Abstract
In this study, as a proof-of-concept, we aim to initiate the development of Radiology Foundation Model, termed as RadFM. We consider three perspectives: dataset construction, model design, and thorough evaluation, concluded as follows: (i), we contribute 4 multimodal datasets with 13M 2D images and 615K 3D scans. When combined with a vast collection of existing datasets, this forms our training dataset, termed as Medical Multi-modal Dataset, MedMD. (ii), we propose an architecture that enables to integrate text input with 2D or 3D medical scans, and generates responses for diverse radiologic tasks, including diagnosis, visual question answering, report generation, and rationale diagnosis; (iii), beyond evaluation on 9 existing datasets, we propose a new benchmark, RadBench, comprising three tasks aiming to assess foundation models comprehensively. We conduct both automatic and human evaluations on RadBench. RadFM outperforms former accessible multi-modal foundation models, including GPT-4V. Additionally, we adapt RadFM for diverse public benchmarks, surpassing various existing SOTAs.
Similar content being viewed by others
Introduction
Generalist foundation models1, the latest generation of artificial intelligence models pretrained on large-scale dataset, have demonstrated remarkable success in various domains, e.g., natural language processing, computer vision2,3. Their ability to address diverse and challenging tasks has also attracted tremendous attention among researchers in the field of AI for Medicine (AI4Medicine)4,5,6,7,8. Despite the promising clinical usage, developing medical foundation models has been fundamentally hindered by three challenges:
-
Lack of multimodal datasets for training: medicine by its nature, requires understanding multimodal data, spanning text (electronic health record, medical reports), 1D signals (ECG), 2D images (ultrasound, X-ray), 3D images (CT or MRI scans), genomics, and more. To support the training of the medical generalist foundation model, a large-scale, diverse, multimodal dataset is desperately required;
-
Lack of general architecture formulation: in the literature of AI4Medicine, various clinical tasks have largely been tackled by following a divide-and-conquer paradigm, i.e., different architectures are designed for the problem of interest, like diagnosis9,10 or report generation11,12. In contrast, developing a medical foundation model requires one general architecture that is capable of tackling a wide spectrum of clinical tasks, by fusing information from a mixture of different modalities;
-
Lack of effective benchmark to monitor progress: benchmarking the models’ clinical knowledge predominantly relies on task-specific datasets with a limited number of testing cases. An high-quality benchmark is yet to be established, to comprehensively measure the progress of the development on medical foundation model across a wide range of clinical tasks.
Considering the abovementioned challenges, in this paper, we take a preliminary, yet realistic step toward developing a generalist medical foundation model for radiology, which has shown to play a vital role in clinical scenarios, for example, disease diagnosis, treatment planning, and monitoring patient progression. Specifically, we present our progress towards building a Radiology Foundation Model (RadFM), that aims to tackle a wide spectrum of clinical radiology tasks, by learning from medical scans (X-ray, CT, MRI, PET, etc.) and corresponding text descriptions/reports.
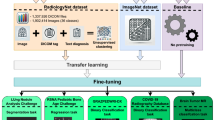
To achieve this, as shown in Fig. 1, we start by constructing four novel medical multimodal datasets, by exploiting the highly specialised, high-quality radiological images on the Internet, where the diagnosis labels have been extensively reviewed by a panel of experienced clinicians, namely, PMC-Inline, RP3D, PMC-CaseReport and MPx, consisting of 13M 2D and 615K 3D radiology scans. Additionally, we combine a vast collection of existing datasets with our collections, resulting in a large-scale Medical Multimodal Dataset, named MedMD, with totally 16M 2D and 3D radiology scans, accompanied with high-quality textual descriptions, for example, radiology reports, visual-language instruction, or crucial disease diagnosis labels. MedMD encompasses a wide range of radiological modalities, covering 17 medical systems, e.g., breast, cardiac, central nervous system, chest, gastrointestinal, gynecology, hematology, head and neck, hepatobiliary, musculoskeletal, obstetrics, oncology, pediatrics, spine, trauma, urogenital and vascular featuring over 5000 diseases, thus potentially serving as the cornerstone for developing foundation models in radiology.
a Dataset demonstration. The colored datasets are constructed by us in this paper. b Our training procedure. For better radiologic performance, we first pre-train our model on the whole medical domain with 16M scans (MedMD), then finetune on a cleaned dataset with 3M radiologic scans (RadMD). c RadFM architecture. Our architecture enables multi-image input interleaving with texts 2D or 3D images. d The human rating comparison of five foundation models under three open-ended task types (medical VQA, report generation, and rationale diagnosis), adding GPT-4V14 into comparison. All evaluations have shown the superiority of RadFM.
Architecturally, RadFM refers to a visually conditioned auto-regressive text generation model, that enables seamless integration of natural language with 2D or 3D medical scans, and address a wide range of medical tasks with natural language as output. The proposed model is initially pretrained on the large MedMD dataset, and subsequently trained with domain-specific visual instruction tuning on a filtered radiology subset, comprising 3M meticulously curated multi-modal samples with only radiologic cases, termed as RadMD, ensuring a high-quality and reliable dataset.
To monitor the developmental progress of the foundation model for radiology, in addition to using the existing benchmarks, we also establish a comprehensive evaluation benchmark, spanning various clinical task types, termed as RadBench, covering a variety of clinical tasks, for example, report generation, and visual question-answering on radiologic modalities and anatomical regions. All samples in RadBench have undergone meticulous manual verification to ensure data quality. We conduct both automatic and human evaluation on RadBench with powerful models that are publicly accessible, for example, Open-flamingo13, MedVInT8, LLaVA-Med4, MedFlamingo6, and GPT-4V14, and observe significant benefits across all considered tasks. In addition, we perform task-specific finetuning of RadFM on several public benchmarks, demonstrating its strong ability for transfer.
Overall, in this work, we demonstrate a preliminary attempt for building a generalist foundation model for radiology, by making contributions from three key aspects: four new large-scale medical multimodal datasets, covering both 2D and 3D medical images, namely, PMC-Inline, RP3D, PMC-CaseReport, and MPx, a superior radiology foundation model (RadFM), and a comprehensive benchmark for radiology (RadBench).
Results
In this section, we start by presenting evaluation results on nine public datasets, comparing them to the existing medical multi-modal foundation models, together with ablation results on our training procedure and our newly collected dataset from the Internet. Considering existing medical datasets cannot comprehensively cover all medical tasks, we further report the results on our proposed RadBench, with three medical tasks, namely, medical VQA, report generation, and rationale diagnosis, as demonstrated in Fig. 2.
We start by comparing the zero-shot prompting results for RadFM with other foundation model baselines (both zero-shot and few-shot settings). Following that, we perform task-specific finetuning experiments to thoroughly evaluate the performance of our model with different state-of-the-art task-specific models. Additionally, to evaluate the model’s generalization ability, we employed a zero-shot evaluation on the unseen classes in the PadChest dataset. It is worth noting that the results on PadChest have not undergone any task-specific finetuning.
Results on existing benchmarks
We compare our model with other foundation models on nine existing benchmarks, e.g., VinDr-Mammo, VinDr-SpineXr, VinDr-PCXR, CXR-Mix, RadChest-CT, PMC-VQA, VQA-RAD, SLAKE, and MIMIC-CXR, covering tasks like diagnosis, medical VQA, and report generation, as shown in Table 1. In detail, we compare with the Open-flamingo13, MedVInT8, LLaVA-Med4, and MedFlamingo6. For the flamingo-series, we adopt the few-shot (three-shot) prompting setting, as the models are supposed to demonstrate better performance under a few-shot scenario, while for MedVInT, LLaVA-Med, and our RadFM, we adopt a zero-shot prompting strategy, as both the two are trained to follow semantic instructions rather than few-shot samples. The zero-shot results for flamingo-series are also included in the Supplementary Table 2.
Comparison with other foundation models
As shown by the results in Table 1, our final model shows superior results on nine publicly available datasets. First, for disease diagnosis, existing foundation models perform poorly, with an accuracy score (ACC) of nearly 50%. Considering that we prompt the problem with a judgment format, i.e., “Does the patient have disease?”, this score is nearly random. In contrast, our proposed RadFM, exhibits evaluation results, with 59.96, 68.82, 56.32, 83.62, and 72.95% ACC scores on the five diagnosis datasets, respectively. Second, for other long sentence generation tasks, i.e., medical VQA and report generation, RadFM also surpasses other models significantly on most metrics.
Ablation studies
In Table 1, we also carry out ablation studies on our methods. First, we dismiss the domain-specific instruction tuning on RadMD. Similar to the observation in language domain15,16,17, we find that domain-specific instruction tuning is a critical step for building up a multi-modal foundation model that can rule various medical tasks with proper instruction prompting. As shown by the results, without domain-specific instruction tuning on RadMD, the model can hardly respond correctly to diverse task instructions. Moreover, we evaluate the effectiveness of our newly collected data, i.e., PMC-Inline, RP3D, PMC-CaseReport, and MPx, by comparing the model trained with and without them for auto-regressive pretraining and domain-specific instruction tuning. As shown by the results, adding our new-collected data can significantly improve the final results regardless of task types, underscoring that, our newly collected datasets, though sourced from Internet, are effective for improving model performance on existing clinical datasets.
Beyond the data-centric ablation studies, we also conducted an additional series of experiments to demonstrate the effectiveness of our training design, as shown in Supplementary Table 3. Given the computational cost of testing these settings across the entire training dataset, we randomly sampled 10% of the RadMD dataset (using a 0.1 sample ratio) for these experiments. We used the default setup of our method—specifically, a 3D ViT model with a patch size of 32 and the prompting strategy described in the Methods section—and systematically varied the following factors: the image-encoder architecture, the patch size, and the prompting strategy.
Specifically, we vary the following factors: (i) to investigate the impact of the image-encoder architecture, we experimented with more recent models, such as those proposed in ref. 18 and ref. 19, (ii) we test the effects of increasing the patch size beyond 32, (iii) we experiment with more complex prompts. For example, we tested prompts with added role-play elements, such as “Assuming you are an experienced radiologist reading modality images, please interpret the following images and answer the related questions. This is a serious clinical case, so please be as careful and disciplined as possible question,” as well as incorporating varied, synonymous prompts generated by GPT-4 and verified by human reviewers.
