Abstract
Identifying receptors for bat coronaviruses is critical for spillover risk assessment, countermeasure development, and pandemic preparedness. While Middle East respiratory syndrome coronavirus (MERS-CoV) uses DPP4 for entry, the receptors of many MERS-related betacoronaviruses remain unknown. The bat merbecovirus HKU5 was previously shown to have an entry restriction in human cells. Using both pseudotyped and full-length virus, we show that HKU5 uses Pipistrellus abramus bat ACE2 but not human ACE2 or DPP4 as a receptor. Cryo-electron microscopy analysis of the virus-receptor complex and structure-guided mutagenesis reveal a spike and ACE2 interaction that is distinct from other ACE2-using coronaviruses. MERS-CoV vaccine sera poorly neutralize HKU5 informing pan-merbecovirus vaccine design. Notably, HKU5 can also engage American mink and stoat ACE2, revealing mustelids as potential intermediate hosts. These findings highlight the versatility of merbecovirus receptor use and underscore the need for continued surveillance of bat and mustelid species.
Similar content being viewed by others
Introduction
Bats are a major reservoir for coronaviruses with pandemic potential1,2. To date, three highly pathogenic betacoronaviruses have spilled over into humans: severe acute respiratory syndrome coronavirus (SARS-CoV), SARS-CoV-2, and Middle East respiratory syndrome coronavirus (MERS-CoV)3,4,5,6. MERS-CoV, a member of the Merbecovirus subgenus, transmits sporadically in humans but has a case-fatality rate of 35%7. Hundreds of other bat merbecoviruses circulate in the wild and thus represent a potential pandemic threat8,9. However, our ability to risk-stratify and develop effective countermeasures against these viruses is precluded by the scarcity of information regarding their receptor use9.
Viral receptors mediate host range, tissue tropism, transmission fitness, and pathogenesis10,11,12. Multiple merbecoviruses, including MERS-CoV and bat coronaviruses HKU4 and 422-CoV, use dipeptidyl peptidase-4 (DPP4) as a receptor13,14,15,16,17. In contrast, many sarbecoviruses (e.g., SARS-CoV, SARS-CoV-2) as well as the alphacoronavirus HCoV-NL63 use angiotensin-converting enzyme 2 (ACE2) for entry18,19,20,21. The coronavirus spike protein is comprised of S1 and S2 subunits, which mediate host receptor attachment and membrane fusion, respectively. S1 contains N-terminal domain (NTD) and C-terminal domain (CTD), with the CTD containing the classical receptor-binding domain (RBD). In addition to receptor binding, cellular proteases such as TMPRSS2 and Cathepsin L prime and activate the spike to enable membrane fusion10,21,22,23.
HKU5-like coronaviruses comprise a clade of group 2c merbecoviruses isolated from Pipistrellus bats in East Asia24,25. Despite being closely related to MERS-CoV and the group 2c bat coronavirus HKU4, the receptor for HKU5 is unknown, hindering initial efforts to rescue a full-length infectious molecular clone of HKU59,23,24,26. Previous studies demonstrated that human cells are permissive to HKU5 but that there was a block to infection at viral entry that could in part be overcome with exogenous trypsin treatment26,27. An HKU5 chimera (HKU5-SE) in which the SARS-CoV spike ectodomain replaced that of HKU5 efficiently replicated in human cells and caused severe disease in aged mice26. This demonstrates the pathogenic potential of the HKU5 viral backbone. African and European bat merbecoviruses (NeoCoV, PDF-2180; MOW15-22, PnNL2018B; respectively) were recently shown to use bat ACE2, albeit by distinct mechanisms9,28,29. Identifying the receptor for HKU5 and related viruses is crucial for understanding their host range, spillover potential, and pandemic risk.
Here, using a panel of receptor orthologs, we identify that a prototypic HKU5 and the related BtPa-BetaCoV/GD2013 (GD2013) use ACE2 from P. abramus bats for entry. We characterize this interaction with genetic, biochemical, and structural studies, including a 4.2 Å cryo-EM structure of the HKU5RBD and P. abramus ACE2 complex. We map key interacting residues via mutagenesis and demonstrate that full-length infectious HKU5 requires P. abramus ACE2. We also identify mustelids as possible intermediate hosts for the HKU5 clade. This work will enable improved surveillance and the development of countermeasures for this group of bat coronaviruses.
Results
Investigation of the receptor usage of diverse bat CoVs
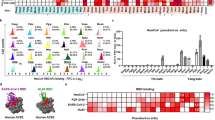
To screen for coronavirus receptors, we synthesized spikes from five diverse bat coronaviruses from three different Betacoronavirus subgenera: bat HKU5-LMH03f (Merbecovirus, HKU5), Erinaceus CoV/2012-174 (Merbecovirus, HHCoV), RoBatCoV/HKU9 (Nobecovirus, HKU9), RoBatCoV/GCCDC1 (Nobecovirus, GCCDC1), and bat Hp-BetaCoV/Zhejiang2013 (Embecovirus, HpCoV) (Fig. 1a and Supplementary Fig. 1a). We generated replication defective vesicular stomatitis virus (VSV) pseudovirus expressing each respective spike protein and a Renilla Luciferase (RLuc) reporter. To test whether ACE2 or DPP4 could mediate entry, we initially synthesized expression constructs for 31 DPP4 (Fig. 1b and Supplementary Table 1) and 48 ACE2 orthologs (Fig. 1c and Supplementary Table 2) from wild, domestic, and peri-domestic species. DPP4 and ACE2 plasmids were transfected into human HEK-293T cells, and expression was confirmed by Western blot (Supplementary Fig. 1b, c). First, we investigated whether any DPP4 ortholog(s) could facilitate viral entry of these orphan coronaviruses, in addition to SARS-CoV-2 and MERS-CoV controls. MERS-CoV spike pseudoviruses (VSV-MERS-CoVspike-RLuc) readily infected cells expressing human DPP4 as well as other mammalian DPP4 orthologs. However, these DPP4s were not sufficient to promote entry of the other pseudoviruses (Fig. 1b). Next, we assessed whether any of these pseudoviruses could efficiently use ACE2 orthologs for entry. The SARS-CoV-2 spike mediated entry of cells expressing the ACE2 of 38/48 ACE2 orthologs, including 19/27 bat species and 19/21 non-bat species, in both HEK-293T and BHK-21 (hamster) cells, the latter of which lacks endogenous ACE2 (Fig. 1c). The ability to engage diverse ACE2 orthologs is consistent with the known broad host range of SARS-CoV-230. Notably, among the pseudoviruses tested, we detected a significant increase in HKU5 pseudovirus (VSV-HKU5spike-Rluc) entry into both Pipistrellus abramus (P. abramus, Japanese house bat) and Neogale vison (N. vison, American mink) ACE2-expressing cells. The four other bat coronavirus spikes did not mediate detectable entry with any tested ACE2 ortholog (Fig. 1c). As expected, sequence alignment revealed high sequence variability in the RBDs of ACE2-using and DPP4-using merbecoviruses31 (Supplementary Figs. 1d and 2).
a Phylogenetic analysis of the full-length amino acid sequences of coronavirus spikes from representative subgenera. Hosts and receptors are indicated. Asterisks (*) highlight bat coronaviruses used in this study. Animal illustrations were sourced from BioRender (https://BioRender.com/d86esp4). b HEK-293T cells were transiently transfected with the indicated animal DPP4 constructs, and infection of each spike pseudovirus was assessed. MERS-CoV pseudovirus uses diverse DPP4 orthologs, but other spike pseudoviruses do not use DPP4. c HEK-293T or BHK-21 cells were transiently transfected with the indicated animal ACE2 constructs. Pseudotyped virus infection with coronavirus spikes revealed that HKU5 uses P. abramus and N. vison ACE2 for entry. In contrast, SARS-CoV-2 uses diverse ACE2 orthologs. The mean of three technical replicates is plotted from one of two independent experiments. RLU relative light units, Sbc sarbecovirus, Merbc merbecovirus, Nobc nobecovirus, Emc embecovirus. Source data are provided as a Source Data file.
Characterization of bat coronavirus HKU5 utilization of P. abramus and N. vison ACE2 for entry
Next, we sought to validate P. abramus ACE2 and N. vison ACE2 as entry receptors for HKU5 from the pseudovirus entry screen. We transfected a targeted panel of ACE2 and DPP4 constructs in HEK-293T cells and used pseudoviruses to assess viral entry. Consistent with our initial screen, HKU5 efficiently used P. abramus and N. vison ACE2 but not DPP4 for entry (Fig. 2a, b). Next, we generated HKU5spike VSV pseudovirus expressing green fluorescence protein (VSV-HKU5spike-eGFP) to assess infection on a per-cell basis. Consistent with the luciferase results, VSV-HKU5spike-eGFP infected HEK-293T cells expressing P. abramus ACE2 and N. vison ACE2 (Fig. 2c, d). At a low multiplicity of infection (MOI), VSV-SARS-CoV-2spike-eGFP infected cells expressing human ACE2 and N. vison ACE2, but not P. abramus ACE2. Next, we tested if murine leukemia virus (MLV) pseudovirus coated with P. abramus ACE2 protein (MLV-P. abramusACE2-Gluc) could infect HEK-293T target cells expressing HKU5spike on their surface (Fig. 2e). MLV bearing P. abramus ACE2 readily infected HEK-293T cells in a HKU5spike- and ACE2-dependent manner, revealing that HKU5spike and P. abramus ACE2 are necessary and sufficient for entry.
HEK-293T cells expressing ACE2 (a) or DPP4 (b) orthologs from H. sapiens, P. abramus, P. pipistrellus, N. vison, or control vectors were infected with VSV pseudoviruses bearing SARS-CoV-2, HKU5, or MERS-CoV spikes. Entry was quantified via luciferase activity. c, d Cells expressing ACE2 orthologs were infected with eGFP-expressing pseudoviruses; GFP-positive cells were counted across 10 fields (100×). e MLV particles pseudotyped with P. abramus ACE2 and Gaussia luciferase infected HKU5 spike-expressing cells. Schematic illustration was generated using BioRender (https://BioRender.com/xige2n2). f Phylogenetic tree of full-length coronavirus spikes related to HKU5. g, h Entry efficiency of HKU5-related viruses in HEK-293T and BHK-21 cells expressing ACE2 orthologs was assessed via Renilla luciferase. i–l Vero81 cells stably expressing ACE2 constructs were infected with full-length HKU5 (FL-HKU5). i Brightfield and j fluorescence microscopy showed syncytia and nucleoprotein-positive cells in P. abramus ACE2-expressing cells. k Spike, ACE2, and nucleocapsid expression were assessed by Western blot at 24 hpi. l Plaque assays measured replication kinetics of FL-HKU5 in Vero81/ACE2 cells. RLU relative light units. Data represent three independent experiments unless otherwise stated. Data in (a, b, g, h) are pooled from two independent experiments with three biological replicates each. Images in (d) are representative of two experiments. Scale bars: 100 µm (d, i); 50 µm (j). Statistical analysis: two-tailed unpaired Student’s t-test and one-way repeated measures ANOVA. Data shown as mean ± s.e.m. ns, not significant; **p = 0.0069, ***p = 0.0022, ****p < 0.0001. Source data are provided as a Source Data file.
