Abstract
The endogenous opioid system provides powerful control over emotions, nociception, and motivation among many other fundamental nervous system functions. Its major components include a panel of opioid peptides that activate four canonical inhibitory opioid receptors. However, its regulatory principles are not fully understood including the existence of additional receptors and other elements. In this study we report the identification of a receptor for the opioid peptide dynorphin. By conducting a screen of a custom library of neuropeptides, we found that orphan receptor GPR139 binds to and is activated by a series of dynorphin peptides. Unlike other opioid receptors, GPR139 couples to Gq/11 and avoids β-arrestin, providing excitatory signaling that homeostatically scales the inhibitory response of neurons to dynorphin. This introduces a non-canonical dynorphin receptor as an essential component of the opioid system.
Similar content being viewed by others
Introduction
The endogenous opioid system plays a critical role in many vital neuronal processes, including nociception1,2, reward3, and mood regulation4,5. Classical opioid receptors, the μ-opioid receptor (MOR), κ-opioid receptor (KOR), and δ-opioid receptor (DOR), share significant homology and belong to the family of G protein coupled receptors (GPCRs)6,7,8. Multiple endogenous opioid peptides derived from three precursors: proenkephalin (PENK), prodynorphin (PDYN), and proopiomelanocortin (POMC), overlap in activating these receptors with varying preferences9,10. In addition, the nociceptin/orphanin FQ receptor is considered to be a more distant member of the opioid family with a dedicated ligand and significant crosstalk with other opioid receptors9,11. All classical opioid receptors invariably signal via Gi/o type of G proteins to inhibit neuronal activity and are blocked by the common antagonist naloxone6,12.
A wealth of evidence also points to non-opioid actions of opioid peptides defined as naloxone-insensitive behavioral reactions and effects on neuronal signaling13,14,15. Furthermore, biochemical and pharmacological studies consistently identify binding and activity of opioid peptides in the neuronal tissues that can’t be explained by their interactions with canonical opioid receptors16,17. Together, these observations strongly suggest the existence of additional receptors for the opioid peptides.
Unbiased genetic screening previously identified evolutionary conserved orphan receptor GPR139 to engage in controlling opioid signaling18. It was shown to uniquely couple to stimulatory Gq/11 proteins, restraining opioid-mediated inhibition of neuronal firing, analgesia, and reward19. However, its endogenous ligand remains unknown.
In this study, we report that opioid peptides derived from prodynorphin serve as endogenous ligands for GPR139. They bind to GPR139 with high affinity in a naloxone insensitive manner and trigger its Gq-mediated effects in neurons. The unique potency range of GPR139-mediated dynorphin signaling allows it to constrain excessive signaling via canonical opioid receptors. These findings establish GPR139 as bonafide non-canonical opioid receptor and an essential component of the endogenous opioid system.
Results
Identification of endogenous peptide ligands for GPR139
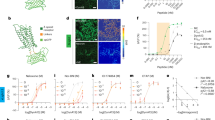
GPR139 exhibits the characteristic features of the peptide family of GPCRs with which it shares significant homology20,21. Given that GPR139 is expressed exclusively in the nervous system22,23,24, we hypothesized that it could be liganded by a neuropeptide. To test this hypothesis, we screened a collection of ~100 neuropeptides. Given that Gq/11 coupling of GPR139 triggers the release of intracellular calcium19, we used a high-throughput Calcium 5 assay to detect responses (Fig. 1a, b). The screen was performed with HEK293 cells transiently transfected with GPR139 and control untransfected cells in parallel and the responses were normalized to those elicited by the synthetic GPR139 agonist JNJ-6353305423. Comparing responses from GPR139-expressing cells to control cells revealed 36 hits that produced higher responses in the presence of GPR139. Fluorescence-based high-throughput screens rely on single replicates conducted in a miniaturized environment and are thus prone to false positives. Therefore, we used an orthogonal approach based on the genetically encoded calcium biosensor CalFluxVTN to validate initial hits (Fig. 1a, c). Because many of the hits from the primary screen contained opioid peptides, we expanded the secondary screen to exhaustively include all peptides derived from precursors of classical opioid peptides as well as collection of related RF amide peptides25,26 (Fig. 1c, Supplementary Fig. 1a–c). Comparing the responses to control cells not expressing GPR139, across 3 independent experiments, we detected significant signals for 8 peptides derived from (PDYN) (Fig. 1c) and two POMC-derived peptides, a-MSH and ACTH (Supplementary Fig. 1b), noted before27. The responses showed characteristic kinetics of Ca2+ waveform (Fig. 1d) and were blocked by GPR139 selective synthetic antagonist JNJ-379216528,29 (Supplementary Fig. 1d), confirming the specificity of GPR139 engagement. Dose response studies showed that dynorphin A peptides were the most efficacious, exhibiting ~70% efficacy relative to reference agonist JNJ-63533054 and had potency in the micromolar range (Fig. 1e, Supplementary Table 1). Analysis of the sequence features revealed that these peptides had characteristic opioid consensus sequence “YGGFL” (Fig. 1f). Interestingly, the N-terminal tyrosine strictly required for activity on classical opioid receptors30,31 was not required for activating GPR139, although it enhanced GPR139 activation. Increasing the length of the peptide also mildly increased the potency of the peptides and so did the amidation of the C terminus (Fig. 1e, f; Supplementary Table 1). Together, these findings reveal that GPR139 is activated by a set of peptide ligands: endogenous PDYN-derived peptides.
a Schematic of screening assay design. b Screening results of ~100 neuropeptides (10 μM) using Calcium 5 assay. Untransfected HEK293T/17 cells were used as negative control. Data were normalized to the 10 μM JNJ-63533054 signal. c Quantification of CalFluxVTN BRET signal of 36 hits at 10 μM. Data were normalized to 10 μM JNJ-63533054. d Traces of BRET signal change in CalFluxVTN assay elicited by 10 μM JNJ-63533054 and 40 μM dynorphin peptides. e The concentration dependence of GPR139 activation by PDYN-derived peptides. Data were normalized to 10 μM JNJ-63533054. f The amino acid sequence of PDYN-derived peptides. Data shown are mean ± SEM of n = 3 independent experiments. Data in (c) were analyzed by two-way ANOVA with Tukey’s test. *p < 0.05, **p < 0.01 and ***p < 0.001.
Dynorphin peptides bind directly and specifically to GPR139 with high affinity
To investigate interaction of PDYN-derived peptides with GPR139, we used a series of orthogonal assays. First, we used a radioligand binding assay (Fig. 2a), a gold standard for determining binding affinity of ligands. In this assay, membranes prepared from the HEK293 cells expressing GPR139 were first incubated with the radioactively labeled [3H]-JNJ-63533054 reference ligand. Titration experiments established specific ligand binding with a Kd of ~20 nM (Supplementary Fig. 2a), consistent with previous reports23. The membranes were then incubated with unlabeled ligands and their ability to displace [3H]-JNJ-63533054 was determined. Indeed, both unlabeled reference ligand JNJ-63533054 and all PDYN-derived peptides tested were able to compete with [3H]-JNJ-63533054 binding to the same extent (Fig. 2b). We next conducted dose-response studies comparing displacing ability of different PDYN peptides. These studies revealed the highest affinity was for Dyn A peptides with Ki in the 12-85 nM range, similar to the ~66 nM Ki of JNJ-63533054 in the same assay (Fig. 2c, Supplementary Fig. 2b, c and Supplementary Table 1). For a reference, we also evaluated POMC-derived active peptides α-MSH and ACTH, as well as inactive Leu-enkephalin as a negative control. These studies revealed no competition from Leu-enkephalin and only modest binding of α-MSH and ACTH with no saturation to discern Ki values (Supplementary Fig. 2b).
a Schematic of radioligand binding competition assays using cell membranes overexpressing GPR139. b Binding of 10 μM JNJ-63533054 and 40 μM dynorphin peptides to the membranes from HEK293T/17 cells transfected with GPR139 following co-incubation with 10 nM [3H]-JNJ-63533054. c Binding competition of [3H]-JNJ-63533054 with increasing concentration of dynorphin peptides. d Schematic of experimental design for detecting FITC-Dyn A13 binding to GPR139 by flow cytometry. e Flow cytometry histogram of cell population distribution after cell sorting. f Quantification of 1 μM FITC-Dyn A13 binding from (e). Median Fluorescence Intensity (MFI) was calculated. g Binding of 10 μM JNJ-63533054 and 40 μM Dyn A17 to the membranes from WT mouse brains in the presence of 10 nM [3H]-JNJ-63533054. h Binding competition of [3H] JNJ-63533054 with increasing concentration of Dyn A17. Data were normalized to specific binding of 10 nM [3H]-JNJ-63533054. i Quantification of the IC50 values from the binding displacement curves in (h). Data are mean ± SEM from n = 3 independent experiments. In (b, g), data were analyzed by one-way ANOVA with Dunnett’s test or unpaired two-tailed t-test for (f, i). *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
We next used intact cells to confirm that GPR139 on a cell surface can directly bind PDYN-derived peptides using the most potent peptides, Dyn A13 and Dyn A17 (Fig. 2c). First, we found that like in the membrane preparation DynA17 was able to displace [3H]-JNJ-63533054 as effectively as reference ligand JNJ-63533054 (Supplementary Fig. 2d, e). Next, we labeled Dyn A13 with fluorescein isothiocyanate (FITC) fluorophore and studied its direct interaction with GPR139 on the surface of intact cells using flow cytometry (Fig. 2d; Supplementary Fig. 2f). This approach showed that cells expressing GPR139 were labeled with DynA13-FITC (Fig. 2e, f). This binding was reduced by both GPR139 agonist JNJ-63533054 and antagonist JNJ-3792165, further confirming the specificity (Supplementary Fig. 2g, h). These observations indicate that dynorphin peptides interact with GPR139 on the surface of intact cells.
