Abstract
Drug-target interaction (DTI) prediction is a crucial component of drug discovery. Recent deep learning methods show great potential in this field but also encounter substantial challenges. These include generating reliable confidence estimates for predictions, enhancing robustness when handling novel, unseen DTIs, and mitigating the tendency toward overconfident and incorrect predictions. To solve these problems, we propose EviDTI, a novel approach utilizing evidential deep learning (EDL) for uncertainty quantification in neural network-based DTI prediction. EviDTI integrates multiple data dimensions, including drug 2D topological graphs and 3D spatial structures, and target sequence features. Through EDL, EviDTI provides uncertainty estimates for its predictions. Experimental results on three benchmark datasets demonstrate the competitiveness of EviDTI against 11 baseline models. In addition, our study shows that EviDTI can calibrate prediction errors. More importantly, well-calibrated uncertainty information enhances the efficiency of drug discovery by prioritizing DTIs with higher confident predictions for experimental validation. In a case study focused on tyrosine kinase modulators, uncertainty-guided predictions identify novel potential modulators targeting tyrosine kinase FAK and FLT3. These results underscore the potential of evidential deep learning as a robust tool for uncertainty quantification in DTI prediction and its broader implications for accelerating drug discovery.
Similar content being viewed by others
Introduction
Drug discovery is the process of discovering drugs that can treat diseases and improve human health. It involves multiple steps1,2, including target identification, compound screening, and lead optimization. Identifying drug-target interactions (DTI) plays a crucial role in compound screening3,4. Although traditional biomedical measurements obtained from in vitro experiments are reliable, they come with significant drawbacks, such as high costs and lengthy development cycles, which significantly limit the pace of drug development5.
In silico methods for predicting DTIs, particularly deep learning (DL) techniques, have received much attention for their potential to reduce drug development costs, shorten time and increase the success rate of new drugs6,7,8,9. These methods can be broadly classified into network-based methods and proteochemometrics (PCM)10. Network-based methods integrate drug-target, drug-drug, and protein-protein interactions, etc. multiple networks into a unified network10,11,12.
Recently, PCM methods have received increasing attention. PCM employs representations of drug and protein information to improve the accuracy of DTI predictions, and its performance largely dependent on the effectiveness of molecular and protein representations13,14. Typically, amino acid sequences of protein are typically used as input for proteins, while molecular graph and SMILES strings are commonly used as input for drugs. To obtain representation of proteins and drugs, convolutional neural networks (CNNs)15, recurrent neural networks (RNNs), graph neural networks (GNNs)16,17,18 and transformer models19,20 are typically applied. A great deal of innovative work has been undertaken to predict interactions more effectively14.To improve model interpretability and capture local interactions of drug-target, the gated cross-attention mechanisms have received more attention21,22,23,24. To address the problem of small dataset sizes and incomplete representations in DTI prediction, pre-training models are emerging as a promising solution25,26,27. These models demonstrate excellent scalability and generalization capabilities across a wide range of prediction tasks28,29,30,31,32. To achieve more comprehensive and nuanced representations, many methods integrate multimodal techniques that combine different types of data33,34.
Despite the significant advances achieved by DL in DTI prediction, its practical application still faces a major challenge: high probability predictions do not necessarily correspond to high confidence35,36. The root of this problem lies in the fundamental difference between DL models and human cognitive models37. Humans are able to dynamically adjust the confidence level according to the knowledge boundary, giving certain answers to familiar questions and explicitly expressing “uncertainty” about unknown domains. In contrast, traditional DL models generate predictions for all inputs, including out-of-distribution and noisy samples. More critically, traditional DL models lack probability calibration ability and may produce high prediction probabilities even in low confidence situations. This phenomenon of “overconfidence” has the tendency to introduce unreliable predictions into downstream processes, including the pushing of false positives into experimental validation, the omission of potentially active compounds in virtual screening, and even the designing of clinical trial protocols based on false predictions. These situations not only lead to inefficient use of resources, but also have the potential to delay the drug discovery process.
Uncertainty quantification (UQ) methods can address above challenge and thus improve the robustness of neural models in scientific applications38,39,40,41,42,43,44, especially in the field of drug discovery. The core value of UQ is to provide a reliable basis for decision-making by distinguishing between plausible predictions and high-risk predictions. Typically UQ45,46 methods include Bayesian neural networks47 and sampling-based48,49 methods. However, these methods typically rely on multiple random sampling to approximate the underlying uncertainty function50,51,52, resulting in high computational costs and extended runtimes. This poses a significant limitation for large-scale DTI prediction. Evidential deep learning (EDL) offers a promising alternative that provides a direct way to learn about uncertainty without relying on random sampling53,54. Furthermore, EDL can be integrated into existing network structures without major architectural modifications. Several approaches have demonstrated the potential of EDL in the field of drug discovery and development43,55,56,57.
In this work, we propose an EDL-based DTI prediction framework (EviDTI). The framework utilizes pre-trained knowledge as well as multi-dimensional representations to enhance the model performance. More importantly, EviDTI provides prediction confidence estimation by introducing EDL to help identify drug candidates that are most likely to be successful, thus reducing the risk and cost associated with false positives. The framework comprises three main components: a protein feature encoder, a drug feature encoder, and an evidence layer. In protein feature encoder, the protein language pre-trained model ProtTrans30 is employed to extract protein sequence features. The light attention mechanism is then employed to provide insights into local interactions at the residue level. For drug features, 2D graph representations are obtained using our previously proposed pre-trained model, MG-BERT58. Additionally, 3D features of drugs are encoded via geometric deep learning. The learned protein and drug representations are concatenated and fed into the evidential layer to obtain DTI probabilities and uncertainties. In a comprehensive evaluation across 11 DTI prediction models, EviDTI demonstrates competitive performance. Moreover, we demonstrate that evidential-based uncertainty can effectively calibrate prediction errors. This uncertainty information can accelerate drug discovery and repositioning by prioritizing DTIs with more confident predictions for experimental validation. By applying uncertainty-guided prediction in the discovery of potential tyrosine kinase modulators, EviDTI identifies potential new modulators targeting tyrosine kinase FAK and FLT3, highlighting the utility of EviDTI in drug discovery. By bridging the gap between prediction and reliability, EviDTI offers a trustworthy solution for DTI prediction. The source code is available at https://github.com/zhaoyanpeng208/EviDTI.
Results
Overview
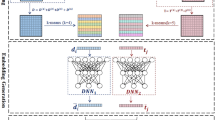
We developed an EDL-based DTI prediction model, EviDTI. As shown in Fig. 1, the EviDTI framework consists of three main components: protein feature encoder, drug feature encoder, and evidential layer. Given a drug-target pair as input, the protein feature encoder utilizes the protein sequence pre-training model ProtTrans as the initial encoder to generate an initial target representation. This representation undergoes further feature extraction through the light attention (LA) module. In the drug feature encoder, both 2D topological information and 3D structural information of the drug are encoded. For the drug 2D topological graph, an initial representation is derived using our previously proposed molecule pre-training model MG-BERT36, which is subsequently processed by a 1DCNN. The 3D spatial structure of the drug is converted into an atom-bond graph and a bond-angle graph, with the representation obtained through the GeoGNN module. The target and drug representation are then concatenated and fed into the evidential layer. The output of the evidential layer is the parameter α, which is used to calculate the prediction probability and the corresponding uncertainty value.
For a given drug-target pair, the protein feature encoder employs the pre-trained ProtTrans for initial target representation, further refined by a light attention (LA) module. The drug feature encoder processes the 2D topology and 3D structure representations. The 2D representation is derived from the pre-trained MG-BERT model and processed by 1D CNN. The 3D structure representation is obtained via the GeoGNN. These representations are concatenated and fed into the evidence layer, which outputs parameter α for prediction probability and uncertainty.
Performance comparison
To evaluate the effectiveness of the EviDTI framework, we validated it on three different experimental datasets: DrugBank, Davis59, and KIBA60. These datasets were randomly divided into training, validation and test sets in a ratio of 8:1:1. We used seven evaluation metrics: accuracy (ACC), recall, precision, Matthews correlation coefficient (MCC), F1 score, area under the ROC curve (AUC), and area under the precision-recall curve (AUPR) to assess model performance. EviDTI is compared with three traditional machine learning methods, including Random Forests (RFs)61, Support Vector Machines (SVMs)62 and Naive Bayesian (NB)63. These methods have been widely used in DTI prediction and are important benchmarks for evaluating the performance of our proposed method. In addition, EviDTI is compared with eight state-of-the-art models in the field, including DeepConv-DTI19, GraphDTA64, MolTrans20, HyperAttention17, TransformerCPI65, GraphormerDTI24, AIGO-DTI66, and DLM-DTI27.
