Abstract
A leading paradigm for understanding the large-scale behavior of tissues is via generalizations of liquid crystal physics; much like liquid crystals, tissues combine fluid-like, viscoelastic behaviors with local orientational order, such as nematic symmetry. Whilst aspects of quantitative agreement have been achieved for flat monolayers, the most striking features of tissue morphogenesis—including symmetry breaking, folding and invagination—concern surfaces with complex curved geometries in three dimensions. As yet, however, characterizing such behaviors has been frustrated due to the absence of proper image analysis methods; current state-of-the-art methods almost exclusively rely on two-dimensional intensity projections of multiple image planes, which superimpose data and lose geometric information that can be crucial. Here, we describe an analysis pipeline that properly captures the nematic order and topological defects associated with tissue surfaces of arbitrary geometry, which we demonstrate in the context of in vitro multicellular aggregates, and in vivo zebrafish hearts.
Similar content being viewed by others
Introduction
Morphogenesis is the striking process by which tissues—large-scale aggregates of cells—adapt and change their shape. It is a ubiquitous phenomenon where cell and molecular biology intersect with mechanics and geometry1. As such, it is central to many important developmental processes, such as wound-healing2, the folding process during gastrulation3,4,5, symmetry breaking events6,7,8, embryonic and organ development4,9, neuronal tube closure10,11, and large-scale regeneration in Hydra12,13.
Notably, such behavior has been the subject of recent intense work, in which tissues are understood via generalizations of liquid crystal physics14,15,16; much like liquid crystals, tissues combine fluid-like, viscoelastic behaviors with local orientational order, such as nematic or hexatic symmetries. This approach has been shown to reproduce several characteristics of flat tissue monolayers17,18,19,20,21,22,23,24, including the apical extrusion of cells as a consequence of topological defects19,25, regeneration in Hydra13,26,27,28,29, and the statistics of active turbulence by which tissues ‘self-stir’30,31,32. Recent theory and experimental work is moreover highly suggestive that such nematic characterizations can describe complex shape changes, with topological defects in particular conjectured to play a significant role in morphogenesis13,26,27,28,29,33,34,35,36.
However, for the latter case, quantitative comparisons with data, as well as the concomitant generation of better, more biologically accurate physico-mathematical models, has been stymied by a lack of appropriate image analysis methods. Specifically, standard algorithms for extracting the nascent nematic order of a tissue monolayer, or exposed interface, rely on those surfaces being flat5,13,26,37,38,39,40,41,42. For curved surfaces, the typical approach is to project the data onto a flat surface and then use standard techniques13,26,27,28,29,34,36. Such projections may be reasonably accurate in regions of low curvature, but (i) will lose information in regions of high curvature, for example, in a protrusion, and (ii) cannot possibly capture the entire surface of a tissue or aggregate in three dimensions (3D) (Fig. 1a, b). Whilst some progress has been made in this area—3D segmentation39,43,44,45 and tracking software46 as well as applied models for 3D reconstructions47,48,49,50,51 are notable advances—the tools to properly capture nematic order on tissue surfaces with arbitrarily complex geometry are still lacking, making experimental investigations of cell and tissue properties difficult as well as leaving experiment and theory unable to verify each other, and therefore breaking the otherwise virtuous feedback cycle.
a Reconstruction of a multicellular MCF10A aggregate. The cell boundaries (dark areas) of the aggregate indicate the fluorescence signal of the actin filament. The signal within a distance of 6 μm to the surface of the southern hemisphere was max-projected onto a 2D plane as shown in (b). b The 2D-projected image is superimposed with the 2D nematic directors (white lines) representing the orientation field and nematic topological defects of charge +1/2 (orange) and −1/2 (cyan) as regions where no prevailing orientation can be found. However, the effects of curvature mean that the full 3D nematic director can have components in the z-direction, as shown in panel (a); this information is lost by projecting into 2D. The size of this effect is moreover proportional to the difference between the normal to the aggregate and the normal to the projection plane—i.e., it is largest at the boundary of the projected image in panel (b). c By contrast, our 3D surface analysis displays the correct orientation field of the cells, especially at the boundary of the aggregate, and thus the correct locations of nematic topological defects, see also Fig. 2g, h.
Here, we present an analysis pipeline to do precisely this: we properly characterize the orientation field on the entire surface of tissues of arbitrary shape, based only on 2D images of a single z-stack acquired with conventional, widely available microscopy (Fig. 1c). At a high level, our method is as follows. We first identify a surface in terms of discrete points, using the images of the acquired z-stack. At a given point, we then construct the tangent plane to the surface. By translating the tangent plane a small distance in the inward normal direction, so that it makes a shallow slice of the surface cells, we are then able to quantify local properties, such as the nematic order parameter. This is then projected back onto the initial surface point. Repeating the procedure for all surface points, therefore, leads to a mapping of surface information that is faithful to the underlying geometry (Fig. 1). In this way, our method is capable of quantifying nematic orientation fields (as well as other quantities, such as fluorescence signals) in a manner independent of a surface’s curvature.
We demonstrate our approach by using it to analyze epithelial MCF10A multicellular aggregates in vitro and zebrafish hearts in vivo. For the former, we identify topological defects and correlate their number to the aggregate’s surface area, as predicted by theory. For the latter, we correlate nematic order with the heart’s curvature and F-actin cytoskeleton signal, seemingly indicating that the system obeys a Laplace law. In addition, we also demonstrate our analysis method on examples of endothelial cells forming a micro-vessel with negative curvature, as well as myoblasts growing on a PDMS sphere, for which the alignment of the nematic director and the underlying actin architecture is visible.
Results
Detecting surface properties using tangent planes
To achieve a full quantification of cell and tissue properties on surfaces of arbitrary shape, a first step is to understand the deficiencies of standard 2D projection methods. We use the illustrative example of a spheroidal MCF10A aggregate of about 100 μm diameter (corresponding to the diameter of about 7 cells) whose cell boundaries are visualized using phalloidin to identify F-actin (Fig. 1a). As shown in Fig. 1a, we restrict our projection range of the xy-images of the z-stack to one of the two hemispheres of the aggregate: the southern hemisphere. The range is defined by the first image of the z-stack, up to the image where the diameter of the cross-section of the aggregate is the largest. Since we are interested in the surface of the aggregate, we project the signal within a distance of 6 μm to the surface onto a flat plane before extracting information (Fig. 1b). The orientation field of the 2D-projected image is then generated using the ImageJ plugin, OrientationJ. However, the resulting flattened image will be only a faithful representation of the cell structure of the aggregate and the nematic order at the pole of the hemisphere, where the normal to the spherical aggregate is aligned with the normal to the flat plane that we have projected the images onto. In other words, this is the area of the aggregate facing the objective of the microscope. Away from this point, the normal direction of the aggregate and the normal to the projection plane have a large discrepancy, which is due to the curvature of the surface. The greater this disparity, the less accurate the analysis, limiting the applicability of the single projections to small areas and making it difficult to analyze the entire curved surface in 3D.
