Abstract
Scavenger receptor CD163 is a marker of M2 type macrophages that play important roles in anti-inflammatory processes. The most extensively studied function of CD163 is related to the elimination of hemoglobin-haptoglobin (Hb-Hp) complexes, to prevent potential oxidative toxicity of the iron-containing heme. However, the structural mechanism of CD163 in ligand binding and internalization remains elusive. Here, we present the cryo-electron microscopy structure of human Hb-Hp recognition by the full ectodomain of CD163. We illuminate that CD163 forms calcium-dependent oligomers and primarily exists as trimeric form under the condition of 2.5 mM calcium. It mainly utilizes two protomers to interact with Hb-Hp complex asymmetrically, while the third protomer of the trimer also has the potential to form calcium-mediated contacts with Hp. Flow cytometry analyses reveal that oligomerization of CD163 significantly enhances the efficiency of ligand endocytosis. These results advance our understanding of the role of CD163 in ligand scavenging.
Similar content being viewed by others
Introduction
Macrophages and monocytes play crucial roles in maintaining homeostasis and regulating inflammatory responses. These mononuclear phagocytes can be divided into two subpopulations, M1 and M2, based on their functional properties. CD163 has been identified as a specific marker of M2 monocytes/macrophages, exhibiting strong anti-inflammatory properties1,2,3,4,5. The protein can transduce signals upon binding of its ligands that lead to the release of anti-inflammatory mediators such as interleukin-10 (IL-10)2. CD163 expression has also been demonstrated to be associated with various diseases, including cancer6,7,8,9.
CD163 is a type I transmembrane protein that belongs to the group B scavenger receptor cysteine-rich (SRCR) superfamily2,10. It comprises nine type B SRCR domains and can be cleaved by ADAM17 to form soluble CD163 (sCD163)11 (Fig. 1a). SRCR6 and 7 are separated by a proline-serine-threonine (PST) rich polypeptide of ~35 amino acids (Fig. 1a). The protein also contains a transmembrane part spanning the plasma membrane once, and a short intracellular tail which contains a hydrophobic internalization motif for mediating endocytosis2. As a multifunctional receptor, CD163 interacts with various ligands and pathogens. It has been identified as a receptor for porcine reproductive and respiratory syndrome virus and African swine fever virus and is suggested to act as an innate immune sensor for bacteria12,13,14. CD163 has been reported as an erythroblast adhesion receptor and has been identified as a receptor for tumor necrosis factor (TNF)-like weak inducer of apoptosis15,16,17,18. However, the most well-known function of CD163 is the elimination of Hb-Hp complexes3.
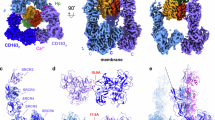
a Domain architecture of CD163, sCD163, Hp1 and Hp2. PST, proline-serine-threonine rich interdomain segment; TM transmembrane, CCP complement control protein, SP serine protease. b Subunit organization of Hp1-1, Hp1-2 and Hp2-2 complexes. c Different views of the cryo-EM map of trimeric CD163 in complex with Hb-Hp1. Three CD163 protomers are shown in blue, cyan, and green, Hb subunits are shown in red (α) and orange (β), Hp1 is shown in pink. d Overall structure of CD163 in complex with Hb-Hp1. e Structure of CD163 SRCR domains from different viewpoints. f Model for CD163-mediated recognition and internalization of Hb-Hp complex.
During extra- or intra-vascular hemolysis, free hemoglobin (Hb) is released into the blood from lysed red blood cells, which can be potentially hazardous due to the oxidative and toxic properties of the iron-containing heme19,20. Haptoglobin (Hp), a plasma stress responding protein, irreversibly captures the free Hb, forming Hb-Hp complexes21,22,23,24. Hp is proteolytically cleaved in vivo after oligomerization into heavy chains (β chains) and light chains (α chains)25,26. In humans, the Hp gene has two major allelic variants, Hp1 and Hp2, leading to three major phenotypes: homozygous Hp1-1, heterozygous Hp1-2, and homozygous Hp2-2 (Fig. 1a)27. Hp2 contains two complement control protein (CCP) domains that can bridge two other Hp CCP domains, leading to the formation of various oligomers (Fig. 1b)28,29. The Hb-Hp complex then forms a neo-epitope that is recognized by CD163 in a calcium- and pH-dependent manner3,30. Binding of Hb-Hp to CD163 leads to the internalization of the Hb-Hp complex and subsequent degradation of the heme subunit, yielding biliverdin, free iron, and the carbon monoxide (CO) molecule31,32.
In this study, we present high-resolution cryogenic electron microscopy (cryo-EM) structures of CD163 bound to the Hb-Hp complex. Previous studies have determined the crystal structure of the porcine Hb-Hp dimer and investigated the structure of CD163-Hb-Hp using small-angle X-ray scattering (SAXS), which indicated that the receptor-binding site is located in the protruding Hp loop 3 (residues 251-275)22. Our cryo-EM structures reveal that the CD163 ectodomain oligomerizes in a calcium-dependent manner through its C-terminal SRCR5-9 domains, while the N-terminal SRCR2-4 domains are crucial for binding to Hb-Hp dimeric complex. Our data suggest that the CD163 oligomer primarily uses two protomers to asymmetrically recognize half of the dimeric Hb-Hp complex, with a third protomer potentially forming calcium-mediated interactions with Lys211 in Hp. We further show that calcium ions are essential for both CD163 oligomerization and ligand binding, and define two distinct dual-point electrostatic interactions induced by calcium-binding. Additionally, our results underscore the significance of CD163 oligomerization in mediating endocytosis. CD163 has been recognized as a marker for many diseases and a target molecule for cell-directed therapies2,33,34,35,36,37. Our study provides valuable insights into the structural basis of CD163 function, facilitating further research and translational application on CD163-positive macrophages and the development of CD163-mediated therapies.
Results
Overall structure of CD163 in complex with Hb-Hp
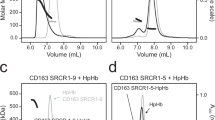
Two Hp isoforms were overexpressed in HEK293F and purified, which resulted in the presence of uncleaved products (Supplementary Fig. 1a, b). The Hb-Hp and CD163-Hb-Hp complexes were then reconstituted and purified in vitro (Supplementary Fig. 1c–f). The chromatogram for Hb-Hp1 shows a peak corresponding to a molecular weight of ~150 kDa, whereas Hb-Hp2 exhibits a peak around 400 kDa, which is an average molecular weight of its various oligomeric forms (Supplementary Fig. 1c, d). CD163 elutes as a peak corresponding to a molecular weight of ~500 kDa, indicating its existence in an oligomeric state (Supplementary Fig. 1f). The CD163-Hb-Hp1 complex is observed at ~650 kDa, representing the molecular mass of oligomeric CD163 in complex with Hb-Hp1. The CD163-Hb-Hp2 complex exhibits a peak about 980 kDa (Supplementary Fig. 1e, f). Notably, the uncleaved Hp also formed complexes with Hb and CD163 (Supplementary Fig. 1a–f). We then examined the particles of CD163-Hb-Hp1 and CD163-Hb-Hp2 complexes using cryo-EM (Supplementary Fig. 1g, h). The CD163-Hb-Hp2 sample contained various oligomerized protein complexes due to the oligomerization property of Hp229 (Fig. 1b), which made it impossible to obtain any distinct 2D classes after preliminary cryo-EM analysis, making it more challenging to solve the structure. Additionally, the oligomerization of Hp2 added complexity to the mixture, complicating the analysis. As a result, we chose the Hp1 isoform for all the assays and Hp2 for a subset of them.