Finally, we examined Dyn A17 binding to GPR139 in the brain tissues. We found that binding of [3H]-JNJ-63533054 to the brain membranes was effectively reduced by preincubation with Dyn A17 or unlabeled JNJ-63533054 (Fig. 2g), suggesting that they share similar binding sites. To better understand the contribution of GPR139 to DynA17 binding, we used brain membranes from GPR139 knockout mice (Gpr139 KO). Titration studies revealed that it took ~4 times more Dyn A17 to displace [3H]-JNJ-63533054 in brain membranes from Gpr139 KO as compared to wild-type membranes, supporting the major contribution of GPR139 to Dyn A17 binding in native tissues (Fig. 2h, i). Interestingly, [3H]-JNJ-63533054 binding was not completely abolished in Gpr139 KO, suggesting that this ligand may also interact with other receptors. Given the overlap in pharmacology of GPR139 with opioid receptors, we tested whether such a target could be opioid receptors. Indeed, we found that JNJ-63533054 could activate KOR with low potency (Supplementary Fig. 3a, b) but not MOR, DOR, or NOR, suggesting that lingering [3H]-JNJ-63533054 binding in Gpr139 KO may be contributed to by KOR. Collectively, ligand binding experiments strongly support direct binding of PDYN-derived peptides to GPR139.
Dynorphin peptides promote G protein activation but not β-arrestin recruitment by GPR139
To understand GPR139 signal transduction induced by dynorphin, we first examined its ability to activate heterotrimeric G proteins. We used a BRET-based GPCR fingerprinting assay that examines activation of unmodified Gα subunits in real time by monitoring the kinetics of Gβγ release (Fig. 3a). With this assay, we examined the activity of GPR139 on an exhaustive set of 15 Gα proteins using a representative potent PDYN peptide: Dyn A13 applied at a saturating concentration. Analysis of response amplitudes and activation kinetics revealed a characteristic profile of G protein activation (Fig. 3b, c). GPR139 activated proteins of Gi/o and Gq/11 classes with varying kinetics and efficacy (Supplementary Fig. 4a–c), but not Gs (Supplementary Fig. 4d) or G12/13 types of proteins (Supplementary Fig. 4e).
a Schematic of G protein BRET fingerprinting assay. G protein coupling activity was measured by the BRET signal between Gβγ-venus and masGRK3ct-Nanoluc upon GPR139 activation by dynorphin peptides. b The maximum amplitude of responses mediated by 15 G proteins upon application of 40 μM Dyn A13. Data are normalized to the response of GαoA. c Initial rate of responses mediated by 15 G proteins upon application of 40 μM Dyn A13. GPR139 mediated activation on GαoA (d) and Gα11 (g) by dynorphin peptides. The maximum amplitude of GαoA (e) and Gα11 (h) activation across different concentration of Dyn A13 and Dyn A17. The EC50 of GαoA (f) and Gα11 (i) coupling. j Schematic of agonist-induced LgBit-β-arrestin recruitment assay in HEK293T/17 cell. k Basal luminescence signal of GPR139-SmBit paired with LgBit-β-arrestin1 or LgBit-β-arrestin2. β-arrestin recruitment kinetics of GPR139-SmBit/LgBit-β-arrestin1 (l) and GPR139-SmBit/LgBit-β-arrestin2 (m) upon application of 40 μM Dyn A13 and 40 μM Dyn A17. Data shown are mean ± SEM of n = 3 independent experiments.
Next, we conducted concentration-response studies comparing peptide-stimulated activities of GPR139 on representative members of two major Gα classes activated by GPR139: GαoA and Gα11(Fig. 3d–i). These studies revealed similar potencies of G protein activation for dynorphin peptides, which were comparable to potencies recorded in the Ca2+ release assays. Interestingly, α-MSH and ACTH did not show any significant coupling activity with either GαoA or Gα11 (Supplementary Fig. 4f, g) in line with their generally weak activity on GPR139. Neither did we observe G protein activation in response to L-Enk, a negative control (Supplementary Fig. 4f, g). Interestingly, we found that naloxone, which antagonizes all canonical opioid receptors, did not block G protein activation by GPR139 (Supplementary Fig. 4h).
In addition to G proteins, GPCRs recruit β-arrestins which could mediate both their signaling and trafficking32,33,34. Therefore, we next investigated the ability of dynorphin peptides to recruit β-arrestin 1 and β-arrestin 2 to GPR139 using a split NLuc complementation assay (Fig. 3j). We employed the same strategy for studying β-arrestin recruitment to MOR35, which served as a positive control for this assay (Supplementary Fig. 5a, b). We detected a significant basal association of both β-arrestins with GPR139 (Fig. 3k; Supplementary Fig. 5c), consistent with our earlier findings that GPR139 can promote heterologous recruitment of β-arrestin to MOR18. However, application of either Dyn A13 or Dyn A17 resulted in no detectable increase in β-arrestin 1 or β-arrestin 2 recruitment to GPR139 (Fig. 3l, m). Because GPCRs can bind either N- or C-termini of β-arrestins36,37, we also tested an alternative configuration placing the LgBit sensor at the C terminus of the molecule (Supplementary Fig. 5d). This configuration resulted in lower signal at the baseline (Supplementary Fig. 5e). However, it similarly resulted in a lack of β-arrestin recruitment upon ligand stimulation (Supplementary Fig. 5f, g). We also did not observe recruitment of β-arrestins in response to application of JNJ-63533054 with either β-arrestin1 or β-arrestin2 in either sensor configuration (Supplementary Fig. 5h). These observations were confirmed by monitoring real time recruitment of β-arrestins to the plasma membrane upon receptor activation. We found no significant changes in localization of either β-arrestin 1-Venus (Supplementary Fig. 6a) or β-arrestin 2-Venus (Supplementary Fig. 6b), which remained cytoplasmic upon activation of SNAP-GPR139 on the plasma membrane by Dyn A13. In contrast, positive control experiments with SNAP-MOR showed robust translocation of β-arrestin 2-Venus to the plasma membrane upon stimulation with DAMGO (Supplementary Fig. 6c). Overall, these results indicate that dynorphins induce GPR139 coupling to multiple G proteins but not β-arrestins.
Dynorphin induces GPR139 internalization in a β-arrestin independent manner
One of the hallmarks of GPCR activation by ligands is their internalization, which physically removes the receptor from the surface of the cell and routes it to intracellular compartments for continued signaling from these locations and/or subsequent recycling back to the plasma membrane33,38,39. Therefore, we next investigated whether dynorphin could induce GPR139 internalization. Upon transfection into HEK293 cells, GPR139 was found mostly localized on the plasma membrane, with some present at the intracellular sites (Fig. 4a). Application of Dyn A13 induced clear internalization of GPR139 as evidenced by its accumulation mostly at intracellular locations and reduced presence on the plasma membrane (Fig. 4a). Indeed, quantification showed significant reduction in GPR139 co-localization with KRas-GFP (Fig. 4b, c).
a Co-expression of KRas-Venus and HA-GPR139 in HEK293T/17 cells. The cells were treated with 40 μM Dyn A13 for 5 min. b HA-GPR139 fluorescence intensity distribution across the line in (a). c Mander’s colocalization coefficient (MOC) analyzed from 11 images (25 cells for control group and 28 cells for Dyn A13 group). d Schematic of luminescence assay for examining cell surface abundance and internalization of GPR139. e Cell surface abundance of GPR139 in HEK293T/17 and β-arrestin1/2 KO cells. Time course of Dyn A13 and Dyn A17 initiated HiBit-GPR139 internalization in HEK293T/17 (f) and in β-arrestin1/2 KO cells (i). Quantification of the internalization of GPR139 in HEK293T/17 (g) and in β-arrestin1/2 KO cells (j). The maximum internalization of GPR139 in HEK293T/17 (h) and β-arrestin1/2 KO cells (k) across different concentrations of Dyn A13 and Dyn A17. l The effect of 40 μM Dyngo-4a, 10 μM PitStop 2 and 4 mM MβCD applied 15 min before the assay on the time course of Hibit-GPR139 internalization induced by 40 μM Dyn A13 in HEK293T/17 cells. m The maximum internalization of GPR139 in HEK293T/17 cells under different treatments. Data are mean ± SEM of n = 3 independent experiments. Data were analyzed by unpaired two-tailed t-test (c, e) or one-way ANOVA with Dunnett’s test (g, j, m). ** p < 0.01, *** p < 0.001, n.s. p > 0.05.
