Abstract
Infections caused by nontuberculous mycobacteria, such as Mycobacterium avium and Mycobacteroides abscessus, are becoming increasingly prevalent, and rising antibiotic resistance poses a significant clinical challenge. However, the mechanisms by which the host defense system controls these infections remain poorly understood. Here we show that the autophagy-related protein ATG7 in innate immune cells plays an essential role in controlling nontuberculous mycobacterial infection and protecting lung tissue from pathological inflammation. Patients with nontuberculous mycobacterial pulmonary disease exhibit reduced ATG7 expression in blood mononuclear cells and decreased ATG7 levels in necrotic lesions at disease sites. Mice lacking Atg7 in innate immune cells display elevated bacterial loads, excessive inflammation, mitochondrial damage, and multiple forms of cell death in the lungs, including pyroptosis, necrosis, and apoptosis. Notably, neutrophil infiltration in the lungs of these mice plays a key role in driving exacerbated inflammation and gasdermin E-associated cell death, which precede bacterial overgrowth. In vitro, Atg7-deficient macrophages exhibit impaired antimicrobial responses and reduced phagolysosomal fusion, but only modest increases in inflammation and cell death. These findings underscore the critical role of ATG7 in innate immune cells in orchestrating an effective host defense against nontuberculous mycobacterial lung infection by mitigating neutrophil-driven pathological inflammation and associated cell death.
Similar content being viewed by others
Introduction
Nontuberculous mycobacteria (NTM) are a diverse group of environmental microorganisms, distinct from Mycobacterium tuberculosis (Mtb) and its close relatives, among them M. leprae 1. NTM pulmonary diseases (PD) can afflict individuals with compromised or robust immune systems, posing a growing clinical concern2. The slowly growing M. avium-intracellulare complex predominates among pathogens responsible for NTM-PD, while rapidly growing Mycobacteroides abscessus (Mabc) complexes present significant multidrug resistance3,4. Existing challenges include treatment non-compliance due to severe adverse effects5, lack of vaccination, and precise diagnostics6. Despite the identification of several host risk factors, the mechanisms governing host-pathogen interactions are unclear3,7. Understanding host-pathogen interactions with Mabc, M. avium (Mav), and M. intracellulare (Mint) could revolutionize vaccination strategies and treatments for susceptible patient populations. However, much less is known about the host factor(s) and the mechanisms underlying protection against NTM-PD, especially in comparison to Mtb, a well-studied tuberculosis pathogen with extensive global research efforts aimed at improving treatment outcomes8.
Autophagy, a fundamental intracellular degradation pathway reliant on lysosomal processes, plays a crucial role in maintaining organelle integrity and quality control in response to various stresses, including infections9,10,11,12,13,14,15. Gene manipulation techniques have been instrumental in dissecting the roles of individual autophagy-related genes (ATGs), which participate in both canonical and non-canonical autophagy16. In the context of Mtb infections, several ATGs play intricate roles in modulating Mtb replication, pathological inflammation, and cell death17,18,19,20,21,22,23,24. Early studies by using Atg5 fl/flLysM-Cre+/− mice demonstrated that autophagy is required for Mtb restriction in primary cultured macrophages17. Additionally, autophagy was shown to mitigate excessive pulmonary inflammation in vivo by limiting neutrophil infiltration and interleukin (IL)−17-driven immune responses18. Further evidence suggests that Atg5 in innate immune cells, particularly polymorphonuclear neutrophils rather than alveolar macrophages, plays a protective role in preventing severe immunopathology driven by neutrophil-induced inflammatory responses during low-dose Mtb infection19. Moreover, Atg5 expression in CD11c+ cells (alveolar macrophages and dendritic cells) regulates the production of proinflammatory cytokines and chemokines during Mtb infection, as its absence leads to excessive neutrophil infiltration20. In a high-dose Mtb infection model that mimics active pulmonary tuberculosis, Atg5 in lung macrophages and dendritic cells modulates susceptibility to infection, prevents the accumulation of myeloid-derived suppressor cells, and safeguards T cell responses21. A recent study employing enhanced genetic depletion of Atg5, Atg16L1, or Atg7 through doubling of the LysMcre copy number revealed increased susceptibility to Mtb infection, impaired control of Mtb growth at the acute phase of infection, and heightened neutrophil infiltration in mice22. Notably, autophagy is required for regulating the mode of cell death during Mtb infection, preventing the transition from apoptosis to necrosis22. Recent studies have also highlighted the role of ATGs in human cells in response to Mtb infection. ATG5 knockout human cells demonstrated that ATG5 is essential for host protection against Mtb infection, not through its canonical autophagy function but through its non-canonical role in lysosomal membrane repair23. Similarly, CRISPR-Cas9-mediated deletion of ATG7 in human induced pluripotent stem cell-derived macrophages impaired intracellular Mtb restriction24. Additionally, the deletion of ATG14 in these cells underscored its critical role in facilitating the fusion of phagosomes with lysosomes24. While the growing body of evidence on ATGs functions in Mtb infection continues to expand, the translational insights, roles, and mechanisms of ATGs in modulating host defense against NTM infections remain largely unexplored.
To identify the role of ATGs in the context of NTM-PD, we revisited the transcriptome data (GSE290289) from our previous work25 and identified that ATG7 levels were significantly reduced in patients with NTM-PD compared to healthy controls (HCs). Additionally, Atg7 levels inversely correlated with proinflammatory cytokines/chemokines in peripheral blood mononuclear cells (PBMCs). In lung tissues from patients with NTM-PD, ATG7 expression was elevated in non-necrotic granulomatous lesions compared to necrotic ones. Using mice with innate immune cell-specific deletion of Atg7 (Atg7 fl/flLysM-Cre+/+, referred to as Atg7 conditional knockout; cKO), we demonstrated that ATG7 innate immune cells is essential for effective in vivo antimicrobial responses against various NTM species. Spatial RNA sequencing and functional assays revealed that Atg7 cKO lungs exhibited heightened inflammation, mitochondrial damage, oxidative stress, neutrophil infiltration, and increased levels of combined cell death during NTM infection. Furthermore, single-cell (sc) RNA sequencing, neutrophil depletion, and early-stage infection assays identified excessive neutrophil infiltration as a major contributor to bacterial growth, exacerbated pathological inflammation, and gasdermin (GSDM)-associated cell death in Atg7 cKO lungs during NTM infection. In vitro, Atg7 cKO macrophages exhibited impaired antimicrobial responses and defective phagolysosomal fusion compared to Atg7 f/f (conditional wild-type; Atg7 cWT) macrophages, although differences in inflammation and cell death were minimal. Our findings highlight the pivotal role of ATG7 in innate immune cells in mediating antimicrobial host defense against NTM-PD in vivo by mitigating neutrophil-driven pathological inflammation and cell death.
Results
ATG7 expression is depressed in PBMCs and necrotic lesions at disease sites of patients with NTM-PD
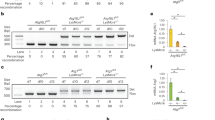
We revisited our previous nCounter assay data (GSE290289) from Mabc-infected patients (n = 11), M. massiliense (Mmass)-infected patients (n = 7), and HCs (n = 6)25. Both ATG10 and ATG7 showed a statistically significant reduction in NTM-PD patients compared to HCs (Supplementary Fig. 1a, b). As previous studies have demonstrated that ATG7 is essential for controlling Mtb pathogenesis in vivo22,24,26, we focused on exploring the involvement of ATG7 in NTM infection. Subsequent quantitative real-time PCR (qRT-PCR) analysis of ATG7 in PBMCs from patients infected with Mabc (n = 31) and Mmass (n = 22), as well as from HCs (n = 39)25 confirmed downregulation of ATG7 in the patients compared to HCs (Fig. 1a and Supplementary Table 1). Notably, ATG7 was negatively correlated with those of inflammatory cytokines/chemokines (IL1B, IL6, CXCL2, and CCL2) (Supplementary Fig. 1c).
a Dot-plot illustrating the relative ATG7 mRNA expression normalized to GAPDH in PBMCs from healthy controls (HCs, n = 39) and NTM patients (M. abscessus subsp. abscessus [Mabc], n = 31; M. abscessus subsp. massiliense [Mmass], n = 22). b, c Immunohistochemical staining of ATG7 in lung tissues from NTM patients (n = 12). ATG7 expression in histocytes (yellow triangle) and giant cells (black triangle) (b). ATG7 immunohistochemical expression in necrotic and non-necrotic region (c). d Atg7 cWT and Atg7 cKO mice were infected with Mabc-R (2 × 106 CFU, n = 4), Mav (1 × 107 CFU, n = 4), or Mint (1 × 107 CFU, Atg7 cWT n = 4; Atg7 cKO n = 3) for 21 days. In vivo bacterial loads in lung tissues were analyzed. e Atg7 cWT and Atg7 cKO mice were infected with ERFP-Mabc-R (2 × 106 CFU) for 7 days. Representative in vivo imaging of lung tissues from mice. f Atg7 cWT and Atg7 cKO mice were infected with Mabc-R (2 × 106 CFU) for 14 days, Mav (1 × 107 CFU), or Mint (1 × 107 CFU) for 21 days. Representative H&E images of lung tissues (left panel). Quantitative analysis of the inflamed area (right panel). e, f are representative of at least three independent experiments performed. Statistical significance was examined by Kruskal–Wallis test with Dunn’s multiple comparison test (a), and two-tailed Student’s t-tests (c, d, f). Data shown (means ± SEM) represent the combined results of triplicate from three independent experiments (c, d, f). Images are representative of at least three independent experiments (b, e, f). Un uninfected. Source data are provided as a Source Data file.
Immunohistochemical staining of ATG7 in tissue blocks from the clinical histopathological archives showed that the lesions contained well-formed granulomas of various sizes and interstitial inflammation (Fig. 1b and Supplementary Table 2). Immunohistochemical staining revealed that the expression level of ATG7 was significantly lower in necrotic areas compared to non-necrotic areas in NTM-PD patients (Fig. 1b, c). In particular, ATG7 expression was evident in epithelioid histiocytes and giant cells, both of which are derived from activated macrophages (Fig. 1b). This suggests decreased ATG7 expression in human PBMCs and at necrotic lesions in NTM-PD patients.
Deletion of Atg7 in innate immune cells suppress antimicrobial responses during NTM infection in vivo
Given the crucial role of macrophages in mycobacterial infection27 and the observed inverse correlation between ATG7 and proinflammatory cytokines/chemokines in NTM-PD patients, we investigated the host protective function of ATG7 in NTM-PD by generating Atg7 cKO mice (Supplementary Fig. 2a, b). Overexposed Western blot analysis for ATG7 expression revealed a significant reduction in ATG7 levels, though not a complete loss. It is estimated that ~3.6% of the ATG7 detected in bone marrow-derived macrophages (BMDMs) from Atg7 cKO mice (Supplementary Fig. 2a).
We first explored the function of ATG7 in innate immune cells in the antimicrobial response to pulmonary infections caused by several strains of NTM, including a smooth strain of Mabc (Mabc-S), a rough strain of Mabc (Mabc-R), Mmass, Mav, or Mint. Following intranasal infection, the NTM loads significantly increased in the lungs of Atg7 cKO mice compared to Atg7 cWT mice (Fig. 1d and Supplementary Fig. 2c), along with elevated bacterial proliferation signals (Fig. 1e) and a higher number of granulomatous lesions (Fig. 1f ). These findings underscore the necessity of ATG7 in innate immune cells for controlling bacterial growth during NTM infection in vivo.
