Abstract
Single-cell RNA sequencing (scRNA-seq) facilitates the study of transcriptome diversity in individual cells. Yet, many existing methods lack sensitivity and accuracy. Here we introduce SCALPEL, a Nextflow-based tool to quantify and characterize transcript isoforms from standard 3’ scRNA-seq data. Using synthetic data, SCALPEL demonstrates higher sensitivity and specificity compared to other tools. In real datasets, SCALPEL predictions have a high agreement with other tools and can be experimentally validated. The use of SCALPEL on real datasets reveals novel cell populations undetectable using single-cell gene expression data, confirms known 3’ UTR length changes during cell differentiation, and identifies cell-type specific miRNA signatures regulating isoform expression. Additionally, we show that SCALPEL improves isoform quantification using paired long- and short-read scRNA-seq data. Overall, SCALPEL expands the current scRNA-seq toolkit to explore post-transcriptional gene regulation across species, tissues, and technologies, advancing our understanding of gene regulatory mechanisms at the single-cell level.
Similar content being viewed by others
Introduction
Alternative polyadenylation (APA) is a general mechanism of post-transcriptional regulation that significantly contributes to the diversification of gene expression patterns under diverse physiological and pathological conditions1. APA defines the end of transcripts by selecting one of the available polyA sites (PAS) at the 3’ end of genes, resulting in the generation of multiple mature RNA isoforms from the same pre-mRNA2. These isoforms may have different coding regions or contain distinct 3’ untranslated regions (3’ UTRs), which contain regulatory elements influencing mRNA stability, localization, and translational efficiency3,4,5. Transcriptomic studies have demonstrated that APA is highly regulated in a tissue specific manner6 and plays a crucial role in various biological processes, including cellular differentiation7, development8,9,10, and response to environmental cues11. Alterations in APA patterns have been linked to various diseases, where they can lead to aberrant gene expression and even cancer12,13.
The development of high-throughput single-cell transcriptomics technologies (scRNA-seq) has led to the emergence of computational methods to characterize the transcriptomic profile of thousands of individual cells in a single experiment14. While these methods are mainly used to quantify gene expression, 3’ tag-based scRNA-seq protocols such as Drop-seq15 or 10x Genomics provide opportunities to study 3’ end isoform diversity. Currently, only a few computational tools allow to study isoform diversity generated by APA in scRNA-seq data and most of them face significant drawbacks. They often fail to detect polyadenylation sites (PAS) with low read coverage due to the sparse nature of single-cell data and they lack the precision needed to accurately pinpoint the exact PAS locations, leading to potential misidentification and incomplete characterization of isoform diversity16,17,18,19. Alternative methods based on isoform quantification such as scUTRquant20 have been shown to be more powerful in quantifying transcript diversity from scRNA-seq data. Yet, the main power of this method relies on an improved curated 3’ end annotation that is not available for most species.
Here, we present SCALPEL, a Nextflow workflow21 to quantify isoform expression using commonly used 3’ tag-based scRNA-seq data. Comparison of SCALPEL to other existing tools using synthetic data shows that SCALPEL predictions have a higher sensitivity than other tools while maintaining a high specificity. On real data, SCALPEL predictions show a strong agreement with other tools and can be validated experimentally. Our analysis revealed that isoform-based analysis of single-cell data recapitulates known biological processes such as 3’ UTR lengthening during mouse spermatogenesis, identify novel cell populations, and reflect post-transcriptional regulatory processes such as microRNA function. Furthermore, we also demonstrate how SCALPEL can be used to improve isoform quantification at the single-cell level using paired long and short scRNA-seq data. Together, our work highlights the versatility of SCALPEL across datasets and highlights the power of isoform-based analysis in single-cell studies.
Results
SCALPEL, a new computational tool for isoform quantification using scRNA-seq data
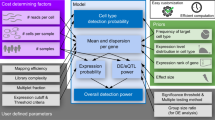
SCALPEL is a new computational Nextflow workflow21 to quantify transcript isoforms from single-cell data. It takes as input the digital gene expression matrix (DGE) generated by a scRNA-seq processing pipeline such as CellRanger or Drop-seq tools and the mapped reads in BAM format and uses them to decompose gene expression into isoform expression data (Fig. 1a). SCALPEL workflow is divided into three main modules (Fig. 1b). In the first module, raw sequencing data and annotation files are processed to perform bulk quantification of the annotated isoforms. These isoforms are then truncated and collapsed, giving rise to a set of distinct isoforms with different 3’ ends for quantification at single-cell resolution. In the second module, scRNA-seq reads are mapped on the set of selected isoforms and reads coming from pre-mRNAs or resulting from internal priming (IP) events are discarded. In the last module, isoforms are quantified in individual cells and an isoform digital gene expression matrix (iDGE) is generated (Fig. S1). The iDGE can be processed to perform downstream single-cell level analyses such as dimensionality reduction, clustering, marker discovery and trajectory inference. Furthermore, it can also be used to study differential isoform usage (DIU) and visualize isoform coverage using additional functions included in SCALPEL repository. The main novelty that SCALPEL brings is the pseudo-assembly of reads with the same cell barcode (CB) and unique molecular identifier (UMI). This approach helps in the assignment of UMIs to individual isoforms by considering the global transcript structure and jointly modeling the distance of the reads with the same UMI to the 3’ end of the transcripts (Fig. S1).
a Schematic representation of SCALPEL function. SCALPEL decomposes conventional scRNA-seq data mapped to a gene to the different isoforms that are expressed. b SCALPEL Nextflow pipeline diagram. SCALPEL is composed of four workflows performing (1) annotation preprocessing (black line); (2) read preprocessing to discard artifacts and reads derived from pre-mRNAs (blue line); (3) quantification of isoforms in individual cells (green line); and (4) characterization of differential isoform usage (orange line).
SCALPEL shows accurate quantification of isoforms at single-cell resolution
To assess the performance of SCALPEL to quantify isoforms in scRNA-seq data, we generated synthetic single-cell isoform expression datasets. First, we simulated single-cell gene expression values for 6000 cells belonging to two different cell populations using Splatter22 v1.28.0. (Fig. 2a). These cells expressed a total of 6560 genes and 12,320 isoforms including genes with changes in expression and/or isoform usage across cell populations (Fig. 2b and Supplementary Data 1). Using this strategy, we generated three datasets with different drop-out rates: one similar to a real 10x dataset used as reference23 and two other datasets with lower coverage (Figs. 2c and S2a). Given that SCALPEL uses as input mapped reads, we developed a new method, scr4eam (https://github.com/plasslab/scr4eam), to generate isoform-aware realistic scRNA-seq reads for the synthetic iDGEs (Fig. S2b, c). We used these three datasets to compare SCALPEL isoform quantifications with simulated isoform quantification. In all datasets, we find a high correlation between simulated isoform abundances and the isoform quantification provided by SCALPEL (Pearson correlation coefficient r ≥ 0.8, Fig. 2d–f).
a UMAP plots depicting the two cell populations, population A (orange) and population B (green), simulated with Splatter. b Graphical representation of the types of genes included in the simulated data. c Violin plots showing the relative number of UMIs per isoform in a reference 10x dataset (gray) and those in the simulated datasets with different dropout rates (high, mid and low depicted in dark, medium and light purple). The real dataset contains information for 40,750 isoforms across 2042 cells. 12,320 isoforms in 6000 cells were simulated for the high, mid and low datasets. Differences in the distribution of UMIs per isoform were tested using Mann–Whitney–Wilcoxon two-sided test. The center of the box plot is denoted by the median, a horizontal line dividing the box into two equal halves. The bounds of the box are defined by the lower quartile (25th percentile) and the upper quartile (75th percentile). The whiskers extend from the box and represent the data points that fall within 1.5 times the interquartile range (IQR) from the lower and upper quartiles. Any data point outside this range is considered an outlier and plotted individually. p-Values for pairwise comparisons (Bonferroni-adjusted) are: High vs. real p-value = 0.86; high vs. mid p-value = 4.40e−259; high vs. low p-value = 2.22e−308; mid vs. low p-value = 2.6e−174. Correlation between simulated isoform abundances (y-axis) and predicted isoform abundances (x-axis) for the high (d), medium (e) and low (f) sequencing depth simulated datasets. g–i Number of correctly identified genes (g), isoforms (h), and DIU genes (i) by each of the sequencing tools in the high (dark purple), medium (medium purple) and low (light purple) simulated datasets. As reference, we provide the number of simulated genes, isoforms and DIU genes simulated (gray bar). j Summary of the performance of the different tools benchmarked on the three synthetic datasets. Source data are provided as a Source Data file.
Benchmark of SCALPEL using synthetic data shows higher sensitivity and specificity than other tools
We used the generated synthetic datasets to benchmark the performance of SCALPEL against existing tools developed to quantify APA in scRNA-seq data16,17,18,20,24,25. Considering the underlying quantification strategy, these methods can be divided into peak-calling based tools (Sierra, scAPA, scAPAtrap, SCAPTURE, and scDaPars), and isoform quantification tools (scUTRquant) (Supplementary Data 2). Following the quantification of peaks or isoforms according to the default parameters of each tool, we performed DIU analysis. In the case of scUTRquant20, which uses an extended curated 3’ UTR annotation (3’ UTRome), we performed the benchmarking using both the 3’ UTRome (scUTRquant) and the same annotation as the other tools (scUTRquant*).