The performance results on the DrugBank dataset are shown in Table 1. EviDTI shows robust overall performance on all metrics, especially in terms of precision (81.90%), and competitive values for Accuracy (82.02%), MCC (64.29%) and F1 score (82.09%). Besides, we evaluated EviDTI on the Davis and KIBA datasets, which are particularly challenging due to class imbalance. The performance results on the Davis and KIBA datasets are presented in Tables 2 and 3, respectively. Specifically, on the KIBA dataset, EviDTI outperformed the best baseline model by 0.6% in accuracy, 0.4% in precision, 0.3% in MCC, 0.4% in F1 score, and 0.1% in AUC. On the Davis dataset, EviDTI exceeds 0.8% in accuracy, 0.6% in precision, 0.9% in MCC, 2% in F1 score, 0.1% in AUC, and 0.3% in AUPR. These results demonstrate the robustness and superior performance of EviDTI when dealing with complex and unbalanced datasets. Overall, EviDTI achieved robust and superior performance on three benchmark datasets, which further validates the effectiveness and competitiveness of EviDTI.
In order to evaluate the effectiveness of the EviDTI in predicting novel DTIs, we introduced a cold-start scenario following the practice established by Wang et al12. Supplementary Table 2 demonstrates the performance of EviDTI under the cold-start scenario. EviDTI outperforms other models in several evaluation metrics, especially in accuracy (79.96%), recall (81.20%), F1 score (79.61%) and MCC value (59.97%). Its AUC value (86.69%) is slightly lower than TransformerCPI’s 86.93%. These results demonstrate that EviDTI is a competitive model in cold start scenarios.
Multidimensional feature fusion and pre-trained models lead to better prediction performance
Two ablation studies were conducted to explored the impact of different combinations of dimensional features and the use of pre-trained model features on DTI prediction.
Firstly, we compare the performance of models using single-dimensional features with models using a multidimensional feature fusion strategy. Supplementary Figs. 1 and 2 illustrate the various feature combination architectures used. Specifically, three multidimensional feature combinations were evaluated on three datasets:
EviDTI: combines the 2D topological representation and the 3D structural representation of small molecules of small molecules, and protein sequence representation.
EviDTI w/o drug 3D: utilizes 2D topological representation of small molecules and the protein sequence representation.
EviDTI w/o drug 2D: utilizes 3D structural representation of small molecules and the protein sequence representation.
As shown in Fig. 2a and Supplementary Table 3, the EviDTI model consistently outperformed the other two models across all three datasets in most evaluation metrics. This result indicates that multidimensional feature fusion significantly enhances DTI prediction performance.
a Performance comparison on the DrugBank, KIBA and Davis datasets using single-dimensional features and multidimensional feature fusion strategies. Five independent replications of each method were performed (n = 5). Data are expressed as means ± std. b Performance comparison of feature extraction with and without pre-trained models on DrugBank, KIBA, and Davis datasets. Five independent replications of each method were performed (n = 5). Data are expressed as means ± std. Source data are provided as a Source Data file.
To explore the benefits of using pre-trained models as initial feature extractors, we compared the performance of architectures with and without pre-trained models. Supplementary Figs. 3 and 4 depict the different ablation architectures examined:
EviDTI-Protein Integer: replaces the protein pre-training model ProtTrans with integer coding and a CNN for feature extraction.
EviDTI-Drug 2D GCN: replaces the small molecule pre-training model MG-BERT with a two-layer GCN.
Figure 2b and Supplementary Table 4 illustrate the performance of these different model frameworks across the three datasets. It is evident that the architectures utilizing pre-trained models for initial feature extraction outperformed those that did not across all metrics. This finding underscores the value of leveraging pre-trained models to enhance DTI prediction performance.
Evidential deep learning provides a reliable measure of uncertainty
Following the evaluation of EviDTI’s prediction performance on the benchmark dataset, it becomes critical to ensure that the model can provide reliable estimates of uncertainty in the DTI prediction task. To assess whether the model can effectively assess prediction uncertainty, two hypotheses are proposed and tested:
-
1.
Samples with correct predictions should have low uncertainty, while samples with incorrect predictions should have high uncertainty.
-
2.
Samples with lower uncertainty should exhibit higher prediction accuracy.
These assumptions are based on the EDL principle, which views learning as a process of acquiring evidence. The more evidence accumulated, the greater the confidence and the higher the probability of prediction.
First, the correlation between uncertainty and prediction results was assessed on the three benchmark datasets. Figure 3a illustrates the relationship between the sample prediction results and the uncertainty values. The horizontal axis divides the sample into true positives (TP, true value 1, predicted value 1), false positives (FP, true value 0, predicted value 1), false negatives (FN, true value 1, predicted value 0) and true negatives (TN, true value 0, predicted value 0). The vertical axis represents the distribution of uncertainty values for each group.
a A Mann–Whitney test was performed on the error distribution of uncertainty in samples classified as TP, FP, FN, TN for three datasets: DrugBank (n = 3312 observations), KIBA (n = 11,639 observations), and Davis (n = 2,583 observations). The central line indicates the median, the box bounds indicate the 25th and 75th percentiles, whiskers extend to the minimum and maximum values (within 1.5× interquartile range), and outliers are shown as individual points. All tests were two-sided, with no adjustments made for multiple comparisons. Asterisks indicate statistically significant differences based on Mann–Whitney U test p-values: ****p ≤ 0.0001. Significance is indicated as follows: For DrugBank dataset, TP vs. FN has a p-value of 1.055e-10, FP vs. TN has a p-value of 4.954e-74, TP vs. FP has a p-value of 1.546e-51, FN vs. TN has a p-value of 1.895e-26. For KIBA dataset, TP vs. FN has a p-value of 9.713e-30, FP vs. TN has a p-value of 4.954e-74, TP vs. FP has a p-value of 1.546e-51, FN vs. TN has a p-value of 1.895e-26. For Davis dataset, TP vs. FN has a p-value of 3.502e-09, FP vs. TN has a p-value of 5.662e-45, TP vs. FP has a p-value of 6.667e-21, FN vs. TN has a p-value of 7.434e-40. b Test data sorted and divided into 20 confidence intervals based on uncertainty. All tests were two-sided with no adjustments made for multiple comparisons. The ACC was calculated for samples within each confidence interval. Five independent replications (n = 5) were performed in each data set. Data are presented as mean ± std. Source data for the figure are shown in Supplementary Data. Source data are provided as a Source Data file.
In all benchmark datasets, the incorrectly predicted samples (FP and FN) typically exhibit higher uncertainty than the correctly predicted samples. However, there were performance differences between the datasets. Fewer outliers were observed in the Davis and KIBA datasets than in the DrugBank dataset, which may be due to the fact that the DrugBank dataset contains a wider range of proteins and small molecules with complex and diverse structures. In contrast, the Davis and KIBA datasets contain a more limited diversity of proteins and small molecules of kinases.
We further analyzed whether the model provides a reliable measure of uncertainty in the cold-start dataset. Supplementary Fig. 5 demonstrates the relationship between sample prediction results and uncertainty values in the cold-start dataset. The incorrectly predicted samples (FP and FN) in the cold-start dataset also typically exhibit higher uncertainty than the correctly predicted samples.
Next, we examined the relationship between sample uncertainty and prediction accuracy to validate the second hypothesis. Samples were ranked according to their uncertainty values and divided into 20 confidence intervals, each containing 5% of the total samples. For example, the first interval includes the top 5% of samples with the lowest uncertainty, while the second interval comprises samples with an uncertainty ranking of 6–10%. The accuracy within each interval was then calculated separately, as shown in Fig. 3b.
Figure 3b shows that within the confidence interval with the lowest uncertainty (top 5 per cent), i.e., the region with the highest confidence in the model’s predictions, the accuracy exceeds 90 per cent for all three benchmark datasets, and is close to 100 per cent for the KIBA and Davis datasets. In contrast, within the confidence interval with the highest uncertainty (96–100%), the accuracy ranged between 0.5 and 0.6. This suggests that the model effectively communicates uncertainty and therefore these predictions should be treated with caution. In conclusion, the predictive accuracy of the model decreases progressively with increasing uncertainty.
These findings suggest that the uncertainty estimates generated by EviDTI can be an important tool for calibrating predictions. It also implies that EviDTI can help reduce the risks and costs associated with incorrect DTI predictions, providing important support for the decision-making process in drug discovery.
Uncertainty prediction accelerates drug discovery
After validating that EviDTI can effectively calibrate predictions, we explored its application in real-world scenarios. In practical settings, model performance in high-confidence intervals is particularly important. Therefore, we aimed for the model to achieve good uncertainty calibration in these intervals and to guide the identification of new DTIs.