The results, summarized in Supplementary Table 3, show that modifying the image-encoder architecture produced some slight gains on certain datasets, but these improvements were not statistically significant. Given that the focus of our work is not on architectural design, we chose to retain the classic 3D ViT architecture, which offers a solid baseline. When we tested the impact of increasing the patch size, we observed that larger patch sizes beyond 32 actually led to a degradation in performance. Notably, here, using smaller patch sizes might be better but they will bring unacceptable computational cost due to the increment of token number in ViT, this confirms our decision to use a patch size of 32. Lastly, variations in prompt complexity, including the use of role-playing or diverse synonymous prompts, had little to no effect on the model’s performance.
These additional ablation studies provide further validation for the choices we made in model architecture, patch size, and prompt design, supporting the robustness of our method.
Results on RadBench
In this section, we further assess the long sentence generation ability for different models on our proposed benchmarks, which compensate for three medical tasks, i.e., medical VQA, report generation, and rationale diagnosis.
Medical visual question answering (VQA)
Medical VQA denotes a comprehensive and versatile challenge in the field of medical image analysis. In a clinical setting, patients and radiologists may pose a wide variety of questions related to medical images, ranging from simple inquiries about image modality to more complex reasoning queries. In contrast to the aforementioned existing medical VQA datasets, on RadBench, the image input is more close to a clinical scenario with 3D scan input.
As shown in Table 2, RadFM generally demonstrates superior performance. Compared to the second best model, MedVInT, which was specifically trained on visual question answering, despite achieving better results on its in-domain PMC-VQA test set, its generalization to real 3D scans is relatively poor, even though the task is still medical visual question answering. MedVInT struggles with real 3D medical scans, which require a model capturing the information from an extra image dimension. In contrast, our RadFM model shows a substantial improvement in UMLS_Precision from 20.12 to 31.77% and UMLS_Recall from 15.82 to 24.93% across the whole test set, demonstrating its proficiency to comprehensively understand the given textual information and flexible adaptation to various complex clinical scenarios.
Report generation
Report generation is a crucial and prominent use case for generative medical foundational models. Unlike Medical VQA, this application generally requires the model to emphasize clinically significant observations based on the image. Considering that, current report generation benchmarks are all concentrated on X-ray; in RadBench, we focus more on testing the report generation ability for other imaging modalities. As shown in Table 2, RadFM shows significant improvement over existing models, across various metrics, particularly in relation to medical-specific terminology. For instance, RadFM improves UMLS_Precision from 9.61 to 22.49%, and UMLS_Recall from 3.66 to 12.07% in the zero-shot setting.
Rationale diagnosis
In addition to basic diagnosis, the ability to scrutinize diagnostic prediction outcomes is crucial, particularly in light of the stringent demands for precision and interpretability within medical contexts. Thus, on RadBench, we further evaluate the ability to generate diagnosis rationale sentences for different models. Much like report generation, this task also requires proficiency in generating supplementary paragraphs and a comprehensive understanding on medical knowledge.
As indicated in Table 2, RadFM is the only model that can effectively respond on this task, outperforming other models on BLEU and ROUGE scores by 16.50 and 12.87%, respectively, even comparing with the few-shot case. Moreover, it exhibits significant improvements in UMLS_Precision and UMLS_Recall scores, showcasing advancements of 21.91 and 16.18%, respectively.
Human rating
In Fig. 3b, we show the human rating results on the three generative tasks for all models. We choose OpenFlamingo to denote the performance of the general-domain multimodal foundation models, MedVInT for zero-shot-based medical multimodal foundation models, MedFlamingo for few-shot-ones, and GPT-4V for best close-sources multimodal foundation models. As shown on the left of the figure, RadFM achieves higher scores on all three generative-based tasks compared with existing open-source models, only falling behind with GPT-4V in rationale diagnosis. On the right, we further show the relative comparison between RadFM and a certain model. In most cases, results from RadFM are preferred by human clinicians. It is worth highlighting that we also show the comparison between RadFM and GPT-4V(ison), which has been widely considered as the strongest foundation model. As GPT-4V can only input up to four 2D pictures per query, we thus ask the radiologists to pick out the most informative slices based on the reference’s answer from 3D volumes. With human prior, answering questions becomes easier than directly inputting original 3D volumes, which is used as the evaluation style for our model. Despite this, RadFM still surpasses GPT-4V in average scores.
a Comparison of RadFM with various foundation models on different subsets under zero-shot evaluation with machine rating scores. The detailed sample sizes for testing are marked as “(n=…)” in the left corner of each bar plot. For the dataset involving diagnosis, like VinDr-Mammo, VinDr-SpineX, VinDr-PCXR, CXR-Mix, RadChest-CT, ”ACC“ scores are plotted in the figure. For the left datasets, “AVG” scores, denoting the average of the four word-overlap-based evaluation metrics, i.e., BLEU, ROUGE, UMLS_Precision, and UMLS_Recall, are plotted. For each bar plot, the error bars represent the 95% confidence interval (CI) via 1000 technical replications, with the center of the error bars indicating the average score across all test cases within each dataset. The exact number for each bar plot can be found in Supplementary Tab. 2. b Comparison of RadFM with other methods on human rating scores. On the left, we show the absolute human rating scores of different methods on the three generative tasks. i.e., VQA, report generation and rationale diagnosis. On the right, we show the relative comparison. Each sub-figure in right shows the number of RadFM win/tie/lose cases when comparing against a certain model. Note that, considering GPT-4V may refuse to answer medical questions for safety, we dismiss such cases when calculating the scores or comparisons relating to GPT-4V. In detail, for 1200 testing cases, 22 cases were dismissed for GPT-4V due to safety.
Results for task-specific finetunine
In Table 3, we treat RadFM as a pretrained model and task-specific finetune it on various downstream datasets. For diagnosis, we use the image-encoder weights as initialization for both 2D and 3D imaging modalities, for VQA and report generation, the whole model is further finetuned on the specific dataset, as shown in the Table 3, our model improves both diagnosis results and text generation quality, based on the automatic metrics. In general, the representation learned in RadFM benefits various clinical tasks across diverse medical imaging modalities, like CT, MRI, X-ray, or even rarely seen PET-CT, regardless of whether they are presented in 2D slices or 3D scans. Consistent improvement can be observed, across various task types, like diagnosis, VQA, and report generation.
Generalization to unseen classes of PadChest
In Fig. 4, we show the results of zero-shot evaluation of RadFM on unseen classes from the PadChest dataset. We modify the task as an induction task, for each disease, we randomly select a prompt sentence like “Is {disease} shown in this image” as input, the network outputs whether the case has this disease. Note that we balance the ratio of “yes” or “no” in the test set, and all the disease classes never appeared in the training set. The prompting utilized here are essentially equivalent to traditional multi-label disease classification methods, with each disease’s performance being evaluated independently, leading to a holistic outcome through sequential assessment of all listed diseases. Following this same rationale, in our context, we can provide comprehensive diagnosis results by iteratively employing the “yes/no” prompting approach.
Qualitative results
In this section, we show the qualitative results for different free-form text generation tasks.
For medical VQA, qualitatively, as shown in Fig. 5, RadFM demonstrates the ability to comprehend the questions and provide answers in a consistent format, accurately addressing the questions. However, in some challenging cases, such as the first example, where the question pertains to the type of abnormality, the model faces difficulty predicting “ectopic ACTH-producing tumor” and mistakenly identifies it as “primary lung neoplasm”, which requires fine-grained discrimination within tumor types.
In Fig. 6, we provide qualitative examples of the radiology reports generation task by RadFM. It can be observed that the model is capable of identifying the underlying diseases and, in some cases, performs exceptionally well. However, the report generated by RadFM may lack specific location information, such as the ‘left’ or “right” of an anatomical region.
We present several examples with reports generated by RadFM and reference reports. The green color highlights accurate keywords, while the red color indicates prediction errors. Additionally, the blue color denotes instances where the model missed this information that has been mentioned in the reference reports.
At last, Fig. 6 shows two rationale diagnosis cases. The first case is a patient with pulmonary embolism and the latter is with subarachnoid haemorrhage. On both cases, RadFM can make accurate diagnosis in free form and give further related radiologic reasoning. However, the limitation can also be observed that the reasoning results are still general and more like background medical knowledge, yet not specific to the input case.
Discussion
RadFM tries to develop a medical foundation model that enables the processing of both 2D and 3D radiologic images with interleaved texts. In the field of radiologic images, one significant challenge in developing a foundation model lies in the disparity of image dimensions, i.e., medical scans are either 2D or 3D, posing challenges in integrating real 3D MRI or CT images alongside 2D images like X-rays or ultrasounds. As a consequence, the development of foundational models has been significantly impeded, with most current models only accommodating 2D images. To overcome these limitations, we propose a new training structure that unifies 2D and 3D images, allowing to process various clinical images. By unifying the training procedure, our model benefits from a more comprehensive understanding of the diverse clinical images, leading to improved performance and versatility. Additionally, to facilitate research and foster collaboration in the field, we collect four medical multimodal datasets, namely, PMC-Inline, RP3D, PMC-CaseReport, and MPx, consisting of 13M 2D and 615K 3D radiology scans with text descriptions or labels.
RadFM unifies the medical tasks with a generative model. While developing AI for medicine, traditional approaches consider a divide-and-conquer idea, that tackles a myriad of specific tasks individually, such as diagnosis, report generation, and medical visual question answering, resulting in separated approaches with limited generalization ability and efficiency. Here, we formulate diverse medical tasks as multi-modal question-answering and develop a generative visual-language model, RadFM, that can answer arbitrary questions or follow instructions. In contrast to existing works with the use of exemplars in prompts, we use zero-shot prompts for all tasks, allowing users to interact with the model without providing any exemplar images, questions, and answers. Training models support zero-shot prompts is certainly more challenging, however, considering the user might be patients without no clinical background, or examplar images, zero-shot prompt would also be indispensable for real-world applications. By unifying the tasks, RadFM achieves promising performance across a wide spectrum of clinical tasks. On the medical VQA task, RadFM surpasses the performance of MedVINT, a pretrained model trained solely on a single Medical VQA dataset.
RadFM supports multiple images as input. Till now, existing multi-modal foundation models in the medical community have been limited to supporting only a single image input per interaction. However, such a design poses critical challenges in medical scenarios where diagnosis and treatment decisions often necessitate longitudinal clinical follow-ups, that comprise a series of radiologic images. To overcome this limitation and pave the way for more comprehensive medical image analysis, our proposed RadFM supports multi-image input. To support the training, our constructed dataset is largely composed of multi-image input data, and our innovative training flow seamlessly accommodates this unique medical scenery, fostering advancements in medical image analysis.
