamic complex known as spliceosome, which is composed of five small nuclear RNAs (U1 snRNA, U2 snRNA, U4 snRNA, U5 snRNA, U6 snRNA) and their corresponding small nuclear ribonucleoproteins (snRNPs)16,17,18. In addition, a number of RNA-binding proteins such as serine/arginine-rich (RS) proteins and heterogenous nuclear ribonucleoproteins, act as auxiliary splicing factors to identify targets and modulate appropriate splicing event18,19. During the process of spliceosome assembly, U1 snRNP initiate recognition of the 5′ splice site (SS) branch point sequence (BPS), followed by the binding of U2 snRNP to 3′SS BPS20,21. The spliceosome orchestrates precise catalysis to generate five major types of AS, including alternative 5′SS (A5SS) and 3′SS (A3SS), intron retention (IR), skipped exon (SE), and mutually exclusive exons19. Emerging evidence showed that light can trigger AS of circadian clock genes22,23. However, the detailed mechanisms regulating AS of circadian clock genes and the physiological consequences remain unclear.
In this study, we identified ELD1 as a photoperiod-dependent flowering time repressor gene in rice. ELD1 encodes a CCHC-type zinc finger protein rich in serine and arginine in its C-terminal region. ELD1 co-localizes and interacts with RS-related protein OsNKAP, as well as core splicing factors. Full-length RNA-seq analyses revealed that ELD1 affects global AS profiles, including the central oscillator gene of the circadian clock, OsCCA1. Molecular and genetic evidence demonstrates that ELD1 regulates flowering time in rice through the OsCCA1-Hd1 module. Additionally, our results revealed that photoactivated phyB interacts with ELD1 in the spliceosome. Red-light modulates the binding activity of ELD1 to OsCCA1, thereby mediating red-light-induced AS of OsCCA1. Overall, our results indicate that ELD1 is involved in light signaling and mediates photoperiodic flowering through regulating AS of OsCCA1 in rice.
Results
ELD1 negatively regulates flowering time in rice under long-day conditions
To explore the regulatory factors influencing flowering time in rice, we isolated the early flowering at long day 1 (eld1) mutant from an ethyl methanesulfonate (EMS) mutation pool of japonica cultivar Ningjing 4. The eld1 mutant exhibited an early flowering phenotype under natural long-day conditions (NLD), flowering 10 days earlier than the wild-type (WT) in Nanjing (E118°46′ N32°03′, ~14.25 h of daylight at transplanting) and 22 days earlier in Beijing (E118°28′ N40°00′, ~15 h of daylight at transplanting). However, the eld1 mutant exhibited no significant phenotype under natural short-day conditions (NSD) in Hainan (E110°18’ N18°34’, ~11 h of daytime at transplanting) (Fig. 1a–c). We crossed the eld1 mutant with WT and generated an F2 population. The early flowering and normal flowering individuals in F2 population segregated in a ratio close to 1:3 (χ² = 0.193, P > 0.05). This demonstrates that the early flowering phenotype of the eld1 mutant is controlled by a single recessive gene (Supplementary Fig. 1a).
a, b Phenotypic comparison of WT and eld1 plants at heading stage under Nanjing natural long-day conditions (NLD, ~14.25 h of daylight at transplanting) and Hainan natural short-day conditions (NSD, ~11 h of daylight at transplanting). White arrows indicate rice panicles. Scale bar, 10 cm. c Days to flowering of WT and eld1 mutant under Beijing NLD conditions (~ 15 h of daylight at transplanting), Nanjing NLD conditions, and Hainan NSD condition. Values are presented as means ± SD (n = 10 biological replicates), two-sided Student’s t-test was used to calculate P values. d Gene structure diagram of ELD1. The white and blue boxes represent UTR and CDS. The sequence and trace file of WT, eld1, and CRISPR-based adenine base editor (ABE) recover edit lines are indicated. e Phenotypic comparison of WT, eld1, and two independent ABE recover edit lines at the heading stage in Nanjing. Scale bars, 10 cm. f Days to flowering of WT, eld1, and two independent ABE recover edit lines at the heading stage in Nanjing. Values are presented as means ± SD (n = 10 biological replicates). g Phenotypic comparison of WT, eld1, and three independent transgenic complemented lines (C1 to C3) at heading stage under Nanjing NLD conditions. Scale bars, 10 cm. h Days to flowering of WT, eld1, and three independent transgenic complemented lines (C1 to C3) grown under Nanjing NLD conditions. Values are presented as means ± SD (n = 10 biological replicates). For (f and h), different letters were used to denote statistically significant differences, which were determined using one-way ANOVA with Tukey’s multiple comparisons test (P < 0.05).
To clone the responsible gene, we selected thirty extremely early-flowering individuals and thirty late flowering individuals from the F2 population to form the early and late bulks, respectively. Subsequently, we performed deep sequencing and employed bulked segregant analysis (BSA) to reveal the causal mutations. A distinct peak containing a cluster of five SNPs was detected on chromosome 9 (Supplementary Fig. 1b), and these SNPs are located on two retrotransposon and three putative protein-encoding genes, according to the rice MSU-RGAP reference genome annotation (Supplementary Table 1). Through sequencing of the individual plants, we confirmed a nonsynonymous nucleotide substitution from C to T in the coding sequence (CDS) of LOC_Os09g01640, resulting in the conversion of proline to leucine at the 39th residue of the encoded protein in the eld1 mutant (Fig. 1d).
To confirm whether this mutation causes the eld1 early flowering phenotype, we employed CRISPR-based adenine base editor (ABE) to restore the mutated T to C in the eld1 mutant, and the edited lines showed a similar flowering time to the WT (Fig. 1d–f). Further, a construct containing the 2.3-kb promoter, 1.4-kb CDS, and 0.8-kb downstream sequence of LOC_Os09g01640 (pELD1:ELD1) fully rescued the early flowering phenotype of eld1 at T1 generation (Fig. 1g, h), suggesting LOC_Os09g01640 corresponds to ELD1. Compared to WT, overexpression of ELD1-GFP fusion driven by the cauliflower mosaic virus 35S promoter (p35S:ELD1-GFP) caused delayed flowering of 3–6 days in the eld1 background (Supplementary Fig. 1c–e). Together, these results suggest ELD1 negatively regulates flowering time in rice under LD conditions.
ELD1 encodes an essential CCHC-type zinc finger protein
ELD1 is predicted to encode a protein that contains the CCHC-type zinc finger (ZCCHC) domain. Phylogenetic analysis showed that homologous proteins containing this domain are widely present across various species but have rarely been studied, particularly in plants. Protein sequence alignment indicates that both ELD1 and its orthologs contain a conserved ZCCHC domain, with the C-terminal region being highly enriched with arginine and serine residues. Notably, the first 39 amino acids of ELD1 are identical among multiple species (Supplementary Fig. 2).
To further investigate the role of ELD1, we attempted to generate ELD1 knockout mutants using CRISPR-Cas9 technology. However, we failed to obtain any homozygous null mutants at T0. Instead, we identified heterozygous lines and two homozygous lines with 6-bp in-frame deletions. We then tried to identify homozygous knockout mutants from the progenies of a heterozygous line, CR#1, which has a 1-bp deletion resulting in a frameshift mutation and premature termination of the encoded protein. However, all T1 progenies were either WT or heterozygous, and the segregation fit a 1:2 ratio (χ² = 0.13, P > 0.05) (Supplementary Fig. 3a). This suggests that ELD1 null mutants are likely embryonically lethal. Consistent with this, the heterozygous CR#1 line has normal pollen fertility, but the seed setting rate is only 71.7%, compared to the 92.5% in the WT (Supplementary Fig. 3b, c).
We further investigated the phenotypes of the two homozygous lines with 6-bp deletions, CR#3 and CR#9. Both lines result in the loss of two conserved amino acids and exhibited early flowering phenotypes similar to the eld1 mutant. Neither line showed any other developmental defects (Supplementary Fig. 3d–f). This suggests that the weak mutation of ELD1 is viable. Additionally, we also employed RNAi technology to generate three ELD1 knock-down transgenic plants. These RNAi lines also showed an early flowering phenotype (5–10 days earlier than WT) (Supplementary Fig. 3g–i). These results suggest that ELD1 is an essential gene and that weak mutations of ELD1 lead to early flowering.
ELD1 is rhythmically expressed and encodes a protein localized in nuclear speckles
To investigate the expression pattern of ELD1, we conducted reverse-transcription quantitative PCR (RT-qPCR) analysis across various tissues. The results revealed that ELD1 was broadly expressed in all tissues, with preferential expression in the leaves, nodes, and panicles (Fig. 2b). This finding was confirmed by β-glucosidase (GUS) staining of the pELD1: GUS transgenic plants (Fig. 2c). Additionally, a 24-h continuous expression analysis under controlled long-day conditions (CLD) revealed a rhythmic expression pattern of ELD1 in both WT and the eld1 mutant, with transcription levels peaking at Zeitgeber time 0 (ZT 0) and then gradually decreasing during the daytime. However, in the eld1 mutant, the expression level of ELD1 is significantly lower compared to the WT (Fig. 2a). This may suggest the involvement of a feedback regulatory mechanism.
a Rhythmic expression of ELD1 over 24 h in WT and eld1 mutant, detected by RT-qPCR in the leaves of 40-day-old plants grown under controlled long-day conditions (CLD, 14 h light/10 h dark). Values are means ± SD (n = 3 biological replicates). b RT-qPCR analysis of ELD1 expression in various tissues at heading stage. Values are means ± SD (n = 3 biological replicates). c GUS staining of the tissues collected from pELD1:GUS transgenic plants at the heading stage. (1) young panicle, (2) (3) panicle, (4) flag leaf, (5) stem node, (6) root, and (7) stem. Scale bars, 2.5 mm. d Subcellular localization of ELD1-GFP and free GFP in root cells of 7-day-old transgenic plants. Scale bars, 10 μm. e Subcellular localization of full-length ELD1 and various truncated forms in rice protoplasts. ELD1 (1–377) represents the full-length ELD1, ELD1-N (1–81) includes the conserved N-terminal, ELD1-M (82–205) contains the conserved ZCCHC domain, and ELD1-C (206–377) comprises the RS-enriched C-terminal region. D53-mCherry was used as a nuclear marker. Scale bars, 5 μm.