Using cryo-EM, we determined the structures of the CD163 ectodomain in complex with Hb-Hp1 (Fig. 1c, d, Supplementary Figs. 2 and 3, Supplementary Table 1). Only the cleaved Hp in the complex was observed in the cryo-EM structures, while the uncleaved Hp was not detected, possibly adopting a flexible conformation. As in previous studies, the Hb-Hp1 formed a complex structure assembled as a dimer (α2β2Hp2)22. In our structures, we observed only half of the Hb-Hp1 complex binding to CD163 with clear density, while the other half displayed much weaker density overall. This structural flexibility, apparent in low-resolution reconstructions derived from 30,000 particles, prevents reliable modeling of the complete complex (Supplementary Fig. 4). We observed two distinct major classes of the complex, with CD163 forming either a trimer or a dimer (Supplementary Fig. 5a). The trimeric CD163 structure resembles a claw with three “toes” when viewed from the side, with the N-terminal SRCR domains capturing the Hb-Hp1 complex (Fig. 1c). The CD163 protomers interact through the C-terminal SRCR5-9 domains, which adopt a stable conformation (Fig. 1d, e). The dimeric CD163 bound to Hb-Hp1 exhibits a structure highly similar to the trimer, except for the absence of protomer C and variations in the SRCR9 domain of protomer B (B/SRCR9) (Supplementary Fig. 5a). In the trimeric CD163-Hb-Hp1 structure, the B/SRCR9 is stably connected to B/SRCR5 (Supplementary Fig. 5b), whereas in the dimeric CD163-Hb-Hp1 structure, the density of B/SRCR9 cannot be resolved in the cryo-EM map (Supplementary Fig. 5c). The direct connection between SRCR6 and SRCR8 appears to be stable in both resolved structures.
The N-terminal SRCR2-4 domains of CD163 protomers directly interact with the ligand (Fig. 1c, Supplementary Fig. 5a). The SRCR2 domains of protomers A and B interact with the Hp1-β subunit, while the SRCR3 and 4 domains contact the Hb subunits. Specifically, protomer A interacts with the Hb-α subunit, while protomer B interacts with the Hb-β subunit (Fig. 1c, d). The protomer C exhibits poor density, but it appears that the SRCR3 domain of protomer C may form direct contacts with Hp (Fig. 1c).
Based on the results, we propose a model for CD163-mediated recognition and internalization of ligands. CD163 forms oligomers through interactions between the SRCR5-9 domains near the cell membrane. The N-terminal SRCR domains are more flexible and play key roles in capturing ligands, such as the Hb-Hp complex. Upon binding to CD163, the ligands are internalized via endocytosis, mediated by the hydrophobic internalization motif YxxΦ (Φ represents a bulky hydrophobic residue)32 (Fig. 1f). This motif is recognized by the AP2 adaptor complex, which is involved in clathrin-dependent endocytosis38.
CD163 can form calcium-dependent oligomers
To investigate the oligomerization of CD163, we first analyzed the protein using size-exclusion chromatography (SEC). The SEC data revealed a shift in the protein peak with increasing calcium concentration, indicating that CD163 underwent conformational changes or oligomerization in a calcium concentration-dependent manner (Fig. 2a). Chromatographic analyses reveal that CD163 exhibits a calcium-dependent molecular weight distribution, transitioning from about 150 kDa in calcium-free conditions to ~500 kDa when the calcium concentration exceeds 2 mM, indicating the formation of an oligomeric species close to a trimer (Fig. 2a). To further explore the oligomerization of CD163, we performed sedimentation velocity-analytical ultracentrifugation (SV-AUC) assays (Fig. 2b, Supplementary Table 2). In the absence of calcium, CD163 remained in a monomeric state, with a peak around 147 kDa, consistent to the SEC results (Fig. 2b, Supplementary Table 2). In contrast, in the presence of calcium, CD163 underwent oligomerization (Fig. 2b). At a calcium concentration of 0.5 mM, two peaks were observed at ~102 and 152 kDa (Fig. 2b, Supplementary Table 2). Considering the effects of frictional ratio on estimated molecular weights, as explained in the Methods section, these peaks likely represent the CD163 monomer and an intermediate state between the monomer and dimer. At 1.0 mM calcium, the protein could form dimers, and at 2.5 mM calcium, which is within the range of serum calcium levels in healthy humans (2.2 to 2.6 mM)39, CD163 exists as a mixture of monomers (18.7%), dimers (26.2%) and trimers (39.1%), with the trimeric form predominating (Fig. 2b, Supplementary Table 2). Additionally, we tested other closely related metal ions to assess whether they could mediate CD163 oligomerization. Our results showed that this concentration-dependent oligomerization was specific to calcium and was not observed with other metal ions (Fig. 2c).
a SEC of CD163 in the presence of different concentrations of calcium. The molecular weights of CD163 estimated from SEC at different calcium concentrations (0–2.5 mM) were 150, 330, 400, 480, 500, and 500 kDa. b SV-AUC results of CD163 ectodomain in the presence of different calcium concentrations. c SEC of CD163 in the presence of 2.5 mM calcium, magnesium, barium, or strontium. d Main 2D classes of purified SRCR5-9. e Cryo-EM map of purified CD163 SRCR5-9. f Cryo-EM map of CD163 SRCR5-9 domains from the complex structure after local refinement. g Structural model of SRCR5-9 trimer. h Interactions between protomers B and C. i Interactions between protomers A and B. j Interactions between protomers A and C.
An expression vector containing the coding sequence for the SRCR5-9 domains of CD163 was constructed, as these domains are responsible for oligomerization according to the structures. The corresponding protein was then expressed and purified from HEK293F cells. To investigate whether the SRCR5-9 domains could form oligomers in the absence of ligand binding, we performed cryo-EM analysis and obtained a low-resolution 3D map. The analysis revealed that SRCR5-9 alone is capable of forming oligomers. The 2D classification and low-resolution cryo-EM map of SRCR5-9 showed that it forms a trimer with an equilateral triangular shape, measuring ~120 Å (Fig. 2d, e). These findings provide additional evidence that CD163 can form oligomers and the SRCR5-9 domains are sufficient for oligomerization.
Calcium-dependent interaction between CD163 protomers
Although we obtained a map of the CD163-Hb-Hp1 complex at a resolution of 3.11 Å, the local resolution, particularly in the triangular head region, is insufficient for detailed interaction analysis (Supplementary Fig. 2b). Therefore, we performed local refinement of the SRCR5-9 domains within the trimeric CD163-Hb-Hp1 complex structure (Supplementary Fig. 2a, h, I). This refinement allowed us to achieve a higher-resolution map of SRCR5-9 at 3.03 Å (Fig. 2f, Supplementary Fig. 2h, I, Supplementary Table 1). In this triangular structure, the length of the side formed by protomers A and C is `127 Å, which differs from that of the purified SRCR5-9 (Fig. 2f). Since the Hb-Hp1 complex is predominately recognized by protomers A and B, ligand binding facilitates the connection between these two protomers, leading to deviations from the purified SRCR5-9 structure.
Application of local refinement generated an enhanced density map suitable for detailed structural interpretation (Supplementary Fig. 2i). Here, we presented the density map and atomic model of SRCR5-9 domains from protomer B (Supplementary Fig. 5d, e). We observed that the SRCR5-9 domains form a shape resembling a pentagon when viewed from the side (Supplementary Fig. 5d). From the top view, they form a corner of a triangular trimer with an angle of ~80° (Supplementary Fig. 5d, e).
