Abstract
Insufficient coverage of cervical cytology screening in resource-limited areas remains a major bottleneck for women’s health, as traditional centralized methods require significant investment and many qualified pathologists. Using consumer-grade electronic hardware and aspherical lenses, we design an ultra-low-cost and compact microscope. Given the microscope’s low resolution, which hinders accurate identification of lesion cells in cervical samples, we train a coarse instance classifier to screen and extract feature sequences of the top 200 instances containing potential lesions from a slide. We further develop Att-Transformer to focus on and integrate the sparse lesion information from these sequences, enabling slide grading. Our model is trained and validated using 3510 low-resolution slides from female patients at four hospitals, and subsequently evaluated on four independent datasets. The system achieves area under the receiver operating characteristic curve values of 0.87 and 0.89 for detecting squamous intraepithelial lesions on 364 slides from female patients at two external primary hospitals, 0.89 on 391 newly collected slides from female patients at the original four hospitals, and 0.85 on 570 human papillomavirus positive slides from female patients. These findings demonstrate the feasibility of our AI-assisted approach for effective detection of high-risk cervical precancer among women in resource-limited regions.
Similar content being viewed by others
Introduction
Cervical cancer ranks as the fourth most common cancer among women worldwide and the fourth leading cause of cancer-related death in women1,2. In low- and middle-income countries such as South Africa, India, China, and Brazil, 84% to 90% of cervical cancer cases occur, imposing a substantial budgetary burden on these regions3,4. While human papillomavirus (HPV) vaccination is critical, cytology screening remains vital for reducing the incidence of cervical cancer. Historically, Pap smear programs have reduced the incidence by up to 80% in developed countries5. In recent years, the World Health Organization (WHO) has recommended HPV screening for high-throughput screening of cervical cancer6,7, and extensive work has been carried out8,9,10,11. However, HPV testing usually suffers from relatively low specificity12,13,14. Cytopathology-based screening, which directly detects cellular morphological changes, continues to serve as a routine and effective screening approach. Despite its importance, widespread implementation of cytopathology screening still encounters challenges, particularly in resource-limited regions, due to the shortage of trained cytologists and the high cost of necessary equipment.
In recent years, cost-efficiency in portable optical instruments has improved significantly. These advancements have propelled them to serve as practical alternatives to commercial systems, addressing diverse demands of bioscience, medical diagnosis15, and other fields16,17,18. For instance, portable microscopes have been effectively utilized in the diagnosis of Malaria, Sickle Cell Anemia, and Tuberculosis19,20,21. Such devices typically integrate Complementary Metal Oxide Semiconductor (CMOS) sensors and compact optical systems, sometimes even utilizing smartphone camera capabilities. In contrast to the large-size and expensive professional microscopes commonly seen in medical facilities, these portable versions dramatically reduce costs while maintaining an acceptable image quality. Concurrently, advanced medical image analysis algorithms have been developed to automate the processing of acquired images. These algorithms can perform tasks such as automated quantification of blood cells18, rapid detection of abnormal morphological targets22, and other diagnostic functions. Consequently, individuals with relatively basic training can conduct these procedures effectively. For cervical cytology, automatic analysis algorithms hold promise in assisting precancer lesion detection. The combination of compact microscopes and artificial intelligence (AI) presents a promising approach for enabling large-scale periodic cervical screening in resource-limited regions.
Over the last few decades, extensive research has focused on cervical cell classification and detection algorithms. Early approaches were primarily based on handcrafted feature extraction, cell or nuclei segmentation23,24, and traditional machine learning classifiers. Subsequent advancements introduced the convolutional neural network25 (CNN) and recent Vision Transformer26, which leverage data-driven methods to identify cervical lesion cells, improving recognition accuracy27,28,29,30,31,32,33,34,35. Despite the progress in cell-level recognition, a discernible gap persists between cell-level and slide-level diagnostic frameworks. When dealing with a gigapixel whole slide images (WSIs) of cervical cytology, which contain tens of thousands of cells or instances (local cell images), researchers have adopted multi-stage or hybrid slide-level analysis methods requiring large-scale lesion cell annotations36,37,38,39,40,41. For example, Zhu et al.39 integrated YOLOv3 for target detection, Xception and Patch-based models for target classification and U-net for nucleus segmentation along with a logical decision tree for slide classifying (using about 1.7 million annotated targets). Cheng et al.36 fused high-resolution (HR) and low-resolution (LR) imaging using CNNs to identify/recommend lesion cells, and then classified WSIs via a recurrent neural network by aggregating deep features of the top 10 recommended lesion instances (using 79,911 annotated lesion cells). Lin et al.37 designed a dual-path network with a synergistic grouping loss function for lesion retrieval, and classified slides by rule-based risk stratification (using 202,557 abnormal cell images). Wang et al.40 utilized RetinaNet to detect the top 20 instances of each class and a random forest algorithm to aggregate the detection results and classify slides. Essentially, these methods utilize high-resolution images to first train an accurate abnormal cell classifier or detector. Then, they aggregate the learned deep features or predicted positive probabilities of the detected abnormal cells in each WSI to represent and classify slides. In these methods, the effectiveness of slide classification depends critically on the initial cell detection precision. This step requires high-resolution images, which provide rich and sufficient texture details, along with large-scale annotation datasets. In contrast, low-resolution WSIs (LRWSIs) captured by a compact microscope have limited resolution and insufficient intricate textures. As a result, they are unable to support accurate cell-level discernment. Consequently, these approaches are not suitable for processing LRWSIs.
In the study, we developed an artificial intelligence-assisted cervical cancer screening system targeting populations at high risk of squamous intraepithelial lesions (SIL) in resource-limited regions. This system integrates a low-cost compact microscope with an artificial intelligence algorithm. By leveraging consumer-grade electronic hardware, our microscope achieves a cost reduction of tens to hundreds of times compared with laboratory-grade high-performance equipment, making it easier to manufacture and maintain. The hardware design employs aspherical lenses to drastically compress the optical path of the system, resulting in an ultra-compact device. The entire system demonstrates excellent usability and supports automated whole-slide scanning. Unlike existing methods that rely on high-quality images and large-scale annotations for precise abnormal cells identification (tens or a few), our approach successfully enables slide classification using low-resolution images from cost-effective hardware. We trained a coarse instance classifier to screen and extracted the feature sequence of the top 200 lesion-candidate instances from a slide and eliminate the majority of negative instances in the first-stage. This step reduces the sequence length of the instance features from several thousand down to hundreds, while still sufficiently covering potential positive cells. We further developed Att-Transformer to focus on and integrate the sparse lesion information from the above sequence, enabling slide grading. The Att-Transformer integrates a Vision Transformer backbone with a Gated-Attention module42, which focuses on the lesion areas and assigns higher weights to these features, achieving excellent feature aggregation and interpretability. In addition, we also employed pretraining and transfer learning strategies for efficient classifier training.