Subcellular localization analysis revealed nuclear speckle localization of the ELD1 protein in the p35S:ELD1-GFP/eld1 transgenic rice root cells (Fig. 2d). To further analyze the key domain responsible for its nuclear speckle localization pattern, we constructed various truncated ELD1 sequences fused in frame with GFP and transformed into rice protoplasts. The results indicated that the RS-enriched C-terminal domain (aa 206-377) is important for the nuclear speckle localization of ELD1 (Fig. 2e).
OsNKAP co-localizes and interacts with ELD1
To investigate the molecular role of ELD1, we performed yeast two-hybrid screening to identify ELD1-interact proteins. We identified OsNKAP and several RNA-binding proteins as interaction partners of ELD1 (Supplementary Table 2). OsNKAP is a putative ortholog of human NKAP (NF-κB activating protein), which is indispensable for the survival of hematopoietic stem cells and for maintaining the functionality and maturation of T cells24,25. MAS2, the Arabidopsis ortholog of OsNKAP, is essential for embryo development, as its null alleles result in embryonic lethality. Interestingly, researchers used MAS2 as a bait protein and similarly identified CXIP4, the homolog of ELD1, but their interaction was not further validated26. Protein alignment revealed that OsNKAP and its orthologous proteins have an RS-enriched region at the N-terminal, a basic region in the middle, and a conserved SynMuv domain at the C-terminal in both animals and plants (Supplementary Fig. 4a). To determine the interact region between ELD1 and OsNKAP, we generated different truncated variants of ELD1 and OsNKAP and tested their interaction using yeast two-hybrid assay. The results showed that the interaction between ELD1 and OsNKAP is highly dependent on the RS-enriched C-terminal region of ELD1 (Fig. 3a) and that the basic region (aa161-349) in the middle of OsNKAP is indispensable for its interaction with ELD1 (Supplementary Fig. 4b). The interaction between ELD1 and OsNKAP was verified through bimolecular fluorescence complementation (BiFC) assays in N. benthamiana leaf epidermal cells and co-immunoprecipitation (Co-IP) assay in rice protoplasts (Fig. 3b, c). Interestingly, we found that the molecular weight of the OsNKAP protein is higher than predicted, possibly due to post-translational modifications or other biochemical alterations. Subcellular localization analysis revealed that OsNKAP was also localized in nuclear speckles in root cells of the p35S: OsNKAP-GFP transgenic plants, mirroring the localization of ELD1 (Supplementary Fig. 5a). The co-localization of ELD1 and OsNKAP was further confirmed by a transit co-expression of ELD1-GFP and OsNKAP-mCherry in rice protoplasts (Fig. 3d).
a The interaction between ELD1 and OsNKAP in yeast cells. The ELD1-N (1–81), ELD1-M (82–205), and ELD1-C regions are indicated; The ZCCHC domain was marked in red. DDO refers to SD/-Trp/-Leu, and QDO refers to SD/-Trp/-Leu/-His/-Ade. The symbol “+” indicates the presence the corresponding construct, whereas “empty” represents the empty construct. b Bimolecular fluorescence complementation (BiFC) analysis revealed the interaction between ELD1 and OsNKAP in N. benthamiana cells. The inset images show the magnified portions of recombinant YFP signal. D53-mCherry serving as nuclear marker. Scale bars, 10 μm. c Co-immunoprecipitation (Co-IP) assays in rice protoplasts show that ELD1 can interact with OsNKAP. Immunoprecipitated samples were detected using anti-HA and anti-FLAG antibodies, respectively. The symbols “+” represent presence corresponding construct; symbols “−” represent free construct. Three independent experiments were performed with similar results. d ELD1-GFP co-localizes with OsNKAP-mCherry in rice protoplasts, and exhibited nuclear speckles localization. Free GFP was used as the control, Scale bars, 5 μm. e RT-qPCR analysis OsNKAP expression level in WT, OsNKAP-CR162, and three OsNKAP-RNAi lines. Samples were collected from 20-day-old seedlings grown under CLD conditions at ZT0. Values are presented as means ± SD (n = 3 biological replicates), two-sided Student’s t-test was used to calculate P values. f Phenotypes of WT, OsNKAP-CR162, and three distinct OsNKAP-RNAi lines at heading stage growth in Nanjing. Scale bars, 10 cm. g Days to flowering of WT, OsNKAP-CR162, and three independent OsNKAP-RNAi lines in Nanjing. Values are presented as means ± SD (n = 10 biological replicates), two-sided Student’s t-test was used to calculate P values.
OsNKAP shares similar biological function with ELD1
To investigate the biological function of OsNKAP in rice, we used CRISPR-Cas9 technology to generate OsNKAP knockout mutants. Similar to the situation with ELD1, we did not discover any homozygous null OsNKAP mutants in T0. In addition, all the identified progenies of a heterozygous 1-bp deletion line OsNKAP-CR18 were either WT or heterozygous, and the segregation fit a 1:2 ratio (χ2 = 0.07, P > 0.05) (Supplementary Fig. 5b), suggesting null mutation of OsNKAP also results in embryo lethality. Intriguingly, we identified a homozygous edited line, OsNKAP-CR162, which has 162 bp deletion, resulting in 54 amino acids missing, and the homozygous OsNKAP-CR162 line flowered 4 days earlier than the WT (Fig. 3f, g). We also knocked down OsNKAP through RNAi technology. Three independent RNAi lines were generated, in which OsNKAP expression was reduced to 15%–34% of WT levels. Under NLD conditions in Nanjing, all these lines exhibited 3–5 days earlier flowering phenotypes than the WT (Fig. 3e–g). However, no obvious phenotypic changes were observed under NSD conditions in Hainan (Supplementary Fig. 5c, d). These findings indicate that OsNKAP is also an essential gene for survival and acts as a flowering suppressor in rice, just like ELD1.
Both ELD1 and OsNKAP interact with the spliceosome complex components
NKAP is an RS-related protein in humans, characterized by a domain rich in arginine and serine residues. Proteins containing RS domains, RS-like, and RS-related proteins have been reported to play important roles in RNA metabolism27,28,29. Additionally, recent studies have reported that NKAP interacts with splicing factors and regulates constitutive splicing29,30. Given that ELD1 interacts with OsNKAP, and both of them are localized in nuclear speckles, we tested potential interactions between ELD1 and core splicing factors. As expected, ELD1 exhibited a strong interaction with U1-70K and U2AF65A, and a weaker interaction with U2AF65B in the yeast two-hybrid assay (Fig. 4a). Additionally, both luciferase complementation imaging (LCI) and BiFC assays confirmed the interaction between ELD1 and U1-70K, U2AF65A, and U2AF65B (Fig. 4c). Notably, the interactions exhibit a nuclear speckle localization (Supplementary Fig. 6a). Furthermore, the in vivo Co-IP assay demonstrated efficient co-precipitation of ELD1-GFP with U1-70K, U2AF65A, and U2AF65B, while the free-GFP control did not exhibit any interaction (Fig. 4b). These results suggest ELD1 physically interacts with core splicing factors. Consistently, ELD1 co-localized with U1-70K and U2AF65A in nuclear speckles in rice protoplasts (Fig. 4d). Similarly, we found that OsNKAP also interacted with U1-70K, U2AF65A, and U2AF65B, just like ELD1. Additionally, OsNKAP interacted with two other core factors, U2AF35B and U2AF35C (Fig. 4a and Supplementary Fig. 6b).
a A yeast-two hybrid assay shows ELD1 and OsNKAP interact with core splicing factors. DDO refers to SD/-Trp/-Leu, and QDO refers to SD/-Trp/-Leu/-His/-Ade. b The in vivo Co-IP assay confirms that ELD1 interacts with U1-70K, U2AF65A, and U2AF65B in N. benthamiana. The immunoprecipitated samples were detected using the anti-GFP and anti-FLAG antibodies. The symbol “+” indicates presence of the corresponding protein, while the symbol “−” represents the empty construct. Three independent experiments were performed with similar results. c The luciferase complementation imaging (LCI) assay confirms that ELD1 interacts with U1-70K, U2AF65A, and U2AF65B. d Co-localization of ELD1-GFP with U1-70K-mCherry and U2AF65A-mCherry in rice protoplasts. Scale bars, 5 μm. e RNA immunoprecipitation followed by qPCR (RIP-qPCR) assay demonstrating the binding affinity of ELD1 protein to snRNAs in vivo. The pUBI:ELD1-FLAG/eld1 transgenic seedlings grown for 10 days under CLD conditions were used. Samples were immunoprecipitated with anti-FLAG Magnetic Beads or the IgG control. UBQ gene serving as an internal control. Values are presented as means ± SD (n = 3 biological replicates). The asterisks indicate significant differences from IgG control using the two-sided student’s t-test (**P < 0.01).