Overall, our analyses show a clear difference in sensitivity between peak- and isoform-based methods. Peak-based methods quantified fewer genes and isoforms than isoform-based methods, which showed similar sensitivity (Fig. 2g, h). Some of the sensitivity differences between peak and isoform tools can be explained by the constraints of the prediction methods. In particular, the low number of isoforms quantified by scDaPars24 could be explained because this tool only predicts PAS in annotated 3’ UTRs. As expected, sequencing depth impacts the quantification of genes and isoforms. Most methods detect fewer genes and isoforms in the datasets with medium and low UMI per isoform (Fig. 2g, h).
We next identified DIU genes across the two cell populations simulated. In all three simulated datasets, SCALPEL recovered the highest number of DIU genes closely followed by scUTRquant* and scUTRquant (Fig. 2i, Supplementary Data 3). Both methods have a superior performance than peak-based tools although SCALPEL has higher sensitivity than scUTRquant (Figs. 2j and S3a–c). The higher sensitivity of SCALPEL is partly explained by a robust performance across expression ranges while all other tools misidentify lowly expressed (bottom 50% expression) DIU genes (Fig. S3d–f). In the low expression dataset, SCALPEL correctly identifies 57% of DIU genes among Q1 genes, while scUTRquant and scUTRquant* identified 19% and 22%, respectively (Supplementary Data 3). We also noticed that true DIU genes predicted by SCALPEL have a high degree of agreement and are co-detected by at least one of the other benchmarked tools (high: 95%, mid: 93%, low: 91%) (Fig. S3g–i), indicating that tool agreement can be used as a metric to assess its performances on real datasets.
Finally, we decided to compare the performance of SCALPEL in terms of execution requirements and runtime. SCALPEL runtime and memory usage are comparable or better than most of the tools analyzed, and only scUTRquant is faster and more memory efficient than SCALPEL. Yet, this can be directly linked to the presence of an already processed annotation as both memory and execution time increase when no 3’ UTRome is provided to scUTRquant (scUTRquant*, Fig. S3j–l, Supplementary Data 4).
SCALPEL predictions on real data are more sensitive and have a high degree of agreement with other tools
We additionally benchmarked SCALPEL performance against the other tools using a publicly available single-cell dataset of mouse sperm cell differentiation generated using 10x Genomics platform23 (Fig. S4a). In this dataset, SCALPEL identified 51,767 isoforms in 17,525 genes that were used for downstream analyses such as dimensionality reduction and clustering. We found three main cell populations using markers from a previous study26: spermatocytes (SC), round spermatids (RS) and elongated spermatids (ES) (Fig. S4b). Pairwise comparison between these populations shows again that the number of predicted DIU genes is generally higher for isoform-based approaches than for peak based methods (Fig. S4c–e, Supplementary Data 5). The higher number of identified DIU genes by SCALPEL is partly explained by a higher prediction among less expressed genes (bottom 50% expression). Here, it is important to note that the number of DIU genes detected by scUTRquant using the standard gene annotation (scUTRquant*) is clearly reduced, indicating that the higher sensitivity of scUTRquant can be directly attributed to the use of an extended annotation (3’ UTRome) and not to the algorithm per se.
Finally, we investigated the agreement in the prediction of genes with differential peak or isoform usage across tools. We observed a substantial overlap in the predictions of SCALPEL with other tools, with more than 70% of SCALPEL predictions supported by one or more tools (Fig. S4f–h). SCALPEL and scUTRquant showed the highest agreement on the identified DIU genes across the cell types (Fig. S4i–n).
We additionally benchmarked the performance of SCALPEL on a shallower single-cell dataset generated in-house with a Drop-seq platform containing human induced pluripotent stem cells (iPSCs) and neural progenitor cells (NPCs) (Fig. 3a, b). In this dataset, SCALPEL quantified 68,813 isoforms in 16,544 genes and more than 10,000 genes with two or more isoforms (Supplementary Data 6–8). When performing the benchmark on the neuronal dataset, SCALPEL and scAPAtrap showed higher sensitivity in the identification of DIU genes while keeping a high degree of agreement with other tools (Fig. S5). In this case, although scUTRquant quantified a high number of isoforms (40,002) and genes with multiple isoforms (10,481), only 246 DIU genes were identified between NPCs and iPSCs (Fig. S5d, e). This drop in the number of detected DIU genes is likely arising from stringent default parameters which discard genes expressed in a few cells (minCellsPerGene = 50).
a Clustering analysis identifies two main populations corresponding to iPSCs (salmon) and NPCs (cyan). b Distribution of the number of UMIs, Genes and % of mitochondrial UMIs (MT) of the two samples analyzed. iPSC violin plots show measurements for 1233 cells and the NPC violin plots show measurements for 1302 cells. Overlaid boxplots indicate the median (center line), the 25th and 75th percentiles (box limits), and the most extreme values within 1.5 times the IQR (whiskers). iPSCs: median = 7047, box = [4166, 11,495], whiskers = [597, 19,973]; NPCs: median = 2952, box = [1513, 5771], whiskers = [414, 19,975]. c SCALPEL identifies a significant change in isoform usage in EIF1 gene between iPSCs and NPCs. Coverage plots show the distribution of filtered reads along isoforms in both conditions. Relative expression of isoform abundance by SCALPEL is provided on the custom tracks. The location of the oligo dT primer (gray) and the gene-specific primer (purple) used for experimental validations are shown on top of the isoform diagrams. d Experimental validation in bulk of the isoform changes predicted by SCALPEL using 3’ RACE. EIF1-204 isoform is more highly expressed in iPSCs than in NPCs while the expression of the EIF1-201 isoform is similar in both conditions. e Quantification of JPT1 isoforms by SCALPEL. f Relative abundance of JPT1−203 in NPCs is higher than in iPSCs while JPT1-207 is more similar across conditions. g Cumulative distribution plot of log2FCs showing the difference in the expression of isoforms from DIU genes where one of them contains miR-128-5p target sites (pink) and the other does not (gray), indicating that changes in isoform usage between iPSCs and NPCs can be attributed to miRNAs known to be implicated in neurogenesis (two-tailed Kolmogorov–Smirnov test, FDR = 0.0000115). Source data are provided as a Source Data file.
Overall, the results from the benchmark show that SCALPEL has higher sensitivity than all other tools while keeping good precision and performance in real datasets generated with 10x and Drop-seq technologies.
SCALPEL predictions can be experimentally validated
Considering that iPSCs and NPCs are easily distinguished experimentally, we decided to use this dataset to experimentally validate SCALPEL predictions. To this end, we selected five genes predicted to have changes in isoform usage by SCALPEL using Chi-square statistical test (false discovery rate, FDR, adjusted p-value < 0.05, see the “Methods” section) across cell types (Supplementary Data 9) and validated the expression of the isoforms quantified by SCALPEL in NPCs and iPSCs using 3’ RACE. With this analysis, we confirmed the 3’ UTR lengthening of two DIU genes predicted by SCALPEL, EIF1 and JPT1. In both examples, SCALPEL quantified two isoforms differing in the length of the last exon and predicted a shift in the expression of the short and the long isoform between iPSCs and NPCs (Fig. 3c, d) that was also validated with the 3’ RACE (Fig. 3e, f). For the other 3 genes tested, we were able to detect all isoforms predicted by SCALPEL in two of the remaining genes (Figs. S6 and S11–13).
Isoform expression at the single-cell level reflects miRNA function
Since it is known that APA changes significantly during neurogenesis27, and that miRNA regulation is very important in this process28,29,30, we investigated if changes in isoform usage in NPCs compared to iPSCs were driven by miRNA function. To address this question, we downloaded the predicted miRNA target sites on the human genome from the MBS database31 and identified all isoforms targeted by miRNAs previously associated to NPCs30 (Supplementary Data 10). Given that isoforms with different 3’ ends could contain different regulatory elements such as miRNA target sites2, and that miRNAs usually downregulate their target RNAs32, we compared the fold change distribution of isoforms containing target sites of miRNAs expressed in NPCs with those of non-targeted isoforms from the same genes. This analysis identified significant downregulation of isoforms targeted by let-7b-5p, miR-9-5p, miR-124-3p, miR-128-5p, miR-128-3p, miR-153-5p, miR-199a-3p, and miR-34a-5p in NPCs compared to iPSCs (FDR < 0.05; Fig. 3g and Supplementary Data 11). This result suggests that miRNAs can explain some of the isoform expression changes predicted by SCALPEL during the differentiation of iPSCs to NPCs.