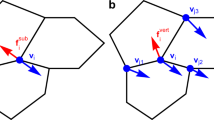
By contrast to such traditional methods, our approach is instead based on the geometrical notion of the tangent plane52 (Fig. 2). It is tantamount to choosing multiple perspectives from which to observe the aggregate and then stitching the information together in a way that is faithful to the surface geometry. The positions of each tangent plane in the ambient 3D space are determined by two criteria: they must each (i) intersect a different boundary surface point (identified from the acquired 2D images of the z-stack) such that (ii) the tangent plane normal coincides with the surface normal at the chosen surface point (Fig. 2a–c, Methods - Surface points and normal vectors). We then use the normal vector to translate the tangent plane in the direction of the bulk of the multicellular system, so that it makes a shallow slice of the outer cell layer (Fig. 2d, e). Ideally, the distance we translate by does not exceed the size of a single cell; we have chosen 5 μm as half the cell diameter. (Distances in the direction of the bulk that exceed the size of a single cell lead to the analysis of cells below the surface layer and possibly to different results). At this stage, there are two options depending on the image quality. Either we directly analyze the fluorescence signal on a single shallow slice, or we first sum-, max-, or mean-project the intensity signal of multiple slices, each at different translation distances. In the following analysis of spheroidal aggregates, we use the former, whilst in our analysis of Zebrafish hearts, we use the latter.
a, b For selected images of the acquired z-stack (Fig. 1a), a black and white mask is created to identify boundary points (yellow dots). c These points are combined to create a discretization of the aggregate’s surface in 3D (yellow dots). To determine the normal (white arrow) at a given point, a set of neighboring points (purple dots) is required to capture the local shape. This is repeated for every surface point (red arrows). d The normal vector (red arrow) at a surface point (yellow dot) defines the tangent plane at that point. This is translated by 5 μm along the normal vector in the direction of the bulk, resulting in a shallow slice (black plane) of the outer cells of the aggregate. e The xy-image of the acquired z-stack at the surface point (yellow dot) in (d) shows the position of the shallow slice (white line) and the 2D component of the 3D normal vector (red arrow). f A continuous 2D image within the shallow slice in (d) is obtained by linear interpolation of the underlying intensity field. Here, dark areas indicate the actin filaments at the cell borders, while lighter areas of the cells are used to perform the orientation analysis using the standard 2D software, OrientationJ. Nematic directors (red lines) are obtained in the shallow slice, with those in a local area ‘beneath’ the surface point (yellow dot) averaged to define n2D (white line). This director is then translated back to the original surface point, where it is expressed in 3D coordinates, n3D (see Methods - 3D nematic director on the surface). g, h The nematic director field (white lines) covers the entire surface of the 3D multicellular MCF10A aggregate and enables the detection of nematic topological defects. Defects of charge ±1/2 are shown here.
For a given shallow slice, we obtain a continuous scalar field by linear interpolation of the acquired intensities. We then perform the orientation analysis using standard flat-surface software, OrientationJ. This involves a local average over nearest neighbors to produce a smooth nematic director field (red lines in Fig. 2f). Whilst this field of directors may be affected by the curvature in a similar way as the 2D intensity projection case, the information in a local region around the translated surface point—i.e., the ‘center’ of the shallow slice—gives a good approximation of the nematic orientation at the aggregate surface. We therefore take an average of the local nematic field (red lines in Fig. 2f) in the small region around the center point. This then defines a nematic director n2D in the plane of the slice (white line in Fig. 2f), which we translate back to the original surface point. We then perform a pull-back to the ambient space and express it in terms of its 3D components, referring to the result as n3D.
Performing the above construction at all surface points yields a nematic director field across the entirety of the aggregate’s surface (Supplementary Fig. S2d in the Supplementary Information - S2 Surface point spacing and coarse-graining). The directors are then coarse-grained on the surface of the aggregate (Fig. 2g, h, Supplementary Fig. S2d–f), see Eq. (11). The larger the coarse-graining radius, the smoother the director field becomes, setting an effective minimum separation for observing defect pairs. In Movie 1, we rotate the view of the aggregate by 360°, showing the entire nematic texture (Supplementary Movie 1). Of note, our method takes care of asymmetric voxels, which are typically longer in the z-direction, as discussed in the Supplementary Information (S1 Signal projection onto shallow slices in 3D space).
Since our method only requires the surface points of a given multicellular system, it allows the analysis of surfaces with arbitrary shapes. Compared to the 2D intensity projection method (Fig. 1b), where the orientation analysis is based on a specific field of view defined by the position of the sample in the microscope—and where the projected nematic directors may differ greatly from the true, geometrically faithful orientation field—our surface analysis method provides a more complete representation (Fig. 1c). As we expected, both methods of orientation analysis give similar results at the centered region of the field of view, where the normal to the aggregate and the normal to the projection plane are similar. However, the two methods give drastically different results near to the depicted aggregate boundary, where the effects of curvature are large. As a result, our method is able to properly identify topological defects on curved surfaces, which are the focus of the next section.
Nematic order and topological defects on multicellular aggregate surfaces
The arrangement of cells within the tissue is crucial for development and disease. On the one hand, an aligned director field has been associated with pattern formation during embryogenesis, convergence and extension in gastrulation, and directional migration during wound-healing processes14,15,16. On the other hand, regions where the director field cannot be defined, also known as topological defects, have been linked to morphogenesis and tissue regeneration13,29,36. These topological defects must occur on the surface of spheres and topologically equivalent systems with a total nematic topological charge of +253. In this context, we used our surface analysis method to (i) identify nematic topological defects within our multicellular aggregates and (ii) ensure that our analysis complies with the mathematical constraint on the total topological charge.
Figure 3a shows a multicellular aggregate superimposed with the nematic director field. The color code of the directors corresponds to the magnitude of the local nematic order parameter, see Eq. (11). Regions with misaligned directors are identified by low values of the nematic order (blue) and high alignments by high values of the nematic order (red). Topological defects are identified as points where the magnitude of the order parameter is zero. These defects are characterized by their positive or negative charge, which is computed by encircling each defect point with a contour and calculating how the director field on that contour rotates around the defect core, see Eq. (13). We performed this analysis by projecting the local nematic directors of interest onto the tangent plane at the location of the defect. Thus, we identified several +1/2 and −1/2 defects on the aggregate surface, with a total defect charge of +2.
a Multicellular MCF10A aggregate, superimposed with the nematic director field (lines) and topological defects (dots). The color code of the nematic directors corresponds to the magnitude of the local nematic order parameter. Red indicates director alignment (with 1 being perfect nematic order) and blue indicates director misalignment (with 0 being the absence of nematic order). The distance between the directors is about 6 μm, which corresponds to the approximate cell radius. Defects of charge +1/2 and −1/2 are shown as orange and cyan dots, respectively. The cell boundaries (dark areas) of the aggregate indicate the fluorescence signal of the actin filament. b Local nematic order parameter visualized as a heatmap. Defects of charge +1/2 and −1/2 are shown as black and white dots, respectively. Panels (a) and (b) show the identical position of the aggregate, while in (b), the aggregate was treated as a sphere for the orientation analysis. c Number of defects per analyzed aggregate (n = 10), including ±1/2 defects, against the aggregate surface area for the coarse-graining factor kcg = 1.2 with a Pearson’s linear correlation coefficient of r = 0.6. The orange line represents a linear regression, indicating the trend of the data points, where m is the slope and d stands for defect. The linear regression intersects the number of 4 defects at 0.63 × 104 μm2. As a note, the plot includes two aggregates with 14 defects and the same surface area, resulting in an overlap of the data points. We separated the points slightly to visualize both. Each aggregate has a total topological charge of +2.
For convenience, we next approximated the aggregate surface by a sphere in order to obtain a regular triangular mesh (see Methods - Surface points and normal vectors). The regular grid with equal spacing between the nematic directors facilitated a more straightforward analysis of the topological charge of the defects (Fig. 3b). This approximation does not materially change either the location of the topological defects or the wider texture of the nematic director field; regions of high nematic order (on the right of the aggregate in this perspective) and regions of disorder (on the left of the aggregate in this perspective) are similarly located between aggregates and spheres (Fig. 3a, b).
We investigated defect proliferation on ten multicellular aggregates of different sizes, limiting ourselves to those with radii < 50 μm, since for larger aggregates, high light absorption and scattering of the fluorescence signal occur in deep layers of the z-stack. In the absence of composite defects with charge ±1, which we did not observe in any of our analyzed aggregates, the topology of the sphere implies the minimal number of defects is four, each with +1/2 charge, for a total charge of +253. Additional defects must come in pairs with charge ±1/2 in order to meet the topological constraint on the total defect charge. Theoretical models have suggested that the number of defects correlates linearly with the surface area of the sphere in a turbulent regime characterized by additional defects54. Accordingly, we plotted the total number of defects against the surface area of the aggregate (Fig. 3c) with a coarse-graining factor of kcg = 1.2 (see Methods - 3D nematic director on the surface). Our data indicate that the number of defects increases with the aggregate surface area, with a slope of m = 63 × 10−5 defects/μm2 of the linear regression, supporting previous theoretical work54. Increasing the coarse-graining radius resulted in a reduction of the number of ±1/2 defect pairs, setting an effective cutoff, in terms of surface area, for observing more than four ±1/2 defects (Fig. S3a, b in the Supplementary Information - S3 Defect density on spheres). All multicellular aggregates analyzed had, as expected, at least four +1/2 defects and a total topological change of +2.
