Abstract
Probing endogenous protein localization and function in vivo remains challenging due to laborious gene targeting and monofunctional alleles. Here, we develop a multifunctional and adaptable toolkit based on genetically encoded affinity reagents (GEARs). GEARs use small epitopes recognized by nanobodies and single chain variable fragments to enable fluorescent visualization, manipulation and degradation of protein targets in vivo. Furthermore, we outline a CRISPR/Cas9-based epitope tagging pipeline to demonstrate its utility for producing knock-in alleles that have broad applications. We use GEARs to examine the native behavior of the pioneer transcription factor Nanog and the planar cell polarity protein Vangl2 during early zebrafish development. Together, this toolkit provides a versatile system for probing and perturbing endogenous protein function while circumventing challenges associated with conventional gene targeting and is broadly available to the model organism community.
Similar content being viewed by others
Introduction
Understanding endogenous protein localization and function in vivo can present challenges, partially due to overexpression approaches, which can lead to artifacts due to non-physiological expression levels1,2,3. Genome engineering has transformed the field of genetics by enabling precise DNA editing across a wide range of organisms4,5,6. This technology enables the generation of tagged fusion proteins to study their endogenous functions7,8,9, overcoming the limitations of traditional antibody-based methods, which are typically restricted to fixed samples and dependent on the availability of specific primary antibodies. Despite this technological advance, low germline transmission rates and complex cloning strategies can make precise gene editing challenging10,11. Additionally, most tags such as fluorescent proteins, optogenetically-inducible domains12 and the auxin-inducible degron (AID)13 have single functions. One exception to this is the green fluorescent protein (GFP), which has targeted binding reagents for degradation14, transcriptional activation15, and proximity interaction mapping16. However, the precise genomic knock-in of GFP is hindered by its relatively large size, which reduces efficiency across different species17,18,19. Moreover, large fusion proteins such as GFP can interfere with native protein function, limiting their utility in functional studies. In contrast, shorter sequences are more efficiently integrated into the genome. When paired with genetically encoded binders, short epitope tags offer a compact, scalable alternative that enables versatile applications across species, highlighting the need for improved small-epitope tools that support multifunctional analysis of endogenous alleles.
To address these limitations, we developed GEARs (Genetically Encoded Affinity Reagents), a modular system composed of short epitopes, their high-affinity binders (single-chain variable fragments (scFvs) or nanobodies (Nb)), and adaptor modules such as fluorophores, degrons, or HaloTags20. This platform enables precise and versatile tagging of endogenous proteins in vivo. We tested the functionality of codon-optimized epitope tags and their binders in both zebrafish and mouse embryos, demonstrating the system’s versatility across species. To streamline genome engineering, we developed a synthetic CRISPR/Cas9-based strategy using single-stranded donor oligonucleotides (ssODNs) for efficient tag insertion, enabling the rapid generation of multifunctional alleles. We applied this approach to several zebrafish gene loci (nanog, vangl2, dicer, and pou5f3), demonstrating its utility by visualizing Nanog dynamics during genome activation and characterizing Vangl2 localization in vivo. This plug-and-play toolkit offers a flexible and scalable platform for endogenously tagging proteins, providing researchers with precise control over protein function in vivo. Moreover, the modular design supports future integration with technologies such as optogenetics, mass spectrometry, and protein relocalization, expanding the potential applications for studying complex biological systems while overcoming many of the limitations of traditional gene targeting approaches.
Results
GEARs function in vivo to detect exogenously expressed protein targets
To develop a platform of genetically encoded affinity reagents (GEARs), we tested a set of small epitopes <20 amino acids and their cognate scFvs or Nbs (Fig. 1a) and determined their functionality in vivo. Specifically, we tested the anti-HA 15F11 scFv21 (FbHA), anti-FLAG scFv22 (FbFLAG), anti-GCN423 scFv (FbSun), anti-ALFA Nb24 (NbALFA), anti-VHH05 Nb25 (NbVHH05), anti-127d01 Nb26 (Nb127d01), and anti-gp41 Nb (NbMoon)27 binders to assess whether they would (1) produce functional protein and localize uniformly within cells, and (2) detect exogenous targets in vivo (Fig. 1b). First, we generated codon-optimized versions of these binders fused to EGFP and synthesized mRNA for embryo injection. Wildtype (WT) zebrafish embryos were injected at the 1-cell stage and imaged at six hours post fertilization (hpf). We observed diffuse cytoplasmic and nuclear fluorescence for all constructs, indicating that these GEARs are well tolerated and fold properly in vivo (Figs. 1c, e, S1a, b, and S2a, b). Despite all binders being injected at the same concentration, we noticed varying fluorescence intensity levels when normalized to an V5-mScarlet-I injection control, suggesting that the scFvs/Nbs have different protein stabilities/half-lives (Figs. 1c, e (“-target” images and curves), S1c, S2c, and S3a). With the exception of NbALFA and FbHA, binders exhibited weak nuclear localization, which was also observed for the V5-mScarlet-I control protein (Nuclear to cytoplasmic ratio >1; Fig. S3b–d). scFv binders (FbSun and FbHA) formed subcellular accumulations, with FbHA accumulating in one or two distinct foci near the nucleus and the mitotic apparatus (Fig. S3e). We attributed the latter to off-target binding of centriolar proteins; a phenomenon observed with cross-reactivity in some commercial antibodies28,29. Importantly, the expression of all GEAR binders did not cause any phenotypic effects at the injected concentrations. Together, these results demonstrate that, different from their initial design (37 °C), the seven tested binders can function in vivo at zebrafish physiological temperatures (24–29 °C) and pH (ranging from pH 6.5–8)30.
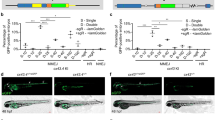
a Overview of genetically encoded probes, their respective sizes, and target epitopes. b Schematic of a binding assay for visualizing GEAR binding in vivo. Nbs or scFvs were fused to EGFP and injected into WT zebrafish embryos, either alone or with tagged versions of nuclear (Nanog) or membrane-bound (Vangl2) targets. Localization of EGFP reflects the in vivo binding ability of these binders. c Single slices of representative microscopy images show EGFP localization when fused to different binders in the absence (top) or presence (middle) of epitope-tagged Nanog. Lines indicate the regions for fluorescence quantification. Line plots (bottom) of normalized EGFP-binder fluorescence intensity in the absence (−target) or presence (+target) of cognate tagged Nanog along lines drawn through cells. Measurements in pixel intervals and centered on the nucleus. d Quantifying and comparing the in vivo binding ability of different GEAR binders to tagged Nanog using the readout of relocalized EGFP fluorescence (nuclear-to-cytoplasmic ratio normalized to a V5-mScarlet-I injection control). Center lines show medians; box limits indicate the 25th and 75th percentiles; whiskers extend to the minimum and maximum value in the data. N = 6 (NbMoon) or 5 embryo data points. Each data point is the mean of n = 12 cells per embryo. Data were analyzed using a nonparametric Kruskal–Wallis test, ∗∗∗ p < 0.001 (p-value = 0.00028875). e As in (c) but for GEAR binders in the absence or presence of epitope-tagged Vangl2. f Quantification and comparison of the in vivo binding ability of GEAR binders to tagged Vangl2 using the readout of relocalized EGFP fluorescence as the ratio of membrane-to-cytoplasm, normalized to the V5-mScarlet-I injection control. Center lines show medians; box limits indicate the 25th and 75th percentiles; whiskers extend to the minimum and maximum value in the data. N = 4 (NbMoon), 3 (NbVHH05) or 5 embryo data points. Each data point is the mean of n = 12 cells per embryo. Data were analyzed using a nonparametric Kruskal–Wallis test, ∗∗∗ p < 0.001 (p-value = 0.00031782). Scale bars: 10 mm (c, e). See also Figs. S1–3.
Next, we evaluated whether GEAR binders recognize their cognate tags in vivo (Fig. 1b). We cloned epitope tags for each GEAR onto the N-terminus of zebrafish nanog and vangl2. Nanog is a maternally deposited transcription factor with pioneering activity in the embryo and regulates genome activation, localizing to the nucleus31. Vangl2, a core component of the planar cell polarity pathway, is localized to the membrane32. Importantly, tagging the N- and C-terminus of Nanog (Fig. S3f), and the N-terminus of Vangl2 did not disrupt their biological functions32. We hypothesized that the translocation of EGFP-GEARs to the nucleus or plasma membrane would serve as a clear indicator of successful binding in vivo (Fig. 1b).
To test the binding efficiency of EGFP-GEARs, we co-injected each EGFP-tagged GEAR mRNA into 1-cell staged embryos along with their cognate-tagged nanog mRNAs and measured the nuclear-to-cytoplasmic fluorescence ratios relative to V5-mScarlet-I control (Figs. 1b, c and S1d–f). All GEARs translocate to the nucleus, though with varying efficacy (Fig. 1c, d). When we quantified the fluorescence intensity relative to the injection control, we observed that the stability of the GEAR binders increased in the presence of their specific antigen, resulting in an overall increased fluorescence relative to the injection control (Figs. 1c and S3a). This revealed lower background fluorescence for the more effective binders, such as ALFA, Sun, and Moon. The most efficient nuclear translocation of fluorescence was observed with the NbALFA and NbMoon, while Nb127d01 showed no significant nuclear enrichment compared to the nontargeted binder (Figs. 1d and S3g, h). Furthermore, tagged Vangl2 induced the EGFP-GEARs to translocate to the membrane, which was quantified as membrane enrichment relative to the cytoplasmic control V5-mScarlet-I (Figs. 1e, f, S2d–f, and S3i–l). Nb127d01, NbVHH05, and FbSun GEARs displayed varying translocation efficiencies depending on the epitope-tagged protein’s cellular localization (Figs. 1d, f and S3h, j). Overall, the NbALFA and NbMoon binders provided the strongest signal for both targets in vivo and exhibited the least background fluorescence (Figs. 1d, f and S3a, h, j). Overall, these data demonstrate the applicability of using GEARs to detect a variety of epitopes and targets during zebrafish development.
We next examined whether nanobody-based GEARs could be combined with other fluorophores. We replaced the EGFP in the NbALFA, NbVHH05, and Nb127d01 GEAR constructs with open reading frames (ORFs) that encode mNeonGreen33, mScarlet-I34, mTagBFP235, and HaloTag20. When these constructs were co-injected with tagged nanog mRNA, all fluorescent protein fusions as well as the HaloTag localized to the nucleus (Fig. S4a). This indicates that GEARs are compatible with different fluorescent adapters.
GEARs can bind and degrade target proteins with high efficiency
Given the variety of adapters compatible with GEARs, we asked whether GEAR binders could be utilized for targeted protein degradation. Recently, several genetic systems have adapted an anti-GFP nanobody for targeted protein destruction in Drosophila14, C. elegans36, zebrafish37, and human cells38. While these strategies rely on binding to GFP-tagged proteins, integrating large tags remains a challenge in several model organisms4,39,40. Therefore, developing degron reagents that rely on shorter, more easily integrated epitope tags, like those offered by GEARs, would be highly valuable.
To test if GEARs could facilitate degradation of tagged proteins, we adapted the zebrafish zGrad GFP nanobody system. zGrad utilizes the zebrafish F-box protein Fbxw11b fused to an anti-GFP Nb for targeting GFP-tagged proteins for proteasomal degradation37. We fused zebrafish fbxw11b to the Nb-based binders, titrated their expression to assess off-target effects in WT embryos and found most were well tolerated at 100 pg (Fig. S4b). We focused on the Nbs due to the observed off-target localization with some of the scFv-based binders (Fig. S3e). To test degradation efficiency, we used a bicistronic reporter in zebrafish embryos that encodes membrane-tdTomato and GEAR epitope-tagged H2B-GFP, separated by a T2A self-cleaving peptide (Fig. 2a, b). This reporter expresses nearly equal amounts of membrane and tagged nuclear fluorescent proteins. We reasoned that the loss of nuclear GFP signal upon degrader expression would indicate specific protein elimination by the GEAR degron (Fig. 2a). Zebrafish embryos were co-injected with the bicistronic mRNA reporter and either zGrad or GEAR degrader mRNAs (referred to as ALFAgrad, VHH05grad, 127d01grad, and MoonGrad) and compared to the no degron control at 10 hpf (Fig. 2b, c). GEARgrads caused a reduction in GFP signal of 89%, 80%, 77%, and 57% for ALFAgrad, VHH05grad, Moongrad, and 127d01grad, respectively (Figs. 2d and S4c). The efficiency of ALFAgrad was comparable to that of zGrad (Fig. 2d), reinforcing the ALFA nanobody system as a highly effective binding platform. Notably, these data suggest that assessing localization alone may not reflect how efficiently these nanobodies perform in degron contexts.
a Degron assay schematic: a split reporter expresses membrane-targeted tdTomato (lyn-tdTomato) and nuclear EGFP (TAG-H2B-EGFP), separated by a T2A peptide. Degradation of nuclear EGFP by GEARs lowers the EGFP/tdTomato ratio. b Zebrafish assay: 1-cell embryos injected with reporters ± GEAR degrader, grown to 10 hpf, and imaged. c Representative images at 10 hpf showing tdTomato (magenta) and EGFP (green) in embryos injected with reporter alone or with degraders (zGrad, ALFAgrad, VHH05grad, Moongrad, 127d01grad). d Quantification of reporter ± degrader assays. Data normalized to control mean = 1. Mean ± SD; N = 3–6 embryos with >188 nuclei analyzed per embryo. e, f Kinetics of ALFAgrad: 1-cell embryos co-injected with ALFA-EGFP and rhodamine dextran; one 2-cell blastomere injected with ALFAgrad. Imaging at 16- and 512-cell stages; degron-injected region marked *. See also Fig. 4d. g–i ALFAgrad kinetics for pre-synthesized proteins: Tg(hsp70:ALFAgrad)/+ embryos injected with ALFA-EGFP and rhodamine dextran, grown to 24 hpf, sorted by transgene status, heatshocked (1 h, 38 °C), and imaged for 5 h. Representative images and quantification show reduced EGFP:rhodamine ratios in degron-expressing embryos. Mean ± SD; N = 3 (control), 5 (degron). j–l Tissue-specific assay: Tg(fli1a:ALFAgrad-P2A-mCherry)/+ embryos injected with ALFA-EGFP or EGFP (control). At 30 hpf, caudal vein plexus (CVP) imaged. Segmentation via mCherry; EGFP:Halo ratios in CVP vs somites assayed. Lower CVP/somite ratios indicate degradation. Mean ± SD; N = 5 embryos. m–o Mouse assay: embryos injected with ALFA split reporter mRNA (1-cell stage) and ALFAgrad mRNA into one 2-cell blastomere. The uninjected cell serves as control. Fixed at various time points. Representative images show EGFP (green), tdTomato (magenta); degron-injected cells indicated by *. Quantification: median ± interquartile range; N = 4 (0 h, 3 h, 6 h), 5 (2 h), 6 (9 h, 12 h). Scale bars: 100 mm (c, f, k), 25 mm (c, inset), 50 mm (h), and 10 mm (n). See also Figs S4 and S5.