SCALPEL isoform quantification recapitulates 3’ UTR shortening during mouse sperm cell differentiation
We decided to use SCALPEL isoform quantification to investigate the benefits of using isoform-based analyses instead of gene-based analyses for clustering and downstream analyses. Using the mouse spermatogenesis dataset, we observed that the use of isoforms instead of genes for clustering analysis results in the identification of the same three main cell populations (Fig. 4a; 94% agreement in cell assignment). In this dataset, most of the genes express a single transcript in individual cells. Yet, we detect on average 917 genes with two or more isoforms per cell, which may be potentially regulated across cell types (Fig. S7a). Given that isoform quantification may be useful to identify new cell populations, we investigated how informative is isoform expression to define cell type identity. For this purpose, we computed for each gene its information content defined as the sum of the information from all its expressed isoforms33. The higher the information content of a gene, the more cell-type-specific the expression of its isoforms is. Using this approach, we noted that most genes had clear cell type-specific expression bias (Fig. 4b). Manual inspection showed that in many cases all isoforms from the same gene showed similar expression changes across cell types, indicating that gene information content mainly reflects transcriptional changes (Fig. 4c). Thus, we used a Chi-square test to identify genes in which isoform usage changes across conditions, reflecting a regulation at the post-transcriptional level. Using this approach, we identified 4196 genes displaying changes in isoform usage across cell types using Chi-square test (FDR adjusted p-value < 0.05, see methods) (Fig. 4b, red dots; Supplementary Data 12). Pairwise DIU analysis showed that most of these genes present changes in isoform usage independent of changes in gene expression, indicating that those events are only regulated at the post-transcriptional level (Fig. 4d). In most cases, changes in APA across cell types involved changes in tandem APA sites, which only affect the length of the 3’ UTR, or complex events including isoforms with changes in the 3’ UTR length and the exon composition (Fig. 4e). These results indicate that the main effect of APA during spermatogenesis is the regulation of the 3’ UTR length. One of these genes is Smg7, a gene that plays a crucial role in male germ cell differentiation in mice through its role in nonsense-mediated mRNA decay25. We observed a switch in isoform usage during cell differentiation where the long isoform of Smg7 gene (Smg7-202) is progressively replaced by a shorter isoform (Smg7-203) (Fig. 4f). Previous studies have shown that APA results in global 3’ UTR shortening during sperm cell differentiation34,35. Thus, we used SCALPEL predictions to assess if the observed changes in 3’ UTR length reflected a coordinated shortening during sperm cell differentiation. To this end, we used the isoform quantification data to order cells by pseudotime and calculated the average 3′ UTR length per cell. In agreement with previous studies, we observed that overall, 3’ UTR length shortens while cells differentiate (Fig. 4g), indicating that SCALPEL predictions recapitulate known changes in 3’ UTR length during mouse spermatogenesis.
a Cell types identified using gene expression (left) and isoform expression estimated by SCALPEL (right). Both analyses identify spermatocytes (SC), round spermatids (RS), and elongated spermatids (ES). Agreement in clustering solutions is shown by a Sankey plot. b Scatter plot showing the cell-type expression specificity relative to gene expression for all genes with only two expressed isoforms. The higher the information content of a gene, the more cell-type-specific its expression is. Colored dots represent genes whose isoform expression usage changes across conditions (Chi-squared test adjusted p-value < 0.05). c SCALPEL quantification of isoform usage from Ighmbp2. Coverage plots show the distribution of filtered reads in SC, RS and ES clusters. SCALPEL quantifications of the different isoforms are shown on top of the custom tracks. d Number of DIU genes identified in each pair of samples that are also differentially expressed (light gray) or not (dark gray). Most genes show changes in isoform usage independently of changes in expression. e Classification of DIU genes depending on the type of APA event identified as alternative last exons (ALE), tandem APA sites (tandem APA), and others. f Quantification of Smg7 isoforms by SCALPEL. Coverage plots show a gradual switch in isoform usage during the differentiation from SC to ES cells. g Pseudotemporal ordering of cells confirms the overall shortening of 3’ UTRs during mouse sperm cell differentiation. h High-resolution clustering using isoform expression data (right) identifies novel cell states (RS6) that cannot be identified using gene expression data (left). Using this resolution, we identified 15 cell populations in the isoform-based analysis including spermatocytes (SC1-5), Round cells (RS1-6), Elongated Spermatids (ES1-2), and Condensing Spermatids (CS1-2). The new RS6 population identified in the isoform-based analysis is composed of cells coming from several RS populations identified using gene expression data. i GO term enrichment analysis using RS6 marker isoforms identified significant terms associated with cilium organization and cell differentiation. In the circular barplot, GO terms are arranged according to their combined enrich R score and colored according to their adjusted p-value. Source data are provided as a Source Data file.
Finally, we investigated if the changes in isoform usage across cell types involved changes in the types of transcripts (i.e. the biotype) expressed in the cells that could further alter cell function by promoting, for instance, the degradation of expressed mRNAs or the production of non-coding isoforms of a gene. We observed that in ~80% of the cases, changes in APA did not involve a change in the type of transcripts expressed (Fig. S7b). In the cases where the biotype of the most expressed isoform changed across cell types, in 82% of the cases this change involved a switch from a mRNA to a non-coding or NMD isoform or vice versa (ES-RS: 429, RS-SC: 406, ES-SC: 690; Fig. S7c). In these cases, APA could alter significantly protein levels and lead to stronger functional changes. Together, these results confirm that SCALPEL robustly captures APA-mediated remodeling of transcriptomes and reveals novel insights into transcript diversity regulation during cell differentiation.
Isoform-based analysis identifies novel cell populations during mouse spermatogenesis
To investigate if isoforms provide additional biological insights in single-cell analyses, we decided to perform a high-resolution clustering analysis of the spermatogenesis dataset. As expected, at higher resolutions both gene and isoform-based analyses identify more cell populations while keeping a high degree of similarity across clustering solutions (Figs. 4h, S8a, Supplementary Data 13, 14). At this higher resolution, we identified a new cell population of RS cells, RS6, in the isoform-based analysis that could not be distinguished using gene-based clustering. This population is composed of cells from other RS cell clusters and is not the result of subclustering existing RS cell populations (Figs. 4h and S8a, b). Compared to other RS populations, the RS6 shows few changes in gene expression (Fig. S8c). Yet, the RS6 cluster presents a clear set of isoform markers (Wilcoxon rank-sum test, FDR adjusted p-value < 0.05, see the “Methods” section) that cannot be identified in the gene-based analysis (Fig. S8d, Supplementary Data 14). GO term enrichment analysis using RS6 isoform markers identified biological processes associated with cilium organization and organelle assembly (Fig. 4i, Supplementary Data 15, Fisher exact test, FDR adjusted p-value < 0.05), which are essential processes for the differentiation and maturation of RS cells36,37. DIU analysis across RS populations identified 543 genes with differential isoform usage in RS6 cells (Supplementary Data 16, Chi-square test, FDR adjusted p-value < 0.05, see the “Methods” section). These isoforms are not the result of changes in gene expression as only 3 DIU genes are differentially expressed. Among them, we identified Dnah3, a structural component of the axoneme essential for the flagellum assembly and the acquisition of sperm motility, and Msi2, an RNA binding protein that plays a central role in coordinating post-transcriptional programs essential for late spermatocytes to early spermatid differentiation38,39,40. These genes exhibited higher expression of longer transcript isoforms in RS6 compared to neighboring RS cells, suggesting that these changes are affecting the resulting proteins and their function within the different cell types (Fig. S8e, f). Together, these results suggest that the RS6 cell population corresponds to elongating spermatids41, an intermediate population state between RS and ES populations previously described morphologically in the literature that had not been identified using single-cell gene expression profiles.
SCALPEL improves isoform quantification of novel isoforms predicted using single-cell long-read sequencing
We investigated if SCALPEL could be used to improve the quantification of isoforms predicted in individual cells using long-read single-cell sequencing data. We reanalyzed two single-cell datasets from the P28 mouse hippocampus and visual cortex containing 10× 3’seq scRNA-seq data and paired PacBio data42 (Fig. S9a, Supplementary Data 17). We performed standard single-cell analysis using gene expression data and subset the data of the main five cell populations: excitatory neurons, astrocytes, NPCs, oligodendrocytes, and microglia (Supplementary Data 18, 19). As expected, a comparison of cell assignments using gene or isoform-based quantification revealed a high degree of agreement between the two approaches (Fig. 5a). Given that SCALPEL can only distinguish isoforms with changes at the 3’ end, we collapsed isoforms and kept only those with changes in the last 600 nt. Then, we compared the abundance per cluster of these isoforms according to isoquant and SCALPEL. The quantification by SCALPEL clearly correlates with that of isoquant although the expression range of SCALPEL is broader as the quantification is done using the single-cell short-read data and not the long-read data (Fig. S9b).
a UMAP plots depicting the main five cell populations selected from Jogleklar et al. identified using gene and isoform-based quantifications including excitatory neurons (EN), neural progenitor cells (NPCs), oligodendrocytes (Olig), astrocytes (Astro) and microglia (MIG). b Comparison of isoform expression quantification for each cell type using isoquant (PacBio data) and SCALPEL. c, d SCALPEL identifies changes in isoform usage in Cdc42 (c) and Dclk1 (d) between astrocytes/microglia and excitatory neurons. Custom tracks on top show the mapping of filtered short reads and SCALPEL quantification for each cell type. On the bottom, sashimi plots show the relative usage of different isoforms in individual cell types and the aligned long-read data supporting them. Source data are provided as a Source Data file.
We also ran SCALPEL to identify genes with changes in isoform usage across cell types. In this dataset, we found that most changes in isoform usage happen between glial cells (microglia, astrocytes, and oligodendrocytes) and the excitatory neurons (Supplementary Data 20). Among these genes, we identified examples in which isoform switch events predicted by SCALPEL were supported by the long-read data. One of those examples is the gene Cdc42. SCALPEL detects a change in isoform usage between excitatory neurons and microglia/astrocytes, which is validated with the PacBio data (Fig. 5c). Dclk1 is another example in which SCALPEL predicts a change in isoform usage between astrocytes/microglia and excitatory neurons. In this case, microglia and astrocytes express only a short version of the gene while excitatory neurons express a much longer isoform (Fig. 5d). Here, it is interesting to highlight that isoquant quantifications disregard changes in the 3’ ends and focus on changes in exon junctions. While SCALPEL quantifies two isoforms with tandem polyA sites (Dclk1-202 and Dclk1-208), isoquant focuses on the changes in exon composition. Together, these analyses demonstrate that SCALPEL can be used to quantify isoform diversity derived from paired long and short-read data with single-cell resolution and that provides additional information to the long-read data that disregards changes in the 3’ ends that do not affect exon composition.
