Abstract
CD4+Foxp3+ regulatory T cells are essential for maintaining immune tolerance and preventing excessive inflammation, making them promising candidates for treating autoimmunity and GvHD. However, the translation of regulatory T cell therapy into clinical practice poses substantial challenges. Here, we show that adoptive regulatory T cell therapy increases IL-6 and TGF-β-dependent pathogenic Th17 cell differentiation in murine models of inflammatory bowel disease and experimental autoimmune encephalomyelitis. Regulatory T cells increase the p-stat3/p-stat5 ratio in effector T cells by suppressing IL-2 secretion and competitively consuming IL-2, thereby promoting Th17 cell differentiation. Notably, IL-2 signaling deficiency not only promotes a Th17 cell-associated transcriptional program, but also enhances the pro-inflammatory properties of Th17 cells. Strikingly, therapeutic blockade of IL-6/STAT3 signaling pathway can reverse pathogenic Th17 cell differentiation and enhance the therapeutic effect of regulatory T cell therapy. Thus, our findings could potentially advance the clinical research progress of adoptive regulatory T cell therapy.
Similar content being viewed by others
Introduction
CD4+Foxp3+ regulatory T (Treg) cells represent a subset of CD4+ T cells dedicated to negative immune regulation and immune homeostatic balance preservation1,2. The absence or insufficient function of Treg cells (Tregs) may over-activate the immune system and inflammatory responses, resulting in autoimmune disease development. The mechanisms by which Tregs exert immune suppression have been extensively elucidated3,4. Tregs release anti-inflammatory cytokines, including transforming growth factor β (TGF-β), IL-10, and IL-35, engage in competitive IL-2 consumption, and express negative regulatory surface receptors such as CTLA-4, CD39, and CD735. These potent immunosuppressive functions make Tregs attractive candidates for treating autoimmune disorders and organ transplant rejection. More than 50 ongoing clinical trials are using Tregs to treat graft-versus-host disease (GvHD), solid organ transplantation, and autoimmune diseases6. These published data confirm the feasibility and potential efficacy of adoptive Treg cell therapy.
In previous clinical trials, polyclonal Tregs were predominantly used7, potentially carrying the risk of nonspecific immunosuppression. Patients receiving polyclonal Treg cell therapy may have an increased risk of viral infection within the first 30 days8. Consequently, there has been a recent shift in research focusing on antigen-specific Tregs that can exert localized and targeted inhibitory effects on only a limited number of cells. In type 1 diabetes (T1D)9, experimental autoimmune encephalomyelitis (EAE)10, and transplantation tolerance11,12, antigen-specific Tregs have demonstrated superior therapeutic efficacy compared with polyclonal Tregs. In addition to the adoptive transfer of Tregs, it is possible to directly induce Treg cell generation in vivo using low-dose IL-213,14,15. Activated Tregs are more sensitive to IL-2 than other T and natural killer (NK) cells because of the constitutive expression of high-affinity IL-2 receptor complexes (CD25, CD122, and CD132)1,16. Furthermore, IL-2 negatively regulates T follicular helper (Tfh) cell development by affecting Bcl6 expression17 and suppresses the T helper 17 (Th17) cell differentiation by inducing STAT5 activation18.
Interleukin-17 (IL-17)-secreting Th17 cells constitute a distinct subset of T helper cells, with retinoic acid receptor-related orphan receptor-γt (RORγt) as the master transcription factor of the TH17 lineage19. RORγt expression initiation and IL-17 transcription involve crosstalk between TGFβ-SMADs signaling and IL6-STAT3 signaling20. As a pivotal component of the adaptive immune system, Th17 cells enhance host protection from extracellular pathogens by fortifying epithelial barrier function. On the other hand, Th17 cells can also exhibit pathogenic behavior and contribute to the development of various autoimmune and inflammatory diseases, including colitis and multiple sclerosis (MS).
Despite the promising results achieved using adoptive transfer and in vivo induction strategies, Treg cell therapy has certain limitations. This is because the therapeutic results manifest differently in various disease types and at distinct disease stages. In a Phase II clinical trial of T1D (NCT02691247), the treatment group did not exhibit significant efficacy following the administration of autologous ex vivo polyclonally expanded Tregs21. In addition, adoptive transfer of Tregs can substantially alleviate symptoms in patients with chronic GvHD22. However, Tregs have shown only short-term efficacy in patients with acute GvHD22. Although Treg cell therapy can improve the condition of an autoimmune gastritis model, a considerable proportion of effector T (Teff) cells persist in the long term23. Inducing T cell depletion before Treg cell therapy in autoimmune diseases and GvHD may achieve better results than Treg cell therapy alone24,25,26. These findings imply that the presence of pro-inflammatory Teff cells (Teffs) may be a critical factor limiting the effectiveness of Treg cell therapy.
Here, our findings suggest that adoptive Treg cell therapy promotes IL-6 and TGF-β-dependent pathogenic Th17 cell accumulation in inflammatory bowel disease (IBD) and EAE models. Specifically, Tregs suppress IL-2 signaling in Teffs via competitive IL-2 consumption and suppression of IL-2 production under inflammatory conditions, thereby increasing the ratio of p-STAT3 to p-STAT5 in Teffs. Furthermore, blocking IL-2 enhances the pro-inflammatory features of Th17 cells. Finally, we determined that concurrent inhibition of IL-6/STAT3 signaling during adoptive Treg cell therapy prevented pathogenic Th17 cell generation, thereby improving the therapeutic effect of Tregs. Our study revealed a previously unrecognized adverse effect of adoptive Treg cell therapy and developed a combination therapy strategy based on mechanistic studies, which could potentially advance the clinical research progress of adoptive Treg cell therapy.
Results
Adoptive polyclonal Treg cell transfer promotes Th17 cell generation in IBD
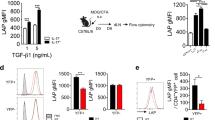
To assess the influence of adoptive Treg cell therapy on the immune microenvironment, we used a classic IBD model (the T cell transfer colitis model) by transferring CD4+CD25-CD45RBhi Naïve T cells alone (IBD group) or in combination with CD4+CD25+Foxp3-eGFP+ Tregs (Treg group) into Rag1-/- mice27. Mice in the IBD group began to lose weight at–5–6 weeks (Fig. 1A) and the infiltration of inflammatory cells into the mucosal layer was severe (Fig. 1B), whereas the Treg group showed no symptoms of colitis. We performed single-cell RNA sequencing (scRNA-seq) on colonic lamina propria cells from six mice (n = 3 per group) when the body weight of the IBD group mice decreased to 80% of their initial weight. Following quality control, we obtained 32898 cells, including 18979 IBD-originating and 13919 Treg-originating cells. Four major cell clusters were constructed using uniform manifold approximation and projection (UMAP): immune cells, fibroblasts, endothelial cells, and epithelial cells (Supplementary Fig. 1A–C and Supplementary Data 1). Consistent with a previous report, immune cells in the Treg group were significantly suppressed (Fig. 1C and Supplementary Fig. 1B)28,29. In our murine model, immune cells (Ptprc+ cells) were classified into seven subgroups: T cells, dendritic cells (DCs), macrophages, NK cells, monocytes, type 2 innate lymphoid cells (ILC2s), and natural killer T (NKT) cells, with T cells representing the predominant component of colonic lamina propria lymphocytes (LPLs) (Fig. 1D and Supplementary Fig. 1D). Similarly, Treg cell therapy significantly reduced the proportion of T cells (Fig. 1E). Notably, IL-4-expressing ILC2s, which play a role in intestinal homeostasis and mucosal tissue repair, were present at high proportions in the Treg group30,31,32. Taken together, these data elucidated the landscape of single-cell subgroups in an IBD model following Treg cell therapy for IBD, providing further confirmation of the immunosuppressive functions of Tregs.
Rag1−/− mice were intravenously injected with CD45.2+CD4+CD25-CD45RBhi T cells, with or without CD45.1+CD4+CD25+ Tregs, and then observed for the development of colitis. A Weight changes of Rag1−/− mice post T cell transfer. B Representative histology images of colon sections. Scale bars, 500 μm. C Total Immune cell numbers in the spleen, mesenteric lymph nodes (MLN) and colon tissues of indicated mice. D Uniform manifold approximation and projection (UMAP) plot of all immune cells colored by cell types. E Fraction of different immune cell types in indicated mice. F UMAP plot of all T cells colored based on cell type. G Fraction of different cell types among all T cells in the indicated mice. H, I Flow cytometry analysis of frequencies of total CD4+IL-17+ Th17 cells. J Frequencies of CD45.1+Th17 cells (black box) and CD45.2+Th17 cells (gray box) among total Th17 cells in the spleen, MLN and colon tissues of the Treg cell therapy group. K, L Flow cytometry to detect the frequencies of CD45.2+CD4+IL-17+ T cells (K) and CD45.2+CD4+RORγt+ T cells (L) in CD45.2+CD4+ cells. Sample sizes: (A, C, I, J, K and L) n = 3 mice per group. Data are representative of two independent experiments. Statistical testing by two-way ANOVA with multiple comparisons test (A) or unpaired two-tailed Student’s t tests (C, I, J, K and L). Summary data are presented as mean ± SEM. Source data are provided as a Source Data file.
By analyzing the data deeply, we identified four T-cell subtypes, including T-helper 0 cells (Th0 cells), T-helper 1 cells (Th1 cells), Th17 cells, and Tregs (Fig. 1F and Supplementary Fig. 1E). Treg cell therapy significantly reduced the proportion of Th1 cells and significantly increased the proportion of Tregs (Fig. 1G). The proportion of Th17 cells in the Treg cell therapy group increased significantly (Fig. 1G), indicating that Th17 cell generation may be a side effect of Treg cell therapy, as an increase in Th17 cells is considered a key factor driving the development of IBD19,20,33. To validate this finding, we established an IBD model using CD45.2+ naïve T cells and CD45.1+ Tregs and conducted flow cytometry analysis of LPLs. Consistently, the frequency of IL-17-producing Th17 cells in the colon, mesenteric lymph nodes (MLN), and spleen was significantly higher in the Treg group than in the IBD group (Fig. 1H, I and Supplementary Fig. 1F). Under inflammatory conditions, some Tregs can be transformed into IL-17- and IFN-γ-secreting pathogenic T cells and lose Foxp3 expression34. However, Th17 cells converted from Tregs constituted only a small fraction of all Th17 cells, with the majority of Th17 cells originating from Teffs (Fig. 1J). Therefore, Th17 cell generation from Teffs rather than Tregs constitutes the main reason for the increase in Th17 cells following Treg cell therapy. As expected, the frequency of CD4+IL17+ T cells within CD45.2+ CD4+ T cells, as well as the key transcription factor for Th17 cells, RORγt, significantly increased in the Treg group (Fig. 1K, L). In contrast, the frequencies of CD45.2+CD4+ T cells producing IFN-γ (Th1 cells), IL-10 (Tr1 cells), and IL-4 (Th2 cells) were significantly decreased (Supplementary Fig. 1G–L). In addition, the overall proportion of Foxp3+ Tregs increased, whereas CD45.2+Foxp3+Tregs showed no significant changes (Supplementary Fig. 1M, N). These data indicate that in the IBD model, polyclonal Treg cell therapy suppressed Th1 and Th2 cell responses in Teffs but selectively promoted Th17 cell generation.