A general evaluation benchmark for radiology foundation models. Evaluating the performance of medical foundation models is challenging, due to the specialized nature of medical tasks. In the pursuit of advancing radiology foundation models, we propose RadBench, a novel benchmark that encompasses a diverse range of medical scenarios. By incorporating both 2D and 3D images, RadBench offers a more comprehensive and realistic evaluation platform compared to existing benchmarks. Combining with the existing medical benchmarks, we comprehensively evaluate models for four medical tasks, namely plain diagnosis, visual question answering, report generation, and rationale diagnosis, covering multiple imaging modalities. Additionally, as existing evaluation metrics are primarily designed for general natural language tasks, which may not adequately capture the intricacies and nuances specific to medical image analysis, thus may not reflect the model’s true capabilities in real-world clinical scenarios. To address this limitation, we propose two new evaluation metrics, namely UMLS_Precision and UMLS_Recall. Unlike conventional metrics, UMLS Precision and Recall are tailored to measure the model’s performance in medical tasks. By leveraging the Unified Medical Language System (UMLS), a comprehensive medical knowledge resource, these metrics provide a more tailored evaluation, ensuring that the model’s outputs align with medical domain expertise.
The superiority of RadFM. As shown in Tables 1, 2 and Fig. 1, while evaluating on our proposed comprehensive benchmark for radiology, namely, RadBench, and nine existing radiological datasets, RadFM outperforms previous methods by a significant margin across all five tasks, showcasing its exceptional capabilities. Notably, RadFM excels in particularly challenging tasks such as medical VQA, report generation, and rationale diagnosis, which demand a profound understanding of both textual information and images. The average human evaluation score for RadFM in these tasks surpasses that of GPT-4V, especially in the medical VQA task, where RadFM achieves a score of 2.87 compared to GPT-4V’s score of 2.13. In medical VQA, the questions can be drastically varying, from simple queries like “What modality is the given image?” to more complex and context-rich questions, such as “Based on the provided images, patient data (age, gender, and medical history), can you identify the disease that is commonly associated with such radiological manifestations?” The complexity of questions makes medical VQA a comprehensive and versatile task. By integrating visual and textual information, RadFM can handle these varying question types, delivering accurate and meaningful answers. Similarly, in report generation, RadFM showcases significant improvement. The model’s ability to discern relevant information from the provided images and weave it cohesively with textual prompts leads to highly informative and contextually rich reports, setting it apart from traditional methods. Overall, the performance of RadFM across these diverse tasks confirms its versatility and transformative potential in radiology image analysis.
Clinical impact. In contrast to all existing medical models, RadFM is the first attempt towards developing foundation models that simultaneously satisfies three important criteria in clinical practice: (i) in support of both 2D and 3D data, for example, 2D chest-X-ray, 3D CT or MRIs; (ii) be able to process multiple scans from various imaging modalities; (iii) to support interleaved data format, for example, allowing the user to freely input additional background information in text form, along with radiology scans, the model enables to accompolish complex clinical decision-making tasks. Overall, RadFM allows users to input 3D multiple scans interleaved with texts per query, which can greatly benefit its clinical usage.
Limitations. Despite our efforts in developing a foundation model for radiology and surpassing former medical foundation models significantly, RadFM is still a proof-of-concept model design towards medical generalist AI (GMAI) and need more efforts for real clinical usage. In detail, it may exhibit the following limitations:
First, the capacity to generate meaningful and accurate long sentences remains underdeveloped, causing the foundation models to be still far from clinically useful. As demonstrated in Supplementary Tab. 2, for rationale diagnosis and report generation, the quantitative results surpass previous works but are still far from practically satisfactory. In human rating, similar results are also observed. As shown in Fig. 3, none of the models gets over score 3 which represents moderately accurate, showing that there is still a long way to go for developing generalist medical foundation models. For real clinical usage, we suggest future improvements, including scaling model sizes, increasing image resolutions, and expanding clinical data, as outlined by the scaling laws20.
Second, the proportion of actual 3D images in the data remains limited. As illustrated in Fig. 7, although we attempt to compensate for the lack of 3D images, 2D images remain to be dominating.
a Overview of Medical Multimodal Dataset (MedMD). Our collected data covers the majority of radiologic modalities and anatomical regions of the human body, such as the brain, head and neck, thorax, spine, abdomen, upper limb, lower limb, and pelvis, etc. The dataset mixes two types of datasets, i.e., interleaved datasets and visual instruction datasets. \({{{\mathcal{T}}}}\) refers to the text of interleaved data, \({{{\mathcal{I}}}}\) refers to the instruction input text, and \({{{\mathcal{R}}}}\) refers to the response text. b The data statistics of RadMD and RadBench. The left image shows the distribution of different modalities of RadMD, and the center image shows the distribution of 2D and 3D sample pairs of RadMD. The right image shows the distribution of the anatomy of the samples in the RadBench.
Third, the automatic evaluation metrics fall short of expectations. Compared to general contexts where the emphasis is placed on the overall coherence and fluency of sentences, medical texts prioritize precision in key statements and contain many synonyms, like “MRI” and “magnetic resonance imaging”, overlooking minor syntax errors. Although we employ UMLS_Precision and UMLS_Recall to mitigate this issue, they do not fully reflect true performance. On the other hand, though human evaluation is flexible and accurate, it is costly and cannot be carried out on a large scale. A robust automatic evaluation metric is essential to guide the construction of reliable and robust medical foundation models.
Fourth, as the 3D images in our dataset are downloaded from the internet, some metadata is missing, for example, the imaging spacing. Such a lack of precise distance measurement makes it impossible to make certain statements, such as “The tumor is 3-cm large”. This specification is crucial for report writing, particularly for tasks requiring precise spatial information, such as tumor size estimation, currently, we acknowledge that our model cannot be used in this way. We propose several potential solutions to address this limitation in future work, for instance, one way is to explore incorporating registration or spacing prediction models to generate the pseudo-spatial information, allowing the model to make more accurate numerical predictions related to physical measurements (e.g., tumor size). Another promising direction is to enhance the model’s ability to interact with external tools or agents. For example, the model could use segmentation models alongside coding functions to calculate physical spacing directly, providing precise numeric feedback to assist in tasks like report generation or anomaly detection.
Lastly, due to the scale of the dataset (16M image-text pairs), model size (14B parameters), investigating the effects of different components becomes increasingly challenging and prohibitively expensive in terms of both time and computational resources. As future work, we will further break down the problem and investigate each component, ultimately enhancing our understanding and refining the model’s performance. This includes, but is not limited to, improved 2D and 3D unified encoding methods, more effective choices for LLM base models, and enhanced training pipelines that address data imbalance arising from data combination.
Related works. With the success of generative language foundation models such as GPT-416 and PaLM-221, there has been a surge of interest in multi-modal foundation models. Significant strides have been made in the realm of natural scenery, as evidenced by BLIP-23 and Flamingo22. Though in the context of the medical language-only models, great steps have been made, like Med-PALM series23,24, PMC-LLaMA25, Meditron-70b26, the development of multimodal medical artificial intelligence is still in its nascent stages5. The relevant research can be bifurcated into two primary areas, namely, dataset construction and model training.
-
Dataset Construction. Contrary to the natural scenery domain, which boasts numerous large-scale multi-modal datasets such as MMC427, Visual Genome28, and LION-5B29, the medical domain is somewhat lacking. The most widely utilized medical multi-modal dataset is MIMIC-CXR30, which only contains chest X-ray images with caption reports, and its quantity (224K) is relatively small. In PMC-OA31, the authors have compiled a dataset containing 1.6M image-caption pairs. Although it encompasses various image modalities, many 3D medical scans are presented as 2D slices since the images are extracted from papers. There are also some medical VQA datasets, such as VQA-RAD32, SLAKE33, and PMC-VQA8, but they are also limited to 2D images. In Med-Flamingo6, they have collected a dataset, MTB, consisting of approximately 0.8M images interleaved with texts while it is not open-source. Consequently, due to the limitations on data availability, existing medical foundation models have concentrated on a narrow range of data modalities. For example, LLaVA-Med4 and MedVInT8 utilize image captions in PubMed Central, which exists a significant domain gap between real-world clinical data. In Med-PaLM M7, the authors amalgamate existing medical images or multi-modal datasets, but the majority of images are X-rays, which are not sufficiently accurate for clinical practice.
-
Model Training. To date, several works have focused on building medical foundation models, yet most of these works are limited to support for 2D images4,6,7,8. An ideal foundation model should exhibit a comprehensive set of capabilities: it should support both 2D and 3D image inputs, multi-image input per case, and images interleaved with text inputs. Currently, there exists no model that can simultaneously support this series of heterogeneous input forms. This paper aims to address these gaps, aligning more closely with clinical practice and we are very glad to see many current works34,35,36,37 have been inspired based on our efforts.
In conclusion, this paper serves as a proof-of-concept initialization for building medical generative vision-language foundation models. Specifically, our work demonstrates:
-
Data collection: Highlighting the importance of leveraging and combining web-scale radiology data sources to ensure robust and comprehensive model training.
-
Architecture formulation: Proposing the unification of all radiology tasks within a generative architecture formulation, carefully designed with task-specific instructions, to develop a versatile and generalist model.
-
Evaluation: Underlining the need to monitor model performance across a diverse range of radiology tasks to ensure comprehensive validation and broader applicability.
While our model significantly outperforms existing open-source multimodal foundation models, we acknowledge that it does not yet meet clinical criteria. Nevertheless, we believe this work provides a starting point for future research and development toward more generalist medical AI models. With the remarkable and rapid advancements in this field, there will be continuous progress in several key areas. For example, more practical and invaluable datasets, such as CT-RATE38, are expected to be released. Similarly, more powerful general multimodal foundation models, such as the latest DeepSeek-VL39 and Qwen2-VL40, will likely emerge, serving as stronger base models along with increasingly sophisticated imaging encoders, like CT-ViT38 and LongViT41. By consistently integrating these distinct impressive advancements in future work, we, together with the entire research community, can drive the evolution of radiology foundation models toward broader adoption in practical clinical applications.. We will release all corresponding data, codes, and models. We believe this can greatly promote the development of medical foundation models.
Methods
In this section, we will detail our method. Notably, our study is based on data obtained from open-source websites, as listed in Supplementary Table 5. Therefore, the relevant ethical regulations are governed by the original data-uploading processes outlined in each dataset’s collection pipeline (please refer to each dataset website in Supplementary Table 5 for more details). Specifically, for the data from Radiopaedia, which forms the main component of our newly proposed dataset, Radiopaedia is a peer-reviewed, open-edit radiology resource collection website. Its mission is to “create the best radiology reference available and to make it available for free, forever, and for all.” We have obtained non-commercial use permission from various uploaders as well as the founder of Radiopaedia. The relevant ethical regulations are governed under Radiopaedia privacy-policy.