Robust internalization of GPR139 amidst undetectable recruitment of β-arrestin, a key driver of internalization for many GPCRs39,40, poses a question about the involvement of β-arrestins in dynorphin-induced GPR139 internalization. We used a nanoluciferase complementation strategy (Fig. 4d) to examine internalization dynamics and probe the influence of β-arrestins. We found that knockout of both β-arrestin1 and β-arrestin 2 had no effect on the cell surface abundance of GPR139 at the baseline (Fig. 4e). In normal cells, Dyn A13 and Dyn A17 induced time-dependent loss of luminescence occurring with similar kinetics (Fig. 4f), indicating internalization of GPR139 and corroborating microscopy data. Concentration-response studies indicated variable potencies of dynorphin peptides in the range of concentrations that induce G protein activation with Dyn A17 showing somewhat greater potency (Fig. 4g, h). We found that in cells lacking both β-arrestin isoforms, both peptides reached a similar extent and kinetics of internalization (Fig. 4I, k). Control experiments confirmed previously noted41 loss of DAMGO-induced internalization of MOR in the same double β-arrestin 1/β-arrestin 2 knockout cell line (Supplementary Fig. 7). To examine internalization mechanisms of GPR139, we blocked several processes known to be involved in GPCR endocytosis38. Indeed, dynamin blocker Dyngo-4a42 and clathrin blocker PitStop 243 significantly inhibited Dyn A13-mediated internalization (Fig. 4l, m). In contrast, blockade of caveolin by methyl-beta-cyclodextrin (MβCD)44 had no effect on GPR139 internalization (Fig. 4l, m). Together, these results indicate that dynorphin peptides are capable of inducing GPR139 internalization in a manner typical to GPCR agonists. This process relies on dynamin and clathrin but occurs in a β-arrestin independent manner.
Co-activation of GPR139 with dynorphin reduces the signaling strength of the canonical opioid receptor MOR
To understand the physiological significance of GPR139 activation by dynorphin peptides that also activate all canonical opioid receptors, we modeled receptor interplay in transfected cells. In these studies, we used a genetically encoded sensor to measure the impact of receptor activation on cAMP (Fig. 5a), one of the central second messengers that crucially shapes cell responses. Interestingly, cAMP is differentially regulated by canonical opioid receptors, which inhibit it by virtue of coupling to Gαi/o G proteins and GPR139, which stimulates cAMP production via Gαq-Ca2+ dependent mechanism19. Since GPR139 has been established as a MOR regulator in an endogenous neuronal setting in vivo18 and because MOR is potently activated by dynorphins45,46, we chose MOR as a model receptor to study the interplay with GPR139.
a Schematic of cAMP signaling platform to study the influence of GPR139 co-expression on MOR activation by different ligands. b Concentration-response curve for DAMGO-mediated MOR activation with or without GPR139 co-expression. c Concentration-response curve for DAMGO-mediated MOR activation with or without GPR139 co-expression in the presence of 10 μM JNJ-63533054. d Maximum efficacy of DAMGO-mediated activation of MOR and MOR co-expressed with GPR139. e The negative logarithm of the EC50 (pEC50) of DAMGO-mediated activation of MOR with and without GPR139 co-expression. f Concentration-response curve for Dyn A13-mediated MOR activation with or without GPR139 co-expression. g Maximum efficacy of Dyn A13-mediated activation of MOR and MOR upon co-expression with GPR139. h The negative logarithm of the EC50 (pEC50) of Dyn A13-mediated activation of MOR and MOR upon co-expression with GPR139. i Schematic of luminescence complementation assay to detect change in cell surface abundance of MOR. Time course of changes in MOR cell surface abundance upon treatment with DAMGO (j) and Dyn A13 (k). Quantification of MOR internalization by DAMGO (l) and Dyn A13 (n). Quantification of MOR internalization rate in response to DAMGO (m) and Dyn A13 (o). Data shown are mean ± SEM of n = 3 independent experiments. Data were analyzed by d two-way ANOVA with Fisher’s least significant difference test, e one-way ANOVA with Dunnett’s test or (g, h, l, m, n, o) unpaired two-tailed t-test. *p < 0.05, **p < 0.01 and ***p < 0.001, n.s. p > 0.05.
In the absence of GPR139 co-activation, application of the MOR-selective agonist DAMGO resulted in a typical dose-dependent inhibition of cAMP production (Fig. 5b). GPR139 did not respond to DAMGO, and its co-expression with MOR at equivalent cDNA concentrations also did not inhibit MOR responses (Fig. 5b, d, e). Next, we activated GPR139 by pre-incubating the cells with its agonist JNJ-63533054 and repeated experiments examining MOR-mediated suppression of cAMP production in response to DAMGO application. While cells expressing MOR alone showed the same response to DAMGO, such response was completely abolished in cells co-expressing MOR and GPR139 (Fig. 5c–e). These experiments confirm that GPR139 exerts a powerful inhibitory influence on MOR-mediated responses in a model system when both receptors are activated by their individual surrogate agonists.
Having established the inhibitory interplay, we next studied the responses of MOR-GPR139 paired system to a common agonist- Dyn A13. For MOR, application of Dyn A13 resulted in a typical dose-dependent inhibition of cAMP production (Fig. 5f). We found that in the presence of GPR139, Dyn A13 inhibition of cAMP mediated by MOR was severely depressed as evidenced by both reduction in the maximal amplitude of the response and nearly ten-fold decrease in potency as compared to responses of the cells expressing MOR alone (Fig. 5g, h).
In addition to signaling interplay at downstream second messenger pathways, receptor internalization is a powerful mechanism to control signal strength. Given the high constitutive level of GPR139 association with β-arrestin that drives MOR internalization and previous findings that GPR139 facilitates β-arrestin recruitment to MOR18, we explored the impact of GPR139 on MOR internalization (Fig. 5i). When only MOR was activated by its selective agonist DAMGO, the presence of GPR139 had no influence on the extent or rate of MOR internalization (Fig. 5j, l, m). However, when MOR was co-activated with GPR139 by a common agonist DynA13 both the rate and extent of MOR internalization significantly increased (Fig. 5k, n, o). Although the degree of this effect was relatively small, it could contribute to reducing MOR content on the surface and thus possibly its signaling capacity over time. Collectively, these results indicate that GPR139 serves to counter-balance signals initiated by MOR when two receptors are activated by the common peptide ligand-dynorphin, thereby allowing GPR139 to provide homeostatic control over the strength of the opioid response.
GPR139 signals in medial habenula neurons to temper dynorphin-induced modulation of firing
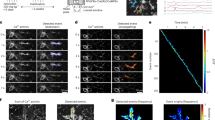
To understand the physiological relevance of GPR139 activation, we examined its contribution to dynorphin responses in the mouse brain. We started our studies in medial habenula (MHb), a region with particularly high expression of GPR13922, where it is co-expressed with MOR18. Using brain slice preparation, we examined Ca2+ responses in MHb neurons transduced with a well-established calcium sensor, jGCaMP7s47 (Fig. 6a). Unlike GPR139, canonical opioid receptors do not couple to Gq types of G proteins and are thus not expected to generate Ca2+ responses. Indeed, application of Dyn A17 resulted in a readily detectable characteristic Ca2+ transient that reached its peak in ~30 s before slowly decaying (Fig. 6b). Importantly, the response was completely blocked by selective GPR139 antagonist JNJ-3792165 (Fig. 6b, c). To further confirm that the response is generated by GPR139 we recorded from MHb neurons in slices from Gpr139 KO mice (Fig. 6b, c). We detected no responses in Gpr139 KO slices, indicating that Dyn A17 selectively engages GPR139 to produce Ca2+ signal.
a Experimental design of recording calcium imaging utilizing jGCaMP7s sensor in medial habenula (MHb) region of mice. b Average of calcium response in MHb neurons of WT and Gpr139 KO mice after puff application with 100 μL of 160 μM Dyn A17 (WT n = 6 cells from 4 mice; WT/JNJ-3792165 n = 7 cells from 4 mice; Gpr139 KO, n = 6 cells from 4 mice). c Quantification of area under curve of calcium response in (b). n = 6 cells from 4 mice for WT; n = 7 from 4 mice for WT/JNJ-3792165, and n = 6 cells from 4 mice for Gpr139 KO. d Representative traces (n = 8 cells from WT mice and n = 11 cells from Gpr139 KO mice) of spontaneous firing activity of MHb neurons in brain slices from WT and Gpr139 KO mice with and without 500 nM Dyn A application. e Time course of normalized firing frequencies of MHb neurons at baseline and following Dyn A application. f Quantification of maximal drug effect as the averaged normalized firing frequency during min 9-10 (WT n = 8 cells from 4 mice; Gpr139 KO n = 11 cells from 5 mice). g Schematic of viral targeting of jGCaMP7s and Flex-ChrimsonR-tdTomato to the nucleus accumbens (NAc) and optogenetic stimulation. ISI inter-stimulation interval. h Average jGCaMP7s responses to optical stimulation in the presence or absence of 10 μM JNJ-3792165 in Pdyn+ that express ChrimsonR-tdTomato (black, n = 4 cells from 3 mice; maroon, n = 3 cells from 3 mice) and in Pdyn- neurons that do not express ChrimsonR-tdTomato (n = 7 cells from 4 mice). i Quantification of the area under the curve calculated from traces in (h). n = 4 cells from 3 mice for Pdyn+ Block; n = 3 cells from 3 mice for Pdyn+ Control. Data are mean ± SEM. Data were analyzed by (c) One-way ANOVA with Dunnett’s post hoc test, (e) Two-way ANOVA with Holm-Šídák’s test, (f) unpaired two-tailed t-test or (i) paired two-tailed t-test. *p < 0.05, **p < 0.01.
