Abstract
Ataxia telangiectasia and Rad3-related kinase (ATR) is a rational radiosensitization target. In this study, we explore the combination of the ATR inhibitor, ceralasertib, and palliative radiotherapy, with primary endpoint the identification of maximum tolerated dose, and secondary endpoints the determination of adverse event causality, pharmacokinetics (PK) and anti-tumor activity. Twenty-seven patients were dosed in escalating dose cohorts from 20 to 80 mg twice daily (BD) with concomitant radiation, 20 Gy in 10 fractions or 30 Gy in 15 fractions. Patients were assessed for acute and late toxicities and response after therapy. A non-tolerated dose was not reached. Maximum administered dose was 80 mg BD ceralasertib over 3 weeks with 30 Gy in 15 fractions, at which 1/6 evaluable patients had dose-limiting toxicities (radiation dermatitis and mucositis). PK was comparable to monotherapy. Of 23 efficacy-evaluable participants, 2 (9%) had complete response (CR), 6 (26%) partial response (PR), 13 (57%) stable disease (SD) and 2 (9%) progressive disease (PD) as best response in irradiated tumors. Response was not clearly linked to genomic aberrations. Increased T and natural killer cell activation as observed in peripheral blood as treatment progressed.
Similar content being viewed by others
Introduction
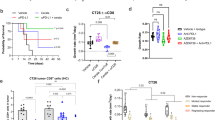
Radiotherapy (RT) is an essential part of cancer treatment. Despite treatment intensification with dose-escalation, acceleration, and concomitant chemotherapy, cure rates require improvement. Inhibitors of the DNA damage response (DDR) improve radiotherapy efficacy in preclinical models1. Ataxia Telangiectasia and Rad3-related (ATR) is a critical kinase in the DDR, being activated by DNA damage and replication stress to cause cell cycle arrest and DNA repair2 (Fig. 1A). ATR inhibition (ATRi) is an attractive combination partner with RT because it offers tumor-selective radiosensitization, mainly targeting the G2/M cell-cycle checkpoint, which is the primary functional checkpoint in G1/S-checkpoint defective cancer cells3 (Fig. 1B). ATRi increases the DNA damage load from radiation and enhances cell death. ATRi combined with RT also significantly modulates the immune tumor microenvironment in preclinical models, augmenting RT-induced inflammation4,5,6.
A Schematic of ATR function. ATR phosphorylates CHK1, on Serine 345, after DNA damage or replication stress. Activated CHK1 causes a predominantly G2 cell cycle arrest. Ceralasertib prevents ATR activating CHK1. B Schematic for tumor selectivity. Normal cells have intact G1 and G2 cell cycle checkpoints, most tumor cells have impaired G1 checkpoint control and will, therefore, be more at risk from G2 checkpoint inhibition when combined with DNA damage. C Study schema for drug and radiation dosing, PD sampling, response and toxicity assessments. Blue boxes: lead-in and post-radiotherapy ceralasertib; green boxes: radiotherapy treatments, grey outline: weekend with ceralasertib and no radiotherapy. D Schema for escalation of ceralasertib and radiotherapy (RT) doses. RT dose in Gy, (2 Gy fractions); ceralasertib dose in mg BD. E Late toxicities identified through medical notes review, AE reporting or LENT-SOMA assessment, compared with radiation dose-volume parameters. All indicated participants received ceralasertib 80 mg BD and 30 Gy in 15 fractions radiation. Density indicates length of available follow-up for that participant, and color indicates toxicity grade. Dmax maximum point dose, Dmean mean organ dose, VxGy volume of organ receiving x Gy, D10cc dose to most irradiated 10 cc of organ. Source data are provided online.
Ceralasertib (AZD6738, AstraZeneca) is an orally bioavailable, potent and selective inhibitor of ATR. Preclinical studies of ceralasertib and radiation have confirmed tumor sensitisation1. We have recently reported monotherapy activity of ceralasertib in the PATRIOT Part A/B study, with modulation of pharmacodynamic markers at the doses used here7. It is currently being investigated in combination with chemotherapy, PARP inhibitors and immunotherapy.
Here, we report the first study of ceralasertib combined with palliative RT8 using 2 Gy-per-fraction schedules typically employed during curative-intent RT. We show that ceralasertib is well tolerated in combination with palliative RT, and that combination therapy results in durable responses in areas irradiated to low doses. We also observe modulation of pharmacodynamic biomarkers, and evidence of systemic and intratumor immunomodulation.
Results
Ceralasertib with palliative radiotherapy was well tolerated
Twenty-seven patients were dosed between January 2016 and December 2020 and included in the safety analysis (6 in the below-diaphragm cohort and 21 in the above-diaphragm cohort). Of these, 22 were considered evaluable for DLT (completed at least 80% of planned treatment) and 23 for response (completed at least one response assessment) (Fig. 1D). Four did not complete planned treatment and were replaced, 2 due to grade 3 thrombocytopenia, 2 due to patient choice with low grade toxicities. A further patient progressed clinically before the first response assessment. Administered radiation dose was 20 Gy in 10 fractions in 13 participants and 30 Gy in 15 fractions in 9 patients. Baseline characteristics are given in Table 1. Radiation details are in Supp. Table 1.
DLT was defined as G4 hematological, G3 non-hematological (see supplementary data for full details) toxicity, G3 or G4 radiation-induced skin or mucosal toxicity or the inability to tolerate one week of combination therapy due to toxicity. There was one DLT at the highest administered dose of 80 mg BD ceralasertib and 30 Gy in 15 fractions RT with all RT fractions sensitized. The DLT was grade 3 mucositis and dermatitis, causing dysphagia, hypokalemia and hypophosphatemia requiring hospital admission in a patient treated for oropharyngeal squamous cell carcinoma.
Adverse events felt possibly, probably or definitely related to ceralasertib and/or radiotherapy are listed in Table 2. The profile of toxicities was mainly hematological. There were mild radiation reactions in the majority of participants, with G1-2 dermatitis, mucositis, esophagitis observed, but these were not felt to be significantly greater than would be observed with radiation alone. At 30 Gy with 3 weeks’ ceralasertib, 3/8 participants had G3 toxicity, however only 1 participant had G3 radiation sensitization (DLT patient above), with the remainder experiencing hematological toxicity. Non-DLT SAEs related to the therapy were G3 radiation-induced mucositis and dermatitis (the DLT at 80 mg/30 Gy), which was associated with G3 dysphagia and hypokalemia, and one episode of febrile neutropenia, with G1 neutropenia and thrombocytopenia (80 mg/30 Gy).
Due to the COVID pandemic and slow recruitment, and the acquisition of sufficient data to support an ongoing, curative-intent phase II study (ADEPT-DDR, EudraCT 2020-001034-35), and evidence of radiosensitization in the AE profile (Fig. 1D, E, see below), further dose levels were not tested beyond 80 mg BD. The exacerbation of radiation-induced normal tissue toxicity in one participant indicates that ATR also plays a role in normal tissue responses to DNA damage, possibly by reducing DNA repair or increasing DNA damage-associated replication stress (Fig. 1A, B).
Median follow-up from start of RT to last clinical review was 151 days (range 31–2054 days). CTCAE and LENT-SOMA toxicities are shown in Fig. 1E and Supp. Fig. 1, which describe the radiation dose metrics to organs-at-risk, the length of available follow-up for late toxicity monitoring and the maximum grade of recorded toxicity. One patient being treated for a lung tumor (with 30 Gy and 80 mg BD, 10 fractions sensitized) developed CTCAE G2 pneumonitis at 3 months after the start of radiation, which responded to steroids. A significant volume of lung was irradiated, with lung V20Gy 33.5% and mean lung dose 11.3 Gy. Eight patients completed at least one year of follow-up and none had severe late toxicity. Nine patients completed at least 1 LENT-SOMA late toxicity assessment9. The relevant domains are shown in Supp. Figs. 2–4. Notable late toxicities included G2-3 salivary dysfunction, and LENT-SOMA G3 lung fibrosis (defined as dense confluent radiographic changes limited to the radiation field) at day 108, which resolved to G2 by day 212, with resolution of symptoms at day 430. Lung V20Gy was 21.5% and mean lung dose 7.3 Gy.
