Abstract
Embryonic stem cells (ESCs) display a distinctive resistance against various viruses, irrespective of any interferon response. Nevertheless, the underlying mechanism of this resistance remains unclear. In this study, we identify vesicle-associated membrane protein 5 (VAMP5) as a potent cell-autonomous defense factor against coronaviruses, including SARS-CoV-2, with high expression levels observed in ESCs and mesoderm. VAMP5 not only exhibits functional conservation in restricting the replication of SARS-CoV-2 and its variants, as well as other highly pathogenic coronaviruses, but also shows efficacy in combating the replication of viruses from other families. Mechanistic investigations reveal that VAMP5 localizes to double membrane vesicles (DMVs) and impedes viral replication by relying on its vesicle-side C-terminal domain to interact with the viral non-structural protein 8 (NSP8), thus inhibiting the synthesis of negative-strand RNA. Our research demonstrates that VAMP5 in ESCs disrupts the protected environment of DMVs, which is essential for viral genome replication, and interacts with RNA replication complexes to defend against viral infection. This provides a novel strategy for developing broad-spectrum antiviral treatments.
Similar content being viewed by others
Introduction
Viral infectious disease outbreaks remain a significant threat to human health in the cross-linked world both historically and into the foreseeable future. Coronaviruses known as crown shape and primarily respiratory transmission caused several pandemics of coronavirus disease. For instance, Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) emerged in Guangdong China and spread worldwide in 20021,2. Also worthy of note is that the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) emerged in Saudi Arabia in 2012 caused respiratory disease in human with a mortality rate of more than 35%3,4. More recently, coronavirus disease 2019 (COVID-19) outbreak, caused by SARS-CoV-2, has had a severe impact on global health5,6,7.
Coronaviruses belong to the Nidovirales order, Coronaviridae family and Orthocoronavirinae subfamily. They are enveloped, positive-sense RNA virus possessing genome sizes ranging from 26 to 32 kilobases (kb) in length8. These RNA viruses exhibit error-prone replication due to their RNA-dependent RNA polymerase (RdRp) conferring rapid mutations to emerge new variants. These mutations help the viruses evolve into more transmittable and adaptable variants, which arouse great concern and are expected to spread worldwide. Notably, SARS-CoV-2 variants of concern (VOC), including Alpha, Beta, Delta, Gamma, Kappa, and Omicron, have challenged the vaccines and neutralizing antibodies, impacting the ongoing COVID-19 pandemic on a global scale9,10,11,12,13.
Due to the potential outbreaks and immune escape mutations, there is a growing effort to investigate broad-spectrum antiviral treatments. These efforts emphasize on understanding the interaction between hosts and viruses to identify specific targets for intervention. One strategy is to develop drugs that target host factors required for viral complete replication cycles14,15. This host factors targeting strategy also impairs the normal function of these factors as a result of side effects. Another strategy to respond to broad antiviral treatment is based on the cell-autonomous immunity to find the pan-viral host restriction factors16. This cell-autonomous immune response, such as interferon (IFN) mediated response, upregulates the expression of IFN stimulated genes (ISGs) to defense against invading pathogens16,17,18. Interestingly, embryonic stem cells (ESCs) do not produce IFN in response to viral infection, but intrinsically express many restriction factors, such as IFN induced transmembrane protein 1(IFITM1). These factors act as central orchestrators of cell-autonomous pan-viral defense to protect stem cells19,20,21. In addition, although ESCs express coronavirus entry dependent factors such as Angiotensin Converting Enzyme-2 (ACE2), transmembrane serine protease 2 (TMPRSS2), furin, and tyrosine-protein kinase receptor UFO (AXL), it is endoderm (END) and ectoderm (ECT) rather than ESCs and mesoderm (MES) that are permissive for SARS-CoV-2 infection22,23,24,25. This observation led us to hypothesize that certain host key restrictive factors that are highly expressed in ESCs and mesodermal cells, may restrict SARS-CoV-2 infection. This intrinsic restrictive activity in stem cells has been shown to confer viral resistance19. Characterizing their activity will shed light on how cell-autonomous antiviral defense to protect ESCs from pathogens and provide the basis for the design and development of broad-spectrum antiviral treatments. Therefore, we set out to identify restriction factors in ESCs against coronavirus infection.
Here, we show vesicle-associated membrane protein 5 (VAMP5) expressed highly in ESCs and mesodermal cells, and present detailed studies on its role in blocking SARS-CoV-2 and other pathogenic coronaviruses, as well as other viral families. This finding shows the different mechanism of stem cells resistance to viral infection and also aid in developing entirely new theory to treat viral infection.
Results
Identification of VAMP5 as a host restriction factor against SARS-CoV-2 infection in ESCs
To identify host restrictive factors conferring viral resistance, we performed RNA sequencing (RNAseq) analysis in ESCs, END, MES and ECT. Consistent with other RNAseq analysis for the expression of host key restrictive factors in different stages of stem cells differentiation, we found that most restrictive factors express highly in ESCs but exhibit different expression patterns during differentiation into END, MES and ECT (Fig. 1a)19. Therefore, the variations in restrictive factor expression in ESCs likely play important roles in their viral resistance capabilities. Next, our focus turned to identify which key factors conferred this effect. The different expression of host genes response to viral replication by RNAseq among END, MES and ECT were analyzed to uncover anti-SARS-CoV-2 restriction factors highly expressed in MES but lowly expressed in END and ECT (Fig. 1b). Interestingly, we then identified 16 genes that were enriched in both MES and ESCs but lowly expressed in END and ECT (Fig. 1c). Upon comparing the expression of these factors before and after SARS-CoV-2 infection, we noted that these factors also highly expressed in ESCs and MES even without the influence of viral infection (data not show), which contrasts with findings from the previously published study where certain host factors were upregulated during SARS-CoV-2 infection26. After analyzing the details, we found that cells with lower expression of ESCs related markers (Nanog and OCT4) expression in their study were permissive for SARS-CoV-2 infection, suggesting that these cells may not have been authentic ESCs, which could potentially explain the discrepancies. These results went on to show that these factors highly intrinsic expression in ESCs may contribute to the restriction of SARS-CoV-2. Next, we used siRNA technology to knock-down these candidates in ESCs which have been less studied in association with viral replication, with CD74 as a positive control (Fig. 1d)27. Notably, VAMP5 is mostly involved in SARS-CoV-2 infection (Fig. 1d). Little is known about the cellular function of VAMP5 as well as its antiviral role in SARS-CoV-2 and even other coronavirus infections. VAMP5, as a vesicle-associated membrane protein, has previously been implicated in vesicle trafficking and docking, fusion and translocation of glucose transport (GLUT4)28,29,30. However, conflicting reports suggest that VAMPs, except for VAMP5, are essential for mediating fusion with plasma membrane target soluble N-ethylmaleimide-sensitive factor attachment protein receptors (t-SNAREs)31, indicating that the precise role of VAMP5 in cells remains unclear. In addition, VAMP5 has been associated with IFN-γ stimulation and responded to influenza virus (IAV) infection32,33. Therefore, based on its relationship with negative strand RNA virus, we aim to investigate its antiviral role in SARS-CoV-2 wild-type (WT) and the variants infections.
a Representative heatmap of ISGs profiles of endoderm (END, n = 3 biological replicates), mesoderm (MES, n = 3 biological replicates) and ectoderm (ECT, n = 3 biological replicates). b Scatterplot of ISGs-wise log2 fold change from this study analysis. c Venn diagram of the top 16 host factors significantly enriched in hESCs and MES but lowly in END and ECT from our own RNAseq data. Selected: selecting 24 ISGs enriched in MES compared to END and ECT. d Screen the influence of top 16 ISGs from (b) by siRNA in hESCs on SARS-CoV-2 replication (MOI = 0.05, 24 hpi). e Representative western blot image of VAMP5 expression in hESCs, END, MES and ECT. The results are representative of three independent experiments. f–k VAMP5 WT and KO (n = 2 cell lines) hESCs infected with SARS-CoV-2 (MOI = 0.05, 24hpi) WT (f) and variants of Alpha (g), Delta (h), Gamma (i), Kappa (j) and Omicron (k) with viral infection in cells at MOI of 1 for 24h. Virus strain is also indicated in the figure. ESC, embryonic stem cells; MES, mesoderm; END, endoderm; ECT, ectoderm; VAMP5, vesicle-associated membrane protein 5; KO, knock-out; OE, overexpression; WT, wild-type; ACE2, Angiotensin-converting enzyme 2. Data are represented as mean ± SEM, n = 3 biological replicates in each group. Student’s two tailed t test was used. *p < 0.05, **p < 0.01, ***p < 0.001.
We next performed western blot to confirm the high expression of VAMP5 in ESCs and MES. As expected, VAMP5 exhibits high expresses in ESCs and MES, while its expression is lower in END and ECT (Fig. 1e). Interestingly, we observed that ACE2 is only significantly higher in END compared to ESCs, MES and ECT. Based on SARS-CoV-2 infection in END and ECT, we speculated that relative lower expression of ACE2 is not the key factor responsible for inhibiting SARS-CoV-2 infection in ESCs and MES.
To investigate whether VAMP5 in ESCs restricts SARS-CoV-2 infection, we used CRISPR-Cas9 technology to generate VAMP5 knock-out (KO) hESC-H9 cell lines to increase the SARS-CoV-2 ability of infection. Verification of VAMP5 KO in these clones was done through genome sequencing and western blot (Supplementary Fig. 1a). We then challenged two independent KO clones to SARS-CoV-2 WT, N501Y (Alpha), Delta, GYKT (Gamma), B.1.617.1(Kappa) and Omicron from the artificial transcription and replication-competent SARS-CoV-2 virus-like-particles (trVLP), which express a reporter gene (GFP) replacing viral nucleocapsid gene (N) and also could complete viral life cycle in the cells when expressing N protein34. Consistent with our siRNA screening results, increased growth of both SARS-CoV-2 WT and its variants was observed in the VAMP5 KO clones (Fig. 1f–k). We next expressed VAMP5 individually in the VAMP5 KO cells to restore its ability to restrict virus infection. As expected, VAMP5 overexpression restored its ability to inhibit SARS-CoV-2 infection (Fig. 1f–k). To ensure that SARS-CoV-2 infection in VAMP5 KO ESCs, we also utilized flow cytometry to detect viral infection in OCT4 positive ESCs. As shown in Supplementary Fig. 1b, the viral infection rate was significantly higher in VAMP5 KO cells with OCT4 expression, indicating that SARS-CoV-2 can replicate in ESCs with VAMP5 KO. Although appearing similar cell viability, VAMP5 KO clones grow relatively slower compared to WT cells (Supplementary Fig. 1c, d). To avoid this influence, we also utilized siRNA to knock down VAMP5 expression in ESCs and challenged the cells to SARS-CoV-2 WT and its variants. As expected, knocking-down VAMP5 expression also enhanced the replication of SARS-CoV-2 WT and its variants (Supplementary Fig. 1e-j).
VAMP5 KO has no impair on stemness of ESCs
While studying SARS-CoV-2 infection in VAMP5 KO ESCs, we noticed an decrease in cell growth compared to WT cells. We hypothesized that this effect may be attributed to VAMP5’s role not only in protecting stem cells from viral infection, but also in facilitating cells growth, stemness and subsequent differentiation. This hypothesis is also supported by recent findings indicating a crucial role for VAMP5 in the differentiation of urinary and respiratory system in mice, without which can make mice died around birth35. To validate this hypothesis, we detected the expression of ESCs related markers in VAMP5 KO ESCs. Indeed, the results, as shown in Fig. 2a, and Supplementary Fig. 2a, revealed the similar rates of SSEA4 and OCT4 expression cells in VAMP5 KO ESCs to WT ESCs, although WT ESCs express relatively higher SSEA4 and OCT4 compared to VAMP5 KO ESCs. Similarly, as mentioned above (Supplementary Fig. 1b), flow cytometry detection revealed that the rates of OCT4 positive cells in VAMP5 ESCs KO cells are similar to ESCs WT cells. This suggests that VAMP5 might has no influence on ESCs stemness. While this does not explain a role for VAMP5 in ESCs functions, further evidence is required to establish it necessity in stem cells differentiation.
a–d Representative western blot images of VAMP5 KO potentially impair the related markers expression in hESCs (a) and also after differentiated into mesoderm (MES) (b), endoderm (END), (c) and ectoderm (ECT) (d). Data are represented as mean ± SEM, n = 3 biological replicates in each group. Student’s two tailed t test was used. No adjustments for multiple comparisons were made, as only two groups were compared. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. e PCA of the transcriptomic comparison of VAMP5 WT and KO in hESCs and followed differentiated three germ layers, n = 3 in each cell type. f Representative heatmap of hESCs and three germ layers related markers comparison of VAMP5 WT and KO in hESCs and followed differentiated three germ layers. g Immunofluorescence staining for lineage differentiation from VAMP5 WT and KO hESCs to mesoderm derived smooth muscles cells (markers of α-SMA and Transgelin), endoderm derived alveolar type II cells (markers of EpCAM and SP-B), and ectoderm derived neuron cells (β3 tubulin and MAP2). The results are representative of three independent experiments. Scale bars, 50 μm. KO, knock-out; WT, wild-type.