Adoptive transfer of antigen-specific Tregs promotes Th17 cell generation in EAE
Compared with polyclonal Tregs, antigen-specific Tregs accumulate in target tissues in an antigen-driven manner and exert antigen-specific immune suppression, reducing the risk of systemic immunosuppression5. Therefore, to verify whether adoptive antigen-specific Treg cell therapy can increase Th17 cell generation, we established EAE as an experimental model of MS and treated EAE mice with myelin oligodendrocyte glycoprotein (MOG)-specific Tregs sorted from 2D2 TCR transgenic mice. C57BL/6 mice were immunized with the MOG35-55 peptide in complete Freund’s adjuvant (CFA) to induce EAE, and MOG-specific Tregs were transferred into the mice on day 9, before the onset of symptoms (Supplementary Fig. 2A). Treg cell-treated mice showed partial improvement in the disease progression of EAE (Fig. 2A), with reduced inflammatory cell infiltration into the central nervous system (CNS) (Fig. 2B). The reduction in immune cell numbers specifically occurred in the drainage lymph nodes (DLNs) and spinal cord, suggesting that systemic immunosuppression did not occur in mice as a result of antigen-specific Treg cell treatment (Fig. 2C). Analysis of T cells from the spleen, DLN, spinal cord, and brain revealed that MOG-specific Treg cell therapy increased the number of Tregs in the spinal cord and brain (Fig. 2D, E). In the meanwhile, the frequency of Th17 cells was significantly increased in the spinal cord and brain (Fig. 2F, G). In addition, the frequency of Th17 cells in the spleen and DLN also increased (Fig. 2F, G). This is most likely because the MOG/CFA injected during modeling expanded the number of antigen-specific T cells in the spleen and DLN, thereby causing MOG-specific Treg cell therapy to have a certain impact on the differentiation of Th17 cells in these organs. In contrast, the frequency of Th1 cells in the spinal cord decreased significantly (Fig. 2H, I), whereas the frequencies of Th2 cells, Tr1 cells, and IFN-γ+CD8+T (Tc1) cells did not exhibit significant changes (Supplementary Fig. 2B–G). Collectively, these data show that the promotion of Th17 cell generation by Treg cell therapy is a common phenomenon and is not limited to the IBD model.
C57BL/6 mice were subcutaneously injected with MOG35-55 peptide emulsified in complete Freund’s adjuvant (CFA) to induce EAE, and MOG-specific Tregs were injected intravenously into mice of the Treg cell therapy group on day 9. A EAE clinical scores of the indicated groups. B Representative histology images of spinal cord sections. Scale bars, 400 μm. C Total Immune cell numbers in the spleen, DLN, brain and spinal cord tissues of indicated mice. D–I Representative flow cytometry plots and bar graphs showing frequencies of CD4+Foxp3+ Tregs (D, E), IL-17+CD4+ Th17 cells (F, G) and IFN-γ+CD4+ T cells (H, I) in the spleen, DLN, brain and spinal cord tissues of indicated mice. Sample sizes: (A) n = 8 mice per group; (C, E, G and I) n = 4 mice per group. Data are pooled from two independent experiments (A) or are representative of two independent experiments (B–I). Statistical testing by two-way ANOVA with multiple comparisons test (A) or unpaired two-tailed Student’s t tests (C, E, G and I). Summary data are presented as mean ± SEM. Source data are provided as a Source Data file.
Adoptive Treg cell therapy-induced Th17 cells exhibit high pathogenicity
To determine the physiological significance of Th17 cells induced by Tregs, we integrated Th17 cells from healthy mouse LPLs using publicly available databases (GSE173887)35 and analyzed the heterogeneity of Th17 cells under three different conditions (healthy, IBD, and Treg groups) (Supplementary Fig. 3A). Exploratory analysis using UMAP and Leiden clustering of all TH17 cells identified two distinct clusters: C0 and C1 (Fig. 3A and Supplementary Data 2). After an unbiased exploration of the available Gene Ontology (GO) terms, the C1 subset of highly expressed genes was highly enriched in immune responses and cell differentiation, suggesting that these cells may be active in IBD (Fig. 3B). Based on differences in gene expression profiles and physiological functions, Th17 cells can be categorized as pathogenic Th17 cells and non-pathogenic (or homeostatic) Th17 cells36. Typically, Th17 cells generated with IL-6 plus TGF-β3 or with IL-6, IL-1β and IL-23 are considered pathogenic Th17 cells37,38. Gene set enrichment analysis (GSEA) revealed that C1 subset is closer to pathogenic Th17 cells (Fig. 3C). Notably, the proportion of Th17 cells represented by the C1 cluster was lowest in the healthy group and highest in the Treg group (Fig. 3D). Furthermore, genes associated with the homeostatic Th17 cell phenotype (Ahr, Ikzf3, and Maf) are specifically accessible in the healthy group (Fig. 3E and Supplementary Fig. 3B), while genes associated with pathogenic Th17 cells (Sata4, Tbx21, Satb1, Nr4a2, Bhe40, Runx1, IL1r1, IL22, Tnfsf11, Nfatc1, S100a4, and Il23r) are specifically accessible in both the IBD and Treg groups (Fig. 3E and Supplementary Fig. 3C), with the expression of pathogenic-associated genes Nfatc1, S100a4, and IL23r higher in the Treg group than that in the IBD group (Supplementary Data 3)39,40,41. These data suggest that Th17 cells generated after adoptive Treg cell therapy may have pathogenic properties.
A UMAP plot of all Th17 cells colored by clusters. B Bubble plot showing the top 15 GO (BP) terms (rows) for C1 upregulated gene enrichment pathways. Functional enrichment analysis was performed using clusterProfiler via over-representation analysis (ORA), which employs the hypergeometric test to calculate enrichment p-values. The Benjamini-Hochberg method controlled the false discovery rate (FDR) across all tested gene sets. C Gene set enrichment analysis (GSEA) of pathogenic Th17-related genes in C1, comparing C0. NES: normalized enrichment score. NES were computed using a weighted enrichment statistic applied to signal-to-noise-ranked genes. P-value for each gene set was estimated through gene set permutation (n = 1000). The BH method controlled the FDR across all tested gene sets. D Fraction of the different clusters in the indicated groups. E Heat map shows the average expression levels of pathogenic/non-pathogenic Th17-related genes (columns; Z normalized per column) in the indicated groups (rows, after batch correction). F Experimental scheme of the IBD model to verify the pathogenicity of Th17 cells induced by Tregs in vivo. G, H CD45.2+CD4+ effector T cells isolated from the IBD group, CD45.2+ CD4+IL17(eGFP)+ Th17 cells and CD45.2+CD4+IL17(eGFP)- effector T cells isolated from the Treg cell therapy group were transferred into Rag1-/- mice and then observed for the development of colitis. G Weight changes of Rag1−/− mice post T cell transfer (n = 3 mice per group); mean ± SD, two-way multiple-range ANOVA test. H Representative histology images of colon sections. Scale bars, 500 μm. Data are representative of two independent experiments (G, H). Source data are provided as a Source Data file.
To determine the pathogenicity of Th17 cells, we established an IBD model using naïve T-cells from IL-17-eGFP (CD45.2) mice and Tregs from CD45.1 mice. Eight weeks later, we sorted CD45.2+CD4+ Teffs from IBD mice, sorted CD45.2+CD4+IL-17-eGFP+ Th17 cells and CD45.2+CD4+IL-17-eGFP- Teffs from Treg cell-treated mice, and transferred these three cell populations into new Rag1-/- mice to re-establish the IBD model (Fig. 3F). The results showed that CD45.2+CD4+IL-17-eGFP+Th17 cells sorted from the Treg cell-treated group caused more severe weight loss in mice than the other two cell populations (Fig. 3G). Moreover, the infiltration of inflammatory cells into the colon and the development of colitis in mice injected with CD45.2+CD4+IL-17-eGFP+Th17 cells were more severe than those in the other two groups (Fig. 3H). Consistently, the frequency of Th17 cells was higher, and the frequency of Tr1 cells was lower in this group (Supplementary Fig. 3D–G). Collectively, these data demonstrate that adoptive Treg cell therapy-induced Th17 cells are pathogenic Th17 cells.
Adoptive Treg cell therapy-induced Th17 cell generation requires IL-6 and TGF-β signaling in vivo
Next, we investigated the molecular mechanisms regulating Treg cell therapy-induced Th17 cell generation. IL-6 production by lamina propria macrophages and CD4 + T cells is higher in an IBD murine model and in patients with IBD, leading to increased levels of IL-6 in both the host bloodstream and intestinal tract42,43. Since IL-6 plays a crucial role in Th17 cell generation, it is vital to identify whether Treg cell therapy-induced Th17 cell generation depends on IL-6 signaling. To evaluate this notion, we established an IBD model using CD4+CD25-CD45RBhi naïve T cells from Il6ra+/+ and Il6ra-/- mice, with or without CD45.1+CD4+CD25+ Tregs. To avoid feedback regulation of the immune microenvironment after the disease occurred, we concluded the experiment 5–6 weeks after T cell transfer, before the mice began to lose weight. Compared to the IBD model established using Il6ra+/+ naïve T cells, the IBD model established with Il6ra-/- naïve T cells showed a significant reduction in the proportion of Th17 cells (Fig. 4A–C). Furthermore, Treg cell treatment did not lead to a substantial increase in Th17 cells in the IBD model established with Il6ra -/- naïve T cells (Fig. 4A–C), indicating that Treg cell therapy-induced Th17 cell generation is dependent on IL-6 signaling. Consistent with a previous report44, the absence of IL-6 signaling increased the frequency of Tregs among CD45.2+ T cells (Supplementary Fig. 4A, B). Meanwhile, the total number of immune cells and the frequency of Th1 cells decreased in the IBD model established with Il6ra-/- naïve T cells (Fig. 4D and Supplementary Fig. 4C, D). Moreover, Treg therapy decreased the total number of immune cells in the IBD model established using Il6ra-/- naïve T cells (Fig. 4D). These results show that Treg cell therapy-induced Th17 cell generation is dependent on IL-6 signaling in vivo.