At the time of data preparation in October 2023, one patient remained alive, with a follow-up of 43 months.
Evidence of target modulation
Ceralasertib PK has been previously reported7 (Supp. Fig. 5). Post-dose plasma ceralasertib concentrations at 40 mg and pre-dose at 80 mg (Fig. 2A) compared favorably to levels required for radiosensitization in vitro1 (radiosensitization of clonogenic assay seen at 0.1 µM, equivalent to 41 ng/mL), and were similar to those observed with monotherapy dosing7.
A Ceralasertib PK. PK samples were taken on the days of PD sampling, pre- and 4–8 h post-dose. Individual points are shown, line = mean, error = 95% confidence interval. B PBMC phospho(S345)Chk1 immunofluorescence intensity, plotted as fold-change vs. baseline levels, pre-fraction 1 (after 3–7 days ceralasertib dosing) and pre-fraction 2 (after a single fraction of 2 Gy radiation), by dose cohort, line = median, box = interquartile range, whisker = range. n = 3 (20 mg BD), n = 4 (40 mg BD), n = 9 (80 mg BD). C PBMC phospho(S345)Chk1 immunofluorescence intensity, individual points as shown in (B). Size of points = dose (mg BD), color indicates corresponding pre-dose plasma ceralasertib level, if available (grey: not available). * = P = 0.035 by two-sided Wilcoxon Signed Rank test, versus a theoretical median of 1. Data includes one-sample where pre-F1 and pre-F2 samples were mixed, this has been included in both pre-F1 and pre-F2 columns. Source data for this figure (A–D) are provided online. D γH2AX foci in skin punch biopsies, absolute count of foci per nucleus, median of 765 (range 72–2204) nuclei were quantified per patient per time point, number of nuclear foci per nucleus displayed. * = P = 0.024 by unpaired t-test (two-tailed), comparing pre-F2 to baseline. E Representative micrographs of skin-punch biopsies stained for γH2AX by IHC. Left = baseline; middle = pre-fraction 1; right = pre-fraction 2. Scale bar = 50 μm. F Tumor pharmacodynamics in paired tumor biopsies before treatment and prior to fraction 2. Left: change in γH2AX % positivity; right: change in phospho(S635)Rad50 % positivity. G Example micrographs for the tumor biopsies quantified in (G). Scale bar = 100 μm.
PBMC were analyzed for pharmacodynamic markers of ATR inhibition. Sampling before the first RT fraction (after 3–7 days of ceralasertib) confirmed reduced phosphorylation of Chk1, a downstream target of ATR, and higher dosing led to more profound suppression (Fig. 2B, C). Before fraction 2 (16–24 h after fraction 1), there was an increase in pChk1 in many participants, likely reflecting DNA damage from radiation (Fig. 2C). Levels of γH2AX on PBMCs, a marker of DNA damage, were not significantly different between groups (Supp. Fig. 6).
Skin punch biopsies were analyzed for γH2AX foci, which are markers of DNA double-strand breaks. Ex-vivo experiments showed that these should approach baseline levels 18 h after irradiation (Supp. Fig. 7A). There was a significant increase in foci per nucleus before fraction 2 (Fig. 2D, Supp. Fig 7B), with a median fold-change of 1.49 before fraction 1 and 1.94 before fraction 2 (example images in Fig. 2E).
One set of paired tumor biopsies was analyzed and showed increased phospho-Rad50, a PD marker of ATR inhibition10, and increased γH2AX before the second RT fraction (Figs. 2F, G). Monotherapy tumor biopsies have previously shown pRad50 modulation at doses above 40 mg BD7.
Durable responses with ATRi + low-dose radiotherapy
Separate response assessments were performed for the irradiated lesions and the overall response, given that the majority of participants had other metastatic disease which was not treated. In irradiated lesions, of 23 evaluable participants (who had at least 1 response assessment), 2 (9%; 95% CI: 2–31%) had RECIST complete response (CR), 6 (26%; 95% CI: 12–49%) partial response (PR), 13 (57%; 95% CI: 35–76%) stable disease (SD) and 2 (9%; 95% CI: 2–31%) progressive disease (PD), with overall best RECIST responses (irradiated and unirradiated lesions) 1 (4%; 95% CI: 1–27%) CR, 5 (22%; 95% CI 9–44%) PR, 13 (57%; 95% CI 35–76%) SD and 4 (17%; 95% CI: 6–40%) PD. Examples of responses are shown in Fig. 3A–E.
A–E example responses with corresponding radiotherapy plans. Legend indicates the isodose levels, expressing the percentage of prescribed radiation dose (either 20 or 30 Gy). Red arrowheads indicate the tumor. A metastatic squamous cell carcinoma of the larynx, mediastinal node treated with 20 Gy and 20 mg BD ceralasertib, best response was PR. B metastatic SCCHN, acetabulum metastasis treated with 20 Gy and 40 mg ceralasertib, best response was PR. C SCCHN (hypopharynx) treated with 20 Gy and 40 mg ceralasertib, best response was PR and later clinical CR. D SCCHN (oropharynx) treated with 30 Gy and 80 mg ceralasertib, best response was PR. E SCCHN treated with 30 Gy and 80 mg ceralasertib, best response was PR. F spider plot of change in diameter, compared to baseline measurement, for irradiated lesions only. G waterfall plot, of change in diameter, compared to baseline measurement, for irradiated lesions only. H Swimmer plot, indicating clinical course for each participant.
Tumor measurements for irradiated and unirradiated lesions are shown in Fig. 3F, G and Supp. Figs. 8 and 9. Measurements prior to starting the study were available in the majority of participants to allow plotting of the kinetics of tumor growth pre-treatment. These data showed a reduction in the rate of tumor growth in the majority of irradiated tumors (Supp. Fig. 10). There were no so-called abscopal responses outside of the treatment field. Four patients had all visible disease encompassed in the RT field, these included:
-
A patient with squamous cell cancer of the hypopharynx (HPV or p16 status unknown) treated with 20 Gy in 10 fractions and 40 mg BD (Fig. 3C). He showed an initial radiological PR and a later clinical CR, with tumor controlled for more than 5 years. He ultimately died of a lung malignancy in the context of heavy smoking history.
-
A patient with sebaceous carcinoma of the maxilla (MMR proficient by IHC, with normal FISH studies for sarcoma breakpoints), treated with 20 Gy in 10 fractions and 20 mg BD. He showed a best response of SD for 4 months and remains alive at the time of data cut-off.
-
A patient with small cell lung cancer (consolidation radiotherapy after second-line chemotherapy), treated with 20 Gy in 10 fractions and 80 mg BD. He showed a best response of SD lasting 2 months.
-
A patient with recurrent adenocarcinoma of maxillary antrum involving skull base and neck, treated with 30 Gy in 15 fractions and 80 mg BD for all fractions. He showed SD for 3 years.
At the last follow-up, 17/22 (77%) of evaluable patients had tumor control within the irradiated field, with a median follow-up of 158 days (range 31–2054). Overall survival is shown in Supp. Fig. 11.