We then tested whether VAMP5 play a role in ESCs differentiating into END, MES and ECT. We differentiated the ESCs with VAMP5 WT and KO independently into three germ layers as previously reported, then independently quantified mRNA encoding END related SOX17 and FoxA2, MES related brachyury and CXCR4, and ECT related nestin and PAX6 by western blot and immunofluorescent staining19. As shown in Fig. 2b–d, and Supplementary Fig. 2b-d, we found that both VAMP5 KO and WT ESCs could be differentiated into three germ layers, although the markers expressions were relatively different. For example, END related markers (SOX17 and FOXA2) and ECT related markers (nestin and PAX6) are relatively higher in VAMP5 KO cells compared to WT cells. In contrast, MES related brachyury and CXCR4 levels were relatively impaired in VAMP5 KO derived cells compared to WT derived cells. These findings indicate that VAMP5 might have no influence on the differentiation of ESCs into three germ layers. To further confirm this hypothesis, we performed RNAseq to assess gene expression profile in ESC WT and VAMP5 KO cells, as well as in their differentiated three multipotent germ layers of END, MES and ECT. We found a similar gene pattern in each stem cell type between VAMP5 WT and KO cells, although with slight differences in expression levels (Fig.2e). However, when we examined specific markers for the three germ layers, we found that ECT derived from VAMP5 KO shows higher NES and PAX6 genes compared to WT cells. Interestingly, END with VAMP5 KO shows increased expression of MES related markers like TBXT (brachyury), MESP1, and ECT related markers like NCAM1 and NES (nestin). Likewise, ESC with VAMP5 KO shows higher expression of MES related marker of MESP1, and ECT related markers of NCAM1 and NES (Fig. 2f). Thus, we hypothesized that VAMP5 might impair three germ layers differentiated into mature tissue cells. We then differentiated these three germ layers into hepatic stellate cells and alveolar type II cells (AT2 cells) (from END)36, Neuron cells (from ECT) and smooth muscle cells (from MED). As shown in Fig. 2g, differentiation into AT2 cells, neurons and smooth muscle cells was not impaired by VAMP5 KO. However, hepatic stellate cell differentiation was disrupted, as evidenced by the lack of desmin expression (Supplementary Fig. 2e), indicating a potential role for VAMP5 involved in hepatic stellate cell differentiation. Together, our data indicate that VAMP5 is not required for maintaining hESCs stemness or general trilineage differentiation capacity, although it might be essential for the differentiation into specific lineages, such as hepatic stellate cells.
VAMP5 is a pan-viruses restriction factor for ESCs in vitro and in vivo
To expand our understanding of VAMP5’s impact, we extended our studies to include other coronaviruses. These coronaviruses we tested include the MERS-CoV, human coronavirus (HCoV)-OC43 and HCoV-NL63, along with Porcine Epidemic Diarrhea virus (PEDV) representing the acute diarrhea and/or vomiting, dehydration and high mortality in neonatal piglets, and mouse hepatitis virus-A59 (MHV-A59). We found that all these viruses replicated more efficiently in VAMP5 KO ESCs than WT cells (Fig. 3a–e). These data demonstrate that VAMP5 serves as a pan-coronavirus restriction factor.
a–e RT-qPCR analysis of VAMP5 WT and KO clones (n = 2 cell lines) in hESCs influence on viral replication of coronavirus, such as MERS-CoV (a), OC43 (MOI = 1, 24 hpi) (b), NL63 (MOI = 1, 24 hpi) (c), PEDV (MOI = 0.1, 24 hpi) (d) and MHV (MOI = 0.1, 24 hpi) (e). f–j RT-qPCR analysis of viral RNA levels from ZIKV (MOI = 0.1, 24 hpi) (f), VSV (MOI = 0.1, 24 hpi) (g), DENV2 (MOI = 0.1, 24 hpi) (h), IAV (MOI = 0.1, 24 hpi) (i) and EV71 (MOI = 0.1, 24 hpi) (j) infecting VAMP5 WT and KO clones (n = 2 cell lines) in hESCs. k, l Luciferase activity analysis of viral replication from AAV2 (MOI = 0.1, 24 hpi) and VSV-G-pseudotyped HIV-1 lentivirus (LV) (MOI = 0.1, 24 hpi) with luciferase as reporter for viral replication in VAMP5 WT and KO clones (n = 2 cell lines) hESCs. m Representative images of VSV-GFP expression in VAMP5 siNC and siRNA induced knocked down hESCs (MOI = 0.1, 24 hpi) (Scale bars, 100 μm) and quantification of GFP fluorescence signal density. n Graphic representation of VAMP5 inhibiting viral infection in vivo. Nude mice were xenografted with ESCs-H9 cells exposed to PEDV and DENV2 (2 × 105 pfu for one mouse), followed by infection of above viruses at set intervals. Created in BioRender. Wang, K. (https://BioRender.com/w02jplv). o Equal amounts of total RNA were obtained from the ESCs clusters to detect PEDV and DENV2 levels by RT-qPCR. WT, wildtype; KO, knock-out; NC, negative control; LV, lentivirus. Data are represented as mean ± SEM, n = 3 biological replicates in each group. For in vivo experiment (o), each group, n = 5 biological replicates as indicated. Student’s two tailed t test was used. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Given that VAMP5 broadly restricts members of the Coronaviridae, we speculated that VAMP5 might also shield stem cells against a wide range of distantly related viruses or unrelated viruses. Based on the VAMP5 KO ESCs, we next also performed similar experiments for a diverse panel of other unrelated viruses, including positive- and negative-strand RNA viruses, retroviruses and DNA viruses, such as Zika virus (ZIKV), Vesicular Stomatitis Virus (VSV), Dengue virus 2 (DENV2), Enterovirus 71 (EV71), IAV, Adeno-associated Virus 2 (AAV2) and VSV-G-pseudotyped HIV-1 lentivirus (LV). Interestingly, we observed enhanced infection of ZIKV, VSV, DENV2, IAV, AAV2 and LV in VAMP5 KO cells compared to WT cells (Fig. 3f–l and Supplementary Fig. 3a, b). We next expressed VAMP5 in the KO cells to restore its ability to inhibit AAV2 and LV infection. As expected, VAMP5 overexpression in KO clones strengthen the ability to restrict viral infection (Fig. 3k, l). However, EV71 infection is effective in both VAMP5 WT and KO cells, suggesting that ESCs are permissive for this virus’ infection, and VAMP5 has no influence on EV71 infection (Fig. 3j). In addition, similar results were observed in VAMP5 knocked-down ESCs using siRNA (Fig. 3m and Supplementary Fig. 3c–g). Hence, VAMP5 exhibits broad antiviral activity against coronavirus as well as other unrelated viruses to protect ESCs.
Next, we tested the in vivo effect of VAMP5 in ESCs on viral infection using nude mice xenografted with ESCs subcutaneously19,37. Considering the spontaneous differentiation of ESCs in mice, we injected these viruses at the inoculation sites of ESCs 48-h post-transplantation (Fig. 3n). After 48-h post-infections of viruses, we harvested the cell clusters and detected viral RNA levels (Fig. 3n). The results demonstrated that VAMP5 KO cells showed similar expression level of stem cell markers consistent with wild-type cells when transplanted in mice for 48 h (Supplementary Fig. 3h). Similar to the in vitro results described above, we found that VAMP5 KO in ESCs enhances DENV2 and PEDV infection (Fig. 3o), suggesting that VAMP5 retained inhibition effects on viral replication after 48-96 hours exposure to in vivo environment.
Collectively, these results indicate that VAMP5 inhibits diverse viruses in vitro and in vivo.
VAMP5 overexpression in tissue cells persists its effect on viral restriction
Given that VAMP5 was found to be highly expressed in ESCs and protects these cells from coronavirus infection, we were curious about its activity in tissue cells regarding SARS-CoV-2 and other viruses. Actually, multiple human epithelial cells serve as major sites of VAMP5 expression, including the respiratory tract related cells and colon tissues (Supplementary Fig. 4a, b). To test it, we generated VAMP5 KO clones in Caco-2-N cells (Supplementary Fig. 5a)34. Overexpression of VAMP5 in these KO cells, doesn’t impair cellular viability and stress (Supplementary Fig. 5b–d), restored their ability to restrict viral infection, including SARS-CoV-2 and other pathogenic coronaviruses (Fig. 4a–n, and Supplementary Fig. 5e). Aside from the coronavirus, we also tested if VAMP5 inhibits other virus in tissue cells. Consistent with earlier findings, VAMP5 overexpression in Vero E6 and HEK293T cells inhibits infection by AAV2, LV, ZIKV, DENV2, VSV, and IAV (Fig. 4o–t). Overexpression of VAMP5 markedly decreased the infectivity of pseudoviruses in a dose-dependent manner (Supplementary Fig. 5f, g). We also investigated VAMP5’s impact on hepatitis B virus (HBV), hepatitis C virus (HCV) and hepatitis D virus (HDV) infection in HepG2-NTCP/CD81/Mir122 and Huh7-NTCP cells38. However, we didn’t find any influence of VAMP5 on HBV, HCV and HDV infection (Supplementary Fig. 5h-j). These data suggest that VAMP5 acts as a broad restriction host factor restricting viral infection in tissue cells except for certain viruses.
a Representative images of the infection of SARS-CoV-2 S pseudotyped VSV-ΔG (MOI = 0.1, 24 hpi) with GFP as reporter in VAMP5 KO Caco-2-N cells with VAMP5 and CD74 overexprssion via transiently transfection system. Scale bars, 100 μm. The results are representative of three independent experiments. b Representative images of SARS-CoV-2 trVLP replication with GFP expression in VAMP5 KO Caco-2-N cells with VAMP5 overexpression via transiently transfection system (MOI = 0.1, 24 hpi). Scale bars, 50 μm. The results are representative of three independent experiments. c Representative images of western blot analysis of SARS-CoV-2 spike protein (S) expression when different concentration of SARS-CoV-2 trVLP infecting VAMP5 KO Caco-2-N cells for 24 h with VAMP5 and negative control plasmids transfection. The results are representative of three independent experiments. d TCID50 analysis of extracellular SARS-CoV-2 titers at different time points after viral infection in Caco-2-N cells with or without VAMP5 knockdown (MOI = 0.05, 24 hpi). e–i RT-qPCR analysis of viral RNA levels from SARS-CoV-2 different variants (MOI = 0.05, 24 hpi) like Alpha (e), Delta (f), Gamma (g), Kappa (h) and Omicron (i) infecting in VAMP5 KO Caco-2-N cells with VAMP5 or negative control (Ctrl) transfection. j-n RT-qPCR analysis of viral RNA levels of other coronaviruses like MERS-CoV (j) in Huh7 cells, OC43 (MOI = 1, 24 hpi) (k) and NL63 (MOI = 1, 24 hpi) (l) in Caco-2-N cells, MHV (MOI = 0.1, 24 hpi) (m) in 17CI-1 cells and PEDV (MOI = 0.1, 24 hpi) (n) in the Vero E6 cells with VAMP5 or negative control (Ctrl) transfection. o, p Luciferase activity analysis of AAV2 (MOI = 0.1, 24 hpi) (o) and VSV-G-pseudotyped HIV-1 lentivirus (LV) (MOI = 0.1, 24 hpi) (p) replication in HEK293T cells with VAMP5 or negative control (Ctrl) transfection. q–t RT-qPCR analysis of viral RNA levels from ZIKV (q), DENV2 (r), VSV (s) and IAV (t) infecting in Vero E6 or HEK293T cells with VAMP5 or negative control (Ctrl) transfection (MOI = 0.1, 24 hpi). Data are represented as mean ± SEM, n = 3 biological replicates in each group. Student’s two tailed t test was used. *p < 0.05, **p < 0.01, ***p < 0.001. MOI, multiplicity of infection; Ctrl, control; NC, negative control.