In eukaryotic organisms, the spliceosome is a dynamic complex composed of small nuclear RNA (snRNA) and large number of RNA-binding proteins31. To confirm whether ELD1 functions as a constituent of the spliceosome and interacts with snRNAs, we performed an RNA immunoprecipitation qPCR (RIP-qPCR) assay using pUBI:ELD1-FLAG/eld1 transgenic plants, which perfectly complement the early flowering phenotype of eld1. Following immunoprecipitation, RT-qPCR was conducted using specific primers to detect the enrichment of snRNA. As expected, the FLAG beads effectively enriched U1, U2, U4, U5, and U6 snRNAs. As a control, no significant enrichment of U3 snRNA was observed (Fig. 4e). This result suggests that ELD1 interacts with mRNA splicing components, including not only the core splicing factors but also the snRNAs.
ELD1 regulates OsCCA1 splicing
We next investigated whether ELD1 regulates mRNA splicing by conducting a deep full-length RNA-seq analysis to compare genome-wide AS between WT and eld1 mutants. Samples from 40-day-old seedlings grown under CLD conditions were collected at ZT0. Among the three independent biological replicates, we identified 1611 significantly different transcripts across 1114 genes loci in the eld1 mutants compared to WT. Gene Ontology (GO) analysis revealed significant enrichment in biological processes such as mRNA processing, RNA splicing, and chromatin organization among the differentially spliced genes (Supplementary Fig. 7). To validate the full-length RNA-seq result, we selected six genes (Ehd4, RSZ21A, UVR8, U2AF65, YTH05, JAZ1) with significantly different AS patterns for RT-qPCR analysis. Using specific primers, we found that RSZ21A, and UVR8 exhibited significantly decreased splicing efficiency in the eld1 mutant compared to the WT, while Ehd4, U2AF65, YTH05, and JAZ1 show a significant increase of splicing efficiency (Supplementary Fig. 8).
OsCCA1 (also known as OsLHY or Nhd1) encodes a conserved MYB domain-containing transcription factor, which acts as the central oscillator of the circadian clock and has been reported to participate in multiple biological processes of plant growth and development, including flowering time7,32,33,34,35. Strikingly, the full-length RNA-seq analysis revealed over 60 splicing isoforms present in the 5′ untranslated region (5′UTR) and CDS of OsCCA1 (Supplementary Fig. 9) and three significant different splicing events between eld1 and WT at the OsCCA1 loci. These include an alternative 3′ splice site, designated as A3SS1, and two IR sites, designated as IRS1 and IRS2, respectively (Fig. 5a). Both A3SS1 and IRS1 are located in the 5′UTR region of OsCCA1 while IRS2 is positioned within the CDS, and IR results in an increase of 30 amino acids in the encoded protein (Supplementary Fig. 10a–d). The differential AS events were validated by semi-quantitative RT-PCR (sqRT-PCR) (Fig. 5b and Supplementary Fig. 10a). RT-qPCR analysis revealed that eld1 mutants and three independent ELD1-RNAi lines exhibited a significantly reduced A3SS1 splicing ratio (2 ~ 5% vs. 8.6%) compared to the WT, while showing a significantly higher IR ratio at IRS1 (70 ~ 90% vs 46%) and IRS2 (23 ~ 36% vs 15%) compared to WT (Supplementary Fig. 11a–d).
a The Sashimi plot from the Integrative Genomics Viewer (IGV) illustrates three distinct alternative splicing sites of OsCCA1, highlighting differences between the WT and eld1 mutant. b sqRT-PCR analysis of A3SS1, IRS1 and IRS2 splicing pattern of OsCCA1 in WT, eld1 and three ELD1-RNAi lines. Using cDNA as a template, the sqRT-PCR primers (ACTIN, A3SS1, and IRS1) amplify the target regions along with adjacent constitutive introns, with genomic DNA (gDNA) serving as a control. All the sqRT-PCR reactions run for 27 cycles, except for A3SS1, which runs for 28 cycles due to its relatively low isoform expression. Samples were collected from 20-day-old seedlings grown under CLD conditions at ZT0. More than three independent experiments were performed with similar results. c Rhythmic expression of OsCCA1 over 24 h in WT and eld1 mutant, detected by RT-qPCR in the leaves of 40-day-old plants grown under CLD conditions. Values are means ± SD (n = 3 biological replicates). d–f The rhythmic splicing pattern of A3SS1 (d), IRS1 (e), and IRS2 (f) was detected by RT-qPCR in both WT and eld1. Total RNA was extracted from 40-day-old plants under CLD conditions. Values are presented as means ± SD (n = 3 biological replicates). g Rhythmic expression of OsCCA1 over 24 h in WT and eld1 mutant, detected by RT-qPCR in the leaves of 27-day-old plants grown under CSD conditions. Values are means ± SD (n = 3 biological replicates). h–j The rhythmic splicing pattern of A3SS1 (h), IRS1 (i) and IRS2 (j) detected by RT-qPCR in WT and eld1. Total RNA was extracted from 27-day-old plants under CSD conditions. Values are presented as means ± SD (n = 3 biological replicates). For (c–f and h), Asterisks indicate significant differences from WT using the two-sided Student’s t-test (*P < 0.01, *P < 0.05).
Given that many other splicing factors also have abnormal splicing pattern in the eld1 mutant, we hypothesize that the observed defect in OsCCA1 splicing in eld1 could be a secondary effect caused by defects in these splicing factors. To test this notion, we employed RIP-qPCR to determine whether ELD1 can bind to OsCCA1 mRNA. The results demonstrated that FLAG beads can effectively enriched OsCCA1 mRNA from the cell homogenate of pUBI:ELD1-FLAG/eld1 transgenic plants (Supplementary Fig. 11e). These findings indicate that ELD1 can directly bind to OsCCA1 mRNA and regulate its AS. Moreover, RT-qPCR analysis showed that the AS patterns of A3SS1 and IRS1, but not IRS2, in three independent OsNKAP-RNAi lines were similar to those observed in eld1 and ELD1-RNAi lines (Supplementary Fig. 11f–h). It is possible that other splicing factors other than OsNKAP that collaborate with ELD1 to regulate the AS of IRS2.
Interestingly, we observed that the splicing efficiency of the three AS sites followed a circadian rhythm in both WT and the eld1 mutant under CLD conditions, with IR peaking around ZT0 and reaching its lowest level around ZT12, aligning with the transcriptional oscillation pattern of OsCCA1 (Fig. 5c–f). Under CSD conditions, the splicing efficiency of IRS1 and IRS2 continued to exhibit rhythmic fluctuations, with IR peaking at ZT16 and reaching its lowest point at ZT8. However, the splicing efficiency of A3SS1 did not display a clear rhythmic pattern under CSD conditions (Fig. 5g–j). In addition, we noticed that the splicing efficiency at the A3SS1 site still showed a significant difference between WT and the eld1 mutant (Fig. 5h). However, the difference in splicing efficiency at IRS1 and IRS2 was eliminated under CSD conditions (Fig. 5i, j). These suggest the ELD1 dependent OsCCA1 splicing is regulated by photoperiod, which may explain why the eld1 mutant does not exhibit a distinct flowering time phenotype under short-day conditions in Hainan.
To investigate whether ELD1 exhibits a rhythmic expression pattern similar to OsCCA1, 24-day-old seedlings grown under CLD conditions were subjected to continuous light treatment. The results showed that ELD1 displayed a rhythmic expression pattern similar to that of OsCCA1. Under free-running conditions, both OsCCA1 and ELD1 maintained rhythmic expression, although the amplitude was significantly reduced (Supplementary Fig. 12). These findings suggest that ELD1 rhythmically regulates the splicing efficiency of OsCCA1.
ELD1 regulates flowering time via the OsCCA1-Hd1 module
Previous studies have elucidated the role of OsCCA1 in regulating flowering time by modulating the rhythm transcription of Hd17,32. To investigate the genetic relationship between ELD1, OsCCA1, and key flowering regulators, we generated oscca1 mutant and eld1 oscca1 double mutant through genome editing. The oscca1 mutant, harboring a frameshift mutation, showed a strong late-flowering time phenotype, delayed by up to 27 days, and the eld1 oscca1 double mutant showed a similar flowering time with the oscca1 single mutant (Fig. 6a, b and Supplementary Fig. 13c), suggesting that OsCCA1 is epistatic to ELD1 in the regulation of flowering time. Consistently, the two florigen genes Hd3a and RFT1 exhibited increased expression in eld1 compared to WT, but decreased expression in oscca1 and eld1 oscca1 backgrounds (Fig. 6c, d). Additionally, we also generated eld1 hd1 and eld1 ghd7 double mutant and eld1 hd1 oscca1 triple mutant, and found that the eld1 hd1 double mutant and eld1 hd1 oscca1 triple mutant exhibited the same flowering time as the hd1 single mutant. However, the eld1 ghd7 double mutant displayed an extremely early flowering phenotype compared to either ghd7 or eld1 single mutant (Fig. 6a, b and Supplementary Fig. 13a–e). These observations suggest ELD1 and Hd1 function in the same genetic pathway, but acts independently of Ghd7 to control flowering time. Consistently, RT-qPCR analyses revealed upregulation of Hd1 expression in the oscca1 mutant and eld1 oscca1 double mutant, suggesting a negative effect of OsCCA1 on Hd1 expression. However, downregulation of Hd1 expression was observed in the eld1 mutant (Fig. 6e), suggesting a positive effect of ELD1 on Hd1 expression. These results, together with the genetic interaction data, strongly support that ELD1 represses the function of OsCCA1 and regulates rice flowering time through modulating the expression of Hd1.
a Phenotypes of WT, eld1, oscca1, eld1 oscca1, hd1, eld1 hd1 and eld1 hd1 oscca1 at heading stage growth in Nanjing NLD conditions. Scale bars, 10 cm. b Days to flowering of WT, eld1, oscca1, eld1 oscca1, hd1, eld1 hd1 and eld1 hd1 oscca1 in Nanjing NLD conditions. Values are presented as means ± SD (n = 12 biological replicates). Different letters were used to denote statistically significant differences, which were determined using one-way ANOVA with Tukey’s multiple comparisons test (P < 0.05). c–e Expression of Hd1 and two florigens genes in WT, eld1, oscca1, and eld1 oscca1 detected by RT-qPCR. Samples were collected from 40-day plants grown under CLD conditions. Values are presented as means ± SD (n = 3 biological replicates).