To assess the kinetics of target degradation using ALFAgrad, we first analyzed the degradation of newly synthesized proteins in a clone of cells (Fig. 2e). To this end, we co-injected mRNA encoding ALFA-EGFP and rhodamine dye at the 1-cell stage. Then, we injected ALFAgrad mRNA into one of the two blastomeres at the 2-cell stage, allowing the uninjected blastomere to serve as a no-degradation control (Fig. 2e). The daughter cells of the degron-injected blastomere did not accumulate GFP compared to the control cells (Figs. 2f and S4d, e), demonstrating that the degradation rate was greater than or equal to the rate of protein synthesis upon ALFAgrad expression.
Second, we examined the kinetics of target degradation using a heatshock-inducible transgene to regulate ALFAgrad expression temporally. Zebrafish expressing the ALFAgrad transgene showed no developmental defects or phenotypic abnormalities (Fig. S4f). We injected ALFA-EGFP mRNA and rhodamine dye into Tg(hsp70:ALFAgrad)/+ and WT sibling embryos (see methods; Fig. 2g). At 28 hpf, embryos were sorted based on transgene status (+ or − myl7:EGFP transgenesis reporter), subjected to heatshock, and imaged over time in the trunk region for GFP and rhodamine fluorescence. Transgene-positive embryos exhibited a significant reduction in EGFP signal over time (51% fluorescent reduction), whereas transgene-negative embryos showed minimal signal loss (Fig. 2h, i). The degradation kinetics post-heatshock induction are comparable with previously benchmarked zGrad degradation kinetics in zebrafish37. These results define the kinetics of degradation for a pre-existing protein pool upon ALFAgrad expression, accounting for the temporal dynamics of transcription, translation, and target degradation. Moreover, they demonstrate that transgenic ALFAgrad expression enables precise temporal control of degron activity.
Finally, we tested whether ALFAgrad could be expressed in a tissue-specific manner, enabling conditional protein loss in targeted cells. To this end, we generated zebrafish expressing ALFAgrad under the control of the vascular-specific fli1a promoter41 and marked by a P2A mCherry bicistronic reporter. We injected either ALFA-EGFP mRNA or untagged EGFP mRNA together with an H2B-Halo injection control into Tg(fli1a:ALFAgrad P2A mCherry)/+ and WT sibling embryos at the 1-cell stage (Fig. 2j). At 28 hpf, we imaged the caudal vein plexus, marked by mCherry expression, and found that ALFA-EGFP was efficiently degraded by 50% in this tissue compared to untagged EGFP (Figs. 2j–l and S5a). These data suggest that ALFAgrad allows tissue-specific protein degradation and loss-of-function analysis.
Since GEARgrads rely on a zebrafish F-box protein, we asked whether it could effectively recruit ubiquitinylation machinery in other model systems. To investigate this, we injected the bicistronic membrane-tdTomato-T2A-ALFA-H2B-GFP mRNA into 1-cell stage mouse zygotes, followed by ALFAgrad injection into one of the two cells at the 2-cell stage, using the uninjected cell as the no degradation control (Fig. 2m). Though ALFA-H2B-EGFP was strongly expressed before degron injection, we observed a significant reduction of nuclear EGFP signal within 2 h of degron injection, with robust clearance of 96% of nuclear EGFP signal in the degron-injected cell by 3 h post-degron injection (Figs. 2n, o and S5b). Using the orthogonal Moongrad revealed efficient Moon-H2B-EGFP degradation by 9 h (Fig. S5c, d). These findings demonstrate the versatility of the GEARs system in targeting proteins across different model organisms and demonstrate the rapid degradation kinetics of ALFAgrad in various biological contexts.
GEARs can be coupled with gene targeting to interrogate endogenous protein function
Given the small size of the epitopes recognized by GEARs (<20 amino acids), we aimed to develop a rapid endogenous tagging method for efficient gene targeting. To achieve this, we employed a knock-in strategy utilizing recombinant Cas9, synthetic single guide RNA (sgRNA) and a ssODN as a donor template (Fig. 3a). This fully synthetic approach allows for a cost-effective and cloning-free method to introduce short epitope tags into endogenous loci by homology-directed repair.
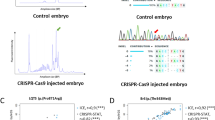
a Schematic of gene targeting using oligo donors and Cas9 to insert epitope tags into target genes. b PCR quantification of germline tag integration in F1 for ALFA−, VHH05−, and 127d01-tagged nanog, dicer, vangl2, and pou5f3. Integration was classified as: partial (one junction+), imprecise full (both junctions+ with indels), or precise full (both junctions+ without indels). N values shown in pie charts. Overall knock-in efficiency across 8 loci reported as mean ± SD. c Representative single plane images of WT, ALFA-nanogKI/KI, and ALFA-vangl2KI/KI embryos at 8 hpf co-injected with memBFP (blue) and EGFP-NbALFA (green). d WT and ALFA-nanogKI/KI embryos, ± ALFAgrad (100 pg, 1-cell stage), compared to MZnanog mutants. Imaged at 6 hpf. e WT and ALFA-vangl2KI/KI embryos, ± ALFAgrad (100 pg, 1-cell stage), compared to vangl2−/− mutants. Imaged at 24 hpf. f Zygotic or maternal-zygotic (MZ) homozygous ALFA-pou5f3KI/KI embryos ± ALFAgrad (150 pg) at the 1-cell stage. Midbrain-hindbrain boundary indicated by black arrowhead. Imaged at 24 hpf. g Homozygous ALFA-dicerKI/KI embryos ± ALFAgrad (150 pg). Embryos were imaged at 24 hpf. h ALFA-dicerKI/KI embryos were injected with 100 pg of the GFP-3xmiR-430 sensor (green) and 100 pg of dsRed (magenta) as an injection control, ± 150 pg of ALFAgrad. Embryos were imaged at 24 hpf. i Western blot comparing endogenous ALFA-Nanog to overexpressed (25 or 50 pg mRNA injection) ALFA-Nanog in WT embryos. Histone H3 used as loading control. N = 25 embryos per condition. j Genome tracks of ALFA-nanogKI/KI and WT ChIP-seq (purple) vs Nanog-Myc overexpression (OE; gray), shown at the bmp2b locus. Scale bars: 20 mm (c) and 250 mm (d–h). See also Figs. S6 and S7.
As a proof-of-principle, we generated eight different knock-in alleles for the nuclear, membrane and cytoplasmic proteins, nanog, vangl2, pou5f3 and dicer, by integrating single epitopes into these loci (ALFA, VHH05, and 127d01 for nanog and vangl2, and ALFA for pou5f3 and dicer; Figs. 3a, b and S6a). Screening F0 founder incrosses identified precisely tagged alleles, confirmed by Sanger sequencing, with an efficiency ranging between 7% and 18% (Table 1 and Figs. 3b and S6a). These efficiencies are comparable to those previously reported with conventional homologous recombination (HR) methods (0.3%–16%), especially without prescreening using secondary reporters, like fluorescent markers10,11,42,43. Maternal homozygous knock-in embryos (derived from homozygous mothers) progressed through gastrulation normally and were phenotypically indistinguishable from WT embryos at 24 hpf, suggesting that the alleles are viable and do not perturb protein function. This efficient method enables the generation of a single tagged allele that can be paired with a wide range of GEARs to allow multifunctional analysis.
To this end, we employed EGFP-tagged ALFA GEAR binders to detect endogenous ALFA-Nanog in the nucleus and ALFA-Vangl2 at the plasma membrane via immunohistochemistry (Fig. 3c), demonstrating the ability of GEARs to bind endogenous proteins in vivo. Next, we used GEARgrads to investigate whether degrading endogenously tagged Nanog, Vangl2, Dicer, and Pou5f3 recapitulate known loss-of-function (LOF) mutant phenotypes. First, injection of ALFAgrad into ALFA-nanogKI/KI homozygous embryos resulted in the arrest of epiboly in 97% of the embryos, phenocopying the maternal-zygotic nanog (MZnanog) mutant phenotype (Fig. 3d)44,45. Similarly, ALFA-vangl2KI/KI homozygous embryos injected with ALFAgrad fully recapitulated the MZvangl2 convergent-extension phenotype (Fig. 3e)46. Injection of VHH05grad or 127d01grad into embryos carrying VHH05- or 127d01-tagged Nanog or Vangl2, respectively, resulted in fewer embryos with an LOF phenotype, consistent with previous findings that these GEARs are less effective at degrading tagged proteins (Fig. S6b, c). Interestingly, the tagged-vangl2 embryos are more sensitive to VHH05grad and 127d01grad than tagged-nanog embryos with 87% and 53% of embryos displaying a LOF-like phenotype (100 pg of GEARgrad), suggesting that smaller reductions in Vangl2 pools are sufficient to induce a mutant-like phenotype (Fig. S6b, c). Together, these data suggest that ALFAgrad is the most efficient at degrading tagged Nanog and Vangl2 and inducing LOF-like phenotypes, followed by VHH05grad, while 127d01grad appears to be the least effective.
Second, injection of ALFAgrad into ALFA-pou5f3KI/KI faithfully recapitulates both the zygotic (loss of midbrain-hindbrain boundary) and the maternal-zygotic (gastrulation defects) LOF phenotypes of pou5f3/spg mutants (Fig. 3f)47,48,49,50. Finally, injection of ALFAgrad into ALFA-dicerKI/KI embryos recapitulated the MZdicer phenotype, apparent by shortened body axis and brain morphogenesis defects (Fig. 3g)51. Dicer is an essential enzyme involved in the processing of mature microRNAs, and MZdicer mutants cannot clear maternally deposited mRNAs, as they cannot generate mature miR-43051. Indeed, injection of a miR-430 sensor showed impaired miR-430 activity in the Dicer-depleted embryos through the expression of GFP, compared to sibling embryos where Dicer was not degraded (Figs. 3h and S6d, e). In contrast, a control reporter for miR-204, which is not expressed during early embryogenesis, was not affected by the degradation of Dicer through ALFAgrad (Fig. S6b, c). These data suggest that ALFAgrad can phenocopy molecular phenotypes and that these effects are specific. Additionally, as a control, injection of ALFAgrad into WT, or heterozygous ALFA-nanogKI/+ or ALFA-vangl2KI/+ embryos yielded wildtype-like embryos, demonstrating the specificity of ALFAgrad for its target (Fig. S6f, g).
Overall, these data demonstrate that ALFAgrad is the most efficient degron in the GEARs toolbox. Furthermore, we show that maternal and zygotic pools of nuclear, membrane, and cytoplasmic proteins can be efficiently degraded with ALFAgrad, effectively mimicking genetic LOF mutants.
GEARs can be utilized to characterize endogenous protein expression
We utilized our ALFA-nanogKI/KI fish line to investigate endogenous Nanog during embryogenesis and performed immunofluorescence (IF), western blotting, and ChIP-seq analysis. First, western blot analysis revealed that Nanog is present from the 4-cell stage and gradually increases over developmental time (Fig. S6h), consistent with its role in genome activation31. We observed an expected band of ~45 kDa representing full-length Nanog, as well as additional lower molecular weight bands that were absent in WT and present in an independent HA-nanog knock-in allele, likely representing splicing variants, truncations or post-translationally modified Nanog species (Fig. S6i). Comparing endogenous Nanog levels to exogenously expressed mRNA (at quantities typically used to rescue nanog mutant embryos31,44,45,52) resulted in 1.76×–4.35× fold higher Nanog levels (Fig. 3i and S6j, k). Although the nature of the shorter Nanog species remains unknown, these experiments demonstrate that ALFA-nanog knock-in fish accurately report the physiological concentration of endogenously produced nanog protein, which is found at lower levels than previously appreciated.
Second, we aimed to define the endogenous Nanog binding profile in the genome by ChIP-seq using the ALFA-Ab and compared it to publicly available data using exogenous nanog expression53. ChIP-seq analysis across two biological replicates of ALFA-nanogKI/KI and two negative control WT replicates identified 13,957 shared peaks between the ALFA-nanogKI/KI replicates that did not overlap with a small set of peaks identified in WT controls (Fig. S7a, c, d). When compared to exogenous nanog-Myc overexpression (OE), 11,949 of those peaks were shared between the two datasets (86% overlap), though OE yielded an additional 46,249 peaks (Fig. S7b). The examination of tracks revealed numerous Nanog target genes in the OE pulldown that contained gene body read clusters not observed in the endogenous pulldown (examples shown in Figs. 3j and S7e). The ALFA-Ab offers a strong signal-to-noise ratio, reflecting the high data quality of these datasets (see y-axis Figs. 3j and S7e). This broad dynamic range allows us to detect weak ChIP-enriched signals that might otherwise be indistinguishable from background noise and which may account partially for the ~ 2000 peaks uniquely identified in the ALFA-nanogKI/KI samples (Fig. S7b). Together, these results suggest that the ALFA-Ab is highly specific in detecting ALFA-tagged protein and a valuable tool for ChIP-seq analysis.
GEARs illuminate the native behavior of Nanog during the earliest transcriptional events in zebrafish embryogenesis
Using the GEARs system, we investigated the spatiotemporal dynamics of endogenous Nanog protein. Nanog is an ideal target for evaluating protein dynamics because (1) it is a pioneer transcription factor with intrinsically disordered regions (IDRs) that engage in concentration-dependent interactions, and (2) exogenous fluorescently-tagged Nanog localizes to subnuclear puncta during the maternal-to-zygotic transition52,54. While live imaging of exogenous Nanog has revealed important molecular behaviors, the localization pattern of endogenous Nanog has not been studied due to the lack of appropriate tools. We performed time-lapse imaging of endogenous ALFA-Nanog during the first 3 h of embryogenesis, using the EGFP-NbALFA GEAR binder for visualization, and compared it to exogenously expressed ALFA-Nanog and the direct fusion protein Nanog-mEmerald54 (Figs. 4a and S7f). When Nanog was exogenously provided by injecting mRNA into WT embryos, subnuclear foci formation was observed starting at the 64-cell stage, with characteristically bright foci forming at the 128-512 cell stages, as previously reported (Figs. 4b and S7g)52,54. While the use of EGFP-NbALFA increases the inherent fluorescence background due to the presence of unbound EGFP-NbALFA in cells, unlike a direct fusion protein that does not have this limitation, we observed no significant difference in the number of Nanog foci when exogenous Nanog was provided (Fig. S7h–j). These results suggest that the increased background of ALFA-nanobody does not hinder the detection of Nanog foci.