A–D Rag1-/- mice were injected with CD45.2+CD4+CD25-CD45RBhi T cells isolated from Il6ra+/+ or Il6ra-/- mice, with or without CD45.1+CD4+CD25+ Tregs from CD45.1 mice. The experiment was ended 5-6 weeks post T cell transfer, before the mice started losing weight (n = 3 mice per group). A–C Representative flow cytometry plots and bar graphs showing frequencies of CD45.2+CD4+IL-17+ Th17 cells (A, B), and CD45.2+ CD4+RORγt+ T cells (C) in spleen, MLN and colon tissues of indicated mice. D Total immune cell numbers in the spleen, MLN and colon tissues of the indicated mice. E–H Rag1-/- mice were injected with CD45.2+CD4+CD25-CD45RBhi T cells isolated from Tgfbr1f/f ER-Cre+ mice treated with tamoxifen (Tgfbr1-/-) or oil (Tgfbr1+/+), with or without CD45.1+CD4+CD25+ Tregs from CD45.1 mice. The experiment was ended 5-6 weeks post T cell transfer, before the mice started to lose weight (n = 4 mice per group). E–G Representative flow cytometry plots and bar graphs showing frequencies of CD45.2+CD4+IL-17+ Th17 cells (E, F), and CD45.2+CD4+RORγt+ T cells (G) in spleen, MLN and colon tissues of indicated mice. H Total Immune cell numbers in the spleen, MLN and colon tissues of indicated mice. Data are representative of two independent experiments. ns, not significant, one-way ANOVA with Tukey’s post hoc test. Summary data are presented as mean ± SEM. Source data are provided as a Source Data file.
Blocking the TGF-β signaling pathway prevents EAE and IBD development in mice, highlighting the essential role of TGF-β in maintaining Th17 cell function and differentiation under pathological conditions33,45. To investigate whether Treg cell therapy-induced Th17 cell generation is dependent on TGF-β signaling, we established the IBD model with CD4+CD25-CD45RBhi naïve T cells from Tgfbr1f/f ER-Cre+ mice treated with tamoxifen (Tgfbr1-/-) or oil (Tgfbr1+/+), with or without CD45.1+CD4+CD25+ Tregs. The experiment was ended 5-6 weeks post T cell transfer, before the mice began to lose weight. The IBD model established with Tgfbr1-/- naïve T cells exhibited a significant reduction in Th17 cell generation, and Treg cell therapy-induced Th17 cell generation was completely suppressed (Fig. 4E–G). However, loss of TGF-β signaling reduced Treg cell differentiation among CD45.2+ T cells (Supplementary Fig. 4E), and the total frequency of Treg cells also decreased in the IBD model established with Tgfbr1-/- naïve T cells (Supplementary Fig. 4F). Besides, since Tregs can suppress Teff cell activation and proliferation by producing TGF-β46, the loss of TGF-β signaling resulted in the reduced capacity of Tregs to suppress the total immune responses (Fig. 4H) and Th1 cells (Supplementary Fig. 4G, H). Therefore, although the loss of TGF-β signaling blocked the generation of Th17 cells, it also reduced the suppression capacity of Treg cell therapy. Taken together, these data indicate that Treg cell therapy-induced Th17 cell generation is dependent on the TGF-β signaling in vivo.
Tregs promote Th17 cell differentiation by reducing IL-2
To further investigate the mechanisms by which Tregs promote Th17 cell generation, we established an in vitro co-culture system in which CD45.2+CD4+CD25-CD62L+ naïve T cells were stimulated with anti-mouse CD3 and antigen-presenting cells (APCs) in the presence or absence of CD45.1+CD4+CD25+ Tregs sorted from the spleen and LNs. Consistent with the in vivo observations, Tregs promoted the differentiation of Th17 cells in CD45.2+ T cells when the Th17-favoring cytokines TGF-β1 and IL-6 were simultaneously present (Fig. 5A, B). In contrast, Th1 cell differentiation in CD45.2+ T cells was significantly suppressed by Tregs (Supplementary Fig. 5A, B). Tregs did not affect the differentiation of Th2 and Tr1 cells but suppressed Treg cell generation in the presence of TGF-β1 (Supplementary Fig. 5C–H). To further confirm the phenotype of Treg cell-induced Th17 cell differentiation, we induced Tregs with TGF-β and IL-2 in vitro, and set up the co-culture with naïve T cells and induced Treg (iTreg) cells, we identified that iTregs induced more Th17 cells in CD45.2+ T cells as well (Supplementary Fig. 5I, J). These data indicate that Tregs promote Th17 cell differentiation in the presence of Th17-favoring cytokines TGF-β1 and IL-6.
Naïve CD4+ T cells isolated from C57BL/6 mice were stimulated with anti-CD3 and antigen-presenting cells (APCs) in the presence or absence of Tregs (1:1 ratio), and cultured in indicated polarization conditions. A, B Representative flow cytometry plots and a bar graph showing frequencies of CD4+IL-17+ Th17 cells among T cells cultured for three days. C, D Representative flow cytometry plots and a bar graph showing frequencies of IL2+CD4+ T cells among T cells cultured for 24 h. E Amount of IL-2 in the culture supernatant was detected using enzyme-linked immunosorbent assay (ELISA) after 24 h of culture. F, G Representative flow cytometry plots and a bar graph showing frequencies of CD4+IL-17+ Th17 cells among T cells cultured in the indicated polarization conditions for three days. Data are pooled from three biological replicates. Summary data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns, not significant, unpaired two-tailed Student’s t tests. Source data are provided as a Source Data file.
Tregs express high-affinity IL-2 receptors (CD25) and can suppress Teffs by competitively consuming IL-24. In contrast, IL-2-STAT5 signaling suppresses Th17 cell differentiation47. Therefore, we expected that Tregs would inhibit IL-2 signaling in effector CD4+ T cells to promote Th17 cell differentiation. Indeed, the secretion of IL-2 by Teffs was significantly inhibited in the presence of Tregs compared to that in the Teff group under Th17 polarizing conditions (Fig. 5C, D). Moreover, Tregs led to a substantial reduction in IL-2 in the culture medium under all culture conditions compared to control conditions (Fig. 5E), indicating that Treg cell-mediated competitive consumption of IL-2 plays a vital role in reducing IL-2. To verify the function of IL-2 in Treg cell-induced Th17 cell differentiation, naïve T cells were cultured in the presence of exogenous IL-2 or IL-2 neutralizing antibody under Th17 polarizing conditions. Neutralizing IL-2 supported Th17 cell differentiation, and Tregs could not further promote Th17 cell differentiation in the absence of IL-2 (Fig. 5F, G). Notably, exogenous IL-2 completely reversed Treg cell-induced Th17 cell generation (Fig. 5F, G). These findings show that Tregs promote Th17 cell differentiation via IL-2 secretion inhibition by Teffs and competitive IL-2 consumption.
Tregs promote Th17 cell differentiation by elevating the p-STAT3/p-STAT5 ratio in Teffs
IL-2 activates STAT5 by inducing STAT5 phosphorylation (p-STAT5), which interferes with the ability of p-STAT3 to bind to the IL-17 gene locus, thereby inhibiting the transcription of IL-1718,47. Therefore, we assessed the levels of p-STAT3 and p-STAT5 in Teffs using a co-culture system. We found that the presence of Tregs significantly decreased the levels of p-STAT5 in Teffs compared to those in the control group (Fig. 6A, B). When T cells were cultured with exogenous IL-2, Treg cell-mediated suppression of p-STAT5 was reversed (Fig. 6A, B). Notably, under Th17 polarizing conditions, Tregs also slightly reduced the level of p-STAT3 in Teffs (Fig. 6C and Supplementary Fig. 6A). However, Tregs significantly increased the ratio of p-STAT3 to p-STAT5 under Th17 polarizing conditions (Fig. 6D). Because p-STAT3 and p-STAT5 share the same binding sites on the Il17a promoter, an increase in the p-STAT3/p-STAT5 ratio results in increased IL-17 transcription level.
A–D Naïve CD4+ T cells isolated from C57BL/6 mice were stimulated with anti-CD3 and APCs in the presence or absence of Tregs (1:1 ratio), and cultured in the indicated polarization conditions. Representative flow cytometry plots and bar graphs showing frequencies of p-STAT5+CD4+ T cells (A, B) and p-STAT3+CD4+ T cells (C) in T cells cultured for 24 hours. D Bar graph showing the ratio of p-STAT3+CD4+cells to p-STAT5+CD4+cells. E–H Naïve CD4+ T cells isolated from C57BL/6 mice were cultured with anti-CD3 and anti-CD28 in the indicated polarization conditions. E Immunoblotting of STAT5, p-STAT5, STAT3, and p-STAT3 in T cells cultured for 24 h. F Density values of the Immunoblot were measured using Image J, and the ratio of p-STAT3 to p-STAT5 was calculated. The samples derive from the same experiment, and the blots were processed in parallel. G Representative flow cytometry plots showing frequencies of p-STAT5+CD4+ T cells after 24 h culture. H Bar graph showing the ratio of p-STAT3+CD4+ T cells to p-STAT5+CD4+ T cells after 24 h culture. Data are pooled from three biological replicates. Data were analyzed by unpaired two-tailed Student’s t tests (B, C and D) or one-way ANOVA with Tukey’s post hoc test (F and H). Summary data are presented as mean ± SEM, ns, not significant. Source data are provided as a Source Data file.
To further confirm that IL-2 inhibition can elevate the p-STAT3/p-STAT5 ratio in Th17 polarizing conditions, we cultured naïve T cells with anti-CD3 and anti-CD28 antibodies, with or without TGF-β, IL-6, and anti-IL-2 antibody. Consistent with the results obtained from the co-culture system, the addition of anti-IL-2 antibody reduced the expression of both p-STAT5 and p-STAT3 in T cells (Fig. 6E and Supplementary Fig. 6B, C) but increased the ratio of p-STAT3 to p-STAT5 (Fig. 6E, F). Moreover, intracellular flow cytometry data showed a substantial increase in the p-STAT3/p-STAT5 ratio in the presence of IL-2 (Fig. 6G, H), accompanied by increased Th17 cell differentiation and decreased Treg cell differentiation (Supplementary Fig. 6D–G). Taken together, these findings indicate that Tregs promote Th17 differentiation in Teffs via an IL-2 inhibition-mediated increase in p-STAT3/p-STAT5 ratio.