The C-terminal transmembrane domain of VAMP5 is responsible for SARS-CoV-2 restriction
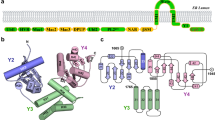
Given that it is difficult to reconstruct ESCs to study VAMP5’s effects on coronavirus39, and tissue cells exhibit a similar phenotype of VAMP5 restriction on SARS-CoV-2 infection, we next performed the mechanic studies in tissue cells like Caco-2-N cells. VAMP5 is anchored to the membrane by the carboxy (C) -terminal transmembrane domain with the proline-rich amino (N)– terminal domain laying in the cytoplasm. To determine which domains of VAMP5 are crucial for restricting SARS-CoV-2 infection, we generated the clones lacking N-terminal or C-terminal domain. A diagram of these VAMP5 constructs is shown in Fig. 5a. We found that only VAMP5 with the full-length sequence and intact C-terminal transmembrane domain decreases viral infection (Fig. 5b). This data suggests that the C-terminal transmembrane domain of VAMP5 may interfere viral infection.
a Graphic representation of VAMP5 constructs with different region deletion. VAMP5-N contains SNARE domain, and VAMP5-C contains TM domain and CTD. b Representative images of western blot analysis of SARS-CoV-2 spike protein (S) expression impaired by transfecting VAMP5 constructs with different region deletion (MOI = 0.05, 24 hpi). The results are representative of three independent experiments. c Graphic representation of VAMP5 sequence comparison among Homo sapiens, Gorilla gorilla gorilla, Macaca mulatta, Mus musculus and Rattus norvegicus. d–i RT-qPCR analysis of viral RNA levels from SARS-CoV-2 WT (d) and variants like Alpha (e), Gamma (f), Kappa (g), Delta (h) and Omicron (i) infection (MOI = 0.05, 24 hpi) in VAMP5 KO Caco-2-N cells rescued with VAMP5 overexpression from different species. j Table shows the frequency of three SNPs in VAMP5 in several human populations. k RT-qPCR analysis of viral RNA levels of SARS-CoV-2 infecting VAMP5 KO Caco-2-N cells rescued with VAMP5 WT and mutants (MOI = 0.05, 24 hpi). l Western blot analysis of SARS-CoV-2-S expression impaired by VAMP5 WT and mutants in VAMP5 KO Caco-2-N cells. The results are representative of three independent experiments. SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptor; TM, transmembrane domain; CTD, C-terminal domain; WT, wild-type; KO, knock-out; Ctrl, control; Hom, homo sapiens; Gor, Gorilla; Mac, macaca mulatta; Mus, mus musculus; Rat, rattus norvegicus. Data are represented as mean ± SEM, n = 3 biological replicates in each group (d–k). Student’s two tailed t test was used. *p < 0.05, **p < 0.01.
C-terminal transmembrane domain of VAMP5 is functionally conserved across mammalian species
It has been noted that VAMP5 functions by anchoring to the membrane to mediate the vesicle trafficking, docking and fusion, implying its importance in membrane fusion during the vesicle trafficking and release of vesicle contents29. This function, combined with its broad antiviral activity, led us to hypothesize that VAMP5 is evolutionarily conserved for viral resistance. After analyzing the amino acid sequences, we observed that Homo Sapiens, Gorilla gorilla gorilla, and Macaca Mulatta share the similar VAMP5 sequence in the C-terminal domain, with differences in only two residue sites (I110T and N116D). However, the sequences of the C-terminal domain in Mus Musculus and Rattus Norvegicus are significant differences, with 14 residues deleted at the end of C-terminal domain compared to Homo Sapiens (Fig. 5c). To determine if these divergent VAMP5 orthologs keep their antiviral activity against SARS-CoV-2 infection, we expressed these VAMP5 orthologs in VAMP5 KO Caco-2-N cells and challenged them with SARS-CoV-2 WT and its variants. Surprisingly, we found that all these VAMP5 orthologs inhibit viral infection, even those from Mus Musculus and Rattus Norvegicus, suggesting that evolutionarily divergent VAMP5 orthologs maintain their antiviral effects (Fig. 5d-i).
Naturally occurring VAMP5 SNPs exhibit no influence on its antiviral activity
As we all known, the interaction between viruses and human can lead to genetic changes in human as they adapt to fight off viral infection. During the long term of strong evolutionary history, naturally single nucleotide polymorphisms (SNPs) can be enriched in human population. As expected, upon analyzing VAMP5 sequences from the database, we discovered several SNPs with extremely low prevalence in human (data not shown). Interestingly, we found only three SNPs are the most prevalent variants. These SNPs are SNP (rs756685424) at amino acid position 82 that results in a Leu82Phe substitution, SNP (rs553284722) resulting in an Ile83Phe substitution and SNP (rs1359803401) as a result of a Gln95Lys substitution. The SNP of Leu82Phe substitution is present at 0.004% in European, Ile83Phe substitution is present at 0.3% in America and Gln95Lys substitution is present at 0.01% in Africa (Fig. 5j). We engineered these amino acid substitutions into a VAMP5 construct independently and expressed them in VAMP5 KO Caco-2-N cells. After challenging these cells with SARS-CoV-2, we found that at 48 h post infection, all the variants exhibit similar restriction activity to WT (Fig. 5k, l). These data suggest that naturally existing variants in VAMP5 may be not related with viral infection pressure and is conserved with their ability to resist viral infection.
Vesicle trafficking function of VAMP5 is not involved in restricting SARS-CoV-2 infection
Previous imaging and proteomics analysis have shown that VAMP5 typically localizes between the cells surface membrane and vesicle membrane, prompting us to hypothesize if VAMP5 interacts with viral spike protein to inhibit viral attachment28. We then conducted Co-Immunoprecipitation experiments and found that VAMP5 does not co-immunoprecipitate with SARS-CoV-2 spike protein, indicating that VAMP5 does not interact with viral spike protein to inhibit viral infection (Fig. 6a and Supplementary Fig. 6a). To further prove this, we next tested if VAMP5 disrupts the attachment of SARS-CoV-2 on the cell surface. We designed the attachment experiment (Fig. 6b)40 and discovered that VAMP5 has no effect on the attachment of SARS-CoV-2 on Caco-2-N cells surface, whereas lactoferrin significantly inhibit SARS-CoV-2 to attach the cell (Fig. 6c)41. Subsequently, we next tested if VAMP5 impairs the entry step of SARS-CoV-2 infection. After performed the special designed viral entry experiment (Fig. 6b), we observed no influence of VAMP5 on viral entry (Fig. 6d). To further confirm this, we used a split-NanoLuc-reporter-based assay to test viral entry and endosome fusion. The reporter displays when self-complementation of luciferase activity upon reaching the cytosol (Fig. 6e). We transduced the LgBiT fragment in Huh7.5 cells and challenged them with a SARS-CoV-2-spike-expressing pseudovirus harboring the HiBiT fragment42. As expected, reconstituted LgBiT-HiBiT bioluminescence is not decreased by VAMP5 overexpression. In contrast, Camostat treatment and CD74 overexpression reduces markedly this response, as previously reported to block the endosomal entry pathway of SARS-CoV-2 (Fig. 6f)43. These data suggest that VAMP5 inhibits SARS-CoV-2 infection beyond the endosomal entry pathway.
a Representative images of analyzing Flag tagged VAMP5 co-immunoprecipitates with SARS-CoV-2 spike protein which recognized by anti-S antibody. The results are representative of three independent experiments. b Graphic representation of designing viral attachment and entry assay in Caco-2-N cells individually with or without VAMP5 overexpression and positive control. c Caco-2-N cells were transfected with VAMP5 or negative control for 24 h, and then infected with SARS-CoV-2 (MOI:0.05). RT-qPCR analysis of viral RNA levels reflecting the influence of VAMP5 on SARS-CoV-2 attachment to Caco-2-N cells, lactoferrin (1 mg/mL) as a positive control. The levels of VAMP5 were detected by western blot (bottom). d Caco-2-N cells were transfected with VAMP5 or negative control for 24 h, and then infected with SARS-CoV-2 (MOI:0.05). RT-qPCR analysis of viral RNA levels reflecting the influence of VAMP5 on SARS-CoV-2 entry into Caco-2-N cells, camostat (10 μM) as a positive control. The levels of VAMP5 were detected by western blot (bottom). e Graphic representation analyzing if virus-cell fusion impaired by VAMP5 in Huh7.5 cells. f Huh7.5 cells were transfected with VAMP5, CD74 or negative control for 24 h, and then infected with pseudoviruses (MOI = 0.1). Nano-luciferase activity was performed to analyze virus-cell fusion in Huh7.5 cells, CD74 and camostat (100 μM) as positive controls. g, h RT-qPCR analysis of VAMP5 influence on SARS-CoV-2 replication (g) and secretion (h) in Caco-2-N cells with or without VAMP5 knockdown by siRNA (MOI = 0.05, 24 hpi). i, Graphic representation of designing specialized primer for detecting SARS-CoV-2 negative strand RNA which reflecting the RNA dependent RNA polymerase (RdRp) activity. j, k RT-qPCR analysis of SARS-CoV-2 negative strand RNA levels in viral infecting Caco-2-N cells with VAMP5 overexpression (j) or knockdown by siRNA (k) (MOI = 0.05, 24 hpi). S, spike protein; NC, negative control; Ctrl, control. Data are represented as mean ± SEM, n = 3 biological replicates in each group. Student’s two tailed t test was used. No adjustments for multiple comparisons were made, as only two groups were compared. *p < 0.05, **p < 0.01, ****p < 0.0001.
After that, we tested if VAMP5 restricts viral secretion. Initially, we knocked down VAMP5 expression in Caco-2-N cells and then challenged them with SARS-CoV-2. As expected, detecting viral RNA revealed that viral infection is enhanced upon knocking down VAMP5 expression during the time course of infection (Fig. 6g). Next, we compared the ratio of extracellular and intracellular viral RNA levels to reflect viral secretion and found that VAMP5 has no impact on viral secretion (Fig. 6h). To prove it, we also designed a new experiment to detect viral secretion by immunofluorescent assay, with the Ankx as the positive control as it prevents protein stop in the endoplasmic reticulum44. We overexpressed VAMP5 and Ankx independently in Caco-2-N cells and then challenged the cells with SARS-CoV-2 and VSV, respectively. We next incubated the cells at 4 °C for 1 h followed by incubation at 37 °C for 4 h. As expected, after 4 h incubation at 37 °C, both VSV glycoprotein and SARS-CoV-2 S protein localized in everywhere of the membrane in VAMP5-expressing cells, whereas the viral proteins are localized in the endoplasmic reticulum vesicle just near the nucleus in the Ankx-expressing cells (Supplementary Fig. 6b, c). These findings demonstrate that VAMP5 also have no influence on the step of viral secretion.
