Abstract
Vertebrate inner ear mechanosensory hair cells detect sound and gravitational forces. Additionally, fishes have homologous lateral line hair cells in the skin that detect water vibrations for orientation and predator avoidance. Hair cells in the lateral line and ear of fishes and other non-mammalian vertebrates regenerate readily after damage, but mammalians lack this ability, causing deafness and vestibular defects. As yet, experimental attempts at hair cell regeneration in mice result in incompletely differentiated and immature hair cells. Despite differences in regeneration capabilities, the gene regulatory networks (GRNs) driving hair cell maturation during development are highly similar across vertebrates. Here, we show that the transcription factor prdm1a plays a key role in the hair cell fate GRN in the zebrafish lateral line. Mutating prdm1a respecifies lateral line hair cells into ear hair cells, altering morphology and transcriptome. Understanding how transcription factors control diverse hair cell fates in zebrafish is crucial for understanding the yet unsolved regeneration of diverse hair cells in mammalian ears to restore hearing and balance.
Similar content being viewed by others
Introduction
Vertebrates possess mechanosensory hair cells in the ear that transduce vibrations into nerve impulses interpreted as sound1,2. Fishes have two separate sensory organ systems with functionally and genetically similar mechanosensory cells; the ear responsible for hearing and orientation, and the lateral line responsible for sensing water vibrations and detecting external physical stimuli (Fig. 1a–c)3. Within the ear, there are several sensory epithelia containing diverse hair cells with specialized functions4,5,6. In the mammalian cochlea, inner hair cells (IHCs) and outer hair cells (OHCs) are responsible for hearing1, while Type I and Type II hair cells in the saccule and utricle detect linear acceleration, and the cristae of the semicircular canals detect rotational acceleration. In the saccule and utricle the two types of hair cells are divided into striolar and extra-striolar regions based on polarity and location within the organs7. Early developmental specification of these different hair cell types is controlled by common gene regulatory networks (GRNs)8,9,10, but control of terminal differentiation is exerted by distinct GRNs that result in different functions and physical characteristics11,12,13. The precise specification of each hair cell type in the ear is essential for hearing and vestibular function, and incorrect developmental fate leads to profound hearing loss1,14,15,16.
a 5 dpf representative zebrafish embryo with hair cells expressing Tg(myo6b:H2B-mScarlet) in magenta and support cells expressing Tg(she:H2B-EGFP) in green, respectively. Scale bar= 100 µm. b Modified schematic of the lateral line with hair cells and support cells in magenta and green, respectively20. c Schematic of the zebrafish ear. AC anterior crista, LC lateral crista, PC posterior crista, AM anterior macula, PM posterior macula. d HCR of lateral line neuromasts and cristae of the zebrafish ear with prdm1a probe and Tg(pou4f3:GAP-EGFP) in hair cells. Representative image of one of three larvae. Scale bars= 20 µm. e prdm1a expression in the lateral line during homeostasis, and during regeneration at 30 min and 5 h post-neomycin, with Tg(pou4f3:GAP-EGFP) in hair cells. HC hair cells, SC support cells. Representative images of one of six larvae. f Schematic of Prdm1a protein with major domains and stop codon in prdm1a mutant (black arrow). g Violin plot of prdm1a expression from sibling scRNA-seq, divided by cell type. h Hair cells labeled with Tg(myo6b:H2B-mScarlet) show developmental defects in prdm1a mutant neuromasts at 2 and 7 dpf compared to siblings. Scale bars= 20 µm. i Quantification of hair cells during development in sibling and prdm1a-/- neuromasts. n = 6–10 fish per timepoint and genotype, 3–4 neuromasts per fish. Dots represent individual neuromasts. n = 27 for 2 dpf sibling, n = 23 for 2 dpf prdm1a-/-, n = 18 for 3 dpf sibling, n = 22 for 3 dpf prdm1a-/-, n = 18 for 5 dpf sibling, n = 38 for 5 dpf prdm1a-/-, n = 8 for 7 dpf sibling, and n = 26 for 7 dpf prdm1a-/-. Two-way Anova with Tukey’s multiple comparisons test, ***p < 0.001, ****p < 0.0001. The box plots show the interquartile range (IQR), with the median (central line) and hinges (25th and 75th percentiles), and the whiskers extending from the box to the minimum and maximum values. j Hair cells labeled with Tg(myo6b:H2B-mScarlet) show regeneration defects in prdm1a-/- neuromasts 48 hours after neomycin treatment; 7 dpf. Scale bars= 20 µm. k Quantification of hair cells during regeneration and homeostasis at the same developmental timepoint, in sibling and prdm1a-/- neuromasts. The percentage of regenerated hair cells compared to homeostasis for each condition is shown above the regeneration counts. n = 6–10 fish per timepoint and genotype, 3–4 neuromasts per fish. n = 8 for 7 dpf sibling, n = 26 for 7 dpf prdm1a-/-, n = 30 for 48 hr post neomycin sibling, and n = 35 for 48 hr post neomycin prdm1a-/-. One-way Anova with Tukey’s multiple comparisons test, ****p < 0.0001. The box plots show the interquartile range (IQR), with the median (central line) and hinges (25th and 75th percentiles), and the whiskers extending from the box to the minimum and maximum values. l EdU (green) incorporation and hair cells expressing Tg(myo6b:H2B-mScarlet) (magenta) during development and regeneration in siblings and mutants. Scale bars= 20 µm. Quantification of EdU incorporation in differentiating divisions (m) and amplifying divisions (n) during regeneration in sibling and prdm1a-/- neuromasts. n = 6–8 fish per treatment and genotype, 3–4 neuromasts per fish. n = 18 for homeostasis sibling, n = 25 for homeostasis prdm1a-/-, n = 16 for neomycin sibling, n = 25 for neomycin prdm1a-/-. One-way Anova with Tukey’s multiple comparisons test, ****p < 0.0001. The box plots show the interquartile range (IQR), with the median (central line) and hinges (25th and 75th percentiles), and the whiskers extending from the box to the minimum and maximum values.
The ears of aquatic vertebrates also have a similar diversity of hair cell types and functions (Fig. 1a, c)17. Like mammals, adult fishes possess vestibular hair cells in cristae, utricles, saccules and the later-developing lagena. Fishes do not possess a cochlea, but the utricle, saccule and lagena also serve an auditory function17. Fish lateral line hair cells share genetic, functional, and structural similarity with ear hair cells of many species6,18, including core gene networks that regulate their early specification8,19,20. Likewise, genes linked to deafness in mammals cause defects in hair cell function when mutated in zebrafish21,22,23.
Despite these similarities, there is a striking disparity of hair cell regeneration between fish and mammals. While hair cells in non-mammalian vertebrates regenerate following death6,24,25, mammalian hair cells lack this ability9,26,27, causing hearing loss and vestibular dysfunction in humans28,29. During development and regeneration of the zebrafish lateral line, hair cells arise from central support cells that turn into hair cell progenitors, then divide and form young hair cells before differentiating into mature hair cells (Fig. 1b)19,30. Attempts at stimulating hair cell regeneration in mammalian cochleae through manipulating signaling pathways or gene therapy result in incomplete differentiation or hair cells of only one type31,32,33,34,35,36,37,38,39,40. Therefore, understanding the unique developmental requirements for each hair cell type is a prerequisite to any attempt to restore proper function. As mammals lack the molecular and genetic programs to regenerate hair cells, studying the process in a robustly regenerating model provides a powerful tool to identify missing components and gain insights for broader applications.
We recently performed a single-cell RNA sequencing (scRNA-seq) time course of lateral line hair cell regeneration, and identified genes and pathways necessary for proliferation and regeneration19. In addition, we and others have characterized single-cell transcriptomic profiles of the zebrafish lateral line and ear during homeostasis and development to provide a comprehensive transcriptional atlas5,20,41. We searched these datasets for genes possibly involved in regeneration and identified the transcription factor prdm1a, which is expressed both during hair cell differentiation and regeneration in the lateral line. prdm1a is not expressed in ear hair cells12, pointing to a unique role in hair cell specification and regeneration in the lateral line. Prdm1a is a direct and indirect transcriptional repressor, but can also activate certain genes42,43,44. In mammals, Prdm1 (Blimp1) directs cell fate decisions in a variety of tissues and organs45,46,47. In zebrafish, prdm1a is a central regulator of neural crest cell proliferation and fate decisions during neurogenesis and specification of slow-twitch muscle fibers48,49,50. prdm1a, therefore, has hallmarks of a gene that controls cell fate, identity, and downstream functional properties across species51 and is a prime candidate for investigations into proliferation and hair cell differentiation and regeneration.
In this work, we mutated prdm1a and observed profound defects in the development and regeneration of lateral line hair cells, and a decrease in support cell proliferation during regeneration. Using scRNA-seq, we characterized transcriptional changes associated with this mutation, and found evidence of a fate switch from lateral line to an ear-like hair cell state, with de-repression of genes normally expressed only in ear hair cells. Morphological characterization of mutant hair cells also indicates a transformation to an ear hair cell-like morphology. We show that the ectopically expressed genes in prdm1a mutants possess enhancers and promoters that respond to direct transcriptional repression by Prdm1a. Lastly, we present a GRN model to describe upstream and downstream genetic interactions of prdm1a in the lateral line that places the gene into the broader context of hair cell specification and regeneration. This GRN shows striking similarities to the core regulatory code shared between lateral line and ear hair cells of fish and mammals. We show that, even though the hair cell GRN is highly similar between the ear and the lateral line, the lack of expression of prdm1a in the ear but continued expression of prdm1a in the lateral line leads to the differentiation of lateral line hair cells instead of ear hair cells.
