Abstract
Arboviruses often employ various strategies to manipulate the behavior of herbivorous vectors and host plants, thereby enhancing their transmission and infection. In this study, we identify a plant lectin protein, OsChtBL1, which possesses chitin-binding activity and specifically accumulates on the stylets of insect vectors. This binding creates a feeding barrier that reduces the vectors’ efficiency in acquiring and transmitting the virus. However, the rice stripe virus, a devastating pathogen in rice, counteracts this defense by utilizing a viral protein to recruit an E3 ubiquitin ligase, leading to the degradation of OsChtBL1. This degradation facilitates vector feeding and enhances viral transmission. Our findings provide insights into how arbovirus-infected host plants improve the performance of herbivorous vectors and offer deeper understanding of the complex interactions among arboviruses, vectors, and host plants.
Similar content being viewed by others
Introduction
In nature, ~80% of plant viruses are known to be transmitted by herbivorous vectors. These arboviruses have established symbiotic relationships with their vectors through co-evolution, allowing them to systematically attack plants and compromise their various immune barriers1. After establishing infection in host plants, arboviruses typically employ diverse strategies to manipulate the host’s physiology to enhance the performance of their insect vectors, thereby amplifying and accelerating viral spread2,3. Extensive research has shown that arboviruses regulated the production of volatile compounds in host plants to attract insect vectors2,4,5. For example, the 2b protein encoded by cucumber mosaic virus (CMV) suppressed jasmonic acid (JA) signaling, making the host plant more attractive to aphid vectors in Arabidopsis6. Additionally, the CMV 1a protein inhibited the biosynthesis of methyl-salicylate (MeSA), reducing the plant’s airborne defense against aphids7. Despite these findings on volatile compounds, the complexity of the relationships among host plants, herbivorous insects, and arboviruses remains poorly understood.
Over the course of their prolonged struggle against herbivorous insects and pathogens in their natural habitats, plants have gradually evolved a sophisticated and effective defense network. Plant lectins, a group of proteins capable of recognizing various carbohydrates based on their molecular structures and biochemical properties without catalyzing them, are widely found across a broad range of plant species8,9. Sixteen plant lectin families, containing lectin domains, have been identified based on amino acid sequence similarities, molecular structure analyses, and the types of carbohydrates they bind. These families are typically named after the species in which the first lectin of the family was discovered. Some of the major families include Galanthus nivalis agglutinin (GNA) lectins, jacalin lectins, Lysin M (LysM) domain lectins, and Hevein domain lectins10. In recent decades, with advancements in high-throughput sequencing, the role of plant lectins has expanded beyond mere carbohydrate recognition. Molecular structure determination and omics analyses have provided deeper insights into the functions of plant lectins, revealing their involvement in multiple critical biological processes11. However, only a limited number of lectin families have been thoroughly studied. For example, GNA lectins exhibit a high degree of affinity for mannose in the gastrointestinal epithelial cells of insects, leading to their poisoning12,13. LysM domain lectins function as receptors for chitin oligosaccharides from pathogens or symbionts, triggering innate immunity mediated by pathogen-associated molecular patterns (PAMPs) and microbe-associated molecular patterns (MAMPs) in plants14,15. Despite these advances, research on Hevein domain lectins, which specifically bind to chitin, remains relatively limited. Chitin, composed mainly of N-acetylglucosamine (GlcNAc), is specifically synthesized by insects, crustaceans, nematodes, and fungi16. Most Hevein domain lectins possess one or more GlcNAc-binding domains and exhibit significant antifungal activity17,18. However, homologs of Hevein domain lectins in rice remain poorly characterized. Furthermore, the mechanisms by which Hevein domain lectins orchestrate arboviruses, herbivorous insects, and host plants' tripartite interactions are largely unexplored.
Rice, as a crucial food crop, serves as the staple food for approximately half of the global population. However, rice production is frequently threatened by numerous agricultural pests and pathogens, which can cause devastating losses. Among these, piercing-sucking insects pose a particularly severe threat, not only through direct damage caused by their feeding activities but also by transmitting viruses that exacerbate the damage19. In nature, the rice stripe virus (RSV), a member of the genus Tenuivirus within the family Phenuiviridae (order Bunyavirales), is a highly destructive pathogen carried and transmitted by the small brown planthopper (SBPH, Laodelphax striatellus Fallén)20. RSV is a single-stranded (ss) negative-sense RNA virus, consisting of four ssRNA segments that encode seven proteins, including a viral nucleocapsid protein (Pc3), a RNA-dependent RNA polymerase (RdRp), and five nonstructural proteins (P2, Pc2, P3, P4, and Pc4)21,22. Considerable research has been explored to understand the RSV-SBPH or RSV-rice interactions. On one hand, RSV enters the gut lumen of SBPH through its mouthparts, overcomes a series of barriers within the insect, reaches the ovaries of female insects for vertical transmission to offspring, and eventually reaches the salivary glands for horizontal transmission to RSV-free rice plants23,24. On the other hand, RSV exploits various cellular processes of the host plant to replicate and spread, thereby facilitating its own infection24,25. However, there has been relatively limited progress in understanding the mechanisms underlying the interactions among RSV, SBPH, and rice, particularly how RSV manipulates host plants to enhance vectors performance, eventually facilitating its own infection and transmission.
In this study, we identified a chitin-binding lectin, OsChtBL1, that binds to and aggregates with the stylets of SBPH, affecting its feeding activity. The accumulation of OsChtBL1 within plants triggers an endoplasmic reticulum (ER) stress-mediated immune response, thereby repressing virus proliferation. Interestingly, the P2 encoded by RSV can degrade OsChtBL1 through the 26S proteasome, suppressing rice resistance to herbivorous vector SBPH and RSV. This degradation process enhances the adaptability of SBPH on host plants and promotes RSV acquisition and transmission. Collectively, our findings provide insights into how arbovirus-infected host plants enhance the performance of herbivorous vectors, facilitating viral infection and transmission.
Results
OsChtBL1 suppress the performance of SBPH
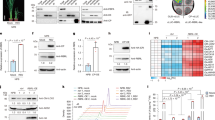
Damage inflicted by herbivorous insects activates multiple immune responses in plants and induces the expression of numerous insect-resistant genes26. Given that insects possess substantial amounts of chitin, we hypothesized that plant chitin-binding lectins may play a crucial role in plant-insect interactions. We thus examined the function of OsChtBL1 in response to SBPH infestation. RT-qPCR assay further demonstrated that OsChtBL1 expression was sharply upregulated within 30 min of SBPH feeding on NIP plants, particularly in the stem tissue (the primary feeding site of SBPH), followed by stabilization over time (Fig. 1a). To explore the function of OsChtBL1 in the rice-feeding process of SBPH, we generated two transgenic rice lines overexpressing OsChtBL1 (OsChtBL1-OE#1, OsChtBL1-OE#6) and two CRISPR-Cas9 mutants (oschtbl1#4, oschtbl1#10) to investigate whether OsChtBL1 influences the performance of SBPH (Supplementary Fig. 1a, b). First, the feeding efficiency of SBPH was evaluated on rice plants. The amount of honeydew, which is a key indicator for SBPH feeding efficiency4,27, produced by SBPH on OsChtBL1-OE plants was significantly lower than that on NIP, while the amount of honeydew was markedly increased on oschtbl1 plants (Fig. 1b). Correspondingly, the mortality rate of SBPH on OsChtBL1-OE plants was significantly higher than that on NIP, while the mortality rate was reduced on oschtbl1 plants after 15 d of SBPH feeding on these plants (Fig. 1c). Using electrical penetration graph (EPG) analysis, we identified four distinct waveform patterns during SBPH feeding on rice plants (Supplementary Fig. 2a). Further data analysis revealed that SBPH fed on OsChtBL1-OE plants exhibited prolonged probing duration, reduced sustained phloem ingestion, and increased probing frequency compared to those fed on NIP plants, whereas SBPH fed on oschtbl1 plants showed the opposite trends (Supplementary Fig. 2b, c). These results suggest that OsChtBL1 might suppress the performance of SBPH by affecting their feeding efficiency.