First, we used the out-of-fold (OFR)45,67 to examine the predictive performance of the model at high confidence intervals. By setting thresholds ranging from 0.1 to 0.01, we compare the performance of the models at different confidence levels. To elucidate the role of evidential uncertainty, we compare two approaches: a probability-based approach that does not use uncertainty and an uncertainty-based approach that uses evidential uncertainty to obtain probabilities. Figure 4a–c shows the OFR (based on five random data splits) for the different methods on each dataset.
a Comparison of OFR between uncertainty-based and probability-based frameworks on DrugBank dataset at different thresholds. Five independent replications of each method were performed (n = 5). Data are expressed as means ± std. b Comparison of OFR between uncertainty-based and probability-based frameworks on the KIBA dataset at different thresholds. Five independent replications of each method were performed (n = 5). Data are expressed as means ± std. c Comparison of OFR between uncertainty-based and probability-based frameworks on the Davis dataset at different thresholds. The line represents the mean OFR, and the shaded area indicates the standard deviation. Five independent replications of each method were performed (n = 5). Data are expressed as means ± std. d Hit rates of the Top20 ranked predictions determined by uncertainty ranking strategies and probability ranking strategies. Five independent replications of each method were performed (n = 5). Data are expressed as means ± std. e Case study. Column interaction is the true label of the DTIs. Column Uncertainty-based is the predicted probability based on the uncertainty method and the predicted label, with the uncertainty given by the model in parentheses. Column Probability-based is the predicted probability of the probability-based method and the predicted labels. Source data are provided as a Source Data file.
According to Fig. 4a–c and Supplementary Table 5, the probability-based approach achieves higher prediction performance at lower thresholds. As the threshold increases, the OFR of both approach decreases, with the uncertainty-based approach decreasing more significantly. The uncertainty-based method has a lower OFR on all three datasets when the threshold is below 0.02. This suggests that the predictive performance of EviDTI in high confidence intervals is higher than that of the method without EDL because it provides better uncertainty calibration.
Next, we conducted several case studies to illustrate how uncertainty can improve prediction reliability. As shown in Fig. 4e, for some DTIs, probability-based methods yield incorrect predictions with high prediction probabilities that may mislead experimental validation. In contrast, uncertainty-based methods identify these predictions as unreliable by assigning a high uncertainty score. This ability to convey prediction uncertainty is critical to avoid misguidance.
While our experiments were trained and validated on the same datasets (with similar sample and feature distributions), real-world challenges involve predicting unseen DTIs, where the distribution differs from the training data. EDL’s ability to assess uncertainty in such cases is essential for making informed decisions about prediction reliability.
To conduct this study, we turned to external datasets to assess the potential of evidence uncertainty in predicting unseen samples. We constructed an independent test set of all new drugs and their targets approved by the US Food and Drug Administration in 202268 to ensure that these drugs did not appear in the training set, as detailed in Methods. A total of 24 pairs of drug-target interactions and 24 pairs of randomly generated negative interactions were collected for model performance testing. These newly reported drugs typically have novel backbones and targets that occupy different regions of chemical space.
We compared two ranking strategies for DTI identification: (1) ranking based on predicted probabilities (probabilities-based ranking), and (2) ranking based on uncertainty scores (uncertainty-based ranking). Hit rates are used to evaluate these strategies, which reflect the proportion of true DTIs correctly identified with minimal false positives. We first select the top-ranked prediction and calculate the hit rate under that ranking. Then, the ranking thresholds are gradually increased and the hit rates of the two ranking strategies are computed at each threshold. Figure 4d shows the hit rate curves determined by these two ranking strategies. Uncertainty-based ranking achieves a 100% hit rate for Top3 predictions and a hit rate of over 80% for Top10 predictions. In contrast, the probability-based ranking achieves a 100% hit rate only for the Top1 prediction and exhibits greater volatility for the Top3 prediction, indicating a lack of robustness in its model predictions. The above results illustrate that uncertainty-based rankings reduce false positives, suggesting that evidential uncertainty can reduce false positives in top DTIs.
It is worth noting that the performance of EviDTI on external datasets (based on standard assessment metrics) is not impressive. In addition, the study investigates whether UQ enhances the overall prediction performance in comparison to non-uncertainty-based models. The performance of model with UQ integration and the model without UQ integration was compared across three benchmark datasets. The model without UQ integration has the same architecture but does not use the evidential layer. As shown in Supplementary Fig. 6, the model with UQ integration performs comparably to the model without UQ integration respect to the standard evaluation metrics. However, uncertainty information remains critical. The primary objective of DTI identification is to maximize the chances of discovering a true DTI by prioritizing high-confidence predictions rather than exhaustively testing all possibilities. Incorporating uncertainty into DTI screening can improve hit rates, reduce false positives, and increase the efficiency of drug discovery by focusing experimental validation on the most reliable predictions.
In summary, these findings demonstrate that EviDTI, by quantifying uncertainty, offers a promising solution for improving the reliability of predictions and the efficiency of screening in drug-target discovery.
Application in multi-target tyrosine kinase modulators research
Tyrosine kinases are a critical class of enzymes that regulate cell signaling and play a central role in cancer therapy by driving proliferation and survival pathways69,70. Tyrosine kinase modulators have been widely used to regulate these kinase by binding to their phosphorylation sites. However, the efficacy of single-target tyrosine kinase modulators is frequently constrained due to the complexity. Additionally, single-target inhibitors frequently induce drug resistance, diminishing their clinical utility71. Multi-target tyrosine kinase modulators offer a promising solution by regulating multiple cancer-associated kinases simultaneously, disrupting the tumor signaling network and improving therapeutic efficacy.
We investigated the potential of EviDTI for the discovery of novel multi-targeted tyrosine kinase regulators. Initially, the efficacy of EviDTI in predicting interactions between tyrosine kinases and tyrosine kinase modulators was validated. Subsequently, EviDTI was applied to screen for new multi-targeted regulators and evaluated them by in vitro experiments.
Firstly, we validated the performance of EviDTI based on patent. Specifically, the interaction data between two Lenvatinib analogs (LYD-2-45 and LYD-2-49) and 11 tyrosine kinase targets was obtained from the patent (Grant No. CN 116751161 A). A total of 22 drug-target pairs were collected, including 20 positive interactions (confirmed interactions) and 2 negative interactions (non-interactions). The probabilities and uncertainties of these drug-target pairs were obtained using EviDTI, which was trained on the DrugBank database. The predicted results are shown in Supplementary Table 6, EviDTI gave correct predictions for 16 drug-target pairs.
Secondly, we validated the performance of EviDTI based on the literature. To perform this validation, we collected 67 known tyrosine kinase targets from literatures and 51 potential tyrosine kinase modulators from the Targetmol-Tyrosine Kinase Modulators Library72 (see METHODS for details). The uncertainty score and probability of interactions between the above kinases and regulators were obtained using the model trained on DrugBank dataset, and the predictions were ranked according to the uncertainty score. The predictive performance of the EviDTIs was validated in three aspects. The first aspect is to validate how many predicted DTIs with the lowest uncertainty score have been reported in literature. As shown in Supplementary Table 7, among the top 10 predicted DTIs, two DTIs were validated in the literature (Interaction between Flumatinib mesylate and c-Kit [Rank 6, Uncertainty value = 0.0419]; Interaction between Flumatinib mesylate and Bcr-Abl [Rank 10, Uncertainty value = 0.0429]73). The second aspect is to validate how many literature-reported DTIs between tyrosine kinase modulators and tyrosine kinase targets are predicted by EviDTI. A total of 27 DTIs were identified between 67 tyrosine kinase targets and 51 tyrosine kinase modulators. The 27 DTIs have been obtained from multiple literature sources. Supplementary Table 8 shows the results of the predictions made using EviDTI. Among the 27 known DTIs, 21 DTIs were predicted by EviDTI. 10 out of 21 predicted DTIs have high confidence (the uncertainty score less than 0.1). The third aspect is to validate how many DTIs between drugs in the top 10 predicted DTIs and their literature-reported targets could be predicted by EviDTI. We collected the drugs in the top 10 predicted DTIs and their all literature-reported targets, totally 10 DTIs between these drugs and their all targets. The results of these predictions are displayed in Supplementary Table 9. Among these 10 DTIs, 7 DTIs were predicted by EviDTI, 4 out of 7 predicted DTIs have high confidence (the uncertainty score less than 0.1). This analysis shows that EviDTI performs well in predicting DTIs reported in the literature, and in particular shows significant potential in the discovery of tyrosine kinase modulators.