We trained and validated our model using 3510 LRWSIs, containing 13 datasets collected from four distinct hospitals over various time periods. In the internal test set, for 194 positive cases and 195 negative cases, we achieved an area under the receiver operating characteristic curve (AUC) of 0.845. Subsequently, the system was further evaluated on four independent test sets for detecting SIL, achieving AUCs of 0.873 and 0.891 on 364 slides from two external primary hospitals (External-A and External-B), 0.889 on 391 newly collected slides from the original four hospitals (NewCohort), and 0.855 on 570 slides from human papillomavirus-positive patients. We compared the optimal pathology WSI analysis algorithms, CLAM and TransMIL on our LRWSI datasets. AUCs below 0.600 on External-A, External-B, and NewCohort indicate that these weakly supervised multiple instance learning (MIL) methods are not suitable for cervical cytology WSIs with sparsely distributed lesion cells. Moreover, our ablation studies demonstrated that: (1) supervised features are more effective and robust in the slide classifier; (2) the proposed Att-Transformer, through its attention module, focuses on sparse lesion areas, achieving better feature aggregation and interpretability.
In this work, we demonstrate that the combination of our algorithm and the miniature microscope offers a cost-effective screening solution for cervical precancer screening in high-risk populations in regions with scarce medical resources. Individuals triaged as high-risk through this screening process are referred to tertiary care centers for further examination and treatment. We substantiate that this cost-effective and easily accessible screening method aligns with the WHO global strategy to eliminate cervical cancer globally.
Results
Compact microscope and cytology LRWSI dataset
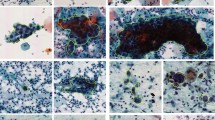
In this paper, we designed a low-cost and compact microscope for resource-limited areas, as shown in Fig. 1a. We use aspherical lenses, which significantly compress the optical path of the system. The optical module contains two lens sets: L1 and L2. L2 is fixed, and L1 is controlled by a voice coil motor (VCM) to support autofocusing. A system on a chip (SoC) is used to control the whole process and store acquired LRWSIs. The entire system has excellent automation and usability. By leveraging a consumer-grade electronic module, the cost of the whole system is only about $300, which is orders of magnitude lower than that of traditional screening systems used in medical centers. More details are shown in Methods (Compact Microscope). We visualized the image quality of a common pathology microscope and our compact microscope in Fig. 1b. For the same target, our imaging results provide supportive evidence for the morphological interpretation of cells, such as the size and contour of the nuclei. However, our images have limited expressiveness regarding details like the texture of the nuclei and cytoplasm. This limitation is evident in Fig. 1b, indicating that our images are relatively constrained in terms of noise levels and contrast. We further analyzed the impact of image quality on interpretation results (Supplementary Fig. 1) and provided standard deviation of noise, percentage of lost focus, saturation probability and optical resolution (1951 USAF resolution calculator) of image quality to illustrate the quality differences between the two imaging modes on 42 pairs of 3000 × 3000 pixel-sized registered images (Supplementary Table 1 and Supplementary Fig. 2).
a The appearance of our compact microscope and the form of an LRWSI. The optical module contains L1 and L2, two lens sets. L2 is fixed, and L1 is controlled by a voice coil motor (VCM) to support autofocusing. We use a CMOS sensor to capture the image and transmitted a light-emitting diode (LED) light source to illuminate the sample between the light source and the objective lens. A system on a chip (SoC) is used to control the whole process and storage acquired LRWSIs. An LRWSI is usually composed of about 90 views of images with 3840 × 3160 pixels and 0.87 μm/pixel. b Imaging quality comparison between a standard microscope and our compact microscope. The left composite image juxtaposes a high-resolution (HR) region (upper-right quadrant, captured by a standard microscope) with a low-resolution (LR) region (lower-left quadrant, obtained using our compact microscope). The right panel statistically quantifies contrast and noise differences: contrast level is calculated as the absolute difference between average nuclear pixel intensity and average cytoplasmic pixel intensity, divided by the average cytoplasmic pixel intensity. The noise level is defined as the standard deviation of the background pixel. Each data point in the contrast level is derived from a single cell in different images, and each data point in the noise level is derived from different images. Statistical significance (Paired samples t test, n = 13, *** denotes p < 0.001) confirms the contrast of LR images is significantly lower than that of HR images, while the noise level is significantly higher. Source data are provided as a Source Data file.
We made our best efforts to collect a diverse and substantial dataset to support our study. A total of 3135 slides from four hospitals were used to train the model, and 375 slides and 389 slides served as the validation and internal test sets, respectively. To better evaluate our model, we tested it on four external test sets: 513 slides from the same hospitals as the training set but from a different time period; 308 and 151 slides collected from another two resource-limited external hospitals; 570 slides from HPV-positive cases. Overall, a total of 5441 slides is used in our experiment. More details can be found in Methods.
Inspiration and algorithm design
Due to the microscope’s low resolution, which hinders accurate identification of lesion cells, we first trained a coarse instance classifier to screen and extract the feature sequence of the top \(k\) instances containing potential lesions from a slide in the first-stage, rather than training an accurate model for detecting several abnormal cells on high-quality images in previous methods. Considering that lesion cells are sparsely distributed in cervical cytopathology slides (Supplementary Figs. 3 and 4), we further developed Att-Transformer to focus and integrate the sparse lesion information from the above sequence, enabling slide grading. We chose 200 as the optimal value of \(k\) through conducting ablation experiments (Supplementary Fig. 5). We named our two-stage method DualCytoNet.
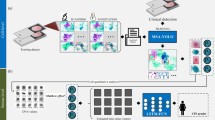
First, we divided each LRWSI into about 5700 instances redundantly, which have a size of 256 × 256 pixels and are resized to 224 × 224 pixels as the input of the first-stage Att-Transformer, which is proposed to handle sparse positive foreground cervical cytopathology images. After that, we obtained the features and positive probability of all the instances as Fig. 2a. We highlighted the instances of top 200 lesion probabilities within a slide, and aggregated their deep features (200 × 768 dimensions) to represent and classify slides by our second-stage Att-Transformer as Fig. 2b. The attention scores output by the two stages can be used separately to identify key regions (16-pixel width) in instances and to the key instances that are decisive for slide interpretation. More details are in Methods (Att-Transformer heatmap).
a Stage one. The input of the first-stage model is instances which are originally cropped from the LRWSI as 256 × 256 pixels and resize to 224 × 224 pixels. The output of the first-stage contains merged features of each instance, attention scores for patches in each instance and positive probability of each instance, which is obtained by mapping the merged feature through a linear layer. The attention scores of each 16 × 16 pixels patch are used to plot the cell-level region of interest (ROI). b Stage two. We choose features of instances which get a top 200 positive probability as input of stage two. The stage two model is similar as the first-stage model, which uses the features of top instances as input. The final output is the attention score of each input feature and the positive probability of a slide. The attention scores of each 224 × 224 pixels instance are used to plot the instance-level ROI. c We train the first-stage model by masked auto-encoder43 (MAE) based self-supervised. The pre-training process has two stages. A standard MAE pretraining is performed, followed by further pretraining with our aggregation module integrated. d The stage one model is finetuned on HR and LR labeled instances after the pretraining process. e The Att-Transformer is composed by a ViT backbone and a Gated-Attention module. The Gated-Attention module applies learnable weighting to the feature sequence output by the ViT backbone and emphasizes the sparse positive cell features in the sequence.