Discussion
In this manuscript, we have presented SCALPEL, a new tool for sensitive isoform quantification and visualization using conventional 3’ based scRNA-seq data. Comparison of SCALPEL to other existing tools using synthetic data shows that SCALPEL correctly identifies a higher fraction of simulated genes and isoforms and has a higher accuracy in the detection of DIU genes than other tools (Fig. 2g–j). Benchmarking of SCALPEL on real data shows that SCALPEL predictions have a high agreement with that of the other tools, which have commonly been used as a measure of accuracy. Additionally, we also show that in contrast to other tools, SCALPEL’s performance is similar in the 10x dataset than in the Drop-seq dataset (Figs. S4 and S5). This is remarkable considering that many other tools perform poorly on the Drop-seq datasets, limiting their usability to high-expression datasets.
We show that SCALPEL predictions can be validated experimentally using 3’ RACE (Figs. 3c–f and S6) and long read sequencing (Fig. 5c, d). While 3’ RACE is not quantitative and can only be used to validate specific isoforms in bulk, we consider that the validation of some of SCALPEL’s predictions using this method confirms the reliability of the predictions. Similarly, while PacBio only provides reliable cell-type specific measurements, the significant correlation between isoquant and SCALPEL using pseudobulk data supports SCALPEL predictions.
We also demonstrate that the iDGE provided by SCALPEL can be used to perform standard single-cell analysis such as dimensionality reduction, clustering, and pseudotime analysis (Fig. 2a). At low resolution, SCALPEL iDGE provides very similar clustering solutions to gene-based analyses (Figs. 4a and 5a). However, at higher resolutions we also show that isoform quantification can be used to gain new insights about cell populations and identify, for instance, novel cell states that cannot distinguished using standard single-cell gene quantification data (Figs. 4h and S8), indicating that we can capture post-transcriptional regulatory events by quantifying isoforms in individual cells.
In agreement with this idea, we noticed that SCALPEL predictions recapitulate known changes in 3’ UTR length during sperm cell differentiation, mainly led by changes in the use of tandem APA sites (Fig. 4). In the human iPSC & NPC data, changes in isoform quantification reflect miRNA function at the single-cell level, as changes in isoform usage across cell types can be directly linked to the presence of cell type-specific miRNAs (Fig. 3g).
Finally, we also show that SCALPEL can be used to improve isoform quantification obtained from paired long and short-read scRNA-seq datasets (Fig. 5). In these cases, SCALPEL can be exploited to quantify the relative abundance of isoforms predicted using long-read data at the individual cell level, which cannot be done with standard scRNA-seq analysis pipelines. Additionally, it provides the quantification of shortening/lengthening events in the last exon, i.e. tandem APA sites, which are usually not considered using long-read data that focuses on events that change exon composition.
Together, our work highlights how SCALPEL expands the current scRNA-seq toolset to explore post-transcriptional gene regulation in individual cells from different species, tissues, and technologies to advance our knowledge on gene regulation from the bulk to the single-cell level.
Methods
Annotation preprocessing
3’ based scRNA-seq protocols use oligo(dT)s to capture polyadenylated RNAs, which introduces a bias in the location of reads towards the 3’ end of the RNAs (Fig. S10). Thus, we truncated the annotated isoforms in the existing annotation and restricted the quantification to isoforms that are different at the 3’ end. Using GENCODE43 annotation as a reference, we truncated all isoforms to include the last 600 nucleotides of spliced sequence from their 3’ end, which is the region that displays coverage by the 3’ tag-based scRNA-seq data (Fig. S10). Then, we collapsed truncated isoforms with exact intron/exon boundaries and fewer than 30 nucleotide differences in their 3’ end coordinates. When multiple isoforms were collapsed, we kept the name of the isoform having the higher expression according to pseudobulk quantification. For this purpose, we used salmon quant v10.144 with default parameters to quantify all isoforms in bulk using as input the scRNA-seq bam files provided. For the analysis of multiple samples, isoform expression was averaged across all samples.
Read preprocessing
We processed all the input BAM files containing aligned and tagged scRNA-seq reads to discard artifacts and reads not supporting annotated transcripts. First, we used samtools v1.19.245 to split the input BAM files by chromosome using the command samtools view and converted them into BED files using bam2bed command from BEDOPS v2.4.4146. We included all reads in the bed file (option –all-reads) and split them into separate entries (option –split) if contained Ns in the CIGAR line (i.e. spliced reads). We used the function findOverlaps from GenomicRanges R package v1.50.047 to overlap the reads with the set of selected isoforms using default parameters. Given that all reads with the same BC and unique molecular identifier UMI are likely generated from the same original RNA molecule, we grouped them into a unique fragment and jointly evaluated them during isoform quantification (Fig. S1). We defined the genomic coordinates of each fragment as the most 5’ and 3’ coordinates of its associated reads. We discarded all the fragments overlapping intronic and intergenic regions except those extending the 3’ end of the gene, and spliced reads not supporting annotated exon-exon junctions, as they were considered to come from pre-mRNAs or unannotated transcripts. To avoid biases in the quantification of the isoforms due to reads mapping to internal priming (IP) locations in the genome, we discarded all the fragments that could arise from these sites. For that purpose, we scanned the whole genome using a custom Perl script and identified putative IP locations as regions containing six consecutive adenosines or seven or more adenosines in a window of 10 nucleotides. We discarded all the fragments located upstream from an IP site. In this case, we only considered IP sites located more than 60 nucleotides upstream of an annotated 3’ end in the transcriptomic space.
Quantification of isoforms at the single-cell level
First, we used all genes with only one detected isoform to assess the empiric distance distribution of scRNA-seq reads with respect to annotated isoform 3’ ends. Considering defined intervals of 30 nt, we calculated the distribution of read 3’ ends relative to annotated 3’ ends by dividing the number of distinct 3’ ends in each bin in the transcriptomic space by the total number of 3’ ends. Using this probability distribution, we assigned to each read a probability to come from a specific isoform based on its relative distance to the 3’ end. Considering that each fragment is composed of one or more reads, we defined the probability of a fragment \(f\) to belong to an isoform \(k\) for a gene \(g\) as
where \({R}_{f}\) is the set of reads associated with the fragment \(f\), \({{\mathbb{P}}}_{r}\left(k|g\right)\) is the probability that read \(r\) is associated with the isoform \(k\), and \({\omega }_{k,g}\) is the weight associated with the isoform \(k\) and gene \(g\) based on the pseudobulk quantification.
We computed the weight \({\omega }_{k,g}\) of an isoform \(k\) and gene \(g\) as
where \({K}_{g}\) is the set of isoforms for the gene \(g\) and \({{{\rm{T}}}{{\rm{P}}}{{\rm{M}}}}_{k,g}\) is the transcript per million counts of isoform \(k\) and gene \(g\) according to Salmon44.
Hence, the probability of a set of fragments \({F}_{g,c}\) for gene \(g\) and cell \(c\) given a fixed set of isoforms \({K}_{g}\) and associated relative abundances values \((\theta )\) is given by
where \({F}_{g,c}\) is a set of fragments for a gene \(g\) and cell \(c\), \({{\mathbb{P}}}_{f}(\varkappa )\) represents the probability of a fragment \(f\) belongs to isoform \(\varkappa\), and \({\theta }_{\varkappa }^{g,c}\) the isoform \(\varkappa\) relative expression for a gene \(g\) and cell \(c\).
Consequently, we estimated the isoform relative expression values \((\theta )\) by maximizing the log-likelihood function using a standard expectation maximization (EM) algorithm. The maximum-likelihood estimators \(\hat{\theta }\) for the isoform relative abundance values are given by:
For each gene \(g\) and cell \(c\), the EM algorithm proceeds as follows:
-
1.
Initialization of value \({\theta }_{k}^{g,c}\) for each isoform \(k\) of the gene \(g\) as \(\frac{1}{{{|K}}_{g}|}\)
-
2.
Iterate until convergence:
-
a.
E-step: Calculation of posterior probability for each fragment \(f\in {F}_{g,c}\) to belong to the isoform \(k\) given that it comes from a gene \(g\) and a cell \(c\) as
$${\mathbb{P}}_f\left(k \mid g \cap c\right)=\frac{{\mathbb{P}}_f\left(k \mid g \cap c\right)\theta_k^{\,g,c}}{\sum_{\varkappa \in K_g}{\mathbb{P}}_f\left(\varkappa \mid g \cap c\right)\theta_\varkappa^{\,g,c}}$$(5) -
b.
M-step: Estimate the isoform relative expressions \({\theta }_{k}^{\, g,c}\)
-
a.
The convergence state of the EM algorithm was settled by a stop criterion condition. This condition was reached when the maximum difference of the estimated relative abundance between two iterations was equal to or lower than 0.001. All the isoforms with a null weight value were discarded from the annotation set. To generate isoform expression values for the iDGE per gene the estimated isoform probabilities were multiplied by the UMI counts assigned to the original DGE (Fig. S1).
