Abstract
The Wnt/β-catenin pathway requires precise regulation for proper development and tissue homeostasis, yet the structural mechanisms enabling its fine-tuned control remain incompletely understood. Here, we reveal how LGR4 achieves differential signaling outcomes through distinct recognition of two key modulators: Norrin and R-spondins (RSPOs). Using cryo-electron microscopy, we determined the structure of full-length LGR4 bound to Norrin in a 2:2 stoichiometry, revealing a molecular bridging mechanism where Norrin dimer connect two LGR4 protomers in a spatial arrangement fundamentally distinct from the LGR4-RSPO2-ZNRF3 complex. Notably, Norrin binding to LGR4 sterically hinders simultaneous interaction with the Frizzled4 receptor, establishing a regulatory checkpoint in Wnt signaling. The partially overlapping binding sites for Norrin and RSPOs on LGR4 enable mutually exclusive interactions that drive distinct signaling outcomes. Disease-linked mutations map to distinct functional regions: those disrupting LGR4 interaction are associated with familial exudative vitreoretinopathy (FEVR), while others impairing Frizzled4 binding are linked to Norrie disease. Furthermore, we developed a high-affinity nanobody that blocks both Norrin and RSPO binding to LGR4, providing a potential tool for therapeutic intervention. These findings elucidate the structural basis of LGR4’s dual signaling roles and lay the groundwork for therapeutic strategies targeting Wnt-related diseases.
Similar content being viewed by others
Introduction
The Wnt/β-catenin signaling pathway orchestrates fundamental biological processes, including embryonic development, tissue homeostasis, and stem cell proliferation1,2,3,4. This pathway is primarily activated when Wnt ligands engage Frizzled receptors and low-density lipoprotein receptor-related proteins 5 and 6 (LRP5/6) on the cell surface5,6,7,8,9,10,11,12,13. Within this complex signaling network, Norrin functions as a distinctive Wnt-like ligand that specifically interacts with the Frizzled-4 (FZD4) receptor, LRP5/6 co-receptors, and the auxiliary transmembrane protein Tetraspanin-12 (TSPAN12) to initiate Wnt signaling14,15,16,17,18,19,20,21,22,23,24.
R-spondins (RSPOs) represent another critical layer of regulation in this pathway. These secreted proteins, comprising four homologs (RSPO1-4), enhance Wnt/β-catenin signaling by binding to leucine-rich repeat-containing G protein-coupled receptors (LGR4/5/6)1,25,26,27. The RSPO-LGR complex modulates E3 ubiquitin ligases ZNRF3/RNF43, which typically promote Frizzled receptor degradation. By inhibiting ZNRF3/RNF43, RSPOs increase Frizzled receptor stability, thereby amplifying cellular sensitivity to Wnt signals28,29,30,31,32,33,34,35,36,37,38,39,40,41,42. Recent studies have revealed that Norrin also acts as a dual-purpose ligand, engaging both FZD4 and LGR4, adding another level of complexity to Wnt/β-catenin signaling regulation14,15. Meanwhile, LGR4 itself serves as a dual-functional receptor, activating distinct signaling pathways in response to binding by Norrin or RSPOs14,25,26,27.
Dysregulation of the Wnt pathway has profound implications for human health, contributing to various diseases including cancer, fibrosis, and neurodegeneration1,43,44,45,46,47,48. Norrin plays a particularly crucial role in retinal and vascular development, where it maintains the integrity of both the blood-retina barrier (BRB) and the cerebellar blood-brain barrier (BBB)15,49,50,51,52,53. Mutations in the Norrin gene are associated with severe clinical conditions, most notably Norrie disease—an X-linked disorder characterized by congenital blindness, progressive hearing loss, and intellectual disability54,55,56,57,58,59. These mutations can also cause familial exudative vitreoretinopathy (FEVR), leading to retinal detachment and vision loss due to impaired Wnt signaling activation54,55,56.
Despite Norrin’s essential role in retinal angiogenesis, the physiological relevance of LGR4/5/6 in retinal vascular development remains elusive. Genetic knockout models in mice and studies in humans carrying LGR4 loss-of-function alleles have failed to reveal overt retinal vascular defects. Similarly, LGR5 or LGR6 knockout mice do not exhibit clear retinal phenotypes. These findings have fueled the perception that LGR4/5/6 are either dispensable or functionally redundant in this context44,60,61,62. Yet, this conclusion is largely based on the absence of phenotypes in single-gene knockouts and does not exclude context-dependent or compensatory roles of these receptors, particularly when considering their co-expression in retinal endothelial cells.
While crystal structures of the Norrin-FZD4 cysteine-rich domain complex and unliganded Norrin have provided initial structural insights19,20,21, they revealed unexpected similarities between Norrin and Wnt proteins in their interaction with Frizzled receptors. Recent studies have further demonstrated that Tspan12 can simultaneously bind Norrin with FZD4, enabling more efficient capture of Norrin at low concentrations63. Moreover, competition between Tspan12 and LRP6 for Norrin binding suggests a precise temporal sequence in pathway activation63.
Our recent structural characterization of LGR4-RSPO2-ZNRF3 assemblies has revealed that LGR4 abundance influences ZNRF3’s conformational states64. Higher LGR4 levels appear to stabilize ZNRF3 in its inactive form, potentially promoting WNT/β-catenin signaling through complex internalization. However, the specific mechanisms by which Norrin regulates LGR4 to modulate Wnt/β-catenin signaling remain unclear. As Norrin interacts with both LGR4 and FZD4, understanding the structural distinctions in these interactions is crucial for elucidating its dual regulatory role.
In this study, we employ cryo-electron microscopy to resolve the structure of the full-length LGR4-Norrin complex, revealing several key insights. First, we demonstrate that Norrin binding to LGR4 sterically prevents formation of the FZD4 CRD-Norrin complex, suggesting distinct regulatory mechanisms. Second, comparison of the 2:2 LGR4-Norrin structure with the 2:2:2 LGR4-RSPO2-ZNRF3 complex reveals overlapping yet distinct binding interfaces, enabling differential signaling outcomes. Additionally, we identify a nanobody (NB18) that effectively blocks both RSPO and Norrin binding to LGR4, presenting a potential therapeutic tool for modulating Wnt signaling pathways in disease contexts.
Results
Structures of LGR4-Norrin complex
To elucidate the structural basis of LGR4-Norrin interaction, we developed a systematic approach for complex assembly and structural determination. First, we generated a high-affinity nanobody (NB52) targeting the LGR4 ectodomain. To optimize particle orientation and improve cryo-EM analysis, NB52 was inserted into a circularly permutated scaffold protein, resulting in the construction of a larger and more rigid “megabody” (MB52)65. Complex formation was achieved by incubating Norrin with lysed LGR4-expressing cells during extraction, followed by the addition of MB52 at a molar ratio of 1.2:1 relative to LGR4. The resulting LGR4-Norrin complexes were then purified for structural studies.
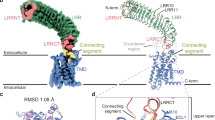
Initial cryo-EM analysis yielded a 2:2 stoichiometric complex between LGR4 and Norrin. Through rigorous 3D classification, we obtained an initial structure at 3.61 Å resolution, which was further improved to 3.05 Å in the extracellular region through local refinement (Supplementary Fig. 1, Supplementary Table 1). This enhanced resolution enabled precise modeling of LGR4-Norrin dimer, revealing clear side-chain densities at their interface (Fig. 1a, b).
a, b Cryo-EM structure of the LGR4-Norrin complex with a 2:2 stoichiometry. Cryo-EM map (a) and atomic model (b) are shown. LGR4 and Norrin are colored light green/cyan and violet/orange, respectively. The nanobody segment of MB52 is shown in gray, with other segments omitted for clarity. The color scheme remains consistent throughout the manuscript unless specified otherwise. c, d Views of the extracellular region of the LGR4-Norrin complex: c bottom view of the Cryo-EM map, d bottom view of the atomic model.