IL-2 deficiency increases Th17 cell pro-inflammatory features
In vivo data from the IBD model demonstrated that adoptive Treg cell-induced Th17 cells are pathogenic (Fig. 3), prompting us to investigate whether IL-2 deficiency increases the pathogenicity of Th17 cells. Accordingly, we performed RNA sequencing on Th17 cells induced by TGF-β1 + IL-6 and anti-IL2 + TGF-β1 + IL-6. A total of 1846 differentially expressed (DE) genes were identified between anti-IL-2 + TGF-β1 + IL-6 induced Th17 cells and TGF-β1 + IL-6 induced Th17 cells (Fig. 7A and Supplementary Data 4). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the upregulated genes in anti-IL-2 + TGF-β1 + IL-6 induced Th17 cells revealed a strong correlation between the deficiency of IL-2 and Th17 cell differentiation (Fig. 7B). Moreover, the deficiency of IL-2 in Th17 cell culture condition significantly upregulated the genes and pathways related to IBD (Fig. 7B). Compared to TGF-β1 + IL-6 induced Th17 cells, anti-IL-2 + TGF-β1 + IL-6 induced Th17 cells exhibited a high expression of genes associated with chronic inflammatory diseases, such as Il23r48,49, Bhlhe4050, S100a451, and Cd24a52 (Fig. 7C). Furthermore, the expression of Rbpj, Nfatc1, Tnfsf11, Casp1, and Cxcr6, which have been identified as part of the pro-inflammatory module within pathogenic Th17 cells38,39,41,53, was significantly upregulated in anti-IL-2 + TGF-β1 + IL-6 induced Th17 cells (Fig. 7C). In contrast, Foxp3, a key transcription factor in Tregs, and Gpr83, a surface marker that plays a key role in the suppression of Tregs54, were notably inhibited (Fig. 7C). Although we did not observe a decrease in Il10 expression at the transcriptional level, a significant reduction of IL-10 at the protein level was identified in anti-IL-2 + TGF-β1 + IL-6 induced Th17 cells by flow cytometry (Supplementary Fig. 7A, B). These findings show that the deficiency of IL-2 may potentially enhance the pro-inflammatory function of Th17 cells.
A–C Naïve CD4+ T cells isolated from C57BL/6 mice were cultured with anti-CD3 and anti-CD28 in ithe ndicated polarization conditions for three days. A Volcano plot showing differentially expressed genes (DEGs) of Anti-IL-2 + TGF-β + IL-6 compared to TGF-β + IL-6 (p < 0.05 and fold change ≥ 1.5). Differential expression analysis was performed using DESeq2. P-values were calculated with the Wald test, and the BH method controlled the FDR. B Bubble plot showing the KEGG terms (rows) of the upregulated gene enrichment pathways in the Anti-IL-2 + TGF-β + IL-6 group (also refer to Supplementary Data 4). Functional enrichment analysis was performed using clusterProfiler via over-representation analysis (ORA), which employs the hypergeometric test to calculate enrichment p-values. The BH method controlled the FDR across all tested gene sets. C Heat map showing the expression levels of pathogenic/non-pathogenic Th17-related genes (rows; Z normalized per row) that are differentially expressed in T cells cultured in the indicated conditions (columns). D–H Naïve CD4+ T cells isolated from OT-II TCR transgenic mice were cultured with anti-CD3 and anti-CD28 in the indicated polarization conditions for three days, and were then adoptively transferred into C57BL/6 mice to establish an acute pulmonary inflammation model. D–F Representative flow cytometry plots (D) and bar graphs (E, F) showing frequencies of CD11b+Ly6G+ neutrophils in BALF and lung tissues. G, H Representative flow cytometry plots (G) and bar graph (H) showing frequencies of F4/80+ Macrophage in lung tissue. Sample sizes: (E, F and H) n = 3 mice per group. Data are representative of two independent experiments. Summary data are presented as mean ± SEM, one-way ANOVA with Tukey’s post hoc test. Source data are provided as a Source Data file.
To further verify the effect of IL-2 deficiency on the function of Th17 cells, we cultured naïve T cells isolated from OT-II TCR transgenic mice under the indicated conditions for three days, adoptively transferred the cells into C57BL/6 mice, and established an acute pulmonary inflammation model with the OVA323-339 peptide by intratracheal instillation53 (Supplementary Fig. 7C, D). Compared with TGF-β1 + IL-6-induced Th17 cells, anti-IL-2 + TGF-β1 + IL-6 induced Th17 cells recruited more CD11b+Ly6G+ neutrophils in both bronchoalveolar lavage fluid (BALF) and the lungs (Fig. 7D–F), indicating that these Th17 cells induced more severe acute pulmonary inflammation. Furthermore, we also observed an increased infiltration of macrophages, which is considered one of the driving factors of lung inflammation55, in the lungs of mice transferred with anti-IL-2 + TGF-β1 + IL-6 induced Th17 cells (Fig. 7G, H). Thus, these data prove that the deficiency of IL-2 increases the pro-inflammatory features of Th17 cells. Taken together, the IL-2 deficiency caused by adoptive Treg cell therapy can not only promote the differentiation of Th17 cells but also enhance the pathogenicity of Th17 cells.
Improving adoptive Treg cell therapy efficacy by reversing pathogenic Th17 cell generation via blocking IL-6/STAT3 signaling pathway
Next, we investigated strategies to reverse Treg cell-induced generation of pathogenic Th17 cells in vivo to improve the efficacy of adoptive Treg cell therapy. We have revealed that Treg cell-mediated IL-2 suppression results in the generation and pathogenicity of Th17 cells, and the generation of these Th17 cells is dependent on TGF-β and IL-6 signaling. On the one hand, the suppression and consumption of IL-2 by Tregs is an important mechanism for Tregs to suppress immune responses6. On the other hand, TGF-β is a key immunosuppressive cytokine that can suppress inflammation and induce immune tolerance56,57. Our data also showed that TGF-β deficiency led to an increase in pro-inflammatory Teffs, especially Th1 cells, in the IBD model (Supplementary Fig. 4G, H). IL-6 is a proinflammatory cytokine that plays an important role in autoimmunity58. Therefore, targeting the IL-6/STAT3 signaling pathway is an ideal strategy for developing a combination treatment to reverse Th17 cell differentiation and enhance the efficacy of Treg cell therapy. To determine the feasibility of the combination treatment, we established an IBD model using naïve T cells from Il6ra+/+ and Il6ra-/- mice, with or without CD45.1+ Tregs. As expected, mice injected with Il6ra+/+ T cells developed severe colitis, whereas mice injected with Il6ra-/- T cells did not develop obvious colitis (Supplementary Fig. 8A, B). In addition, mice injected with Il6ra-/- T cells together with Tregs did not develop colitis, and the generation of Th17 cells was completely inhibited (Supplementary Fig. 8C). Moreover, more Tregs were generated in mice transferred with Il6ra-/- T cells together with Tregs, and the total immune cells and Th1 cells were significantly suppressed in these mice compared to those in the other groups (Supplementary Fig. 8D–F). These data show that the combination of adoptive Treg cell therapy and IL-6 deficiency is a promising strategy to reverse Th17 cell differentiation and enhance the efficacy of Treg cell therapy.
To develop a physiological therapeutic strategy, we used an IL-6 neutralizing antibody together with Tregs for a combination therapy study in the IBD model (Fig. 8A). Consistent with the findings in the IBD model set up with Il6ra+/+ and Il6ra-/- T cells, colitis development in both the anti-IL6 antibody treatment group and the Treg cell therapy group was significantly suppressed compared to that in the control group (Fig. 8B, C). Moreover, the combination therapy of Tregs and anti-IL-6 antibody not only suppressed IBD development but also inhibited Th17 cell generation (Fig. 8B–E). Th1 and total immune cells were significantly suppressed in the combination therapy group (Fig. 8F, G). In addition, the frequency of Tregs significantly increased in the combination therapy group, which was due to the differentiation of Tregs in the CD45.2+ T cell population (Fig. 8H and Supplementary Fig. 8G, H). However, both the anti-IL6 antibody treatment group and Treg cell therapy group were used ideal experimental settings; they had very well therapeutic effects even when used alone, resulting in no further pathological improvement in the combination therapy group. To address this limitation, we optimized the therapeutic regimen by utilizing an increased number of naïve T cells for disease modeling and by administering a low dose of IL-6 neutralizing antibody and a low number of Tregs. Notably, treatment with a low dose of IL-6 neutralizing antibody (Low anti-IL-6) delayed the onset of IBD without achieving complete disease prevention (Fig. 8I, J). Similarly, treatment with a low number of Tregs (Low Treg) exhibited progressively attenuated efficacy over time, with limited control over pathological progression (Fig. 8I, J). In contrast, the combination therapy group demonstrated markedly improved therapeutic outcomes (Fig. 8I, J), accompanied by significant reductions in the frequencies of both Th17 and Th1 cells (Fig. 8K, L and Supplementary Fig. 8I, J). Therefore, the combination therapy with Tregs and anti-IL-6 antibody can improve the efficiency of IBD treatment by reversing Treg cell-induced Th17 generation and increasing Treg cell generation.
A–H Rag1-/- mice were injected with 4 × 105 CD45.2+CD4+CD25-CD45RBhi T cells isolated from C57BL/6 mice, with or without 1 × 105 CD45.1+CD4+CD25+ Tregs from CD45.1 mice and anti-IL-6 neutralizing antibody (300 μg/mouse every three days), and then observed for the development of colitis (n = 4 mice per group). A Experimental scheme of the combination therapy of the IBD model. B Weight changes of Rag1-/- mice post T cell transfer. C Representative histology images of colon sections. Scale bars, 500 μm. D–F Representative flow cytometry plots and bar graphs showing frequencies of CD45.2+CD4+IL-17+ Th17 cells (D, E), and CD45.2+CD4+IFN-γ+ Th1 cells (F) in the spleen, MLN and colon tissues of indicated mice. G A bar graph showing total Immune cell numbers in the spleen, MLN and colon tissues of the indicated mice. H A bar graph showing frequencies of total CD4+Foxp3+ Tregs in the spleen, MLN and colon tissues of indicated mice. I–L Rag1-/- mice were injected with 6 × 105 CD45.2+CD4+CD25-CD45RBhi T cells isolated from C57BL/6 mice, with or without 0.5 × 105 CD45.1+CD4+CD25+ Tregs from CD45.1 mice and anti-IL-6 neutralizing antibody (100 μg/mouse every four days), and then observed for the development of colitis (n = 4 mice per group). I Weight changes of Rag1-/- mice post T cell transfer. J Representative histology images of colon sections. Scale bars, 250 μm. (K and L) Representative flow cytometry plots and a bar graph showing frequencies of CD45.2+CD4+IL-17+ Th17 cells. Data were analyzed by two-way multiple-range ANOVA test (B, I), one-way ANOVA with Tukey’s post hoc test (E–H and L). Data are representative of two independent experiments. Summary data are presented as mean ± SEM, ns, not significant. Source data are provided as a Source Data file.