VAMP5 inhibits coronavirus replication to synthesize negative strand RNA
As shown in Fig. 4, we observed that VAMP5 not only decreases SARS-CoV-2 RNA, as detected with primers designed at the 5’ terminal of viral genome, but also decreases viral S and N protein expression (reflected by GFP in trVLP system). Combing this information with the lack of influence of VAMP5 on viral attachment, entry and secretion, we hypothesized that VAMP5 restricts viral replication process. Viral genomic replication initiates with the synthesis of full-length negative-sense genomic copies, which serve as templates for generating new positive-sense genomic RNA and can thus be used as a marker to assess viral replication activity45. To better test if VAMP5 inhibits viral replication, we designed a new pair of primers to detect viral negative-strand RNA, which is transcribed by viral RdRp and its level can reflect the RdRp activity of SARS-CoV-2. The diagram of negative-strand RNA is shown in Fig. 6i. Surprisingly, viral negative-strand RNA levels are inhibited significantly by overexpressing VAMP5 in Caco-2-N cells compared to control group, but enhanced by knocking down the expression of VAMP5 (Fig. 6j, k).
VAMP5 targets SARS-CoV-2 non-structural protein 8 (NSP8) of RdRp
Next, we assessed whether the cellular localization of VAMP5 changed upon viral infection. The HAtag-VAMP5 expression was utilized to monitor VAMP5 cellular localization prior to and during SARS-CoV-2 infection. Previous imaging studies have shown that VAMP5 localized to the cell surface and some vesicular structures in Golgi and peripheral regions28,46. In our study, we observed that HAtag-VAMP5 expressing in Caco-2-N cells and ESCs are consistent with the cell surface and some vesicular structures localization (Fig. 4b, and Supplementary Fig. 6d). Interestingly, we didn’t see any viral replication in the Caco-2-N cells overexpressing HAtag-VAMP5 (Fig. 4b). HAtag-VAMP5 in these cells localizes is consistent with the cells without any viral infection, indicating that VAMP5 effectively inhibits viral replication to the extent that co-localizations between virus and VAMP5 is difficult to detect within a cell. Given that VAMP5 restricts the negative-strand RNA formation as mentioned above, we hypothesized that VAMP5 may interact with RdRp of SARS-CoV-2 to restrict viral replication. The structure of SARS-CoV-2 RdRp consists of NSP12 and its cofactors of NSP7 and NSP8. Therefore, we engineered the Flag-NSP7, Flag-NSP8 and Flag-NSP12 constructs with the tagFlag fused to the N terminus of these proteins. We then co-transfected these plasmids independently with HA-VAMP5 in HEK293T cells. We observed a co-localization of HA-VAMP5 with viral nonstructural protein of Flag-NSP8, whereas Flag-NSP7 and Flag-NSP12 show less co-localization possibility with HA-VAMP5 (Fig. 7a).
a Representative immunofluorescent images of analyzing VAMP5 co-localization with NSP7, NSP8 and NSP12 in HEK293T cells. Yellow color in the merged column shows the co-localization of VAMP5 with viral antigens. DAPI stains cell nuclei. Scale bars, 10 μm. The results are representative of three independent experiments. b, c Representative western blot images of analyzing HA tagged VAMP5 immunoprecipitates Flag tagged NSP8 (b) and Flag tagged NSP8 immunoprecipitates GFP tagged VAMP5 WT and its special domain (c). The results are representative of three independent experiments. d Representative image of predicting VAMP5 key residues may interacts with NSP8 by the protein structure interaction prediction tool of ZDOCK. e Graphic image of constructing VAMP5 variants which the mutation engineered as prediction from (d). f Representative western blot images of analyzing the potential mutants impair VAMP5 immunoprecipitates NSP8. The results are representative of three independent experiments. g Representative images of analyzing the influence of VAMP5 on viral RNA binding to NSP8 when SARS-CoV-2 trVLP infecting the VAMP5 expressing Caco-2-N cells (MOI = 0.05, 24 hpi). Viral RNA was reverse transcribed into DNA, amplified by PCR and then displayed by agarose gel electrophoresis. NSP8 was detected by western blot. The results are representative of three independent experiments. h RT-qPCR analysis of VAMP5 WT and L82A mutation potentially impair viral negative strand RNA synthesis in Caco-2-N cells when SARS-CoV-2 trVLP infecting the VAMP5 expressing Caco-2-N cells (MOI = 0.05, 24 hpi). n = 3 biological replicates. i Representative western blot images of analyzing VAMP5 WT and L82A mutation potentially impair SARS-CoV-2 spike protein (S) expression in Caco-2-N cells when SARS-CoV-2 trVLP infecting the VAMP5 expressing Caco-2-N cells (MOI = 0.05, 24 hpi). The results are representative of three independent experiments. j, k Representative image of analyzing VAMP5 potentially impair RNA dependent RNA polymerase (RdRp) activity for RNA synthesis in vitro (j) and quantification of RNA signal (k). n = 2 biological replicates. WT, wild-type; Ctrl, control; N, N-terminus; C, C-terminus; RIP, RNA immunoprecipitation. Data are represented as mean ± SEM. Student’s two tailed t test was used. **p < 0.001.
To further confirm the co-localization of VAMP5 with NSP8, we performed Co-Immunoprecipitation experiments using anti-HA or anti-Flag antibody to immunoprecipitate VAMP5 or NSP8. We found that HA-VAMP5 (Fig. 7b) and endogenous VAMP5 (Supplementary Fig. 7a) co-immunoprecipitates with Flag-NSP8, whereas NSP7 and NSP12 were not detected in the lysates co-immunoprecipitated with VAMP5 (Supplementary Fig. 7b, c). In this study, we probed for the flag to confirm the similar amounts of lysate were used as input. Since the C-terminal domain of VAMP5 plays a key role in SARS-CoV-2 restriction, we tested if this domain interacts with NSP8 to inhibit the RdRp activity. After performing the Co-Immunoprecipitation experiment, we found that the C-terminal domain of VAMP5 indeed interacts with NSP8 (Fig. 7c). We then determined which specific sites in the C-terminal domain may interact with NSP8. Based on the protein structure interaction prediction tool of ZDOCK, we found residues of R68, G74, G79, V77, L82, D98 and A102 as potential interaction sites with NSP8 (Fig. 7d, e). To validate these predictions, we engineered this artificial amino acid substitution into a VAMP5 expression construct with HA tag fused to the N-terminus. Western blot analysis confirmed that HA-VAMP5 WT and the variants (R68A, G74A/G79A, V77A, L82A, D98A and A102S) are expressed at similar levels in HEK293 cells (Fig. 7f). Subsequently, we performed the Co-Immunoprecipitation experiments and found that all these variants decrease the binding ability of VAMP5 to NSP8, particularly the L82A variant, which may explain the transmembrane region in the C-terminal domain play the key role in viral restriction (Fig. 7f). In addition, we also performed an immunofluorescent assay to examine the influence of these variants on VAMP5 co-localization with NSP8. As expected, VAMP5 variants showed altered localization in the cells, with most of them localizing in the nucleus (Supplementary Fig. 7d). As the predicted residues mentioned above are not entirely reliable, we also engineered VAMP5 variants with substitution in the remaining residues (P94A, Q95A, S96A, S97A, and S99-101A) and performed the Co-Immunoprecipitation experiment (Supplementary Fig. 8a). We found that P94A and S97A also impact the interaction of VAMP5 with NSP8, further highlighting the importance of the C-terminal domain in VAMP5’s interaction with NSP8 (Supplementary Fig. 8b). Similarly, expanding our analysis to other viruses, we found that predicted structural interaction analysis showed that VAMP5 also interacts with RdRps from other coronaviruses like MERS-CoV, OC43, NL63, PEDV and MHV (Supplementary Fig. 8c). In addition, we also performed the Co-Immunoprecipitation experiment to test the potential interaction between VAMP5 with RdRps of DENV2 and ZIKV. Unfortunately, we found no interaction of VAMP5 with these RdRps (NS5), indicating that VAMP5 restrict ZIKV and DENV2 infection through other pathways that require further study (Supplementary Fig. 8d). Collectively, these results suggest that VAMP5 restricts SARS-CoV-2 replication by interacting with NSP8.
VAMP5 disrupts the replication and transcription complex at the double membrane vesicles (DMVs)
As mentioned above, we hypothesized that the C-terminal domain enables VAMP5 to target SARS-CoV-2 NSP8, potentially interfering the formation of replication and transcription complex (RTC) and subsequent the synthesis of negative-strand genomic RNA. As shown in Fig. 6F, the critical importance of VAMP5’s C terminal domain is likely determined by residues R68, G74/G79, V77, L82, D98 and A102, with L82 playing a key role in VAMP5’s binding to NSP8. Firstly, we sought to determine if VAMP5 targets NSP8 to disrupt the interaction between RdRp and viral genomic RNA. We performed the RNA immunoprecipitation (RIP) experiment using the anti-NSP8 antibody to immunoprecipitate NSP8 protein and PCR assay to detect the RNA bound by RdRp. As expected, the viral RNA bound to RdRp decreases upon overexpressing VAMP5 WT, as well as the truncated VAMP5 (C) and VAMP5 (ΔTM) in Caco-2-N cells. This suggests that the C-terminal domain and ΔTM of VAMP5 disrupts the binding of RdRp with viral genomic RNA, halting the negative RNA synthesis process (Fig. 7g). However, the VAMP5 L82A variant did not disrupt RNA binding to RdRp. To exclude the reduction in viral RNA immunoprecipitated is not due to the general reduction in viral RNA in cells overexpressing VAMP5, we designed the experiments to prove that VAMP5 overexpression does not affect the expression of RdRp (Supplementary Fig. 9a). In addition, we used the remdesivir, which blocks viral replication, to prove whether the RNA levels prior to binding with RdRp remain consistent across different treatments with VAMP5 WT and mutants. As expected, the RNA levels prior to RdRp binding were consistent (Supplementary Fig. 9b), indicating that the observed reduction of RNA levels may result from VAMP5 disrupting the binding ability of RdRp to viral RNA. To further prove it, we expressed VAMP5 WT and L82A variant in the Caco-2 cells and challenged them with SARS-CoV-2. We observed that viral negative RNA levels are relatively increased in cells expressing the L82A variant compared to WT, indicating that L82A substitution disrupts VAMP5’s restriction on viral replication (Fig. 7h). To our surprise, we found that VAMP5 L82A still inhibits SARS-CoV-2 S protein expression, which conflicts with the negative RNA results (Fig. 7i). We speculated that L82A substitution impairs VAMP5’s ability to bind to NSP8 and subsequent disrupts negative RNA synthesis, whereas it still keeps its ability to inhibit mRNA formation and S protein expression. The detail mechanism of this phenomenon will need further investigation. To directly understand that VAMP5 disrupts RdRp activity to inhibit SARS-CoV-2 replication, we also did the in vitro RdRp activity experiments. We mixed the NSP7, NSP8 and NSP12 together to form the RdRp with VAMP5 WT or fragment in the buffer, with suramin as positive control. We found that the RNA fragments synthesized by RdRp are disrupted upon adding recombined VAMP5 at 100 μM, whereas the deleted fragment of VAMP5 has no influence on RdRp activity (Fig. 7j, k), indicating that VAMP5 disrupt the interaction between RdRp and viral genomic RNA resulting in stopped synthesis of viral RNA by RdRp.
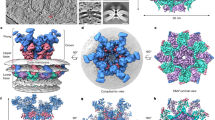
SARS-CoV-2, like other coronaviruses, establishes a virus-induced membrane-bound replication organelle to enable RNA replication. This organelle comprises double membrane vesicles (DMVs), serving as optimal platforms for viral RNA synthesis by concentrating viral replicative proteins to form RTC and relevant host factors, and for hiding replication intermediates to help virus evade innate immune sensors that can detect viral RNA47,48. However, the mechanism by which VAMP5 targets viral RdRp to impede viral replication remains unclear. We hypothesized that this restriction activity may stem from VAMP5’s presence on the DMVs, where its C-terminal domain within the vesicle interacts with NSP8 to inhibit negative-strand RNA synthesis. With VAMP5 located in the DMVs, viral double stranded RNA intermediates protected by DMVs from host innate immune sensors that survey the cytosol for pathogen-associated molecular patterns (PAMPs) can be still sensed by host factors like VAMP5 to eliminate viruses replication48. To test the localization of VAMP5 in DMVs, we transfected Caco-2-N cells with HA-VAMP5 and challenged them with SARS-CoV-2. After 48 h of infection, we detected the localization of VAMP5 on the surface of DMVs by immunoelectron microscopy (IEM). As expected, the anti-HA labeling VAMP5 WT and C terminal domain rather than VAMP5 N terminal domain in Supplementary Fig. 10 shows gold particles decorating DMVs interacting with NSP8, indicating that VAMP5 also localizes on the DMVs, where its C-terminal domain interacting NSP8 to inhibit viral replication.