Because the Gαq/11-Ca2+ signal generated by GPR139 counteracts Gαi/o-mediated responses initiated by canonical opioid receptors including MOR, which was observed in transfected HEK cells, we hypothesized that simultaneous engagement of GPR139 and MOR by dynorphin in MHb neurons would dampen MOR inhibitory signaling, and that genetic deletion of GPR139 should result in amplified dynorphin-mediated inhibition due to the loss of GPR139 constraint over MOR. To test this, we used patch clamp electrophysiology in acute MHb brain slices performing cell-attached recordings of MHb tonic neuronal firing (Fig. 6d). We detected significant reduction in the baseline firing rates in Gpr139 KO neurons (Supplementary Fig. 8a), suggesting that GPR139 constitutive activity contributes to constraining tonic opioidergic inhibition of MHb neurons, consistent with earlier findings18. Next, we bath applied Dyn A17 and detected a robust reduction in the tonic firing frequency of MHb neurons (Fig. 6e). Importantly, the inhibitory effect of Dyn A17 (Fig. 6e, f) was significantly greater in neurons from slices from Gpr139 KO mice relative to slices from WT mice (Fig. 6e, f). The effect of Dyn A17 on neuronal firing was blocked by naloxone (Supplementary Fig. 8b, c), indicating that it is mediated by canonical opioid receptors other than KOR as the KOR-specific agonist U-50488 failed to elicit changes in firing (Supplementary Fig. 8d, e). Indeed, MHb neurons are highly enriched with MOR and lack detectable KOR expression48,49. These data demonstrate that GPR139 significantly contributes to shaping MHb neuronal activity by several mechanisms that include signaling in response to dynorphin as well as changes in the basal activity, among other possibilities.
To better assess the physiological relevance of dynorphin action on GPR139 in intact circuits, we adopted an optogenetic approach to trigger dynorphin release. In these studies, we focused on the nucleus accumbens (NAc), another area of the brain with significant GPR139 expression also noted for the highest density of dynorphin release50,51,52. Interestingly, previous studies have shown high expression of GPR139 in the dynorphinergic neurons of the NAc53,54, suggesting possible autocrine action of dynorphin upon its release. To trigger dynorphin release, we selectively excited dynorphinergic neurons in the NAc by optogenetic stimulation of a ChrimsonR driver virally expressed in neurons of Pdyn-Cre mice (Fig. 6g). To validate the release of dynorphin, we used recently developed dynorphin sensor κLight1.352. We found that our optogenetic protocol reliably produced the signal with similar characteristics upon repeated stimulations, indicating that our protocol evokes dynorphin release in the NAc (Supplementary Fig. 9). We then virally expressed the jGCaMP7s sensor that reports Gq/11 signaling of GPR139 in the same NAc region. Optogenetic stimulation produced significant fluorescence increases that reached peak after about 200 s in Pdyn+ neurons (Fig. 6h), matching the kinetics of dynorphin release recorded by the κLight1.3 sensor. In contrast, we detected no significant responses in Pdyn− neurons. Importantly, notable jGCaMP7s response was also detected with repeated stimulation, but it was completely abolished upon application of GPR139 antagonist JNJ-3792165 (Fig. 6h, i), indicating that it is mediated by GPR139. These experiments support that dynorphin release upon neuronal excitation is capable of activating GPR139 in endogenous circuits.
Discussion
In this study, we report the identification of an endogenous ligand for the orphan receptor GPR139. Orphan GPCRs have attracted considerable attention for their promise in uncovering previously unrecognized biology and untapped potential as drug targets. GPR139 has been a subject of intense interest for its involvement in neuropsychiatric processes55 and noted roles in controlling opioidergic and dopaminergic neuromodulation in key neural circuits18,53,56. In particular, it has been shown to negatively regulate MOR signaling, serving as an “anti-opioid” receptor18,19. Thus, unbiased identification of the opioid peptide dynorphin as a ligand for GPR139 retrospectively makes logical sense and supports its role as an essential component of the opioid system.
We propose that GPR139 be considered a non-canonical member of the opioid receptor family. Importantly, it endows the system with an additional signaling modality. All canonical opioid receptors couple to Gi/o type of G proteins and exert net inhibitory effect on neuronal activity. In contrast, by virtue of Gq/11 coupling GPR139 mobilizes excitatory Ca2+ signaling pathway. This establishes a pivotal interplay between GPR139 and canonical opioid receptors as concurrent activation of GPR139 by a shared ligand acts to diminish the inhibitory influence of canonical opioid receptors. Despite the high binding affinity of dynorphin to GPR139, it exhibits modest signaling potency. Such dissociation is not uncommon in GPCRs and in previously documented cases45,57,58 is thought to drive differential engagement of transducers, e.g., G proteins vs β-arrestin, a phenomenon known as ligand bias. In these cases, the low intrinsic efficacy of an agonist, perhaps due to conformational flexibility of its orthosteric site in the receptor, could be overcome by higher ligand concentrations to shift the equilibrium towards the activated receptor state. GPR139's low functional potency may be a feature that exploits the conformational bias of the receptor to deliberately set up the range of GPR139 engagement by dynorphin outside of canonical opioid receptors. It is estimated that the concentration of opioid peptides, including dynorphin, at the sites of release could reach tens of micromolar which would clearly influence GPR139 activity59,60,61,62,63. Our results showed that calcium signal evoked by optogenetic stimulation of dynorphin expressing neurons is mediated by GPR139. However, these experiments do not directly establish that the responses are solely driven by dynorphin leaving a possibility that they may be influenced by other mediators co-released with dynorphin upon stimulation.
We therefore propose a model where a physiological function of GPR139 would be to homeostatically scale the strength of opioid mediated inhibition (Fig. 7). At a low level of dynorphin release, it will primarily trigger canonical opioid receptors, first KOR and then MOR and DOR to inhibit neuronal activity. As the dynorphin concentration increases further, it engages GPR139 gradually dampening MOR/KOR/DOR signaling to avoid extreme silencing of neuronal activity and maintain the operational range of the responses. Basal activity of GPR139 likely also contributes to tonic inhibition of the canonical opioid receptors signaling at the low concentration of dynorphin insufficient for triggering GPR139 activity. These effects may become exacerbated upon prolonged exposure to the dynorphin, which would cause the internalization of MOR/KOR/DOR but not GPR139 further strengthening homeostatic influence. The counteracting effect of GPR139 on canonical opioid receptor actions likely involves multiple mechanisms that include changes in β-arrestin recruitment and opposing influence of Gαq on Gαi/o signaling. Based on the extent of the effects we observed in this study, the latter mechanism likely dominates the influence, consistent with previous observations19. As proof of principle, our studies focused on MOR given the physiologically established significance of GPR139-MOR interplay and their close overlap in expression. However, GPR139 may similarly cross-talk with KOR and DOR and exploring this avenue will be an exciting future direction.
At a low level of dynorphin release, only canonical opioid receptors (MOR/KOR/DOR) are engaged to inhibit neuronal activity via Gi/o signaling. More dynorphin release upon increase in the activity of dynorphinergic neurons progressively engages GPR139, which gradually counteracts MOR/KOR/DOR thereby avoiding excessive silencing of neuronal activity to maintain operating range of the circuit. Created in BioRender. Martemyanov, K. (2025) https://BioRender.com/mlzlbxs.
Previous studies showed that PDYN-derived peptides can modulate several non-opioid targets, including the acid-sensing ion channels (ASICs)64, nicotinic acetylcholine receptor65, N-methyl-d-aspartate (NMDA) receptors63, and bradykinin receptor66. For example, Dyn big67,68 antagonizes ASIC decreasing the proton-sensitivity of its steady state desensitization while activation of bradykinin receptor by Dyn A peptides exacerbates pain, an effect opposite from canonical actions of opioid peptides66. Our studies introduce GPR139 as another intriguing target of dynorphin peptides, broadening our understanding of their biological effects.
GPR139 is an evolutionary ancient receptor, which, unlike canonical opioid receptors, is widely conserved and present even in invertebrates69. It was likely co-opted to influence opioid system when it appeared, but its interaction with dynorphin and related peptides possibly predated the emergence of the canonical opioid receptors. From this perspective, it is interesting to note the possibility of broader ligand tuning of GPR139. This receptor has also been noted to be activated by the aromatic amino acids tryptophan and phenylalanine23, which we did not explore in this study, and peptides derived from the same precursor as opioid peptides, such as adrenocorticotropic hormone (ACTH), and α- and β-melanocyte-stimulating hormone (MSH), which we found to have extremely low potency and binding affinity to GPR139. Thus, GPR139 engagement by opioid peptides and possibly other ligands serves to provide homeostatic scaling of opioid responses, calibrating their strength for the operational tuning of neural circuits.
