Abstract
Many wild relatives of wheat in the Triticeae tribe provide important genetic resources for crop improvement, but their complex, polyploid genomes shaped by hybridization remain poorly understood. Here, we assemble and analyze the genomes of Thinopyrum intermedium and Roegneria kamoji, two species commonly used in wheat hybridization. We show that Th. intermedium contains genomic contributions from Pseudoroegneria (St), Dasypyrum (V), and Aegilops (J), while R. kamoji contains subgenomes related to Pseudoroegneria (St), Dasypyrum (V-related “Y”), and Hordeum (H). Phylogenomic evidence indicates that both species underwent independent polyploidization events, with Pseudoroegneria serving as the original maternal donor. R. kamoji likely evolved from tetraploid Roegneria. We also identify two Fhb7 homologs in the St and H subgenomes of R. kamoji that enhance Fusarium head blight resistance in a dosage-dependent manner. These findings refine the understanding of Triticeae polyploid evolution and offer valuable genomic resources for wheat improvement and forage breeding.
Similar content being viewed by others
Introduction
Bread wheat (Triticum aestivum L.) is one of the most crucial staple crops globally, occupying the largest cultivated area and contributing approximately one-fifth of the daily caloric and protein intake for humans1. Similar to other domesticated crops, wheat originated from a small subset of wild plants selected as founder stock2. This selection, combined with harsh environmental conditions and repeated cycles of cultivation, results in a genetic bottleneck that constrains productivity gains3,4. Wild relatives of wheat represent valuable genetic reservoirs, offering beneficial traits such as higher yield potential, improved quality, and enhanced resistance or tolerance to biotic and abiotic stresses5. Introducing advantageous alleles from these wild relatives into cultivated wheat through distant hybridization has proven to be an effective strategy for developing improved cultivars5. Intermediate wheatgrass (Thinopyrum intermedium (Host) Barkworth & D.R. Dewey, syn. Trichopyrum intermedium (Host) Á.Löve; 2n = 6x = 42) and Roegneria kamoji (Ohwi) Ohwi ex Keng (syn. Campeiostachys kamoji (Ohwi) B.R. Baum, J.L. Yang & C. Yen; Elymus kamoji (Ohwi) S.L.Chen; 2n = 6x = 42) are allohexaploid perennial species within the Triticeae tribe and are considered key genetic resources for wheat improvement and forage breeding6,7. These species contribute beneficial traits, including resistance to leaf rust, stripe rust, stem rust, powdery mildew, Fusarium head blight (FHB), and yellow dwarf virus, as well as tolerance to abiotic stress and desirable quality attributes8,9,10,11,12. Additionally, recent breeding efforts in the United States have domesticated Th. intermedium as a perennial grain crop, resulting in the development of Kernza®—an innovative step toward sustainable agriculture13.
Th. intermedium is native to Europe and Central Asia, and has been introduced to other temperate regions as a forage grass14. It features extensively creeping rhizomes with erect, robust culms that reach heights of 70–100 cm. The leaves exhibit a stiff texture, and the inflorescence is a straight, linear spike measuring 11–17 cm in length and ~5 mm in width15. The chromosome composition of Th. intermedium remains contentious owing to the high prevalence of repetitive sequences, extensive hybridization events, and the evolutionary complexity of Triticeae species. Despite extensive studies, the genome constitution of Th. intermedium remains unresolved. Based solely on genomic in situ hybridization (GISH) analyses, the genomic formula of Th. intermedium was designated as EeEeEbEbStSt, which represents two closely related genomes similar to those of Thinopyrum elongatum (Host) D.R.Dewey (EeEe or JeJe, 2n = 2x = 14) and Thinopyrum bessarabicum (Săvul. & Rayss) Á.Löve (EbEb or JbJb, 2n = 2x = 14), along with a more distantly related genome similar to Pseudoroegneria strigosa (M. Bieb.) Á.Löve (StSt, 2n = 2x = 14)16. Subsequent phylogenetic analyses using Expressed Sequence Tag-Simple Sequence Repeat (EST-SSR) markers further support this designation17 and re-designate the genomic formula as JvsJvsJrJrStSt, where Jvs and Jr represent ancestral genomes of the present-day Jb of Th. bessarabicum and Je of Th. elongatum, respectively. The St genome is similar to the present-day St found in diploid species of Pseudoroegneria native to Eurasia. A recent study employing Oligo-fluorescence in situ hybridization (FISH) indicated that Jvs was largely derived from Dasypyrum breviaristatum (H. Lindb.) Fred (VVVbVb, 2n = 4x = 28), while Jr was related to either Th. elongatum or Th. bessarabicum18. However, an alternative perspective suggested that two of the subgenomes of Th. intermedium originated from Pseudoroegneria and Dasypyrum, while the third subgenome likely arose through hybridization involving contributions from Thinopyrum and Aegilops19,20. Collectively, while the involvement of the Pseudoroegneria subgenome in Th. intermedium has been demonstrated, the origins of the other subgenomes remain unclear. This ambiguity, particularly concerning contributions from Aegilops, Thinopyrum, or Dasypyrum, continues to complicate the taxonomic classification of Th. intermedium.
Roegneria, primarily distributed in the warm-temperate and humid regions of Central and East Asia, is a polyploid perennial genus within the Triticeae tribe and serves as a valuable genetic resource for wheat disease-resistance breeding, particularly against FHB8,21,22. The allohexaploid species R. kamoji is characterized by densely tufted culms (30–100 cm tall), with 4–5 nodes and a slightly geniculate base. Its spicate inflorescence (7.5–8.5 cm long) features densely arranged spikelets (12–14 mm), with a single spikelet per rachis node23. Although Roegneria was established by Koch in 184824 and has been supported by numerous taxonomists based on genomic and morphological criteria23,25,26, some North American botanists often synonymize Roegneria with Elymus sensu lato, owing to morphological overlap and limited characterization of the Y subgenome prevalent in Asian Roegneria species15,27. Cytogenetic studies have distinguished tetraploid Roegneria (StStYY) from Elymus genus, but the classification of hexaploid Roegneria species (StStYYHH), also designated as Campeiostachys Drobow, remains unresolved and requires further elucidation of taxonomic treatment. Currently, the presence of the St genome of Pseudoroegneria and the H genome of Hordeum genus is well-established in hexaploid Roegneria species. However, the origin of the Y genome remains uncertain28,29,30. Two primary hypotheses have been proposed regarding the origin of the Y genome. The first hypothesis postulates that the Y genome evolved from the St genome, supported by nuclear ribosomal internal transcribed spacer sequence analyses and chromosome-specific painting studies31,32. The second hypothesis posits that the Y genome originated independently. Phylogenetic reconstructions based on disrupted meiotic cDNA (DMC1) sequences indicate that although the Y subgenome is closely related to the St genome, it likely originated from a distinct diploid species33,34. Additional analyses of nuclear and chloroplast DNA regions further suggest that the Y subgenome may have been contributed by an independent diploid donor, potentially involving the Dasypyrum and Peridictyon (Xp) genomes in its formation and evolution28. Resolving the enigmatic origin of the Y subgenome requires further investigation into the genome constitution of Roegneria species. This knowledge will enhance our understanding of their evolutionary history.
In this work, we generate high-quality whole-genome assemblies of Th. intermedium and R. kamoji and resolve their phylogenetic placements within the Triticeae tribe. We also functionally characterize the previously identified FHB resistance gene Fhb7 in both species35. These findings offer genomic resources to support future studies on polyploidization and facilitate trait introgression in wheat breeding.