Results
prdm1a expression is integral to the lateral line and hair cells during development and regeneration
prdm1a is expressed in central support cells during the early stages of regeneration and subsequently in differentiating hair cells (Fig. 1g)19. To analyze hair cell expression in more detail, we performed scRNA-seq on 5 days post fertilization (dpf) homeostatic prdm1a mutants (described below) and siblings (Supplementary Fig. 1g). We confirmed that prdm1a partially overlaps with the hair cell specifying gene atoh1a and is strongly expressed in young and mature hair cells and sometimes in a few support cells (Supplementary Fig. 1h, i). prdm1a truncated transcript is also detected in mutant cells, even though it is non-functional. We validated the expression results with in situ hybridization chain reaction (HCR) in larvae, showing that prdm1a is expressed in all hair cells of the lateral line. prdm1a, however, is excluded from hair cells in the ear, which still express other hair cell marker genes such as pou4f3 (Fig. 1d, e). To characterize prdm1a’s spatiotemporal expression during regeneration, we killed hair cells using neomycin52 and performed HCR. We observed a strong upregulation of prdm1a expression in central support cells 30 min and 1 h after killing hair cells (Fig. 1e and Supplementary Fig. 1a). Some of these central support cells will proliferate and differentiate into regenerated hair cells19,30. As regeneration progresses, prdm1a expression is once again restricted to newly regenerated hair cells 5 h after hair cell killing, overlapping with pou4f3:EGFP expression.
We previously identified a mutation in prdm1a in an unpublished chemical mutagenesis screen, allowing us to test its function in vivo. The mutation causes a premature stop codon in prdm1a, truncating the zinc finger DNA-binding domain, rendering the protein unable to bind to DNA and regulate target genes (Fig. 1f, black arrow). Consistent with previous findings, we see defects in slow muscle fibers along the myoseptum and in melanocytes48,53,54. The defect in myoseptum formation causes the random or stalled migration of the posterior lateral line primordium, leading to irregularly deposited neuromasts along the trunk (Supplementary Fig. 1d, e)55. The larvae also have small, deformed pectoral fins, but look morphologically relatively normal.
We assayed lateral line hair cell development in prdm1a mutants, from 2 dpf through neuromast maturation and homeostasis at 7 dpf. We find a failure of prdm1a mutant hair cells to increase in number starting at 2 dpf (Fig. 1h, i; Supplementary Fig. 1f). The result of this developmental stalling in hair cell formation is a permanent reduction in the number of hair cells compared to sibling embryos. To evaluate the impact of the prdm1a mutation on regenerative ability, we killed the hair cells and assayed regeneration in siblings and mutants 48 h later. In siblings, we observed an average of 11–12 regenerated hair cells, or 55.3% of homeostasis, and four hair cells, or 52.0% of homeostasis on average in mutants (Fig. 1j, k). Since prdm1a mutant neuromasts possess on average 6 or 7 hair cells before neomycin treatment and the regeneration rates are very similar, we conclude that regeneration of hair cells is unaffected beyond the developmental defects we observed.
Hair cell regeneration in the lateral line also crucially depends on proliferation of two different support cell types. Central support cells undergo differentiating divisions and develop into regenerated hair cells, and peripheral support cells undergo amplifying divisions to replenish the central support cell pool19,30. As a prdm1a mutation decreases proliferation in other zebrafish tissues50,56, we performed EdU incorporation experiments to assay proliferation in siblings and prdm1a mutants. Differentiating divisions result in EdU-positive hair cells, while amplifying divisions result in EdU-positive support cells. During both development and regeneration, the ratio of peripheral, amplifying support cells marked by EdU is significantly lower in the prdm1a mutant embryos (Fig. 1l, n). This indicates a non-cell autonomous role for prdm1a in proliferation, as it is not expressed in these amplifying, peripheral support cells. We did not, however, observe a difference in EdU in hair cells themselves (Fig. 1l, m), indicating that differentiating divisions are not affected by loss of prdm1a, and that prdm1a plays a proliferation-independent role in hair cells, consistent with our observations about similar rates of regeneration overall.
Together, these results point to dual functions for prdm1a in the lateral line. prdm1a plays an indirect, non-cell autonomous role in amplifying support cell proliferation, which likely underlies the defect in the development of a normal number of hair cells. While the role of prdm1a in proliferation requires further investigation, in this manuscript, we explore the previously unknown cell-autonomous role that prdm1a plays in postmitotic hair cells.
scRNA-seq indicates a cell fate switch in prdm1a mutant hair cells
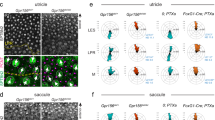
The hair cell-specific and regeneration-responsive expression of prdm1a in the lateral line led us to conduct a scRNA-seq experiment with prdm1a mutants and siblings to determine what role it might be playing in hair cell differentiation. We FAC-sorted 5 dpf zebrafish embryos using a transgenic line driving mCherry in all lateral line cells (Tg(she:mCherry)), separately sorting prdm1a mutant and sibling embryos, and performed scRNA-seq using the 10X Chromium platform. The resulting datasets were subsequently integrated for downstream analysis. All expected lateral line cell types were present, including sibling and mutant hair cells along our previously established developmental trajectory19 (Supplementary Fig. 1g, Supplementary Data 1). We subsetted the integrated hair cell lineage from central support cells to mature hair cells, as these are the cell types that express prdm1a in the lateral line. Only after submitting, we observed a split in the hair cell developmental trajectory at the young hair cell stage (Fig. 2a, green cells), with hair cells located in two transcriptionally distinct clusters after the split. Strikingly, when we color the cells by genotype, the prdm1a mutant hair cells segregate from sibling hair cells (Fig. 2b). The split in hair cell trajectory occurs when prdm1a is first expressed (Fig. 2c, black arrow), suggesting that the activity of prdm1a is integral to hair cell fate. We performed a differential gene expression analysis between sibling and mutant hair cell clusters to determine the transcriptional changes driving the separate trajectory and clustering of mutant hair cells and identified 1589 upregulated and 344 downregulated genes (Supplemental Data 2). As prdm1a acts mostly as a repressor, we focused on the larger set of upregulated genes as likely targets of direct repression, while acknowledging the possibility of indirect interactions as well. These ectopic genes are upregulated primarily in mutant hair cells after they differentiate (Fig. 2d–p; Supplementary Fig. 2a–h). The prdm1a mutant hair cells still express canonical hair cell markers, such as myo6b, pou4f3, gfi1aa/ab, and atoh1a/b (Fig. 2d) and they also express genes typical for mature hair cells, such as fgf10b and the tip link component pcdh15a (Supplementary Fig. 2m, n), suggesting that they are not delayed in their development. Many of the upregulated genes cause auditory dysfunction in mice and/or humans when mutated, pointing to their function in ear hair cells (mutant phenotypes reviewed in Supplementary Table 1A)14,57,58,59,60,61,62,63,64,65,66,67,68,69. We subsetted scRNA-seq data for all other neuromast cell types, but found no additional separate clustering based on prdm1a genotype, supporting a hair cell-specific effect of prdm1a on gene regulation.
a UMAP plot of prdm1a-/- and sibling scRNA-seq, from central support cells to mature hair cells, colored by cell type. Black arrows indicate developmental trajectory. b UMAP plot of prdm1a-/- and sibling scRNA-seq, from central support cells to mature hair cells, colored by genotype. c Feature plot of prdm1a expression in the scRNA-seq hair cell lineage sub-clustering. d Heatmap of selected genes upregulated in prdm1a-/- hair cells, and non-differentially expressed hair cell marker genes. e–h Feature plots of pvalb9, ckbb, s100a1 and tmc2a from the hair cell trajectory sub-clustered scRNA-seq. i–p HCRs of pvalb9, ckbb, s100a1 and tmc2a in 5 dpf sibling and prdm1a-/- neuromasts. Representative images of two independent experiments. q–t HCRs of pvalb9, ckbb, s100a1 and tmc2a in the lateral crista of the ear in Tg(myo6b: H2B-mScarlet) larvae, with hair cells are labeled in magenta. Scale bars= 10 µm. Representative images of two independent experiments.
To validate the expression of genes upregulated in prdm1a mutants, we performed HCRs for pvalb9, ckbb, s100a1, tmc2a, strc, kncn, lhfpl5a, tbx2a, and tbx2b and found them to all be strongly expressed in the lateral line hair cells of prdm1a mutants, but not sibling hair cells (Fig. 2i–p, Supplementary Fig. 2e–h). On the other hand, all these genes are strongly expressed in sibling hair cells in the ear (Fig. 2q–t and Supplementary Fig. 2i, j). Of particular interest are the family of Tmc genes, which are members of the mechanotransduction complex, as they are differentially expressed in lateral line and ear hair cells70,71,72,73. In the sibling lateral line, tmc2a is only expressed in a few sibling lateral line hair cells, but is expressed in all ear (lateral crista) hair cells (Fig. 2h, o, p, t)73. In prdm1a mutants all lateral line hair cells express tmc2a, suggesting that mutant lateral line hair cells acquired an ear hair cell fate (Fig. 2p). On the other hand, the paralog tmc2b is highly expressed in the sibling and prdm1a mutant lateral line hair cells, suggesting that it is not regulated by prdm1a (Supplementary Fig. 2l). Also, in the lateral crista in the ear tmc2b is only expressed in a few hair cells that do not appear to express tmc2a (Supplementary Fig. 2l, t).