Furthermore, we tested our 3D surface analysis method on C2C12 mouse myoblasts grown on a PDMS sphere with a diameter of 300 μm (Fig. S4 in the Supplementary Information - S4 Other examples), showing a highly organized F-actin network with spindle-like morphology. The nematic directors align with the F-actin signal and form 5 × +1/2 and 1 × −1/2 defects. Compared to the multicellular aggregates with different radii (Fig. 3c), the number of detected defects is much lower in the myoblast monolayer grown on the sphere. This is probably due to their larger nematic correlation length compared to epithelial cells, such as MCF10A cells55,56.
Taken together, our surface analysis method is able to identify the nematic director field of multicellular aggregates and myoblasts grown on spheres, and provides information for further analysis, such as the detection of topological defects. Our ability to determine quantities such as the total number of nematic defects and to investigate the dependence of these quantities on material properties such as aggregate surface area, then facilitates quantitative comparisons between experimental systems and mathematical models of active nematics.
Spatio-temporal correlations in the Zebrafish Heart
To demonstrate the robustness of our surface analysis method, we applied it to a complex in vivo system with distinctly different shapes and curvature than the spheroidal multicellular aggregates: zebrafish hearts at different stages of development. We imaged the ventricular myocardium of zebrafish hearts at 72 and 120 hours post-fertilization (hpf) using fluorescent reporters for the cell membrane and actin cytoskeleton. At these stages, the myocardium is composed of an outer compact layer and an inner trabecular layer. Here, we focused on the outer compact layer (CL), where actomyosin remodeling has previously been shown to allow cells to stretch, therefore increasing ventricle size57. To apply our surface analysis method to only the compact layer, we set the distance of the tangent plane to the surface to be 3 μm, which is about the thickness of the outer cell layer, and then max-projected the intensity signal of the membrane from four generated planes with distance of 1 μm in this range (see Methods) in order to increase the signal of the thin cell layer. We then performed the orientation analysis on these tangent planes and projected the nematic directors back onto the surface of the ventricle, exactly as described above for the multicellular aggregates (Fig. 2). Compared to the tissue orientation analysis of 2D-projected images, the nematic orientation field obtained from our method is much more accurate at regions, e.g., the lateral surface of the ventricle, where the surface strongly curved out of the projection plane (as shown in the 2D perspective in Fig. 4a, b) and an analysis based on 2D projections is not valid.
a 2D image of the ventricle at 120 hpf. The signal within a distance of 8 μm to the surface was max-projected onto a 2D plane. The cell boundaries (dark areas) of the heart indicate the fluorescence signal of the membrane (myl7:BFP-CAAX). The image is superimposed with the 2D nematic directors (red lines) representing the orientation field of the cells. b Result of our 3D surface analysis that displays the correct orientation field of the cells. c, d Ventricles at 72 and 120 hpf, superimposed with nematic directors, see also Supplementary Movie 2 and Supplementary Movie 3. The color code of the nematic directors corresponds to the local nematic order parameter. Red indicates high alignment of cells (i.e., order of 1) and blue misalignment (i.e., order of 0). The distance between the directors is about 10 μm. Region I and II (cyan dots, encircled in cyan) were used for further analysis. e, f Distribution of the local nematic order parameter and the absolute value of the Gaussian curvature of the two different regions of zebrafish hearts, as shown in panels (c, d). Each box plot contains data points of these regions of zebrafish hearts at 72 and 120 hpf, NI,72hpf = 66, NII,72hpf = 59, NI,120hpf = 62, NII,120hpf = 63. In each case, we use data from five different hearts. The black dots are the mean values of selected data points per analyzed heart. Each box shows the median (red line), 25th and 75th percentiles (box), maximum and minimum without outliers (whiskers), and 95% confidence interval of the median (notches). P-values were calculated from Dunn’s test of multiple comparisons after a significant Kruskal-Wallis test.
The myocardium is under constant tensile stress due to the fluid pressure in the ventricle lumen while the heart is pumping. It has been shown that cells elongate and align in the direction of tension57,58. To investigate this, we calculated the local nematic order parameter and selected data points within a radius of 15 μm from two different regions on the surface of a total of 5 hearts at 72 and 120 hpf for comparison (Fig. 4c, d, Supplementary Movie 2 and Supplementary Movie 3). Region I was located in the outer curvature of the ventricle, i.e., the opposite to where blood flows in from the atrium. Due to the geometry of the ventricle and the contraction to regulate the blood flow, we expect the cells to experience the highest tension here. Region II was located in the apex, i.e., on the opposite side where the blood exits. For both developmental stages, we measured a significantly higher local nematic order of the cells in region I with 0.937 ±0.020 (mean ±s.d.) at 72 hpf and 0.949 ±0.016 at 120 hpf compared to region II with 0.86 ±0.05 at 72 hpf and 0.87 ±0.13 at 120 hpf (Fig. 4e). In addition, we did not observe any nematic topological defects in region I at either 72 or 120 hpf. However, defects appeared in region II, where the local nematic order was lower (Supplementary Movie 2 and Supplementary Movie 3). The difference in director alignment and distribution of defects suggests57,58 that cells were indeed under higher tension in region I. Moreover, the local nematic order significantly increased from 72 to 120 hpf in both regions, indicating an increase in tension.
Previous work has suggested that regions where the absolute value of Gaussian curvature is large correlate with regions of lower nematic order35,59,60,61. Accordingly, we measured a significantly lower absolute value of Gaussian curvature in region I with (7 ±8) × 10−4 μm−2 (mean ±s.d.) at 72 hpf and (3.6 ±3.0) × 10−4 μm−2 at 120 hpf compared to region II with (16 ±15) × 10−4 μm−2 at 72 hpf and (8 ±7) × 10−4 μm−2 at 120 hpf (Fig. 4f). In addition, the absolute value of the Gaussian curvature decreased between 72 and 120 hpf in both regions, reflecting the shape change and enlargement of the heart during development.
The major player in the contraction of the heart is the sarcomeric network composed of actomyosin. We used our surface analysis method to measure the intensity distribution of F-actin labeled by LifeAct (-0.2myl7:Lifeact-mNeongreen) in the CL of the ventricle, using the same imaged zebrafish hearts and tangent plane positions as described above. The intensity was averaged over the four tangent planes at each surface point and related to the background signal (see Methods - Intensity analysis on the plane). We compared the mean signal-to-background ratio of the fluorescence signal (Fig. 5a–c, Supplementary Movie 4 and Supplementary Movie 5) between the two regions at 72 and 120 hpf. For both developmental stages, we measured a significantly higher mean intensity of F-actin in region I with 8.8 ±2.5 (mean ±s.d.) at 72 hpf and 9.1 ±2.3 at 120 hpf compared to region II with 6.6 ±1.6 at 72 hpf and 6.8 ±1.6 at 120 hpf (Fig. 5c). We speculated above that cells align in the direction of tension, and therefore the stronger alignment of cells in region I compared to region II (Fig. 4e) suggests a positive correlation with tissue tension. Our measurement of a dominant presence of F-actin in region I supports this idea.
a, b 3D reconstruction of the ventricle at 72 and 120 hpf. The signal within a distance of 3 μm to the surface is shown. Bright areas of the heart show the fluorescence signal of F-actin (-0.2myl7:Lifeact-mNeongreen). The ventricle is superimposed with nematic directors with a distance of about 10 μm, determined based on the membrane signal shown in (d, e), whose color code corresponds to the mean intensity signal in respect to the background, see also Supplementary Movie 4 and Supplementary Movie 5. Region I and II (red dots, encircled in red) were used for quantification and comparisons in (c). d, e 3D reconstruction of the ventricle at 72 and 120 hpf. Dark areas of the heart indicate the fluorescence signal of the membrane (myl7:BFP-CAAX). The color code of the ventricle is superimposed with nematic directors, whose color code corresponds to the mean curvature, see also Supplementary Movie 6 and Supplementary Movie 7. Region I and II (white dots, encircled in white) were used for quantification and comparisons in (f). c, f Each box plot contains data points of these regions of zebrafish hearts at 72 and 120 hpf, five analyzed hearts each, NI,72hpf = 66, NII,72hpf = 59, NI,120hpf = 62, NII,120hpf = 63. The black dots are the mean values of selected data points per analyzed heart. Each box shows the median (red line), 25th and 75th percentiles (box), maximum and minimum without outliers (whiskers), and 95% confidence interval of the median (notches). P-values were calculated from Dunn’s test of multiple comparisons after a significant Kruskal-Wallis test.