Isoform entropy and gene information content calculation
In order to deconvolute the DGE into an iDGE, SCALPEL estimates the isoform relative expression distributions for each gene and cell (θ, see above). Considering a fragment \(\bar{f}\) across the set of all the fragments \(F\), we can define the conditional probability of fragment \(\bar{f}\) to originate from an isoform \(k\) given it comes from a gene \(g\) and cell \(c\) as:
where \({\theta }_{k}^{g,c}\) is the isoform \(k\) relative expression estimated by SCALPEL for gene \(g\) in cell \(c\). Taking this into account, the probability of fragment \(\bar{f}\) to originate from isoform \(k\) of gene \(g\) given that it originates from cell \(c\) can be derived as
where the probability of fragment \(\bar{f}\) to originate from any isoform of gene \(g\) given it originates from cell \(c\) can be calculated from the iDGE as
where \({{{\rm{D}}}{{\rm{G}}}{{\rm{E}}}}_{g,c}\) denotes the number of UMIs assigned to gene \(g\) in cell c, \(G\) the set of all the genes, \({i{{\rm{D}}}{{\rm{G}}}{{\rm{E}}}}_{\varkappa,c}\) the number of UMIs assigned to isoform \(\varkappa\) in cell \(c\), \(K\) the set of all the isoforms across all the genes.
Applying Bayes’ theorem, the probability of fragment \(\bar{f}\) to originate from cell \(c\) given it originates from isoform \(k\) can be derived as
where the probabilities \({P}_{\bar{f}}(c)\) of fragment \(\bar{f}\) to originate from cell \(c\) and \({P}_{\bar{f}}(k)\) of fragment \(\bar{f}\) to originate from isoform \(k\) can be estimated from the iDGE corresponding column and row sum fractions, respectively.
where \(C\) is the set of all the cells, and
Given the cell-to-cell cluster mapping ϕ:C↦L, where \(L\) is the set of all cell clusters, the probability of fragment \(\bar{f}\) to originate from a cell in cluster \(l\), given it originates from isoform \(k\) can be derived by summing up the corresponding cell probabilities:
where \({C}_{l}\) is the set of the cells associated to the cluster \(l\).
Based on those conditional probabilities, each isoform’s entropy across cell cluster is defined as:
If all fragments originating from a given isoform originate from cells of the same cell type, the corresponding entropy is minimal, at a value of
bits. If an isoform \(k\) is expressed perfectly equal across all clusters, the maximum isoform entropy
is reached, where |L| denotes the number of cells clusters.
The isoform-level entropies were summarized to the gene level to quantify the randomness of a gene’s isoform distribution across cell types:
While the minimal gene entropy
is gene-independent, its upper bound depends on the number of expressed isoforms \(|{K}_{g}|\) and thus differs across genes:
The difference from that maximum entropy defines a gene’s information content across isoforms and cell types/clusters
Detection of differential isoform usage between cell clusters
For the identification of differential isoform expression between clusters of cells, we implemented the function FindIsoforms. This function aggregates the isoform counts using AggregateExpression from Seurat and retains only genes with two or more isoforms. All isoforms that represent < 10% of the total gene expression in at least one of the conditions were discarded. Additionally, to reduce the number of false positives, we also discarded genes whose isoforms changed across conditions but had a very low expression variance (< 5% standard deviation), which would otherwise be likely identified as significant DIU genes. For the remaining genes, the function performs a Chi-squared test to assess if the read distribution across isoforms is the same between clusters. We selected significant DIU genes with an FDR < 0.05.
Average 3’ UTR length calculation
We extracted from the iDGE all protein-coding isoforms and computed the lengths of the corresponding 3’ UTR regions from the reference annotation. For each gene \(g\), we calculated the gene average 3’ UTR length \({\tau }_{g}\) in a cell \(c\) as
where \({\tau }_{k}\) is the 3’ UTR length of isoform \(k\), \({i{{\rm{D}}}{{\rm{G}}}{{\rm{E}}}}_{k,c}\) are the UMI counts of isoform \(k\).
Next, we calculated the average weighted 3’ UTR isoform length within each cell \({\tau }_{c}\) for all the expressed isoforms as
where \({\mathbb{G}}\) is the set of all genes with protein-coding isoforms expressed.
Generation of read coverage plots
SCALPEL outputs a BAM file including the reads used for isoform quantification. Within the SCALPEL framework, we have implemented the function CoveragePlot in R to visualize the read coverage on the isoforms using a transcriptome annotation in GTF format and BAM files. We generated the visualization tracks using the R Gviz library v1.46.048.
Generation of synthetic data for benchmark analysis
We used Splatter22 to simulate 12,320 isoforms across 6000 cells representing two equally probable cell populations (group.prob = 0.5) using the function splatSimulatedGroups. For these cells, we generated 3 datasets with distinct sequencing depth: low (lib.loc: 29,000 counts, lib.scale: 0.5), medium (lib.loc: 39,000 counts, lib.scale: 0.5), high (lib.loc: 120,000 counts, lib.scale: 0.5). Dropout effects were modeled using the experimental dropout function with the parameters shape and midpoint adjusted to reflect 10x genomic profiles in the 10x scRNA-seq mouse dataset (shape: 0.5; midpoint: 0.4). For each simulation, genes were stratified into five regulatory categories based on their number of isoforms and their expression dynamics: unique isoform with no change, unique isoform with expression change, multi-isoform with no change, multi-isoform with coordinated expression change, and multi-isoform with isoform-specific regulation (DIU) (Supplementary Data 1 and Fig. 2b). Furthermore, genes were sampled across five expression quantiles to ensure even coverage across expression levels.
To complement the synthetic iDGE matrices, we generated synthetic FASTQ files that reflect realistic read structure and position distribution observed in the mouse 10x scRNA-seq data. Considering the mapped data, we extracted the genes, isoforms, and fragments, as well as the distance to the transcript’s 3’ end for all reads assigned to at least one transcript. Next, we used a custom bash script to sort these data and extract the following three (empirical) distributions: (i) the number of reads per fragment; (ii) their distance to the fragment’s 3’ end; and (iii) the distance to the transcript’s 3’ end for all fragments that were unambiguously mapped to a gene expressing a single isoform. From these distributions and the counts from the corresponding synthetic iDGE, fragments and reads were sampled using a custom R script. Then, a second custom awk script was used to extract the corresponding synthetic read sequenced from a FASTA file providing the transcripts’ spliced sequences. This single FASTA file was finally converted to a pair of FASTQ files adding ‘@’ quality scores and transforming the placeholder cell and fragment indices to valid 10x barcodes using a custom Python script. All custom scripts used to generate the synthetic FASTQ files are implemented within the scr4eam pipeline (see the “Code availability” section).
Analysis of mouse spermatogenesis 10x scRNA-seq data
We downloaded the 10x scRNA-seq samples from male mouse germline23 from the GEO database (accession number GSE104556) and processed them using Cell Ranger v7.1.049, using mm10 mouse genome assembly50 and GENCODE M2143 as reference annotation. We merged the processed data and analyzed them jointly using Seurat v5.0.051. We restricted the analysis to the cells with a UMI count superior to 500, inferior to 80,000 and < 5% of mitochondrial genes. The final Seurat object contained 2174 cells and 22,158 genes. We performed dimensionality reduction on the DGE using 2000 genes as variable features and used the first 9 principal components (PCs) to build the kNN graph and compute a UMAP. We used the function FindClusters with a resolution of 0.02 to identify 3 clusters. We compared our clustering results with the cell clustering from previous analysis on the same dataset by Schulman et al.16 and annotated the defined cell clusters in our analysis as ES, RS, and SC according to their previous corresponding annotation. Next, we filtered the input BAM file to retain the 2,174 filtered cells using samtools view -D CB from samtools v1.19.245 and extracted the DGE matrix for SCALPEL analysis. Following the execution of SCALPEL, we generated a Seurat object containing the same 2,042 cells, 23,151 genes, and 76,419 isoforms. For downstream analyses, we discarded all isoforms expressed in less than four cells and cells with less than three isoforms expressed using CreateSeuratObject function (option min.cells = 4, min.features = 3). The final Seurat object included 51,767 isoforms of 17,525 genes. We performed dimensionality reduction on the iDGE using 2000 isoforms as variable features, and we used 9 PCs for the generation of the kNN graph for the UMAP. We used a resolution of 0.02 to perform the clustering and identified three cell populations. We used the approach described above to annotate the cell clusters. We compared the iDGE clusters with the DGE clusters using a Jaccard index.