To stabilize this conformation, SRCR6 directly interacts with SRCR8, and SRCR5 contacts SRCR9 (Supplementary Fig. 5f). These interactions are mediated by Arg647, Ser907, Tyr580, and Gln904 between SRCR6 and SRCR8, while hydrophobic interactions also play a critical role in maintaining the stable conformation (Supplementary Fig. 5g, h). Within SRCR8, a cavity is primarily composed of valine, leucine, and tryptophan residues, forming a hydrophobic core, into which a leucine residue from SRCR6 inserts (Supplementary Fig. 5h, i). The interaction between SRCR5 and SRCR9 is driven by the contacts between Asp504 and Gly989, as well as the interactions between Arg571 with Glu933, Gln986, and Gly987 (Supplementary Fig. 5j).
In the complex structure, the interface between protomers A and C differs from the other two interfaces (Supplementary Fig. 6a–c). A/SRCR7 interacts with C/SRCR9, while in the other two interfaces, SRCR7 interacts with SRCR5 and SRCR9 from the other protomer (Supplementary Fig. 6d–f). This variation is induced by ligand binding to protomers A and B, resulting in conformational changes in the SRCR5-9 domains. A/SRCR7 and C/SRCR9 rotate in opposite directions, and the two SRCR domains interact with each other, exhibiting weaker contacts (Supplementary Fig. 6g, h).
The structure reveals the critical role of calcium ions in trimer formation (Fig. 2g–j, Supplementary Fig. 3a). The interface between protomers involves two calcium-binding sites, including a one-calcium-binding site in SRCR9, formed by residues Asp955-Asp956-Glu1022, and a two-calcium-binding site in SRCR7, formed by residues Asp745-Asp746-Asp-784-Glu785-Asn-807-Glu812, from different protomers, such as B/SRCR9 and C/SRCR7 (Fig. 2h, i). These acidic residues from the calcium-binding sites form highly negatively charged patches that are not fully compensated by calcium ions and need to be stabilized by positively charged residues. Two lysine residues, Lys811 in C/SRCR7 and Lys1021 in B/SRCR9, interact with the two calcium-binding sites, respectively, forming stable interface interactions (Fig. 2h, i). In addition, two residues in SRCR5, A/Pro561 and B/Glu564, form interactions with B/Gln805 and C/His801 in SRCR7, respectively (Fig. 2h, i). However, due to the conformational changes induced by ligand binding, there is a difference between protomers A and C (Fig. 2j). In the interface between protomers A and C, only the two-calcium-binding site in A/SRCR7 is involved in the interaction (Fig. 2j). The Lys977 residue from C/SRCR9 contacts with A/Asp746 (Fig. 2j). Besides, the residues C/Asp961 and A/Arg809 interact with each other (Fig. 2j).
The high-resolution structure demonstrated the significance of calcium ions in the oligomerization of CD163. To investigate whether this oligomerization is conserved across species, we analyzed the protein sequences of CD163 from different organisms (Supplementary Fig. 7a). Multiple sequences alignment revealed a high degree of identity in the SRCR5-9 domains, with the calcium-binding residues being highly conserved among mammals (Supplementary Fig. 7a). The two lysine residues that interact with the calcium-binding sites are also found to be conserved in these CD163 proteins (Supplementary Fig. 7a). Furthermore, the structures of CD163 SRCR5-9 domains from various species, predicted by Alphafold, exhibited a conformation strikingly similar to the solved structure (Supplementary Fig. 7b). This suggests that calcium-dependent oligomerization is a conserved mechanism among mammalian CD163.
Calcium-dependent interaction between CD163 and Hb-Hp
To investigate the molecular mechanism underlying the recognition of the Hb-Hp1 complex by CD163, a mask was created to enclose the Hb-Hp1 complex along with the SRCR2-4 domains of protomers A and B. The mask was then applied for local refinement, yielding a final map at a resolution of 2.94 Å (Fig. 3a, Supplementary Fig. 2a, j, k, Supplementary Table 1).
a Cryo-EM map of Hb-Hp1 binding to SRCR2-4 of two CD163 protomers after local refinement. b Structural model of CD163 protomers bound to Hb-Hp1. c Interactions between CD163 and Hb-Hp1 complex. d NHS-pull down analyses of Hb-Hp binding activity of recombinant CD163 variants, with each experiment has been repeated at least twice. Overexpressed and purified CD163 variants were incubated with ligand-bound beads in the presence of 2.5 mM calcium. Source data are provided as a Source Data file.
The recognition of Hb-Hp1 by CD163 is also dependent on calcium3. There are two calcium-binding sites located in SRCR2 and SRCR3, respectively (Fig. 3b, c, Supplementary Fig. 3b), consistent with previous studies40,41. This calcium-dependent ligand binding involves a one-calcium-binding site in SRCR3, formed by residues Asp292-Asp293-Glu359, as well as a two-calcium-binding site in SRCR2, formed by residues Asp185-Asp186-Asp224-Asn247-Glu252 (Fig. 3c, Supplementary Fig. 3b, 8a). The SRCR2 domains of the two protomers are responsible for interacting with Hp1 subunit (Fig. 3b, Supplementary Fig. 8a). Specifically, Hp1/Lys262 binds to residues Asp186 and Glu252 of the calcium-binding site in A/SRCR2, and Hp1/Lys94 binds to Asp186 and Asn247 of the calcium-binding site in B/SRCR2 (Fig. 3c, Supplementary Fig. 3b, 8a). In addition to calcium-mediated contacts, A/Glu216, A/Asp249, and B/N247 contribute to the recognition of Hp1 (Fig. 3c, Supplementary Fig. 8a). The SRCR3 domains are responsible for recognizing the Hb subunits (Fig. 3b, Supplementary Fig. 8a). Protomer A interacts with the Hb-α subunit, where the residue Hb-α/Lys11 binds to Asp293 and Glu359 of the calcium-binding site in A/SRCR3 (Fig. 3c, Supplementary Fig. 3b, 8a). Protomer B interacts with the Hb-β subunit, with the residue Hb-β/Lys82 binding to Asp293 and Glu359 of the calcium-binding site in B/SRCR3 (Fig. 3c, Supplementary Fig. 3b, 8a). Additionally, A/Tyr354 contacts Hb-α/Asp74, while B/Gly294, B/Lys352, as well as B/Tyr354 form interactions with Hb-β (Fig. 3c, Supplementary Fig. 8a). In addition, SRCR4 of protomer B also directly binds to Hb-β subunit through interactions between Hb-β/Asp79 with B/Gly401 and B/Tyr465 (Supplementary Fig. 8a). Although we cannot draw the direct interactions between protomer C and Hb-Hp1 due to the poor density, we observed that residue Lys211 from Hp1 is in close proximity to C/Asp293 and C/Glu359 (Supplementary Fig. 8b). The residues Asp292-Asp293-Glu359 can form a calcium-binding site, suggesting that there may also be a calcium-mediated interaction between protomer C and Hp1 (Supplementary Fig. 8b). This potential interaction could enhance the trimer’s selectivity and affinity for the Hb-Hp1 complex.
Previous studies have demonstrated the calcium-dependent recognition of the Hb-Hp by CD163, and the calcium-binding sites in SRCR2-3 domains play crucial roles in enabling SRCR1-5 to recognize Hb-Hp3,41. To investigate the roles of these residues in Hb-Hp binding by the full CD163 ectodomain, we generated several CD163 mutants and successfully purified some of the proteins in HEK293F cells. Using these protein mutants, we conducted NHS-pull down assays to assess the impact of the calcium-binding sites on ligand binding (Fig. 3d). The results indicated that mutations in the calcium-coordinated residues of SRCR3, such as E359A and D292A/D293A, did not prevent CD163 from binding to the Hb-Hp complex (Fig. 3d). However, when any residue in the two-calcium-coordinated site in SRCR2 was mutated to alanine, the interaction between CD163 and the Hb-Hp complex was disrupted (Fig. 3d). This disruption was observed in both the multi-site mutants and the single-site mutants, including D185A, D186A, N247A, and E252A (Fig. 3d). These findings suggest that the two-calcium-binding site in SRCR2 is essential for the binding of Hb-Hp complex by CD163.
