Abstract
Neonatal meningitis Escherichia coli (NMEC) breaching the blood-brain barrier (BBB) is a critical event in the development of E. coli meningitis. Brain microvascular endothelial cells (BMECs), the primary structural component of the BBB, play a central role in defending against pathogen invasion. In this study, we employ the NMEC strain RS218 (O18:K1:H7) to investigate the molecular mechanisms of cell death in BMECs and its pivotal contribution to BBB disruption. The study reveals that RS218 infection promotes assembly of the Ripoptosome complex. This leads to the coordinated activation of apoptosis, pyroptosis, and necroptosis. Notably, necroptosis can also occur through RIPK1-independent pathways. By generating Ripk1 D138N/D138N, Gsdmd -/-, and Casp8 -/- Ripk3 -/- mice, we demonstrate that the regulation of BMECs death was an important factor in BBB resistance to infection. Among these models, kinase-dead Ripk1 D138N/D138N mice exhibit the most effective BBB protection, independent of innate immune responses. Inhibition of RIPK1 kinase significantly preserves BBB integrity, and lowers RS218 invasion and neuroinflammation. Moreover, the combination of RIPK1 inhibition with antibiotics yields additive therapeutic effects. Our study advances the understanding of NMEC pathogenesis and supports the potential of RIPK1 as a therapeutic target for E. coli meningitis.
Similar content being viewed by others
Introduction
Neonatal meningitis Escherichia coli (NMEC) is the second most prevalent bacterial pathogen responsible for meningitis, posing a substantial threat to neonates and accounting for approximately 35% of clinical cases1,2. Current antibiotic therapies include ampicillin/penicillin, gentamicin, and cefotaxime, administered alone or in combination3. Although combinations of two or more antibiotics can have additive or synergistic effects, the increasing prevalence of antibiotic resistance has led to mortality rates for E. coli meningitis remaining between 15% and 40% annually. In the absence of treatment, mortality approaches 100%1,4. The NMEC strain RS218 (O18:K1:H7) is the prototypical model in many studies, with its capacity to traverse the blood-brain barrier (BBB) playing a pivotal role in the pathogenesis of meningitis4. Brain microvascular endothelial cells (BMECs), critical components of the BBB, maintain its integrity through tight junctions that effectively block the paracellular transport of small molecules5,6. The survival of BMECs during bacterial invasion is thus intimately linked to the preservation of BBB integrity. Understanding the mechanisms by which RS218 crosses the BBB and developing novel therapeutic approaches remain critical challenges for treating neonatal meningitis.
Programmed cell death (PCD) is a tightly regulated biological process essential for maintaining host homeostasis and modulating disease progression7,8. The principal modes of cell death in host cells during infection include caspase-8-dependent apoptosis, receptor-interacting protein kinase 1 (RIPK1) and RIPK3-dependent necroptosis, and caspase-1/caspase-11-dependent pyroptosis9. Recent studies have revealed complex interactions among these distinct pathways. Inactivation of caspase-8 can trigger necroptosis and inflammation10. Although MLKL deficiency can block this process, it instead activates caspase-1/ASC signaling and leads to fatal pyroptosis11,12. Notably, necroptosis can promote caspase-1 activation by triggering the NLRP3 inflammasome13,14. These interactions make it challenging to define the precise roles of each pathway in disease pathogenesis. As a key regulatory factor in both apoptosis and necroptosis, caspase-8 has recently been shown to cleave gasdermin D, thereby directly initiating pyroptosis15. These findings position caspase-8 at the center of the regulatory network governing cell death. The cross-regulation of different cell death pathways may serve multifaceted roles in disease progression. For example, during Yersinia infection, switching between death pathways can disrupt intracellular niches and facilitate bacterial clearance16.
RIPK1 serves as a pivotal mediator of cell death and inflammation, fundamentally influencing cell fate. RIPK1 is activated by signals from various death receptors (TNFR1, TRAILR, and FasR), TLR3/4, and ZBP1. It exhibits kinase activity while also serving scaffolding functions independent of this activity. Numerous inflammatory and infectious diseases are associated with the kinase activity of RIPK117. To date, RIPK1 has been identified as the sole substrate for its own kinase activity, which primarily facilitates autophosphorylation18. This autophosphorylation induces conformational changes in RIPK1, allowing interactions with downstream effectors such as RIPK3 and FADD to initiate cell death18. Identified autophosphorylation sites in humans include S14/15, S20, S161, and S166, and phosphorylation of S161 and S166 is critical for downstream signaling18,19,20. In addition, RIPK1’s scaffold function contributes to cell survival by stabilizing pro-survival NF-κB signaling pathways. Deficiency in RIPK1 resulted in diminished NF-κB-mediated transcription of pro-survival proteins. Loss of RIPK1 in mice led to systemic inflammation and death shortly after birth21,22. However, mice expressing kinase-dead RIPK1 (D138N) alone did not exhibit the same detrimental phenotype and showed a reduced incidence of disease23,24,25. This observation underscores the delicate balance between the stabilization of RIPK1’s pro-survival scaffolding and the activation of its pro-death kinase activity, which may critically influence disease prognosis.
In this study, we delineated the mechanisms by which the NMEC strain RS218 induced cell death in BMECs. RS218 activated multiple signaling pathways, leading to distinct forms of programmed cell death. Gene-editing models revealed a central role for RIPK1 kinase in mediating BBB disruption and driving meningitis pathogenesis. These findings identified RIPK1 as a promising therapeutic target in E. coli meningitis and provided a foundation for the development of targeted intervention strategies.
Results
RS218 activated multiple cell death pathways in hBMECs
To assess the potential induction of cell death in human BMECs (hBMECs) by NMEC RS218 infection, we performed live cell imaging analysis. As the infection progressed, dying cells became shrunken and exhibited plasma membrane protrusions (Fig. 1a and Supplementary Movie 1). Furthermore, cytotoxicity (Fig. 1b) and degree of cell membrane rupture (Fig. 1c) increased in a time- and dose-dependent manner, indicating that hBMECs underwent membrane rupture-associated cell death during infection.
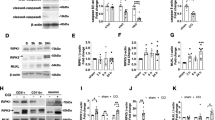
a Time-lapse microscopy of PI-stained (red) hBMECs infected with RS218 (MOI = 10), showing 8 min interval images taken at 60 × magnification. The 10 frames demonstrated the progression of cell death within 72 min before membrane rupture. White dashed lines indicate the boundaries of the cell membrane. Scale bar, 20 μm. b, c hBMECs stimulated with RS218 (MOI = 1 or 10) were used to determine cytotoxicity by LDH release (b, n = 3 biologically independent samples) and kinetics of cell death of PI penetration (c, n = 5 biologically independent samples). d, e hBMECs were pretreated with NSA or zVAD alone or in combination for 0.5 h, and cytotoxicity (d, n = 6 biologically independent samples) and cell death kinetics of PI penetration (e, n = 3 biologically independent samples) were assayed. f Flow cytometry of Annexin V-FITC and PI staining in hBMECs infected with RS218 (MOI = 1). Representative images of immunofluorescence represented morphological analysis of Annexin V-FITC and PI-labeled cells for the indicated times postinfection. Scale bar, 20 μm. g Immunoblot analysis of MLKL phosphorylation, GSDMD, caspase-3, and PARP1 cleavage in hBMECs infected with RS218 (MOI = 1) at the indicated times. Blots for GAPDH served as loading controls. +, indicated pharmacologically-induced death of hBMECs. FL, full-length. h Pro caspase-8 was co-immunoprecipitated with RIPK1, RIPK3, ZBP1 and ASC in uninfected or RS218-infected hBMECs (MOI = 1) for 3 h. i Immunofluorescence representative images of 2-day-old mice brain tissue sections 18 h after injection. The nucleus was labeled in blue, pMLKL or GSDMD-N or caspase-3 p17 in green, and endothelial cell marker CD31 in red. Scale bar, 20 μm. All experiments were representative of at least three independent experiments with similar results. Bars indicated the mean plus SD. Statistical significance was determined by one-way ANOVA with Tukey’s post-hoc test, with P-values denoted as follows: **** P < 0.0001, *** P < 0.0005, ** P < 0.005, * P < 0.05, n.s., no significant difference. Exact P-values were provided in the source data. Source data are provided as a Source Data file.
hBMECs were treated with the pan-caspases inhibitor z-VAD-FMK (zVAD) (Supplementary Fig. S1a, b) or the necroptosis and pyroptosis inhibitor necrosulfonamide (NSA) (Supplementary Fig. S1c, d), with 20 μM identified as the optimal inhibitory concentration. Compared to the RS218-treated group, both inhibitors significantly reduced cytotoxicity (Fig. 1d) and membrane rupture (Fig. 1e) 6 h post-infection at MOIs of 1 or 10. Notably, NSA exhibited a more pronounced protective effect (Fig. 1d, e). These findings suggested the differential effects of the two inhibitors reflected the involvement of multiple pathways in RS218-induced hBMECs death. Annexin V-FITC and propidium iodide (PI) labeling revealed that Annexin V-FITC⁺/PI⁺ cells comprised the majority of the infected cell population, increasing in a time-dependent manner (Fig. 1f). Peak levels of Annexin V-FITC⁺/PI⁻ cells were observed at 3 h (Fig. 1f), indicating significant early apoptotic activity in hBMECs.
We further detected activation of necroptotic, pyroptotic and apoptotic signature proteins, MLKL, GSDMD, caspase-3, and PARP1 by Western blot. The results showed phosphorylation of MLKL (pMLKL) and cleavage of GSDMD into its N-terminal fragment (GSDMD-N) were detected during infection, similar to the positive control for pharmacologically induced hBMECs death (Fig. 1g). Cleavage of the effector caspase-3 into its active p17 form (cl. CASP3), along with cleavage of its substrate PARP1 (cl. PARP1), was predominantly observed at 3 h post-infection (Fig. 1g). This time point corresponded with the Annexin V-FITC/PI staining (Fig. 1f) and was selected as the optimal window for subsequent experiments. These results suggested that necroptosis, pyroptosis and apoptosis were activated in hBMECs. Furthermore, co-immunoprecipitation (Co-IP) assays revealed that the formation of a cytoplasmic death complex composed of caspase-8, RIPK1, RIPK3, ZBP1, and ASC at 3 h post-infection (Fig. 1h), confirming the coordinated activation of multiple PCD pathways. In addition, a neonatal meningitis model by intraperitoneal infection of 2-day-old mice. Active forms of caspase-3 (p17), GSDMD (GSDMD-N), and MLKL (pMLKL) were detected in the BMECs of neonatal mice (Fig. 1i), indicating that RS218 activated apoptosis, pyroptosis, and necroptosis in BMECs in vivo.
RIPK1 kinase drove the assembly of Ripoptosome
To investigate whether RIPK1 functioned upstream of the death complex, we generated RIPK1 knockout (KO) cells using the CRISPR/Cas9 gene-editing system (Supplementary Fig. S2a). Co-IP analyses revealed that the loss of RIPK1 inhibited the complex formation (Fig. 2a), confirming the assembly of the Ripoptosome. Although ZBP1 was involved in Ripoptosome assembly, its deletion did not substantially disrupt the formation of the complex (Fig. 2a and Supplementary Fig. S2b). The knockout of RIPK1 resulted in the simultaneous loss of scaffold and kinase functions. To differentiate between the scaffold and kinase functions of RIPK1, we reintroduced a kinase-dead RIPK1 mutant (D138N) into RIPK1 KO cells (Supplementary Fig. S2c). Despite retaining scaffold function, RIPK1 D138N failed to restore Ripoptosome formation (Fig. 2b), indicating that the kinase activity of RIPK1 preceded its scaffold function in driving Ripoptosome assembly.
a Co-IP of pro-caspase-8 with RIPK1, RIPK3, ZBP1, and ASC in WT, RIPK1 KO, or ZBP1 KO cells infected with RS218 (MOI = 1). b Co-IP of pro-caspase-8 with RIPK1, RIPK3, ZBP1, and ASC in hBMECs expressing RIPK1 WT or D138N under RS218 infection (MOI = 1). c Immunoblot of RIPK1 total protein and phosphorylation levels at S161 and S166 in WT, TNFR1 KO, TLR4 KO, or double KO cells post RS218 infection (MOI = 1). Blots for GAPDH served as loading controls. d Co-IP of pro-caspase-8 with RIPK1, RIPK3, ZBP1, and ASC in hBMECs expressing RIPK1 WT, S161N, or S166N cells (MOI = 1). e, f In WT, TNFR1 KO, TLR4 KO, or double KO cells, the cytotoxicity was assessed by LDH release (e, n = 3 biologically independent samples), and cell death kinetics were measured by PI uptake (f, n = 3 biologically independent samples). g Co-IP analysis of RIPK1 with TNFR1 and TLR4 interactions in WT and ZBP1 KO cells. h Co-IP of pro-caspase-8 with RIPK1, RIPK3, ZBP1, and ASC in WT, TNFR1 KO, TLR4 KO or double KO cells (MOI = 1). i Immunoblot of RIPK1 total protein and phosphorylation levels at S161 in WT or ZBP1 KO cells. Blots for GAPDH served as loading controls. All experiments were representative of at least three independent experiments with similar results. Bars indicated the mean plus SD. Statistical significance was determined by one-way ANOVA with Tukey’s post-hoc test, with P-values denoted as follows: **** P < 0.0001, *** P < 0.0005, ** P < 0.005, * P < 0.05, n.s., no significant difference. Exact P-values were provided in the source data. Source data are provided as a Source Data file.
