Abstract
Eating disorders arise from a complex interaction of genetic and environmental influences. Here we provide comprehensive population-level estimates of the heritability of eating disorders and their genetic relationships with various mental health and cardiometabolic disorders (CMDs), expanding beyond genome-wide association studies. We examined the heritability of three eating disorders—anorexia nervosa (AN), bulimia nervosa (BN), and other eating disorders (OED)—and investigated shared familial and genetic risk factors with mental health disorders and CMDs. Using national register data from Denmark and Sweden (1972–2016), we analysed clinical diagnoses for over 67,000 individuals with eating disorders, their first-degree relatives, and matched controls from populations totalling 17 million. Heritability estimates were moderate, h2AN = 36%, h2BN = 39%, and h2OED = 30% and genetic correlations revealed substantial overlap between AN and obsessive-compulsive disorder (rg = 0.65) and moderate correlations with other mental health disorders such as autism (rg = 0.36). Significant genetic associations were also identified between eating disorders and CMDs, showing strong replication across both countries. These findings emphasise the genetic foundations of eating disorders and their shared genetic architecture with mental health and CMDs. This research enhances our understanding of comorbidity patterns and has important implications for developing integrated treatment approaches.
Similar content being viewed by others
Introduction
Eating disorders (ED) such as anorexia nervosa (AN), bulimia nervosa (BN), binge-eating disorder (BED), and other eating disorders (OED), are often chronic and debilitating disorders occurring in 5–10% of the general population1. As with all psychiatric disorders, both genes and the environment contribute to the risk of developing an ED. The role of genetics in eating disorders has been well-established using several study designs (e.g. family- and twin studies)2, and more recently via molecular techniques such as genome-wide association studies (GWAS)3,4,5. These different study designs have provided ample evidence of the heritability of eating disorders, the occurrence within families, the potential involvement of specific gene variants, and the genetic correlations with other disorders. Heritability estimates ascertained from twin studies vary across eating disorders6. For instance, of these three disorders, AN has the highest reported twin-based heritability but also a broad range of estimates (0.28–0.74)7,8,9, followed by BN (0.55–0.62)9,10,11,12, and BED (0.39–0.45)11,13,14. Although not the focus of this study, a recent study has shown high twin-based heritability (0.79; 95% CI: 0.70–0.85) of broad avoidant restrictive food intake disorder (ARFID)15. Moderate heritability estimates for individual symptoms of eating disorders have also been shown, such as weight and shape concern (0.43)16,17, binge eating (0.49)16,17, and self-induced vomiting (0.72)18, with some variability across studies19.
Historically, genetic studies, such as GWAS, have focused heavily on AN3,4,20,21,22. However, a recent GWAS identified multiple common genetic risk variants associated with a BED phenotype obtained via machine-learning23. The discovery of more risk variants is likely given that a larger AN GWAS and the first binge-eating GWAS are currently underway and nearing completion24. Both twin studies and GWAS (specifically those of AN) have demonstrated the genetic overlap across eating disorders10, and between eating- and other psychiatric disorders3,4,25,26. The strongest positive genetic correlations with AN have been identified for obsessive-compulsive disorder (OCD), major depressive disorder (MDD), schizophrenia, and anxiety. In contrast, negative genetic correlations have been shown with cardiometabolic- and anthropometric traits (e.g. body fat percentage, insulin resistance, and leptin)3,4. In a recent study, we showed that whilst AN, BN, and BED share genetic risk with other psychiatric disorders, they differ in their shared genetic risk with cardiometabolic and anthropometric traits5. For example, whereas BED has positive genetic similarity with waist circumference and obesity, opposite patterns were observed for AN.
Twin studies and GWAS are useful in estimating the heritability and genomic architecture of complex traits and diseases; however, they have methodological limitations. First, both methods are often applied to highly self-selected clinical samples or rely on self-reported data, leading to a study sample that is not representative of the general population and thus potentially over- or underestimating heritability. Second, single-nucleotide polymorphism (SNP)-based heritability estimates are consistently lower than twin-based ones27. This is in large part because GWAS exclusively assesses the association between common additive genetic variants and a disorder, and thus does not capture the effect of gene-environment interactions, rare variants, and other non-additive contributions. The greatest limitation of twin-based studies is reliance on the equal environment assumption, which assumes that mono- and dizygotic twins are similarly exposed to relevant environmental factors, biasing heritability estimates upwards. Indeed, a recent register-based study in Denmark supported the non-representativeness of twins as it showed that compared to singletons, twins carry a 40% increased risk, whilst triplets/quadruplets carry a 92% increased risk for developing AN28. Therefore, the estimates using twin populations only may not reflect the genomic architecture of the general population, as they are likely to be biased upwards.
Building on existing knowledge and recent methodological advances29,30, we leveraged nationwide healthcare registers and near-complete population genealogies of ~17 million individuals spanning four generations across Denmark and Sweden, to: (a) estimate the risk of eating disorders in first-degree relatives of probands with eating disorders; (b) determine the heritability of three eating disorders; and (c) investigate co-heritability (i.e. the genetic correlation) amongst specific eating disorders and, between eating disorders and multiple psychiatric disorders and cardiometabolic diseases (CMD).