Dataset
Here, we describe the procedure for constructing the datasets and benchmark. In the section “Medical Multimodal Dataset (MedMD)”, we present several medical multimodal datasets and merge them with an extensive collection of preexisting datasets, resulting Medical Multimodal Dataset (MedMD). MedMD is a large-scale, high-quality medical vision-language dataset, covering a wide range of anatomies with over 5000 diseases, as shown in Fig. 7a. We further construct a filtered radiology subset Radiology Multimodal Dataset (RadMD). In the section “Radiology Evaluation Benchmark (RadBench)”, we introduce a new Radiology Benchmark for evaluation, termed RadBench, with three distinct tasks, e.g., visual question answering, report generation and rationale diagnosis, aiming to monitor the progress of developing foundation models.
Medical multimodal dataset (MedMD)
To start, we construct a candidate data pool by pulling a variety of existing visual-language medical datasets together, for example, MIMIC-CXR30 and PMC-OA31. Despite the scale of these high-quality datasets, they are fundamentally limited in several aspects: (i) Data format. These datasets are only composed of 2D medical images, which do not fully capture the complexities in clinical use cases, for example, 3D medical imaging modalities, like CT, MRI; (ii) Modality diversity. A noteworthy limitation arises from the fact only chest X-ray images are provided with medical reports, training models on such data will clearly pose limitation on the generalizability to a broader range of imaging modalities and anatomical regions; (iii) Report quality. Another critical limitation lies in the use of data extracted from figures and captions from research papers. The gap between research-oriented data and real-world clinical scenarios may not support accurate and reliable clinical diagnoses. Therefore, to support the training of our proposed Radiology Foundation Model (RadFM), we augment the dataset with four new ones, including PMC-Inline, PMC-CaseReport, RP3D-Series, and MPx-Series, resulting in MedMD. MedMD has a total of 16M 2D image-text pairs, including around 15.5M 2D images and 500k 3D scans with corresponding captions or diagnosis labels, as shown in Supplementary Table 3. More detailed introduction of different data sources can be found in the Supplementary Section “Detailed Introduction for Different Data Sources”.
Generally speaking, we split the candidate data pool into two parts (i) interleaved image-language data that is collected from academic papers and (ii) image-language data constructed for visual-language instruction tuning, as detailed below.
Interleaved dataset
PMC-Inline. PMC-Inline contains 11M 2D radiology images that are collected from PubMed Central papers. In contrast to existing work, for example, PMC-OA31, that only contains figures and corresponding captions, here, we focus on the inline reference from the main body of papers. For example, one paper may contain many sentences like “As shown in Fig. 2, we can see …”, we localise the keyword “Fig. 2” and link its corresponding figure back into sentences, ending up with interleaved images and texts, with rich context. This dataset shares the same format as MMC427, which has shown to be effective in training foundation models in the computer vision community, for example, Flamingo22.
Visual-language instruction tuning dataset
PMC-CaseReport. Inspired by former works leveraging clinical case reports42, PMC-CaseReports is a filtered subset of PMC-Inline with around 103K case reports, where the doctors typically document the valuable clinical cases, based on their contact with the patients, such as family medical history, preliminary diagnosis, radiographic exam results, surgical records, etc., together with critical radiologic scans, that generally follows the real timeline.
Similar to PMC-VQA8 that generates VQA pairs by querying ChatGPT with image captions, we also generate 1.1M question-answer pairs by querying ChatGPT with the sentences containing inline references in case reports. However, in contrast to PMC-VQA, we keep background information of the patients to simulate the clinical diagnosis scenario, thus can be seen as a medical contextual VQA dataset. For example, a question-answer pair may like “Question: A 58-year-old woman presented to the emergency department …Postoperatively, her pain significantly relieved. What did the MRI indicate? Answer: The MRI indicated tumor recurrence at L2 and S1-S2.”
RP3D. RP3D (RadioPaedia 3D) is a novel dataset with 3D radiology scans, sourced from the Radiopaedia website (https://radiopaedia.org/). All privacy issues have already been resolved by the clinician who uploaded the case. Specifically, each patient case comprises one or more images from the same or different modalities, accompanied by high-quality captions that have been meticulously peer-reviewed by experts in the Radiopaedia Editorial Board (https://radiopaedia.org/editors). We have included a response letter from Radiopaedia, with the agreement for us to use the dataset for training under non-commercial cases. In addition, for each disease, we can get corresponding radiological features across different modalities. We convert the image-caption pairs into a variety of formats, namely, RP3D-Caption, RP3D-Modality, RP3D-Rationale, and RP3D-VQA, depending on their corresponding text content. Specifically, RP3D-Caption denotes the images paired with their corresponding captions; RP3D-Modality refers to images with modality labels; RP3D-Rationale incorporates radiological features with disease labels for each case; RP3D-VQA involves visual question-answering pairs generated from captions by querying ChatGPT, as illustrated in Supplementary Fig. 1.
MPx. MPx is collected from the MedPix website (https://medpix.nlm.nih.gov/) and organized by cases. Each case contains multiple radiologic scans, along with general clinical findings, discussions, and diagnostic results. In addition, MPx also provides annotations on the scan level, including information such as image modality, shooting plane, and captions for each scan. Thus, we separate it into MPx-Single and MPx-Multi, containing annotations on the case-level and scan-level, respectively.
Radiology multimodal dataset (RadMD)
For domain-specific finetuning, we filter out the non-radiology images from MedMD, and construct a clean subset, named Radiology Multimodal Dataset (RadMD), dedicating to supervised visual instruction tuning. It contains a total of 3M images, spanning various data formats, modalities, and tasks, featuring over 5000 diseases, as shown in Fig. 7b.
In general, we have conducted the following filtering process: (i) remove non-radiologic images; (ii) remove the entire PMC-OA and PMC-Inline datasets, as the images in PubMed are 2D-only, thus differ from real clinical cases, additionally, the writing styles between academic papers and real clinical reports are inconsistent; (iii) remove a large portion of 2D image cases from PMC-Series, to emphasize the 3D images in training. (iv) filter out the information about patient age or structure size, as the image spacing and patient background information are not provided. Specifically, we applied string matching techniques using Python’s regular expressions to remove any sentences containing terms related to physical measurements, such as “mm”, “cm”, or decimal numbers (e.g., “2.5 cm”), as these are indicative of missing or incomplete metadata related to patient age, structure size, or image spacing. This step primarily addresses the problem in the report generation tasks, where such metadata would otherwise cause incorrect or unpredictable descriptions.; (v) balance the number of normal and abnormal patients in the diagnosis datasets, as generative models are sensitive to data imbalances. More comprehensive details regarding the filtering process and the resulting dataset sizes can be found in Supplementary Table 3.
Radiology evaluation benchmark (RadBench)
In addition to the training set, we also introduce RadBench, a comprehensive evaluation benchmark for monitoring progress in the development of radiology foundation model for generative tasks. Considering that most existing medical benchmarks may only include a plain label (like disease categories), that are not suitable to assess the models’ long sentence generation ability, our RadBench is targeted at compensating for this.
In detail, RadBench is first randomly sampled from the RP3D dataset. Then, We further carry out meticulous manual verification to ensure data quality on all the samples. Specifically, we developed a human evaluation interface, visually presenting the data source, image, question, and answer of each case. Eight human annotators were asked to assess the quality of these cases by addressing the following criteria:
-
Image types: remove the images that do not fall in radiology.
-
Question reasonability: keep the questions that can be answered from the given radiology image, for example, on visual question answering, remove the question related to size; on report generation, remove cases containing sentences like “Compared with previous cases”; on rationale diagnosis, remove cases lacking corresponding radiological features are filtered out.
-
Answer correctness: keep those with correct answers based on the given text reports.
As a result, we have obtained 4229 for visual question answering, 1468 for report generation, and 1000 for rationale diagnosis. Additionally, we also consider nine existing tasks for our evaluation, which include plain diagnosis and medical VQA tasks. A detailed breakdown of each dataset, including task descriptions and modalities, is provided in Supplementary Table 4. Combining them with our RadBench, in evaluation, we will comprehensively assess models for four tasks, i.e., disease diagnosis, medical VQA, report generation, and rationale diagnosis. The details of the four evaluation tasks and metrics are introduced in the following.
Disease diagnosis
This task involves analyzing the radiology images to determine the likelihood of specific diseases. Here, we modify this task to an induction task, which uses introductory text explaining the classification task and providing the name of the queried disease at the beginning of the prompt. Given a medical image, we randomly select a disease and a prompt sentence like “Is {disease} shown in this image” as input, querying the model to determine the existence of a certain disease. Due to this being formulated as a generation task, “AUC” cannot be calculated, so we match the output with ground-truth to calculate the ACC and F1 score. Similarly, we match the output with a closed ground-truth list {“yes”, “no”} using difflib.SequenceMatcher, and choosing the most similar one as the prediction of the model. Considering ACC scores may suffer from data unbalancing, we keep the same ratio to sample positive and negative cases. In our dataset, we do not put prior on the disease, and over 5000 diseases are considered, with a balanced ratio of “yes” or “no” responses.
Medical visual question answering
This task is a combination of popular visual question-answering challenges. Given a medical image and a clinically relevant question in natural language as a prompt, the medical VQA system is expected to predict a plausible and convincing answer.
Radiology report generation
This task focuses on the automatic generation of reports, i.e., summarizing the radiologic findings based on radiology images, such as X-rays, CT scans, and MRI scans. Given a medical image, we randomly select a prompt sentence like “Please caption this scan with findings” as input.
Rationale diagnosis
This task involves analyzing radiology images to predict both the underlying disease and the typical radiologic features of different modalities, such as X-rays, CT scans, and MRI scans associated with that disease. Specifically, we randomly select a prompt sentence like “Determine the disease that corresponds to the given radiographic images, starting with the established radiological features and concluding with the ultimate diagnosis.” Since we have evaluated disease diagnosis accuracy in the common “Disease Diagnosis” setting, for rational diagnosis, we mainly focus on how well the foundation model can give reasons.
Building generalist foundation model for radiology
In this section, we start by describing the paradigm for unifying different medical tasks into a generative framework, followed by detailing the proposed RadFM model, and its training details. Our training adopts two types of datasets, namely, interleaved datasets and visual instruction datasets. It is worth noting that their training objectives differ slightly, which will be detailed in the following.