a RT-qPCR assay showing the relative expression level of OsChtBL1 within 48 h of SBPH feeding on NIP plants. b The quantity of honeydew collected from NIP, OsChtBL1-OE and oschtbl1 plants after SBPH feeding for 48 h. n = 12 individual SBPH. c The mortality rate of SBPH fed with NIP, OsChtBL1-OE and oschtbl1 plants after 15 d. d The predicted structural domain of OsChtBL1. e, f IP assay showing OsChtBL1 exhibits a strong binding affinity with chitin in N. benthamiana and rice. g The protein degradation assay showing chitin enhanced the protein stability of OsChtBL1. The concentration of chitin used was 10 μg/ml. h The survival rate of SBPH feeding on purified GST or GST-OsChtBL1 in vitro for 48 h. i Detailed Schematic diagram of the stylet of SBPH. Mg midgut, Sg salivary gland, Fc food canal, Sc salivary canal, Lmx left maxillary stylet, Rmx right maxillary stylet, Lmd left mandibular stylet, Rmd right mandibular stylet, * dendritic canal. j IF assay showing OsChtBL1 binds and aggregates on the stylets of SBPH. Scale bar, 50 μm. k. The food canal of SBPH feeding on purified GST and GST-OsChtBL1 in vitro observed by TEM. Scale bar, 200 nm. l. IEM assay showing OsChtBL1 accumulation in the food canal of SBPH. Scale bar, 200 nm. SBPH fed on purified GST and GST-OsChtBL1 (50 μg/ml) for 36 h were subjected to IF, TEM, and IEM assays (j–l). OsUBQ5 serves as an internal reference gene (a). n = 3 (a, c, h) independent biological replicates. Error bars represent SD, and values are means ± SD. All the statistical analysis data were performed using a two-tailed Student’s t test. *At the top of columns indicates significant differences with the control group at P < 0.05. Rubisco serves as an internal reference protein for Western blot analysis and detected by anti-RUB antibody (g). Experiments in (e–g) were repeated three times with similar results.
OsChtBL1 establishes a feeding barrier for SBPH by accumulating within their stylets
Structural domain analysis predicted that OsChtBL1 contains four tandem chitin-binding domains (Fig. 1d). To determine whether OsChtBL1 possesses chitin-binding activity, we performed an immunoprecipitation assay using chitin-coupled magnetic beads. The results showed that OsChtBL1 binds strongly to chitin, regardless of whether OsChtBL1 was transiently expressed in N. benthamiana or overexpressed in transgenic rice (Fig. 1e, f). Co-immunoprecipitation (Co-IP) assay and bimolecular fluorescence complementation (BiFC) assay demonstrated that OsChtBL1 exhibits a strong self-association (Supplementary Fig. 2d, e). When the purified GST or GST-OsChtBL1 was separately incubated with water or chitin at 4 °C for 4 h, transmission electron microscopy (TEM) revealed that GST-OsChtBL1 formed large aggregates in the presence of chitin, while GST alone did not (Supplementary Fig. 2f). In addition, protein degradation assays revealed that OsChtBL1 exhibited enhanced stability in the presence of chitin compared to control buffer, regardless of cycloheximide (CHX) treatment (Fig. 1g and Supplementary Fig. 2g). To validate the specificity of this effect, we conducted parallel degradation assays using OsHDA706, a rice histone deacetylase lacking a chitin-binding domain. Notably, chitin had no discernible impact on the stability of OsHDA706, demonstrating that the chitin-mediated stabilization of OsChtBL1 is specific (Supplementary Fig. 2h). These results suggest that OsChtBL1 can bind to chitin, resulting in a more stable complex.
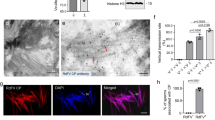
Considering that insect mouthpart and stylets is composed largely of chitin16,28, we hypothesize that OsChtBL1 might directly bind to stylets of SBPH, reducing feeding efficiency and increasing mortality rates. To test this, we first feed SBPH with a mixture of 4% sucrose and purified GST or GST-OsChtBL1. While the mixture of 4% sucrose and GST (50 μg/ml) had no significant impact on the performance of SBPH, the mixtures of 4% sucrose and either low-dose (10 μg/ml) or high-dose (50 μg/ml) GST-OsChtBL1 caused a substantial increase in abnormal SBPH deaths. Notably, the high-dose GST-OsChtBL1 resulted in a mortality rate exceeding 50% within 48 h (Fig. 1h). We then dissected the SBPH that died in the in vitro parafilm feeding assay (Fig. 1h, i) to examine their stylets using an immunofluorescence (IF) assay and TEM. The results of IF assay showed that multiple types of protein aggregations and obstructions were observed on the stylets of SBPH fed with GST-OsChtBL1, but not on those of SBPH fed with GST (Fig. 1j). TEM revealed that the food canal of SBPH stylets fed with GST-OsChtBL1 were densely filled with mesh-like substances, whereas those fed with GST alone appeared largely transparent (Fig. 1k). Immunoelectron microscopy (IEM) further confirmed that these structures were specifically labeled by anti-GST antibodies, demonstrating their identity as accumulated GST-OsChtBL1 within the food canal of SBPH stylets (Fig. 1l). We further performed TEM and IEM analyses on food canal of SBPH after 7 days of feeding on NIP, OsChtBL1-OE, and oschtbl1 plants. Our observations revealed substantial mesh-like substance accumulation in the food canal of SBPH fed on OsChtBL1-OE plants, minimal accumulation in those fed on NIP plants, and no detectable accumulation in those fed on oschtbl1 plants (Supplementary Fig 2i). Statistical analysis of 50 food canal samples confirmed these observations (Supplementary Fig. 2j). Further IEM analyses confirmed that the abundant mesh-like substances in food canal of SBPH fed on OsChtBL1-OE plants were indeed OsChtBL1 (Supplementary Fig. 2k). Collectively, these results demonstrate that OsChtBL1 binds to chitin in the SBPH food canal, forming stable aggregates that establish a feeding barrier.
OsChtBL1 suppresses the efficiency of virus acquisition and transmission by SBPH
The acquisition and transmission of arboviruses by herbivorous insects are crucial processes in the life cycle of arboviruses29. To investigate whether OsChtBL1 affects the efficiency of SBPH in acquiring viruses, we employed a mixture of 4% sucrose, purified GST or GST-OsChtBL1 (20 μg/ml), and RSV virus particles crudely extracted from infected rice as feed and enabled SBPH to feed through in vitro parafilm feeding for 2 days, followed by a 5-day latent period on fresh rice seedlings (Fig. 2a). Both the quantity of RSV acquired by SBPH and the proportion of infected SBPH were significantly lower in those fed with GST-OsChtBL1 compared to those fed with GST alone (Fig. 2b, c). These results indicate that OsChtBL1 reduces the efficiency of acquiring viruses by SBPH. To further investigate whether OsChtBL1 affects the efficiency of virus transmission by SBPH, we allowed RSV-free SBPH to feed on RSV-infected rice for 3 days and to spend a 10-day latent period on healthy rice seedlings in order to obtain an adequate quantity of RSV. Subsequently, these RSV-infected SBPH fed on healthy NIP, OsChtBL1-OE, and oschtbl1 plants at the same proportion (Fig. 2d). We first evaluated the early-stage viral transmission efficiency of RSV-infected SBPH feeding on rice. Our results showed that RSV was detectable in rice plants 72 h after feeding by RSV-infected SBPH (Supplementary Fig. 3a). Furthermore, when the ratio of RSV CP mRNA to OsUBQ5 mRNA reached ≥0.0005, significantly higher numbers of RSV-infected individuals and elevated viral loads were observed in oschtbl1 plants compared to NIP plants, while the opposite trend was observed in OsChtBL1-OE plants (Supplementary Fig. 3b and Fig. 2e). Subsequently, we quantified RSV CP protein levels in secreted saliva from RSV-free/infected SBPH using an in vitro parafilm feeding system (Supplementary Fig. 3c). After 24 h of feeding on protein extracts from NIP and OsChtBL1-OE plants, western blot analysis revealed significantly reduced accumulation of RSV CP in salivary secretions of RSV-infected SBPH fed on OsChtBL1-OE plant extracts compared to those fed on NIP plant extracts (Supplementary Fig. 3d). Following the protocol outlined in Fig. 2d, we inoculated NIP, OsChtBL1-OE, and oschtbl1 plants with RSV and assessed incidence rate 30 days post-inoculation. The results revealed that OsChtBL1-OE plants displayed a lower incidence rate compared to NIP plants, whereas oschtbl1 plants exhibited a significantly higher incidence rate (Fig. 2f). These results indicate that OsChtBL1 significantly inhibits the efficiency of SBPH in acquiring and transmitting RSV. Apart from the extremely low incidence rate, we observed milder symptoms on RSV-infected OsChtBL1-OE plants compared to RSV-infected NIP (Fig. 2g). Whereas RSV-infected oschtbl1 plants not only had an extremely high incidence rate but also exhibited more severe symptoms (Fig. 2j). Analysis of the mRNA and protein levels of RSV CP demonstrated that the viral accumulation in OsChtBL1-OE plants was considerably lower than that in NIP (Fig. 2h, i), whereas the viral accumulation in oschtbl1 plants was significantly higher than that in NIP (Fig. 2k, l). We speculate that, in addition to OsChtBL1 inhibiting the feeding activity of SBPH and causing a low initial amount of RSV invading into the plants, OsChtBL1 may also activate antiviral immune responses of rice.
