Abstract
Lack of standardization in biofoundries limits the scalability and efficiency of synthetic biology research. Here, we propose an abstraction hierarchy that organizes biofoundry activities into four interoperable levels: Project, Service/Capability, Workflow, and Unit Operation, effectively streamlining the Design‑Build‑Test‑Learn (DBTL) cycle. This framework enables more modular, flexible, and automated experimental workflows. It improves communication between researchers and systems, supports reproducibility, and facilitates better integration of software tools and artificial intelligence. Our approach lays the foundation for a globally interoperable biofoundry network, advancing collaborative synthetic biology and accelerating innovation in response to scientific and societal challenges.
Similar content being viewed by others
Introduction and motivations
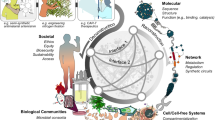
In June 2018, 15 noncommercial biofoundries from four continents gathered in London and agreed to establish the Global Biofoundry Alliance1, a collaborative effort to share experiences and resources while addressing common challenges and unmet scientific and engineering needs. Following the experience of the pandemic2, the importance of biofoundries as a main workforce of biomanufacturing and a sustainable bioeconomy has become even more highlighted. Biofoundries are more than facilities for conducting experiments using automated equipment; they are structured Research and Development (R&D) systems where biological design, validated construction, functional assessment, and mathematical modeling are performed following the Design-Build-Test-Learn (DBTL) engineering cycle1. A biofoundry can be used for conducting many heterologous experiments, necessitating the analysis of a wide range of different experimental protocols and biological assays. In synthetic biology and engineering biology various terms may be used interchangeably (and occasionally inappropriately), such as “protocols”, “Standard Operating Procedures”, “workflows”, and “tasks”. Or, for example, the term “protein design” sometimes refers only to the design step, but at other times it can refer to the entire DBTL process of protein design and engineering. For the operation of automated systems like biofoundries, it is essential to precisely define these concepts and the scope of terms used to describe different biofoundry activities. Synthetic biology is an applied field that merges disciplines from the life sciences and engineering, including molecular biology, chemical biology, genetics, bioinformatics, chemical and computer engineering. The experiments conducted in biofoundries extend beyond normal molecular and cell biology experiments and encompass a wide range of application-driven protocols and methods. This diversity and complexity underscore the need for a unified framework that not only standardizes terminologies and methodologies but also facilitates the exchange of best practices across biofoundries3. Therefore, it is timely to build an international collaborative network for sharing biofoundry methodologies and applications using common terminology and standardized methods.
Given that biofoundry workflows span from low‑throughput manual protocols to high‑throughput operations using 96‑, 384‑, and 1536‑well plates, quantitative metrics are crucial for benchmarking performance improvements, ensuring reproducibility, and maintaining operational quality across scales. These metrics also enable performance comparisons across different biofoundries, whether the processes involve semi-automated workflows with manual plate transfers between instruments or fully automated workflows using robotic arms4. However, developing such quantitative metrics requires a foundational framework based on standardized protocols. Once standardized workflows are established, biofoundries can create reference materials and calibration tools to assess reproducibility and quality levels, enabling measurement comparisons across different instruments. Prioritizing the standardization of workflows as a prerequisite for metric development enhances the reliability and interoperability of biofoundry operations. This approach not only ensures consistent performance across facilities but also mitigates the adverse effects of monopolies by equipment manufacturers, fostering a more collaborative and equitable biofoundry ecosystem.
Shifting to a biofoundry environment introduces challenges in adapting experimental protocols. Many existing lab-based synthetic biology protocols are optimized for manual execution and often omit details that are assumed to be obvious to trained researchers. When these protocols are directly applied to automated biofoundry platforms, which typically operate in 96/384-well plate formats and use liquid-handling robots, differences in sample volumes, concentrations, and equipment specifications can result in deviations from expected outcomes. In other words, protocols that work reliably in manual settings may yield inconsistent results in automated environments unless they are explicitly adapted for such systems. Additionally, human-executed protocols often omit obvious steps in publications or laboratory manuals, such as sample preparation. Automated workflows, however, require precise definitions of the location, state, quantity, and behavior of all materials used. The same equipment is used differently depending on the application, and equipment turnover, in which older instruments are replaced by new ones, further complicates reproducibility. These challenges underscore the need for highly abstracted workflows that encapsulate biofoundry-specific processes while accommodating automation variability.