Results
Descriptive statistics
The prevalence of AN and BN was close to 0.5% in both countries, but was higher in Denmark than in Sweden for individuals born between 1968 and 2000 (AN: 0.59% vs 0.53% and BN: 0.48% vs 0.32%). These differences were more pronounced within the younger (1985–2000) cohort (AN: 0.80% vs 0.72% and BN: 0.55% vs 0.37%). Conversely, the prevalence of OED in Sweden was higher than that of Denmark in both the older (1.02% vs 0.6%) and younger cohort (1.40% vs 0.79%). As expected, in both countries individuals who received an eating disorder diagnosis in were substantially more likely to be female than male (13:1). However, we observed small differences in sex-specific eating disorder prevalence between Denmark and Sweden; with BN having a higher prevalence in males in Sweden (3.4% vs 2.5%, p = 1.19 × 10−3) and OED being observed more commonly in Danish males (9.3% vs 7.6%, p = 1.01 × 10−8). We observed a significant difference in the use of in- and outpatient registration between the two countries (Table 1). For instance, Danish individuals with an AN or OED diagnosis were more likely to be treated exclusively as inpatients compared to Swedish individuals with the same diagnosis (ORAN = 3.83, OROED = 1.19), who were more likely to be treated via the outpatient hospital system (Supplementary Data 2A). The opposite relationship was observed for BN. Overall, Danish individuals were significantly more likely to have both an in- and outpatient recording for all three EDs, potentially suggesting a hospitalisation trajectory. Individuals in Sweden were significantly younger when receiving their first AN diagnosis (median age in years: 17.8 vs 18.6, p = 9.74 × 10−11) or OED (20 vs 21, p = 7.09 × 10−17), whereas Danish individuals received a BN diagnosis at a younger age (24.3 vs 26.3, p = 1.30 × 10−47). Across all eating disorder diagnoses, Danish individuals were, on average, born earlier than Swedish individuals and diagnosed earlier, based on calendar years (Table 1, Supplementary Figs. 1–3, and Supplementary Note 3). These differences in prevalence, age at first diagnosis, year of birth, and year of diagnosis were too small to be clinically impactful but statistically significant due to the large sample size.
We observed a statistically significant difference between the number of eating disorder diagnoses per individual in each country. For instance, individuals diagnosed in Denmark were substantially more likely to have an exclusive diagnosis of AN (OR = 1.88, p < 1 × 10−99) or BN (OR = 2.54, p < 1 × 10−99), whilst individuals diagnosed with an eating disorder in Sweden were more likely to have exclusively an OED diagnosis (OR = 0.41, p < 1 × 10−99). If multiple eating disorder diagnoses were given, Swedish individuals were more likely than Danish individuals to receive a diagnosis of AN (OR = 0.65, p = 4.74 × 10−75) or BN (OR = 0.84, p = 4.27 × 10−7) together with an OED diagnosis (Fig. 1), whereas Danish individuals were more likely to have both a diagnosis of AN and BN (OR = 3.06, p = 1.22 × 10−83) compared with individuals in Sweden. More can be found in Supplementary Data 2A, B.
Familial risk
To assess the familial risk of eating disorders, we compared CIFs between individuals with a first-degree family member (i.e. parents and siblings) diagnosed with an eating disorder relative to the risk in the general population. The incidences were calculated from the number of new cases occurring for each age year. We calculated cumulative incidences up to a maximum of age 49 for birth cohort 1968–2000 and age 32 for birth cohort 1985–2000. We observed that individuals were substantially more likely to receive an eating disorder diagnosis if they had a first-degree relative (i.e. parent or full sibling) diagnosed with the same disorder (Fig. 2). In general, the probability of an eating disorder increased on average three-fold for all disorders when having either a parent or full sibling with an eating disorder diagnosis. However, in Sweden, children were less likely to receive a BN diagnosis if they had a parent with a BN diagnosis, regardless of birth cohort, compared to Danish individuals, who had an increased risk of 1.5- to 8.3-fold. This might, in part, be due to differences in recording time and left-sided censoring of medical data, as well as children of BN-diagnosed individuals in Sweden being too young to be diagnosed themselves. All estimates can be found in Supplementary Data 3.
Cumulative incidences (%) and 95% confidence intervals (reported as cumulative incidence estimate ± 1.96 × standard error) for eating disorders were calculated for different birth cohorts a 1985–2000 (nDenmark = 1,026,609, nSweden = 1,698,409) and b 1968–2000 (nDenmark = 2,124,093, nSweden = 3,435,832) using medical records up to 2016. Cumulative incidences were calculated for: the general population (all individuals born in the birth window), individuals with at least one full sibling diagnosed with the same eating disorder, and individuals with at least one parent diagnosed with the same eating disorder. Individuals diagnosed before age 10 were excluded. Risk estimates were taken, if possible, at age 49 (1968–2000) and 32 (1985–2000). General population confidence intervals are narrow due to the large sample size and may appear overlapping or not visible.
Heritability estimates (h 2)
To estimate the heritability of eating disorders, we combined familial risk estimates as described in the methods section. We calculated \({h}_{{AN}}^{2}\) as 0.36 (95% CI: 0.30–0.41), \({h}_{{BN}}^{2}\) as 0.39 (95% CI: 0.32–0.46), and \({h}_{{OED}}^{2}\) as 0.30 (95% CI: 0.20–0.40, Fig. 3). The h2 of AN and BN were generally lower than previously reported estimates, however it should be noted that these estimates varied widely (0.25–0.8)2,12,31. The heritability of OED as defined in this study has, to our knowledge, not been studied enough to make an adequate comparison. We observed no statistically significant difference between country-specific estimates (Bonferroni p < 3.33 × 10−3) (Supplementary Data 4).
All within-country (Denmark and Sweden) h2 estimates reported per birth year were first meta-analysed using random effect inverse variance weighting, followed by a between-country meta-analysis using the same method. Confidence intervals are reported as a meta-analysis of Danish and Swedish narrow-sense h2 estimates per birth year ± 1.96 × standard error.