To investigate whether other merbecoviruses closely related to the prototypic HKU5 isolate LMH03f could use P. abramus and N. vison ACE2, we synthesized five additional bat coronavirus spike proteins: BatCoV HKU5r isolate BY140568 (HKU5r BY140568), GD2013, BatCoV HKU25 isolate NL140462 (HKU25), BtVs-BetaCoV/SC2013 (SC2013), and BatCoV/H.savii/Italy/206645-40/2011 (Italy/2011)32,33,34,35,36 (Fig. 2f). HKU5r BY140568, GD2013, and HKU25 were isolated from Pipistrellus bats while SC2013 and Italy/2011 were isolated from Vespertilio superans and Hypsugo savii bats, respectively32,33,34,35,36. GD2013 could use both P. abramus and N. vison ACE2 for entry, while the four other coronaviruses could not use any of the tested ACE2 orthologs (Fig. 2g, h). Together, this confirms that HKU5 and GD2013 use P. abramus and N. vison ACE2 as receptors, while the receptors for genetically similar coronaviruses remain unknown.
To confirm the pseudovirus data with full-length infectious HKU5, we inoculated Vero-CCL-81 (Vero81) cells stably expressing P. abramus ACE2 (Vero81/P. abramus ACE2) with a full-length HKU5 infectious recombinant virus (FL-HKU5). We observed virus-induced cytopathic effects at 72 h post-infection (hpi) in Vero81 cells expressing P. abramus ACE2 but not human ACE2 (Fig. 2i). FL-HKU5 readily infected Vero81/P. abramus ACE2 cells as demonstrated by the presence of nucleoprotein antigen-positive cells and syncytia formation at 24 hpi (Fig. 2j). Next, we challenged HEK-293T cells transiently expressing ACE2 orthologs with FL-HKU5. The HKU5 spike and nucleocapsid proteins were detected by Western blot from cells expressing P. abramus ACE2 and N. vison ACE2 but not in cells expressing human or other bat (Pteropus alecto and Rhinolophus pusillus) ACE2s (Fig. 2k). Next, we performed a viral growth curve with FL-HKU5 on Vero81 cells stably expressing human ACE2, P. abramus ACE2, or N. vison ACE2. Vero81/P. abramus ACE2 cells infected with FL-HKU5 exhibited a greater than one-log increase in virus titer at 24 hpi and nearly four-log increase in viral titers by 72 hpi (Fig. 2l). In contrast, the viral titers in Vero81/N. vison ACE2 cells did not show an increase at 24 hpi, but rapidly caught up by 48 hpi, resulting in a nearly four-log increase by 72 hpi (Fig. 2l). These findings indicate that P. abramus ACE2 or N. vison ACE2 expression is sufficient to facilitate productive infection and spread of native HKU5 (Fig. 2l).
Evaluation of the HKU5 receptor-binding domain for targeted ACE2 binding
Next, we sought to determine the mechanism by which HKU5spike interacts with ACE2. We generated mouse immunoglobulin G (IgG) Fc-fusion proteins containing the RBD (RBD-Fc) from either SARS-CoV-2 (SARS-CoV-2RBD-Fc), MERS-CoV (MERS-CoVRBD-Fc), PDF-2180 (PDF-2180RBD-Fc), or HKU5 (HKU5RBD-Fc) spike proteins (Supplementary Fig. 3a, b). We transiently expressed human ACE2, P. abramus ACE2, N. vison ACE2, or human DPP4 on HEK-293T cells and assessed RBD-Fc binding to these cells by flow cytometry. SARS-CoV-2RBD-Fc readily bound human and N. vison ACE2 but not P. abramus ACE2 or human DPP4 (Fig. 3a), consistent with our data and prior studies35. MERS-CoVRBD-Fc bound human DPP4 but not to any tested ACE2 ortholog (Fig. 3b). The PDF-2180RBD-Fc bound to P. abramus and N. vison ACE2 but exhibited low levels of binding to human ACE2 (Fig. 3c). The HKU5RBD-Fc showed robust binding to cells displaying P. abramus ACE2 (Fig. 3d). However, binding of HKU5RBD-Fc to N. vison ACE2 was below the limit of detection (Fig. 3d).
Flow cytometry analysis of SARS-CoV-2RBD-Fc (a), MERS-CoVRBD-Fc (b), PDF-2180RBD-Fc (c), and HKU5RBD-Fc-fusion protein (d) binding to HEK-293T cells expressing the indicated receptors. The mean fluorescence intensity (MFI) was calculated. Data are plotted from two independent experiments with two biological replicates per experiment. The same mock samples were used in (a–d), as the experiments were processed together. HKU5RBD-Fc directly binds the P. abramus ACE2. e BLI analysis reveals binding kinetics of HKU5RBD with P. abramus ACE2. The reported KD values correspond to avidities due to the use of dimeric ACE2 constructs. Analysis was conducted using curve-fitting kinetic global fitting (1:1 binding model). f–h Neutralization assays using SARS-CoV-2 vaccine sera, MERS-CoV-S2P mouse vaccine sera, and a monoclonal antibody against MERS-CoV (MERS-CoV-27) in BHK-21 cells revealed that HKU5 is resistant to SARS-CoV-2- and MERS-CoVelicited antibodies. Each point represents the mean neutralization value of two biologically independent infection replicates. Nonlinear regression (4-parameter logistic model) was performed using Python, with curve fitting implemented via scipy.optimize.curve_fit, and graphs were generated using GraphPad Prism v10.4.2. For panel f, the SARS-CoV-2 IC₅₀ is presented from a single biological experiment for clarity in presentation. #Not reliable: The confidence intervals could not be reliably estimated in some cases due to poor curve fit, likely resulting from flat or noisy responses. a–d Statistical analyses were performed using two-tailed unpaired Student’s t-tests and one-way ANOVA. Data are represented as mean ± s.e.m. ns not significant, *p = 0.0365, ***p = 0.0003, ****p < 0.0001. Source data are provided as a Source Data file.
To quantify the affinity and kinetics of the spike-receptor interactions, we performed biolayer interferometry (BLI). Recombinant HKU5RBD bound to recombinant P. abramus ACE2, with a binding affinity (Kd) of 120 nm (Fig. 3e, Supplementary Fig. 3c), whereas binding to N. vison ACE2 was below the limit of detection (Supplementary Data Fig. 3d). Consistent with the flow cytometry binding assay, SARS-CoV-2RBD bound to N. vison ACE2 (Kd = 1.5 nm) while SARS-CoV-2RBD binding to P. abramus ACE2 was undetectable (Supplementary Data Fig. 3d, e). These results demonstrate direct physical interaction between the RBD of HKU5spike and P. abramus ACE2.
The identification of P. abramus ACE2 as a receptor for HKU5 enables assessment of antibody-mediated protection. To evaluate antibody-mediated cross-protection between HKU5, MERS-CoV, and SARS-CoV-2, we tested neutralizing activity of SARS-CoV-2 human vaccine sera, MERS-CoV-S2P mouse vaccine sera, and a MERS-CoV-targeting monoclonal antibody (MERS-27 mAb)37,38 against VSV-HKU5spike-RLuc (Fig. 3f–h). SARS-CoV-2 vaccine sera nor the MERS-27 mAb neutralized VSV-HKU5spike-RLuc (Fig. 3f–h). The MERS-CoV-S2P mouse sera had detectable but limited neutralizing activity against VSV-HKU5spike-RLuc (Fig. 3g). Our results highlight antigenic differences between HKU5, SARS-CoV-2, and MERS-CoV and underscore the need for novel mAbs and vaccines that cross-react with HKU5 viruses.
Structural basis of HKU5 recognition of P. abramus ACE2
To understand HKU5RBD engagement with P. abramus ACE2, we performed cryo-EM analysis of HKU5RBD bound to dimeric P. abramus ACE2. Data processing yielded a structure at 4.2 Å resolution, revealing the ACE2 dimer with HKU5RBD bound to each peptidase domain (Fig. 4a, Supplementary Figs. 4 and 5, and Supplementary Table 3). The binding interface buried a surface of ~1000 Å2 on both sides of the interaction area (Fig. 4b and Supplementary Table 4). The 1040 Å2 ACE2 binding surface on HKU5RBD is formed by regions on α-helices 4 and 5, β strands 5, 6 and 7, and the loop region exiting β5 (Fig. 4b). HKU5RBD binds to ACE2 on a surface boarded by the inner side of helix α1, tip of helix α3, glycan on N387ACE2 and the R328ACE2 helix (Fig. 4b).
a Cryo-EM density (left) and refined model (right) for the HKU5RBD in complex with P. abramus ACE2. The ACE2 is colored in green, and the RBD is colored in orange. b Footprints of the P. abramus ACE2 and HKU5RBD complex are shown in open-book view. c Interactions at the interface. The complex is shown in the 45-degree rotated view compared to the orientation in (a). The interactions are divided into two patches. HKU5RBD tyrosines provide more than 500 Å2 binding surface for the P. abramus ACE2. Tyrosines in patch 1 form extensive interactions with a helical region on ACE2 that spans P324ACE2, R328ACE2, and D329ACE2. Patch 2 interactions involve hydrogen bonding between HKU5RBD tyrosines and A385ACE2, glycan 387ACE2, K352ACE2, as well salt bridge between E510RBD and K352ACE2. The Y517RBD is wedged between I92ACE2 and I93ACE2 and interacts with R26ACE2 and D90ACE2. d Comparison of binding modes of the RBDs of HKU5, SARS-CoV-2, NeoCoV and PDF-2180 CoV to ACE2. All structures (PDB ID: 6M0J, 7WPO, and 7WPZ) were superimposed onto the HKU5RBD-bound to P. abramus ACE2. Footprints of the HKU5RBD, SARS-CoV-2RBD, PDF-2180RBD, and NeoCoVRBD are marked on the surface representation of P. abramus ACE2 and colored in the same color as their respective RBDs in the left panel. A zoomed-in view is shown to the lower right to compare the binding difference of HKU5RBD and SARS-CoV-2RBD relative to the ACE2 α1 helix. e, f Structural conservation map of the RBDs from the spike proteins of MERS-CoV, PDF-2180, and HKU5. Sequence conservation between HKU5, NeoCoV, PDF-2180, and MERS-CoV is mapped along the HKU5-P. abramus ACE2 binding footprint (outlined in black) on the HKU5 RBD (burnt orange) e. Pairwise sequence conservation between HKU5 and the RBD of NeoCoV (sand), PDF-2180 (pink), or MERS-CoV (salmon) is mapped to their respective binding footprints (outlined in black) f. Amino acid sequence with conservation colored blue and divergence red.