a Schematic diagram of the virus acquisition system for SBPH through in vitro parafilm feeding. b OsChtBL1 inhibiting the quantity of RSV acquired by SBPH. The SBPH obtained following the process in (a). The mRNA ratio of RSV CP/LsGADPH is used to determine the quantity of RSV carried by SBPH. LsGADPH serves as an internal reference gene of SBPH. The numbers above the scatter plots indicate the ratio of RSV-infected SBPH individuals (left of the slash) to the total number of SBPH samples tested (right of the slash). c OsChtBL1 reducing the rate of SBPH infected with RSV. The column chart represents the ratio of SBPH infected with RSV in (b). d Schematic diagram of virus transmission system for SBPH. e Ratio of rice infected with RSV after RSV-infected SBPH fed on NIP, OsChtBL1-OE, and oschtbl1 plants for 72 h. Quantification of SBPH viral transmission efficiency in Supplementary Fig. 3b. f The incidence rate of NIP, OsChtBL1-OE, and oschtbl1 plants under field conditions. These plants are inoculated with RSV following the process in (d). g, j Phenotypes of Mock- or RSV-infected NIP, OsChtBL1-OE, and oschtbl1 plants after RSV inoculation for 30 days. Scale bar, 10 cm. h, k RT-qPCR assay showing the mRNA relative expression level of RSV CP in RSV-infected NIP, OsChtBL1-OE, and oschtbl1 plants. i, l Western blot assay showing protein accumulation level of RSV CP in Mock- or RSV-infected NIP, OsChtBL1-OE, and oschtbl1 plants. OsUBQ5 serves as an internal reference gene (h, k). n = 3 (c, e, f, h, k) independent biological replicates. Error bars represent SD, and values are means ± SD. All the statistical analysis data were performed using a two-tailed Student’s t test. *At the top of columns indicates significant differences with the control group at P < 0.05. Rubisco serves as an internal reference protein for Western blot analysis and detected by anti-RUB antibody (i, l). Experiments in (i, l) were repeated three times with similar results.
OsChtBL1 positively regulate endoplasmic reticulum stress
To further explore whether OsChtBL1 is involved in activating immune response of rice, we conducted an in-depth transcriptome analysis of NIP and OsChtBL1-OE plants. The transcriptome analysis identified a total of 2042 differentially expressed genes (DEGs) altogether, including 1051 upregulated DEGs and 991 downregulated DEGs (Fig. 3a). The gene ontology (GO) enrichment analysis demonstrated that these DEGs were mainly concentrated in pathways like “protein folding in endoplasmic reticulum”, “regulation of salicylic acid biosynthetic process”, “hydrogen peroxide catabolic process” and “defense response” (Fig. 3b). External biotic or abiotic stresses increase the load of misfolded or unfolded proteins in the endoplasmic reticulum (ER) of plants, stimulating ER stress and conferring resistance of plants to adverse external factors30,31. Therefore, we carried out a more profound hierarchical cluster analysis of the ER stress-related genes within all the DEGs and discovered that the ER stress-related genes in OsChtBL1-OE#1 plants were significantly upregulated compared to NIP plants (Fig. 3c). Then, we detected the expression of previously reported ER stress-related genes in transgenic plants32,33. RT-qPCR assay showed that the ER stress-related genes were significantly upregulated in OsChtBL1-OE plants but markedly downregulated in oschtbl1 plants compared to NIP plants (Fig. 3d), which was consistent with the results of transcriptome analysis. GO enrichment analysis further implicated OsChtBL1 in the salicylic acid (SA) pathway, which has been shown to positively regulate rice resistance to RSV34. Correspondingly, RT-qPCR confirmed that SA pathway-related genes were significantly upregulated in OsChtBL1-OE plants and downregulated in oschtbl1 plants relative to NIP plants (Fig. 3e). Activation of ER stress destabilizes the redox environment on the ER membrane, inducing the production of reactive oxygen species (ROS)35,36. In addition, GO enrichment analysis also showed that DEGs are involved in “response to oxidative stress” and “hydrogen peroxide catabolic process”, suggesting a close relationship between OsChtBL1 and ROS levels (Fig. 3b). Therefore, we carried out histochemical staining of NIP, OsChtBL1-OE and oschtbl1 plants with 3, 3'-diaminobenzidine (DAB), and found that the ROS level of OsChtBL1-OE plants was markedly higher than that of NIP plants, whereas the ROS level of oschtbl1 plants was significantly lower (Supplementary Fig. 4a, b). Together, these results suggest that OsChtBL1 likely activates rice antiviral immunity by modulating ER stress, SA pathway, and ROS production.
a The volcano plot showing the overall distribution of DEGs through transcriptome analysis of NIP and OsChtBL1-OE#1 plants. The threshold criteria for screening DEGs are | log2(fold change) | ≥2 and a q-value < 0.05 (q-value is the adjusted value of P value). b GO enrichment analysis of the DEGs in OsChtBL1-OE#1 versus NIP plants. c Hierarchical clustering analysis showing ER stress-related genes in OsChtBL1-OE#1 plants are significantly upregulated compared to those in NIP plants. d, e RT-qPCR assay showing mRNA relative expression level of ER stress and SA-related genes in NIP, OsChtBL1-OE, and oschtbl1 plants. OsUBQ5 serves as an internal reference gene (d, e). n = 3 (d, e) independent biological replicates. Error bars represent SD, and values are means ± SD. All the statistical analysis data were performed using a two-tailed Student’s t test. *At the top of columns indicates significant differences with the control group at P < 0.05.
OsChtBL1 specifically Interacts with RSV P2
For the purpose of thoroughly dissecting the function of OsChtBL1 during RSV infection, we detected the protein level of OsChtBL1 in OsChtBL1-OE#1 plants after RSV infection. Western blot assay showed that the protein level of OsChtBL1 in RSV-infected OsChtBL1-OE#1 plants was markedly decreased compared with that in Mock-OsChtBL1-OE#1 plants (Fig. 4a). Correspondingly, the quantity of honeydew collected from RSV-infected OsChtBL1-OE#1 plants was greater than that from Mock-OsChtBL1-OE#1 plants at 48 h after SBPH feeding (Fig. 4b). These results imply that the reduced protein accumulation of OsChtBL1 caused by RSV infection promotes the behavioral performance of SBPH.
a Western blot showing RSV infection reduces the protein accumulation level of OsChtBL1. b The quantity of honeydew collected from Mock- and RSV-infected OsChtBL1-OE#1 plants after SBPH feeding for 48 h. n = 19 individual SBPH. c Y2H assay showing P2 specifically interacts with OsChtBL1 in yeast cells. These yeast cells containing different combinations were observed after growing on the SD-LT (lacking leucine and tryptophan) and SD-AHLT (lacking alanine, histidine, leucine, and tryptophan) medium at 30 °C for 3 days. d Co-IP assay showing P2 interacts with OsChtBL1 in N. benthamiana. e LCI assay showing the combination of P2 and OsChtBL1 generates a strong fluorescent signal. f BiFC assay showing P2 interacts with OsChtBL1 in the cytoplasm of N. benthamiana. Scale bar, 50 μm. g Co-IP showing P2 interacts with OsChtBL1 during RSV infection. h Co-IP showing P2 interacts with OsChtBL1 in the hybrid rice line. i The protein degradation assay showing P2 promotes the degradation of OsChtBL1 via the 26S proteasome in rice. DMSO or MG132 and Purified GST or GST-P2 were added, respectively, to the total protein extract of OsChtBL1-OE plants. Samples were collected every 30 min at 28 °C and analyzed by Western blot using anti-MYC antibody. j The protein accumulation level of OsChtBL1 in OsChtBL1-OE#1 and OsChtBL1-OE#1/P2-OX plants. Error bars represent SD, and values are means ± SD. All the statistical analysis data were performed using a two-tailed Student’s t test. *At the top of columns indicates significant differences with the control group at P < 0.05. Rubisco serves as an internal reference protein for western blot analysis and detected by anti-RUB antibody (a, i, j). Experiments in (d–j) were repeated three times with similar results.