Results
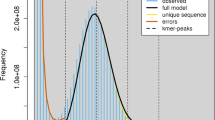
Genome assembly and annotation
We selected accessions of Th. intermedium (PI 440031) and R. kamoji (Pr 87-88 353) for whole-genome sequencing and assembly. Genome size estimates derived from k-mer frequency analysis yielded values of 11.75 Gb for Th. intermedium and 11.26 Gb for R. kamoji (Supplementary Figs. 1 and 2), consistent with flow cytometry measurements of 11.84 Gb and 11.16 Gb per 1C, respectively, using the 14.51 Gb telomere-to-telomere genome assembly of wheat cultivar “Chinese Spring” as a reference36 (Supplementary Fig. 3). PacBio HiFi sequencing generated ~349 Gb and 551 Gb of data, yielding assembled genome sizes of 10.89 Gb and 11.14 Gb, closely aligning with the initial estimates (Supplementary Data 1, Supplementary Figs. 1 and 2). To further improve the assemblies, high-throughput chromosome conformation capture (Hi-C) data were generated at ~136× and 149× coverage for Th. intermedium and R. kamoji, respectively. These data enabled the organization and anchoring of 10.65 Gb and 11.07 Gb of contigs onto 21 pseudo-chromosomes per species, representing 97.8% of the Th. intermedium assembly and 99.4% of the R. kamoji assembly (Supplementary Fig. 4, Supplementary Data 2 and 3). The remaining unanchored sequences totaled 240 Mb for Th. intermedium and 64.5 Mb for R. kamoji. Both assemblies demonstrated exceptional genic completeness, with 99.5% of Embryophyta genes identified based on Benchmarking Universal Single-Copy Orthologs (BUSCO) analysis37 (Table 1, Supplementary Data 4). Of these, 98.9% and 99.3% were classified as complete. Assessment of repeat sequence integrity using the long terminal repeat (LTR) assembly index (LAI)38, yielded scores ≥15 in both assemblies, with Th. intermedium slightly outperforming R. kamoji (Supplementary Data 5). Subgenome partitioning using Subphaser software39 effectively clustered and assigned the 21 pseudo-chromosomes into three subgenomes for each species (Supplementary Fig. 5), further reinforcing the high quality of the assemblies and establishing a solid foundation for downstream phylogenetic and comparative genomic analyses.
Transposable element (TE) annotation revealed that TEs occupied 77.15% and 78.49% of the genome sequences in Th. intermedium and R. kamoji, respectively (Supplementary Fig. 6, Supplementary Data 6 and 7). As observed in other Triticeae species, Gypsy and Copia retrotransposons were the most abundant TE families. Gypsy elements were predominantly concentrated in centromeric regions, whereas Copia elements exhibited a more dispersed, contrasting distribution (Fig. 1a, b). The density patterns of Cereba and Quinta LTR retrotransposons across the pseudo-chromosomes confirmed the successful assembly of centromeric regions (Supplementary Figs. 7 and 8). No evidence of recent LTR retrotransposon bursts was detected in either species, suggesting relatively stable genomes (Supplementary Fig. 9). This genomic stability may help explain the compatibility of these species in distant hybridization with common wheat. TE activity was higher in the subgenomes of Th. intermedium than in R. kamoji subgenomes, potentially explaining the higher LAI score observed in Th. intermedium (Supplementary Fig. 9 and Supplementary Data 5). For comparative analysis, TE annotation was also performed on selected diploid species from genera likely involved in the ancestry of the studied hexaploids, including Aegilops tauschii Coss. (DD, 2n = 2x = 14)40, Pseudoroegneria libanotica (Hack.) D.R.Dewey (StSt, 2n = 2x = 14)41, Dasypyrum villosum (L.) P.Candargy (VV, 2n = 2x = 14)42, and Hordeum marinum Huds. (XaXa, 2n = 2x = 14)43 (Supplementary Data 8). TEs accounted for ~85.09%, 70.92%, 83.07% and 82.34% of the total genome content in these species, respectively, consistent with their original annotation results.
a Th. intermedium; b R. kamoji. Lanes in the circular diagram, from outside to inside, are as follows: (A) chromosome names with tick marks placed at 25-Mb intervals; (B) GC content percentage (40%–50%); (C) density of high-confidence genes (0%–15%); (D) Copia-like retrotransposon density (0%–30%); (E) Gypsy-like retrotransposon density (0%–80%); (F) CACTA DNA transposon density (0%–30%). Connecting lines in the center represent synteny (gray lines) and translocated regions (red lines) among chromosomes of different subgenomes.
We annotated 133,758 and 135,450 high-confidence protein-coding genes in the Th. intermedium and R. kamoji assemblies, respectively, using an integrated approach combining homology-based, ab initio, and transcriptome-based prediction methods. Gene distribution across subgenomes revealed 43,284, 42,646, and 43,020 genes in Th. intermedium and 43,153, 39,845, and 45,456 genes in R. kamoji (Table 1 and Supplementary Data 9). Functional annotation was achieved for ~98.2% of the predicted genes in Th. intermedium and 99% of the predicted genes in R. kamoji, indicating high quality and completeness of the genome assemblies (Supplementary Data 10). Genome-wide visualization of key structural features, including TEs, Copia, and Gypsy retrotransposons, and annotated genes, was performed across all pseudo-chromosomes, providing a detailed representation of the genomic architecture and functional elements (Fig. 1a, b).
Molecular phylogeny and genome constitution of Th. intermedium
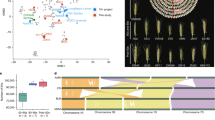
To elucidate the phylogeny and genome constitution of hexaploid Th. intermedium and R. kamoji, we employed two distinct methodological approaches to reconstruct phylogenetic relationships within the Triticeae tribe. First, we conducted an integrated analysis using transcriptomic, exomic, and genomic datasets to construct a phylogeny based on 150,514 shared SNP loci (Fig. 2a and Supplementary Data 11). Second, we performed a complementary phylogenomic reconstruction using single-copy orthologous genes extracted from publicly available Poaceae genomes and the assemblies generated in this study (Supplementary Fig. 10). Both approaches yielded concordant topologies. In Th. Intermedium, the three subgenomes clustered with: St-clade of Pseudoroegneria, V-clade of Dasypyrum, and a heterogeneous clade of Aegilops, which we designated as ThinSt, ThinV, and ThinJ, respectively. Reciprocal mapping, conducted using 30× simulated whole-genome reads from the St genome of Pse. libanotica, the V genome of D. villosum, and the D genome of Ae. tauschii, further validated the phylogenetic placement of these subgenomes (Fig. 2b and Supplementary Fig. 11). Notably, the ThinJ subgenome was found to be a sister to the D- and S-genome Aegilops species, with an estimated divergence time of ~3.8 million years. Analysis of TE composition revealed notable similarities between ThinJ and Ae. tauschii, particularly in the CACTA DNA transposon family, which constituted 20.02% of the ThinJ subgenome and 21.51% of the Ae. tauschii genome (Supplementary Data 6 and 8). Furthermore, the 10 most abundant LTR retrotransposon subfamilies revealed no significant differences between ThinJ and Ae. tauschii (Supplementary Fig. 12). These findings collectively confirmed that ThinJ originated from the Aegilops genus. Comparative structural analysis of Th. intermedium subgenomes and their corresponding diploid relatives revealed no major chromosomal translocations or inversions, with the exception of a V04 to V07 translocation between ThinV and the V genome of D. villosum. This translocation, also observed in other Triticeae species, appeared to be restricted to the Dasypyrum lineage42 (Fig. 2c).
a Maximum-likelihood phylogeny of Triticeae species constructed using whole-genome shared SNPs. Branch support values displayed at nodes represent 1000 standard bootstrap replicates. Evolutionary nodes annotated with red ellipses indicate divergence times and 95% Highest Posterior Density Intervals. b Whole-genome mappings among closely related diploid species, Th. intermedium, and R. kamoji. Xa: H. marinum; V: D. villosum; St: Pse. libanotica; D: Ae. tauschii; E: Th. elongatum; Thin: Th. intermedium; Rka: R. kamoji. c Synteny analysis among Th. intermedium, R. kamoji, and closely related diploid species. Blue lines indicate the 4V-5V-7V translocation events among V-clade species; red lines indicate inversion events; gray lines represent synteny blocks. d Inferred speciation of Th. intermedium and R. kamoji.