HKU5RBD and P. abramus ACE2 form potential hydrogen bonds, salt bridges, polar and stacking interactions (Supplementary Table 4). These interactions can be categorized into two major patches. Patch 1 involves five tyrosine residues (Y463RBD, Y507RBD, Y544RBD, Y557RBD, Y559RBD) and M460RBD. These residues form a pocket to wrap around the R328ACE2 helix with polar and stacking interactions (Fig. 4c). Patch 2 is mainly formed by β5 and its exiting loop, β6, and β7. Despite the 4.2 Å resolution, hydrogen bonds form between NAG386ACE2 and Y523RBD, A386ACE2 and Y521RBD, K352ACE2 and Y512RBD, N353ACE2 and E510RBD, N353ACE2 and Y544RBD. Y517RBD on the tip of the β5-exiting loop makes extensive interactions with R26ACE2, D90ACE2, I92ACE2, and I93ACE2. A prominent feature of the ACE2 interface on RBD is that tyrosine-rich interactions provide 50% of the receptor-binding surface (Supplementary Table 4).
To determine whether dimeric ACE2 can interact with two HKU5 spikes simultaneously, we modeled a complex between an ACE2 dimer and two spike trimers by superposing the HKU5 RBDs onto the RBDs of two MERS-CoV spikes (Supplementary Fig. 6). The MERS-CoV spike structure was used because, unlike the HKU5 spike, it contains one RBD in the raised position, which is necessary for RBD-ACE2 interaction. The model also incorporated the structure of B0AT2, another known binding partner of ACE2. This modeling revealed no steric clashes between the two spikes, suggesting that such a complex is structurally possible.
Comparison of the structures of ACE2-bound HKU5RBD, SARS-CoV-2RBD, PDF-2180RBD, and NeoCoVRBD shows that HKU5RBD binds to ACE2 in a different binding mode (Fig. 4d). Both PDF-2180RBD and NeoCoVRBD bind to ACE2 mainly through their β5-exiting loops and the β-hairpins between strand β6 and β7 with smaller footprints. In contrast, HKU5RBD and SARS-CoV-2RBD recognize much larger areas on ACE2. While SARS-CoV-2RBD grabs both sides of ACE2 α1 helix, HKU5 only interacts with the inner side of the α1 helix, shifting its footprint to the center region of ACE2 peptidase domain (Fig. 4d). Despite the common usage of ACE2 as a receptor and the similar overall folding of their RBDs, subtle sequence differences in the receptor-binding footprints may result in distinct binding modes to ACE2. To explore this sequence variation, we mapped the sequence conservation between HKU5, NeoCoV, PDF-2180, and MERS-CoV onto the HKU5 RBD-ACE2 binding footprint (Fig. 4e). This revealed divergence along this receptor-binding interface. We then mapped sequence conservation as a pairwise comparison between HKU5 and NeoCoV, PDF-2180, and MERS-CoV onto their respective binding footprints, revealing differences between HKU5 and each of these at their binding footprint (Fig. 4f). Together, this illustrates sequence divergence along the binding footprints, which likely drives distinct binding modes.
P. abramus ACE2 determinants of HKU5 spike interaction
To functionally validate residues in the binding interface, we generated both combinatorial and individual substitutions in P. abramus ACE2, transfected these constructs into BHK-21 cells, and used VSV-HKU5spike-RLuc or VSV-SARS2-CoV-2spike-RLuc to assess the ability of these constructs to support viral entry. We tested four combinations of substitutions: R26K/V30D/N38D/H41YACE2, D90N/I92T/I93VACE2, P324Q/R328E/D329NACE2, and N353GACE2. Of these four ACE2 combinations, only P324Q/R328E/D329NACE2 (patch 1 in Fig. 4C) completely abrogated HKU5 entry, while N353GACE2 partially reduced entry (Fig. 5a, b). Each individual substitution in P324Q/R328E/D329NACE2 also modestly reduced HKU5 entry.
a Schematic of the ACE2 gene highlighting substituted residues and domains. Positions in gray had minimal effect on HKU5 entry, while substitutions that reduced entry are shown in red. b Wild-type P. abramus ACE2, individual, and combinatorial ACE2 mutants were transiently transfected into BHK-21 cells and then infected with VSV-HKU5spike-RLuc or VSV-SARS-CoVspike-RLuc. c We generated two patches of substitutions (1, 2) on the HKU5RBD and tested each for infectivity on BHK-21 cells stably expressing P. abramus ACE2 (Patch 1: Y507A/Y544Q/Y557SRBD; patch 2: E510K/Y512S/Y517S/Y521E/Y523ARBD. Patches 1 and 2 of the HKU5RBD were necessary for P. abramus ACE2 utilization, while individual substitutions Y507ARBD and E510KRBD were sufficient to block infection. d We generated four patches of substitutions (3, 4, 5, 6) on the HKU5RBD (orange) and tested each for infectivity on BHK-21 cells stably expressing P. abramus ACE2 (green). Patch 3: S457P/D459S/Y463D/A471PRBD; patch 4: T509N/E510K/Y512S/T514L/S515L/A516F/Y517D/G518D/K519R/Y521ERBD; patch 5: 542-548→EDGDYYRKQLSPLEGRBD; patch 6: T553A/T555S/Y557S/I558T/Y559VRBD). HKU5 patches 4, 5, and 6 were required for P. abramus ACE2 use, with individual substitutions K519RRBD and Q545PRBD sufficient to block infection. Dots represent the mean from each of three independent experiments, each done in technical triplicate. Statistical analyses were performed using two-tailed unpaired Student’s t-tests and one-way ANOVA. Data are mean ± s.e.m. b *D90N-I92T-I93V: p = 0.0426, D90N: p = 0.0317, **V30D: p = 0.0089, **I93V: p = 0.0094, **R328E: p = 0.0020, ****p < 0.0001. c *p = 0.0445, ***p = 0005, ****p < 0.0001. d *Y463D: p = 0.0294, *T514L: p = 0.0473, *S515L: p = 0.0176, *A516F: p = 0.0158, **Patch 1: p = 0.0060, **K543D: p = 0.0047, ****p < 0.0001. e HKU5RBD patches 3, 4, 5, and 6. Residue positions for each set of substitutions are shown in stick and dot representations. K519RBD and Q545RBD, which showed infection-blocking effects when mutated, are labeled along with other residues for positional reference. Source data are provided as a Source Data file.
Although we did not previously observe detectable SARS-CoV-2 entry in P. abramus ACE2-expressing cells (Figs. 1 and 2), we hypothesized that a higher MOI using a concentrated VSV-SARS-CoV-2spike-RLuc stock could potentially overcome the barrier to infection. Indeed, our higher titer VSV-SARS-CoV-2spike-RLuc demonstrated viral entry with P. abramus ACE2 (Fig. 5b). Of note, none of the P. abramus ACE2 substitutions reduced SARS-CoV-2 entry, demonstrating that each ACE2 construct was functionally expressed on the cell surface.
An earlier study examining interactions between P. pipistrellus ACE2 and NeoCoV/PDF-2180 revealed a glycan-dependent ACE2 binding mechanism, which mediates species-specific ACE2 use28. Specifically, P. pipistrellus residues 337–342ACE2 (SDGRQV) mediated entry of both NeoCoV and PDF-218028. These residues are conserved in P. abramus ACE2 but differ between human and P. abramus ACE2 orthologs (Supplementary Fig. 7a, b). To determine whether these ACE2 determinants can modulate HKU5 entry, we generated six human ACE2-P. abramus ACE2 chimeras (Supplementary Fig. 8) and expressed the ACE2 chimeras in HEK-293T cells. VSV-HKU5spike-RLuc and VSV-SARS-CoV-2spike-RLuc were used to assess the ability of these chimeras to support entry. Notably, the introduction of the N-terminus of P. abramus ACE2 (1–83 aa, chimera #1) into human ACE2 enabled HKU5 entry; however, the reciprocal insertion of the human ACE2 N-terminus into P. abramus ACE2 (chimera #2) did not support entry (Supplementary Fig. 8a, b). Insertion of P. abramus residues 337–342ACE2 (SDGRQV) also did not facilitate HKU5 entry (chimera #3). Insertion of human residues 337–342ACE2 (GNVQKA, chimera #4) into P. abramus ACE2 modestly reduced but did not eliminate HKU5 entry. Next, we tested VSV-GD2013spike-RLuc to determine whether these interactions were generalized to other HKU5-like viruses. VSV-GD2013spike-RLuc revealed a similar pattern to HKU5 amongst the ACE2 chimeras on HEK-293T cells (Supplementary Fig. 8c). We validated ACE2 chimera expression by Western blot (Supplementary Fig. 8d). In contrast to HKU5 and GD2013, SARS-CoV-2 was much more tolerant of ACE2 chimeras and substitutions. Both insertion of the P. abramus ACE2 N-terminus into human ACE2 (chimera #1), as well as the reciprocal insertion (chimera #2), were sufficient for SARS-CoV-2 entry (Supplementary Fig. 8c). Substitution of residues 337–342ACE2 had no effect on SARS-CoV-2 entry in cells expressing human or P. abramus ACE2, consistent with our structural analysis (Supplementary Fig. 8c).
To further investigate spike-ACE2 interactions, we conducted a flow cytometry-based binding assay with RBD-Fc-fusion proteins on HEK-293T cells expressing the ACE2 chimeras. Consistent with the pseudovirus results, HKU5RBD strongly bound both wild-type P. abramus ACE2 and the P. abramus ACE2 N-terminus on human ACE2 (chimera #1) (Supplementary Fig. 9a). Substitutions of residues 337–342ACE2 had no effect on HKU5RBD-Fc binding. In contrast to HKU5, SARS-CoV-2RBD-Fc bound to all ACE2 chimeras except chimera #4, phenocopying VSV-SARS-CoV-2spike-RLuc results. Next, we directly compared PDF-2180RBD-Fc binding relative to HKU5 with the ACE2 chimeras. PDF-2180 exhibited a broad but distinct pattern of ACE2 use relative to HKU5, consistent with different binding mechanisms and specificities (Fig. 4d). Consistent with previous results28, insertion of human ACE2 residues 337–342ACE2 (chimera #5) into P. abramus ACE2 impaired PDF-2180RBD-Fc binding (Supplementary Fig. 9c). Our structural analysis and mutagenesis results confirm that the N-terminus of P. abramus ACE2 is important for interaction with the HKU5RBD and reveal distinct mechanisms of binding and entry between SARS-CoV-2, PDF-2180, and HKU5.