To explore this idea further, and considering that the cell layer is under pressure due to blood flow, we thought to correlate our results with Laplace’s law. According to Laplace’s law, the pressure difference across a surface, ΔP, is proportional to the surface tension, γ, multiplied by the surface’s mean curvature, H (i.e., ΔP ∝ γH). This implies that, at constant pressure difference, when comparing region I and II for each developmental stage, surface tension and mean curvature should be anti-correlated. To investigate whether the F-actin signal is related to surface tension, we measured the mean curvature of the ventricle (Fig. 5d–f, Supplementary Movie 6 and Supplementary Movie 7). The mean curvature in region I, with 0.037 ±0.046 μm−1 (mean ±s.d.) at 72 hpf and 0.022 ±0.042 μm−1 at 120 hpf was significantly lower when compared with region II, with 0.06 ±0.07 μm−1 at 72 hpf and 0.04 ±0.037 μm−1 at 120 hpf (Fig. 5f). For each individual developmental stage, where pressure is assumed to be constant, the intensity of F-actin anti-correlates with the mean curvature. This indicates that F-actin may represent a proxy for surface tension. When we compared the two different developmental stages, we did not measure a significant difference of the F-actin signal. Since we only analyzed the CL of the ventricle, we do not want to speculate whether the significant decrease in mean curvature in both regions indicates a decrease in blood pressure from 72 to 120 hpf.
Using the zebrafish heart as an example, we have demonstrated how our surface analysis method can be used to investigate and spatio-temporally correlate various tissue properties of complex multicellular systems in development, such as local tissue orientation and curvature. We identify regions on the surface of the heart with different orders of cell alignment, possibly in the direction of tension, which we find to be anti-correlated with the absolute value of the Gaussian curvature of the surface. In addition, we have used our method to measure and quantify the intensity of fluorescence signals from cells, which can be spatio-temporally correlated with the tissue alignment and mean curvature of the ventricle. Our results show that the correlation of these properties follows Laplace’s law, linking biological properties with physical interpretations.
Discussion
In this work, we have demonstrated how to properly analyze local, tensor-based descriptors of tissue surfaces, with the nematic director our primary motivating example. This moves beyond the standard, and widely-used projection methods that we have shown to be insufficient to meet the needs of contemporary mechano-biological understanding of morphogenesis, much of which is rooted in the physics of liquid crystals.
We used our method to detect the nematic ordering on the surface of spheroidal epithelial aggregates, identifying nematic topological defects in regions with misaligned directors (Fig. 3). As expected for spheres and topologically equivalent shapes, all analyzed aggregates had a total topological charge of +253. The identification of defects allows the comparison of experimental data with predictions from theory and simulations. By counting the defects on multicellular aggregates of different sizes, we showed that, over and above a minimum area set by the choice of a surface coarse-graining length scale, the number of defects appears to scale linearly with the aggregate surface area (Fig. 3c). This correlation has been suggested by simulations in a turbulent regime characterized by additional ±1/2 defect pairs beyond the four +1/2 defects54. It would be interesting to study this for larger aggregates; other microscopy techniques, such as light-sheet microscopes and with larger working distance objectives, might allow going beyond our measurement range.
In addition, we have analyzed a monolayer of myoblasts that have been cultured on a sphere. This is shown to have six defects, which is fewer than on the surface of epithelial aggregates of similar size. We argue that this is likely due to a significantly larger nematic correlation length as compared to epithelial cells55,56. In this context, it would be interesting to investigate the length scale of orientational order for different cell types grown on surfaces of different geometries, how the curvature impacts the result, and whether other defects, such as integer defects, occur.
For completeness, we have also analyzed one example of negative mean curvature in Supplementary Fig. S5, where the nematic orientation field aligns with the stress fibers of HUVECs forming a micro-vessel in the longitudinal direction, which is consistent with previous work58.
More generally, the spatio-temporal quantification of topological defects on curved surfaces offered by our 3D surface analysis method enables the investigation of dynamic systems, such as defect migration on curved surfaces or on more complex 3D systems. It provides their quantification beyond 2D projections (Fig. 1), local analysis of relatively flat regions, or simplifying multicellular aggregates as perfect spheres62,63,64,65, and can be used to further investigate the role of topological defects in morphogenesis13,26,27,28,29,36. It would also be interesting to link computational input parameters, such as activity and energy terms33,35,54,61,66,67 with experimental results to strengthen interdisciplinary research between active soft matter physics and tissue biology.
We tested our approach on a complex in vivo system: the zebrafish heart. The nematic analysis of the heart’s ventricle uncovered regions of lower and higher alignment (Fig. 4), which we were able to correlate with the ventricle’s curvature as a geometrical property and the fluorescence expression of F-actin as a molecular signal (Fig. 5). The regions of the ventricle on the outer curvature, defined here as region I, showed a strong alignment of cells perpendicular to the long axis of the ventricle57. It has been suggested that the local nematic order will align with the direction of tension as the heart contracts58,68. Indeed, we found a greater F-actin signal in region I compared with the region at the apex, defined here as region II. These results are anti-correlated with the absolute value of Gaussian and mean curvature and, assuming F-actin can be used as a proxy for the surface tension due to actomyosin contractility, are indicative of a Laplace law for a constant pressure. However, it would be interesting to combine our 3D surface analysis with other techniques such as stress inference43 or flipper probes69,70 to measure tissue tension.
Our 3D surface analysis enables multiscale correlations of surface curvature and nematic alignment at the tissue scale with molecular responses at the subcellular scale. It opens the possibility to investigate curvature-dependent cell and tissue organizations beyond regular structures such as cylinders and spheres71,72,73, and lays the foundation to better characterize developmental processes in three dimensions.
Methods
All animal research complies with all relevant ethical regulations. All regulated procedures were performed under the UK project license PP8356093, according to institutional (The Francis Crick Institute) and national (UK Home Office) requirements as per the Animals (Scientific Procedures) Act of 1986.
Preparation of PDMS spheres
PDMS spheres were fabricated using a homemade microfluidic system. Briefly, PDMS droplets were generated by ejecting uncured PDMS through fine needles into a 2% (w/v) sodium alginate (SA) aqueous solution. To prevent droplet coalescence, the SA solution was continuously stirred using a magnetic stirrer. Once the PDMS droplets were uniformly dispersed in the SA matrix, a 1% (w/v) calcium chloride (CaCl2) solution was added to induce ionic crosslinking of the SA, forming a stable hydrogel network around the PDMS droplets. The mixture was then transferred to an oven and heated at 80 °C for 2 h to cure the PDMS droplets. After curing, ethylenediaminetetraacetic acid (EDTA) solution was introduced to chelate the calcium ions and disrupt the alginate crosslinking, thereby releasing the PDMS spheres. The spheres were subsequently collected by centrifugation, washed three times with deionized water, and dried at 80 °C.
Cell culture
MCF10A cells (CRL-10317) were obtained from ATCC and cultured in DMEM/F-12 (Thermo Fisher Scientific, 10565-018) supplemented with 5% horse serum (Sigma Aldrich, H1138, USA), 10 μg/ml insulin (Sigma Aldrich, I1884), 0.5 μg/ml hydrocortisone (Sigma Aldrich, H0888), 100 ng/ml chlorotoxin (DC Chemicals, DC23913), 20 ng/ml epidermal growth factor (Proteintech Group, HZ-1326), and 100 units/ml penicillin/streptomycin, 37 °C, 5% CO2. For multicellular aggregate experiments, single MCF10A cells were seeded in a 50% Matrigel matrix (Corning, 354230) supplied with 0.5 mg/ml collagen type 1 (Corning, 354236, rat tail, 3.2 mg/ml), 6 mM NaOH (Merck, 106498), and 15.6 mM HEPES (Thermo Fisher Scientific, 15630-080) in culture medium. Drops of 20 μl per well of an 8-well cell culture chamber on coverglass (Sarstedt. 94.6190.802) were solidified upside-down (hanging drop) at 37 °C for 30 min before adding 500 μl cell culture medium. Medium was refreshed every three to four days.