The refined structure reveals a striking architecture of the LGR4-Norrin complex characterized by twofold symmetry. A central Norrin dimer serves as a molecular bridge between two horseshoe-shaped LGR4 protomers, binding to their extracellular domains (Fig. 1c, d). The complex adopts a distinctive spatial arrangement where the LGR4 ectodomains face opposite directions without direct contact. Norrin binds near the upper regions of the LGR4 ectodomains, inducing a specific geometric configuration: the LGR4 heads are tilted at a 38.4° angle and separated by approximately 47.5 Å (Supplementary Fig. 2a, b). This arrangement positions the transmembrane domains at a considerable distance of 184 Å apart (Supplementary Fig. 2a), suggesting potential implications for signal transduction.
Norrin recognition of LGR4
Having established the overall architecture of the LGR4-Norrin complex, we next investigated the molecular details of their interaction to understand how this binding interface enables LGR4’s unique regulatory functions. Our structural analysis reveals that Norrin forms an elongated dimer reminiscent of its previously characterized conformations, but adopts a distinct binding mode with LGR4. The interaction occurs along LGR4’s concave surface, spanning from the LRRNT to the LRR10 region, and is mediated through specific structural elements of Norrin: the β1 and β2 strands, their connecting loop, and the segment between β5 and β6 (Fig. 2a, b).
a The binding interface between the LGR4 ectodomain and Norrin in the LGR4-Norrin complex, LGR4 transmembrane is omitted for clarity, LGR4 is shown in light green, while Norrin is depicted in violet and orange. b Detailed view of the specific interactions between LGR4 (light green) and Norrin (violet), highlighting key binding residues. c The LGR4 double mutant (H207N/H209T) was transiently expressed in LGR4-knockout HEK293T/17 cells and stimulated with 100 nM Norrin-Fc. Wnt/β-catenin signaling activity was assessed using the TOPFlash luciferase reporter assay. The H207N/H209T mutation significantly impaired Norrin-induced signaling compared to wild-type LGR4, n = 3, p < 0.001 (***). Error bars represent the S.E.M. Source data are provided as a Source Data file. d Superimposed structures of the LGR4-Norrin and FZD4 CRD-Norrin complexes (PDB code: 5CL1), aligned based on the Norrin dimer. This alignment reveals a steric clash between FZD4 CRD and LGR4 when both attempt to bind Norrin. For clarity, transmembrane regions of LGR4 are omitted. LGR4 is colored light green and cyan, Norrin in violet and orange, and FZD4 CRD in pink. e Table summarizing Norrin binding residues, their variants, and associated disease implications with the LGR4 and FZD4 receptors57,66,67,68,69.
The extensive interaction surface covers approximately 1483 Ų, establishing a robust complex between the two proteins. Importantly, the identified Norrin-LGR4 binding interface is spatially and functionally distinct from Norrin’s dimerization interface. The binding surface exhibits a sophisticated organization with three distinct interaction zones: two hydrophilic regions flanking a central hydrophobic core (Fig. 2a, b).
Detailed examination of these interaction zones reveals precise molecular complementarity. At the N-terminal end, Norrin residues R41, Y122, and L124 form an intricate network of salt bridges and polar interactions with LGR4 residues R40 and E85. The opposite end features hydrophilic interactions between Norrin residues K54 and S57 and LGR4 residues H209, Y234, S257, and D281. The central hydrophobic core is stabilized through specific contacts between Norrin residues S47, M59, and L61 and LGR4 residues V109, H157, W159, A181, T183, and H207 (Fig. 2b). LGR4 double mutant (H207N/H209T) significantly impaired Norrin-induced Wnt signaling, supporting the structural predictions and confirming the functional importance of these residues in mediating the LGR4-Norrin interaction (Fig. 2c).
The functional significance of this binding interface extends beyond LGR4 alone. We find that the LGR4 residues critical for Norrin binding are highly conserved in LGR5 and LGR6, providing a structural explanation for Norrin’s ability to recognize all three receptors (Supplementary Fig. 2c). Most notably, many of the Norrin residues involved in this interface correspond to known mutation sites associated with genetic retinal disorders (Fig. 2e), suggesting that disruption of this specific binding mode may contribute to disease pathogenesis.
Structural mechanisms of Norrin-related diseases
The complex interplay between Norrin and its receptors is crucial for retinal vascular development and neuronal health18,49,50,51,52. While Norrin’s interaction with FZD4-LRP5/6 has been well-characterized19,20,21, our structural analysis of the LGR4-Norrin complex provides insights into how different mutations lead to distinct retinal vascular diseases54,55,56,57,58,59.
Through comparative analysis of the LGR4-Norrin and FZD4 CRD-Norrin complexes, we found that FZD4 CRD and LGR4 cannot simultaneously bind to Norrin due to steric interference (Fig. 2d). This mutually exclusive binding is explained by overlapping interaction surfaces on Norrin, particularly at residues M59, L61, and Y122. Mutations affecting these shared binding regions or adjacent areas correlate with both Norrie disease (ND) and familial exudative vitreoretinopathy type 2 (FEVR2), suggesting a molecular basis for the spectrum of disease manifestations (Fig. 2e)19,21,57,66,67.
Our structural analysis reveals distinct sets of Norrin residues that preferentially interact with either LGR4 or FZD4. For LGR4-specific interactions, we identified K54 in the β1-β2 loop, R41, which forms a salt bridge with LGR4’s E85, and L124, which hydrogen bonds with LGR4’s R40 (Fig. 2e). Notably, these residues do not participate in FZD4 binding19,21, and their mutations are specifically associated with FEVR2 (Fig. 2e)67,68. This finding suggests that disruption of LGR4-specific interactions leads to the FEVR2 phenotype.
Conversely, FZD4-specific interactions involve residues such as K104, which forms a hydrogen bond with N152 of FZD4, and V45, which engages in hydrophobic contacts with multiple FZD4 residues (M105, T107, K109, I110, and M157) (Supplementary Fig. 3)19. Mutations in these FZD4-specific interaction residues (K104 and V45) correlate exclusively with Norrie disease (Fig. 2e)66,69.
Interestingly, H43 appears to engage both FZD4 and LGR4. In our structure, the pyridine-like nitrogen (4-position) of the H43 imidazole ring forms a 3.1 Å contact with the side-chain carboxyl group of LGR4 E85. However, since both atoms carry lone electron pairs, this interaction is predicted to be weak. The H43R mutation replaces histidine with arginine, whose guanidinium group can potentially form a stronger salt bridge with E85, possibly enhancing the Norrin-LGR4 interaction. Therefore, H43R is unlikely to impair LGR4 binding and would not be expected to contribute to FEVR2. In contrast, H43 makes multiple stabilizing contacts with FZD4, including hydrogen bonding with G57 and hydrophobic interactions with F96, M105, and M159, explaining why the H43R mutation is exclusively linked to Norrie disease.
Structural basis for differential LGR4 regulation by Norrin and RSPOs
Having established the molecular details of LGR4-Norrin interaction, we next investigated how this binding mode differs from LGR4’s engagement with RSPOs, as these distinct interactions are known to activate different signaling pathways14,25,26,27. This comparison is particularly important because while Norrin-LGR4 signaling is crucial for retinal vascular development, RSPO-LGR4 interaction more broadly regulates Wnt pathway activation through ZNRF3/RNF43 modulation16,70.