HKU5 spike determinants of P. abramus and N. vison ACE2 interaction
We next sought to determine the HKU5spike residues that interact with P. abramus and N. vison ACE2. We tested the infectivity of VSV-HKU5spike-RLuc in BHK-21 cells expressing either wild-type P. abramus ACE2 or N. vison ACE2. We introduced substitutions from MERS-CoVspike on HKU5spike at sites residing in the P. abramus ACE2 contact interface. We tested two mutant sets depicted in Fig. 4c: Patch 1 (Y507A/Y544Q/Y557S/Y559VRBD) and Patch 2 (E510K/Y512S/Y517S/Y521E/Y523ARBD). Both Patch 1 and Patch 2 substitutions effectively blocked infection in P. abramus ACE2, with individual substitutions Y507ARBD and E510KRBD being sufficient to confer this restriction (Fig. 5c). Additionally, HKU5spike substitutions S457P/D459S/Y463D/A471PRBD (Patch 3) did not affect HKU5 entry in cells expressing P. abramus ACE2. Whereas HKU5spike patch 4 (T509N/E510K/Y512S/T514L/S515L/A516F/Y517D/G518D/K519R/Y521ERBD), patch 5 (542-548→EDGDYYRKQLSPLEGRBD), and patch 6 (T553A/T555S/Y557S/I558T/Y559VRBD) substitutions restricted infection with P. abramus ACE2. We further identified that the individual substitutions K519RRBD (from patch 4) and Q545PRBD (from patch 5) were sufficient to reduce P. abramus ACE2 use (Fig. 5d, e). In contrast, all four patches (3, 4, 5, 6) blocked infection of N. vison ACE2-expressing cells, with individual substitutions Y463DRBD, S515LRBD, A516FRBD, K519RRBD, K543DRBD, Q545PRBD, and S546LRBD each being sufficient to restrict N. vison ACE2-dependent entry (Supplementary Fig. 10). These findings suggest that N. vison ACE2 is more susceptible to HKU5spike perturbations, which is consistent with the reduced HKU5 binding and entry efficiency relative to P. abramus ACE2.
N. vison ACE2 determinants of HKU5 spike interaction
Our screen revealed that ACE2 from American mink (N. vison) but not ferrets (Mustela putorius) could support HKU5 entry (Fig. 1). Therefore, we asked which other mustelid species could serve as potential intermediate hosts for HKU5. To test this, we synthesized ACE2 constructs from nine additional mustelids including European badgers (Meles meles), greater hog badgers (Arctonyx collaris), Chinese ferret badger (Melogale moschata), North American river otters (Lontra canadensis), Eurasian otters (Lutra lutra), wolverines (Gulo gulo), stoats (Mustela erminea), black-footed ferrets (Mustela nigripes), and European mink (Mustela lutreola) (Fig. 6a and Supplementary Table 2). Of note, mink ACE2 contains an insertion which results in the alteration of residue positions compared to P. abramus ACE2 (Fig. 6a). VSV-HKU5spike-RLuc and VSV-SARS-CoV-2spike-RLuc were used to assess the ability of wild-type mustelid ACE2s to support viral entry. N. vison and M. erminea ACE2 but not that of the other tested mustelids allowed for HKU5 entry (Fig. 6b). In contrast to VSV-HKU5spike-RLuc, VSV-SARS-CoV-2spike-RLuc similarly used all mustelid ACE2s (Fig. 6b). These findings suggest that N. vison and M. erminea could serve as intermediate hosts for HKU5-related viruses.
a Phylogenetic tree of mustelid species for which ACE2 constructs were tested. Shown are the nine amino acid positions that differ between N. vison (susceptible to HKU5) and M. putorius (resistant to HKU5). The figure was created using PhyloPic, an open-access database licensed under Creative Commons. b Wild-type mustelid ACE2 constructs were transiently transfected into BHK-21 cells, and VSV-HKU5-spike-RLuc and VSV-SARS-CoV-spike-RLuc were used to assess entry. N. vison (American mink) and M. erminea (stoat) ACE2, but not the ACE2 of other mustelids, were sufficient for HKU5 entry. SARS-CoV-2 could use all tested mustelid ACE2s. c Wild-type and mutant N. vison ACE2 and M. putorius ACE2s were transiently transfected into BHK-21 cells, and entry was assessed using VSV-HKU5-spike-RLuc and VSV-SARS-CoV-spike-RLuc. N. vison ACE2 residues at A387ACE2 and R548ACE2 (highlighted in red) are necessary for HKU5 use of N. vison ACE2. No individual substitutions were sufficient for HKU5 use of M. putorius ACE2. Data are means from three independent experiments, each done in triplicate. d Key amino acids are mapped onto the P. abramus ACE2 structure. Positions 387ACE2 and 548ACE2 are necessary for HKU5 to utilize mink ACE2. Statistical analyses were performed using two-tailed unpaired Student’s t-tests and one-way ANOVA. Data are mean ± s.e.m. b *M. erminea-HKU5: p = 0.0412, **N. vison ACE2-HKU5: p = 0.0032, *A. collaris ACE2-SARS-CoV-2: p = 0.0109, *M. nigripes-ACE2: p = 0.0172, **M. meles ACE2-SARS-CoV-2: p = 0.0067, **L. canadensis ACE2-SARS-CoV-2: p = 0.0089, **G. gulo ACE2-SARS-CoV-2: p = 0.0052, **L. lutra ACE2-SARS-CoV-2: p = 0.0016, **M. moschata ACE2-SARS-CoV-2: p = 0.0065, **M. putorius ACE2-SARS-CoV-2: p = 0.0013, **M. lutreola ACE2-SARS-CoV-2: p = 0.0042, ***p = 0.0002. c N. vison ACE2-HKU5: **p = 0.0024, ****p < 0.0001; M. putorius ACE2-HKU5: *p = 0.0207, **p = 0.0061 ***p = 0.0009; N. vison ACE2-SARS-CoV-2: *K309E, p = 0.0245, *K313T, p = 0.0284; *H354R, p = 0.0202; ***p = 0.0002; M. putorius ACE2-SARS-CoV-2: ****p < 0.0001. Source data are provided as a Source Data file.
To gain mechanistic insight into the N. vison ACE2 determinants of HKU5 entry, we leveraged the fact that N. vison and M. putorius have similar ACE2 sequences but dissimilar HKU5 susceptibilities. N. vison (susceptible) and M. putorius (resistant) differ at only nine sites across the ACE2 coding sequence (Fig. 6a). We induced reciprocal single amino acid substitutions at these sites in N. vison and M. putorius ACE2 in an attempt to modify the observed susceptibility of the wild-type ACE2 (Fig. 6a). Substitutions A387E and R548S on N. vison ACE2 reduced VSV-HKU5spike-RLuc entry, while no tested substitutions on M. putorius ACE2 conferred HKU5 susceptibility (Fig. 6c). VSV-SARS-CoV-2spike-RLuc entered all constructs for both species, underscoring the broad ACE2 host range of SARS-CoV-2 (Fig. 6c). Position 387ACE2 represents an N-linked glycan site on P. abramus (N387ACE2) but not on that of N. vison (A387ACE2), M. putorius (E387ACE2), or human (A387ACE2). We identified that the glycan at N387ACE2 forms a hydrogen bond with Y523RBD (Fig. 4c); however, the role of 387ACE2 in mediating differences between mink and ferret ACE2 is independent of this glycan, as neither N. vison nor M putorius possesses an N-linked glycan site at this position.
Our finding that 548ACE2 is required for HKU5 use of N. vison ACE2 was unexpected, given that this residue is remote from the binding interface, consistent with an allosteric effect. In addition, 548ACE2 was not identified as critical for HKU5 infection of P. abramus ACE2, PDF-2180 infection of P. pipistrellus ACE2, or SARS-CoV-2 infection of human or N. vison ACE29,39,40 (Fig. 6). Position 548ACE2 was also not interrogated in previous ACE2 deep mutational scanning studies41,42,43. This position 548 represents the third residue in the NxS/T consensus sequence for a putative N-linked glycan site at 546ACE2 for P. abramus (T548ACE2), human (T548ACE2), and M. putorius (S548ACE2) but not N. vison (R548ACE2) nor M. erminea (R548ACE2) ACE2. Consistent with the distinct mechanism of interaction of HKU5-ACE2 relative to other coronaviruses, residues 337–342ACE2, which mediated PDF-2180 binding and entry, did not have an effect on N. vison ACE2 use by HKU5 (Supplementary Fig. 11). This suggests that differential ACE2 glycosylation plays an important role in mediating mustelid ACE2 use by HKU5 and highlights that HKU5 may interact with N. vison ACE2 in a distinct manner relative to P. abramus ACE2.
Discussion
Identifying viral receptors is critical for pandemic preparedness as receptor usage determines host range, cell and tissue tropism, pathogenesis, and potential for cross-species transmission44. Understanding virus-receptor interactions also informs the development of vaccines, therapeutic monoclonal antibodies, antiviral drugs, and diagnostics. In this study, we employed a diverse panel of receptor orthologs from 57 wild, domestic, and peri-domestic species to demonstrate that HKU5 efficiently utilizes ACE2 from P. abramus (Japanese house bat), N. vison (American mink), and M. erminea (Eurasian ermine, stoat) as a receptor. This was confirmed using both pseudovirus and full-length HKU5 virus. Furthermore, we observed that the related merbecovirus GD2013 also engages P. abramus ACE2. Our findings that HKU5 uses P. abramus ACE2 were simultaneously confirmed by two additional groups45,46. Additionally, a distinct bat HKU5 coronavirus lineage (HKU5-CoV-2) has been reported to use human ACE2 as a functional receptor, further underscoring the potential zoonotic risk of HKU547.
Cryo-EM analysis, coupled with mutagenesis studies, identified residues in P. abramus ACE2 that mediate HKU5spike binding and entry. These residues are distinct from those involved in ACE2 interactions with SARS-CoV-2, PDF-2180, and NeoCoV, consistent with convergent evolution28,29,48. Notably, we observed that the critical ACE2 residues facilitating HKU5 entry in N. vison differ from those in P. abramus, revealing multiple, distinct mechanisms of ACE2 engagement among receptor orthologs. This observation further underscores the evolutionary plasticity of coronavirus spike-ACE2 interactions49.