Abstraction hierarchy for biofoundry operations
To address the issues of biofoundry interoperability, we have designed a flexible abstraction hierarchy for the operation of a biofoundry (Fig. 1). Level 0 refers to the Project that is to be carried out in the biofoundry. This represents a series of tasks to fulfill the requirements of external users who wish to use the biofoundry. Level 1 Service/Capability, refers to the functions that external users require from the biofoundry and/or that the biofoundry can provide. Level 2, Workflow, refers to the DBTL-based sequence of tasks needed to deliver the Service/Capability. Each workflow is intentionally assigned to a single stage of the DBTL cycle to ensure modularity and clarity in execution. Level 3 is Unit-operations which represents the actual hardware or software that will perform the tasks required to fulfill the desired workflow. Engineers or biologists working at the highest abstraction level do not need to understand the lowest Level 3 operations.
Level 1: services and capabilities
Researchers and companies in the field of biotechnology can leverage the specialized services and capabilities provided by biofoundries to achieve their R&D project goals. Examples include modular long-DNA assembly or Artificial Intelligence (AI) driven protein engineering. In this report, a biofoundry capability refers to the specialized processes or activities conducted by biofoundries where clients can be from both academia and industry (including startups/spinouts, SMEs and larger organizations). Biofoundry services can be divided into various tiers—these range from simply providing access to specialist equipment to offering a fully comprehensive support package from project conception to commercialization and scale-up. We can categorize these tiers of services/capabilities in relation to the synthetic biology DBTL cycle (Table 1).
Level 2: workflows
A service/capability consists of sequentially and logically interconnected multiple workflows. Workflows are designed to be highly abstracted and modularized for clarity and reconfigurability. Although Workflow has been used to describe the entire DBTL cycle, here we introduce functionally modular workflows for each stage of the DBTL cycle. Table S1 shows 58 biofoundry workflows with short descriptions. Each workflow is assigned to one of the specific Design, Build, Test, or Learn stages. These workflows encompass the diversity and complexity of synthetic biology experiments, allowing the reconfiguration and reuse of workflows to achieve different functional and executable outcomes. For example, the DNA Oligomer Assembly workflow could be understood to indicate the entire DBTL process for constructing a complete target gene sequence. However, here we use it specifically to define the DNA assembly step where DNA oligomers are assembled. This allows for the development of an ontology of specific actions (workflows) that define the individual steps required to fulfill the entire synthetic biology DBTL cycle. The modularized workflows can be arranged sequentially to perform arbitrary services. Figure S1 represents an example of a protein library construction service.
Level 3: unit-operations
We define unit operations as the lowest abstraction hierarchy level. Unit operations indicate individual experimental or computational tasks. These tasks can be conducted by automated instruments or software tools. By combining unit operations in a sequential manner, workflows can be designed for specific biological tasks. Table S2 and Table S3 show unit operations for hardware and software, respectively. A hardware unit operation can be considered the smallest unit of operation for an experiment corresponding to one or more pieces of equipment. For example, the Liquid Transfer unit operation is an experiment that can be performed by a single liquid handling robot, including PCR setup, dilution, and dispensing. For software unit operations, they are defined based on a software application or package as the smallest unit of operation for an experiment. For example, Protein Structure Generation unit operation is performed for example by RFdiffusion5 software application. We propose an initial set of 42 unit operations for hardware (Table S2) and 37 unit operations for software (Table S3). As an example, DNA Oligomer Assembly (WB010) workflow can be represented by 14 unit operations as described in a protocol for synthetic genome synthesis6 (Table S4, Fig. S2).