To obtain structural information, we prepared the CD163-Hb-Hp1 complex with a limited amount of CD163, at a molar ratio of 1:1 (monomeric CD163:Hb-Hp1 complex), which facilitated easier separation by SEC of the free Hb-Hp1 complex from the CD163-Hb-Hp1 complex (Supplementary Fig. 1e). To investigate whether CD163 can recognize both sides of the Hb-Hp1 dimer, we incubated CD163 with Hb-Hp1 at a molar ratio of 10:1 and analyzed the mixture by SEC, resulting in two overlapped peaks (Supplementary Fig. 9a). We then examined the particles from peak1 and collected some data using cryo-EM (Supplementary Fig. 9b). 2D classification revealed that a small fraction of particles (about 9.9%) showed two CD163 oligomers recognizing both sides of the Hb-Hp1 dimer (Supplementary Fig. 9c). We then performed SV-AUC in the presence of 5 mM calcium (Supplementary Fig. 9d–g). CD163 exists as monomers, dimers, and trimers, while Hb-Hp1 remains in monomeric state with a molecular weight ~128 kDa in the presence of calcium (Supplementary Fig. 9d, e). When mixed with Hb-Hp1 at a 1:1 molar ratio, the four observed peaks likely correspond to Hb-Hp1, monomeric CD163 bound to Hb-Hp1, dimeric CD163 bound to Hb-Hp1, and trimeric CD163 bound to Hb-Hp1 (Supplementary Fig. 9f). When the molar ratio was adjusted to 10:1, most of the proteins were observed in a peak ~420 kDa (60% of the mixture) (Supplementary Fig. 9g). Considering the effects of frictional ratio on molecular weights estimation, this peak likely represents the interaction of CD163 with Hb-Hp1 at a 3:1 molar ratio (monomeric CD163:Hb-Hp1), with CD163 corresponding to either a single trimer or a combination of one monomer and one dimer. Additionally, a peak around 616 kDa was observed in SV-AUC (10.5% of the mixture), corresponding to a 5:1 molar ratio interaction (Supplementary Fig. 9g). In this case, the Hb-Hp1 should interact with a CD163 trimer and a dimer. However, we did not observe two CD163 trimers binding to the Hb-Hp1 complex.
CD163 oligomerization is more efficient for endocytosis
To assess the importance of the oligomeric form of CD163 for its function, we purified an oligomer disruption mutant, CD163TriMut, which contains four mutations (745 A/746 A/955 A/956 A). We then performed SEC assays, and the results showed that the peak of CD163TriMut did not shift in the presence of 2.5 mM calcium (Fig. 4a). SV-AUC analysis further confirmed that the mutant maintained a monomeric form, regardless of the calcium presence (Fig. 4b). Furthermore, we mixed the CD163TriMut with Hb-Hp1 complex and analyzed the mixture using SV-AUC. The resulting peak, representing the complex of CD163 with Hb-Hp1, indicated that these two components interact (Fig. 4c). We also conducted NHS-pull down assays with both Hp isoforms, which demonstrated that CD163TriMut retained its ability to interact with the Hb-Hp complex (Fig. 4d).
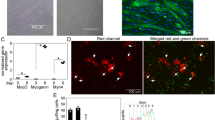
a SEC of CD163TriMut in the presence of 0 and 2.5 mM calcium. b SV-AUC of CD163TriMut in the presence of 2.5 mM calcium. The molecular weight of the peak in SV-AUC is ~139 kDa. c SV-AUC of CD163TriMut mixed with Hb-Hp1 at a molar ratio of 1:2. The frictional ratio is 1.51812. The molecular weights of the peaks in SV-AUC are ~138 and ~251 kDa. d Pull down analyses of Hb-Hp binding activity of recombinant CD163TriMut in the presence of 2.5 mM calcium, with each experiment repeated at least twice. e Western blot analyses of CD163 expression in HEK293T cells and HEK293T cells transfected with plasmids overexpressing full-length CD163. This experiment has been repeated twice. Uncropped blots in Supplementary Fig. 13. f Flow cytometry and imaging analyses of HEK293T cells binding to and internalizing Hb-Hp. g Flow cytometry and imaging analyses of HEK293T cells overexpressing CD163, showing binding to and internalizing Hb-Hp. h Flow cytometry and imaging analyses of HEK293T cells overexpressing CD163TriMut, showing binding to and internalizing Hb-Hp. Each flow cytometry experiment was repeated three times individually (marked with black dots on the bars). Data was plotted as the mean with SD. Statistical analysis is based on the two-tailed unpaired t-test (****, p < 0.0001; **, p = 0.0068). Source data are provided as a Source Data file.
To investigate the functional implications of CD163TriMut, we performed flow cytometry assays using HEK293T cells. We first confirmed that these cells did not express any endogenous CD163, and that cells transfected with plasmids were able to produce CD163 (Fig. 4e). Flow cytometry and imaging flow cytometry analyses demonstrated that HEK293T cells could not facilitate the internalization of Hb-Hp (Fig. 4f, Supplementary Fig. 10a). In contrast, both CD163 and CD163TriMut were capable of binding to the Hb-Hp complexes and mediating the endocytosis of ligands (Fig. 4g, h, Supplementary Fig. 10b, c). Interestingly, the efficiency of CD163TriMut-mediated recognition and internalization of Hb-Hp was nearly ten-fold lower than that of CD163 (Fig. 4g, h). These results demonstrate the oligomeric form is efficient and significant for CD163-mediated endocytosis of Hb-Hp complex. Additionally, the efficiency of Hb-Hp2 recognition and internalization by CD163 or CD163TriMut was higher than that of the Hb-Hp1 complex (Fig. 4g, h). Due to its duplicated CCP domain, Hp2 can form various oligomers, resulting in enhanced avidity in the Hb-Hp2 complex that facilitates interaction with CD163. This contributes to an enhanced functional affinity of the ligand, which is consistent with the higher affinity observed in a previous study3.
Discussion
Our results elucidate how CD163 recognizes the Hb-Hp complex. Contrary to the previous model, which suggested that two monomeric CD163 chains bind symmetrically to the dimeric Hb-Hp1 complex, our data suggest that CD163 oligomers interact with half SIDE of the dimeric ligand in an asymmetric manner. This interaction involves binding of the Hb and heavy chain of Hp1 to the SRCR2-4 domains of CD163. In humans, the Hb and the heavy chain of Hp interact similarly in both Hb-Hp1 and Hb-Hp2 complexes28, indicating that CD163 recognizes these complexes using a common molecular mechanism. Differently, flow cytometry showed that the Hb-Hp2 complex exhibits a higher affinity for CD163 than Hb-Hp1, which facilitates a more efficient elimination of free Hb in humans, consistent with previous studies3. Notably, a recent study, consistent with our findings, reported the analysis of the CD163 and Hb-Hp1 complex structure42. They also obtained results for the binding of both dimeric and trimeric CD163 to Hb-Hp1. In our study, Hp was recombinantly produced and contained uncleaved products, while Hb was obtained from a commercial source, which may introduce potential limitations. However, our results are consistent with the published work, in which the Hb-Hp1 complex was directly purified from human blood42.