A unified learning paradigm
In both of our proposed multimodal datasets, i.e., MedMD and RadMD, each training sample is essentially consisting of two elements, i.e., \({{{\mathcal{X}}}}=\{{{{\mathcal{T}}}},{{{\mathcal{V}}}}\}\), where \({{{\mathcal{T}}}}\) refers to the language part in the case, with special placeholder tokens for images, e.g., “The patient is 47-year-old. 〈image-1〉 〈image-2〉 We can see opacity on the X-ray”. \({{{\mathcal{V}}}}\) refer to the visual parts containing a set of 2D or 3D image scans, i.e., \({{{\mathcal{V}}}}=\{{v}_{1},{v}_{2},\ldots,{v}_{N}\}\), \({v}_{i}\in {{\mathbb{R}}}^{H\times W\times C}\) or \({v}_{i}\in {{\mathbb{R}}}^{H\times W\times D\times C}\), H, W, D, C are height, width, depth, and channel, respectively, corresponding to the “〈image-i〉 ” token in \({{{\mathcal{T}}}}\). In general, \({{{\mathcal{T}}}}\) and \({{{\mathcal{V}}}}\) can be considered as prompts input to model with interleaved language and image.
The goal is to model the likelihood of generated text tokens in \({{{\mathcal{T}}}}\), conditioned on interleaved scans as:
where \({{{{\mathcal{T}}}}}_{l}\) represents the l-th token in \({{{\mathcal{T}}}}\) and \({{{{\mathcal{V}}}}}_{ < l}\), \({{{{\mathcal{T}}}}}_{ < l}\) represent the image and language text appearing before the l-th token. We use a generative model (ΦRadFM) to parameterize the probability p, and our final training objective can be expressed as the negative log-likelihood of the correct next token in the text sequence:
where wl refers to a per-token weighting, aiming to either emphasize key tokens or skip special tokens. Its value differs for different datasets and we detail this in the following.
Interleaved datasets. For samples in visual-language interleaved dataset, i.e., PMC-Inline, there are no strong question-and-answer relationships between contexts, we extract medical-related words in each sentence by using unified medical language system (UMLS)43, and give them a high loss weights. Additionally, we avoid calculate loss on the image placeholder token. Overall, wl can be formulated as,
Note that, PMC-Inline is the only dataset fit in this case.
Visual instruction datasets. For samples from visual instruction datasets like PMC-VQA8 or PMC-CaseReport, they are often in the format of dialogue, for example, “What can you see from the image? 〈image-1〉 I can see lesions.” or “Please describe the scans 〈image-1〉. The scan is …”, we further separate the language part \({{{\mathcal{T}}}}\) into instruction and response, denoted as \({{{\mathcal{I}}}}\) and \({{{\mathcal{R}}}}\) respectively. For example, as in the former two cases, \({{{\mathcal{I}}}}\) refers to “What can you see from the image? 〈image-1〉 ” and “Please describe the scans 〈image-1〉 ”. In a practical scenario, \({{{\mathcal{I}}}}\) is expected to be given by users, and the model is only required to output correct responses. Overall, wl can be formulated as,
Most samples from MedMD fit the weighting formulation. All prompts used for instruction tuning are listed in the Supplementary Tables 8–11. We describe the detailed prompting for different problem settings:
-
Modality recognition. Here, we adopt two types of prompts, (i) we use inductive prompts, and the 2D or 3D medical scan as input, for example, “〈image-1〉 Is this image captured by {modality}?”, and the modality category is randomly sampled from the modality set, forming the text input \({{{\mathcal{I}}}}\) and if the modality matches the ground truth labels we set the \({{{\mathcal{R}}}}\) as “yes” otherwise “no”. (ii) we use open prompts, like “What’s the modality of the input scan 〈image-1〉 ?” to form the \({{{\mathcal{I}}}}\), and translate the corresponding modality label into \({{{\mathcal{R}}}}\). Samples for training such functionality are from RP3D-Modality and MPx-Single, with modality annotations available.
-
Disease diagnosis. All the datasets listed as “image data” in Supplementary Table 3 are built for diagnosis, they only have binary labels for diseases. Similarly to modality recognition, we use two prompts to transform them into our desired format, (i) we use inductive prompts, like “〈image-1〉 Does the patient have {disease}?” and the disease category is randomly sampled from a disease set, forming the text input \({{{\mathcal{I}}}}\) and if the disease matches the ground truth labels we set the \({{{\mathcal{R}}}}\) as “yes” otherwise “no”, note that, during sampling, we balance the positive and negative ratio, (ii) we use open diagnosis prompts, like “Please make diagnosis based on the images 〈image-1〉 〈image-2〉.” to construct the instruction (\({{{\mathcal{I}}}}\)), and translate the positive disease labels into response (\({{{\mathcal{R}}}}\)), by simply using their category names. A simple example is, \({{{\mathcal{I}}}}\)="Please make diagnosis based on the image 〈image-1〉.” with \({{{\mathcal{R}}}}\) = “Edema, pneumothorax.”. With such instruction, the model is thus required to complete a difficult task, i.e., directly outputting the disease name.
-
Visual question answering. Beyond the abovementioned task formulation, there are more complex questions that can be asked, such as those about the spatial relationships among objects ("What is the location of the lesion?”) and common sense reasoning questions ("Given the image context and patient history, what is likely to be the cause of the observed symptoms?”). A robust medical VQA system must be capable of solving a wide range of classic medical diagnosis tasks, as well as the ability to reason about images. Existing medical VQA datasets like VQA-RAD32, SLAKE33, PMC-VQA8 and RP3D-VQA naturally fit into this paradigm. They contain a mixture of question types, thus the language questions can naturally be treated as text instruction (\({{{\mathcal{I}}}}\)) and the corresponding answer as response (\({{{\mathcal{R}}}}\)). It is worth noting that, our constructed PMC-CaseReport dataset also falls into this category, with more contextual information available for instruction, for example, history diagnosis, is also available, thus providing critical information for answering the question.
-
Report generation. MIMIC-CXR30, RP3D-Caption, PMC-OA31, MPx-Multi, and MPx-Single are all captioning datasets, the task is to write a long caption or report given one or a set of images. The language instruction for this task are like “What can you find from the scans 〈image-1〉 〈image-2〉?”.
-
Rationale diagnosis. We construct RP3D-Rationale based on the RP3D dataset. This task encompasses disease prediction and the generation of typical radiological features associated with the diagnosed disease. Specifically, we design some prompts like “What disease can be diagnosed from these radiological images and what specific features are typically observed on the images? 〈image-1〉 〈image-2〉 ” as instruction (\({{{\mathcal{I}}}}\)), and response (\({{{\mathcal{R}}}}\)) refers to the disease label along with radiological features collected from the Radiopaedia website.
Architecture detail
In this section, we aim to describe the proposed model in detail. As shown in Fig. 1c, our proposed RadFM model consists of a visual encoder Φvis, that can process both 2D and 3D medical scans; a perceiver44 module Φper for aggregating a sequence of scans into a fixed number of tokens, for example, taken with different modalities (CT, MRI) or various time point; and a large language model (LLM) Φllm that enables to generate free-form text responses, based on the input visual-language information.
Visual encoding. Given one sample instance from our dataset, denoted as \({{{\mathcal{X}}}}=\{{{{\mathcal{T}}}},{{{\mathcal{V}}}}\}\), where \({{{\mathcal{V}}}}=\{{v}_{1},{v}_{2},\ldots,{v}_{N}\}\), we first encode each input image separately with an image-encoder Φvis. Specifically, we adopt 3D ViT here to be compatible with both 2D and 3D image input. For 2D images, we expand a new dimension for depth by replicating the slices. Therefore, each image scan can be denoted as \({v}_{i}\in {{\mathbb{R}}}^{H\times W\times {D}_{i}\times C}\), where C denotes the image channels and H, W, Di are the height, width, and depth of the image, respectively. The rationale behind this design choice is as follows: (i) increasingly more radiology diagnosis rely on 3D scans, for example, CT, MRI, the foundation model should certainly be able to process 3D data input; (ii) in 3D data, consecutive slices are highly similar, thus padding 2D into 3D, on the one hand, does not lead information loss, on the other hand, resembles a good approximation of 3D data; (iii) padding 2D images will only affects the tokenization layer, i.e., converting image patches into continuous embedding, while still keep the rest of model shared with 3D scans, thus facilitating knowledge share.
Note that, comparing to the typical visual encoding scenario that assumes different images have unified shape, we do not normalize the depth dimension into an exact size, only round into a factor of 4, depending on their original resolution. Note that, all the 2D images are padded into four slices on the depth channel. We convert the image into 3D patches, embed them into a token sequence, and feed into the encoder (Φvis). To retain the 3D position of these tokens, we adopt learnable 3D position embeddings, the detailed procedure can be formulated as:
where vi is the output embedding for image vi, encoded with 3D ViT, Pi is the total number of tokens, and d is the feature dimension. Due to the inconsistency in depth dimension, Pi varies across 2D and 3D images, and the model can get to know the original image size by positional encoding.
Aggregation with perceiver. After visual encoding, we adopt a perceiver44 module Φper to aggregate visual representation. Specifically, Φper follows the classical perceiver architecture with a fix number of learnable queries as the latent array input, and the visual embedding vi is treated as the byte array input, so that the final output embeddings will be normalized into the same length with the pre-defined learnable query sequence. The aggregation procedure can be formulated as:
where ui refers to the aggregated visual embedding, P denotes the number of learnable queries. Leveraging perceiver architecture, we can map an arbitrary number of patch tokens into the same length, such that images of different sizes can be treated equally in the following fusion flow.
Multimodal fusion. To fuse the visual-language information, we interleave the visual embedding with text embeddings from tokenization, where the special image placeholder token is simply replaced with the corresponding visual embedding. The resulting interleaved sequence is then passed into a decoder-only large language model (Φllm), the self-attention transformer layers in LLM can thus naturally be reused as multi-modal fusion modules:
where ti, ui refer to the text and visual embeddings, p is the probability distribution for the next token.
Training procedure
Our training procedure includes two stages, namely, pretraining, and domain-specific finetuning, as shown in Fig. 1b. Note that, all training settings remain identical at two stages, with the only distinction lying in the training data, from generalist to radiologic-specific.
Generally, all the data used for model training is listed in Supplementary Table 2 with citations indicating their sources (those without citations denoting the data are contributed by this work). For pretraining, all the listed data are employed. While for domain-specific instruction tuning, we further filter out some relatively low-quality data, i.e., generated data without human verification or non-radiology data, focusing more on high-quality question-answering pairs. Next, we will describe this in detail.