ELD1 is involved in phytochrome signaling to mediate AS of OsCCA1
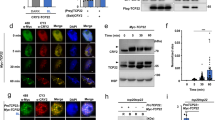
It has been reported that RNA splicing and mRNA processing GO terms were significantly enriched among phytochrome-mediated AS genes during the early light response in Arabidopsis22,36. The observation that these GO terms are also enriched in the ELD1-mediated AS events suggests a potential correlation between ELD1 and the phytochrome-mediated AS process (Supplementary Fig. 7). We thus tested whether there is direct interaction between ELD1 and phyB. BiFC assays demonstrated that ELD1 interacts with phyB in N. benthamiana cells and colocalizes with the splicing factor U1-70K (Fig. 7a). Additionally, the in vivo Co-IP assay also showed an efficient co-precipitation of ELD1-GFP with phyB-FLAG (Fig. 7b). Furthermore, the LCI assay revealed that the interaction between ELD1 and phyB is light-dependent (Supplementary Fig. 14a).
a BiFC analysis revealed the interaction between ELD1 and phyB in N. benthamiana cells. The inset images display the magnified portions of YFP signal. U1-70K mCherry serving as nuclear speckles marker, Scale bars, 10 μm. b The in vivo Co-IP assay confirms the interaction between ELD1 and phyB in N. benthamiana. The immunoprecipitated samples were detected using the anti-GFP and anti-FLAG antibodies. The symbols “+” represent presence corresponding protein; symbols “−” represent empty construct. Three independent experiments were performed with similar results. c The AS pattern of A3SS1, IRS1, and IRS2 detected by sqRT-PCR after 2 h of red-light treatment in WT and phyBT822*. Continuous dark treatment serves as control. More than three independent experiments were performed with similar results. d–f The AS pattern of A3SS1 (d), IRS1 (e), and IRS2 (f) detected by RT-qPCR after 2 h of red-light treatment in WT and eld1 mutant. The indicated genotypes of 10-day-old seedlings were grown under CLD conditions, pretreated with darkness for 48 h, and then exposed to red light for 2 h. Continuous dark treatment serves as a control. Values are presented as means ± SD (n = 3 biological replicates). g RIP-qPCR assay shows the in vivo binding affinity of ELD1 to OsCCA1 mRNA. pUBI:ELD1-FLAG/eld1 transgenic seedlings grown for 10 days under CLD conditions were pretreated with darkness for 48 h, and then exposed to red light for 2 h. Continuous dark treatment serves as the control. Samples were immunoprecipitated with anti-FLAG Magnetic Beads. UBQ gene serves as an internal control. Values are presented as means ± SD (n = 3 biological replicates). For (d–g), two-sided Student’s t-test was used to calculate P values.
To determine whether phyB is involved in the AS of OsCCA1, we examined the 10-day-old WT and a phyB stop-gain mutant phyBT822* grown under 28 °C CLD conditions. The seedlings were pretreated in darkness for 48 h, and then exposed to red light, with continuous dark treatment serving as a control to eliminate circadian clock interference. RT-qPCR and sqRT-PCR results indicated that a 2-h red-light treatment significantly increased the IR ratio at the IRS1 and IRS2 sites of OsCCA1 (Fig. 7c–f), while decreasing the AS ratio at A3SS1 site in WT (Fig. 7d). This effect was not observed in phyBT822* mutant, suggesting that phyB also regulates the red-light dependent AS of OsCCA1 (Fig. 7c and Supplementary Fig. 14b–d).
To investigate whether this red-light regulated AS of OsCCA1 is dependent on ELD1, we detected the AS of OsCCA1 at the A3SS1, IRS1, and IRS2 in both WT and eld1 mutant after red-light treatment. RT-qPCR analysis revealed that, after 2 h red-light treatment, the AS ratio at A3SS1 significantly decreased, and the IRS2 IR ratio significantly increased compared to continuous darkness in WT, but similar changes were not observed in eld1 (Fig. 7d, f). These findings suggest that ELD1 plays an important role in phyB mediated AS of OsCCA1. Notably, the IR ratio at IRS1 site was significantly increased after red-light treatment both in WT and eld1, but the fold change in eld1 was notably lower (Fig. 7e), suggesting that other factors may also mediate red-light regulated AS of OsCCA1 at specific sites.
The change of OsCCA1 AS pattern after red-light treatment is similar to that of eld1 mutant, suggesting that phyB may exert a repressive effect on the regulatory function of ELD1. To determine how red-light affects the function of ELD1, we conducted immunoblot analysis to detect protein accumulation of ELD1 in pUBI:ELD1-FLAG/eld1 transgenic seedling exposed to red light for different durations, and red-light seems not to affect the stability of the ELD1 protein (Supplementary Fig. 14e). Further, ELD1 protein still forms nuclear speckles in root cells of dark grown etiolated p35S:ELD1-GFP transgenic seedlings (Supplementary Fig. 14f), suggesting red-light also not affects the subcellular localization of ELD1. However, the RIP-qPCR assay showed that ELD1 co-immunoprecipitated less OsCCA1 mRNA after exposed to red light 2 h compared to darkness (Fig. 7g). These results indicate that phyB may repress the ELD1 RNA-binding activity to regulate OsCCA1 AS.
To examine the genetic relationship between ELD1 and phyB, we crossed eld1 with the phyBT822* mutant to generate eld1 phyBT822* double mutant. Phenotypic analyses showed that phyBT822* exhibited pronounced dwarfism compared to WT at the 15-day-old seedling stage growing under CLD conditions. However, the plant height of eld1 phyB T822* double mutant showed remarkable recovery (Supplementary Fig. 15a, b), demonstrating that ELD1 is epistatic to phyB regarding seedling phenotype. Surprisingly, the eld1 phyBT822* double mutant displayed an extremely early flowering phenotype that exceeded the combined additive effects of eld1 and phyBT822* under NLD conditions (Supplementary Fig. 15c, d). These findings suggest that phyB genetically interacts with ELD1 to regulate rice flowering, and may control flowering through multiple pathways in addition to ELD1.
Manipulating ELD1 activity is advantageous for rice breeding
Earlier flowering is desired trait in rice breeding for expanding the elite cultivars to higher latitudes. To evaluate the value of the eld1 weak mutant alleles in rice breeding, we thoroughly investigated its agronomic traits. Compared to WT, eld1 showed a normal seed setting rate, a 5.7% reduction in plant height, and a 14% reduction in grain number, but grain length and 1000-grain weight remarkably increased. Ultimately, the yield of eld1 plot was only reduced by 5.9% compared to WT. However, when the yields were adjusted for their growth durations, there is no significant difference in daily grain production between eld1 and WT (Supplementary Fig. 16a–c and Table 3). We also investigated the agronomic traits of the ELD1-RNAi lines, which show similar results to the eld1 mutant (Supplementary Table 3). This suggests a promising prospect for utilizing ELD1 in rice breeding.
Thus, we employed the CRISPR-based cytosine base editors (CBE) to mimic the C to T mutation found in the eld1 mutant. We successfully generated four distinct editing lines harboring the designed C to T mutation with different accessory mutations within the editing window (CBE#2 to CBE#5), resulting in two types of amino acid variations. These four transgenic lines exhibited a phenotypic similarity to the eld1 mutant, characterized by early flowering and a normal seed setting rate in Nanjing (Supplementary Fig. 16d–f). To evaluate the field performance of eld1 and four CBE lines in higher latitudes, we planted these lines at Beijing (E118°28’ N40°00’). The WT plants flowered too late, encountered frost stress at the grain-filling stage, resulting in failure to mature and nearly no harvest. The eld1 and four CBE lines flowered approximately 22 days earlier than the WT, they were able to mature and be harvested normally (Fig. 8a–d). The same strategy was applied on another elite cultivar, Ningjing 7. As expected, we successfully generated five distinct editing lines that flowered 13-15 days earlier than Ningjing 7 in Nanjing (E118°46′ N32°03′) (Supplementary Fig. 16g–i).
a Phenotypes of WT, eld1 and four independent CBE editing lines at heading stage during growth in Beijing NLD conditions. Scale bars, 10 cm. b Panicle maturity of WT, eld1, and four independent CBE editing lines at the time of harvest in Beijing. Scale bars, 2 cm. c Days to flowering of WT, eld1 and the CBE editing lines in Beijing NLD conditions. Values are presented as means ± SD (n = 12 biological replicates). Different letters were used to denote statistically significant differences, which were determined using one-way ANOVA with Tukey’s multiple comparisons test (P < 0.05). d Brown rice of WT, eld1, and CBE editing lines at the time of harvest in Beijing. The WT failed to mature and nearly no harvest, while eld1 and four CBE lines were able to mature and be harvested normally. e The working model of ELD1 in regulating photoperiodic flowering. ELD1 interacts OsNKAP and core splicing factors to regulate global AS. The ELD1-containing spliceosome complex directly binds and regulates the rhythmic AS of OsCCA1. Additionally, phyB physically interacts with ELD1 in a light dependent manner, inhibiting its binding to OsCCA1 pre-mRNA. This regulation ensures appropriate OsCCA1 activity and a properly tuned circadian clock, resulting in high Hd1 transcription and prolonged flowering at LD. In the eld1 mutant, the OsCCA1 has aberrant splicing pattern at A3SS1, IRS1, and IRS2 sites. These aberrantly spliced forms of OsCCA1 strongly suppress Hd1 transcription, leading to the activation of downstream florigen genes and ultimately causing early flowering.