No subgenome of Th. intermedium exhibited close genetic similarity to Th. elongatum, previously proposed as an ancestral donor18. Simulated reads from the E genome of Th. elongatum could not be mapped to any of the Th. intermedium subgenomes, effectively ruling out Th. elongatum as a diploid contributor (Fig. 2b). To validate this result, we sequenced six additional Th. intermedium accessions at an average coverage of 14× (Supplementary Data 12). When mapped to the combined Th. intermedium and Th. elongatum genome assemblies, the sequenced reads showed a significantly lower alignment rate to Th. elongatum than to the Th. intermedium subgenomes (Supplementary Fig. 13), further excluding Th. elongatum as a genomic contributor to Th. intermedium.
To cytogenetically validate the genome composition of Th. intermedium, we performed sequential GISH and FISH analyses. FISH with the V-genome-specific probe Oligo-pDb12H44 and the Aegilops species abundant tandom repeat probe Oligo-Ae584 we designed in this study revealed distinct hybridization patterns: 14 chromosomes displayed strong V genome signals, consistent with a previous study18, while Oligo-Ae584 predominantly labeled the ThinJ chromosomes (Supplementary Fig. 14a, b, Supplementary Data 13). Sequential GISH using the total genomic DNA of Pse. spicata (St) as probe confirmed 14 chromosomes of St origin (Supplementary Fig. 14c). These cytogenetic results definitively support the genome composition inferred from phylogenetic analyses (Supplementary Fig. 14d). Accordingly, the genome of Th. intermedium is derived from progenitors corresponding to present-day genera Pseudoroegneria, Dasypyrum, and Aegilops.
Molecular phylogeny and genome constitution of R. kamoji
Phylogenetic analysis of Triticeae species revealed that two subgenomes of R. kamoji (hereafter referred to as RkaSt and RkaH) clustered with the St-clade of Pseudoroegneria and H-clade of Hordeum species, respectively, consistent with a previous report33 (Fig. 2a). The third subgenome of R. kamoji clustered with the V-clade of Dasypyrum (hereafter referred to as RkaY), rather than the previously presumed St-clade of Pseudoroegneria31 (Fig. 2a). This subgenome exhibited an ancient divergence (~6.0 million years ago) from other V-clade species. Whole-genome mapping of simulated reads from the V genome of D. villosum to R. kamoji showed no significant alignment signals with RkaY, suggesting a more ancient phylogenetic relationship between this subgenome and extant diploid species (Fig. 2b). The presence of a distinct V04-V05 translocation, characteristic of V-clade species, was also detected in RkaY, with breakpoints precisely matching those observed in the V genome of D. villosum and ThinV (Fig. 2c and Supplementary Fig. 15).
GISH and FISH analyses were further performed to validate the phylogenetic divergence of the RkaY subgenome. The 14 chromosomes belonging to the RkaSt subgenome were distinguished using Pse. spicata (St) genomic DNA (Supplementary Fig. 16a). Chromosomes of RkaH were also identified using the H-genome-specific probe Oligo-HvCSR45 and the tandem repeat probe Oligo-pTa535, which is highly amplified in wild Hordeum46 (Supplementary Fig. 16b). However, no hybridization signals were detected for the RkaY subgenome using either the total genomic DNA of D. villosum or the V-genome-specific probe Oligo-pDb12H (Supplementary Fig. 16a). The absence of detectable hybridization signals in RkaY supports our phylogenetic and comparative analyses, suggesting that this subgenome represents an ancient V-clade lineage that has diverged beyond the genomic recognition threshold of extant diploid Dasypyrum species. Collectively, R. kamoji exhibits a tripartite genome structure comprising three distinct subgenomes derived from the St-clade of Pseudoroegneria, the H-clade of Hordeum, and an evolutionarily divergent lineage closely related to the V-clade of Dasypyrum.
Speciation of Th. intermedium and R. kamoji
Although Th. intermedium and R. kamoji share St- and V(Y)-related subgenomes, their overall genome sizes differ significantly (Table 1; ThinSt: 3.04 Gb, RkaSt: 3.98 Gb; ThinV: 3.8 Gb, RkaY: 3.21 Gb). This variation is mainly attributed to differences in repetitive sequences, particularly retrotransposons, as revealed by consistent TE annotation pipelines (Supplementary Fig. 12, Supplementary Data 6 and 7). The size differences corresponded with the more distant phylogenetic divergence observed between ThinSt and RkaSt and between ThinV and RkaY (Fig. 2a), indicating that these subgenomes underwent independent polyploidization events. Notably, the closer affinity of Pse. strigosa to RkaSt and Pse. stipifolia to ThinSt further supports this hypothesis (Fig. 2a).
To elucidate the maternal origins of these two hexaploid species, we generated complete circularized chloroplast genomes using de novo assembly of PacBio HiFi sequencing data. Phylogenetic reconstruction based on these chloroplast assemblies and representative Triticeae chloroplast sequences definitively identified Pseudoroegneria (St) as the maternal donor for both species (Supplementary Fig. 17, Supplementary Data 14), consistent with the established pattern of maternal inheritance in hexaploids20,28. Moreover, the chloroplast phylogeny revealed tetraploid Roegneria species as the closest relatives of the hexaploid Roegneria lineage. Combined with nuclear genome evidence, these findings suggest that the hexaploidization of R. kamoji likely resulted from hybridization between a tetraploid Roegneria ancestor and a diploid Hordeum-related paternal donor (Fig. 2d), and the origin time was estimated to be at least 2.8 million years ago based on current phylogenetic relationships.
Functional evaluation of Fhb7 in Th. intermedium and R. kamoji
FHB, caused by Fusarium species, is a devastating fungal disease affecting wheat globally. Despite over five decades of research, only a few major resistance loci have been identified in common wheat47. Certain R. kamoji germplasms have previously demonstrated excellent resistance to FHB48. In our genome assemblies, we identified two copies of Fhb7 homologs located on the St07 and H07 chromosomes of R. kamoji, designated as Fhb7RkaSt and Fhb7RkaH, respectively. No Fhb7 homolog was detected on the ThinSt subgenome of Th. intermedium; however, one copy (Fhb7ThinJ) was found on J07 chromosome of ThinJ, containing a 19-bp insertion that disrupts protein translation (Supplementary Fig. 18). Comparative genomic analysis indicated that only limited portions of the gene body and its flanking regions of Fhb7 homologs are conserved among Th. intermedium, R. kamoji, Th. elongatum, and their potential donor, Epichloë (Fig. 3a). These results suggest that this horizontal gene transfer of Fhb7 into the Triticeae tribe likely occurred before their divergence, resulting in the integration of a small genomic fragment into the plant genome35.
a Comparison analysis among Fhb7 homologs. b Inoculated wheat spikes including the susceptible cultivar “Fielder,” and resistant transgenic lines T3_Fhb7RkaH, T3_Fhb7RkaSt, and F3_Fhb7RkaSt + Fhb7RkaH. c Statistics of inoculated spikes from wheat cultivar “Fielder,” transgenic lines T3_Fhb7RkaH, T3_Fhb7RkaSt, and F3_Fhb7RkaSt + Fhb7RkaH. The central box denotes the interquartile range (25th to 75th percentiles), with the median marked by an internal line. Whiskers indicate the standard deviation. Group differences were assessed using a two-sided Wilcoxon test, with significance levels: P < 0.05. Sample sizes: Fielder (n = 10), T3_Fhb7RkaH (n = 11), T3_Fhb7RkaSt (n = 11) and F3_Fhb7RkaSt + Fhb7RkaH (n = 36). The letter at the top of the boxes indicates significant differences among groups. d Predicated protein structure of Fhb7 homologs (Fhb7RkaSt and Fhb7RkaH) in R. kamoji. e Enzymatic activity (gray squares) and thermal stability (half-life, t1/2; red circles) of two engineered Fhb7RkaSt missense mutants (P65L and L217M) and their wild-type homologs. Data represent mean ± standard deviation from three independent biological replicates, with error bars indicating variability across measurements. Source data are provided as a Source data file.