Pretraining. At this stage, we use all available data in MedMD as listed in Supplementary Table 3, the main components of the data are PMC-Inline and PMC-OA31, which are all collected from 2.4M PMC papers. These two datasets contain diverse medical vocabularies and images with cutting-edge medical knowledge, however, they are relatively noisy, so we only use them during pretraining in the hope that the network can accumulate enough knowledge about medical-specific terminologies and images. Additionally, we also include other VQA, captioning, and diagnosis datasets, as they are much cleaner.
Domain-specific Instruction Tuning. At this stage, we adopt RadMD for domain-specific instruction tuning, which contains over 3M radiologic images, with high-quality language instructions and responses. In this stage, we utilize RadMD for domain-specific instruction tuning, which includes over 3M radiological images accompanied by high-quality language instructions and responses. Notably, we filter out PMC-Inline and PMC-OA, as these datasets are not derived from real clinical scenarios. For the remaining data sources, we primarily filter out non-radiology-related content. Specifically, the filtering process targets the MPx-series, RP3D-series, and PMC-CaseReport datasets. For both MPx-series and RP3D-series, the filtering is straightforward since the original websites provide related imaging modalities for each case. For PMC-CaseReport, which is generated from the case reports subset of PMC-Inline using ChatGPT, we rely on the image captions to filter the cases. Only those with captions explicitly mentioning radiology-related terms—such as “MRI”, “CT”, “X-ray”, “ultrasound”, or “mammography”—are retained. We acknowledge that some noisy cases may still remain in the dataset. Therefore, in our evaluation dataset, RadBench, the selected test cases undergo additional manual inspection to further ensure quality.
Training details
Image preprocessing. To dismiss the differences of medical images in different modalities, certain preprocessing steps are applied. Specifically, (i) to align the intensity distributions, we employ min-max normalization of all images; (ii) given that medical images can exist in either 3D or 2D formats (such as MRI being 3D and X-ray being 2D), we convert all 2D images to 3D simply by expanding an extra dimension. Consequently, all images, irrespective of their original format, can be processed uniformly as 3D images; (iii) to ensure consistent sizes across all images, we resize them using the torchvision.transforms.Resize function. For height and weight dimensions, we resize them to 512 × 512 for 2D images and 256 × 256 for 3D images because 3D data has more slices, thus taking more computational memorization. For the depth dimension, since our visual encoder, a 3D vision transformer (ViT), requires the input image sizes to be divisible by the patch size of 32 × 32 × 4, we resize the depth dimension to the nearest multiple of 4 and will not surpass 64. Please check the Supplementary Table 6 to obtain more details.
A detailed forward example. To better illustrate our model architecture, we present a simple instruction tuning example: a radiology image paired with the text prompt “Does the case 〈image〉 have pneumonia?”, with the ground truth response “Yes.” The model forward procedure will include three main steps, i.e., visual encoding, text fusion, and loss calculation. Visual encoding: A 2D image is first expanded into a pseudo-3D format by adding an extra dimension of size 4. It is then processed by a 3D Vision Transformer (ViT) to produce visual tokens. These are compressed to a fixed length of 32 using a perceiver module, ensuring consistent input regardless of image size. Text fusion: The text prompt is tokenized using the LLM’s embedding layer, and the “〈image〉 ” placeholder is replaced with the visual tokens. This fused sequence is input to the LLM’s causal self-attention layers for multimodal understanding. Loss calculation: The model predicts the next tokens auto-regressively, and the loss is computed against the ground truth “Yes”. During pretraining, the same forward process is used, but the loss is calculated over all text tokens except the image placeholder, following GPT-style training.
Implementation. For the visual encoder, we adopt a 12-layer 3D ViT with 768 feature dimensions and the perceiver is chosen as a six-layer transformer decoder with a learnable latent array in 32 × 5120 dimensions, so that all images will be embedded as a 32 × 5120 feature embedding after passing visual encoding and perceiver aggregation. When inserting them into the text embedding, we will add two extra special tokens 〈image〉, 〈/image〉 at the beginning and ending, respectively, to distinguish them from common text tokens. For the large language model, we initialize it with the MedLLaMA-13B model introduced by PMC-LLaMA25, which has further finetuned the LLaMA-13B2 model on the medical corpus. Our final model has 14B parameters.
In training, we vary the batch size, i.e., one batch size per device for 3D images and four batch size per device for 2D images with four-step gradient accumulation, and the max token length is set to be 2048. We totally train the model for eight epochs, four epochs for pretraining and four epochs for instruction tuning. In the first one epoch, we freeze the language model to align image embedding space with that of texts, in the following epochs, all parameters are updated. To improve the training speed, we adopt FSDP acceleration strategy45, together with automatic mixed precision (AMP) and gradient checkpointing46. All models are implemented in PyTorch and trained on 32 NVIDIA A100 GPUs with 80 GB memory.
Evaluation
In this section, we introduce three evaluation settings, i.e., zero-shot, few-shot and task-specific evaluation, together with the models in comparison. Note that, the first two evaluations require no further training, while the last requires additional finetuning on specific tasks. Afterward, we introduce the automatic metrics and human rating progress.
Zero-shot and few-shot evaluation
Foundation models, as a generalist model, the most appealing characteristic is that they can be applied to various tasks just with proper prompting strategies, like zero-shot or few-shot prompting, without any specific training. In the zero-shot setting, models will be given task-related semantic instructions to indicate which task it is expected to perform, and in the few-shot prompting scenario, some similar cases related to the task will be given instead. The insight of both is to use appropriate textual instructions to prompt the model on what tasks to perform, while which one is more suitable for a certain model depends on its training approach.
Baselines. For our RadFM, we mainly adopt zero-shot evaluation, as in the instruction tuning step, we focus on promoting the model to understand diverse zero-shot instructions. For other baselines, we compare with the following publicly accessible foundation models under these two settings, as follows:
-
OpenFlamingo13. This is an open-source implementation of the prior state-of-the-art generalist visual-language model Flamingo22, that was trained on large-scale data from general visual-language domain. We utilized the released checkpoint for zero-shot and few-shot evaluation in our study.
-
MedVInT8. This is a visual instruction-tuned visual-language model based on LLaMA2, which was trained on PMC-VQA8. Considering that the PMC-VQA data does not contain any few-shot cases, mainly targeting at zero-shot prompting cases, we directly use the released checkpoint of the MedVInT-TD model with PMC-LLaMA and PMC-CLIP backbone for zero-shot evaluation.
-
LLaVA-Med4. LLaVA-Med is a medical-specifical vision-language foundation model trained based on LLaVA47 leveraging zero-shot instruction tuning dataset generated from pubmed image-caption pairs. Similar to MedVInT, it also mainly targets zero-shot prompting cases and we directly use the released checkpoint LLaVA-Med-v1.5 for zero-shot evaluation.
-
Med-Flamingo6. This is a multimodal model developed based on OpenFlamingo-9B13, that can handles multi-image input interleaving with texts. We use the released checkpoint for zero-shot and few-shot evaluation.
-
GPT-4V14. GPT-4V is widely considered as the most powerful multi-modal foundation model, released by OpenAI. Since until our submission, GPT-4V can only input 4 images which can hardly allow few-shot cases with multiple images, thus we evaluate it in zero-shot cases Besides, GPT-4V can be only accessed through the online chatting website, therefore, large-scale auto-evaluation is not feasible. In this paper, we only use it for evaluation under the human rating setting.
For OpenFlamingo and Med-Flamingo, we perform both zero-shot and few-shot evaluations in our study. Specifically, we follow the prompts derived from the official Med-Flamingo repository. The example prompt for zero-shot evaluation: ‘You are a helpful medical assistant. Please answer the question about the given image. 〈image〉 Question: the query question. Answer:”. In the few-shot setting, we expand upon this format by supplying the models with additional examples to guide their responses. This is structured as follows: “You are a helpful medical assistant. You are being provided with images, a question about the image, and an answer. Follow the examples and answer the last question. 〈image〉 Question: [the first question]. Answer: [the first answer]. 〈 —endofchunk—〉 〈image〉 Question: [the second question]. Answer: [the second answer]. 〈 —endofchunk—〉 〈image〉 Question: the query question. Answer:”.
To our knowledge, there are currently no existing foundation models that can effectively handle both 2D and 3D radiology images. For comparison, we have strong baseline models that are publicly accessible, for example, OpenFlamingo13, MedVInT8, LLaVA-Med4, and Med-Flamingo6, which have demonstrated efficacy in processing slices and making predictions. In addition, we also compare with GPT-4V(ision)14 use its online chatting website version.
Datasets. We evaluate the above foundation models on RadBench and 9 exising datasets as introduced in section “Radiology evaluation benchmark (RadBench)”. Additionally, we also evaluate them on PadChest48. It is a labeled large-scale, high-resolution chest x-ray dataset including 160,000 images obtained from 67,000 patients, with 174 different radiographic finding labels. We dismiss the classes with cases fewer than 10 together with the seen classes appearing in our training set, resulting in 163 totally unseen classes. We therefore ensure that not only images, but also categories in the texts never appear in the training, which requires more generalization ability of models.
Task-specific evaluation
In addition to directly evaluating different foundation models using zero-shot or few-shot prompting, without any training, our model can also serve as a pretrained model, that can be adapted to different specific tasks by further finetuning on its corresponding training set, giving up the ability to generalize between tasks, but getting better performance on a specific task. In such a case, we compare our final results with different task-specific state-of-the-arts (SOTAs) according to the related datasets. In detail, we use the following datasets, and the corresponding SOTAs for comparison are listed in Table 3 with citations:
-
VinDr-Mammo49 is a mammography diagnosis dataset comprising 20,000 images (5000 four-view scans). Each scan was manually annotated with a five-level BI-RADS score. We view this as a multi-class classification task with the official split following the BenchMD50.
-
CXR1451 is a widely-used chest X-ray diagnosis dataset containing 112,120 frontal-view X-ray images of 30,805 (collected from the year of 1992 to 2015) unique patients with 14 finding labels. We follow its official split and evaluate the SOTA52 on the split.
-
LDCT53 Low dose computed tomography (LDCT) is a procedure that uses an x-ray machine linked with a computer to create 3D images of a patient’s tissues and organs. LIDC-IDRI53 dataset is used here, containing 1018 low-dose lung CTs, where each CT has small/large/no nodule labels. We follow BenchMD50 to set this dataset as a 3D diagnosis task and split it follow BenchMD.
-
MosMedData54 is a set of 1110 3D CT cases labeled with COVID-19 related findings, as well as without such findings. We view it as a classification task and split it randomly with 8:2 for training and testing following54.