To further investigate how RSV regulates the protein level of OsChtBL1, we employed the yeast two-hybrid (Y2H) assay to screen whether any of the six proteins (P2, P3, P4, Pc2, Pc3, and Pc4) encoded by RSV directly interacted with OsChtBL1. Surprisingly, OsChtBL1 specifically interacts only with P2 (Fig. 4c). Moreover, mutant analysis of OsChtBL1 showed that P2 interacts with the full length of OsChtBL1 (Supplementary Fig. 4c, d). The interaction of OsChtBL1 with P2 was further confirmed by Co-IP assay and luciferase complementation imaging (LCI) assay (Fig. 4d, e). Subcellular localization assay indicated that OsChtBL1 was predominantly localized in the endoplasmic reticulum, and P2 did not alter the localization of OsChtBL1 (Supplementary Fig. 4e). BiFC assay showed that OsChtBL1 interacts with P2 mainly in the cytoplasm (Fig. 4f). To further verify whether OsChtBL1 and P2 interact in RSV-infected rice plants, we utilized the specific anti-P2 antibody to immunoprecipitate the P2 protein from RSV-infected NIP and OsChtBL1-OE#1 plants, as well as healthy OsChtBL1-OE#1 plants. Western blot assay showed that the endogenous P2 in RSV-infected OsChtBL1-OE#1 plants co-precipitated with OsChtBL1-MYC (Fig. 4g). Furthermore, we also constructed a transgenic rice line overexpressing P2-FLAG (P2-OX) and hybridized it with OsChtBL1-OE plants, generating a double overexpression line (OsChtBL1-OE#1/P2-OX) of P2-FLAG and OsChtBL1-MYC. We utilized MYC magnetic beads to immunoprecipitate OsChtBL1-MYC from P2-OX, OsChtBL1-OE#1, and OsChtBL1-OE#1/P2-OX plants. Western blot assay showed that OsChtBL1-MYC co-precipitated with P2-FLAG in OsChtBL1-OE#1/P2-OX plants (Fig. 4h). These results strongly suggest that P2 specifically interacts with OsChtBL1.
P2 mediates the degradation of OsChtBL1 through 26S proteasome
Based on these findings, we speculated that the decrease in the OsChtBL1 protein level mediated by RSV might be attributed to P2. We transiently co-expressed OsChtBL1-FLAG, GFP, and P2-MYC. Western blot assay indicated that with the increase in the protein level of P2-MYC, the accumulation of OsChtBL1-FLAG decreased, while the accumulation of GFP, which served as an internal reference protein, did not change significantly (Supplementary Fig. 5a). In addition, RT-qPCR assay showed that there was no significant change in the transcriptional level of OsChtBL1-FLAG (Supplementary Fig. 5b). Meanwhile, upon the treatment with MG132, the degradation of OsChtBL1-FLAG mediated by P2-MYC was recovered (Supplementary Fig. 5c). To further verify that P2 degrades OsChtBL1, we purified GST or GST-P2 and added to the protein extract of OsChtBL1-OE#1 plants. The results showed that the degradation rate of OsChtBL1-MYC markedly accelerated in the presence of GST-P2 compared to the control GST protein, and this degradation rate of OsChtBL1 clearly slowed down with the treatment of MG132 (Fig. 4i). Furthermore, we also detected the protein level of OsChtBL1-MYC in OsChtBL1-OE#1 and OsChtBL1-OE#1/P2-OX plants. Western blot assay showed that the protein level of OsChtBL1-MYC significantly decreased in OsChtBL1-OE#1/P2-OX plants (Fig. 4j), but RT-qPCR assay showed that the transcriptional level of OsChtBL1-MYC in OsChtBL1-OE#1 and OsChtBL1-OE#1/P2-OX plants did not change significantly (Supplementary Fig. 5d). Ubiquitination assay showed that OsChtBL1 undergoes specific ubiquitination, and the presence of the P2 significantly enhanced the accumulation of ubiquitinated OsChtBL1 (Supplementary Fig. 5e). Together, these results indicate that P2 promotes the degradation of OsChtBL1 through 26S proteasome.
P2 attenuates OsChtBL1-mediated resistance to SBPH and RSV
Arboviruses often manipulate both herbivorous insects and host plants to facilitate their own transmission2. To determine P2’s role in this complex interaction network, we first analyzed whether P2 influences the binding to chitin by the OsChtBL1. IP assays showed that P2 protein did not interfere with the recognition of chitin by OsChtBL1-GFP (Supplementary Fig. 6a). However, further research has indicated that GST-P2 attenuated the stabilizing effect of chitin on the OsChtBL1-MYC protein level (Fig. 5a). Meanwhile, SBPH feeding on OsChtBL1-OE#1/P2-OX plants exhibited significantly increased honeydew production relative to those on OsChtBL1-OE#1 plants, while SBPH on P2-OX plants produced more honeydew than those on NIP plants (Fig. 5b). Consistently, SBPH mortality was significantly reduced on OsChtBL1-OE#1/P2-OX compared to OsChtBL1-OE#1 plants and on P2-OX relative to NIP plants (Fig. 5c). These results suggest that P2 accelerates the degradation of OsChtBL1 via the 26S proteasome, reducing OsChtBL1’s binding to the stylets of SBPH and thereby enhancing SBPH performance.
a The protein degradation assay showing P2 attenuates the protein stability of OsChtBL1 conferred by chitin. Water or 10 μg/ml Chitin and Purified GST or GST-P2 were added respectively to the total protein extract of OsChtBL1-OE plants, treated with 100 μg/mL CHX, and DMSO or MG132, Samples were collected every 30 min at 28 °C and analyzed by western blot using anti-MYC antibody. b The quantity of honeydew collected from NIP, OsChtBL1-OE#1, OsChtBL1-OE#1/P2-OX and P2-OX plants after SBPH feeding for 48 h. n = 19 individual SBPH. c The mortality rate of SBPH fed with NIP, OsChtBL1-OE#1, OsChtBL1-OE#1/P2-OX, and P2-OX plants after 15 days. d Phenotypes of Mock- or RSV-infected NIP, OsChtBL1-OE#1, OsChtBL1-OE#1/P2-OX, and P2-OX plants after RSV inoculation for 30 days. Scale bar, 10 cm. e The incidence rate of NIP, OsChtBL1-OE#1, OsChtBL1-OE#1/P2-OX, and P2-OX plants under field conditions at 30 days. f RT-qPCR assay showing the mRNA relative expression level of RSV CP in RSV-infected NIP, OsChtBL1-OE#1, OsChtBL1-OE#1/P2-OX, and P2-OX plants. g Western blot assay showing protein accumulation level of RSV CP in Mock- or RSV-infected NIP, OsChtBL1-OE#1, OsChtBL1-OE#1/P2-OX, and P2-OX plants. OsUBQ5 serves as an internal reference gene (f). n = 3 (b, c, e, f) independent biological replicates. Error bars represent SD, and values are means ± SD. All the statistical analysis data were performed using a two-tailed Student’s t test. *At the top of columns indicates significant differences with the control group at P < 0.05, and ns at the top of columns indicates no significant differences with the control group at P > 0.05. Rubisco serves as an internal reference protein for western blot analysis and detected by anti-RUB antibody (a, g). Experiments in (a, g) were repeated three times with similar results.
We further performed DAB staining on NIP, OsChtBL1-OE#1, OsChtBL1-OE#1/P2-OX, and P2-OX plants. The ROS level observed was significantly lower in OsChtBL1-OE#1/P2-OX plants versus OsChtBL1-OE#1 plants, and in P2-OX versus NIP plants (Supplementary Fig. 6b, c). When RSV was inoculated onto these plants, OsChtBL1-OE#1/P2-OX plants displayed enhanced disease severity and higher RSV infection rates compared to OsChtBL1-OE#1 plants, with P2-OX plants similarly showing exacerbated symptoms and elevated infection incidence relative to NIP plants (Fig. 5d, e). RT-qPCR and Western blot assays confirmed significantly increased accumulation of RSV CP mRNA and protein in OsChtBL1-OE#1/P2-OX versus OsChtBL1-OE#1 plants, and in P2-OX versus NIP plants (Fig. 5f, g). These results collectively indicate that P2 manipulates the performance of SBPH on rice by degrading OsChtBL1 via the 26S proteasome, facilitating RSV infection and proliferation.