Thirdly, based on EviDTI’s predictions for interactions between 67 tyrosine kinase targets and 51 potential modulators, the most promising interactions (lowest uncertainty scores) were selected for experimental validation. We focused on two key tyrosine kinases targets (FAK and FLT3). For each kinase, the seven interactions with the highest confidence (lowest uncertainty score) between these targets and potential modulators were prioritized for experimental validation. These interactions were validated using ADP-Glo™ Kinase Assay Kit. For the FAK kinase experiment, we selected PF-562271 as the positive control. In our experimental system, the 50% effective concentration of this compound was 2.91 ± 0.47 nM, which is comparable to the previously reported kinase inhibition activity (FAK: 1.5 nM)74. As shown in Fig. 5b, Tyrphostin 975 and Vodobatinib76 exhibited an inhibition of FAK activity with the 50% effective concentration of 35.7 ± 3.4 nM and 85.7 ± 8.2 nM, respectively. Also, Flumatinib mesylate73 inhibited FAK activity with a 50% effective concentration of 14.9 ± 2.1 nM (Supplementary Table 10a and Supplementary Fig. 7a–d). For the FLT3 kinase experiment, sorafenib are used as the positive control. The 50% effective concentration of sorafenib in our system was 51.7 ± 8.47 nM, which is comparable to literature value (FLT3: 58 ± 20 nM)77. As shown in Fig. 5c, Tyrphostin 975 and Vodobatinib76 activated FLT3 activity with the 50% effective concentration 1265.9 ± 244.6 nM and 406.8 ± 74.5 nM, respectively (Supplementary Table 10b and Supplementary Fig. 7e–g).
a The validation framework on multi-target tyrosine kinase modulators. Initially, the validation was carried out using the data reported in the patents. Two lenvatinib analogs with 11 known targets were collected from the patents to conduct this validation. Subsequently, the validation was carried out using the data reported in the literature. The uncertainty score and probability of interactions between the 67 tyrosine kinase targets and 51 tyrosine kinase modulators were predicted via EviDTI. Finally, two targets of interest were selected from the 67 targets collected above. Based on the uncertainty for these two targets and 51 modulators, the interactions with lowest uncertainty between these two targets and the seven modulators were validated experimentally. b The 50% effective concentration of Tyrphostin 9, Vodobatinib, Flumatinib and PF-562271 in the FAK kinase ADP-Glo assays, respectively. PF-562271 as positive control. Mean ± SEM of three independent experiments is shown (n = 3). c The 50% effective concentration of Vodobatinib, Tyrphostin 9 and sorafenib in the FLT3 kinase ADP-Glo assays, respectively. Sorafenib as positive control. Mean ± SEM of three independent experiments is shown (n = 3). Source data are provided as a Source Data file.
In summary, EviDTI demonstrated its potential in identifying novel multi-target tyrosine kinase modulators, not only by predictively validating the targets of existing drugs, but also by experimentally validating its effectiveness in improving the efficiency of drug discovery and reducing the risk of development.
Interpretability of light attention
EviDTI provides an additional advantage by offering residue-level insights and elucidating key factors in drug design. The light attention module facilitates this capability by enabling visualization of the contribution of each amino acid to the final predicted outcome.
Firstly, a visualization case study demonstrated the relationship between attention weights and binding residues. Four DTI pairs were randomly selected from the DrugBank dataset, and their tertiary structures were downloaded from the PDB database. The PDB IDs of these complexes are 1y91, 1d6n, 1czh, and 1z83, respectively. Attention values were mapped to these structures and visualized using PyMOL (Fig. 6). In Fig. 6, the binding pockets of proteins and small molecules are visualized, showcasing residues with high attentional values. Across all cases, residues with high attention values coincide with the binding site, underscoring its importance in predicting and validating the attention mechanism’s efficacy.
a–d The correctly predicted amino acid residues surrounding the corresponding ligands (sticks) are highlighted. The residues around the corresponding ligands that were correctly predicted are highlighted in the figure. Their color indicates the degree of contribution of these residues to the prediction results. 3D representations of all structures were visualized using Pymol software.
Overall, the case study findings demonstrate the interpretability of the model and are expected to reveal previously unexplored local interactions, providing opportunities to mine hidden knowledge. These findings could guide strategies to help develop drug targets.
To further explore the interpretability of the model, all known DTI complex structures appearing in the DrugBank dataset were collected from the PDB database. The degree of correspondence between the residues with the highest attention value and the binding site was also evaluated using the binding sites-hit ratio.
Specifically, the residues were first ranked according to their attention value, and the top N% residues were selected to calculate the binding sites hit ratio. A true binding site is defined as residue whose Euclidean distance from the drug is within 4 Å of any atom.
The binding sites hit ratio at different thresholds is reported in Table 4 and Supplementary Fig. 8. When the number of true binding sites was used as the threshold value, the binding sites hit ratio was 0.125, and more than 70% of the binding sites ranked in the top 50% of the attention values. These results suggest that confidence in the interpretability of protein sequences may be lacking because 1D protein sequences (used as inputs for protein information in our model) do not necessarily indicate the 3D conformation and location of the binding pocket. However, the results for primary protein sequences are encouraging, and it is reasonable to assume that further incorporation of 3D protein information into the modeling framework will ultimately improve the model’s interpretability of the drug-target interaction network.
Discussion
Although current deep learning models have made significant progress in DTI prediction, they typically lack the ability to provide confidence estimates for predictions. This limitation seriously affects the effectiveness of these models in real-world applications, where decision making often requires a clear understanding of the reliability of the predicted results. To bridge the gap between predictive models and practical applications, we present EviDTI, a novel framework that integrates evidential-based deep learning. This method not only predicts DTI with high accuracy, but also provides reliable quantification of uncertainty, which enhances the practicality and reliability of the model. This is important for advancing the field of drug discovery as it is directly related to the success and cost-effectiveness of experiments. EviDTI leverages 2D topological graphs, 3D molecular geometries of drug molecules, and target protein sequences for DTI prediction. Additionally, EDL is employed to estimate uncertainty, providing a robust measure of prediction confidence. The performance comparison across three datasets demonstrates that EviDTI achieves competitive performance compared to 11 DTI prediction models. This study demonstrates that EDL can provide a reliable uncertainty quantification for DTIs prediction. Importantly, we showed that uncertainty-guided predictions can prioritize experiments in drug discovery and drug repositioning studies. The experimental validation of interactions between two tyrosine kinase targets and seven potential tyrosine kinase modulators (totally 14 potential DTIs) was performed. Among those predicted DTIs, 5 DTIs showed binding affinity, and 4 of them reached the nanomolar level of binding affinity. These results demonstrate that EviDTI is a powerful tool that can accelerate the translation process from theoretical models to actual drug development.
However, the model still has some limitations. Although EviDTI was designed with the objective of identifying potential DTIs, it is currently cannot determine whether the molecule has an activating or inhibitory effect on the target. To overcome this limitation, future directions for improvement include introducing more molecular biochemical properties into the model, developing a multi-task learning framework capable of predicting activation and inhibition simultaneously.
In addition, EviDTI has difficulty in providing discriminating probabilities and uncertainties for interactions between compounds and the target and its mutants. The development of effective drugs that specifically targeting mutants is essential to improve treatment outcomes, especially in diseases driven by specific genetic alterations such as acute myeloid leukemia (AML). However, EviDTI relies on one-dimensional protein sequence information, its ability to capture the nuanced effects of mutations may be limited. We believe in incorporating more informative representations such as protein 3D structural information and pharmacologically perturbed transcriptomics data can more effectively address these limitations.
In the future, the model can be extended as follows to enhance its performance and generalizability. First, with the rapid advancements in 3D protein structure prediction methods29,78, incorporating 3D protein structure information into DTI predictions has the potential to significantly enhance prediction accuracy. However, modeling 3D protein structures remains complex and considerably increases computational time; therefore, this study did not utilize such information in the modeling process. Nevertheless, we believe that integrating 3D structural data—particularly when combined with pocket information79,80—can yield deeper insights into interaction mechanisms, ultimately leading to more accurate DTI predictions and improved interpretation of these interactions.
Furthermore, this study uses LA module to obtain amino acid attention weights, providing molecular-level insights into DTIs. However, the EviDTI framework still faces limitations in explicitly learning the interactions between drug and protein local structures. Several approaches have been developed to visualize the contributions of protein sequences and drug substructures to the final prediction results through a cross-attention mechanism, thus enhancing the interpretability of the model. Combining this cross-attention mechanism with evidential-based deep learning can improve model accuracy and interpretability while effectively calibrating uncertainty.