The hypothesis that loss of prdm1a leads to a fate switch is supported by our analysis of the genes upregulated in prdm1a mutant lateral line hair cells in published zebrafish ear and mammalian ear scRNA-seq data5,12,13,74,75. We used the gEAR data exploration tool76 to compile a table of expression of these genes, and show that they are normally expressed in zebrafish and mammalian ear hair cells (Supplementary Table 1). Together, the expression analyses strongly suggest that loss of prdm1a causes lateral line hair cells to adopt an ear hair cell fate.
prdm1a regulates the expression of target genes as a transcription factor
prdm1a is a strong repressor with a DNA-binding zinc finger domain, and a large number of genes are upregulated specifically in hair cells when it is mutated. Therefore, we asked if Prdm1a regulates these genes by binding to their enhancers and promoters. We performed ATAC-seq on FAC-sorted wild-type lateral line cells during homeostasis. We then performed a motif enrichment analysis specifically on the putative regulatory regions and promoters of genes upregulated in the prdm1a mutant hair cells. We find that the Prdm1 binding site is highly enriched in these putative enhancers and promoters (Fig. 3a and Supplementary Fig. 3a), suggesting that Prdm1a directly controls these genes. To test these interactions, we cloned putative regulatory elements of the prdm1a target s100a1 into a construct driving EGFP (Fig. 3a). In wild-type embryos, the reporter construct for regulatory elements of s100a1 drives EGFP strongly in ear hair cells, but not lateral line hair cells (Fig. 3b–d), recapitulating endogenous ear expression and demonstrating that these regulatory elements are ear hair cell-specific. We also injected the s100a1 reporter construct into prdm1a mutant embryos and observed ectopic expression in mutant lateral line hair cells (Fig. 3e). These results show that Prdm1a interacts with the regulatory elements of at least some of its putative target genes, and that loss of Prdm1a binding to these elements in reporter constructs leads to ectopic expression in lateral line cells, recapitulating the de-repression of the genes in the mutants.
a Schematic of the s100a1 locus showing lateral line (gray) or ear (black) ATAC-seq reads with location of Prdm1 binding motifs in red. Exons and introns in purple. Cloned enhancer-promoter fusion of Tg(s100a1:H2B-EGFP) in green. s100a1 HCR (yellow) and H2B-GFP expression (cyan) in 5 dpf wild type embryos driven by Tg(s100a1:H2B-EGFP) in the posterior macula (b) and lateral crista (c). 5 dpf sibling neuromast (d) and prdm1a-/- dorsal neuromast (e). Scale bars= 10 µm. Representative images of two independent experiments. f Illustration of the zebrafish ear from a ventral view and orientation of the larva during imaging. AC anterior crista, LC lateral crista, PC posterior crista, AM anterior macula, PM posterior macula, NM neuromast. Ventral view of the zebrafish ear. Created in BioRender. Piotrowski, T. (2025) https://BioRender.com/vl2w553 (g), HCR with a prdm1a probe (magenta) in a Tg(myo6b:prdm1a) larva (h). Generated using Biorender. Representative images of six independent experiments. Scale bars= 20 µm (i–k) HCR signals for prdm1a (yellow), pvalb9, s100a1, and ckbb (cyan) in the lateral crista of sibling and the Tg(myo6:prdm1a; cryaa:mTurqoise2) embryos, respectively. Scale bars= 10 µm. Images are representative of embryos from two independent experiments. l Quantification of fluorescent signal from HCRs of pvalb9 (p = 0.0081), s100a1 (p = 0.0018), ckbb (p = 0.0003), tbx2b (p = 0.3699), and tmc2b (p = 0.2497). n = 8 embryos (two ears each) imaged per probe and genotype. Multiple unpaired two-sided t-test, ns>0.05, **p < 0.01, ***p < 0.001. Data are presented as mean ± SD. m HCRs of ckbb in sibling and prdm1a mutant neuromasts. Images representative of two independent experiments. n Expression of ckbb in prdm1a-/- hair cells using the Tg(myo6:prdm1a; cryaa:mTurqoise2) line downregulates the expression of ckbb. Scale bars= 10 µm. Images representative of two independent experiments. o Quantification of ckbb expression in the various conditions. n = 12 for sibling, n = 11 for prdm1a-/-, n = 15 for ectopic prdm1a-expressing sibling and n = 8 for ectopic prdm1a-expressing prdm1a-/-. Two-way Anova with Tukey’s multiple comparisons test, *p < 0.05, **p < 0.01(**p = 0.0069), ***p < 0.001, ****p < 0.0001. Data are presented as mean ± SD. p Quantification of tmc2a HCR fluorescence intensity in the various conditions. n = 21 for sibling, n = 12 for prdm1a-/-, n = 23 for ectopic prdm1a-expressing sibling and n = 23 for ectopic prdm1a-expressing prdm1a-/-. Two-way Anova with Tukey’s multiple comparisons test, *p < 0.05(*p = 0.0262), **p < 0.01, ***p < 0.001, ****p < 0.0001. Data are presented as mean ± SD.
We also investigated the ability of Prdm1a to regulate its putative target genes by ectopically expressing it in all ear hair cells using a myo6b:prdm1a driver (Fig. 3i–k). We performed HCRs for three genes upregulated in mutant lateral line hair cells and quantified expression in the lateral cristae of the ear in ectopic prdm1a-expressing larvae and siblings. The cristae were imaged from a ventral view (Fig. 3f–h). The putative prdm1a target genes pvalb9, s100a1, and ckbb are repressed in the ear in the presence of ectopic prdm1a as indicated by reduced HCR signal intensity (Fig. 3i–l). tbx2b, the ortholog of mammalian Tbx2, is an essential transcription factor for hair cell fate and development in ear hair cells14,77, and is expressed in ear hair cells and support cells (Supplementary Fig. 2n). After ectopic expression of prdm1a in ear hair cells, tbx2b is not significantly repressed in hair cells. As tmc2b is expressed in both sibling and prdm1a mutant lateral line hair cells (Supplementary Fig. 2l) it served as a control and was indeed not affected by the ectopic expression of prdm1a in the ear (Fig. 3l). We also tested if overexpression of prdm1a represses expression of selected ectopically expressed ear genes in prdm1a mutant hair cells and indeed observed significant downregulation of ckbb and tmc2a (Fig. 3m–p).
Taken together, the derepression of prdm1a targets in the lateral line, the repression of these targets upon ectopic prdm1a expression in the ear, and the activity of prdm1a target gene enhancers all demonstrate a direct genetic interaction between prdm1a and at least some of its targets and highlight the central role that prdm1a plays in hair cell fate.
prdm1a mutant hair cells have structural changes concordant with expression changes
Different zebrafish hair cell populations exhibit different physical characteristics that are molecularly determined, including length of kinocilia16,78,79,80. Microtubule-based kinocilia are components of the mechanosensory apparatus at the apex of the hair cell. To test if the prdm1a mutation impacts these features, we used a pou4f3:gap43-EGFP transgenic line and measured kinocilia length in sibling and mutant lateral line hair cells. Sibling kinocilia are 23 µm in length, and mutant kinocilia are on average 12 µm long. Thus, prdm1a mutant hair cells show a significant reduction in kinocilia length (Fig. 4a–c). Macula and cristae in the ear have kinocilia lengths of 8 µm and 30 µm, respectively16,78,79. Therefore, a shortening of lateral line kinocilia resembles a macular hair cell morphology, as macula hair cells possess the shortest kinocilia of zebrafish hair cell populations. We also measured the kinocilia lengths of hair cells of the ear lateral crista in the myo6b:prdm1a larvae, and observed a reduction in length from 30 µm to 23 µm in the presence of ectopic prdm1a (Fig. 4d, e). This shortening of kinocilia is concordant with the repression of prdm1a target genes observed in the same transgenic embryos (Figs. 3i-l), and a partial fate shift toward lateral line characteristics of shorter kinocilia. Mutant lateral line hair cells also possess a higher cell volume (Fig. 4f, g) and an enlarged cuticular plate area (Fig. 4h, i). However, hair cell polarity is not affected (Fig. 4h, j). Mutant lateral line hair cells are likely functional as they take up Daspei, and they can be killed by neomycin, which requires functional mechanosensory channels (Fig. 4k, l).