Our recent structural studies of LGR4-RSPO2-ZNRF3 assemblies provide a foundation for this comparison64. These studies revealed that LGR4 and RSPO2 together regulate ZNRF3 through distinct conformational states. In its active form, ZNRF3 exists as a dimer with coiled-coil transmembrane helices and dimerized RING domains. However, the presence of additional LGR4 molecules induces an inactive ZNRF3 state, characterized by widely separated transmembrane helices and RING domains. This conformational plasticity highlights LGR4’s ability to fine-tune ZNRF3 activity through different molecular assemblies.
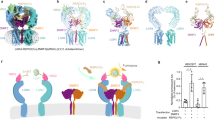
Comparative analysis of the LGR4-Norrin (2:2) and LGR4-RSPO2-ZNRF3 (2:2:2) complexes reveals striking differences in their molecular architecture (Fig. 3)64. While both complexes show face-to-face orientation of LGR4 protomers bridged by their respective ligands (Fig. 3a, c), the spatial arrangement differs significantly. The LGR4-Norrin complex exhibits a 101.5 Å greater separation between LGR4 protomers compared to the LGR4-RSPO2-ZNRF3 complex (Fig. 3b, d), with distinct tilting of the protomers. These architectural differences suggest that Norrin activates LGR4 through a mechanism distinct from RSPO-mediated signaling (Fig. 3e).
a Structure of the LGR4-RSPO2-ZNRF3 complex in a 2:2:2 stoichiometry. LGR4 is shown in light blue and pink, ZNRF3 in purple, and RSPO2 in brown (PDB code: 8Y69). b Front view of the arrangement of two LGR4 protomers in the LGR4-RSPO2-ZNRF3 complex, with LGR4 colored cyan and pink. c Structure of the LGR4-Norrin complex in a 2:2 stoichiometry, with LGR4 and Norrin displayed in light green/cyan and violet/orange, respectively. d Front view showing the arrangement of two LGR4 protomers in the LGR4-Norrin complex in a 2:2 stoichiometry, with LGR4 colored light green and cyan. e Superimposition of LGR4 from the LGR4-Norrin complex (light green/cyan) and the LGR4-RSPO2-ZNRF3 complex (cyan/pink), with Norrin, RSPO2, and ZNRF3 omitted for clarity. f Superposition of the LGR4 ectodomains in the LGR4-Norrin and LGR4-RSPO2-ZNRF3 complexes. LGR4 is colored light green, Norrin in violet, and RSPO2 in blue; other segments are omitted for clarity. g, h Binding affinity measurements between LGR4 and RSPO2-Fc (g) and between LGR4 and Norrin-Fc (h) using biolayer interferometry (BLI). The binding affinity between LGR4 and RSPO2-Fc was significantly higher than that reported for monomeric RSPO2, and the values may not be directly comparable due to differences in oligomeric state. i, j Dose-dependent TOPFlash activity induced by LGR4 after stimulation with Norrin-Fc (i) or RSPO2-Fc (j). RSPO2-Fc and Norrin-Fc were serially diluted and applied to transiently transfected HEK293T/17 cells (plasmid ratio: receptor: TOPFlash: pRL = 1:1:0.1; within the receptor plasmids, LGR4:LRP6 = 3:1). Firefly luciferase activity was normalized to Renilla luciferase activity (Rlu). The EC50 and EC80 values were determined as follows: Norrin-Fc EC50 = 2.807 × 10−8M and EC80 = 7.557 × 10−8M, Emax = 47.91%; RSPO2-Fc EC50 = 1.251 × 10−11M and EC80 = 4 × 10−10M, Emax = 91.67%, n = 3, Error bars represent the S.E.M. k Dose-dependent TOPFlash activity induced by LGR4 in HEK293T cells upon stimulation with RSPO4-Fc alone (black) or co-stimulation with RSPO4-Fc and Norrin-Fc (red), n = 3. Error bars represent the S.E.M. l Knockdown of ZNRF3 in HEK293T/17 cells enhances Norrin-Fc (100 nM) induced Wnt signaling, as measured by normalized luciferase activity. m Co-transfection of LGR4 and LRP6 into HEK293T/17 cells significantly enhances Norrin-Fc (100 nM) induced signaling activity, relative to the expression of either receptor individually. l, m: n = 3, Error bars represent the S.E.M. p < 0.01 (**), p < 0.001 (***), p < 0.0001 (****). Source data are provided as a Source Data file.
The molecular basis for these distinct signaling outcomes becomes apparent when examining the binding interfaces. RSPO2 and Norrin binding regions on LGR4 partially overlap, creating steric hindrance that prevents simultaneous binding (Fig. 3f). RSPO2-Fc exhibits markedly higher binding affinity for LGR4 than Norrin-Fc (Fig. 3g, h), and induces significantly stronger Wnt/β-catenin signaling in the TOPFlash assay, reflecting its greater potency (Fig. 3i, j).
The partial overlap in binding sites raises the possibility that Norrin may antagonize RSPO activity. Given that RSPO4 signaling is fully dependent on LGR471, we conducted antagonist assays using Norrin in the presence of increasing RSPO4 concentrations. In co-transfection experiments with LGR4 and LRP6, RSPO4 and Norrin exhibited additive effects. However, we observed an inhibition of RSPO4-induced signaling, evidenced by a rightward shift in the EC50, indicative of competitive binding for LGR4 (Fig. 3k). Interestingly, the maximal signaling response was significantly increased when both ligands were present, suggesting that Norrin and RSPO4 can synergize under certain conditions to enhance Wnt pathway activation.
Collectively, these findings demonstrate that Norrin and RSPOs engage LGR4 through distinct mechanisms to modulate β-catenin signaling. While their binding sites overlap, enabling Norrin to competitively inhibit RSPO signaling at high concentrations, the ability of Norrin to enhance maximal signaling in combination with RSPO4 also points to potential synergy. This implies a finely tuned regulatory mechanism wherein the shared binding interface on LGR4 allows mutually exclusive ligand engagement and context-dependent switching between distinct signaling modes.
To further dissect the mechanism of Norrin-LGR4 signaling, we performed siRNA-mediated knockdown of ZNRF3 in HEK293 cells. Remarkably, Norrin retained its ability to activate Wnt signaling in the absence of ZNRF3, indicating that its effect is not dependent on E3 ligase inhibition (Fig. 3l). Moreover, co-expression of LGR4 and LRP6 further potentiated Norrin-induced signaling, supporting a model in which Norrin promotes the formation or stabilization of a signaling-competent complex involving LGR4 and LRP6, functionally analogous to the canonical Wnt-Frizzled-LRP5/6 complex (Fig. 3m).