Upregulation of proinflammatory cytokines/chemokines in lung tissues from Atg7 cKO mice during NTM infection
Observing heightened bacterial load and exacerbated lung pathology in Atg7 cKO mice compared to Atg7 cWT mice during infection, we investigated the inflammatory target gene(s) modulated by ATG7 in NTM infection. Spatial transcriptomic analysis on formalin-fixed paraffin embedded (FFPEs) tissues from Atg7 cWT and Atg7 cKO mice at 21 days post-infection (dpi) following Mav infection was performed using the 10× Genomics Visium platform (Fig. 2a and Supplementary Fig. 3a). Unsupervised Louvain clustering yielded 18 clusters, which were partitioned into 8 major nodal clusters. Clusters 1–4 exhibited enrichment in Atg7 cWT lungs, and clusters 5–8 in Atg7 cKO lungs. Clusters 2 and 5 accounted for the largest proportions in any given sample (Fig. 2b,c). Cluster 2 revealed high expression of Pecam1, a canonical marker of endothelial cells, and cluster 5 indicated upregulation of innate immune marker genes such as Cd68 and Itgam (Fig. 2d). A pathway enrichment analysis (GO Biological Process) analysis confirmed that cluster 5 is significantly enriched in innate-immunity signaling associated with the induction of inflammation (Supplementary Fig. 3b). To identify cell types within the clusters, particularly in cluster 5, which is inferred to be innate immune cells, we conducted a deconvolution analysis using scRNA sequencing data (GSE151974)28 and compared the probabilities of the presence of 13 cell types across the 8 major nodal clusters (Fig. 2e and Supplementary Fig. 3c). According to the heatmap, the cell-type composition of cluster 5, which distinguished it from other clusters, included alveolar macrophages (Alv macrophages), interstitial macrophages (Int macrophages), neutrophils, and monocytes (Fig. 2e). These four cell types were present in greater numbers in Atg7 cKO lungs than in Atg7 cWT lungs after Mav infection. Specifically, neutrophils were not visible in Atg7 cWT lungs, but were present in significantly higher numbers in Atg7 cKO lungs, suggesting heightened neutrophil infiltration (Supplementary Fig. 3d).
a–g Spatial transcriptomics data obtained lung tissues from Mav-infected (1 × 107 CFU) Atg7 cWT and Atg7 cKO mice at 21 dpi. a Representative H&E staining images of spatial transcriptomics data generated using the 10× Genomics Visium platform. b UMAP plot of Louvain clustering with 6971 spots from 2 spatial transcriptomics samples. Hierarchical clustering tree divided into major nodes, each represented by a distinct color. c UMAP plot of 8 major nodal clusters and comparison of cluster composition in each sample. UMAP clusters are mapped into spatial context. d Heatmap of the expression of cell type specific gene signatures of epithelial cells, endothelial cells, fibroblasts, myeloid cells, and lymphoid cells among 8 major nodal clusters. e Heatmap comparing expression profiles between the 8 major nodal clusters and the 13 cell types resulting from deconvolution with scRNA sequencing data (GSE151974). f Pathway enrichment analysis (GO Biological Process) comparing total spatial transcriptomic spots between Atg7 cWT (n = 3152 spots) and Atg7 cKO (n = 3798 spots), showing the top enriched pathways per group. Enriched GO terms were identified using one-sided hypergeometric tests, and p-values were adjusted for multiple comparisons using the Benjamini-Hochberg method. g UMAP plots of the module scores for the inflammatory pathway (HALLMARK: INFLAMMATORY_ RESPONSE), mapped to both the total and cluster five regions within the spatial context, comparing Atg7 cWT and Atg7 cKO. Violin plot shows a significant difference in cluster 5, assessed using a two-sided unpaired Welch’s t-test (P < 2.2 × 10−16; Atg7 cWT: n = 490 spots; Atg7 cKO: n = 1,275 spots). a is representative of at least three independent experiments performed. Alv Alveolar, Int interstitial, NK natural killer. Source data are provided as a Source Data file.
Pathway enrichment analysis (GO biological process) comparing Atg7 cWT and Atg7 cKO lungs revealed a significant upregulation of cytokine/chemokine activity and related signaling pathways in Atg7 cKO lungs, indicating hyperactivation of proinflammatory responses (Fig. 2f ). Module scores confirmed higher values for the inflammatory pathway in total clusters and cluster 5, indicative of an innate immune cell area, in Atg7 cKO lungs compared to Atg7 cWT lungs (Fig. 2g). Further analysis of cluster 5 spots in the spatial transcriptomes corroborated these findings, showing significantly elevated module scores for the inflammatory pathway in Atg7 cKO lungs (P < 0.0001; Fig. 2g). These results were further supported by cytokine/chemokine gene expression analyses and the module scores of three inflammatory pathways (HALLMARK) (Supplementary Fig. 4a–c).
Bulk RNA sequencing analysis demonstrated significant differences in the levels of inflammatory cytokines/chemokines between Atg7 cWT and Atg7 cKO lungs after Mav infection (Fig. 3a). qRT-PCR confirmed a substantial increase in the mRNA levels of these inflammatory cytokines/chemokines and mediators in the lung tissues of Atg7 cKO compared to Atg7 cWT mice (Fig. 3b-d). Consistently, the levels of IL-1β and IL-6 in lung lysate supernatants were significantly elevated in Atg7 cKO mice compared Atg7 cWT mice (Fig. 3e, f). These findings implicate ATG7 in controlling the expansion of innate immune cells, including neutrophils, and pathological inflammation in lung tissues, during pulmonary NTM infections.
a Atg7 cWT and Atg7 cKO mice were infected with Mabc-R (2 × 106 CFU) for 3 days. A heatmap analysis was performed to visualize the expression of inflammatory genes in the mouse lung tissues. The color scale bar of the heatmap represents z-scores of normalized gene counts. b, c Atg7 cWT and Atg7 cKO mice were infected with Mabc-R (2 × 106 CFU) for 3 days (n = 3 for Ninjurin 1; n = 6 for Tnf, Il1b, and Mmp12). Relative mRNA expression of various inflammatory chemokines/cytokines in the lung tissues (b), and of S100a8 and S100a9 (c), were measured (d) Atg7 cWT and Atg7 cKO mice were infected with Mav (1 × 107 CFU) for 3 days (n = 6 mice per group). Relative mRNA expression of various inflammatory chemokines/cytokines in the lung tissues. e, f Atg7 cWT and Atg7 cKO mice were infected with Mabc-R (2 × 106 CFU, 21 dpi, n = 4), Mabc-S (1 × 107 CFU, 7 dpi, n = 3), Mav (1 × 107 CFU, 21 dpi, n = 4), or Mint (1 × 107 CFU, 21 dpi, Atg7 cWT: n = 4; Atg7 cKO: n = 2). The protein levels of IL-1β (e) and IL-6 (f) in lung homogenates were analyzed by ELISA. Statistical significance was determined by two-sided unpaired t-test with Mann–Whitney U-test (b; Ccl2, Cxcl2, Ccl5, Cxcl5, Tnf, Il1b, and Mmp12), two-sided unpaired Welch’s t-test (b; Ninjurin1), and two-tailed student’s t-test (c–f). Data shown (means ± SEM) represent the combined results of three independent experiments (b–f). dpi days post-infection. Source data are provided as a Source Data file.
Atg7 deficiency in innate immune cells heightens oxidative stress and mitochondrial damage in the lungs during NTM infection
Excessive mitochondrial damage and dysregulated redox homeostasis exacerbate lung inflammation during severe infections29. Given the amplified inflammatory responses in Atg7 cKO lungs, as compared to Atg7 cWT lungs, we investigated whether oxidative stress and mitochondrial damage were heightened in the Atg7 cKO lungs. To pinpoint the spatial location of oxidative stress, we computed module scores for the reactive oxygen species (ROS) pathway (HALLMARK) and assessed them by means of UMAP clusters and spatial transcriptome spots (Fig. 4a). High ROS pathway module scores were predominantly present in cluster 1 (likely epithelial area) in Atg7 cWT lungs, whereas in Atg7 cKO lungs, scores were elevated in cluster 5 (inferred innate immune cell area). Subsequent analysis of cluster 5 confirmed a significant increase of the ROS pathway module score in Atg7 cKO lungs (P < 0.0001; Fig. 4a).
a Atg7 cWT and Atg7 cKO mice were infected with Mav (1 × 107 CFU) for 21 days. UMAP plots of the module scores for the ROS pathway (HALLMARK_ REACTIVE_ OXYGEN_SPECIES_PATHWAY), mapped to both the total and cluster 5 regions within the spatial context, comparing lung tissues from mice. Violin plot shows a significant difference in cluster 5, assessed using a two-sided unpaired Welch’s t-test (P < 2.2 × 10−16; Atg7 cWT: n = 490 spots; Atg7 cKO: n = 1275 spots). b Atg7 cWT and Atg7 cKO mice were infected with Mav (1 × 107 CFU) for 21 days. Lung tissues were stained with MitoSOX (red) and DAPI (for nuclei; blue). Representative immunofluorescence images. c, d Atg7 cWT and Atg7 cKO mice were infected with Mabc-R (2 × 106 CFU) for 14 days or Mav (1 × 107 CFU) for 21 days. ROS levels in lung tissues were visualized using DHE (red). Representative immunofluorescence images (c) and quantitative analysis of DHE relative intensity (d, n = 12; independent biological replicates). e, f Atg7 cWT and Atg7 cKO mice were infected with Mabc-R (2 × 106 CFU) or Mav (1 × 107 CFU) for 10 days. Representative TEM images of damaged mitochondria in lung tissues from mice (e). Red stars indicate damaged mitochondria. The percentage of damaged mitochondria (Mav: n = 10; Mabc-R: n = 17; independent biological replicates) and the quantification of circularity of the mitochondria (Mav: n = 27; Mabc-R: n = 35; independent biological replicates) (f). Statistical significance was determined by two-sided unpaired t-test with Mann-Whitney U-test (d, f). Data shown (means ± SEM) represent the combined results of triplicate experiments from three independent experiments (c, f). Images are representative of three independent experiments (b, c, e). Un uninfected, arb. units arbitrary unit. Source data are provided as a Source Data file.
Alarmins such as S100A8/A9 are accompanied by vascular inflammation and oxidative stresses30. S100a8/a9 expression was significantly increased in areas with elevated ROS pathway module scores in Atg7 cKO lungs (Supplementary Fig. 5a). Because heightened oxidative stress induces endoplasmic reticulum (ER) stress31, we analyzed the expression levels of genes related to ER stress. The expression levels of the heat shock protein family A (Hsp70) member 5 (Hspa5) marker gene (a central regulator of ER stress) and most ER stress genes were increased in the innate immune cell cluster 5 of Atg7 cKO lungs (Supplementary Fig. 5b).
We next assessed mitochondrial superoxide and total ROS levels in lung tissues from Atg7 cWT and Atg7 cKO mice during infection. MitoSOX Red—a fluorescent dye specific for mitochondrial (mt) ROS32—staining revealed a significant increase in mtROS levels in Atg7 cKO lungs compared to Atg7 cWT lungs after infections with Mav or Mabc (Fig. 4b and Supplementary Fig. 5c). Dihydroethidium (DHE) staining showed upregulated total ROS levels in Atg7 cKO lungs post-infection (Fig. 4c, d). Excessive mtROS levels, driven by a cycle of ROS-induced ROS release, accelerate mitochondrial destruction, often triggering a hyperinflammatory state and disrupting intracellular homeostasis in pathological conditions33,34. Transmission electron microscopy (TEM) depicted notable mitochondrial damage, including mitochondrial swelling, lengthening, and disrupted cell membranes35, in lung tissues from Atg7 cKO mice after Mabc-R or Mav infection (Fig. 4e, f and Supplementary Fig. 5d). These findings suggest that Atg7 cKO lungs undergo significant mitochondrial damage and oxidative stress, accompanied by neutrophil infiltration during NTM infections.