C2C12 cells (CRL-1772) were obtained from ATCC. To facilitate myoblast attachment on PDMS spheres, the surfaces were first coated with fibronectin. PDMS spheres were plasma-treated for 3 min to enhance surface hydrophilicity, then incubated in a fibronectin-containing solution (Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 5% fibronectin) for 2 h at room temperature. Following coating, C2C12 cells were suspended in the fibronectin solution and incubated with the PDMS spheres for 30 min to allow initial cell attachment. The spheres with attached cells were then transferred into a 1% agarose gel matrix to support 3D suspension and cultured in growth medium (DMEM +10% FBS) for several days to allow the formation of a confluent cell layer on the PDMS surface.
Human umbilical venous endothelial cells (HUVECs) from a single donor (Lonza, CC-2935), authenticated and tested for contamination by Lonza, were cultured in Endothelial Cell Basal Media-Plus supplemented with SingleQuots Bullet Kit (EGM-Plus, CC-5035 Lonza) until passage 2-3.
Immunostaining
Cells in Matrigel were cultured for seven to eight days, forming multicellular aggregates. After cell fixation with 4% paraformaldehyde (43368; Alfa Aesar) for 15 min, aggregates were permeabilized with 0.1% Triton-X 100 for 10 min, blocked with 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) for 1 h. F-actin was visualized with Alexa Fluor 546 Phalloidin (1:100 ratio; A22283, Invitrogen).
C2C12 cells were fixed with 4% paraformaldehyde for 10 min at room temperature, followed by permeabilization with 0.1% Triton X-100 in PBS for 5 min. Samples were then washed three times (5 min each) in PBS. Blocking was performed in PBS containing 1% BSA and 10% FBS for 1 h at room temperature. The actin cytoskeleton was stained using Alexa Fluor 568 Phalloidin (1:200; A12380, Invitrogen) overnight at 4 °C.
HUVECs were cultured for 3 days as 3D micro-vessel74 using specialized extracellular matrix consisting of 1.2 mg/ml polyisocyanopeptide (PIC) hybridized with 2 mg/ml type 1 atelo bovine collagen (Advanced Biomatrix, 5010)75. The micro-vessel was fixed and incubated overnight with Actin-Stain 555 Phalloidin (1:400, PHDH1-A, Cytoskeleton) to label F-actin.
Zebrafish husbandry
Zebrafish (Danio rerio) were reared at 28.0 °C according to standard practice, in fresh water with pH 7.5 and conductivity 500 μS, on a 15 h on 9 h off light cycle.
Imaging
Multicellular aggregates were imaged on an Andor Dragonfly spinning-disc confocal microscope attached to a Nikon Ti2 stand, equipped with a 514 nm laser and Zyla 4.2 sCMOS camera using the 40 μm pinhole disc, z-stacks were acquired using a 40x water immersion objective (Nikon, MRD77410, 1.15 N.A.) with 0.5 μm spacing. Data were collected using Fusion software.
Micro-vessel z-stack images were imaged on a Zeiss Axiovert 200 Inverted Microscope Stand with LSM 880 Confocal Scanner, equipped with a 514 nm laser. The z-stack was acquired using a 40x/N.A. 1.3 oil immersion objective with 1 μm spacing. Data were collected using Zen Black.
C2C12 cells on PDMS spheres were imaged using a Nikon CSU-W1 spinning disk confocal microscope. Illumination was performed using a 568 nm laser. Cells were inspected with a 20x 0.75 objective. Images were taken in z-stack focal planes with distances of 1 μm. Data were collected using NIH-Elements software.
Live zebrafish embryos were imaged at either 72 hpf or 120 hpf, prior to sexual differentiation, so no selection was made for embryo sex. Tg(myl7:BFP-CAAX)bns193;Tg(-0.2myl7:Lifeact-mNeongreen)fci611 embryos57,76 were screened for positive BFP and mNeongreen fluorescence using a Leica SMZ18 fluorescence stereomicroscope at 72 hpf. Selected embryos were mounted in 1% (w/v) low-melting-point agarose containing 2 μg/μl tricaine in a 35 mm diameter glass-bottomed dish. Once set, the dish was filled with egg water containing 4 μg/μl tricaine. All imaging was performed within 10 min of the heart stopping, given that prolonged anesthesia causes the heart to collapse and lose its shape. All imaging was performed using a Zeiss LSM 980 Axio Examiner confocal microscope equipped with a Zeiss W Plan-Apochromat 40x/1.0 DIC M27 water immersion dipping objective. A 1024 × 1024-pixel scan field was used at 1x optical zoom, resulting in a pixel size of 0.21 μm. Data were collected using Zen Blue software.
Analysis
The analysis was performed using Fiji/ImageJ and MATLAB R2021b. Codes are openly available on GitHub.
Z-stacks and image pre-processing
Orientation analysis was performed for the structure of interest. The structures of interest were the F-actin signal for multicellular aggregates, the HUVEC micro-vessel, and C2C12 cells on spheres, as well as the membrane signal for zebrafish hearts. The channel containing the structures’ information was pre-processed in Fiji/ImageJ. (1) The background signal, i.e., low intensity values from the constant signal not coming from the structure, was subtracted, (2) the structure was smoothed by using the Gaussian Blur filter to locally smooth the intensity values, (3) the intensity of each image was uniformized using the Normalized Local Contrast plugin, and (4) uniformized across the z-stack using the Stack Contrast Adjustment plugin to equalize the details in the same image and across the z-stack.
For intensity analysis, the raw images of the acquired z-stack of the lifeAct signal of zebrafish hearts were used. No image pre-processing was performed.
Mask
The black-and-white mask (cells: white, background: black) of each image was created using an appropriate threshold method in Fiji/ImageJ. Depending on the quality of the image, i.e., the strength of the intensity signal compared to the background, the background was subtracted and/or the image was smoothed by the Gaussian Blur filter in Fiji/ImageJ. In addition, the created mask was manually corrected in certain instances.
Surface points and normal vectors
To get the surface points, we first determined the boundary of the mask created in each image plane of the z-stack. In a given plane, the bwboundaries() function in MATLAB provided the xy-pixel positions of each boundary, which we converted to units of μm using the known camera resolution. Starting from an initial point, only positions with regular spacings of size dP along the boundary were considered as xy-boundary points, i.e., xyz-surface points, for further calculations. If the last point of the boundary was within a distance of dP/2 from the initial point, then it was removed. This process was repeated for a subset of z-stack planes with separation dP, whereby the starting point within each alternating plane was shifted by dP/2. This ensured a hexagonal-like configuration on the surface in 3D space instead of a square configuration. The result is a set of surface points whose characteristic separation is dP, up to variations due to curvature. The following values were used in the analysis presented here. dP = 6 μm for multicellular aggregates and HUVEC cells forming a micro-vessel. dP = 30 μm for C2C12 cells grown on a sphere. dP = 10 μm for zebrafish hearts. In these cases, dP was chosen to be between the radius and the diameter of a representative cell. Larger distances are discussed in Supplementary Fig. S2h, i in the Supplementary Information (S2 Surface point spacing and coarse-graining).
To obtain the normal N (rP) at a given surface point, a local set of points within a ball of a radius of 2dP were selected. These were used to find the so-called ‘best-fitting’ plane that minimizes the least squares distances to the points. This is achieved by singular value decomposition using the svd() function in MATLAB (purple dots in Fig. 2c). The direction of the normal vector was inverted if it pointed in the direction of the mask. Of note, changes to dP can affect the normal by altering the neighboring set of points and therefore their representation of the local geometry (Supplementary Fig. S2a–d in the Supplementary Information - S2 Surface point spacing and coarse-graining). We do not recommend increasing dP beyond the characteristic size of a cell.