In RS218-infected hBMECs, phosphorylation at both the S161 and S166 sites of RIPK1 was detected (Fig. 2c). To identify key residues regulating kinase activity, we introduced S161N and S166N mutants into RIPK1 KO cells (Supplementary Fig. S2d). The results demonstrated that RIPK1 S161N effectively inhibited Ripoptosome assembly, while RIPK1 S166N exhibited a limited inhibitory effect on its formation (Fig. 2d). These findings suggested that the S161 residue was pivotal for the regulation of RIPK1 kinase-driven Ripoptosome assembly.
Next, we aimed to elucidate the activation mechanism of RIPK1, focusing on TNFR1 and TLR4 as important receptors involved in this process. In knockout cells (Supplementary Fig. S2e), the deletion of either receptor significantly reduced cytotoxicity (Fig. 2e) and membrane rupture (Fig. 2f) in hBMECs. However, autophosphorylation at RIPK1 S161 and S166 was inhibited specifically by TNFR1 knockout (Fig. 2c), suggesting that the activation of RIPK1 was related to the TNFR1 signaling pathway. Co-IP further found that RIPK1 activation occurs through recruitment to TNFR1, independently of TLR4 (Fig. 2g). Deletion of TNFR1 prevented Ripoptosome assembly (Fig. 2h), indicating that TNFR1 was the activating receptor for RIPK1-driven death. In addition, Ripoptosome formation was diminished in ZBP1 KO (Fig. 2a). Subsequent investigations revealed that ZBP1 enhanced the assembly of RIPK1 with TNFR1 (Fig. 2g) and facilitated S161 autophosphorylation (Fig. 2i). This suggested that ZBP1, as a non-essential component of Ripoptosome, was able to promote the recruitment of RIPK1 to TNFR1. Collectively, these findings indicated that RIPK1 was recruited to TNFR1 with the aid of ZBP1, mediating S161 autophosphorylation to activate its kinase activity and promote the Ripoptosome assembly.
RS218 mediated necroptosis through RIPK1-dependent and independent pathways activation of RIPK3
Mechanistically, a mutation at RIPK1 S161 was able to inhibit downstream cell death by restricting the assembly of Ripoptosome. To investigate this hypothesis, we compared RIPK1 WT and S161N activation differences in apoptosis, pyroptosis, and necroptosis under infection conditions. Compared to RIPK1 WT, the latter significantly reduced the cleavage of apoptosis-related caspase-3, PARP1, and pyroptosis-related GSDMD in response to RS218 infection (Fig. 3a). This reflected the dependence of apoptosis and pyroptosis on RIPK1. For necroptosis, while RIPK1 S161 was not phosphorylated, the downstream effectors RIPK3 (pRIPK3) and MLKL (pMLKL) still exhibited the presence of partial phosphorylation. (Fig. 3a). These findings suggested that RIPK3-MLKL activation might not have been entirely reliant on RIPK1 kinase. To further investigate whether RIPK1’s non-kinase function contributed to RIPK3 activation, we conducted assays in RIPK1 KO. The results showed that cleaved caspase-3, PARP1, and GSDMD were significantly inhibited in RIPK1 KO (Fig. 3b), demonstrating that the knockout of RIPK1 similarly inhibited pyroptosis and apoptosis. Intriguingly, unlike the results for RIPK1 S161N, RIPK3 and MLKL exhibited increased phosphorylation in RIPK1 KO, and only the knockout of RIPK3 (Supplementary Fig. S2f) completely abrogated MLKL activation (Fig. 3b). This finding indicated that the RIPK3-MLKL pathway could be activated independently of RIPK1, while the presence of RIPK1’s non-kinase function appeared to inhibit this pathway to some extent.
a, b Immunoblot analysis of cleaved caspase-3, PARP1, GSDMD, and phosphorylated RIPK1, RIPK3, and MLKL in RIPK1 WT and RIPK1 S161N cells (a) or in WT, RIPK1 KO, and RIPK3 KO cells (b) after RS218 infection (MOI = 1). Blots for GAPDH served as loading controls. c Immunoblot analysis of total and phosphorylated levels of RIPK1, RIPK3, and MLKL in WT, TNFR1 KO, TLR4 KO, or double KO hBMECs expressing RIPK1 WT or S161N. Blots for GAPDH served as loading controls. d TLR4 was co-immunoprecipitated with RIPK3 and TRIF in RIPK1 WT and S161N cells. e In RIPK1 S161N background, TLR4 was co-immunoprecipitated with RIPK3 and TRIF in TRIF WT, TRIF KO, and TRIF mRHIM cells. f In the RIPK1 S161N background, immunoblot analysis of total and phosphorylated levels of RIPK1, RIPK3, and MLKL in TRIF WT, TRIF KO, and TRIF mRHIM cells. Blots for GAPDH served as loading controls. All experiments were representative of at least three independent experiments with similar results. Source data are provided as a Source Data file.
We next sought to elucidate the activation mechanism of additional RIPK3-MLKL signaling. TNFR1 functioned as RIPK1-activating receptor, and the observed decrease in RIPK3 and MLKL phosphorylation following TNFR1 deletion was anticipated (Fig. 3c). However, TLR4 knockout effectively reduced the phosphorylation of RIPK3 and MLKL without affecting RIPK1 phosphorylation (Fig. 3c). Co-treatment of cells abrogated RIPK3 and MLKL activation (Fig. 3c) while minimizing cytotoxicity and membrane rupture (Fig. 2e, f). This suggested that TLR4 acted as a receptor that independently activated the RIPK3-MLKL pathway. To further clarify this speculation, we performed assays in RIPK1 S161N to exclude interference from RIPK1-dependent RIPK3 activation. In this context, the presence or absence of TNFR1 did not affect RIPK3 phosphorylation (Fig. 3c). In contrast, only TLR4 deletion suppressed the phosphorylation of RIPK3 and downstream MLKL (Fig. 3c). These findings demonstrated that the TLR4-activated RIPK3-MLKL pathway was not regulated by the Ripoptosome. Moreover, the formation of TLR4-TRIF-RIPK3 complex was observed in both RIPK1 WT and S161N (Fig. 3d). Loss of TRIF (Supplementary Fig. S2g) disrupted the formation of this complex (Fig. 3e) and the RIPK1-independent activation of RIPK3 (Fig. 3f). When VQLG, the critical domain of RIP homotypic interaction motif (RHIM) utilized by TRIF to bind RIPK3, was mutated to AAAA (TRIF mRHIM) (Supplementary Fig. S2h), TLR4-TRIF-RIPK3 complex formation (Fig. 3e) and the phosphorylation of RIPK3 (Fig. 3f) were no longer detected in RIPK1 S161N. This suggested that TRIF interaction with RIPK3 via the RHIM domain was critical for triggering TLR4-mediated necroptosis. Collectively, these results indicated that RS218 induced TLR4 to activate the RIPK3-MLKL pathway, mediating necroptosis in a manner independent of the Ripoptosome. This explained the continued activation of RIPK3 and MLKL following inhibition of RIPK1 kinase activity.
Compared to WT, cytotoxicity (Supplementary Fig. S3a) and membrane rupture (Supplementary Fig. S3b) were significantly reduced in RIPK1 D138N and S161N due to the inhibition of cell death. However, no significant differences were observed between RIPK1 KO and WT (Supplementary Fig. S3a, b). While RIPK1 depletion suppressed pyroptosis and apoptosis, the exacerbation of necroptosis likely contributed to this observation (Fig. 3b). We hypothesized that the RHIM domain of RIPK1 might inhibit TLR4-TRIF-mediated phosphorylation by competitively binding to RIPK3, thereby blocking its activation within the signaling cascade. To exclude the confounding effects of RIPK1-dependent RIPK3 activation, we introduced an IQIG-to-AAAA mutation in the RHIM domain (RIPK1 mRHIM) on the RIPK1 S161N background (Supplementary Fig. S2i). The results showed that RIPK1 mRHIM induced significantly elevated cytotoxicity (Supplementary Fig. S3c) and membrane rupture (Supplementary Fig. S3d) compared to RIPK1 with the wild-type RHIM domain (RIPK1 RHIM). Further analysis revealed that the RHIM mutation in RIPK1 enhanced RIPK3 recruitment to TLR4 (Supplementary Fig. S3e), leading to markedly increased phosphorylation of RIPK3 and MLKL (Supplementary Fig. S3f). These findings suggested that the RHIM domain of RIPK1 inhibited TLR4 signaling by binding to RIPK3, making RIPK1 the essential protein for cell survival, while its kinase activity was critical for reducing cell death in hBMECs.
Next, we analyzed the dynamics of necroptosis in hBMECs. Continuous monitoring of the phosphorylation of RIPK1, RIPK3, and MLKL revealed that RIPK1 phosphorylation occurred 2 h post-infection and progressively increased, whereas phosphorylation of RIPK3 and MLKL was detectable as early as 1 h post-infection (Supplementary Fig. S3g). This indicated that during the early stages of infection, the activation of RIPK3 and MLKL preceded that of RIPK1. Compared to the death signal induced by TNFR1, necroptosis mediated by TLR4 elicited a more rapid response and played a critical role in early cell death in hBMECs. In addition, RIPK1 activation gradually increased at later time points (Supplementary Fig. S3g), implying that RIPK1 and RIPK3 may regulate distinct cell death pathways at different stages during infection.
Caspase-8 was critical to balancing cell death in hBMECs
Caspase-8 was found to be involved in the assembly of the Ripoptosome in RS218-infected hBMECs, and exhibiting time-dependent activation of its enzymatic activity during infection (Supplementary Fig. S4a), suggesting that caspase-8 may play a role in the regulation of multiple death pathways. To test this hypothesis, we compared cell death activation between caspase-8 deficient cells (Supplementary Fig. S2j) and WT. RS218 infection induced greater cytotoxicity (Supplementary Fig. S4b) and cellular membrane disruption (Supplementary Fig. S4c) in CASP8 KO cells compared to WT. At 3 h post-infection, the number of PI-labeled cells was significantly greater in CASP8 KO than in WT (Supplementary Fig. S4d). These findings demonstrated that caspase-8 knockout exacerbated RS218-induced cell death in hBMECs. Western blot analysis revealed that cleavage of caspase-3 and PARP1 was undetectable in CASP8 KO (Fig. 4a), suggesting that apoptosis was inhibited following loss of caspase-8, and the observed exacerbation of cell death in CASP8 KO resulted from increased necroptosis or pyroptosis.
a Immunoblot analysis of the processing of caspase-3, PARP1, caspase-1, and GSDMD, as well as total and phosphorylated levels of RIPK1, RIPK3, and MLKL in WT or CASP8 KO cells. Blots for GAPDH served as loading controls. FL, full-length. b, c Bar graphs represent the relative expression levels of RIPK1 (b, n = 3 biologically independent samples) and RIPK3 (c, n = 3 biologically independent samples) based on (a). d, e Cytotoxicity was assessed by LDH release (d, n = 3 biologically independent samples), and cell death kinetics were detected by PI penetration (e, n = 5 biologically independent samples) in RIPK1 WT and D324A cells stimulated with RS218 (MOI = 1). f In RIPK1 WT or S161N backgrounds, immunoblot analysis of total and phosphorylated levels of RIPK1, RIPK3, and MLKL in RIPK1 D324 (non-mutated) or RIPK1 D324A (point-mutated) cells. Blots for GAPDH served as loading controls. g Immunoblot analysis of the expression of pro-caspase-1 and its cleaved fragment (p10). Blots for GAPDH served as loading controls. h Representative immunofluorescence images showing co-localization of caspase-1 or caspase-8 with ASC in hBMECs. Scale bar, 20 μm. Line plots show fluorescence intensity profiles over 250 pixels for co-localization analysis in each image. i Immunoblot analysis of GSDMD, caspase-3, and PARP1 processing in WT, CASP1 KO, or CASP1/8 KO cells. Blots for GAPDH served as loading controls. FL, full-length. j Co-IP of ASC with pro-caspase-8 or pro-caspase-1 in WT or CASP8 KO cells. k Schematic representation of the modes of RS218-induced hBMECs death. All experiments were representative of at least three independent experiments with similar results. Bars indicated the mean plus SD. Statistical significance was determined by two-tailed unpaired Student t tests, with P-values denoted as follows: **** P < 0.0001, *** P < 0.0005, ** P < 0.005, * P < 0.05, n.s., no significant difference. Exact P-values were provided in the source data. Source data are provided as a Source Data file.