Moreover, while EDL can calibrate prediction uncertainty for unknown samples, it does not inherently improve prediction performance for out-of-distribution samples (i.e., cross-domain generalization). Transfer learning can address this limitation by aligning the feature distributions of the training set with new samples and learning transferable representations. Combining EDL with transfer learning could significantly enhance the efficiency of drug discovery and drug repositioning.
Finally, the EviDTI framework, with its focus on generalizability, holds the potential to be extended for application to other interaction prediction problems. This includes, but is not limited, to enzymatic reaction kinetic parameters prediction and drug-drug interaction prediction. The adaptability and versatility of this framework open up new avenues for research and application in the field of drug-target interaction prediction.
Methods
Datasets
Three datasets DrugBank68, Davis59, and KIBA60 were used for model evaluation. Supplementary Table 1 summarizes these datasets.
DrugBank: the data used in this study were published on January 3, 2020 (version 5.1.5). Drugs with inorganic compounds, very small molecule compounds [e.g., iron (DB01592), zinc (DB01593)], or SMILES strings that the RDKit Python package could not recognize were removed. Drugs with a SMILES length greater than 300 were also removed to comply with the maximum input of MG-BERT. Ultimately, 6148 drugs and 4085 targets, totaling 16,531 pairs of known drug-target interactions, were collected, and these known drug-target interactions were used as positive samples for training. We randomly disrupted these positive pairs to generate negative samples, specifically by randomly selecting a drug-target interaction pair and forming a negative drug-target interaction pair between the target in that pair and another drug with a known drug-target interaction pair. Negative samples with the same number of positive samples, totaling 16,531 negative sample pairs, were generated through the above steps. Thus, a total of 33,062 pairs of samples were collected in the DrugBank dataset, with a ratio of positive to negative samples of 1:1.
Davis and KIBA: The Davis and KIBA dataset records wet-lab values for determining the binding affinity between drugs and proteins. We refer to previous work and divide positive and negative samples using 5.0 and 12.1 as thresholds to construct a binary classification dataset.
In order to assess the ability of the model to predict novel compound interactions in real-world drug discovery, we implemented a cold-start scenario following the practice established by Wang et al.12 Selected 10% of drugs from DrugBank dataset, and then all the DTI pairs associated with these drugs were used as a test set. Resulting in 3170 cold-start test samples VS. 29,894 training samples.
To perform the unseen data prediction (see “Uncertainty prediction accelerates drug discovery”), we collected all new drugs approved by the FDA in 2022 and their targets from the DrugBank dataset. In 2022, the FDA approved 37 new drugs for marketing, including 22 new molecular entities and 15 new biologic applications. Only molecular drugs were selected among these new approvals, while drugs already appearing in the training set were removed. Ultimately, we collected 13 drugs, 22 targets, and 24 pairs of interactions. We used the same method for constructing the negative samples as for the DrugBank training set, randomly generating as many negative samples as positive samples. Thus, we collected 48 pairs of samples for out-of-domain performance testing. In these samples, the ratio of positive to negative samples was 1:1.
Framework architecture
Protein feature encoder
The protein feature encoder consists of an initial encoding module and a LA module81.
In the initial coding module, we extracted sequence information using the widely used protein language model ProtTrans30 (denoted as ProtT5), which is a self-supervised autocoder based on the transformer model. A 1024-dimensional sequence embedding for each residue was extracted using ProtT5. For each protein, we obtained a \(L\times 1024\)-dimensional feature matrix, where L is the length of the protein chain. We did not train ProtT5, but used its locked parameters to obtain the representations.
The protein feature matrix is encoded using light attention. The input to light attention is a protein embedding \(x\) of size \(L\times {d}_{{in}}\), where \(L\) is the length of the protein chain, i.e., the number of amino acids, and \({d}_{{in}}\) is the length of the initial representation of each amino acid, in this case, 1024. Two independent 1D convolutions transform the input. Convolution is used in the length dimension to generate attention coefficients and values \(e,v\).
where \(b\) is a learned bias, \({s}\) is filter sizes, \({W}^{(e)}\) is the weights of the 1D convolutions.
To obtain the attention weights, softmax normalization is performed on \({e}_{{ij}}\). The attention weight \({\alpha }_{{ij}}\) for the j-th residue and the i-th feature dimension is computed as:
The weight distribution of each feature dimension i is independent, and they can produce different attention patterns. The attention distribution calculates a weighted sum of the transformed residual embeddings \({v}_{{ij}}\). Thus, we obtain a representation \(x{\prime}\) independent of protein length.
Drug feature encoder
The drug feature encoder consists of a 2D topology encoding module and a 3D structure encoding module.
In the 2D topology encoding module, the small molecule pre-training model MG-BERT58 is used to extract drugs’ 2D topological graph information. MG-BERT is a pre-trained model based on a self-supervised learning approach for learning atomic-level representations of drug molecules. The model uses the transformer architecture and employs a mask-based self-supervised learning approach. Again, we did not train MG-BERT but used its frozen parameters to obtain an initial representation of the drug. In this work, the representation obtained by MG-BERT is extracted as an initial representation of the drug molecule, which is used as input to 1DCNN to obtain a final representation of the 2D information of the drug.
In the 3D structure encoding module, given a 3D structure graph, an atom-bond graph and a bond-angle graph are constructed, under which the representation vectors of atoms and bonds are learned iteratively82. In the atom-bond graph, the nodes of the graph are atoms, and the edges of the graph are covalent bonds of atomic bonds. In the bond-angle graph, the nodes of the graph are the bonds, and the edges are the bond angles between the two bonds. The scalar atomic distances and angles are mapped into high-dimensional vectors using the Gaussian kernel function. In a more concrete sense, the representation vectors of atom \(u\) and bond \((u,v)\) for the kth iteration are denoted as \({h}_{u}\) and \({h}_{{uv}}\), respectively. The initialization is set as \({h}_{u}^{(0)}={x}_{u}\) and \({h}_{{uv}}^{(0)}={x}_{{uv}}\).
Given the bond \((u,v)\), its representation vector \({h}_{{uv}}^{(k)}\) at the kth iteration is formulated by
\(N(u)\) and \(N(v)\) represent the adjacent atoms of u and \(\,v\),\(\{(u,w):w\in N(u)\}\cup \{(v,w):w\in N(v)\}\) are the neighboring bonds of \((u,v)\).
The function “Aggregate” aggregates messages, while “Combine” updates the bond-angle graph.
The information from adjacent bonds and corresponding bond angles is aggregated into \({a}_{{uv}}^{\left(k\right)}\), and subsequently, the bond representation \(\left(u,v\right)\) is updated based on the aggregated information.
For an atom \(u\), at the kth iteration, its representation vector \({{h}}_{u}^{\left(k\right)}\) can be expressed as:
The messages from the bonds are learned from the bond-angle graph. The aggregated messages then update the representation vector of atom u.
The atom’s representation vectors at the final iteration are integrated to obtain the molecular representation vector \({h}_{G}\) through the READOUT function.
Evidential layer
The learned drug and protein representations are concatenated and serve as inputs to the evidential layer. The evidence layer is a fully connected neural network with N layers. Unlike classical neural networks, the softplus layer is used to ascertain non-negative output. In the deep evidential classification model, the joint evidence distribution is presented as a Dirichlet distribution, which is used to represent the interaction prediction and the degree of evidence associated with that prediction. The outputs of the evidence layer are used as parameters of the predicted Dirichlet distribution. For the DTI prediction task, the network has two outputs.
Evidential learning for classification
In subjective logic (SL), for each class \(k=1,\,.\,..,K\), SL will provide a belief mass bk and an overall uncertainty mass u to compose a framework for evaluating the K classification. These mass values are all non-negative and sum to 1, i.e.,
where \({{\rm{u}}}\ge \,0\) and\(\,{{{\rm{b}}}}_{k}\,\ge \,0\) for \({k}=1,\,.\,..,K\).
The bk value is computed from the amount of evidence for the kth class. Let \({{{\rm{e}}}}_{k}\,\ge \,0\) be the evidence for the kth class, then the belief bk and uncertainty u are calculated as
where\(\,S=\,{\sum }_{i=1}^{K}{e}_{i}-1\).
Here, the evidence is defined as a measure of the support from the data collection to a certain class to support the sample classification. A belief mass assignment, i.e., subjective opinion, corresponds to a Dirichlet distribution with parameters \({\alpha }_{k}={e}_{k}+1\).
That is:
Where \({{\rm{S}}}=\,{\sum }_{i=1}^{K}{\alpha }_{i}\)
The expected probability of the Kth class is the mean of the corresponding Dirichlet distribution, calculated as
The EviDTI is trained end-to-end using a multi-objective loss function \({{\boldsymbol{L}}}(\varTheta )\) to achieve the maximum fit and minimum incorrectly predicted evidence for the model.