Radiosensitization is independent of genetic background
Tumor profiling was possible for 21 participants (Fig. 4A). Tumor DNA sequencing, either whole exome or a targeted panel of ATR-relevant genes (Supp. Table 2), was available for 8 participants (4 with CR/PR and 4 with SD). Plasma sequencing for a panel of relevant genes (Supp. Table 3) was available for a further 13 (4 PR, 8 SD and 1 PD). TP53 mutation was the only abnormality in the patient with CR. Of 7 patients with PR, 6 had a genomic aberration of interest. The participant with PD had a TP53-mutant tumor. Overall, there were no clear biomarkers of response based on tumor sequencing. Protein expression for ATM, Ki67, Cyclin E and ARID1A was only available for a limited number of patients (Supp. Table 4): no ATM or ARID1A loss was detected, 2 tumors (1 PR and 1 SD) had high cyclin E expression.
A Oncoprint of DNA sequencing data for 20 participants. Tumor: whole exome sequencing (“exome”) or customized panel (“panel”; gene list in Supp. Table 2). Plasma: customized panel (gene list in Supp. Table 3). SLD: change in sum of longest diameter of irradiated lesion. Histology: H&N other: other non-squamous head and neck tumors. Ceralasertib dose in mg BD. RT: radiation dose in Gy. BOR: best overall response (by RECIST: CR complete response, PR partial response, SD stable disease, PD progressive disease). G1: genes involved in G1 cell-cycle checkpoint control. DDR: genes involved in DNA damage repair pathways. B Quantification of tumor-infiltrating lymphocytes (TIL) in H&E sections of available tumors. TIL density was quantified in tumor and stromal regions: 3 tumors with CR/PR and 5 with SD or PD were quantified, whole tumor area lymphocyte count. * = P = 0.036 by two-sided Mann–Whitney test. Source data are provided online. C Volcano plot of differentially expressed genes in on-treatment (pre-fraction 2) vs. baseline tumor biopsies for n = 2 paired tumors. Fold-change of treatment versus control: effect size calculated in DESeq2 R package; p-value: Wald test p-value with Benjamini–Hochberg adjustment. Genes of interest indicated. D Gene set enrichment analysis performed for the data shown in (C) using the “hallmarks” gene set. NES normalized enrichment score, FDR Q False discovery rate Q value, EMT epithelial-mesenchymal transition. E Cell-type deconvolution of RNAseq data for the 2 available paired biopsies using CIBERSORTx. Mean fold-change vs. baseline cell-type scores is shown. Source data are provided online. F Cell-type deconvolution of cell type infiltration using mMCPcounter at day 3 and day 10 after irradiation in a mouse model of head and neck cancer. Animals were treated with 4 fractions of 2 Gy and ATRi 25 mg/kg/day for 7 days, starting 2 h before the first fraction of radiation, 3 animals per group. Mean fold-change in cell-type scores vs. vehicle-treated animals for the same time-point is shown. Error bars = SEM. Source data are provided online.
We have previously observed that tumors with higher baseline inflammation are more likely to respond to ATRi monotherapy. Tumor-infiltrating lymphocytes (TIL) were quantified on H&E sections for 8 participants, 3 with PR (including 1 with durable clinical CR), 4 with SD and 1 with PD. There was significantly more stromal TIL infiltration in responding patients (Fig. 4B).
TME modulation by ATRi-RT
Two paired tumors were available for RNA sequencing (RNAseq), with biopsies taken at baseline and prior to the second fraction of RT (16–24 h after the first fraction). These were from one participant with oropharyngeal SCC and a PR, and one with recurrent laryngeal SCC and CR. In contrast to data from ATRi monotherapy, there was downregulation of a number of transcripts at this early time point (prior to the second RT fraction) (Fig. 4C). These included G2/M checkpoint, as anticipated with this combination, and downregulation of a number of immune pathways, including interferon signaling, inflammatory response and allograft rejection signature, all of which are shown to be upregulated at later timepoints in preclinical models of this combination4,6. Of note, there was upregulation of pathways involved in TNF-alpha signaling, unfolded protein response, and p53 signaling (Fig. 4D), as well as a number of matrix metalloproteinases and chemokines. Cell-type deconvolution indicated an increase in natural killer (NK) and some macrophages at this early time-point (Fig. 4E). RNAseq from the same combination in a mouse model of head and neck cancer, from sampling at day 3 after completion of therapy6, indicated that there was significant inflammatory infiltrate. Those data demonstrated an abundance of macrophages and NK cells compared with controls, and that adaptive immune cells do not infiltrate until a later time (Fig. 4F and Supp. Fig. 12). The mouse data also share similarities with the human samples, including upregulation of TNF-alpha signaling, epithelial-mesenchymal transition and downregulation of G2/M cell cycle checkpoint, and suggest that stromal factors may be upregulated at early time points following combination treatment (Supp. Figs. 13–15). Taken together, these data support further studies of the microenvironmental response to ATRi-RT, in particular early stress responses, innate immune activation and adaptive immune cell depletion, before a subsequent repopulation with adaptive immune cells.
Immunomodulation with ATRi-RT
Due to multiple lines of preclinical data suggesting that ATRi with RT can modulate the tumor-immune microenvironment, and clinical evidence of immunomodulation, we profiled immune changes in peripheral blood with therapy in 7 trial participants, all of whom underwent treatment with 80 mg BD ceralasertib and 30 Gy (all fractions sensitized). Blood samples were analyzed by flow cytometry before treatment, prior to RT fraction 1 (after 3–7 days of ceralasertib), and prior to fractions 6 and 11 (after 1 and 2 weeks of RT, respectively). Of the 7 subjects, there were 3 with best response of PR/CR, 3 with SD and 1 not evaluable for response.
The total proportion of circulating immune cells in the lymphocyte compartment did not significantly change, with a trend toward increased CD8-positive and unconventional T-cells (Fig. 5A and Supp. Fig 16A). When markers of activation were examined, there was evidence of activation of NK cells and both CD8-positive and unconventional T-cells (Fig. 5B). Specifically, as treatment proceeded: CD69 expression progressively and significantly increased on CD8-positive T-cells and NK cells (Fig. 5C, D); CD95 (Fas) was progressively elevated on CD8-positive T-cells and NK cells (Fig. 5C); there was a trend towards increased NKG2A expression on NK cells (Fig. 5E). The modulation of some immune cells seemed dependent on ceralasertib, and others dependent on ceralasertib with RT: when compared with samples from participants treated with ceralasertib monotherapy (160 mg BD from PATRIOT Part A/B), combination with RT led to a significant increase in activated CD8 T-cells, as measured by CD69 expression (Fig. 5C) and there was a trend towards activation of NK cells, as measured by NKG2A expression (Fig. 5E). Increased expression of TIM-3 was observed on unconventional T-cells (Supp. Fig. 16D). Nonsignificant changes were observed in the frequencies of multiple activation markers on T and NK cells (Supp. Fig. 16B–E) as well as monocyte subpopulations (Supp. Fig. 16F). We detected significant upregulation of PD-L1 on circulating monocytes (Supp. Fig. 16E). When analyzed by response, patients with tumor shrinkage (CR and PR) had a nonsignificant trend towards increased frequency of NKG2A and PD-1 double-positive circulating CD4+ and CD8 + T-cells compared with those who did not have a shrinkage (RECIST SD, Fig. 5F, G).