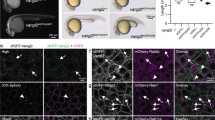
a Live imaging schematic: EGFP-NbALFA mRNA (75 pg) was injected into ALFA-nanogKI/KI embryos to visualize endogenous Nanog, or imaging was done directly from ALFA-nanogKI/KI; Tg(actb2:EGFP-NbALFA)/+ crosses. For exogenous Nanog, WT embryos were co-injected with ALFA-nanog mRNA (25 pg) and EGFP-NbALFA. b Max-projected stills comparing endogenous and exogenous ALFA-Nanog. Endogenous Nanog was visualized via EGFP-NbALFA mRNA injection (row 1) or transgene expression (row 2); exogenous Nanog via co-injection of ALFA-nanog and EGFP-NbALFA mRNAs into WT embryos (row 3). Stills show matched cell stages (E = early, L = late); brightness and contrast adjusted per image for visibility. Insets highlight Nanog foci (arrowheads). c Quantification of the number of visible Nanog foci during the 256-cell stage; mean ± SD of N = 3 embryos with n = 6 (endogenous) and 7 (exogenous) nuclei. d Quantification of Nanog foci from 50% to 75% of the 256-cell stage; n = 37 (endogenous) and 35 (exogenous) data points from N = 3 embryos. Box limits indicate the 25th and 75th percentiles; whiskers extend to the minimum and maximum value. Mann–Whitney test, ∗∗∗∗ p < 0.0001 (p-value < 1 × 10−11). e Live imaging setup: mScarlet-i3-NbALFA and EGFP-fused Pol-II Ser2-P mintbody (recognizing Ser2P in C-terminal domain (CTD)) were co-injected into ALFA-nanogKI/KI embryos at the 1-cell stage to visualize endogenous Nanog and elongating RNA Pol II. f Stills and 3D rendering of a 512-cell stage nucleus show Nanog foci (magenta, white arrows) appearing before and near miR-430 Pol-II transcription sites (green, blue arrows). Example 3D distances: 0.56 and 2.80 μm³. g Quantification from (f); n = 15 Nanog/Pol-II measurements at 512- and 1k-cell stages from N = 3 embryos. Bright Nanog foci closely associate with two large Pol-II transcription foci (mean = 1.80 μm³). Box plots show median; limits indicate the 25th and 75th percentiles; whiskers extend to the minimum and maximum value in the data. Scale bars: 10 mm (b), 5 mm (f), and 2 mm (b and f insets). See also Fig. S7.
Strikingly, endogenous Nanog accumulates in fewer fluorescent foci beginning at the 64-cell stage (Figs. 4b–d and S7h), likely reflecting the lower protein abundance observed by IF and western blot and indicative of concentration-dependent foci formation. Interestingly, two bright fluorescent foci were robustly detected during the early phases of the 64–512 cell stages (Video 1, Figs. 4b and S7h), strongly suggesting that these regions represent the priming of miR-430 transcription sites as recently shown in the literature52. The miR-430 locus, the first zygotically transcribed region in the zebrafish genome, can be detected as transcriptionally active starting at the 64-cell stage52,55,56. Additionally, we generated a transgenic zebrafish line that ubiquitously expresses EGFP-NbALFA under the control of a beta-actin promoter in an ALFA-nanogKI/KI background, allowing us to visualize endogenous Nanog protein behavior directly without the need of mRNA injection (Figs. 4b; S7h and Video 1). Similar results were observed when the EGFP-NbALFA GEAR binder was supplied via transgene expression versus mRNA injection. While we quantified Nanog clusters, additional signal can be observed in the nucleus (Fig. 4b inset) that we cannot quantify, given the spatial resolution of these data. Overall, these findings indicate that when Nanog is overexpressed, it can either seed additional foci or enlarge existing ones due to excess Nanog molecules, possibly aided by its IDRs.
Endogenous Nanog foci prime the formation of large transcription bodies
In zebrafish embryos, elongating RNA Polymerase II (Pol-II) molecules are initially concentrated in two large, long-lived transcription bodies corresponding to the transcription of the miR-430 gene cluster52,55. Exogenously provided Nanog transcription factor foci have been shown to precede the formation of these transcription bodies, consistent with Nanog’s role as a pioneer transcription factor31. To investigate whether endogenous Nanog behaves similarly and to demonstrate the utility of GEAR reagents in studying biological phenomena, we used a fast-maturing mScarlet-i357-NbALFA GEAR to detect Nanog protein, alongside a genetically encoded Pol-II Ser2-EGFP reporter54,58 (Mintbody detecting RNA Pol II phosphorylated on Serine 2) to visualize transcriptional elongation (Fig. 4e). As observed in studies with exogenous Nanog, we found that Nanog foci precede the formation of transcription bodies, appearing in close proximity but dissolving rapidly while the transcription bodies persist (Fig. 4f, g and Video 2)52. These data support the model where Nanog binds to promoters and enhancers of miR-430, forming foci that dissolve at the onset of transcription. Together, these results highlight the rapid mobility of Nanog protein during embryogenesis and demonstrate the effectiveness of GEARs to visualize the behavior of endogenous proteins to gain biological insights.
Discussion
We present the GEARs toolkit and a streamlined CRISPR/Cas9 knock-in strategy for creating multifunctional, adaptable alleles. GEARs is a tripartite system consisting of a short epitope and a binder fused to assay-specific adapter proteins. After evaluating seven epitope-binder pairs, we identified those with the strongest in vivo performance. We demonstrate diverse applications of GEARs—including live imaging, targeted degradation, western blotting, immunostaining, and ChIP—highlighting the broad utility of the engineered alleles.
The GEARs toolbox concept is based on epitope tagging, a well-established technique for protein detection that typically does not disrupt protein function59,60. For instance, HA-tagging Wnt3 enabled visualization of its endogenous morphogen gradient, a challenge with larger tags61. Epitope tagging also allows detection when primary antibodies are unavailable60. Recent advances in Drosophila and cell culture62,63 show the growing potential and broad applicability of in vivo detection tools. In this study, we applied in vivo binding tools to vertebrate systems, systematically assessing their performance in binding assays. Testing nuclear and membrane-targeted proteins, we found ALFA and Moon tag-binder pairs showed the strongest binding efficiency and fluorescence localization. Interestingly, the Sun and VHH05 tags performed better in the nucleus, while Nb127d01 exhibited higher efficiency in targeting plasma membrane protein (Fig. 1e, f). We favor the Nb-based binders over scFv-binders due to their higher in vivo specificity and efficiency.
A critical aspect in protein tagging is ensuring that the resulting fusion protein retains its biological function. While genes tested here tolerated N-terminal fusions and had suitable Cas9 target sites, the NGG PAM requirement of S. Pyogenes Cas9 can limit the availability of genomic regions for targeting. Using other nucleases (Cas9 variants64 or Cas12a65) with different cleavage site preferences, could expand the targeting space. Our streamlined cloning-free genome editing protocol demonstrated improved integration efficiency from 0.3 to 16% (after fluorescent marker selection)11,42 to 7–18% without pre-selection. The increased efficiency likely stems from the ease of inserting short DNA fragments via non-HR repair, which zebrafish favor during early embryogenesis66,67. Based on the frequency of germline transmission, we hypothesize that oligo-based tagging likely occurs at earlier developmental stages, thereby increasing the likelihood of recovering germline-transmitting events. Future optimizations in choosing tag insertion sites, nuclease selection and germline screening methods will further enhance the throughput and efficiency of allele generation.
In this study, we introduced an F-box-based degron into the GEARs toolbox, inspired by the zGrad system, which uses the F-box-NbGFP fusion protein37. The degrons developed—ALFAgrad, Moongrad, VHH05grad, and 127d01grad—demonstrated over 50% degradation efficiency, with ALFAgrad achieving the highest performance, clearing ≥90% of nuclear protein in both zebrafish and mouse embryos. Furthermore, ALFAgrad reduced previously synthesized cytoplasmic protein, comparable to zGrad (Fig. 2d, I, n)37. In addition to our use of ALFAgrad in zebrafish and mouse, protein depletion using NbALFA was recently shown in Drosophila, further highlighting the cross-species functionality of these reagents68. GEARgrads offer several key advantages over other targeted protein depletion systems. Unlike other degron systems such as dTAG69 and HaloPROTAC70, which require large protein domain insertions into the gene of interest and the addition of compounds to induce degradation, GEARgrads utilize smaller tags without the need for small molecules. Finally, traditional degron alleles cannot be repurposed for applications beyond protein depletion. In contrast, the modular design of the GEARs toolbox enables the use of epitope-tagged lines for both degrons and other biological applications.
While GEARs use small epitopes to generate multifunctional alleles, any sequence addition could impair protein function. Short epitopes are more likely to be tolerated than large sequence insertions, yet it is important to experimentally validate the functionality of tagged proteins, such as through genetic rescue experiments. Furthermore, the interaction between the epitope and the GEAR binder-adaptor may interfere with protein function, and careful analysis of protein localization and ectopic phenotypes should be considered. Despite these potential limitations, we believe the usefulness of rapid swapping of nanobody fusions significantly outweighs these potential drawbacks. Background fluorescence from unbound fluorophore-GEAR binders could mask the true signal, potentially hindering detection, especially for low-abundance proteins. However, we observe that the fluorescence of binders in the absence of the epitope is significantly reduced, possibly through destabilization of the GEAR binders in the absence of the epitope. To further lower background, a recently developed conditionally destabilized ALFA nanobody68 could enable a higher turnover of unbound fluorophore-NbALFA and, therefore, effectively reduce background fluorescence while enhancing signal detection. Here, we have demonstrated protein degradation using four endogenous targets as well as cytoplasmic GFP and H2B-GFP. It remains to be determined whether there are proteins that are refractory to protein degradation, either by masking of the epitope, inaccessible subcellular localization, alternative splicing that excludes the epitope or degron adaptor specificity. To overcome this, using an alternative established degron system (such as the Ab-SPOP degradation system71) or using a degron adapter may expand the degradation abilities of this system to its maximal capabilities. Finally, the use of tissue-specific ALFAgrad expression in the vasculature (transgenic fli1a:ALFAgrad P2A mCherry) demonstrates that GEARs can be used for targeted protein degradation with spatiotemporal control. This approach opens many avenues for using ALFAgrad for conditional protein loss-of-function analysis.
Using GEARs and time-lapse imaging in zebrafish embryos, we visualized endogenous Nanog as two bright nuclear foci near transcription bodies, previously shown to correspond to transcription of the miR-430 gene locus52,55, consistent with the region’s high density of Nanog binding sites31. In contrast, Nanog overexpression leads to the formation of ectopic foci, with only two seeding at the miR-430 locus52,54. This phenomenon may stem from IDR-mediated recruitment of excess, unbound Nanog or its binding to low-affinity or non-canonical sites. This could also partially explain why ChIP-seq analysis reveals over 30,000 additional peaks—approximately a two-fold increase—when comparing ectopically expressed Nanog-Myc to endogenously expressed ALFA-Nanog. Consistent with this, previous work has shown that exogenous Nanog foci are DNA-bound52,54, suggesting they reflect altered binding rather than DNA-independent protein accumulation. Given the growing interest in phase transitions and nuclear foci formation by transcription factors72, it is important to note that overexpression can cause concentration-dependent ectopic foci. This underscores the need to study proteins at physiological levels—something the GEARs toolbox can enable.
A key advantage of the GEARs toolbox is its flexibility in integrating new components. We have shown this with various fluorophores and degron adapters; additional binders or epitopes can be easily added. For example, future introduction of chemical or light-inducible elements could enable spatiotemporal control over GEAR activity. As new intracellular probes emerge, GEARs can rapidly adapt, maintaining its versatility for future applications.
Methods
Zebrafish husbandry and maintenance
Danio rerio (zebrafish) embryos were obtained from natural matings of adult fish of mixed wild-type backgrounds (TU-AB and TLF strains) of mixed ages, ranging from 5 to 18 months. Zebrafish were maintained and used in accordance with the Yale University AAALAC guidelines under a protocol approved by the Yale University IACUC (protocol number 2021-11109). Embryos were grown and staged according to published standards73 and all zebrafish and live cell embryo experiments were performed at 28 °C.
Mouse husbandry and maintenance
Mouse experiments complied with ethical protocols approved by the Yale University Institutional Animal Care and Use Committee (IACUC) under protocol #2023-20324. The housing used was consistent with the Guide for the Care and Use of Laboratory Animals and compliant with the Animal Welfare Act and Regulations. Mice were housed on ventilated Tecniplast lixit racks with an ambient temperature of 22 °C and 50 ± 10% humidity with a 12-h light–dark cycle (lights on 7:00–19:00) and fed ad libitum. Mouse embryos were generated by inducing hyperovulation in 4-week-old B6D2F1 females (Jackson Laboratory), which were mated with 8-week to 6-month-old B6D2F1 males (Jackson Laboratory). To induce hyperovulation, 5 IU of pregnant mare serum gonadotropin (PMSG; BioVendor R&D, Cat# RP1782725000) was injected intraperitoneally, followed by 7.5 IU of human chorionic gonadotropin (hCG; Sigma Aldrich, Cat# CG10-1VL; CAS: 9002-61-3) 47 h later. Zygotes were collected 20 h post-hCG and cumulus cells were removed using hyaluronidase in M2 media (Sigma Aldrich, Cat# MR-051-F). Embryos were cultured in 25 µL drops of KSOM Mouse Embryo Media (Sigma, MR-106) covered with cell-culture grade paraffin oil (Copper Surgical, Cat# ART-4008-5P) in a cell culture incubator maintained at 37 °C and 5% CO2.
Molecular cloning
GEAR binders
DNA fragments containing the ORFs for FbHA, FbFLAG, NbALFA, NbVHH05, Nb127d01, NbMoon, and FbSun were codon-optimized for zebrafish using iCodon74 and purchased from IDT as GBlocks. Fragments were cloned into a pCS2+ EGFP expression vector as C-terminal fusions (EGFP-GEAR binder) using the InFusion HD Cloning Kit (TaKaRa, Cat# 639650) and sequence verified. For mRNA production, vectors were linearized with NotI-HF restriction enzyme (New England Biolabs, Cat# R3189S) and in vitro transcribed using the mMessage mMachine SP6 kit (Thermo Fisher Scientific, Cat# AM1340). Injection control V5-mScarlet-I34 was codon-optimized, purchased as a GBlock from IDT, cloned using InFusion enzyme (TaKaRa, Cat# 639650), sequence verified, and linearized/transcribed using the same procedure.