When multicellular aggregates were considered as spheres, the surface points in a regular triangle configuration and the corresponding normal vectors were determined using the icosphere() function in MATLAB77 (Fig. S3c in the Supplementary Information - S3 Defect density on spheres). The number of subdivisions of the edge into equal segments was chosen in respect to the sphere’s surface area to match the number of surface points with those obtained by the surface method above.
Tangent plane and shallow slice
The tangent planes were obtained by using the surf() function in MATLAB. For each surface point, rP, we generate a plane with a resolution of 100 × 100 μm, which corresponds to 1000 × 1000 pixels, with 0.1 μm/pixel. The plane has a normal N(rP), which corresponds to polar and azimuthal angles
and
respectively, with ez the Cartesian unit vector in the z-direction. By translating the plane in the direction of the bulk of the multicellular system, a shallow slice at position rP − dsN (rP) is obtained. The slice() function in MATLAB was used to generate a linearly interpolated image, i.e., a cross-section of the 3D volumetric data.
2D nematic director in a slice
The shallow slice had a distance of ds = 5 μm for multicellular aggregates, which is half the cell diameter to capture the outer cell layer. For the zebrafish hearts, C2C12 on the sphere, and the HUVEC micro-vessel, the signal of multiple planes of distance ds ∈ {0,1,2,3} μm, ds ∈ {− 5,− 3,− 1,1,3} μm, ds ∈ {− 3,− 2,− 1,0,1,2,3} μm, respectively, was max-projected. The max-projection increased the fluorescence signal for orientation analysis and captured thin structures that may not be at ds = 0 due to deviations in mask creation. The orientation field of the generated 2D image was determined using the ImageJ plugin OrientationJ implemented in MATLAB and using the MIJ MATLAB-ImageJ connector78. OrientationJ computes the largest eigendirection of the structure tensor for each pixel, i.e., its intensity orientation, of the image. The Gaussian-shaped window size was set to σ = 3.
The orientation of each pixel, n, at position rn was transferred into a complex order parameter
with p = 2 being the signature of the nematic of the p − fold symmetry, α the angle of the local intensity orientation in respect to an arbitrary axis obtained by OrientationJ.
The 2D local nematic order parameter was obtained by calculating the mean order parameter of pixels within the distance, W = dP/2, to the grid point, rw, with an overlap of 1/4 of W and can be expressed by
The N grid points rw are spaced by W. The nematic order parameter was further coarse-grained within a distance of 2W:
to smoothen the orientation field. The spacing between rw is smaller than dP in order to increase the local set of nematic directors for coarse-graining in Eq. (5) and to increase the local accuracy. The nematic director in the flat plane at the grid point rw = rgp corresponding to the 3D surface point rP was calculated by
with β = Arg\(\left({\Psi }_{{{{\rm{cg}}}}}\left({{{r}}}_{{{{\rm{gp}}}}}\right)\right)/2\) being the phase of the coarse-grained 2D order parameter.
3D nematic director on the surface
The 2D nematic director in Eq. (6) was transferred onto the 3D surface of the multicellular system using the unit vectors in spherical coordinates
where
rS is the plane position rP in spherical coordinates, and ϑ and ϕ the polar and azimuthal angles as calculated in Eq. (1) and Eq. (2), respectively.
The nematic director n3D(rP) is then coarse-grained. Each nematic director at the surface point, rP, determines the local order parameter in the form of a traceless rank-2 tensor,
with i and j being the xyz-coordinates of n(rP) at point rP. The coarse-grained rank-2 tensor results in
where rB are the M surface points within the distance dcg = dPkcg to rP, with kcg being a coarse-graining factor. Unless otherwise specified, kcg = 1.7, which provides a coarse-graining over about 7 surface points (see Supplementary Information - S2 Surface point spacing and coarse-graining) and accounts for slight variations of surface point distances due to local curvature. Here, we coarse-grain on the multicellular’s surface over a distance of the cell diameter.
The 3D coarse-grained nematic director is the eigendirection of Eq. (11) corresponding to the largest eigenvalue.
Nematic order parameter
The nematic order parameter, i.e., the local nematic alignment parameter, is the positive eigenvalue of the traceless rank-2 tensor \({\overline{{{{\bf{Q}}}}}}_{ij}^{P}\) in Eq. (11), using the coarse-grained director field. dcg = dPkcg with kcg = 1.7 to calculate the order with respect to the nearest directors.
Nematic defects
Topological defects were identified by projecting the 3D nematic directors onto the tangent plane and computing the winding number along a closed contour, C, or more precisely, along the N nearest neighbors of each 2D nematic director. Thus, the topological charge can be expressed as
with \(\theta=\arctan ({n}_{y}/{n}_{x})\) being the angle of the 2D-projected nematic director at position rn. The parameter c = −π for \(\left({\theta }_{j+1}-{\theta }_{j}\right) > \pi /2\), c = +π for \(\left({\theta }_{j+1}-{\theta }_{j}\right) < -\pi /2\), and c = 0 for all other angular differences79.
Intensity analysis on the plane
As in the orientation analysis above, the slice() function in MATLAB was used to get the 2D image of the zebrafish heart at the tangent plane with a linear interpolation method. The intensity signal of four planes, positioned with a distance of 1 μm between the surface and 3 μm towards the bulk, was averaged. As the center point, rgp, of the generated image corresponds to the 3D surface point, rP, of the multicellular system, the intensity signal was averaged within a radius of dP/2 in respect to rgp. This value determined the mean intensity at rP. In addition, we calculated the median intensity of the background of the generated 2D image that was not covered by the mask. The signal-to-background ratio was used for the analysis.
Mean and Gaussian curvature
To compute the scalar curvatures of the zebrafish hearts, we first construct a triangular mesh of the surface points using Spyder IDE and Python. At a given surface point ri, we then compute the Gaussian curvature Gi at this surface point using the Gauss–Bonnet Theorem80. For each triangular face of the mesh Tj having ri as a vertex, we compute the angle θj of the triangle at vertex ri. Then the Gaussian curvature is
where the sum is taken over triangles with ri as a vertex. Here, Ai is an appropriate approximation to the area of the surface patch centered on ri whose boundary consists of the edges of the triangles Tj. We use the barycentric cell area approximation to this area, which is one-third of the sum of the areas of all the triangular faces,
We compute the mean curvature at the surface point ri by computing the discrete Laplacian80. For each surface point rj connected to ri by an edge of the triangular mesh, we consider the two triangular faces of the mesh which have ri and rj as two of their vertices. The cotangent weights αij, βij are the internal angles of the triangles at the third vertex, which is not ri or rj. Then we approximate the discrete Laplacian by ref. 80
where the sum is over all surface points rj connected to ri by an edge of the mesh. This discrete Laplacian is related to the mean curvature Hi and normal vector N (ri) at ri by
By taking the magnitude, we obtain the absolute value of the mean curvature; by dotting with the normal, we determine the sign.
This approach cannot accurately determine the mean and Gaussian curvatures at points that lie on the boundary of the surface. Thus, for our analysis of the correlation between nematic order and surface curvature we only use the points in the interior of the surface, disregarding points on the boundary.
Statistics
In total, ten multicellular aggregates and five zebrafish hearts at 72 hpf and 120 hpf were analyzed. We did not consider biological replicates as the focus of this article is on our method for quantifying surfaces of multicellular systems, and the type of analysis is dependent on the geometry rather than biological quantification.
P-values between two groups were calculated using the non-parametric two-sided Wilcoxon rank sum test in MATLAB, as they were non-normally distributed. The null hypothesis is fulfilled if the medians are equal. For comparisons of more than two groups, p-values were calculated using Dunn’s test of multiple comparisons after first performing a Kruskal-Wallis significance test in R.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Imaging data and MATLAB files containing analyzed data that support the findings of this study have been deposited in Zenodo [https://doi.org/10.5281/zenodo.15509350]. The raw image showing myoblasts on the PDMS sphere is available from the corresponding author upon request. Source data are provided in this paper.