A model was manually built based on the resolved density map, with two full-length CD163 trimers bound to the Hb-Hp1 dimer. This model showed that protomer C of one CD163 trimer is positioned too close to protomer B or C of the other CD163, leading to possible steric hindrance. This may explain why we did not observe Hb-Hp1 interacting with two CD163 trimers simultaneously in SV-AUC (Supplementary Fig. 11a). A few 2D class averages observed in cryo-EM and SV-AUC appear to show a trimer and a dimer (lacking protomer C) binding to both sides of the Hb-Hp1 dimer (Supplementary Fig. 9). However, in the recent published work, based on modelling, the authors did not exclude the possibility of two trimeric CD163 binding to the Hb-Hp1 complex, but this complex was not found in their cryo-EM data42. In contrast, we found that the N-terminal SRCR domain of one CD163 protomer is positioned as SRCR2 in our model (Supplementary Fig. 11b). Based on our high-resolution map, we are confident that this domain corresponds to SRCR2, not SRCR1 (Supplementary Fig. 11c). Comparison of the Hb-Hp1 structures revealed that the Hb and the SP of Hp maintain a highly rigid conformation, while the CCP exhibits significant conformational changes, leading to a wobbling structure of Hb-Hp1 (Supplementary Fig. 11d, e). This flexibility may explain the poor density observed for the other half of the Hb-Hp1 dimer in the cryo-EM map.
Our study provides insights into the specific interaction between CD163 and the Hb-Hp complex. The adjacent SRCR domains 2 and 3 contain two calcium-binding sites, which is consistent with previous studies suggesting that calcium-dependent two-point electrostatic pairing is a common mechanism for coupling ligands to endocytic receptors41. Additionally, we confirmed that the Hp loop 3, which has been demonstrated to be crucial for recognition of Hb-Hp41, plays a key role in this interaction in our structures. The residue Lys262 in loop 3 forms a significant interaction with the calcium-binding site in SRCR2. Besides, residues Arg252 and Gln249 in loop 3 also contribute to the interaction (Fig. 4c, Supplementary Fig. 9a). The SRCR2 domain of CD163 contains a two-calcium-coordinated acidic cluster which is critical for the recognition of the Hb-Hp complex. Mutations in this site abolish the interaction, consistent with previous findings involving mutations in the acidic triad of SRCR domain 2 (D185A, D186A, E252A)41. In previous studies, single (E359A) and double (D292A, D293A) mutations in the one-calcium-binding site in SRCR3 were shown to completely abrogate interactions between CD163 SRCR1-5 and the Hb-Hp complex41. However, our results revealed that CD163 ectodomain carrying these mutations still retained the ability to recognize the Hb-Hp complex. This suggests that the ability of the CD163 ectodomain, which can form oligomers, to capture the Hb-Hp complex is less affected by these mutations. In contrast, the mutations in SRCR3 appear to have a more significant impact on the recognition of the ligand by SRCR1-5, which exists as a monomer. The monomeric state likely exhibits reduced ligand-binding affinity, making it more susceptible to mutation-induced perturbations.
SRCR domains 7 and 9 also contain conserved calcium-binding sites, which have been suggested to be possibly involved in the binding of free Hb as well as the recognition of other ligands41. However, our results revealed that the two calcium-coordinated acidic clusters in SRCR7 and 9 can form a two-point interaction with two lysine residues, playing crucial roles in forming CD163 oligomers. Furthermore, we found that CD163 oligomerization is specifically dependent on calcium concentration. Since calcium concentration has been measured to be less than 10 μM in endosomes43, our findings indicate that CD163 oligomerizes on the cell surface to bind ligands and dissociates into monomers in endosomes. This behavior is appropriate for the segregation of ligands and the recycling of CD163. The recent study also experimentally confirmed the critical role of calcium ions in the oligomerization of CD16342. In addition to the calcium-binding sites involved in Hb-Hp recognition and CD163 oligomerization, we identified at least three calcium-binding sites within SRCR6, 7, and 9 (Supplementary Fig. 3c). These calcium-binding residues may have the potential to recognize other ligands.
A crystal structure of SRCR5-9 from wild boar has been solved at a resolution of 2.5 Å and deposited recently (PDB: 8H7J). The crystal was grown in the absence of calcium, and the structure was found to be a monomer. The conformation of the SRCR5-9 from the crystal structure closely resembles the SRCR5-9 domains in our structure, suggesting that calcium binding does not induce significant conformational changes in the SRCR5-9 domains (Supplementary Fig. 12a, b). Surface electrostatic potential analysis revealed that electrostatic interactions, particularly those involving calcium-coordinated acidic clusters interacting with basic amino acid residues, are the dominant forces driving both oligomerization and Hb-Hp recognition (Fig. 5a, b). Two models illustrate the differences between the two dual-point electrostatic interactions induced by calcium binding (Fig. 5c, d). These calcium-dependent interactions open up additional possibilities for SRCR domain-mediated protein-protein interactions, including homomeric aggregation and ligand recognition.
a, b Surface electrostatic potential of CD163 and Hb-Hp showed the surfaces in the interface are highly charged, especially the calcium-coordinated acidic clusters. Main electrostatic interactions are circled and connected with dotted lines. c Model for calcium-dependent interaction between CD163 protomers A and B. d Model for calcium-dependent interaction between two CD163 protomers and Hb-Hp complex.
In summary, our study elucidates the molecular mechanism of CD163 oligomerization and Hb-Hp recognition, while also highlighting the functional significance of the oligomeric form of CD163 in facilitating the endocytosis of the Hb-Hp complex. Mutational analysis of CD163TriMut revealed a significantly reduced efficiency in mediating the internalization of the Hb-Hp complex. These findings deepen our understanding of CD163 function and have important implications for the study of CD163-positive monocytes/macrophages and the development of novel therapeutic approaches targeting CD163-mediated processes.
Methods
Cell culture
HEK293F and HEK293T (ATCC, CRL-3216) cells were originally purchased from the American Type Culture Collection. HEK293F cells were cultured using FreeStyle 293 expression medium (Gibco) in a humidified shaker at 37 °C with 5% CO2. HEK293T cells were cultured in DMEM/HIGH GLUCOSE (HyClone) supplemented with 1% penicillin–streptomycin (Gibco) and 10% FBS (YEASEN) in a humidified incubator at 37 °C with 5% CO2.
Protein expression and purification
The gene of human CD163 (residue 1-1050), CD163 SRCR5-9 (residue 1–41 and residue 473–1050), human haptoglobin Hp1 (residue 1-347), as well as human haptoglobin Hp2 (residue 1–406) were cloned and inserted into BamHI and XhoI sites of pBMCL1 vector with a C-terminal 6×His tag and a C-terminal twin-strep-tag44. The CD163 mutated variants (D185A, D186A, D224A, N247A, E252A, E359A, D185A/D186A, D292A/D293A, D186A/D292A/D293A, D185A/D186A/D292A/D293A, D745A/D746A/D955A/D956A) were generated by site-directed mutagenesis using pBMCL1-CD163 (residue 1–1050) as a template using the Q5 Site-Directed Mutagenesis Kit (E0554S, New England Biolabs). Primers used for site-directed mutagenesis are provided in the Supplementary Data. Plasmids were transfected into HEK293F cells (cell density between 1 and 1.5 million cells/mL) using polyethylenimine (Polysciences). Cells were cultured in a humidified shaker for four days after transfection. Then the conditioned media were collected by centrifugation at 1000 × g, concentrated using a Hydrosart Ultrafilter (Sartorius), and exchanged into the binding buffer (25 mM Tris-HCl, pH 8.0, 150 mM NaCl). The proteins were purified using a Ni-NTA affinity column (GE Life Sciences) and eluted with 25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 250 mM imidazole. Then the proteins were purified using Superose 6 increase column (GE Life Sciences) in the final buffer (20 mM HEPES, pH 7.2, 150 mM NaCl). Hp1 was used for all the assays in this study and Hp2 was used to perform pull-down and flow cytometry assays.