To ensure the robustness of our feature extraction, we introduced Att-Transformer, which represents an improvement over the ViT model as Fig. 2e. By integrating an attention module after the ViT backbone, Att-Transformer achieves a weighted aggregation of the sequential features generated by the original ViT. This augmentation effectively highlights the sparse positive cell features in the sequence, allowing them to dominate the classification results. Moreover, in comparison to the original ViT, this design promotes more seamless parameter transfer from pretraining to downstream tasks (Supplementary Fig. 6). For the first-stage model, we employed masked auto-encoder43 (MAE) based self-supervised learning for training and subsequently fine-tuned it using annotated instances, as illustrated in Figs 2c, d. The pretraining process helps the model acquire a more reliable and robust feature extraction capability across a broader range of data distributions, simultaneously diminishing its reliance on the scale of annotated data. We used data augmentation focused on color change and a false positive mining strategy to further enhance the performance of the first-stage model. More details about Att-Transformer, pretraining and finetuning can be seen in Methods.
Cervical precancerous screening for SIL cases
To evaluate the effectiveness and generalization of our DualCytoNet, we compared it with state-of-the-art WSI classifier methods CLAM44 and TransMIL45. The two methods achieved optimal outcomes on histopathology datasets through aggregating pretrained features (ResNet50 pretrained on ImageNet) of all instances within a slide to represent the slide. Initially, we trained and validated the models for classifying slides into positives and negatives. In the internal test set, our method, CLAM, and TransMIL achieved AUC of 0.845, 0.654 and 0.670, respectively. Subsequently, we further evaluated all methods for detecting SIL cases on three independent test sets: NewCohorts (n = 391), External A (n = 253) and External B (n = 111). Figure 3 shows receiver operating characteristic (ROC) curves and the sensitivity and specificity (threshold = 0.5). On the three independent test sets, CLAM and TransMIL achieved poor results of AUC < = 0.567. Our method achieved AUCs of 0.889, 0.873 and 0.891. Similarly, both the sensitivity and specificity of our method far exceed CLAM and TransMIL on the three independent test sets. We also evaluated the screening performance for detecting positive cases (atypical squamous cells, ASC; low-grade squamous intraepithelial lesion, LSIL; high-grade squamous intraepithelial lesion, HSIL) on the three test sets. Our method achieved overall average AUC of 0.780 whereas AUCs of the comparison methods are lower than 0.525 (Supplementary Fig. 7). The analysis of the false cases shows low false rate on SIL and high false rate on ASC (Supplementary Fig. 10). Besides, we found that partial misclassification attributes to several image quality problems such as under-staining or fading, bubble artifacts, insufficient cell count and inflammatory reaction (Supplementary Fig. 11). Therefore, our method can effectively identify SIL types, which aligns with our goal of screening in resource-limited areas, but its ability to identify ASC is limited.
Top figures illustrate the ROC curves of CLAM, TransMIL and our methods (DualCytoNet) on internal test (InTest), NewCohorts, External-A and External-B sets. AUC and sample size are added in the legend and above the figure, respectively. The bottom figures show the sensitivity and specificity (threshold = 0.5) of three methods for each test set. Source data are provided as a Source Data file.
Considering the relatively low specificity of the HPV test, we additionally carried out a sub-study focused on HPV-positive cases to investigate the feasibility of integrating our method with the current HPV screening approach recommended by the WHO. On 570 HPV-positive slides, we achieved an AUC of 0.850 (Supplementary Fig. 12). The results reveal that our method can effectively identify SIL cases among cases who have tested positive for HPV, improving the specificity of co-testing.
Feature representation of instances
To verify the effectiveness of the feature representations by the instance classifier, we utilized t-SNE46 to visualize the instance features within the instance test set. Ordinarily, due to the limited sampling of real-world data in datasets and the innate constraints of deep learning models, the classification boundaries modeled by the classifier lack a high degree of precision and accuracy. In the t-SNE dimensionality reduction plot, samples located at the cluster boundaries are regarded as hard samples.
As shown in Fig. 4, while there exist certain hard instances on the classification boundary that extend into the opposite class, the majority of positive and negative instances are well distinguished. In addition, our model effectively clusters the morphological characteristics of cells. For example, we can observe that the far-right area shows the negative instances with few cells, whereas the left area shows the characteristics of perinuclear halos and nuclear enlargement. Given that the LR images possess insufficient textures, some lesion cells cannot be recognized by the model and thus manifest as hard samples. These results indicate that the instance classifier based on SSL and transfer learning is effective and meets the design goal of the first-stage, which is to eliminate the vast majority of negative instances within a slide.
767 positive instances and 767 negative instances are randomly selected from the LR instance test set, and are used to get feature embedding by our first-stage model. The embeddings are reduced to two dimensions by t-SNE, and the corresponding instances and relative positions are plotted to visualize the feature space. We only show one image in one place in map to avoid overlapping. The color of the frame around the instances indicates the annotation category of them.
Supervised feature is more effective and robust for slide classifier
To further demonstrate the effectiveness of our feature extraction, we compared three different types of instance features: Supervised, Pretrained, and Pretrained on ImageNet. “Supervised” refers to the features of top-200 lesion probability instances extracted by our first-stage model. “Pretrained” denotes features of the same top-200 instances extracted by the model pretrained solely on unlabeled cervical images, while “Pretrained on ImageNet” pertains to the features of those instances obtained from a model pretrained on ImageNet. Notably, we did not use any instance-level augmentation in these experiments to reduce computation cost, thus, the results are different from that in other sections (see Train for stage two in Methods).
Figure 5 shows the ROC curves of slide classification results. For the internal test set, Supervised, Pretrained and Pretrained on ImageNet achieved high AUC of 0.921, 0.914 and 0.868 for detecting positive, respectively. In External-A, External-B, and NewCohorts test sets, Pretrained and Pretrained on ImageNet achieved average AUC of 0.557 and 0.457 for detecting SIL, respectively, while Supervised achieved an average AUC of 0.862. We can see that Pretrained and Pretrained on ImageNet features have high AUC values in the internal test set, but they decrease significantly on the independent test sets, while supervised features maintain a highly consistent level. This indicates that features of Pretrained and Pretrained on ImageNet are overfitted during slide classification, whereas supervised features are capable of identifying true lesion information among the top 200 instances and thus support a robust slide classifier. These experimental results indicate that supervised representation learning is more effective and robust compared to self-supervised representation and ImageNet-pretrained representation.