Fluorescent GEAR adapters
DNA fragments coding for mNeonGreen (gift from Ken-Ichi Takemaru; Addgene plasmid #128144; RRID:Addgene_128144), mScarlet-I (gift from Dorus Gadella; Addgene plasmid #85044; RRID:Addgene_85044), mTagBFP2 (gift from Michael Davidson; Addgene plasmid #55295; RRID:Addgene_55295) and HaloTag (gift from Marvin Tanenbaum; Addgene plasmid #128603; RRID:Addgene_128603) were amplified, cloned into pCS2-EGFP-NbALFA to replace the EGFP ORF using InFusion enzyme (TaKaRa, Cat#639650) and sequence verified. For mRNA production, vectors were linearized with NotI-HF restriction enzyme (New England Biolabs, Cat# R3189S) and in vitro transcribed using mMessage mMachine SP6 kit (Thermo Fisher Scientific, Cat#AM1340). A DNA fragment for mScarlet-i357 was codon-optimized using iCodon74, purchased from IDT as GBlock and cloned into pCS2-EGFP-NbALFA, replacing the EGFP ORF, as described above.
Degron GEAR adapters and degron reporters
Zebrafish fbxw11b ORF was amplified from the pCS2+ zGrad plasmid (gift from Holger Knaut; Addgene plasmid # 119716; RRID:Addgene_119716) and cloned into pCS2+ GEAR Nb-binder clones to replace the EGFP ORF using InFusion enzyme (TaKaRa, Cat# 639650), creating GEARdegrons that we call ALFAgrad, Moongrad, VHH05grad, and 127d01grad. Split fluorescent reporters were cloned by introducing ALFA, VHH05, Moon, and 127d01 epitope tags in-frame to the H2B-EGFP ORF in pCS2+TAG (TdTomato-2A-H2B-GFP-SV40pA; gift from Shankar Srinivas; Addgene plasmid # 26772; RRID:Addgene_26772). The ALFA-EGFP reporter was cloned by introducing an N-terminal ALFA epitope in frame to a pCS2+ EGFP vector. Vectors were sequence verified, linearized with NotI-HF restriction enzyme (New England Biolabs, Cat# R3189S) and in vitro transcribed using mMessage mMachine SP6 kit (Thermo Fisher Scientific, Cat#AM1340 for mRNA injection.
Nanog and Vangl2 fusions
Tandem HA tags were cloned as N-terminal or C-terminal fusions using InFusion enzyme (TaKaRa, Cat# 639650) into a pCS2+nanog expression vector31. vangl2 was amplified from cDNA and cloned into the pCS2+ expression vector to generate pCS2+vangl2. Single HA, FLAG, ALFA, VHH05, 127d01, Sun/SunGb1 and Moon epitope tags were cloned as N-terminal fusions using InFusion enzyme (TaKaRa, Cat# 639650) into a pCS2+nanog and pCS2+ vangl2 expression vector. Vectors was sequence verified, linearized with NotI-HF restriction enzyme (New England Biolabs, Cat# R3189S) and in vitro transcribed using mMessage mMachine SP6 kit (Thermo Fisher Scientific, Cat#AM1340 for mRNA injection.
Live imaging reagents
Expression vectors containing nanog-mEmerald54 and the Pol-II PhosphoSer2 Mintbody-EGFP54,58 were linearized using NotI-HF restriction enzyme (New England Biolabs, Cat# R3189S) and in vitro transcribed using mMessage mMachine SP6 kit (Thermo Fisher Scientific, Cat#AM1340) for mRNA injection.
Transgenesis
Tol2-based transgenes75 were assembled using InFusion cloning (TaKaRa, Cat# 639650). To generate Tg(actb2:EGFP-NbALFA) zebrafish, an actb2 promoter, EGFP-NbALFA open reading frame and a prrg2 3′UTR were amplified and assembled into a pDEST Tol2 pA2 transgenesis vector. To generate Tg(hsp70l:ALFAgrad) zebrafish, a hsp70l promoter, ALFAgrad open reading frame and a prrg2 3′UTR were amplified and assembled into a pDEST Tol2 CG2 transgenesis vector containing myl7:EGFP transgenesis reporter. To generate Tg(fli1a:ALFAgrad P2A mCherry) zebrafish, a fli1a promoter, ALFAgrad open reading frame and a P2A mCherry polyA cassette were assembled into a pDEST Tol2 pA2 transgenesis vector. Transgenes were fully sequence verified before injection. Embryos were injected at the one-cell stage with 25 pg of assembled transgene and 25 pg of Tol2 mRNA. Embryos were sorted at 24 hpf for ubiquitous EGFP reporter expression (actb2:EGFP-NbALFA transgene), heart-specific myl7:EGFP (hsp70l:ALFAgrad transgene) or vascular-specific mCherry expression and were subsequently grown to adulthood. Individual fish were crossed to AB wild-type zebrafish to generate stable F1 lines. Two independent lines were established for each transgene and verified to exhibit similar behavior. All stable transgenic lines used were hemizygous for the respective transgenes. To evaluate potential off-target effects of heatshock promoter-driven ALFAgrad expression, embryos from a hemizygous male Tg(hsp70l:ALFAgrad) outcross were screened at 24 hpf for the presence or absence of the myl7:EGFP transgene reporter. Based on their expression status, embryos were sorted into transgene-positive and transgene-negative sibling groups. Both groups were subjected to heat shock at 24 hpf at 38 °C for 1 h. Phenotypes were assessed and imaged at 1 dpf, 3 dpf, and 5 dpf, with representative images presented in Fig. S4f.
Epitope tag knock-in
sgRNA design and synthesis
To target the N-terminus of Nanog, Vangl2, Dicer, and Pou5f3 sgRNAs were designed to target as close to the start ATG codon as possible using CRISPRscan76. The sgRNA sequences were ordered from Synthego, resuspended to 500 ng/µL stocks in RNase-free H2O, and stored at −80 °C until use.
Nuclease test
A 5 µl mix containing 500 ng recombinant Engen Spy Cas9 NLS (New England Biolabs; Cat#M0646T), 250 ng sgRNA and 300 mM KCl was incubated at 37 °C for 5 min to form ribonucleoprotein complexes (RNPs). Embryos were injected at the 1-cell stage into the cell with the RNPs and grown to 48 hpf. Uninjected and injected embryos were lysed in a 1× PCR buffer (GoTaq; Promega, Cat# M7123) and 1 µg/µL Proteinase K (Thermo Fisher Scientific, EO0492) solution for 1 h at 55 °C followed by inactivation for 10 min at 95 °C. 1 µL of this lysis was used in a PCR reaction that overlapped the target site listed below. The reactions were cleaned up using Zymo DNA Purification columns (Zymo Research, Cat#D4014) and Sanger sequenced. The resulting chromatograms were used for ICE analysis (Synthego; https://ice.synthego.com/) to determine cleavage efficiency. Each sgRNA was confirmed to have >80% cleavage efficiency across 3 independent F0 injected embryos. The targeting sgRNA sequences were as follows: nanog TTTATCTAACGGCGAAATGG, vangl2TGCGACTCGTTATCCATGTC, dicer TATCAGTCTCTAAGCATGGC, and pou5f3 CCTTTTTAGCGGAAAGATGA.
Genotyping for nanog was performed using the following primers, Fwd: GTTGTAGGACAGAAAGAGCCGT, Rev: CACCTGGCAATATAAATCAGCA.
Genotyping for vangl2 was performed using the following primers, Fwd: CCGCGCTCTCCAGTCCGTCA, Rev: CGAGAGCTGCGTGAGTGTGAA.
Genotyping for dicer was performed using the following primers, Fwd: CAGTAACCCGCCTGATCCTG, Rev: AATTTTCTGGGGGTGCCACT.
Genotyping for pou5f3 was performed using the following primers,
Fwd: GGAAGAGTTGGAGGTGGTGA, Rev: AAACGGGTACCAGTGTTTGG.
Tag knock-in design
Single-stranded oligo donor nucleotides (ssODNs) were designed by inserting epitope sequences in frame with nanog, vangl2, dicer, and pou5f3 coding sequences, respectively, directly proceeding the ATG start codon. Homology arms of 20 bp were appended to epitope tag sequences on the sense strand and ordered as PAGE-purified oligos (Sigma Aldrich). Oligos were resuspended in RNase-free H2O at a concentration of 50 µM and stored at −20 °C until use.
Donor ssODNs for nanog were the following:
ALFA: GAGTTTATCTAACGGCGAAATGCCATCACGTCTGGAGGAAGAGCTGCGTCGTCGCCTGACCGAACCTGCGGACTGGAAGATGCCA
VHH05: GAGTTTATCTAACGGCGAAATGCCGCAGGCTGATCAGGAGGCTAAAGAGCTGGCAAGACAGATTAGCCCCGCGGACTGGAAGATGCCA
127d01: GAGTTTATCTAACGGCGAAATGCCATCCTTCGAAGATTTCTGGAAGGGTGAGGATCCTGCGGACTGGAAGATGCCA
Donor ssODNs for vangl2 were the following:
ALFA: CCCGCCCACTGGCCCCCGACATGCCATCACGTCTGGAGGAAGAGCTGCGTCGTCGCCTGACCGAACCTGATAACGAGTCGCAGTACTC
VHH05: CCCGCCCACTGGCCCCCGACATGCCGCAGGCTGATCAGGAGGCTAAAGAGCTGGCAAGACAGATTAGCCCCGATAACGAGTCGCAGTACTC
127d01: CCCGCCCACTGGCCCCCGACATGCCATCCTTCGAAGATTTCTGGAAGGGTGAGGATCCTGATAACGAGTCGCAGTACTC
Donor ssODN for dicer was the following:
ALFA: CTTATCAGTCTCTAAGCATGCCATCACGTCTGGAGGAAGAGCTGCGTCGTCGCCTGACCGAACCTGCTGGCCTACAGCTGGTGAC
Donor ssODN for pou5f3 was the following:
ALFA: ACCTTTTTAGCGGAAAGATGCCATCACGTCTGGAGGAAGAGCTGCGTCGTCGCCTGACCGAACCTACGGAGAGAGCGCAGAGCCC
Tag knock-in injection
A 5 µL mix containing 2 µM ssODN in RNase-free H2O was prepared. A second 5 µl mix containing 500 ng recombinant Engen Spy Cas9 NLS (New England Biolabs; Cat#M0646T), 250 ng sgRNA and 300 mM KCl was incubated at 37 °C for 5 min to form RNPs. Wildtype embryos were injected at the 1-cell stage twice: first, embryos were injected in the yolk with 1 nL of the donor oligo solution, and then second, embryos were re-injected into the cell with the RNPs. Uninjected, donor-only injected, and dual-injected embryos were grown to 48 hpf. 5 embryos per condition (uninjected, donor-only and dual injection) were lysed in a 1× PCR buffer (GoTaq; Promega, Cat# M7123) and 1 µg/µL Proteinase K (Thermo Fisher Scientific, EO0492) solution for 1 h at 55 °C followed by inactivation for 10 min at 95 °C. 1 µL of this lysis was used in a PCR reaction that used nanog, vangl2 or dicer genotyping primers with tag-specific primers, such that two junction PCRs were run for each sample. Clutches that showed positive junction PCR amplification were grown to adulthood for germline screening. Genotyping for ALFA integration was performed using the following primers, Fwd: CACGTCTGGAGGAAGAGCTG, Rev: GTTCGGTCAGGCGACGAC. Genotyping for VHH05 integration was performed using the following primers, Fwd: TGGCAAGACAGATTAGCCCC, Rev: TTAGCCTCCTGATCAGCCTG. Genotyping for 127d01 integration was performed using the following primers, Fwd: TCTGGAAGGGTGAGGATCCT, Rev: TCACCCTTCCAGAAATCTTCGA.
Knock-in line establishment
Adult F0 animals were incrossed to increase throughput of screening. Pools of 8 embryos were lysed in 50 µL 1× PCR buffer (GoTaq; Promega, Cat# M7123) and 1 µg/µL Proteinase K (Thermo Fisher Scientific, EO0492) solution for 1 h at 55 °C followed by inactivation for 10 min at 95 °C. 12 pools of embryos per pair were screened by PCR to maximize screening efficiency. F0 pairs that had both positive 5′ and 3′ junction PCRs were then outcrossed to wildtype fish, and 24 individual embryos from those crosses were re-screened by PCR. Embryos that were positive for both 5′ and 3′ junction PCRs were Sanger sequenced to assess tag integration. Those with precise integrations were grown to adulthood, genotyped for tag integration, and incrossed to generate homozygous knock-in animals. Homozygous animals were further confirmed for precise tag integration by PCR using genotyping primers listed above. Homozygous animals were maintained for all knock-in alleles and exhibited normal development.
GEAR localization experiments
Injections
Wildtype 1-cell staged embryos were dechorionated in 0.83 mg/ml Protease from Streptomyces griseus (pronase; Sigma Aldrich, Cat# P5147, CAS: 9036-06-0) and injected with 25 pg of a V5-mScarlet-I control for normalization and 50 pg of EGFP-GEAR mRNA alone or co-injected with 50 pg of epitope-tagged nanog mRNA or 100 pg tagged vangl2 mRNA. Embryos were incubated at 28 °C until they reached 30% epiboly (~5 hpf) and mounted in 0.8% low-melt agarose (GPG/LE AmericanBio, CAS: 9012-36-6) in system water against a No. 1.5 cover slip (Thermo Fisher Scientific, Cat# 22-050-246).
Imaging
Embryos were imaged on a temperature-controlled (28 °C) upright Zeiss LSM 980 confocal microscope with an Airyscan 2 detector (RRID:SCR_025048) and a W Plan-Apochromat 40×/1.0 DIC VIS-IR M27 objective with bidirectional line scanning at a format of 2532 × 2532 pixels, equivalent to an imaging field of 150.81 × 150.81 µm and 1.4× optical zoom with 2× averaging. All images were collected at 16 Bit aryscan mode, and optical stacks were acquired at 0.35 µm spacing. EGFP fluorophores were excited using the 488 nm laser line at 4% laser power and mScarlet-I fluorophores were excited using the 561 nm laser line at 7% laser power. Raw images were deconvolved using the Airyscan software. Individual representative slices are shown in Fig. 1 and S1 + S2.