Transcriptomic data mapping showed that Fhb7RkaH and Fhb7RkaSt were mainly expressed in the root and grain tissues of R. kamoji, with Fhb7RkaSt exhibiting a higher expression level than Fhb7RkaH (Supplementary Fig. 19). No expression was detected for the disrupted Fhb7ThinJ copy. To evaluate the functional role of Fhb7RkaH and Fhb7RkaSt in FHB resistance, two genomic DNA fragments containing the complete coding sequences and flanking regions were introduced into common wheat. The resulting T3 transgenic lines carrying either homolog exhibited significantly greater FHB resistance than wild-type Fielder (Fig. 3b, c and Supplementary Fig. 20). Furthermore, two transgenic lines verified to contain a single copy of Fhb7 homolog (T1_Fhb7RkaH-5 and T1_Fhb7RkaSt-11) were crossed to combine both homologs (Supplementary Fig. 21, Supplementary Data 15). The resulting homozygous plants displayed slightly enhanced FHB resistance than transgenic plants carrying a single Fhb7 homolog (Fig. 3b, c). Liquid chromatography-high resolution mass spectrometry (LC-HRMS) analysis revealed that both Fhb7RkaH-GST and Fhb7RkaSt-GST could catalyze the formation of deoxynivalenol-glutathione (DON-GSH) adducts, thereby detoxifying DON35. However, Fhb7RkaSt-GST produced more DON-GSH than Fhb7RkaH-GST over the same period, while the pyramid line with both Fhb7RkaH-GST and Fhb7RkaSt-GST showed higher detoxification efficiency than their parental lines (Supplementary Fig. 22).
DNA sequence differences between Fhb7RkaH and Fhb7RkaSt resulted in 12 amino acid substitutions (Fig. 3d and Supplementary Fig. 18). One substitution (P65L) occurred within the predicted N-terminal glutathione binding site (G-site), while the remaining substitutions occurred within the C-terminal hydrophobic substrate binding site (H-site; Fig. 3d). Mutations at positions I127-P128-R129 in Fhb7RkaH-GST formed a loop structure, whereas the corresponding region (M127-H128-M129) in Fhb7RkaSt-GST formed a helix, which was hypothesized to enhance the thermal stability of Fhb7RkaSt-GST. Site mutations of these amino acid substitutions with Fhb7RkaSt as template verified that the relative enzyme activities and protein half-life (t1/2) of P65L and L217M were significantly reduced (Fig. 3e and Supplementary Fig. 23). This mechanistic insight explains the better FHB resistance observed in transgenic lines expressing Fhb7RkaSt-GST compared with those expressing Fhb7RkaH-GST. Overall, these results suggested that Fhb7RkaH and Fhb7RkaSt confer varying degrees of FHB resistance, and their combination can exert a synergistic effect to enhance overall resistance.
Discussion
Deciphering the phylogeny and genome composition of polyploid species within the Triticeae tribe is challenging owing to the presence of excessive repetitive DNA sequences, frequent hybridization events, and complex evolutionary histories49. Addressing these complexities requires advanced genomic tools to unravel the relationships among subgenomes and trace their evolutionary origins. In this study, comparative genomic analyses revealed that the genome constitution of Th. intermedium was contributed by progenitors corresponding to present-day genera Pseudoroegneria, Dasypyrum, and Aegilops. These findings were inconsistent with some earlier hypotheses suggesting that diploid Thinopyrum species directly contributed to the genome of Th. intermedium17,18. Moreover, the ThinJ subgenome of Th. intermedium appeared to represent an ancient lineage closely related to Aegilops, corroborating previous findings19. This relationship might explain the high interspecific compatibility of Th. intermedium with wheat and its extensive use in wheat genetic improvement programs. Nevertheless, comprehensive surveys of Th. intermedium germplasm collections were needed to validate the genome constitution reported, particularly the contributions from the Aegilops lineage. The absence of Thinopyrum-derived subgenomic components in Th. intermedium might necessitate a re-evaluation of its taxonomic placement within the Thinopyrum genus.
The genome constitution of R. kamoji was resolved as comprising three distinct subgenomes derived from Pseudoroegneria, Hordeum, and an evolutionarily divergent lineage closely related to Dasypyrum. The ancient phylogenetic relationship between the RkaY subgenome and the V-clade of Dasypyrum, along with its close association with the St-clade of Pseudoroegneria, has long confounded researchers relying on cytogenetic and limited DNA sequence analyses. Despite sharing two nominally shared subgenomes (St- and V(Y)-related), the divergent evolutionary trajectories of Th. intermedium and R. kamoji reflect distinct polyploidization histories. In particular, the St and V(Y) subgenomes within each species have separate origins, indicating that Th. intermedium and R. kamoji underwent independent polyploidization events. These differences highlighted the unique evolutionary paths that shaped the adaptations and functionalities of Th. intermedium and R. kamoji. Chloroplast phylogenomics identified Pseudoroegneria as the maternal progenitor for both species, consistent with known maternal inheritance patterns. The hexaploidization of R. kamoji likely originated through hybridization between a tetraploid Roegneria ancestor and a diploid Hordeum-related paternal donor.
Our previous study demonstrated that Fhb7 was likely introduced into the ancestor of Thinopyrum through horizontal gene transfer from Epichloë35. In this study, we identified Fhb7 homologs in the Hordeum, Aegilops, and Pseudoroegneria related subgenomes of both Th. intermedium and R. kamoji, suggesting that this HGT event dates back to an earlier time before the speciation of the Triticeae tribe. Further examination of publicly available Poaceae genomes also revealed conserved Fhb7 homologs in non-Triticeae Pooideae species (Alopecurus aequalis, Bromus sterilis, Bromus tectorum, and Hierochloe odorata), while no homologs were detected in other Poaceae subfamilies. Given the limited sampling of sequenced genomes, the occurrence time of this HGT event might be updated in future studies. Fhb7 tends to be predominantly retained in perennial species, such as Thinopyrum, Elymus, Roegneria, and Pseudoroegneria. This retention pattern indicates that perennial species, which are exposed to prolonged biotic stress during Fusarium infections, particularly in the crown region, likely exhibited greater evolutionary pressure to maintain Fhb7 than annual species.
In this study, the presence of functional Fhb7 homologs in R. kamoji and their demonstrated dosage-dependent effects on FHB resistance highlight the potential of wild relatives in addressing critical agricultural challenges. Variations in the stability and enzymatic activity of Fhb7-GST directly influence the FHB resistance. Therefore, enhancing the activity and environmental suitability of this enzyme through rational engineering or directed evolution could improve its application value in the development of FHB-resistant wheat varieties and in mycotoxin detoxification for the feed industry. These findings provide a framework for leveraging wild relatives to enhance wheat resilience by introducing beneficial traits related to biotic and abiotic stresses.