Interestingly, HKU5 displayed a highly restricted ACE2 host range, successfully using only P. abramus, N. vison, and M. erminea ACE2 among the 57 orthologs tested. In contrast, other ACE2-using coronaviruses, including most sarbecoviruses (e.g., SARS-CoV and SARS-CoV-2), the alphacoronavirus HCoV-NL63, and the merbecoviruses PDF-2180 and NeoCoV, exhibit much broader ACE2 usage across mammalian species18,19,20,21,28,50,51. The narrow ACE2 host range of HKU5 is reflected in the distinct molecular interactions with P. abramus ACE2, differing substantially from those of SARS-CoV-2, PDF-2180, and NeoCoV. Importantly, sequence-based predictions alone would not have identified mink and stoat as potential intermediate hosts, as their ACE2 sequences do not phylogenetically cluster with P. abramus. This highlights the necessity of functional, unbiased experimental approaches for assessing coronavirus host susceptibility.
The narrow receptor usage of lineage 1 HKU5 raises an important evolutionary question as to why the HKU5 receptor host range is so constrained relative to other coronaviruses. One possibility is that HKU5 has evolved a highly specific adaptation to P. abramus ACE2, optimizing replication in its natural reservoir at the cost of reduced cross-species transmissibility. It is also possible that the co-existence of other coronaviruses in P. abramus bats may have induced antibodies that forced HKU5 to employ an alternative mode for binding ACE2. Given the evolutionary trajectory of other coronaviruses, HKU5 may acquire mutations over time that expand its ACE2 host range. Recent studies have shown that a related lineage 1 HKU5 spike can use bovine ACE2 in addition to Pipistrellus ACE245, and that newly discovered lineage 2 HKU5 spikes can use ACE2 receptors from a broader range of mammalian species47. Identifying the potential evolutionary pathways that could enable HKU5 to infect additional species, including humans, remains a critical area for future investigation.
P. abramus bats, commonly known as Japanese house bats, are ~1.5-inch-long insectivorous bats abundant in East Asia that thrive in human-altered landscapes, often roosting in barns, bridges, and other man-made shelters. This places P. abramus bats in frequent contact with humans and farmed animals. While N. vison is native to only North America, they are routinely farmed and invasive in parts of Europe and Asia52,53 enabling interactions between P. abramus bats. Our findings here that HKU5 can use P. abramus and N. vison ACE2 have proven foreboding, as during the preparation of this manuscript, HKU5 was reported in two diseased minks on a Chinese mink farm54. This was the first description of HKU5 outside of Pipistrellus bats and was accurately predicted by our study. HKU5 infection of farmed American mink is of particular concern given multiple events of SARS-CoV-2 spillover from humans to mink and spillback to humans have been described, highlighting mink as an effective intermediate host for coroanviruses52,53,55,56. The bunyavirus SFTSV, which can cause hemorrhagic fever, has also been transmitted from mink to mink farmers, further highlighting the zoonotic risk of farmed mink57.
Bats of the larger Pipistrellus genus, which are likely susceptible to HKU5, are distributed throughout Asia, Europe, and North Africa, thus presenting the opportunity for expanded HKU5 geographical range and further contacts with susceptible mustelids. It is possible that other mustelids beyond American mink and stoat will be susceptible to HKU5 viruses, which may facilitate HKU5 evolution and host range expansion, perhaps even to humans. This underscores the need for increased surveillance of farmed and wild mustelids, particularly in areas with Pipistrellus bats58,59.
Our study identifies ACE2 as a previously unrecognized receptor for the merbecoviruses HKU5 and GD2013 and underscores mustelids, including mink and stoats, as potential intermediate hosts. Group 2c merbecoviruses, including HKU5, have pandemic potential and should be considered in the design of merbecovirus vaccines. Our findings demonstrate that SARS-CoV-2 and MERS-CoV vaccine sera poorly neutralize HKU5, underscoring the need for pan-merbecovirus vaccine strategies. The identification of P. abramus ACE2 as a receptor for HKU5 will facilitate these efforts. These findings have important implications for zoonotic spillover risk and underscore the urgent need for continued surveillance of coronaviruses in both their natural reservoirs and potential bridging hosts.
Methods
Receptor and virus sequences
The sequence information of the 31 mammalian DPP4s and 57 mammalian ACE2s, directly retrieved from GenBank, is described in Supplementary Tables 1 and 2. The whole genome sequences of selected coronaviruses were retrieved the following GenBank accessions: SARS-CoV-2 (MT461669), bat HKU5-LMH03f (NC_009020.1)60, PDF-2180 (NC_034440.1)61, NeoCoV (KC869678.4)62, bat Hp-BetaCoV/Zheijiang2013 (NC_025217.1)63, RoBatCoV HKU9 (NC_009021.1)25, RoBatCoV GCCDC1 (NC_030886.1)64, Erinaceus CoV/2012-174 (NC_039207.1)65, Erinaceus CoV/HKU31strainF6 (MK907286.1)66, BatCoV HKU5 isolate BY140568 (MN611520.1)33, BtPa-BetaCoV/GD2013 (KJ473820.1)34, BatCoV HK25 isolate NL140462 (KX442565.1), BatCoV HKU25 isolate YD131305 (KX442564.1), BtVs-BetaCoV/SC2013 (KJ473821.1)35, BtCoV/133/2005 (Q0Q4F2.1)67, BtTP-BetaCoV/GX2012 (KJ473822.1)34, BatCoV/H. savii/Italy/206645-40/2011 (MG596802.1)36, BatCoV HKU4r isolate GZ131656 (MN611519.1)33, BatCoV HKU4 (NC_009019.1)17, BatCoV HKU4r isolate SM3A (MW218395.1), Bat-SL-CoVZC45 (MG772933.1)68, human NL63 (JX504050.1)69, human 229E (MT797634.1), camel MERS KFU-HKU19Dam (KJ650296.1)70, and human MERS-CoV (NC_019843.3)6. Genes synthesized in this study were produced by GenScript or Twist.
Plasmids
Human codon-optimized sequences of DPP4s, ACE2s, ACE2 chimeras, and ACE2 mutants with a C-terminal HA-tag were commercially synthesized and subcloned into an expression vector (pUC57) through the BsmBI restriction site. The DNA sequences of human codon-optimized coronavirus spikes and HKU5 mutants with a C-terminal Flag tag were commercially synthesized and subcloned into the pCAGGS vector through the EcoRI and NotI restriction sites. Plasmids expressing coronavirus spike NTD-IgG-mFc and RBD-IgG-mFc fusion proteins were commercially synthesized and generated by inserting the putative coding sequence of HKU5NTD (amino acids 22–359), SARS-CoV-2RBD (amino acids 316–527), HKU5RBD (amino acids 376–586), MERS-CoVRBD (amino acids 368–588), and PDF-2180RBD (amino acids 372–584) into a modified pFuse-mIgG1-Fc2 vector (InvivoGen) through the EcoRI and NheI restriction sites. An IL2 signal sequence was included for protein secretion.
Cells lines
HEK-293T and BHK-21 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) with 10% heat-inactivated fetal bovine serum (FBS) and 1% Penicillin/Streptomycin (Pen/Strep). Vero-CCL-81 cells were cultured in DMEM containing 5% FBS, 1% Pen/Strep, and 1% NEAA. For the Vero-CCL-81 ACE2 stable cell lines, 10 µg/ml of puromycin (Gibco) was added. Cells were cultured at 37 °C in 5% CO2 with regular passaging every 2–3 days. Expi293F cells used for protein production were cultured in Expi293 Expression Medium (Thermo Fisher Scientific), according to the manufacturer’s instructions. Expi293F cells were maintained at 37 °C at 8% CO2 in an orbital shaker (125 rpm) and split at 0.3–0.5 × 106 viable cells/ml every 3–4 days. BHK-21 cells were stably transduced with a lentiviral vector encoding P. abramus ACE2 for pseudovirus experiments with HKU5 mutants.
Production of VSV-ΔG pseudoviruses
VSV-based pseudotyped viruses were produced as described previously71,72,73,74. pCAGGS containing different coronavirus spikes were synthesized by GenScript. Briefly, HEK-293T cells were transfected with the pCAGGS vector expressing the different spikes using polyethylenimine (PEI), and after 24 h, were infected with a replication-deficient VSV vector containing expression cassettes for RLuc or eGFP in lieu of the VSV-G open reading frame. After an infection period of 1 h at 37 °C, the inoculum was removed, and cells were washed with 1X PBS prior to the addition of media supplemented with anti-VSV-G clone I4 (Kerafast) to neutralize residual input virus75. Pseudotyped particles were harvested at 24 and 48 hpi, clarified from cellular debris by centrifugation, and stored at −80 °C prior to use.
Production and purification of spike RBD and NTD proteins
RBD- and NTD-mFc fusion proteins (SARS-CoV-2RBD (amino acids 316–527), HKU5RBD (amino acids 376–586), MERS-CoVRBD (amino acids 368–588), PDF-2180RBD (amino acids 372–584), and HKU5NTD (amino acids 22–359)) were expressed in Expi293F cells by transfecting the corresponding plasmids using the Expi293 expression system kit (Thermo Fisher Scientific) following the manufacturer’s instructions. Briefly, Expi293F cells were subcultured in 125-ml shaker flasks. Once the cell density reached 2.5 × 106 viable cells/ml in a 45 ml culture, 45 µg of plasmids were transfected using the Expifectamine 293 transfection kit. After 20 h post-transfection (hpt), Enhancer 1 and 2 from the kit were added, and transfected cells were incubated for an additional 4 days in an orbital shaker at 125 rpm at 37 °C with humidified 8% CO2. The supernatant was subsequently harvested by centrifugation at 2000 rpm for 2 min. The protein-containing supernatant was transferred to a sterile 50-ml Falcon tube and mixed with pre-washed Protein A agarose resin in 1X PBS (GoldBio). The mixture was incubated at room temperature with gentle rocking for 2 h. The protein-bound agarose resin was purified through a gravity flow column (BioRad) and washed with 1X PBS. Before storing at −80 °C, RBD proteins were eluted with 100 mM glycine (pH 3.2) and 5 M NaCl and neutralized with 100 mM Tris-HCl (pH 8). The purified RBD and NTD proteins were visualized on SDS-PAGE gel by Coomassie Blue staining using the BioRad ChemiDoc Imaging System.