Where \({\lambda }_{t}\) is the annealing coefficient, t is the index of the current training epoch, \({D}({p}_{i}\left|\left\langle 1,...,1\right\rangle \right.)\) is the uniform Dirichlet distribution, and \({\widetilde{\alpha }}_{i}={y}_{i}+(1-{y}_{i})\odot {\alpha }_{i}\) is the Dirichlet parameters after removal of the non-misleading evidence from predicted parameters \({\alpha }_{i}\) for sample i. More details about the Evidential Deep Learning to Quantify Classification Uncertainty.
Comparison methods
For the DrugBank, Davis and KIBA dataset, we retrained and tuned all models based on the parameters provided in their respective GitHub repositories. All methods were conducted with five repeated experiments, and the results are reported as the mean and variance.
DeepConv-DTI19
DeepConv-DTI utilizes CNNs to extract features from proteins and fully connected networks (FCNs) to extract features from drugs, followed by making predictions.
GraphDTA64
GraphDTA employs GNN and CNN to represent drugs and proteins, respectively. As Nguyen et al. reported, the GAT_GCN combination is chosen as the feature extractor in subsequent experimental comparisons.
For a fair comparison, GraphDTA were adapted according to the following steps:(1) A Sigmoid activation function was added after the last layer to convert the model output to binary probabilities. (2) A binary cross-entropy was used as the loss function instead of the original mean square error (MSE) loss. (3) A grid search was performed to optimise key hyperparameters such as learning rate and batch size, and a validation set was used to determine the best configuration.
MolTrans20
MolTrans employs the transformer model to encode protein and drug sequences into feature embeddings. These embeddings are used to construct an interaction matrix, which is subsequently processed by CNNs and FCNs to make predictions.
TransformerCPI65
TransformerCPI utilizes the Transformer architecture, treating drugs and proteins as distinct sequences. By employing CNN and GCN, the model generates representations for protein sequences and atom structures, respectively. Subsequently, TransformerCPI extracts interaction features through the transformer decoder and employs linear layers to produce the final output, representing the interaction probability.
HyperAttentionDTI17
HyperAttention DTI is an end-to-end bio-inspired model built upon the CNN and attention mechanism. Deep CNNs are employed to learn feature matrices for both drugs and proteins adeptly. The attention mechanism was leveraged on these feature matrices to capture intricate non-covalent intermolecular interactions among atoms and amino acids, assigning an attention vector to each atom or amino acid.
AIGO-DTI66
AIGO-DTI proposes an Adaptive Iterative Graph Optimization (AIGO)-DTI prediction framework. This framework integrates atomic cluster information and enhances molecular features through the design of functional group prompts and graph encoders, thereby optimizing the construction of DTI association networks. Furthermore, the optimization of graph structure is transformed into a node similarity learning problem, utilizing multi-head similarity metric functions to iteratively update the network structure to improve the quality of DTI information.
GraphormerDTI24
GraphormerDTI integrates the Graph Transformer neural network with a 1D-CNN to extract the representations of drug and target, respectively. Specifically, GraphormerDTI embeds molecular graphs into vector representations through iterative Transformer-based message passing. The structural representations of the molecules are encoded through node centrality encoding, node spatial encoding and edge encoding. The attentional mechanisms are used to model interactions.
DLM-DTI27
DLM-DTI was comprised of three primary components: the drug encoder, the target encoder, and the interaction prediction head. The target encoder incorporates both the teacher and student models of language models for protein sequences. The teacher model employed for target sequence encoding was the ProtBERT model. The student model was designed to be consistent with the original teacher model with fewer layers to conduct knowledge distillation. The function of the drug encoder is to convert SMILES sequences to molecular representations, and this is achieved by employing the ChemBERTa encoder. The interaction prediction header predicts interaction probabilities by concatenating drug and target representations.
Statistical analysis of binding sites
To verify the correlation between residues with higher attention values in LA and drug-target binding sites, we obtained the real three-dimensional structures of the DTI interaction pairs in the DrugBank dataset from the PDB database for statistical analysis of binding sites.
First, drug-target pairs lacking the true structure of the protein were excluded. Next, for each protein, only structures containing small molecules that interact with them in the PDB file were retained. The binding sites of the drugs were then calculated, defining a binding residue as any atom of that residue within a 4 Å Euclidean distance of the drug molecule. Finally, to ensure accurate mapping of attention values to residues, the amino acid sequence indexes in the PDB were aligned with those in UniProt, which were used during training.
In the Light Attention network, for a given amino acid sequence of length L, the network will output an \(L\times {d}_{{in}}\) dimensional attention vector, where \({d}_{{in}}\) denotes the dimension of the attention vector. We get the attention value for each amino acid by taking the average of this matrix in the\(\,{d}_{{in}}\) dimension. A larger attention value means that the amino acid contributes more to DTI prediction.
Experimental settings
EviDTI is implemented in Python 3.8 and PyTorch 1.12.0, as well as functions in PYG 2.2.0, Scikit-learn 1.2.2, Numpy 1.24.3, Pandas 1.4.2, and RDKit 2020.09.1. The batch size was set to 32, and the Adam optimizer was used with a learning rate of 5e-4 and a weight decay of 1e-5. We allowed the model to run for a maximum of 500 epochs for all datasets, and if the model’s accuracy on the validation set did not decrease within 30, training would stop. The parameters of all neural networks are listed in the Supplementary Table 11 and Supplementary Fig. 9.
Compounds collection from Targetmol-Tyrosine kinase modulator library (L2200)
There were 746 potential tyrosine kinase modulators collected from the Targetmol-Tyrosine kinase modulator library (L2200). To ensure the credibility of the experiments, we excluded kinase modulators with drug similarity greater than 0.7 to the DrugBank dataset and ultimately retained 52 potential modulators. Drug similarity was calculated based on the Tanimoto coefficient.
In vitro kinase activity assays
The FAK (Cat#V1971) and FLT3 (Cat#V4064) were purchased from Promega. Compounds Tyrphostin 9 (Cat#T2479), GSK180736A(Cat#3513), Vodobatinib (Cat# T8882), BIX02188 (Cat# T1744), PKG drug G1 (Cat# T4661), ST271 (Cat# T4511), Flumatinib mesylate (Cat# T7861), Mubritinib (Cat# T6124), PF-562271 (Cat# T6124) and Sorafenib (Cat# T0093L) were purchased from Targetmol, USA. All compounds were tested from 1000 nM, with 3-fold dilution for 10 points.
In vitro kinase activity assays were conducted through ADP-Glo assays (#V9101) provided by Promega. The protocol for the FAK and Flt3 assay is described as follows: Enzyme, substrate, ATP, and compounds were diluted in 1X reaction buffer composed of 40 mM Tris (pH 7.5), 2 mM MnCl, 100 µM sodium vanadate. In a 384-well low-volume plate, l μl of the compound at indicated doses or 5% dimethyl sulfoxide (DMSO), 2 μl of FAK or FLT3 enzyme (15 ng/well), and 2 ul of substrate/ATP mix (final concentration: 20 μM ATP) were added to each well. The plate was then incubated at 25 °C for 60 min to allow for kinase activity. Following the enzymatic reaction, 5 μl of ADP-GloT Reagent was added to each well, and the plate was incubated at 25 °C for an additional 40 min. Subsequently, 10 μl of Kinase Detection Reagent was added to convert ADP to ATP and introduce luciferase and luciferin to detect ATP, and the plate was incubated for a final 30 min at 25 °C. Luminescence was recorded with an integration time of 0.5 s using the SpectraMax iD3 instrument (Molecular Devices).
The 50% effective concentration was calculated using Prism 8 by fitting the following equation:
where X is a log of concentration, Y is a response, and top and bottom are the responses of controls, each assay was repeated at least three times, and we computed the mean and standard deviation for the values.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The source data of three datasets used to train and evaluate the model is provided at https://github.com/zhaoyanpeng208/EviDTI/tree/main/dataset and https://zenodo.org/records/14056305. The source data of 67 targets with 51 drugs used for literature validation of tyrosine kinase modulators is provided in https://github.com/zhaoyanpeng208/EviDTI/tree/main/dataset/len_v_s_dataset and https://zenodo.org/records/14056305. The source data of two Lenvatinib analogs (LYD-2-45 and LYD-2-49) with 11 tyrosine kinase targets used for patent validation of tyrosine kinase modulators is provided in https://github.com/zhaoyanpeng208/EviDTI/tree/main/dataset/len_case_dataset and https://zenodo.org/records/14056305. Source data are provided with this paper through Figshare https://doi.org/10.6084/m9.figshare.28816634. Source data are provided with this paper.