Flexibility for general applicability
The modular workflows and unit operations defined here describe various synthetic biology experiments through the reconfiguration and reuse of these elements. However, due to the diversity of biological experiments and the continuous development of improved equipment and software, detailed protocols may vary, which can limit the general applicability of fixed workflows and unit operations. For example, the Liquid Media Cell Culture (WB140) workflow could refer to simple liquid culture for DNA amplification or could include a culture process involving cell-based enzyme assays. In other words, the same workflow or unit operation name can encompass different experimental processes depending on the objectives of the biological experiments. Additionally, workflows or unit operations may differ among laboratories depending on the functionality of their available equipment. For instance, the DNA Extraction (WB045) workflow involves sequential unit operations such as cell lysis and centrifugation. However, some automated equipment can perform the entire DNA purification process in a single operation, so the Nucleic Acid Extraction (UH250) unit operation has been separately added to account for such cases. Similarly, some automated parallel fermenters with functionalities like HT Aerobic Fermentation (UH180) and Microbioreactor Fermentation (UH200) may integrate Microplate Reading (UH370) or simple metabolic/sugar detection functionalities.
These challenges highlight the importance of establishing data standards and methodologies for protocol exchange. Existing standards such as Synthetic Biology Open Language (SBOL)7 and Laboratory Operation Ontology (LabOp)8 provide good starting points for describing protocols and workflows in a standardized format. In particular, SBOL’s data model is well-suited to represent each stage of the Design, Build, Test, and Learn cycle, and it offers a range of tools9 that support data sharing between users, making it compatible with the workflow abstraction proposed in this study. Developing and collecting biofoundry-specific protocols tailored to diverse workflows will be crucial for achieving greater interoperability and reproducibility across biofoundries. This initial version of workflows and unit operations proposed here focuses more on a conceptual framework, definition and classification for biofoundry operations rather than precise definitions. Additionally, a set of unit operations can often resemble familiar protocols with slight variations in methods and naming conventions across laboratories. For example, Golden Gate Assembly, a well-known assembly protocol in synthetic biology, can be viewed as the sequential use of unit operations such as Liquid Handling for DNA part preparation and Thermocycling for enzyme reactions and annealing. This set of unit operations could be named as a distinct Golden Gate Assembly workflow, though further discussions would be required to formalize this classification. However, our proposed conceptual framework allows biofoundry operations to be classified and shared, leading to more standardized operations and the development of calibrants and measurands to allow comparison and interoperability.
Software tools and data management
Ensuring that biofoundry-generated protocols and data are reusable, interoperable, and accessible across diverse systems and institutions will require alignment with the Findable, Accessible, Interoperable, and Reusable (FAIR) principles10, which are essential for effective biofoundry design and software integration. The workflows and unit operations proposed here, for each stage of the DBTL cycle, need to be supported by software tools on multiple levels. For example, the Design step requires Computer-Aided Design (CAD) tools; the Build step requires simulation of laboratory operations and translation of protocols into robotic instructions, via files or application programming interfaces (APIs). The Test stage requires bioinformatics pipelines for data analysis, and finally, the Learn stage is supported by mathematical and other computational modeling tools.
Due to limitations of hardware drivers, a soft integration approach that consolidates data is one of the best options for early-stage biofoundries. Using an integrated database as a single source of truth aligns well with the FAIR principles. However, each unit operation generates a variety of metadata such as operational logs, experimental conditions11, and biological raw data12,13 requiring careful curation and integration of relevant information. To address this, implementing an API service that runs independently on the computer controlling each piece of equipment, as part of a distributed data management system, would allow seamless accessibility from anywhere.
Software tools for biofoundries must efficiently analyze large volumes of biological data and manage a wide variety of diverse experiments. Laboratory Information Management Systems (LIMS) and Electronic Lab Notebooks (ELNs) are essential for comprehensive data management, working in tandem with specialized tools tailored to specific experiments or analytical tasks. Well-known open-source ELN-LIMS solutions include openBIS14, Aquarium15, Leaf-LIMS16, and Galaxy-SynBioCAD17, while Teselagen Operating System18 and Benchling19 are recognized end-to-end commercial solutions. To enable the configurability and flexibility of the workflow approach proposed here, the software tools are best implemented using a modular architecture. This approach accommodates the unique setup of individual biofoundries and makes it easier to add new features or tools to support novel projects. A microservices architecture consisting of smaller, independently functioning applications simplifies adding or modifying services to adapt to specific workflows. This architecture is flexible, scalable, and adaptable to meet diverse biofoundry needs. A microservice architecture with multiple applications specialized for different workflows is more suitable for diverse biofoundry operations than an all-encompassing solution. These applications should be developed with separate front-end and back-end components, adhere to Representational State Transfer (REST) principles20, and be deployed using containerization technologies like Docker and Kubernetes.