Atg7 deficiency in innate immune cells is associated with cell death in vivo at a time of increased inflammation and NTM burden
Deletion of several ATGs is associated with increased cell death in human and murine cells infected with Mtb22,24. To verify this, we compared the module scores of three cell death pathways—pyroptosis, necrosis, and apoptosis—between Atg7 cWT and Atg7 cKO lungs using spatial transcriptome profiles. Specifically, cluster 5, representing an innate immune cell area, showed significantly higher cell death levels in Atg7 cKO lungs compared to Atg7 cWT lungs, as demonstrated by statistical analysis and the spatial mapping of pathway module scores and gene expression at individual spots (Fig. 5a and Supplementary Fig. 6a, b). Necroptosis was also confirmed through pathway analysis and the expression of key necroptosis-related genes, yielding consistent findings (Supplementary Fig. 7a, b). Propidium iodide (PI) staining revealed increased cell death in the inflammatory lesion lung tissues of Atg7 cKO compared to Atg7 cWT lungs during Mav or Mabc-R infection (Fig. 5b and Supplementary Fig. 8a).
a Atg7 cWT and Atg7 cKO mice were infected with Mav (1 × 107 CFU) for 21 days. Violin plots of module scores for three cell death pathways (REACTOME: PYROPTOSIS; REGULATED NECROSIS; KEGG: APOPTOSIS), comparing lung tissues from mice in cluster 5 between Atg7 cWT (n = 490 spots) and Atg7 cKO (n = 1275 spots). Statistical significance was assessed using a two-sided unpaired Welch’s t-test (P < 2.2 × 10−16). b Atg7 cWT and Atg7 cKO mice were infected with Mav (1 × 107 CFU) for 21 days. Immunofluorescence analysis of Ly6G+PI+ cells in the lung tissues of mice. Atg7 cWT and Atg7 cKO mice were infected with Mabc-R (2 × 106 CFU) for 21 days (c) or Mabc-S (1 × 106 CFU) for 7 days (d). Representative immunoblotting of IL-1β, caspase-1, GSDMD, caspase-3, and GSDME in the lung tissues of mice. e Relative quantification of GSDMD-NT and GSDME-NT, normalized to GSDMD-FL and GSDME-FL, respectively. (uninfected samples: n = 6; infected samples: n = 9) f Atg7 cWT and Atg7 cKO mice were infected with Mabc-R (2 × 106 CFU) or Mav (1 × 107 CFU) for 10 days. Representative TEM images of pyroptotic cell death in mouse lung tissues. Yellow arrows indicate cell swelling and membrane rupture. g Atg7 cWT and Atg7 cKO mice were infected with Mav (1 × 107 CFU) or Mabc-R (2 × 106 CFU) for 10 days. Representative TEM images of neutrophil cell death in mouse lung tissues. Yellow circles indicate granule,s and red star indicates nuclear membrane break down. Statistical significance was determined by one-way ANOVA (e). Data shown (means ± SEM) represent the combined results of duplicate from three independent experiments (e). Images are representative of three independent experiments (b–d, f, g). Un uninfected, N nucleus, NT N-terminal, FL full length. Source data are provided as a Source Data file.
Disrupted mitochondrial and redox balance can trigger cell death pathways mediated by GSDM-related signaling36,37,38. Given the involvement of caspases, a family of conserved cysteine proteases, in the regulation of apoptosis and pyroptosis36,37,38, we measured the levels of caspases (caspase-1 and -3), pro-IL-1β, GSDMD, and GSDME in lung tissues of Atg7 cWT and Atg7 cKO mice after Mabc or Mav infection. Western blotting revealed that Atg7 cKO lungs upregulated the expression levels of GSDME-N-terminal (NT)/GSDME-full length (FL) and GSDMD-NT/GSDMD-FL compared to Atg7 cWT lungs during Mabc or Mav infection (Fig. 5c–e and Supplementary Fig. 8b). Moreover, TEM showed morphological changes characteristic of cell death in the Atg7 cKO lung tissues (Fig. 5f, g), such as balloon-like bubbles, cell swelling, and membrane rupture, which are typical features of pyroptosis39,40,41. These findings suggest that Atg7 deficiency in innate immune cells is associated with heightened multiple cell death pathways, with a notable increase in GSDM-associated pyroptotic cell death in lung tissues during NTM infections.
Neutrophils in innate immune cell cluster of Atg7 cKO lungs constitute a major population associated with hyperinflammation and combined cell death during NTM infection
To identify the innate immune cell types involved in antimicrobial host defense against NTM infection in Atg7 cKO lungs, we performed scRNA sequencing using 10X Genomics platform on mouse lung cells under experimental conditions identical to those used for spatial transcriptomic analysis. After filtering out low-quality cells and processing the data with Seurat, we obtained a total of 25,040 cells, which were grouped into 37 unsupervised Louvain clusters (Supplementary Fig. 9a). These clusters were classified into 22 global cell types based on the expression of canonical marker genes, including five innate immune cell types: alveolar macrophages, interstitial macrophages, neutrophils, monocytes, and dendritic cells. The identities of these innate immune cell populations were further validated using additional canonical markers (Fig. 6a, b and Supplementary Fig. 9b, c).
a–e Atg7 cWT and Atg7 cKO mice infected with Mav (1 × 107 CFU) for 21 days. scRNA sequencing data were obtained under the same experimental conditions as the spatial transcriptomics data from lung tissues. a UMAP plot of all scRNA sequencing data, comprising a total of 25,040 cells from two samples: Atg7 cWT and Atg7 cKO. The plot shows cells from 22 distinct cell types (left), with an enlarged view of 5 innate immune cell types (right). b Dot Plot of marker gene expression for innate immune cell populations. c, d Violin plots of the module scores for the inflammatory pathway and three cell death pathways (same as Figs. 2g and 5a) in innate immune cell populations, comparing Atg7 cWT (n = 3457 cells) and Atg7 cKO (n = 3389 cells). The distribution of module scores across all cell types is shown on the UMAP plot. Statistical significance was assessed using a two-sided unpaired Welch’s t-test for each plot (P = 7.7 × 10−5, P = 5.8 × 10−8, P = 0.025, P = 3.5 × 10−12, listed in the order of plots). e Violin plots across innate immune cells in Atg7 cKO (Int Macrophages, n = 1512 cells; Alv Macrophages, n = 268 cells; Neutrophils, n = 419 cells; Monocytes, n = 753 cells; DCs, n = 437 cells), showing the module scores for the pathways from Fig. 6c, d. For multi-group comparisons, normality was assessed using the Shapiro-Wilk test, and homogeneity of variance was evaluated using Levene’s test. One-way ANOVA followed by Tukey’s HSD test was used for post-hoc pairwise comparisons. Exact p-values for each plot are provided in the Source data file. AT alveolar type cell, Ciliated ciliated cell, Cap capillary endothelial cell, Vein venous endothelial cell, Lymph lymphatic endothelial cell, Int interstitial, Alv alveolar, DC dendritic cell. Source data are provided as a Source Data file.
scRNA sequencing analysis revealed a significant increases in module scores for both inflammatory responses and three cell death pathways—pyroptosis, necrosis, and apoptosis—in innate immune cells from Atg7 cKO lungs compared to Atg7 cWT lungs (Fig. 6c, d). Neutrophils formed the most prominent cluster on the UMAP (Fig. 6c, d). Additionally, Gsdme, a key gene associated with neutrophil-lytic pyroptosis42, was upregulated in neutrophils (Supplementary Fig. 10a). Among the five distinct innate immune cell populations, neutrophils exhibited the highest levels of inflammatory response and cell death pathway activation (Fig. 6e). Additionally, ROS levels were elevated in both neutrophils and alveolar macrophages (Supplementary Fig. 10b). These findings suggest that neutrophils, among innate immune cells, are primarily associated with hyperinflammation and increased combined cell death, including Gsdme-associated cell death, in Atg7 cKO lung tissues during infection.
Neutrophils in Atg7 cKO lungs contribute to defective antimicrobial responses, exaggerated pathological inflammation, and cell death during NTM infection
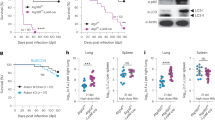
Based on the findings of scRNA sequencing analysis, we then examine whether neutrophil infiltration was increased in Atg7 cKO lung tissues during infection. Flow cytometry and immunofluorescence microscopy revealed a notable increase in the infiltration of Ly6G+ neutrophils in the lung tissues of Atg7 cKO mice compared to Atg7 cWT mice (Fig. 7a, Supplementary Fig. 11a, b). During infection, neutrophils infiltrating lung tissue undergo degranulation and phagocytosis, releasing neutrophil extracellular traps (NETs) composed of decondensed chromatin, myeloperoxidase (MPO), and antibacterial proteins43,44. Additionally, Atg7 cKO lungs exhibited a significant increase in the expression of citrullinated histone H3 and MPO compared to Atg7 cWT mice during NTM infection (Fig. 7b, c). Furthermore, the upregulated level of chromatin structures was visualized by TEM analysis in Atg7 cKO lung cells during Mav or Mabc-R infection (Fig. 7d).
a Atg7 cWT and Atg7 cKO mice were infected with Mabc-R (2 × 106 CFU) for 10 days. Representative flow cytometry plots (left) show live CD45+MerTK-CD64- cells gated for Ly6G+ neutrophils in lung tissues. Quantification of Ly6G+ neutrophils is shown on the right (n = 4). b, c Atg7 cWT and Atg7 cKO mice were infected with Mabc-R (2 × 106 CFU) for 14 days. Representative 3D images of CitH3 (green), MPO (red), and DAPI (blue) in lung tissues were generated using Aivia (b). The percentage of Ly6G+ cells that were also CitH3+ (n = 21, independent biological replicates) (c). d Atg7 cWT and Atg7 cKO mice were infected with Mabc-R (2 × 106 CFU) or Mav (1 × 107 CFU) for 10 days. Representative TEM images of decondensed chromatin in lung tissues. e–h Atg7 cWT and Atg7 cKO mice were infected with Mabc-S (1 × 107 CFU) for 7 days. Mice were administered with anti-IgG2a or anti-Ly6G antibodies (Ab) via daily intraperitoneal injection until sacrifice. In vivo bacterial loads in lung tissues (e Atg7 cWT: n = 3; Atg7 cKO: n = 4). IL-6 (f IgG2a: n = 6, Ly6G: n = 8) and TNF (g n = 4) levels in lung tissues were measured by ELISA. Expression of GSDME was assessed by immunoblotting in lung tissue lysates. Dot plots show quantification of the GSDME-NT normalized to GSDME-FL, based on Western blot (h). Statistical significance was determined by a two-sided unpaired t-test with Mann–Whitney U-test (a, c), and two-tailed t-test (e–g). Data shown (means ± SEM) represent the combined results of two or three independent experiments (a, c, e, f, g). Images are representative of three independent experiments (b, d). dpi days post infection, CitH3 citrullinated histone H3, NTN-terminal, FL full length, ONM outer nuclear membrane, INM inner nuclear membrane. Source data are provided as a Source Data file.
To examine whether neutrophils were the most prominent contributors to increased inflammation and cell death in Atg7 cKO lung tissues, we next conducted neutrophil depletion experiments and observed that increased neutrophil infiltration exacerbated inflammatory and cell death responses in Atg7 cKO lungs during infection. Treatment of Atg7 cKO mice with anti-neutrophil antibodies (anti-Ly6G Ab) significantly reduced Mabc and Mav loads (Fig. 7e and Supplementary Fig. 12a), granulomatous lesions (Supplementary Fig. 12b), and IL-6 and TNF-α levels (Fig. 7f, g and Supplementary Fig. 12c) in lung tissues. In contrast, Atg7 cWT mice showed no reduction in bacterial burden, histological changes, or inflammatory cytokine production following the same treatment. Notably, anti-Ly6G Ab treatment significantly suppressed the cleavage of GSDME in Atg7 cKO lungs, but not in Atg7 cWT lungs, during Mabc-S infection (Fig. 7h). These findings suggest that excessive neutrophil responses play a pivotal role in the impaired host defense observed in Atg7 cKO mice.
Given these findings, we further questioned whether Atg7 is directly required for neutrophil-induced immunopathology, which could influence host defense against NTM infection. To explore this, we performed bacterial load assay, cell death analysis, and inflammatory response analyses at early stages of infection (1–3 dpi). By 3 dpi, Atg7 cKO mice exhibited significantly higher bacterial loads compared to Atg7 cWT mice (Supplementary Fig. 13a). However, at 1 dpi, no significant differences in Mav growth were observed between Atg7 cKO and Atg7 cWT mice (Supplementary Fig. 13b). Interestingly, inflammatory cytokine levels (Supplementary Fig. 13c), neutrophil infiltration (Supplementary Fig. 13d), and GSDME-associated cell death (Supplementary Fig. 13e) were already elevated in Atg7 cKO lungs at 1 dpi following Mav infection. These findings suggest that the exaggerated neutrophil infiltration, which drives inflammation and cell death in Atg7 cKO mice, precedes bacterial overgrowth. This indicates that early immune dysregulation may be a key driver of the phenotypes observed during NTM infection in vivo.