To further determine the effectiveness of the combined therapy with antigen-specific Tregs and IL-6 signaling blockade, we established an EAE model. Because antibodies cannot effectively cross the blood-brain barrier, we used Stattic59, a specific inhibitor of STAT3, to block the IL-6/STAT3 signaling pathway. Naïve T cells were cultured in vitro to verify the ability of Stattic to inhibit Th17 cell differentiation (Supplementary Fig. 9A, B). Next, we established the EAE model and treated the mice with or without MOG-specific Tregs on day 9, then treated the mice with or without Stattic every other day from day 11 (Fig. 9A). Consistent with our earlier findings, the adoptive transfer of MOG-specific Tregs had an inhibitory effect on the pathogenesis of EAE (Fig. 9B, C). However, treatment with Stattic alone was not effective in suppressing EAE progression (Fig. 9B, C), although it did suppress the generation of Th17 cells in the spleen and brain (Fig. 9D, E and Supplementary Fig. 9C, D). Despite this, the combination treatment with Stattic and MOG-specific Tregs exhibited a notable synergistic effect in suppressing EAE and had no marked impact on systemic immunity (Fig. 9B–G), indicating that blocking the IL-6/STAT3 signaling pathway can indeed enhance the efficacy of adoptive Treg cell therapy. Moreover, Stattic treatment not only reversed the generation of Th17 cells induced by Treg cell therapy (Fig. 9D, E and Supplementary Fig. 9C, D) but also reduced the frequency of Th1 cells in the CNS (Fig. 9H, I and Supplementary Fig. 9E, F). Besides, Stattic treatment resulted in an increase in the number of Tregs in the CNS and DLN of EAE mice (Fig. 9J, K and Supplementary Fig. 9G, H). Taken together, these data indicate that blocking IL-6/STAT3 signaling can improve the efficacy of adoptive Treg cell therapy by reversing the generation of Th17 cells and inducing stronger immune tolerance.
C57BL/6 mice were subcutaneously injected with MOG35-55 peptide emulsified in CFA to induce EAE, with or without MOG-specific Tregs and Stattic treatment, and then observed for the development of EAE. A Experimental scheme of the combination therapy of the EAE model. B EAE clinical scores of indicated groups (n = 8 mice per group). C Representative histology images of spinal cord sections. Scale bars, 500 μm. D, E Representative flow cytometry plots and a bar graph showing frequencies of CD4+IL-17+ Th17 cells in the brain and spinal cord tissues of the indicated mice. F, G Bar graphs showing total Immune cell numbers in the spleen, DLN, spinal cord and brain tissues of the indicated mice. H–K Representative flow cytometry plots and bar graphs showing frequencies of CD4+IFN-γ+ Th1 cells (H, I) and CD4+ Foxp3+ Tregs (J and K) in the brain and spinal cord tissues of indicated mice. Sample sizes: (B) n = 8 mice per group; (E–G, I and K) n = 4 mice per group. Data were analyzed by unpaired two-tailed Student’s t tests (B and I) or one-way ANOVA with Tukey’s post hoc test (E–G and K). Data are representative of two independent experiments (C–K) or are pooled from two independent experiments (B). Summary data are presented as mean ± SEM, ns, not significant. Source data are provided as a Source Data file.
Discussion
Several preclinical investigations and clinical trials have demonstrated that adoptive Tregs represent a promising and attractive treatment strategy for treating autoimmune disorders and GvHD5,6. Current perspectives acknowledge that one of the obstacles in Treg therapy is their plasticity34,60,61. Whether used for treating GvHD or autoimmune diseases, Tregs are exposed to inflammatory microenvironments, which may lead to the generation of Tregs with pro-inflammatory phenotypes or the polarization of Teffs from the conversion of Tregs. However, some studies have suggested that the expression of plasticity and transcription factors may enable Tregs to track and suppress inflammation mediated by Th1 and Th17 cells62,63. In this study, we discovered that adoptive Treg cell therapy specifically induced Th17 cell generation in both IBD and EAE models. Moreover, we identified that the majority of these adoptive Treg cell therapy-induced Th17 cells were derived from Teffs. Only a small proportion of Th17 cells were generated from the conversion of Tregs.
IL-17-producing Th17 cells play a crucial role in the pathogenesis of autoimmune diseases, including IBD, rheumatoid arthritis, asthma, MS, and psoriasis36. Our investigation indicated that adoptive Treg cell therapy-induced Th17 cells exhibit a more pathogenic transcriptional profile and can trigger severe intestinal inflammation in an IBD model. Tregs are necessary for the early initiation of Th17 cells in vivo64. In vitro, Tregs can promote Th17 cell differentiation by producing TGF-β in the presence of LPS-stimulated DCs65. These investigations are in line with our observation that the presence of Tregs under Th17-polarizing conditions (TGF-β1 + IL-6) resulted in an increased differentiation of Th17 cells from Teffs. Treg cell-mediated Th17 cell generation occurs during acute infection in vivo, and these Th17 cells are important for controlling the infection66. However, in chronic inflammatory responses, Tregs are believed to exert inhibitory effects on the differentiation of Th17 cells66. However, our data showed that in the chronic inflammatory microenvironment of IBD and EAE, the adoptive transfer of Tregs induces pathogenic Th17 cell generation, and these Th17 cells can persist at the inflammatory site for a long time. Other researchers have shown similar phenomena of Th17 cell generation in adoptive Treg cell therapy studies using an IBD model67,68,69. However, these studies have not focused on this crucial phenotype. Moreover, in a clinical trial involving the adoptive transfer of Tregs for the treatment of systemic lupus erythematosus (SLE), patients experienced a concurrent increase in Th17 cells within the local inflamed sit after 12 weeks of receiving 1 × 108 Treg cell infusions70. In another clinical trial, some patients with amyotrophic lateral sclerosis (ALS) who received Treg cell therapy exhibited elevated levels of IL-17F, a pro-inflammatory cytokine secreted by Th17 cells, in their peripheral blood71. Therefore, it is likely that adoptive Treg cell therapy in patients may also drive the differentiation of pathogenic Th17 cells in autoimmune contexts, which could chronically persist and accumulate at inflammatory sites. However, due to the limited availability of clinical data, further experimental and clinical investigations are required to confirm these observations. Moreover, whether adoptive Treg cell therapy can induce Th17 cell generation in different autoimmune diseases may also be related to the microenvironment of organs or tissues.
Although IL-21 can somewhat replace IL-6 and synergistically drive Th17 differentiation together with TGF-β72, IL-6 plus TGF-β are still considered to be essential for the maintenance of Th17 in both uninflamed and inflamed intestine45,73. Our experiments further validate this perspective as T cells lacking IL-6 and TGF-β signaling exhibit a dramatic reduction in Th17 cell numbers and cannot induce colitis development effectively. Meanwhile, the reverse of adoptive Treg cell therapy-induced Th17 cell differentiation in the absence of IL-6 signaling or TGF-β signaling proves that the generation of these Th17 cells is also dependent on IL-6 and TGF-β signaling in vivo. In contrast, Tregs express high levels of high-affinity IL-2 trimers, and their binding affinity for IL-2 is approximately 1000 times stronger than that of IL-2 receptor heterodimers expressed on Teffs74. Therefore, the ability of Tregs to capture IL-2 is considered a crucial mechanism for suppressing immune responses, as the deficiency of IL-2 leads to the death of Teffs75,76. Moreover, the absence of IL-2 reduces STAT5 activation, thereby reducing its inhibition of the IL-6/STAT3 signaling pathway18, leading to increased Th17 cell differentiation. Tregs modulate the balance between p-STAT5 and p-STAT3 in Teffs by inhibiting IL-2, thereby promoting Th17 cell differentiation64,66, which is consistent with our findings. Furthermore, we demonstrated that under Th17-polarizing conditions, Tregs could directly inhibit the production of IL-2 in Teffs. This observation may partially explain why Tregs still upregulate the p-STAT3/p-STAT5 ratio under Th17-polarizing conditions in the presence of an IL-2 neutralizing antibody. IL-2 induces the binding of STAT5 to the regulatory element CNS2 of Foxp3 and promotes the expression of Foxp3 to maintain Treg cell function77. We discovered that IL-2 deficiency leads to the downregulation of immune regulatory genes, including Foxp3 and Gpr83, and the upregulation of pathogen-related genes, such as Il23r, S100a4, Tnfsf11, and Bhlhe40, thereby enhancing the pro-inflammatory properties of Th17 cells. Collectively, these results show that adoptive Treg cell therapy-mediated IL-2 inhibition leads to increased differentiation and enhanced pathogenicity of Th17 cells in autoimmunity.
Based on phenotypic and mechanistic studies, we investigated a combination therapeutic strategy to improve the efficacy of Tregs by inhibiting pathogenic Th17 cell generation. As we mentioned earlier, TGF-β signaling negatively regulates the immune response by inhibiting the proliferation and function of Teffs and antigen-presenting cells78. Therefore, we believe that targeting TGF-β is not an ideal strategy for developing combination therapy. Consistently, we found that although Rag1-/- mice transferred with Tgfbr1-/- T cells failed to generate Th17 cells, the differentiation of Th1 cells and the accumulation of Teffs in the colon of these mice could not be controlled by Treg cell therapy. In contrast, although IL-2 can suppress Th17 cell differentiation and support Treg cell expansion, it can also promote Teff cell activation, survival, and proliferation. Owing to the pleiotropic nature of IL-2, precise dosage control is crucial during clinical administration. Low-dose IL-2 therapy enhances the function of Tregs in patients with T1D and GvHD. In adition, it is also associated with an increase in the numbers of NK cells and CD8+ T cells79,80. Therefore, there is a potential risk of activating immune cells with pro-inflammatory potential even with low-dose IL-2 therapy. Based on these considerations, we combined the adoptive transfer of Tregs with the blockade of the IL-6/STAT3 signaling pathway to develop a combination therapeutic strategy. Our data demonstrate that this combination therapy can effectively inhibit the generation of Treg cell-induced Th17 cells without compromising their function. Moreover, the combination therapeutic strategy increased the generation of Tregs and further suppressed the generation of Th1 cells and the infiltration of total immune cells.