Quantification
Nanog: The nuclear-to-cytoplasmic ratio in samples expressing EGFP-GEAR binders without an epitope target present was compared to samples expressing EGFP-GEAR binders with epitope-tagged Nanog. In each confocal image, lines (width of 1) were drawn through 12 cells in ImageJ and assigned to the region of interest (ROI) manager. Lines were drawn through cells below the enveloping layer in single Z-slices. Raw values of fluorescence intensity (arb. unit) were extracted for both channels (EGFP and mScarlet-I) along the line at every pixel (1px = 59.9 nm) and further processed in R-studio (Version 4.2.3; RRID:SCR_001905). In detail, the center of the nucleus was determined for each cell and aligned at 0 µm for further analysis. Next, the mean V5-mScarlet-I fluorescence was used to normalize all fluorescence values within a cell (Mean V5-mScarlet-I fluorescence = 1 arb. unit). The nuclear region was defined as −2.442 µm to +2.442 µm, which includes 83 data points per cell and contains the “center/middle” of each nucleus to account for small variations in nuclear size between cells (shaded region, Fig. S3g). The mean fluorescence intensity for each measured position throughout the nucleus was determined across the 12 nuclei. The median of all 83 nuclear measurements per image was recorded and used in the nuclear-to-cytoplasmic ratio calculation. The cytoplasm was defined as the regions of −8.0407 µm to −7.0282 µm and +7.0282 µm to +8.0407 µm, which contain 36 data points per cell (shaded region, Fig. S3g). The mean fluorescence intensity (arb. units) for each data point position in the cytoplasm was determined across the 12 cells investigated and the median of all 36 averaged cytoplasmic measurements per image was recorded for each embryo replicate and used to calculate the nuclear-to-cytoplasmic ratio (shown in Figs. 1d and S3h). The area under the curve or total fluorescence intensity was determined by quantifying the sum fluorescence (nucleus + cytoplasm) for each cell, results shown in Fig. S3a.
Vangl2: The membrane-to-cytoplasmic ratio in samples expressing EGFP-GEAR binders without an epitope target present was compared to samples expressing EGFP-GEAR binders with epitope tagged Vangl2. Raw fluorescence intensity measurements were extracted from single Z-slices of 12 cells per image as described above and further processed in R-studio. The line was drawn to cross the plasma membrane in two instances and to avoid the nucleus. In summary, the mean V5-mScarlet-I fluorescence was used to normalize all fluorescence values within a cell (Mean V5-mScarlet-I fluorescence = 1 arb. unit). Next, the two regions where the ROI line crossed the plasma membrane were isolated and the center of the membrane was aligned at 0 µm for further analysis (Both peaks shown in Fig. S3i). The membrane was defined as the region surrounding the peak membrane data point as −0.1191 µm to +0.1191 µm, which includes 5 data points per membrane crossing and, therefore, 10 data points per cell (Fig. S3i). The mean fluorescence intensity for each position was determined across the 12 nuclei measured per biological replicate and the median of all 10 plasma membrane measurements per image was recorded and used in the membrane-to-cytoplasmic ratio calculation for each embryo (shown in Figs. 1f and S3j). The cytoplasm was defined as the region of +2.384 µm to +10.132 µm, where the first plasma membrane position determined the position measurements. The cytoplasm data includes 131 data points in total per nucleus analyzed. The mean fluorescence intensity for each data point position in the cytoplasm was determined across the 12 cells investigated and the median of all 131 averaged cytoplasmic measurements per image was recorded and used to calculate the plasma membrane-to-cytoplasm ratio (shown in Figs. 1f and S3j).
GEAR multicolor adapter experiments
Injections
Wildtype 1-cell staged embryos were dechorionated in 0.83 mg/ml pronase (Sigma Aldrich, Cat# P5147, CAS: 9036-06-0) and injected with 50 pg of ALFA tagged nanog mRNA alone or with 50 pg of mNeonGreen-NbALFA, 50 pg of mScarlet-I- NbALFA, 50 pg of mTagBFP2-NbALFA or 50 pg of HaloTag-NbALFA mRNA. Embryos were incubated at 28 °C until they reached 30% epiboly (~5 hpf) and mounted in 0.8% low-melt agarose (GPG/LE AmericanBio, CAS: 9012-36-6) in system water against a No. 1.5 cover slip (Thermo Fisher Scientific, Cat# 22-050-246). For HaloTag imaging, JFX650 (Janelia Fluor®; Promega, Cat# HT1070) HaloTag ligand was directly added at a concentration of 10 nM to low-melt agarose and mixed thoroughly before mounting.
Imaging
Embryos were imaged on a temperature-controlled (28 °C) upright Zeiss LSM 980 confocal microscope with an Airyscan 2 detector (RRID:SCR_025048) and a Plan-Apochromat 20×/0.8 M2 objective with bidirectional line scanning at a format of 4084 × 4084 pixels and 1.4× optical zoom in aryscan mode. All images were collected at 16 Bit, and optical stacks were acquired at 0.35 µm spacing. Fluorophores were excited using the 408 laser line (mTagBFP2) at 2% laser power, 488 laser line (mNeonGreen) at 1% laser power, 561 laser line (mScarlet-I) at 8% laser power and the 639 laser line (Halo/JFX650) at 0.6% laser power. Raw images were deconvolved using the Airyscan software. Individual representative slices of embryonic cells are shown in Fig. S2.
Degron experiments
Zebrafish toxicity titrations
Wildtype 1-cell staged embryos were dechorionated in 0.83 mg/ml pronase (Sigma Aldrich, Cat# P5147, CAS: 9036-06-0) and injected with 25, 50, 75, or 100 pg of fbxw11b-GEAR degron mRNA. Embryos were scored at 24 hpf for morphological defects relative to wildtype uninjected controls.
Zebrafish reporter injections
Wildtype 1-cell staged embryos were dechorionated in 0.83 mg/ml pronase (Sigma Aldrich, Cat# P5147, CAS: 9036-06-0) and injected with 50 pg of split fluorescent reporter (TdTomato-2A-epitopeTag-H2B-EGFP-SV40pA, containing the ALFA, Moon, VHH05 or 127d01 epitope tag) mRNA alone or with fbxw11b-GEAR degron mRNA. As a positive control, embryos were injected with tdTomato-2A-H2B-EGFP alone or with 100 pg of zGrad37. For imaging, embryos were incubated at 28 °C until they reached tailbud stage and mounted in 0.8% low-melt agarose (GPG/LE AmericanBio, CAS: 9012-36-6) in system water against a no. 1.5 cover slip (Thermo Fisher Scientific, Cat# 22-050-246).
Zebrafish reporter imaging
Embryos were imaged at 10 hpf on a temperature-controlled (28 °C) upright LSM 980 confocal microscope (RRID:SCR_025048) with an Airyscan 2 detector and an EC Plan-Neofluar 10×/0.3 M27 objective with bidirectional line scanning at a format of 4084 × 4084 pixels and 1.0× optical zoom. All images were collected at 16 Bit in aryscan mode, and optical stacks were acquired at 3.08 µm spacing. Fluorophores were excited using the 488 laser line (EGFP) at 5% laser power and the 561 laser line (tdTomato) at 8% laser power. Raw images were deconvolved using the Airyscan software. Representative maximum projections of individual embryos are shown in Fig. 2.
Quantification of zebrafish reporters
3D image analysis was performed in the IMARIS software (Bitplane, Oxford Instruments, Concord MA; Version: 10.0; RRID:SCR_007370). Nuclei were identified using the ‘spot’ function and an object with a constant size of 5 μm in xy and a z-axis point spread function of 8μm was placed in the center of nuclei. The membrane fluorescence of a cell was quantified by sampling the membrane circumference in multiple regions using the ‘spots’ function. Smaller spots with a size of 2 μm xy and 4 μm in the z-dimension were added based on the tdTomato staining. A screenshot visualizing the analysis setup is shown in Fig. S4c. To determine the background fluorescence in both channels, sets of ‘spots’ of the same volume as the nuclei and membrane spots were generated outside the embryo for background correction. Statistics for all spot objects were exported and membrane spots were linked to their closest nucleus in 3D using a published Python script (https://github.com/TMinchington/sass, RRID:SCR_018797)77. Median background fluorescence was subtracted from each channel measurement, and the median nuclear sum fluorescence was divided by the median membrane sum fluorescence for each cell. The resulting mean of the control embryo condition was set to 1, and all other values were adjusted using the conversion factor accordingly.
Zebrafish tissue-specific ALFAgrad expression and imaging
Embryos derived from a male Tg(fli1a:ALFAgrad P2A mCherry) hemizygous outcross were injected at the 1-cell stage with 1 nL of a mixture of either 20 pg ALFA-EGFP or 20 pg EGFP mRNA with 20 pg H2B-Halo mRNA as an injection control. Embryos were screened at 24 hpf for the presence or absence of fli1a mCherry transgene reporter expression, sorted, and incubated in JFX650 dye (Promega, Cat#HT1070) for 30 min at 28 °C. Embryos were washed 3 times in embryo water. At 29 hpf, embryos were mounted in 0.8% low-melt agarose (GPG/LE AmericanBio, CAS: 9012-36-6) in system water and placed in a preheated (28 °C) confocal chamber. Two uninjected WT embryos were mounted and imaged for background autofluorescence normalization. Embryos were imaged on an inverted Leica SP8 confocal microscope with a white light laser (70% laser power) and an HC PL APO CS2, 20×/0.75 DRY objective. Images were acquired using bidirectional line scanning at a scan speed of 400 Hz and a format of 3288 × 1696 pixels, equivalent to a pixel size of 35.37 nm × 35.37 nm and an image size of 116.25 µm × 59.95 µm. All images were collected at 16-bit, 1× optical zoom, 2× frame accumulation, and frame sequential scanning with optical stacks acquired at 1.5 µm spacing. Fluorophores were excited using excitation wavelengths of 472 nm (EGFP; laser power: 16%, pinhole = 1.2 Airy Units, HyD detector, collection window: 493–564 nm, time gating: 0.5 ns–8 ns), 666 nm (mCherry laser power; 16%, pinhole = 2 Airy Units, HyD detector, collection window: 569–658 nm, time gating: 0.5 ns–9 ns) and a 523 nm (Halo; laser power: 10.5%, pinhole = 1 Airy Units, PMT detector, collection window: 658–776 nm). Images were deconvolved using the Huygens Deconvolution software (Scientific Volume Imaging).
Zebrafish tissue-specific ALFAgrad quantification
The EGFP/Halo ratio quantification was performed in 3D using the IMARIS software (Bitplane, Oxford Instruments, Concord, MA; Version 10.0; RRID:SCR_007370). The mCherry fluorescence of the transgene was used to segment out the vasculature using the surface function. For better segmentation, a Gaussian blur of 1 was applied to the mCherry channel prior to segmentation. The sum EGFP and Halo fluorescence within the segmented vascular tissue was exported. Two control regions in the somite tissue were segmented manually in each embryo using the surface function, and sum EGFP and Halo fluorescence within the segmented regions was extracted. Two uninjected WT embryos were imaged to quantify tissue autofluorescence in each channel. The embryo was segmented manually, excluding the yolk region, which has higher inherent autofluorescence. The average background fluorescence per µm³ was calculated using the segmented volume and subtracted from all vasculature and somite measurements according to their respective volume measurements. After background correction, the EGFP-to-Halo ratio was calculated for the vasculature and somite tissues. The average of the two somite tissue ratios was used to calculate the vasculature-to-somite ratio of EGFP:Halo fluorescence. Embryos that expressed ALFAgrad in the vasculature and injected with ALFA-EGFP, EGFP degradation occurred as expected, resulting in a reduced vasculature-to-somite ratio.
Mouse injections
Mouse embryos were injected using a digital injection system (Xenoworks, Sutter Instruments) and borosilicate filamented glass needles (Sutter Instruments, Cat# NC9955576) that were prepared in-house. The microinjection system comprises a DMi8 microscope (Leica) with micromanipulators (Sutter Instruments). Embryos were injected in glass-bottomed dishes (MatTek, Cat# P35GC-1.5-14-C) in 25 µL drops of M2 media (Sigma Aldrich, Cat# MR-015-D) and covered in cell-culture grade paraffin oil (Copper Surgical, Cat# ART-4008-5P). First, 50 ng/µL of split fluorescent ALFA mRNA reporter (TdTomato-2A-ALFA-H2B-EGFP) was injected into the zygote immediately after isolation, with 100 ng/µL of ALFAgrad (Fbxw11b-NbALFA) mRNA or Moongrad (Fbxw11b-NbMoon) injected into one cell of the two-cell embryos 24 h later. Embryos were fixed at 0, 2, 3, 6, 9, and 12 h post-degron injection for ALFAgrad and at 6, 9, and 12 h for Moongrad. For fixation, the zona pellucida was first removed using acidified Tyrode solution (Sigma Aldrich, Cat# T1788) and embryos were attached to a glass bottom dish (MatTek, Cat# P35GC-1.5-14-C). A solution of 4% Paraformaldehyde (PFA; Electron Microscopy Sciences, Cat# 50-980-487) in 1× Phosphate Buffered Saline (PBS; American bio, Cat# AB11072-01000) was added to embryos for 15 min at room temperature, followed by nuclear staining using 5 µg/ml DAPI (Thermo Fisher Scientific, Cat# D1306) in 1× PBS for 15 min, 3× washes in 1 × PBS and imaging.
Mouse imaging
Mouse degron images were acquired using an inverted Leica SP8 confocal microscope with a white light laser (70% laser power) and an HC PL APO CS2, 40×/1.30 oil objective. Images were acquired using bidirectional line scanning at a scan speed of 400 Hz and a format of 1024 × 1024 pixels, 3× (ALFAgrad) or 2.8× (Moongrad) optical zoom, 2× line averaging, and a pinhole diameter of 1 Airy Units. All images were collected at 8 Bit in sequential mode, and optical stacks were acquired at 1 µm (ALFAgrad) or 1.5 µm (Moongrad) spacing. Fluorophores were excited using the 405 laser line (DAPI) at 11% (ALFAgrad) or 32.93% (Moongrad) laser power, the 561 laser line (tdTomato) at 12.95% (ALFAgrad) or 30.2% (Moongrad) laser power, and the 488 laser line (EGFP) at 5.5% (ALFAgrad) or 9.75% (Moongrad) laser power. Raw images were deconvolved using the Airyscan software. Representative images of individual embryos are shown in Figs. 2 and S5.
Quantification of mouse reporters
Two-cell mouse embryos were microinjected, with one cell serving as a direct control to the second cell that was injected with the degrader. Image analysis was performed using the IMARIS software (Bitplane, Oxford Instruments, Concord MA; Version 10.0; RRID:SCR_007370). Nuclei were segmented using the ‘spots’ module based on DAPI signal, with a diameter of 10 μm. Membrane fluorescence was sampled for each cell by creating 2 μm (xy) × 4 μm (z) spots based on the tdTomato fluorescence. Shared membrane regions between two or four cells were excluded, leaving only unique membrane regions for each cell for analysis. To correct for background fluorescence, spots of equivalent volume to nuclei and membrane spots were placed outside the cells in both channels. The median background fluorescence in both fluorescence channels was subtracted from all measurements. At the time of fixation, most embryos remained at the 2-cell stage, although a few from the 12-h time point had reached the 4-cell stage. For embryos analyzed at the 2-cell stage, the nuclear sum fluorescence for each cell was divided by its median membrane sum fluorescence. The nuclear-to-membrane ratio of the control cell was set to 1, with the degrader-injected cell ratio adjusted relative to this value. For embryos at the 4-cell stage, the nuclear sum fluorescence of each cell was divided by its median membrane sum fluorescence. The average ratio of the two control cells was set to 1, and the degrader-injected cell ratios were adjusted accordingly. To assess the variation in nuclear GFP levels at the 2-cell stage (following zygote injection with the split reporter but prior to ALFAgrad injection), fluorescence was quantified as described above for the time point 0. The cell with the higher nuclear-to-membrane ratio was designated as Cell A, while the cell with the lower ratio was designated as Cell B (see Fig. 2o). This approach allowed us to determine the maximum potential bias that could influence downstream measurements (assuming every cell injected with degron corresponded to “Cell B”). However, this value is likely an overestimate, as the degron injection was performed randomly.