Methods
Reagents and constructs
Lipofectamine LTX was from Invitrogen. Dulbecco Modified Eagle Medium (DMEM), MEM non-essential amino acids (NEAA), and sodium pyruvate were from Life Technologies (Grand Island, NY). Fetal bovine serum (FBS) was purchased from Millipore-Sigma (Burlington, MA). NanoGlo® reagent and NanoGlo® HiBiT Extracellular Detection System was purchased from Promega (Madison, WI). Peptides were purchased from Phoenix Pharmaceuticals (Burlingame, CA) and synthesized by Genscript (Piscataway, New Jersey). [3H]-JNJ-63533054 was purchased from Moravek (Brea, CA). The GPR139 cDNA used for all experiments (HASP-HA-GPR139) contained an N-terminal HA signal peptide (HASP), followed by an HA tag and human GPR139 (NM_001002911.4)19.
Animal studies
All studies involving mice were carried out in accordance with the National Institutes of Health guidelines and were granted formal approval by the Institutional Animal Care and Use Committee of the Herbert Wertheim UF Scripps Institute for Biomedical Innovation & Technology, University of Florida. Wild-type (WT) C57Bl6 mice were purchased from Charles River. Generation and characterization of GPR139 knockout mice (Gpr139 KO) on C57Bl6 background was reported before18. Pdyn-Cre mice (B6;129S-Pdyntm1.1(cre)Mjkr/LowlJ) were purchased from the Jackson Laboratory. All mice were housed in groups on a 12:12 h light-dark cycle with food and water available ad libitum.
Cell culture and transfection
β-arrestin1/2 knockout cell70 was kindly provided by Dr. Asuka Inoue. HEK239T/17 cells (ATCC, VA) and β-arrestin1/2 knockout cells were maintained in culture medium (DMEM supplemented with 10% FBS, NEAA, 1 mM sodium pyruvate and antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin)) at 37 °C in a humidified incubator containing 5% CO2. Transfections were performed as described71. Briefly, cells were seeded into 60-mm dishes (coated with 1 μg/ml Matrigel (Corning)) at a density of 3.2 × 106 cells. After 3–4 h, expression constructs (total 10 μg/dish) were transfected into the cells using PLUS reagent (10 μl/dish) and Lipofectamine LTX (12 μl/dish) reagents. All transfected DNAs were balanced to 10 μg using pcDNA3.1 and the transfection mixture was added dropwise according to the manufacturer’s instructions. Cells were incubated for 16–24 h at 37 °C before experiments.
Calcium 5 assay
HEK293T/17 cells were transfected with 25 μg pCDNA3.1-HASP-HA-GPR139/ml at 1 × 108 cells using Maxcyte electroporation system (Rockville, MD). After transfected, cells were incubated overnight at 37 °C, 5% CO2 before freezing down cells. On the day for experiment, thawed cells, resuspended in assay buffer (HBSS, 20 mM HEPES, 1% DMSO), and seed 3 μl/well (3000/well) in 1536 black wall clear bottom plate. Calcium dye (Molecular Device) working solution was made in assay buffer and dispensed 3 μl/well, then incubation 60 min at 37 °C. Fluorescence recording were done with FLIPR-Tetra (Molecular Devices). Basically, 5 s basal readings were done before adding 30 nL compounds followed by 2 min fluorescence reading.
CalFluxVTN BRET assay
HEK293T/17 cells were transfected with 0.42 μg pCDNA3.1-HASP-HA-GPR139 and 0.42 μg pCDNA3.1-CalFluxVTN. The CalFluxVTN sensor was described before72. The assay was performed as described73. About 16 h post-transfection, cells were washed once with Dulbecco’s Phosphate-Buffered Saline (PBS) and treated with 1 mM EDTA/PBS until cells were detached from dishes. Approximately 80,000 cells per well were distributed in Matrigel pre-coated 96-well flat-bottomed white microplates (Greiner Bio-One) and cultured for ~3 h in CO2 incubator. Before BRET measurements were made, the medium was replaced with HBSS (CaCl2, MgCl2) (Gibco). The signal reading was performed using the PHERAstar FSX plate reader POLARstar and Omega plate reader (BMG Labtech). Briefly, the Nano-Glo substrate was injected at 0 s and ligands were injected at 20 s. Signal reading started at 15 s and time interval was 0.1 s. The BRET signal is determined by calculating the ratio of the signal emitted by the Venus (535 nm with a 30 nm band path width) over the light emitted by the Nluc (475 nm with a 30 nm band path width). The average baseline BRET ratio recorded prior to agonist stimulation was subtracted from the experimental BRET signal values to obtain ΔBRET ratio.
Radioligand binding assay
For cell membrane radioligand binding assay, membranes were prepared from GPR139 expressing HEK293T/17 cells as follows: 48 h after transfection, cells were detached from plates by pipetting with PBS, centrifuged at 500 × g for 5 min, and then the fluid was aspirated away; the cell pellets were frozen with liquid nitrogen and stored at −80 °C. Pellets were homogenized in TE buffer (50 mM Tris-HCl, pH 7.4, 5 mM EDTA)23 and spin down at 1000 × g for 5 min at 4 °C. Supernatant was collected and recentrifuged at 30,000 × g for 30 min at 4 °C. The pellet was re-homogenized in TE buffer and incubated in 5 ml tube with eight concentrations of [3H]-JNJ-63533054 (specific activity 24.7 Ci/mmol) ranging from 0.024 to 400 nM for 60 min at room temperature (total volume 2 ml). The binding reaction was terminated by filtration through GF/C filter (Waterman) followed by washes with cold TE buffer. Filters were dried in a hood overnight followed by the addition of scintillation fluid. Then, bound radioactivity was counted on a Hidex 600 liquid scintillation counter. For inhibition of radioligand binding (10 nM [3H]-JNJ-63533054), peptides were added to the cell membrane at seven concentrations. Nonspecific binding was determined with 10 μM JNJ-63533054. The Kd values of the radioligands and IC50 of the antagonist were calculated by Graphpad Prism. Ki values were calculated as Ki = IC50/(1 + C/Kd), where C is the concentration of the radioligand. Whole cell radioligand binding assay used human GPR139 expressing HEK293T/17 cells. Both male and female mice 4–12 weeks were used for the brain membrane collection. For the preparation of brain tissue membranes, mice were anesthetized using isoflurane followed by decapitation; then the brains were collected and immediately transferred in liquid nitrogen and subjected to the same procedure as above.
Flow cytometry binding assay
Flow cytometry binding assay was performs as described74. HEK293T/17 cells were cultured in 6-well plates at a density of 8 × 105 cells/ per well. After 4–6 h, cells were transfected with 2 μg of GPR139-mCherry using Metafectene Pro. After ~36 h, transfected cells were washed with PBS and blocked using PBS-BSA (0.1%) for 1 hr at 4 °C Dynorphin A-FITC was incubated with 1 × 106 cells in the dark, rotating for 2 h at 4 °C Agonist JNJ-63533054 and antagonist JNJ-3792165 were added 1 h before Dynorphin A-FITC. After washing, cells were analyzed in BD Symphony flow cytometer. Acquired data were analyzed using FlowJo software.
G protein BRET assay
Agonist-dependent cellular measurements of BRET between Venus-Gβ1γ2 and masGRK3ct-Nluc were performed to examine activation of G protein signaling in living cells71. GRP139, G protein (Gi1, Gi2, Gi3, GoA, GoB, Gz, Ga11, Gq, G14, G15, Gs short, Gs long, Golf, G12, G13), venus-1-155 Gγ2, venus-155-239 Gβ1, masGRK3ct-Nluc-HA constructs were used at a 2:2:1:1:1 ratio (ratio 1 = 0.42 μg of plasmid DNA). Gα14/15 and Gαolf were transfected with Flag-Ric-8A (ratio 1), respectively. A construct carrying catalytic subunit of pertussis toxin (PTX-S1) was transfected with Gα subunits except for Gαi and Gαo to inhibit the possible coupling of endogenous Gi/o to GPCRs. 16–24 h post-transfection, cells were seeded into 96-well flat-bottomed white microplates (Greiner Bio-One) and cultured for ~3 h in CO2 incubator. Before signaling reading, cells were washed twice and replaced with BRET buffer (PBS containing 0.5 mM MgCl2 and 0.1% glucose). The signal reading was performed using the PHERAstar FSX and POLARstar Omega plate reader (BMG Labtech).
NanoBiT β-arrestin recruitment assay
Assays were performed as described18,75 by using the Promega NanoBiT Protein Protein Interaction assay system. cells were transfected with pcDNA3.1-GPR139-SmBiT (0.42 μg), 0.42 μg of Lgbit tagged β-arrestin1 or 2 (pcDNA3.1-β-arrestin1-LgBiT, pcDNA3.1-LgBiT-β-arrestin1, pcDNA3.1-β-arrestin2-LgBiT, pcDNA3.1-LgBiT-β-arrestin2). About 16 h after transfection, cells were detached and seeded in 96-well white plate. Before reading, 50 μl substrate working solution was added to each well. After 5 min basal reading, 50 μl agonist was manually added to each well followed by reading for another 20 min.