a,b Tg(pou4f3:EGFP) in sibling and prdm1a-/- neuromast hair cells. c Quantification of neuromast kinocilia length in sibling and prdm1a-/- hair cells. n = 11 sibling neuromasts, 89 kinocilia; n = 21 prdm1a-/- neuromasts, 117 kinocilia. Student’s two-sided t-test, ****p = 8.165*10-73. Data are presented as mean ± SD. d Tg(pou4f3:EGFP) expression in sibling and Tg(myo6b:prdm1a) cristae. Scale bars= 10 µm. e Quantification of Tg(myo6b:prdm1a) cristae kinocilia length. n = 15 sibling cristae, 226 kinocilia; n = 15 Tg(myo6b:prdm1a) cristae, 258 kinocilia. Student’s two-sided t-test, ****p = 7.314*10-54. Data are presented as mean ± SD. f Tg(pou4f3:EGFP) labeling cell bodies in sibling and prdm1a-/- lateral line hair cells. g Quantification of cell body volume in sibling and prdm1a-/- lateral line hair cells. n = 12 sibling neuromasts, 37 cell bodies; n = 11 prdm1a-/- neuromasts, 24 cell bodies. Student’s two-sided t test, **p = 0.0026. Data are presented as mean ± SD. h Phalloidin staining of sibling and prdm1a-/- neuromasts. i Quantification of cuticular plate area in sibling and prdm1a-/- neuromasts. n = 14 sibling neuromasts, 29 cuticular plates; n = 10 prdm1a-/- neuromasts, 23 cuticular plates. Student’s two-sided t-test, ****p = 6.296*10-5. Data are presented as mean ± SD. j Quantification of the percentage of anterior-polarized hair cells per neuromast. n = 14 sibling neuromasts and n = 10 prdm1a-/- neuromasts. Student’s two-sided t-test p = 0.2916. Data are presented as mean ± SD. k DASPEI uptake in sibling and prdm1a-/- lateral line hair cells. Representative of two independent experiments with similar results. l Tg(pou4f3:EGFP) in sibling and prdm1a-/- lateral line hair cells during homeostasis and 2 h after neomycin treatment. Representative of two independent experiments with similar results. m Tg(myo6b:prdm1a) overexpression in prdm1a-/- hair cells. n Quantification of kinocilia length in Tg(myo6b:prdm1a) positive and negative prdm1a-/- hair cells. n = 12 Tg(myo6b:prdm1a) negative neuromasts, 12 kinocilia; n = 19 Tg(myo6b:prdm1a) positive neuromasts. Student’s two-sided t-test p = 0.0174. Data are presented as mean ± SD. All Scale bars = 10 µm.
As prdm1a is also expressed in support cells during early stages of regeneration, we wondered in the hair cell defect is possibly non-cell autonomous. However, overexpression of prdm1a with the hair cell-specific myo6b-promoter increases the kinocilia length, demonstrating that the kinocilium defect in prdm1a mutant hair cells is, in part, cell-autonomous (Fig. 4m, n).
Taken together, the changes in kinocilia length when manipulating prdm1a expression in different hair cell types provide evidence that the transcriptional changes we observe correlate with physical differences in hair cells, reinforcing the idea of prdm1a controlling hair cell fate in the zebrafish.
Integrated scRNA-seq analyses suggest prdm1a mutant hair cell fate switch to striolar hair cells
While our data provide convincing evidence that prdm1a is central to zebrafish lateral line hair cell fate, we asked how complete the conversion to an ear hair cell fate is in the absence of prdm1a. We therefore integrated our lateral line scRNA-seq data with two published scRNA-seq studies of zebrafish ear hair cells. Specifically, we integrated an otic subset from a whole-embryo developmental atlas and an ear-specific dataset5,75, which we term “embryo otic subset” and “ear scRNA-seq” respectively (Fig. 5a–e, Supplementary Data 3). We observed clustering of the prdm1a mutant lateral line hair cells with wildtype ear hair cells from both ear datasets, indicating that prdm1a mutant lateral line hair cells share transcriptional similarity with ear hair cells (Fig. 5e, Supplemental Data 4). When we label the ear hair cell clusters by annotated cell type5, the prdm1a mutant hair cells overlap most with striolar hair cells of the macula (Fig. 5f, Supplementary Data 4), consistent with the shortening of kinocilia. In addition, a differential gene expression analysis comparing prdm1a mutant lateral line, sibling lateral line, and ear hair cells shows that a large group of genes is shared between the mutant lateral line and ear hair cells, but not sibling lateral line (Fig. 5g, Supplementary Data 5). While the fate switch is not complete based on transcriptional changes, as some lateral line-specific hair cell genes in the prdm1a mutant hair cells are not completely downregulated (Supplementary Data 4 and 5), the fate change is nevertheless dramatic.
a–e UMAPs of integrated scRNA-seq, showing sibling lateral line, prdm1a-/- lateral line, embryo otic subset, ear scRNA-seq, and complete integration, respectively. f Integrated ear prdm1a-/- hair cell cluster with cells color coded by cell type from the ear scRNA-seq annotation5. prdm1a-/- hair cells are in gray. g Heatmap of mature hair cell genes of the triple integrated scRNA-seq data set, divided into prdm1a-/- lateral line cells, ear hair cells of the embryo otic subset and ear scRNA-seq data sets. h GRN of zebrafish lateral line hair cell specification and development. i GRN of zebrafish ear hair cell specification and development. j GRN of mouse inner hair cell specification and development. k GRN of mouse outer hair cell specification and development. Numbers in brackets indicate citations for the individual interactions.
Here, we show that within the lateral line, prdm1a promotes the proliferation of support cells and the development and regeneration of lateral line hair cells. prdm1a also suppresses an ear hair cell fate through its role as a transcriptional repressor. Loss of prdm1a in the lateral line leads to de-repression of a large group of ear-specific hair cell genes, resulting in physical changes in lateral line hair cells consistent with an ear macula hair cell identity. We provide multiple lines of evidence—through scRNA-seq, ATAC-seq, reporters of regulatory element activities, and ectopic prdm1a expression experiments—that prdm1a represses ear fate in lateral line hair cells. Importantly, our integrated scRNA-seq analysis of prdm1a mutant and wild-type ear hair cells shows that the fate conversion is almost complete. Therefore, the expression of prdm1a during hair cell maturation reflects an inflection point in the determination of hair cell fate between the lateral line and the ear. The differential regulation of prdm1a in the lateral line and the ear likely played an important role in the evolution of these two hair cell types and also demonstrates that individual transcription factors play key roles in determining hair cell subtypes, also during regeneration.
Discussion
Questions of how individual genes and gene regulatory networks determine cell fate are central to developmental biology, and foundational works in the field have broad influence over research to this day81,82,83,84,85. We investigated the role of prdm1a, central to cell fate decisions, with wide implications for linking evolution and specification of cell fate with regenerative genetics to restore hearing. We uncovered two roles for prdm1a in the lateral line: determining hair cell fate and promoting proliferation during development and regeneration.
We focused on the significant role of prdm1a in determining hair cell fate. Our integration of prdm1a mutant scRNA-seq data with two zebrafish ear hair cell scRNA-seq datasets shows how a single transcription factor can shift cell fate with consequences for transcriptional identity and physical characteristics that affect cell function. When comparing all three zebrafish hair cell types (sibling, prdm1a mutant lateral line, and the wildtype ear), it is important to note that although there are transcriptional and morphological differences between the three populations, they are all hair cells. All three hair cell types share genes along the differentiation trajectories, with more genes in common than different (Supplementary Data 3 and 4). Therefore, the three datasets present an ideal foundation from which to build GRNs describing hair cell development and fate.
Just as there are transcriptionally and morphologically distinct hair cells in the zebrafish ear and lateral line5,79, there are also unique hair cell types in the mammalian cochlea and vestibular organs86. In both the ear and lateral line, the genes that initiate specification and differentiation of hair cell progenitors have been well characterized87,88,89,90,91,92,93. Specifically, Atoh1 and Insm1 are key transcription factors for hair cell specification and differentiation across species, and their epistatic interactions have been investigated in mice94,95. By using our ATAC-seq data to identify conserved transcription factor binding sites and incorporating the developmental timing of expression, we infer the existence of a coherent feed forward subcircuit96 in the lateral line involving atoh1a/b and insm1a/b activating downstream hair cell genes, including prdm1a (Fig. 5h, i and Supplementary Fig. 3b). The presence or absence of prdm1a in this subcircuit determines lateral line or ear hair cell fate.
As prdm1a is the key gene in determining ear versus lateral line hair cell fate in the zebrafish, we compared its place in the GRN to another hair cell fate specification process; the inner versus outer hair cell fate decision in the mammalian cochlea. The network of Insm1, Ikzf2, and Tbx2 in cochlear hair cells determines if they will develop into inner or OHCs, each with different morphologies and functions in hearing14,58,60,77,94,97. While these studies each uncovered key aspects of this fate decision, a unified network has not yet been constructed. We built a GRN underlying the inner-, versus outer hair cell fate decision in the mouse cochlea (Fig. 5j, k) based on published data and compared it to the model zebrafish GRNs for the lateral line and ear (Fig. 5h, i).
The zebrafish GRNs possess striking similarities to the mouse ear hair cell fate GRN. The same coherent feedforward loop with Atoh1 and Insm1 appears to be present in both models, and the expression of either Tbx2 or Ikzf2 determines the fate of developing hair cells60,94. This similarity between hair cell GRNs in the zebrafish ear, and mouse ear, and lateral line reveals a shared architecture of the hair cell determination and differentiation network, implying deeply conserved systems at the heart of hair cell development and fate.