Degradation kinetics
Heatshock transgene
Embryos derived from a male Tg(hsp70l:ALFAgrad) hemizygous outcross were injected at the 1-cell stage with 1 nL of a mixture of 25 pg ALFA-EGFP mRNA and 0.04% Tetramethylrhodamine-Dextran (Invitrogen, Cat# D1816). Embryos were screened at 24 hpf for presence or absence of myl7:EGFP transgene reporter expression and sorted. Transgene-positive and negative embryos were heatshocked at 38 °C for 1 h, mounted in 0.7% low-melt agarose (GPG/LE AmericanBio, CAS: 9012-36-6) against a no. 1.5 cover slip and placed in a preheated (28 °C) confocal chamber. Two uninjected embryos were mounted for background autofluorescence normalization. Embryos were imaged simultaneously on an upright Zeiss LSM 980 confocal microscope with an Airyscan 2 detector (RRID:SCR_025048) and an EC Plan-Neofluar 10×/0.3 M27 objective with bidirectional line scanning at a format of 1699 × 1699 pixels and 2.5× optical zoom resulting in an image size of 337.62 × 337.62 µm. Z-stacks were taken starting at 20 min post-heatshock and captured every 10 min for 5 h using the multiple-position feature. All images were collected at 16 Bit in aryscan mode, and optical stacks were acquired at 3.4 µm spacing and 34 slices for a total depth of 112.2 µm; LSM Scan Speed 9 (pixel dwell time 0.31 μs) and a 131 ms frame time. Fluorophores were excited using the 488 laser line (EGFP) at 8% laser power and 561 laser line (Tetramethylrhodamine-Dextran) at 3% laser power. Raw images were deconvolved using the Airyscan software. Representative maximum projections of individual embryos are shown in Fig. 2.
Quantification
The quantification of signal intensities was performed as previously described37 using FIJI. In summary, the Rhodamine dextran channel was duplicated, a Gaussian Blur (sigma = 1) was applied and a mask was generated using the FIJI Default Thresholding. This mask was applied to the EGFP and the Rhodamine dextran channel to extract the signal intensities of the masked regions. The extracted fluorescence signal was sum projected and the mean intensity of each time point was recorded. The average background signal was quantified from two uninjected embryos using the same method as described, and their mean fluorescence intensities were subtracted from the mean intensity values obtained from injected embryos at each time point. The EGFP to Rhodamine dextran ratio was calculated for each time point and graphed on a scale between 1 and 0.
2-cell live imaging degradation kinetics
Wildtype embryos were injected at the 1-cell stage with 1 nL of a 25 pg ALFA-EGFP mRNA mixture and 0.04% Tetramethylrhodamine-Dextran (Invitrogen, Cat# D1816). At the 2-cell stage, 1 of the 2 cells was reinjected with 50 pg ALFAgrad mRNA. Embryos were mounted at the 4-cell stage in 0.7% low-melt agarose (GPG/LE AmericanBio, CAS: 9012-36-6) and placed in a preheated (28 °C) confocal chamber. Time-lapse images were acquired using an upright Zeiss LSM 980 confocal microscope with an Airyscan 2 detector (RRID:SCR_025048) and an EC Plan-Neofluar 10×/0.3 M27 objective with bidirectional line scanning and 1.0× optical zoom. In detail, images were obtained at 16 Bit in Aryscan mode, LSM scan speed 6 (pixel dwell time 0.51 μs), and using an image size of 4086 × 4086 pixel corresponding to an image size of 825.74 × 825.74 μm. EGFP was excited using the 488 nm laser line at 4% laser power and the Tetramethylrhodamine-Dextran was excited using the 561 laser line at 8% laser power. Three-dimensional optical sections were acquired at 2 μm distance, a final depth of 130 μm and a final temporal resolution of 9 min in between time frames. Representative maximum projections of individual embryos are shown in Figs. 2 and S4.
2-cell live imaging degradation kinetics quantification
The fluorescence intensity was quantified in 3D using the IMARIS software (Bitplane, Oxford Instruments, Concord, MA; Version 10.0; RRID:SCR_007370) with the ‘spot’ function. Multiple spheres with a diameter of 12 μm were placed within the degron and control cell regions in the embryo, and their total fluorescence in the Rhodamine dextran and EGFP channels were measured. The extracted fluorescence data were exported to R-Studio for further analysis. The average Rhodamine dextran fluorescence was calculated for each time point and all measurements at that time point were normalized accordingly using a normalization factor. The median fluorescence intensity was then determined for the degron and control regions at each time point for Rhodamine dextran and EGFP channels. Subsequently, the EGFP:Rhodamine dextran fluorescence ratio was calculated for each time point in both areas. To facilitate comparison, the starting value of each replicate dataset was set to 1, and all ratios were normalized accordingly.
Endogenous ALFA-tagged protein imaging
Wildtype, homozygous ALFA-nanogKI/KI and ALFA-vangl2KI/KI embryos were dechorionated in 0.83 mg/ml pronase (Sigma Aldrich, Cat# P5147, CAS: 9036-06-0) and injected at the 1-cell stage with 25 pg membrane TagBFP2 (Addgene #55295) and 50 pg EGFP-NbALFA. Embryos were incubated at 28 °C until they reached 30% epiboly (~5 hpf) and mounted in 0.8% low-melt agarose (GPG/LE AmericanBio, CAS: 9012-36-6) in system water against a no. 1.5 cover slip (Thermo Fisher Scientific, Cat# 22-050-246). Embryos were imaged on a temperature controlled (28 °C) upright Zeiss LSM 980 confocal microscope with an Airyscan 2 detector (RRID:SCR_025048) and a Plan-Apochromat 20×/0.8 M2 objective with, bidirectional line scanning at a format of 4060 × 4060 pixels and 1.4× optical zoom. All images were collected at 16 Bit in aryscan mode, and optical stacks were acquired at 0.3 µm spacing. Fluorophores were excited using the 408 laser line (mTagBFP2) at 2% laser power and 488 laser line (EGFP) at 1% laser power. Raw images were deconvolved using the Airyscan software. Individual representative slices of the enveloping layer are shown in Fig. 3.
Endogenous ALFA-tagged protein depletion
Wildtype, ALFA-nanogKI/KI, ALFA-nanogKI/+, ALFA-vangl2KI/KI, ALFA-vangl2KI/+, VHH05-nanogKI/KI, VHH05-vangl2KI/KI, 127d01-nanogKI/KI, 127d01-vangl2KI/KI, ALFA-dicerKI/KI and zygotic as well as maternal-zygotic ALFA-pou5f3KI/KI embryos were dechorionated in 0.83 mg/ml pronase (Sigma Aldrich, Cat# P5147, CAS: 9036-06-0) and injected at the 1-cell stage with 100 pg (for ALFA-nanog and ALFA-vangl2), 150 pg (for ALFA-pou5f3 and ALFA-dicer) ALFAgrad mRNA, 50–150 pg VHH05grad mRNA (for VHH05-nanog and VHH05-vangl2) or 50–150 pg 127d01grad mRNA (for 127d01-nanog and 127d01-vangl2). ALFA-nanogKI/KI, embryos from ALFA-nanogKI/KI incrosses or outcrosses (male knock-ins crossed to wildtype for heterozygous controls) were incubated at 28 °C and scored at 6 hpf for the nanog loss-of-function phenotype (gastrulation failure44,45). For ALFA-vangl2KI/KI, embryos from ALFA-vangl2KI/KI incrosses or outcrosses (male knock-ins crossed to wildtype for heterozygous controls) were incubated at 28 °C and scored at 24 hpf for the vangl2 loss-of-function phenotype (shortened body axis46). For ALFA-dicerKI/KI, embryos from ALFA-dicer/KI incrosses were incubated at 28 °C and scored at 24 hpf for the dicer loss-of-function phenotype (shortened body axis, brain morphogenesis defects51). For ALFA-pou5f3KI/KI, embryos from female ALFA-pou5f3KI/+ to male ALFA-pou5f3KI/KI (to generate 50% zygotic (Z) clutches) or ALFA-pou5f3KI/KI incrosses (to generate maternal-zygotic (MZ) clutches) or were incubated at 28 °C and scored at 24 hpf for the Zpou5f3 loss-of-function phenotype (failure to form midbrain-hindbrain boundary47,48) or the MZpou5f3 phenotype (gastrulation and axis formation defects49,50). Zygotic knock-in embryos in Fig. 3f were imaged and subsequently genotyped to confirm the correct genotype.
microRNA sensors
Uninjected and ALFAgrad-injected ALFA-dicerKI/KI embryos were injected at the 1-cell stage with either (1) 100 pg control non-targeted GFP-F-3 × Perfect Target miR-204 sensor and 100 pg dsRed mRNA, or (2) GFP-F-3 × Imperfect Target miR-430 sensor and 100 pg dsRed mRNA (as previously described by Giraldez et al. 2005). Embryos were incubated at 28 °C and imaged side-by-side at 24 hpf for GFP and dsRed fluorescence on an Olympus MVX10 fluorescence microscope with a DP74 camera with identical exposures for all images.
Western blot
Comparison to nanog overexpression
Wildtype embryos were injected with 25 pg or 50 pg of ALFA-nanog mRNA at the 1-cell stage and incubated at 28 °C until they reached 50% epiboly. 25 embryos from uninjected, 25 pg injected, 50 pg injected and stage-matched ALFA-nanogKI/KI were batch deyolked in deyolking buffer (55 mM NaCl, 1.8 mM KCl, 1.25 mM NaHCO3) and snap frozen in liquid nitrogen. The embryos were boiled in sample buffer (1× NuPAGE™ LDS Sample Buffer; Thermo Fisher Scientific, Cat# NP0007 and 1× 10× NuPAGE™ Sample Reducing Agent; Thermo Fisher Scientific, Cat# NP0009) for 10 min. Samples were resolved on a 4–12% NuPAGE Bis-Tris gel (Thermo Fisher Scientific, Cat# NP0335BOX) in NuPAGE MOPS Running Buffer (Thermo Fisher Scientific, Cat# NP0001) and transferred to a nitrocellulose membrane with the iBlot 2 Gel Transfer Device (Thermo Fisher Scientific, Cat# IB21001). The membrane was blocked in 5% milk (American bio, Cat# AB10109-01000) in 1 × PBS with 0.1% Tween-20 (PBS-Tw) and then cut at ~25 kDa. The membranes were incubated with primary antibody solution (1:1000 anti-ALFA antibody (NanoTag Biotechnologies, Cat# N1583; RRID:AB_3075998) for the top (>25 kDa) membrane, 1:5000 anti-H3 antibody (Abcam, Cat# ab1791; RRID:AB_302613) for the bottom (<25 kDa) membrane) prepared in blocking solution, and then incubated with secondary antibody solution (1:10000 of horseradish peroxidase-conjugated anti-rabbit antibody (Abcam, #Cat ab671; RRID:AB_955447)). The Super-Signal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific, Cat# 34580) was used for protein detection. FIJI (Fiji is just ImageJ; RRID:SCR_002285)78 software was used for densitometry to assess levels of ALFA-tagged Nanog in different samples normalized to histone H3 loading control.
Timecourse
For each time point, 15 ALFA-nanogKI/KI or HA-nanogKI/KI embryos were dechorionated by hand in Ringer’s solution (116 mM NaCl, 2.9 mM KCl, 1.8 mM CaCl2, 5.0 mM HEPES, pH 7.2) and rinsed in 1× PBS. The embryos were boiled in sample buffer (1× NuPAGE™ LDS Sample Buffer; Thermo Fisher Scientific, Cat# NP0007 and 1× 10× NuPAGE™ Sample Reducing Agent; Thermo Fisher Scientific, Cat# NP0009) for 10 min. The samples were resolved on a 4–12% NuPAGE Bis-Tris gel (Thermo Fisher Scientific, Cat# NP0335BOX) in NuPAGE MOPS Running Buffer (Thermo Fisher Scientific, Cat# NP0001) and transferred to a nitrocellulose membrane with the iBlot 2 Gel Transfer Device (Thermo Fisher Scientific, Cat# IB21001). The membrane was blocked in 5% milk (American bio, Cat# AB10109-01000) in 1× PBS-Tw and then cut at ~25 kDa. The membranes were incubated with primary antibody solution prepared in block solution (1:1000 anti-ALFA antibody (NanoTag Biotechnologies, Cat# N1583; RRID:AB_3075998) or anti-HA antibody (Cell Signaling Technology, Cat# 3724; RRID:AB_1549585) for the top (>25 kDa) membrane, 1:5000 anti-H3 antibody (Abcam, Cat# ab1791; RRID:AB_302613) for the bottom (<25 kDa) membrane), and then incubated with secondary antibody solution (1:10000 of horseradish peroxidase-conjugated anti-rabbit antibody (Abcam, #Cat ab671; RRID:AB_955447)). The Super-Signal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific, Cat# 34580) was used for protein detection.