Differentiation and characterization of iPSCs and NPCs using Drop-seq technology
We differentiated human iPSCs (Ic-Ctrl3-F-iPSC-4F-1 from Spanish Stem Cell Bank) towards NPCs using a protocol previously established in the lab52,53. Briefly, we differentiated iPSCs to neuroepithelial cells over a period of 10 days by dual SMAD inhibition using neural maintenance medium, 1:1 ratio of DMEM/F-12 GlutaMAX (Gibco, #10565018) and neurobasal (Gibco, #21103049) medium complemented with 0.5x N-2 (Gibco, #17502048), 0.5x B-27 (Gibco, #17504044), 2.5 μg/ml insulin (Sigma, #I9278), 100 mM L-glutamine (Gibco, #35050061), 50 μM non-essential amino acids (Lonza, #BE13-114E), 50 μM 2-mercaptoethanol (Gibco, #31350010), 50 U/ml penicillin and 50 mg/ml streptomycin (Gibco, #15140122), supplemented with 500 ng/ml noggin (R&D Systems, # 3344-NG-050), 1 μM Dorsomorphin (StemCell technologies, #72102) and 10 μM SB431542 (Calbiochem, #616461). After the initial neural induction step, we differentiated the cells to NPCs by neural maintenance medium replacement up to day 22. We cryopreserved iPSCs and NPCs in fetal bovine serum (Cytiva, #SV30160.03) or neural maintenance medium supplemented with 10% of DMSO (Sigma, #D2438) for iPSC or NPCs respectively. Before single-cell encapsulation using the NADIA instrument (Dolomite Bio), we thawed, filtered, and counted the samples. We retrotranscribed RNAs captured with the oligo(dT) for cDNA library preparation. Finally, we sequenced the final Illumina tagged libraries on an Illumina NextSeq 550 sequencer using the NextSeq 550 High Output v2 Kit (75 cycles) (Illumina, #20024906) in pared-end mode; read 1 of 20 bp with custom primer Read1CustSeqB54 (5’-GCCTGTCCGCGGAAGCAGTGGTATCAACGCAGAGTAC-3’) read 2 of 64 and 8 bp for i7 index.
scRNA-seq analysis of Drop-seq data
We processed the iPSC and NPC scRNA-seq libraries using Drop-seq tools v2.5.154 pipeline to generate DGE matrices. We merged the FASTQ files containing paired-end reads into a single unaligned BAM file using Picard tools v2.27.455. We tagged the reads with cell and the molecular barcodes, trimmed them at the 5’ end to remove adapter sequences and at the 3’ end to remove polyA tails, and mapped them to the human genome (GRCh38) with STAR v2.7.1056. We tagged the resulting BAM files with the annotation metadata using the human GENCODE v4157 annotation as a reference. Finally, we performed the cell barcode correction using the programs DetectBeadSubstitutionError and DetectBeadSynthesisErrors with default parameters. To estimate the number of cells obtained, we used a knee plot considering the top 3,000 cell barcodes and generated a DGE count matrix for each sample.
We used Seurat v5.0.051 and R v4.3.258 to merge the DGEs and preprocess the scRNA-seq data. We discarded all genes expressed in less than four cells and all cells with less than three genes expressed. We also discarded low-quality cells with less than 300 UMIs, less than 300 genes, and more than 5% mitochondrial gene, and cell artifacts with more than 20,000 UMIs and 7000 genes. The final Seurat object contained 2535 cells and 19,103 genes. We performed a dimensionality reduction analysis using the 2000 most variable genes to calculate 50 PCs. We used the ElbowPlot function to manually inspect the amount of variability explained by each PC and selected the first 9 PCs to build the kNN graph and compute the UMAP plot.
Benchmark analysis
We downloaded each of the benchmarked tools from their respective GitHub repository. For each tool, all commands for its default execution were integrated into Nextflow workflows and were executed using the default parameters indicated by the authors. We performed the benchmark analysis on the preprocessed mouse spermatogenesis scRNA-seq23 data using the GENCODE vM2157 GTF and transcriptome FASTA files as reference annotation. Additionally, we performed a second benchmark analysis on the preprocessed neuronal cell differentiation scRNA-seq Drop-seq data using the analogous reference files obtained from GENCODE v4157. As scAPA only considers disjoint 3’ UTR regions annotation from the hg19 version of the human genome for the annotation of its quantified peaks, we generated a new annotation of non-overlapping 3’ UTR regions using GENCODE v41 and GenomicRanges R package. We intersected all the peaks detected by scAPA16 following its peak calling process to this new reference annotation. Following Dapars2 v2.159 default procedure24, we downloaded the gene region annotation reference for the human and mouse genome (GRCh38, mm10) using the UCSC Table browser. Then, we extracted 3’ UTR regions from the gene annotation using Dapars2 script DaPars_Extract_Anno. Finally, we calculated the raw percentage of distal PAS sites usage index (PDUI) values using DaPars2 script DaPars2_Multi_Sample_Multi_Chr and provided them to scDaPars24 to infer their expression at single-cell level. scUTRquant20 was executed using the target transcriptome annotation files provided in the GitHub repository for the human and mouse genomes (hg38 and mm10). These files included high-confidence cleavage sites called from the Human Cell Landscape and Mouse Cell Atlas dataset. Additionally, we re-ran scUTRquant20 using a custom target transcriptomic annotation generated from the input genome annotation using the Bioconductor package txcutr v1.8.060. For each tool, we performed a differential peak or isoform usage analysis using the default parameters and statistical test (Sierra: DEXSeq FDR adjusted p-value; scAPA: Chi-square FDR adjusted p-value; scAPAtrap: Wilcoxon Rank-Sum Test FDR adjusted p-value; SCAPTURE: Wilcoxon rank-sum test FDR adjusted p-value; scDapars: two-sided t-test FDR adjusted p-value, scUTRquant: two-sample hypothesis testing with a bootstrap using weighted utr expression index FDR adjusted p-value). An adjusted p-value threshold of 0.05 was used for all genes with at least two peaks or isoforms detected. For each comparison test, we generated UpSet plots using the R library UpSetR v1.4.061 for the set of DIU genes co-detected by at least two tools. We applied a cutoff threshold of 10 genes for each intersection set.
Isoform validation using nested PCRs
To validate the changes of isoform usage between the iPSCs and NPCs predicted by SCALPEL, we performed nested PCR as previously described62,63. 1 µg of total mRNA extracted using Maxwell® RSC simplyRNA Cells kit protocol (Promega Corporation, #AS1340) was used as input RNA for the cDNA synthesis using an oligo(dT)-adapter sequence TAP-VN as a primer for the reverse transcription. For the first nested PCR, 1 µL of 1:10 cDNA dilution was used, with a gene-specific primer (GSP) which is shared by all isoforms and an adapter primer (AP) as a reverse primer. The second nested PCR was performed with 1 µL of a 1:10 dilution of the first PCR using a second gene-specific primer and a second adapter primer (MAP) as a reverse primer. This second nested PCR reaction that anneals 3’ to the first GSP is essential to reduce the amplification of undesired products64. The resulting nested PCR products are resolved by an agarose gel. Primer sequences are provided in Supplementary Data 21.
Identification of miRNA signatures in differentially regulated isoforms
To obtain isoform-level identification of miRNA target sites, we overlapped the genome-wide miRNA target site annotation included in the MBS database31 with the GENCODE annotation reference v4157 (hg38). Target sites of different miRNAs were grouped by their seed sequences according to miRBase v22.165 The seed–target isoform pairs were used for downstream analyses. Using a set of neurogenesis-related miRNA30, we filtered genes displaying changes in isoform usage between iPSCs and NPCs as predicted by SCALPEL with at least one isoform targeted by a neurogenesis-related miRNA and one non-targeted isoform. We normalized the isoform abundances by the number of cells in NPCs and iPSCs and used these values to calculate their log2 fold changes (log2FC). For each miRNA, we used a two-tailed Kolmogorov–Smirnov test to check for differences in the cumulative distribution of log2FC between targeted and non-targeted isoforms from the same set of genes. The resulting p-values were FDR-adjusted using Benjamini–Hochberg correction.
Identification of novel cell populations in the mouse 10x dataset
We increased the clustering resolution in the gene and the isoform-based analyses to identify new cell states. We tested a different number of variable features in the DGE and iDGE datasets and assessed the resulting clustering using a range of resolutions. For each parameter set, we evaluated the concordance between the DGE and iDGE clustering by calculating the Jaccard index between the cell barcodes in each cluster. Based on this analysis, we selected 4000 variable features and a clustering resolution of 0.5 for the DGE, and 6000 variable features with a resolution of 0.9 for the iDGE. Using the markers from Lukassen26, we annotated the cell clusters of the gene-based analysis. For the isoform analysis, we calculated a Jaccard index score between all gene and isoform clusters (Fig. S8a) and assigned to each isoform-based cluster the identity of the most similar cluster. We performed differential isoform analysis using FindAllMarkers function with a two-tailed Wilcoxon rank-sum statistical test (option min.pct = 3, adjusted p-value < 0.05) on the iDGE to identify isoform markers for each cluster. We visualized the top 100 isoform markers' average expression within each cluster in the iDGE using the Heatmap function from the ComplexHeatmap66 package along with the average expression of their corresponding genes in the DGE clusters (Fig. S8d).
Differential gene expression analysis between RS populations
We performed a differential gene expression analysis between the RS6 and other RS populations using a pseudo-bulk approach. We aggregated gene expression values across cell clusters using the Seurat function AggregateExpression to generate a pseudo-bulk count matrix. After removing genes with zero counts, we defined a design matrix indicating the condition for each sample (RS6-RS cells). Using DESeq2 R library67, we constructed a DESeq2 object with DESeqDataSetMatrix function and performed differential expression analysis with DESeq function with a two-tailed Wald test. We identified significantly differentially expressed genes with an FDR-adjusted p-value inferior to 0.05 and an absolute log2 fold-change value superior to 0.5.
GO term enrichment analysis
We performed GO term enrichment analysis using the R package enrichR v3.268. The reference used for the enrichment analysis was the GO_Biological_Process_202369. We selected all the GO terms using a one-sided Fisher exact test with an FDR adjusted p-value inferior to 0.05 and visualized the associated adjusted p-value and enrichR combined scores using ggplot267.
Trajectory inference analysis
We used the iDGE-based Seurat object to derive a pseudo-temporal ordering of the cells using Monocle3 v1.3.470. First, we converted the Seurat object into a CellDataSet object which includes the cluster annotation and the UMAP embedding previously computed. Next, we fit the principal trajectory graph within each cluster partition using the function learn_graph. Finally, we calculated the pseudotime values using the function pseudotime and the ES cells as root cells.