Discussion
As the key resources for growth and regeneration, stem cells are essential for tissue maintenance and repair. Without any immune system protection, except for the protection from maternal origin, the embryo at the early development stages are often at risk of viral infection. The ESCs have to evolve their own protection system against viral infection related damage. Thus, it is important to study how ESCs protects themselves from viral infection.
We have identified VAMP5 as a crucial cell-intrinsic host defense factor for ESCs against SARS-CoV-2 infection. Notably, VAMP5 is highly expressed in ESCs and mesodermal cells rather than in END and ECT, independent of any viral infection or IFN treatment response. Knocking out VAMP5 renders ESCs susceptible to SARS-CoV-2 as well as a variety of viruses’ infection, whereas overexpressing VAMP5 in the KO cells restores the antiviral activity. In addition, VAMP5 inhibits not only coronavirus (human related SARS-CoV-2, MERS-CoV, CoV-OC43, CoV-NL63, and mammalian related PEDV and MHV-A59) but also other related or unrelated virus infection (ZIKV, VSV, DENV2, AAV2, IAV and LV). These findings suggest that VAMP5 is an important cellular determinant shaping the viral resistance of stem cells. However, it is unlikely that only a single gene can provide resistance against all viruses, as VAMP5 does not restrict EV71, HBV, HCV and HDV. Given that IFITMs are also highly expressed in stem cells and inhibit most viral infection whereas enhancing coronavirus infection like CoV-OC4349,50, we hypothesized that stem cells require a combination of genes to inhibit different stages of viral replication cycle. Our studies uncovered that the combined overexpression of VAMP5 with CD74 enhances the antiviral activity of ESCs (data not shown). Therefore, it is urgent to identify more host restriction factors in stem cells. This will enable us to understand different mechanisms of stem cells resistance to viral infection and also aid in developing entirely new theory to treat viral infection.
In our follow-up studies, we discovered that orthologues of VAMP5 in rats and mice also restrict SARS-CoV-2 and its variants, despite notable differences in residues, particularly in the C-terminus. Moreover, human VAMP5 effectively inhibits swine and mouse coronaviruses, such as PEDV and MHV-A59. These findings suggest that VAMP5 is functionally conserved during mammalian evolution. Along with these situations, it is noteworthy that the SNP presence in VAMP5 is very low in humans. We detected only three SNPs with relative high rates: Leu82Phe substitution present at 0.004% in European, Ile83Phe substitution present at 0.3% in America, and Gln95Lys substitution present at 0.01% in Africa. Of these, only Ile83Phe SNP affects the restrictive activity against SARS-CoV-2. The reason why the Ile83Phe SNP has evolved in these population remains unclear. Given that VAMP5 is also involved in other cellular functions, we hypothesize that these other functions may drive the selection of this SNP.
As VAMP5 usually involves in vesicle trafficking, docking and fusion, we asked if this function of VAMP5 is responsible for restricting SARS-CoV-2 infection. Our findings suggest that these vesicle-related functions of VAMP5 do not influence SARS-CoV-2 restriction under various conditions of viral entry and secretion. This suggests that the high expression of VAMP5 in ESCs cannot solely be explained by its traditional vesicle trafficking and fusion roles. This led us to hypothesize that VAMP5 might localize to the membrane and utilize a novel pathway to restrict viral infection. Together these results highlight the multi-functions of host factors in the microenvironment of ESCs to maximize the maintenance of stem cells against essential damage.
Viruses continually evolve to survive in their environment, resulting in single nucleotide polymorphism variants and numerous lineages. This ongoing evolution poses accurately determining SARS-CoV-2 infection, such as designing effective neutralizing antibodies and developing vaccines. In instance, SARS-CoV-2 omicron continuously evolves during the pandemic leading to various new subvariants. These subvariants exhibit strong neutralizing evasion activity against neutralizing antibody drug and convalescent plasma, presenting a challenge to the efficacy of existing humoral immunity established from vaccination or prior infection10. These challenges have prompted us to question the necessity of developing neutralizing antibody drugs and to reconsider alternative treatment approaches, such as exploring broad-spectrum antiviral treatment to circumvent immune escape mutations. Our study has uncovered that VAMP5’s antiviral restriction effect appears to be unaffected by existed viral mutations, as it significantly inhibits all major variants, including alpha, beta, gamma, delta and omicron. This contrasts the concept that resistance escape barriers are higher when relying on host restrictive factors compared to targeting neutralizing antibodies and the virus directly.
Viruses usually exploit cellular resources to support their replication when infecting eukaryotes. A striking example of this is the virus-induced membrane modification known as viral replication organelles (ROs). These ROs are subcellular structures that serves the invading virus by concealing viral replication intermediates, thus evading detection by innate immune sensors that typically detect viral RNA. Interestingly, SARS-CoV-2 ROs are composed of DMVs48. Host factors such as VMP1 and TMEM41B are essential for DMVs formation during SARS-CoV-2 infection51. TMEM41B deficient cells initiate a strong innate immune response when exposed to viral infection14. However, our findings revealed that VAMP5 is also present within DMVs and inhibits viral replication. This suggests that host cells can also restrict viral replication by interacting with viral replication intermediates within DMVs. At this point, although host innate immune sensors may not directly surveil viral infection within DMVs, other host factors like VAMP5 can access the protective membrane to inhibit viral replication without triggering an innate immune response.
VAMP5, anchored at the membrane of DMVs, relies on its C-terminal domain to disrupt SARS-CoV-2 replication29. Structurally, VAMP5 is relatively simple, featuring an α-helix in its N-terminal domain, a transmembrane (TM) region, and a coil structure in its C-terminal region. The TM and C-terminal domain are positioned on the vesicle side of the DMVs, suggesting their potential to interact with viral replication intermediates and mediated viral restriction. Along with this line, our research indicates that the C-terminal domain of VAMP5 potentially interacts with SARS-CoV-2 NSP8 protein. This interaction leads to the disruption of the RTC. Consequently, the synthesis activity of RdRp is compromised, hindering the synthesis of negative-strand RNA, mRNA, and viral proteins. This disruption ultimately halts the infectious replication cycle. It highlights the VAMP5 TM and C-terminal domain as a major determinant of stem cell intrinsic resistance against coronavirus. They also shed light on how stem cells protect themselves from pathogens damage without relying on human IFN response.
It’s notable that there are some limitations of the in vivo transplantation and viruses’ infection experiments. Short duration of transplantation restricts the effects of in vivo environment on the transplanted cells and subcutaneous transplantation site is different from natural conditions in which hESCs reside (Fig. 3n). However, extended exposure to in vivo environment would influence ESCs pluripotency and induce differentiation, making it difficult to inspect the exact role of in vivo environment. Moreover, technical limitations also make it hard to mimic the hESCs natural conditions. Thereby, the protective role of VAMP5 in vivo should be verified in a new animal model in future.
In conclusion, we recently identified intrinsic VAMP5 highly expresses in ESCs, protecting embryonic stem cells from SARS-CoV-2 and several related or unrelated viruses’ infection. This discovery positions VAMP5 as a potential target for designing broad spectrum antiviral treatment capable of inhibiting a broad range of viral infection.
Methods
Our research complies with all relevant ethical regulations, following protocols approved by the Animal Care and Use Review Committee of Peking University Health Science Center (LA2021093).
Cell culture
Caco-2-N, HEK293T, HepG2-NTCP/CD81/Mir122, Huh7-NTCP, Huh7, Huh7.5, 17CI-1, A549, and Vero E6 cells were cultured and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 U/mL penicillin, and 100 mg/mL streptomycin in a humidified 37 °C, 5% CO2 incubator. The hESC lines WA09 (H9) obtained from female embryonic stem cells were cultured on growth-factor-reduced Matrigel according to the manufacturer’s recommendation in mTeSR1 medium. The cells were replenished with fresh medium every day.
Viruses and infections
The SARS-CoV-2 trVLP and their mutants of N501Y(alpha), P.1 (gamma), B.1.617.1 (kappa), B.1.617.2 (Delta) and Omicron expressing GFP replacing viral nucleocapsid gene (N) were used in this study52. Caco-2-N cells were infected with trVLP to amplify viruses34. ZIKV53 (strain SPH2015) was obtained from Prof. Qiang Ding (Tsinghua University, China) and passaged on Vero E6 cells. IAV strain A/WSN/33 (H1N1) was obtained from Prof. Demin Zhou54 (Peking University, China) and passaged on Vero E6 cells. DENV237 was obtained from Prof. Gong Cheng (Tsinghua University, China) and passaged on Vero E6 cells. MHV-A5955 was obtained from Prof. Dan Lu (Peking University, China) and passaged on 17CI-1 cells. EV7156 (strain Isehara) was obtained from Prof. Yihong Peng (Peking University, China) passaged on HEK293T cells. Infectious virus stocks were obtained for the viruses VSV-GFP (based on strain Indiana), PEDV (CV777), AAV2, LV, HBV, HCV and HDV. All infections were performed by incubation of virus inoculum with cells for 2 hr before the cells were washed and changed to the medium appropriate for the specific cell type and differentiation stage.
WT and VAMP5 KO Caco-2-N cells were infected with SARS-CoV-2 trVLP, their mutants at MOI = 0.05 PFU/cell for 24 h and were infected with HCoV-OC43, HCoV-NL63 at MOI = 1 PFU/cell for 24 h. Vero-E6 were infected with IAV (strain A/WSN/33), DENV2, ZIKV (strain SPH2015) and PEDV (CV777) at MOI = 0.1 PFU/cell for 24 h. HEK293T were infected with VSV-GFP (strain Indiana), EV71 (strain Isehara), AAV2-Luc and LV-Luc at MOI = 0.1 PFU/cell for 24 h. 17CI-1 were infected with MHV-A59 at MOI = 0.1 PFU/cell for 24 h. HepG2-NTCP/CD81/Mir122 were infected with HBV and HCV at MOI = 0.1 PFU/cell for 24 h. Huh7-NTCP were infected with HDV at MOI = 0.1 PFU/cell for 24 h. Viral infection was performed using MOI = 0.1 (as determined of the cell type used to titrate each virus), unless stated otherwise in the figure legend.
Plasmids
VAMP5 plasmid was constructed to pLVX-AcGFP1-N1 plasmid and pcDNA3.1 (+) with hemagglutinin (HA) -tag, Flag-tag at the N terminal region or GFP at the C terminal region. VAMP5 mutations with deletion of 1-70, 71-116, 94-116, TM and point mutation of R68A, G74A/G79A, V77A, L82F, I83F, P94A, Q95K, S96A, S97A, D98A, S99A/S100A/S101A, and A102S were cloned into the vector of pLVX-AcGFP1-N1 with HA-tag at the N terminal region or GFP at the C terminal region respectively. SARS-CoV-2 ORF7b, M, and spike protein were cloned into pLVX-AcGFP-N1 vector. The plasmids of pcDNA3.1 (+)-nsp12, pcDNA3.1 (+)-nsp7, pcDNA3.1 (+)-nsp8, and pcDNA-nsp8-152 were used to express codon-optimized Flag-nsp12, Flag-nsp7, Flag-nsp8 respectively, all of which contain a Flag tag at the N-terminus. CD74 from human and VAMP5 sequences from Garillia gorilla gorilla, Macaca mulatta, Mus musculus, and Rattus norvegicus were synthesis artificially and inserted into pLVX-AcGFP1-N1 vector. split-NanoLuc fragments of LgBiT, and HiBit fused with CypA were cloned into pLVX-AcGFP1-N1 and pcDNA3.1, respectively. ZIKV NS5 and DENV2 NS5 fragments were cloned into pcDNA3.1 with Flag-tag at the C terminal. The primers used for plasmids construction abovementioned are shown in Supplementary table 1.