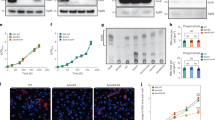
ATG7 is involved in phagolysosomal maturation and controls the intracellular NTM growth and inflammation in macrophages in vitro
Because ATG7 is a core ATG protein45, we investigated whether Atg7 deficiency in macrophages affects the trafficking of phagosomes into autophagosomal and lysosomal structures during Mabc infection. Upon infecting Atg7 cWT and Atg7 cKO BMDMs with green fluorescence protein (GFP)-expressing Mabc-S or Mabc-R, the colocalization level of Mabc-R/-S with autophagosomes was significantly reduced in Atg7 cKO BMDMs compared to Atg7 cWT BMDMs (Fig. 8a–c). Notably, LC3 puncta formation decreased in Atg7 cKO BMDMs during Mabc-R or Mabc-S infection (Fig. 8a–c). TEM supported these observations, demonstrating a decreased presence of Mabc in autophagosomes in Atg7 cKO BMDMs (Fig. 8d). Next, we investigated whether ATG7 is essential for phagolysosomal fusion of Mabc-S or Mabc-R in macrophages during infection. The co-localization of Mabc-R with LAMP1-positive lysosomes was significantly decreased in Atg7 cKO BMDMs compared to Atg7 cWT BMDMs (Fig. 8a, c, e). A representative image of LC3 and LAMP1 in uninfected control Atg7 cWT and Atg7 cKO BMDMs is shown in Supplementary Fig. 14.
a–d BMDMs from Atg7 cWT and Atg7 cKO mice were infected with GFP-Mabc-S (MOI of 5) or GFP-Mabc-R (MOI of 5) for 6 h (a–c, e) or 18 h (d). Cells were stained with LC3 (red) and LAMP1 (blue) and visualized by confocal microscopy. Representative confocal images (a, c). Mabc and LC3 colocalization was assessed by calculating Pearson’s coefficient (b Un: n = 10; Mabc-S and Mabc-R: n = 12; independent biological replicates). Mabc and LAMP1 colocalization was assessed by calculating Pearson’s coefficient (e Un: n = 10, Mabc-S and Mabc-R: n = 12, independent biological replicates). Representative TEM images showing autophagosome in of BMDMs (d). Statistical significance was determined by two-tailed unpaired Student’s t-test or unpaired Student’s t-test with Welch’s correction (b, e). Data shown (means ± SEM) represent the combined results of triplicates from three independent experiments (b, e). Images are representative of three independent experiments (a, c, d). Un uninfected. Source data are provided as a Source Data file.
We performed an intracellular colony-forming unit (CFU) assay to assess the host-defense responses of Atg7 cWT and Atg7 cKO macrophages to Mabc-R, Mabc-S, Mav, and Mint. The number of NTM was significantly higher in Atg7 cKO BMDMs compared to Atg7 cWT BMDMs (Fig. 9a–d). Additionally, Mabc-induced IL-6 levels were slightly elevated in Atg7 cKO BMDMs compared to Atg7 cWT BMDMs (Fig. 9e). Furthermore, when primed with supernatants from lung lysates of Mabc-R-infected mice or Pam3CSK4, a TLR2 ligand, the increase in proinflammatory cytokine production was greater in Atg7 cKO macrophages compared to Atg7 cWT (Fig. 9f and Supplementary Fig. 15a, b). These findings were further validated through knockdown experiments using human primary monocyte-derived macrophages (hMDMs), which were transduced with lentiviral small hairpin RNA (shRNA) targeting hATG7 (shATG7) or a nonspecific shRNA control (shNS) prior to infection with the Mabc-R or Mabc-S. Knocking down ATG7 significantly increased the intracellular survival of Mabc-R or Mabc-S (Fig. 9g, h). Additionally, ATG7 knockdown in hMDMs led to a marked increase in the production of IL-6 and IL-1β, both in uninfected cells and during infection with Mabc-R (Fig. 9i) or Mav (Fig. 9j). Overall, these findings suggest that ATG7 is required for LC3+ autophagosome formation and phagolysosomal fusion, and regulation of intracellular NTM survival and inflammatory responses in macrophages during infection in vitro.
a–d Atg7 cWT and Atg7 cKO BMDMs were infected with Mabc-R (MOI of 1 or 3, a), Mabc-S (MOI of 1 or 5, b), Mav (MOI of 1 or 3, c), or Mint (MOI of 1 or 3, d) for the indicated times. Intracellular survival of NTM was determined by CFU assay. e Atg7 cWT and Atg7 cKO BMDMs were infected with Mabc-R (MOI of 3) for the indicated times. IL-6 levels were measured by ELISA. f Atg7 cWT and Atg7 cKO BMDMs were pretreated with lung homogenate supernatants (lung sup) for 4 h, followed by infection with Mabc-S (MOI of 1) for the indicated times. IL-6 levels were measured by ELISA. g Human primary MDMs were infected with Mabc-R (MOI of 3) for 1 day and then transduced with lentivirus expressing shNS or shATG7 for 36 h. Transduction efficiency was evaluated by qRT-PCR. h Human primary MDMs were transduced with lentivirus expressing shNS or shATG7, followed by infection with Mabc-R (MOI of 3; left) or Mabc-S (MOI of 3; right). Bacterial burden was assessed by CFU assay at 1 or 2 dpi, respectively. i, j Human primary MDMs were transduced with lentivirus expressing shNS or shATG7 and then infected with Mabc-R (MOI of 5, i) or Mav (MOI of 5, j) for 12 h or 24 h, respectively. The concentrations of IL-1β and TNF were measured using ELISA. Statistical significance was determined by two-sided one-way ANOVA (a–d, f, j) or two-tailed Student’s t tests (e, g, h). Data shown (means ± SEM) represent the combined results of triplicates from three independent experiments. dpi days post infection, NS non-specific, sup supernatant. Source data are provided as a Source Data file.
Atg7 deficiency in macrophages marginally affects mitochondrial function and host-cell survival during NTM infection in vitro
We next assessed the impact of Atg7 deficiency on mitochondrial mitophagy, mtROS generation, and cell death in macrophages during NTM infection. Mitochondrial colocalization with LC3 was significantly decreased in Atg7 cKO BMDMs compared to Atg7 cWT BMDMs during Mabc infection (Fig. 10a, b and Supplementary Fig. 16a, b). However, mitochondrial colocalization with lysosomes was still detectable in Atg7 cKO BMDMs, though it was substantially reduced compared to Atg7 cWT BMDMs during Mabc-S and Mabc-R infections (Fig. 10a, b and Supplementary Fig. 16c, d).
a, b BMDMs from Atg7 cWT and Atg7 cKO mice were infected with Mabc-R (MOI of 5) for 6 h. Cells were stained with mitotracker (red), LC3 (green), and LAMP1 (green). Representative confocal images (a). Mitotracker/LC3 and mitotracker/LAMP1 colocalization were assessed by calculating Pearson’s coefficient (b, n = 10; independent biological replicates). c, d BMDMs from Atg7 cWT and Atg7 cKO mice were infected with Mabc-S (MOI of 5) or Mabc-R (MOI of 5) for 6 h. Cells were stained with MitoSOX (red) and DAPI (for nuclei). Representative confocal images (c). Average mean fluorescence intensities (MFI) of MitoSOX (d, n = 12; independent biological replicates). e Atg7 cWT and Atg7 cKO BMDMs were infected with Mabc-R (MOI of 1) for 96 h. Annexin V/PI analysis was measured by flow cytometry. Statistical significance was determined by two-tailed unpaired Student’s t-test or unpaired Student’s t-test with Welch’s correction (b, d), Welch’s ANOVA (P < 0.0001), followed by Dunnett’s T3 multiple comparisons test (d), and a two-tailed Student’s t-test (e). Data shown (means ± SEM) represent the combined results of triplicates from three independent experiments (b, d, e). Images are representative of three independent experiments (a, c). Un uninfected. Source data are provided as a Source Data file.
We further assessed mtROS generation in Atg7 cWT and Atg7 cKO macrophages during Mabc infection and observed a greater increase in mtROS levels in Atg7 cKO macrophages both before and after Mabc-S and Mabc-R infections (Fig. 10c, d). Regarding cell death, no significant differences were observed between Atg7 cWT and Atg7 cKO BMDMs until 72 h of infection. However, at 96 h of Mabc-R infection, cell death (PI+) was slightly but significantly increased in Atg7 cKO BMDMs compared to Atg7 cWT BMDMs, indicating increased cell death at later time points (Fig. 10e). No cleavage of GSDME-NT or caspase-3 was detected in either Atg7 cWT or Atg7 cKO BMDMs up to 72 h after Mabc-R infection (Supplementary Fig. 17). Overall, our findings suggest that Atg7 deficiency in macrophages has a marginal effect on mitochondrial function and host-cell survival during NTM infections in vitro.
Discussion
Our findings highlight the essential role of ATG7 in innate immune cells in defending against NTM-PD (Supplementary Fig. 18). Previously, we observed increased inflammatory cytokine/chemokine expression in PBMCs from NTM-PD patients25. In this study, ATG7 expression was significantly reduced in PBMCs from NTM-PD patients compared to HCs and was inversely correlated with inflammatory cytokine and chemokine levels. Additionally, ATG7 expression was diminished in necrotic lesions at disease sites in patients with NTM-PD. Further investigations in a mouse infection model underscore the critical role of ATG7 in innate immune cells in controlling bacterial loads and limiting excessive pathological inflammation during NTM infections. Atg7 cKO mice exhibited elevated levels of IL-6, IL-1β, chemokines, and S100a8/a9 in lung tissues, particularly within innate immune cell clusters, during infections. The role of ATG7 in regulating inflammatory responses has been explored in various disease models. Studies have shown that ATG7 modulates inflammation in sepsis-induced acute lung injury46, experimental colitis47, and Klebsiella pneumonia-induced inflammatory responses in macrophages48. In the gut, deletion of Atg5 or Atg7 in intestinal antigen-presenting cells exacerbates intestinal inflammation and enhances Th17 responses49. However, in neuronal cells, Atg7 deletion protects against neuronal death and brain injury following neonatal hypoxia-ischemia50. Additionally, ATG7 has been implicated in ultraviolet irradiation-induced inflammation in keratinocytes and skin tumorigenesis51. These findings suggest that ATG7 regulates inflammation in a cell type-specific and context-dependent manner.
Additionally, Atg7 cKO mice displayed exacerbated mitochondrial damage, as well as increased oxidative and ER stresses, in lung tissues during various NTM infections. While mtROS typically aids anti-mycobacterial defense52, excessive generation due to mitochondrial damage can worsen infection and trigger harmful inflammatory and cytotoxic responses53. Our data suggest that dysfunctional mitochondria in Atg7 cKO lungs lead to elevated oxidative stress, manifested by excessive production of mtROS and cellular ROS. mtROS, when combined with cellular ROS, exacerbates tissue pathologies, inflammation, and cell death processes34,54. We previously demonstrated that scavenging mtROS significantly reduces the in vivo growth of Mabc-R and mitigates excessive pathological inflammation and mitochondrial damage in the lung tissues of infected mice55. Although the current study does not directly establish a link between increased oxidative stress and impaired antimicrobial defense in Atg7 cKO mice, our findings, together with previous data55, suggest that ATG7 in innate immune cells plays a crucial role in maintaining mitochondrial homeostasis, which may contribute to host defense against NTM-PD.