Live cell imaging of β-arrestin recruitment
HEK293T/17 cells were co-transfected with 0.2 μg of SNAP-GPR139 or SNAP-MOR and 0.01 μg β-arrestin 1-Venus or β-arrestin -Venus in 35 mm dish. Twenty-four hours after transfection, cells were plated on poly D-lysine coated No. 1.5 coverglass bottom 35 mm dishes in complete DMEM overnight. Prior to imaging, the SNAP-tag of surface expressed SNAP-GPR139 or SNAP-MOR was labeled by incubation with SNAP surface 594 (New England Biolabs) diluted to 1 µM in complete media at 37 °C for 30 min to 1 h. Cells were imaged in HBSS (CaCl2, MgCl2) supplemented with 10 mM HEPES in a temperature controlled chamber at 37 °C under ambient atmosphere. Imaging was performed using an Olympus FV3000 LSM confocal microscope through the ×100, 1.50 NA objective. For live cell β-arrestin recruitment, a single focus on a single optical section was maintained using ZDC autofocus, and images were collected at a rate of 0.1 Hz with simultaneous excitation with the 488 and 561 laser lines and detection at 500–548 nm and 584–656 nm for Venus and SNAP-594, respectively. After a 1 min baseline, agonist was added, and cells were recorded for an additional 20 min. For Z-stacks of cells were taken before and after agonist addition.
Immunocytochemistry
HEK293T/17 cells were transfected with 0.42 μg pCDNA3.1-HASP-HA-GPR139 and 0.42 μg pCDNA3.1-kRas-venus. After 24 h, cells were detached and reseeded on coverslips coated with poly D-lysine in 24-well plates. The next day, cells were treated with 40 μM Dyn A13 and incubation stopped by aspirating Dyn A13 following by adding ice-cold PBS. Cells were fixed in 4% PFA in PBS for 20 min and then permeabilized in 0.1% Triton X-100 in PBS for 5 min. Cells were blocked with 10% donkey serum in 0.1% Triton X-100 in PBS for 1 h at room temperature. The anti-HA-AlexFluo 594 antibody (1:500) was diluted in 2% donkey serum in 0.1% Triton X-100 in PBS. Cells were incubated with antibody for 2 h at room temperature and washed with 0.1% Triton X-100 in PBS for 15 min. Then cells were mounted onto slides using Fluoromount G containing DAPI.
Imaging and analysis
Imaging of HEK293T/17 cells was performed on Olympus FV3000 confocal microscope using a 100x objective. Co-localization analysis was performed with ImageJ (https://imagej.nih.gov/ij/) and 11 images for each condition. Lines were drawn across cells, avoiding the nucleus, and the fluorescence values from the plasma membrane (confirmed by KRas-venus) and intracellular membranes were quantified. Co-localization analysis using the entire cell was done using ImageJ and the Pearson’s correlation coefficient was reported.
Surface abundance assay
Cell surface GPR139 was measured using Promega’s NanoGlo® HiBiT Extracellular Detection System18. HEK293T/17 cells were transfected with 0.42 μg HiBit-GPR139. For testing MOR internalization, cells were transfected with 0.42 μg HiBit-MOR with or without 0.21 μg pCDNA3.1-HASP-HA-GPR139. The next day, cells were reseed into white-walled-96-well plate at about 80,000 cells per well and incubated for 3 h in CO2 incubator. Medium was then aspirated and 50 µl of BRET buffer containing Nano-Glo® HiBit extracellular substrate and LgBiT protein was added to each well. Plates were loaded into PHERAstar FSX plate reader set at 37 °C and luminescence was measured every 150 s or 90 s. To measure internalization, surface abundance was first measured as described, then PDYN-derived peptides were applied, and luminescence was measured for 30 min. The final time point was recorded as surface abundance after internalization and compared with the surface abundance before the addition of peptides.
cAMP measurements
cAMP assays were performed as described76,77. Briefly, HEK293T/17 cells were transfected with 2.52 μg of Promega-22F cAMP pGloSensor, 0.42 μg pCDNA3.1-MOR-FLAG, with or without 0.42 μg pCDNA3.1-HASP-HA-GPR139. The next day, cells were incubated in Promega GLO reagent for ~2 h at 37 °C and cooled down to room temperature before reading. Luminescence signal was read on PHERAstar FSX plate reader. Cells were treated with various concentration of DAMGO or Dyn A13. After 5 min basal reading, cells were treated with 1 μM of forskolin (FSK) and readings were continued for up to 20 min. The FSK-mediated increase in luminescence in the absence of agonist was used to normalize the data, where a decrease in luminescence signaling was attributed to MOR activity.
Live imaging of calcium dynamics
Adeno-associated virus (AAV) serotype 9 encoding the calcium indicator jGCaMP7s driven by synapsin promoter (pGP-AAV-Syn-jGCaMP7s-WPRE (AAV9)) was obtained from Addgene (104487-AAV9). Male mice 4–8 weeks old were anesthetized with vaporized isoflurane and mounted on a stereotaxic frame. Gpr139 KO and their WT littermates received a 300 nl bilateral stereotaxic guided injection targeted to the medial habenula (AP −1.7, ML ± 0.25 relative to bregma, DV −2.25 relative to dura). The injection volume and flow rate were controlled with an injection pump (World Precision Instruments, Sarasota, USA). Calcium imaging experiments were performed after a minimum of 2 weeks post-surgery. On the day of the experiment, mice were anesthetized with isoflurane and decapitated. The brain was quickly removed and rested for 30 s in ice-cold oxygenated NMDG cutting solution containing (in mM): 93 N-Methyl-D-glucamine (NMDG), 2.5 KCl, 1.2 NaH2PO4, 30 NaHCO3, 20 HEPES, 25 glucose, 2 thiourea, 5 Na-ascorbate, 3 Na-pyruvate, 0.5 CaCl2·2H2O, 10 MgCl2 (adjusted to 7.2–7.4 pH with HCl). Coronal slices (300 μm thick) containing the habenula were cut on a vibratome (VT1200S, Leica)18. Slices were recovered at room temperature in recording buffer containing (in mM): 103 NaCl, 3 KCl, 5 TES, 8 trehalose, 10 glucose, 26 NaHCO3, 1 NaH2PO4 (adjusted to 7.3 PH with NaOH). The experimenter was blind to the mouse genotypes. Excitation was performed with a 488 nm laser. The acquisition of XYZ image stacks at 1.5 s intervals was collected through a HyD detector tuned to 495–600 nm. Quantification of fluorescence intensity was performed on neuronal cell bodies using ImageJ. One hundred microliters of 160 μM Dyn A17 was added in phasic puffs in recording buffer after ~50 s basal signal recording.
For calcium imaging experiments assessing optogenetic release of endogenous dynorphin in slices, 4–8 weeks male Pdyn-Cre mice were used for viral injection targeted at nucleus accumbens (AP + 1.4, ML ± 1 relative to bregma, DV −4.5 relative to dura). Adeno-associated virus (AAV) serotype 5 encoding the ChrimsonR-tdTomato driven by synapsin promoter (pAAV-Syn-FLEX-rc[ChrimsonR-tdTomato] (AAV5)) (Addgene, 62723-AAV5) and AAV9-Syn-jGCaMP7s-WPRE virus were mixed at 1: 1 dilution (400 nl final volume) before injection. Imaging was carried out at room temperature in oxygenated ACSF containing the following (in mM): 126 NaCl, 2.5 KCl, 2.4 CaCl2, 1.3 MgCl2, 18 NaHCO3, 1.2 NaH2PO4, 10 glucose. Imaging was performed with excitation by a 488 nm laser and the acquisition of XYZ image stacks at 1.5 s intervals was collected through a HyD detector tuned to 500–570 nm. A fiber optic (CFM14L20, Thorlabs) coupled to a LED (M617F2, Thorlabs) was used for optogenetic stimulation with 20 trains of 1 s stimulation followed by 0.5 s inter-stimulation interval52. Quantification of the fluorescence intensity from cell bodies was performed using ImageJ. For κLight 1.3 imaging experiments assessing dynorphin release by optogenetic stimulation in slices, 8–12 weeks male Pdyn-Cre mice were used for viral injection at NAc as above. Adeno-associated virus (AAV) serotype 5 encoding the ChrimsonR-tdTomato driven by synapsin promoter (pAAV-Syn-FLEX-rc[ChrimsonR-tdTomato] (AAV5)) (Addgene, 62723-AAV5) and AAV-CAG-DIO-κLight1.3 (UNC Neurotools) were mixed at 1:1 dilution (400 nl final volume) before injection. Slice preparation and optogenetic stimulation were done as above. Imaging was performed with excitation by a 488 nm laser and the acquisition of XYT mode at 300 ms intervals was collected through a HyD detector tuned to 500–550 nm. Quantification of the fluorescence intensity was performed using ImageJ.