Antibodies and chemicals
Primary antibodies used for western blot and immunofluorescent staining include rabbit polyclonal anti-VAMP5 (Proteintech: 11822-1-AP; 1:1000), rabbit polyclonal anti-SARS-CoV-2 spike (Abcam: ab272504; 1:1000), mouse monoclonal anti-β-actin (Beyotime: AA128; 1:1000), mouse monoclonal anti-GAPDH (Proteintech: 60004-1-Ig; 1:1000), mouse monoclonal anti-HA (Cell Signaling Technology: 2367S; 1:1000), rabbit monoclonal anti-HA (Cell Signaling Technology: 3724S; 1:1000), rabbit polyclonal anti-GFP (GeneTex: GTX113617; 1:5000), mouse monoclonal anti-Flag (Sigma-Aldrich: F1804; 1:1000), rabbit polyclonal anti-OCT4 (Proteintech: 11263-1-AP; 1:1000), mouse monoclonal anti-SSEA-4 (STEMCELL: 60062Fl.1; 1:1000), mouse monoclonal anti-SOX17 (R&D systems: MAB1924; 1:1000), rabbit monoclonal anti-FoxA2 (Cell Signaling Technology: 8186 T; 1:1000), rabbit monoclonal anti-Branchyury (Cell Signaling Technology: 81694; 1:1000), mouse monoclonal anti-CXCR4 (Proteintech: 60042-1-Ig; 1:1000), rabbit polyclonal anti-PAX6 (Proteintech: 12323-1-AP; 1:1000), mouse monoclonal anti-Nestin (R&D systems: MAB1259; 1:1000), rabbit polyclonal anti-ACE2 (Abcam: ab65863; 1:1000), rabbit monoclonal anti-VSV-G tag antibody (Abcam: ab309106; 1:1000), rabbit monoclonal anti-SARS-CoV-2 nsp8 (GeneTex: GTX636997; 1:1000), mouse monoclonal anti-SMA (Santa Cruz: sc-32251; 1:500), rabbit polyclonal anti-Transgelin (Abcam: ab14106; 1:500), rabbit polyclonal anti-MAP2 (Abcam: ab32454; 1:500), mouse monoclonal anti-Tubulin β3 (BioLegend: 801202; 1:500), rabbit monoclonal anti-surfactant protein B (SP-B; Abcam: ab271345; 1:500), mouse monoclonal anti-EpCAM (Invitrogen: 9326-82; 1:500).
Primary antibodies used for immunoprecipitation include rabbit monoclonal anti-HA (Cell Signaling Technology: 3724S; 1:50), mouse monoclonal anti-Flag (Sigma-Aldrich: F1804; 1:200), rabbit polyclonal anti-GFP (GeneTex: GTX113617; 1:200), rabbit polyclonal anti-desmin (Proteintech: 16520-1-AP), rabbit monoclonal anti-calponin (Abcam: ab46794), mouse monoclonal anti-calnexin (Proteintech: 66903-1-Ig). Secondary antibodies, proteins and staining solutions used for western blot, EM and IF include goat anti-rabbit IgG conjugated to HRP (EASYBIO: BE0101-100; 1:10000), goat anti-mouse IgG conjugated to HRP (EASYBIO: BE0102-100; 1:10000), goat anti-rabbit IgG conjugated to Alexa Fluor 594 (Life Technologies: A-11012; 1:1000), goat anti-mouse IgG conjugated to Alexa Fluor 594 (Life Technologies: A-11005; 1:1000) goat anti-rabbit conjugated to Alexa Fluor 488 (Life Technologies: A-11008; 1:1000) goat anti-mouse conjugated to Alexa Fluor 488 (Life Technologies: A-11001; 1:1000), Nanogold goat anti-rabbit IgG conjugated Fab’ fragment (Nanoprobes: NNP-2004; 1:50).
Drugs and antibiotics were used at concentrations indicated in figure legends and methods, and include puromycin (Abcam: Ab141453), polybrene (R&D Systems: 7711), Suramin (Sigma: S2671), Remdesivir (Aladdin: R287694), Camostat (MedChemExpress, HY-13512), Lactoferrin (Sigma, 61327). Reactive Oxygen Species (ROS) Assay were detected with 2,7-dichlorofluorescein diacetate (DCFH-DA) according to the manufacturers’ manuals (Beyotime, S0033).
Generation of VAMP5-knockout ESCs and Caco-2
Two guide RNA (gRNA) targeting VAMP5 exon were designed using the online CRISPR design tool (http://crispr.mit.edu/) to generate VAMP5-knockout cells (Supplementary table 2). The gRNA was cloned into the lentiCRISPR v2 vector using standard techniques. Lentiviral particles were generated as reported previously57. For generation of VAMP5-knockout in ESCs, cells were dissociated into single cells using Accutase and plated onto Matrigel-coated plates with ROCK inhibitor (Y-27632) to a confluency of approximately 20–30%. Cells were transduced twice in mTeSR1 medium with concentrated lentiviral particles and puromycin (1 μg/ml) was added to culture medium at 48 hr post transduction. Clones were expanded and screened by western blot analysis and homozygous VAMP5 knockout clones were chosen for further characterization. Pluripotency was evaluated by checking expression of pluripotent markers and differentiation capability into different germ layers, using the conditions described above.
Lentivirus production and transduction
Lentiviral particles were generated as reported previously57. Briefly, HEK293T cells plated on a 10-cm dish with 9 μg pLVX vector expressing the ORF of interest, 4.5 μg psPAX2 and 3 μg VSV-G by Lipofectamine 2000 according to the manufacturer’s directions. Medium containing the lentiviral particles was harvested at 48 h post transfection and filtered through 0.22 μm filter. For lentiviral transduction, cells were transferred to 24-well plates in suspension and transduced with several lentivirus particles in medium containing 3% FBS, 20 mM HEPES, and 4 μg/mL polybrene.
MERS-CoV replicon reporter assay
An amount of 100 ng of pBAC-CMV-MERS replicon58 (generously provided by Zhengli Shi, Wuhan Institute of Virology, Chinese Academy of Science, China) was transfected into Huh7 that had been grown to 70% confluence in a 96-well plate. Transfection was conducted by using Lipofectamine 3000 (Invitrogen) in Opti-MEM Medium (Gibco) according to the manufacturer’s instructions. Amount of 400 ng of pBAC-CMV-MERS replicon were added to a 96-well plate containing 0.1 mL of ESCs-H9 cells (5 × 104) in Electroporation Solution (Thermo). Single electrical pulse was given with a Neon (Invitrogen) with setting of 1200 V for 30 ms. After 5 min recovery at room temperature, the electroporated cells were seeded into 96-well plate and incubated at 37 °C with 5% CO2. After incubation for 6 h, the culture medium was replaced with DMEM containing 10% FBS. For the Nano-Glo assay, aspirate medium from cells in 96-well plates and replace with 100 µl of buffered cell culture medium. Add 25 µl of Nano-Glo Live Cell Reagent to each well, and gently mix the plate by hand or with an orbital shaker. Measure luminescence immediately after adding the Nano-Glo Live Cell Reagent.
Antiviral test of VAMP5 in nude mice
Female BALB/c nude mice was used for ESCs transplantation. Mice aged 6–8 weeks were randomly distributed into two distinct groups (five per group). All mice subjected to different stimuli were maintained in the same environmental conditions for growth. Female mice were housed in groups of two, with 12 h light and 12 h dark conditions, feeding on a standard diet. The room temperature was 22 °C and humidity was 50%. All animal protocols were approved by the Animal Care and Use Review Committee of Peking University Health Science Center (LA2021093). ESCs-H9 cells control or VAMP5 KO ESCs mixed with matrigel were xenografted subcutaneously. After 48-h post xenograft, PEDV or DENV2 (2 × 105 pfu for one mouse) was injected at the subcutaneous injection site (Fig. 3n). After 48 h of viral infection, the mice were sacrificed and the ESCs clusters were harvested for the detection of viral replication by Quantitative real-time PCR (RT-qPCR).
RNA interference (RNAi)
Small interfering RNAs (siRNAs) targeting candidate genes and negative control (NC) siRNA were synthesized by Genepharma (Shanghai, China). The sequences of the siRNAs are indicated in Supplementary table 2. According to the manufacturer’s instructions, cells grown to 60–70% confluence were treated with siRNA using Lipofectamine RNAi MAX.
RT-qPCR
The RNA extraction and quantification were performed as previously described34. Briefly, Total RNA of cells was isolated with the RNAprep pure Cell Culture/Bacterial total RNA extraction kit. The reverse transcription primer tag-F for negative-strand viral RNA was 5’-TAATACGACTCACTATAGGG CGAAAGGTAAGATGGAGAGCC-3’ and the other reverse transcription primers were the general oligo-dT. The negative-strand viral RNA primer tag-F was designed to contain the SARS-CoV-2-targeting sequence F in 3’- part and the non-SARS-CoV-2 sequence tag in 5’ -part. The cDNA was synthesized by RevertAid first strand cDNA synthesis kit. The qPCR for SARS-CoV-2 RNA was performed using the 2× RealStar Green Power Mixture according to the instruction. The primers were shown in the Supplementary table 3. GAPDH was detected and used as an internal control.
Western blot analysis
Western blotting was performed as described previously59. Briefly, cells were lysed with commercial RIPA buffer [50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate, 1% NP-40 with protease inhibitor] and centrifuged at 12,000 × g at 4 °C for 30 min to extract the total protein. Cell lysates were loaded on the 12% SDS-PAGE gel and transferred to a polyvinylidene fluoride membrane. After blocking with blocking buffer with 5% milk, the membranes were incubated with antibodies. The β-actin was used as loading control. Primary antibodies used for western blot analysis are listed in the Key Resources Table. The membranes were exposed using ECL substrate and the readout was detected using the BioRad ChemiDoc MP system.
Immunoprecipitation assay
HEK293T cells were lysed with radioimmunoprecipitation assay (RIPA) lysis buffer IP lysis buffer [10 mM HEPES (pH 7.5), 150 mM KCl, 3 mM MgCl2, 0.5% NP-40, 0.1 mM phenylmethylsulfonyl fluoride (PMSF) and protease inhibitor] for 30 min on ice and then subjected to centrifugation at 12,000 × g for 10 min at 4 °C. Supernatants were immunoprecipitated with the appropriate antibodies at 4 °C overnight. The immune complexes were incubated with protein A/G-agarose beads for 2 h, washed five times with lysis buffer, and eluted in 1×SDS-PAGE sample buffer western blot analysis.
Viral attachment assay
The viral attachment assay was performed as previously reported41. Briefly, Caco-2-N were seed in 24-well plates with 500,000 cells/well one day before the infection. SARS-CoV-2 trVLP (MOI = 1) was added into the cells and put at 4 °C for 2 hr to allow viral attachment to cells. After washing out of free virus, cell surface trVLP was extracted and quantified by RT-qPCR.
Viral entry assay
The viral entry assay was performed as previously described41. Briefly, Caco-2-N were seeded in a 24-well plate with 500,000 cells/well one day before the infection. Cells were incubated with trVLP (MOI = 1) at 37 °C for 2 hr to allow viral internalization into cells. Then, the cells were washed with PBS for 3 times. The mRNA of trVLP was measured by RT-qPCR.
Viral release assay
Caco2-N cells were infected with SARS-CoV-2 trVLP in 24-well plates. The culture fluid was harvested and filtered through a 0.2-μm pore filter at different infection times. Finally, the viral RNA in cells (intracellular) and culture fluid (extracellular) were determined by real-time RT-qPCR at different infection times. The ratio of extracellular to intracellular viral yields represents the virion release efficiency.