Significant increases in the phosphorylation of RIPK1 and its downstream effectors, RIPK3 and MLKL (Fig. 4a), confirmed that caspase-8 knockout triggered enhanced necroptosis. Intriguingly, intracellular RIPK1 levels decreased by approximately 51% following WT infection with RS218, but returned to baseline levels in caspase-8 knockout conditions (Fig. 4a, b). In contrast, RIPK3 did not exhibit a similar trend (Fig. 4a, c). These findings suggested that caspase-8 might regulate necroptosis by cleaving RIPK1. To test this hypothesis, we generated RIPK1 D324A cells with mutations at the caspase-8 cleavage site (Supplementary Fig. S2k). Compared to RIPK1 WT, cytotoxicity (Fig. 4d) and membrane disruption (Fig. 4e) were significantly increased upon infection in RIPK1 D324A. Western blot analysis revealed that blocking RIPK1 cleavage by caspase-8 led to a significant increase in the phosphorylation of RIPK1, RIPK3, and MLKL (Fig. 4f). These results demonstrated that caspase-8 inhibited necroptosis in the RIPK1-dependent RIPK3-MLKL pathway by cleaving RIPK1. To examine the effect of RIPK1 cleavage on necroptosis within the TLR4-RIPK3-MLKL pathway, we introduced the D324A mutation into RIPK1 S161N. The results showed that blocking caspase-8-mediated cleavage of RIPK1 reduced phosphorylation of RIPK3 and MLKL (Fig. 4f), indicating that the non-kinase function of RIPK1 suppressed necroptosis in the TLR4-RIPK3-MLKL pathway. These findings suggested that caspase-8 cleavage of RIPK1 inhibited RIPK1-associated necroptosis while promoting necroptosis through the TLR4-RIPK3-MLKL pathway.
We next investigated the potential involvement of caspase-8 in pyroptosis. While GSDMD was typically activated via pro-caspase-1 cleavage, RS218 infection of hBMECs failed to produce detectable levels of the cleavage fragment p10 of pro-caspase-1, despite a time-dependent increase in pro-caspase-1 expression (Fig. 4g). Activation of pro-caspase-1 required inflammasome formation with ASC. Colocalization analysis revealed that pro-caspase-1 did not form specks with ASC (Fig. 4h and Supplementary Fig. S4e, f), confirming that pro-caspase-1 was not incorporated into RIPK1 death complex and was not activated during associated pyroptosis. In contrast, caspase-8 exhibited strong colocalization with ASC (Fig. 4h and Supplementary Fig. S4e, f). Correspondingly, the absence of pro-caspase-1 (Supplementary Fig. S2l) did not affect GSDMD cleavage, whereas the additional knockout of caspase-8 significantly inhibited GSDMD cleavage (Fig. 4i). These findings suggested that hBMECs undergo caspase-8-dependent pyroptosis, independent of pro-caspase-1-ASC inflammasome.
Based on these findings, we hypothesized that GSDMD cleavage in hBMECs would be abolished upon caspase-8 knockout. Contrary to expectations, the GSDMD-N level was significantly increased in CASP8 KO compared to WT (Fig. 4a). Further analysis revealed that pro-caspase-1 expression was markedly elevated in CASP8 KO, accompanied by the appearance of its active p10 fragment (Fig. 4a). This unexpected observation indicated that caspase-8 deficiency activated pro-caspase-1. Co-IP assays confirmed the formation of pro-caspase-1-ASC complexes in CASP8 KO (Fig. 4j), and colocalization analysis detected pro-caspase-1-ASC specks (Supplementary Fig. S4g). These results demonstrated that caspase-8 suppressed excessive pyroptosis by inhibiting the formation of pro-caspase-1-ASC inflammasome and the activation of pro-caspase-1. In summary, Caspase-8 played a crucial role in restraining excessive pyroptosis and necroptosis, acting as an essential regulator for maintaining the balance of cell death.
We next investigated the upstream events governing caspase-8 activation. Western blot analysis revealed a significant reduction in the caspase-8 p18 fragment in RIPK1 KO compared to WT following RS218 infection (Supplementary Fig. S4h), accompanied by a loss of caspase-8 enzymatic activity (Supplementary Fig. S4i). Similarly, mutation of RIPK1 S161 inhibited both caspase-8 cleavage (Supplementary Fig. S4j) and enzymatic activity (Supplementary Fig. S4i). These findings indicated that caspase-8 activation was regulated by RIPK1 kinase. In summary, caspase-8 was activated by RIPK1 and, in turn, cleaved RIPK1 in a negative feedback loop to maintain its levels at low abundance. This regulatory mechanism mitigated excessive necroptosis and pyroptosis, highlighting caspase-8 as a critical regulator in preserving the balance of cell death in hBMECs (Fig. 4k).
RIPK1 kinase drove BBB disruption through endothelial cell death in E. coli meningitis
To investigate how RIPK1, caspase-8, and RIPK3 affect BBB function through the regulation of BMECs death, we employed a Transwell-based in vitro model (Supplementary Fig. S5a). The permeability of the BBB was evaluated by measuring transendothelial electrical resistance (TEER), and the efficiency of paracellular transport was assessed using fluorescent tracer NaFI permeability. The results showed a time-dependent reduction in TEER with increasing infection duration (Supplementary Fig. S5b). RIPK1 knockout failed to reverse the TEER reduction (Supplementary Fig. S5b) or limit NaFI permeability (Supplementary Fig. S5c). In contrast, RIPK1 kinase-inactive mutants (D138N and S161N) significantly reduced TEER loss and NaFI leakage (Supplementary Fig. S5b, c), highlighting the specific role of RIPK1 kinase activity in maintaining BBB integrity. Although RIPK3 deletion blocked MLKL phosphorylation, it promoted the transition of hBMECs toward pyroptosis and apoptosis (Fig. 3b), leading to increased cytotoxicity (Supplementary Fig. S3a) and membrane rupture (Supplementary Fig. S3b). As a result, RIPK3 deficiency did not protect the BBB (Supplementary Fig. S5b, c), suggesting limited therapeutic value as a standalone target during RS218 infection. Similarly, the loss of caspase-8 exacerbated cell death and showed a trend toward impaired BBB integrity (Supplementary Fig. S5b, c), emphasizing its essential role in hBMECs. These results indicated that RIPK1 kinase activity was a key molecular switch driving BBB disruption and increased paracellular permeability, while targeting RIPK3 or caspase-8 alone did not provide protection.
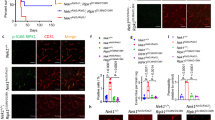
Building on in vitro findings, we generated Ripk1 D138N/D138N, Gsdmd -/-, and Casp8 -/- Ripk3 -/- mice to examine regulatory mechanisms in vivo. Casp8 and Ripk3 were co-deleted to avoid embryonic lethality associated with caspase-8 deficiency. In the neonatal meningitis model, Gsdmd -/- mice exhibited a trend towards improved survival compared to WT, though not statistically significant (Fig. 5a). In contrast, both Ripk1 D138N/D138N and Casp8 -/- Ripk3 -/- mice showed a marked survival benefit (Fig. 5a). RIPK1 kinase mutation conferred stronger protection, with recovery of body weight observed at 30 h post-infection (Fig. 5b). These results suggested that pyroptosis was not the primary lethal mechanism in this context, and that RIPK1 kinase and necroptosis played central roles in meningitis pathogenesis. Histological analysis revealed extensive erythrocyte extravasation in the WT brain (Fig. 5c), consistent with BBB disruption. This pathology was still evident in Gsdmd -/- and Casp8 -/- Ripk3 -/- mice. In contrast, Ripk1 D138N/D138N mice exhibited a marked reduction in hemorrhage, although vascular congestion remained (Fig. 5c). These findings indicated mechanistic differences underlying host protection between kinase-inactive RIPK1 and combined Casp8/Ripk3 deficiency. Bacterial loads were subsequently assessed across major organs. Brain bacterial loads were significantly reduced in both Ripk1 D138N/D138N and Casp8 -/- Ripk3 -/- mice compared to WT, while levels in Gsdmd -/- mice remained comparable to WT (Fig. 5d). Similarly, bacterial loads in the liver and lung were markedly reduced in Ripk1 D138N/D138N and Casp8 -/- Ripk3 -/- mice, with no significant differences observed in the spleen (Supplementary Fig. S5d). In contrast, peripheral blood bacterial loads did not differ across genotypes (Supplementary Fig. S5e). These findings indicated that the reduced bacterial loads in the brain, liver, and lung of Ripk1 D138N/D138N and Casp8 -/- Ripk3 -/- mice could not be attributed to variations in systemic bacterial burden.
a, b Two-day-old mice of indicated genotypes were challenged with 1 × 105 CFU of RS218. Survival curve (a, n = 12 biologically independent samples) and body weight change (b, n = 6 biologically independent samples) of mice were recorded. c H&E-stained brain sections. Black arrows showed erythrocytes within vessels, red arrows showed extravasated erythrocytes, and orange arrows showed vascular congestion. Scale bars, 20 μm. d Bacterial load in brain tissue with or without immune exhaustion (n = 6 biologically independent samples). e, f In the background of immune exhaustion, survival curve (e, n = 12 biologically independent samples) and body weight change (f, n = 6 biologically independent samples) of mice were recorded. g H&E staining of brain sections under immune exhaustion. Black arrows showed erythrocytes within vessels, red arrows showed extravasated erythrocytes, and orange arrows showed vascular congestion. Scale bars, 20 μm. h, i Activation of BMECs death in the brain detected by immunofluorescence. The nucleus was labeled in blue, endothelial cell marker CD31 in red, and pMLKL or GSDMD-N or caspase-3 p17 in green. Scale bars, 20 μm. j, k Detection of tight junction proteins in the brain. The nucleus was labeled in blue, CD31 in red, and VE-cadherin or claudin-5 in green. Scale bars, 20 μm. All experiments were representative of at least three independent experiments with similar results. Bars indicate the mean plus SD. Statistical significance was determined by Log-rank (Mantel-Cox) test (a, e), two-tailed unpaired Student t tests (b, f) and one-way ANOVA with Tukey’s post-hoc test (d), with P-values denoted as follows: **** P < 0.0001, *** P < 0.0005, ** P < 0.005, * P < 0.05, n.s., no significant difference. Exact P-values were provided in the source data. Source data are provided as a Source Data file.