Due to the large sex difference in eating disorder prevalence (>90% female) and small sample size in the Danish cohort, the heritability of eating disorders in females was nearly identical to the non-sex-stratified heritability. In comparison, the male estimates presented substantially large confidence intervals, or no heritability could be calculated (BN). We did not observe significant differences in sex-stratified h2 estimates of eating disorders in the Swedish sample (Supplementary Data 4). Estimates of h2 of other mental health disorders and CMD, as well as all meta-analysed estimates of h2 including SE, CIs, and p-values, are reported in Supplementary Data 4.
We performed a sensitivity analysis to assess the effect of exclusion criteria on the heritability (Supplementary Data 4). First, we assessed the effect of inpatient and outpatient recording by estimating the h2 separately. The \({{{{\rm{h}}}}}_{{{{\rm{OED}}}}}^{2}\) using inpatient ICD-10 diagnoses was 0.6 (95% CI: 0.42–0.77), significantly higher (Bonferroni p < 1.15 × 10−3) than the \({{{{\rm{h}}}}}_{{{{\rm{OED}}}}}^{2}\) using outpatient ICD-10 diagnoses: 0.27 (95% CI: 0.19–0.36) using Danish individuals, and we observed no such difference using Swedish data. No significant differences between in- and outpatient heritability estimates of AN and BN were observed. Furthermore, no significant difference was observed between the \({{{{\rm{h}}}}}_{{{{\rm{AN}}}}}^{2}\) diagnosed under ICD-8 or ICD-9 (exclusively inpatient) and the \({{{{\rm{h}}}}}_{{{{\rm{AN}}}}}^{2}\) diagnosed as inpatient using ICD-10 (p = 0.26). All estimates of h2 using various exclusion criteria can be found in Supplementary Data 5 and Supplementary Note 4.
Genetic correlations (r g)
We first quantified the genetic relationship across the three eating disorders via genetic correlation analyses. Next, we calculated genetic correlations across the three eating disorders by performing multiple between-disorder comparisons using (a) seven other mental disorders, and (c) six CMDs. All analyses were conducted using data from Denmark and Sweden using both full-sibling and parent-offspring comparisons. All estimates of rg are reported in Supplementary Data 6 and 7.
Between eating disorders
Genetic correlation analysis between eating disorders using full-sibling information showed a large shared genetic contribution between eating disorders, ranging from rg = 0.62 to rg = 0.96 (Supplementary Data 6). Meta-analyses of Danish and Swedish estimates did not significantly change the estimates. The analysis was repeated by stratifying individuals by inpatient and outpatient eating disorder hospitalisation; we observed no significant change in genetic correlation estimates.
Eating disorders and other mental health disorders
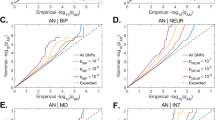
We estimated genetic correlations for each of the 21-eating disorder and other mental disorder diagnostic pairs using full-sibling comparisons and observed, again, similar estimates between countries (Fig. 4). After meta-analysis, 25 genetic correlations were statistically significant (Bonferroni p < 1.39 × 10−3) (Fig. 4). Genetic correlations for all the ED-other mental disorder pairs were positive and statistically significant except AN-schizophrenia. In line with previous literature32, OCD showed the largest genetic correlation with both AN (rg = 0.65, 95% CI: 0.56–0.73) and OED (rg = 0.66, 95% CI: 0.53–0.79). Other large genetic correlations were observed between OED and other mental disorders such as MDD (rg = 0.57, p = 1.01 × 10−131), ADHD (rg = 0.43, p = 2.03 × 10−96), anxiety disorders (rg = 0.57, p = 1.59 × 10−79), and ASD (rg = 0.46, p = 6.30 × 10−65). Genetic correlations between AN and ADHD, and between OED and ASD, were not replicated using parent-offspring comparisons. Overall, estimates from Denmark replicated well with Swedish estimates, with one genetic correlation showing a significant difference between countries using full-sibling information and five using parent-offspring data. In general, our results suggest that the genetic architecture of eating disorders is widely shared (between 0.14 and 0.69) with other mental disorders.
All within-country (Denmark and Sweden) rg estimates reported per birth year were first meta-analysed using random effect inverse variance weighting, followed by a between-country meta-analysis using the same method. Top row panels rg estimates based on full-sibling comparison, bottom row panels rg based on parent-offspring comparisons. Bonferroni p = 1.28 × 10−3. Confidence intervals are reported as a meta-analysis of Danish and Swedish rg estimates ± 1.96 × standard error.
Eating disorders and CMD
We observed significant positive genetic correlations between AN, BN and two CMDs (heart failure and peripheral artery disease) in full-sibling comparisons (Fig. 4), and five estimates were significantly different between Denmark and Sweden. The observed significant genetic correlations between AN, BN and two CMDs did not replicate using parent-offspring comparisons. Finally, no significant associations were observed between obesity and AN and BN. However, obesity shares a substantial genetic overlap with OED (rg = 0.13, p = 5.46 × 10−7), which was replicated using parent-offspring comparisons (rg = 0.16, p = 1.71 × 10−5). All but one rg (OED and obesity) using parental information were similar between countries.
As sex stratification did not seem to affect the heritability, or no (reliable) heritability could be estimated using the Danish data, we opted not to perform any sex stratified genetic correlation analysis.