Development and structural characterization of an LGR4 inhibitory nanobody
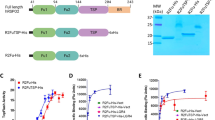
Having established the distinct mechanisms by which Norrin and RSPOs regulate LGR4, we sought to develop a molecular tool that could modulate both signaling pathways. Through camel immunization with recombinant LGR4 ectodomain and subsequent phage display screening, we identified several high-affinity nanobodies specific to LGR4. Among these, NB18 emerged as a particularly promising candidate, demonstrating exceptional binding affinity (0.15 nM) to LGR4 (Fig. 4a).
a Binding affinity measurements between LGR4 and NB18 using biolayer interferometry. b, c Cryo-EM structure of the LGR4-NB18 complex, showing the Cryo-EM map (b) and corresponding atomic model (c). LGR4 is displayed in light green, NB18 in salmon, with the nanobody portion of MB52 in gray (remaining segments are omitted for clarity). d Close-up view of the LGR4-NB18 binding interface, highlighting key residues involved in specific interactions. e, f Superimposition of LGR4 from the LGR4-NB18 complex with LGR4 from the LGR4-RSPO2-ZNRF3 and LGR4-Norrin complexes. LGR4 is shown in light green, with Norrin in magenta and RSPO2 in blue; non-essential segments are omitted for clarity. g TOPFlash reporter assay in HEK293T cells demonstrate that NB18 (5 μM) inhibits LGR4-mediated Wnt signaling activity upon RSPO2-Fc stimulation. h, i Dose-dependent inhibition of Wnt signaling activity by NB18 in response to Norrin-Fc (75 nM) (h) and RSPO2-Fc (0.2 nM) (i) stimulation, as measured by TOPFlash reporter assay in HEK293T cells. NB18 was tested in a concentration-dependent manner to generate full dose-response curves. g–i: n = 3, all error bars represent the S.E.M. Source data are provided as a Source Data file.
To understand the molecular basis of NB18’s interaction with LGR4, we determined their complex structure using cryo-EM to 2.7 Å resolution. The resulting high-quality density map enabled precise modeling of both main chain and side-chain conformations (Supplementary Fig. 4, Supplementary Table 2). The structure reveals that NB18 engages the concave inner surface near the top of the LGR4 ectodomain through its complementarity-determining regions (CDRs), forming an extensive interface that covers 2018.64 Ų of solvent-accessible surface area (Fig. 4b, c). Notably, NB18 adopts an orientation parallel to the cell surface, making specific contacts with multiple leucine-rich repeat domains (LRR5, LRR6, LRR8, LRR10, and LRR11) while preserving LGR4’s native conformation.
Detailed analysis of the binding interface reveals a sophisticated network of interactions. The primary contact points involve hydrophilic interactions between the CDR3 region of NB18 (residues L122, R125, and W132) and LGR4 residues (D231, N233, D281, and S300), forming an extensive network of hydrogen bonds and salt bridges (Fig. 4d). A secondary interaction network is established by NB18’s N-terminus (Q23) and CDR1 region (Y51 and R53) with LGR4 residues (H157, D161, Q180, T183, and H209). These hydrophilic interactions frame a central hydrophobic core formed by Y49 and Y51 of NB18 and W159 and H207 from LGR4 (Fig. 4d). Importantly, the LGR4 residues involved in these interactions are conserved across LGR5 and LGR6, suggesting NB18’s potential as a pan-LGR4/5/6 inhibitor (Supplementary Fig. 5).
Comparison of the LGR4-NB18 complex with our previously determined LGR4-Norrin and LGR4-RSPO2 structures revealed a critical insight: NB18’s binding site directly overlaps with both Norrin and RSPOs binding regions on LGR4 (Fig. 4e, f). This structural overlap provides a clear mechanism for NB18’s inhibitory function; by occupying these shared binding surfaces, NB18 prevents both Norrin and RSPO engagement with LGR4.
Functionally, NB18 exhibited dose-dependent inhibition of Wnt signaling in luciferase reporter assays (Fig. 4g). When Norrin was used as the stimulus, NB18 had an IC₅₀ of 5.940 × 10−¹⁰M and a corresponding Ki of 1.618 × 10−¹⁰M. In contrast, when RSPO2-Fc was used, the IC₅₀ was higher at 4.305 × 10−9M, with a similar Ki of 2.5 × 10−¹⁰M. Notably, the lower IC₅₀ for Norrin indicates that NB18 more effectively inhibits Norrin-induced signaling, likely due to Norrin-Fc’s weaker affinity for LGR4, making it more susceptible to competitive displacement. In contrast, RSPO2-Fc binds LGR4 with substantially higher affinity, which reduces the effectiveness of NB18 competition, resulting in a higher IC₅₀ (Fig. 4h, i). Given that these ligand-receptor interactions are critical for normal Wnt pathway regulation, NB18 effectively inhibits both Norrin-Fc and RSPO2-Fc-induced signaling, highlighting its potential as a competitive antagonist of LGR4 function.
Discussion
The Wnt/β-catenin pathway orchestrates crucial biological processes through complex interactions between multiple components, including Wnt ligands, Frizzled receptors, LRP5/6 co-receptors, and regulatory proteins such as LGR4/5/6, RSPOs, and Norrin1,14,15,25,26,27. Dysregulation of this pathway underlies various pathological conditions, from cancer to retinal vascular disorders2,3,4,5,6,7,8,9,10,11,12,13.
Despite this, previous genetic studies involving LGR4 and LGR5 knockout mice, as well as individuals carrying a putative loss-of-function mutation in LGR4, have not shown clear retinal vascular abnormalities. These findings have contributed to the prevailing notion that LGR family members play a limited or redundant role in retinal vascular disease44,60,61,62. However, our structural and functional analyses of the LGR4-Norrin complex provide an important piece of evidence that LGR4 contributes to retinal vascular development and pathology, and offer additional structural mechanistic insights into how this pathway is regulated.
First, our cryo-EM structure of the LGR4-Norrin complex demonstrates a previously uncharacterized 2:2 stoichiometry with distinct architectural features. The complex shows Norrin bridging two LGR4 ectodomains with a characteristic 38.4° tilt and a substantial transmembrane separation of 184 Å (Fig. 1, Supplementary Fig. 1). This large inter-receptor distance raises the possibility that the two LGR4 molecules may originate from adjacent regions of a highly curved membrane, or potentially from the opposing membranes of neighboring cells. This architecture contrasts sharply with the previously described LGR4-RSPO2-ZNRF3 complex, suggesting that Norrin and RSPOs engage LGR4 in fundamentally distinct ways (Fig. 3a–e).
While RSPO2 activates Wnt signaling by binding to both LGR4 and the E3 ligases RNF43/ZNRF3, leading to their internalization and degradation, our data indicate that Norrin utilizes a different mechanism. siRNA-mediated knockdown of ZNRF3 in HEK293 cells did not impair Norrin-induced Wnt activation, suggesting that Norrin-LGR4 signaling is independent of E3 ligase inhibition (Fig. 3l). Additionally, we found that co-transfection of LGR4 and LRP6 significantly enhanced Norrin-induced Wnt signaling, suggesting that Norrin-LGR4 may recruit or stabilize LRP6 to form an active signaling complex (Fig. 3m). This behavior closely mirrors the canonical Wnt-FZD-LRP6 system, where ligand-induced complex formation leads to LRP6 phosphorylation and β-catenin stabilization. Taken together, our findings support a model in which Norrin-LGR4 activates Wnt/β-catenin signaling through a mechanism more akin to canonical Wnt-FZD-LRP6 signaling, rather than RSPO-like modulation of E3 ligase activity.
A key finding from our structural analysis is the identification of partially overlapping binding regions for Norrin and RSPOs on LGR4 (Fig. 3f). This overlap creates a molecular switch mechanism where LGR4 can alternate between two distinct signaling modes: Norrin-mediated signaling for retinal vascular development and RSPO-mediated signaling for broader Wnt pathway regulation.