Pseudotyped virus entry assay
1.5 × 104 HEK-293T or BHK-21 cells were seeded in 100 µl total volume in each well of a black-walled, clear-bottom 96-well plate. The following day, cells were transfected with 0.1 µl of either DPP4- or ACE2-expressing plasmids (1 µg/µl stock), using 0.3 µl PEI in 9 µl of Opti-MEM (Gibco) per well. The following day, spike-expressing VSV-pseudotyped viruses were added at a 1:10 final concentration volume/volume (v/v) in DMEM with 2% FBS and incubated for 24 h. After 24 h, cells were lysed using 5X Renilla lysis buffer (Promega) diluted using 1X PBS. 25 µl of 1X lysis buffer was added to each well prior to incubating for 20 min with shaking at room temperature. RLuc substrate (Promega) was mixed with Renilla assay buffer (10 µL of RLuc substrate per ml of assay buffer) and 25 µl was added to the lysed cells. The cells were incubated for 10 min with shaking in the dark. Each infection condition was performed in at least duplicate in each independent experiment. Luciferase activity was measured using a microplate reader (BioTek Synergy) and analyzed with Gen5 software. Relative light units (RLU) were plotted and normalized in GraphPad Prism (v10.2)
MLV-ACE2 entry into spike-expressing cells
Murine leukemia virus luciferase reporter virions bearing P. abramus ACE2 (MLV-P. abramusACE2-Gluc) were tested for entry into spike-expressing HEK-293T cells. MLV-ACE2 virions were produced by transfecting 0.4 µg of MLV-GagPol plasmid (pMDoldGag-Pol, Richard Mulligan, Harvard Medical School, Boston, MA, USA), 0.4 µg of ACE2 plasmid or empty vector, and 0.2 µg of an intron-regulated MLV-based Gaussia Luciferase (GLuc) reporter gene (Dave Derse, NCI, Frederick, MD, USA) into a 24-well plate of HEK-293T cells using PEI76. MLV-P. abramusACE2-GLuc containing supernatants were purified with a 0.45 µm syringe filter 2 days post-transfection (dpt) and used fresh. HEK-293T cells were 0.2 µg spike plasmids or empty vector using PEI into a 48-well plate 1 day prior to the assay. Purified MLV-P. abramusACE2-Gluc supernatants (100 µl) were added to spike-expressing target cells in quadruplicate and incubated at 37 °C. After two days, 100 µl of assay supernatant was mixed with 30 µl Gluc substrate (Thermo Fisher Scientific) containing 100 mM NaI in a white 96-well plate and subsequently read for luminescence with a Promega GloMax Explorer plate reader.
Spike RBD-Fc and NTD-Fc-protein binding assay
HEK-293T cells transiently transfected with P. abramus ACE2 were trypsinized and incubated with 50 µg/ml of recombinant RBD- or NTD-mFc protein diluted in DMEM with 2% FBS. The cell and Fc-protein mix was incubated for 2 h in a roller at 4 °C (cold room). Unbound spike RBD- or NTD-mFc proteins were removed, and cells were washed with cold 1X PBS. For flow cytometry analysis, spike RBD- or NTD-mFc bound cells were incubated with a live/dead stain (Zombie Violet, Biolegend) in 1X PBS for 10 min at 4 °C and fixed in 4% paraformaldehyde (PFA) for 10 min at room temperature. Following cell fixation, 100 µl (1:500) of Alexa Fluor 488-conjugated goat anti-mouse IgG (Invitrogen) was added to the spike RBD-mFc or NTD-fc bound cells and incubated for 30 min at 4 °C. The cells were then washed and resuspended with cold FACS buffer (1X PBS, 2% FBS, 1 mM EDTA). Dead cells, as indicated by SSC/FSC, were excluded by gating. The spike RBD-mFc or NTD-Fc bound cells were assessed by the fluorescence intensity compared to HEK-293T control cells without the receptor expression. Flow cytometry data were collected on a Beckman Coulter CytoFlex S and analyzed using FlowJo (v.10).
Infection with the authentic full-length HKU5 virus
P. abramus and N. vison ACE2-expressing cells were infected with full-length-HKU5 infectious clone virus (FL-HKU5, MOI: 0.01) for 72 h26,77,78. CPE was monitored, and images were taken using an inverted optical microscope (Olympus, IX73). For the multistep growth curve analysis, cells were infected at an MOI of 0.01 and incubated at 37 °C with 5% CO2 for 1 h. After removing the inoculum, the monolayer was washed with 1X PBS. Complete growth media was added back to each well, and samples of the infected culture supernatant were collected at 0, 4, 24, 48, and 72 hpi. Samples were stored at −80 °C for viral titration, which was performed by plaque assay on Vero81 cells expressing P. abramus ACE2 or N. vison ACE2 (Vero81-PaACE2 and -NvACE2, respectively). 10-fold serial dilutions of virus in 1X PBS were prepared and used to inoculate Vero81-PaACE2 and -NvACE2 cells, as described above. After 1 h of viral absorption, the monolayers were overlaid with 0.8% agarose in Eagle minimum essential medium (MEM). Plaques were visualized and manually quantified at 4 days post-infection (dpi) using neutral red stain. All work with full-length HKU5 virus were performed under biosafety level 3 conditions, at negative pressure and with personal powered-air purifying respirators. All full-length HKU5 experiments were approved by the UNC Institutional Biosafety Committee.
Western blot
HEK-293T cells transfected with ACE2 or coronavirus spikes were washed with 1X PBS and lysed with NP-40 lysis buffer (Thermo Fisher Scientific) on ice for 10 min. Cell lysates were clarified by centrifugation at 10,000 × g at 4 °C for 5 min. The lysate was mixed with 4X Laemmli sample buffer (BioRad) at a 1:4 (v/v) ratio of buffer to lysate and incubated at 95 °C for 5 min. For HEK-293T cells transfected with DPP4 orthologs, cells were lysed using RIPA buffer on ice for 10 min, mixed with 4X Laemmli sample buffer (BioRad) at a 1:4 (v/v) ratio of buffer to lysate, and loaded without boiling. To detect the Flag tag on pseudotyped viruses, the virus-containing supernatant was mixed with 4X Laemmli sample buffer. After SDS-PAGE and PVDF membrane transfer, the blots were blocked with 5% milk in 1X TBS containing 0.1% Tween-20 (TBST) at room temperature for 1 h. Primary antibodies against Flag (Sigma-Aldrich), HA (Biolegend), and GAPDH (Biolegend) were added at a 1:1000 dilution in TBST with 5% milk and incubated on a shaker at 4 °C overnight. After three washes with TBST, the blots were incubated with horseradish peroxidase (HRP) conjugated secondary antibody, goat anti-mouse IgG (H+L) (Jackson Immuno Research, 1:5000 dilution) in 1X TBST for 1 h on a shaker at room temperature. The blots were washed three times with 1X TBST and visualized using the BioRad ChemiDoc Imaging system.
A full-length molecular clone for HKU5 (FL-HKU5) has been previously reported26,27. To detect the nucleocapsid in FL-HKU5 virus infection, HEK-293T cells were transiently transfected with ACE2. Following 24 hpi, the cells were exposed to FL-HKU5 (MOI: 1.0), and the infected cells were collected at 24 hpi. These cells were then lysed using 8 M urea, combined with Laemmli sample buffer (BioRad), and heated at 60 °C for 10 min. After SDS-PAGE and PVDF membrane transfer, the blots were blocked with 5% milk in 1X PBS at room temperature for 1 h. Primary antibodies targeting spike (1:2000, gift from David Veesler, University of Washington, clone 76E1)79, HA (1:2000, CST catalog number: 3724), nucleocapsid (1:2000, Sino Biological 40068-RP02), and GAPDH (1:5000, CST catalog number: 2118) were diluted in PBS with 5% BSA and 0.1% Tween 20 and left to incubate on a shaker at 4 °C overnight. Following three washes with 1X PBS, blots were exposed to HRP conjugated secondary antibodies goat anti-human (1:5000, SeraCare catalog number: 5220-0330) and goat anti-rabbit (1:5000, CST catalog number: 7074S) in 1X PBS with 5% BSA and 0.1% Tween 20 for 1 h on a shaker at room temperature. Finally, the blots were washed three times with 1X PBS and visualized using the iBright 1500 Imaging system (Invitrogen).
Immunofluorescence assay
For pseudovirus experiments, 2 × 105 HEK-293T or BHK-21 cells were plated per well in 8-well chamber slides and transfected with 0.5 µl ACE2 (1 µg/µl stock), infected with pseudotyped-Spike-eGFP virus at 1:10 final concentration v/v in DMEM with 2% FBS, and incubated for 24 h. Cells were subsequently fixed with 4% PFA at room temperature for 10 min. The nuclei were stained with Hoechst 33342 (1:10,000 dilution in 1X PBS, Thermo Fisher Scientific). Images were captured with a fluorescence microscope (Zeiss Axio Imager) and analyzed using the Zeiss Zen Pro microscopy software.
For FL-HKU5 experiments, Vero81-PaACE2 cells were seeded on coverslips in 6-well plates and subsequently infected with FL-HKU5 virus (MOI: 0.01). At 24 hpi, cells were fixed with 4% PFA for 15 min at room temperature. Cells were then blocked and permeabilized in 10% normal goat serum and 0.3% Triton-X 100 for 1 h at room temperature, followed by incubation with rabbit anti-MERS polyclonal sera (1:2000; Sino Biological; 40068-RP02) diluted in 5% normal goat sera for 16 h at 4 °C. Cells were then washed three times with 1X PBS and incubated with goat anti-rabbit conjugated to Alexa488 (1:3000) diluted in 5% normal goat sera at room temperature for 2 h. Nuclei were stained with Hoechst (1:10,000), and the cells were washed three times prior to mounting on slides with ProLong Gold antifade reagent (Invitrogen). HKU5 antigen-positive cells were observed under a fluorescence microscope (Keyence, BZ-X810), and images were captured for further analysis.
Bioinformatic and computational analyses
Protein sequence alignment was performed using the MUSCLE algorithm by MEGA-X software (v.10.17)80,81. For phylogenetic analysis, amino acid sequences of receptors or spikes were first aligned using MUSCLE, and phylogenetic trees were generated using the maximum-likelihood method in MEGA-X (1000 bootstraps). The model and parameters utilized for the phylogenetic analysis were implemented based on the recommended optimal protein model using the MEGA-X software. SimPlot (v.3.5.1) was used to analyze nucleotide similarities with a sliding window size of 200 nucleotides and a step size of 20 nucleotides using a gap-stripped alignment and Kimura (2-parameter) distance model.