P2 facilitates the degradation of OsChtBL1 mediated by OsRING18
In order to explored how P2 promotes the degradation of OsChtBL1, we utilized the Y2H assays and employed OsChtBL1 as the bait to screen out a RING-type E3 ubiquitin ligase OsRING18, which interacted with OsChtBL1 (Fig. 6a). Co-IP assay further revealed that OsRING18 specifically interacts with OsChtBL1 (Fig. 6b). To determine whether OsRING18 is capable of leading to the degradation of OsChtBL1, we transiently co-expressed OsChtBL1-FLAG, GFP and OsRING18-MYC with different protein levels in plants. Western blot assay demonstrated that with the increase in the protein level of OsRING18-MYC, the protein accumulation of OsChtBL1-FLAG decreased, while the protein level of the internal control protein GFP remained unaffected (Supplementary Fig. 7a). Meanwhile, the transcriptional level of OsChtBL1-FLAG was also not affected by OsRING18-MYC (Supplementary Fig. 7b), and the degradation of OsChtBL1-FLAG mediated by OsRING18-MYC was recovered with the treatment with MG132 (Supplementary Fig. 7c). Protein degradation assay indicated that GST-OsRING18 significantly facilitated the degradation of OsChtBL1-MYC, while MG132 slowed this process (Supplementary Fig. 7d). We further employed the CRISPR-Cas9 technology to establish two OsRING18 knocked out mutant lines (osring18#5 and osring18#8) (Supplementary Fig. 7e). Adding purified GST-OsChtBL1 to the protein extracts of ZH11 and osring18#5 plants. The results revealed that GST-OsChtBL1 is more stable in the osring18#5 plants than in the control ZH11 plants (Supplementary Fig. 7f). These results indicate that OsRING18 mediates the degradation of OsChtBL1 via the 26S proteasome.
a, c Y2H assay showing OsRING18 interacts with OsChtBL1 and P2 in yeast cells. These yeast cells containing different combinations were observed after growing on the SD-LT and SD-AHLT medium at 30 °C for 3 days. b, d Co-IP assay showing OsRING18 interacts with OsChtBL1 and P2 in N. benthamiana. e The protein degradation assay showing P2 synergizes with OsRING18 to promote the degradation of OsChtBL1. f The protein degradation assay showing the degradation of OsChtBL1 mediated by P2 is dependent on OsRING18. g The quantity of honeydew collected from ZH11 and osring18 plants after SBPH feeding for 48 h. n = 17 individual SBPH. h Phenotypes of Mock- or RSV-infected ZH11 and osring18 plants after RSV inoculation for 30 days. Scale bar, 10 cm. i The incidence rate of ZH11 and osring18 plants under field conditions at 30 days. j RT-qPCR assay showing the mRNA relative expression level of RSV CP in RSV-infected ZH11 and osring18 plants. k Western blot assay showing protein accumulation level of RSV CP in Mock- or RSV-infected ZH11 and osring18 plants. OsUBQ5 serves as an internal reference gene (j). n = 3 (g, i, j) independent biological replicates. Error bars represent SD, and values are means ± SD. All the statistical analysis data were performed using a two-tailed Student’s t test. *At the top of columns indicates significant differences with the control group at P < 0.05. Rubisco serves as an internal reference protein for Western blot analysis and detected by anti-RUB antibody (e, f, k). Experiments in (b, d–f, k) were repeated three times with similar results.
Surprisingly, Y2H and Co-IP assays also demonstrated that OsRING18 interacts with P2 (Fig. 6c, d). Competitive Co-IP experiment showed that HIS-P2 did not affect the co-precipitation level of GST-OsRING18 and OsChtBL1-MYC (Supplementary Fig. 7g). To explore whether P2-mediated OsChtBL1 degradation depends on OsRING18, we added the purified HIS-P2 and GST-OsRING18 either together or individually to the protein extract of OsChtBL1-OE plants. Western blot assay revealed that the degradation rate of OsChtBL1-MYC was strikingly elevated when both HIS-P2 and GST-OsRING18 were present compared to either HIS-P2 or GST-OsRING18 alone (Fig. 6e). Furthermore, in OsRING18-knockout mutant plants (osring18#5), P2 failed to significantly promote OsChtBL1 degradation (Fig. 6f). These results indicate that the degradation of OsChtBL1 mediated by P2 depends on OsRING18.
We further explored whether OsRING18 deficiency indirectly enhances rice resistance to SBPH and RSV. The quantity of honeydew produced by SBPH on osring18 plants was lower than on ZH11 plants (Fig. 6g). When RSV-infected SBPH were on ZH11 and osring18 plants, osring18 plants displayed milder disease symptoms and lower incidence rates compared to ZH11 plants (Fig. 6h, i). Western blot assay and RT-qPCR assay indicated that osring18 plants had lower mRNA levels and protein accumulation of RSV CP than ZH11 plants (Fig. 6j, k). Collectively, these results suggest that OsRING18 negatively regulates the rice resistance to SBPH and RSV by degrading OsChtBL1.
Based on the above findings, we propose a model describing OsChtBL1’s role in the interaction among SBPH, RSV, and rice (Fig. 7). When SBPH feed on RSV-infected rice, the RSV-encoded P2 protein hijacks OsRING18 to mediate OsChtBL1 degradation via the 26S proteasome. This process suppresses rice antiviral immunity while reducing OsChtBL1 accumulation in the SBPH food canal, thereby enhancing viral acquisition by improving insect feeding performance. Subsequently, when RSV-acquired SBPH feed on healthy rice, OsChtBL1 is strongly induced and accumulates in the SBPH’s food canal, blocking viral transmission by inhibiting insect performance and simultaneously activating rice antiviral immunity. In conclusion, our work reveals insights into how host plants defend against dual attacks from arboviruses and herbivorous insects, and how arboviruses manipulate the performance of herbivorous insects and host plants to promote infection and transmission.
In RSV-infected rice, the RSV-encoded P2 protein recruits OsRING18 to promote OsChtBL1 degradation via the 26S proteasome, thereby suppressing OsChtBL1-mediated dual resistance against both SBPH and RSV, while enhancing SBPH performance and virus acquisition. In healthy rice, OsChtBL1 accumulates in the SBPH food canal, inhibiting SBPH performance and virus transmission. Concurrently, OsChtBL1 activates rice antiviral immunity.
Discussion
The interaction network among arboviruses, herbivorous insects, and host plants is highly complex and diverse. Arboviruses inhibit the innate immune response of host plants to attract herbivorous insects, which in turn can use the virus to weaken plant defenses, facilitating their feeding and reproduction. In response, host plants have evolved sophisticated defense systems to counter the dual threat of arboviruses and herbivorous insects2,29,37. Previous studies have shown that arboviruses and herbivorous insects coevolve to jointly launch a comprehensive attack on host plants, targeting multiple immune pathways, particularly those regulated by plant hormones7,38. For example, while the attack of piercing-sucking herbivorous insects typically evades jasmonic acid (JA)-mediated defense responses, it triggers an increase in salicylic acid (SA) levels in the host plant37. Our earlier research has demonstrated that many arboviruses employ strategies to suppress JA- and SA-mediated broad-spectrum antiviral immune responses34,39,40,41. These findings suggest that the mechanisms through which arboviruses and herbivorous insects jointly target host plants are highly diverse and complex. Although the broad-spectrum resistance mediated by plant hormones against arboviruses and herbivorous insects is effective, it often revolves around changes in hormone levels that lead to the production of volatiles, altering the attractiveness of host plants to herbivorous insects1,5,7. In our study, we identified a plant lectin protein, OsChtBL1, that accumulates at the stylets of SBPH, creating a feeding barrier (Fig. 1j–l). We also found that P2 protein encoded by RSV that recruits an E3 ubiquitin ligase, OsRING18, to mediate the degradation of OsChtBL1 via the 26S proteasome, thereby enhancing SBPH performance and adaptability on rice (Fig. 4a, b, i, j). These findings provide perspectives on how arboviruses manipulate host plant defense to promote their own transmission through herbivorous vectors.
Protein folding within ER is particularly sensitive to external stress factors, which can activate ER stress responses, allowing eukaryotes to quickly sense and respond to environmental changes31,42. Recent studies have found that ER stress activated by a HVA22 family protein OsHLP1 positively regulates rice resistance to blast disease30,43. In our study, we found that OsChtBL1 activates ER stress and positively regulates rice antiviral immunity (Figs. 2f–l and 3c, d). These results indicate that ER stress-mediated immune responses in rice may have broad-spectrum effects against diverse pathogens. In mammals, ER stress is closely linked to the production of ROS. External environmental stresses can disrupt ER redox homeostasis, leading to electron transfer and Ca2+ signaling, which in turn increases ROS production35,36. In yeast, heat shock responses mitigate ER stress-induced damage by regulating cytoplasmic processes and suppressing ROS generation44. Our findings indicate that OsChtBL1 not only activates ER stress but also likely promotes ROS production in rice (Supplementary Fig. 4a, b). These findings suggest that the link between ER stress and ROS generation is highly conserved across eukaryotes. Numerous studies have indicated that ER stress and ROS production possess broad-spectrum resistance to herbivorous insects and various pathogens45,46. Recent studies suggest that rice disulfide isomerase-like proteins (OsPDILs) regulate ER stress and interact with OsRBOHb (a ROS-generating NADPH oxidase) to modulate ROS bursts under stress47. This indicates that OsPDILs in rice may serve as critical mediators between ER stress and ROS production. Our RT-qPCR data further demonstrate that expression of OsPDIL2-3 is strongly induced in OsChtBL1-OE plants (Fig. 3d), suggesting that OsChtBL1 may indirectly activate ER stress and ROS production via OsPDILs. Together, our findings demonstrate that OsChtBL1 not only directly suppresses herbivorous insect performance by establishing feeding barriers but also enhances rice resistance by activating ER stress and ROS production.