Our data uniquely demonstrate that innate immune cells from Atg7 cKO lungs undergo combined cell death, including pyroptosis, necrosis, and apoptosis during NTM infection—an aspect not previously reported in the context of Mtb infection. A previous study showed that depletion of Atg5, Atg7, or Atg16L1 in macrophages shifts the mode of cell death from apoptosis to a necrotic-like pattern —but not pyroptosis—during Mtb infection22. Notably, we did not observe significant mortality in either Atg7 cKO or control mice up to 60 days post-infection with Mabc or Mav. These differences in mortality may reflect the distinct virulence factors and pathologies associated with Mtb versus NTM strains. Our findings partially align with this study22 in that neutrophil infiltration and pathological inflammation were increased in Atg7 cKO lung tissues during infection. However, our study provides distinct evidence that Atg7 cKO lung tissues exhibit a heightened degree of combined cell death, particularly GSDME-associated cell death, across various NTM infections. GSDME-mediated cell death is linked to amplified inflammatory responses, contributing to the pathogenesis of infectious and inflammatory diseases. For instance, Ma et al. demonstrated that GSDME functions in neutrophil lytic death and amplifies inflammatory responses, thereby promoting pulmonary inflammation and lung injury 42. Additionally, cigarette smoking and lipopolysaccharide co-stimulation trigger caspase-3-induced GSDME cleavage, releasing damage-associated molecular patterns like S100A8/A9, which impairs defense against infections56. In brain cells, herpes simplex virus-2 infection induces ER stress-mediated GSDME cell death, releasing alarmins and triggering inflammatory responses57. Due to the increased ER stress and elevated alarmin S100a8/S100a9 observed in Atg7 cKO lungs, these factors may collectively contribute to drive enhanced GSDME-associated cell death during NTM infection.
Using scRNA sequencing analysis and neutrophil depletion experiments, we identified neutrophils as the primary drivers of increased bacterial loads, inflammatory responses, and GSDME-associated cell death in Atg7 cKO lungs during NTM infection. These findings partially align with previous studies on Mtb infection using various gene-deficient mouse models, further underscoring the role of neutrophils in immunopathology. For example, innate immune cells lacking Atg5 exhibit reduced control of Mtb infection due to excessive neutrophil-driven inflammation19. Similarly, neutrophil depletion in Sirtuin 3-deficient mice enhances host protection and mitigates lung pathological inflammation during Mtb infection55. In chronic tuberculosis models, neutrophil depletion improves Mtb control, although it has no significant effect during acute infection58. Additionally, immune-responsive gene 1 expression protects against Mtb infection by mitigating neutrophil-mediated immunopathology59. In a primate model, neutrophils play a key role in shaping granuloma dynamics, facilitating local dissemination and Mtb spread60. A recent study further demonstrated that, upon encountering type I interferon, neutrophils release NETs, promoting necrosis and granulomatous caseation61. These findings, along with our data, highlight the critical role of neutrophil-mediated immunopathology in the pathogenesis of both Mtb and NTM infections in a context-dependent manner. Moreover, our results raise important questions about causality—specifically, the potential links between bacterial growth, neutrophil infiltration, excessive inflammation, and cell death in the context of Atg7 deficiency in innate immune cells during NTM infection. Notably, even at 1 dpi, when no significant differences in NTM growth were observed, Atg7 cKO lungs exhibited increased neutrophil infiltration, heightened inflammation, and GSDME-associated cell death. These results strongly suggest that the neutrophil-induced exaggeration of immunopathology in Atg7 cKO mice precedes bacterial overgrowth, further implicating early immune dysregulation as a key driver of the observed phenotypes during NTM infection in vivo.
Compared to Mtb, our understanding of autophagy activation during NTM infection remains limited, highlighting the need for further research. Our data demonstrate a significant reduction in Mabc-R within LC3+ autophagosomal structures in Atg7 cKO BMDMs compared to Atg7 cWT BMDMs, suggesting that Atg7 deficiency markedly suppresses canonical autophagy activation in macrophages. Nonetheless, a substantial proportion of Mabc-R colocalized with LAMP1-positive lysosomes in Atg7 cKO BMDMs, indicating that canonical autophagy-independent phagosomal maturation controls both Mabc-R and Mabc-S infections in Atg7-deficient macrophages. Furthermore, Atg7 cKO macrophages exhibit only minor disruptions in mitochondrial function, cell death, and inflammatory responses compared to Atg7 cWT macrophages during NTM infection. However, distinct phenotypic differences in Atg7-deficient macrophages became evident upon priming with lung lysate supernatants, which mimic the infection microenvironment. Thus, the discrepancy between in vivo and in vitro findings underscores the importance of considering the broader cellular and tissue context. In vivo, Atg7 depletion in innate immune cells exacerbates hyperinflammation and increases cell death in response to excessive danger signals, likely driven by neutrophil infiltration at infection sites in Atg7 cKO mice. Collectively, our findings highlight the essential role of ATG7 in innate immune cells in promoting antimicrobial host defense by mitigating excessive neutrophil infiltration, pathological inflammation, and the activation of multiple cell death pathways during NTM-PD.
Methods
Ethics statement
Human samples and clinical data in this study were obtained from studies approved by the Samsung Medical Center Institutional Review Board (clinicaltrials.gov identifier: NCT00970801) and the Institutional Research and Ethics Committee at Chungnam National University Hospital (approval numbers: CNUH 2019-04-046, CNUH 2020-07-082, CNUH 2020-09-015; Daejeon, Korea). All patients provided informed consent, and the protocol adhered to the guidelines of the Declaration of Helsinki.
Patients and clinical samples
ATG7 and inflammatory cytokines/chemokines (IL1B, IL6, CXCL2, and CCL2) expression in PBMCs was evaluated in NTM patients (Mabc, n = 31; Mmass, n = 22) and HCs (n = 39) from the previously studied cohort25. Thirteen surgical lung resection patients were selected, and their NTM infection was confirmed through microbiological evaluation. These patients were diagnosed at Chungnam National University Hospital (Daejeon, Korea) between January 2018 and December 2022. Sputum samples were collected for acid-fast bacilli smears and cultures. TB/NTM genotyping was performed using real-time PCR with the PANA qPCR™ assay (Panagene, Korea). Lung tissues were obtained from FFPE tissue blocks.
Data analysis of the human PBMC samples
nCounter assay results for human PBMCs from NTM cohort were obtained from a previous study (GSE290289)25. Raw counts were normalized using positive controls and housekeeping genes with nSolver 4.0 software. The normalized counts for each gene were then compared between the NTM-PD patients and HCs using t-tests. For the heatmap, the normalized counts were converted to Z-scores, which were calculated based on the mean and standard deviation of each gene. ATG7 mRNA levels were measured by qRT-PCR and normalized to GAPDH expression using the 2−ΔΔCt (delta-delta Ct) method. The primer sequences are listed in Supplementary Table 3.
Mice
The mice used in individual experiments were age- (6–8 weeks old) and sex-matched. To generate transgenic in innate immune cell-specific Atg7-deficient mice (Atg7 fl/flLysM-Cre+/+) using the Cre/loxP recombination system, Atg7-floxed mice (Atg7 fl/fl, C57BL/6 background)62 were crossed with B6.129P2-Lyz2 tm1(cre)Ifo/J mice (004781, The Jackson Laboratory) expressing Cre recombinase under the control of a Lyz2 promoter. F1 progeny from Atg7 fl/flLysM-Cre+/+ × Atg7 fl/fl crosses were genotyped for the presence (LysM-Cre+) or absence (LysM-Cre−) of the LysM-Cre allele. Cre-mediated recombination was confirmed by PCR using genomic DNA from mouse tails. The genotyping primers were as follows: IMR3066 (Mutant reverse, 5′-CCCAGAAATGCCAGATTACG-3′); IMR3067 (Common, 5′-CTTGGGCTGCCAGAATTTCTC-3′); IMR3068 (WT reverse, 5′-TTACAGTCGGCCAGGCTGAC-3′). The absence of expression of ATG7 in BMDMs was confirmed by immunoblot analysis using an ATG7 antibody (8558, 1:1000, Cell Signaling). Mice were housed at 21–22 °C, with 30–70% humidity on a 12 h light/dark cycle with ad libitum access to food and water. All procedures involving animals were reviewed and approved by the Institutional Animal Care and Use Committee, Chungnam National University College of Medicine (202109A-CNU-180; Daejeon, South Korea). These procedures were conducted in compliance with the guidelines of the Korean Food and Drug Administration.
Cells
The isolation of PBMCs from heparinized venous blood was conducted using the density gradient centrifugation method using Ficoll-Hypaque (Lympoprep; 1114545, axis-sheid). Plastic adherent monocytes derived from PBMCs were cultured in complete RPMI 1640 (10-040-CV, Corning) supplemented with recombinant human macrophage-colony stimulating factor (M-CSF; M6518, Sigma Aldrich) for 7 days to differentiate MDMs63 at placed at 3 × 106 cells per 96-well plate. Primary BMDMs isolated from Atg7 cWT and Atg7 cKO mice were cultured in Dulbecco’s modified Eagle’s medium (DMEM; 10-013-CVRC, Corning) supplemented with 10% fetal bovine serum (FBS; 16000-044, Gibco), 1% Antibiotic-Antimycotic (15240062, Gibco). BMDMs were differentiated for 5 days in the presence of M-CSF (416-ML-010, R&D Systems). HEK293T (ATCC, CRL-3216) cells were cultured in DMEM and maintained at 37 °C in a humidified incubator with 5% CO2.
Mycobacterial strains and culture
The smooth and rough ATCC19977 strain of Mabc, red fluorescent protein (RFP)-labeled Mabc-R, and GFP-labeled Mabc-S, Mint (ATCC 13950), Mav (ATCC 25291), and Mmass (KMRC-00136-13018) were used for this study. Mabc CIP 104536 T R and S morphotype carrying a pMV262-mWasabi or RFP plasmid enabling the expression of mWasabi or RFP were employed. Mabc CIP 104,536 S- and R-type strains were generously provided by Laurent Kremer (Université de Montpellier, Montpellier, France). Each Mycobacterium strain was cultured on 7H10 (Difco, 262710) plates supplemented with 10% oleic albumin dextrose catalase (OADC; 212240, BD Bioscience) for 4 days or 2–4 weeks to confirm colony formation and the absence of contamination. A single colony was selected using a loop and inoculated into 25 ml of 7H9 liquid medium without added surfactant but supplemented with 10% OADC. This pre-culture was incubated at 37 °C for 3 days or 2–4 weeks before initiating the main culture. Once the absorbance reached an OD600nm of 0.5 or higher, 10 ml of the seed culture was transferred to 1 l of the same main culture medium composition and incubated at 37 °C with rotary shaking. The culture was terminated when the absorbance approached an OD600nm of 0.6. The bacterial culture was then centrifuged to pellet the cells, and the medium components were removed by resuspending and centrifuging the cells three times in PBS buffer. Following centrifugation, an equal volume of glass beads was added to the pelleted bacterial cells, and twice their volume of PBS was used to homogenize the mixture using a Mixer Mill 400 (Retsch, Haan, Germany) at 30 Hz for 4 min, resulting in the separation of the mycobacteria into single-cell units. The homogenized mycobacteria were then aliquoted into cryogenic vials for storage. Upon thawing, the strains were diluted and quantitatively cultured on agar plates to verify the colony-forming units per milliliter (CFU/mL).
Antibodies and reagents
Anti-IL-1β (1:1000, CST#12242), anti-Caspase-1 (1:1000, CST#2225), anti-Caspase-3 (1:1000, CST#9662), anti-cleaved Caspase-3 (1:1000, CST#9661), anti-GSDMD (1:1000, CST#39754), anti-ATG7 (1:1000, CST#8558), anti-ACTB (1:4000, CST#4970), anti-mouse IgG (1:4000, CST#7076) and anti-rabbit IgG (1:4000, CST#7074) were purchased from Cell Signaling Technology for western blotting. Anti-DFNA5/GSDME [EPR19859]-N-terminal (1:1000, ab2159) were purchased from Abcam. Anti-LC3A/B (1:400, PM036) was purchased from MBL International. Anti-LAMP1 (1:400, sc-19992) was purchased from Santa Cruz Biotechnology. Alexa Fluor 405-conjugated anti-rat IgG (1:400, A48261), Alexa Fluor 488-conjugated anti-rabbit IgG (1:400, A17041), Alexa Fluor 594-conjugated anti-rabbit IgG (1:400, A21207), Pam3CSK4 (tlrl-pms), and Dihydroethidium (1:200, D11347) were purchased from Invitrogen. MitoTracker Deep Red FM (M22426) was purchased from Thermo Fisher Scientific. RIPA buffer (CBR002) was purchased from LPS solution and 1× blocking solution (OP105-500) was from Biofact. Protease inhibitor (4693116001) and phosphatase inhibitor (4906837001) cocktails were purchased from Roche. MPO (1:200, AF3667) was purchased from R&D System. Anti-Histone H3 [Citruline Arg17, Citruline Arg2, Citrulline Arg8] (1:200, NB100-57135) was purchased from Novus Biologicals. InVivoMAb mouse IgG2a isotype control (BE0085), InVivoMAb anti-mouse Ly6G (BE0075-1) were purchased BioXCell. Alexa Fluor™ 700 rat anti-mouse CD45 (1:250, 56-0451-82), PE-Cyanine7 rat anti-Ly6G (25-9668-82) were purchased from Invitrogen. Zombie Violet™ Fixable Viability Kit (423114), APC anti-mouse CD64 (FcγRI) (1:250, 161006), and PE anti-mouse MERTK (Mer) (151506) were purchased from BioLegend. APC-Cy™7 rat anti-CD11b (1:250, 557657) and Annexin V-PI staining kit (556547) were purchased from BD Biosciences.