Interpretable slide classification based on Att-Transformer
We designed Att-Transformer. This model not only capitalizes on the transformer’s ability to model long and flexible feature sequences, but also leverages gated attention to weighting and aggregate instance features to represent slides. Instance features containing discriminative information are emphasized, and the attention scores provide interpretability. To better understand the model’s decision-making process, we made use of the attention scores output by our model to generate the Attention Score Map. We also generated CAMs based on the feature maps from the penultimate multi-head self-attention (MHSA) layer. We chose GradCAMElementWise47 and HiResCAM48 to plot the map. Figure 6 shows the two-stage attention mechanism. All three heatmaps consistently focus on the lesion regions within the images, and the results demonstrate similarity between the two CAMs and the attention map generated using our attention scores, indicating that the lesion areas emphasized by our model contain reliable semantic information. The heatmaps generated by our method achieve the best performance in identifying sensitive lesion regions. Here, the lesion regions primarily consist of cells with abnormally enlarged nuclei.
Cell-level ROI and instance-level ROI are acquired by the attention score of first-stage model and second-stage model. We frame out instances whose positive probability is in the top 200 as instance-level ROI in a view of a slide using a yellow square (i.e., a and b) and plot the heat map of our attention score in stage one. In addition, class activation maps (CAM) generated from the output of the penultimate multi-head self-attention layer of our model is plotted, specifically for the positive class. Two types of CAM are shown: GradCAMElementWise47 and HiResCAM48. We have augmented the dataset in Supplementary Fig. 16 with seven additional experimental replicates to demonstrate the method’s performance.
Further, we compared the classification performance of different instance aggregation methods on the same top-200 instance features extracted by our method. Figure 7 shows AUC, sensitivity and specificity for SIL and negative cases by these methods in External-A, External-B, and NewCohorts test sets. We evaluated eight different aggregation methods: MaxPool, MeanPool, ABMIL49, GatedAtt49, CLAM44, TransMIL45, and our Att-Transformer. We can see that Att-Transformer achieves optimal overall performance. In the feature sequence output by the model backbone, our attention scores assign higher weights to the tokens corresponding to the lesion areas, allowing the final output features of the model to be primarily determined by the lesion regions. All these findings underscore the interpretability and superiority of Att-Transformer as an instance feature aggregator.
Discussion
The WHO has set the goal of eliminating cervical cancer worldwide. However, the challenge remains in achieving widespread screening coverage in resource-limited regions. We have designed an extremely low-cost and compact microscope by leveraging consumer-grade electronic hardware and aspherical lenses, and developing complementary artificial intelligence algorithms for low-resolution cervical cytopathology slides with sparse lesion cells. The experimental results on multiple external datasets have demonstrated that our system can effectively screen SIL cases.
The lesion areas are small and dispersed in cytopathology, while the lesion areas in histopathology are larger and more concentrated. In our statistical results, the proportion of positive foregrounds to all foregrounds is on the order of 0.01 at its highest, and more commonly on the order of 0.001 (Supplementary Fig. 4). The sparsity of lesion cells in cytopathology makes it challenging to apply weakly supervised classification methods (used effectively in histopathology) to our LRWSIs. The poor performance of TransMIL and CLAM on our LRWSIs indicate this point (Fig. 3). These weakly supervised methods generally aggregate the long sequence of features (extracted by a pretrained model) from all instances in a slide to represent and classify slides. The proportion of positive instance features in the sequence affects the distinction between positive and negative slide feature sequences, which in turn impacts classification difficulty. Generally, a higher proportion of positive features in the sequence corresponds to greater the specificity differential between positive slides and negative slides. Through ablation experiments, we observed the performance of CLAM and TransMIL progressively degrades as the proportion of positive foreground decreases, ultimately failing below a critical threshold (Supplementary Fig. 13). On the contrary, our method excludes the majority of negative instances in the first-stage, focus on sparse positive information through Att-Transformer to better represent slide, and ultimately enables effective classification of slides.
We focus on SIL cases with a high risk of cervical cancer, and our method can effectively identify SIL, which aligns with our goal of screening in resource-limited areas. Although ASC is also a common type of cervical abnormality, its cancer risk is lower. Therefore, it is not our primary research focus. The AUC of our method decreased by 0.145, 0.088, and 0.118 on three external test sets after adding ASC (Supplementary Fig. 7). We found the majority of false negative slides are misclassified ASC slides (Supplementary Fig. 10). Distinguishing ASC in clinical practice is inherently challenging due to its atypical or subtle cytomorphological features, unlike the distinct changes observed in SIL. Low-resolution (LR) images exacerbate this challenge by losing partial texture details, and the scarcity of abnormal cells in ASC slides further complicates slide-level interpretation. In clinical practice, SIL is the most important type for screening, as it shows clear cytomorphological changes that indicate a higher risk of cervical cancer50. ASC cases also exhibit significant inter-observer variability51,52, with a relatively lower risk of malignancy. Further examination or follow-up is typically recommended for ASC. Consequently, it is reasonable for us to prioritize the identification of SIL as our main screening target. In the future, we will improve the recognition of ASC by optimizing both the imaging quality of our microscope and the performance of our algorithms.
As the WHO has recommended HPV screening for high-throughput screening of cervical cancer, joint screening with the HPV testing is an important potential application of our system. We tested our method on a dataset (n = 570) that was already identified as HPV-positive (Supplementary Fig. 12). Our overall AUC was 0.850 and 0.755 when excluding ASC or when not, respectively. Our method is still able to effectively distinguish SIL in HPV-positive slides. Considering the relatively low specificity of the HPV testing, our model can effectively exclude a large number of cytopathology-negative cases, thereby improving the specificity of co-testing. These results indicate that our system can still play an important role in the co-testing or HPV-first screening scenarios.
We have demonstrated a complete, fully automated workflow, but there is still significant room for improvement in real-world clinical practice, such as increasing scanning speed and integrating hardware with software. In addition, the lack of standardization in slide preparation and staining processes in resource-limited areas affects the generalizability of AI algorithms. Standardizing and implementing data quality control are critical for the application of our screening system. In the future, we plan to promote our system in more resource-limited regions.
Methods
Ethical statement
This study was approved by the Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology (2020-S353). Informed consent was waived for using anonymized, retrospectively collected cervical slides solely for imaging and analysis.