Virus-cell membrane fusion assay
The virus-cell membrane fusion assay was performed as previously described42. Briefly, HIV-based pseudoviral particles containing CypA-HiBiT were prepared by transfecting HEK293T cells plated on a 10-cm dish with 9 μg pLVX-AcGFP1-N1, 4.5 μg psPAX2, 2 μg CypA-HiBiT and 3 μg pCMV-spike (encoding the spike protein from SARS-CoV-2) by Lipofectamine 2000. At 48 h after transfection, the supernatant was filtered through a 0.22-μm filter, laid onto a 20% sucrose (w/v in 1 × HBSS) cushion and centrifuged using a Sorvall TH-641 rotor at 100,000 g for 2 h. The supernatant was discarded and the pseudoviral particles concentrated in the pellet were resuspended with 500 μl of DMEM + 10% FBS medium. Huh7.5 cells were stably transduced with pLVX-LgBiT, followed by puromycin selection for 7 days. Huh7.5 expressing the LgBiT fragment were plated in a 96-well dish. One day after plating, each well of cells was infected with 100 μl of pseudoviruses containing CypA-HiBiT at 1,000 g and 4 °C for 30 min, followed by incubation at 37 °C for 2 h. The medium was removed and Nano-Glo assay was performed to detect SARS-CoV-2 pseudovirus infection.
In vitro RdRp activity assay
The in vitro RNA dependent RNA polymerase (RdRp) activity of SARS-CoV-2 were performed as described previously60. Briefly, 1uM of nsp12-nsp7-nsp8 were mixed with 1uM annealed RNA and 0.5 mM NTP in a reaction buffer containing 10 mM Tris-HCl (pH 8.0), 10 mM KCl, 1 mM betamercaptoethanol and 2 mM MgCl2 (freshly added prior usage). Then, the mixture was incubated at 30 °C for 30 min. The annealed RNA was mixed by a 40-nt template RNA (5’-CUAUCCCCAUGUGAUUUUAAUAGCUUCUUAGGAGAAUGAC-3’, Takara) corresponding to the 3’ end of the SARS-CoV2 genome and a 5’-fluorescein label (5’FAM- GUCAUUCUCCUAAGAAGCUA-3’, Takara). The products were denatured by boiling (100 °C, 10 min) in the presence of formamide and separated by 20% PAGE containing 9 M urea run with 0.5 × TBE buffer. Images were taken using a Vilber Fusion system
Differentiation into three germ layers
For endoderm, ESCs (H9, Wicell Research Institute) were dissociated into single cells using Accutase and plated in the Matrigel-coated plates with 10 mM ROCK inhibitor (Y-27632). Differentiation was induced by using the STEMdiff definitive endoderm kit following the manufacturer’s instructions19. For mesoderm, differentiation of ESCs into mesoderm was performed accordingly. Briefly, ESCs were dissociated into single cells using Accutase and plated onto Matrigel-coated plates with ROCK inhibitor (Y27632) to reach approximately 60% confluence. Differentiation was induced by the STEMdiff mesoderm induction medium kit following the manufacturer’s instructions19. For ectoderm, ESCs were differentiated into neuroectoderm cells as described previously. Briefly, ESCs were cultured in mTeSR1 medium with ROCK inhibitor (Y27632). The next day, the cells were differentiated with SRM (DMEM/F12, 10% knockout serum replacement (KOSR), and 1% GlutaMax) supplemented with 10 mM SB431542 and 200 nM Noggin for 5 days19.
The hESCs induced stellate cells
To generate stellate cells, endoderm was dissociated into single cells using Accutase and plated onto Matrigel coated plates with Y-27632 in basal medium (DMEM/F12 + 10% KOSR + 1% GlutaMax +1% penicillin-streptomycin; Life Technologies, USA) supplemented with 10 ng/mL FGF2 (Peprotech, USA), 5 ng/mL BMP4 (R&D, USA), 10 ng/mL VEGF (Peprotech, USA), and 10 ng/mL HGF (Peprotech, USA). The medium was replaced every 2 days for 4 days. Then, the cells were exposed to BM containing 1 × B27 (Life Technologies, USA), 1 × insulin-transferrin-selenium-ethanolamine (ITS-X; Life Technologies, USA), 1 mM retinoic acid (RA; Sigma-Aldrich, USA), 20 mM rosiglitazone (Selleck Chemicals, USA), and 2.5 mM dexamethasone (Sigma Aldrich, USA) for 6 days36.
The hESCs induced alveolar type II cells (AT2 cells)
To generate AT2 cells, ESCs were seeded at 4 × 104 cells/ cm2 on Matrigel coated 6 well plates at day 0. The endoderm differentiation basal medium consisted of 11.6 g/L MCDB131, 0.44 g/L D-Glucose, 2.68 g/L NaHCO3, 5 g/L BSA, 80 μg/mL vitamin C, 1×GlutaMAX. 24 h after passage, the medium was changed into basal medium containing 100 ng/mL activin A and 3 μM CHIR99021 for 24 h and then 100 ng/mL activin A for another 24 h to differentiate ESCs into definitive endoderm.
The lung progenitor’s basal medium consisted of DMEM/F12 supplemented with 0.5 mg/mL bovine serum albumin, 50 μg/mL L-ascorbic acid, 1×GlutaMAX, 1×N2, 1×B27, 0.04 μl/mL β-mercaptoethanol. 2 μM dorsomorphin, and 10 μM SB431542 were added for 3 days and 3 μM CHIR99021, 10 ng/mL BMP4, and 50 nM retinoic acid were added for 9 days to differentiate into lung progenitor cells. The CPM+ lung progenitor cells was isolated with mojoSor anti-PE nanobeads (Biolegend). To generate AT2 cells, the isolated lung progenitor cells were treated with 3 μM CHIR99021, 10 ng/mL KGF, 50 nM dexamethasone, 100 μM IBMX and 100 μM cAMP was added for 10 days61.
The hESCs induced smooth muscle cells
To generate smooth muscle cells (iSMC), hESCs need to be induced to mesoderm. hESCs were seeded at 2×104 cells/ cm2 on Matrigel coated 6 well plates. 24 h after passage, the medium was changed into LaSR medium consists of DMEM/F12 supplement with 1% Glutamax, 60 μg/mL L-ascorbic acid and 1% penicillin-streptomycin, supplement with 8 μM CHIR99021 for 3 days with medium changed daily. Cells were then reseeded at 6.25×104 cells/cm2 on Matrigel coated plates in SMC medium for 3 day with medium changed daily. SMC medium was LaSR supplement with 2 ng/mL Activin-A and 10 ng/mL PDGF-BB. After three days of differentiation, the induced SMC can be passaged using 0.05% Trypsin-EDTA treated for 1 min at 37 °C62.
The hESCs induced neurons cells
To generate neurons, ESCs were dissociated with TrypLE and plated at 1 × 105 cells/cm2 on Matrigel coated plates. 24 h after passage, the medium was replaced with DMEM/F12 supplemented with 1% Glutamax, 1 × NEAA, 1 × B27 w/o Vitamin A, 1 × N2 Max, 10 μM DAPT, 10 μM SB431543, 100 ng/mL Noggin, 2 μM XAV939, and 1 μg/mL doxycycline for 6 days with medium changed daily63.
RNA-sequencing (RNA-seq)
ESCs and three germ layers were analyzed by RNA-seq. RNA-seq was performed at Novogene Co. Ltd. Total RNA was isolated from ESCs and three germ layers. Total RNA was extracted using Trizol and sent to the Novogene Company (China) for RNA-seq analysis. RNA sequencing libraries were prepared by using the NEB Next Ultra TM RNA library Prep kit for Illumina (NEB) following the manufacturer’s instructions. The randomly primed 150-bp paired-end libraries were sequenced on Illumina Hiseq 4000 platform. The reads were aligned to the human genome (hg38) using STAR v2.7.9a64 and gene expression levels were estimated using feature Counts65. DESeq266 was used to perform differential gene expression analysis. R package cluster Profiler was used to perform gene ontology (GO) pathway analysis67. Principal component analysis (PCA) was generated with R PCA function using scaled gene expression. Heatmaps were generated by the pheatmap package.
Immunofluorescence assay
Cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.1% Triton X-100 for 15 min, and blocked with phosphate-buffered saline (PBS) containing 10% horse serum plus 1% bovine serum albumin (BSA) for 2 h. The cells were washed three times with PBS and incubated with diluted primary antibodies overnight at 4 °C. Cells were washed with PBS three times at room temperature, then incubated with fluorochrome-conjugated secondary antibodies in the dark for 1 h at room temperature. The cells were then rinsed and incubated with DAPI for 5 min, Images were taken by Echo Laboratories Revolve FL fluorescent microscopy and processed with ImageJ.
Immunogold staining and electron microscopy
For pre-embedding IEM, the cells were fixed, permeabilized and labeled as described previously. In brief, the cells were fixed with a mixture of 4% PFA and 0.05% glutaraldehyde prepared in 0.2 M HEPES buffer for 10 min (room temperature) and then with 4% PFA alone for 30 min (room temperature), followed by incubation with blocking/permeabilizing solution (0.5% BSA, 0.1% saponin and 50 mM NH4Cl in PBS) for 30 min. Cells were incubated with a primary anti-HA monoclonal antibody and anti-NSP8 antibody diluted in blocking/permeabilizing solution overnight and then a secondary anti-rabbit antibody (coated with15 nm gold particles against VAMP5 and 5 nm gold particles against NSP8) was added for 2 hr. The GoldEnhance EM kit (from Nanoprobes) was used to enhance ultrasmall gold particles. Cells prepared for IEM were scraped, pelleted, post-fixed in OsO4 and uranyl acetate, dehydrated, embedded in Epon and polymerized at 60 °C for 72 hr. For each sample, thin sections were cut using a Leica EM UC7 ultramicrotome (Leica Microsystems). The samples were observed using TEM (FEI Tecnai G2) operating at a 75-kV acceleration voltage. All samples preparation and immunocolloidal gold transmission electron microscopy were performed at the Center for Biological Imaging (CBI, http://www.ibp.cas.cn/cbi/), Institute of Biophysics, Chinese Academy of Sciences68.
RNA immunoprecipitation and RT-PCR
Lysates from SARS-CoV-2 trVLP infected Caco-2-N cells in immunoprecipitation assays were collected 24 hpi and preincubated with protein A/G-agarose on ice for 1 h. Non-specific complexes were pelleted by centrifugation at 12,000 × g for 10 min at 4 °C. The supernatants were recovered, and 100 µL samples were each diluted with 450 µL of lysis buffer and then added to either NSP8 antibody or no antibody, and then incubated on ice for 2 h. Prewashed protein A/G-agarose was added to each sample, then incubated on ice for 1 h. RNA-protein coimmunoprecipitation complexes were pelleted by centrifugation at 1000 × g at 4 °C for 5 min and washed three times with lysis buffer. Each pellet was resuspended in 400 µL of proteinase K buffer [100 mM Tris-HCl (pH 8.0), 12.5 mM EDTA, 150 mM NaCl, 1% SDS] and incubated with 100 µg predigested proteinase K for 30 min at 37 °C. RNA was extracted and subsequently RT-PCR amplified. RT-PCR was performed using a PrimeScript one-step RT-PCR kit and primers specific to SARS-CoV-2 S. Primers specific to actin were used as control. The primers shown in the Key Resources Table. PCR products were resolved in a 1% agarose gel pre-stained with Gel Red nucleic acid gel stain.
Flow cytometry
Differentiated cells were dissociated into a single-cell suspension and stained with antibodies for SSEA4, SARS-CoV-2-S, ZIKV-NS5. Stained cells were analyzed with FACSCanto II cytomerter and FlowJo VX software. FACS plot showing populations were usually 10,000 and the positive gates were defined based on the isotype control.
Cell Viability Assay
Cells in 96-well plates were equilibrated at room temperature for approximately 30 min. Add 100 µl CellTiter-Glo® Reagent (Promega) equal to the volume of cell culture medium present in each well. Mix contents for 2 minutes on shaker to induce cell lysis. Allow the plate to incubate at room temperature for 10 min to stabilize luminescent signal. Record luminescence.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 8. The two-tailed Student’s t test was used comparing two conditions. Statistical significance was defined as: p < 0.05 (*), p < 0.01 (**), p < 0.001 (***) and p < .00001 (****).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNAseq data generated in this study have been deposited in the NCBI database under accession code PRJNA1097823 (https://www.ncbi.nlm.nih.gov/bioproject/). All other data are available in the article and its supplementary files. Source data are provided with this paper.