Code availability
Our 3D surface analysis MATLAB codes are available on GitHub [https://github.com/Julia-Eckert/3D_SurfProps], and archived in Zenodo [https://doi.org/10.5281/zenodo.15509350].
References
Thompson, D. W.On Growth and Form. (Cambridge University Press, 1992).
Tetley, R. J. et al. Tissue fluidity promotes epithelial wound healing. Nat. Phys. 15, 1195–1203 (2019).
Paluch, E. & Heisenberg, C.-P. Biology and physics of cell shape changes in development. Curr. Biol. 19, R790–9 (2009).
Lemke, S. B. & Nelson, C. M. Dynamic changes in epithelial cell packing during tissue morphogenesis. Curr. Biol. 31, R1098–R1110 (2021).
Smits, C. M., Dutta, S., Jain-Sharma, V., Streichan, S. J. & Shvartsman, S. Y. Maintaining symmetry during body axis elongation. Curr. Biol. 33, 3536–3543.e6 (2023).
Ishihara, K. & Tanaka, E. M. Spontaneous symmetry breaking and pattern formation of organoids. Curr. Opin. Syst. Biol. 11, 123–128 (2018).
Katsuno-Kambe, H. et al. Collagen polarization promotes epithelial elongation by stimulating locoregional cell proliferation. Elife 10, e67915 (2021).
Gsell, S., Tlili, S., Merkel, M. & Lenne, P.-F. Marangoni-like tissue flows enhance symmetry breaking of embryonic organoids. Nat. Phys. 21, 644–653 (2025).
Andrews, T. G. R. & Priya, R. The mechanics of building functional organs. Cold Spring Harb. Perspect. Biol. 17, https://doi.org/10.1101/cshperspect.a041520 (2024).
Moon, L. D. & Xiong, F. Mechanics of neural tube morphogenesis. Semin. Cell Dev. Biol. 130, 56–69 (2022).
Alvarez, Y. D. et al. A lifeact-egfp quail for studying actin dynamics in vivo. J. Cell Biol. 223, e202404066 (2024).
GIERER, A. et al. Regeneration of hydra from reaggregated cells. Nat. New Biol. 239, 98–101 (1972).
Maroudas-Sacks, Y. et al. Topological defects in the nematic order of actin fibres as organization centres of hydra morphogenesis. Nat. Phys. 17, 251–259 (2021).
Doostmohammadi, A., Ignés-Mullol, J., Yeomans, J. M. & Sagués, F. Active nematics. Nat. Commun. 9, 3246 (2018).
Saw, T. B., Xi, W., Ladoux, B. & Lim, C. T. Biological tissues as active nematic liquid crystals. Adv. Mater. 30, e1802579 (2018).
Balasubramaniam, L., Mège, R.-M. & Ladoux, B. Active nematics across scales from cytoskeleton organization to tissue morphogenesis. Curr. Opin. Genet. Dev. 73, 101897 (2022).
Duclos, G., Erlenkämper, C., Joanny, J.-F. & Silberzan, P. Topological defects in confined populations of spindle-shaped cells. Nat. Phys. 13, 58–62 (2016).
Kawaguchi, K., Kageyama, R. & Sano, M. Topological defects control collective dynamics in neural progenitor cell cultures. Nature 545, 327–331 (2017).
Saw, T. B. et al. Topological defects in epithelia govern cell death and extrusion. Nature 544, 212–216 (2017).
Balasubramaniam, L. et al. Investigating the nature of active forces in tissues reveals how contractile cells can form extensile monolayers. Nat. Mater 20, 1156–1166 (2021).
Endresen, K. D., Kim, M., Pittman, M., Chen, Y. & Serra, F. Topological defects of integer charge in cell monolayers. Soft Matter. 17, 5878–5887 (2021).
Armengol-Collado, J.-M., Carenza, L. N., Eckert, J., Krommydas, D. & Giomi, L. Epithelia are multiscale active liquid crystals. Nat. Phys. 19, 1773–1779 (2023).
Eckert, J., Ladoux, B., Mège, R.-M., Giomi, L. & Schmidt, T. Hexanematic crossover in epithelial monolayers depends on cell adhesion and cell density. Nat. Commun. 14, 5762 (2023).
Happel, L. et al. Quantifying the shape of cells - from minkowski tensors to p-atic order. ELife 14, RP105680 (2025).
Monfared, S., Ravichandran, G., Andrade, J. & Doostmohammadi, A. Mechanical basis and topological routes to cell elimination. ELife 12, e82435 (2023).
Bailles, A. et al. Anisotropic stretch biases the self-organization of actin fibers in multicellular Hydra aggregates. Proc. Natl Acad. Sci. USA 122, e2423437122 (2025).
Maroudas-Sacks, Y., Garion, L., Suganthan, S., Popović, M. & Keren, K. Confinement modulates axial patterning in regenerating hydra. PRX Life 2, 043007 (2024).
Maroudas-Sacks, Y. et al. Mechanical strain focusing at topological defect sites in regenerating hydra. Development 152, DEV204514 (2025).
Ravichandran, Y., Vogg, M., Kruse, K., Pearce, D. J. G. & Roux, A. Topology changes of hydra define actin orientation defects as organizers of morphogenesis. Sci. Adv. 11, eadr9855 (2025).
Sanchez, T., Chen, D. T. N., DeCamp, S. J., Heymann, M. & Dogic, Z. Spontaneous motion in hierarchically assembled active matter. Nature 491, 431–434 (2012).
Thampi, S. P., Golestanian, R. & Yeomans, J. M. Vorticity, defects and correlations in active turbulence. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 372, 20130366 (2014).
Alert, R., Casademunt, J. & Joanny, J.-F. Active turbulence. Annu. Rev. Condens. Matter Phys. 13, 143–170 (2022).
Metselaar, L., Yeomans, J. M. & Doostmohammadi, A. Topology and morphology of self-deforming active shells. Phys. Rev. Lett. 123, 208001 (2019).
Hoffmann, L. A., Carenza, L. N., Eckert, J. & Giomi, L. Theory of defect-mediated morphogenesis. Sci. Adv. 8, eabk2712 (2022).
Hoffmann, L. A., Carenza, L. N. & Giomi, L. Tuneable defect-curvature coupling and topological transitions in active shells. Soft Matter 19, 3423–3435 (2023).
Guillamat, P., Blanch-Mercader, C., Pernollet, G., Kruse, K. & Roux, A. Integer topological defects organize stresses driving tissue morphogenesis. Nat. Mater. 21, 588–597 (2022).
Farrell, D. L., Weitz, O., Magnasco, M. O. & Zallen, J. A. Segga: a toolset for rapid automated analysis of epithelial cell polarity and dynamics. Development 144, 1725–1734 (2017).
Chen, D.-Y., Crest, J., Streichan, S. J. & Bilder, D. Extracellular matrix stiffness cues junctional remodeling for 3d tissue elongation. Nat. Commun. 10, 3339 (2019).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021).
Aigouy, B. & Prud’homme, B. Segmentation and quantitative analysis of epithelial tissues. Methods Mol. Biol. 2540, 387–399 (2022).
Shroff, N. P. et al. Proliferation-driven mechanical compression induces signalling centre formation during mammalian organ development. Nat. Cell Biol. 26, 519–529 (2024).
Pérez-Verdugo, F., Maniou, E., Galea, G. L. & Banerjee, S. Self-organized cell patterning via mechanical feedback in hindbrain neuropore morphogenesis. Preprint at https://doi.org/10.1101/2024.11.21.624679 (2024).
Ichbiah, S., Delbary, F., McDougall, A., Dumollard, R. & Turlier, H. Embryo mechanics cartography: inference of 3d force atlases from fluorescence microscopy. Nat. Methods 20, 1989–1999 (2023).
Jiang, J. et al. Segmentation, tracking, and sub-cellular feature extraction in 3d time-lapse images. Sci. Rep. 13, 3483 (2023).
Zhou, F. Y. et al. A general algorithm for consensus 3d cell segmentation from 2d segmented stacks. Preprint at https://doi.org/10.1101/2024.05.03.592249 (2024).