The expression of prdm1a is a key difference between lateral line-, and ear hair cells. However, prdm1a and Prdm1 are both expressed in embryonic-developing zebrafish ears (Supplementary Fig. 1b, c) and mammalian ears, respectively98, but repressed in sensory epithelia and completely eliminated from ears by the time hair cells mature and function. We therefore suggest that ancestral hair cells diverged into ear and lateral line fates based on the differential regulation of prdm1a.
It is well understood that transcriptional repression of a GRN branch or node is a powerful tool to direct cell type specification in developing tissues99,100,101. The repeated deployment of Prdm1/prdm1a in cell fate decisions in hematopoiesis and photoreceptors in mice45,46,47, neural crest-, muscle-, and hair cells in zebrafish48,49,50 highlights the potency of its activity as a transcriptional regulator across tissues and species. In all these cases, the mutation of Prdm1/prmd1a leads to a derepression of a large number of genes and a shift in cell fate. Our study cements prdm1a in this role as a master regulator of cell fate and the possible conservation of its function across evolution. The results not only inform our understanding of how hair cell types might have evolved but also serve as a template to induce correct hair cell differentiation via the manipulation of key transcription factors during regeneration in the cochlea.
Methods
Work with zebrafish followed standard procedures and was overseen by the IACUC at the Stowers Institute for Medical Research.
Zebrafish
The following transgenic lines were used: Tg(she:H2A-mCherry)psi56Tg, Tg(she:H2B-EGFP)psi59Tg, Tg(myo6:H2B-mScarlet-I)psi66Tg 102, Tg(pou4f3:Gap43-EGFP) 103 and Tg(sqet20Et:EGFP) 104. The prdm1apsi91 mutant was identified in an ENU-induced mutagenesis screen.
Confocal microscopy
Embryos were embedded in 0.8% low-melting-point agarose (Promega) and mounted on 35 mm glass-bottom dishes (Mat Tek). Imaging was performed on a Nikon Ti2 Yokogawa CSU-W1 spinning disk microscope with a Hamamatsu Orca Fusion sCMOS camera. Images were acquired with either 10x CFI Apo LWD or 40x WI 1.15 NA Lambda Objectives. Images were processed using Imaris versions 8.3.1 and 9.6.1 (Bitplane) and Fiji105.
HCR in situ hybridization
All HCRs were performed using probes and reagents from Molecular Instruments according to standard zebrafish embryo protocols106, with the following changes: Embryos were permeabilized in 80% acetone at −20 °C for 30 min, followed by washing in PBST 3 × 10 min prior to probe pre-hybridization. All reaction volumes were reduced by half, but probe and amplifier concentrations maintained with protocol recommendations; amplification step was extended to 48 h. All probes were custom-synthesized by Molecular Instruments.
HCR signal quantification
The HCR signal was normalized with a 50 μm rolling ball background subtraction in Fiji. The signal was then quantified by drawing ROIs around cells of interest, and using the Measure function to measure mean signal intensity.
Kinocilia length quantification
Using images of Tg(pou4f3:EGFP), maximum intensity projections of z-stacks were created in Fiji and lengths of individual kinocilia were measured using the Measure function.
Cuticular plate measurements
Cuticular plate measurements were obtained by manually drawing ROIs around the phalloidin signal of stained cuticular plates and using the measure function in Fiji.
Daspei staining
To visualize hair cells embryos were placed in a 0.006% solution of DASPEI (2-4-(dimethylamino)steryl)-N-ethylpyridinium iodide, [Invitrogen, USA]) diluted in embryo media for 10 min. Embryos were then washed and anesthetized with MS-222 for imaging on a spinning disk microscope.
Phalloidin staining
Phalloidin staining was performed using Alexa Fluor 488-conjugated phalloidin (Thermo Fisher Scientific). Zebrafish were fixed in 4% paraformaldehyde with 1% PBS for 4 h at 4 °C, then washed in PBS containing 1% DMSO, 0.5% Triton X- 100, and 0.1% Tween-20 (PBDTT buffer). Phalloidin (1:40, #A12379, Thermo Fisher Scientific) and DAPI (1:1000) were then added in PBDTT buffer supplemented with 1% BSA and incubated at room temperature for 2 h.
Cell body measurements
To measure hair cell body volumes, masks were drawn manually around the body of hair cells, excluding the kinocilia, in Fiji. Using the measure function in Fiji, the hair cell body volume was estimated as an ellipsoid from the major and minor axes of the hand-drawn ROI output by Fiji.
Hair cell ablation
Lateral line sensory hair cells were ablated in 5 dpf larvae by incubation in 300 μM neomycin (Sigma-Aldrich) in standard embryo media for 30 min at 28 °C52. Neomycin was washed out thoroughly, and embryos returned to 0.5x E2 embryo media until analysis timepoint.
Cell proliferation assay
Proliferating cells were assayed as previously described107. 5 dpf larvae were incubated in 3.3 mM EdU (Carbosynth) in 1% DMSO in standard embryo medium for 48 h following hair cell ablation. Larvae were then fixed in 4% paraformaldehyde overnight at 4 °C. All subsequent steps were performed at room temperature. Larvae were washed 3 × 10 min in PBS/0.8% Trition-X (PBSTX), blocked in 3% BSA in PBSTX for 1 h, and washed 3 × 5 min in PBS. To stain, embryos were incubated in staining cocktail consisting of 1x TBS, 2 mM CuSO4, 2.5 μM Alexa-647-Azide (Invitrogen), 50 mM ascorbic acid, and 0.4% Triton-x for 30 min. After staining, larvae were washed 3 × 10 min in PBSTX, stained with 5 μg/ml DAPI at RT for 5 min, and imaged as described above.
scRNA-seq
Larva dissociation and FACS
600 each prdm1a mutant and sibling embryos with the Tg(she:H2A-mCherry)psi56Tg lateral line marker were separately dissociated in 4.5 ml 0.25% Trypsin-EDTA (Gibco) for 3.5 min on ice by triturating with a 1 ml pipette repeatedly. Supernatant containing cells was filtered through a 70 μm Cup Filcon (BD Biosciences), and cells were pelleted by centrifugation at 720 × g for 5 min at 4 °C. Cells were washed in cold PBST by resuspending the pellet and pelleted again at 720 g for five minutes at 4 °C. Cells were filtered through a 35 μm strainer (Falcon-Corning), and stained with Draq5 (BioStatus) at a dilution of 1:500. Cells were then sorted on a BD Influx Cell Sorter (BD Biosciences). mCherry and Draq5 double-positive cells were sorted into 90% MeOH for fixation and stored at 4 °C until rehydration.
10x Chromium scRNA-seq library prep
FAC-sorted MeOH fixed cells prepared as above were processed according to 10X Genomics standard protocols. Cells were rehydrated in 1% BSA and 0.5 U/μl RNase-inhibitor in ice-cold DPBS. A volume of 34ul was loaded onto the Chromium Controller, targeting ~12,000 cells per sample. Library prep was performed using the Chromium Next GEM Single Cell 3’ GEM, Library & Gel Bead Kit v3.1. Two independent replicates were sequenced for both prdm1a mutant and sibling conditions. Libraries were pooled and sequenced on an Illumina NextSeq 500 in High Output mode, with 28 bp, 8 bp, and 91 bp for read 1, i7 index, and read 2, respectively.
Single-cell RNA-seq data analysis
Data were processed using 10X Genomics Cellranger v4.0, first by demultiplexing using cellranger mkfastq with default parameters, then cellranger count was used to align reads to the GRCz11 genome (Ensembl release 98) and generate a feature count matrix. The matrix was then loaded into R package Seurat (v4.3)108 for further bioinformatics analysis. First, the top 2000 variable genes were selected using the loess span parameter for principal component analysis. Based on elbow plot, 30 PCs were selected to generate the nearest neighbor graph, clustering, as well as the UMAP visualization were done using Seurat default parameters. External scRNA-seq data used for integration were downloaded from GEO. All replicates and samples were then merged and integrated using Seurat’s default CCA method for downstream analysis. For marker gene detection, we used the Seurat FindAllMarkers function (logfc.threshold = 0.25) to generate differentially expressed genes for every cluster and annotated based on ZFIN database knowledge. Heatmaps were then generated using the DoHeatmap function. av
ATAC-seq
Larva dissociation and FACS
ATAC-seq was performed generally following a previously established protocol109, with some modifications. 600–1200 Tg(she:H2A-mCherry)psi56Tg embryos were fixed in 1% PFA in PBS at 4 °C for 30 min. Glycine was added to a final concentration of 125 mM for 5 min to quench formaldehyde. Embryos were moved to 4.5 ml 0.25% Trypsin-EDTA (Gibco) and dissociated for ~8 min on ice by triturating with a 1 ml pipette repeatedly. Supernatant containing cells was filtered through a 70 μm Cup Filcon (BD Biosciences), and cells were pelleted and washed twice by centrifugation at 720 × g for 5 min at 4 °C and resuspending in cold PBST. Cells was filtered through a 70μm Cup Filcon (BD Biosciences) one additional time and pelleted by centrifugation at 720 × g for 5 min at 4 °C. Cells were resuspended in 4 ml PBST and stained with Draq5 at a dilution of 1:500. mCherry and Draq5 double-positive cells were then sorted into pools of 50,000 on a BD Melody Cell Sorter (BD Biosciences) into chilled 1.5 ml centrifuge tubes.