Cryo-EM grid preparation, data collection, and structure determination
The complex of P. abramus ACE2-ACT66266.1 and HKU5-RBD-10lnQQAVI was assembled by mixing ACE2 and HKU5-RBD at a molar ratio of 1:1.2 to a final concentration of the complex of 3.7 mg/ml, followed by incubating for 1 h at room temperature. Before vitrification, 6 mM 3-[(3-cholamidopropyl) dimethylammonio]-2-hydroxy-1-propanesulfonate (CHAPSO) was added to the sample. A sample volume of 2.8 µl was applied onto a freshly glow-discharged Quantifoil R 2/2 gold grid and plunge-frozen using a Thermo Scientific Vitrobot Mark IV plunger with the following parameters: chamber humidity of 95%, chamber temperature of 4 °C, blotting force of −5, and blotting time between 1 and 2.5 s.
The data were acquired using SerialEM82 on an FEI Titan Krios G1 electron microscope equipped with a Direct Electron Apollo direct electron detector. A total of 10,702 movies were collected at a nominal magnification of 47,000× with a pixel size of 0.5 Å in super-resolution mode. Movie frames were aligned with MotionCor283, and CTF parameters were estimated using the cryoSPARC 4.484 module patch CTF estimation. 9529 micrographs were selected for downstream processing after curating in cryoSPARC based on full-frame motion, CTF fit resolution, and relative ice thickness. Particles were picked from a subset of 500 micrographs using crYOLO85. After several rounds of 2D classification, selected 2D templates and corresponding particles were used for template picking and Topaz training86. The particles obtained by template-based and Topaz picking were merged, followed by the elimination of duplicates. Ab initio reconstruction in cryoSPARC was used to generate initial models, followed by multiple rounds of heterogeneous refinement to remove low-quality particles. The resulting 242,675 particles representing the complex dimer were subjected to non-uniform refinement in cryoSPARC with C2 symmetry imposed to produce the final map of ACE2-HKU5RBD. The reported resolution of 4.2 Å was determined based on the “gold standard” criterion at the FSC curve threshold of 0.14387. Further steps included symmetry expansion followed by 3D classification and refinement, as well as focused refinement of one complex monomer within the dimer. These measures did not improve the cryo-EM map. We also performed non-uniform refinement for the monomer of the complex (184,339 particles), which produced a map with a resolution of 4.8 Å (Supplementary Fig. 4). While this resolution was too low for molecular interpretation, docking of the monomer from the structure of the complex dimer demonstrated an exact fit (correlation: 0.89), indicating that no global structural changes were induced by dimerization.
To build the atomic model of the complex, an initial model of P. abramus ACE2-ACT66266.1 was generated using SWISS-MODEL88 based on the structure from PDB entry 8WBY, while the initial model of HKU5-RBD-10lnQQAVI was obtained using ColabFold89. These models were docked into the cryo-EM map using UCSF Chimera90 and refined in Coot91 using the sharpened map generated by cryoSPARC and map post-processed using EMReady92. The improved model was refined using real-space refinement in Phenix93 against the original cryoSPARC map. Molprobity94 was used to validate the final model. The refinement statistics are summarized in the Supplementary Table 3.
Production and purification of soluble ACE2 ectodomain proteins and biotinylated RBD proteins
N-terminus HRV3C cleavable single-chain Fc-tagged P. abramus ACE2 ectodomain proteins, and N-terminus HRV3C cleavable single-chain Fc-tagged and C-terminus AVI-tagged HKU5 RBD and SARS-CoV-2 RBD proteins were expressed in Expi293F cells by transfecting the corresponding plasmids using the Turbo293 transfection reagent (Speed BioSystems). Briefly, pre-mixed 1 mg plasmid in 20 ml Opti-MEM (Thermo Fisher Scientific) and 3 ml of Turbo293 transfection reagent in 20 ml Opti-MEM were added to 0.8 liter of Expi293F cells at a cell density of 2.5 × 106 viable cells/ml. The transfected cells were incubated for 6 days in an orbital shaker at 125 rpm at 37 °C in a humidified 9% CO2 incubator before the supernatant was harvested by centrifugation and filtration. Subsequently, the supernatant was incubated with 5 ml of PBS-equilibrated protein A resin for 1–2 h. The ACE2-bound resin was then collected, washed with 1X PBS, and cleaved overnight at 4 °C with 200 μg of HRV3C. The RBD-bound resin was collected, washed with 1X PBS, and subjected to an overnight incubation in a 3-ml BirA biotin-protein ligase mixture (Avidity) and 200 μg of HRV3C at 4 °C. The liberated, untagged, and soluble ACE2 proteins and biotinylated RBD proteins were applied to a Superdex 200 16/600 gel filtration column equilibrated with PBS the next day. Peak fractions corresponding to the target proteins were pooled for subsequent analysis.
Binding analysis using biolayer interferometry (BLI)
Binding affinities were assessed using BLI assays conducted on the Octet HTK instrument. Briefly, biotinylated recombinant RBD proteins at a concentration of 3 µg/ml were immobilized on streptavidin biosensors for 300 s. Following this, the biosensors were immersed in kinetic buffer for 180 s to remove any unbound biotinylated RBD or NTD proteins. Subsequently, the biosensors were immersed in kinetic buffer containing soluble P. abramus ACE2 ectodomain proteins at concentrations ranging from 0 to 500 nm for 120 s to capture association kinetics and then placed in kinetic buffer for 300 s to capture dissociation kinetics. A kinetic buffer lacking ACE2 was used as a reference for background determination. The affinities were then calculated using Octet Data Analysis software (v.12.2.0.20), employing curve-fitting kinetic analysis or steady-state analysis with global fitting. Kd, app values were reported due to the utilization of dimeric ACE2.
Neutralization assay
For pseudotyped virus neutralization, BHK-21 cells were transfected with plasmids containing the full-length human ACE2, human DPP4, and P. abramus ACE2-using PEI as described above. The following day, two-fold serial dilutions of sera and antibody were prepared in DMEM. SARS-CoV-2 (Moderna) vaccine sera were pooled from healthy humans (BEI Cat# NRH-21747). MERS-CoV-27 monoclonal antibody was described previously36,37. MERS-CoV-S2P serum was harvested (Day 60) from mice primed (Day 0) and boosted (Day 30) with MERS-CoV-S2P (full-length spike) and pooled from 10 mice. Subsequently, 5 µl of the corresponding pseudovirus was mixed with 20 µl of DMEM with 2% FBS and 25 µl of each serum/antibody dilution, and the mixtures were incubated for 45 min at 37 °C. Transfected BHK-21 cells were trypsinized and seeded into 96-well plates at a density of 50,000 cells per well with the pseudovirus-sera/antibody mixtures. After 24 h, the cells were lysed, and RLuc activity was measured as described above.
Statistical analysis
Infection and binding assay experiments were repeated at least 2–3 independent times, with 2–4 technical replicates per experiment. Data are presented as mean ± s.e.m., as specified in the figure legends. IC₅₀ values and 95% confidence intervals for the neutralization assay were calculated using Python, with curve fitting implemented via scipy.optimize.curve_fit. Statistical analyses were conducted using GraphPad Prism (v10.2) using unpaired two-tailed Student’s tests or One-way ANOVA, as stated in the figure legends. P < 0.05 was considered significant; *P < 0.05, **P < 0.01, *** P < 0.001, ****P < 0.0001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The structural data generated in this study are available in the Protein Data Bank (PDB) under accession number 9MV0 and in the Electron Microscopy Data Bank (EMDB) with an entry ID EMD-48650. Information about the key resources used in this study is listed in Supplementary Table 5. The data generated in this study are provided in the Source Data File. Source data are provided with this paper.
References
Cohen, L. E., Fagre, A. C., Chen, B., Carlson, C. J. & Becker, D. J. Coronavirus sampling and surveillance in bats from 1996-2019: a systematic review and meta-analysis. Nat. Microbiol. 8, 1176–1186 (2023).
Keusch, G. T. et al. Pandemic origins and a One Health approach to preparedness and prevention: solutions based on SARS-CoV-2 and other RNA viruses. Proc. Natl. Acad. Sci. USA 119, e2202871119 (2022).
Zhou, H. et al. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell 184, 4380–4391.e14 (2021).
Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020).
Ksiazek, T. G. et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1953–1966 (2003).
Zaki, A. M., van Boheemen, S., Bestebroer, T. M., Osterhaus, A. D. & Fouchier, R. A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820 (2012).
Hui, D. S. et al. Middle East respiratory syndrome coronavirus: risk factors and determinants of primary, household, and nosocomial transmission. Lancet Infect. Dis. 18, e217–e227 (2018).
Letko, M., Seifert, S. N., Olival, K. J., Plowright, R. K. & Munster, V. J. Bat-borne virus diversity, spillover and emergence. Nat. Rev. Microbiol. 18, 461–471 (2020).
Xiong, Q. et al. ACE2-using merbecoviruses: Further evidence of convergent evolution of ACE2 recognition by NeoCoV and other MERS-CoV related viruses. Cell Insight 3, 100145 (2024).
Li, F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 3, 237–261 (2016).
Lu, G., Wang, Q. & Gao, G. F. Bat-to-human: spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 23, 468–478 (2015).
Graziano, V. R., Wei, J. & Wilen, C. B. Norovirus attachment and entry. Viruses 11, 495 (2019).
Raj, V. S. et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495, 251–254 (2013).
Lau, S. K. P. et al. Isolation of MERS-related coronavirus from lesser bamboo bats that uses DPP4 and infects human-DPP4-transgenic mice. Nat. Commun. 12, 216 (2021).
Wang, Q. et al. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe 16, 328–337 (2014).
Yang, Y. et al. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc. Natl. Acad. Sci. USA 111, 12516–12521 (2014).
Luo, C. M. et al. Discovery of novel bat coronaviruses in South China that use the same receptor as middle east respiratory syndrome coronavirus. J. Virol. 92, e00116–e00118 (2018).
Hofmann, H. et al. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. USA 102, 7988–7993 (2005).
Li, W. et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454 (2003).
Letko, M., Marzi, A. & Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol 5, 562–569 (2020).
Hoffmann, M. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8 (2020).
Simmons, G., Zmora, P., Gierer, S., Heurich, A. & Pöhlmann, S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antivir. Res. 100, 605–614 (2013).
Fehr, A. R. & Perlman, S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 1282, 1–23 (2015).
Woo, P. C. et al. Molecular diversity of coronaviruses in bats. Virology 351, 180–187 (2006).
Woo, P. C. et al. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J. Virol. 81, 1574–1585 (2007).
Agnihothram, S. et al. A mouse model for Betacoronavirus subgroup 2c using a bat coronavirus strain HKU5 variant. mBio 5, e00047–14 (2014).