To clarify the source of resistance in the two mouse genotypes, we employed clodronate liposomes to systemically deplete phagocytes, thereby eliminating potential confounding effects of innate immune cell-mediated bacterial clearance linked to genetic modifications. Following depletion, neutrophil infiltration in the brain was virtually absent (Supplementary Fig. S5f). Under these conditions, all WT mice succumbed within 12 h post-infection (Fig. 5e), indicating that the loss of innate immune defense precipitates rapid progression to lethal systemic infection. Although Ripk1 D138N/D138N and Casp8 -/- Ripk3 -/- mice exhibited significantly prolonged survival, none survived beyond 24 h (Fig. 5e). Notably, Gsdmd -/- mice demonstrated a distinct delay in mortality under immunodeficient conditions (Fig. 5e), suggesting that in the absence of phagocytes, pyroptosis played a more pronounced role in controlling infection progression. Body weight loss trajectories did not differ significantly between infection groups (Fig. 5f). Histological analysis revealed extensive erythrocyte extravasation in the brains of immunodeficient WT mice (Fig. 5g), whereas Ripk1 D138N/D138N, Gsdmd -/- and Casp8 -/- Ripk3 -/- mice exhibited attenuated pathology to varying degrees, with the most prominent protection observed in Ripk1 D138N/D138N mice (Fig. 5g). These findings suggested that genetic inhibition of cell death pathways could partially preserve BBB integrity even in the context of impaired innate immunity. Furthermore, while immune depletion markedly increased bacterial burdens in the brains of WT mice, bacterial loads in Ripk1 D138N/D138N, Gsdmd -/-, and Casp8 -/- Ripk3 -/- mice remained comparable to those observed under immunocompetent conditions (Fig. 5d). The shift in statistical significance for Gsdmd -/- mice from non-significant to significant (Fig. 5d) underscores the heightened contribution of pyroptosis to infection control in the absence of phagocytic clearance. These results indicated that all three mutant mice retained resistance to meningitis under immunocompromised conditions, highlighting a non-immune cell-dependent role of the BBB in preserving brain homeostasis. In contrast, no differences in bacterial clearance were observed among groups in peripheral organs following immune depletion, including the liver, lung, spleen and blood (Supplementary Fig. S5d, e), indicating that the antibacterial effects of Ripk1 D138N/D138N and Casp8 -/- Ripk3 -/- in peripheral tissues were primarily dependent on phagocyte-mediated immunity. In addition, the regulatory role of pyroptosis on the BBB was amplified under specific immune conditions, underscoring its secondary yet non-negligible role in host defense.
We hypothesized that the varying degrees of meningitis resistance conferred to the BBB in mice were related to the regulation of BMECs death. Therefore, we focused on delineating the impact of BMECs death modalities on BBB structural integrity. In WT mice, the presence of significant activation of pMLKL, GSDMD-N and cleaved caspase-3 (p17) was detected in BMECs (Fig. 5h), with all three forms of cell death further exacerbated under conditions of immune depletion (Fig. 5i). Concordantly, tight junction protein expression declined markedly following immune suppression (Fig. 5j, k). Under immunocompetent conditions, GSDMD deficiency abrogated pyroptosis in BMECs, while necroptosis and apoptosis remained unaffected (Fig. 5h). Correspondingly, tight junction protein levels in Gsdmd -/- mice were comparable to those in WT mice (Fig. 5j), indicating that pyroptosis contributed modestly to BBB impairment in the context of an intact immune system. However, in the setting of immune deficiency, the protective effect of GSDMD deletion became more pronounced, as evidenced by increased expression of tight junction proteins compared with WT mice (Fig. 5k). This correlated with a reduction in bacterial burden in the brain (Fig. 5d), collectively suggesting that GSDMD loss conferred a selective advantage in preserving BBB integrity under immunosuppressed conditions. In Casp8 -/- Ripk3 -/- mice, both necroptosis and apoptosis were effectively suppressed in BMECs irrespective of immune status (Fig. 5h, i). In fact, these cells exhibited heightened GSDMD-N cleavage (Fig. 5h, i) and specific caspase-1 activation (Supplementary Fig. S5g), consistent with in vitro findings. In contrast, Ripk1 D138N/D138N mice displayed simultaneous suppression of necroptosis, pyroptosis and apoptosis (Fig. 5h, i). This divergence in BMECs death regulation was reflected in greater tight junction protein expression in Ripk1 D138N/D138N mice relative to Casp8 -/- Ripk3 -/- mice (Fig. 5j), which also explained their differences in meningitis resistance (Fig. 5c). To further quantify BBB function, we assessed NaFI permeability in brain tissue. Gsdmd -/- mice exhibited significant barrier protection only under immune-depleted conditions (Supplementary Fig. S5h). In contrast, Ripk1 D138N/D138N and Casp8 -/- Ripk3 -/- mice demonstrated consistent and robust BBB preservation regardless of immune status (Supplementary Fig. S5h). Nevertheless, Casp8 -/- Ripk3 -/- mice showed a non-significant trend toward increased BBB permeability following immune depletion (Supplementary Fig. S5h), potentially attributable to excessive pyroptotic activation. Collectively, these data indicated that the extent and nature of BMECs death directly govern BBB integrity. Among the cell death pathways examined, inhibition of RIPK1 kinase activity conferred the most effective protection against BBB disruption induced by E. coli.
Intervention with RIPK1 kinase improved treatment outcomes in neonatal mouse E. coli meningitis
The above results provided a rationale and potential for RIPK1 kinase to serve as a therapeutic target for E. coli meningitis. To explore this approach, necrostatin-1 stable (Nec-1s), a selective RIPK1 kinase inhibitor targeting the activation loop at S161, was chosen. Initially, the efficacy of Nec-1s was tested in vitro to determine whether its effects were consistent with the experimental outcomes of RIPK1 D138N and S161N mutations. As expected, Nec-1s treatment significantly suppressed Ripoptosome formation (Supplementary Fig. S6a) and downstream cell death, including apoptosis (cl. caspase-3 and cl. PARP1), pyroptosis (GSDMD-N) and necroptosis (pRIPK1, pRIPK3 and pMLKL) (Supplementary Fig. S6b). The inhibition of caspase-8 activation downstream of Ripoptosome by Nec-1s resulted in the rescue of cleaved RIPK1, thereby increasing cellular RIPK1 abundance (Supplementary Fig. S6b), which subsequently enhanced the inhibitory effect of RIPK1 on the TRIF-RIPK3 complex (Supplementary Fig. S6c). Nec-1s did not interfere with RIPK1 recruitment to TNFR1 (Supplementary Fig. S6d). These factors were finally reflected in significant inhibition of membrane rupture (Supplementary Fig. S6e) and cytotoxicity (Supplementary Fig. S6f). Furthermore, Nec-1s intervention prevented the reduction in TEER of the BBB (Supplementary Fig. S6g) and NaFI permeability (Supplementary Fig. S6h), thereby preserving BBB integrity. For comparison, the caspase-8 inhibitor Z-IETD-FMK (IETD) and the RIPK3 inhibitor GSK’872 were also included. In line with findings from knockout models, neither inhibitor alone conferred a protective effect on the BBB (Supplementary Fig. S6g, h). In addition, we excluded the potential impact Nec-1s, IETD, and GSK'872 could have on the viability of RS218 (Supplementary Fig. S7a).
Based on these findings, we assessed these interventions in 2-day-old CD1 mice. Mice were infected with RS218, followed by intraperitoneal administration of Nec-1s, IETD, or GSK'872 6 h post-infection. PBS was exclusively administered to the blank control group (Fig. 6a). Over the 48 h observation period, mice in the RS218, IETD, and GSK'872 groups exhibited progressive weight loss until mortality, whereas Nec-1s treated mice showed gradual weight recovery beginning 30 h post-infection (Fig. 6b). Notably, Nec-1s treatment significantly improved survival rates in meningitis model, achieving 40% survival, while mice in the IETD and GSK'872 groups succumbed within 18 h (Fig. 6c). Histopathological examination unveiled pronounced erythrocyte extravasation (Fig. 6d) and neutrophil infiltration (Fig. 6e) within the meninges of mice from the RS218, IETD, and GSK'872 groups, but these features were absent in the Nec-1s group (Fig. 6d, e). These observations indicated that IETD and GSK'872-mediated inhibition of caspase-8 and RIPK3 failed to protect mice from E. coli meningitis, in contrast to the effects seen with Nec-1s.
a Schematic of the neonatal mouse E. coli meningitis model and treatments. b, c Two-day-old mice were challenged with 1 × 105 CFU of RS218 and treated with PBS or Nec-1s or IETD or GSK’872. Body weight (b, n = 6 biologically independent samples) and survival curve (c, n = 12 biologically independent samples) were recorded. d H&E-stained brain sections. Black arrows showed erythrocytes within vessels, and red arrows showed extravasated erythrocytes. Scale bars, 20 μm. e Immunofluorescence analysis of neutrophil infiltration. The nucleus was labeled in blue, MPO in red, and GFAP in green. Scale bars, 20 μm. f Bacterial load in brain, liver, lung, spleen, and blood. n = 6 biologically independent samples. g Activation of BMECs death in the brain detected by immunofluorescence. The nucleus was labeled in blue, endothelial cell marker CD31 in red, and pMLKL or GSDMD-N or caspase-3 p17 in green. Scale bars, 20 μm. h Detection of tight junction proteins. The nucleus was labeled in blue, CD31 in red, and VE-cadherin or claudin-5 in green. Scale bars, 20 μm. i Immunoblotting of brain tissue. GAPDH served as a loading control. j The analysis of CNS injury. The nucleus was labeled in blue, and GFAP in green. Scale bars, 20 μm. k Cytokine levels in brain tissue. n = 6 biologically independent samples. l Survival curve of mice was recorded. All experiments were representative of at least three independent experiments with similar results. Bars indicate the mean plus SD. Statistical significance was determined by one-way ANOVA with Tukey’s post-hoc test (c, f, k, and 6–24 h for b), two-tailed unpaired Student t-tests (30–48 h for b), and Log-rank (Mantel-Cox) test (l), with P-alues denoted as follows: **** P < 0.0001, *** P < 0.0005, ** P < 0.005, * P < 0.05, n.s., no significant difference. Exact P-values were provided in the source data. Source data are provided as a Source Data file.
Mice subjected to Nec-1s treatment exhibited significantly reduced bacterial loads in the brain, liver, and lung compared to the RS218 group, with the most substantial reduction observed in the brain (Fig. 6f). In contrast, mice treated with IETD showed markedly higher bacterial loads in both the brain and liver (Fig. 6f). GSK'872 group did not exhibit a significant difference from the RS218 group in other assessed organs, except for a notable decrease in bacterial loads in the lung (Fig. 6f). In peripheral blood, bacterial load tended to decrease in the Nec-1s-treated mice (Fig. 6f), suggesting that Nec-1s may facilitate bacterial clearance through additional mechanisms. To explore whether the inhibitors enhanced the phagocytic activity of macrophages, we treated differentiated THP-1 and RAW 264.7 cells with inhibitors, respectively. The results showed that neither IETD nor GSK'872 significantly affected macrophage phagocytosis, while Nec-1s appeared to a certain extent to enhance phagocytic activity (Supplementary Fig. S7b), suggesting that macrophage activation contributed to bacterial clearance in the blood to a certain extent.
Compared to the modest reduction in bacterial load in the blood, a dramatic decrease was observed in the brain (Fig. 6f), indicating that Nec-1s treatment primarily reduced brain bacterial load through mechanisms other than blood bacterial clearance. Therefore, we further investigated the mechanisms underlying Nec-1s-induced bacterial clearance in the brain. In the RS218 group, clear evidence of pMLKL, GSDMD-N, and caspase-3 p17 was observed, which was nearly completely suppressed following Nec-1s administration (Fig. 6g). In contrast, the levels of pMLKL in the IETD group remained similar to those in the RS218 group (Fig. 6g). GSK'872 treatment also failed to inhibit GSDMD and caspase-3 activation in BMECs (Fig. 6g). These findings indicated that Nec-1s-mediated inhibition of RIPK1 kinase significantly reduced RS218-induced cell death in BMECs, whereas an effect not observed with IETD or GSK'872. To further explore the impact of BMECs survival on BBB integrity in mice, the expression levels of the tight junction proteins VE-cadherin and claudin-5 were examined. In comparison to the PBS group, VE-cadherin and claudin-5 expression were significantly reduced in BMECs in the RS218 group (Fig. 6h). This reduction was exacerbated in the IETD and GSK'872 groups but was recovered following treatment with Nec-1s (Fig. 6h). These results demonstrated that Nec-1s-inhibited BMECs death was essential for the restoration of BBB integrity. Coupled with the significant reduction in bacterial load in the brain of the Nec-1s group (Fig. 6f), these findings indicated that increased BBB permeability due to BMEC death was a critical factor in RS218 invasion of the brain.