Methods
Sampling
The Th. intermedium accession PI 440031 used for sequencing was originally obtained from the USDA National Plant Germplasm System (NPGS). To minimize risks of seed mix-ups, this accession underwent multiple generations of self-pollination to ensure genetic homogeneity. The R. kamoji accession Pr 87-88 353 was obtained from the Triticeae Research Institute of Sichuan Agricultural University (Ya’an, Sichuan, China), an authoritative institution for Triticeae systematics. This accession was originally collected and taxonomically validated by Prof. Chi Yen and Prof. Junliang Yang, pioneering scholars in Triticeae systematics. Additionally, six Th. intermedium accessions were obtained from the Chinese Crop Germplasm Resources Information System (https://www.cgris.net/) for genome re-sequencing. All accessions were cultivated in greenhouse conditions at Shandong Agricultural University.
Genome survey
Genomic DNA was extracted from young leaves of Th. intermedium and R. kamoji through the CTAB method50. Size-selected paired-end libraries (~500 bp) were constructed according to the standard manufacturer protocol (BGI Inc., Shenzhen, China) and sequenced on the DNBSEQ-T7 platform. For Th. intermedium and R. kamoji, 1270 Gb and 913 Gb of paired-end reads were generated, providing ~108× and 81× coverage of their respective estimated genome sizes. The genome size of Th. intermedium was estimated as 11.75 Gb, with a heterozygosity rate of 1.35% and a repeat content of 85.8%. By contrast, R. kamoji had an estimated genome size of 11.26 Gb, a heterozygosity rate of 0.17%, and a repeat content of 87.42%, as determined using Jellyfish (v. 2.3.1) software51.
The genome sizes of Th. intermedium and R. kamoji were also determined via flow cytometry with wheat cultivar “Chinese Spring” as an internal reference standard. Fresh leaf tissues (~0.5 cm2 per sample) were rapidly excised and immersed in 500 μL of CyStain™ PI Absolute P nuclei extraction buffer (Sysmex Partec, Görlitz, Germany). Tissue homogenization was performed by chopping samples with a sterile razor blade for 30 s in a Petri dish containing the extraction buffer, followed by a 30-s incubation to facilitate nuclei release. The resulting lysate was filtered through a 50 μm CellTrics® nylon mesh (Sysmex Partec) to remove cellular debris. The nuclear suspension was then mixed with 2 mL of CyStain™ PI Absolute P staining buffer containing propidium iodide (Sysmex Partec) and RNase A, followed by 30-min dark incubation at 4 °C for chromatin stabilization and stoichiometric DNA staining. Nuclear DNA content was analyzed using a BD FACSAria™ III flow cytometer (BD Biosciences, San Jose, CA, USA) equipped with a 488 nm laser. Three biological replicates were analyzed for each species to ensure measurement reproducibility. Genome size = mean peak value/peak value (CK) × genome size (CK).
PacBio HiFi sequencing
High-molecular-weight genomic DNA was sheared to a target size of ~20 kb. Library preparation was performed using the SMRTbell Express Template Prep Kit 2.0 (Pacific Biosciences, Menlo Park, CA, USA). The resulting libraries were sequenced on a Pacific Biosciences Sequel II platform in CCS mode. Raw data were processed into high-precision HiFi reads at Novogene Co., Ltd. (Tianjin, China). Sequencing yielded ~349 Gb of raw reads for Th. intermedium and 551 Gb for R. kamoji, with N50 read lengths of ~16 kb and 18 kb, respectively. These datasets provided genome coverage of ~30× for Th. intermedium and 49× for R. kamoji.
Hi-C library construction and sequencing
Hi-C libraries were prepared following a previously described protocol35. In brief, fresh leaf tissues from Th. intermedium and R. kamoji were fixed in 2% formaldehyde solution prepared in MS buffer. Nuclei were isolated by homogenizing the tissue in liquid nitrogen, followed by filtration through a cell strainer. The cross-linked DNA was subjected to digestion using the restriction enzyme DpnII at 37 °C. Biotin-labeled adapters were ligated to the sticky ends of the digested fragments to form chimeric junctions. These fragments were enriched and trimmed to ~450 bp. Quality-controlled Hi-C libraries were sequenced on the DNBSEQ-T7 platform at Novogene Co. Ltd. (Tianjin, China). Sequencing produced ~1600 Gb and 1681 Gb of Hi-C reads, supporting pseudo-chromosome assembly for Th. intermedium and R. kamoji, respectively.
Genome assembly and evaluation
PacBio HiFi reads generated for Th. intermedium and R. kamoji were initially assembled into contigs by using hifiasm (v. 0.13-r30852)52, with default parameters. Hi-C reads were mapped to the assembled contigs with HiCUP (v. 0.7.3-1)53. Contig clustering was performed using ALLHiC54 with the parameter “ALLHiC_partition -e GATC -k 21.” The resulting draft assemblies were manually refined and corrected using Juicebox (v. 1.11.08)55. Genome assembly quality was evaluated using BUSCO (v. 5.8.1)37 to assess gene completeness against 1,614 embryophyte genes from the “Embryophyta_odb10” dataset. In addition, repeat sequence completeness was assessed through the LAI38, which quantifies full-length LTR retrotransposons.
Subgenome phasing and assignment
Protein-coding gene sequences from diploid Th. elongatum35 were aligned to the genome sequences of Th. intermedium and R. kamoji using Gmap (v. 2023-04-28)56. Pseudo-chromosomes were assigned to homoeologous chromosome sets based on the whole-genome protein sequences alignment results, with each group consisting of three pseudo-molecules. Subphaser (v. 1.2)39 was employed to phase the assembled Th. intermedium and R. kamoji genomes by identifying subgenome-specific k-mers and TEs.
TE annotation
TEs in the Th. intermedium and R. kamoji genome assemblies were annotated using a homology-based prediction approach. The ClariTeRep TE databank (https://github.com/jdaron/CLARI-TE), containing 3050 high-quality wheat TE sequences, was combined with the TREP nucleotide sequence database, which includes 4162 complete plant TE sequences (https://trep-db.uzh.ch/downloads/trep-db_complete_Rel-19.fasta.gz), to create a universal library. TE annotation was performed using RepeatMasker (v. 4.0.5)57, with the merged datasets serving as the search library. The same method was applied to annotate TE sequences in the genome assemblies of potential diploid progenitors, including Ae. tauschii (NCBI accession GCA_002575655.2), Pse. libanotica (PRJNA940619), D. villosum (GWH accession GWHBJXA00000000), and H. marinum (GWH accession GWHBJBH00000000).
Gypsy family members RLG_famc8.3 (Cereba), RLG_famc8.1 (Quinta), and RLG_famc8.2 (Quinta) were extracted from the annotated TE datasets of Th. intermedium and R. kamoji. Their density distribution across each pseudo-chromosome was calculated and plotted at a resolution of 1 Mb, which provided a suitable balance between resolution and signal stability for identifying centromere-associated enrichment.
Full-length LTR-RTs were extracted from the evaluation results generated using LTR_retriever (v. 2.9.0)58. The left and right LTR sequences for each LTR-RT were aligned using MUSCLE (v. 3.7)59, and the accumulated divergence (K) between the LTRs was calculated with the distmat utility based on the Kimura substitution correction algorithm in the EMBOSS60 software package. The insertion age of each LTR-RT was estimated using the formula
where r represents the plant TE-specific mutation rate of 1.3 × 10−8 per site per year61.
Transcriptome sequencing
To annotate protein-coding genes in the genomes of Th. intermedium and R. kamoji, total RNA was extracted from various tissues. For Th. intermedium, RNA was collected from the root, shoot, leaf, and spike tissues, whereas for R. kamoji, RNA was extracted from the root, shoot, leaf, spike, and grain tissues. All fresh tissues were immediately frozen in liquid nitrogen to preserve RNA integrity. RNA extraction was performed for each sample using TRIzol Reagent (Invitrogen), following the manufacturer’s protocol. Paired-end RNA sequencing (RNA-seq) libraries were prepared according to the standard manufacturer’s instructions and sequenced on the DNBSEQ-T7 platform at Novogene Co. Ltd. (Tianjin, China). In addition, ISO-seq libraries with an insert size range of 0–10 kb were constructed using the same tissue samples for each species. These libraries were sequenced on the PacBio SMRT Sequel platform at Novogene Co. Ltd.