A Percentage of indicated cell types in lymphocyte gate at baseline, before fraction 1 (“Pre-F1”, after 3–7 days’ ceralasertib); fraction 6 (“Pre-F6”, after 1 week of radiation with ceralasertib); fraction 11 (“Pre-F11” after 2 weeks of radiation with ceralasertib). B average Log2 fold-change versus baseline sample of indicated activation markers on CD8, CD4, unconventional (CD3+CD4−CD8−) T cells and NK cells. C Change in percentages of CD8-positive T-cells and NK cells positive for indicated activation markers, CD95 and CD69. Each point represents an individual patient (n = 6–7 patients per group), percent positivity of indicated subpopulations of lymphocytes. Statistical significance: mixed effects model with Geisser-Greenhouse correction and Fisher’s Least Significance Difference test to compare time points without multiple comparisons testing. D Fold-change versus baseline in percent CD69-positive CD8 cells. “Day 14” is fold-change vs. baseline for participants treated with ceralasertib 160 mg BD monotherapy in PATRIOT Part A/B study (n = 8). Each point represents an individual patient (n = 6–7 patients per time point), fold-change percent positivity versus individual baseline sample. Two-sided p-values: one-sample t-test against hypothetical median of 1. Line: mean; error bars: SEM. E Fold-change versus baseline in percent NKG2A-positive NK cells. “Day 14” is fold-change vs. baseline for participants treated with ceralasertib 160 mg BD monotherapy in PATRIOT Part A/B study (n = 8). Each point represents an individual patient (n = 6–7 patients per time point). Two-sided p-values: one-sample t-test against a hypothetical median of 1. Line: mean; error bars: SEM. F Representative flow cytometry plot showing change in NKG2A- and PD-1-positive CD8 T-cells with treatment (baseline and before the 6th fraction of radiotherapy). G Upper row: change in mean percentage of NKG2A- and PD-1-positive CD8 T-cells (upper left) and CD4 T cells (upper right). Lower row: fold-change in proportion of NKG2A- and PD-1-positive CD8 (lower left) and CD4 (lower right) T-cells. Error bars: SEM. Source data for this figure (A, B–E, G, H) are provided online. H Plasma cytokines were measured at baseline, prior to fraction 1 and prior to fraction 2. Fold-change versus baseline is plotted, color indicates best overall response of irradiated lesions. * = two-sided p = 0.013 by Wilcoxon Test vs. a hypothetical median of 1.
Analysis of circulating cytokines in 7 participants (4 with PR/CR, 2 with SD and one not evaluable) showed an increase in CCL3 with combination treatment and modulation of other cytokines, without a clear correlation with response (Fig. 5H).
Discussion
In this work, we have established that ceralasertib 80 mg BD is a tolerable dose in combination with 30 Gy in 15 fractions of 2 Gy. This dose effectively modulates ATR signaling and is associated with durable responses, including cure of an HNSCC with 20 Gy in 10 fractions. There was little unexpected toxicity, with mainly hematological toxicities related to ceralasertib. We did observe increased normal tissue reactions to radiotherapy in some patients, together with persistent γH2AX foci in normal skin within the radiation field, in keeping with the status of ceralasertib as a radiosensitizer and indicating a potential increase in acute toxicity, but no severe late toxicities were identified in patients followed for a median of about 6 months.
Limitations of our data include the lack of identification of MTD, and the small numbers of patients with extended long-term follow-up to establish a better estimate of late toxicity. Recruitment of participants undergoing palliative RT was challenging. Studies currently recruiting with RT and ATRi are taking advantage of newer study designs, such as the time-to-event continual reassessment model which formally build late toxicities into dose-escalation decisions.
RECIST response rates to palliative RT are not well described, given that the goal is symptom control rather than tumor volume change. As a result, there are no good studies evaluating RECIST response rates from RT doses equivalent to those employed in this trial.
Monotherapy activity of ATRi has been associated with oncogene activation, mutations in DDR genes, and loss of proteins such as ATM and ARID1A11,12. There are some preclinical data suggesting that ATM-deficient, but not BRCA1-deficient tumors may show greater radiosensitization13. We did not find any clear signals of predictive biomarkers in combination with RT in this relatively small population of patients, most likely because the mechanism of sensitization to exogenous DNA damage is different from monotherapy ATR inhibition. It seems likely that those tumors with more reliance on G2 cell-cycle checkpoint would be more at risk, but given that this is a near-universal feature of cancers, it may be difficult to identify a definitive biomarker. The cure of a patient with head and neck cancer with no clear mutations and p53-wild-type status is a good example of the ability of the RT-ATRi combination to achieve remarkable results without a molecularly-defined basis for the response. The consistent signal that more inflamed tumors seem to benefit in both the monotherapy and combination settings is intriguing and demands further investigation. Higher numbers of TILs in responding tumors could represent a genomically unstable tumor (potentially susceptible to RT/RT-drug combination) or could indicate an immune-related mechanism of action of this combination.
It is increasingly recognized that RT has an immunomodulatory effect and that outcomes from treatment are influenced by the immune microenvironment14,15. Modulation of the DDR results in amplification of nucleic acid sensing pathway effects and the resultant downstream influence on immune infiltration. In preclinical models, increases in inflammation after ATRi-RT have been demonstrated4,5,16,17. In our data, we observed immune modulation in circulating immune cells after at least one week of treatment, and the effect increased with time. We noted an increase in T-cells positive for the activation/exhaustion marker, PD-1, and the inhibitory receptor NKG2A, which were only elevated in responding participants. Crucially, this has led to in vivo studies showing the benefit of dual NKG2A-PD-1 targeting as adjuvant therapy after ATRi-RT17.
In tumor biopsies after just one fraction of RT, we observed a reduction in many adaptive immune-related transcripts and an upregulation of stromal and myeloid factors, suggesting this may be an important part in the initial response to ATRi-RT. Few studies have examined tumor biopsies at early time points after irradiation, with available data suggesting that conventionally-fractionated RT leads to a depletion of immune cells18, with inconsistent results from high dose-per-fraction radiation19,20. Window-of-opportunity studies have also suggested an early suppression of T-cell proliferation, followed by a rebound repopulation with enhanced activity after ATRi monotherapy21, suggesting this may be a feature in both ATRi and ATRi-RT treatment. Whether the extensive changes in immune populations suggested by our data lead to a favorable immune repopulation requires further investigation with larger sample numbers but it is reasonable to hypothesize, based on other preclinical and clinical data showing ATRi effect on immune populations22, as well as the data from our study that the ATRi-RT combination may deplete resident anergic/exhausted or suppressive populations and make space for repopulation with new, vigorous T-cells. The increase in peripheral immune activation as treatment progressed suggests that this may be the case. There is also evidence that the scheduling of these therapies will be crucial to maximizing the effect on the immune microenvironment, with prolonged treatment associated with more immune cell depletion23. We and others have demonstrated that ATRi monotherapy modulates both tumor and peripheral immune cells, and preclinical data suggested that combination with radiation further enhances this phenomenon4. Critically, we also observed an increase in NK-related transcripts. When taken together with the activation seen in circulating NK cells with treatment, and preclinical data suggesting NK targeting together with ATRi-RT may be beneficial6, we believe this maybe a promising immune combination target for future studies.
A number of studies are treating patients with combination ATRi and RT, including the ADEPT-DDR study of ceralasertib with curative-intent radiotherapy in platinum non-eligible HNSCC patients (EudraCT 2020-001034-35), the CONCORDE study of ceralasertib with radical radiotherapy in lung cancer24, and the CHARIOT study with palliative chemotherapy and radiotherapy in esophageal cancer25,26. A phase I study of the intravenous ATRi, berzosertib, with radical chemoradiotherapy in HNSCC has recently completed27. ATRi are also being tested in combination with cytotoxic chemotherapies28, targeted therapies29 and immunotherapy30,31.
Other drugs targeting the DDR are in various stages of clinical development with radiotherapy, including those targeting PARP, ATM, DNA-PK, and WEE1. It remains to be seen which, if any, of these options is the most viable. Targeting ATR has, in theory, advantages through its predominant G2 checkpoint-targeted effect, although the increased normal tissue reactions seen here indicate that the selectivity may not be absolute.
In summary, ceralasertib is a promising radiation combination partner. The next wave of studies with this combination is currently combining with radical doses of RT alone. Future studies need to assess the potential of combining with standard-of-care chemoradiotherapy regimens. Future translational work should be directed at providing a deeper understanding of the immunomodulatory effects of ATRi as single agents and alongside RT, chemoradiotherapy and combinatorial regimens incorporating immune checkpoint inhibitors.