Chromatin immunoprecipitation sequencing
Spike-in Chromatin Immunoprecipitation Sequencing (ChIP-seq) was performed with modifications based on previously established protocols31,79,80. Briefly, wildtype and ALFA-nanogKI/KI embryos were dechorionated at the 1-cell stage (2 wildtype replicates and 2 ALFA-nanogKI/KI replicates). At 4 hpf, ~600 embryos per group were fixed with 1.9% paraformaldehyde (Electron Microscopy Sciences, Cat# 50-980-487) for 15 min at room temperature, quenched with 0.125 M glycine for 5 min, washed 3 times with cold 1× PBS, and snap-frozen with liquid nitrogen. The embryos were homogenized and lysed for 15 min in cell lysis buffer (10 mM Tris–HCl pH 7.5, 10 mM NaCl, 0.5% IGEPAL (Sigma Aldrich, Cat# I8896; CAS: 9002-93-1), protease inhibitor cocktail (Sigma Aldrich, Cat# 11873580001)) on ice. Nuclei were pelleted by centrifugation for 5 min at 1200 × g at 4 °C. Nuclei were then lysed with nuclei lysis buffer (50 mM Tris–HCl pH 7.5, 10 mM EDTA, 1% SDS, protease inhibitor cocktail (Sigma Aldrich, Cat# 11873580001)) on ice, diluted with IP dilution buffer (16.7 mM Tris–HCl pH 7.5, 167 mM NaCl, 1.2 mM EDTA, 0.01% SDS, protease inhibitor cocktail (Sigma Aldrich, Cat# 11873580001)), and sonicated (15 cycles of sonication with 30 s ON and 30 s OFF, 15 mins in ice, and another 15 cycles of sonication; Bioruptor Pico sonication device (Diagenode, Cat# B01080010). 8 μl of 10% Triton X-100 (Sigma Aldrich, Cat# X100; CAS: 9036-19-5) was added per 100 μl of sonicated chromatin to the chromatin suspension, which was then centrifuged for 10 min at 18500 × g at 4 °C. 5% of the supernatant was taken as input and stored at −80 °C until use. 50 ng spike-in chromatin (Active Motif, Cat# 53083) was mixed with the chromatin supernatant from each sample. 25 µl of Protein G Dynabeads (Thermo Fisher Scientific, Cat# 10003D) were washed with 0.5% Bovine Serum Albumin (BSA; Sigma Aldrich, Cat# A9647; CAS:9048-46-8) in 1 × PBS, incubated with 4 μg ALFA antibody (Nanotag Biotechnologies, Cat# N1583; RRID:AB_3075998) and 2 μg spike-in antibody (Active Motif, Cat# 61686; RRID:AB_2737370) overnight at 4 °C, washed three times with cold 0.5% BSA/PBS, and added to the chromatin mix to incubate overnight at 4 °C. The beads were washed five times with cold RIPA wash buffer (50 mM HEPES pH 7.6, 1 mM EDTA, 0.7% DOC, 1% IGEPAL (Sigma Aldrich, Cat# I8896; CAS: 9002-93-1), 0.5 M LiCl), two times with Tris-buffered saline (50 mM Tris pH 7.5, 150 mM NaCl) before eluted with elution buffer at 65 °C for 15 min (50 mM NaHCO3, 1% SDS). Both the input and ChIP samples were purified for sequencing: reverse-crosslinking at 65 °C overnight, RNase A (0.33 µg/µl; Thermo Fisher Scientific, Cat# EN0531) treatment at 37 °C for 2 h, Proteinase K (0.2 µg/µl; Thermo Fisher Scientific, Cat# EO0492) treatment at 55 °C for 2 h, and purification with ChIP DNA Clean & Concentrator (Zymo Research, Cat# D5205). Library preparation (Illumina TruSeq protocol) and sequencing (Illumina NovaSeq 6000 System, pair-end) were performed by the Yale Center for Genome Analysis.
Spike-in ChIP-seq analysis
The Spike-in ChIP-seq data were managed with the LabxDB81 and analyzed as previously described82. Raw reads were mapped to the combined genome (zebrafish GRCz11 genome + fly BDGP6.32 genome) sequence83 with LabxPipe (https://github.com/vejnar/LabxPipe) using Bowtie2 (option: –no-unal, –no-discordant, –no-mixed, version: 2.5.1; RRID:SCR_016368)84 after adapter-trimming (using ReadKnead, https://github.com/vejnar/ReadKnead). Reads were filtered (deduplicated, and only uniquely mapped reads (MAPQ ≥ 30) were kept) using SAMtools (RRID:SCR_002105)85 before any downstream analysis. Filtered BAM files were split into four BAM files using split_bam.py80, and zebrafish.bam was used to call ALFA-Nanog binding peaks in zebrafish. Narrow peaks were called with input data as controls from the filtered BAM files (significance cut-off: q = 0.05; options: “-f BAMPE”) using MACS386. Peaks were filtered by removing the ones that overlap with peaks called from their cognate input data (narrow peaks; significance cut-off: q = 0.05); overlapping peaks are defined using BEDTools (RRID:SCR_006646)87 with the default minimum overlap. The two ALFA-nanogKI/KI replicates were merged, and then peaks were called from the merged data (significance cut-off: q = 0.05). Peaks called from the merged data that overlap with peaks called from the two biological replicates individually were defined as confident binding peaks. To plot the correlation between replicates, the spike-in normalized average ChIP-seq signal (normalized using spike-in reads as previously described82 was calculated across each 5 kb window of the zebrafish genome; the Pearson correlation was then calculated on all genomic windows. Genomic tracks were created using the LabxPipe trackhub option, which uses GeneAbacus (https://github.com/vejnar/GeneAbacus) to compute fragment coverage, as well as utilities from the UCSC genome browser88. Signal intensity in spike-in normalized tracks is in spike-in scaling factor corrected RPM82.
To further validate our endogenous ALFA-Nanog ChIP-seq (4 hpf), we also analyzed the 4.5 hpf Nanog-Myc ChIP-seq that was done by overexpressing Myc-tagged Nanog in WT embryos53. Raw reads were mapped to the zebrafish GRCz11 genome sequence83 with LabxPipe (https://github.com/vejnar/LabxPipe) using Bowtie2 (option: –no-unal, –no-discordant, –no-mixed, version: 2.5.1; RRID:SCR_016368)84 after adapter trimming (using ReadKnead, https://github.com/vejnar/ReadKnead). Reads were filtered (deduplicated, and only uniquely mapped reads (MAPQ ≥ 30) were kept) using SAMtools (RRID:SCR_002105)85 before any downstream analysis. Reads from the single-end Nanog-Myc ChIP-seq53 were extended to 200 nt for all downstream analyses. Genomic tracks were created using the LabxPipe trackhub option, which uses GeneAbacus (https://github.com/vejnar/GeneAbacus) to compute fragment coverage (normalized to the total fragments per million fragments). Narrow peaks were called with input data as controls from the filtered BAM files (significance cut-off: q = 0.05; options: “-f BAM –nomodel –extsize 200”) using MACS386. Raw reads generated in this study are publicly available in the Sequence Read Archive (SRP547050).
Immunofluorescence of ALFA-Nanog
Wildtype and ALFA-nanogKI/KI embryos were dechorionated in 0.83 mg/ml pronase (Sigma Aldrich, Cat# P5147, CAS: 9036-06-0) at the 1-cell stage. Wildtype embryos were injected with 50 pg ALFA-nanog mRNA. All embryos were grown at 28 °C until they reached sphere stage (~4 hpf). Embryos were fixed in 4% PFA (Electron Microscopy Sciences, Cat# 50-980-487) in 1 × PBS overnight at 4 °C, followed by 3 washes in 1 × PBS with 0.1% Triton-X (PBS-Tx; Sigma Aldrich, Cat# X100; CAS: 9036-19-5) for 10 min at room temperature. Embryos were blocked in 10% Normal Goat Serum (NGS) (Thermo Fisher Scientific, Cat# 50062Z) for 2 h with gentle rotation and incubated with primary antibody solution (1:1000 anti-ALFA antibody; NanoTag Biotechnologies, Cat# N1583; RRID:AB_3075998) prepared in 10% NGS overnight at 4 °C. Embryos were washed 3 times for 30 min in 0.1% PBS-Tx, followed by incubation with secondary antibody solution (1:1000 Goat anti-Rabbit Alexa Fluor 488; Thermo Fisher Scientific, Cat# A-11008; RRID:AB_143165) and DAPI (5 ng/ml; Thermo Fisher Scientific, Cat# D1306) with gentle rotation for 1 h at room temperature protected from light. Embryos were washed 3 times for 30 min in PBS-Tx and mounted in 0.8% low-melt agarose (GPG/LE AmericanBio, CAS: 9012-36-6) on glass-bottom dishes (MatTek, Cat# P35GC-1.5-14-C). Embryos were imaged on an upright Zeiss LSM 980 confocal microscope with an Airyscan 2 detector (RRID:SCR_025048) and a Plan-Apochromat 20×/0.8 M2 objective with bidirectional line scanning at a format of 4084 × 4084 pixels and 1.3× optical zoom. All images were collected at 16 Bit in aryscan mode, and optical stacks were acquired at 3.08 µm spacing. Fluorophores were excited using the 405 laser line (DAPI) at 0.5% laser power and 488 laser line (EGFP) at 7.5% laser power. Raw images were deconvolved using the Airyscan software. Representative maximum intensity projections of individual embryos are shown in Fig. S6.
Quantification
Nuclear Nanog concentration was determined based on ALFA-Ab immunofluorescence staining. Nuclei were segmented in the IMARIS software (Bitplane, Oxford Instruments, Concord, MA; Version 10.0; RRID:SCR_007370) using the ‘surface’ module based on the DAPI channel. The sum fluorescence in the Ab-ALFA channel of each nucleus was adjusted for the respective nuclear volume and background corrected to account for background variation between experiments. The median background fluorescence for each embryo was determined by placing 3 × 3 µm spheres using the ‘spot’ function randomly within the embryo (excluding nuclear regions).
Live image acquisition
For live imaging of Nanog, ALFA-nanogKI/KI, ALFA-nanogKI/KI; Tg(actb2:EGFP-NbALFA)/+ and wildtype embryos were dechorionated in 0.83 mg/ml pronase (Sigma Aldrich, Cat# P5147, CAS: 9036-06-0). Dechorionated ALFA-nanogKI/KI embryos were injected with 75 pg of the EGFP-NbALFA GEAR-binder at the 1-cell stage. ALFA-nanogKI/KI; Tg(actb2:EGFP-NbALFA)/+ embryos did not require any microinjection prior to imaging. Wiltype embryos were injected with 25 pg exogenous ALFA-nanog mRNA together with 75 pg of the EGFP-NbALFA GEAR-binder or 25 pg exogenous nanog-mEmerald54 reporter. Embryos were grown at 28 °C and mounted in 0.8% low-melt agarose (GPG/LE AmericanBio, CAS: 9012-36-6) in system water at the 4-cell stage against a no. 1.5 cover slip (Thermo Fisher Scientific, Cat# 22-050-246). Temperature-controlled live imaging (28 °C) was performed using an upright Zeiss LSM 980 confocal microscope with an Airyscan 2 detector (RRID:SCR_025048) and a LD LCI Plan-Apochromat 40×/1.2 Imm Corr DIC M2 objective with water immersion. Time series were acquired starting at the 8-cell stage and confocal stacks were readjusted during mitosis of every cleavage cycle. In detail, images were obtained at 16 Bit in aryscan mode with 2× line averaging, bidirectional scanning, LSM scan speed 8 (pixel dwell time 0.37 μs), 2× optical zoom and using an image size of 2124 × 2124 pixel corresponding to an image size of 105.47 × 105.47 μm. EGFP was excited using the 488 nm laser line at 2.8% laser power. Three-dimensional optical sections were acquired at 1 μm distance, a final depth of 31 μm and a final temporal resolution of 32 s per time frame.
Maximum intensity projections of 3D stacks are shown in Figs. 4 and S7. Embryos were imaged for 120–180 min, which included the beginning of the 1k cell stage.
Imaging of Nanog in conjunction with elongating Pol II (Pol II pSer2 mintbody54,58) was carried out using the microscope setup described above with the following alterations. Injection of 75 pg mScarlet-i3-NbALFA GEAR-binder and 25 pg pSer2-EGFP mintbody into ALFA-nanogKI/KI embryos was performed after dechorionation at the 1-cell stage. In total, 61 planes were acquired with 1 μm spacing (total 60 μm), optical zoom of 1.5, a z-stack acquisition time of ∼147 s and a format of 2848 × 2848 pixels corresponding to an image size of 140.83 × 140.83 μm. mScarlet-i3 was excited by the 561 nm laser line at 1.1% laser power and the mintbody by the 488 nm laser line at 2.4%.
Live imaging analysis
During image analysis, all datasets were adjusted in time to account for slight temperature differences during imaging that can alter the speed of development. Therefore, each cell cycle was aligned to begin with the completion of telophase and end with chromatin decondensation.
Nanog foci quantification
Nanog foci segmentation from live imaging data was conducted using IMARIS software (Bitplane, Oxford Instruments, Concord, MA; Version 10.0; RRID:SCR_007370). Foci were defined by an x-y diameter of 0.5 µm and a z-length of 1 µm and identified via the ‘spot’ function. Thresholding was applied to distinguish spots based on fluorescence intensity relative to the background. To quantify the total number of Nanog clusters (Figs. 4d and S7j) we report the number of total clusters detected at time points from 50 to 75% of the cell cycle (Note: If the same clusters are present in consecutive imaging frames, they are counted more than once, leading to an additive effect). We chose this time window as this is where we observe the maximum number of Nanog clusters (time span indicated by shading in Fig. S7i).
Nanog foci distance to active transcription sites
Analysis was performed in IMARIS software (Bitplane, Oxford Instruments, Concord, MA; Version 10.0; RRID:SCR_007370). First, nuclei were segmented using the’ surface’ module based on their nuclear fluorescence intensity. Each nucleus was then “immobilized” through drift correction in x/y/z dimensions based on its centroid. Nanog foci and transcription foci were segmented using the ‘spots’ module and their x/y/z centroid position was extracted. The shortest distance in 3D between Nanog foci and their respective closest transcription spot was computed.
Statistical analysis
Statistical comparisons were performed using nonparametric Kruskal–Wallis tests with and without Dunn’s multiple comparisons test and two-tailed nonparametric Mann–Whitney tests in GraphPad Prism 10. ChIP-seq analysis and Pearson Correlation was performed using Python (Version 3.11; RRID:SCR_008394). Statistical test and sample sizes can be found in Figure legends. Statistical significance was assumed by p < 0.05. Individual p-value ranges are indicated in the Figure legends. The total number of embryos analyzed is represented by N. Individual data points such as cells or nuclei are represented by n numbers. The detailed number of embryos (N) and number of cells/nuclei (n) for each experiment can be found in the respective figure legend.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the findings of this study are available from the corresponding authors upon request or in the Source Data file. Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Curtis W. Boswell (curtis.boswell@yale.edu). All plasmids will be available on Addgene. Zebrafish lines generated in this study are available from the Lead Contact on request. Raw ChIP sequence reads generated in this study are available in the Sequence Read Archive (SRP547050). Source data are provided with this paper.
References
Moriya, H. Quantitative nature of overexpression experiments. Mol. Biol. Cell 26, 3932–3939 (2015).
Eguchi, Y. et al. Estimating the protein burden limit of yeast cells by measuring the expression limits of glycolytic proteins. Elife 7, https://doi.org/10.7554/eLife.34595 (2018).
Xu, R. & Du, S. Overexpression of lifeact-GFP disrupts F-actin organization in cardiomyocytes and impairs cardiac function. Front. Cell Dev. Biol. 9, 746818 (2021).
Peng, Y. et al. Making designer mutants in model organisms. Development 141, 4042–4054 (2014).