Human hemoglobin (H7379, Sigma) and Hp were mixed at a molar ratio of 1.5:1 and incubated at 4 °C for 1 h. The Hb-Hp complex were then purified using Superose 6 increase column in the final buffer. Then CD163 and Hb-Hp complex were mixed with a molar ratio (monomeric CD163:Hb-Hp1 complex) of 1:1 for Hp1, or 5:1 for Hp2 in 25 mM Tris, pH 8.0, 150 mM NaCl, 2.5 mM CaCl2 and incubated at 4 °C for 1 h. The CD163-Hb-Hp complex subsequently purified using Superose 6 increase column (GE Life Sciences) in the final buffer containing 2.5 mM CaCl2. All peak fractions containing the target protein and protein complex were collected and analyzed by SDS-PAGE.
Cryo-EM sample preparation and data collection
The purified CD163-Hb-Hp1 complex was diluted to 0.4 mg/mL and loaded onto a glow-discharged Holey-carbon gold grid (Quantifoil, R1.2/1.3). After incubation for 10 s, the grids were then blotted for 2 s with a blot force of −1 at 4 °C and 100% humidity and plunge frozen in liquid ethane using a Vitrobot Mark IV System (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The grids were first screened using a 200 kV Talos Arctica transmission electron microscope equipped with a Ceta camera (Thermo Fisher Scientific, Inc.). Data collection was carried out using a Titan Krios electron microscope (Thermo Fisher Scientific, Inc.) operated at 300 kV. Movies were recorded on a K3 Summit direct electron detector (Gatan Inc.) using EPU (E Pluribus Unum, Thermo Scientific), in the super-resolution mode at a nominal magnification of ×81,000 with a defocus range of –1.0 to –2.0 μm, resulting in a pixel size of 1.07 Å per pixel. A dose rate of 17.9 electrons per pixel per second and an exposure time of 3.84 s were used, generating 32 movie frames with a total dose of ~60 e/Å2. A total of 5919 movie stacks were collected for CD163-Hb-Hp1 complex.
Cryo-EM data processing
Movie frames were motion-corrected and dose-weighted using the MotionCor245. The defocus values were estimated with the Gctf program46. Around 2000 particles were manually picked without a template to generate 2D averages for subsequent template-based auto-picking. 2,739,731 particles were auto-picked and extracted from the dose-weighted micrographs using RELION47 and subjected to 2D classification without symmetry restriction using cryoSPARC48. 1,032,569 good particles were selected and used to generate initial models using the ab initio reconstruction with no symmetry function in CryoSPARC. The 3D maps were used as references for running heterogeneous refinement using cryoSPARC and generated two classes of CD163-Hb-Hp1 complex. 432 933 particles from the best class were used for non-uniform refinement in cryoSPARC to obtain the final 3D reconstruction of trimeric CD163-Hb-Hp1 complex with an overall resolution of 3.11 Å. 332,640 particles were used for final non-uniform refinement of dimeric CD163-Hb-Hp1 complex, yielding a final reconstruction with an overall resolution of 3.21 Å. Reported resolutions are based on the gold-standard Fourier shell correlation (FSC) of 0.143 criterion49. Local resolution estimation and filtering was carried out using CryoSPARC. One mask that encases the SRCR5-9 domains and another mask that encases the Hb-Hp1 complex and SRCR2-4 domains of protomers A and B were generated and used to perform local refinement in cryoSPARC to obtain final local 3D maps with higher resolution at 3.03 Å and 2.95 Å, respectively. 3D reconstruction and local resolution were summarized in Supplementary information Fig. S2.
Model building and refinement
The reported crystal structure of human Hb-Hp1 complex (PDB: 4WJG) and Alphafold50 predicted CD163 structure were used for the model building of our CD163-Hb-Hp1 complexes. The corresponding structures were roughly fitted into our cryo-EM maps using UCSF ChimeraX51. The structure models were then adjusted and refined manually in Coot52. Further real-space refinements were carried out using phenix.real_space_refine53. Statistics associated with data collection, 3D reconstruction, and model building were summarized in Supplementary Information Table S1.
Sequence alignment
Sequence alignment was performed using Clustal Omega54 and the results were visualized and modified with Esprint 355.
Size exclusion chromatography
Size exclusion chromatography was carried out using an AKTA Purifier system (GE Healthcare). Purified CD163 was divided and incubated in the gel filtration buffer (20 mM HEPES, pH 7.2, 150 mM NaCl) with different concentration of calcium, as well as different metal ions at 4 °C for 30 min. The samples were loaded onto a Superose 6 increase column (GE Life Sciences).
SV-AUC
Protein samples were diluted to 1 mg/mL and incubated in buffer (20 mM HEPES, pH 7.2, 150 mM NaCl) with different concentrations of calcium. For WT CD163 and Hb-Hp1, they were incubated with different molar ratios of 1:1 and 10:1 for 1 h and then used for SV-AUC analysis. And for CD163TriMut and Hb-Hp1, they were incubated at a 1:2 molar ratio. SV-AUC experiments were carried out using a 12-mm charcoal-filled Epon centerpieces (Beckman, 306493) and a four-hole An50 Ti rotor at 40,000 rpm (Optima AUC analytical-ultracentrifuge, Beckman Coulter) with absorbance detection at 280 nm at 20 °C. The data were analyzed with SEDFIT software to obtain sedimentation coefficient distribution C (S)56. Samples were analyzed at the same time using the same equipment.
During the analysis of a mixed sample, the software automatically computes a frictional ratio for all components, which may not accurately reflect the properties of each individual component, potentially leading to errors in the estimation of molecular weights. The frictional ratio (f/f₀) serves as a critical parameter characterizing molecular shape and size. Defined as the ratio of a molecule’s actual frictional coefficient (f) in solution to that of an ideal spherical particle with equivalent mass and density (f₀), this parameter reflects molecular asymmetry57. The Svedberg equation is helpful for evaluating and comparing the estimated molecular weights: M = (RTs0)/[D(1-ῡρ)]×(f/f0), where M: molecular weight, R: gas constant, T: temperature, s0: theoretical sedimentation coefficient for a sphere of same mass and density, D: diffusion coefficient, ῡ: partial specific volume, and ρ: solvent density57.
NHS-pull down assay
NHS-Activated Beads (Smart Lifescience) were washed three times with 1 mM HCl and one time with 200 mM NaHCO3, pH 8.0, 500 mM NaCl before use. Hb-Hp1 or Hb-Hp2 complex at concentration of 10 mg/mL was incubated with the beads at room temperature for 2 h. The beads were washed three times and blocked with 100 mM Tris, pH 8.5 at room temperature for 2 h. Then the beads were washed three times and stored in PBS. Purified CD163 and its variants at a concentration of 0.4 mg/mL were incubated with beads in binding buffer (20 mM HEPES, pH 7.2, 150 mM NaCl, 2.5 mM CaCl2) for 1 h at 4 °C. The beads were spun down and then washed three times using Binding buffer. Proteins retained on the beads were suspended and directly used for SDS-PAGE. The results were analyzed by SDS-PAGE and Coomassie staining.