Menachery, V. D. et al. Trypsin treatment unlocks barrier for zoonotic bat coronavirus infection. J. Virol. 94, e01774–19 (2020).
Xiong, Q. et al. Close relatives of MERS-CoV in bats use ACE2 as their functional receptors. Nature 612, 748–757 (2022).
Ma, C. B. et al. Multiple independent acquisitions of ACE2 usage in MERS-related coronaviruses. Cell 188, 1693–1710.e18 (2025).
Yan, H. et al. ACE2 receptor usage reveals variation in susceptibility to SARS-CoV and SARS-CoV-2 infection among bat species. Nat. Ecol. Evol. 5, 600–608 (2021).
Tolentino, J. E., Lytras, S., Ito, J. & Sato, K. Recombination analysis on the receptor switching event of MERS-CoV and its close relatives: implications for the emergence of MERS-CoV. Virol. J. 21, 84 (2024).
Lau, S. K. P. et al. Receptor usage of a novel bat lineage c betacoronavirus reveals evolution of middle east respiratory syndrome-related coronavirus spike proteins for human dipeptidyl peptidase 4 binding. J. Infect. Dis. 218, 197–207 (2018).
Li, B. et al. Discovery of bat coronaviruses through surveillance and probe capture-based next-generation sequencing. mSphere 5, e00807–e00819 (2020).
Wu, Z. et al. ORF8-related genetic evidence for Chinese horseshoe bats as the source of human severe acute respiratory syndrome coronavirus. J. Infect. Dis. 213, 579–583 (2016).
Yang, L. et al. MERS-related betacoronavirus in Vespertilio superans bats, China. Emerg. Infect. Dis. 20, 1260–1262 (2014).
Moreno, A. et al. Detection and full genome characterization of two beta CoV viruses related to Middle East respiratory syndrome from bats in Italy. Virol. J. 14, 239 (2017).
Du, L. et al. A conformation-dependent neutralizing monoclonal antibody specifically targeting receptor-binding domain in Middle East respiratory syndrome coronavirus spike protein. J. Virol. 88, 7045–7053 (2014).
Jiang, L. et al. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci. Transl. Med. 6, 234ra59 (2014).
Lan, J. et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020).
Wang, Q. et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181, 894–904.e9 (2020).
Chan, K. K. et al. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science 369, 1261–1265 (2020).
Starr, T. N. et al. Deep mutational scans for ACE2 binding, RBD expression, and antibody escape in the SARS-CoV-2 Omicron BA.1 and BA.2 receptor-binding domains. PLoS Pathog. 18, e1010951 (2022).
Starr, T. N. et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 182, 1295–1310.e20 (2020).
Dufloo, J., Andreu-Moreno, I., Moreno-García, J., Valero-Rello, A. & Sanjuán, R. Receptor-binding proteins from animal viruses are broadly compatible with human cell entry factors. Nat. Microbiol 10, 405–419 (2025).
Park, Y. J. et al. Molecular basis of convergent evolution of ACE2 receptor utilization among HKU5 coronaviruses. Cell 188, 1711–1728.e21 (2025).
Catanzaro, N. J. et al. ACE2 from Pipistrellus abramus bats is a receptor for coronaviruses. Nat. Commun. 16, 4932 (2025).
Chen, J. et al. Bat-infecting merbecovirus HKU5-CoV lineage 2 can use human ACE2 as a cell entry receptor. Cell 188, 1729–1742.e16 (2025).
Yan, R. et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448 (2020).
Starr, T. N. et al. ACE2 binding is an ancestral and evolvable trait of sarbecoviruses. Nature 603, 913–918 (2022).
Ma, C. et al. Broad host tropism of ACE2-using MERS-related coronaviruses and determinants restricting viral recognition. Cell Discov. 9, 57 (2023).
Peña-Hernández, M. A. et al. SARS-CoV-2-related bat viruses evade human intrinsic immunity but lack efficient transmission capacity. Nat. Microbiol 9, 2038–2050 (2024).
Wang, Y., Yang, M., Zhou, H., Quan, C. & Kang, H. Outbreak of natural severe fever with thrombocytopenia syndrome virus infection in farmed minks, China. Emerg. Infect. Dis. 30, 1299–1301 (2024).
Fenollar, F. et al. Mink, SARS-CoV-2, and the human-animal interface. Front. Microbiol. 12, 663815 (2021).
Zhao, J. et al. Farmed fur animals harbour viruses with zoonotic spillover potential. Nature 634, 228–233 (2024).
Oreshkova, N. et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Euro Surveill. 25, 2001005 (2020).
Koopmans, M. SARS-CoV-2 and the human-animal interface: outbreaks on mink farms. Lancet Infect. Dis. 21, 18–19 (2021).
Li, J. et al. Direct transmission of severe fever with thrombocytopenia syndrome virus from farm-raised fur animals to workers in Weihai, China. Virol. J. 21, 113 (2024).
Guan, Y. et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302, 276–278 (2003).
Worobey, M. et al. The Huanan Seafood Wholesale Market in Wuhan was the early epicenter of the COVID-19 pandemic. Science 377, 951–959 (2022).
Lau, S. K. et al. Genetic characterization of Betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications for the origin of the novel Middle East respiratory syndrome coronavirus. J. Virol. 87, 8638–8650 (2013).
Anthony, S. J. et al. Further evidence for bats as the evolutionary source of Middle East respiratory syndrome coronavirus. mBio 8, e00373-17 (2017).
Ithete, N. L. et al. Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerg. Infect. Dis. 19, 1697–1699 (2013).
Wu, Z. et al. Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J. 10, 609–620 (2016).
Obameso, J. O. et al. The persistent prevalence and evolution of cross-family recombinant coronavirus GCCDC1 among a bat population: a two-year follow-up. Sci. China Life Sci. 60, 1357–1363 (2017).
Corman, V. M. et al. Characterization of a novel betacoronavirus related to Middle East respiratory syndrome coronavirus in European hedgehogs. J. Virol. 88, 717–724 (2014).
Lau, S. K. P. et al. Identification of a novel betacoronavirus. Viruses 11, 980 (2019).
Tang, X. C. et al. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 80, 7481–7490 (2006).
Hu, D. et al. Genomic characterization and infectivity of a novel SARS-like coronavirus in Chinese bats. Emerg. Microbes Infect. 7, 154 (2018).
Lednicky, J. A. et al. Isolation and genetic characterization of human coronavirus NL63 in primary human renal proximal tubular epithelial cells obtained from a commercial supplier, and confirmation of its replication in two different types of human primary kidney cells. Virol. J. 10, 213 (2013).
Hemida, M. G. et al. MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerg. Infect. Dis. 20, 1231–1234 (2014).
Wei, J. et al. The KDM6A-KMT2D-p300 axis regulates susceptibility to diverse coronaviruses by mediating viral receptor expression. PLoS Pathog. 19, e1011351 (2023).
Wei, J. et al. A chemically-targetable transcription factor-chromatin remodeler interaction underlies SARS-CoV-2 susceptibility. Nat. Genet. 55, 471–483 (2023).
Wei, J. et al. Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell 184, 76–91.e13 (2021).
Strine, M. S. et al. DYRK1A promotes viral entry of highly pathogenic human coronaviruses in a kinase-independent manner. PLoS Biol. 21, e3002097 (2023).
Lyles, D. S., Puddington, L. & McCreedy, B. J. Vesicular stomatitis virus M protein in the nuclei of infected cells. J. Virol. 62, 4387–4392 (1988).
Jin, J., Sherer, N. M., Heidecker, G., Derse, D. & Mothes, W. Assembly of the murine leukemia virus is directed towards sites of cell-cell contact. PLoS Biol. 7, e1000163 (2009).
Becker, M. M. et al. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc. Natl. Acad. Sci. USA 105, 19944–19949 (2008).
Donaldson, E. F. et al. Systematic assembly of a full-length infectious clone of human coronavirus NL63. J. Virol. 82, 11948–11957 (2008).
Sun, X. et al. Neutralization mechanism of a human antibody with pan-coronavirus reactivity including SARS-CoV-2. Nat. Microbiol. 7, 1063–1074 (2022).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Wagner, T. et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 (2019).
Bepler, T. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019).
Henderson, R. et al. Outcome of the first electron microscopy validation task force meeting. Structure 20, 205–214 (2012).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
He, J., Li, T. & Huang, S. Y. Improvement of cryo-EM maps by simultaneous local and non-local deep learning. Nat. Commun. 14, 3217 (2023).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D. Struct. Biol. 75, 861–877 (2019).
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Acknowledgements
This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Prime Contract No. 75N91019D00024, Task Order No. 75N91023F00016. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This work was funded in part by Burroughs Wellcome Fund (C.B.W.), NIH R01 AI148467 (C.B.W.), NIH R21 AI173821 (S.L. and C.B.W.), NIH PO1 AI167966 and CEIRS Contract 75N93021C00014 (R.S.B.), R01 AI183155 (S.L.), NIH grant R01 AI163395 (W.M.), Hanna H. Gray Fellowship from the Howard Hughes Medical Institute (D.R.M.), NIH grant F31 AI176650 (M.W.G.), and Metropolitan AntiViral Drug Accelerator (C.B.W.). B.L.M. was supported by NIH T32 HL007974. AKS was supported by the Fulbright Junior Research Award Scholarship (PL/2023/21/JR). A.K. is in part supported by Open Philanthropy and the Life Sciences Research Foundation. M.W.G. is a recipient of the Gruber Science Fellowship and was supported by NIH T32 AI055403. We thank Peter Cresswell, Matthew Spencer, and Leo Phan for helpful discussions and technical guidance.
Author information
Authors and Affiliations
Contributions
Conceptualization: C.B.W., D.C.D., T.Z., M.M.A.; methodology: C.B.W., M.M.A., E.L.K., T.Z.; investigation: M.M.A., E.L.K., N.L., N.J.C., I.T.T., Z.Z., M.W.G., B.Y., A.S., D.W., A.S.K., A.L.S., M.P.H., S.Z., R.A., R.K., B.L.M., J.W., G.A.R., J.H., J.S., A.R., A.I., R.K.J., S.L., D.R.M., W.M., P.D.U., J.G.D., A.B.S., R.S.B., L.S., Y.T.; funding acquisition: C.B.W., D.C.D., T.Z.; writing—original draft, reviewing and editing: C.B.W., M.M.A., E.L.K., D.C.D., T.Z.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Kenneth Matreyek and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Madel Alfajaro, M., Keeler, E.L., Li, N. et al. HKU5 bat merbecoviruses engage bat and mink ACE2 as entry receptors. Nat Commun 16, 6822 (2025). https://doi.org/10.1038/s41467-025-61583-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-61583-7