In summary, we determined that adoptive Treg cell therapy leads to an increased differentiation of pathogenic Th17 cells in autoimmune disorders. Mechanistically, adoptive Treg cell therapy-mediated IL-2 inhibition led to increased differentiation and enhanced pathogenicity of Th17 cells in autoimmunity. This previously overlooked side effect may explain the suboptimal outcomes observed in clinical trials. By blocking this unemphasized side effect, the efficacy of adoptive Treg cell therapy can be improved, which is expected to promote translational research of adoptive Treg cell therapy.
Methods
Mice
The mice used in this study had a C57BL/6 background and were housed in a specific pathogen-free animal facility at Sichuan University. C57BL/6 (Cat#000664), Rag1-/- (Cat#002216), 2D2 TCR-transgenic (Cat#006912), CD45.1 (Cat#002014), IL-17-eGFP (Cat#018472), Foxp3-eGFP (Cat#006772), ER-Cre (Cat#007001), Tgfbr1f/f (Cat#028701) and OT-II TCR-transgenic mice (Cat#004194) were all obtained from Jackson Lab. Tgfbr1f/f (Cat#NM-CKO-200142) and Il6ra-/- (Cat#NM-KO-18042) mice were obtained from Shanghai Model Organisms. All mice used in this study were on a C57BL/6 background. All animal studies were performed in accordance with the guidelines of the Animal Care and Use Committee of West China Hospital, Sichuan University and were approved by the Animal Care and Use Committee of West China Hospital, Sichuan University (2021736 A, 20211212 A).
T cell-transfer colitis
Rag1-/- mice, aged 6–12 weeks, were intravenously injected with 4 × 105 CD4+CD25-CD45RBhi T cells sorted from C57BL/6 mice, Tgfbr1f/f ER-Cre+ mice (oil-treated or tamoxifen-treated), Il6ra+/+ mice, or Il6ra-/- mice alone or together with 1 × 105 CD45.1+CD4+CD25+ Tregs isolated from CD45.1 mice. For some experiments, CD45.1+CD4+CD25-CD45RBhi T cells and CD4+CD25+Foxp3 (eGFP)+ Tregs were used to establish the IBD model. For the IL-6 neutralization experiment, anti-IL6 antibody (300 μg/mouse, MP5-20F3, Bio X Cell) or rat IgG1 isotype control antibody (300 μg/mouse, HRPN, Bio X Cell) was injected intraperitoneally every three days. In the low-dose combination therapy experiment involving Tregs and anti-IL-6 antibody, Rag1−/− mice were intravenously injected with 6 × 105 CD4+CD25-CD45RBhi T cells alone or together with 0.5 × 105 Tregs, followed by intraperitoneal administration of anti-IL6 antibody (100 μg/mouse) or rat IgG1 isotype control antibody (100 μg/mouse) every four days. Body weights were measured every 3–4 days. The colon, spleen, and MLN tissues were collected at the end of the experiment for histopathological and immunological analyses.
EAE
Eight-week-old male C57BL/6 mice were utilized for establishing the EAE model. Briefly, MOG35-55 peptide (200 µg; QYAOBIO, #163913-87-9) was emulsified in complete Freund’s adjuvant (Difco, # 263910) containing 300 μg of inactivated Mycobacterium tuberculosis H37Ra (Difco, #231141), followed by subcutaneous injection in mice. On the day of immunization and two days later, pertussis toxin (Absin, # 70323-44-3) was intraperitoneally administered at 200 ng per mouse. MOG-specific Tregs (3 × 105 cells/mouse) sorted from 2D2 TCR-transgenic mice were injected intravenously into EAE mice on day 9. Stattic (5 mg/kg weight; sellcek, #S7024) was injected intraperitoneally into EAE mice every other day from day11. EAE scores were as follows: 0, no disease; 1, limp tail; 2, limp tail and hind limb weakness; 3, hind limb paralysis; 4, forelimb and hind limb paralysis; and 5, moribund or dead. After the experiment, the spleen, DLN, brain, and spinal cord tissues were collected for histopathological and immunological analyses.
Cell cultures
Cells were cultured at 37 °C, 5% CO2 in complete Dulbecco’s Modified Eagle’s Medium - high glucose (cDMEM), which was DMEM (Gibco, #11960044) supplemented with 10% fetal bovine serum (Gibco, #10099141 C), 50 μM β-mercaptoethanol (Sigma, #516732), 100 U/mL Penicillin-Streptomycin (Gibco, #15140122), 10 mM HEPES buffer solution(Gibco, #15630080), 1 × nonessential amino acids (Gibco, #15630080), 1 mM pyruvic acid (Gibco, #11360070), and 2 mM L-glutamine (Gibco, #25030081). Naïve CD4+ T cells and CD45.1+ Tregs were isolated from C57BL/6 and CD45.1 + mice, respectively, using a magnetic cell sorting kit (Miltenyi Biotec, #130-106-643). For in vitro co-culture of T cells, naïve T cells were cultured in 48-well plates at a density of 2 × 106 cells per well and were stimulated with APCs (1:5), with or without CD45.1+ Tregs (1:1 ratio), recombinant mouse IL-6 (50 ng/ml; R&D systems, #406-ML), recombinant human TGF-β1 (2 ng/mL; PeproTech, #100-21 C), recombinant human IL-2 (10 ng/mL; PeproTech, #200-02), and anti-mouse IL-2 antibody (50 μg/mL; Bio X Cell, #BE0042). For T cell culture in vitro, naïve T cells were cultured at 0.2 × 106 cells per well in 48-well plates, with plate-bound anti-mouse CD3 (1.5 μg/mL; Bio X Cell, #BE0001) and soluble anti-mouse CD28 (1.5 μg/mL; Bio X Cell, #BE0015), with or without recombinant mouse IL-6 (50 ng/mL), recombinant human TGF- β1 (2 ng/mL), recombinant human IL-2 (10 ng/mL), and anti-mouse IL-2 antibody (50 μg/mL). Cells were collected for flow cytometry analysis 24 h or three days later.
Acute lung inflammation model
Naïve CD4+ OVA-specific T cells were isolated from the spleen and peripheral lymph nodes of OT-II TCR transgenic mice using a magnetic sorting kit (Miltenyi Biotec). Cells cultured at a density of 0.4 × 106 cells per well on 24-well plates with plate-bound anti-CD3 (1.5 µg/mL) and soluble anti-CD28 (1.5 µg/mL), with or without TGF-β1 (2 ng/mL), IL-6 (50 ng/mL) and anti-IL-2 antibody, at 37 °C, 5% CO2 for three days. Cultured OVA-specific T cells (1 × 106 cells/mouse) were transferred into the recipient mice, followed by intratracheal instillation of OVA323-339 peptide (25 μg/mouse; QYAOBIO, # 92915-79-2)53. After the mice were euthanized, their lungs and bronchoalveolar BALF were collected for further analysis.
Cell isolation from colon and CNS
To isolate single cells from the colon, the colon was washed and cut into 5 mm pieces, then shaken in cDMEM containing 5 mM EDTA (Solarbio, #E1170) and 1 mM DTT (Thermo, #R0862) for 20 min at 37 °C. Tissues were then transferred to PBS containing 5 mM EDTA and shaken vigorously by hand for 45 s. The wash step was repeated three times to completely remove the intraepithelial lymphocytes. The remaining tissue was minced and digested in PBS containing 0.2 mg/mL Liberase TL (Roche, #05401020001) and 0.2 mg/mL DNase I (Aladdin, #D106200) and shaken continuously at 200 rpm, 37 °C for 20 min. The digested tissue was mashed and filtered through a 70 μm cell strainer to obtain LPLs.
For single-cell isolation from the brain and spinal cord, tissues were cut into small pieces and digested with 4 mg/mL collagenase IV (Worthington Biochem, # LS004188) and 1 mg/mL DNase I at 37 °C for 30–40 min. The digested tissues were then mashed and filtered through a 70 μm mesh to remove impurities, and finally, single-nucleus cells were separated using a 30%/70% Percoll gradient (GE Healthcare, #17089109).
Enzyme-linked immunosorbent assay (ELISA)
IL-2 in the cell culture supernatants was detected using the Mouse IL-2 ELISA MAX kit (BioLegend, #431004), according to the manufacturer’s instructions. Briefly, capture antibodies (1:200) were added to the plate and incubated at 2–8 °C overnight. After sealing the plate, the samples were incubated at room temperature for 2 h, and the detection antibody (1:200) was added and incubated for 1 h. Subsequently, avidin-HRP (1:1000) was added, and the substrate was added after 30 min. Finally, the absorbance values were measured using an ELISA Reader. The cytokine concentration in the sample was calculated using a standard curve.
Droplet-based scRNA-seq
Single cells were captured via the GemCode Single Cell Platform using the GemCode Gel Bead, Chip and 3ʹ-end Library Kits (10X Genomics), according to the manufacturer’s protocol. Briefly, the intestinal cells were loaded at a density of 30,000 cells/channel. All cells were loaded at 8000 cells per channel. The cells were then partitioned into GemCode instruments in which individual cells were lysed and mixed with beads carrying unique barcodes in individual oil droplets. The products were then subjected to reverse transcription, emulsion breaking, cDNA amplification, and sample index attachment. Libraries were paired-end (150 + 150 bp) and sequenced using a NovaSeq (Illumina).
scRNA-seq data processing
Initial processing and estimation of gene expression were performed using a cell ranger count (version 7.1.0) with the refdata-gex-mm10-2020-A reference from 10X Genomics for alignment. The unique molecular identifier (UMI) count matrix was converted into Seurat objects using the R package Seurat (version 4.3.0) in R (version 4.2.2). Cells with more than two or without tags were excluded. After further removing low-quality cells with more than 20% of UMIs mapped to mitochondrial genes, we obtained 32898 cells (18979 cells from the IBD group, 13919 cells from the Treg therapy group). The filtered gene expression matrix was normalized using Seurat’s NormalizeData function, and 2000 highly variable genes were identified per dataset using Seurat’s FindVariableFeatures function. After scaling the normalized data, highly variable genes were selected for principal component analysis (PCA). The first 15 principal components (PCs) were used for Uniform Manifold Approximation and Projection (UMAP) for visualization and graph-based clustering with a resolution of 1.0. Using normalized expression values, marker genes for each cluster were inferred using the Wilcoxon rank-sum test, as implemented in the FindAllMarkers function of Seurat. Immune cell clusters were used as a subset for downstream analysis based on the identified marker genes. Subsequently, we reran the PCA and used the first 15 PCs for UMAP and clustering. Marker genes for new clusters were identified using the FindAllMarkers function.