Methods
Ethics statement
The study was ethically approved by the NHS NRES Committee London—City and East, reference 14/LO/0465. All patients gave written informed consent. The study design and conduct complied with all relevant regulations regarding the use of human study participants and was conducted in accordance with the criteria set by the Declaration of Helsinki. The study was registered with clinicaltrials.gov in August 2014 (NCT02223923), all participants were enrolled after this date. The full study protocol is available with this manuscript in supplementary note 2.
Patient population
Eligible participants were aged 18 or over, with advanced solid tumors and ECOG performance status of 2 or better, adequate organ function and an indication for palliative RT. Full inclusion and exclusion criteria are detailed in supplementary materials.
Study design
This was part of a 3-part study of ceralasertib. Dose escalation and expansion of ceralasertib monotherapy have been previously reported (parts A and B)7. Part C is the RT combination component of the study which ran in parallel, at least two dose cohorts behind the monotherapy phase. Primary objective was feasibility and safety of ceralasertib in combination with RT. Secondary objectives were to guide dose and schedule selection for future studies. This was a multi-center, open-label, phase I dose-escalation study, using a 3 + 3 design (Fig. 1A), where 3 participants were recruited per dose cohort, with dose escalation if there were no dose-limiting toxicities (DLT), and expansion to 6 participants if there was 1 DLT, with the maximum administered dose defined as the dose level at which 2 participants experienced a DLT, and maximum tolerated dose the dose level below, where no more than 1/6 participants had DLT. There was escalation of both ceralasertib and RT doses. Two separate arms were opened, dose-escalating separate cohorts by site of irradiation: either above or below the diaphragm, to separately define toxicity by irradiated site. The data from both arms have been presented together, due to no clear site-specific DLT and poor recruitment below the diaphragm. The dose-limiting toxicity (DLT) window ended 2 weeks after completion of RT. DLT definitions are given in the study protocol (supplementary note 2).
Study treatments
Ceralasertib
Ceralasertib was administered orally using a continuous twice-daily (BD) dosing schedule. A starting dose of 20 mg BD was selected for both monotherapy and combination arms. Ceralasertib dosing commenced a minimum of 3 and a maximum of 7 days prior to RT. Ceralasertib dosing finished 48 h after the final sensitized fraction of RT (Fig. 1C). Dose escalation used a modified Fibonacci method with the same dose cohorts used for monotherapy.
Radiotherapy
RT starting dose was 20 Gy in 10 daily fractions, restricted to weekdays and starting on a Monday. This dose was selected as comparable to standard palliative doses (8 Gy in 1 fraction, equivalent dose in 2 Gy fractions (EQD2) of 17.6Gy3 for normal tissues and 12 Gy10 for tumors; 20 Gy in 5 fractions EQD2 of 28 Gy3 and 23.3 Gy10) but divided into 2 Gy fractions, with a view to future studies of curative-intent RT. Radiation dose level 2 was 30 Gy in 15 daily fractions (Fig. 1D). As the duration of the two RT regimens was different, after radiotherapy dose escalation to 30 Gy, the first cohort received the first 20 Gy with ceralasertib and the final week of RT (10 Gy) without ceralasertib, while the final cohort received all 30 Gy with ceralasertib concomitantly (Fig. 1A). All RT delivery techniques were permissible, and standard departmental palliative RT protocols were used. All radiotherapy was CT-planned. Minimum field size was 7.5 × 7.5 cm and maximum field size was 15 × 15 cm, except for cases involving irradiation of the pelvis where 20 × 20 cm was permitted. Dose to normal structures in the field was recorded.
Study assessments
Pharmacodynamics and immunohistochemistry
Tumor biopsies were analyzed for phospho-Rad50 by IHC. Formalin-fixed peripheral blood mononuclear cells (PBMC) were analyzed for phospho-Chk1 and γH2AX by immunofluorescence as previously described7.
Skin punch from within the radiation field was taken at baseline, prior to fraction 1 and 16–24 h after fraction 1, prior to fraction 2. This was immediately fixed in formalin and embedded in paraffin after 24 h.
Bloods were taken at the same time points, and PBMC formalin-fixed for 20 min after isolation from Cell Preparation Tubes (Sodium Citrate, BD). PBMCs were analyzed by immunofluorescence for γH2AX (Abcam ab11174, 1:5000), phospho-(S345)Chk1 (Cell Signaling 2438, 1:50) and total Chk1 (Abcam ab40866, 1:100). Paired tumor biopsies were formalin-fixed and analyzed for nuclear phospho-(S635)Rad50 by IHC (Cell Signaling 14223, 1 μg/mL), using a previously published method10. H-score was calculated after pathologist assessment. Tumor samples were also analyzed by a pathologist for γH2AX (S139, Cell Signaling 9718) positivity by IHC. A positive nucleus was defined as one with >5 nuclear foci or pan-nuclear staining. Nuclear ATM (Abcam 32420) staining was assessed by H-score, but samples were only deemed acceptable if lymphocyte staining (internal control) was at least moderate. Ki67 (Mib-1, Dako M7240) was scored by percent nuclear positivity using a global unweighted method, scoring 4 areas of 100 cells. ARID1A IHC was performed as previously published32, using EPR13501 antibody (Abcam) and scored using H-score for nuclear positivity. Quantification of TIL was done using a semi-automated method in QuPath33. Scanned H&E sections were first segmented into tumor and stroma, using a random trees machine-learning classifier, for each tumor section. Nuclei were segmented using Stardist34, then stromal nuclei were sorted by size (area <20 μm2) and circularity (> 0.85), using small circular nuclei to approximate lymphocyte count, where tumor cells were small, the mean nuclear hematoxylin intensity was added to separate lymphocytes (mean > 0.235). For skin punch γH2AX quantification, nuclei were counted automatically in QuPath and nuclear foci were counted manually.
Pharmacokinetics
Blood for pharmacokinetic (PK) analysis was taken pre- and 4–8 h post-dose on the days of pharmacodynamic (PD) sampling.
Plasma samples (50 μL) were analyzed using validated bioanalytical methods for ceralasertib and its metabolite AZ13368982, after the addition of deuterated internal standards, by protein precipitation followed by reversed-phase high-performance liquid chromatography with tandem mass spectrometric detection (HPLC-MS/MS). Concentrations of each analyte were calculated with reference to a calibration series covering the concentration ranges 41.3–41,300 ng/mL and 3.98–3980 ng/mL for ceralasertib and AZ13368982, respectively, constructed by adding known amounts of each to control human plasma and processing these standards in parallel with the trial samples. Both pre- and in-study validation was successfully conducted according to the FDA’s Guidance for Industry Bioanalytical Method Validation35. Evaluated PK parameters for ceralasertib included area under the plasma concentration–time curve (AUC), maximum observed plasma concentration (Cmax), time to Cmax (tmax), and apparent terminal half-life (t1/2) calculated as ln/lambda-z where lambda-z (λz) is the apparent terminal phase rate-constant estimated by linear regression of logarithmically-transformed concentration-versus-time data. A minimum of three data points were used in calculating λz as per AstraZeneca standard operating procedures. Data from multiple dosing were used to derive accumulation ratios based on Cmax (Race Cmax) and AUC(0–8) (Rac AUC0-8) defined as the ratio between Cmax or AUC(0–8) after multiple dose and Cmax and AUC(0–8) at day 1. All PK parameters for ceralasertib were derived using non-compartmental analysis method in Phoenix WinNonLin v8.3 software or higher where the “linear up/log down trapezoidal rule” for AUC was applied. All PK concentrations and parameters were listed and summarized as per AstraZeneca standard operating procedures by dose level and by treatment (single or multiple dose).