Discussion
Here we have shown that the h2 estimates were substantial, i.e. \({h}_{{AN}}^{2}\) = 0.36, \({h}_{{BN}}^{2}\) = 0.39 and \({h}_{{OED}}^{2}\) = 0.30, and not significantly different across eating disorders. Reducing phenotypic heterogeneity by excluding individuals with prior eating disorder diagnoses other than the eating disorder of interest did not affect the h2. The h2 of ICD-10 OED in Denmark for individuals receiving inpatient care was two times larger (0.60) than the h2 of ICD-10 OED diagnosed in outpatient care (0.27), suggesting stronger genetic underpinnings for eating disorders with greater severity33.
The observed pattern of genetic correlations with mental disorders and CMD advances understanding of the underlying aetiology, comorbidities, and complexity of different eating disorders. The largest positive genetic correlation observed between AN and OCD (rg = 0.65, p = 6.68 × 10−48) is in line with previous family34,35, register26, and genetic studies32. Moreover, previous analyses grouped mental disorders based on shared genomics and clustered AN, OCD, and Tourette syndrome into one category36. These findings therefore point towards a higher-level genetic structure that underlies disorders characterised by compulsivity32 compared to other mental disorders. In addition, eating disorders are often comorbid with internalising disorders, e.g. multiple anxiety phenotypes37,38 and mood disorders39,40, which were also observed in this study.
The positive genetic correlations seen in this study between AN and ASD (rg = 0.36, p = 2.50 × 10−22) support the increasing hypothesis of a substantial genetic overlap between these disorders41. Several clinical studies have hinted at an overlap between eating disorders (predominantly AN) and ASD, given the overlapping features such as limited emotional expression, reduced social contacts, and cognitive rigidity42,43, yet no significant genetic associations have been observed previously4,44. Differences in full-sibling and parent-offspring genetic correlations are likely due to increased awareness and diagnosis of neurodevelopmental disorders.
In line with recent work5, we did not observe a significant negative genetic correlation between AN and type-2 diabetes using parent-offspring comparisons (rg = -0.06, p = 2.50 × 10−3) and when using sibling comparisons (rg = 0.02, p = 0.37). However, this warrants further investigation as other large GWAS using linkage-disequilibrium score regression have observed significant negative genetic correlations between the two outcomes3,4. Nor did we observe a significant negative genetic correlation between AN and obesity (rg = -0.11, p = 0.02)4; however this is likely due to the nature of our sample, as obesity is likely to be diagnosed in the registers only if relevant to pathological findings in the healthcare context, and will be under-represented in this context. The discrepancy between previously observed negative genetic correlations between AN and type-2 diabetes can partially be explained by differences in sampling of individuals across different types of studies. In general, GWAS are performed using non-population representative samples, whereas a national register-based study contains all individuals with diagnoses that have been in contact with the hospital at any time and therefore is mostly free from generally occurring biases (e.g. volunteering bias and selection bias). In this case, it is entirely plausible that observed negative genetic correlation using GWAS data may in fact be influenced by other hidden and co-occurring disorders. Our finding of a relationship between AN, BN and two CMDs (heart failure and peripheral artery disease) using (young) full-sibling comparisons is novel and warrants further investigation. The large overlap between OED and obesity potentially hints at an enrichment of binge-eating disorder diagnoses in OED, which is not defined as a separate diagnostic entity in ICD-10. Our findings overall highlight the need to implement current diagnostic manuals (ICD-11) in research that includes BED as a separate diagnosis, given the impact this has on a large group of the population. In contrast to the genetic correlations calculated between EDs and other disorders, the genetic correlations between the eating disorders were substantially larger and often not significantly different from one, suggesting a high degree of genetic similarity across all EDs. Stratification based on hospital admission type (i.e. in or outpatient) did not affect the genetic correlation estimates, which may rule out disorder severity affecting these genetic correlations.
Although the genetic correlations were comparable between the two familial relationships in our study, we observed substantially larger confidence intervals between specific clusters of disorder constellations related to differences in age of onset. For example, most mental disorders have a relatively early age of onset, whereas CMDs tend to be diagnosed later in life. This, in part, explains why the confidence intervals of the genetic correlations between eating disorders and CMDs are substantially smaller using parent-offspring information compared to full-sibling information, as parents generally have lived long enough to develop a CMD. The opposite pattern was observed for eating disorders and other mental disorders, apart from schizophrenia, which has an older average age at onset than other mental disorders. We believe that our findings might reflect differences in the underlying genetic architecture of disorders. For example, it is entirely plausible that individuals that receive a diagnosis of CMD (i.e. disorders that generally onset later in life) at a relatively young age might have a specific phenotype that has a higher genetic liability than individuals that develop the same disorder at an older age (with a larger environmental liability, related to lifestyle factors).