Slice electrophysiology
For acute ex vivo brain slice preparation, both male and female mice were used. Mice were anesthetized using isoflurane followed by decapitation and brains collected and immediately transferred to ice-cold and oxygenated cutting solution consisting of (mM): 93 NMDG, 2.5 KCl, 20 HEPES, 10 MgSO4·7H2O, 1.2 NaH2PO4, 30 NaHCO3, 0.5 CaCl2·2H2O, 25 glucose, 3 Na-pyruvate, 5 ascorbic acid, and 5 N-acetylcysteine. Two hundred and fifty micrometers coronal slices containing the medial habenula were prepared on a vibrating microtome using standard procedures. Following the collection of coronal sections, the brain slices were transferred to a 32 °C chamber containing oxygenated cutting solution for a 10–15 min recovery period. Slices were then transferred to a 25 °C holding chamber with solution consisting of (mM): 92 NaCl, 2.5 KCl, 20 HEPES, 2 MgSO4·7H2O, 1.2 NaH2PO4, 30 NaHCO3, 2 CaCl2·2H2O, 25 glucose, 3 Na-pyruvate, 5 ascorbic acid, 5 N-acetylcysteine, and were allowed to recover for ≥30 min. For recording, slices were placed in a perfusion chamber and continuously perfused with oxygenated artificial cerebrospinal fluid (ACSF; 31–33 °C) consisting of (in mM): 113 NaCl, 2.5 KCl, 1.2 MgSO4·7H2O, 2.5 CaCl2·2H2O, 1 NaH2PO4, 26 NaHCO3, 20 glucose, 3 Na-pyruvate, 1 ascorbic acid, at a flow rate of 2–3 ml/min.
Medial habenula neurons were identified at ×40 magnification with an immersion objective with differential interference contrast microscopy. Cells for recording were selected from the lateral portion of the medial habenula due to the high density of mu opioid receptor and GPR139 expression in this area18. All recordings were carried out in loose-seal cell-attached configuration with borosilicate glass pipettes (3–6 MΩ) filled with extracellular ACSF solution. Tonic action potential firing experiments were carried out in voltage clamp configuration with the holding potential set such that baseline current was at or near I = 0 pA. Baseline firing was recorded for 5 min, followed by 5 min perfusion of 500 nM Dyn A17. Signals were filtered at 1 kHz and sampled at 10 kHz using Patchmaster software (HEKA Instruments) with an EPC10 amplifier (HEKA Instruments). To reliably detect pharmacological effects, cells with a basal firing frequency <1 Hz were labeled as silent and excluded from recordings. Data files were then converted to Axon Text File (.atf) format using Stimfit software and analyzed using Clampfit 10.7 software (Molecular Devices).
Statistics
Statistical significance was analyzed using GraphPad Prism 10.1.1 software. Concentration-response curves were fitted to four-parameter sigmoidal function. Data are displayed as mean ± SEM from three or four independent experiments. Analysis of variance (ANOVA) with appropriate post-hoc test and two-tailed Student’s t-test were used for statistical analysis as indicated in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data generated during the study are deposited at Figshare and are accessible via the https://doi.org/10.6084/m9.figshare.29533004. Source data are provided with this paper.
References
D’Amato, F. R. & Pavone, F. Modulation of nociception by social factors in rodents: contribution of the opioid system. Psychopharmacology 224, 189–200 (2012).
Gavériaux-Ruff, C. & Kieffer, B. L. Opioid receptor genes inactivated in mice: the highlights. Neuropeptides 36, 62–71 (2002).
Darcq, E. & Kieffer, B. L. Opioid receptors: drivers to addiction? Nat. Rev. Neurosci. 19, 499–514 (2018).
Nummenmaa, L. & Tuominen, L. Opioid system and human emotions. Br. J. Pharmacol. 175, 2737–2749 (2018).
Lutz, P.-E. & Kieffer, B. L. Opioid receptors: distinct roles in mood disorders. Trends Neurosci. 36, 195–206 (2013).
McDonald, J. & Lambert, D. G. Opioid mechanisms and opioid drugs. Anaesth. Intensive Care Med. 14, 505–509 (2013).
Che, T. & Roth, B. L. Molecular basis of opioid receptor signaling. Cell 186, 5203–5219 (2023).
Valentino, R. J. & Volkow, N. D. Untangling the complexity of opioid receptor function. Neuropsychopharmacology 43, 2514–2520 (2018).
Fricker, L. D., Margolis, E. B., Gomes, I. & Devi, L. A. Five decades of research on opioid peptides: current knowledge and unanswered questions. Mol. Pharm. 98, 96–108 (2020).
Benarroch, E. E. Endogenous opioid systems: current concepts and clinical correlations. Neurology 79, 807–814 (2012).
James, A. & Williams, J. Basic opioid pharmacology - an update. Br. J. Pain. 14, 115–121 (2020).
van Dorp, E., Yassen, A. & Dahan, A. Naloxone treatment in opioid addiction: the risks and benefits. Expert Opin. Drug Saf. 6, 125–132 (2007).
Przewłocki, R., Shearman, G. T. & Herz, A. Mixed opioid/nonopioid effects of dynorphin and dynorphin related peptides after their intrathecal injection in rats. Neuropeptides 3, 233–240 (1983).
Wollemann, M. & Benyhe, S. Non-opioid actions of opioid peptides. Life Sci. 75, 257–270 (2004).
Walker, J. M., Moises, H. C., Coy, D. H., Baldrighi, G. & Akil, H. Nonopiate effects of dynorphin and des-Tyr-dynorphin. Science 218, 1136–1138 (1982).
Tang, Q., Lynch, R. M., Porreca, F. & Lai, J. Dynorphin A elicits an increase in intracellular calcium in cultured neurons via a non-opioid, non-NMDA mechanism. J. Neurophysiol. 83, 2610–2615 (2000).
Shukla, V. K. & Lemaire, S. Central non-opioid physiological and pathophysiological effects of dynorphin A and related peptides. J. Psychiatry Neurosci. 17, 106–119 (1992).
Wang, D. et al. Genetic behavioral screen identifies an orphan anti-opioid system. Science 365, 1267–1273 (2019).
Stoveken, H. M., Zucca, S., Masuho, I., Grill, B. & Martemyanov, K. A. The orphan receptor GPR139 signals via G(q/11) to oppose opioid effects. J. Biol. Chem. 295, 10822–10830 (2020).
Foster, S. R. et al. Discovery of human signaling systems: pairing peptides to G protein-coupled receptors. Cell 179, 895–908.e821 (2019).
Vassilatis, D. K. et al. The G protein-coupled receptor repertoires of human and mouse. Proc. Natl. Acad. Sci. USA 100, 4903–4908 (2003).
Vedel, L., Nøhr, A. C., Gloriam, D. E. & Bräuner-Osborne, H. Pharmacology and function of the orphan GPR139 G protein-coupled receptor. Basic Clin. Pharmacol. Toxicol. 126, 35–46 (2020).
Liu, C. et al. GPR139, an orphan receptor highly enriched in the Habenula and septum, is activated by the essential amino acids L-tryptophan and L-phenylalanine. Mol. Pharmacol. 88, 911–925 (2015).
Süsens, U., Hermans-Borgmeyer, I., Urny, J. & Schaller, H. C. Characterisation and differential expression of two very closely related G-protein-coupled receptors, GPR139 and GPR142, in mouse tissue and during mouse development. Neuropharmacology 50, 512–520 (2006).
Nässel, D. R. & Winther, A. M. Drosophila neuropeptides in regulation of physiology and behavior. Prog. Neurobiol. 92, 42–104 (2010).
Rőszer, T. & Bánfalvi, G. FMRFamide-related peptides: anti-opiate transmitters acting in apoptosis. Peptides 34, 177–185 (2012).
Nøhr, A. C. et al. The orphan G protein-coupled receptor GPR139 is activated by the peptides: adrenocorticotropic hormone (ACTH), α-, and β-melanocyte stimulating hormone (α-MSH, and β-MSH), and the conserved core motif HFRW. Neurochem. Int. 102, 105–113 (2017).
Zhou, Y. et al. Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139. Cell Res. 32, 210–213 (2022).
Nepomuceno, D. et al. Re-evaluation of adrenocorticotropic hormone and melanocyte stimulating hormone activation of GPR139 in vitro. Front. Pharm. 9, 157 (2018).
Hiramatsu, M. & Inoue, K. Des-tyrosine(1) dynorphin A-(2-13) improves carbon monoxide-induced impairment of learning and memory in mice. Brain Res. 859, 303–310 (2000).
Gardell, L. R. et al. Sustained morphine exposure induces a spinal dynorphin-dependent enhancement of excitatory transmitter release from primary afferent fibers. J. Neurosci. 22, 6747–6755 (2002).
Wang, W., Qiao, Y. & Li, Z. New insights into modes of GPCR activation. Trends Pharmacol. Sci. 39, 367–386 (2018).
Pavlos, N. J. & Friedman, P. A. GPCR signaling and trafficking: the long and short of It. Trends Endocrinol. Metab. 28, 213–226 (2017).
Luttrell, L. M. & Lefkowitz, R. J. The role of beta-arrestins in the termination and transduction of G-protein-coupled receptor signals. J. Cell Sci. 115, 455–465 (2002).
Miess, E. et al. Multisite phosphorylation is required for sustained interaction with GRKs and arrestins during rapid μ-opioid receptor desensitization. Sci Signal. 11, eaas9609 (2018).
Kumari, P. et al. Functional competence of a partially engaged GPCR-β-arrestin complex. Nat. Commun. 7, 13416 (2016).
Lee, Y. et al. Molecular basis of β-arrestin coupling to formoterol-bound β(1)-adrenoceptor. Nature 583, 862–866 (2020).