Piercing-sucking herbivorous insects possess highly refined mouthparts, which are integral components of their exoskeleton, allowing them to feed on plant sap by puncturing the phloem or xylem—a process often associated with the transmission of symbiotic viruses48. The mouthparts of these insects are composed mainly of cuticular proteins with chitin-binding domains, lipids, and a cross-linked aromatic matrix rich in chitin28,49. Our study found that OsChtBL1, which contains four chitin-binding domains, specifically binds to chitin in the stylets of SBPH, resulting in feeding impairment (Fig. 1). However, SBPH feeding on uninfected rice plants remains highly efficient in natural environments. We hypothesize that this efficiency may result from evolutionary adaptations in SBPH to counteract plant defense mechanisms developed through long-term co-evolution. When feeding on uninfected plants, SBPH likely secretes effector proteins to suppress lectin-mediated resistance, thereby partially overcoming the toxicity of OsChtBL1 and enabling efficient feeding. In addition, as the global food crisis intensifies due to climate change and population growth, genetically modified crops offer a promising solution50. Given that chitin is uniquely biosynthesized and metabolized by arthropods, nematodes, crustaceans, and fungi but not by mammals16,49. Hence, OsChtBL1, a plant-derived protein that binds to chitin and is toxic to herbivorous insects, represents a potentially safe and effective transgenic tool for addressing future agricultural challenges.
Methods
Insect vectors and plant materials
The SBPH was captured from the rice field in Zhejiang Province, China, and then fed and bred in the laboratory. N. benthamiana plants were used in the transient expression system of Agrobacterium tumefacien (GV3101) infiltration. Rice (Oryza sativa) seeds of cv. Nipponbare (NIP) Zhonghua 11 (ZH11) were used as the wild-type. Transgenic rice lines OsChtBL1-OE (OsChtBL1 fusing MYC tag) and P2-OX (P2 fusing FLAG tag), and CRISPR-Cas9 mutants oschtbl1 were constructed using NIP as the background. CRISPR-Cas9 mutants osring18 were constructed using ZH11 as the background. SBPH and all rice plants were maintained at 28–30 °C under a light/dark photoperiod of 14/10 h. N. benthamiana plants were maintained at 25 °C under a light/dark photoperiod of 16/8 h.
Insect bioassays
For honeydew measurement, the parafilm (Cat# PM-996, Amcor, USA) was fixed on the stem of the rice plants with different treatments, and the fifth-instar SBPH was placed into the sealed space bound by parafilm. After 48 h of feeding, an electronic balance (1 mg, METTLER TOLEDO, Switzerland) was used to weigh the parafilm before and after feeding to calculate the amount of honeydew produced by SBPH. For the mortality rate of SBPH measurement, 50 third-instar SBPH were placed into a beaker containing 10 growing rice plants, and sealed the outlet with gauze netting. After 15 days of feeding, the mortality rate of SBPH was counted, and at least three biological replicates were performed for each treatment.
Western blot assay
The leaves of rice and N. benthamiana were thoroughly ground in liquid nitrogen and then fully lysed in SDS lysis buffer (1 M Tris-HCl pH 6.8, 10% SDS and water) or IP buffer (40 mM Tris-HCl pH 7.5, 100 mM NaCl, 2% glycerol, 1% TritonX-100, 1 mM DDT, 1 mM MgCl2, 1 μM ZnCl2, 50 μM MG132, 1× Protease inhibitor cocktail and water) for 10 min. The mixture was centrifuged at 12,000 × g for 10 min, and the supernatant was aspirated. Then, 5× SDS loading buffer (250 mM Tris-HCl pH 6.8, 50% glycerol, 10% SDS, 0.5% bromophenol blue, 2% β-mercaptoethanol, and water) was added to the supernatant and boiled in water for 5 min. Finally, the boiled supernatant was separated by SDS-PAGE gel and transferred to a PVDF membrane for detection using the corresponding antibodies. Anti-MYC/FLAG/GFP/GST/HIS antibody (1:5000 dilution, TransGen Biotech, China), Anti-RSV-CP antibody34, Anti-RSV-P2 antibody34, Goat anti-mouse/rabbit antibody (1:10,000 dilution, TransGen Biotech, China).
Chitin-binding assay
The full length of OsChtBL1 and FLAG (negative control) were cloned into the pCAMBIA1300-GFP vector. The leaves of N. benthamiana transiently expressing OsChtBL1-GFP or GFP-FLAG for 48 h, and OsChtBL1-OE plants were ground thoroughly in liquid nitrogen. In total, 0.5 g of ground tissue was fully lysed with IP buffer, and the supernatant was collected and incubated with chitin magnetic beads (Cat# E8036S, NEW England Biolabs, USA) at 4 °C for 4 h. Then, the chitin magnetic beads were adsorbed using a magnetic grate and washed three times with 1× PBS for 10 min each time. After adding 1× SDS loading buffer to the washed chitin magnetic beads and boiled in water for 5 min, the supernatant was aspirated and used for western blot analysis.
Immunofluorescence assay (IF)
SBPH was fed with 4% sucrose and purified GST or GST-OsChtBL1 through parafilm for 36 h, and the stylets of dead SBPH were dissected out and fixed in 4% paraformaldehyde at room temperature for 2 h. The fixed stylets were washed three times with 1× PBS, incubated with anti-GST antibody (1:50 dilution) at 37 °C for 2 h, washed three times with 1× PBS. These stylets were labeled with goat anti-mouse fluorescent antibody conjugated with green fluorescence (1:100 dilution, Cat# F0257, Sigma, USA) at 37 °C for 1 h, washed three times with 1× PBS, and observed using a fluorescence microscope (Leica TCS SP8 X, Germany).
Transmission electron microscopy assay
The stylets of SBPH were dissected in the same way as in the IF assay. According to the previously described methods, these stylets were fixed, dehydrated, embedded, and observed using TEM (Hitachi HT-7800, Japan) after ultrathin sectioning51,52. For IEM, these stylets of SBPH were labeled with anti-GST antibody (1:200 dilution) as the primary antibody and goat anti-mouse antibody conjugated with 10 nm gold particles (1:200 dilution, Sigma, USA) as the secondary antibody, and observed using TEM (Hitachi HT-7800, Japan) after ultrathin sectioning.
RNA extraction and quantitative RT-qPCR analysis
SBPH, the leaves of rice plants, and N. benthamiana were placed in 2 ml RNA-free EP tubes and thoroughly ground in liquid nitrogen using a high-speed grinder. TRIzol reagent and chloroform were added to the 2-ml EP tubes, centrifuged at 12,800 × g, washed two times with 75% RNA-free ethanol, and DEPC water was added for the extraction of total RNA. According to the manufacturer’s instructions, Evo M-MLV RT Mix Kit with gDNA Clean (Cat# AG11728, Accurate Biology) is used for total RNA reverse transcription and SYBR Green Premi × Pro Tag HS qPCR Kit (Cat# AG11718, Accurate Biology) is used for RT-qPCR reactions, which were performed by ABI QuantStudio5 (Thermo Fisher Scientific). The entire process was carried out on ice. At least two technical replicates and three biological replicates were performed, and the 2-ΔΔCt method was used for data processing. LsGADPH, OsUBQ5, and NbACTIN were respectively used as reference genes for SBPH, rice, and N. benthamiana, and all the primers used in this study are listed in Supplementary Table 1.
RNA sequencing and analysis
The seeds of NIP and OsChtBL1-OE plants (with three biological replicates, 30 seeds for each replicate) were grown in hydroponic solution for 15 days. These plant samples were collected and sent to LC-Bio Technologies Company (Hangzhou, China) for RNA sequencing and analysis, as described in our previous work53,54.
Yeast two-hybrid assay (Y2H)
The full length of OsChtBL1 and OsRING18 were cloned into the pGADT7 vector containing the activation domain. The full length of viral protein (P2, P3, P4, Pc2, Pc3, and Pc4) and OsChtBL1 were cloned into the pGBDT7 vector containing a DNA-binding domain. According to the manufacturer’s instructions, different plasmid combinations were co-transformed into yeast cells (AH109, Cat# YC1010, WEI DI, China). The transformed yeast cells grew on SD-LT lacking leucine and tryptophan (Cat# PM2220, COOLABER, China) and SD-AHLT lacking alanine, histidine, leucine, and tryptophan (Cat# PM2110, COOLABER, China) medium at 30 °C for 3 d, then were photographed and recorded.