Compact microscope
We first completed the design of the prototype of the small microscope and acquired the data collection for experiments. For the prototype version, the schematic diagram is shown as Fig. 1a. We used a compact microscope module (dimensions: 48 mm × 38 mm × 20 mm) from Tinyphoton Technology Co., Ltd. (Wuhan, China). Sony IMX 258 (1/3.06 “, pixel size 1.12 μm) image sensor and two aspherical lens sets are integrated inside the module. The two lens sets are with focal lengths of 3.6 mm and 4.7 mm, serving as objective and tube lenses, respectively. This configuration achieves an approximate magnification of 1.3 for the entire microscopy system, providing a pixel size of 0.87 μm/pixel. The module also integrates a voice coil motor (VCM) module with a travel range of 200μm and a step precision of less than 8 μm, which is supported by an integrated autofocus algorithm. A diffuse LED light panel serves as a transmitted illumination source. Effective microscope illumination can be acquired by adjusting the distance between the panel and the sample. The structural components are constructed using 3D-printed materials to form the framework of the entire system. In order to meet the demand for edge computing, we have selected the Hi3519 SoC for image acquisition and storage. This SoC incorporates a highly efficient neural network inference engine, capable of delivering up to 2.5 trillion operations per second (TOPS) in neural network computations. The SoC is also equipped with abundant peripheral interfaces, including high-definition multimedia interface (HDMI) and USB, which streamline the connectivity process with devices such as microscope modules and display units. The entire system is designed with energy efficiency, ensuring that the power consumption remains below 5 watts. When powered by a 6000 mAh battery, the device is projected to offer over 4 h of continuous operation. Parameters are shown in Supplementary Table 2, and a picture of our prototype microscope and more details can be seen in Supplementary Fig. 14. After validating our method on the prototype, we further improved our compact microscope to enhance its usability. These optimizations include improved image quality, enhanced human-computer interaction, and fully automated whole-slide scanning as shown in Supplementary Fig. 15. And we have a demo movie to show the system as Supplementary movie 1.
LRWSI imaging and preprocessing
A low-resolution WSI is composed by about 90 fields of view, and each field of view has the size of 3840 × 2160 pixels. Due to uneven exposure at the field of view boundaries, we set the camera movement step size slightly smaller than the field of view width, ensuring each cell is imaged at least once within the central 1600 pixels region of the field. After acquiring the original images, we crop the central 1600 pixels region. We then apply a sliding window operation with a step size of 196 pixels and a window size of 256 pixels to convert the entire field of view into a set of instances. Each instance is resized to 224 pixels before being inputted into the model using linear interpolation. Given a dataset contains multiple slides \(\{{X}_{1},{X}_{2},\ldots,{X}_{N}\}\), \(N\) is the number of slides, and each slide \({X}_{{\mbox{i}}}\) contains multiple instances \(\{{x}_{i,1},{x}_{i,2},\ldots,{x}_{i,n}\}\), and a corresponding label \({Y}_{i}\). \(n\) is the number of instances in a slide. In our setting, an instance is \({x}_{i,j}\in {R}^{3\times 224\times 224}\), and its label is \({y}_{i,j}\).
Typically, the annotation of whole slides is assumed to be a multi-instance problem. Given a slide \({X}_{i}\) and its corresponding label \({Y}_{i}\), if the label is negative, all instances are considered negative; if the label is positive, at least one instance among them is positive.
LRWSI datasets
Table 1 shows the LRWSI datasets used in our paper for training, validating and testing. In our study, we focus on the screening of intraepithelial lesion, which mainly consists of ASC, LSIL and HSIL samples (POS), while negative samples (NEG) are the same as negative for intraepithelial lesion or malignancy (NILM) samples. For the external dataset, we further annotated its subcategories: ASC, LSIL, and HSIL, to evaluate the model’s performance on each subclass. In contrast to the scenario in histopathology where a patient may correspond to multiple slides, in our dataset, each patient corresponds to a single slide.
All experimental data were collected from female participants through self-reporting. For the internal dataset, we first collect 3,899 LRWSIs from four different hospitals at different periods: A1–A5, B1-B2, C1-C2, and D1–D4. The A and D datasets are slides from the same hospital, which is Tongji Hospital of Huazhong University of Science and Technology, but they differ in slide preparation and staining scheme. Slides made by Wuhan Union Hospital of Huazhong University of Science and Technology is referred as B. C is slides made by Maternal and Child Hospital of Hubei Province. We randomly separated the whole internal dataset to train, validate and test set by 8:1:1. For the external dataset, we collect 972 LRWSIs. E1 and F1 are named External-A and External-B in the paper, respectively. E is from Wuhan Landing Institute for Artificial Intelligence Cancer Diagnosis Industry Development. F is slides from Duodao People’s Hospital, Jingmen. NewCohorts is composed by C3, D5 and D6, which is new cohorts from the same hospital of internal datasets. It is worth noting that E and F are sourced from enterprises and primary hospitals in a resource-limited region. Compared to central hospitals with specialized equipment and experienced pathology experts, these data are more aligned with our application scenario.
To demonstrate the performance of our method on HPV-positive samples, we additionally collected a dataset of 570 slides from HPV-positive cases. These slides were obtained using our improved compact microscope. All slides in the dataset were derived from cases with HPV-positive test results. Details of the dataset are presented in Table 1. The HPV-positive dataset is from Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China.
Two groups of pathology experts were involved in annotating our dataset, each consisting of two junior pathologists and one senior pathologist. The two junior pathologists independently completed the slide annotations, and in cases where their annotations differed, the senior pathologist reviewed and resolved the discrepancies. The same slides were scanned once using a standard microscope and once using our compact microscope, with the annotation process conducted on the images from the standard microscope. We abandoned questionable annotations.
Instance datasets
In the SSL training step of the stage one model, we utilized 239,000 unlabeled instances, which were acquired by randomly cropping instances from slides in train set. During the fine-tuning step, we used labeled instances. As shown in Supplementary Table 3, these labeled data include HR instances imaged using a standard microscope and LR instances imaged using our compact microscope. We annotated a total of 133,447 positive HR instances, which were randomly divided into training and testing sets. In addition, we collected ~ 6 million negative HR instances, which were also randomly split into training and testing sets. The large number of negative HR instances was made possible by randomly cropping them from negative slides. All annotations were achieved through consensus among three pathology experts. To obtain annotations for the LR instances, we used both a standard microscope and a portable microscope to scan the same slide. Pathology experts annotated the HR instances, and we acquired the annotation information for the LR instances through registration. We got 7599 positive LR instances (contain at least one positive cell) and 65,700 negative LR instances for training, since negative samples were easy to be acquired by cropping from negative slides. We have also obtained another LR instance dataset specifically designed for testing purposes, which contains 767 positive instances and 767 negative instances. The LR test set also served as an essential foundation for creating t-SNE maps in Fig. 3. Before inputting into the model, the HR instances are down-sampled to match the pixel size of LR instances.
Att-Transformer
We propose the Att-Transformer as shown in Fig. 2e, which is composed of three consecutive modules. Initially, an embedding layer converts an instance into a sequence of tokens. Subsequently, a ViT-based backbone processes the token sequence and extracts a deep feature representation. Ultimately, a GatedAttention-based aggregation head consolidates all output tokens after the backbone. Given a input \(x\in {{\mbox{R}}}^{3\times 224\times 224}\). The embedding layer (\({\mbox{Conv}}2{\mbox{d}}\)) is a convolutional network with a stride and kernel size equal to the patch size. In our model, the patch size is set to 16 and the kernel number is 768. The output is \(T=\{{t}_{1},{t}_{2},\ldots,{t}_{196}\}\), \({t}_{i}\in {{\mbox{R}}}^{768},T\in {{\mbox{R}}}^{196\times 768}\).
\({{\mathrm{Reshape}}}\) refers to flattening the width and height dimensions of the output feature.