-
COVID-CT55 is a set of 349 2D CT slices labeled with COVID-19 collected from 216 patients. We split it randomly with an 8:2 ratio for training and testing.
-
BraTs201932 is an MRI dataset with four MRI modalities T1WI, T2WI, T2FLAIR, and T1 contrast-enhanced(T1CE). There are 259 volumes of high-grade glioma (HGG) and 73 volumes of low-grade glioma (LGG). We follow the setting as DSM56 that uses T1CE to diagnose the HGG or LGG. Due to the original paper did not release their splits we randomly split the dataset following 7:3 for training and testing and re-tested the SOTA on it.
-
ADNI (Alzheimer’s disease neuroimaging initiative)57 is a large collection alzheimer’s disease dataset with 3D brain MRI scans. We follow the setting introduced in ref. 58 and split it randomly 8:2 for training and testing.
-
BTM-17 (Brain-tumor-17)59 is a challenge about classifying an MRI case into 17 tumor types, with 4449 real images. We adopt its official split.
-
Lung-PET-CT-Dx60 consists of CT and PET-CT DICOM images of 355 lung cancer subjects. We treat it as a diagnosis dataset to further distinguish whether one patient is diagnosed with Adenocarcinoma, small cell carcinoma, large cell carcinoma, or squamous cell carcinoma. Considering its limited case number, we split it with 7:3 (train:test) to ensure enough cases for evaluation.
-
VQA-RAD32 is a radiology VQA dataset containing 3515 questions with 517 possible answers. We follow the official dataset split for our evaluation.
-
SLAKE33 is an English-Chinese medical VQA dataset composed of 642 images and 14K questions. There are 224 possible answers in total. We only use the “English” part, and follow the official split.
-
PMC-VQA8 is an English medical VQA dataset generated with auto-nlp methods containing 149K images with 227K questions. Its answers are diverse for different questions. Considering its test set is also auto-generated, we have manually cleaned it as mentioned in section “Radiology Evaluation Benchmark (RadBench)” and retest the SOTA MedVInt8 checkpoint on the cleaned test set.
-
MedDiffVQA61 is a large-scale dataset for difference medical VQA (involving historical comparison) in medical chest x-ray images with 700,703 pairs of question-answer. We follow its official split.
-
IU-X-ray62 is a set of chest X-ray images paired with clinical reports. The dataset contains 7470 pairs of images and reports. We follow the setting and split as CDGPT263 where we use a single-view image to generate the reports.
Evaluation metrics
Machine rating. We evaluate on four distinct tasks, e.g., disease diagnosis, visual question answering, report generation and rationale diagnosis. The details of the four tasks and automatic metrics are introduced in section “Radiology Evaluation Benchmark (RadBench)”. To evaluate the model’s performance across a range of tasks, distinct evaluation metrics are employed based on the task type. For tasks with pre-defined answer choices, such as disease diagnosis, we adopted standard metrics developed in the community, for example, F1 stands for “F1 score”, and ACC stands for “Accuracy”. Conversely, for tasks involving open-ended responses, like report generation and visual question answering (VQA) and rationale diagnosis, alternative evaluation metrics, like BLEU, ROUGE and BERT-sim are employed. BLEU stands for “BiLingual Evaluation Understudy”64, ROUGE stands for “Recall-Oriented Understudy for Gisting Evaluation”65. BERT-sim stands for “BERT similarity score”, the F1 BERT score between the generated answer and the correct answer66. For BLEU and ROUGE, if not specific pointing, we all use 1-gram by default.
In addition, inspired by the score RadCliQ12 designed specifically for evaluating generated chest X-ray reports, we also propose two new metrics, UMLS_Precision and UMLS_Recall, which aim to measure the overlapping ratio of medical-related words between ground truth and predicted response. Specifically, given a pair of ground-truth and prediction, we extract the medical-related words from them by using unified medical language system (UMLS)43, and count the overlap words as true-positive. UMLS_Precision is defined with the classical precision concept, i.e., the number of true-positive divides the whole generated medical-related word number. On the other hand, UMLS_Recall also follows the recall concept, i.e., the number of true-positive words divides the total number of medical-related words in the ground truth.
Discussion on automatic metrics. Despite these automatic metrics have been widely adopted by the community, they often struggle to capture the semantic accuracy in generative tasks, for example, question answering, report generation, and rationale generation. To address these limitations and ensure a more accurate evaluation of system performance, we incorporate human evaluation, leveraging the expertise of radiologists, to get a professional evaluation on the quality of generated answers.
Human rating. For the sake of clinical utility, we further involve manual checking in the evaluation stage and compute the human rating score. Three radiologists were asked to rate the quality of the generated answers using a 0–5 scale. Each radiologist has five years of clinical experience in radiology departments. One is affiliated with Shanghai General Hospital, and the other two are from Shanghai Sixth People’s Hospital. All three completed their studies in “Medical imaging and nuclear medicine” at Shanghai Jiao Tong University. Here are the specifics of each rating:
-
1.
Garbled - The content is incomprehensible and lacks any readability.
-
2.
Inaccurate - While readable, the content is entirely incorrect and lacks meaningful information.
-
3.
Partially informative - The content holds some reference value, yet its correctness is subpar.
-
4.
Moderately accurate - The content provides reference points, with approximately half of the information being correct, but containing several errors.
-
5.
Mostly accurate - The content is almost entirely correct, with only a few omissions or errors present.
-
6.
Completely correct - The content is accurate in its entirety, without any mistakes.
To facilitate this assessment, we have developed a human evaluation interface, visually presenting the generative instances with images, as depicted in Supplementary Fig. 2. Prior to the full evaluation, we conducted a preliminary exam involving 20 randomly sampled test cases. This exam was designed to ensure that the radiologists understood the evaluation criteria. All three radiologists showed consistent results, with one exception: for one case, one radiologist rated the answer as 2 while the others rated it as 3. This indicates that our five-point rating system was sufficiently clear for evaluating the model’s outputs. The exam results were also reviewed by a senior radiologist with over 10 years of experience from the radiology department of Shanghai Sixth People’s Hospital, further confirming the validity of the evaluation process. In the evaluation, raters are provided with images, the question, the correct answer, and a set of generated responses from different models, arranged in a randomized order. The evaluation score given by the professional radiologists differs from the automatic evaluation metrics, offering greater accuracy and flexibility. In the context of the report generation example shown in the figure, they focus on the most crucial aspects, rather than solely on word matching, recall or precision.
Note that, human rating is only performed for the open-ended tasks, i.e., medical VQA, report generation and rationale diagnosis. As for disease diagnosis, their answers are fixed without confusion; thus, the automatic metrics can already well reflect the performance. Considering the cost for human rating, for each open-ended task, we randomly sample 400 test cases from RadBench, as they are generally collected from clinical practice across the world, and can represent real scenarios, resulting in 1.2K cases for human rating in total.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data used in this study can be downloaded via the links provided in Supplementary Table 5. Most datasets can be directly downloaded from the listed websites and used for non-commercial purposes. For the RP3D and MPx datasets, due to licensing restrictions, we cannot release the original data directly. However, links to the official websites are provided. For MPx, please ensure you review the official MedPix license and mail us to request the download link to the data. For RP3D, users can contact the official Radiopaedia licensing team at license@radiopaedia.org to obtain usage approval. Once approved, the confirmation can be shared with us, and we will provide the detailed data download link. Commonly, we will respond to inquiries regarding the two datasets within 3–5 business days. Most figures in this paper include detailed numerical annotations; the only exception is Fig. 4, for which the data is provided in the Source Data file. Source data are provided with this paper.
Code availability
The code is available on GitHub at https://github.com/chaoyi-wu/RadFM67.
References
Bommasani, R. et al. On the opportunities and risks of foundation models. Preprint at arXiv:2108.07258 (2021).
Touvron, H. et al. Llama: Open and efficient foundation language models. Preprint at arXiv:2302.13971 (2023).
Li, J., Li, D., Savarese, S. & Hoi, S. Blip-2: bootstrapping language-image pre-training with frozen image encoders and large language models. In Proc. 40th International Conference on Machine Learning 19730–19742 (PMLR, 2023).
Li, C. et al. Llava-med: Training a large language-and-vision assistant for biomedicine in one day. In Proc. 37th International Conference on Neural Information Processing Systems 28541–28564 (NeurIPS, 2023).
Moor, M. et al. Foundation models for generalist medical artificial intelligence. Nature 616, 259–265 (2023).
Moor, M. et al. Med-flamingo: a multimodal medical few-shot learner. In Proc. 3rd Machine Learning for Health Symposium 353–367 (PMLR, 2023).
Tu, T. et al. Towards generalist biomedical AI. NEJM AI 1, AIoa2300138 (2024).
Zhang, X. et al. Development of a large-scale medical visual question-answering dataset. Commun. Med. 4, 277 (2024).
Tiu, E. et al. Expert-level detection of pathologies from unannotated chest x-ray images via self-supervised learning. Nat. Biomed. Eng. 6, 1399–1406 (2022).
Zhang, X., Wu, C., Zhang, Y., Wang, Y. & Xie, W. Knowledge-enhanced pre-training for auto-diagnosis of chest radiology images. Nat. Commun. 14, 4542 (2023).
Monshi, MaramMahmoudA., Poon, J. & Chung, V. Deep learning in generating radiology reports: a survey. Artif. Intell. Med. 106, 101878 (2020).
Yu, F. et al. Evaluating progress in automatic chest x-ray radiology report generation. Patterns 4, 100802 (2023).
Awadalla, A. et al. OpenFlamingo: an open-source framework for training large autoregressive vision-language models. Preprint at arXiv 2308.01390 (2023).
OpenAI (2023). Chatgpt can now see, hear, and speak. https://openai.com/blog/chatgpt-can-now-see-hear-and-speak (2023).
Peng, B, Li, C., He, P., Galley, M. & Gao, J. Instruction tuning with gpt-4. Preprint at arXiv:2304.03277 (2023).
OpenAI. Gpt-4 technical report. Preprint at arXiv abs/2303.08774 (2023).
Wang, Y. et al. Super-naturalinstructions: generalization via declarative instructions on 1600+ NLP tasks. In Proc. 2022 Conference on Empirical Methods in Natural Language Processing 5085–5109 (Association for Computational Linguistics, 2022).
Hamamci, I. E. et al. Generatect: text-conditional generation of 3D chest CT volumes. In Proc. European Conference on Computer Vision (ECCV) 126–143 (Springer, 2024).
Wang, W. et al. When an image is worth 1,024 x 1,024 words: a case study in computational pathology. Preprint at arXiv:2312.03558 (2023).