Our study has some limitations. First, we utilised secondary care hospital diagnoses from the 1960s onwards since the establishment of the inpatient registers in both countries. Therefore, some relatively newer diagnoses (e.g. BN) will be under-ascertained in older individuals and may skew the age at onset estimations. This underrepresentation might also result in misclassification (e.g. in Denmark) due to the non-adoption of ICD-9 and therefore, the “late” introduction of BN as a substantive diagnostic category. Second, the outpatient diagnoses were recorded decades later than the inpatient diagnoses in both countries, and diagnostic practices might have varied over time and between countries. However, we hypothesise that our calendar year-stratified analyses partially account for these time trends within and between countries. However, while we attempt to compensate for within-country differences by calculating the genetic correlation per birth-year and meta-analysing these estimates, we believe that some of the observed differences in rg between Denmark and Sweden are due to strong country-specific effects, such as differences in diagnostic and referral practices (temporal effects). Another limitation concerns undiagnosed individuals, for instance, only ~20-30% of individuals with an eating disorder in the general population seek or access medical services45,46,47. Therefore, our cases likely represent more severely affected individuals who sought treatment and, therefore, are not fully representative of the entire population of individuals affected by eating disorders, which in turn may have inflated our h2 and rg estimates. Due to diagnostic manuals used in the healthcare systems in Denmark and Sweden, we were not able to differentiate other eating disorders outside of AN and BN, therefore, our OED category is likely to be a heterogeneous group and might comprise multiple eating disorders that are undefined up to ICD-10 (e.g. BED). We did not account for the influence of shared environmental factors among close relatives on heritability and genetic correlation estimates. As a result, these estimates may be slightly inflated, although prior research suggests that such effects are minimal29. Regarding ancestry, in this study, to minimise the impact of left censoring, we excluded individuals born outside of Denmark and Sweden, as their medical diagnoses may have been recorded in other countries to which we did not have access. This exclusion applied both to Danish and Swedish citizens born abroad and to individuals who migrated to these countries. While no data on race, ancestry, or ethnicity were recorded in the registers, it is important to acknowledge that Denmark and Sweden have historically been predominantly composed of individuals of white individuals of European ancestry. However, both countries have experienced increasing migration from non-European regions in recent decades. As a result, by restricting our sample to individuals born in Denmark and Sweden, we likely captured a population primarily of white individuals of European ancestry while inadvertently reducing the representation of individuals from more recent immigrant backgrounds. While this study likely includes more individuals of diverse backgrounds, our sample is likely to be representative of individuals of white-European ancestry. This should be considered when interpreting the generalisability of our findings beyond those of European ancestry.
Nonetheless, our study has several important implications for research and clinical practice. By quantifying heritability in a non-twin, near-complete total population sample of two countries, we provide a more comprehensive estimate of the contribution of genetic factors to the variance of eating disorders. Second, we show an increased familial risk of eating disorders, namely that there is, on average, a threefold higher risk among individuals who have a first-degree relative with an ED compared to the general population. Quantification of familial risk can help prevention and early identification. Although the intergenerational transmission of eating disorders has been studied less than other mental disorders, ongoing studies will help understand risk markers and developmental trajectories. The identified genetic correlations between eating disorders and other somatic diseases and psychiatric disorders have supported both a deeper investigation of the pathophysiology of disorders5, and a reconceptualisation of eating disorders as disorders encompassing brain and body48. This study confirms the strong genetic relationship between eating disorders and other psychiatric disorders, particularly internalising disorders (e.g. OCD, anxiety, and depressive disorders). Clinically, these disorders are highly comorbid with eating disorders, and clarifying the nature of this overlap can improve clinical management and therapeutic approaches to comorbidities. For example, the overlap between ASD and eating disorders in clinical samples has received much attention recently49, leading to the development of specific care pathways, and this paper is the first to provide a likely genetic basis for this observed overlap50,51.
In summary, we derived population-wide genetic estimates by harnessing the power of data containing almost all hospital records and genealogical information of two entire Scandinavian countries (n = 17 million). Our results provide a unique perspective on the complex heritable nature of eating disorders, as well as the overlap in genetic aetiology underlying psychiatric and CMD.
Methods
Case definition
This study was based on the Danish and Swedish civil and national health registries, which have been previously described in detail in refs. 52,53,54,55,56,57,58,59 (Supplementary Notes 1 and 2). Eating disorder cases included all individuals born in Denmark or Sweden who were clinically diagnosed with either AN, BN, and/or OED (Supplementary Data 1) according to inpatient and outpatient discharge records from all Danish and Swedish hospitals up to December 31, 2016, obtained from the Danish and Swedish National Patient Register or Psychiatric Central Research Register (for details about years of coverage of both registers see)59. We applied the same criteria to identify cases for attention-deficit/hyperactivity disorder (ADHD), anxiety disorders, autism spectrum disorders (ASD), bipolar disorder, MDD, obsessive compulsive disorder (OCD), schizophrenia, type 2 diabetes, heart failure, hypertension, coronary artery disease, obesity, and peripheral artery disease cases. The total number of cases is less than the sum of all individual disorder diagnoses because many individuals have more than one diagnosis. Most AN, BN, OED, MDD, schizophrenia, and bipolar disorder diagnostic codes translate poorly to individuals below the age of 10, therefore, we excluded individuals who received a diagnosis before this age. Family members (non-twin full-siblings and parents) were identified using the near-complete population genealogies derived from the multi-generation registers60. Information about being male or female was obtained from the civil registers, and for most individuals born in Denmark and Sweden refers to sex, which was assigned at birth. However, for some individuals, this information should be referred to as gender, as individuals are allowed to request a change to this information based on their preferred gender identity. Details regarding exclusion criteria and ICD-8, 9, and 10 codes used in this study are reported in Supplementary Data 1.
Statistical analysis
Primary analyses were cumulative incidence functions (CIF) with age at first hospital contact as an inpatient or outpatient for all eating disorders, other mental health disorders, and CMD. CIFs were estimated for: (a) the general population, and for individuals with any (b) full sibling or (c) parent diagnosed with the studied disorder, and finally for individuals with any (d) full sibling or (e) parent diagnosed with a diagnosis different than their own disorder (e.g. all offspring with AN with at least one parent diagnoses with MDD). All CIFs were calculated using the Nelson-Aalen estimator for individuals born in the same calendar year to account for substantial changes over time in the underlying incidence, as well as censoring and competing risks.