In addition to BMECs, we also examined the damage in brain parenchymal cells. The results demonstrated that in comparison to the RS218 group, IETD inhibited the cleavage of GSDMD-N, caspase-3, and PARP1, but high levels of pRIPK1, pRIPK3 and pMLKL caused necroptosis in the brain (Fig. 6i). GSK'872 treatment detected significant cleavage of caspase-3, PARP1 and GSDMD, maintaining high levels of apoptosis and pyroptosis (Fig. 6i). Pro-caspase-1 expression in the brain increased under RS218 infection compared to the PBS group. IETD treatment suppressed this increase (Fig. 6i), likely due to the inhibitory effect of caspase-8 on pro-caspase-1. A small amount of activated pro-caspase-1 was only detected following GSK'872 treatment (Fig. 6i), suggesting that pro-caspase-1 contributed to pyroptosis in the brain within the GSK'872 group. In contrast, Nec-1s reduced necroptosis without activating pro-caspase-1 and simultaneously inhibited both apoptosis and pyroptosis (Fig. 6i). GFAP expression in the brain was significantly downregulated in the Nec-1s group (Fig. 6j). These findings indicated that Nec-1s targeted RIPK1 to alleviate cell death in the brain, thereby mitigating neurological damage. In addition, the IETD group displayed similar levels of CXCL-1, IL-1β, and IL-6 as the RS218 group, while TNF-α exhibited a significant increase (Fig. 6k). IL-1β was also increased in the GSK'872 group (Fig. 6k). Nec-1s treatment reduced the release of CXCL-1, IL-1β, TNF-α, and IL-6 in the brain (Fig. 6k), which correlated with lower bacterial loads. Collectively, these findings suggested that Nec-1s attenuated meningitis-induced brain damage by limiting excessive inflammation and reducing neurological injury.
Treatment with Nec-1s significantly reduced RS218 counts in the brain but did not substantially decrease bacterial load in peripheral blood (Fig. 6f). This reflected the therapeutic limitations of targeting BMECs death inhibition for bacterial clearance. Cefotaxime (CTX), a third-generation cephalosporin antibiotic, was a primary treatment for E. coli meningitis3,26. We evaluated the effects of Nec-1s and CTX, both individually and in combination. Over the 48 h observation period, mice treated with Nec-1s or CTX, either alone or in combination, began to regain body weight 30 h post-infection in comparison to the RS218 group (Supplementary Fig. S8a). Survival rates were comparable between the Nec-1s and CTX monotherapy groups, while combination therapy rescued a greater proportion of mice (Fig. 6l), indicating a synergistic effect between Nec-1s and CTX. Although the Nec-1s and CTX monotherapy mice showed similar changes in body weight and survival, the brain from the latter mice exhibited distinct hemorrhagic spots, similar to those observed in RS218-infected mice (Supplementary Fig. S8b). Histopathological analysis showed reduced congestion in the treatment groups compared to the RS218-infected group (Supplementary Fig. S8c). Moreover, none of the treatment groups exhibited the severe hemorrhagic phenomena observed in the IETD or GSK'872 treatment groups (Fig. 6d). CTX monotherapy did not significantly alleviate the RS218-induced reduction in VE-cadherin and claudin-5 expression as evidenced by tight junction protein labeling (Supplementary Fig. S8d). However, Nec-1s alone or in combination with CTX reversed these changes (Supplementary Fig. S8d). These findings underscore the critical role of BMECs death inhibition in maintaining BBB integrity and revealed the limitations of antibiotic therapy alone in preventing cell death.
Compared to the RS218 group, CTX-treated mice exhibited significantly reduced bacterial loads in the brain, liver, spleen, lung, and particularly in peripheral blood (Supplementary Fig. S8e), indicating the efficacy of CTX in bacterial clearance. The combination of Nec-1s and CTX further reduced bacterial loads in brain tissue (Supplementary Fig. S8e), with CTX enhancing bacterial clearance in other organs compared with Nec-1s monotherapy. (Supplementary Fig. S8e). Cytokine levels in the brain showed no significant inhibition in the CTX group compared to the RS218 group (Supplementary Fig. S8f), and GFAP expression remained elevated (Supplementary Fig. S8g), indicating the limitations of CTX monotherapy in controlling brain inflammation and neurological damage. However, improved outcomes were observed with the combined treatment of Nec-1s and CTX (Supplementary Fig. S8f, g). These results suggested that targeting BMECs death via RIPK1 kinase inhibition, in combination with bacterial clearance, may offer enhanced therapeutic efficacy in E. coli meningitis.
Discussion
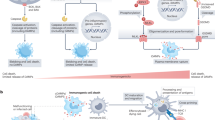
It is well known that infection with NMEC compromises the structural integrity of the BBB27,28. Here, we identify a key mechanism by which the NMEC strain RS218 disrupts BBB function through intricate crosstalk among multiple PCD pathways in BMECs (Fig. 7). The absence of a single death effector molecule resulted in a shift in the type and extent of cell death, thereby breaking cellular equilibrium. This dysregulation was detrimental to the maintenance of BBB function and neural homeostasis during E. coli meningitis. Cell death increases the paracellular translocation of RS218 into brain tissue, representing a key mechanism underlying bacterial invasion. Previous studies have indicated that mice deficient in caspases-1, − 11, − 12, − 8, and RIPK3 exhibit impaired bacterial clearance following infection with low doses of attenuated Salmonella strains, ultimately resulting in mortality. Host defense was facilitated by the activation of apoptosis, pyroptosis and necroptosis, which promoted bacterial clearance and the release of inflammatory mediators, yielding beneficial outcomes29. This comparison underscores that the advantages and disadvantages of PCD activation varied across different cell types and infectious agents. Thus, a comprehensive investigation of the molecular mechanisms governing PCD crosstalk is essential to advancing understanding and developing intervention strategies for infectious diseases.
Under physiological conditions, the integrity of the BBB is maintained by endothelial cells, tight junctions, pericytes, the basement membrane, and astrocytes, effectively preventing macromolecules and pathogens from translocating from the bloodstream (top) into the brain (bottom). In E. coli meningitis, RS218 induced BMECs death through RIPK1 kinase-dependent and independent pathways. While caspase-8 limited programmed necrosis and pyroptosis by cleaving RIPK1 and suppressing caspase-1-ASC inflammasome formation, the death of membrane-compromised BMECs remained inevitable. This detrimental effect on BMECs directly compromised BBB integrity, facilitating RS218 penetration into brain tissue via the paracellular pathway. Pharmacological inhibition of RIPK1 kinase activity with Nec-1s effectively suppressed BMECs death and preserved RIPK1. The scaffolding function of RIPK1 was thus maintained, enhancing its ability to inhibit RIPK3-MLKL-mediated necroptosis. Ultimately, this mitigated BMECs death and BBB disruption.
Although RS218 induces caspase-8-mediated apoptosis in hBMECs by secreting the effector Hcp1 through its type VI secretion system (T6SS)30, the precise mechanism underlying cell death remained unclear. In this study, we provide direct evidence that RS218 triggers BMECs death and underscore the central role of caspase-8 in maintaining cell death homeostasis. The cleavage of RIPK1 and RIPK3 represents a key mechanism through which caspase-8 prevents excessive cell death12,31. Our findings indicate that RIPK1, rather than RIPK3, serves as the major substrate of caspase-8 during RS218 infection. The expression of a cleavage-resistant RIPK1 mutant (D324A) further supports the importance of RIPK1 processing in limiting necroptosis. This observation aligns with studies in RIPK1 D325A knock-in mice, in which midgestational lethality was rescued by combined deletion of RIPK3 and FADD11,32,33. Moreover, caspase-8 cleaves GSDMD independently of canonical caspase-1 activity. This mechanism resembles that observed in L. pneumophila infection, where caspase-8 was recruited by Naip5/NLRC4/ASC inflammasomes in the absence of caspase-1, resulting in GSDMD cleavage and pyroptosis in bone marrow-derived macrophages (BMDMs). The study also suggested that caspase-8 activation occurred only when caspase-1 was inhibited15,34. In contrast, our data reveal that caspase-8 remains active despite the presence of caspase-1 during RS218 infection in BMECs. Indeed, caspase-1-ASC inflammasome formation and caspase-1 activation were detectable only in the absence of caspase-8. Given that caspase-1 cleaves GSDMD more efficiently than caspase-815,35, its activation resulted in heightened pyroptosis. This demonstrated the role of caspase-8 in inhibiting caspase-1 activation and preventing excessive pyroptosis. Elevated pyroptosis levels in BMECs exert a more pronounced disruptive effect on the BBB. The complex regulatory relationship between caspase-8 and caspase-1 under specific stimuli was revealed.
In this study, we demonstrated that RS218-induced BMECs death is critically dependent on RIPK1 kinase activity. The serine S161 is a critical site for the kinase to function, with its mutation producing effects equivalent to those of the kinase-dead D138N variant. While a mutation at S166 affected RIPK1 activity, it did not completely block Ripoptosome assembly. It had been pointed out that the S161 site played a crucial role in determining the binding of downstream effector molecules by altering the conformation of RIPK119,20,36. In contrast, S166 enhanced kinase activity only in the presence of RIPK1 activation, but was insufficient to initiate activity independently37. Mechanistically, targeting the RIPK1 S161 locus appeared more aligned with effective therapeutic strategies. We demonstrated the efficacy of inhibiting autophosphorylation at RIPK1 S161 in preserving BBB integrity in BMECs in vitro and in vivo. A recent study indicated that blocking RIPK1 kinase activity with Nec-1s mitigated macrophage migration inhibitory factor (MIF)-promoted apoptosis and necroptosis in BMECs, thereby reducing BBB permeability38. Together with the infection resistance observed in Ripk1 D138N/D138N mice in this study, these findings collectively supported the concept that the RIPK1 kinase activity was involved in BBB disruption due to BMECs death in inflammatory and infectious diseases33. Furthermore, increasing evidence suggests that RIPK1 promotes cell survival signals through its scaffolding function and inhibits downstream cell death checkpoints in TNFR1 signaling39,40,41. Concurrently, RIPK1 competitively inhibited RIPK3-mediated necroptosis by interacting with the RHIM domain in TRIF and ZBP1, independently of its kinase activity40,42. In this study, we also acknowledged the importance of RIPK1 scaffolding function in BMECs during E. coli meningitis. Although RIPK1 knockout blocked caspase-8-mediated apoptosis and pyroptosis, it relieved RIPK1 inhibition of RIPK3 and promoted necroptosis of the RIPK1-independent pathway. This alteration negatively impacted the in vitro BBB model constructed by BMECs. The necessity of the scaffold function for the BBB could not be confirmed by constructing RIPK1-null mice, as loss of this function proved lethal during embryonic development21,43. This underscored the indispensable role of RIPK1 scaffolding in cell death regulation. Notably, RIPK1 (D138N) mice expressing loss of kinase activity did not exhibit a spontaneous phenotype and significantly reduced pathogenesis in experimental models of inflammatory and degenerative diseases in mice23,24,25. This suggested that inhibiting RIPK1 kinase activity may represent a potential therapeutic target to mitigate or prevent tissue damage, provided it did not adversely affect RIPK1 expression levels or its prosurvival scaffolding function under ideal circumstances.