Sequencing
Formalin-fixed samples were assessed by a pathologist for tumor-rich areas, which were marked for microdissection at extraction, and tumor content estimated. Tumor and normal tissue (DNA extracted from buffy coats) were analyzed where possible (and for all non-archival biopsies) to remove germline variants.
Formalin-fixed paraffin embedded (FFPE) tissue slides, fresh frozen material, and frozen buffy coat samples were extracted for this study.
FFPE slides were reviewed for tumor content by a pathologist and tumor rich areas were marked for macrodissection at extraction. DNA was extracted using from five 10-micron sections of FFPE tumor samples using QIAamp DNA FFPE tissue kit (56404 QIAGEN).
FF samples were extracted using QIAamp DNA Mini kit (51304 QIAGEN).
Frozen buffy coat samples were extracted on Qiagen QIAsymphony SP instrument, using QIAsymphony DNA_Blood_400_V6_DSP protocol, and QIAsymphony DNA
Midi Kit (931255, QIAGEN).
DNA was quantified using the Qubit dsDNA High Sensitivity Assay Kit with the Qubit 3.0 Fluorometer (Invitrogen, Carlsbad, CA).
NGS libraries were prepared from 25 to 400 ng DNA using the KAPA HyperPlus Kit (Kapa Biosystems, Wilmington, MA, USA) and IDT UDI 8 bp adapters (Integrated DNA Technologies, Coralville, USA), following the manufacturer’s protocol, including dual-SPRI size selection of the libraries (250–450 bp). To optimize enrichment and reduce off-target capture, pooled, multiplexed, amplified pre-capture libraries (up to 20 samples per hybridization) were hybridized overnight using 1 µg of total DNA to a custom design of DNA baits complementary to the genomic regions of interest (NimbleGen SeqCap EZ library, Roche, Madison, WI, USA). Hybridized DNA was PCR amplified and products purified using AMPure XP beads (Beckman Coulter, Danvers, MA, USA) and quantified using Qubit dsDNA High Sensitivity Assay Kit with the Qubit 3.0 Fluorometer (Invitrogen, Carlsbad, CA), and High Sensitivity D1000 TapeStation (Agilent, Santa Clara, USA).
Samples were captured using a targeted capture panel (DDR panel) consisting of 173 genes, including multiple potential sensitizers to ATRi, such as DDR genes, and oncogenes (Supplementary Table 2).
Sequencing was performed on a NextSeq (Illumina, San Diego, CA, USA) with 75 bp paired-end reads and v2 chemistry, or NovaSeq6000 with 100 or 150 bp paired-end reads and v1 chemistry, according to the manufacturer’s instructions.
Sequencing runs were analyzed using an in-house pipeline. For the demultiplexing, Illumina bcl2fastq was used to assign reads for each sample based on the sequencing of 8-bp unique dual indexes. The reads were aligned to the reference genome build GRCh37/Hg19 using Burrows–Wheeler Aligner (BWA-MEM), followed by the marking of PCR duplicates and calculation of various quality control (QC) metrics using Picard. Genome Analysis Toolkit was used for realigning around known indels to improve indel calling and base quality score recalibration for adjusting systematic errors made by the sequencer when estimating quality scores of each base call. HaplotypeCaller is used for variant calling in germline sample (limit of detection ~10%) and Mutect2 is used for tumor-normal paired somatic analysis (limit of detection ~5%). Mutect2 tumor only mode is used for tumor only somatic analysis (limit of detection ~5%). VCF files were then annotated using oncotator (for samples pre-2019) and Personal Cancer Genome Reporter (for post 2019 samples). The potential mutations identified by in-house pipeline were further checked manually on IGV. Copy number variant was estimated by generalizing the coverage expected for a copy of any given targeted region (i.e.,an exon), taking the average coverage across all captured regions to estimate the average coverage of one targeted region. Any ratio below 0.5-fold was defined as a potential deletion, whereas a ratio above 2.4 was flagged as a potential amplification if 80% of the target regions had exceeded the thresholds. Manta and Pindel was used for the detection of structural variants including large indels, potential fusions and ITDs.
For whole exome sequencing, genomic DNA was extracted from Buffy Coat using the Qiagen DNeasy Blood & Tissue Kit, from Frozen Solid Tumors using the Qiagen All Prep DNA/RNA Micro Kit, and from FFPE samples using the Covaris truXTRAC FFPE DNA Kit.
All genomic DNA (200–1000 ng) was fragmented to 200 bp using a Covaris E Series and the resultant libraries were subjected to DNA Capture using SureSelect XT Human All Exon v5 or v6 kit (Agilent) following the manufacturer’s instructions.
Final libraries were quantified using qPCR and clustered at a molarity of 14.5 pM; sequencing was performed on an Illumina HiSeq 2500 using 2 × 101 cycles of version 2 RAPID SBS chemistry. Tumor samples were sequenced at 100–150× depth and germline samples at 40–60×. Tumor mutational burden was defined as total number of somatic mutations, including synonymous mutations, divided by the library panel size. Tumor mutational burden was defined as high (≥ 20 mutations/Mb), intermediate (10–20) or low (< 10).
For plasma DNA sequencing, 17 frozen plasma samples were extracted from average of 9.3 ml of available plasma volume (5–16 ml), using QIAsymphony SP Instrument, and QIAsymphony SP Circulating DNA kit (Qiagen). cfDNA was eluted in 60 μl and quantified using High Sensitivity Qubit dsDNA kit (Invitrogen), yielding an average of 5.3 ng/μl of cfDNA (0.954–21.4 ng/μl). Three samples had to undergo size selection to eliminate high molecular weight DNA. Libraries were then generated using Cell3Target library preparation kit (Nonacus), with UDI-UMI adapters, following manufacturer’s protocol. All samples generated sufficient libraries (average library concentration 35 ng/ul, range 3.3–55 ng/μl), and were subsequently pooled by equal mass into separate capture pools. Those pooled captures were then hybridised, one with ctPARP and one with ctATR custom-designed panels (see supp. Table 3 for genes), and sequenced in separate NovaSeq6000 SP 2 × 100PE (Illumina) sequencing runs. A custom bioinformatics pipeline was developed, involving demultiplexing of raw sequencing data, alignment, QC, and variant calling. Demultiplexing was performed using Bcl2fastq2version 2.20.0. Sequenced reads were aligned to the hg19 genome, and consensus BAM files were generated using the fulcrumgenomics bio (fgbio) suite of tools including consensus read generation determined by UMI and clipping of overlapping reads. Variant calling was performed with VarDictJava, and the output was filtered to remove low quality, synonymous and intronic mutations, as well as any variants below a defined depth or allele frequency (0.1% VAF in UMI consensus reads). High (> 90%) on-target rates are chiefly a consequence of the error-correction process. Variants were then manually curated using IGV software to exclude false positives. Read uniqueness was based on (i) a minimum of 5 UMI reads for novel SNV calling, (ii) clearly unique UMIs and unique genomic alignment positions for read pairs, (iii) absence of clear strand bias (iv) absence of mapping to a different genomic location. The established the limit of detection for known variants to be 0.125%.
Tumor RNAseq
RNA samples were quality-controlled and sequenced by the ICR Genomics Facility. RNA polyA method was used for mRNA selection. Strand-specific libraries were generated using the NEB ultra II directional kit. Illumina paired-end libraries were sequenced on a NovaSeq (Illumina) using Novaseq chemistry acquiring 100 bp paired-end (PE) reads. Bcl2fastq software (v2.2.20, Illumina) was used for converting the raw base calls to fastqs and to de-multiplex further the sequencing data. The PE fastq files were used for further analysis. The STAR alignment software (v.2.7.6a) was used to align reads to the reference genome (GRCh38). Once the reads were aligned, HTSeq-count (HTSeq v0.12.4) was used to count the number of reads mapping unambiguously to genomic features in each sample.