Our structural comparisons between LGR4-Norrin and FZD4 CRD-Norrin complexes reveal another layer of regulation through competitive binding. The finding that Norrin cannot simultaneously engage both LGR4 and FZD4 due to steric hindrance suggests that this competition may contribute to the distinct phenotypes observed in Norrin-related disorders. Furthermore, our detailed mapping of the binding interfaces has revealed a clear structure-function relationship in disease manifestation: mutations affecting LGR4-specific binding residues lead to FEVR2, while mutations in FZD4-specific interaction sites result in Norrie disease57,66,67,68,69,72. These structure-based findings support a strong genotype-phenotype correlation, providing mechanistic insight into how different Norrin mutations lead to distinct disease outcomes. Together, these results offer a structural framework for understanding Norrin-mediated signaling specificity and its implications in human disease, potentially guiding future therapeutic strategies. We note, however, that our conclusions are primarily based on structural analysis and previously reported patient mutations. While they offer a mechanistic hypothesis, further experimental validation will be essential to establish definitive functional links between Norrin mutations, receptor binding, and disease manifestation.
The lack of reported retinal vascular phenotypes in LGR4 and LGR5 knockout models may be explained by two factors44,60,61,62. First, LGR5 knockout leads to neonatal lethality, potentially obscuring its role in postnatal retinal vascular development in the absence of conditional or tissue-specific models. Second, functional redundancy among LGR4, LGR5, and LGR6 due to their potential co-expression in retinal endothelial cells could result in compensatory signaling that masks the effects of individual gene deletions. To definitively clarify the roles of these receptors in Norrin signaling and retinal vascular development, combinatorial and tissue-specific knockout studies targeting LGR4-6 are essential.
To further explore this regulatory axis, we developed NB18, a high-affinity nanobody that effectively blocks both Norrin and RSPO binding to LGR4. The conserved nature of NB18’s binding interface across LGR4, LGR5, and LGR6 suggests its potential as a pan-LGR4/5/6 inhibitor. This therapeutic tool could be particularly valuable for diseases involving Wnt pathway dysregulation, as it can simultaneously inhibit multiple signaling modes through these receptors.
Looking forward, our findings have significant implications for therapeutic development. The distinct conformational states adopted by LGR4 when binding different ligands suggest the possibility of developing ligand-specific modulators. Such agents could selectively target either the Norrin-LGR4 or RSPO-LGR4 signaling axis, potentially enabling more precise therapeutic interventions for retinal vascular disorders or other Wnt-related diseases.
In conclusion, our structural and functional analyses provide a comprehensive framework for understanding LGR4’s dual regulatory roles in Wnt signaling. The molecular mechanisms we have uncovered—from the stoichiometry of the LGR4-Norrin complex to the competitive binding between different ligands—explain how mutations in a single protein can lead to distinct disease phenotypes. These insights not only advance our understanding of Wnt pathway regulation but also provide a foundation for developing targeted therapeutic strategies for diseases marked by Wnt signaling dysregulation.
Methods
Construct design and cloning
Human LGR4 (residues A25-Q833) was cloned into a modified pEG BacMam vector, incorporating an N-terminal influenza hemagglutinin (HA) signal peptide and a C-terminal Flag tag. A synthetic gene encoding human Norrin (K25-S133, UniProt Q00604) was cloned into the pFBDM vector with an N-terminal gp67 secretion signal. The Norrin sequence was followed by a human rhinovirus 3C protease site, a human IgG1 Fc (hFc) fragment and a C-terminal FLAG tag for downstream applications. The Furin-like (FU) domain of RSPO2 (residues N37-E143) and RSPO4 (residues G32-G142) was cloned into the pEG BacMam vector, incorporating an N-terminal Gaussia signal peptide for secretion and a C-terminal HRV-3C protease cleavage site, followed by a human IgG Fc domain to enhance solubility and facilitate purification. To support functional studies, the full-length human LGR4 sequence was also cloned into the pcDNA3.1(+) vector for use in cell-based assays, ensuring efficient expression in mammalian cells.
NB18 and MB52 generation, expression, and purification
Camel immunizations and nanobody library generation were performed as described previously58. In brief, two camels were immunized subcutaneously with approximately 1 mg human LGR4 protein combined with an equal volume of Gerbu FAMA adjuvant once a week for seven consecutive weeks. Three days after the final boost, peripheral blood lymphocytes (PBLs) were isolated from the whole blood using Ficoll-Paque Plus according to the manufacturer’s instructions. Total RNA from the PBLs was extracted and reverse transcribed into cDNA using a Super-Script III FIRST-Strand SUPERMIX Kit (Invitrogen). The VHH encoding sequences were amplified with two-step enriched-nested PCR using VHH-specific primers and cloned between PstI and BsteII sites of the pMECS vector. Electro-competent E. coli TG1 cells (Lucigen) were transformed.
E. coli strain TG1 cells containing the VHH library were superinfected with M13KO7 helper phages to obtain a library of VHH-presenting phages. Phages presenting LGR4-specific VHHs were enriched after two rounds of bio-panning. Periplasmic extracts were made and analyzed using ELISA screens. NB52 was cloned into a pMECS vector (NTCC) that contains a PelB signal peptide and a hemagglutinin (HA) tag followed by a His6 tag at the C-terminus. It was expressed in the periplasm of E. coli strain TOP10F’ cells.
The VHH gene of NB52 was expanded by fusion to the circular permutated extracellular adhesin domain of Helicobacter pylori (HopQ, 45 kDa) to generate the megabody referred to as MB52. MB52 was expressed as a periplasmic protein in E. coli strain TOP10F’ cells and purified using previously described protocols.
Expression and purification of RSPOs-Fc
The plasmid of the RSPO2-Fc domain protein was transiently transfected into the HEK293F cells using Polyethyleneimine (PEI) at a cell density of 2 × 106 cells/mL. Twenty-four hours post-transfection, sodium butyrate was added to a final concentration of 10 mM. Cells were incubated at 30 °C for 72 h, and then the cell supernatant was harvested by centrifugation at 5000×g for 30 min. The RSPO2-Fc protein was isolated from the cell supernatant by protein A affinity chromatography. The eluted protein was concentrated to 5 mL and further purified by size-exclusion chromatography on a HiLoad 16/600 Superdex 200 pg column. The purification method for RSPO4-Fc was the same as that used for RSPO2-Fc.
Expression and purification of Norrin-Fc protein
Norrin-Fc was cloned into the pFBDM vector to enable recombinant protein production in insect cells via the baculovirus system. Hi5 cells, at a density of 2 × 106 cells/mL, were infected with Norrin-Fc P3 viruses and cultured at 27 °C for 72 h. The cell supernatant was collected by centrifugation at 5000×g for 30 min. Norrin-Fc protein was then isolated from the supernatant using anti-Flag M2 antibody affinity chromatography. Elution was performed with 20 mM HEPES (pH 7.4), 500 mM NaCl, 10% glycerol, 0.5% CHPAS, 1 mM TCEP, 1 mM CaCl2, and 0.3 mg/mL Flag-peptide. The protein was concentrated and further purified by a HiLoad™ 16/600 Superdex™ 200 pg size-exclusion chromatography (SEC) column (Cytiva) equilibrated in a buffer containing 20 mM HEPES (pH 7.5), 500 mM NaCl, 10% glycerol, 0.002% GDN, and 0.0002% CHS to assemble the complexes with LGR4.