An example is the Edinburgh Genome Foundry’s software suite21 that enables in silico sequence design, modification and cloning; simulation of protocols by modeling microplates and liquid transfers; and QC through design and analysis of sequencing data. The suite is made up of several independent libraries (packages of the Python programming language) that, for each workflow, can be operated individually via a graphical interface (web apps) or are linked together with a shell script. Using scripts to utilize software to perform the required steps, as opposed to a manual procedure, is preferable as it has the same advantages as laboratory automation protocols, namely: batch processing, self-documentation, precision, reproducibility and speed22. Ideally, these tools, and the scripts (which represent protocols), are distributed under a free and open-source license, which is both cost-efficient and allows quick and immediate sharing of expertise and developments between biofoundries and other users.
ELNs play a crucial role in integrating various applications and databases, consolidating the planning and results of experiments, and providing a central source of information. Flexibility can be maximized by using natural language-based software tools, such as ELNs, to conduct actual biofoundry experiments. Incorporating natural language to describe experiments enhances the flexibility of workflows and unit operations. A recently proposed approach based on literate programming23, which integrates text and computer code, offers new possibilities for future ELN development. The ability to embed computer code in ELNs is crucial for extending their functionality and interacting with other biofoundry applications. In this regard, open-source programming editors like Jupyter Notebook, RStudio(with Quarto), and VSCode are among the best options for use as a biofoundry ELN. Each of these editors can also be leveraged in cloud environments such as Google Colab, Posit Workbench and GitHub Codespaces, respectively. However, it is important to note that many institutions and companies require their data to remain outside the cloud due to security concerns. Furthermore, as data volumes grow and project durations extend, the high cost of cloud storage can pose a financial burden for biofoundry operations. Therefore, adopting a strategy that combines the advantages of local storage and cloud environments is essential to balance cost and accessibility effectively.
For compatibility with ELNs, we illustrate a Tier 3-level Service/Capability example (Supplementary Information) focused on Part DNA Assembly workflows. This example shows the design of workflows (Table S5, Fig. S3), provides corresponding experimental records structured according to modular unit operations (Table S6) and its rendered screenshot (Fig. S4). Each modular unit operation is documented in Markdown format using natural language, with explicit specifications for title, metadata, inputs, outputs, equipment, reagents, and sample IDs, thereby ensuring full traceability across the workflow. This example illustrates the possibility of how biofoundry experiments built on an abstraction hierarchy framework, can contribute to improved reusability, modularity, and enhanced interoperability across different biofoundries.
Discussions and future directions
Compared to a regular laboratory, a biofoundry must comprehensively manage a significantly larger number of equipment, materials, data, experiments, and operations. This necessitates a robust operational framework that ensures seamless functionality, including equipment accessibility, consistent material supply, and rapid analysis of collected data to guide subsequent experimental designs. Biofoundries integrate various automated equipment that should be cohesively connected and substituted with devices from different manufacturers, emphasizing the need for a standardized operational platform. This platform should independently manage user-designed workflows and data, separate from vendor-dependent hardware. RESTful APIs might be useful for effectively translating information exchanged between these workflows and automated equipment. By developing an open lexicon and ontology, multiple public-funded biofoundries can foster cooperation and collaboration on an international scale. While private-sector biofoundries often employ proprietary toolchains that limit broader interoperability, our proposed standardization efforts primarily target public-sector and newly emerging biofoundries that require accessible and flexible operational frameworks. Rather than attempting to encompass all proprietary systems, we emphasize the use of community-driven open-source standards, such as SBOL and LabOp, to overcome technical barriers and accelerate the establishment of interoperable biofoundry infrastructures. A recent report highlighted the need for the development of technical standards and metrics for engineering biology3, and biofoundries could play a leading role in enabling such developments.