Differential expression analysis of the count data was performed in R using the Bioconductor package DESeq2 (v1.34.0). Dispersion estimations were corrected using the fdrtool (v1.2.17). Gene Set Enrichment and Pathway Analysis was carried out in R using DOSE (v3.20.1), pathview (v1.34.0) and clusterProfiler (v4.2.2) packages, and using GSEA (v4.3.2)36. Volcano plots were generated using EnhancedVolcano v1.16.0. Heatmaps were constructed using the pheatmap package (v1.0.12) using euclidean clustering and scaling by row; gene signatures for heatmap construction were used from MSigDB37 and cell-type-related transcripts from NanoString annotations and Bindea et al.38. Cell-type deconvolution was performed using CIBERSORTx39 for human samples and mMCPcounter40 for murine samples.
Plasma cytokine analysis
Samples were run in duplicate using the Bio-Plex Pro Human Cytokine 27-plex panel (Bio-Rad) as per manufacturer’s instructions.
Flow cytometry
Peripheral blood was drawn into 8 mL EDTA tubes (Vacutainer, BD) and analyzed within 24 h. ACK-lysed whole blood (for myeloid panel) or PBMC from density-gradient centrifugation (for lymphocyte panel) were used. Samples were surface-stained with antibodies for 30 min at 4 °C. The full antibody list used for these studies is available in Supp. Table 5. Samples were analyzed on a LSR Fortessa (BD Biosciences). FACS analyses were performed in FlowJo v10. Gating strategy is shown in Supp. Fig. 17.
Response assessment
Participants underwent RECIST v1.1 response assessment imaging 4 and 8 weeks after the completion of RT, and then at the treating physician’s discretion.
Follow-up
Participants were followed up until resolution of acute toxicities, and then every 3 months until 1 year, to assess for late toxicities.
Statistics and reproducibility
Descriptive statistical data analysis methods were used to summarize the data. Continuous data were described using mean and standard deviation for approximately normally distributed measures (assessed by inspection of histograms), median and interquartile ranges where there were marked departures from normality and numbers and percentages were reported for categorical variables. Where appropriate, 95% confidence intervals were estimated using appropriate methods. Statistical analyses were conducted using STATA version 18.0. Statistical testing for genomic and laboratory data was performed in Prism v9 (GraphPad). Data were graphed using Prism v9 or ggplot2 R package. Statistical tests used are indicated in the figure legends. Significance was taken as a P value of <0.05 and no adjustment for multiple testing was made. Missing data was reported as missing with no imputation conducted. No data were excluded from the analyses. The Investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Individual participant data that underlie the figures in this article will be made available, upon de- identification, to researchers who provide a methodologically sound proposal. Requests should be submitted to magnus.dillon@icr.ac.uk, a signed data access agreement will be required. Tumor profiling, flow cytometry and clinical annotations can be provided. All other data supporting this work are provided in the main article, supplementary information or source data file. Source data are provided with this paper.
References
Dillon, M. T. et al. Radiosensitization by the ATR Inhibitor AZD6738 through Generation of Acentric Micronuclei. Mol. Cancer Ther. 16, 25–34 (2017).
Cimprich, K. A. & Cortez, D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9, 616–627 (2008).
Dillon, M. T., Good, J. S. & Harrington, K. J. Selective targeting of the G2/M cell cycle checkpoint to improve the therapeutic index of radiotherapy. Clin. Oncol. (R. Coll. Radio.) 26, 257–265 (2014).
Dillon, M. T. et al. ATR inhibition potentiates the radiation-induced inflammatory tumor microenvironment. Clin. Cancer Res. 25, 3392–3403 (2019).
Vendetti, F. P. et al. ATR kinase inhibitor AZD6738 potentiates CD8+ T cell-dependent antitumor activity following radiation. J. Clin. Investig. 128, 3926–3940 (2018).
Patin, E. C. et al. Harnessing radiotherapy-induced NK-cell activity by combining DNA damage-response inhibition and immune checkpoint blockade. J. Immunother. Cancer 10, e004306 (2022).
Dillon, M. T. et al. Durable responses to ATR inhibition with ceralasertib in tumors with genomic defects and high inflammation. J. Clin. Invest. 134, e175369 (2024).
Dillon, M. T. et al. PATRIOT: a phase I study to assess the tolerability, safety and biological effects of a specific ataxia telangiectasia and Rad3-related (ATR) inhibitor (AZD6738) as a single agent and in combination with palliative radiation therapy in patients with solid tumours. Clin. Transl. Radiat. Oncol. 12, 16–20 (2018).
Lent soma scales for all anatomic sites. Int. J. Radiat. Oncol. Biol. Phys. 31, 1049–1091 (1995).
Jones, G. N. et al. pRAD50: a novel and clinically applicable pharmacodynamic biomarker of both ATM and ATR inhibition identified using mass spectrometry and immunohistochemistry. Br. J. Cancer 119, 1233–1243 (2018).
Yap, T. A. et al. First-in-human trial of the oral ataxia telangiectasia and RAD3-Related (ATR) inhibitor BAY 1895344 in patients with advanced solid tumors. Cancer Discov. 11, 80–91 (2021).
Dillon, M. T. & Harrington, K. J. Targeting ATR for cancer therapy: ATR-targeted drug candidates. In Targeting the DNA Damage Response for Anti-Cancer Therapy (eds. Pollard J. & Curtin N. J.) 99–127 (London: Springer; 2018).
Ng, V. et al. Genotype-directed synthetic cytotoxicity of ATR inhibition with radiotherapy. Clin. Cancer Res. 30, 5643–5656 (2024).
McLaughlin, M. et al. Inflammatory microenvironment remodelling by tumour cells after radiotherapy. Nat. Rev. Cancer 20, 203–217 (2020).
Wilkins, A. C., Patin, E. C., Harrington, K. J. & Melcher, A. A. The immunological consequences of radiation-induced DNA damage. J. Pathol. 247, 606–614 (2019).
Sheng, H. et al. ATR inhibitor AZD6738 enhances the antitumor activity of radiotherapy and immune checkpoint inhibitors by potentiating the tumor immune microenvironment in hepatocellular carcinoma. J. Immunother. Cancer 8, e000340 (2020).
Patin, E. C. et al. Sculpting the tumour microenvironment by combining radiotherapy and ATR inhibition for curative-intent adjuvant immunotherapy. Nat. Commun. 15, 6923 (2024).
Yoneyama, M. et al. Longitudinal assessment of tumor-infiltrating lymphocytes in primary breast cancer following neoadjuvant radiation therapy. Int. J. Radiat. Oncol. Biol. Phys. 120, 862–874 (2024).
Steverink, J. G. et al. Early tissue effects of stereotactic body radiation therapy for spinal metastases. Int. J. Radiat. Oncol. Biol. Phys. 100, 1254–1258 (2018).
Kotecha, R. et al. Evaluation of the impact of pre-operative stereotactic radiotherapy on the acute changes in histopathologic and immune marker profiles of brain metastases. Sci. Rep. 12, 4567 (2022).
Jones, G. N. et al. Abstract CT198: immunomodulatory effects of the ATR inhibitor ceralasertib in a window of opportunity biomarker trial in patients with head and neck squamous cell carcinoma. Cancer Res. 83, CT198–CT198 (2023).
Hardaker, E. L. et al. The ATR inhibitor ceralasertib potentiates cancer checkpoint immunotherapy by regulating the tumor microenvironment. Nat. Commun. 15, 1700 (2024).
& Vendetti, F. P. et al. The schedule of ATR inhibitor AZD6738 can potentiate or abolish antitumor immune responses to radiotherapy. JCI Insight 8, e165615 (2023).