Expression and purification of LGR4-Norrin complex
Human LGR4 was expressed in HEK293 GnTI- cells cultured in FreeStyle™ 293 medium (Gibco). HEK293 GnTI- cells at a density of 2 × 106 cells/mL were infected with LGR4 P3 viruses. After 12 h, 10 mM sodium butyrate was added to enhance protein expression, and the cells were incubated at 30 °C for an additional 72 h. After 72 h of expression, the cell pellet (~2.2 g) was collected and lysed using a homogenizer in lysis buffer containing 20 mM HEPES (pH 7.5), 5 mM MgCl₂, 5 mM CaCl₂, 10% glycerol, supplemented with a protease inhibitor cocktail (EDTA-free) and 1 mg of Norrin-Fc protein. NaCl was then added to a final concentration of 150 mM, and membranes were solubilized by adding 1% Lauryl maltose neopentyl glycol (LMNG) and 0.1% cholesteryl hemisuccinate (CHS) at 4 °C for 2 h. The supernatant was obtained by centrifugation at 40,000 rpm for 1 h and applied to an anti-Flag M2 antibody affinity chromatography column. Elution was performed with 20 mM HEPES (pH 7.4), 150 mM NaCl, 5% glycerol, 2 mM MgCl2, 2 mM CaCl2, 0.02% GDN, 0.002% CHS, and 0.3 mg/mL Flag-peptide. MB52 was added at a molar ratio of 1:1.2 to form the complex. The protein complex was concentrated and further purified by size-exclusion chromatography on a Superose™ 6 Increase 10/300 GL column in 20 mM HEPES (pH 7.4), 100 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 0.004% GDN, and 0.0004% CHS.
Biolayer interferometry (BLI) analysis
We employed the Octet RED96 system to assess protein-protein affinity. Streptavidin Biosensors (SA) were first immersed in a 5 μg/mL LGR4-ECD solution to immobilize LGR4. The biosensors were then transferred to PBS containing 0.2% BSA to confirm stable immobilization. Subsequently, Norrin-Fc protein was serially diluted in a twofold concentration gradient, and the biosensors with immobilized LGR4 were placed in wells containing different concentrations of Norrin-Fc for approximately 300 s until saturation. Dissociation was then measured by transferring the biosensors to PBS with 0.2% BSA for approximately 200 s. The binding kinetics and dissociation rates were analyzed using ForteBio Data Analysis Software V3.34.
The same procedure was applied to determine the affinity between LGR4 and NB18. For LGR4-RSPO2-Fc interactions, Octet® ProA Biosensors were used to immobilize RSPO2-Fc, while the subsequent measurement steps remained the same.
Cryo-EM data collection
For the preparation of cryo-EM grids, 3 μL of purified complexes at a concentration of 2–3 mg/mL was applied onto the freshly glow-discharged 300 mesh R1.2/R1.3 UltrAufoil holey gold grids (Quantifoil) or ANTcryoTM Au300-1.2/1.3 (Nanodim) under 100% humidity at 4 °C. The grid was blotted with a wait time of 5 s, and a blot time for 3 s, and plunged-frozen into liquid ethane using the Vitrobot Mark IV (Thermo Fisher Scientific, FEI), and cooled by liquid nitrogen.
Final datasets were collected on a Titan Krios with a K3 detector. All micrographs were acquired at a calibrated pixel size of 0.5355 Å with a dose rate of 23.3 electrons per pixel per second, and defocus values range from −1.5 to −2.5 μm. The micrograph has 36 frames each and was collected over a 3 s exposure and resulting in a total dose of 70 electrons per Å2. At these settings, a total of 5859 movies for the LGR4-Norrin complex and 5303 movies for the LGR4-NB18 complex were collected.
Cryo-EM image processing
All datasets were imported into cryoSPARC for image processing. Motion correction was performed using the Patch Motion Correction module, and the image stacks were binned 2×, corresponding to the pixel size of 1.071 Å. Patch CTF Estimation was applied to each non-dose-weighted micrograph to determine contrast transfer function (CTF) parameters. Micrographs were selected based on estimated CTF fit resolution (better than 4 Å) and relative ice thickness to ensure high-quality input for further processing.
Initial particle picking was performed manually to generate templates for automated particle picking. Auto-picked particles were subjected to two rounds of 2D classification to remove junk and select high-quality particle classes. These particles were then used for Ab initio Reconstruction and multiple rounds of 3D Heterogeneous Refinement to eliminate low-quality classes. High-quality particles were further refined through Ab initio Reconstruction, Homogeneous Refinement, and Non-Uniform Refinement to generate an initial model and improve particle quality.
Subsequently, global and local CTF refinement was conducted. Reference-based motion correction was applied, and particles were stacked using UCSF PyEM and RELION, followed by additional processing in CryoSieve. The final set of screened particles was re-imported into cryoSPARC for a final round of Ab initio Reconstruction, Homogeneous Refinement, and Non-Uniform Refinement, resulting in the final electron density map.
For the LGR4-Norrin complex, a total of 1,244,888 particles were initially picked and subjected to reference-free 2D classification, resulting in 1,005,139 high-quality particles retained for Ab initio Reconstruction into two classes. After multiple rounds of Heterogeneous Refinement, 598,305 particles were subjected to Homogeneous Refinement and Non-Uniform Refinement, yielding a map at 3.60 Å resolution. Applying C2 symmetry slightly reduced the resolution to 3.61 Å. This C2-symmetrized map was used to build the atomic model (PDB: 9KHH).
To improve interpretability, the C2-symmetrized map was processed with emready, a deep-learning-based tool for map enhancement. For detailed visualization of side-chain interactions at the LGR4-Norrin interface, local refinement was performed on the extracellular region of the LGR4-Norrin dimer, focusing on the higher-resolution side. This yielded a local map at 3.05 Å, which enabled the construction of a higher-resolution model (PDB: 9UOK).
For the LGR4-NB18 complex, an initial set of 1,003,958 particles was extracted, of which 490,220 high-quality particles were retained following reference-free 2D classification. These selected particles were then used to generate an ab initio model. Following Heterogeneous Refinement, 420,577 particles were further processed through Homogeneous Refinement and Non-Uniform Refinement, resulting in a reconstruction at 2.97 Å. Reference-based motion correction was then applied, and particles were stacked using UCSF PyEM and RELION, followed by further processing in CryoSieve.
The final subset of 70,544 particles was re-imported into cryoSPARC for another round of Ab initio Reconstruction, Homogeneous Refinement, and Non-Uniform Refinement, yielding a high-resolution map at 2.63 Å. This final map was used to build the atomic model (PDB: 9KGK).
Model building and refinement
The initial model of LGR4, NB18, and MB52 were all generated using AlphaFold, and Norrin from 5BQB. The above structures were fit into the electron density map using Chimera and combined into a single PDB file. The E2GMM module in EMAN2 was used to optimize the PDB, followed by multiple rounds of refinement and manual adjustment using Phenix and Coot on the optimized result. All models were validated using Phenix. Structural figures were prepared in PyMOL (PyMOL | pymol.org).
Luciferase reporter assay
HEK293T/17 cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS). HEK293T/17-LGR4-KO cells, which were maintained in DMEM with 10% FBS and 2 μg/mL puromycin, were generated using CRISPR-Cas9-mediated knockout. The guide sequences used to target LGR4 were cloned into the Lenti-CRISPRv2 vector, with sequences TACCCAGTGAAGCCATTCGA and AGGGTCAGCGCCTGTAGGGT. Cells were grown in 6 cm culture dishes until reaching over 90% confluence. Transient transfection was carried out in 293T/17 cells using Lipofectamine® 3000 (Invitrogen), with plasmid mixtures at molar ratios of 1:1:0.1 for the receptor complex (LGR4:LRP6 = 3:1), Super 8×TOPFlash Wnt reporter, and pRL-SV40 normalization plasmid, respectively. Ten hours post-transfection, cells were seeded into 96-well white plates at a density of 0.7 × 106 cells per well. After allowing 8 h for attachment, cells were treated for an additional 16 h in media with or without Norrin-Fc protein, RSPO2/4-Fc, or NB18. Luciferase activity was measured using the Dual-Luciferase-Reporter Assay Kit (Yeasen) following the manufacturer’s instructions, with data normalized to Renilla luciferase activity. Dose-response curves (log(agonist) versus response (three parameters)) were fitted using GraphPad Prism, and EC50 was retrieved. All experiments were repeated at least three times with duplicates or triplicates in each experiment.