References
Stadler, K. et al. SARS - Beginning to understand a new virus. Nat. Rev. Microbiol 1, 209–218 (2003).
Zhong, N. S. et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 362, 1353–1358 (2003).
de Wit, E., van Doremalen, N., Falzarano, D. & Munster, V. J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol 14, 523–534 (2016).
Lim, P. L., Lee, T. H. & Rowe, E. K. Middle East Respiratory Syndrome coronavirus (MERS CoV): update 2013. Curr. Infect. Dis. Rep. 15, 295–298 (2013).
Guan, W. J. et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med 382, 1708–1720 (2020).
Ma, J. X. et al. Coronavirus disease 2019 patients in earlier stages exhaled millions of severe acute respiratory syndrome coronavirus 2 per hour. Clin. Infect. Dis. 72, E652–E654 (2021).
Huang, C. L. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020).
Cui, J., Li, F. & Shi, Z. L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol 17, 181–192 (2019).
Carabelli, A. M. et al. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat. Rev. Microbiol 21, 162–177 (2023).
Cao, Y. L. et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature 614, 521–529 (2023).
Plante, J. A. et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 592, 116–121 (2021).
Garcia-Beltran, W. F. et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 184, 2523–2523 (2021). (vol 184, pg 2372, 2021).
Zhang, L. et al. SARS-CoV-2 BA.2.86 enters lung cells and evades neutralizing antibodies with high efficiency. Cell 187, 596–608 (2024).
Hoffmann, H. H. et al. TMEM41B is a pan-flavivirus host factor. Cell 184, 133–148 (2021).
Zhang, S. J. et al. Comparison of viral RNA-host protein interactomes across pathogenic RNA viruses informs rapid antiviral drug discovery for SARS-CoV-2. Cell Res. 32, 9–23 (2022).
Schneider, W. M., Chevillotte, M. D. & Rice, C. M. Interferon-stimulated genes: a complex web of host defenses. Annu Rev. Immunol. 32, 513–545 (2014).
Schoggins, J. W. et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 (2011).
Tian, X. et al. TRIM56 impairs HBV infection and replication by inhibiting HBV core promoter activity. Antivir. Res 207, 105406 (2022).
Wu, X. et al. Intrinsic immunity shapes viral resistance of stem cells. Cell 172, 423–438 (2018).
Wolf, D. & Goff, S. P. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458, 1201–1204 (2009).
Wu, X. F., Kwong, A. C. & Rice, C. M. Antiviral resistance of stem cells. Curr. Opin. Immunol. 56, 50–59 (2019).
Hong, B. et al. SARS-CoV-2 and Malayan pangolin coronavirus infect human endoderm, ectoderm and induced lung progenitor cells. Preprint at bioRxiv, https://doi.org/10.1101/2020.09.25.313270 (2020).
Jackson, C. B., Farzan, M., Chen, B. & Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Bio 23, 3–20 (2022).
Wang, S. et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 31, 126–140 (2021).
Lim, S., Zhang, M. & Chang, T. L. ACE2-independent alternative receptors for SARS-CoV-2. Viruses-Basel 14, 2535 (2022).
Zeng, W. J. et al. Evidence of infection of human embryonic stem cells by SARS-CoV-2. Front Cell Infect. Mi 12, 911313 (2022).
Wells, A. I. & Coyne, C. B. Inhibiting Ebola virus and SARS-CoV-2 entry Screening identifies CD74 as a cellular antiviral protein against Ebola virus and SARS-CoV-2. Science 370, 167–168 (2020).
Tran, T. H. T., Zeng, Q. & Hong, W. J. VAMP4 cycles from the cell surface to the trans-Golgi network via sorting and recycling endosomes. J. Cell Sci. 120, 1028–1041 (2007).
Chen, Y. A. & Scheller, R. H. Snare-mediated membrane fusion. Nat. Rev. Mol. Cell Bio 2, 98–106 (2001).
Rose, A. J., Jeppesen, J., Kiens, B. & Richter, E. A. Effects of contraction on localization of GLUT4 and v-SNARE isoforms in rat skeletal muscle. Am. J. Physiol. -Reg. I 297, R1228–R1237 (2009).
Hasan, N., Corbin, D. & Hu, C. A. Fusogenic pairings of vesicle-associated membrane proteins (VAMPs) and plasma membrane t-SNAREs-VAMP5 as the exception. Plos One 5, e14238 (2010).
Rhein, B. A. et al. Interferon-γ Inhibits Ebola Virus Infection. Plos Pathogens 11, https://doi.org/10.1371/journal.ppat.1005263 (2015).
Forst, C. V. et al. Integrative gene network analysis identifies key signatures, intrinsic networks and host factors for influenza virus A infections. Npj Syst. Biol. Appl 3, 35 (2017).
Ju, X. et al. A novel cell culture system modeling the SARS-CoV-2 life cycle. PLoS Pathog. 17, e1009439 (2021).
Ikezawa, M. et al. Loss of VAMP5 in mice results in duplication of the ureter and insufficient expansion of the lung. Dev. Dynam 247, 754–762 (2018).
Lai, X. et al. Generation of functionally competent hepatic stellate cells from human stem cells to model liver fibrosis in vitro. Stem Cell Rep. 17, 2531–2547 (2022).
Zhu, Y. B. et al. Host serum iron modulates dengue virus acquisition by mosquitoes. Nat. Microbiol. 4, 2405–2415 (2019).
Zhang, K. et al. A novel cell culture model reveals the viral interference during hepatitis B and C virus coinfection. Antivir. Res 189, 105061 (2021).
Martin, R. M. et al. Highly efficient and marker-free genome editing of human pluripotent stem cells by CRISPR-Cas9 RNP and AAV6 donor-mediated homologous recombination. Cell Stem Cell 24, 821–828 (2019).
Lai, X. Y., Zhuang, H., Li, T. & Xiang, K. H. Protocol for characterizing the inhibition of SARS-CoV-2 infection by a protein of interest in cultured cells. Star Protoc 3, https://doi.org/10.1016/j.xpro.2022.101802 (2022).
Lai, X. et al. Identified human breast milk compositions effectively inhibit SARS-CoV-2 and variants infection and replication. iScience 25, 104136 (2022).
Xu, D. J. et al. PLSCR1 is a cell-autonomous defence factor against SARS-CoV-2 infection. Nature 619, 819–827 (2023).
Bruchez, A. et al. MHC class II transactivator CIITA induces cell resistance to Ebola virus and SARS-like coronaviruses. Science 370, 241–247 (2020).
Allgood, S. C. et al. Effector AnkX disrupts host cell endocytic recycling in a phosphocholination-dependent manner. Front Cell Infect. Mi 7, 397 (2017).
V’kovski, P., Kratzel, A., Steiner, S., Stalder, H. & Thiel, V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol 19, 155–170 (2021).
Matsui, T., Sakamaki, Y., Hiragi, S. & Fukuda, M. VAMP5 and distinct sets of cognate Q-SNAREs mediate exosome release. Cell Struct. Funct. 48, 187–198 (2023).
Ricciardi, S. et al. The role of NSP6 in the biogenesis of the SARS-CoV-2 replication organelle. Nature 606, 761–768 (2022).
Wolff, G., Melia, C. E., Snijder, E. J. & Bárcena, M. Double-membrane vesicles as platforms for viral replication. Trends Microbiol 28, 1022–1033 (2020).
Zhao, X. S. et al. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. P Natl. Acad. Sci. USA 111, 6756–6761 (2014).
Shi, G. L. et al. Opposing activities of IFITM proteins in SARS-CoV-2 infection. Embo J. 40, e106501 (2021).
Ji, M. M. et al. VMP1 and TMEM41B are essential for DMV formation during β-coronavirus infection. J. Cell Biol. 221, e202112081 (2022).
Ren, W. L. et al. Mutation Y453F in the spike protein of SARS-CoV-2 enhances interaction with the mink ACE2 receptor for host adaption. PLoS Pathog. https://doi.org/10.1371/journal.ppat.1010053 (2021).
Ding, Q. et al. Species-specific disruption of STING-dependent antiviral cellular defenses by the Zika virus NS2B3 protease. P Natl. Acad. Sci. USA 115, E6310–E6318 (2018).
Si, L. L. et al. Generation of influenza A viruses as live but replication-incompetent virus vaccines. Science 354, 1170–1173 (2016).
Sun, Y. Z. et al. A Gradient pH-sensitive polymer-based antiviral strategy via viroporin-induced membrane acidification. Adv. Mater. 34, e2109580 (2022).
Zhu, M., Duan, H., Gao, M., Zhang, H. & Peng, Y. H. Both ERK1 and ERK2 Are Required for Enterovirus 71 (EV71) Efficient Replication. Viruses-Basel 7, 1344–1356 (2015).
Zhang, Y. J. & Stefanovic, B. mTORC1 phosphorylates LARP6 to stimulate type I collagen expression. Sci Rep. https://doi.org/10.1038/srep41173 (2017).
Chen, J. et al. Development of a MERS-CoV replicon cell line for antiviral screening. Virol. Sin. 36, 730–735 (2021).
Fan, H. et al. The effect of whey protein on viral infection and replication of SARS-CoV-2 and pangolin coronavirus in vitro. Signal Transduct. Target Ther. 5, 1849–1851 (2020).
Fan, H. et al. The effect of whey protein on viral infection and replication of SARS-CoV-2 and pangolin coronavirus in vitro. Signal Transduct. Target Ther. 5, 275 (2020).
Burgess, C. L. et al. Generation of human alveolar epithelial type I cells from pluripotent stem cells. Cell Stem Cell https://doi.org/10.1016/j.stem.2024.03.017 (2024).
Patsch, C. et al. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat. Cell Biol. 17, 994–U294 (2015).
Zhang, Y. S. et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78, 785–798 (2013).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Wu, T. Z. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021).
Liu, W. et al. E3 ubiquitin ligase ANKIB1 attenuates antiviral immune responses by promoting K48-linked polyubiquitination of MAVS. Cell Rep. 43, 114687 (2024).
Acknowledgements
We declare grants from Peking University Health Science Center Fund (BMU2023YJ016 to K.X.), Young Elite Scientists Sponsorship Program by BAST (QNRCNRC001 to K.X), the National Natural Science Foundation of China [grant No. 81873579 (to K.X.)], and Emerging Engineering Interdisciplinary-Young Scholars Project, Peking University, the Fundamental Research Funds for Central Universities (to K.X.). We gratefully thank Prof. Demin Zhou (Peking University, China) for sharing the IAV, Prof. Gong Cheng (Tsinghua University, China) for sharing Dengue Virus, Prof. Yihong Peng (Peking University, China) for sharing EV71, Prof. Zhengli Shi (Wuhan Institute of Virology, Chinese Academy of Sciences, China) for sharing the plasmid of MERS-CoV, Prof. Dan Lu (Peking University, China) for sharing MHV-A59 and 17CI-1 cells, Prof. Xiaoyun Liu (Peking University, China) for sharing the plasmid of Ankx, Prof. Stephan Urban (Heidelberg University, Germany) for sharing the Myrcludex B. We also thank Prof. Ling Liang (Peking University, China), Prof. Ye Xiang (Tsinghua University, China), and Prof. Xianfang Wu (Cleveland Clinic Foundation) for the helpful discussion.
Author information
Authors and Affiliations
Contributions
Conceptualization, K.X.; methodology, H.D., Z.P., P.J., F.Y., X.L., H.L., Z.G., J.D., Q.P., Y.Y.; data analysis, H.D., K.X., Z.P., J.P.; writing-original draft, K.X., H.D.; writing-review and editing, K.X., Q.D., J.L., Y.S., W.T., T.L., H.Z., T.S.; supervision and funding acquisition, K.X.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Xianfang Wu, Chengjiang Gao, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Dong, H., Pan, Z., Jiao, P. et al. Vesicle-associated membrane protein 5 is an intrinsic defense factor for embryonic stem cells against coronaviruses. Nat Commun 16, 6241 (2025). https://doi.org/10.1038/s41467-025-61655-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-61655-8
This article is cited by
-
VAMP proteins: molecular architects in disease pathogenesis and therapeutic innovation
Medical Oncology (2025)