We also examined the natural variation of ELD1 in rice germplasms, SNP data of 844 accessions were obtained from previously published studies37. Based on two nonsynonymous SNPs (SNP1, SNP3) and one synonymous SNP (SNP2) in the coding region, ELD1 can be classified into four haplotypes (Hap1-Hap4). Among these haplotypes, Hap2 carries a natural variation at SNP3, resulting in an amino acid substitution from Arginine (Arg) to Lysine (Lys) at position 280 (Supplementary Fig. 17a). Amino acid alignment analysis revealed that this Arg is highly conserved across different species (Supplementary Fig. 2b). Flowering time analysis indicated that Hap2 exhibited significantly delayed flowering compared to the other haplotypes under long-day conditions in Nanjing. However, under short-day conditions in Hainan, no significant difference in flowering time was observed between Hap2 and Hap1, although Hap2 exhibits a slightly delayed flowering time compared to Hap3 and Hap4 (Supplementary Fig. 17b). These findings align well with the absence of a significant phenotype in the eld1 mutant under short-day conditions, further supporting the conclusion that ELD1 regulates rice flowering time specifically under long-day conditions. Therefore, manipulating ELD1 activity through targeted mutations at key amino acid sites, such as the mutated site in eld1 or SNP3 in natural variation, holds promise for crop improvement and provides an approach for utilizing essential genes in breeding.
Discussion
AS is an important post-transcriptional regulatory process that can diversify the transcriptome and proteome in eukaryotes. Although AS of flowering time genes has been observed in Arabidopsis38,39,40, the mechanisms by which AS is regulated and how it integrates various developmental and environmental signals into flowering pathways remain unclear. In this study, we identified ELD1 as a CCHC-type zinc finger protein, and its partial loss of function leads to early flowering at LD in rice. ELD1 interacts with OsNKAP and core splicing factors to regulate global AS. The ELD1-containing spliceosome complex directly binds to and regulates the rhythmic AS of OsCCA1. Additionally, phyB physically interacts with ELD1 in a light-dependent manner, and inhibiting its binding to OsCCA1 pre-mRNA. This phyB-ELD1-OsCCA1 module represents a previously unrecognized photoperiodic flowering pathway, through which ELD1 mediates light signaling to modulate the circadian clock via AS of central clock gene.
In mammals, NKAP has been reported as a multifunctional protein that regulates numerous physiological processes and diseases. It participates in pre-mRNA splicing, chromosome alignment, and transcript regulation25,29,41. However, the function of NKAP family in rice remain unclear. Here, we found that knockout of OsNKAP in rice leads to embryonic lethality, consistent with observations in mammals and Arabidopsis, where deletion of NKAP in hematopoietic stem cells and Treg cells leads to cell death24,42,43, and null alleles of MAS2 (the Arabidopsis ortholog of NKAP) cause embryonic lethality26. The ZCCHC superfamily proteins were widely found in various species, but most of their biological functions remain unclear44,45. In this study, we show that ELD1 interacts with the core splicing factors to regulate AS in rice (Fig. 4a–c), and null allele of ELD1 also results in embryonic lethality. Consistent with these findings, a recent study showing that depletion of CXIP4, the Arabidopsis ortholog of ELD1, leads to lethality and pre-mRNA missplicing46. This suggests an essential role for OsNKAP and ELD1 in plant viability. Interestingly, both the eld1 and the osnkap weak alleles obtained in this study exhibit early flowering phenotypes, providing an opportunity to shed light another layer of their functions on the photoperiod flowering and adapt to the environment changes. Recent research has revealed cigarette smoke condensate enhanced NKAP binding to the pri-miR-25 m6A site and promoted its maturation, ultimately facilitating pancreatic cancer progression47. Similarly, NKAP also has been reported safeguards glioblastoma cells against ferroptosis by regulates SLC7A11 mRNA splicing dependent m6A modification30. It’s worth exploring whether the ELD1 and OsNKAP complex regulate pre-mRNA splicing in rice is an m6A-dependent manner in the future.
Circadian clocks in numerous organisms predominantly function through intricately interconnected transcriptional regulatory feedback loops48,49. Meanwhile, diverse post-transcriptional regulatory processes, including AS, are also important for maintaining circadian homeostasis. AS of clock genes has been extensively observed in Arabidopsis48,50, but the biological significance and the regulatory mechanism remain largely unknown. In the eld1 mutant, we identified three significantly defective splicing events at the OsCCA1 locus (Fig. 5a). The altered splicing pattern of OsCCA1 disrupts the rhythmic expression of Hd1, resulting in the early flowering phenotype observed in eld1. These findings offer valuable insights into how specific splicing regulators modulate the AS of clock genes. Both A3SS1 and IRS1 sites are located in the 5′UTR, which encompasses a multitude of elements, such as internal ribosome entry sites, upstream open reading frames, and structural components involved in regulating mRNA stability51,52,53. The IRS2 site is located in the coding region, and its retention results in an additional 30 amino acid residues of the encoded protein. Our full-length RNA-seq analysis revealed abundant splicing isoforms present in OsCCA1, making it challenging to dissect the functions of individual isoforms (Supplementary Fig. 9). The molecular mechanism by which AS of OsCCA1 affects its function needs further investigation in the future. Precisely editing these AS sites using CRISPR-Cas systems may be an effective approach.
Light signals induce physiological responses by regulating gene expression at both transcriptional and post-transcriptional levels. Recent studies have shown that light can trigger pre-mRNA AS cascades. Deep RNA-seq analysis has also revealed that early red-light treatment induces AS of circadian clock genes22. However, there are only scattered clues about how light regulates AS in plants. For example, SUPPRESSOR-OF-WHITE-APRICOT1 (SWAP1)-SPLICING FACTOR FOR PHYTOCHROME SIGNALING (SFPS)-REDUCED RED LIGHT RESPONSES IN CRY1CRY2 BACKGROUND 1 (RRC1) splicing factor complex facilitates red-light-induced AS by physically interacting with phyB54,55,56. Additionally, the interaction between CRY2 INTERACTING SPLICING FACTOR 1 (CIS1) and the blue-light receptor cryptochrome 2 (CRY2) has been shown to regulate the AS of FLOWERING LOCUS M (FLM)57. AtPRMT5, UAP56, and DCS1 have been reported to work in concert with the E3 ubiquitin ligase CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) to regulate pre-mRNA splicing during red-light response36,58,59. In this study, we obtained several pieces of evidence supporting that ELD1 is a splicing factor involved in phytochrome signaling. Firstly, although quite a lot of AS sites were discovered in OsCCA1, the AS sites regulated by red light align with those of ELD1 (Fig. 7d–f), indicating a close functional relationship between them. Secondly, photoactivated phyB physically interacts with ELD1 and represses its RNA-binding activity. Thirdly, GO terms associated with RNA processing and RNA splicing were enriched in ELD1-mediated AS genes (Supplementary Fig. 7), consistent with the previously reported enriched GO terms in light-induced AS22,36,60. These results suggest that ELD1 may function as a potential splicing factor in mediating pre-mRNA splicing during light responses in rice.
Notably, ELD1 is epistatic to phyB at the seedling stage, supporting its role in mediating red light signaling during this phase. However, the eld1 phyBT822* double mutant exhibited an exceptionally early flowering phenotype that surpassed the combined additive effects of eld1 and phyBT822*, indicating a genetic interaction distinct from that observed at the seedling stage (Supplementary Fig. 15a–d). This phenomenon can be attributed to phyB’s involvement in regulating flowering time through ELD1-independent pathways. For example, phyB has been shown to positively regulate Ghd7 at the transcriptional level and directly interact with, and stabilize GHD7 at the protein level61,62. This notion is further supported by the genetic evidence suggesting that ELD1 represses rice flowering independently of Ghd7 in this study (Supplementary Fig. 13a, b).
Essential genes are crucial for an organism’s survival, playing pivotal roles in fundamental cellular processes. A significant number of essential genes have been identified in the genomes of plants and animals63,64. In crops, null mutants of these genes often result in lethality or poor agronomic traits, which makes it challenging to utilize essential genes in crop breeding. In this study, we found that knocking out either ELD1 or OsNKAP in rice leads to embryonic lethality, indicating both are essential for rice survival. An interesting finding is that the eld1 in our study, which is a partial loss of function mutant, exhibits normal development except for early flowering. This suggests that the residual activity in eld1 may be sufficient to support most biological processes. The eld1 mutant flowers 10 days earlier, with slightly reduced plant height and grain number, but increased grain length and weight. Ultimately, this results in only a 5.9% reduction in plot yield, leading to a daily yield comparable to that of WT in Nanjing. This offers an effective way to utilize essential genes in crop breeding. Indeed, we successfully mimicked the eld1 allele in two elite cultivars (Ningjing 4 and Ningjing 7) using CRISPR-based CBE, and both exhibited early flowering without significant agronomic drawbacks (Supplementary Fig. 16d–i). These edited lines can flower and set seeds before low temperatures arrive in autumn, allowing them to be planted in higher latitudes, thereby expanding the cultivation range of these elite cultivars. Therefore, manipulating the activity of ELD1 holds promise for crop improvement and provides insights into essential genes function research.