Kawahira, N., Ohtsuka, D., Kida, N., Hironaka, K.-i & Morishita, Y. Quantitative analysis of 3d tissue deformation reveals key cellular mechanism associated with initial heart looping. Cell Rep. 30, 3889–3903. (2020).
Mitchell, N. P. & Cislo, D. J. Tubular: tracking in toto deformations of dynamic tissues via constrained maps. Nat. Methods 20, 1980–1988 (2023).
Kuang, X., Guan, G., Tang, C. & Zhang, L. Morphosim: an efficient and scalable phase-field framework for accurately simulating multicellular morphologies. Npj Syst. Biol. Appl. 9, 6 (2023).
Zhou, F. Y. et al. Surface-guided computing to analyze subcellular morphology and membrane-associated signals in 3d. Preprint at 12:arXiv:2304.06176v1. (2023).
Fuhrmann, J. F. et al. Active shape programming drives <i>drosophila</i> wing disc eversion. Sci. Adv. 10, eadp0860 (2024).
Runser, S., Vetter, R. & Iber, D. Simucell3d: three-dimensional simulation of tissue mechanics with cell polarization. Nat. Comput. Sci. 4, 299–309 (2024).
doCarmo, M. P. Differential Geometry of Curves and Surfaces (Dover Publications, 2016).
Eisenberg, M. & Guy, R. A proof of the hairy ball theorem. Am Math Mon.y 86, 571–574 (1979).
Henkes, S., Marchetti, M. C. & Sknepnek, R. Dynamical patterns in nematic active matter on a sphere. Phys. Rev. E 97, 042605 (2018).
Skillin, N. P. et al. Stiffness anisotropy coordinates supracellular contractility driving long-range myotube-ecm alignment. Sci. Adv. 10, eadn0235 (2024).
Carenza, L. N., Armengol-Collado, J.-M., Krommydas, D. & Giomi, L. Quasi-long-ranged order in two-dimensional active liquid crystals. Phys. Rev. Lett. 134, 128304 (2025).
Andrews, T. G. R. et al. Mechanochemical coupling of cell shape and organ function optimizes heart size and contractile efficiency in zebrafish. Dev. Cell 60, 1–18 (2025).
Dessalles, C. A. et al. Interplay of actin nematodynamics and anisotropic tension controls endothelial mechanics. Nat. Phys. 21, 999–1008 (2025).
Alaimo, F., Köhler, C. & Voigt, A. Curvature controlled defect dynamics in topological active nematics. Sci. Rep. 7, 5211 (2017).
Napoli, G. & Vergori, L. Cooling a spherical nematic shell. Phys. Rev. E 104, L022701 (2021).
Mesarec, L., Iglič, A. & Kralj, S. Spatial manipulation of topological defects in nematic shells. Eur. Phys. J. E 45, 62 (2022).
Tan, T. H. et al. Emergent chirality in active solid rotation of pancreas spheres. PRX Life 2, 033006 (2024).
Tang, W. et al. Collective curvature sensing and fluidity in three-dimensional multicellular systems. Nat. Phys. 18, 1371–1378 (2022).
Ruske, L. J. & Yeomans, J. M. Activity-driven tissue alignment in proliferating spheroids. Soft Matter 19, 921–931 (2023).
Tang, W. et al. Topology and nuclear size determine cell packing on growing lung spheroids. Phys. Rev. X 15, 011067 (2025).
Wang, Z., Marchetti, M. C. & Brauns, F. Patterning of morphogenetic anisotropy fields. Proc. Natl. Acad. Sci. USA 120, e2220167120 (2023).
Firouznia, M. & Saintillan, D. Self-organized dynamics of a viscous drop with interfacial nematic activity. Phys. Rev. Res. 7, L012054 (2025).
López-Gay, J. M. et al. Apical stress fibers enable a scaling between cell mechanical response and area in epithelial tissue. Science 370, eabb2169 (2020).
Colom, A. et al. A fluorescent membrane tension probe. Nat. Chem. 10, 1118–1125 (2018).
Roffay, C. et al. Tutorial: fluorescence lifetime microscopy of membrane mechanosensitive flipper probes. Nat. Protoc. 19, 3457–3469 (2024).
Bade, N. D., Kamien, R. D., Assoian, R. K. & Stebe, K. J. Curvature and rho activation differentially control the alignment of cells and stress fibers. Sci. Adv. 3, e1700150 (2017).
Callens, S. J. P., Uyttendaele, R. J. C., Fratila-Apachitei, L. E. & Zadpoor, A. A. Substrate curvature as a cue to guide spatiotemporal cell and tissue organization. Biomaterials 232, 119739 (2020).
Luciano, M., Tomba, C., Roux, A. & Gabriele, S. How multiscale curvature couples forces to cellular functions. Nat. Rev. Phys. 6, 246–268 (2024).
Polacheck, W. J., Kutys, M. L., Tefft, J. B. & Chen, C. S. Microfabricated blood vessels for modeling the vascular transport barrier. Nat. Protoc. 14, 1425–1454 (2019).
Enriquez Martinez, M. A. et al. Tuning collagen nonlinear mechanics with interpenetrating networks drives adaptive cellular phenotypes in three dimensions. Sci. Adv. 11, eadt3352 (2025).
Guerra, A. et al. Distinct myocardial lineages break atrial symmetry during cardiogenesis in zebrafish. ELife 7, e32833 (2018).
wil. icosphere. https://www.mathworks.com/matlabcentral/fileexchange/50105-icosphere. MATLAB Central File Exchange. (2023).
Sage, D. Mij: Running imagej and fiji within matlab. https://www.mathworks.com/matlabcentral/fileexchange/47545-mij-running-imagej-and-fiji-within-matlab. MATLAB Central File Exchange. (2023).
Huterer, D. & Vachaspati, T. Distribution of singularities in the cosmic microwave background polarization. Phys. Rev. D 72, 043004 (2005).
Meyer, M., Desbrun, M., Schröder, P. & Barr, A. H. Visualization and Mathematics III. (Springer, Berlin, Heidelberg, 2003).
Acknowledgements
J.E. and A.S.Y. were supported by the Australian Research Council (FL230100100 and DP220103951). J.E. was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) - 553948485. Microscopy of aggregates and microvessels were performed at the ACRF/IMB Cancer Research Imaging Facility, created with the generous support of the Australian Cancer Research Foundation. R.G.M. and J.P. acknowledge the EMBL Australia Program. R.G.M. acknowledges support from the Australian Research Council Center of Excellence for Mathematical Analysis of Cellular Systems (MACSYS, CE230100001) and the National Health and Medical Research Council (NHMRC) of Australia (GNT2003832). Work in R.P.’s laboratory is supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC011160), the UK Medical Research Council (FC011160), the Wellcome Trust (FC011160), and the British Heart Foundation (SP/F/20/150014). This work was supported by the European Research Council (Grant No. Adv-101019835 “DeadorAlive” to B.L.) and the Alexander von Humboldt Foundation (Alexander von Humboldt Professorship to B.L.). Y.S. would like to acknowledge the Human Frontier Science Program (grant number LT0007/2023-C) for funding. P.E. was supported by a Wainwright Diversity and Inclusion Scholarship (Institute for Molecular Bioscience). P.E. and A.K.L. were supported by an Australian Research Council Discovery Project (DP230100393).
Author information
Authors and Affiliations
Contributions
J.E., A.S.Y., and R.G.M. defined the project. J.E. performed analytic work and the experiments on cell aggregates, wrote the 3D surface analysis software, and analyzed the data. J.P. wrote the code for analyzing the Gaussian and mean curvature. T.G.R.A. and R.P. performed experimental imaging of zebrafish hearts. Y.S. and B.L. performed experimental imaging of myoblasts grown on the sphere. P.E. and A.K.L. provided the images of HUVECs forming microvessels. J.E. wrote the manuscript with help from J.P., A.S.Y., and R.G.M., along with selected contributions from T.G.R.A. and R.P., as well as feedback from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Claire Dessalles, and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Eckert, J., Andrews, T.G.R., Pollard, J. et al. Capturing nematic order on tissue surfaces of arbitrary geometry. Nat Commun 16, 7596 (2025). https://doi.org/10.1038/s41467-025-62694-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-62694-x