Protein-coding gene annotation
Protein-coding genes in the Th. intermedium and R. kamoji genomes were annotated using a previously established pipeline35. In brief, annotation results from homology-based, ab initio, and RNA-seq-based prediction methods were integrated through a comprehensive strategy. High-confidence protein-coding genes from annotations of T. urartu62, Ae. tauschi40, T. turgidum63, T. aestivum64, Th. elongatum35, D. villosum42, H. vulgare65, Brachypodium distachyon66, Oryza sativa67, and Zea mays68 were aligned to the Th. intermedium and R. kamoji genome assemblies by using WUblast (v. 2.0)69. The genomic regions identified through alignment were further analyzed using GeneWise (v. 2.4.1)70 to refine gene structures. Ab initio gene prediction was conducted using multiple software tools, including Augustus (v. 2.5.5)71, SNAP (v. 2013-02-16)72, Genscan (v. 1.0)73, Geneid (v. 1.4)74, and GlimmerHMM (v. 3.0.1)75, on repeat-masked Th. intermedium and R. kamoji genome assemblies. RNA-seq data from earlier experiments were mapped to the genome assemblies using HISAT2 (v. 2.2.1)76, and transcripts were assembled using StringTie (v. 2.2.3)77. The gene expression value FPKM was also calculated using this software based on the final annotated genes. ISO-seq reads were mapped to the genome using BLAT (v. 36)78, and gene models were assembled with PASA (v. 2.2.0)79. The resulting gene models were used as training data for further ab initio predictions. Gene models obtained using the homology-based, ab initio, and RNA-seq-based methods were integrated into a nonredundant gene dataset using EvidenceModeler (EVM) (v. 1.1.1)79. Low-confidence gene models were filtered out based on the following criteria: (1) coding sequences shorter than 150 bp and (2) supported only through ab initio predictions. The final predicted protein-coding genes were functionally annotated by searching the SwissProt80, KEGG81, and NR82 databases using BLASTN (v. 2.6.0+)83, with an e-value cutoff of 1e-5. Protein domains were identified by querying the InterPro (v. 32.0)84 and Gene Ontology databases through InterProScan (v. 5.51)85, using the following parameters: “-f TSV -dp -goterms –iprlookup.”
Phylogenetic analysis of the Triticeae species
We employed two complementary approaches to reconstruct phylogenetic relationships within the Triticeae tribe. The first strategy utilized single-copy orthologous genes identified across publicly available Poaceae genomes and our assemblies. Using OrthoFinder (v. 2.5.4)86, we analyzed genome annotations from 15 species: T. aestivum64, T. urartu62, Ae. tauschii40, Ae. sharonensis87, Ae. longissima87, Ae. bicornis87, Ae. searsii87, Ae. speltoides87, Th. elongatum35, S. cereale88, Pse. libanotica41, D. villosum42, H. marinum43, H. vulgare65, B. distachyon66, and the Th. intermedium and R. kamoji assemblies generated in this study. To ensure phylogenetic resolution, polyploid subgenomes (T. aestivum, Th. intermedium, and R. kamoji) were treated as independent diploid entities. This analysis identified 129 single-copy genes for phylogenetic reconstruction using RAxML (v.8.2.12)89 under the maximum-likelihood framework, with subsequent amino acid substitution modeling performed through CodeML and MCMCTree in PAML (v. 4.9)90. Branch support values were based on 1000 standard bootstrap replicates.
The second approach leveraged shared SNP loci derived from transcriptomic, exomic, and genomic datasets. For non-sequenced species, we obtained NCBI datasets including Aegilops caudata (CC, 2n = 2x = 14), Ae. comosa (MM, 2n = 2x = 14), Ae. mutica (TT, 2n = 2x = 14), Ae. umbellulata (UU, 2n = 2x = 14), Hordeum pusillum (II, 2n = 2x = 14), H. chilense (II, 2n = 2x = 14), H. bulbosum (II, 2n = 2x = 14), H. bogdanii (II, 2n = 2x = 14), H. brevisubulatum (II, 2n = 2x = 14), H. murinum (II, 2n = 2x = 14), Pseudoroegneria spicata (StSt, 2n = 2x = 14), Pse. stipifolia (StSt, 2n = 2x = 14), Pse. strigosa (StSt, 2n = 2x = 14), Pse. cognata (StSt, 2n = 2x = 14), Eremopyrum triticeum (FF, 2n = 2x = 14), Crithopsis delileana (KK, 2n = 2x = 14), Psathyrostachys juncea (NsNs, 2n = 2x = 14), Henrardia persica (OO, 2n = 2x = 14), Agropyron cristatum (PP, 2n = 2x = 14), Heteranthelium piliferum (QQ, 2n = 2x = 14), Taeniatherum caput-medusae (TaTa, 2n = 2x = 14), Australopyrum retrofractum (WW, 2n = 2x = 14) and Peridictyon sanctum (XpXp, 2n = 2x = 14). For species with available assemblies, we simulated 30× whole-genome paired-end reads using wgsim (https://github.com/lh3/wgsim), including T. aestivum, T. urartu, Ae. tauschii, Ae. sharonensis, Ae. longissima, Ae. bicornis, Ae. searsii, Ae. speltoides, Thinopyrum elongatum, Secale cereale, Pse. libanotica, Dasypyrum villosum, H. marinum, H. vulgare, Brachypodium distachyon, and the assemblies generated in this study. Subgenomes of polyploids were treated as diploid species.
All datasets underwent standardized processing: Reads were aligned to the Th. intermedium J-subgenome using BWA-MEM (v. 0.7.17-r1188)91 with the parameters “-t 4 -k 32 –M.” Subsequently, PCR duplicate removal was performed using SAMtools (v. 1.17)92. Variant calling was performed via GATK HaplotypeCaller (v 4.6.0.0)93, and population-level genotypes were obtained after filtering with VCFtools (v. 2.31.0)94 using the parameter: “--max-missing 1.” Phylogenetic tree was reconstructed using IQ-TREE295 software with parameter: “-T 20 -m MFP -b 1000.” Finally, the MCMCtree program implemented in PAML (v. 4.9)90 was applied to infer the divergence time with the following parameters: “burn-in: 10,000, sample-number: 1,000,000, sample-frequency: 2.” Divergence time estimates were calibrated using established TimeTree (http://www.timetree.org/) benchmarks: Hordeum-Triticum (8.7–11.1 Mya) and Ae. tauschii-Th. elongatum (4.7–5.0 Mya).
Whole-genome synteny analysis
Protein sequences of annotated genes from Ae. tauschii, D. villosum, Pse. libanotica, H. marinum, and our assemblies were reciprocally aligned using BLASTP (v. 2.6.0+) with an e-value cutoff of 1e−05. The reciprocal best hit for each alignment was used to build and illustrate the whole-genome collinearity among these species using jcvi software (https://github.com/tanghaibao/jcvi/wiki).
Whole-genome reads sequencing, simulating, and mapping
To further clarify the genome constitution of Th. intermedium and R. kamoji, high-molecular-weight genomic DNA was extracted from six Th. intermedium accessions and sequenced on the DNBSEQ-T7 platform (Novogene Co. Ltd., Tianjin, China). All reads were mapped to the combined Th. intermedium and Th. elongatum assemblies using BWA-MEM (v. 0.7.17-r1188)91. The coverage of mapped reads was calculated for each pseudo-chromosome using bedtools (v. 2.31.0)96 at a resolution of 1 Mb. Simulated reads from Ae. tauschii, D. villosum, Pse. libanotica, and Th. elongatum were mapped to the Th. intermedium genome assembly, whereas reads from H. marinum, D. villosum, and Pse. libanotica were mapped to the R. kamoji genome assembly. Reads from Th. intermedium were mapped to the combined Ae. tauschii, D. villosum, and Pse. libanotica genome assemblies, and reads from R. kamoji were mapped to the combined H. marinum, D. villosum, and Pse. libanotica assemblies. The mapped read coverage was calculated within a 1-Mb window using bedtools96.