Luciferase complementation imaging assay (LCI)
The full-length of OsChtBL1 and GUS (negative control) were cloned into the pCAMBIA1300-nLuc vector, and the full length of P2 and GUS (negative control) were cloned into the pCAMBIA1300-cLuc vector. These plasmids were transformed into Agrobacterium tumefacien (GV3101) and co-expressed in N. benthamiana with different combination. After 48 hours of transient expression, 0.1 mM luciferase substrate was injected into the same position of N. benthamiana in the dark for 10 min, and observed by using In-Vivo Imaging System (Lumazone SOPHIA 2048B, Singapore).
Bimolecular fluorescence complementation assay (BiFC)
The full length of OsChtBL1 and GUS (negative control) were cloned into the pCAMBIA1300-nYFP (N terminus of YFP) vector, and the full length of OsChtBL1, P2 and GUS (negative control) were cloned into the pCAMBIA1300-cYFP (C terminus of YFP) vector. These plasmids were transformed into Agrobacterium tumefacien (GV3101) and co-expressed in N. benthamiana with different combinations for 48 h, and observed by using a fluorescence microscope (Nikon A1R, Japan).
Subcellular localization assay
The effect of P2 on OsChtBL1 localization, OsChtBL1-GFP with P2-MYC or without P2-MYC were co-expressed in N. benthamiana for 48 h, and observed by using a fluorescence microscope (Nikon A1R, Japan). AtWAK2 was used as a maker protein of the endoplasmic reticulum55.
Co-immunoprecipitation assay (Co-IP)
For N. benthamiana, the full length of OsChtBL1 and P2 was cloned into the pCAMBIA1300-FLAG vector, and the full length of P2 and OsRING18 was cloned into the pCAMBIA1300-MYC vector. These plasmids were transformed into Agrobacterium tumefacien (GV3101) and co-expressed in N. benthamiana with different combination. After 48 hours of transient expression, the leaf samples were collected and ground thoroughly in liquid nitrogen. The ground tissue (0.2 g) was fully lysed with IP buffer. The total protein extracts with FLAG magnetic beads (Cat# L00835, GenScript, USA) or MYC magnetic beads (Cat# AB-2631370, Proteintech, USA) at 4 °C for 2 h, washed three times with 1× PBS, added 1× SDS loading buffer, and boiled in water for 5 min, the supernatant was aspirated and used for western blot analysis. For rice, ground tissue (0.2 g) from Mock- or RSV-infected NIP, OsChtBL1-OE#1, OsChtBL1-OE#1/P2-OX, P2-OX plants were fully lysed with IP buffer, then the total protein extract was treated in the same way as for N. benthamiana.
Protein degradation assay
For N. benthamiana, OsChtBL1-FLAG and GFP (negative control) were transiently co-expressed with different protein-abundance P2-MYC or OsRING18-MYC in N. benthamiana for 48 h. Equal-weight N. benthamiana leaves (3–4 pieces) were collected using a punch (Diameter 5 mm) and fully lysed with SDS lysis buffer, centrifuged at 12,000× g for 10 min, extracted the supernatant, added 1× SDS loading buffer, boiled in water for 5 min, and analyzed by western blot using anti-FLAG antibody. MG132 was injected into the same position of N. benthamiana leaves 8–10 h before sampling for analysis. For rice, ground tissue (0.2 g) from OsChtBL1-OE, ZH11, or osring18 plants were fully lysed with IP buffer without MG132, centrifuged at 12,000 × g for 10 min with centrifuge at 4 °C and aspirated the supernatant. Selectively added MG132 and purified protein to the obtained supernatant, incubated on a rotating shaker at 28 °C, and collected samples every 30 min. The collected samples were added to 5× SDS loading buffer, boiled in water for 5 min, and analyzed by western blot using corresponding antibodies.
Ubiquitination assay
OsChtBL1-FLAG or GFP-FLAG was expressed in N. benthamiana leaves for 48 h. Leaf samples were collected and ground thoroughly in liquid nitrogen. 2 g of ground tissue was lysed in IP buffer (40 mM Tris-HCl pH 7.5, 100 mM NaCl, 2% glycerol, 1% TritonX-100, 1 mM DDT, 1 mM MgCl2, 1 μM ZnCl2, 1× Protease inhibitor cocktail, 50 μM MG132, and water) at 4 °C for 20 min to extract active total proteins. Purified GST or GST-P2 was added to the protein extracts and co-incubated with FLAG magnetic beads at 4 °C for 4 h. Beads were washed three times with 1× PBS buffer, then boiled for 5 min in SDS loading buffer. Proteins were resolved by SDS-PAGE, and ubiquitination of OsChtBL1 was detected using anti-Ub antibody (1:1000 dilution, Cat# B1422, Santa Cruz Biotechnology, USA).
Statistical analysis
All statistical significance analyses were performed using a two-tailed Student’s t test. Each experiment included at least three biological replicates. Error bars represent SD, and values are means ± SD. *At the top of columns indicates significant differences with the control group at P < 0.05 and ns at the top of columns indicates no significant differences with the control group at P > 0.05. All statistical analyses were conducted using software Microsoft Office Excel 2007 software. For immunoblot quantification and LCI assay, band intensities and fluorescent signal were measured with ImageJ software.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All the RNA-seq data generated in this study have been deposited in the Genome Sequence Archive (GSA) public repository under accession number [CRA019755]. The data are fully accessible via the following link: [https://bigd.big.ac.cn/gsa/browse/CRA019755]. Source data are provided with this paper.
References
Eigenbrode, S. D., Bosque-Pérez, N. A. & Davis, T. S. Insect-borne plant pathogens and their vectors: ecology, evolution, and complex interactions. Annu. Rev. Entomol. 63, 169–191 (2018).
Ye, J., Zhang, L., Zhang, X., Wu, X. & Fang, R. Plant defense networks against insect-borne pathogens. Trends Plant Sci. 26, 272–287 (2021).
Turlings, T. C. J. & Erb, M. Tritrophic interactions mediated by herbivore-induced plant volatiles: mechanisms, ecological relevance, and application potential. Annu. Rev. Entomol. 63, 433–452 (2018).
Xia, J. et al. Whitefly hijacks a plant detoxification gene that neutralizes plant toxins. Cell 184, 1693–1705.e1617 (2021).
Joo, Y. et al. Herbivory elicits changes in green leaf volatile production via jasmonate signaling and the circadian clock. Plant Cell Environ. 42, 972–982 (2018).
Wu, D. et al. Viral effector protein manipulates host hormone signaling to attract insect vectors. Cell Res. 27, 402–415 (2017).
Gong, Q. et al. Molecular basis of methyl-salicylate-mediated plant airborne defence. Nature 622, 139–148 (2023).
Van Damme, E. J. M. 35 years in plant lectin research: a journey from basic science to applications in agriculture and medicine. Glycoconj. J. 39, 83–97 (2021).
Tsaneva, M. & Van Damme, E. J. M. 130 years of plant lectin research. Glycoconj. J. 37, 533–551 (2020).
Petrova, N. V., Aglyamova, A. R., Mokshina, N. E. & Gorshkova, T. A. Current state of plant lectinology. Russian J. Plant Physiol. 71, 50 (2024).
De Coninck, T. & Van Damme, E. J. M. Review: the multiple roles of plant lectins. Plant Sci. 313, 111096 (2021).
Walski, T., De Schutter, K., Cappelle, K., Van Damme, E. J. M. & Smagghe, G. Distribution of glycan motifs at the surface of midgut cells in the cotton leafworm (Spodoptera littoralis) demonstrated by lectin binding. Front. Physiol. 8, 1020 (2017).
Vandenborre, G., Smagghe, G. & Van Damme, E. J. M. Plant lectins as defense proteins against phytophagous insects. Phytochemistry 72, 1538–1550 (2011).
Desaki, Y., Miyata, K., Suzuki, M., Shibuya, N. & Kaku, H. Plant immunity and symbiosis signaling mediated by LysM receptors. Innate Immun. 24, 92–100 (2017).
De Coninck, T. & Van Damme, E. J. M. Plant lectins: handymen at the cell surface. Cell Surf. 8, 100091 (2022).
Muthukrishnan, S., Mun, S., Noh, Y. M., Geisbrecht, R. E. & Arakane, Y. Insect cuticular chitin contributes to form and function. Curr. Pharm. Des. 26, 3530–3545 (2020).
Itakura, Y., Nakamura-Tsuruta, S., Kominami, J., Tateno, H. & Hirabayashi, J. Sugar-binding profiles of chitin-binding lectins from the hevein family: a comprehensive study. Int. J. Mol. Sci. 18, 1160 (2017).