RIPK1 has been the focus of numerous small-molecule inhibitors. These kinase inhibitors were classified based on their binding sites and mechanisms of action into three categories: type I (bound the active DFG/DLF-in conformation), type II (bound the inactive DFG/DLF-out conformation), and type III (bound the allosteric pocket)44. Nec-1 (the prototype of Nec-1s) was the first identified type III RIPK1 inhibitor19,45. The nitrogen atom in the indole ring of Nec-1 inhibited autophosphorylation at Ser161 and prevented conformational changes in RIPK1 by binding to the hydroxyl oxygen in the activation loop44. While Nec-1 exhibited moderate potency in inhibiting RIPK1 kinase, it demonstrated significant therapeutic efficacy in various mouse models, including those for cardiovascular, neurodegenerative, renal, hepatic, systemic, and intestinal inflammatory diseases46. As more RIPK1 inhibitors were designed, multiple potent kinase inhibitors were discovered. Type II inhibitors GSK'414 and GSK'157 were reported to be 10-100 times more effective than Nec-1 in inhibiting RIPK1 kinase and proved more effective in blocking TNF-induced RIPK1-dependent cell death47,48. However, it was important to note that greater inhibitor potency did not necessarily correlate with improved therapeutic outcomes. The type III inhibitor 6E11, although less effective than Nec-1 in TNF-induced cell death and exhibiting lower binding affinity for human RIPK1, provided better protection for human aortic endothelial cells during hypothermic hypoxia/reoxidation49. This suggested the importance of selecting appropriate agents for targeted therapy among the diverse array of RIPK1 inhibitors. Further studies were warranted to evaluate whether Nec-1s was the most potent inhibitor for targeting RIPK1 kinase to mitigate BMECs death in E. coli meningitis. Caspase-8 and RIPK3 were also significant targets for small-molecule drug development. The multiple roles of caspase-8 made it an attractive target for therapeutic interventions. However, as caspase-8 functioned both as an inducer and an inhibitor of cell death, targeted therapy presents challenges12,50. Our findings indicated that inhibiting caspase-8 enhanced necroptosis and pyroptosis of the caspase-1 pathway in BMECs, leading to increased BBB permeability and exacerbation of RS218 infection. Similarly, inhibition of RIPK3 did not confer benefits in E. coli meningitis in mice, suggesting that targeting RIPK3 alone was not a viable strategy, consistent with the observations of Kim Newton et al.51. To date, no RIPK3 inhibitor has been selected for the treatment of RIPK3-driven inflammatory diseases44,52, likely due to the minimal benefits observed in disease progression following RIPK3 inhibition. These results highlighted the limitations of targeting caspase-8 and RIPK3, suggesting that they may not represent optimal therapeutic strategies for E. coli meningitis. Taken together, given the pivotal role of RIPK1 kinase in orchestrating cellular responses, prioritizing upstream RIPK1 kinase activity emerges as a more promising avenue for drug development and therapy in E. coli meningitis.
The contribution of cell death to E. coli meningitis has remained largely underappreciated. Here, we elucidate the regulatory mechanism by which RS218 induced cell death in BMECs and identify RIPK1 kinase as a therapeutically actionable target within the intricate network of PCD crosstalk. In mice, the D138N mutation in RIPK1 attenuates BMECs death, and enhances resistance to RS218 invasion by preserving BBB integrity. Pharmacological inhibition of RIPK1 kinase recapitulates this protective effect and stabilizes RIPK1 protein levels, thereby augmenting its scaffold function to suppress RIPK1-independent necroptotic signaling. Furthermore, preventing BMECs death mitigated excessive neutrophil infiltration into the brain, which arose from the release of cellular contents, leading to heightened inflammatory responses and central nervous system damage. The combination of RIPK1 kinase inhibition and antibiotic treatment resulted in improved outcomes. Therefore, RIPK1 warrants further investigation as a potential therapeutic target for E. coli meningitis.
Methods
Ethics statement
All animals were housed individually in the Laboratory Animal Center of Nanjing Agricultural University. All animal studies were performed according to protocols approved by the Animal Ethics Committee of Nanjing Agricultural University, procedure number SYXK (SU) 2011-0036, Nanjing. All experiments were performed according to the International Code of Practice for the Care and Use of Animals for Scientific Purposes. Temperature and humidity were maintained at 21–23 °C and 60–65%, respectively.
Animal infections
Gsdmd -/- mice (Strain NO. T010437) were purchased from GemPharmatech (Nanjing, China). Ripk1 D138N/D138N mice were generously provided by Junying Yuan lab (Harvard Medical School) and Daichao Xu lab (Chinese Academy of Sciences). Casp8 -/- Ripk3 -/- mice were generously provided by Xin Lin lab (Tsinghua University). All genetically modified strains were maintained on a C57BL/6 J background. Two-day-old female neonates were genotyped and randomly assigned to experimental groups. To establish the E. coli meningitis model, RS218 was inoculated intraperitoneally at a dose of 1 × 105 CFU in a volume of 20 μL per mouse, as previously described53. For phagocyte depletion, neonatal mice received daily intraperitoneal injections of 10 μL of Clodronate Liposomes (YEASEN, 40337ES08) or PBS Liposomes (YEASEN, 40338ES08) from day 0 to day 2. For pharmacological intervention, 2-day-old female CD1 mice (Specific pathogen free, from Sipeifu, Beijing, China) treated intraperitoneally with Nec-1s (5 mg/kg) or IETD (5 mg/kg) or GSK’872 (5 mg/kg) or cefotaxime (30 mg/kg) at 6 h post-infection. Treatments were administered every 12 h thereafter. The control group was injected intraperitoneally with PBS. The dose was 20 μL per mouse. All mice were monitored every 6 h. The neonates were euthanized humanely if they showed signs of severe illness. The euthanasia was performed by Cervical Dislocation (CD) and counted as death. The monitoring of body weight and survival curves in the experiment continued until 48 h post-infection. For downstream analyses, tissue samples were collected at 12 h post-infection or 12 h after the first drug intervention. Bacterial loads were quantified by plating tissue homogenates on Luria-Bertani (LB) agar. Cytokine concentrations in serum or brain tissue were measured by ELISA. Histopathological assessment was performed using hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC) of brain tissue. For assessment of BBB integrity, NaFI infiltration into brain tissue was quantified as previously described54. Relative permeability was calculated by normalizing to the control group.
Bacterial strains and growth conditions
NMEC strain RS218 were stored at − 80 °C, melted in a 37 °C water bath, and streaked on sheep blood plates. On the following day, single colonies were picked up and cultured in LB medium at 37 °C and 120 rpm for 8 h. They were inoculated into LB medium at 1:100 and subcultured at 37 °C and 120 rpm until the log phase of growth for subsequent tests.
Cell culture
The hBMECs used in this study were purchased from ScienCell Research Laboratories (Catalog #1000). HEK293T cells (ATCC® CRL-3216), THP-1 cells (ATCC® TIB-202), RAW 264.7 cells (ATCC® TIB-71), and U-118MG cells (ATCC® HTB-15) were obtained from the American Type Culture Collection. RIPK1 KO, ZBP1 KO, RIPK3 KO, TRIF KO, CASP8 KO, CASP1 KO, CASP1/8 KO, TLR4 KO, TNFR1 KO, and TLR4/TNFR1 KO cell lines were obtained by the CRISPR/Cas9 gene editing system. The mutant lines were generated by introducing the Ripk1 gene with the D138N, S161N or S166N or D324A or RHIM mutated site into RIPK1 KO and the Trif gene with the RHIM mutated site into TRIF KO via the lentiviral system. The cell lines of hBMECs, U-118MG, HEK293T, and RAW 264.7 were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, Cat# C11995500BT) supplemented with 10% fetal bovine serum (FBS, Gibco, Cat# 10270-106) and 1% (v/v) penicillin/streptomycin (Solarbio, Cat# P1400) at 37 °C with 5% CO2. THP-1 was cultured in RPMI-1640 supplemented with 10% fetal bovine serum and 1% (v/v) penicillin/streptomycin at 37 °C with 5% CO2. Before the experiment, THP-1 cells were differentiated into macrophages by treatment with 30 ng/mL phorbol 12-myristate 13-acetate (PMA) for 48 h. Following this treatment, the medium was replaced, and the cells were cultured for an additional 24 h to prepare for subsequent experiments.
Generation of knockout hBMECs by CRISPR/Cas9
sgRNAs targeting proteins of interest or non-targeting sgRNAs were cloned into lentiCRISPRv2 puro (Addgene, 98290) according to the instructions given by Zhang Feng’s laboratory55,56, and an additional G was added at the beginning of the sgRNA sequence. LentiCRISPRv2 puro, pCMV-VSV-G (Addgene, 8454), and psPAX2 (Addgene, 12260) were transfected into HEK293T packaging lentiviruses at a ratio of 4:2:3 by LipofectamineTM3000 Transfection Reagent (ThermoFisher Scientific, L3000008). After incubation for 6 h at 37 °C, 5% CO2, the medium was replaced with fresh medium, and the incubation was continued until 60 h. Lentiviruses were obtained after filtration of the collected supernatants. Lentiviral titers were detected, and hBMECs containing 5 μg/mL polybrene (MedChemExpress, HY-112735) were infected at an MOI of 20. After 6 h, the supernatant was replaced with fresh medium. Stable stress-resistant cells were then selected with 1 μg/mL puromycin (MedChemExpress, HY-B1743A), and positive cell lines were obtained by limiting dilution.
The most effective sequences among multiple sgRNA sequences were selected for further experiments by Western blot. Specific sgRNAs used were listed in Table 1.
Construction of the mutant cell lines
RIPK1 mRNA sequence (GenBank ID: NM_003804.6) or TRIF mRNA sequence (GenBank ID: NM_001385678.1) was obtained from the NCBI database. The gene sequence with aspartic acid mutation to alanine at position 324 or aspartic acid mutation to asparagine at position 138 or serine mutation to asparagine at position 161 and 166 or IQIG of RHIM mutation to AAAA for RIPK1 and the gene sequence with VQLG of RHIM mutation to AAAA for TRIF were synthesized and ligated into GL208 pcSLenti-EF1-EGFP-F2A-BSR-CMV-MCS-3×FLAG-WPRE vector (purchased from Obiosh) by EcoR I (Takara, 1040 A) and Xba I (Takara, 1093 A). The products were amplified by CMV-F: CGCAAATGGGCGGTAGGCGTG and WPRE-R: CATAGCGTAAAAGGAGCAACA, and sequenced. Lentiviruses were generated as described, and RIPK1 KO cells or TRIF KO cells were infected at an MOI of 20, and fresh medium containing blasticidin S (10 μg/mL, Merck, SBR00022) was replaced 12 h later to select stable stress-resistant cell populations. Flag tag expression was verified by Western blot. Gene sequence analysis and editing were performed using SnapGene software.
Cell stimulations
hBMECs, or knockout cell or mutant cell lines, were plated at a concentration of 1 × 105 cells/cm2 and cultured until reaching a confluent monolayer. Cells were counted before all infection assays to ensure an accurate MOI. For assays associated with inhibitor treatment, cells were pretreated with 20 μM Z-VAD-FMK (Medchem Express, HY-16658B) or 20 μM necrosulfonamide (Medchem Express, HY-100573) or 20 μM necrostatin-1 stable (Nec-1s, Merck, 5.04297) or 5 μM GSK'872 (Medchem Express, HY-101872) or 20 μM Z-IETD-FMK (Medchem Express, HY-101297) or DMSO (Sigma, D1435) for 0.5 h at 37 °C, 5% CO2, and incubated with RS218 at an MOI of 1 or an MOI of 10. For drug-induced positive assays, apoptosis was triggered by treatment with 10 μM Raptinal (Sigma, SML1745) for 0.5 h; necroptosis was obtained by treating hBMECs with Z-VAD-FAM at 20 μM for 0.5 h and then adding TNF-α (Sigma, H8916) at a final concentration of 20 ng/mL and SM-164 (Medchem Express, HY-15989) at 100 nM for an additional 8 h; pyroptosis was obtained by treating hBMECs with 200 ng/mL LPS (E. coli O55: B5, Sigma) for 6 h and then adding nigericin at a final concentration of 20 μM for an additional 45 min as previously described57. Supernatants were used for LDH release assays or cytokine assays, RNA was extracted for analysis of transcriptional levels of the indicated genes, and cell extracts were used for expression level detections of given proteins by western blot. These inhibitors were not significantly cytotoxicity to hBMECs or mice at the indicated concentrations.
Time-lapse microscopy analysis
Time-lapse microscopy analysis was performed as previously described58. Briefly, hBMECs were seeded overnight in a 0.17 mm thick cell culture dish, and DMEM containing a final concentration of 10 μg/mL PI (Solarbio, C0080) was replaced before testing. RS218 was inoculated at an MOI of 10, and kinetic imaging analysis of viable cells was performed using a cell observer automated microscope at 37 °C and 5% CO2. Cell morphology and PI uptake were photographed at 60-fold magnification every minute with natural light and excitation at 535 nm and emission at 615 nm. Real-time images of cell death were processed with ZEISS ZEN 2.3 lite software. The images shown represent three randomly selected regions.