RNA extraction and qRT-PCR
Total RNA from cells and tissues was isolated using TRIzol reagent (15596018, Invitrogen) according to the manufacturer’s instructions. cDNA synthesis was performed using Reverse Transcriptase Premix (EBT 1515C, ELPIS-BIOTECH). qRT-PCR was performed using SYBR Green Master Mix (208056, Qiagen) and primers for the indicated genes in a Rotor-Gene Q 2plex system (Qiagen). Data were analyzed using the 2–ΔΔCt threshold cycle method; Gapdh was used for normalization. Primer sequences (mouse, human) are shown in Supplementary Table 3.
Intracellular CFU assay
For the assessment of intracellular bacterial viability, Mabc, Mav, Mmass, or Mint-infected cells were lysed in distilled water to release the intracellular bacteria. Serial dilutions of the lysates were plated on 7H10 agar plates and incubated at 37 °C. After 3–4 days (Mabc and Mmass) or 21 days (Mav, Mint) of incubation, the colonies were counted to measure the bacterial loads.
Mouse in vivo infection model
Atg7 cWT and Atg7 cKO (8–10 weeks old) mice were anesthetized and challenged intranasally with Mabc-S (1 × 106 or 1 × 107 CFU/mouse), Mabc-R (2 × 106 CFU/mouse), Mav (1 × 106 or 1 × 107 CFU/mouse), Mint (1 × 107 CFU/mouse), or Mmass (1 × 107 CFU/mouse). At the indicated times points post-infection, the mice were euthanized and the lungs were harvested to measure the bacterial burden. Lung tissues were homogenized using a tissue homogenizer (OMNI TH; OMNI Inc.) in PBS with detergent (PBST), and serial dilutions of the homogenates were plated on 7H10 agar plates. After 3–4 days (Mabc and Mmass), or 21 days (Mav and Mint) of incubation the colonies formed in the plates were counted.
Lentiviral shRNA production and transduction
shRNA plasmids (ATG7, sc-41447-SH, Santa Cruz Biotechnology) were used for silencing of human ATG7. For lentivirus production, ATG7 shRNA plasmid, pRSV-Rev (12253, Addgene), pMDLg/pRRE (12251, Addgene), and pMD2.G (12259, Addgene) were transfected into HEK293T cells (crl-3216, ATCC) via the Lipofectamine 3000 (L300015, Invitrogen) for 72 h. After transfection, the supernatants were collected and filtered through the filter with 0.45 μm pore (Millipore) and then centrifuged. The collected supernatants were stored at −80 °C. For lentiviral infection, human primary monocytes and MDMs cultured in 96-well plates were infected with a lentiviral vector (MOI of 10) for 36 h, followed by subsequent NTM infection.
Neutrophil depletion
Atg7 cWT and Atg7 cKO mice were infected intranasally with Mabc-S or Mav (1 × 106 CFU/mouse). For in vivo neutrophil depletion, mice were treated with 0.5 mg of anti-Ly6G mAb (neutrophil-depleting, clone 1A8; BE0075-1, BioXcell) or anti-IgG2a Ab (isotype control, clone 2A3; BE0085, BioXcell) via intraperitoneal injection every 1 or 2 days throughout the indicated depletion periods.
Immunohistochemical staining and analysis
For histological analysis in NTM patients, whole FFPE tissues were sectioned onto coated slides, deparaffinized with xylene, and hydrated in a series of alcohol solutions. The sections underwent antigen retrieval by heating in a pressure cooker (containing 10 mmol/L sodium citrate [pH 6.0]) for 3 min. Endogenous peroxidase blocking was achieved using 0.03% hydrogen peroxide for 10 min. The sections were then incubated at room temperature for 1 h with the following primary antibodies: ATG7 (1:500, ab54272, Abcam). Liver hepatocytes served as positive controls, and tonsil tissues as negative controls. For automated staining quantification, a two-step computational algorithm (based on color deconvolution) was validated using QuantCenter software (available on 3DHistech image analysis platform, Sysmex, Budapest, Hungary). The DensitoQuant module was trained for the recognition of immunohistochemical staining.
Lung tissues dissected from Atg7 cWT and Atg7 cKO mice were fixed with 10% formaldehyde solution before paraffin embedding. The sectioned tissues (thickness of 4μm) were stained with Hematoxylin and Eosin (H&E) and then scanned using the PANNORAMIC 300 Flash DX device (3DHISTECH Ltd, Budapest, Hungary). To quantify the inflamed area from stained lung tissues, red-stained areas with high thresholds in three representative lungs were measured as a percentage of the total area using FIJI software. For immunofluorescence, lung paraffin blocks were sectioned and stained with specific targets: anti-MPO (1:200, R&D systems), anti-Histone H3 (1:200, Novus biologicals), Dihydroethidium (1:200, Invitrogen), anti-mouse Ly6G (1:200, BioXcell), and PI (1:200, P3566, Thermo Fisher Scientific). The sections were then washed three times in PBS (CBP007B, LPS solution), and then incubated in fluorescent antibodies: Alexa Fluor 488-conjugated Goat anti-rabbit IgG (1:400), Alexa Fluor 594-conjugated Donkey anti-rabbit IgG (1:400). Coverslips were then mounted onto the slides c using Prolong™ Gold Antifade Mountant with DNA Stain DAPI (P36931, Invitrogen). Immunofluorescence images were acquired with Zeiss LSM 900. 3D reconstruction image acquisition by Aivia (Leica microsystems).
Enzyme-linked immunosorbent assay (ELISA)
Cell supernatants and tissue homogenates were collected and stored at −80 °C. Supernatants were used to measure mouse IL-1β (88-7013-22, Invitrogen), mouse IL-6 (555240, BD Bioscience), mouse TNF (560478, BD Bioscience), human IL-1β (557953, BD Bioscience), and human IL-6 (555220, BD Bioscience) secretion according to the manufacturer’s instructions.
Western blotting
Cell lysates from Atg7 cWT and Atg7 cKO mice BMDMs and lung homogenates were prepared in RIPA buffer (CBR002, LPS solution) containing protease inhibitor (4693116001, Roche) and phosphatase inhibitor (04906845001, Roche). Samples were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to nitrocellulose membrane. Membranes were blocked in 1x blocking solution (OP105-500, Biofact) for 30 min at room temperature. The membranes were probed overnight at 4 °C with primary antibodies as described: Anti-pro-IL-1β, anti-Caspase-1, anti-Caspase-3, anti-cleaved Caspase-3, anti-GSDMD, anti-ATG7, anti-ACTB, and anti-DFNA5/GSDME. The membranes were washed using TBS-T buffer and further incubated for 1 h at room temperature with appropriate secondary antibodies: anti-mouse IgG (Cell Signaling Technology, #7076, 1:4000) and anti-rabbit IgG (Cell Signaling Technology, #7074, 1:4000). The immune-reactive proteins were detected by using Immobilon western chemiluminescent HRP substrate (WBKLS0500, Merck), and were visualized by iBrightTM Image system (CL750, Invitrogen). The intensity value of target protein bands was normalized by using ImageJ software (National Institutes of Health, USA, 1.52a) for densitometry analysis.
Tissue processing and spatial transcriptomic generation
Following the resection of lungs from Mav-infected Atg7 cWT and Atg7 cKO at 21 dpi, tissues underwent fixation in 10% formalin and subsequent embedding in paraffin. RNA extraction was performed on FFPE samples utilizing the RNeasy FFPE Kit (#73504, Qiagen), followed by DV 200 evaluation using an Agilent TapeStation. FFPE tissue samples were then prepared according to the Visium Spatial Gene Expression for FFPE protocol (#CG000409, 10xGenomics Pleasanton, CA, USA). Libraries were constructed utilizing the Visium Spatial Gene Expression for FFPE Kit, Mouse Transcriptome v.1 (PN-1000521, 10× Genomics) along with the Dual Index Kit TS Set A (PN-1000251, 10xGenomics). Subsequent sequencing was conducted on an Illumina NovaSeq6000 S1 Rgt Kit v1.5 200 cycles, employing paired-end 200 bp FlowCells. For the data analysis pipeline, reads were aligned to the mouse reference genome (mm10-2020) using Space Ranger (10× Genomics). Matrix and H&E image analysis were performed using Seurat v.4.0 (R 4.1) for further analysis.
Spatial transcriptomic data pre-processing
The aggregated HDF5matrix was imported into R and split by sample. Feature-barcode matrices for each sample were imported into the R package “Seurat” (Version 5.0.1) for normalization, quality control, batch effect correction, dimensionality reduction, and Louvain clustering. Spots expressing less than 300 features were excluded from downstream analysis.
scRNA sequencing and data analysis
Following the resection of lungs from Mav-infected Atg7 cWT and Atg7 cKO at 21 dpi, tissues were dissociated using Liberase TL (#05401020001, Roche). Cell viability and number were assessed using the LUNA-FX7™ Automated Cell Counter (Logos Biosystems) with acridine orange/PI stain. Single-cell suspensions were generated using the Chromium Next GEM Single Cell 3’ Kit v3.1 (16 rxns, PN: 1000268) in conjunction with the Chromium Next GEM Chip G Single Cell Kit (48 rxns, PN: 1000120) and the Dual Index Kit TT Set A (96 rxns, PN: 1000215), following the manufacturer’s protocol (10xGenomics). Libraries were sequenced on an Illumina NovaSeq 6000 platform using the NovaSeq 5000/6000 S2 Reagent Kit (200 cycles) v1.5 (PN: 20028315). The sequencing was performed in paired-end mode with Read1: 28 cycles, Read2: 90 cycles, i7 Index: 10 cycles, and i5 Index: 10 cycles. Reads were aligned to the mouse reference genome (mm10-2020-A) using Cell Ranger v8.0.1 (10× Genomics) and gene expression matrices were generated for downstream analysis. Low-quality cells expressing fewer than 200 genes were removed from the dataset. After quality control, 25,040 cells were retained. Seurat’s standard preprocessing workflow was applied, including normalization, data scaling, Harmony integration for batch effect correction, dimensionality reduction, and clustering based on the first 30 principal components. Thirty-seven cell clusters were annotated into 22 cell types based on differentially expressed genes and canonical cell type markers, with references from the PanglaoDB, including AT1 (Hopx, Akap5), AT2 (Sftpc, Stfpa1), Ciliated (Scgb1a1, Sec14l3), Capillary (Cd93, Hpgd), Vein (Vwf), Lymph (Fbln2, Ccl21a, Mmrn1), Fibroblast (Col14a1, Col13a1), Pericyte (Acta2, Gucy1a1), Myeloids (Cd68, Itgam, Fcgr3), T cells (Cd3d, Cd4, Cd8a), NK cells (Nkg7, Gzma, Ccl5), B/Plasma cells (Cd19, Igkc, Ighm, Mzb1), and five innate immune cell types: Interstitial macrophage (Cd68, Fcgr1, Stab1, Aoah, Sdc3), alveolar macrophage (Marco, Siglecf, Itgax, Olr1), neutrophil (Csf3r, Ly6g, Il1r2, S100a8, Trem1), monocyte (Ccr2, Ifitm3, Tet2, Sirpb1c, Klra2, Gpr141), and dendritic cell (Zbtb46, Slamf7, Traf1).