The backbone is composed of multiple stacked standard multi-head self-attention layers (\({{\mathrm{MSA}}}\)) and feed-forward networks (\({{\mathrm{FFN}}}\)). MSA is an extension of self-attention (\({\mbox{SA}}\)). For a \(T\) as an input sequence, we first compute \(q,k,v\) by a linear layer.
The scalers\(\,{{\mbox{n}}}^{q},{{\mbox{n}}}^{k},{{\mbox{n}}}^{v}\) represent the dimensions of \(q,k,v\) respectively, and scaler \({\mbox{d}}\) is equal to \({{\mbox{n}}}^{q}\) and \({{\mbox{n}}}^{k}\). MSA refers to the process of applying multiple SA modules to the input during forward propagation. The outputs of all SA modules are concatenated (\({\mbox{Concatenate}}\)), and a linear layer is used to map the concatenated result back to the input feature space.
m represents the number of SA module. In addition, residual connections are employed during the feature forward process. Then the output features \(Z\) will be further encoded through the \({{\mathrm{FFN}}}\) network. The \({{\mathrm{FFN}}}\) is defined as follows which is a nonlinear network using GELU53 as activation function and b as bias.
Our model’s parameters are fully inherited from the ViT-based architecture, and thus will not be further elaborated here. The difference between our backbone and the ViT45 architecture lies in the absence of the CLS token as the representative of the model’s merged feature. Instead, each token output by the model is mapped to an attention score \({a}_{i}\) using a GatedAttention42 network. We normalize the attention scores of all tokens through a \({\mbox{softmax}}\) function and compute a weighted sum of all tokens using these scores to obtain the backbone’s output. Given an output of a token from the backbone \({h}_{i}\in {R}^{768}\) and merged feature \({h}_{o}\in {R}^{768}\).
Here, \({{\mbox{W}}}^{{\mbox{s}}},{{\mbox{W}}}^{{\mbox{t}}},{{\mbox{W}}}^{{\mbox{m}}}\) is learnable parameters, \({{\mbox{W}}}^{s}\in {R}^{768\times {\mbox{r}}},{{\mbox{W}}}^{{\mbox{t}}}\in {R}^{768\times {\mbox{r}}},{{\mbox{W}}}^{{\mbox{m}}}\in {R}^{{\mbox{r}}\times 1}\), scaler \({\mbox{r}}\) represents the dimension of intermediate features.
In stage one, we draw on the position embedding operation from ViT to modify the feature sequence fed into the model. In stage two, we do not apply this modification, as the sequences in this stage do not carry relative order semantics.
SSL for stage one
We employ self-supervised learning (SSL) to leverage the vast number of unlabeled instances in slides, enabling a more reasonable initial feature space for the model during fine-tuning. We utilize the Masked Autoencoder45 (MAE) pretraining framework as c) in Fig. 2. We primarily utilize the MAE pretraining framework with some modifications. The entire pretraining process consists of two steps, starting with the standard MAE pretraining procedure.
For step one, given a sequence of token \(T=\{{t}_{1},{t}_{2},\ldots,{t}_{{\mbox{d}}}\}\) with d elements, \({t}_{i}\in {R}^{768},T\in {{\mbox{R}}}^{d\times 768}\). \({{\mbox{MASK}}}_{i,d\to n}\) represents randomly deleting a subset of elements in the sequence, while \({{\mbox{MASK}}}_{i,d\to n}^{-1}\) refers to filling in the randomly deleted elements using learnable parameters \({h}_{o}\). \(n\) represents the number of left elements. \(i\) represents the \(i\)-th iteration of the random process.
The loss for MEA training is defined as follows:
\({\mbox{Encoder}}\) and \({\mbox{Decoder}}\) represent two ViT backbones act as encoder and decoder. MSE represents mean squared error. We used 50% mask rate and ImageNet pretrain weight to initialize the model. We pretrained the model on 239k unlabeled instances for 100 epochs.
For step two, we modified the original MAE. We observed that during the MAE pretraining process, each token has an equal probability of contributing to the final loss calculation in the random process. Therefore, we aim to merge all features together as the final output, rather than relying solely on the CLS token while inevitably discarding other features. We achieved this by inserting a GatedAttention module between the encoder and decoder to aggregate all tokens from the encoder into a merged feature, and the loss is defined as follows:
The number of epochs for step two is also 100. Finally, we used the trained encoder as the backbone for the next stage.
Finetune for stage one
We use a linear layer to map the merged features output by the Att-Transformer to one-hot encoded class labels, allowing us to fine-tune the stage one model using our HR and LR instance dataset with cross-entropy loss. The optimizer is Adam54 with 5% warm up and cosine like learning rate decay. Before fine-tuning, the model needs to load the parameters from our SSL pretrained model. When training, all learnable parameters are involved in the training process. To enhance the robustness of the first-stage model, we employed data augmentation and false positive mining training techniques.
We use similar augmentation in our former work36 for two pretrain steps and fine-tuning, which can refer to https://github.com/ShenghuaCheng/Aided-Diagnosis-System-for-Cervical-Cancer-Screening/blob/main/core/data/preprocess.py. These augmentations primarily include color jittering in the RGB and HSV color spaces, Gaussian blur, contrast enhancement, as well as rotation and symmetry operations. We do not use any crop and resize style augmentation as the relative size of the nucleus to the cytoplasm is a critical morphological feature for detecting abnormal cervical cells.
Typically, the annotation of whole slides is assumed to be a multi-instance problem. If a slide \({X}_{{\mbox{i}}}=\{{x}_{i,1},{x}_{i,2},\ldots,{x}_{i,n}\}\) has a corresponding label \({Y}_{i}\,=0\), then all labels of instances \({y}_{i,j}=0\). Based on this, we used the preliminarily trained model to infer all instances from the negative slides in the training set, identifying instances with high positive prediction values (> 0.5) within each negative slide. These hard instances were then incorporated into the model’s fine-tuning process to better delineate the classification boundary. Specifically, we used the initially trained model to infer instances from all negative slides in the training set. These instances were divided into 10 intervals based on the positive probability output by the model, with a step size of 0.1. Instances with a positive probability greater than 0.5 are considered hard instances for the current model, as a negative slide should not contain positive instances. We selected instances from each interval to add to the original training set, updating the training dataset. Next, the model was trained on the new training set until the loss converged. This process was repeated continuously.
We dynamically adjusted the proportion of the training data to obtain a stable model. We kept the balance between number of positive and negative instances in a training batch, and controlled the proportion of hard instances less than 5% to prevent the model from collapsing. We refer to the above approach as false positive mining training (FPMT).
Train for stage two
Given a slide \({X}_{{\mbox{i}}}=\{{x}_{i,1},{x}_{i,2},\ldots,{x}_{i,n}\}\), we use the trained first-stage model to infer all instances, resulting in a set of merged features \({H}_{i}=\{{h}_{i,1},{h}_{{i,2}},\ldots,{h}_{{i,n}}\}\) and a positive probability prediction value for each instance \({P}_{i}=\{{p}_{i,1},{p}_{i,2},\ldots,{p}_{i,n}\}\). We select the features of the top k instances with the highest positive probabilities as the representation for the entire slide. Then, we train our whole slide classifier in an end-to-end manner.