LDH release assay
hBMECs were seeded into 96-well plates and cultured until reaching a confluent monolayer at 37 °C, 5% CO2. Fresh DMEM medium containing RS218 (MOI of 1 or MOI of 10) was added by washing three times with PBS before the assay. Supernatants were collected at indicated times postinfection, centrifuged at 1000 × g for 10 min at 4 °C to remove cell debris and bacteria, and the release of LDH was detected using the Lactate Dehydrogenase (LDH) Cytotoxicity Colorimetric Assay Kit (Sigma, MAK529) according to the manufacturer’s instructions. Expressed as a percentage of total cellular LDH (100% lysis), the culture medium without cells was used for background subtraction.
PI analysis of cell membrane integrity
hBMECs were seeded into 96-well plates on the bottom of black transparent glass and cultured until reaching a confluent monolayer at 37 °C, 5% CO2. RS218 or 0.5% Triton X-100 was inoculated as infected or positive wells. DMEM medium containing PI (final concentration of 10 μg/mL) was taken as control wells. Cell-free culture medium containing PI served as blank wells for background subtraction. Each group contained 3–5 replicate wells. Fluorescence intensity was detected by using a multimode microplate reader (TECAN, Spark) for 8 h at excitation 535 nm and emission 615 nm, and measurements were expressed as a percentage of total cellular PI uptake, as previously described32. For microscopic analysis, 60-fold images were taken from at least three randomly selected areas under the given conditions, capturing approximately 100 cells per field of view, and the percentage of PI-positive cells was calculated.
Flow cytometric analysis
hBMECs were transferred to dishes and cultured until reaching a confluent monolayer at 37 °C, 5% CO2. The following day, cells were washed three times with PBS, and inoculated with DMEM supplemented with RS218 and supernatants were collected at given times. Cells were then washed three times with an equal volume of PBS, and the PBS was transferred to a centrifuge tube. Cells were treated with EDTA-free trypsinization and sedimented by centrifugation at 350 g for 5 min. Cell sediments were labeled using Annexin V-FITC/PI Apoptosis Kit (Elabscience, E-CK-A211) according to the manufacturer’s instructions and detected by BD Accuri C6 (BD Biosciences). The results were processed by FlowJo V10 software.
Immunoblotting and co-immunoprecipitation
During all RS218 infections, unless otherwise indicated, hBMECs lysates were collected for testing at an MOI of 1 for 3 h of infection. hBMECs were first cleaved in RIPA (Proteintech, PR20001) supplemented with 1 mM protease inhibitor PMSF (Solarbio, P0100). For detection of phosphorylated proteins,hBMECs were treated with RIPA combined with phosphatase inhibitor cocktail 2 (Apexbio, K1013). Lysates were mixed with SDS-PAGE polyclonal loading buffer (Solarbio, P1040), treated at 95 °C for 10 min, centrifuged at 12,000 × g for 5 min at 4 °C. 20 μg of lysate supernatants were loaded onto 4–20% FuturePAGE (ACE Biotechnology, ET15420LGel), protein gel electrophoresis was performed by an electrophoresis system (Tanon, EPS 300), and proteins were then transferred to 0.2 μm or 0.45 μm PVDF membranes (Millipore, ISEQ00010 and IPVH00010) according to target protein size. The PVDF membranes then blotted with rabbit anti-caspase 3/p17 antibody (Proteintech, 19677-1-AP, 1:2000), mouse anti-PARP1 Monoclonal antibody (Proteintech, 66520-1-Ig, 1:20000), rabbit anti-GSDMD-NT polyclonal antibody (Bioworld, BS67358, 1:2000), rabbit anti-MLKL Antibody (ABclonal, A19685, 1:2000), rabbit anti-Phospho-MLKL (Ser358) Antibody (Affinity, AF7420, 1:2000), rabbit anti-RIPK1-Specific Polyclonal antibody (ABclonal, A19580, 1:2000), rabbit anti-Phospho-RIPK1 (Ser161) Antibody (Abmart, TA7377, 1:2000), rabbit anti-Phospho-RIPK1 (Ser166) Antibody (Abmart, TA2398S, 1:2000), rabbit anti-RIP3 Polyclonal antibody (Proteintech, 17563-1-AP, 1:2000), rabbit anti-RIP3 (phospho S227) antibody (abcam, ab209384, 1:2000), mouse anti-caspase-1/p10 antibody (Santa Cruz Biotechnology, sc-56036, 1:500), rabbit anti-caspase 8/p43/p18 Polyclonal antibody (Proteintech, 13423-1-AP, 1:1000), rabbit anti-ASC/TMS1 Polyclonal antibody (Proteintech, 10500-1-AP, 1:20000) or rabbit anti-TRIF (TICAM1) antibody (abcam, ab302562, 1:1000) or rabbit anti-ZBP1 Polyclonal antibody (Proteintech, 13285-1-AP, 1:5000) or rabbit anti-TLR4 Polyclonal antibody (Proteintech, 19811-1-AP, 1:2000) or rabbit anti-TNF Receptor I antibody (abcam, ab223352, 1:1000) or rabbit anti-GAPDH polyclonal antibody (Bioworld, AP0063, 1:10000), followed by incubation overnight at 4 °C. Finally, the PVDF membranes were incubated with secondary antibodies, goat anti-rabbit (Bioworld, BS13278, 1:50000) or goat anti-mouse (Bioworld, BS12478, 1:50000) at room temperature for 1 h, followed by 5 times wash for 10 min in PBST before addition of ECL reagent (Millipore, WBKLS0100). Chemiluminescence was detected by an automated chemiluminescence image analysis system (Tanon, Tanon 5200).
In co-immunoprecipitation experiments, cells were then harvested and lysed in NP-40 lysis buffer (Solarbio, N8032) containing protease inhibitors. The lysate was incubated with protein A/G immunoprecipitation beads (Bimake, B23202) combined with 2 μg of polyclonal antibody for 15 min at room temperature. After washing three times with NP-40 lysis buffer, antibody-coated beads were pelleted to ASC complexes from cell lysates at 4 °C. The precipitates were treated in SDS-PAGE loading buffer at 95 °C for 10 min and eluted from the beads. Western blot was then performed as described above. Secondary antibodies were horseradish peroxidase-conjugated goat anti-rabbit IgG heavy chain (H) antibody (Abbkine, A25222) or goat anti-mouse IgG light chain (L) antibody (Abbkine, A25012). Images were grayscale measured by ImageJ and analyzed for changes in expression levels of the indicated proteins in comparison to the internal reference GAPDH. The images shown were representative of three independent replicates.
Immunofluorescence
Cells were seeded into confocal dishes and cultured until reaching a confluent monolayer. Supernatants were discarded and washed three times with PBS. DMEM medium containing RS218 at an MOI of 1 was added and incubated for 3 h at 37 °C in 5% CO2. After washing 3 times with PBS, cells were fixed with 5% paraformaldehyde at room temperature for 20 min and washed three times with PBS for 5 min. Cell membranes were punched for 5 min at room temperature with 0.1% Triton-100 and washed three times with PBS for 5 min. Cells were then blocked with 5% BSA for 1 h at room temperature and labeled overnight in blocking buffer containing the following antibodies: rabbit anti-ASC/TMS1 Polyclonal antibody (Proteintech, 10500-1-AP, 1:2000), mouse anti-caspase 8 antibody (Santa Cruz Biotechnology, sc-56070, 1:200), or mouse anti-caspase-1/p10 antibody (Santa Cruz Biotechnology, sc-56036, 1:200). Next, Cells were incubated with secondary antibodies Goat Anti-Rabbit IgG H&L (Alexa Fluor ® 647) antibody (Abcam, ab150083, 1:1000) and Goat Anti-Mouse IgG H&L (Alexa Fluor ® 488) antibody (Abcam, ab150113, 1:1000) for 1 h at room temperature in the dark. Finally, DAPI was added to label the nuclei. Images were obtained using confocal laser scanning microscopy (ZEISS, LSM 800) and analyzed using ZEISS ZEN 2.3 lite software and ImageJ software. The images shown were representative of three independent replicates.
Caspase-8 activity assay
According to the manufacturer’s instructions, caspase-8 activity assay was performed using FAM-FLICA® Caspase-8 Assay Kit (ImmunoChemistry Technologies, 99). Briefly, hBMECs were seeded into 96-well plates on the bottom of black transparent glass and cultured until reaching a confluent monolayer at 37 °C, 5% CO2. After washing thrice with PBS, RS218 at MOI of 1 was added. At indicated times postinfection, FAM-FLICA working solution was added to each well and incubated for 1 h, during which the culture was gently mixed every 20 min. The supernatant was discarded and washed three times for 10 min each with Apoptosis Wash Buffer (AWB). Finally, FAM-FLICA fluorescence intensity was detected by a multimode microplate reader (TECAN, Spark) at excitation wavelength 488 nm and emission wavelength 530 nm. For image acquisition, Hoechst 33342 was added at an additional ratio of 1:200 per well for 10 min at 37 °C. Cells were washed 3 times with AWB, fixed for 15 min at room temperature with fixative buffer, and collected fluorescence images as described above. All experiments were performed with at least three independent replicates.
Cytokine release detection by ELISA
For tissue samples, fresh tissue was collected and homogenized in 1 mL PBS, and the supernatant was obtained by centrifugation at 5000 g for 10 min at 4 °C. Mouse IL-1β (E-EL-M0037c), IL-6 (E-EL-M0044c), CXCL-1 (E-EL-M0018c), TNF-α (E-EL-M3063) ELISA kits were purchased from Elabscience, and the detections of these cytokines were performed according to the manufacturer’s instructions.
Growth curve
Growth curves were assessed within LB medium. RS218 was inoculated at a 1:100 dilution into 20mL of fresh LB medium, with or without 20 μM Nec-1s or 5 μM GSK'872 or 20 μM Z-IETD-FMK, and growth was monitored using an automated biological growth detection system, RTS1 (BioSan, Riga, Latvia). Growth curves were generated from a minimum of three independent experiments.
Transwell culture and analysis
The in vitro BBB model was established according to the previously described59, with minor optimizations. In brief, cell culture inserts (Corning, CLS3470) pre-coated with collagen (Sigma, 11179179001) were conditioned in medium at 37 °C for 4 h. The inserts were then inverted and placed in a 12-well plate, ensuring that the co-culture medium was level with the bottom of the inserts. U-118MG cells were seeded on the underside of the inserts at a density of 1 × 105 cells/cm2 and incubated overnight at 37 °C and 5% CO2. Subsequently, the inserts were returned to their upright position and transferred to a 24-well plate containing 500 µL of co-culture medium. hBMECs were seeded on the upper surface of the inserts at a density of 1 × 105 cells/cm2 and cultured for 5 days. TEER was measured using an EVOM2 epithelial voltohmmeter with STX2 electrodes to assess the resistance between the luminal (upper) and abluminal (lower) chambers. TEER values from cell-free inserts were used as blanks and subtracted from the experimental readings. The measured resistance (Ω) was multiplied by the membrane surface area (cm2) to obtain the TEER value (Ω•cm2). For permeability assays, 1% NaFI was added to the luminal chamber, followed by incubation at 37 °C with 5% CO2 for 0.5 h. The sample was collected from the abluminal chamber, and fluorescence was measured at excitation and emission wavelengths of 460 nm and 520 nm, respectively. The calculation of the permeability coefficient (Papp) was calculated based on the in vitro BBB model, as described60. All experiments were performed with at least three independent replicates.
Phagocytic function assessment
The phagocytic activity of cells against RS218 was assessed according to established protocols61. Bacterial cultures were added to RAW 264.7 or differentiated THP-1 cells at an MOI of 10. The mixture was centrifuged at 200 g for 5 min and subsequently incubated for 1 h at 37 °C with 5% CO2. Following three washes with PBS, the cells were treated with 200 µg/mL gentamicin for 1 h to eliminate extracellular bacteria. After an additional three washes with PBS, the cells were lysed with 0.1% (v/v) Triton X-100 for 15 min. Cell lysates were serially diluted in PBS and plated onto LB agar, followed by incubation at 37 °C for 12 h. The number of bacterial colonies was enumerated. Phagocytic activity was calculated as the ratio of internalized bacteria at the designated time point to the initial bacterial inoculum. The experiment was conducted with a minimum of three biological replicates.
Statistical analyses
The data in this study were representative of at least three independent experiments. All statistical analyses were performed using GraphPad Prism for two-tailed unpaired Student t tests, one-way ANOVA with Tukey’s post-hoc test or Log-rank (Mantel-Cox) test. All data were represented as mean ± standard deviation (mean ± SD). Differences were considered statistically significant when P < 0.05, where * P < 0.05, ** P < 0.005, *** P < 0.0005, **** P < 0.0001, n,s., no significant difference.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The relevant data supporting the results of this study can be found in the article, the Supplementary Information and the source data. Source data are provided in this paper.