Genomic in situ hybridization and Fluorescence in situ hybridization analysis
Mitotic metaphase chromosomes were prepared from the sample root tips following the previous protocol with modifications97. In brief, seeds were germinated on moist filter paper in Petri dishes at 23 °C in the dark until the roots reached 1.5–2.0 cm in length. Root tips were excised and treated with nitrous oxide gas under 1 MPa pressure for 2 h, fixed in ice-cold 90% acetic acid for 10 min, and then stored in 70% ethanol at −20 °C. For cell dissociation, root tip sections containing dividing cells were washed in ice-cold water, digested in 20 μL of enzymatic solution (1% pectolyase Y23 and 2% cellulose Onozuka R-10; Yakult Pharmaceutical, Tokyo) at 37 °C for 55 min, and rinsed sequentially with 70% and 100% ethanol. The meristematic region was mechanically disrupted using a needle, vortexed in 100% ethanol for 30 s to separate cells, and centrifuged. Pelleted cells were resuspended in 90% acetic acid, and the suspension was dropped onto glass slides in a humidity-controlled chamber. For GISH, genomic DNA from Pse. spicata (St) and D. villosum (V) was labeled with Fluorescein-12-dUTP (NEL 413, Perkin Elmer) and Texas Red-5-dATP (NEL 471, Perkin Elmer), respectively, via the nick translation method (Roche Diagnostics, Indianapolis, IN, USA). For FISH, publicly available Oligo probes, including Oligo-pDb12H (TCAGAATTTTTAGGATAGCAGAAGTATTCGAAATACCCAGATTGCTACAG), Oligo-HvCSR (ACAACGACAACAACGACAATGACGAGA), and Oligo-pTa535 (AAAAACTTGACGCACGTCACGTACAAATTGGACAAACTCTTTCGGAGTATCAGGGTTTC), were collected, and Non-denaturing FISH experiments were performed. The Oligo-Ae584 probe sequence was designed based on the genome sequences of Aegilops species87 using Tandem Repeat Finder software (v 4.09)98 with default parameters. Publicly available probes and custom oligonucleotides (Tsingke Biological Technology, Beijing) were 5′-end labeled with 6-carboxyfluorescein (green fluorescence) or tetramethylrhodamine (red fluorescence). Hybridized slides were imaged using an Olympus BX-53 fluorescence microscope equipped with a DP-80 CCD camera. Sequential GISH and FISH analyses were performed to confirm genomic composition and chromosomal localization of signals. Probe specificity and hybridization efficiency were validated through repeated experiments under standardized conditions.
Chloroplast genome assembly and phylogeny analysis
To elucidate the maternal origins of Th. intermedium and R. kamoji, complete circularized chloroplast genomes were generated by de novo assembly of PacBio HiFi sequencing data using the oatk software (v. 1.0)99 with the parameter “-k 1001 -c 200.” The assemblies, along with representative Triticeae chloroplast sequences, were merged and aligned using MAFFT (v. 7.520) software100. A maximum-likelihood phylogenetic tree was constructed with IQ-TREE295 under the parameters “-m MFP -b 1000.”
Wheat transformation and copy number assay evaluation
To evaluate the functions of Fhb7RkaH and Fhb7RkaSt, a 9281-bp genomic DNA fragment containing the entire coding region (849 bp) of Fhb7RkaH, along with 4586 bp upstream of the start codon and 3846 bp downstream of the stop codon, was amplified from R. kamoji using 2-x Phanta Max Master Mix (Vazyme, Nanjing, China). Similarly, a 5566-bp genomic DNA fragment containing the full coding region (852 bp) of Fhb7RkaSt, 1346 bp upstream of the start codon, and 3368 bp downstream of the stop codon, was amplified. The amplified fragments were inserted into the linearized binary vector pCAMBIA3301 by using LightNing DNA Assembly Mix Plus (iScience, Jiangsu, China). The resulting plasmids, pCAMBIA3301-Fhb7RkaH and pCAMBIA3301-Fhb7RkaSt, were introduced into the susceptible hexaploid wheat line Fielder through Agrobacterium tumefaciens (strain EHA105)-mediated transformation. Transgene integration was confirmed using the primer pairs Fhb7RkaHF1R1 and Fhb7RkaStF1R2. The number of integrated Fhb7RkaH and Fhb7RkaSt transgenes in each transgenic line was estimated based on the segregation ratios of T1 plants and validated through a droplet digital PCR copy number assay101.
FHB evaluation of Th. intermedium, R. kamoji and wheat lines
Wheat lines, including the T3_Fhb7RkaH-5 line, T3_Fhb7RkaSt-11 line, F3 line of combined Fhb7RkaH and Fhb7RkaSt, and the non-transgenic wheat line Fielder, were cultivated in a growth chamber at Shandong Agricultural University. Growth conditions were maintained at 25/20 °C (day/night), with a 16/8 h light/dark photoperiod and 60%–80% relative humidity.
At the flowering stage, 20 spikes from each wheat line were selected for inoculation. A single basal floret from the fifth spikelet at the tip of each spike was injected with 10 μL of Fusarium graminearum (PH-1) conidia suspension (50,000 conidia mL−1) by using a syringe. After inoculation, the spikes were enclosed in transparent plastic bags to maintain high humidity for 48–96 h. The bags were removed once water-soaked lesions appeared at the base of the glumes in susceptible control spikelets. Disease severity was assessed 21 days post-inoculation (dpi) by counting the number of diseased spikelets per spike. Disease severity is expressed as the percentage of infected spikelets relative to the total number of spikelets per spike.
For leaf inoculation assays, leaves were collected at the one-leaf stage from Th. intermedium, R. kamoji, and wheat lines with uniform morphological characteristics (including the T3_Fhb7RkaH-5 line, the T3_Fhb7RkaSt-11 line, and an F3 line carrying both Fhb7RkaH and Fhb7RkaSt). The leaves were excised into 3- to 4-cm segments, and circular lesions were created on the adaxial surfaces by using a hole punch. A 2.5 μL aliquot of F. graminearum conidia suspension (50,000 conidia mL−1) was applied to each wound site. The inoculated leaves were incubated at 25 °C in a controlled humid environment for 3–4 days. Disease progression was monitored, and lesion areas were quantified using ImageJ software.
Quantification of deoxynivalenol and DON-GSH
To quantify the content of DON-GSH, wheat lines were inoculated with deoxynivalenol at the one-leaf stage. Leaf samples from each treatment group, with at least eight biological replicates, were collected 12 h post-inoculation, flash-frozen in liquid nitrogen, and ground to a fine powder for LC-HRMS. LC-HRMS was conducted using a Thermo Scientific Q Exactive instrument (Accela, Thermo Fisher Scientific, San Jose, CA, USA)35. Prior to analysis, the samples were filtered through a 0.22-μm filter and injected into the LC-HRMS system. The injection volume was 3 μL, with a flow rate of 300 μL/min, and the column temperature was maintained at 35 °C. The mobile phase consisted of 0.1% acetic acid (phase A) and acetonitrile (phase B). The gradient program was as follows: 0–0.5 min: 90% phase A, 0.5–3.5 min: 90% to 10% phase A, 3.5–6.5 min: 10% phase A, 6.5–6.6 min: 10% to 90% phase A, and 6.6–10 min: 90% phase A. Parallel reaction monitoring mode was used to quantify the relative levels of deoxynivalenol and its derivatives in enzymatic solutions. Quantification was achieved by rapid switching between positive and negative ion modes, with a resolution of 17,500 and an automatic gain control target value of 2 × 105. The MS/MS acquisition rate was set at 2.5 spectra per second, with normalized collision energies of 15, 30, and 45 eV applied depending on the analyte102. Data acquisition and processing were performed using Xcalibur v. 2.1.0 (Thermo Fisher Scientific).