The model architecture in stage two is consistent with that of the first-stage, with the distinction that the model directly takes the feature sequence as input. Similarly, we attach a linear layer after the Att-Transformer to enable the model to output one-hot encoded classes, and we use cross-entropy loss to train the model. The Att-Transformer here has no position embedding as there is no specific order in feature sequence.
During the training process of the model in the second-stage, data augmentation can also be applied. We first inferred all instances without augmentation using the model from the first-stage and recorded the top k instances within the slide. Subsequently, we performed data augmentation on the selected instances before extracting the features, thus indirectly enhancing the input to the model in the second-stage. In the results shown in Figs. 3 and 7, the second-stage model utilized data augmentation.
Att-Transformer heatmap and ROI
In our Att-Transformer architecture, we use a GA module to aggregate the output token sequence. Given an input token sequence \(T=\{{t}_{1},{t}_{2},\ldots,{t}_{n}\}\), we can get the attention scores \(W=\{{w}_{1},{w}_{2},\ldots,{w}_{n}\}\) by using formulas (7), (8) and (9). We map each token’s attention score \({w}_{i}\) back to the corresponding input, obtaining a heatmap, where regions with higher attention scores represent our region of interest (ROI). In the first-stage model, each token corresponds to a patch in Transformer (16 × 16 pixels), while in the second-stage model, each token corresponds to one of the top k instances (224 × 224 pixels).
Computational hardware and software
The operation system of the server is Linux version 5.4.0−150-generic. The server is equipped with four NVIDIA TITAN V graphics processing unit (GPU) and Intel(R) Core (TM) i9-9940X CPU @ 3.30 GHz. CUDA Version is 11.4. The Python version is 3.8.16. Other libraries mainly are torch 2.0.1, torchvision 0.15.2, opencv-Python 1.2.0, pillow 9.5.0 and openslide-Python 1.2.0.
Comparison methods
We compared our method with CALM (GitHub - mahmoodlab/CLAM: Data-efficient and weakly supervised computational pathology on whole slide images - Nature Biomedical Engineering) and TransMIL (GitHub - szc19990412/TransMIL: TransMIL: Transformer based Correlated Multiple Instance Learning for Whole Slide Image Classification). For CALM, we choose the single-attention-branch version for experiments. We also make use of the encoder on the CLAM project to extract the ImageNet feature of instances.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Due to personal information protection, patient privacy regulations, and medical institutional data policies, slide images have not been publicly deposited. However, we have made all supporting resources publicly available to ensure reproducibility of the technical pipeline and analyses, including the source code, software methods, and supplementary information. In addition, a de-identified demonstration dataset is accessible at: ShenghuaCheng/high-risk-Cervical-Precancerous-Screening (github.com). All data supporting the findings of this study are provided within the paper’s Source Data files. Source data are provided in this paper.
Code availability
We release code, demonstration data and well-trained weights for the first and second-stage models in Python using PyTorch as the primary deep-learning library, which is available at https://github.com/ShenghuaCheng/high-risk-Cervical-Precancerous-Screening. All source codes have been released under the GNU GPLv3 free software license. Source Data are provided in this paper.
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
World Cancer Report: Cancer Research for Cancer Prevention – IARC. https://www.iarc.who.int/featured-news/new-world-cancer-report/ (2020).
Hull, R. et al. Cervical cancer in low and middle‑income countries (Review). Oncol. Lett. 20, 2058–2074 (2020).
Small, W. Jr et al. Cervical cancer: A global health crisis. Cancer 123, 2404–2412 (2017).
Vu, M., Yu, J., Awolude, O. A. & Chuang, L. Cervical cancer worldwide. Curr. Probl. Cancer 42, 457–465 (2018).
Canfell, K. Towards the global elimination of cervical cancer. Papillomavirus Res. 8, 100170 (2019).
Shastri, S. S. et al. Secondary prevention of cervical cancer: ASCO resource–stratified guideline update. JCO Glob. Oncol. https://doi.org/10.1200/go.22.00217 (2022).
de Sanjose, S. & Holme, F. What is needed now for successful scale-up of screening?. Papillomavirus Res. 7, 173–175 (2019).
Hu, L. et al. An observational sudy of deep learning and automated evaluation of cervical images for cancer screening. J. Natl. Cancer Inst. 111, 923–932 (2019).
Serrano, B. et al. Worldwide use of HPV self-sampling for cervical cancer screening. Prev. Med. 154, 106900 (2022).
Cox, B. & Sneyd, M. J. HPV screening, invasive cervical cancer and screening policy in Australia. J. Am. Soc. Cytopathol. 7, 292–299 (2018).
Arbyn, M. et al. Chapter 9: Clinical applications of HPV testing: A summary of meta-analyses. Vaccine 24, S78–S89 (2006).
Denton, K. J. et al. The sensitivity and specificity of p16INK4a cytology vs HPV testing for detecting high-grade cervical disease in the triage of ASC-US and LSIL Pap cytology results. Am. J. Clin. Pathol. 134, 12–21 (2010).
Benevolo, M. et al. Sensitivity, specificity, and clinical value of human Papillomavirus (HPV) E6/E7 mRNA assay as a triage test for cervical cytology and HPV DNA test. J. Clin. Microbiol. 49, 2643–2650 (2020).
Richards-Kortum, R., Lorenzoni, C., Bagnato, V. S. & Schmeler, K. Optical imaging for screening and early cancer diagnosis in low-resource settings. Nat. Rev. Bioeng. 2, 25–43 (2023).
Diederich, B. et al. A versatile and customizable low-cost 3D-printed open standard for microscopic imaging. Nat. Commun. 11, 5979 (2020).
Ozcan, A. Mobile phones democratize and cultivate next-generation imaging, diagnostics and measurement tools. Lab Chip 14, 3187–3194 (2014).
Chen, D. et al. Multiparameter mobile blood analysis for complete blood count using contrast-enhanced defocusing imaging and machine vision. Analyst 148, 2021–2034 (2023).
Hernández-Neuta, I. et al. Smartphone-based clinical diagnostics: towards democratization of evidence-based health care. J. Intern. Med. 285, 19–39 (2019).
de Haan, K. et al. Automated screening of sickle cells using a smartphone-based microscope and deep learning. Npj Digit. Med. 3, 76 (2020).
Tapley, A. et al. Mobile digital fluorescence microscopy for diagnosis of tuberculosis. J. Clin. Microbiol. 51, 1774–1778 (2013).
D’Ambrosio, M. V. et al. Point-of-care quantification of blood-borne filarial parasites with a mobile phone microscope. Sci. Transl. Med. 7, https://doi.org/10.1126/scitranslmed.aaa3480 (2015).