Rasys, A. M. et al. CRISPR-Cas9 gene editing in lizards through microinjection of unfertilized oocytes. Cell Rep. 28, 2288–2292.e3 (2019).
Momose, T. et al. High doses of CRISPR/Cas9 ribonucleoprotein efficiently induce gene knockout with low mosaicism in the hydrozoan Clytia hemisphaerica through microhomology-mediated deletion. Sci. Rep. 8, 11734 (2018).
Dickinson, D. J., Ward, J. D., Reiner, D. J. & Goldstein, B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10, 1028–1034 (2013).
Hisano, Y. et al. Precise in-frame integration of exogenous DNA mediated by CRISPR/Cas9 system in zebrafish. Sci. Rep. 5, 8841 (2015).
Gu, B., Posfai, E. & Rossant, J. Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos. Nat. Biotechnol. 36, 632–637 (2018).
Boel, A. et al. CRISPR/Cas9-mediated homology-directed repair by ssODNs in zebrafish induces complex mutational patterns resulting from genomic integration of repair-template fragments. Dis. Model. Mech. 11, https://doi.org/10.1242/dmm.035352 (2018).
Hoshijima, K., Jurynec, M. J. & Grunwald, D. J. Precise editing of the zebrafish genome made simple and efficient. Dev. Cell 36, 654–667 (2016).
Strickland, D. et al. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat. Methods 9, 379–384 (2012).
Nishimura, K., Fukagawa, T., Takisawa, H., Kakimoto, T. & Kanemaki, M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6, 917–922 (2009).
Caussinus, E., Kanca, O. & Affolter, M. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat. Struct. Mol. Biol. 19, 117–121 (2012).
Tang, J. C. Y. et al. A nanobody-based system using fluorescent proteins as scaffolds for cell-specific gene manipulation. Cell 154, 928–939 (2013).
Xiong, Z. et al. In vivo proteomic mapping through GFP-directed proximity-dependent biotin labelling in zebrafish. Elife 10, https://doi.org/10.7554/eLife.64631 (2021).
Zhang, Z. Generation of epitope tag knock-in mice with CRISPR-Cas9 to study the function of endogenous proteins. STAR Protoc. 4, 102518 (2023).
Carrington, B., Ramanagoudr-Bhojappa, R., Bresciani, E., Han, T.-U. & Sood, R. A robust pipeline for efficient knock-in of point mutations and epitope tags in zebrafish using fluorescent PCR based screening. BMC Genomics 23, 810 (2022).
Paix, A. et al. Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Proc. Natl. Acad. Sci. USA 114, E10745–E10754 (2017).
Los, G. V. et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 3, 373–382 (2008).
Zhao, N. et al. A genetically encoded probe for imaging nascent and mature HA-tagged proteins in vivo. Nat. Commun. 10, 2947 (2019).
Liu, Y. et al. Visualizing looping of two endogenous genomic loci using synthetic zinc-finger proteins with anti-FLAG and anti-HA frankenbodies in living cells. Genes Cells 26, 905–926 (2021).
Tanenbaum, M. E., Gilbert, L. A., Qi, L. S., Weissman, J. S. & Vale, R. D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646 (2014).
Götzke, H. et al. The ALFA-tag is a highly versatile tool for nanobody-based bioscience applications. Nat. Commun. 10, 4403 (2019).
Ling, J. et al. A nanobody that recognizes a 14-residue peptide epitope in the E2 ubiquitin-conjugating enzyme UBC6e modulates its activity. Mol. Immunol. 114, 513–523 (2019).
Bradley, M. E. et al. Potent and efficacious inhibition of CXCR2 signaling by biparatopic nanobodies combining two distinct modes of action. Mol. Pharmacol. 87, 251–262 (2015).
Boersma, S. et al. Multi-color single-molecule imaging uncovers extensive heterogeneity in mRNA decoding. Cell 178, 458–472.e19 (2019).
Friedman, N., Barzily-Rokni, M., Isaac, S. & Eden, A. The histone H2A variant macroH2A1 does not localize to the centrosome. PLoS ONE 6, e17262 (2011).
Lukinavičius, G., Lavogina, D., Gönczy, P. & Johnsson, K. [Letter to the editor]: Commercial Cdk1 antibodies recognize the centrosomal protein Cep152. Biotechniques 55, 111–114 (2013).
Aleström, P. et al. Zebrafish: housing and husbandry recommendations. Lab. Anim. 54, 213–224 (2020).
Miao, L. et al. The landscape of pioneer factor activity reveals the mechanisms of chromatin reprogramming and genome activation. Mol. Cell 82, 986–1002.e9 (2022).
Jussila, M., Boswell, C. W., Griffiths, N. W., Pumputis, P. G. & Ciruna, B. Live imaging and conditional disruption of native PCP activity using endogenously tagged zebrafish sfGFP-Vangl2. Nat. Commun. 13, 5598 (2022).
Shaner, N. C. et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10, 407–409 (2013).
Bindels, D. S. et al. mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat. Methods 14, 53–56 (2017).
Subach, O. M., Cranfill, P. J., Davidson, M. W. & Verkhusha, V. V. An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PLoS ONE 6, e28674 (2011).
Wang, S. et al. A toolkit for GFP-mediated tissue-specific protein degradation in C. elegans. Development 144, 2694–2701 (2017).
Yamaguchi, N., Colak-Champollion, T. & Knaut, H. zGrad is a nanobody-based degron system that inactivates proteins in zebrafish. Elife 8, e43125 (2019).
Arnold, B. et al. hGRAD: A versatile “one-fits-all” system to acutely deplete RNA binding proteins from condensates. J. Cell Biol. 223, e202304030 (2024).
Rozov, S. M., Permyakova, N. V. & Deineko, E. V. The problem of the low rates of CRISPR/Cas9-mediated knock-ins in plants: approaches and solutions. Int. J. Mol. Sci. 20, 1–12 (2019).
Yao, X. et al. Generation of knock-in cynomolgus monkey via CRISPR/Cas9 editing. Cell Res. 28, 379–382 (2018).
Lawson, N. D. & Weinstein, B. M. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307–318 (2002).
Shin, J., Chen, J. & Solnica-Krezel, L. Efficient homologous recombination-mediated genome engineering in zebrafish using TALE nucleases. Development 141, 3807–3818 (2014).
DiNapoli, S. E. et al. Synthetic CRISPR/Cas9 reagents facilitate genome editing and homology directed repair. Nucleic Acids Res. 48, e38–e38 (2020).
Gagnon, J. A., Obbad, K. & Schier, A. F. The primary role of zebrafish nanog is in extra-embryonic tissue. Development 145, https://doi.org/10.1242/dev.147793 (2018).
Veil, M. et al. Maternal Nanog is required for zebrafish embryo architecture and for cell viability during gastrulation. Development 145, https://doi.org/10.1242/dev.155366 (2018).
Ciruna, B., Jenny, A., Lee, D., Mlodzik, M. & Schier, A. F. Planar cell polarity signalling couples cell division and morphogenesis during neurulation. Nature 439, 220–224 (2006).
Belting, H.-G. et al. Spiel ohne Grenzen/pou2 is required during establishment of the zebrafish midbrain-hindbrain boundary organizer. Development 128, 4165–4176 (2001).
Burgess, S., Reim, G., Chen, W., Hopkins, N. & Brand, M. The zebrafish spiel-ohne-grenzen (spg) gene encodes the POU domain protein Pou2 related to mammalian Oct4 and is essential for formation of the midbrain and hindbrain, and for pre-gastrula morphogenesis. Development 129, 905–916 (2002).
Lunde, K., Belting, H.-G. & Driever, W. Zebrafish pou5f1/pou2, homolog of mammalian Oct4, functions in the endoderm specification cascade. Curr. Biol. 14, 48–55 (2004).
Reim, G. & Brand, M. Maternal control of vertebrate dorsoventral axis formation and epiboly by the POU domain protein Spg/Pou2/Oct4. Development 133, 2757–2770 (2006).
Giraldez, A. J. et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science308, 833–838 (2005).
Kuznetsova, K. et al. Nanog organizes transcription bodies. Curr. Biol. 33, 164–173.e5 (2023).
Xu, C. et al. Nanog-like regulates endoderm formation through the Mxtx2-nodal pathway. Dev. Cell 22, 625–638 (2012).
Pownall, M. E. et al. Chromatin expansion microscopy reveals nanoscale organization of transcription and chromatin. Science 381, 92–100 (2023).
Hadzhiev, Y. et al. The miR-430 locus with extreme promoter density forms a transcription body during the minor wave of zygotic genome activation. Dev. Cell 58, 155–170.e8 (2023).
Chan, S. H. et al. Brd4 and P300 confer transcriptional competency during zygotic genome activation. Dev. Cell 49, 867–881.e8 (2019).
Gadella, T. W. J. et al. mScarlet3: a brilliant and fast-maturing red fluorescent protein. Nat. Methods 20, 541–545 (2023).
Uchino, S. et al. Live imaging of transcription sites using an elongating RNA polymerase II-specific probe. J. Cell Biol. 221, https://doi.org/10.1083/jcb.202104134 (2022).
Hammond, J. W., Blasius, T. L., Soppina, V., Cai, D. & Verhey, K. J. Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J. Cell Biol. 189, 1013–1025 (2010).
Burg, L. et al. Internal epitope tagging informed by relative lack of sequence conservation. Sci. Rep. 6, 1–8 (2016).
Farin, H. F. et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature 530, 340–343 (2016).
Xu, J. et al. Protein visualization and manipulation in Drosophila through the use of epitope tags recognized by nanobodies. Elife 11, e74326 (2022).
Vigano, M. A. et al. Protein manipulation using single copies of short peptide tags in cultured cells and in Drosophila melanogaster. Development 148, https://doi.org/10.1242/dev.191700 (2021).
Vicencio, J. et al. Genome editing in animals with minimal PAM CRISPR-Cas9 enzymes. Nat. Commun. 13, 2601 (2022).
Moreno-Mateos, M. A. et al. CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nat. Commun. 8, 2024 (2017).
Thyme, S. B. & Schier, A. F. Polq-mediated end joining is essential for surviving DNA double-strand breaks during early zebrafish development. Cell Rep. 15, 707–714 (2016).
Ata, H. et al. Robust activation of microhomology-mediated end joining for precision gene editing applications. PLoS Genet. 14, 1–22 (2018).
Aguilar, G. et al. Seamless knockins in Drosophila via CRISPR-triggered single-strand annealing. Dev. Cell 59, 2672–2686.e5 (2024).
Nabet, B. et al. The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 14, 431–441 (2018).
Tovell, H. et al. Rapid and reversible knockdown of endogenously tagged endosomal proteins via an optimized HaloPROTAC degrader. ACS Chem. Biol. 14, 882–892 (2019).
Shin, Y. J. et al. Nanobody-targeted E3-ubiquitin ligase complex degrades nuclear proteins. Sci. Rep. 5, 14269 (2015).
Hayward-Lara, G., Fischer, M. D. & Mir, M. Dynamic microenvironments shape nuclear organization and gene expression. Curr. Opin. Genet. Dev. 86, 102177 (2024).
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B. & Schilling, T. F. Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 (1995).
Diez, M. et al. iCodon customizes gene expression based on the codon composition. Sci. Rep.12, 1–16 (2022).
Kwan, K. M. et al. The Tol2kit: a multisite gateway-based construction kit forTol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088–3099 (2007).
Moreno-Mateos, M. A. et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 12, 982–988 (2015).
Hoppe, C. et al. Modulation of the promoter activation rate dictates the transcriptional response to graded BMP signaling levels in the Drosophila embryo. Dev. Cell 54, 727–741.e7 (2020).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Bogdanović, O., Fernández-Miñán, A., Tena, J. J., de la Calle-Mustienes, E. & Gómez-Skarmeta, J. L. The developmental epigenomics toolbox: ChIP-seq and MethylCap-seq profiling of early zebrafish embryos. Methods 62, 207–215 (2013).
Wu, D., Wang, L. & Huang, H. Protocol to apply spike-in ChIP-seq to capture massive histone acetylation in human cells. STAR Protoc. 2, https://doi.org/10.1016/j.xpro.2021.100681 (2021).
Vejnar, C. E. & Giraldez, A. J. LabxDB: versatile databases for genomic sequencing and lab management. Bioinformatics 36, 4530–4531 (2020).
Li, X. Y., Harrison, M. M., Villalta, J. E., Kaplan, T. & Eisen, M. B. Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. Elife 3, 1–20 (2014).
Yates, A. D. et al. Ensembl 2020. Nucleic Acids Res. 48, D682–D688 (2019).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H., and Subgroup, 1000 Genome Project Data Processing et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Lee, C. M. et al. UCSC genome browser enters 20th year. Nucleic Acids Res. 48, D756–D761 (2020).
Acknowledgements
We thank Sarah Dube, Timothy Gerson, and Damilola Olowookere for zebrafish husbandry, Srikar Krishna, Scott Youlten, Elizabeth Eck, Dié Tang, Linnea Weiss, Odelya Kaufman, Valerie Tornini, Maria Benitez, Ethan Strayer, Haejeong Lee, Fiona Sievers, Gal Jaschek, Ayushi Hegde, Miguel Ángel Delgado Toscano, Ella Callahan, Evie Boswell, and Juniper Mark for research and intellectual support. We thank Christopher Castaldi, Irina Tikhonova, and Bryan Szewczyk from the Yale Center for Genome Analysis for sequencing support. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number 1S10OD030363-01A1. This project was directly supported by the Canadian Institutes of Health Research (CIHR) postdoctoral fellowship to C.W.B., the EMBO (ALTF 794-2021) and HFSP (LT0037/2022-L) postdoctoral fellowships to C.H., A.S. is supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under Award Number K99HD112607. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, National Institutes of Health grant R01 HD100035, and National Institutes of Health grant R35 GM122580 to A.J.G.
Author information
Authors and Affiliations
Contributions
Conceptualization: C.W.B., C.H., A.J.G.; Formal analysis: C.W.B., C.H., L.M., M.K; Investigation: C.W.B., C.H., A.S., L.M., M.L.K.; Resources: N.Z., T.J.S., P.M., S.N.; Writing - Original Draft: C.W.B., C.H., A.J.G; Visualization: C.W.B., C.H., L.M., M.L.K.; Supervision: C.W.B., A.J.G.; Funding acquisition: A.J.G.
Corresponding authors
Ethics declarations
Competing interests
A.J.G. is founder of and has an equity interest in RESA Therapeutics, Inc. All other authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Boswell, C.W., Hoppe, C., Sherrard, A. et al. Genetically encoded affinity reagents are a toolkit for visualizing and manipulating endogenous protein function in vivo. Nat Commun 16, 5503 (2025). https://doi.org/10.1038/s41467-025-61003-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-61003-w