Asensio, J. L. et al. Structural basis for chitin recognition by defense proteins: GlcNAc residues are bound in a multivalent fashion by extended binding sites in hevein domains. Chem. Biol. 7, 529–543 (2000).
Cho, W. K., Lian, S., Kim, S. M., Park, S. H. & Kim, K. H. Current insights into research on rice stripe virus. Plant Pathol. J. 29, 223–233 (2013).
Zhu, J. et al. Genome sequence of the small brown planthopper, Laodelphax striatellus. GigaScience 6, 1–12 (2017).
Yang, Z. H. et al. Insights into the effect of rice stripe virus P2 on rice defense by comparative proteomic analysis. Front. Microbiol. 13, 897589 (2022).
Ma, Y. et al. Membrane association of importin α facilitates viral entry into salivary gland cells of vector insects. Proc. Natl. Acad. Sci. USA 118, e2103393118 (2021).
Liu, Q. et al. Insect-transmitted plant virus balances its vertical transmission through regulating Rab1-mediated receptor localization. Cell Rep. 43, 114571 (2024).
Xu, Y., Fu, S., Tao, X. & Zhou, X. Rice stripe virus: exploring molecular weapons in the arsenal of a negative-sense RNA virus. Annu. Rev. Phytopathol. 59, 351–371 (2021).
Li, L., Chen, J. & Sun, Z. Exploring the shared pathogenic strategies of independently evolved effectors across distinct plant viruses. Trends Microbiol. 32, 1021–1033 (2024).
Jing, T. et al. Herbivore-induced DMNT catalyzed by CYP82D47 plays an important role in the induction of JA-dependent herbivore resistance of neighboring tea plants. Plant Cell Environ. 44, 1178–1191 (2020).
Li, L. et al. A class of independently evolved transcriptional repressors in plant RNA viruses facilitates viral infection and vector feeding. Proc. Natl. Acad. Sci. USA 118, e2016673118 (2021).
Guschinskaya, N. et al. Insect mouthpart transcriptome unveils extension of cuticular protein repertoire and complex organization. iScience 23, 100828 (2020).
Wei, T. & Li, Y. Rice reoviruses in insect vectors. Annu. Rev. Phytopathol. 54, 99–120 (2016).
Meng, F. et al. A rice protein modulates endoplasmic reticulum homeostasis and coordinates with a transcription factor to initiate blast disease resistance. Cell Rep. 39, 110941 (2022).
Howell, S. H. Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 64, 477–499 (2013).
Wakasa, Y., Hayashi, S. & Takaiwa, F. Expression of OsBiP4 and OsBiP5 is highly correlated with the endoplasmic reticulum stress response in rice. Planta 236, 1519–1527 (2012).
Hayashi, S., Wakasa, Y., Takahashi, H., Kawakatsu, T. & Takaiwa, F. Signal transduction by IRE1-mediated splicing of bZIP50 and other stress sensors in the endoplasmic reticulum stress response of rice. Plant J. 69, 946–956 (2011).
Zhang, H. et al. Different viral effectors suppress hormone-mediated antiviral immunity of rice coordinated by OsNPR1. Nat. Commun. 14, 3011 (2023).
Zeeshan, H., Lee, G., Kim, H.-R. & Chae, H.-J. Endoplasmic reticulum stress and associated ROS. Int. J. Mol. Sci. 17, 327 (2016).
Bassot, A. et al. The endoplasmic reticulum kinase PERK interacts with the oxidoreductase ERO1 to metabolically adapt mitochondria. Cell Rep. 42, 111899 (2023).
Cao, H.-H., Liu, H.-R., Zhang, Z.-F. & Liu, T.-X. The green peach aphid Myzus persicae perform better on pre-infested Chinese cabbage Brassica pekinensis by enhancing host plant nutritional quality. Sci. Rep. 6, 21954 (2016).
Zhao, Y. et al. A viral protein orchestrates rice ethylene signaling to coordinate viral infection and insect vector-mediated transmission. Mol. Plant 15, 689–705 (2022).
He, Y. et al. The OsGSK2 kinase integrates brassinosteroid and jasmonic acid signaling by interacting with OsJAZ4. Plant Cell 32, 2806–2822 (2020).
Tan, X. X. et al. NF-YA transcription factors suppress jasmonic acid-mediated antiviral defense and facilitate viral infection in rice. PLoS Pathog. 18, e1010548 (2022).
Li, L. et al. Independently evolved viral effectors convergently suppress DELLA protein SLR1-mediated broad-spectrum antiviral immunity in rice. Nat. Commun. 13, 6920 (2022).
Celik, C., Lee, S. Y. T., Yap, W. S. & Thibault, G. Endoplasmic reticulum stress and lipids in health and diseases. Prog. Lipid Res. 89, 101198 (2023).
Liang, Y., Meng, F., Zhao, X., He, X. & Liu, J. OsHLP1 is an endoplasmic-reticulum-phagy receptor in rice plants. Cell Rep. 42, 113480 (2023).
Hou, J. et al. Management of the endoplasmic reticulum stress by activation of the heat shock response in yeast. FEMS Yeast Res. 14, 481–494 (2014).
Mittler, R., Zandalinas, S. I., Fichman, Y. & Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 23, 663–679 (2022).
Ribeiro, B. et al. Interference between ER stress-related bZIP-type and jasmonate-inducible bHLH-type transcription factors in the regulation of triterpene saponin biosynthesis in Medicago truncatula. Front. Plant Sci. 13, 903793 (2022).
Zhao, Q. et al. OsPDIL1-1 controls ROS generation by modulating NADPH oxidase in developing anthers to alter the susceptibility of floret fertility to heat for rice. Environ. Exp. Bot. 205, 105103 (2023).
Zhai, Z.-C., Wang, J.-J., Dietrich, C. H. & Huang, M. SEM study of the mouthparts of Nacolus tuberculatus (Walker) (Hemiptera: Cicadellidae) with comparative notes on other Hemiptera. Zoomorphology 142, 35–49 (2022).
Zhu, K. Y., Merzendorfer, H., Zhang, W., Zhang, J. & Muthukrishnan, S. Biosynthesis, turnover, and functions of chitin in insects. Annu. Rev. Entomol. 61, 177–196 (2016).
Nicolia, A., Manzo, A., Veronesi, F. & Rosellini, D. An overview of the last 10 years of genetically engineered crop safety research. Crit. Rev. Biotechnol. 34, 77–88 (2013).
Wu, W., Yi, G., Lv, X., Mao, Q. & Wei, T. A leafhopper saliva protein mediates horizontal transmission of viral pathogens from insect vectors into rice phloem. Commun. Biol. 5, 204 (2022).
Mao, Q. et al. Filamentous structures induced by a phytoreovirus mediate viral release from salivary glands in its insect vector. J. Virol. 91, e00265–17 (2017).
Yang, Z. et al. Histone deacetylase OsHDA706 orchestrates rice broad-spectrum antiviral immunity and is impeded by a viral effector. Cell Rep. 43, 113838 (2024).
Wei, Z. et al. Transcriptional profiling reveals a critical role of GmFT2a in soybean staygreen syndrome caused by the pest Riptortus pedestris. N. Phytol. 237, 1876–1890 (2023).
Dangol, S., Singh, R., Chen, Y. & Jwa, N.-S. Visualization of multicolored in vivo organelle markers for co-localization studies in Oryza sativa. Molecules Cells 40, 828–836 (2017).
Acknowledgements
We thank Prof. Mike Adams for critically reading and improving the manuscript. The TEM data were collected at the Bioimaging Center, State Key Laboratory of Agricultural Products Safety, Ningbo University, and we are deeply grateful to Mao Qianzhuo, Lei Jianing, and Ma Yige for their substantial assistance in this work. This work was funded by the National Key Research and Development Program of China (2021YFD1400500), National Natural Science Foundation of China (U23A6006, 32272555, 32470148), Zhejiang Provincial Natural Science Foundation (LZ22C140001), and Ningbo Major Research and Development Plan Project (2023Z124).
Author information
Authors and Affiliations
Contributions
Z.Y. and Z.S. conceived and designed the experiments. Z.Y., W.W., and Z.X. performed the experiments; Z.Y. and Z.S. analyzed the data with the help of Y.L., H.Z., L.L., and B.S.; Z.Y. and Z.S. wrote the manuscript; Z.Y., J.C., and Z.S. revised the manuscript. All authors have discussed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Yunjing Wang, Murad Ghanim and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, Z., Wu, W., Xu, Z. et al. Arbovirus suppression of a lectin protein-mediated broad-spectrum resistance enhances herbivorous vector performance and viral transmission. Nat Commun 16, 6873 (2025). https://doi.org/10.1038/s41467-025-62233-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-62233-8