We calculated the additive heritability (h2) under the liability threshold model based on the cumulative incidence as a function of pedigree relatedness. Briefly, we used the general population and full sibling CIFs at the last overlapping observed time point as estimates of the proportion of the population born in the same calendar year that were affected in their lifetime. Using these estimates, we calculated h2 estimates as described by Wray and Gottesman29,61,62 per given calendar year. All within-country h2 estimates were first meta-analysed using random effect inverse variance weighting, followed by a between-country meta-analysis using the same method.
We calculated the genetic correlation (rg) between two traits by deriving the general population cumulative incidence-based risk for both disorders (e.g. AN and SCZ) and full siblings’ cross disorder familial risk (i.e. the cumulative incidence risk for AN when having a full sibling with SCZ). We extracted the last possible time point for each birth-year-stratified CIF and calculated the rg by inputting the previously calculated h2 estimates using the formula described by Wray and Gottesman29,61,62. In line with the h2 calculations, all birth-year specific genetic correlation estimates were meta-analysed first within-country and followed by a between-country meta-analysis. We repeated the analyses using parent-offspring comparisons. Danish register data was stored on a PostgreSQL 13.3 database server information was extracted using the psql 16.2 database client. All register-based analyses were done in R v4.2.1 using the cmprsk v2.2 package.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
This work is based on Danish register data that are not publicly available due to privacy protection, including the General Data Protection Regulation (GDRP). Only Danish research environments are granted authorisation. Foreign researchers can, however, get access to data under Danish research environment authorisation. Further information on data access can be found at https://www.dst.dk/en/TilSalg/Forskningsservice or by contacting Thomas Werge (thomas.werge@regionh.dk). The use of Swedish data was approved by the regional ethics review board in Stockholm, Sweden, with DNR 2012/1814-31/4. Data from Swedish registers is not available for sharing due to policies and regulations in Sweden. Swedish register data are available to all researchers through applications at Statistics Sweden (SCB, https://www.scb.se/en/) and the National Board of Health and Welfare (Socialstyrelsen, https://www.socialstyrelsen.se/). By both Danish and Swedish law, individual consent to use register data for register-based studies is not required.
References
Stice, E., Nathan Marti, C. & Rohde, P. Prevalence, incidence, impairment, and course of the proposed DSM-5 eating disorder diagnoses in an 8-year prospective community study of young women. J. Abnorm. Psychol. 122, 445–457 (2013).
Thornton, L. M., Mazzeo, S. E. & Bulik, C. M. The heritability of eating disorders: methods and current findings. Curr. Top. Behav. Neurosci. 6, 141–156 (2011).
Duncan, L. et al. Significant locus and metabolic genetic correlations revealed in genome-wide association study of anorexia nervosa. Am. J. Psychiatry 174, 850–858 (2017).
Watson, H. J. et al. Genome-wide association study identifies eight risk loci and implicates metabo-psychiatric origins for anorexia nervosa. Nat. Genet. 51, 1207–1214 (2019).
Hübel, C. et al. One size does not fit all. Genomics differentiates among anorexia nervosa, bulimia nervosa, and binge-eating disorder. Int J. Eat. Disord. 54, 785–793 (2021).
Bulik, C., Yilmaz, Z. & Hardaway, A. Genetics and epigenetics of eating disorders. Adv. Genomics Genet. 5, 131 (2015).
Bulik, C. M. et al. Prevalence, heritability, and prospective risk factors for anorexia nervosa. Arch. Gen. Psychiatry 63, 305–312 (2006).
Klump, K. L., Miller, K. B., Keel, P. K., McGue, M. & Iacono, W. G. Genetic and environmental influences on anorexia nervosa syndromes in a population-based twin sample. Psychol. Med. 31, 737–740 (2001).
Kortegaard, L. S., Hoerder, K., Joergensen, J., Gillberg, C. & Kyvik, K. O. A preliminary population-based twin study of self-reported eating disorder. Psychol. Med. 31, 361–365 (2001).
Bulik, C. M. et al. Understanding the relation between Anorexia Nervosa and Bulimia Nervosa in a Swedish National Twin Sample. Biol. Psychiatry 67, 71–77 (2010).
Bulik, C. M., Sullivan, P. F. & Kendler, K. S. Heritability of binge-eating and broadly defined Bulimia nervosa. Biol. Psychiatry 44, 1210–1218 (1998).
Trace, S. E., Baker, J. H., Peñas-Lledó, E. & Bulik, C. M. The genetics of eating disorders. Annu. Rev. Clin. Psychol. 9, 589–620 (2013).
Javaras, K. N. et al. Co-occurrence of binge eating disorder with psychiatric and medical disorders. J. Clin. Psychiatry 69, 266–273 (2008).
Mitchell, K. S. et al. Binge eating disorder: a symptom-level investigation of genetic and environmental influences on liability. Psychol. Med. 40, 1899–1906 (2010).
Dinkler, L. et al. Etiology of the broad avoidant restrictive food intake disorder phenotype in Swedish twins aged 6 to 12 years. JAMA Psychiatry 80, 260–269 (2023).
Munn, M. A. et al. Bivariate analysis of disordered eating characteristics in adolescence and young adulthood. Int J. Eat. Disord. 43, 751–761 (2010).
Root, T. L. et al. Shared and unique genetic and environmental influences on binge eating and night eating: a Swedish twin study. Eat. Behav. 11, 92–98 (2010).
Sullivan, P. F., Bulik, C. M. & Kendler, K. S. Genetic epidemiology of binging and vomiting. Br. J. Psychiatry 173, 75–79 (1998).
Mazzeo, S. E. et al. A twin study of specific bulimia nervosa symptoms. Psychol. Med. 40, 1203–1213 (2010).