Analysis of paired short and long scRNA-seq datasets
We downloaded the FASTQ files for Illumina short-read and PacBio long-read datasets of four samples from the P28 developmental stagepoint from Jogleklar et al.42 from the Knowledge Brain Map database (RRID:SCR_016152). These datasets correspond to two samples from the hippocampus (M1-HIPP, M2-HIPP) and two samples from the visual cortex (M1-VIS, M2-VIS).
We processed the Illumina short reads FASTQ files using Cell Ranger v7.1.049 with the mm10 mouse genome assembly50 and generated Gene × Cell count matrices. We merged the hippocampus and visual datasets and analyzed them jointly using Seurat v5.0.051. We filtered out cells with < 1000 and more than 25,000 UMI counts and cells with more than 15% mitochondrial gene content. After quality control, we retained 43,094 cells for 32,285 genes. We performed dimensionality reduction on the DGE using 2000 genes as variable features and integrated the data using Harmony71 to correct for sample-specific batch effects72. Next, we used the first 30 principal components (PCs) to build the kNN graph and compute a UMAP. We applied the Seurat function FindClusters with a resolution of 0.05 to identify 14 clusters and annotated them using known marker genes from the CellMarkers database73. Then, we reduced the dataset by selecting the set of barcodes corresponding to excitatory neurons, astrocytes, neural progenitor cells (NPCs), microglia, and oligodendrocytes to retain 32,356 cells. We repeated the dimensionality reduction analysis using 2000 variable genes and 7 PCs and applied a clustering resolution of 0.03 to generate five distinct clusters.
We processed the PacBio long-read FASTQ files using the scisorseqr package v5.0.142. First, we deconvoluted the samples with the 32,356 cell barcodes from the short read single-cell analysis using the function GetBarcodes. Then, we aligned the filtered reads to the mm10 mouse reference genome using the function MMalign, to generate a BAM file for each cell type. We used the function MapAndFilter to identify full-length, spliced reads supported by both CAGE and polyA peaks using annotation references from FANTOM572 and polyASite74. We set the filterFullLength parameter to TRUE to retain only 5′- and 3′-complete reads. Finally, we quantified isoform usage for cell types using the IsoQuant function (Iso = TRUE, TSS = TRUE, PolyA = TRUE).
We applied SCALPEL on the filtered 10x short-read single-cell dataset, using mouse GENCODE M24 annotation75 reference to generate an isoform-based count matrix (iDGE). We filtered all the isoforms expressed in less than 4 cells to retain 43,140 isoforms across 32,356 cells. Using Seurat51, we performed a dimensionality reduction analysis by selecting 9500 variable features and 10 principal components. We integrated the samples with Harmony71 with default parameters to correct for batch effects, then clustered the iDGE data with a resolution of 0.05. We identified gene and isoform markers for each cell-type within the data using the Seurat function FindMarkers with a two-tailed Wilcoxon rank-sum test, an adjusted p-value < 0.05, and avg.log2 fold-change cutoff of 0.1 (Supplementary Data 18 and 19. Finally using SCALPEL function FindIsoforms with a two-tail Chi-square statistical test and default parameters (FDR Adjusted p-value = 0.05, threshold.var = 0.05, threshold.abund = 0.1), we identified DIU genes between the distinct cell types (Supplementary Data 20).
Statistical analyses
We performed all the statistical tests in this manuscript using R v4.3.258.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The 10x scRNA-seq samples from male mouse germline23 have been downloaded from GEO database under the accession number GSE104556. The Drop-seq data from iPSCs and NPCs generated in this study have been deposited in the GEO database under accession number GSE268222. FASTQ files from the 10x Chromium 3’seq and PacBio platform for the mouse hippocampus and visual cortex have been downloaded from the Knowledge Brain Map database under the data repository RRID RRID:SCR_016152. Full scans of gels included in the main article are provided in the Source Data file. Full scans of gels included in the supplementary data are provided at the end of the supplementary data file. Raw/intermediate data including the single-cell datasets analyzed and the simulated reads generated in this paper are provided on Figshare (project: 249509). The data used for Fig. 2, S2, S3 are available at https://doi.org/10.6084/m9.figshare.29197175, https://doi.org/10.6084/m9.figshare.29107757; Fig. 3, S5, S6, S10a are available at https://doi.org/10.6084/m9.figshare.29180207; Fig. 4, S4, S7, S8, S10b are available at https://doi.org/10.6084/m9.figshare.29179352, https://doi.org/10.6084/m9.figshare.29195165; Fig. 5, S9 are available at https://doi.org/10.6084/m9.figshare.29191262.
Code availability
SCALPEL is implemented in R. The code of SCALPEL is publicly available and has been deposited in GitHub (https://github.com/plasslab/SCALPEL) under the GNU Affero General Public License (version 3 or later)76. The code of scr4eam is publicly available and has been deposited in GitHub (https://github.com/plasslab/scr4eam) under the GNU Affero General Public License (version 3 or later)77. All the scripts used in this study for the analysis and benchmarking are deposited in GitHub (https://github.com/Franzx7/SCALPEL_analysis_repo)78.
References
Gruber, A. J. & Zavolan, M. Alternative cleavage and polyadenylation in health and disease. Nat. Rev. Genet. 20, 599–614 (2019).
Tian, B. & Manley, J. L. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 18, 18–30 (2016).
Mayr, C. Regulation by 3′-untranslated regions. Annu. Rev. Genet. 51, 171–194 https://doi.org/10.1146/annurev-genet-120116-024704 (2017).
Mayr, C. Evolution and biological roles of alternative 3’UTRs. Trends Cell Biol. 26, 227–237 (2016).
Mayr, C. What are 3’ UTRs doing? Cold Spring Harb. Perspect. Biol. 11, a034728 (2019).
Lianoglou, S., Garg, V., Yang, J. L., Leslie, C. S. & Mayr, C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 27, 2380 (2013).
Sommerkamp, P., Cabezas-Wallscheid, N. & Trumpp, A. Alternative polyadenylation in stem cell self-renewal and differentiation. Trends Mol. Med. 27, 660–672 (2021).
Agarwal, V., Lopez-Darwin, S., Kelley, D. R. & Shendure, J. The landscape of alternative polyadenylation in single cells of the developing mouse embryo. Nat. Commun. 12, 5101 (2021).
Berry, C. W. et al. Developmentally regulated alternate 3′ end cleavage of nascent transcripts controls dynamic changes in protein expression in an adult stem cell lineage. Genes Dev. 36, 916–935 (2022).
Vallejos Baier, R., Picao-Osorio, J. & Alonso, C. R. Molecular regulation of alternative polyadenylation (APA) within the Drosophila nervous system. J. Mol. Biol. 429, 3290 (2017).
Mitschka, S. & Mayr, C. Context-specific regulation and function of mRNA alternative polyadenylation. Nat. Rev. Mol. Cell Biol. 23, 779–796 (2022).
Chen, H. et al. A distinct class of pan-cancer susceptibility genes revealed by an alternative polyadenylation transcriptome-wide association study. Nat. Commun. 15, 1729 (2024).
Gruber, A. J. et al. Discovery of physiological and cancer-related regulators of 3′ UTR processing with KAPAC. Genome Biol. 19, 44 (2018).
Jovic, D. et al. Single-cell RNA sequencing technologies and applications: A brief overview. Clin. Transl. Med. 12, e694 (2022).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Shulman, E. D. & Elkon, R. Cell-type-specific analysis of alternative polyadenylation using single-cell transcriptomics data. Nucleic Acids Res. 47, 10027–10039 (2019).
Patrick, R. et al. Sierra: Discovery of differential transcript usage from polyA-captured single-cell RNA-seq data. Genome Biol. 21, 1–27 (2020).
Wu, X., Liu, T., Ye, C., Ye, W. & Ji, G. scAPAtrap: identification and quantification of alternative polyadenylation sites from single-cell RNA-seq data. Brief Bioinform. 22, bbaa273 (2021).
Kang, B. et al. Infernape uncovers cell type-specific and spatially resolved alternative polyadenylation in the brain. Genome Res. 33, 1774 (2023).
Fansler, M. M., Mitschka, S. & Mayr, C. Quantifying 3′UTR length from scRNA-seq data reveals changes independent of gene expression. Nat. Commun. 15, 1–18 (2024).
Di Tommaso, P. et al. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 35, 316–319 (2017).
Zappia, L., Phipson, B. & Oshlack, A. Splatter: simulation of single-cell RNA sequencing data. Genome Biol. 18, 1–15 (2017).
Lukassen, S., Bosch, E., Ekici, A. B. & Winterpacht, A. Single-cell RNA sequencing of adult mouse testes. Sci. Data 5, 180192 (2018).
Gao, Y., Li, L., Amos, C. I. & Li, W. Analysis of alternative polyadenylation from single-cell RNA-seq using scDaPars reveals cell subpopulations invisible to gene expression. Genome Res. 31, 1856–1866 (2021).
Li, G. W. et al. SCAPTURE: a deep learning-embedded pipeline that captures polyadenylation information from 3′ tag-based RNA-seq of single cells. Genome Biol. 22, 1–24 (2021).
Lukassen, S., Bosch, E., Ekici, A. B. & Winterpacht, A. Characterization of germ cell differentiation in the male mouse through single-cell RNA sequencing. Sci. Rep. 8, 1–7 (2018).