Methods
Plant materials and growth conditions
The eld1 mutant was identified from an EMS pool of japonica rice cultivar Ningjing 4. To observe their flowering phenotypes, plants were grown in paddy fields under natural long-day (NLD) conditions in Beijing (E118°28′ N40°00′) and Nanjing (E118°46′ N32°03′) and natural short-day (NSD) conditions in Hainan (E110°18′ N18°34′). For molecular experiments, plants were grown in light incubators under controlled long-day (CLD, 14 h light /10 h darkness at 28 °C) and controlled short-day (CSD, 10 h light /14 h darkness at 28 °C) conditions, light intensity of approximately 800 µmol/s/m² and a relative humidity of 70%. For red-light treatment, approximately 120 µmol/s/m² red light intensity. To assess agronomic traits, the WT, eld1 mutants and three ELD1-RNAi lines were grown in paddy fields under NLD conditions in Nanjing. The yield per plot was measured after harvesting and drying the seeds. Six plots, each containing 20 plants, were established for both the WT and eld1 mutant to assess plot yield.
Bulked segregant analysis to cloning ELD1
To identify the candidate gene causing the early flowering time phenotype, we crossed the eld1 mutant with WT to generate an F2 population. After assessing the flowering time of each individual, 30 plants exhibiting extremely early flowering and 30 plants with extremely late flowering were selected from a total of 171 F2 individuals to form the early and late bulks. Genomic DNA was extracted from each bulk using the CTAB method after the leaves were ground in liquid nitrogen. Deep sequencing was performed to elucidate the causal mutation, generating 8 Gb of clean data for each bulk. The causal mutation was identified using a modified MutMap pipeline65.
Vector construction and plant transformation
The 3.9-kb genomic DNA fragment, including a 2.5-kb promoter, the complete transcribed region of ELD1, and 0.3-kb downstream region, was cloned into the pCAMBIA1390 vector to generate a complementation construct. The full-length CDS of ELD1 was cloned into the pCUBI1390-FLAG and pCAMBIA1305-GFP vectors to generate FLAG-tagged ELD1 and GFP-tagged ELD constructs, respectively. These constructs were then introduced into Agrobacterium tumefaciens strain EHA105, and subsequently transferred into eld1 mutant. Similarly, the CDS of OsNKAP was cloned into the pCAMBIA1305-GFP vector to generate a GFP-tagged OsNKAP construct, which was then transferred into the WT background. To generate the ELD1-RNAi construct, both the sense and antisense versions of a 203-bp specific fragment from the CDS of the ELD1 were amplified with the primer pairs ELD1-RNAi-F and ELD1-RNAi-R, sequentially cloned into the pCUbi1390-∆FAD2 vector. In the same way, a 196-bp specific fragment from the CDS of OsNKAP was cloned into pCUbi1390-∆FAD2 vector to generate OsNKAP-RNAi construct. The CRISPR-based adenine base editor (ABE) was used to revert the eld1 mutant site to WT (T116 to C116). The specific sgRNA was cloned into pH-ABE8e-SpRY-esgRNA vector66 and transformed into eld1 mutant. The CRISPR-based cytosine base editor (CBE) was used to generate the eld1 alleles in breeding early flowering cultivars. The specific sgRNA was cloning into pH-CBEA3A-SpRY-esgRNA vector and transformed into WT (Ningjing 4) and Ningjing 7, respectively. Primers used to construct the vectors are listed in Supplementary Data 2.
Phylogenetic analysis
The orthologous sequences of ELD1 in Arabidopsis, corn, sorghum, and other species were downloaded from National Center for Biotechnology Information (NCBI). The ClustalW program was used to perform multiple sequence alignment of these proteins. The phylogenetic tree was generated using the neighbor-joining method with 1,000 bootstrap replicates in MEGA 6 software. The amino acid sequences of ELD1 and OsNKAP homologous proteins were aligned and displayed using DNAMAN software.
Subcellular localization
To analyze the subcellular location of ELD1 and OsNKAP in plants, the CDS of both genes were cloned into the pCAMBIA1305-GFP vector, and subsequently transformed into the eld1 mutant and WT, respectively. A confocal laser scanning microscope (Leica TCS SP8) was used to detect GFP fluorescence signal in young root (5-day seedlings) of transgenic plants, with the same genotypes transformed with free-GFP as a control. The full-length and various truncated CDS of ELD1 were cloned into the PAN580-GFP vector to determine the subcellular localization of different domains. All plasmids were co-expressed with the nuclear marker D53-mCherry in rice protoplasts for 16 h before the fluorescence signals were detected using the confocal microscope. For colocalization analysis, the genes OsNKAP, U1-70K, and U2AF65A were inserted into the PAN583-mCherry vector and individually co-expressed with ELD1-GFP in rice protoplasts.
RNA extraction and RT-qPCR
In this study, RNA used for gene expression and AS analysis was extracted using the Tiangen RNA extraction kit (Tiangen, Beijing, China), which contains DNase I to digest genomic DNA. Reverse transcription was performed using oligo-dT primer and PrimeScript II reverse transcriptase (TaKaRa, Shiga, Japan). RT-qPCR was conducted on an ABI prism 7500 Real-Time PCR System with the SYBR Premix Ex Taq Kit (TaKaRa, Shiga, Japan). The RT-qPCR primers are listed in the Supplementary Data 2, with the rice UBQ gene serving as an internal control.
Full-length RNA-seq and data analysis
WT and eld1 mutant were grown in a light incubator under CLD conditions (14 h light / 10 h darkness). Three independent samples were collected from each genotype during the ZT0 time point of 40-day-old plants. Total RNA extracted from each sample was treated with RNase-free DNase I to eliminate genomic DNA. Full-length cDNA was sequenced using Nanopore’s PromethION platform, generating 15 Gb clean data to ensure a high standard AS analysis. After removing adapter sequences and filtering low-quality reads, over 98% full-length reads were aligned to the NCBI rice reference genome (Genome assembly IRGSP-1.0) using Minimap2 software67. rMATS-long software [https://github.com/Xinglab/rMATS-long] was used to detect the significant differences (P < 0.05) AS genes between WT and eld1 mutant. The significant differences transcripts list in Supplementary Data 1. GO analysis was conducted online using Gene Ontology Resource [https://www.geneontology.org/].
Yeast two-hybrid assay
The full-length CDS of ELD1 was cloned into the pGBKT7 vector and transformed into the yeast strain AH109. The mating method was employed to screen for interacting proteins following the Matchmaker GAL4 Two-Hybrid System manual (Clontech). A cDNA library in the Y187 yeast strain was prepared from 50-day-old (under CLD conditions) and 27-day-old (under CSD conditions) mixed rice leaves (ZT0, ZT4, ZT8, ZT12, ZT16, and ZT20). To confirm the interaction between ELD1 and OsNKAP, the truncated variants of ELD1 were cloned into pGBKT7 vector, and the full-length OsNKAP cloned into pGADT7 vector were co-transformed into yeast strain AH109 individually. To further explore potential interactions with splicing factors, ELD1 and OsNKAP were cloned into the pGADT7 vector, while U1-70K, U1A, U1C, U2AF35A, U2AF35B, U2AF35C, U2AF65A, U2AF65B were cloned into pGBKT7 vector. Combinations with empty pGBKT7 or pGADT7 vectors were employed as negative controls. Yeast strains carrying these plasmid combinations were spread evenly onto SD/-Trp/-Leu plates and subsequently incubated at 30 °C for 3 days. The colonies were then transferred to SD/-Trp/-Leu/-His/Ade plates and incubated for additional 3–4 days to verify interactions.
Bimolecular fluorescence complementation (BiFC) assay
To verify the interaction between ELD1 and OsNKAP, the full-length CDS of ELD1 was fused in-frame with the N-terminal of yellow fluorescent protein (YFP) in the p2YN vector to generate ELD1-YN, while the full-length CDS of OsNKAP was fused in-frame with the C-terminal of YFP in the p2YC vector, resulting in OsNKAP-YC construct. To assess whether ELD1 and OsNKAP interact with splicing factors, ELD1 and OsNKAP were cloned into p2YN vector, the full-length CDS of U1-70K, U2AF65A, and U2AF65B were cloned into the p2YC vector to generate the U1-70K-YC, U2AF65A-YC, U2AF65B-YC constructs. These constructs were then transformed into A. tumefaciens strain EHA105. Various combinations and the nuclear marker strains D53-mCherry were co-infiltrated into young tobacco leaves (N. benthamiana). After 36–48 h, confocal laser scanning microscope was used to detect the fluorescence signals. To confirm the location of the interaction between ELD1 and phyB, we used U1-70K-mCherry as a nuclear speckle marker.