Single-cell sequencing (scRNA-seq) data from the wild-type mouse gut were obtained from the GSE173887 dataset of the Gene Expression Omnibus (GEO) database. The analysis process and clustering method were consistent with a previous description, defining the subsets with high expression of the Cd4, Rorc, and Il17a genes as Th17 cells, and integrating them with Th17 cells. The batch effects were corrected using the Harmony R package (version 1.2.0). Differentially expressed genes (DEGs) in different clusters were calculated using the FindAllMarkers function (default parameters), and the gene ontology (GO) pathways were enriched and analyzed using the clusterProfiler package (version 4.7.2) and the org.Mm.eg.db package (version 3.16.0). The normalized matrix was extracted for GSEA analysis, which was conducted using the GSEA software (version 4.3.2), and it was also exported to GraphPad Prism 9 for variance analysis. The heatmaps were generated using the pheatmap package (version 1.0.12) and the ClusterGVis Package (version 0.0.6). Data cleaning was performed using the dplyr package (version 1.0.2), and the ggplot2 package (version 3.3.3) was employed for the typical adjustment of figures.
RNA-Seq data analyses
RNA-Seq experiments were performed using total RNAs from T cells. Raw sequencing data were filtered using Trimmomatic (version 0.36); low-quality reads were discarded, and reads contaminated with adapter sequences were trimmed. Clean data were mapped to the reference genome of mm10 using the STAR software (version 2.5.3a) with default parameters. The reads mapped to the exon regions of each gene were counted using featureCounts (Subread-1.5.1; Bioconductor). DESeq2 (version 1.30.1) was used to determine the DEGs using the criteria of false discovery rate (FDR) < 0.05 (the p-value adjusted by the Benjamini–Hochberg method), log2(fc) ≥ 0.58. KEGG pathways were obtained by enrichment analysis of the differentially expressed genes using the clusterProfiler package (version 4.7.2) and the org.Mm.eg.db package (version 3.16.0). The heatmaps were generated using the pheatmap package (version 1.0.12). Data cleaning was performed using the dplyr package (version 1.0.2), and the ggplot2 package (version 3.3.3) was employed for the typical adjustment of figures.
Flow cytometry
The Zombie Yellow Fixable Viability Kit (BioLegend, #423104) was used to label the dead cells. For cell surface staining, appropriate fluorescent-conjugated antibodies were incubated with cells at 4 °C for 15–20 min. For intracellular cytokine staining, the cells were restimulated with PMA (5 ng/mL; Sigma, #P8139) and ionomycin (1 µg/mL; BD Biosciences, #554722) in the presence of GolgiPlug (1:1000 dilution; BD Biosciences, #555029) for 4 h. The cells were fixed and permeabilized for 20 min using a fixation/permeabilization solution kit (BD Biosciences, #554722) before intracellular staining. For transcription factor staining, the cells were fixed and permeabilized for 1 h using the Foxp3 / Transcription Factor Staining Buffer Set (Thermo, #00-5523-00) before intracellular staining. The following antibodies were used: PerCP-Cy5.5-conjugated anti-CD4 (RM4-5, #45-0042-82), FITC-conjugated anti-CD8α (53-6.7, #11-0081-85), FITC-conjugated anti-CD8β (H35-17.2, #11-0083-85), FITC-conjugated anti-CD45.1 (A20, #11-0453-85), APC-eFluor 780-conjugated anti-CD45.2 (104, #47-0454-82), PE-Cy7-conjugated anti-IL-17 (eBio17B7, #25-7177-82), PE-conjugated anti-IL-4 (11B11, #12-7041-82), APC-conjugated anti-IL-10 (JES5-16E3, #12-7101-82), eFluor 450-conjugated anti-IFNγ (XMG1.2, #48-7311-82), APC-eFluor 780-conjugated anti-TCRβ (H57-597, #47-5961-82), eFluor 450-conjugated anti-Foxp3 (FJK-16s, #48-5773-82), PE-conjugated anti-T-bet (4B10, #12-5825-82), APC-conjugated anti-RORγt (B2D, #17-6981-82), FITC-conjugated anti-CD11b (M1/70, #12-0112-83), and PerCP-Cy5.5-conjugated anti-F4/80 (BM8, #45-4801-82) were all from Thermo Fisher Scientific; APC-conjugated anti-Ly6G (1A8, #5310796) was from BD Pharmingen. Data were collected on a BD LSR Fortessa or Beckman CYTOFLEX S and analyzed using FlowJo 10.6.2 software.
Immunoblot analysis
T cells were washed twice with PBS after cell culture, resuspended in cell lysis buffer (50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM ethylenediaminetetraacetic acid (EDTA), and 1 × protease inhibitor), and lysed on ice for 30 min. Proteins were separated using sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gel electrophoresis and transferred onto polyvinylidene difluoride (PVDF) membranes (Merck Millipore, #IPVH00005). The PVDF membrane was blocked in a 5% bovine serum albumin (BSA) solution to prevent nonspecific binding, followed by three washes with TBST. The membranes were then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies. Detection was performed using an ECL chemiluminescence substrate (Yeasen, #36208ES60). The primary antibodies used included anti-β-Actin (13E5), anti-STAT5 (D2O6Y, #94205), anti-p-STAT5 (Tyr694, #9351), anti-STAT3 (124H6, #9319), and anti-p-STAT3 (Tyr705, #9145). Horseradish peroxidase-conjugated anti-mouse (#7076) and anti-rabbit (#7074) secondary antibodies were used. All antibodies were purchased from Cell Signaling Technology.
Statistics and reproducibility
For animal experiments, mice were randomly assigned to different groups, with at least two independent experiments performed. For cell culture experiments, each group was assayed in at least three biological replicates. No statistical methods were used to predetermine sample sizes. Instead, sample sizes were preselected based on standard practices described in the literature for the stated experiments, with clear specifications provided for each experimental set. Statistical analyses were performed using GraphPad Prism (version 8.4.1). The results are presented as mean ± SD or mean ± SEM. Unless otherwise specified, comparisons between two groups were performed using unpaired two-tailed Student’s t tests, and comparisons between more than two groups were performed using one-way ANOVA (with Tukey’s multiple-comparison post-tests). Results with p < 0.05 were considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The scRNA-seq and RNA-seq data generated in this study have been deposited in the National Genomics Data Center database under accession code CRA018819. The scRNA-seq data from the wild-type mouse used in this study are publicly available in the Gene Expression Omnibus database under accession code GSE173887. Source data are provided in this paper.
References
Sakaguchi, S., Sakaguchi, N., Asano, M., Itoh, M. & Toda, M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol. 155, 1151–1164 (1995).
Fontenot, J. D., Gavin, M. A. & Rudensky, A. Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4, 330–336 (2003).
Sakaguchi, S., Wing, K., Onishi, Y., Prieto-Martin, P. & Yamaguchi, T. Regulatory T cells: how do they suppress immune responses?. Int. Immunol. 21, 1105–1111 (2009).
Vignali, D. A. A., Collison, L. W. & Workman, C. J. How regulatory T cells work. Nat. Rev. Immunol. 8, 523–532 (2008).
Raffin, C., Vo, L. T. & Bluestone, J. A. T(reg) cell-based therapies: challenges and perspectives. Nat. Rev. Immunol. 20, 158–172 (2020).
Ferreira, L. M. R., Muller, Y. D., Bluestone, J. A. & Tang, Q. Next-generation regulatory T cell therapy. Nat. Rev. Drug Discov. 18, 749–769 (2019).
Zhang, Q. et al. Chimeric Antigen Receptor (CAR) treg: A promising approach to inducing immunological tolerance. Front. Immunol. 9, 2359 (2018).
Brunstein, C. G. et al. Adoptive transfer of umbilical cord blood-derived regulatory T cells and early viral reactivation. Biol. Blood Marrow Transpl. 19, 1271–1273 (2013).
Tarbell, K. V., Yamazaki, S., Olson, K., Toy, P. & Steinman, R. M. CD25+ CD4+ T cells, expanded with dendritic cells presenting a single autoantigenic peptide, suppress autoimmune diabetes. J. Exp. Med. 199, 1467–1477 (2004).
Stephens, L. A., Malpass, K. H. & Anderton, S. M. Curing CNS autoimmune disease with myelin-reactive Foxp3+ Treg. Eur. J. Immunol. 39, 1108–1117 (2009).
Sagoo, P. et al. Human regulatory T cells with alloantigen specificity are more potent inhibitors of alloimmune skin graft damage than polyclonal regulatory T cells. Sci. Transl. Med. 3, 83ra42 (2011).
Noyan, F. et al. Donor-specific regulatory T cells generated on donor B cells are superior to CD4+CD25high cells in controlling alloimmune responses in humanized mice. Transpl. Proc. 45, 1832–1837 (2013).
Humrich, J. Y. et al. Homeostatic imbalance of regulatory and effector T cells due to IL-2 deprivation amplifies murine lupus. Proc. Natl. Acad. Sci. USA 107, 204–209 (2010).
Koreth, J. et al. Interleukin-2 and regulatory T cells in graft-versus-host disease. N. Engl. J. Med. 365, 2055–2066 (2011).
Klatzmann, D. & Abbas, A. K. The promise of low-dose interleukin-2 therapy for autoimmune and inflammatory diseases. Nat. Rev. Immunol. 15, 283–294 (2015).
Shevach, E. M. Application of IL-2 therapy to target T regulatory cell function. Trends Immunol. 33, 626–632 (2012).
Ballesteros-Tato, A. et al. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity 36, 847–856 (2012).
Yang, X. P. et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat. Immunol. 12, 247–254 (2011).
Chi, X. et al. RORgammat expression in mature T(H)17 cells safeguards their lineage specification by inhibiting conversion to T(H)2 cells. Sci. Adv. 8, eabn7774 (2022).
Weaver, C. T., Elson, C. O., Fouser, L. A. & Kolls, J. K. The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annu. Rev. Pathol. 8, 477–512 (2013).
Bender, C. et al. A phase 2 randomized trial with autologous polyclonal expanded regulatory T cells in children with new-onset type 1 diabetes. Sci. Transl. Med. 16, https://doi.org/10.1126/scitranslmed.adn2404 (2024).
Trzonkowski, P. et al. First-in-man clinical results of the treatment of patients with graft versus host disease with human ex vivo expanded CD4+CD25+CD127- T regulatory cells. Clin. Immunol. 133, 22–26 (2009).
Tu, E., Bourges, D., Gleeson, P. A., Ang, D. K. & van Driel, I. R. Pathogenic T cells persist after reversal of autoimmune disease by immunosuppression with regulatory T cells. Eur. J. Immunol. 43, 1286–1296 (2013).