AI is essential for enhancing the operational efficiency of biofoundries. High construction and operational costs have been identified as significant challenges, with operational expenses particularly threatening the sustainability of biofoundries. AI models capable of analyzing biological and equipment log data generated in biofoundries will be critical for mitigating these risks. The operational efficiency of a biofoundry is directly related to the efficiency of the workflows, such as minimizing consumable usage and saving time and labor within workflows. Optimizing overall biofoundry operations requires a scheduling algorithm that allows multiple workflows to run simultaneously which minimizes interference between them. To optimize the use of limited equipment, it is crucial to continuously monitor the availability of both equipment and materials, maximize the utilization of available time, and effectively coordinate the workflows of various users. AI models are also indispensable for predicting errors and equipment failures during experiments, which helps minimize idle time. This involves collecting data from equipment log files and using additional edge devices to monitor each piece of equipment. Combining AI for real-time task scheduling with predictive modeling for potential failures creates a resilient and adaptive system. Furthermore, biofoundries are uniquely positioned to provide highly curated and quality-assured datasets, which are critical for the development of robust AI/ML models. By leveraging their ability to generate standardized, high-quality data, biofoundries can significantly accelerate advancements in AI/ML-driven R&D. Text-based descriptions of workflows and unit operations in ELNs (Table S6) will be comprehensively extended by large language models, bringing innovative changes to R&D processes in biofoundries.
As a follow-up study, developing quantitative metrics to compare workflow performance comparison and evaluate QC is essential for enhancing reproducibility and maintaining high-quality performance in a biofoundry. For example, quality metrics such as cloning success rates can be compared between traditional manual vector construction and automated equipment outcomes. Throughput metrics can measure the workload completed within the same time frame and scale by manual researchers versus automated systems. Capacity metrics could include the number of DNA, plasmids, or RNA synthesized within a given timeframe, as well as the number of strains constructed. Strain construction metrics, often derived from multiple workflows, serve as a representative indicator of overall biofoundry performance. Establishing such metrics requires clear definitions, precise explanations, and measurable formulas. Collaboration within international partner institutions is essential, not only for building workflows but also for gathering input on metric development and selection. Such collaboration will facilitate the identification and adoption of appropriate metrics that accurately reflect biofoundry performance.
The abstraction hierarchy framework proposed here will streamline the integration of diverse protocols and serve as a foundation for standardization efforts, ensuring reproducibility and facilitating interoperability across biofoundries. These advancements will enhance the flexibility of workflow management and establish a strong foundation for distributed biofoundry networks. Such networks, supported by AI, standardized data, and workflows, represent a transformative step toward a sustainable bioeconomy and the capacity to address complex global challenges, including pandemics24.
References
Hillson, N. et al. Building a global alliance of biofoundries. Nat. Commun. 10, 2040 (2019).
Crone, M. A. et al. A role for Biofoundries in rapid development and validation of automated SARS-CoV-2 clinical diagnostics. Nat. Commun. 11, 4464 (2020).
Freemont, P. Engineering Biology Metrics and Technical Standards for the Global Bioeconomy. https://spiral.imperial.ac.uk/handle/10044/1/110822https://doi.org/10.25561/110822 (2024).
Martin, H. G. et al. Perspectives for self-driving labs in synthetic biology. Curr. Opin. Biotechnol. 79, 102881 (2023).
Watson, J. L. et al. De novo design of protein structure and function with RFdiffusion. Nature 620, 1089–1100 (2023).
Hutchison, C. A. et al. Design and synthesis of a minimal bacterial genome. Science 351, aad6253 (2016).
Buecherl, L. et al. Synthetic biology open language (SBOL) version 3.1.0. J. Integr. Bioinform. 20, 20220058 (2023).
Bartley, B. et al. Building an open representation for biological protocols. J. Emerg. Technol. Comput. Syst. 19, 1–21 (2023).
SynBioDex/SBOL-utilities. Command-line utilities and scripts for manipulating SBOL data. https://github.com/SynBioDex/SBOL-utilities.
Wilkinson, M. D. et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3, 1–9 (2016).
Barrett, T. et al. BioProject and biosample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res. 40, D57–D63 (2012).
Ham, T. S. et al. Design, implementation and practice of JBEI-ICE: an open source biological part registry platform and tools. Nucleic acids Res. 40, e141–e141 (2012).
McLaughlin, J. A. et al. SynBioHub: a standards-enabled design repository for synthetic biology. ACS Synth. Biol. 7, 682–688 (2018).
Barillari, C. et al. openBIS ELN-LIMS: an open-source database for academic laboratories. Bioinformatics 32, 638–640 (2016).