Immunoblotting assay
293T cells, and 293T cells that transfected with pBMCL1 plasmids44 overexpressing fusion proteins composed of full length CD163 and C-terminal GFP were lysed with Lysis Buffer (20 mM HEPES, pH 7.2, 150 mM NaCl, 5 mM CaCl2, 1×protease inhibitor cocktail, 0.5% LMNG) at 4 °C for 2 h. At the same time, HRV 3 C protease was added to remove the GFP. The precipitation was then removed by centrifugation at 10,000 × g. The protein concentration of the supernatant was measured by the Bicinchoninic Acid Assay (BCA). Twenty microgram of total proteins were mixed with 6× protein buffer (TransGen Biotech) and heated to 100 °C for 2 min. After fractionated by 12% SDS-PAGE, the proteins were transferred to nitrocellulose membranes (GE Healthcare). The membranes were incubated in TBST buffer containing 5% skim milk for 2 h at room temperature and then incubated with the specific primary antibodies at 4 °C overnight. After three washes in TBST, each time for 5 min, the membranes were incubated with HRP-conjugated goat anti-rabbit IgG antibodies (1:50000; HA1001, HuaBio) for 2 h at room temperature. After three washes in TBST, the membranes were incubated with the Super ECL Detection Reagent (YEASEN) and imaged by the Tanon-5200 Chemiluminescent Imaging System (Tanon). The primary rabbit monoclonal antibody anti-CD163 (1:500; JA51-30, HuaBio) and anti-β-actin antibody (1:100,000; AC026, ABclonal) were used in this study. Uncropped and unprocessed scans of the blots are provided in the Source Data file.
Flow cytometry
To examine the endocytosis of Hb-Hp complex mediated by CD163, HEK293T cells were transfected with pBMCL1 plasmids44 encoding full-length CD163-GFP, and full-length CD163TriMut-GFP for 48 h. Twin-strep-tagged Hb-Hp1 and twin-strep-tagged Hb-Hp2 purified from HEK293F cells were incubated with PE-Streptavidin (BioLegend) at 4 °C for 1 h firstly. Then the cells were incubated with the marked twin-strep-tagged Hb-Hp1 and Hb-Hp2 at a concentration of 6.5 µg/mL and 7.4 µg/mL at 4 °C for 30 min, respectively. An additional 30 min incubation at 37 °C was followed before the cells were subsequently collected and washed with DPBS (HyClone) for 3 times. Then the cells were incubated with 7-AAD Viability Staining Solution (BioLegend) to exclude dead cells on ice for 5 min in the dark before being analyzed with BD LSRFortessa flow cytometry. The data was analyzed using FlowJo software.
Imaging flow cytometry
The incubation method was the same as flow cytometry, but twin-strep-tagged Hb-Hp complexes were marked with APC-Streptavidin (Invitrogen) and nuclei were stained with Hoechst 33342 (Beyotime) before being trypsinized instead of 7-AAD staining. Finally, cells were resuspended in 50 µl DPBS supplemented with 5% FBS. Images were acquired in the INSPIRE™ software on the Aminis ImagestreamX Mark II imaging flow cytometer at ×60 magnification. Images were processed with IDEAS 6.2 software.
Statistics and reproducibility
All statistical analyses were conducted with GraphPad Prism version 8. Each group was repeated three times individually for the flow cytometry experiment and analyzed equally. All the flow cytometry data were presented as mean ± SD, and the significance was evaluated using the two-tailed unpaired Student’s t-test. The statistical comparisons were considered significant if the p-values were below 0.05. No statistical method was used to predetermine sample size. No data were excluded from the analyses. The randomization and blinding were not required for the flow cytometry experiment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Cryo-EM density maps of trimeric CD163 in complex with Hb-Hp, dimeric CD163 in complex with Hb-Hp, local SRCR5-9 domains, and local Hb-Hp binding to CD163 have been deposited in the Electron Microscopy Data Bank under accession codes EMD-38485, EMD-38486, EMD-38480, and EMD-38490. Structural coordinates have been deposited in the Protein Data Bank under the accession codes 8XMP, 8XMQ, 8XMK, and 8XMW. Source data are provided with this paper.
References
Law, S. A. et al. A new macrophage differentiation antigen which is a member of the scavenger receptor superfamily. Eur. J. Immunol. 23, 2320–2325 (1993).
Van Gorp, H., Delputte, P. L. & Nauwynck, H. J. Scavenger receptor CD163, a Jack-of-all-trades and potential target for cell-directed therapy. Mol. Immunol. 47, 1650–1660 (2010).
Kristiansen, M. et al. Identification of the haemoglobin scavenger receptor. Nature 409, 198–201 (2001).
Geissmann, F. et al. Development of monocytes, macrophages, and dendritic cells. Science 327, 656–661 (2010).
Yona, S. & Jung, S. Monocytes: subsets, origins, fates and functions. Curr. Opin. Hematol. 17, 53–59 (2010).
Hazzaa, H. H. et al. Expression of CD 163 in hereditary gingival fibromatosis: a possible association with TGF-β1. J. Oral. Pathol. Med. 47, 286–292 (2018).
Jones, K. et al. Serum CD163 and TARC as disease response biomarkers in classical Hodgkin lymphoma. Clin. Cancer Res. 19, 731–742 (2013).
Ye, H., Wang, L. Y., Zhao, J. & Wang, K. Increased CD163 expression is associated with acute-on-chronic hepatitis B liver failure. World J. Gastroenterol. 19, 2818 (2013).
Li, J., Liu, C. H., Xu, D. L. & Gao, B. Significance of CD163-positive macrophages in proliferative glomerulonephritis. Am. J. Med. Sci. 350, 387–392 (2015).
Ritter, M., Buechler, C., Langmann, T. & Schmitz, G. Genomic organization and chromosomal localization of the human CD163 (M130) gene: a member of the scavenger receptor cysteine-rich superfamily. Biochem. Biophys. Res. Commun. 260, 466–474 (1999).
Etzerodt, A., Maniecki, M. B., Møller, K., Møller, H. J. & Moestrup, S. K. Tumor necrosis factor α-converting enzyme (TACE/ADAM17) mediates ectodomain shedding of the scavenger receptor CD163. J. Leukoc. Biol. 88, 1201–1205 (2010).
Fabriek, B. O. et al. The macrophage scavenger receptor CD163 functions as an innate immune sensor for bacteria. Blood 113, 887–892 (2009).
Calvert, J. G. et al. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J. Virol. 81, 7371–7379 (2007).
Sánchez-Torres, C. et al. Expression of porcine CD163 on monocytes/macrophages correlates with permissiveness to African swine fever infection. Arch. Virol. 148, 2307–2323 (2003).
Barbe, E., Huitinga, I., Döpp, E. A., Bauer, J. & Dijkstra, C. D. A novel bone marrow frozen section assay for studying hematopoietic interactions in situ: the role of stromal bone marrow macrophages in erythroblast binding. J. Cell Sci. 109, 2937–2945 (1996).
Fabriek, B. O. et al. The macrophage CD163 surface glycoprotein is an erythroblast adhesion receptor. Blood 109, 5223–5229 (2007).
Bover, L. C. et al. A previously unrecognized protein-protein interaction between TWEAK and CD163: potential biological implications. J. Immunol. 178, 8183–8194 (2007).
Maecker, H. et al. TWEAK attenuates the transition from innate to adaptive immunity. Cell 123, 931–944 (2005).
Sadrzadeh, S. M. et al. Hemoglobin. A biologic fenton reagent. J. Biol. Chem. 259, 14354–14356 (1984).
Ascenzi, P. et al. Hemoglobin and heme scavenging. IUBMB Life 57, 749–759 (2005).