References
Hasbun, R. Progress and challenges in bacterial meningitis: A review. JAMA 328, 2147–2154 (2022).
Liu, Y. et al. Escherichia coli causing neonatal meningitis during 2001–2020: A study in eastern China. Int. J. Gen. Med. 14, 3007–3016 (2021).
Readman, J. B. et al. Cefotaxime/sulbactam plus gentamicin as a potential carbapenem- and amikacin-sparing first-line combination for neonatal sepsis in high ESBL prevalence settings. J. Antimicrob. Chemother. 78, 1882–1890 (2023).
Kim, K. S. Human Meningitis-Associated Escherichia coli. EcoSal Plus 7, https://doi.org/10.1128/ecosalplus.esp-0015-2015 (2016).
Galea, I. The blood-brain barrier in systemic infection and inflammation. Cell. Mol. Immunol. 18, 2489–2501 (2021).
Sweeney, M. D., Zhao, Z., Montagne, A., Nelson, A. R. & Zlokovic, B. V. Blood-brain barrier: From physiology to disease and back. Physiol. Rev. 99, 21–78 (2019).
Tong, X. et al. Targeting cell death pathways for cancer therapy: recent developments in necroptosis, pyroptosis, ferroptosis, and cuproptosis research. J. Hematol. Oncol. J. Hematol. Oncol. 15, 174 (2022).
Wang, Y. & Kanneganti, T.-D. From pyroptosis, apoptosis and necroptosis to PANoptosis: A mechanistic compendium of programmed cell death pathways. Comput. Struct. Biotechnol. J. 19, 4641–4657 (2021).
Lamkanfi, M. & Dixit, V. M. Manipulation of host cell death pathways during microbial infections. Cell Host Microbe 8, 44–54 (2010).
Lalaoui, N. et al. Mutations that prevent caspase cleavage of RIPK1 cause autoinflammatory disease. Nature 577, 103–108 (2020).
Newton, K. et al. Activity of caspase-8 determines plasticity between cell death pathways. Nature 575, 679–682 (2019).
Fritsch, M. et al. Caspase-8 is the molecular switch for apoptosis, necroptosis and pyroptosis. Nature 575, 683–687 (2019).
Conos, S. A. et al. Active MLKL triggers the NLRP3 inflammasome in a cell-intrinsic manner. Proc. Natl, Acad. Sci. USA 114, E961–E969 (2017).
Polykratis, A. et al. A20 prevents inflammasome-dependent arthritis by inhibiting macrophage necroptosis through its ZnF7 ubiquitin-binding domain. Nat. Cell Biol. 21, 731–742 (2019).
Orning, P. et al. Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science 362, 1064–1069 (2018).
Demarco, B. et al. Caspase-8-dependent gasdermin D cleavage promotes antimicrobial defense but confers susceptibility to TNF-induced lethality. Sci. Adv. 6, eabc3465 (2020).
Degterev, A., Ofengeim, D. & Yuan, J. Targeting RIPK1 for the treatment of human diseases. Proc. Natl. Acad. Sci. USA 116, 9714–9722 (2019).
Delanghe, T., Dondelinger, Y. & Bertrand, M. J. M. RIPK1 Kinase-dependent death: A symphony of phosphorylation events. Trends Cell Biol. 30, 189–200 (2020).
Degterev, A. et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 4, 313–321 (2008).
Zhang, Y. et al. RIP1 autophosphorylation is promoted by mitochondrial ROS and is essential for RIP3 recruitment into necrosome. Nat. Commun. 8, 14329 (2017).
Kelliher, M. A. et al. The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity 8, 297–303 (1998).
Shan, B., Pan, H., Najafov, A. & Yuan, J. Necroptosis in development and diseases. Genes Dev. 32, 327–340 (2018).
Takahashi, N. et al. RIPK1 ensures intestinal homeostasis by protecting the epithelium against apoptosis. Nature 513, 95–99 (2014).
Newton, K. et al. RIPK3 deficiency or catalytically inactive RIPK1 provides greater benefit than MLKL deficiency in mouse models of inflammation and tissue injury. Cell Death Differ. 23, 1565–1576 (2016).
Polykratis, A. et al. Cutting edge: RIPK1 Kinase inactive mice are viable and protected from TNF-induced necroptosis in vivo. J. Immunol. Baltim. Md 1950 193, 1539–1543 (2014).
Gillet, Y. et al. Antibiotic treatment of neuro-meningeal infections. Infect. Dis. Now. 53, 104788 (2023).
du Moulin, G. C., Paterson, D., Hedley-Whyte, J. & Broitman, S. A. E. coli peritonitis and bacteremia cause increased blood-brain barrier permeability. Brain Res. 340, 261–268 (1985).
Fu, J. et al. Meningitic Escherichia coli α-hemolysin aggravates blood-brain barrier disruption via targeting TGFβ1-triggered hedgehog signaling. Mol. Brain 14, 116 (2021).
Doerflinger, M. et al. Flexible usage and interconnectivity of diverse cell death pathways protect against intracellular infection. Immunity 53, 533–547 (2020).
Zhou, Y. et al. Hcp family proteins secreted via the type VI secretion system coordinately regulate Escherichia coli K1 interaction with human brain microvascular endothelial cells. Infect. Immun. 80, 1243–1251 (2012).
Kaiser, W. J. et al. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature 471, 368–372 (2011).
Philip, N. H. et al. Caspase-8 mediates caspase-1 processing and innate immune defense in response to bacterial blockade of NF-κB and MAPK signaling. Proc. Natl. Acad. Sci. USA 111, 7385–7390 (2014).
Schwarzer, R., Laurien, L. & Pasparakis, M. New insights into the regulation of apoptosis, necroptosis, and pyroptosis by receptor interacting protein kinase 1 and caspase-8. Curr. Opin. Cell Biol. 63, 186–193 (2020).
Mascarenhas, D. P. A. et al. Inhibition of caspase-1 or gasdermin-D enable caspase-8 activation in the Naip5/NLRC4/ASC inflammasome. PLoS Pathog. 13, e1006502 (2017).
Taabazuing, C. Y., Okondo, M. C. & Bachovchin, D. A. Pyroptosis and apoptosis pathways engage in bidirectional crosstalk in monocytes and macrophages. Cell Chem. Biol. 24, 507–514.e4 (2017).
McQuade, T., Cho, Y. & Chan, F. K.-M. Positive and negative phosphorylation regulates RIP1- and RIP3-induced programmed necrosis. Biochem. J. 456, 409–415 (2013).
Laurien, L. et al. Autophosphorylation at serine 166 regulates RIP kinase 1-mediated cell death and inflammation. Nat. Commun. 11, 1747 (2020).
Li, Y. et al. Myeloid-derived MIF drives RIPK1-mediated cerebromicrovascular endothelial cell death to exacerbate ischemic brain injury. Proc. Natl. Acad. Sci. USA 120, e2219091120 (2023).
Samir, P., Malireddi, R. K. S. & Kanneganti, T.-D. The PANoptosome: A deadly protein complex driving pyroptosis, apoptosis, and necroptosis (PANoptosis). Front. Cell. Infect. Microbiol. 10, 238 (2020).
Kearney, C. J., Cullen, S. P., Clancy, D. & Martin, S. J. RIPK1 can function as an inhibitor rather than an initiator of RIPK3-dependent necroptosis. FEBS J. 281, 4921–4934 (2014).
Orozco, S. et al. RIPK1 both positively and negatively regulates RIPK3 oligomerization and necroptosis. Cell Death Differ. 21, 1511–1521 (2014).
Lin, J. et al. RIPK1 counteracts ZBP1-mediated necroptosis to inhibit inflammation. Nature 540, 124–128 (2016).
Dillon, C. P. et al. RIPK1 blocks early postnatal lethality mediated by caspase-8 and RIPK3. Cell 157, 1189–1202 (2014).
Martens, S., Hofmans, S., Declercq, W., Augustyns, K. & Vandenabeele, P. Inhibitors Targeting RIPK1/RIPK3: Old and New Drugs. Trends Pharmacol. Sci. 41, 209–224 (2020).
Teng, X. et al. Structure-activity relationship study of novel necroptosis inhibitors. Bioorg. Med. Chem. Lett. 15, 5039–5044 (2005).
Cao, L. & Mu, W. Necrostatin-1 and necroptosis inhibition: Pathophysiology and therapeutic implications. Pharmacol. Res. 163, 105297 (2021).
Klück, V. et al. Dapansutrile, an oral selective NLRP3 inflammasome inhibitor, for treatment of gout flares: an open-label, dose-adaptive, proof-of-concept, phase 2a trial. Lancet Rheumatol. 2, e270–e280 (2020).
Madurka, I. et al. DFV890: a new oral NLRP3 inhibitor-tested in an early phase 2a randomised clinical trial in patients with COVID-19 pneumonia and impaired respiratory function. Infection 51, 641–654 (2023).
Delehouzé, C. et al. 6E11, a highly selective inhibitor of Receptor-Interacting Protein Kinase 1, protects cells against cold hypoxia-reoxygenation injury. Sci. Rep. 7, 12931 (2017).
Karki, R. et al. Synergism of TNF-α and IFN-γ triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell 184, 149–168 (2021).
Newton, K. et al. Cleavage of RIPK1 by caspase-8 is crucial for limiting apoptosis and necroptosis. Nature 574, 428–431 (2019).
Mandal, P. et al. RIP3 induces apoptosis independent of pronecrotic kinase activity. Mol. Cell 56, 481–495 (2014).
Pons, S. et al. A high-throughput sequencing approach identifies immunotherapeutic targets for bacterial meningitis in neonates. EBioMedicine 88, 104439 (2023).
Kim, Y.-E. et al. Endothelial SHANK3 regulates tight junctions in the neonatal mouse blood-brain barrier through β-Catenin signaling. Nat. Commun. 16, 1407 (2025).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Shalem, O. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
Tweedell, R. E., Malireddi, R. K. S. & Kanneganti, T.-D. A comprehensive guide to studying inflammasome activation and cell death. Nat. Protoc. 15, 3284–3333 (2020).
Wang, Y. et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature 547, 99–103 (2017).
Bayir, E. & Sendemir, A. In Vitro Human Blood-Brain Barrier Model for Drug Permeability Testing. Methods Mol. Biol. 2367, 73–85 (2021).
Hanada, S. et al. Cell-based in vitro blood-brain barrier model can rapidly evaluate nanoparticles’ brain permeability in association with particle size and surface modification. Int. J. Mol. Sci. 15, 1812–1825 (2014).
Sun, Y. et al. Extraintestinal Pathogenic Escherichia coli Utilizes Surface-Located Elongation Factor G to Acquire Iron from Holo-Transferrin. Microbiol. Spectr. 10, e0166221 (2022).
Acknowledgements
This work was supported by Outstanding Youth Fund of Jiangsu Provincial Natural Science Foundation (BK20240090 to F.T.) and the National Natural Science Foundation of China (32172858 to F.T.). We sincerely thank Prof. Junying Yuan and Prof. Daichao Xu for generously providing the Ripk1 D138N/D138N mice, and Prof. Xin Lin for generously providing the Casp8 -/- Ripk3 -/- mice used in this study. We sincerely appreciate the technical support and valuable guidance provided by Prof. Zhe Ma from Nanjing Agricultural University for this study.
Author information
Authors and Affiliations
Contributions
X.W. conceptualized the study, performed most experiments, curated data, conducted formal analysis, created visualizations, and wrote the original manuscript. J.D. and F.T. contributed to conceptualization, project administration, supervision, and writing review/editing. X.C., J.W., and Y.S. participated in data curation and validated results. Y.Z. and K.F. assisted with formal analysis and methodology, while J.R. and F.X. contributed to investigation and resources. J.C. helped with methodology. All authors have read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, X., Zhang, Y., Chen, X. et al. RIPK1 kinase drove brain microvascular endothelial cells death and blood-brain barrier disruption in neonatal Escherichia coli meningitis. Nat Commun 16, 7309 (2025). https://doi.org/10.1038/s41467-025-62760-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-62760-4