Vrana, J. et al. Aquarium: open-source laboratory software for design, execution and data management. Synth. Biol. 6, ysab006 (2021).
Craig, T. et al. Leaf LIMS: a flexible laboratory information management system with a synthetic biology focus. ACS Synth. Biol. 6, 2273–2280 (2017).
Hérisson, J. et al. The automated Galaxy-SynBioCAD pipeline for synthetic biology design and engineering. Nat. Commun. 13, 5082 (2022).
TeselaGen Operating System Overview-Teselagen. https://teselagen.com/teselagen-operating-system-overview/ (2021).
Cloud-based platform for biotech R&D | Benchling. https://www.benchling.com.
Fielding, R. T. REST: Architectural Styles and the Design of Network-based Software Architectures. Doctoral dissertation, University of California (2000).
Edinburgh Genome Foundry. GitHub https://github.com/Edinburgh-Genome-Foundry.
Zhang, X. & Jonassen, I. RASflow: an RNA-Seq analysis workflow with Snakemake. BMC Bioinform. 21, 110 (2020).
Petersen, S. D. et al. teemi: An open-source literate programming approach for iterative design-build-test-learn cycles in bioengineering. PLoS Comput. Biol. 20, e1011929 (2024).
Vickers, C. E. & Freemont, P. S. Pandemic preparedness: synthetic biology and publicly funded biofoundries can rapidly accelerate response time. Nat. Commun. 13, 453 (2022).
Dauparas, J. et al. Robust deep learning–based protein sequence design using ProteinMPNN. Science 378, 49–56 (2022).
Robinson, C. J. et al. Rapid prototyping of microbial production strains for the biomanufacture of potential materials monomers. Metab. Eng. 60, 168–182 (2020).
Acknowledgments
We thank all members of the SBLab at KRIBB for their valuable advice and support throughout this research. We also gratefully acknowledge the use of the KRIBB Biofoundry-beta facility. This research was funded by the Bio & Medical Technology Development Program (grant number RS-2024-00335300, RS-2024-00508861, RS-2024-00509115, and RS-2024-00445145) of the National Research Foundation, funded by the Ministry of Science and ICT of the Republic of Korea, and the KRIBB Research Initiative Program. P.S.F. thanks UKRI EPSRC (EP/5001859/1) and Schmidt Sciences for funding. This work was part of the Agile BioFoundry (agilebiofoundry.org) supported by the U.S. Department of Energy, Energy Efficiency and Renewable Energy, Bioenergy Technologies Office through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy. The views and opinions of the authors expressed herein do not necessarily state or reflect those of the U.S. Government or any agency thereof. Neither the U.S. Government, nor any agency thereof, nor any of their employees makes any warranty, expressed or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed or represents that its use would not infringe privately owned rights. The U.S. Government retains and the publisher, by accepting the article for publication, acknowledges that the U.S. Government retains a nonexclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allow others to do so, for U.S. Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan). Funding for open access charge: U.S. Department of Energy.
Author information
Authors and Affiliations
Contributions
H.K. S.-G.L., and P.S.F. conceptualized the study. H.K., D.-H.L., W.S. and B.H.S. developed the abstraction hierarchy and drafted the manuscript. H.K., N.J.H. and M.S. curated and organized the workflow and unit operation dataset. B.-K.C., D.-M.K., M.-K.O., M.W.C. and Y.-S.J. contributed domain-specific insights into biofoundry operations and synthetic biology protocols. S.J.R., P.V., R.F., R.L.F. and N.S.S. provided critical feedback on interoperability and standardization frameworks. All authors reviewed and edited the manuscript. S.-G.L. and P.S.F. supervised the project and secured funding.
Corresponding authors
Ethics declarations
Competing interests
The authors declare the following competing financial interest(s): N.J.H. has financial interests in TeselaGen Biotechnologies and Ansa Biotechnologies. All other authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Chris Myers and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kim, H., Hillson, N.J., Cho, BK. et al. Abstraction hierarchy to define biofoundry workflows and operations for interoperable synthetic biology research and applications. Nat Commun 16, 6056 (2025). https://doi.org/10.1038/s41467-025-61263-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-61263-6