Hwang, P. K. & Greer, J. Interaction between hemoglobin subunits in the hemoglobin. Haptoglobin complex. J. Biol. Chem. 255, 3038–3041 (1980).
Andersen, C. B. F. et al. Structure of the haptoglobin–haemoglobin complex. Nature 489, 456–459 (2012).
Nagel, R. L. & Gibson, Q. H. The binding of hemoglobin to haptoglobin and its relation to subunit dissociation of hemoglobin. J. Biol. Chem. 246, 69–73 (1971).
Shim, B. S., Lee, T. H. & Kang, Y. S. Immunological and biochemical investigations of human serum haptoglobin: composition of haptoglobin–haemoglobin intermediate, haemoglobin-binding sites and presence of additional alleles for β-chain. Nature 207, 1264–1267 (1965).
Bowman, B. H. & Kurosky, A. Haptoglobin: the evolutionary product of duplication, unequal crossing over, and point mutation. Adv. Hum. Genet. 12, 189–261 (1982).
Kurosky, A. et al. Covalent structure of human haptoglobin: a serine protease homolog. Proc. Natl. Acad. Sci. USA 77, 3388–3392 (1980).
Maeda, N., Yang, F., Barnett, D. R., Bowman, B. H. & Smithies, O. Duplication within the haptoglobin Hp2 gene. Nature 309, 131–135 (1984).
Andersen, C. B. F. et al. Haptoglobin. Antioxid. Redox Signal. 26, 814–831 (2017).
Wejman, J. C., Hovsepian, D., Wall, J. S., Hainfeld, J. F. & Greer, J. Structure and assembly of haptoglobin polymers by electron microscopy. J. Mol. Biol. 174, 343–368 (1984).
Madsen, M. et al. Molecular characterization of the haptoglobin· hemoglobin receptor CD163: Ligand binding properties of the scavenger receptor cysteine-rich domain region. J. Biol. Chem. 279, 51561–51567 (2004).
Schaer, C. A., Schoedon, G., Imhof, A., Kurrer, M. O. & Schaer, D. J. Constitutive endocytosis of CD163 mediates hemoglobin-heme uptake and determines the noninflammatory and protective transcriptional response of macrophages to hemoglobin. Circ. Res. 99, 943–950 (2006).
Nielsen, M. J., Madsen, M., Møller, H. J. & Moestrup, S. K. The macrophage scavenger receptor CD163: endocytic properties of cytoplasmic tail variants. J. Leukoc. Biol. 79, 837–845 (2006).
Møller, H. J. et al. Soluble CD163: a marker molecule for monocyte/macrophage activity in disease. Scand. J. Clin. Lab. Investig. 62, 29–33 (2002).
Schaer, D. J. et al. Soluble hemoglobin–haptoglobin scavenger receptor CD163 as a lineage-specific marker in the reactive hemophagocytic syndrome. Eur. J. Haematol. 74, 6–10 (2005).
Etzerodt, A. et al. Specific targeting of CD163+ TAMs mobilizes inflammatory monocytes and promotes T cell-mediated tumor regression. J. Exp. Med. 216, 2394–2411 (2019).
Bachli, E. B., Schaer, D. J., Walter, R. B., Fehr, J. & Schoedon, G. Functional expression of the CD163 scavenger receptor on acute myeloid leukemia cells of monocytic lineage. J. Leukoc. Biol. 79, 312–318 (2006).
Cheng, Z., Zhang, D., Gong, B., Wang, P. & Liu, F. CD163 as a novel target gene of STAT3 is a potential therapeutic target for gastric cancer. Oncotarget 8, 87244 (2017).
Boll, W. et al. Sequence requirements for the recognition of tyrosine-based endocytic signals by clathrin AP-2 complexes. EMBO J. 15, 5789–5795 (1996).
Robertson, W. G., Marshall, R. W. & Walser, M. Calcium measurements in serum and plasma—total and ionized. Crit. Rev. Clin. Lab Sci. 11, 271–304 (1979).
Ojala, J. R., Pikkarainen, T., Tuuttila, A., Sandalova, T. & Tryggvason, K. Crystal structure of the cysteine-rich domain of scavenger receptor MARCO reveals the presence of a basic and an acidic cluster that both contribute to ligand recognition. J. Biol. Chem. 282, 16654–16666 (2007).
Nielsen, M. J., Andersen, C. B. F. & Moestrup, S. K. CD163 binding to haptoglobin-hemoglobin complexes involves a dual-point electrostatic receptor-ligand pairing. J. Biol. Chem. 288, 18834–18841 (2013).
Etzerodt, A. et al. The Cryo-EM structure of human CD163 bound to haptoglobin-hemoglobin reveals molecular mechanisms of hemoglobin scavenging. Nat. Commun. 15, 10871 (2024).
Gerasimenko, J. V., Tepikin, A. V., Petersen, O. H. & Gerasimenko, O. V. Calcium uptake via endocytosis with rapid release from acidifying endosomes. Curr. Biol. 8, 1335–1338 (1998).
Guo, W., Wang, M. & Chen, L. A co-expression vector for baculovirus-mediated protein expression in mammalian cells. Biochem. Biophys. Res. Commun. 594, 69–73 (2022).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. elife 7, e42166 (2018).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019).
Madeira, F. et al. (2024). The EMBL-EBI job dispatcher sequence analysis tools framework in 2024. Nucleic Acids Res. 52, W521–W525 (2024).
Robert, X. & Gouet, P. “Deciphering key features in protein structures with the new ENDscript server”. Nucleic Acids Res. 42, W320–W324 (2014).
Gabrielson, J. P. et al. Precision of protein aggregation measurements by sedimentation velocity analytical ultracentrifugation in biopharmaceutical applications. Anal. Biochem. 396, 231–241 (2010).
Lebowitz, J., Lewis, M. S. & Schuck, P. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 11, 2067–2079 (2002).
Acknowledgements
We thank the Cryo-EM Platform of Peking University and Changping Laboratory for their support with data collection; the High-performance Computing Platform of Peking University for help with computation; we thank the Center for Quantitative Biology at Peking University for assistance with flow cytometry and Ms. Fei Wang for help with Imaging flow cytometry; We thank Dr. Yilan E from State Key Laboratory of Membrane Biology, Peking University for the assistance of SV-AUC assay; we thank the Protein Preparation and Identification Core at National Center for Protein Sciences at Peking University; we thank Mr. Zhaoxing Wang for providing facility support at the Protein Preparation and Identification Facilities at Technology Center for Protein Science, Tsinghua University. We specially thank Prof. Xiangyu Liu in Tsinghua University for the help with SV-AUC assay. We thank Prof. Zhe Zhang in Peking University for help with full-length CD163 expression and purification. We thank Dr. Bo Wang for suggestions in revision progress. This work was partly supported by the National Key Research and Development Program of China (2021YFC2301400) to X.D.S. and by China Postdoctoral Science Foundation (8206400138) to H.X.
Author information
Authors and Affiliations
Contributions
H.X. and X.H.S. contributed equally to this work. H.X. carried out the cryo-EM studies, as well as pull-down and SV-AUC assays. X.H.S. carried out the immunoblotting, flow cytometry, and imaging assays. X.D.S. and H.X. conceived and supervised the project and wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xu, H., Song, X. & Su, Xd. Calcium-dependent oligomerization of scavenger receptor CD163 facilitates the endocytosis of ligands. Nat Commun 16, 6679 (2025). https://doi.org/10.1038/s41467-025-62013-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-62013-4
This article is cited by
-
Strength in numbers: how multimerization drives HpHb uptake by CD163 scavenging receptor
Nature Communications (2025)