Code availability
The source data and codes of EviDTI are available on GitHub at https://github.com/zhaoyanpeng208/EviDTI, which has also been deposited in the Zenodo under accession code https://zenodo.org/records/1576047183. Prottrans (https://github.com/agemagician/ProtTrans) is used for calculating target protein representations. Data are analyzed using numpy v1.24.3 (https://numpy.org/), pandas v1.4.2 (https://pandas.pydata.org/), Seaborn V0.11.2 (https://seaborn.pydata.org/) and Matplotlib v3.4.3 (https://matplotlib.org/). Structures are visualized by Pymol v2.5.2 (https://www.pymol.org/).
References
Miteva, M. A. et al. FAF-Drugs: free ADME/tox filtering of compound collections. Nucleic Acids Res. 34, W738–W744 (2006).
Bagherian, M. et al. Machine learning approaches and databases for prediction of drug–target interaction: a survey paper. Brief. Bioinforma. 22, 247–269 (2021).
Yang, Z., Zeng, X., Zhao, Y. & Chen, R. AlphaFold2 and its applications in the fields of biology and medicine. Signal Transduct. Target. Ther. 8, 115 (2023).
Liu, T. et al. Applying high-performance computing in drug discovery and molecular simulation. Natl. Sci. Rev. 3, 49–63 (2016).
Zitnik, M. et al. Machine learning for integrating data in biology and medicine: principles, practice, and opportunities. Inf. Fusion 50, 71–91 (2019).
Zahra, N. & Mohammad Keyvanpour, R. A Comparative Analytical Review on Machine Learning Methods in Drugtarget Interactions Prediction. Curr. Comput.Aided Drug Des. 19, 325–355 (2023).
Sydow, D. et al. Advances and challenges in computational target prediction. J. Chem. Inf. Model. 59, 1728–1742 (2019).
Sieg, J., Flachsenberg, F. & Rarey, M. In need of bias control: evaluating chemical data for machine learning in structure-based virtual screening. J. Chem. Inf. Model. 59, 947–961 (2019).
Lim, S. et al. A review on compound-protein interaction prediction methods: data, format, representation and model. Comput. Struct. Biotechnol. J. 19, 1541–1556 (2021).
Ye, Q. et al. A unified drug–target interaction prediction framework based on knowledge graph and recommendation system. Nat. Commun. 12, 6775 (2021).
Yamanishi, Y., Araki, M., Gutteridge, A., Honda, W. & Kanehisa, M. Prediction of drug–target interaction networks from the integration of chemical and genomic spaces. Bioinformatics 24, i232–i240 (2008).
Wang, X. et al. Multitask joint strategies of self-supervised representation learning on biomedical networks for drug discovery. Nat. Mach. Intell. 5, 445–456 (2023).
van Westen, G. J. P., Wegner, J. K., IJzerman, A. P., van Vlijmen, H. W. T. & Bender, A. Proteochemometric modeling as a tool to design selective compounds and for extrapolating to novel targets. Med. Chem. Commun. 2, 16–30 (2011).
de la Fuente, J. et al. Towards a more inductive world for drug repurposing approaches. Nat. Mach. Intell. 7, 495–508 (2025).
Lee, I. & Nam, H. Sequence-based prediction of protein binding regions and drug–target interactions. J. Cheminform. 14, 5 (2022).
Tsubaki, M., Tomii, K. & Sese, J. Compound–protein interaction prediction with end-to-end learning of neural networks for graphs and sequences. Bioinformatics 35, 309–318 (2019).
Zhao, Q., Zhao, H., Zheng, K. & Wang, J. HyperAttentionDTI: improving drug–protein interaction prediction by sequence-based deep learning with attention mechanism. Bioinformatics 38, 655–662 (2022).
Bai, P., Miljković, F., John, B. & Lu, H. Interpretable bilinear attention network with domain adaptation improves drug–target prediction. Nat. Mach. Intell. 5, 126–136 (2023).
Lee, I., Keum, J. & Nam, H. DeepConv-DTI: prediction of drug-target interactions via deep learning with convolution on protein sequences. PLOS Comput. Biol. 15, e1007129 (2019).
Huang, K., Xiao, C., Glass, L. M. & Sun, J. MolTrans: molecular interaction transformer for drug–target interaction prediction. Bioinformatics 37, 830–836 (2021).
Bian, J., Zhang, X., Zhang, X., Xu, D. & Wang, G. MCANet: shared-weight-based MultiheadCrossAttention network for drug–target interaction prediction. Brief. Bioinforma. 24, bbad082 (2023).
He, Y. et al. Flexible drug-target interaction prediction with interactive information extraction and trade-off. Expert Syst. Appl. 249, 123821 (2024).
Hadipour, H. et al. GraphBAN: an inductive graph-based approach for enhanced prediction of compound-protein interactions. Nat. Commun. 16, 2541 (2025).
Gao, M. et al. GraphormerDTI: a graph transformer-based approach for drug-target interaction prediction. Comput. Biol. Med. 173, 108339 (2024).
Sun, Y., Li, Y. Y., Leung, C. K. & Hu, P. iNGNN-DTI: prediction of drug–target interaction with interpretable nested graph neural network and pretrained molecule models. Bioinformatics 40, btae135 (2024).
Lu, Z. et al. DTIAM: a unified framework for predicting drug-target interactions, binding affinities and drug mechanisms. Nat. Commun. 16, 2548 (2025).
Lee, J., Jun, D. W., Song, I. & Kim, Y. DLM-DTI: a dual language model for the prediction of drug-target interaction with hint-based learning. J. Cheminform 16, 14 (2024).
Hayes, T. et al. Simulating 500 million years of evolution with a language model. Science 387, 850–858 (2025).
Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023).
Elnaggar, A. et al. ProtTrans: toward understanding the language of life through self-supervised learning. IEEE Trans. Pattern Anal. Mach. Intell. 44, 7112–7127 (2022).
Fang, Y. et al. Knowledge graph-enhanced molecular contrastive learning with functional prompt. Nat. Mach. Intell. 5, 542–553 (2023).
Li, H. et al. A knowledge-guided pre-training framework for improving molecular representation learning. Nat. Commun. 14, 7568 (2023).
He, C., Yang, C., Zhang, H., Long, Y. & Zhao, X. DTI-MPFM: a multi-perspective fusion model for predicting potential drug–target interactions. Expert Syst. Appl. 264, 125740 (2025).
Zhao, W. et al. MSI-DTI: predicting drug-target interaction based on multi-source information and multi-head self-attention. Brief. Bioinforma. 25, bbae238 (2024).
Dalkıran, A. et al. Transfer learning for drug–target interaction prediction. Bioinformatics 39, i103–i110 (2023).
Abbasi, K. et al. DeepCDA: deep cross-domain compound–protein affinity prediction through LSTM and convolutional neural networks. Bioinformatics 36, 4633–4642 (2020).
Collins, K. M. et al. Building machines that learn and think with people. Nat. Hum. Behav. 8, 1851–1863 (2024).
Nix, D. A. & Weigend, A. S. Estimating the mean and variance of the target probability distribution. Proc. 1994 IEEE Int. Conf. Neural Netw. (ICNN’94) 1, 55–60 (1994).
Amini, A., Soleimany, A. P., Schwarting, W., Bhatia, S. N. & Rus, D. Uncovering and mitigating algorithmic bias through learned latent structure. in Proceedings of the 2019 AAAI/ACM Conference on AI, Ethics, and Society 289–295 (Association for Computing Machinery, 2019). https://doi.org/10.1145/3306618.3314243.
Luo, Y., Liu, Y. & Peng, J. Calibrated geometric deep learning improves kinase–drug binding predictions. Nat. Mach. Intell. 5, 1390–1401 (2023).
Fan, Z. et al. Reducing overconfident errors in molecular property classification using Posterior Network. Patterns 5, 100991 (2024).
Xu, H. et al. Deep active learning with high structural discriminability for molecular mutagenicity prediction. Commun. Biol. 7, 1071 (2024).
Tan, H. S., Wang, K. & McBeth, R. Deep evidential learning for radiotherapy dose prediction. Comput. Biol. Med. 182, 109172 (2024).
Lin, Y. et al. Uncertainty quantification via a memristor Bayesian deep neural network for risk-sensitive reinforcement learning. Nat. Mach. Intell. 5, 714–723 (2023).
Janet, J. P., Duan, C., Yang, T., Nandy, A. & Kulik, H. J. A quantitative uncertainty metric controls error in neural network-driven chemical discovery. Chem. Sci. 10, 7913–7922 (2019).
Tran, K. et al. Methods for comparing uncertainty quantifications for material property predictions. Mach. Learn. Sci. Technol. 1, 025006 (2020).