Kaplan, J. et al. Scaling laws for neural language models. Preprint at arXiv:2001.08361 (2020).
Anil, R. et al. PaLM 2 technical report. Preprint at arXiv:2305.10403 (2023).
Alayrac, J.-B. et al. Flamingo: a visual language model for few-shot learning. In Proc. 36th International Conference on Neural Information Processing Systems 23716–23736 (NeurIPS, 2022).
Singhal, K. et al. Towards expert-level medical question answering with large language models. Nat. Med. 31, 943–950 (2025).
Singhal, K. et al. Large language models encode clinical knowledge. Nature 620, 172–180 (2023).
Wu, C., Zhang, X., Zhang, Y., Xie, W. & Wang, Y. PMC-LLaMA: further finetuning LLaMA on medical papers. J. Am. Med. Inform. Assoc. (2023).
Chen, Z. et al. Meditron-70b: scaling medical pretraining for large language models. Preprint at arXiv:2311.16079 (2023).
Zhu, W. et al. Multimodal C4: an open, billion-scale corpus of images interleaved with text. In Proc. 37th International Conference on Neural Information Processing Systems 8958–8974 (NeurIPS, 2023).
Krishna, R. et al. Visual genome: connecting language and vision using crowdsourced dense image annotations. Int. J. Comput. Vis. 123, 32–73 (2017).
Schuhmann, C. et al. Laion-5b: an open large-scale dataset for training next generation image-text models. Adv. Neural Inf. Process. Syst. 35, 25278–25294 (2022).
Johnson, AlistairE. W. et al. Mimic-cxr, a de-identified publicly available database of chest radiographs with free-text reports. Sci. Data 6, 317 (2019).
Lin, W. et al. Pmc-clip: contrastive language-image pre-training using biomedical documents. In Medical Image Computing and Computer Assisted Intervention (MICCAI) 2023. (Springer, 2023).
Lau, J. J., Gayen, S., Ben Abacha, A. & Demner-Fushman, D. A dataset of clinically generated visual questions and answers about radiology images. Sci. Data 5, 1–10 (2018).
Liu, B. et al. Slake: a semantically-labeled knowledge-enhanced dataset for medical visual question answering. In 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI) 1650–1654 (IEEE, 2021).
Zhang, K. et al. A generalist vision–language foundation model for diverse biomedical tasks. Nat. Med. 30, 3129–3141 (2024).
Chen, Z. et al. Chexagent: towards a foundation model for chest x-ray interpretation. Preprint at arXiv:2401.12208 (2024).
Chen, J. et al. Huatuogpt-vision, towards injecting medical visual knowledge into multimodal llms at scale. Preprint at arXiv:2406.19280 (2024).
He, S. et al. Meddr: diagnosis-guided bootstrapping for large-scale medical vision-language learning. Preprint at arXiv:2404.1527 (2024).
Hamamci, I. E. et al. A foundation model utilizing chest CT volumes and radiology reports for supervised-level zero-shot detection of abnormalities. Preprint at arXiv:2403.17834v1 (2024).
Lu, H. et al. Deepseek-vl: towards real-world vision-language understanding. Preprint at arXiv:2403.05525 (2024).
Wang, P. et al. Qwen2-vl: enhancing vision-language model’s perception of the world at any resolution. Preprint at arXiv:2409.12191 (2024).
Ding, J. et al. Longnet: scaling transformers to 1,000,000,000 tokens. In Proc. 10th International Conference on Learning Representations (ICLR, 2023).
Zhao, Z., Jin, Q., Chen, F., Peng, T. & Yu, S. A large-scale dataset of patient summaries for retrieval-based clinical decision support systems. Sci. Data 10, 909 (2023).
Bodenreider, O. The unified medical language system (umls): integrating biomedical terminology. Nucleic Acids Res. 32, D267–D270 (2004).
Jaegle, A. et al. Perceiver: general perception with iterative attention. In International Conference on Machine Learning 4651–4664 (PMLR, 2021).
Zhao, Y. et al. Pytorch fsdp: experiences on scaling fully sharded data parallel. Proc. VLDB Endow. 16, 3848–3860 (2023).
Chen, T., Xu, B., Zhang, C. & Guestrin, C. Training deep nets with sublinear memory cost. Preprint at arXiv:1604.06174 (2016).
Liu, H., Li, C., Wu, Q. & Lee, Y. J. Visual instruction tuning. In Proc. 37th International Conference on Neural Information Processing Systems 34892–34916 (NeurIPS, 2023).
Bustos, A., Pertusa, A., Salinas, JoseeMaría & de la Iglesia-Vayá, María Padchest: a large chest x-ray image dataset with multi-label annotated reports. Med. Image Anal. 66, 101797 (2019).
Nguyen, H. T. et al. Vindr-mammo: a large-scale benchmark dataset for computer-aided diagnosis in full-field digital mammography. Sci. Data 10, 277 (2023).
Wantlin, K. et al. Benchmd: a benchmark for modality-agnostic learning on medical images and sensors. Preprint at arXiv:2304.08486v2 (2023).
Wang, X. et al. Chestx-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 2097–2106 (IEEE, 2017).
Wu, C., Zhang, X., Zhang, Y., Wang, Y. & Xie, W. Medklip: medical knowledge enhanced language-image pre-training. In Proc. IEEE International Conference on Computer Vision (ICCV) (IEEE, 2023).
Armato III, S. G. et al. The lung image database consortium (lidc) and image database resource initiative (idri): a completed reference database of lung nodules on ct scans. Med. Phys. 38, 915–931 (2011).
Morozov, S. P. et al. Mosmeddata: chest CT scans with covid-19 related findings dataset. Preprint at arXiv:2005.06465 (2020).
Zhao, J., Zhang, Y., He, X. & Xie, P. Covid-ct-dataset: a ct scan dataset about covid-19. Preprint at arXiv:2003.13865 (2020).
Dosovitskiy, A. et al. An image is worth 16 × 16 words: transformers for image recognition at scale. In Proc. 9th International Conference on Learning Representations (ICLR, 2021).
Petersen, RonaldCarl et al. Alzheimer’s disease neuroimaging initiative (adni) clinical characterization. Neurology 74, 201–209 (2010).
Korolev, S., Safiullin, A., Belyaev, M. & Dodonova, Y. Residual and plain convolutional neural networks for 3d brain mri classification. In 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017) 835–838 (IEEE, 2017).
Feltrin, F. Brain tumor MRI images 17 classes. Kaggle https://www.kaggle.com/datasets/fernando2rad/brain-tumor-mri-images-17-classes (2024).
Pam, A. & Tracy, N. A large-scale ct and pet/ct dataset for lung cancer diagnosis (lung-pet-ct-dx). Cancer Imaging Archive (2021).
Hu, X. et al. Expert knowledge-aware image difference graph representation learning for difference-aware medical visual question answering. In Proc. 29th ACM SIGKDD Conference on Knowledge Discovery and Data Mining 4156–4165 (Association for Computing Machinery, 2023).
Demner-Fushman, D. et al. Preparing a collection of radiology examinations for distribution and retrieval. J. Am. Med. Inform. Assoc. 23, 304–310 (2016).
Alfarghaly, O., Khaled, R., Elkorany, A., Helal, M. & Fahmy, A. Automated radiology report generation using conditioned transformers. Inform. Med. Unlocked 24, 100557 (2021).
Papineni, K., Roukos, S., Ward, T. & Zhu, W.-J. Bleu: a method for automatic evaluation of machine translation. In Proc. 40th annual meeting of the Association for Computational Linguistics 311–318 (Association for Computational Linguistics, 2002).
Lin, C.-Y. Rouge: a package for automatic evaluation of summaries. In Text Summarization Branches Out 74–81 (Association for Computational Linguistics, 2004).
Zhang, T., Kishore, V., Wu, F., Weinberger, K. Q. & Artzi, Y. Bertscore: evaluating text generation with bert. In Proc. 8th International Conference on Learning Representations (ICLR, 2020).
Wu, C. & Zhang, X. chaoyi-wu/radfm: Radfm_official_code (2025).
Chatterjee, S., Nizamani, FarazAhmed, Nürnberger, A. & Speck, O. Classification of brain tumours in mr images using deep spatiospatial models. Sci. Rep. 12, 1505 (2022).
Bazi, Y., Rahhal, MohamadMahmoudAl, Bashmal, L. & Zuair, M. Vision–language model for visual question answering in medical imagery. Bioengineering 10, 380 (2023).
van Sonsbeek, T., Derakhshani, M. M., Najdenkoska, I., Snoek, C. G. M. & Worring, M. Open-ended medical visual question answering through prefix tuning of language models. In Proc. Medical Image Computing and Computer Assisted Intervention (MICCAI) 726–736 (Springer, 2023).
Acknowledgements
This work is supported by the National Key R&D Program of China (No. 2022ZD0160702), STCSM (No. 22511106101, No. 18DZ2270700, and No. 21DZ1100100), 111 plan (No. BP0719010), and State Key Laboratory of UHD Video and Audio Production and Presentation. All the radiology examples used in our figures are obtained from the Radiopaedia website. We sincerely acknowledge their invaluable efforts.
Author information
Authors and Affiliations
Contributions
All listed authors clearly meet the ICMJE 4 criteria. C.W. and X.Z. contribute equally to this work, and Y.W. and W.X. are the corresponding authors. Specifically, C.W., X.Z., Y.Z., H.H., Y.W., and W.X. all make contributions to the conception or design of the work, and C.W. and X.Z. further perform acquisition, analysis, or interpretation of data for the work. In writing, C.W. and X.Z. draft the work and Y.Z., H.H., Y.W., and W.X. review it critically for important intellectual content. All authors approve of the version to be published and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks, Synho Do, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, C., Zhang, X., Zhang, Y. et al. Towards generalist foundation model for radiology by leveraging web-scale 2D&3D medical data. Nat Commun 16, 7866 (2025). https://doi.org/10.1038/s41467-025-62385-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-62385-7
This article is cited by
-
A data-efficient 3D medical vision-language model using only a 2D encoder
Scientific Reports (2026)
-
Bridging modalities with AI: a review of AI advances in multimodal biomedical imaging
Communications Engineering (2026)
-
Towards Automated Reporting: A Bronchoscopy Report Dataset for Enhancing Multimodality Large Language Models
Scientific Data (2026)
-
Foundation models in radiology: a primer for pediatric radiologists
Pediatric Radiology (2026)
-
Implementing generative artificial intelligence in precision oncology: safety, governance, and significance
Journal of Hematology & Oncology (2026)