Tareef, A. et al. Multi-pass fast watershed for accurate segmentation of overlapping cervical cells. IEEE Trans. Med. Imaging 37, 2044–2059 (2018).
Zhao, L. et al. Automatic cytoplasm and nuclei segmentation for color cervical smear image using an efficient gap-search MRF. Comput. Biol. Med. 71, 46–56 (2016).
Kim, Y. Convolutional neural networks for sentence classification. Proc. 2014 Conf. Empir. Methods Nat. Lang. Process. 2014, 1746–1751 (2014).
Dosovitskiy, A. et al. An image is worth 16x16 words: transformers for image recognition at scale. Int. Conf. Learn. Represent. (2021).
Hussain, E., Mahanta, L. B., Das, C. R., Choudhury, M. & Chowdhury, M. A shape context fully convolutional neural network for segmentation and classification of cervical nuclei in Pap smear images. Artif. Intell. Med. 107, 101897 (2020).
Shi, J. et al. Cervical cell classification using multi-scale feature fusion and channel-wise cross-attention. In 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI) 1–5 (IEEE, Cartagena, Colombia, 2023).
K, H., V, V., Dhandapani, M. & A, A. G. CervixFuzzyFusion for cervical cancer cell image classification. Biomed. Signal Process. Control 85, 104920 (2023).
Liu, W. et al. CVM-Cervix: A hybrid cervical Pap-smear image classification framework using CNN, visual transformer and multilayer perceptron. Pattern Recognit. 130, 108829 (2022).
Shinde, S., Kalbhor, M. & Wajire, P. DeepCyto: a hybrid framework for cervical cancer classification by using deep feature fusion of cytology images. Math. Biosci. Eng. 19, 6415–6434 (2022).
Du, Li, X. & Li, Q. Detection and classification of cervical exfoliated cells based on faster R-CNN. In 2019 IEEE 11th International Conference on Advanced Infocomm Technology (ICAIT) 52–57 (IEEE, Jinan, China, 2019).
Liu, S. & Zeng, S. Portable Device based Cervical Lesion Cell Detection Using Knowledge Distillation on Paired High-Low Quality Images. IEEE TRANSACTIONS ON MEDICAL IMAGING (2022).
Cao, L. et al. A novel attention-guided convolutional network for the detection of abnormal cervical cells in cervical cancer screening. Med. Image Anal. 73, 102197 (2021).
Wang, C.-W. et al. Artificial intelligence-assisted fast screening cervical high grade squamous intraepithelial lesion and squamous cell carcinoma diagnosis and treatment planning. Sci. Rep. 11, 16244 (2021).
Cheng, S. et al. Robust whole slide image analysis for cervical cancer screening using deep learning. Nat. Commun. 12, 5639 (2021).
Lin, H. et al. Dual-path network with synergistic grouping loss and evidence driven risk stratification for whole slide cervical image analysis. Med. Image Anal. 69, 101955 (2021).
Cao, M. et al. Patch-to-Sample Reasoning for Cervical Cancer Screening of Whole Slide Image. IEEE Trans. Artif. Intell. 5, 2779–2789 (2023).
Zhu, X. et al. Hybrid AI-assistive diagnostic model permits rapid TBS classification of cervical liquid-based thin-layer cell smears. Nat. Commun. 12, 3541 (2021).
Wang, J. et al. Artificial intelligence enables precision diagnosis of cervical cytology grades and cervical cancer. Nat. Commun. 15, 4369 (2024).
Zhang, Z. et al. SCAC: A Semi-Supervised Learning Approach for Cervical Abnormal Cell Detection. IEEE J. Biomed. Health Inform. 28, 3501–3512 (2024).
Dauphin, Y. N., Fan, A., Auli, M. & Grangier, D. Language modeling with gated convolutional networks. Proc. Mach. Learn. Res. 70, 933–941 (2017).
He, K. et al. Masked autoencoders are scalable vision learners. Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit. 15979–15988 (2022).
Lu, M. Y. et al. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 5, 555–570 (2021).
Shao, Z. et al. TransMIL: Transformer based correlated multiple instance learning for whole slide image classification. Adv. Neural Inf. Process. Syst. 34, 2136–2147 (2021).
Maaten, L. vander & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Jacob Gildenblat & contributors. PyTorch library for CAM methods. (2021).
Draelos, R. L. & Carin, L. Use HiResCAM instead of Grad-CAM for faithful explanations of convolutional neural networks. Preprint at https://doi.org/10.48550/arXiv.2011.08891 (2021).
lse, M., Tomczak,. M. & Welling, M. Attention-based deep multiple instance learning. Proc. Mach. Learn. Res. 80, 2127–2136 (2018).
Nayar R. & Wilbur, D.C.The Bethesda System for Reporting Cervical Cytology: Definitions, Criteria, and Explanatory Notes. (Springer International Publishing, Cham, 2015).
Balulescu, I. & Badea, M. Interobserver variability in the interpretation of cervical smears - a must for developing an internal laboratory quality control system. Gineco.eu 9, 178–183 (2013).
Ndifon, C. O. & Al-Eyd, G. Atypical Squamous Cells of Undetermined Significance. in StatPearls (StatPearls Publishing, Treasure Island (FL), 2024).
Hendrycks, D. & Gimpel, K. Gaussian error linear units (GELUs). Preprint at https://doi.org/10.48550/arXiv.1606.08415 (2023).
Kingma, D. P. & Ba, J. Adam: A method for stochastic optimization. Preprint at https://doi.org/10.48550/arXiv.1412.6980 (2017).
Acknowledgements
This work is supported by the NSFC projects (grant 62375100, 62471212, 62201221), China Postdoctoral Science Foundation (grant 2021M701320, 2022T150237), Guangzhou Science and Technology Project (grant 2024A04J4960) and Southern Medical University Research Start-up Foundation.
Author information
Authors and Affiliations
Contributions
X.L. (Xiuli Liu), S.C., J.C., and S.Z. conceived of the project. N.L., Q.H., X.L., and X.L. (Xiaohua Lv) designed the miniature microscope and the hardware system. J.B., H.Y., S.C., and X.L. (Xu Li) developed the algorithms. J.C., G.R., L.C., C.L., S.S., and J.H. annotated the lesion cells. J.C., L.C., J.H., B.P., and X.C. provided the cervix cytology glass slides. X.L., G.R., and N.L. scanned the slides. S.C., J.B., H.Y., and X.L. (Xu Li) managed the data. J.B., H.Y., S.C., X.L. (Xu Li), and S.L. performed the image analysis and processing. S.C., J.B., X.L. (Xiuli Liu), and J.C. mainly wrote and revised the manuscript, and all the authors participated in revising the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Weimiao Yu, who co-reviewed with KokHaur ONG, Qionghai Dai and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bai, J., Li, N., Ye, H. et al. AI-assisted cervical cytology precancerous screening for high-risk population in resource-limited regions using a compact microscope. Nat Commun 16, 7429 (2025). https://doi.org/10.1038/s41467-025-62589-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-62589-x