Jospin, L. V., Buntine, W. L., Boussaid, F., Laga, H. & Bennamoun Hands-on Bayesian neural networks—a tutorial for deep learning users. IEEE Comput. Intell. Mag. 17, 29–48 (2020).
Lakshminarayanan, B., Pritzel, A. & Blundell, C. Simple and scalable predictive uncertainty estimation using deep ensembles. in Proc. 31st International Conference on Neural Information Processing Systems 6405–6416 (Curran Associates Inc., 2017).
Y. Gal & Ghahramani, Z. Dropout as a Bayesian approximation: representing model uncertainty in deep learning. in Proc. 33rd International Conference on Machine Learning (eds Balcan, M. F. & Weinberger, K. Q.) Vol. 48, 1050–1059 (PMLR, 2016).
Eyke, N. S., Green, W. H. & Jensen, K. F. Iterative experimental design based on active machine learning reduces the experimental burden associated with reaction screening. React. Chem. Eng. 5, 1963–1972 (2020).
Hirschfeld, L., Swanson, K., Yang, K., Barzilay, R. & Coley, C. W. Uncertainty quantification using neural networks for molecular property prediction. J. Chem. Inf. Model. 60, 3770–3780 (2020).
Smith, J. S., Nebgen, B., Lubbers, N., Isayev, O. & Roitberg, A. E. Less is more: sampling chemical space with active learning. J. Chem. Phys. 148, 241733 (2018).
Sensoy, M., Kaplan, L. & Kandemir, M. Evidential deep learning to quantify classification uncertainty. in Proc. 32nd International Conference on Neural Information Processing Systems 3183–3193 (Curran Associates Inc., 2018).
Amini, A., Schwarting, W., Soleimany, A. & Rus, D. Deep evidential regression. in Proc. 34th International Conference on Neural Information Processing Systems (Curran Associates Inc., 2020).
Soleimany, A. P. et al. Evidential deep learning for guided molecular property prediction and discovery. ACS Cent. Sci. 7, 1356–1367 (2021).
Wang, R. et al. An uncertainty-guided deep learning method facilitates rapid screening of CYP3A4 inhibitors. J. Chem. Inf. Model. 63, 7699–7710 (2023).
Wang, D. et al. Learning with uncertainty to accelerate the discovery of histone lysine-specific demethylase 1A (KDM1A/LSD1) inhibitors. Brief. Bioinforma. 24, bbac592 (2023).
Zhang, X.-C. et al. MG-BERT: leveraging unsupervised atomic representation learning for molecular property prediction. Brief. Bioinforma. 22, bbab152 (2021).
Davis, M. I. et al. Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 29, 1046–1051 (2011).
Tang, J. et al. Making sense of large-scale kinase inhibitor bioactivity data sets: a comparative and integrative analysis. J. Chem. Inf. Model. 54, 735–743 (2014).
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001).
Cortes, C. & Vapnik, V. Support-vector networks. Mach. Learn. 20, 273–297 (1995).
Bhuvaneswari, R. & Kalaiselvi, K. Naive Bayesian Classification Approach in Healthcare Applications. International Journal of Computer Science and Telecommunications. 3, 106–112 (2012).
Nguyen, T. et al. GraphDTA: predicting drug–target binding affinity with graph neural networks. Bioinformatics 37, 1140–1147 (2021).
Chen, L. et al. TransformerCPI: improving compound–protein interaction prediction by sequence-based deep learning with self-attention mechanism and label reversal experiments. Bioinformatics 36, 4406–4414 (2020).
Zhang, S. et al. 111AIGO-DTI: predicting drug–target interactions based on improved drug properties combined with adaptive iterative algorithms. J. Chem. Inf. Model. 64, 4373–4384 (2024).
Han, K., Lakshminarayanan, B. & Liu, J. Reliable Graph Neural Networks for Drug Discovery Under Distributional Shift. Arxiv https://doi.org/10.48550/arXiv.2111.12951 (2021)
Law, V. et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 42, D1091–D1097 (2014).
Zhao, Y. et al. Tyrosine kinase inhibitors and their unique therapeutic potentialities to combat cancer. Int. J. Biol. Macromol. 168, 22–37 (2021).
Huang, L., Jiang, S. & Shi, Y. Tyrosine kinase inhibitors for solid tumors in the past 20 years (2001–2020). J. Hematol. Oncol. 13, 143 (2020).
Yang, Y., Li, S., Wang, Y., Zhao, Y. & Li, Q. Protein tyrosine kinase inhibitor resistance in malignant tumors: molecular mechanisms and future perspective. Signal Transduct. Target. Ther. 7, 329 (2022).
Manning, G., Whyte, D. B., Martinez, R., Hunter, T. & Sudarsanam, S. The protein kinase complement of the human genome. Science 298, 1912–1934 (2002).
Zhao, J. et al. Flumatinib, a selective inhibitor of BCR-ABL/PDGFR/KIT, effectively overcomes drug resistance of certain KIT mutants. Cancer Sci. 105, 117–125 (2014).
Sidwell, R. W., Bailey, K. W., Wong, M.-H., Barnard, D. L. & Smee, D. F. In vitro and in vivo influenza virus-inhibitory effects of viramidine. Antivir. Res. 68, 10–17 (2005).
Kandandapani, S., Ridzwan, N. F. W., Mohamad, S. B. & Tayyab, S. Exploring the interaction between tyrphostin 9 and human serum albumin using biophysical and computational methods. J. Biomol. Struct. Dyn. 38, 4134–4142 (2020).
Walsh, R. R. et al. Plasma and cerebrospinal fluid pharmacokinetics of vodobatinib, a neuroprotective c-Abl tyrosine kinase inhibitor for the treatment of Parkinson’s disease. Parkinsonism Relat. Disord. 108, 105281 (2023).
Wilhelm, S. et al. BAY 43-9006 Exhibits Broad Spectrum Anti-Tumor Activity and Targets Raf/MEK/ERK Pathway and Receptor Tyrosine Kinases Involved in Tumor Progression and Angiogenesis. Clin. Cancer Res. 9, 6088S (2003).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Zhao, Y. et al. A point cloud graph neural network for protein–ligand binding site prediction. Int. J. Mol. Sci. 25, 9280 (2024).
Zhao, Y. et al. Identification of metal ion-binding sites in RNA structures using deep learning method. Brief. Bioinforma. 24, bbad049 (2023).
Stärk, H., Dallago, C., Heinzinger, M. & Rost, B. Light attention predicts protein location from the language of life. Bioinforma. Adv. 1, vbab035 (2021).
Fang, X. et al. Geometry-enhanced molecular representation learning for property prediction. Nat. Mach. Intell. 4, 127–134 (2022).
Zhao, Y. Evidential deep learning-based drug-target interaction prediction. zhaoyanpeng208/EviDTI. Zenodo https://zenodo.org/records/15760471 (2025).
Acknowledgements
The authors would like to thank Jingjing Wang for her help. The authors sincerely appreciate the support of the Partners. All individuals have consented to the acknowledgment. This work was supported by the National Key R&D Program of China (Grant No. 2023YFC2604400, 2024YFA1307700) and Natural Science Foundation of Shanghai (Grant No. 25ZR1402171). Funding was provided to S.H. (2023YFC2604400), X.B. (2024YFA1307700), and Y. Zhao (25ZR1402171).
Author information
Authors and Affiliations
Contributions
Y. Zhao, Y.X., C.W., S.H., D.Z., and X.B. conceived the study; Y. Zhao, Y.X., Y. Zhang, D.Y., M.W., H.X., and S.L. performed the experiments; Y. Zhao, H.Z., Z.L., G.Z., M.L, Z.C., and X.W. conducted the surveys; Y. Zhao, Y.X., L.W., and R.L. collated the data; Y. Zhao, Y.X., Y.W. and Y. Zhang performed the writing-primer preparation; S.H., D.Z., and X.B. performed the writing-reviewing and editing, and S.H., D.Z., P.Z., and X.B. supervised the study. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Brad Haubrich, Guishen Wang, and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhao, Y., Xing, Y., Zhang, Y. et al. Evidential deep learning-based drug-target interaction prediction. Nat Commun 16, 6915 (2025). https://doi.org/10.1038/s41467-025-62235-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-62235-6
This article is cited by
-
An Improved Machine Learning Model for Screening and Activity Prediction of Receptor Tyrosine Kinase
Journal of Bionic Engineering (2026)
-
An uncertainty-driven gated feature selection network (UGFS-Net) for TG level prediction: linking high-altitude exposure to lipid metabolism disorder via elevated TG
Lipids in Health and Disease (2025)
-
Applications of artificial intelligence in non–small cell lung cancer: from precision diagnosis to personalized prognosis and therapy
Journal of Translational Medicine (2025)