Quantitative RT-PCR
Two siRNAs directed at human ZNRF3 were made by GenePharma. Sense sequences: ZNRF3-1 sense: GGACAGAUGGGGUGAAAUAUU; ZNRF3-2 sense: GCUCUAGAGAAGAUGGAAAUU; N.A. Non-Targeting Pool from GenePharma as a negative control. The total RNAs were isolated using an RNA extraction kit (Vazyme) and eluted with RNase-free, DEPC-treated water before treatment with DNase. After reverse transcription using the Sensiscript RT kit (Vazyme), quantitative PCR was performed in triplicate using the iTaq SYBR Green Supermix kit (Bio-Rad). Expression levels for ZNRF3 were normalized to GAPDH levels. The q-PCR primers were: forward sequence: TCCTGGCTTTCTTCGTCGTGGT, reverse sequence: TGCTCTTGGAGTTGAACTTTCTGG.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All cryo-EM structural models and maps have been deposited in the Protein Data Bank (PDB) and Electron Microscopy Data Bank (EMDB). The PDB codes are 9UOK, 9KHH and 9KGK. The EMDB codes are EMD-64380, EMD-62340 and EMD-62321. These data are publicly available as of the date of publication. The data that support the conclusions of this study are either presented in the paper or in its Supplementary Information. Source data are provided with this paper.
Materials availability
Recombinant DNA plasmids for constructs generated in this study are available upon request.
References
Nusse, R. & Clevers, H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017).
Steinhart, Z. & Angers, S. Wnt signaling in development and tissue homeostasis. Development 145, dev146589 (2018).
Wang, Y., Chang, H., Rattner, A. & Nathans, J. Frizzled receptors in development and disease. Curr. Top. Dev. Biol. 117, 113–139 (2016).
Rim, E. Y., Clevers, H. & Nusse, R. The Wnt pathway: from signaling mechanisms to synthetic modulators. Annu. Rev. Biochem. 91, 571–598 (2022).
Eubelen, M. et al. A molecular mechanism for Wnt ligand-specific signaling. Science 361, eaat1178 (2018).
Janda, C. Y., Waghray, D., Levin, A. M., Thomas, C. & Garcia, K. C. Structural basis of Wnt recognition by Frizzled. Science 337, 59–64 (2012).
Mahoney, J. P. et al. PI(4,5)P(2)-stimulated positive feedback drives the recruitment of Dishevelled to Frizzled in Wnt-β-catenin signaling. Sci. Signal. 15, eabo2820 (2022).
Ahn, V. E. et al. Structural basis of Wnt signaling inhibition by Dickkopf binding to LRP5/6. Dev. Cell 21, 862–873 (2011).
Cho, C., Smallwood, P. M. & Nathans, J. Reck and Gpr124 are essential receptor cofactors for Wnt7a/Wnt7b-specific signaling in mammalian CNS angiogenesis and blood-brain barrier regulation. Neuron 95, 1221–1225 (2017).
Cho, C., Wang, Y., Smallwood, P. M., Williams, J. & Nathans, J. Molecular determinants in Frizzled, Reck, and Wnt7a for ligand-specific signaling in neurovascular development. Elife 8, e47300 (2019).
Dijksterhuis, J. P. et al. Systematic mapping of WNT-FZD protein interactions reveals functional selectivity by distinct WNT-FZD pairs. J. Biol. Chem. 290, 6789–6798 (2015).
Tsutsumi, N. et al. Structure of the Wnt-Frizzled-LRP6 initiation complex reveals the basis for coreceptor discrimination. Proc. Natl. Acad. Sci. USA 120, e2218238120 (2023).
Bourhis, E. et al. Reconstitution of a frizzled8.Wnt3a.LRP6 signaling complex reveals multiple Wnt and Dkk1 binding sites on LRP6. J. Biol. Chem. 285, 9172–9179 (2010).
Deng, C. et al. Multi-functional norrin is a ligand for the LGR4 receptor. J. Cell Sci. 126, 2060–2068 (2013).
Xu, Q. et al. Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell 116, 883–895 (2004).
Ye, X. et al. Norrin, frizzled-4, and Lrp5 signaling in endothelial cells controls a genetic program for retinal vascularization. Cell 139, 285–298 (2009).
Zhang, C. et al. Norrin-induced Frizzled4 endocytosis and endo-lysosomal trafficking control retinal angiogenesis and barrier function. Nat. Commun. 8, 16050 (2017).
Wang, Y. et al. Interplay of the Norrin and Wnt7a/Wnt7b signaling systems in blood-brain barrier and blood-retina barrier development and maintenance. Proc. Natl. Acad. Sci. USA 115, E11827–E11836 (2018).
Chang, T. H. et al. Structure and functional properties of Norrin mimic Wnt for signalling with Frizzled4, Lrp5/6, and proteoglycan. Elife 4, e06554 (2015).
Ke, J. et al. Structure and function of Norrin in assembly and activation of a Frizzled 4-Lrp5/6 complex. Genes Dev. 27, 2305–2319 (2013).
Shen, G. et al. Structural basis of the Norrin-Frizzled 4 interaction. Cell Res. 25, 1078–1081 (2015).
Junge, H. J. et al. TSPAN12 regulates retinal vascular development by promoting Norrin- but not Wnt-induced FZD4/beta-catenin signaling. Cell 139, 299–311 (2009).
Lai, M. B. et al. TSPAN12 Is a Norrin co-receptor that amplifies Frizzled4 ligand selectivity and signaling. Cell Rep. 19, 2809–2822 (2017).
Bang, I. et al. Biophysical and functional characterization of Norrin signaling through Frizzled4. Proc. Natl. Acad. Sci. USA 115, 8787–8792 (2018).
Hao, H. X. et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 485, 195–200 (2012).
Koo, B. K. et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 488, 665–669 (2012).
de Lau, W. et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–297 (2011).
Carmon, K. S., Lin, Q., Gong, X., Thomas, A. & Liu, Q. LGR5 interacts and cointernalizes with Wnt receptors to modulate Wnt/β-catenin signaling. Mol. Cell Biol. 32, 2054–2064 (2012).
Carmon, K. S., Gong, X., Yi, J., Thomas, A. & Liu, Q. RSPO-LGR4 functions via IQGAP1 to potentiate Wnt signaling. Proc. Natl. Acad. Sci. USA 111, E1221–E1229 (2014).
Binnerts, M. E. et al. R-Spondin1 regulates Wnt signaling by inhibiting internalization of LRP6. Proc. Natl. Acad. Sci. USA 104, 14700–14705 (2007).
Glinka, A. et al. LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and Wnt/PCP signalling. EMBO Rep. 12, 1055–1061 (2011).
Carmon, K. S., Gong, X., Lin, Q., Thomas, A. & Liu, Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc. Natl. Acad. Sci. USA 108, 11452–11457 (2011).
Huch, M. et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494, 247–250 (2013).
Park, S. et al. Unlike LGR4, LGR5 potentiates Wnt-β-catenin signaling without sequestering E3 ligases. Sci. Signal. 13, eaaz4051 (2020).
Xu, K., Xu, Y., Rajashankar, K. R., Robev, D. & Nikolov, D. B. Crystal structures of Lgr4 and its complex with R-spondin1. Structure 21, 1683–1689 (2013).