Tagmentation
Cells were pelleted by spinning in a microcentrifuge for 5 min at 4 °C and 1000 g. Supernatant was removed carefully by pipette, and cells were resuspended in tagmentation reaction mix (22.5 μl H2O, 25 μl TD Buffer (Illumina), and 2.5 μl TD Enzyme (Illumina)) and incubated in a ThermoMixer (Eppendorf) at 37 °C for 30 min, 1000 RPM. 50 μl reverse crosslinking buffer (final concentration of 50 mM TRIS pH 8, 1 mM EDTA, 1% SDS, 200 mM NaCl, 5 ng/ml Proteinase K (Roche)) was added, and the mixture was incubated at 65 °C overnight in a ThermoMixer. Tagmented DNA was purified with a DNA Clean & Concentrator 5 kit (Zymo Research) and eluted in 20 μl H2O. Library PCR amplification was performed with 20 μl eluted DNA, 25 μl NEBNext Ultra II Q5 Master Mix, 2.5 μl forward primer Ad1 25 mM stock, 2.5 μl reverse barcode primer110 Ad 2.x 25 mM stock. PCR conditions as follows: 72 °C 5 min, 98 °C 30 s, 11 cycles of 98 °C 10 s, 63 °C 30 s, 72 °C 1 min, 4 °C hold. The Library was size selected with SPRIselect beads (Beckman Colter) with the following conditions: 0.8× volume of beads first selection, the supernatant was retained, and an additional 1.0× volume of beads was added (total volume 1.8×). The bead pellet was retained and rinsed with 85% EtOH, dried, and DNA was eluted in 30 μl H2O. Libraries were analyzed on a Bioanalyzer 2100 (Agilent) to evaluate tagmentation and quantified on a Qubit 3.0 (Thermo Fisher).
Sequencing and read alignment
ATAC libraries were pooled and sequenced on an Illumina NextSeq 500, 2x75bp paired-end mode. The zebrafish reference genome and transcriptome were obtained from Ensembl (version: GRCz11-102)111. It was segmented into contiguous windows at 50 bp resolution using the makewindows function of the bedtools package112. These windows were used as a basis for this analysis. Sequencing reads for each sample were mapped to the genome using the STAR aligner (v 2.7.9)113 (using parameters—alignIntronMax -1—alignIntronMin −1—outFilterMultimapNmax 1—outFilterMultimapScoreRange 1—outFilterMismatchNmax 3), producing corresponding BAM files.
Peak calling and motif search
A peak set (in narrowPeak format) was determined separately for each experimental group using MACS2 to analyze the group’s replicate BAM files (with parameters “ --shift–100 --extsize 200 --nomodel–f BAM -q 0.1”). A single Ensembl gene was assigned to each peak, called as its putative target based on proximity, as follows. It was classified as a promoter targeting the closest protein coding gene to which it is within 250 bp upstream or 100 bp downstream, if any; Using the list of peak regions called in ATAC-seq peaks described above, Samtools114 were used to extract the genomic sequence under to the peak region and the MEME suite tool FIMO to scan for enriched TF motifs, with a dynamic programming algorithm to convert log-odds scores into p-values, assuming a zero-order background model. Genomic tracks were then produced using the bigwig files, highlighting the enriched MOTIF regions with the Python package pyGenomeTracks.
Generation of transgenic reporter lines
We identified ATAC peaks associated with s100a1 and designed primers to clone the gene’s enhancer and promoter. To clone the 3’UTR peak primers s100a1: F-5’ACTTTCATGTGTTACTTCATTTGAAGGAAAATG’3, R- 5’TTTAACCATTTTATTTTGCTTCTGTCAAAATGTACATC’3 were used. To clone the upstream promoter primers F- 5’CAGAATTCAGCCAAAACGTCTTAAAGCATGG’3, R- 5’GTTAGAAAATATTATCTGCAAGATAAATAATTAAATACAAGTC’3 were used. These regions were then inserted together into a miniTol2115 plasmid containing a beta-actin minimal promoter upstream of H2B-GFP, followed by the SV40 polyadenylation signal via Gibson assembly116. This reporter construct, s100a1:H2B-EGFP, was then injected at a final concentration of 30 ng/μl with tol2 mRNA into one-cell stage zebrafish embryos for Tol2-mediated integration117,118. Injected embryos were screened for EGFP expression at 5 dpf and raised for outcrossing with wild-type fish to establish stable transgenic lines. The myo6:prdm1a; cryaa:mTurqoise2 construct was created using Gateway assembly and the Tol2Kit integration117,118. An existing p5E entry vector with the myo6b enhancer was used119. The prdm1a cDNA was cloned into the pME entry vector, with the SV40 poly-A signal. cDNA was synthesized using RNA extracted from 5 dpf zebrafish embryos with TRIzol (Invitrogen), following standard protocols. RNA was precipitated with isopropanol, washed with 75% EtOH, dried, and resuspended in RNAse-free dH2O. cDNA was synthesized using ProtoScript II Reverse Transcriptase (NEB) with 1 μg total RNA and oligo d(T)23VN as a primer, following standard protocols. Following reverse transcription, the reaction volume of 20ul was diluted up to 100ul with dH2O. prdm1a cDNA was amplified with the following primers: F-5’ ATCTCAGGCACTTGCAGGAGAAGTC 3’, R-5’ CTAGGTATCCATGGCCTCCTCTGTCTC 3’. The cryaa promoter was cloned into the p3E entry vector driving mTurquoise using cDNA prepared above using the following primers: F-5’ GTGGAGACCCCTGATTAATAAAGGGACTTA 3’, R-5’ AATGTCAGACCTGGTAACTCCTTACTGTAA 3’. The final plasmid was assembled by the gateway reaction using all three entry vectors described above and the pDestTol2pA2 destination vector.
Statistical analysis
Statistical tests were performed using GraphPad Prism 10 (Version 10.2.3). When comparing data from more than two groups, statistical significance was calculated using one-way ANOVA with Tukey’s post hoc test. Data from two groups were compared using two-tailed unpaired t-test. p-values smaller than 0.05 were considered to be statistically significant. Statistical details of experiments, such as p-values and sample size, are specified individually for each experiment in the Fig. legends. Plots were made in GraphPad Prism 10.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data are provided as a Source Data file. Raw data from images used in this manuscript can be accessed from the Stowers Institute for Medical Research Original Data Repository at http://www.stowers.org/research/publications/libpb-2495. All raw and processed scRNA-seq and ATAC-seq data have been deposited into the NCBI Gene Expression Omnibus (GEO) database with accession number GSE268538.
References
Corwin, J. T. & Warchol, M. E. Auditory hair cells: structure, function, development, and regeneration. Annu. Rev. Neurosci. 14, 301–333 (1991).
Hudspeth, A. J. How the ear’s works work. Nature 341, 397–404 (1989).
Coombs S., Görner P., Münz H., Universität Bielefeld. Zentrum für Interdisziplinäire Forschung. The Mechanosensory Lateral Line: Neurobiology and Evolution. (Springer-Verlag, 1989).
McPherson, D. R. Sensory hair cells: an introduction to structure and physiology. Integr. Comp. Biol. 58, 282–300 (2018).
Shi, T. et al. Single-cell transcriptomic profiling of the zebrafish inner ear reveals molecularly distinct hair cell and supporting cell subtypes. Elife 12, e82978 (2023).
Brignull, H. R., Raible, D. W. & Stone, J. S. Feathers and fins: non-mammalian models for hair cell regeneration. Brain Res. 1277, 12–23 (2009).
Burns, J. C. & Stone, J. S. Development and regeneration of vestibular hair cells in mammals. Semin. Cell Dev. Biol. 65, 96–105 (2017).
Fritzsch, B., Jahan, I., Pan, N. & Elliott, K. L. Evolving gene regulatory networks into cellular networks guiding adaptive behavior: an outline how single cells could have evolved into a centralized neurosensory system. Cell Tissue Res. 359, 295–313 (2015).
Atkinson, P. J., Huarcaya Najarro, E., Sayyid, Z. N. & Cheng, A. G. Sensory hair cell development and regeneration: similarities and differences. Development 142, 1561–1571 (2015).
Groves, A. K., Zhang, K. D. & Fekete, D. M. The genetics of hair cell development and regeneration. Annu. Rev. Neurosci. 36, 361–381 (2013).
Jahan, I., Pan, N., Kersigo, J. & Fritzsch, B. Beyond generalized hair cells: molecular cues for hair cell types. Hear Res. 297, 30–41 (2013).
Kolla, L. et al. Characterization of the development of the mouse cochlear epithelium at the single-cell level. Nat. Commun. 11, 2389 (2020).
Elkon, R. et al. RFX transcription factors are essential for hearing in mice. Nat. Commun. 6, 8549 (2015).
Garcia-Anoveros, J. et al. Tbx2 is a master regulator of inner versus outer hair cell differentiation. Nature 605, 298–303 (2022).
Fettiplace, R. & Fuchs, P. A. Mechanisms of hair cell tuning. Annu. Rev. Physiol. 61, 809–834 (1999).
Nicolson, T. The genetics of hair-cell function in zebrafish. J. Neurogenet. 31, 102–112 (2017).
Baeza-Loya, S. & Raible, D. W. Vestibular physiology and function in zebrafish. Front. Cell Dev. Biol. 11, 1172933 (2023).