Kasagi, S. et al. In vivo-generated antigen-specific regulatory T cells treat autoimmunity without compromising antibacterial immune response. Sci. Transl. Med. 6, 241ra278 (2014).
Xia, G., He, J. & Leventhal, J. R. Ex vivo-expanded natural CD4+CD25+ regulatory T cells synergize with host T-cell depletion to promote long-term survival of allografts. Am. J. Transpl. 8, 298–306 (2008).
Lee, K., Nguyen, V., Lee, K. M., Kang, S. M. & Tang, Q. Attenuation of donor-reactive T cells allows effective control of allograft rejection using regulatory T cell therapy. Am. J. Transpl. 14, 27–38 (2014).
Wang, Y., Su, M. A. & Wan, Y. Y. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity 35, 337–348 (2011).
Singh, B. et al. Control of intestinal inflammation by regulatory T cells. Immunol. Rev. 182, 190–200 (2001).
Hori, S., Nomura, T. & Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003).
Monticelli, L. A. et al. IL-33 promotes an innate immune pathway of intestinal tissue protection dependent on amphiregulin-EGFR interactions. Proc. Natl. Acad. Sci. USA 112, 10762–10767 (2015).
Tsou, A. M. et al. Neuropeptide regulation of non-redundant ILC2 responses at barrier surfaces. Nature 611, 787–793 (2022).
Cui, W. et al. Diet-mediated constitutive induction of novel IL-4+ ILC2 cells maintains intestinal homeostasis in mice. J. Exp. Med. 220, https://doi.org/10.1084/jem.20221773 (2023).
Zhang, D. et al. High glucose intake exacerbates autoimmunity through reactive-oxygen-species-mediated TGF-beta cytokine activation. Immunity 51, 671–681 (2019).
Zhou, X. et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat. Immunol. 10, 1000–1007 (2009).
Ma, S. Y. et al. RORγt phosphorylation protects against T cell-mediated inflammation. Cell Rep. 38, 110520 (2022).
Wang, C. et al. CD5L/AIM Regulates lipid biosynthesis and restrains Th17 cell pathogenicity. Cell 163, 1413–1427 (2015).
Ghoreschi, K. et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature 467, 967–971 (2010).
Lee, Y. et al. Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 13, 991–999 (2012).
Schnell, A. et al. Stem-like intestinal Th17 cells give rise to pathogenic effector T cells during autoimmunity. Cell 184, 6281–6298 (2021).
Yasuda, K. et al. Satb1 regulates the effector program of encephalitogenic tissue Th17 cells in chronic inflammation. Nat. Commun. 10, 549 (2019).
Meyer Zu Horste, G. et al. RBPJ Controls development of pathogenic Th17 cells by regulating IL-23 receptor expression. Cell Rep. 16, 392–404 (2016).
Atreya, R. et al. Blockade of interleukin 6 trans signaling suppresses T-cell resistance against apoptosis in chronic intestinal inflammation: evidence in crohn disease and experimental colitis in vivo. Nat. Med. 6, 583–588 (2000).
Kai, Y. et al. Colitis in mice lacking the common cytokine receptor gamma chain is mediated by IL-6-producing CD4+ T cells. Gastroenterology 128, 922–934 (2005).
Chandran, S. et al. Interleukin-6 blockade with tocilizumab increases Tregs and reduces T effector cytokines in renal graft inflammation: A randomized controlled trial. Am. J. Transpl. 21, 2543–2554 (2021).
Veldhoen, M., Hocking, R. J., Flavell, R. A. & Stockinger, B. Signals mediated by transforming growth factor-beta initiate autoimmune encephalomyelitis, but chronic inflammation is needed to sustain disease. Nat. Immunol. 7, 1151–1156 (2006).
Konkel, J. E. et al. Transforming growth factor-β signaling in regulatory T cells controls T helper-17 cells and tissue-specific immune responses. Immunity 46, 660–674 (2017).
Laurence, A. et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity 26, 371–381 (2007).
Duerr, R. H. et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314, 1461–1463 (2006).
Gaffen, S. L., Jain, R., Garg, A. V. & Cua, D. J. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat. Rev. Immunol. 14, 585–600 (2014).
Lin, C. C. et al. IL-1-induced Bhlhe40 identifies pathogenic T helper cells in a model of autoimmune neuroinflammation. J. Exp. Med. 213, 251–271 (2016).
Oslejskova, L. et al. Metastasis-inducing S100A4 protein is associated with the disease activity of rheumatoid arthritis. Rheumatology48, 1590–1594 (2009).
Bai, X. F. et al. The heat-stable antigen determines pathogenicity of self-reactive T cells in experimental autoimmune encephalomyelitis. J. Clin. Invest. 105, 1227–1232 (2000).
Wang, X. et al. Febrile temperature critically controls the differentiation and pathogenicity of T helper 17 cells. Immunity 52, 328–341 (2020).
Hansen, W. et al. G protein-coupled receptor 83 overexpression in naive CD4+CD25- T cells leads to the induction of Foxp3+ regulatory T cells in vivo. J. Immunol. 177, 209–215 (2006).
Wang, S. et al. Sphingosine-1-phosphate receptor 4 attenuates neutrophilic airway inflammation in experimental asthma via repressing proinflammatory macrophage activation. Int. J. Biol. Sci. 19, 1597–1615 (2023).
Chen, W. J. TGF-β Regulation of T cells. Annu. Rev. Immunol. 41, 483–512 (2023).
Chen, W. J. & ten Dijke, P. Immunoregulation by members of the TGFβ superfamily. Nat. Rev. Immunol. 16, 723–740 (2016).
Hunter, C. A. & Jones, S. A. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 16, 448–457 (2015).
Schust, J., Sperl, B., Hollis, A., Mayer, T. U. & Berg, T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem. Biol. 13, 1235–1242 (2006).
Goschl, L., Scheinecker, C. & Bonelli, M. Treg cells in autoimmunity: from identification to Treg-based therapies. Semin. Immunopathol. 41, 301–314 (2019).
Rubtsov, Y. P. et al. Stability of the regulatory T cell lineage in vivo. Science 329, 1667–1671 (2010).
Levine, A. G. et al. Stability and function of regulatory T cells expressing the transcription factor T-bet. Nature 546, 421–425 (2017).
Beriou, G. et al. IL-17-producing human peripheral regulatory T cells retain suppressive function. Blood 113, 4240–4249 (2009).
Chen, Y. et al. Foxp3(+) regulatory T cells promote T helper 17 cell development in vivo through regulation of interleukin-2. Immunity 34, 409–421 (2011).
Veldhoen, M., Hocking, R. J., Atkins, C. J., Locksley, R. M. & Stockinger, B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 24, 179–189 (2006).
Pandiyan, P. et al. CD4(+)CD25(+)Foxp3(+) regulatory T cells promote Th17 cells in vitro and enhance host resistance in mouse Candida albicans Th17 cell infection model. Immunity 34, 422–434 (2011).
Liu, Q. et al. Non-oxidative pentose phosphate pathway controls regulatory T cell function by integrating metabolism and epigenetics. Nat. Metab. 4, 559–574 (2022).
Lee, J. eeH., Elly, C., Park, Y. & Liu, Y.-C. E3 Ubiquitin Ligase VHL Regulates Hypoxia-Inducible Factor-1α to Maintain Regulatory T Cell Stability and Suppressive Capacity. Immunity 42, 1062–1074 (2015).
Essig, K. et al. Roquin suppresses the PI3K-mTOR signaling pathway to inhibit T helper cell differentiation and conversion of treg to Tfr cells. Immunity 47, 1067–1082.e1012 (2017).
Dall’Era, M. et al. Adoptive treg cell therapy in a patient with systemic Lupus erythematosus. Arthritis Rheumatol. 71, 431–440 (2019).
Thonhoff, J. R. et al. Combined regulatory T-lymphocyte and IL-2 treatment is safe, tolerable, and biologically active for 1 year in persons with amyotrophic lateral sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 9, https://doi.org/10.1212/nxi.0000000000200019 (2022).
Korn, T. et al. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature 448, 484–487 (2007).
Harbour, S. N. et al. T(H)17 cells require ongoing classic IL-6 receptor signaling to retain transcriptional and functional identity. Sci. Immunol. 5, https://doi.org/10.1126/sciimmunol.aaw2262 (2020).
Waldmann, T. A. The multi-subunit interleukin-2 receptor. Annu. Rev. Biochem. 58, 875–911 (1989).
Pandiyan, P., Zheng, L., Ishihara, S., Reed, J. & Lenardo, M. J. CD4+CD25+Foxp3+ regulatory T cells induce cytokine deprivation-mediated apoptosis of effector CD4+ T cells. Nat. Immunol. 8, 1353–1362 (2007).
Tang, Q. & Vincenti, F. Transplant trials with Tregs: perils and promises. J. Clin. Invest. 127, 2505–2512 (2017).
Feng, Y. et al. Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell 158, 749–763 (2014).
Batlle, E. & Massague, J. Transforming growth factor-beta signaling in immunity and cancer. Immunity 50, 924–940 (2019).
Dong, S.,et al. The effect of low-dose IL-2 and Treg adoptive cell therapy in patients with type 1 diabetes. JCI Insight 6, https://doi.org/10.1172/jci.insight.147474 (2021).
Hirakawa, M. et al. Low-dose IL-2 selectively activates subsets of CD4(+) Tregs and NK cells. JCI Insight 1, e89278 (2016).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (NO. 82171829 to D.Z.), the Key Project of the Science and Technology Department of Sichuan Province (NO. 2025YFHZ0205 to D.Z.), and the 1·3·5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (NO. ZYYC25010 to D.Z.). D.Z. sincerely wants to commemorate Dr. Sang-A Park, who passed away suddenly in an auto accident in Bethesda, MD, USA.
Author information
Authors and Affiliations
Contributions
H.C. designed and performed experiments, analyzed data, and drafted the manuscript. F.N. and N.J. designed and performed experiments, analyzed data and edited the manuscript. X.M., J.Z., H.L., and W.Z. performed experiments. H.N., H.Z., W.J., H.J., J.T., X.Z., N.L., Q.Z., H.H., and W.J.C. provided critical scientific inputs. H.X. designed experiments, analyzed data, and wrote the manuscript. D.Z. conceptualized, supervised and supported the whole study, designed experiments, analyzed data, and wrote the manuscript. All authors have read and approved the final manuscript for publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Wayne Hancock, and the other anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cheng, H., Nan, F., Ji, N. et al. Regulatory T cell therapy promotes TGF-β and IL-6-dependent pro-inflammatory Th17 cell generation by reducing IL-2. Nat Commun 16, 7644 (2025). https://doi.org/10.1038/s41467-025-62628-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-62628-7