Cell type population heatmap with deconvolution
We conducted deconvolution of cell types within spatial transcriptomic spots using the python package “Cell2location”, with the scRNA sequencing data from GSE151974 serving as the reference28. Subsequently, we integrated the results using Seurat object metadata into the R package “Seurat” for downstream analysis. Mean average expression in each spot was calculated based on unsupervised clusters, and the results were visualized using the Heatmap function in the R package “pheatmap” (Ver 1.0.12).
Differential expression analysis to pathway enrichment analysis (GO Biological Process)
Differentially expressed genes were identified using the “FindMarkers”, “MAST” and “FindAllMarkers” function in the “Seurat”. Pathway enrichment analysis was performed using the R package “ClusterProfiler” (Ver 4.10.0) with gene identifiers mapped using the R package “org.Mm.eg.db” package (Ver 3.18.0). Statistical significance for enrichment was assessed using the hypergeometric test with Benjamini-Hochberg correction for multiple comparisons, as implemented in the “ClusterProfiler” package, with a p-value threshold of <0.05. The selected GO terms represent biological processes. As enrichment testing is inherently one-sided, the test directionality was not explicitly stated.
Statistical approach for comparing gene set scores
The average expression of selected gene set signatures from signaling pathways in the databases HALLMARK (HALLMARK_INFLAMMATORY_RESPONSE, HALLMARK_IL6_JAK_STAT3_SIGNALING, HALLMARK_IL2_STAT5_SIGNALING, HALLMARK_TNFA_SIGNALING_VIA_NFKB, HALLMARK_REACTIVE_OXIGEN_SPECIES_PATHWAY), REACTOME (R-MMU-5620971, R-MMU-5218859), KEGG (mmu04215), and GO_BP (GO:0097527) was calculated for each spot and cell in the Spatial Transcriptomes and scRNA sequencing datasets using the “AddModuleScore” function. Data visualization was performed using “DimPlot”, “Featureplot”, and “SpatialFeaturePlot” function in the “Seurat”.
Transmission electron microscopy
For transmission electron microscopy analysis, lung tissues or BMDMs from Atg7 cWT and Atg7 cKO were sequentially fixed with 2.5% glutaraldehyde and 1% osmium tetroxide on ice for 2 h and washed with PBS. The cells were then dehydrated in ethanol and propylene oxide series, embedded in Epon 812 mixture, and polymerized in an oven at 60 °C for 24 h. The 70 nm sections acquired from polymerized blocks were collected on 150 mesh copper grids, counterstained with 4% uranyl acetate for 10 min and lead citrate for 10 min, and examined with a KBSI Bio-HVEM system (JEM-1400Plus at 120 kV and JEM-1000BEF at 1000 kV, JEOL). Data were collected using a OneView camera with DM software (Image size: 4 × 4k, Gatan Inc., Pleasanton, CA, USA). ImageJ software was used for the quantification of damaged mitochondria.
Flow cytometric analysis
To analyze apoptotic cells, Mabc-R-infected BMDMs were harvested and stained with FITC-conjugated Annexin V and PI as provided by the Annexin V-PI staining kit (556547, BD Biosciences). For neutrophil infiltration staining, Mabc-R or Mav-infected Atg7 cWT and Atg7 cKO lung samples were harvested using a mouse lung dissociation kit (130-095-927, Miltenyi Biotech). Briefly, the lungs of the mice were transferred to a C tube containing the 1× S buffer with the mix solution (enzyme D and A in S buffer). The GentleMACS programs were run with a 30 min incubation on the MACSmix™ Tube Rotator at 37 °C. The cell suspension was subsequently passed through a 70-µm strainer, and the cell pellets were resuspended in Zombie-violet Live-Dead stain (1:1000) and incubated for 15 min in the dark. Cells were then washed with PBS containing 2% FBS (v/v). Cells were then stained for 30 min at 4 °C with following specific antibodies: Alexa Fluor™ 700 rat anti-mouse CD45 (1:250), APC anti-mouse CD64 (FcγRI) (1:250), PE anti-mouse MERTK (Mer) (1:250), PE-Cyanine7 Rat Anti-Ly6G (1:250), and APC-Cy™7 Rat Anti-CD11b (1:250). All stains were carried out in PBS containing 2% FBS (v/v). After washing twice with PBS containing 2% FBS, stained cells were resuspended in 2% paraformaldehyde and assayed immediately on the BD FACSDiscover™ S8 Cell Sorter (NFEC-2025-02-303724, BD Biosciences). Data were collected and analyzed using BD Flow Jo (ver10.8.1, BD Biosciences), BD FACSChorus™ software (BD Biosciences).
Immunofluorescence and confocal microscopy analysis
Cells were cultured on coverslips and infected with NTM. Subsequently, the cells were washed three times with PBS, fixed with 4% paraformaldehyde for 15 min, and permeabilized with 0.25% Triton X-100 (Sigma-Aldrich) for 10 min. After a 2 h incubation with primary antibodies at room temperature, cells were washed with PBS to remove excess primary antibodies. The samples were incubated with secondary antibodies for 2 h and nuclei were stained with DAPI for 5 min at room temperature. After mounting, fluorescence images were captured using confocal laser microscope (TCS SP8, Leica).
To quantify the colocalization of mycobacteria with LC3/LAMP1 and mitotracker with LC3/LAMP1, images were captured using confocal laser-scanning microscopy. Each experiment was performed on triplicate coverslips, and the results are expressed as the mean and standard deviation. Pearson’s correlation coefficient was measured by ImageJ software.
Measurement of mtROS
To quantify mtROS, BMDMs and lung tissues were incubated with 2.5 μM MitoSOXTM Red Mitochondrial Superoxide Indicator (M36007, Invitrogen). After 20 min, the cells were washed and measured by confocal microscope (LSM900, Zeiss). MitoSOX intensity was analyzed using ImageJ software.
RNA sequencing analysis
Paired-end sequencing reads were generated on the Illumina sequencing NovaSeq platform. Trimmomatic v0.38 was used to remove adapter sequences and trim bases with poor base quality. The cleaned reads were aligned to the Mus musculus (mm10) using HISAT v2.1.0, based on the HISAT and Bowtie2 implementations. Aligned data (SAM file format) were sorted and indexed using SAMtools v 1.9. After alignment, the transcripts were assembled and quantified using StringTie v2.1.3b. Gene-level and Transcript-level quantification was calculated as raw read count, fragments per kilobase of transcript per million mapped reads (FPKM) and transcripts per million. Differential expressed gene (DEG) analysis and RLE normalization were performed by DESeq2 v 1.38.3. Statistical significance of differential expression gene was determined using DESeq2 nbinom WaldTest. Fold change and P-value were extracted from the result of WaldTest. All P-values are adjusted by Benjamini–Hochberg algorithm to control false discovery rate. The significant gene list was filtered by |fold change| ≥ 2 & raw P-value < 0.05. Gene-enrichment and functional annotation analysis for significant gene list were performed based on gProfiler (https://biit.cs.ut.ee/gprofiler/orth), and KEGG pathway database (https://www.genome.jp/kegg/) respectively. The heatmap data was produced utilizing the heatmap (version 1.0.12). All data analysis and visualization of differentially expressed genes was conducted using R 4.2.2 www.r-project.org.
Optical bioluminescence imaging
For optical fluorescence imaging, Atg7 cWT and Atg7 cKO were intranasally infected with ERFP-Mabc-R (2 × 106 CFU/mouse) for 7 days. To check bacterial colonization in the lung, ex vivo fluorescence imaging was performed using an optical imaging system (Night OWL LB983, Berthold Technologies, USA). At 7 days after bacterial infection, mice were sacrificed and lungs were excised for ex vivo imaging. ERFP-Mabc-R were detected with excitation/emission wavelength at 580/620 nm with an exposure of 7 s. Data were analyzed with IndiGo software (Berthold Technologies).
Statistics and reproducibility
For statistical analysis, data obtained from at least three independent experiments [mean ± standard deviation (SD) or standard error of the mean (SEM)] were analyzed using a two-tailed Student’s t-test, Mann–Whitney U-test, and one-way analysis of variance (ANOVA) with Tukey’s or Dunnett’s multiple comparison tests. The sample size was determined based on biological/technical replicates with sufficient reproducibility. For analysis of the human PBMC sample, 3 groups (NTM patients with Mabc and NTM patients with Mmass as experimental groups and healthy group as the control group) were examined by Kruskal-Wallis test with post hoc using Dunn’s multiple comparison test. For the Visium spatial transcriptomics and scRNA sequencing data, each group (Atg7 cKO as the experimental group and Atg7 cWT as the control group) consisted of one mouse. The unit of analysis was “spot” for Visium and “cell” for scRNA sequencing. Sample sizes (n) for all plots are indicated in the corresponding figure legends. Two-group comparisons of module scores between Atg7 cKO and Atg7 cWT were performed using two-sided, unpaired Welch’s t-tests (R packages ggpubr v0.6.0 and ggplot2 v3.4.4). For comparisons among more than two groups, normality was assessed using the Shapiro-Wilk test (stats package, v4.3.2), and homogeneity of variances was evaluated using Levene’s test (car package, v3.1-2). If both assumptions were met (P > 0.05), a one-way ANOVA (stats package, v4.3.2) was performed, followed by Tukey’s Honestly significant difference (HSD) post hoc test for pairwise comparisons. Effect sizes (Cohen’s d) and their 95% confidence intervals were calculated using the effsize package (v0.8.0) and are provided in the Source Data file. Statistical significance was defined as P < 0.05. Exact p-values are reported in the figures or figure legends; p-values below the numerical precision limit of R (double-precision floating point) are reported as being less than the corresponding threshold.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNA sequencing, Visium, and scRNA sequencing datasets generated during this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE267176,GSE297621, and GSE297620. The public nCounter assay datasets analyzed in this study have been deposited in GEO under accession code: GSE29028925. The public scRNA sequencing datasets reused in this study are available in GEO under accession code: GSE15197428. Source data are provided with this paper.
Code availability
The analysis scripts are accessible at Github: https://github.com/YEONJU-L/ATG7_NTM-infections.git with identifier https://doi.org/10.5281/zenodo.1536850664. Source data are provided with this paper.
References
Haworth, C. S. et al. British Thoracic Society Guideline for the management of non-tuberculous mycobacterial pulmonary disease (NTM-PD). BMJ Open Respir. Res. 4, e000242 (2017).
Kang, Y. A. & Koh, W. J. Antibiotic treatment for nontuberculous mycobacterial lung disease. Expert Rev. Respir. Med. 10, 557–568 (2016).
Lopez-Roa, P., Esteban, J. & Munoz-Egea, M. C. Updated review on the mechanisms of pathogenicity in Mycobacterium abscessus, a rapidly growing emerging pathogen. Microorganisms 11, 90 (2022).
Luo, X. et al. Risk factors for microbiological persistence after 6 months of treatment for Mycobacterium intracellulare and its impact on the drug-resistance profile. Microbiol. Spectr. 11, e0080523 (2023).
Ku, J. H. et al. Treatment of nontuberculous mycobacterial (NTM) pulmonary infection in the US bronchiectasis and NTM registry: treatment patterns, adverse events, and adherence to American Thoracic Society/Infectious Disease Society of America treatment guidelines. Clin. Infect. Dis. 76, 338–341 (2023).
Abdelaal, H. F. M., Chan, E. D., Young, L., Baldwin, S. L. & Coler, R. N. Mycobacterium abscessus: it’s complex. Microorganisms 10, 1454 (2022).
Parmar, S. & Tocheva, E. I. The cell envelope of Mycobacterium abscessus and its role in pathogenesis. PLoS Pathog. 19, e1011318 (2023).
Garcia-Prats, A. J. et al. Characteristics of children and adolescents with multidrug-resistant and rifampicin-resistant tuberculosis and their association with treatment outcomes: a systematic review and individual participant data meta-analysis. Lancet Child Adolesc. Health 9, 100–111 (2025).
Wang, Y. T. et al. Select autophagy genes maintain quiescence of tissue-resident macrophages and increase susceptibility to Listeria monocytogenes. Nat. Microbiol. 5, 272–281 (2020).