Luciferase complementation imaging (LCI) assay
To confirm the interaction between ELD1 and splicing factors, we cloned the CDS of ELD1 was cloned in-frame with the N-terminal region of firefly luciferase (nLUC), while the CDS of U1-70K, U2AF65A, and U2AF65B were cloned in-frame with the sequence encoding the C-terminal of firefly luciferase (cLUC). These constructs were transformed into A. tumefaciens strain EHA105, followed by co-infiltration of various combinations of strains into young tobacco leaves. After 36–48 h, the leaves were harvested and incubated in darkness with 1 mM luciferin (adding 1% Triton X-100) for 5 min. LUC images were captured using a low-light cooled CCD imaging system (Tanon 5200). To investigate the light-dependent interaction between ELD1 and phyB, we constructed the phyB-cLUC plasmid by fusing the CDS of phyB with the C-terminus of LUC. Various combinations of strains were co-infiltrated into two distinct leaves of the same tobacco plant. After infiltration, one leaf was exposed to light while the other was completely shielded from light using aluminum foil as the control.
Co-immunoprecipitation (Co-IP)
To confirm the interaction between ELD1 and OsNKAP, we cloned the full-length CDS of ELD1 into the PAN580-FLAG vector to generate the ELD1-FLAG plasmid, while the full-length CDS of OsNKAP was cloned into the PAN580-HA vector to generate the OsNKAP-HA plasmid. The OsNKAP-HA construct was co-transferred with ELD1-FLAG or the PAN580-FLAG empty vector into rice protoplasts, respectively. The Co-IP assay was performed as previously described68. To investigate whether ELD1 interacts with U1-70K, U2AF65A, U2AF65B or phyB, the CDS of ELD1 was cloned into the pCAMBIA1305-GFP vector to generate the ELD1-GFP plasmid. Similarly, the CDS of U1-70K, U2AF65A, U2AF65B, and PHYB were cloned into the pCAMBIA1300-FLAG vector to generate the U1-70K-FLAG, U2AF65A-FLAG, U2AF65B-FLAG, and phyB-FLAG plasmid, respectively. These constructs were introduced into the A. tumefaciens strain EHA105, followed by co-infiltration of various combinations into young tobacco leaves. After 36 h, the leaves were harvested and ground in liquid nitrogen. Samples were homogenized in extraction buffer [50 mM Tris–MES (pH 8.0), 0.5 M Sucrose, 1 mM MgCl2, 10 mM EDTA, 0.1% NP40, 5 mM DTT and protease inhibitor cocktail tablets (Roche, 11836170001)], and then incubated with rotation at 4 °C for 30 min to promote nuclear lysis. After centrifuged at 13,000 × g for 10 min at 4 °C, the supernatant was collected and added with 80 μL protein A/G mix, rotating at 4 °C for 1 h to remove non-specific proteins and reduce background. Following the removal of the protein A/G beads, 20 μL of GFP magnetic beads (MBL, D153-10) were added and incubated at 4 °C for 2 h. The beads were then collected and washed with 2 mL lysis buffer for three times. Then the beads were boiled in SDS loading buffer at 100 °C for 10 min. Sample proteins were separated on a 12% SDS-PAGE gel (GenScript, M00656) and then detected by immunoblotting using anti-FLAG (MBL; M185-7; 1:5000) and anti-GFP (Abcam; ab6556; 1:5000) antibodies.
RNA immunoprecipitation (RIP) assay
The RIP experiment was carried out as previously described with some modifications57. Seedlings of ELD1-flag complement lines in eld1 background were grown in climate chamber under LD conditions. Samples were harvested, cut into small pieces, and cross-linked with 1% (v/v) formaldehyde for 15 min under vacuum. Cross-linking was stopped by adding glycine to the final concentration of 125 mM. After rinsing three times with DEPC water, samples were dried with absorbent paper and ground into a fine powder in liquid nitrogen. Then the samples were thoroughly incubated in Nuclear Extraction Buffer [10 mM Tris–HCl (pH 8.0), 10 mM MgCl2, 0.4 M sucrose, protease inhibitor cocktail tablets, 0.1 mM PMSF, and 40 U/mL RNase inhibitor], and then filtered with Miracloth (Millipore). The filtrate was then centrifuged at 2000 g to collect the nuclei, and then lysed with the nuclei lysis buffer [50 mM Tris–MES (pH 8.0), 0.5 M Sucrose, 1 mM MgCl2, 10 mM EDTA, 0.1% SDS, 1% NP40, 5 mM DTT, protease inhibitor cocktail tablet, 0.1 mM PMSF and 160 U/mL RNase inhibitor] with rotation at 4 °C for 30 min. After centrifuging at 13,000 × g for 10 min at 4 °C, the supernatant was collected and added with protein A/G mix, rotating at 4 °C for 1 h to remove non-specific proteins. Following discarding protein A/G beads, the FLAG magnetic beads (Sigma, M8823) or the control protein A/G beads were added and incubated at 4 °C for an additional 2 h. The beads were then collected and washed with lysis buffer for three times. RNA from the input and bound in beads was extracted using the Trizol regent (Invitrogen), genomic DNA was removed with DNase I, and reverse transcription was performed with random hexamers. RT-qPCR was conducted on an ABI Prism 7500 Real-Time PCR System with the SYBR Premix Ex Taq Kit (TaKaRa).
Accession numbers
The sequences obtained from this study can be downloaded from the rice genome annotation project [http://rice.uga.edu] using the following accession numbers: ELD1 (LOC_Os09g01640); OsNKAP (LOC_Os09g28220); U1A (LOC_Os05g06280); U1C (LOC_Os02g16640); U1-70K (LOC_Os10g02630); U2AF35A (LOC_Os02g35150); U2AF35B (LOC_Os05g48960); U2AF35C (LOC_Os09g31482); U2AF65A (LOC_Os11g41820); U2AF65B (LOC_Os11g45590); OsCCA1 (LOC_Os08g06110); Hd1 (LOC_Os06g16370); Ghd7 (LOC_Os07g15770); PHYB (LOC_Os03g19590).
Statistics and reproducibility
Statistical analysis was assessed as described in the figure legends. P values were calculated by two-sided Student’s t-tests or by one-way ANOVA with Tukey’s multiple comparisons tests using Microsoft Excel and GraphPad Prism9. No statistical methods were used to predetermine sample size. The experiments were not randomized, and investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All other data are included in the manuscript and/ or supporting information. The full-length RNA-seq data generated in this study have been deposited in the National Center for Biotechnology Information under accession numbers PRJNA1181295. Source data are provided with this paper.
References
Yano, M. et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2483 (2000).
Hu, Z. et al. Autophagy targets Hd1 for vacuolar degradation to regulate rice flowering. Mol. Plant 15, 1137–1156 (2022).
Doi, K. et al. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Gene Dev. 18, 926–936 (2004).
Zhou, S. et al. Transcriptional and post-transcriptional regulation of heading date in rice. N. Phytol. 230, 943–956 (2021).
Lee, S. J., Kang, K., Lim, J. H. & Paek, N. C. Natural alleles of CIRCADIAN CLOCK ASSOCIATED1 contribute to rice cultivation by fine-tuning flowering time. Plant Physiol. 190, 640–656 (2022).
Song, Y. H., Shim, J. S., Kinmonth-Schultz, H. A. & Imaizumi, T. Photoperiodic flowering: time measurement mechanisms in leaves. Annu. Rev. Plant Physiol. 66, 441–464 (2015).
Sun, C. et al. Dual function of clock component OsLHY sets critical day length for photoperiodic flowering in rice. Plant Biotechnol. J. 19, 1644–1657 (2021).
Huang, W. et al. Mapping the core of the circadian clock defines the network structure of the oscillator. Science 336, 75–79 (2012).
Ahn, G. et al. HOS15 represses flowering by promoting GIGANTEA degradation in response to low temperature in Arabidopsis. Plant Commun. 4, 100570 (2023).
Cao, S. H. et al. Genetic architecture underlying light and temperature mediated flowering in Arabidopsis, rice, and temperate cereals. N. Phytol. 230, 1731–1745 (2021).
Greenwood, M. & Locke, J. C. W. The circadian clock coordinates plant development through specificity at the tissue and cellular level. Curr. Opin. Plant Biol. 53, 65–72 (2020).
Wang, X. et al. A photoregulatory mechanism of the circadian clock in Arabidopsis. Nat. Plants 7, 1397–1408 (2021).
Wu, J. F. et al. LWD-TCP complex activates the morning gene CCA1 in Arabidopsis. Nat. Commun. 7, 13181 (2016).
Liu, Y. et al. Transcription factors FHY3 and FAR1 regulate light-induced CIRCADIAN CLOCK ASSOCIATED1 gene expression in Arabidopsis. Plant Cell 32, 1464–1478 (2020).
Yeom, M. et al. How do phytochromes transmit the light quality information to the circadian clock in Arabidopsis?. Mol. Plant 7, 1701–1704 (2014).
Shi, Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 18, 655–670 (2017).
Hoskins, A. A. & Moore, M. J. The spliceosome: a flexible, reversible macromolecular machine. Trends Biochem. Sci. 37, 179–188 (2012).
Lee, Y. & Rio, D. C. Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 84, 291–323 (2015).
Kathare, P. K. & Huq, E. Light-regulated pre-mRNA splicing in plants. Curr. Opin. Plant Biol. 63, 102037 (2021).
Michaud, S. & Reed, R. A functional association between the 5’ and 3’ splice sites is established in the earhest presphceosome complex (E) in mammals. Gene Dev. 7, 1008–1020 (1993).
Wan, R. X., Bai, R., Zhan, X. C. & Shi, Y. G. How is precursor messenger RNA spliced by the spliceosome?. Annu. Rev. Biochem. 89, 333–358 (2020).
Shikata, H. et al. Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proc. Natl. Acad. Sci. USA 111, 18781–18786 (2014).