Guvenek, A. & Tian, B. Analysis of alternative cleavage and polyadenylation in mature and differentiating neurons using RNA-seq data. Quant. Biol. 6, 253–266 (2018).
Kawahara, H., Imai, T. & Okano, H. MicroRNAs in neural stem cells and neurogenesis. Front. Neurosci. 6, 1–13 (2012).
Lang, M. F. & Shi, Y. Dynamic roles of microRNAs in neurogenesis. Front. Neurosci. 6, 22665 (2012).
Tsujimura, K., Shiohama, T. & Takahashi, E. microRNA Biology on Brain Development and Neuroimaging Approach. Brain Sci. 12, 1366 (2022).
Arancio, W., Sciaraffa, N. & Coronnello, C. MBS: a genome browser annotation track for high-confident microRNA binding sites in whole human transcriptome. Database 2023, 1–7 (2023).
Jonas, S. & Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 16, 421–433 (2015).
Ameri, A. J. & Lewis, Z. A. Shannon entropy as a metric for conditional gene expression in Neurospora crassa. G3 Genes|Genomes|Genetics 11, jkab055 (2021).
Li, W. et al. Alternative cleavage and polyadenylation in spermatogenesis connects chromatin regulation with post-transcriptional control. BMC Biol. 14, 1–17 (2016).
Liu, D. et al. Systematic variation in mRNA 3′-processing signals during mouse spermatogenesis. Nucleic Acids Res. 35, 234–246 (2007).
Liu, H. et al. IFT25, an intraflagellar transporter protein dispensable for ciliogenesis in somatic cells, is essential for sperm flagella formation†. Biol. Reprod. 96, 993–1006 (2017).
Sironen, A. et al. Loss of SPEF2 function in mice results in spermatogenesis defects and primary ciliary Dyskinesia1. Biol. Reprod. 85, 690–701 (2011).
Yogo, K. Molecular basis of the morphogenesis of sperm head and tail in mice. Reprod. Med. Biol. 21, e12466 (2022).
Meng, G.-Q. et al. Bi-allelic variants in DNAH3 cause male infertility with asthenoteratozoospermia in humans and mice. Hum. Reprod. Open 2024, hoae003 (2024).
Sutherland, J. M. et al. RNA binding protein Musashi-1 directly targets Msi2 and Erh during early testis germ cell development and interacts with IPO5 upon translocation to the nucleus. FASEB J. 29, 2759–2768 (2015).
Vanderzwalmen, P. et al. The problems of spermatid microinjection in the human: the need for an accurate morphological approach and selective methods for viable and normal cells. Hum. Reprod. 13, 515–519 (1998).
Joglekar, A. et al. Single-cell long-read sequencing-based mapping reveals specialized splicing patterns in developing and adult mouse and human brain. Nat. Neurosci.27, 1051–1063 (2024).
Frankish, A. et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773 (2019).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, 1–4 (2021).
Neph, S. et al. BEDOPS: high-performance genomic feature operations. Bioinformatics 28, 1919–1920 (2012).
Lawrence, M. et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 9, e1003118 (2013).
Hahne, F. & Ivanek, R. Visualizing genomic data using Gviz and bioconductor. Methods Mol. Biol. 1418, 335–351 (2016).
Zheng, G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 1–12 (2017).
Church, D. M. et al. Modernizing reference genome assemblies. PLoS Biol. 9, e1001091 (2011).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Shi, Y., Kirwan, P. & Livesey, F. J. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat. Protoc. 7, 1836–1846 (2012).
Gutiérrez-Franco, A. et al. Methanol fixation is the method of choice for droplet-based single-cell transcriptomics of neural cells. Commun. Biol.6, 1–12 (2023).
Macosko, E. Z. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Broad Institute. Picard toolkit (Broad Institute, GitHub repository, 2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Frankish, A. et al. GENCODE 2021. Nucleic Acids Res. 49, D916–D923 (2021).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2021).
Feng, X., Li, L., Wagner, E. J. & Li, W. TC3A: the Cancer 3′ UTR Atlas. Nucleic Acids Res. 46, D1027 (2018).
Fansler, M. M. Txcutr: transcriptome CUTteR. https://doi.org/10.18129/B9.bioc.txcutr (2025).
Conway, J. R., Lex, A. & Gehlenborg, N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940 (2017).
Diag, A., Schilling, M., Klironomos, F., Ayoub, S. & Rajewsky, N. Spatiotemporal m(i)RNA architecture and 3′ UTR regulation in the C. elegans germline. Dev. Cell 47, 785–800.e8 (2018).
Green, M. R. & Sambrook, J. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 2012).
Scotto-Lavino, E., Du, G. & Frohman, M. A. 3’ end cDNA amplification using classic RACE. Nat. Protoc. 1, 2742–2745 (2006).
Kozomara, A., Birgaoanu, M. & Griffiths-Jones, S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 47, D155–D162 (2019).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York, 2016).
Xie, Z. et al. Gene set knowledge discovery with Enrichr. Curr. Protoc. 1, e90 (2021).
Ashburner, M. et al. Gene Ontology: tool for the unification of biology. Nat. Genet 25, 25–29 (2000).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Abugessaisa, I., Noguchi, S., Carninci, P. & Kasukawa, T. The FANTOM5 computation ecosystem: genomic information hub for promoters and active enhancers. Methods Mol. Biol. 1611, 199–217 (2017).
Zhang, X. et al. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 47, D721–D728 (2019).
Moon, Y., Herrmann, C. J., Mironov, A. & Zavolan, M. PolyASite v3.0: a multi-species atlas of polyadenylation sites inferred from single-cell RNA-sequencing data. Nucleic Acids Res. 53, D197–D204 (2025).
Frankish, A. et al. GENCODE: reference annotation for the human and mouse genomes in 2023. Nucleic Acids Res. 51, D942 (2022).
Ake, F., Schilling, M. & Plass, M. Quantification of Transcript Isoforms at Single-cell Level Using SCALPEL https://github.com/plasslab/SCALPELhttps://doi.org/10.5281/zenodo.15553480 (2025).
Schilling, M., Ake, F. & Plass, M. Single-cell RNA-seq Realistic Random Read Emitting Awk Mess. https://github.com/plasslab/scr4eamhttps://doi.org/10.5281/zenodo.15553161 (2025).
Ake, F., Schilling, M. & Plass, M. Code for the Analysis and Bechmarking of SCALPEL https://github.com/Franzx7/SCALPEL_analysis_repohttps://doi.org/10.5281/zenodo.15553554 (2025).
Acknowledgements
The authors thank all members of the Plass Lab for useful comments and critical discussions. We thank Loris Mularoni from the Regenerative Medicine Program for his support with the cluster management. We also thank Nicole Grieger for her help setting up 3’ RACE and Yvonne Richaud-Patin and Dr. Zomeño from the Regenerative Medicine Program and IDIBELL’s Advanced Cell and Tissue Culture platform respectively for their help with iPSC cell culture. This research was funded by research projects awarded to M. P. from the State R&D Program Research Challenges from the Spanish Ministry of Science, Innovation and Universities, PID2019-108580RA-I00 funded by MICIU/AEI /10.13039/501100011033 and PID2022-139580OB-I00 funded by MICIU/AEI /10.13039/501100011033 and FEDER and by “ERDF A way of making Europe”, by the European Union and by grant CNS2023-144872 funded by MICIU/AEI /10.13039/501100011033 and by “European Union NextGenerationEU/PRTR”. M.P. work was supported by a Ramón y Cajal contract of the Spanish Ministry of Science, Innovation and Universities (Grant: RYC2018-024564-I funded by MICIU/AEI /10.13039/501100011033 and by “El FSE invierte en tu futuro”). L.L. work was supported by the National Natural Science Foundation of China (no. 32370721). F.A. work was supported by a predoctoral contract of the Spanish Ministry of Science, Innovation and Universities (Grant: PRE2020-094049 funded by MICIU/AEI /10.13039/501100011033 and by “FSE invierte en tu futuro”). A.J.G. work was supported by the predoctoral program AGAUR-FI ajuts (2024 FI-1 00072) Joan Oró, which is backed by the Secretariat of Universities and Research of the Department of Research and Universities of the Generalitat of Catalonia, as well as the European Social Plus Fund. We thank the CERCA Program/Generalitat de Catalunya for IDIBELL institutional support.
Author information
Authors and Affiliations
Contributions
F.A. developed SCALPEL. F.A., M.S., A.J.G., L.L. and M.P. designed the computational analyses. F.A., M.S. and A.J.G. performed computational analyses. F.A., M.S., A.J.G., and M.P. interpreted the data. A.G.F. generated scRNA-seq data. S.M.F.M., F.A., and M.P. selected candidate genes for validation. S.M.F.M. performed experimental validations. M.P. designed the project, acquired funding, supervised, and coordinated the work. F.A. and M.P. wrote the initial draft with input from all authors. All authors reviewed the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare the following competing interests: M.P. is an editorial board member of NPJ Systems Biology and Applications. F.A. is a Nextflow Ambassador of Seqera. The remaining authors declare no competing interests.
Ethics
The use of hiPSCs in this work was approved by the Spanish National Commission of Guarantees concerning the donation and use of human cells and tissues from the Carlos III National Institute of Health.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ake, F., Schilling, M., Fernández-Moya, S.M. et al. Quantification of transcript isoforms at the single-cell level using SCALPEL. Nat Commun 16, 6402 (2025). https://doi.org/10.1038/s41467-025-61118-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-61118-0