Walls, G. M. et al. CONCORDE: a phase I platform study of novel agents in combination with conventional radiotherapy in non-small-cell lung cancer. Clin. Transl. Radiat. Oncol. 25, 61–66 (2020).
Javed, S. R. et al. CHARIOT: a phase I study of berzosertib with chemoradiotherapy in oesophageal and other solid cancers using time to event continual reassessment method. Br. J. Cancer 130, 467–475 (2024).
El Badri, S. et al. 484P CHARIOT trial (cohort A2): a phase I dose-escalation study combining the ATR inhibitor berzosertib with cisplatin and capecitabine. Ann. Oncol. 33, S761 (2022).
Bhatia, A. et al. 656MO Phase I study of M6620 (VX-970, berzosertib) in combination with cisplatin and XRT in patients with locally advanced head and neck squamous cell carcinoma. Ann. Oncol. 33, S842 (2022).
Yap, T. A. et al. Ceralasertib (AZD6738), an oral ATR kinase inhibitor, in combination with carboplatin in patients with advanced solid tumors: a phase I study. Clin. Cancer Res. 27, 5213–5224 (2021).
Banerjee, S. et al. ATARI trial: ATR inhibitor in combination with olaparib in gynecological cancers with ARID1A loss or no loss (ENGOT/GYN1/NCRI). Int. J. Gynecol. Cancer 31, 1471–1475 (2021).
Kim, R. et al. Phase II study of ceralasertib (AZD6738) in combination with durvalumab in patients with advanced/metastatic melanoma who have failed prior anti-PD-1 therapy. Ann. Oncol. 33, 193–203 (2022).
Kwon, M. et al. Phase II study of ceralasertib (AZD6738) in combination with durvalumab in patients with advanced gastric cancer. J. Immunother. Cancer 10, e005041 (2022).
Khalique, S. et al. Optimised ARID1A immunohistochemistry is an accurate predictor of ARID1A mutational status in gynaecological cancers. J. Pathol. Clin. Res. 4, 154–166 (2018).
Bankhead, P. et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
Schmidt, U., Weigert, M., Broaddus, C. & Myers, G. Cell detection with star-convex polygons. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2018 (eds. Frangi, A. F. et al.) 265–273 (Cham: Springer International Publishing, 2018).
Guidance for Industry, Bioanalytical Method Validation, U.S. Department of Health and Human Services, Food and Drug Administration, Center for Drug Evaluation and Research (CDER), Center for Veterinary Medicine (CMV) (Rockville, MD: Food and Drug Administration, 2018).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Bindea, G. et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39, 782–795 (2013).
Newman, A. M. et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 37, 773–782 (2019).
Petitprez, F. et al. The murine Microenvironment Cell Population counter method to estimate abundance of tissue-infiltrating immune and stromal cell populations in murine samples using gene expression. Genome Med. 12, 86 (2020).
Acknowledgements
The authors would like to thank all study participants and their loved ones. We acknowledge the assistance of the ICR flow cytometry and genomics facilities, and the Breast Cancer Now histopathology core facility. We acknowledge Dr Anna Wilkins for scoring Ki67 staining. We acknowledge Gemma Jones and Sophie Willis (AstraZeneca) for histopathology assistance. We acknowledge Anton Patrikeev for RNAseq analysis. The authors acknowledge support from the ICR/RM CRUK RadNet Centre of Excellence and the ICR Centre for Translational Immunotherapy (CTI). This study was co-sponsored by The Royal Marsden and The Institute of Cancer Research. Financial and in-kind drug support was provided by AstraZeneca and Cancer Research UK, through the CRUK Combinations Alliance. The authors acknowledge additional financial support from the UK Department of Health and Cancer Research UK via Experimental Cancer Medicine Centre and NIHR Biomedical Research Centre grants to The Institute of Cancer Research/Royal Marsden Hospital, and University College London/UCL Hospital NHS Trust. This project represents independent research supported by the National Institute for Health and Care Research (NIHR). The views expressed are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care. The funding institution had no role in study design, data collection, data interpretation, or writing of the report. Pharmacokinetic data interpretation and some tumor IHC staining were performed by AstraZeneca. Cancer Research UK C7224/A23275, CRUKD/14/007 (K.J.H.). Cancer Research UK C347/A18077, C309/A25144, CTRQQR-2021\100009 (K.S.). Cancer Research UK (M.T.D.). UK Department of Health (National Institute for Health Research) NIHR202438 (K.J.H.). UK Department of Health (National Institute for Health Research) (M.T.D., M.D.F.). Rosetrees Trust (K.J.H., M.T.D.). Taylor Family Foundation (M.T.D.). CRIS Cancer Foundation (P.N.). Experimental Cancer Medicine Centre. AstraZeneca, Cancer Research UK, National Institute of Health Research, Paul and Kathy Beaumont, Rosetrees Trust. Clinicaltrials.gov: NCT02223923, EudraCT: 2013-003994-84.
Author information
Authors and Affiliations
Contributions
Conceptualization: K.J.H., M.T.D., M.D.F., S.A.S. Methodology: M.T.D., E.C.P., S.A.S., M.D.F. Investigation: M.T.D., E.C.P., H.S., P.N., M.R., D.N., L.G., C.J.L., S.P., M.T., K.S., M.D.F., K.J.H. Funding acquisition: K.J.H. Project administration: J.G., M.T.D., K.M. Supervision: K.J.H., M.S., U.B., M.D.F. Writing–original draft: M.T.D., K.M. Writing–review and editing: M.T.D., E.C.P., K.M., J.G., S.A.S., E.D., H.S., P.N., M.R., D.N., N.S., L.G., C.J.L., S.P., M.T., K.E.S., U.B., A.A.M., M.D.F., K.J.H.
Corresponding author
Ethics declarations
Competing interests
K.E.S is an employee of The Institute of Cancer Research, which is involved in the development of PI3K, HSP90, HDAC, AKT, ROCK, RAF, CHK1, and HSF1 inhibitors. C.J.L. makes the following disclosures: receives and/or has received research funding from: AstraZeneca, Merck KGaA, Artios, Neophore. Received consultancy, SAB membership or honoraria payments from: Syncona, Sun Pharma, Gerson Lehrman Group, Merck KGaA, Vertex, AstraZeneca, Tango, 3rd Rock, Ono Pharma, Artios, Abingworth, Tesselate, Dark Blue Therapeutics, Pontifax, Astex, Neophore, Glaxo Smith Kline, Dawn Bioventures. Has stock in: Tango, Ovibio, Hysplex, Tesselate. U.B. reports research funding, honoraria and advisory board membership for Chugai Pharmaceutical, Verastem Inc., and Carrick Therapeutics K.J.H. reports honoraria (inst.): ALX Pharma, Arch Oncology, AstraZeneca, Bicara Therapeutics, BMS, Boehringer Ingelheim, Codiak Biosciences, F-Star Therapeutics, Inzen Therapeutics, Merck Serono, Merus, MSD, Oncolys Biopharma, Pfizer, Replimune, VacV Biotherapeutics; Consulting or Advisory Role (inst.): Arch Oncology, AstraZeneca, BMS, Boehringer Ingelheim, Inzen Therapeutics, Merck Serono, MSD, Oncolys BioPharma, Replimune; Speakers’ Bureau (inst.): BMS, Merck Serono, MSD; Research Funding (inst.): AstraZeneca, Boehringer Ingelheim, Merck Sharp & Dohme, Replimune. E.D., S.A.S., N.S. are employees of AstraZeneca with stock ownership. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Timothy Yap, Toshimitsu Hamasaki, Xianming Tan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Dillon, M.T., Patin, E.C., Mohammed, K. et al. Tumor control and immune activation through palliative irradiation and ATR inhibition, PATRIOT Part C: a phase Ib trial. Nat Commun 16, 7064 (2025). https://doi.org/10.1038/s41467-025-62249-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-62249-0