Fritzsch, B. & Straka, H. Evolution of vertebrate mechanosensory hair cells and inner ears: toward identifying stimuli that select mutation-driven altered morphologies. J. Comp. Physiol. A Neuroethol. Sens Neural Behav. Physiol. 200, 5–18 (2014).
Baek, S. et al. Single-cell transcriptome analysis reveals three sequential phases of gene expression during zebrafish sensory hair cell regeneration. Dev. Cell 57, 799–819.e796 (2022).
Lush, M. E. et al. scRNA-Seq reveals distinct stem cell populations that drive hair cell regeneration after loss of Fgf and Notch signaling. Elife 8, e44431 (2019).
Mulvaney, J. F. et al. Kremen1 regulates mechanosensory hair cell development in the mammalian cochlea and the zebrafish lateral line. Sci. Rep. 6, 31668 (2016).
Nicolson, T. The genetics of hearing and balance in zebrafish. Annu. Rev. Genet. 39, 9–22 (2005).
Whitfield, T. T. Zebrafish as a model for hearing and deafness. J. Neurobiol. 53, 157–171 (2002).
Kniss, J. S., Jiang, L. & Piotrowski, T. Insights into sensory hair cell regeneration from the zebrafish lateral line. Curr. Opin. Genet. Dev. 40, 32–40 (2016).
Cruz, I. A. et al. Robust regeneration of adult zebrafish lateral line hair cells reflects continued precursor pool maintenance. Dev. Biol. 402, 229–238 (2015).
Rubel, E. W., Furrer, S. A. & Stone, J. S. A brief history of hair cell regeneration research and speculations on the future. Hear Res. 297, 42–51 (2013).
Gonzalez-Garrido, A. et al. The differentiation status of hair cells that regenerate naturally in the vestibular inner ear of the adult mouse. J. Neurosci. 41, 7779–7796 (2021).
Agrawal, Y., Carey, J. P., Della Santina, C. C., Schubert, M. C. & Minor, L. B. Disorders of balance and vestibular function in US adults: data from the National Health and Nutrition Examination Survey, 2001-2004. Arch. Intern. Med. 169, 938–944 (2009).
Wilson, B. S., Tucci, D. L., Merson, M. H. & O’Donoghue, G. M. Global hearing health care: new findings and perspectives. Lancet 390, 2503–2515 (2017).
Romero-Carvajal, A. et al. Regeneration of sensory hair cells requires localized interactions between the Notch and Wnt pathways. Dev. Cell 34, 267–282 (2015).
Samarajeewa, A., Jacques, B. E. & Dabdoub, A. Therapeutic potential of Wnt and Notch signaling and epigenetic regulation in mammalian sensory hair cell regeneration. Mol. Ther. 27, 904–911 (2019).
Zheng, J. L. & Gao, W. Q. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat. Neurosci. 3, 580–586 (2000).
Atkinson, P. J., Wise, A. K., Flynn, B. O., Nayagam, B. A. & Richardson, R. T. Hair cell regeneration after ATOH1 gene therapy in the cochlea of profoundly deaf adult guinea pigs. PLoS One 9, e102077 (2014).
White, P. M., Doetzlhofer, A., Lee, Y. S., Groves, A. K. & Segil, N. Mammalian cochlear supporting cells can divide and trans-differentiate into hair cells. Nature 441, 984–987 (2006).
Izumikawa, M. et al. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat. Med. 11, 271–276 (2005).
Kuo, B. R., Baldwin, E. M., Layman, W. S., Taketo, M. M. & Zuo, J. In vivo cochlear hair cell generation and survival by coactivation of beta-Catenin and Atoh1. J. Neurosci. 35, 10786–10798 (2015).
Ikeda, R., Pak, K., Chavez, E. & Ryan, A. F. Transcription factors with conserved binding sites near ATOH1 on the POU4F3 gene enhance the induction of cochlear hair cells. Mol. Neurobiol. 51, 672–684 (2015).
Costa, A. et al. Generation of sensory hair cells by genetic programming with a combination of transcription factors. Development 142, 1948–1959 (2015).
Samarajeewa, A. et al. Transcriptional response to Wnt activation regulates the regenerative capacity of the mammalian cochlea. Development 145, dev166579 (2018).
Iyer, A. A. et al. Cellular reprogramming with ATOH1, GFI1, and POU4F3 implicate epigenetic changes and cell-cell signaling as obstacles to hair cell regeneration in mature mammals. Elife 11, e79712 (2022).
Jimenez, E. et al. A regulatory network of Sox and Six transcription factors initiate a cell fate transformation during hearing regeneration in adult zebrafish. Cell Genom. 2, 100170 (2022).
Bikoff, E. K., Morgan, M. A. & Robertson, E. J. An expanding job description for Blimp-1/PRDM1. Curr. Opin. Genet Dev. 19, 379–385 (2009).
Doody, G. M. et al. An extended set of PRDM1/BLIMP1 target genes links binding motif type to dynamic repression. Nucleic Acids Res. 38, 5336–5350 (2010).
Powell, D. R., Hernandez-Lagunas, L., LaMonica, K. & Artinger, K. B. Prdm1a directly activates foxd3 and tfap2a during zebrafish neural crest specification. Development 140, 3445–3455 (2013).
JAt, B. rzezinski, Lamba, D. A. & Reh, T. A. Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development 137, 619–629 (2010).
Nutt, S. L., Fairfax, K. A. & Kallies, A. BLIMP1 guides the fate of effector B and T cells. Nat. Rev. Immunol. 7, 923–927 (2007).
Turner, C. A. Jr, Mack, D. H. & Davis, M. M. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell 77, 297–306 (1994).
Hernandez-Lagunas, L. et al. Zebrafish narrowminded disrupts the transcription factor prdm1 and is required for neural crest and sensory neuron specification. Dev. Biol. 278, 347–357 (2005).
Prajapati R. S., Hintze M., Streit A. PRDM1 controls the sequential activation of neural, neural crest and sensory progenitor determinants by regulating histone modification. Development 146, dev181107 (2019).
LaMonica, K., Ding, H. L. & Artinger, K. B. prdm1a functions upstream of itga5 in zebrafish craniofacial development. Genesis 53, 270–277 (2015).
Spitz, F. & Furlong, E. E. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626 (2012).
Venero Galanternik, M., Navajas Acedo, J., Romero-Carvajal, A. & Piotrowski, T. Imaging collective cell migration and hair cell regeneration in the sensory lateral line. Methods Cell Biol. 134, 211–256 (2016).
Wilm, T. P. & Solnica-Krezel, L. Essential roles of a zebrafish prdm1/blimp1 homolog in embryo patterning and organogenesis. Development 132, 393–404 (2005).
Iwanaga, R. et al. Loss of prdm1a accelerates melanoma onset and progression. Mol. Carcinog. 59, 1052–1063 (2020).
David, N. B. et al. Molecular basis of cell migration in the fish lateral line: role of the chemokine receptor CXCR4 and of its ligand, SDF1. Proc. Natl. Acad. Sci. USA 99, 16297–16302 (2002).
Birkholz, D. A., Olesnicky Killian, E. C., George, K. M. & Artinger, K. B. Prdm1a is necessary for posterior pharyngeal arch development in zebrafish. Dev. Dyn. 238, 2575–2587 (2009).
Yasunaga, S. et al. A mutation in OTOF, encoding otoferlin, a FER-1-like protein, causes DFNB9, a nonsyndromic form of deafness. Nat. Genet. 21, 363–369 (1999).
Bi, Z. et al. Revisiting the potency of Tbx2 expression in transforming outer hair cells into inner hair cells at multiple ages in vivo. J. Neurosci. 44, e1751232024 (2024).
Hu, Q. et al. Deletion of Kncn does not affect Kinocilium and Stereocilia bundle morphogenesis and mechanotransduction in cochlear hair cells. Front. Mol. Neurosci. 11, 326 (2018).
Chessum, L. et al. Helios is a key transcriptional regulator of outer hair cell maturation. Nature 563, 696–700 (2018).
Tong, B. et al. Oncomodulin, an EF-hand Ca2+ buffer, is critical for maintaining cochlear function in mice. J. Neurosci. 36, 1631–1635 (2016).
Kawashima, Y., Kurima, K., Pan, B., Griffith, A. J. & Holt, J. R. Transmembrane channel-like (TMC) genes are required for auditory and vestibular mechanosensation. Pflug. Arch. 467, 85–94 (2015).
Francis, S. P. et al. A short splice form of Xin-actin binding repeat containing 2 (XIRP2) lacking the Xin repeats is required for maintenance of stereocilia morphology and hearing function. J. Neurosci. 35, 1999–2014 (2015).
Nakanishi, H., Kurima, K., Kawashima, Y. & Griffith, A. J. Mutations of TMC1 cause deafness by disrupting mechanoelectrical transduction. Auris Nasus Larynx 41, 399–408 (2014).
Francey, L. J. et al. Genome-wide SNP genotyping identifies the Stereocilin (STRC) gene as a major contributor to pediatric bilateral sensorineural hearing impairment. Am. J. Med. Genet. A 158A, 298–308 (2012).
Lin, Y. S. et al. Dysregulated brain creatine kinase is associated with hearing impairment in mouse models of Huntington disease. J. Clin. Investig. 121, 1519–1523 (2011).