Moo, E. V., van Senten, J. R., Bräuner-Osborne, H. & Møller, T. C. Arrestin-dependent and -independent internalization of G protein-coupled receptors: methods, mechanisms, and implications on cell signaling. Mol. Pharm. 99, 242–255 (2021).
Rajagopal, S. & Shenoy, S. K. GPCR desensitization: acute and prolonged phases. Cell Signal. 41, 9–16 (2018).
Miller, W. E. & Lefkowitz, R. J. Expanding roles for beta-arrestins as scaffolds and adapters in GPCR signaling and trafficking. Curr. Opin. Cell Biol. 13, 139–145 (2001).
Groer, C. E., Schmid, C. L., Jaeger, A. M. & Bohn, L. M. Agonist-directed interactions with specific beta-arrestins determine mu-opioid receptor trafficking, ubiquitination, and dephosphorylation. J. Biol. Chem. 286, 31731–31741 (2011).
Pergel, E., Veres, I., Csigi, G.I. & Czirják, G. Translocation of TMEM175 lysosomal potassium channel to the plasma membrane by dynasore compounds. Int. J. Mol. Sci. 22, 10515 (2021).
Dutta, D., Williamson, C. D., Cole, N. B. & Donaldson, J. G. Pitstop 2 is a potent inhibitor of clathrin-independent endocytosis. PLoS ONE 7, e45799 (2012).
Reppetti, J. et al. Intact caveolae are required for proper extravillous trophoblast migration and differentiation. J. Cell Physiol. 235, 3382–3392 (2020).
Gomes, I. et al. Biased signaling by endogenous opioid peptides. Proc. Natl. Acad. Sci. USA 117, 11820–11828 (2020).
Wang, Y. et al. Structures of the entire human opioid receptor family. Cell 186, 413–427.e417 (2023).
Dana, H. et al. High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat. Methods 16, 649–657 (2019).
Hashikawa, Y. et al. Transcriptional and spatial resolution of cell types in the mammalian habenula. Neuron 106, 743–758.e745 (2020).
Chen, C. et al. Characterization of a knock-in mouse line expressing a fusion protein of κ opioid receptor conjugated with tdTomato: 3-dimensional brain imaging via CLARITY. eNeuro 7, ENEURO.0028-20.2020 (2020).
Castro, D. C. & Berridge, K. C. Opioid hedonic hotspot in nucleus accumbens shell: mu, delta, and kappa maps for enhancement of sweetness “liking” and “wanting”. J. Neurosci. 34, 4239–4250 (2014).
Al-Hasani, R. et al. Distinct subpopulations of nucleus accumbens dynorphin neurons drive aversion and reward. Neuron 87, 1063–1077 (2015).
Dong, C. et al. Unlocking opioid neuropeptide dynamics with genetically encoded biosensors. Nat. Neurosci. 27, 1844–1857 (2024).
Wang, L. et al. GPR139 and dopamine D2 receptor co-express in the same cells of the brain and may functionally interact. Front. Neurosci. 13, 281 (2019).
Sarin, S. et al. Role for Wnt signaling in retinal neuropil development: analysis via RNA-seq and in vivo somatic CRISPR mutagenesis. Neuron 98, 109–126.e108 (2018).
Dao, M., Stoveken, H. M., Cao, Y. & Martemyanov, K. A. The role of orphan receptor GPR139 in neuropsychiatric behavior. Neuropsychopharmacology 47, 902–913 (2022).
Rabiner, E. A. et al. Endogenous dopamine release in the human brain as a pharmacodynamic biomarker: evaluation of the new GPR139 agonist TAK-041 with [(11)C]PHNO PET. Neuropsychopharmacology 47, 1405–1412 (2022).
Eiger, D. S., Pham, U., Gardner, J., Hicks, C. & Rajagopal, S. GPCR systems pharmacology: a different perspective on the development of biased therapeutics. Am. J. Physiol. Cell Physiol. 322, C887–c895 (2022).
Wootten, D., Christopoulos, A., Marti-Solano, M., Babu, M. M. & Sexton, P. M. Mechanisms of signalling and biased agonism in G protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 19, 638–653 (2018).
Viveros, O. H., Diliberto, E. J. Jr., Hazum, E. & Chang, K. J. Opiate-like materials in the adrenal medulla: evidence for storage and secretion with catecholamines. Mol. Pharm. 16, 1101–1108 (1979).
Winkler, H. & Westhead, E. The molecular organization of adrenal chromaffin granules. Neuroscience 5, 1803–1823 (1980).
Klein, R. L., Wilson, S. P., Dzielak, D. J., Yang, W. H. & Viveros, O. H. Opioid peptides and noradrenaline co-exist in large dense-cored vesicles from sympathetic nerve. Neuroscience 7, 2255–2261 (1982).
Chen, L., Gu, Y. & Huang, L. Y. The mechanism of action for the block of NMDA receptor channels by the opioid peptide dynorphin. J. Neurosci. 15, 4602–4611 (1995).
Chen, L. & Huang, L. Y. Dynorphin block of N-methyl-D-aspartate channels increases with the peptide length. J. Pharm. Exp. Ther. 284, 826–831 (1998).
Vick, J. S. & Askwith, C. C. ASICs and neuropeptides. Neuropharmacology 94, 36–41 (2015).
Lioudyno, M. I. et al. The alpha9/alpha10-containing nicotinic ACh receptor is directly modulated by opioid peptides, endomorphin-1, and dynorphin B, proposed efferent cotransmitters in the inner ear. Mol. Cell Neurosci. 20, 695–711 (2002).
Lai, J. et al. Dynorphin A activates bradykinin receptors to maintain neuropathic pain. Nat. Neurosci. 9, 1534–1540 (2006).
Borg, C. B. et al. Mechanism and site of action of big dynorphin on ASIC1a. Proc. Natl Acad. Sci. USA 117, 7447–7454 (2020).
Leisle, L. et al. Dynorphin Neuropeptides Decrease Apparent Proton Affinity of ASIC1a by Occluding the Acidic Pocket. J. Med Chem. 64, 13299–13311 (2021).
Gloriam, D. E. I., Schiöth, H. B. & Fredriksson, R. Nine new human Rhodopsin family G-protein coupled receptors: identification, sequence characterisation and evolutionary relationship. Biochim. et Biophys. Acta 1722, 235–246 (2005).
Luttrell, L.M. et al. Manifold roles of β-arrestins in GPCR signaling elucidated with siRNA and CRISPR/Cas9. Sci Signal 11, eaat7650 (2018).
Masuho, I., Martemyanov, K. A. & Lambert, N. A. Monitoring G protein activation in cells with BRET. Methods Mol. Biol. 1335, 107–113 (2015).
Yang, J. et al. Coupling optogenetic stimulation with NanoLuc-based luminescence (BRET) Ca(++) sensing. Nat. Commun. 7, 13268 (2016).
Masuho, I. et al. Molecular deconvolution platform to establish disease mechanisms by surveying GPCR signaling. Cell Rep. 24, 557–568.e555 (2018).
Laboute, T. et al. Orphan receptor GPR158 serves as a metabotropic glycine receptor: mGlyR. Science 379, 1352–1358 (2023).
Maza, N. et al. Ptchd1 mediates opioid tolerance via cholesterol-dependent effects on μ-opioid receptor trafficking. Nat. Neurosci. 25, 1179–1190 (2022).
Dunn, H. A., Patil, D. N., Cao, Y., Orlandi, C. & Martemyanov, K. A. Synaptic adhesion protein ELFN1 is a selective allosteric modulator of group III metabotropic glutamate receptors in trans. Proc. Natl. Acad. Sci. USA 115, 5022–5027 (2018).
Dunn, H. A., Zucca, S., Dao, M., Orlandi, C. & Martemyanov, K. A. ELFN2 is a postsynaptic cell adhesion molecule with essential roles in controlling group III mGluRs in the brain and neuropsychiatric behavior. Mol. Psychiatry 24, 1902–1919 (2019).
Acknowledgements
We thank Natalia Martemyanova for husbandry, maintenance, and genotyping of all the mice examined in this study. This work was supported by NIH grant DA036596 (K.A.M.).
Author information
Authors and Affiliations
Contributions
X.L. and K.A.M. conceived the project; X.L. performed all functional experiments and calcium imaging; N.W. performed electrophysiology experiments; S.P. performed studies exploring internalization mechanisms; C.L. performed live cell imaging experiments; H.S., E.S., L.S., and T.S. conducted and analyzed the peptide screening experiments; C.C. performed and analyzed flow cytometry experiments; S.Z. performed viral experiments in mice; X.L. and K.A.M. wrote the manuscript with input from all other authors; and K.A.M. supervised the project.
Corresponding author
Ethics declarations
Competing interests
K.A.M. is a consultant and stakeholder in EvoDenovo, Inc. a company commercializing the development of innovative treatments for opioid use disorder. All other authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Christopher Evans and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, X., Winters, N.D., Pandey, S. et al. Homeostatic scaling of dynorphin signaling by a non-canonical opioid receptor. Nat Commun 16, 6786 (2025). https://doi.org/10.1038/s41467-025-62133-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-62133-x