Zhang, H., Lin, C. & Gu, L. Light regulation of alternative Pre-mRNA splicing in plants. Photochem. Photobiol. 93, 159–165 (2017).
Pajerowski, A. G. et al. Adult hematopoietic stem cells require NKAP for maintenance and survival. Blood 116, 2684–2693 (2010).
Pajerowski, A. G., Nguyen, C., Aghajanian, H., Shapiro, M. J. & Shapiro, V. S. NKAP, a novel modulator of Notch signaling, is required for T cell development. Immunity 30, 696–707 (2009).
Sanchez-Garcia, A. B., Aguilera, V., Micol-Ponce, R., Jover-Gil, S. & Ponce, M. R. Arabidopsis MAS2, an essential gene that encodes a homolog of animal NF-κB activating protein, is involved in 45S ribosomal DNA silencing. Plant Cell 27, 1999–2015 (2015).
Long, J. C. & Caceres, J. F. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 417, 15–27 (2009).
Barta, A., Kalyna, M. & Reddy, A. S. Implementing a rational and consistent nomenclature for serine/ arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell 22, 2926–2929 (2010).
Burgute, B. D. et al. NKAP is a novel RS-related protein that interacts with RNA and RNA binding proteins. Nucleic Acids Res. 42, 3177–3193 (2014).
Sun, S. et al. RNA binding protein NKAP protects glioblastoma cells from ferroptosis by promoting SLC7A11 mRNA splicing in an m6A-dependent manner. Cell Death Dis. 13, 73 (2022).
Karijolich, J. & Yu, Y. T. Spliceosomal snRNA modifications and their function. RNA Biol. 7, 192–204 (2010).
Li, C. et al. OsLHY is involved in regulating flowering through the Hd1- and Ehd1- mediated pathways in rice (Oryza sativa L.). Plant Sci. 315, 111145 (2022).
Wang, F. et al. The rice circadian clock regulates tiller growth and panicle development through strigolactone signaling and sugar sensing. Plant Cell 32, 3124–3138 (2020).
Zhang, S. N. et al. Nitrogen mediates flowering time and nitrogen use efficiency via floral regulators in rice. Curr. Biol. 31, 671–683 (2021).
Yang, L. et al. A central circadian oscillator confers defense heterosis in hybrids without growth vigor costs. Nat. Commun. 12, 2317 (2021).
Li, Y., Du, Y., Huai, J., Jing, Y. & Lin, R. The RNA helicase UAP56 and the E3 ubiquitin ligase COP1 coordinately regulate alternative splicing to repress photomorphogenesis in Arabidopsis. Plant Cell 34, 4191–4212 (2022).
Lei, J. et al. Plastidic pyruvate dehydrogenase complex E1 component subunit Alpha1 is involved in galactolipid biosynthesis required for amyloplast development in rice. Plant Biotechnol. J. 20, 437–453 (2022).
Marquardt, S. et al. Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol. Cell 54, 156–165 (2014).
Chang, P., Hsieh, H. Y. & Tu, S. L. The U1 snRNP component RBP45d regulates temperature-responsive flowering in Arabidopsis. Plant Cell 34, 834–851 (2021).
Gil, K. E. et al. Alternative splicing provides a proactive mechanism for the diurnal CONSTANS dynamics in Arabidopsis photoperiodic flowering. Plant J. 89, 128–140 (2017).
Li, T. et al. SUMOylated NKAP is essential for chromosome alignment by anchoring CENP-E to kinetochores. Nat. Commun. 7, 12969 (2016).
Dash, B., Shapiro, M. J., Chung, J. Y., Arocha, S. R. & Shapiro, V. S. Treg-specific deletion of NKAP results in severe, systemic autoimmunity due to peripheral loss of Tregs. J. Autoimmun. 89, 139–148 (2018).
Shapiro, M. J. et al. NKAP regulates senescence and cell death pathways in hematopoietic progenitors. Front. Cell Dev. Biol. 7, 214 (2019).
Aceituno-Valenzuela, U., Micol-Ponce, R. & Ponce, M. R. Genome-wide analysis of CCHC-type zinc finger (ZCCHC) proteins in yeast, Arabidopsis, and humans. Cell Mol. Life Sci. 77, 3991–4014 (2020).
Wang, Y. et al. The distinct roles of zinc finger CCHC-type (ZCCHC) superfamily proteins in the regulation of RNA metabolism. RNA Biol. 18, 2107–2126 (2021).
Aceituno-Valenzuela, U. et al. CAX-INTERACTING PROTEIN4 depletion causes early lethality and pre-mRNA missplicing in Arabidopsis. Plant Physiol. 197, kiae641 (2024).
Zhang, J. et al. Excessive miR-25-3p maturation via N6-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat. Commun. 10, 1858 (2019).
Filichkin, S. A. et al. Environmental stresses modulate abundance and timing of alternatively spliced circadian transcripts in Arabidopsis. Mol. Plant 8, 207–227 (2015).
Pruneda-Paz, J. L. & Kay, S. A. An expanding universe of circadian networks in higher plants. Trends Plant Sci. 15, 259–265 (2010).
James, A. B. et al. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 24, 961–981 (2012).
Leppek, K., Das, R. & Barna, M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Bio. 19, 673–673 (2018).
Ryczek, N., Lys, A. & Makalowska, I. The functional meaning of 5′UTR in protein-coding genes. Int. J. Mol. Sci. 24, 2976 (2023).
Dong, J., Chen, H., Deng, X. W., Irish, V. F. & Wei, N. Phytochrome B induces intron retention and translational inhibition of PHYTOCHROME-INTERACTING FACTOR3. Plant Physiol. 182, 159–166 (2020).
Kathare, P. K. et al. SWAP1-SFPS-RRC1 splicing factor complex modulates pre-mRNA splicing to promote photomorphogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 119, e2214565119 (2022).
Xin, R. et al. SPF45-related splicing factor for phytochrome signaling promotes photomorphogenesis by regulating pre-mRNA splicing in Arabidopsis. Proc. Natl. Acad. Sci. USA 114, E7018–E7027 (2017).
Xin, R., Kathare, P. K. & Huq, E. Coordinated regulation of Pre-mRNA splicing by the SFPS-RRC1 complex to promote photomorphogenesis. Plant Cell 31, 2052–2069 (2019).
Zhao, Z. et al. CRY2 interacts with CIS1 to regulate thermosensory flowering via FLM alternative splicing. Nat. Commun. 13, 7045 (2022).
Yan, Y. et al. Light controls mesophyll-specific post-transcriptional splicing of photoregulatory genes by AtPRMT5. Proc. Natl. Acad. Sci. USA 121, e2317408121 (2024).
Zhou, H. et al. Light regulates nuclear detainment of intron-retained transcripts through COP1-spliceosome to modulate photomorphogenesis. Nat. Commun. 15, 5130 (2024).
Godoy Herz, M. A. et al. Light regulates plant alternative splicing through the control of transcriptional elongation. Mol. Cell 73, 1066–1074 e1063 (2019).
Zheng, T. et al. Post-transcriptional regulation of Ghd7 protein stability by phytochrome and OsGI in photoperiodic control of flowering in rice. N. Phytol. 224, 306–320 (2019).
Osugi, A., Itoh, H., Ikeda-Kawakatsu, K., Takano, M. & Izawa, T. Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice. Plant Physiol. 157, 1128–1137 (2011).
Lloyd, J. P., Seddon, A. E., Moghe, G. D., Simenc, M. C. & Shiu, S. H. Characteristics of plant essential genes allow for within- and between-species prediction of lethal mutant phenotypes. Plant Cell 27, 2133–2147 (2015).
Wang, T. et al. Identification and characterization of essential genes in the human genome. Science 350, 1096–1101 (2015).
Fekih, R. et al. MutMap plus: genetic mapping and mutant identification without crossing in rice. PLoS ONE 8, e68529 (2013).
Li, C. et al. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat. Biotechnol. 38, 875–U866 (2020).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Liu, T. et al. Dwarf and High Tillering1 represses rice tillering through mediating the splicing of D14 pre-mRNA. Plant Cell 34, 3301–3318 (2022).
Acknowledgements
This work was supported by the Key Laboratory of Biology, Genetics and Breeding of Japonica Rice in Mid-lower Yangtze River, Ministry of Agriculture and Rural Affairs, China, and the Jiangsu Collaborative Innovation Center for Modern Crop Production, China. Funding for this work was provided by the National Natural Science Foundation of China (32272115), and Jiangsu Provincial Key Research and Development Program (BE2023362), and the Foundation of Biological Breeding Zhongshan Lab (BM2022008-03) and the Biological Breeding-National Science and Technology Major Project (2022ZD04001).
Author information
Authors and Affiliations
Contributions
L.C., J.W., and S.Z. designed the study. L.C. performed most of the experiments with the help of B.H., Z.X., S.C., Q.W., H.H., Y.H., L.Z., J.W., W.L., K.C., W.S., and S.S.Z. performed part of the work. C.L., L.J., Y.T., X.L., S.L., and L.M.C. provided technical assistance. J.L. and X.G. performed bioinformatic analysis. L.C., S.Z., and H.W. wrote the manuscript with comments from all authors. J.W. conceived the project and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Paula Duque and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. [A peer review file is available.]
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cai, L., Hao, B., Xu, Z. et al. ELD1 mediates photoperiodic flowering via OsCCA1 alternative splicing and interacts with phytochrome signaling in rice. Nat Commun 16, 5329 (2025). https://doi.org/10.1038/s41467-025-60839-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-60839-6