Nakabayashi, K. et al. Identification of novel candidate loci for anorexia nervosa at 1q41 and 11q22 in Japanese by a genome-wide association analysis with microsatellite markers. J. Hum. Genet. 54, 531–537 (2009).
Boraska, V. et al. A genome-wide association study of anorexia nervosa. Mol. Psychiatry 19, 1085–1094 (2014).
Wang, K. et al. A genome-wide association study on common SNPs and rare CNVs in anorexia nervosa. Mol. Psychiatry 16, 949–959 (2011).
Burstein, D. et al. Genome-wide analysis of a model-derived binge eating disorder phenotype identifies risk loci and implicates iron metabolism. Nat. Genet. 55, 1462–1470 (2023).
Termorshuizen, J. D. Genetic and Enviromental Factors Influencing the Risk an Course of Eating Disorders (Karolinska Institutet, 2023, accessed 9 April 2024); https://openarchive.ki.se/xmlui/handle/10616/48881.
Wade, T. D., Bulik, C. M., Neale, M. & Kendler, K. S. Anorexia nervosa and major depression: Shared genetic and environmental risk factors. Am. J. Psychiatry 157, 469–471 (2000).
Cederlöf, M. et al. Etiological overlap between obsessive-compulsive disorder and anorexia nervosa: a longitudinal cohort, multigenerational family and twin study. World Psychiatry 14, 333–338 (2015).
Mayhew, A. J. & Meyre, D. Assessing the heritability of complex traits in humans: methodological challenges and opportunities. Curr. Genomics https://doi.org/10.2174/1389202918666170307161450 (2017).
Larsen, J. T., Bulik, C. M., Thornton, L. M., Koch, S. V. & Petersen, L. Prenatal and perinatal factors and risk of eating disorders. Psychol. Med. 51, 870–880 (2021).
Wray, N. R., Gottesman II. Using summary data from the Danish National Registers to estimate heritabilities for schizophrenia, bipolar disorder, and major depressive disorder. Front. Genet. https://doi.org/10.3389/fgene.2012.00118 (2012).
Meijsen, J. et al. Quantifying the relative importance of genetics and environment on the comorbidity between mental and cardiometabolic disorders using 17 million Scandinavians. Nat. Commun. 15, 1–10 (2024).
Bulik, C. M., Blake, L. & Austin, J. Genetics of eating disorders: what the clinician needs to know. Psychiatr. Clin. North Am. 42, 59–73 (2019).
Lee, P. H. et al. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell 179, 1469–1482.e11 (2019).
Dellava, J. E., Thornton, L. M., Lichtenstein, P., Pedersen, N. L. & Bulik, C. M. Impact of broadening definitions of anorexia nervosa on sample characteristics. J. Psychiatr. Res. 45, 691–698 (2011).
Lilenfeld, L. R. et al. A controlled family study of anorexia nervosa and bulimia nervosa: psychiatric disorders in first-degree relatives and effects of proband comorbidity. Arch. Gen. Psychiatry 55, 603–610 (1998).
Bellodi, L. et al. Morbidity risk for obsessive-compulsive spectrum disorders in first-degree relatives of patients with eating disorders. Am. J. Psychiatry 158, 563–569 (2001).
Yang, Z. et al. Investigating shared genetic basis across tourette syndrome and comorbid neurodevelopmental disorders along the impulsivity-compulsivity spectrum. Biol. Psychiatry 90, 317–327 (2021).
Thornton, L. M., Dellava, J. E., Root, T. L., Lichtenstein, P. & Bulik, C. M. Anorexia nervosa and generalized anxiety disorder: further explorations of the relation between anxiety and body mass index. J. Anxiety Disord. 25, 727–730 (2011).
Kerr-Gaffney, J., Harrison, A. & Tchanturia, K. Social anxiety in the eating disorders: a systematic review and meta-analysis. Psychol. Med 48, 2477–2491 (2018).
Hambleton, A. et al. Psychiatric and medical comorbidities of eating disorders: findings from a rapid review of the literature. J. Eat. Disord. 10, 1–23 (2022).
Ulfvebrand, S., Birgegård, A., Norring, C., Högdahl, L. & von Hausswolff-Juhlin, Y. Psychiatric comorbidity in women and men with eating disorders results from a large clinical database. Psychiatry Res. 230, 294–299 (2015).
Koch, S. V. et al. Autism spectrum disorder in individuals with anorexia nervosa and in their first- and second-degree relatives: Danish nationwide register-based cohort-study. Br. J. Psychiatry 206, 401–407 (2015).
Nickel, K. et al. Systematic review: overlap between eating, autism spectrum, and attention-deficit/hyperactivity disorder. Front. Psychiatry. https://doi.org/10.3389/fpsyt.2019.00708 (2019).
Zucker, N. L. et al. Anorexia nervosa and autism spectrum disorders: guided investigation of social cognitive endophenotypes. Psychol. Bull. 133, 976–1006 (2007).
Grove, J. et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. https://doi.org/10.1038/s41588-019-0344-8 (2019).
Hudson, J. I., Hiripi, E., Pope, H. G. & Kessler, R. C. The prevalence and correlates of eating disorders in the National Comorbidity Survey Replication. Biol. Psychiatry 61, 348–358 (2007).
Eisenberg, D., Nicklett, E. J., Roeder, K. & Kirz, N. E. Eating disorder symptoms among college students: prevalence, persistence, correlates, and treatment-seeking. J. Am. Coll. Heal 59, 700–707 (2011).
Micali, N. et al. Lifetime and 12-month prevalence of eating disorders amongst women in mid-life: a population-based study of diagnoses and risk factors. BMC Med. 15, 1–10 (2017).