Zhang, Y. et al. Sensorineural deafness and male infertility: a contiguous gene deletion syndrome. J. Med. Genet. 44, 233–240 (2007).
Kalay, E. et al. Mutations in the lipoma HMGIC fusion partner-like 5 (LHFPL5) gene cause autosomal recessive nonsyndromic hearing loss. Hum. Mutat. 27, 633–639 (2006).
Strathdee, G. et al. Molecular characterization of patients with 18q23 deletions. Am. J. Hum. Genet. 60, 860–868 (1997).
Sun P., Smith E., Nicolson T. Transmembrane Channel-Like (TMC) subunits contribute to frequency sensitivity in the zebrafish utricle. J. Neurosci. 44, e1298232023 (2024).
Smith, E. T., Pacentine, I., Shipman, A., Hill, M. & Nicolson, T. Disruption of tmc1/2a/2b genes in zebrafish reveals subunit requirements in subtypes of inner ear hair cells. J. Neurosci. 40, 4457–4468 (2020).
Zhu, S., Chen, Z., Wang, H. & McDermott, B. M. Jr Tmc reliance is biased by the hair cell subtype and position within the ear. Front. Cell Dev. Biol. 8, 570486 (2020).
Kindig, K., Stepanyan, R., Kindt, K. S. & McDermott, B. M. Jr Asymmetric mechanotransduction by hair cells of the zebrafish lateral line. Curr. Biol. 33, 1295–1307.e1293 (2023).
Cai, T. et al. Characterization of the transcriptome of nascent hair cells and identification of direct targets of the Atoh1 transcription factor. J. Neurosci. 35, 5870–5883 (2015).
Sur, A. et al. Single-cell analysis of shared signatures and transcriptional diversity during zebrafish development. Dev. Cell 58, 3028–3047.e3012 (2023).
Orvis, J. et al. gEAR: Gene Expression Analysis Resource portal for community-driven, multi-omic data exploration. Nat. Methods 18, 843–844 (2021).
Kaiser, M. et al. TBX2 specifies and maintains inner hair and supporting cell fate in the Organ of Corti. Nat. Commun. 13, 7628 (2022).
Whitfield, T. T. Cilia in the developing zebrafish ear. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190163 (2020).
Tanimoto, M., Ota, Y., Inoue, M. & Oda, Y. Origin of inner ear hair cells: morphological and functional differentiation from ciliary cells into hair cells in zebrafish inner ear. J. Neurosci. 31, 3784–3794 (2011).
Yu, X., Lau, D., Ng, C. P. & Roy, S. Cilia-driven fluid flow as an epigenetic cue for otolith biomineralization on sensory hair cells of the inner ear. Development 138, 487–494 (2011).
Waddington C. H. The Strategy of the Genes: A Discussion of Some Aspects of Theoretical Biology. (Allen & Unwin, 1957).
Britten, R. J. & Davidson, E. H. Gene regulation for higher cells: a theory. Science 165, 349–357 (1969).
Levine, M. & Davidson, E. H. Gene regulatory networks for development. Proc. Natl. Acad. Sci. USA 102, 4936–4942 (2005).
Haghverdi, L. & Ludwig, L. S. Single-cell multi-omics and lineage tracing to dissect cell fate decision-making. Stem Cell Rep. 18, 13–25 (2023).
Badia, I. M. P. et al. Gene regulatory network inference in the era of single-cell multi-omics. Nat. Rev. Genet. 24, 739–754 (2023).
Khan, A. et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46, D260–D266 (2018).
Cafaro, J., Lee, G. S. & Stone, J. S. Atoh1 expression defines activated progenitors and differentiating hair cells during avian hair cell regeneration. Dev. Dyn. 236, 156–170 (2007).
Costa, A., Powell, L. M., Lowell, S. & Jarman, A. P. Atoh1 in sensory hair cell development: constraints and cofactors. Semin. Cell Dev. Biol. 65, 60–68 (2017).
Chonko, K. T. et al. Atoh1 directs hair cell differentiation and survival in the late embryonic mouse inner ear. Dev. Biol. 381, 401–410 (2013).
Millimaki, B. B., Sweet, E. M., Dhason, M. S. & Riley, B. B. Zebrafish atoh1 genes: classic proneural activity in the inner ear and regulation by Fgf and Notch. Development 134, 295–305 (2007).
Lorenzen, S. M., Duggan, A., Osipovich, A. B., Magnuson, M. A. & Garcia-Anoveros, J. Insm1 promotes neurogenic proliferation in delaminated otic progenitors. Mech. Dev. 138, 233–245 (2015).
Bermingham, N. A. et al. Math1: an essential gene for the generation of inner ear hair cells. Science 284, 1837–1841 (1999).
Chen, P., Johnson, J. E., Zoghbi, H. Y. & Segil, N. The role of Math1 in inner ear development: uncoupling the establishment of the sensory primordium from hair cell fate determination. Development 129, 2495–2505 (2002).
Li, S., He, S., Lu, Y., Jia, S. & Liu, Z. Epistatic genetic interactions between Insm1 and Ikzf2 during cochlear outer hair cell development. Cell Rep. 42, 112504 (2023).
Sun, S. et al. Dual expression of Atoh1 and Ikzf2 promotes transformation of adult cochlear supporting cells into outer hair cells. Elife 10, e66547 (2021).
Peter, I. S. & Davidson, E. H. Assessing regulatory information in developmental gene regulatory networks. Proc. Natl. Acad. Sci. USA 114, 5862–5869 (2017).
Wiwatpanit, T. et al. Trans-differentiation of outer hair cells into inner hair cells in the absence of INSM1. Nature 563, 691–695 (2018).
Urness, L. D. et al. Spatial and temporal inhibition of FGFR2b ligands reveals continuous requirements and novel targets in mouse inner ear morphogenesis. Development 145, dev170142 (2018).
Lim, B., Domsch, K., Mall, M. & Lohmann, I. Canalizing cell fate by transcriptional repression. Mol. Syst. Biol. 20, 144–161 (2024).
Kutejova, E., Sasai, N., Shah, A., Gouti, M. & Briscoe, J. Neural progenitors adopt specific identities by directly repressing all alternative progenitor transcriptional programs. Dev. Cell 36, 639–653 (2016).
Davidson, E. H. Emerging properties of animal gene regulatory networks. Nature 468, 911–920 (2010).
Peloggia, J. et al. Adaptive cell invasion maintains lateral line organ homeostasis in response to environmental changes. Dev. Cell 56, 1296–1312.e1297 (2021).
Xiao, T., Roeser, T., Staub, W. & Baier, H. A GFP-based genetic screen reveals mutations that disrupt the architecture of the zebrafish retinotectal projection. Development 132, 2955–2967 (2005).
Parinov, S., Kondrichin, I., Korzh, V. & Emelyanov, A. Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev. Dyn. 231, 449–459 (2004).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Choi, H. M. T. et al. Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753 (2018).
Lush, M. E. & Piotrowski, T. Sensory hair cell regeneration in the zebrafish lateral line. Dev. Dyn. 243, 1187–1202 (2014).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Chen, X. et al. ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat. Methods 13, 1013–1020 (2016).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Martin, F. J. et al. Ensembl 2023. Nucleic Acids Res 51, D933–D941 (2023).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021).
Balciunas, D. et al. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2, e169 (2006).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Kwan, K. M. et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088–3099 (2007).
Kawakami, K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 8, S7 (2007).
Kindt, K. S., Finch, G. & Nicolson, T. Kinocilia mediate mechanosensitivity in developing zebrafish hair cells. Dev. Cell 23, 329–341 (2012).
Acknowledgements
We thank Jillian Blanck, Kevin Ferro, KeyongMin Bae, Fang Liu, and Jeffery Haug for technical assistance and advice on performing FAC-sorting. We thank Michael Peterson, Kate Hall, and Anoja Perera for technical assistance and advice in sequencing libraries on NGS instruments. We thank Mark Miller for the graphic design. We thank Tatjana Sauka-Spengler and Robb Krumlauf for thoughtful discussions and Piotrowski Lab members Daniela Münch, Julia Peloggia and Aurelie Hintermann for thorough readings of the manuscript. This research was supported by NIH (NIDCD) award 1R01DC015488-01A1 to T.P., NIH (NIDCD) award 1F32DC019023-01A1 to J.E.S., National Institutes of Health grant ZIAHD008997 to J.A.F., funding from the Hearing Health Foundation to T.P., and institutional support from the Stowers Institute for Medical Research to T.P.
Author information
Authors and Affiliations
Contributions
J.E.S., Y.-Y.T. and T.P. conceived the project and designed experiments. JES, Y.-Y.T., L.S., M.E.L., E.E., A.R.S., and J.S.K. performed experiments. J.E.S., Y.-Y.T., L.S. and M.E.L. analyzed experimental data. S.C., N.T.T.T., and M.C. performed computational and biostatistical analysis. J.E.S., Y.-Y.T. and T.P. wrote the manuscript with input from all authors. A.S. and J.A.F. provided the data for otic subset of zebrafish scRNA-seq analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Katie Kindt, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sandler, J.E., Tsai, YY., Chen, S. et al. prdm1a drives a fate switch between hair cells of different mechanosensory organs. Nat Commun 16, 7662 (2025). https://doi.org/10.1038/s41467-025-62942-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-62942-0