Xu, J. G. et al. Crystal structure of LGR4-Rspo1 complex: insights into the divergent mechanisms of ligand recognition by leucine-rich repeat G-protein-coupled receptors (LGRs). J. Biol. Chem. 290, 2455–2465 (2015).
Chen, P. H., Chen, X., Lin, Z., Fang, D. & He, X. The structural basis of R-spondin recognition by LGR5 and RNF43. Genes Dev. 27, 1345–1350 (2013).
Peng, W. C. et al. Structure of stem cell growth factor R-spondin 1 in complex with the ectodomain of its receptor LGR5. Cell Rep. 3, 1885–1892 (2013).
Zebisch, M. et al. Structural and molecular basis of ZNRF3/RNF43 transmembrane ubiquitin ligase inhibition by the Wnt agonist R-spondin. Nat. Commun. 4, 2787 (2013).
Zebisch, M. & Jones, E. Y. Crystal structure of R-spondin 2 in complex with the ectodomains of its receptors LGR5 and ZNRF3. J. Struct. Biol. 191, 149–155 (2015).
Wang, D. et al. Structural basis for R-spondin recognition by LGR4/5/6 receptors. Genes Dev. 27, 1339–1344 (2013).
Peng, W. C. et al. Structures of Wnt-antagonist ZNRF3 and its complex with R-spondin 1 and implications for signaling. PLoS ONE 8, e83110 (2013).
Seshagiri, S. et al. Recurrent R-spondin fusions in colon cancer. Nature 488, 660–664 (2012).
Styrkarsdottir, U. et al. Nonsense mutation in the LGR4 gene is associated with several human diseases and other traits. Nature 497, 517–520 (2013).
van Andel, H. et al. Aberrantly expressed LGR4 empowers Wnt signaling in multiple myeloma by hijacking osteoblast-derived R-spondins. Proc. Natl. Acad. Sci. USA 114, 376–381 (2017).
Giannakis, M. et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat. Genet. 46, 1264–1266 (2014).
Salik, B. et al. Targeting RSPO3-LGR4 signaling for leukemia stem cell eradication in acute myeloid leukemia. Cancer Cell 38, 263–278.e266 (2020).
Jiang, X. et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc. Natl. Acad. Sci. USA 110, 12649–12654 (2013).
Wang, Y. et al. Norrin/Frizzled4 signaling in retinal vascular development and blood brain barrier plasticity. Cell 151, 1332–1344 (2012).
Zhou, Y. et al. Canonical WNT signaling components in vascular development and barrier formation. J. Clin. Invest. 124, 3825–3846 (2014).
Ye, X., Wang, Y. & Nathans, J. The Norrin/Frizzled4 signaling pathway in retinal vascular development and disease. Trends Mol. Med. 16, 417–425 (2010).
Miller, S. J. et al. Molecularly defined cortical astroglia subpopulation modulates neurons via secretion of Norrin. Nat. Neurosci. 22, 741–752 (2019).
Ding, J. et al. Therapeutic blood-brain barrier modulation and stroke treatment by a bioengineered FZD(4)-selective WNT surrogate in mice. Nat. Commun. 14, 2947 (2023).
Smallwood, P. M., Williams, J., Xu, Q., Leahy, D. J. & Nathans, J. Mutational analysis of Norrin-Frizzled4 recognition. J. Biol. Chem. 282, 4057–4068 (2007).
Gilmour, D. F. Familial exudative vitreoretinopathy and related retinopathies. Eye 29, 1–14 (2015).
Panagiotou, E. S. et al. Defects in the cell signaling mediator β-catenin cause the retinal vascular condition FEVR. Am. J. Hum. Genet. 100, 960–968 (2017).
Berger, W. et al. Mutations in the candidate gene for Norrie disease. Hum. Mol. Genet. 1, 461–465 (1992).
Meitinger, T. et al. Molecular modelling of the Norrie disease protein predicts a cystine knot growth factor tertiary structure. Nat. Genet. 5, 376–380 (1993).
Rehm, H. L. et al. Vascular defects and sensorineural deafness in a mouse model of Norrie disease. J. Neurosci. 22, 4286–4292 (2002).
Morita, H. et al. Neonatal lethality of LGR5 null mice is associated with ankyloglossia and gastrointestinal distension. Mol. Cell Biol. 24, 9736–9743 (2004).
Zhu, J. et al. Targeted deletion of the murine Lgr4 gene decreases lens epithelial cell resistance to oxidative stress and induces age-related cataract formation. PLoS ONE 10, e0119599 (2015).
Siwko, S., Lai, L., Weng, J. & Liu, M. Lgr4 in ocular development and glaucoma. J. Ophthalmol. 2013, 987494 (2013).
Bruguera, E. S., Mahoney, J. P. & Weis, W. I. The co-receptor Tetraspanin12 directly captures Norrin to promote ligand-specific β-catenin signaling. Elife 13, RP96743 (2025).
Wang, L. et al. Structural insights into the LGR4-RSPO2-ZNRF3 complexes regulating WNT/β-catenin signaling. Nat. Commun. 16, 362 (2025).
Uchański, T. et al. Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM. Nat. Methods 18, 60–68 (2021).
Royer, G. et al. NDP gene mutations in 14 French families with Norrie disease. Hum. Mutat. 22, 499 (2003).
Shastry, B. S., Hejtmancik, J. F. & Trese, M. T. Identification of novel missense mutations in the Norrie disease gene associated with one X-linked and four sporadic cases of familial exudative vitreoretinopathy. Hum. Mutat. 9, 396–401 (1997).
Kondo, H. et al. Novel mutations in Norrie disease gene in Japanese patients with Norrie disease and familial exudative vitreoretinopathy. Invest. Ophthalmol. Vis. Sci. 48, 1276–1282 (2007).
Riveiro-Alvarez, R. et al. Gene symbol: NDP. Disease: Norrie disease. Hum. Genet. 124, 308 (2008).
de Lau, W., Peng, W. C., Gros, P. & Clevers, H. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev. 28, 305–316 (2014).
Park, S. et al. Differential activities and mechanisms of the four R-spondins in potentiating Wnt/beta-catenin signaling. J. Biol. Chem. 293, 9759–9769 (2018).
Dickinson, J. L. et al. Mutations in the NDP gene: contribution to Norrie disease, familial exudative vitreoretinopathy and retinopathy of prematurity. Clin. Exp. Ophthalmol. 34, 682–688 (2006).
Acknowledgements
The Cryo-EM data were collected at the Cryo-Electron Microscopy Research Center, Shanghai Institute of Materia Medica, Chinese Academy of Sciences. Camel immunization, nanobody (Nb) library generation, screening, and identification of nanobody were performed at Shanghai Kailuo Biotechnology Co., Ltd. This work is supported by Shanghai Kailuo Biotechnology Co., Ltd.
Author information
Authors and Affiliations
Contributions
Y.G. conceived and designed the study. H.R.Q., F.Z.H., Y.A.W., L.W., S.Y.Z., S.J.G., Y.W.X., J.F.X., Q.Q.C. and Q.L.Y. performed all experiments. H.R.Q., F.Z.H. and Y.A.W. participated in manuscript preparation. Y.G., J.W.Z. and H.E.X. supervised the project, analyzed the structures, and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Qiao, H., Hu, F., Wang, Y. et al. Distinct structural mechanisms of LGR4 modulation by Norrin and RSPOs in Wnt/β-catenin signaling. Nat Commun 16, 6256 (2025). https://doi.org/10.1038/s41467-025-61545-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-61545-z