Bulik, C. M., Carroll, I. M. & Mehler, P. Reframing anorexia nervosa as a metabo-psychiatric disorder. Trends Endocrinol. Metab. 32, 752–761 (2021).
Råstam, M. et al. Eating problems and overlap with ADHD and autism spectrum disorders in a nationwide twin study of 9- and 12-year-old children. Sci. World J. https://doi.org/10.1155/2013/315429 (2013).
Tchanturia, K. et al. A novel approach for autism spectrum condition patients with eating disorders: analysis of treatment cost-savings. Eur. Eat. Disord. Rev. 29, 514–518 (2021).
Tchanturia, K., Smith, K., Glennon, D. & Burhouse, A. Towards an improved understanding of the anorexia nervosa and autism spectrum comorbidity: PEACE pathway implementation. Front Psychiatry 11, 640 (2020).
Pedersen, C. B. The Danish civil registration system. Scand. J. Public Health. https://doi.org/10.1177/1403494810387965 (2011).
Pedersen, C. B., Gøtzsche, H., Møller, JØ, Mortensen, P. B. The Danish Civil Registration System. A cohort of eight million persons. Dan. Med. Bull. 53, 441–449 (2006).
Schmidt, M. et al. The Danish National patient registry: a review of content, data quality, and research potential. Clin. Epidemiol. https://doi.org/10.2147/CLEP.S91125 (2015).
Mors, O., Perto, G. P. & Mortensen, P. B. The Danish psychiatric central research register. Scand. J. Public Health https://doi.org/10.1177/1403494810395825 (2011).
Ludvigsson, J. F. et al. Registers of the Swedish total population and their use in medical research. Eur. J. Epidemiol. 31, 125–136 (2016).
Ekbom, A. The Swedish multi-generation register. Methods Mol. Biol. 675, 215–220 (2011).
Ludvigsson, J. F. et al. External review and validation of the Swedish national inpatient register. BMC Public Health 11, 450 (2011).
Pasman, J. A. et al. Epidemiological overview of major depressive disorder in Scandinavia using nationwide registers. Lancet Reg. Health 29, 100621 (2023).
Athanasiadis, G. et al. A comprehensive map of genetic relationships among diagnostic categories based on 48.6 million relative pairs from the Danish genealogy. Proc. Natl. Acad. Sci. USA 119, e2118688119 (2022).
Reich, T., James, J. W. & Morris, C. A. The use of multiple thresholds in determining the mode of transmission of semi-continuous traits. Ann. Hum. Genet. 36, 163–184 (1972).
Falconer, D. S. The inheritance of liability to certain diseases, estimated from the incidence among relatives. Ann. Hum. Genet. 29, 51–76 (1965).
Acknowledgements
J.M., A.B., T.W., and F.F. received funding from the European Union’s Horizon 2020 Research and Innovation Programme: the “Predicting Comorbid Cardiovascular Disease in Individuals with Mental Disorder by Decoding Disease Mechanisms” project (CoMorMent, grant number 847776, to Ole Andreassen). J.M., J.P., T.W., and A.B. were supported by the US National Institutes of Health study on extreme MDD (R01 MH123724, to Patrick Sullivan). J.M., N.M., S.A., and H.D. were supported by the Laureate Grant Award from the Novo Nordisk Foundation (grant no: NNF22OC0071010, to N.M.). R.Z. was supported by the Swedish Research Council (Vetenskapsrådet, grant no. 2022-00242); J.P. received funding from the European Research Council (grant number 101042183). C.B. was supported by the National Institute of Mental Health (R56MH129437; R01MH120170; R01MH124871; and R01MH124871); Swedish Research Council (Vetenskapsrådet, award: 538-2013-8864); and Lundbeck Foundation (grant no. R276-2018-4581). A.B., F.F., and T.W. were supported by the Nordforsk grant 164218.
Author information
Authors and Affiliations
Contributions
J.M., A.B., and N.M. conceived the study. J.M., A.B., C.B., R. Zhang, and N.M. contributed to the study design. J.M., S.A., and H.L.D., performed the literature search, and J.M., K.H., D.W., M.L., J.P., and R. Zetterberg performed programming and/or data analyses. J.M., A.B., R. Zhang, N.M., F.F., and T.W. contributed to data interpretation. F.F., W.Y., A.B., and T.W. provided access to data. J.M., C.B., and N.M. wrote the initial draft. N.M., F.F., and T.W. obtained the funding.
Corresponding authors
Ethics declarations
Competing interests
N.M. receives an honorarium as associate editor on the European Eating Disorders Review. C.B. receives royalties from Pearson Education Inc. and has served as a consultant with Orbimed. All other authors declare no competing interests.
Ethical approval
The use of Danish data was approved by the Danish Health Data Authority (project no. FSEID-00003339) and the Danish Data Protection Agency. By Danish law, informed consent is not required for register-based studies. The use of Swedish data was approved by the regional ethics review board in Stockholm, Sweden, with DNR 2012/1814-31/4.
Peer review
Peer review information
Nature Communications thanks Shahram Bahrami and Sarah Cohen-Woods for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Meijsen, J., Hu, K., Wei, D. et al. Shared genetic architecture between eating disorders, mental health conditions, and cardiometabolic diseases: a comprehensive population-wide study across two countries. Nat Commun 16, 6193 (2025). https://doi.org/10.1038/s41467-025-61496-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-61496-5
This article is cited by
-
‘Will They Have It Too?’ Mothers’ Perspectives of Familial Risk for Eating Disorders
Child Psychiatry & Human Development (2025)