Gomez-Virgilio, L. et al. Autophagy: a key regulator of homeostasis and disease: an overview of molecular mechanisms and modulators. Cells 11, 2262 (2022).
Yamamoto, H., Zhang, S. & Mizushima, N. Autophagy genes in biology and disease. Nat. Rev. Genet. 24, 382–400 (2023).
Levine, B., Mizushima, N. & Virgin, H. W. Autophagy in immunity and inflammation. Nature 469, 323–335 (2011).
Deretic, V., Saitoh, T. & Akira, S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 13, 722–737 (2013).
Deretic, V. Autophagy in inflammation, infection, and immunometabolism. Immunity 54, 437–453 (2021).
Cadwell, K. Crosstalk between autophagy and inflammatory signalling pathways: balancing defence and homeostasis. Nat. Rev. Immunol. 16, 661–675 (2016).
Kuma, A., Komatsu, M. & Mizushima, N. Autophagy-monitoring and autophagy-deficient mice. Autophagy 13, 1619–1628 (2017).
Watson, R. O., Manzanillo, P. S. & Cox, J. S. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 150, 803–815 (2012).
Castillo, E. F. et al. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc. Natl. Acad. Sci. USA 109, E3168–E3176 (2012).
Kimmey, J. M. et al. Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature 528, 565–569 (2015).
Kinsella, R. L. et al. Autophagy prevents early proinflammatory responses and neutrophil recruitment during Mycobacterium tuberculosis infection without affecting pathogen burden in macrophages. PLoS Biol. 21, e3002159 (2023).
Feng, S. et al. Autophagy promotes efficient T cell responses to restrict high-dose Mycobacterium tuberculosis infection in mice. Nat. Microbiol. 9, 684–697 (2024).
Golovkine, G. R. et al. Autophagy restricts Mycobacterium tuberculosis during acute infection in mice. Nat. Microbiol. 8, 819–832 (2023).
Wang, F. et al. ATG5 provides host protection acting as a switch in the atg8ylation cascade between autophagy and secretion. Dev. Cell 58, 866–884 e868 (2023).
Aylan, B. et al. ATG7 and ATG14 restrict cytosolic and phagosomal Mycobacterium tuberculosis replication in human macrophages. Nat. Microbiol. 8, 803–818 (2023).
Kim, H. J. et al. MiR-144-3p is associated with pathological inflammation in patients infected with Mycobacteroides abscessus. Exp. Mol. Med. 53, 136–149 (2021).
Mittal, E. et al. Mycobacterium tuberculosis virulence lipid PDIM inhibits autophagy in mice. Nat. Microbiol. 9, 2970–2984 (2024).
Gatfield, J. & Pieters, J. Molecular mechanisms of host-pathogen interaction: entry and survival of mycobacteria in macrophages. Adv. Immunol. 81, 45–96 (2003).
Hurskainen, M. et al. Single cell transcriptomic analysis of murine lung development on hyperoxia-induced damage. Nat. Commun. 12, 1565 (2021).
Taylor, J. P. & Tse, H. M. The role of NADPH oxidases in infectious and inflammatory diseases. Redox Biol. 48, 102159 (2021).
Xiao, X. et al. S100 proteins in atherosclerosis. Clin. Chim. Acta 502, 293–304 (2020).
Bhattarai, K. R., Riaz, T. A., Kim, H. R. & Chae, H. J. The aftermath of the interplay between the endoplasmic reticulum stress response and redox signaling. Exp. Mol. Med. 53, 151–167 (2021).
Bulua, A. C. et al. Mitochondrial reactive oxygen species promote production of proinflammatory cytokines and are elevated in TNFR1-associated periodic syndrome (TRAPS). J. Exp. Med. 208, 519–533 (2011).
Zorov, D. B., Juhaszova, M. & Sollott, S. J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 94, 909–950 (2014).
Lira Chavez, F. M. et al. Restoring the infected powerhouse: mitochondrial quality control in sepsis. Redox Biol. 68, 102968 (2023).
Avdoshina, V. et al. The HIV protein gp120 alters mitochondrial dynamics in neurons. Neurotox. Res. 29, 583–593 (2016).
Liu, J. et al. cGAS-STING, inflammasomes and pyroptosis: an overview of crosstalk mechanism of activation and regulation. Cell Commun. Signal 22, 22 (2024).
Liang, X., Qin, Y., Wu, D., Wang, Q. & Wu, H. Pyroptosis: a double-edged sword in lung cancer and other respiratory diseases. Cell Commun. Signal 22, 40 (2024).
Yuan, J. & Ofengeim, D. A guide to cell death pathways. Nat. Rev. Mol. Cell Biol. 25, 379–395(2023).
Wu, J. et al. Gasdermin D silencing alleviates airway inflammation and remodeling in an ovalbumin-induced asthmatic mouse model. Cell Death Dis. 15, 400 (2024).
He, B. et al. Single-walled carbon-nanohorns improve biocompatibility over nanotubes by triggering less protein-initiated pyroptosis and apoptosis in macrophages. Nat. Commun. 9, 2393 (2018).
Zhao, P. et al. Neutrophil extracellular traps induce pyroptosis of pulmonary microvascular endothelial cells by activating the NLRP3 inflammasome. Clin. Exp. Immunol. 217, 89–98 (2024).
Ma, F. et al. Gasdermin E dictates inflammatory responses by controlling the mode of neutrophil death. Nat. Commun. 15, 386 (2024).
Brinkmann, V. et al. Neutrophil extracellular traps kill bacteria. Science 303, 1532–1535 (2004).
Papayannopoulos, V. Neutrophil extracellular traps in immunity and disease. Nat. Rev. Immunol. 18, 134–147 (2018).
Collier, J. J., Suomi, F., Olahova, M., McWilliams, T. G. & Taylor, R. W. Emerging roles of ATG7 in human health and disease. EMBO Mol. Med. 13, e14824 (2021).
Li, G. et al. Plasma extracellular vesicle delivery of miR-210-3p by targeting ATG7 to promote sepsis-induced acute lung injury by regulating autophagy and activating inflammation. Exp. Mol. Med. 53, 1180–1191 (2021).
Lee, H. Y. et al. Autophagy deficiency in myeloid cells increases susceptibility to obesity-induced diabetes and experimental colitis. Autophagy 12, 1390–1403 (2016).
Ye, Y. et al. Inhibition of p-IkappaBalpha ubiquitylation by autophagy-related gene 7 to regulate inflammatory responses to bacterial infection. J. Infect. Dis. 212, 1816–1826 (2015).
Ravindran, R. et al. The amino acid sensor GCN2 controls gut inflammation by inhibiting inflammasome activation. Nature 531, 523–527 (2016).
Xie, C. et al. Neuroprotection by selective neuronal deletion of Atg7 in neonatal brain injury. Autophagy 12, 410–423 (2016).
Qiang, L. et al. Autophagy gene ATG7 regulates ultraviolet radiation-induced inflammation and skin tumorigenesis. Autophagy 13, 2086–2103 (2017).
Silwal, P. et al. Mitofusin-2 boosts innate immunity through the maintenance of aerobic glycolysis and activation of xenophagy in mice. Commun. Biol. 4, 548 (2021).
Roca, F. J. & Ramakrishnan, L. TNF dually mediates resistance and susceptibility to mycobacteria via mitochondrial reactive oxygen species. Cell 153, 521–534 (2013).
Ellzey, L. M., Patrick, K. L. & Watson, R. O. Mitochondrial reactive oxygen species: double agents in Mycobacterium tuberculosis infection. Curr. Opin. Immunol. 84, 102366 (2023).
Kim, Y. J. et al. Sirtuin 3 is essential for host defense against Mycobacterium abscessus infection through regulation of mitochondrial homeostasis. Virulence 11, 1225–1239 (2020).
Cristaldi, M. et al. Caspase-8 activation by cigarette smoke induces pro-inflammatory cell death of human macrophages exposed to lipopolysaccharide. Cell Death Dis. 14, 773 (2023).
Ren, F. et al. ER stress induces caspase-2-tBID-GSDME-dependent cell death in neurons lytically infected with herpes simplex virus type 2. EMBO J. 42, e113118 (2023).
Scott, N. R. et al. S100A8/A9 regulates CD11b expression and neutrophil recruitment during chronic tuberculosis. J. Clin. Investig. 130, 3098–3112 (2020).
Nair, S. et al. Irg1 expression in myeloid cells prevents immunopathology during M. tuberculosis infection. J. Exp. Med. 215, 1035–1045 (2018).
Hult, C., Mattila, J. T., Gideon, H. P., Linderman, J. J. & Kirschner, D. E. Neutrophil dynamics affect Mycobacterium tuberculosis granuloma outcomes and dissemination. Front. Immunol. 12, 712457 (2021).
Chowdhury, C. S. et al. Type I IFN-mediated NET release promotes Mycobacterium tuberculosis replication and is associated with granuloma caseation. Cell Host Microbe 32, 2092–2111 e2097 (2024).
Komatsu, M. et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 (2005).
Drexhage, L. Z. et al. Apoptosis-mediated ADAM10 activation removes a mucin barrier promoting T cell efferocytosis. Nat. Commun. 15, 541 (2024).
Sang Min Jeon, et al. ATG7 in innate immune cells is required for host defense against nontuberculous mycobacterial pulmonary infections. Github. https://doi.org/10.5281/zenodo.15368506. (2025).
Acknowledgements
We thank the patients and healthy donors for participating in the study. We thank Dr. L. Kremer (Université de Montpellier, Montpellier, France) for kind provision of mycobacterial strains; Dr. S.Y. for his support in bioinformatic analysis and helpful discussions; S.O.K. for kind support with sample inventory and tracking; K.A.R. for assistance with RNA work; J.W.Y. for assistance with breeding and maintaining of mice; B.K.J. (Chungnam National University College of Medicine) for technical assistance. This research was supported by the Korea Basic Science Institute under the R&D program (Project No. C523311) supervised by the Ministry of Science and ICT (S.-H.L.). This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (RS-2023-00255021) (E.-K.J.), and the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (RS-2023-00227274) (E.-K.J.).
Author information
Authors and Affiliations
Contributions
E.-K.J. initiated and conceived the project. E.-K.J., W.Y.P., and J.K.K. designed and supervised the study. E.-K.J., W.Y.P., J.K.K., S.M.J., Y.J.L., S.-H.L., and S.I.K. contributed to writing—original draft, review, and editing of the manuscript. S.M.J., Y.J.L., S.-H.L., and S.I.K. analyzed and interpreted the data. S.M.J., S.I.K., I.S.K., H.J.K., M.-K.Y., J.-M.K., J.W., S.-Y.K., J.-J.M., J.J., D.H.K., B.W.J., C.C., and M.K. organized sample acquisition and collection. I.S.K., D.H.K., B.W.J., and C.C. recruited participants and collected samples and clinical metadata. S.I.K., B.L., T.R., H.J.K., and I.S.K. collected immune cells from blood. S.M.J., S.I.K., B.L., Y.J.K., H.J.K., and J.K.K. performed and analyzed the in vitro experiments. S.M.J., S.I.K., and B.L. performed flow cytometry and analyzed the data. S.M.J., S.I.K., Y.J.K., J.W., and J.J. performed the mycobacterial infection. S.M.J., S.I.K., and Y.J.K. performed the in vivo experiments. Y.J.L. analyzed single-cell and spatial RNA sequencing data. Y.J.L., I.S.K., and T.R. analyzed the transcriptomic sequencing data. S.-H.L. performed transmission electron microscopy. J.W. and J.J. prepared the mycobacterial strains. M.-K.Y. and J.-M.K. analyzed and interpreted the clinical data. S.-Y.K. and J.-J.M. supported optical biofluorescence imaging and data analysis. M.K. generated and prepared Atg7 knockout mice. All authors contributed to the critical review, finalized, and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
W.-Y.P. is a current employee of Geninus Inc. and GxD Inc. The work reported here was not funded by either company. All other authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Jingjing Ling, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jeon, S.M., Lee, Y.J., Lee, SH. et al. ATG7 in innate immune cells is required for host defense against nontuberculous mycobacterial pulmonary infections. Nat Commun 16, 6966 (2025). https://doi.org/10.1038/s41467-025-61791-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-61791-1