Plasmid construction, protein purification, and activity analysis
For protein expression and purification, the full-length coding sequences of Fhb7RkaH and Fhb7RkaSt were cloned into the pET28a(+) vector, adjacent to a His-tag-encoding sequence, using the ClonExpress MultiS One Step Cloning Kit. The resulting constructs were transformed into the Escherichia coli strain BL21 (DE3). Recombinant protein production was induced by the addition of 0.25 mM IPTG, followed by incubation at 20 °C for 12 h. Expressed Fhb7RkaH-GST and Fhb7RkaSt-GST proteins were purified using BeyoGold His-tag Purification Resin (P2218, Beyotime, Shanghai, China). To remove the 6xHis tag, TEV protease was added to the purified samples, and the mixture was incubated overnight with gentle shaking. The digested samples were passed through a gravity column containing Ni-NTA resin to eliminate the 6xHis tag and TEV enzyme. Fractions containing the purified Fhb7RkaH-GST or Fhb7RkaSt-GST proteins were concentrated to a final concentration of 1 mg/mL at 4 °C by using an Amicon Ultra tube (Millipore; molecular weight cutoff: 10 kDa) for subsequent activity analysis.
To determine the relative activity of Fhb7RkaH-GST, Fhb7RkaSt-GST, and their mutant variants, reactions were performed in a final volume of 100 μL. Each reaction contained 5 μg of purified protein, 100 μM of deoxynivalenol, and 1 mM of freshly prepared GSH at pH 8.0 and 30 °C for 6 min. The reactions were terminated by incubation at 65 °C for 3 min. The supernatants were analyzed via LC-HRMS as mentioned above. To determine the half-lives (t1/2) of Fhb7RkaH-GST, Fhb7RkaSt-GST, and their mutant variants, the residual activities of enzymes incubated at 30 °C for varying durations were measured. For each assay, 5 µg of protein was added to 100 µL of enzyme buffer (20 mM NaH2PO4–Na2HPO4, 0.2 M NaCl, pH 8.0) and incubated at 30 °C for different times (0–20 min). Samples were then immediately placed on ice. After a 5-min incubation on ice, the reaction was initiated by adding 100 µM DON and 1 mM fresh GSH. Reactions proceeded at 30 °C for 6 min, after which aliquots were quenched with 100 µL of ice-cold acetonitrile and centrifuged at 15,000 × g, 4 °C for 30 min. Supernatants were analyzed by reverse-phase LC-HRMS. Residual activity was defined as the activity of the enzyme incubated for a given time relative to that of the enzyme incubated for 0 min.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw PacBio HiFi, Hi-C, RNA-seq, re-sequencing reads, and genome assemblies of Th. intermedium and R. kamoji generated in this study have been deposited in the NCBI database under accession PRJNA1235488 and PRJNA1235489. The complete chloroplast genomes of these two species generated in this study have been deposited in the NCBI database under accession PV435186.1 [https://www.ncbi.nlm.nih.gov/nuccore/PV435186.1/] and PV435187.1 [https://www.ncbi.nlm.nih.gov/nuccore/PV435187.1/], respectively. The genome assemblies and annotations of these two species were also deposited to Zenodo [https://doi.org/10.5281/zenodo.15412758]. The seeds of these two sequenced accessions could be obtained upon request to the corresponding author to facilitate independent verification. Source data are provided with this paper.
References
Shiferaw, B. et al. Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Secur. 5, 291–317 (2013).
Doebley, J. F., Gaut, B. S. & Smith, B. D. The molecular genetics of crop domestication. Cell 127, 1309–1321 (2006).
Pour-Aboughadareh, A., Kianersi, F., Poczai, P. & Moradkhani, H. Potential of wild relatives of wheat: ideal genetic resources for future breeding programs. Agronomy 11, 1656 (2021).
Stetter, M. G., Gates, D. J., Mei, W. & Ross-Ibarra, J. How to make a domesticate. Curr. Biol. 27, R896–R900 (2017).
Tekin, M. et al. Wild relatives and their contributions to wheat breeding. in Ancient Wheats (eds Zencirci, N., Ulukan, H., Baloch, F. S., Mansoor, S. & Rasheed, A.) 197–233 (Springer International Publishing, 2022).
Pototskaya, I. V., Shamanin, V. P., Aydarov, A. N. & Morgounov, A. I. The use of wheatgrass (Thinopyrum intermedium) in breeding. Vavilov J. Genet. Breed. 26, 413–421 (2022).
Weng, Y. & Liu, D. J. Morphology, scab resistance and cytogenetics of intergeneric hybrids of Triticum aestivum L. with Roegneria C. Koch (Agropyron) species. Sci. Agric. Sin. 22, 1–7 (1989).
Song, R. et al. Transferring a new Fusarium head blight resistance locus FhbRc1 from Roegneria ciliaris into wheat by developing alien translocation lines. Theor. Appl. Genet. 136, 36 (2023).
Zhao, F. et al. Resistance of Roegneria kamoji (Poaceae: Triticeae) populations to stripe rust and powdery mildew. Acta Prataculturae Sin 25, 149–158 (2016).
Salina, E. A. et al. A Thinopyrum intermedium chromosome in bread wheat cultivars as a source of genes conferring resistance to fungal diseases. Euphytica 204, 91–101 (2015).
Ayala-Navarrete, L., Thompson, N., Ohm, H. & Anderson, J. Molecular markers show a complex mosaic pattern of wheat-Thinopyrum intermedium translocations carrying resistance to YDV. Theor. Appl. Genet. 121, 961–970 (2010).
Bao, Y., Li, X., Liu, S., Cui, F. & Wang, H. Molecular cytogenetic characterization of a new wheat-Thinopyrum intermedium partial amphiploid resistant to powdery mildew and stripe rust. Cytogenet. Genome Res. 126, 390–395 (2009).
Ginot, C. et al. Introducing intermediate wheatgrass as a perennial grain crop into farming systems: insights into the decision-making process of pioneer farmers. Agron. Sustain. Dev. 44, 58 (2024).
de Oliveira, G., Brunsell, N. A., Crews, T. E., DeHaan, L. R. & Vico, G. Carbon and water relations in perennial Kernza (Thinopyrum intermedium): an overview. Plant Sci 295, 110279 (2020).
Löve, Á. Conspectus of the Triticeae. Feddes Repert 95, 425–521 (1984).
Ji, W. et al. GISH analysis of Thinopyrum intermedium. Acta Bot. Boreali occident. Sin. 21, 401–405 (2000).
Wang, R. R. et al. Genome evolution of intermediate wheatgrass as revealed by EST-SSR markers developed from its three progenitor diploid species. Genome 58, 63–70 (2015).
Qi, F. et al. Genome analysis of Thinopyrum intermedium and its potential progenitor species using Oligo-FISH. Plants 12, 3705 (2023).
Mahelka, V., Kopecký, D. & Baum, B. R. Contrasting patterns of evolution of 45S and 5S rDNA families uncover new aspects in the genome constitution of the agronomically important grass Thinopyrum intermedium (Triticeae). Mol. Biol. Evol. 30, 2065–2086 (2013).
Mahelka, V., Kopecký, D. & Paštová, L. On the genome constitution and evolution of intermediate wheatgrass (Thinopyrum intermedium: Poaceae, Triticeae). BMC Evol. Biol. 11, 127 (2011).
Song, R. et al. Identification and transferring of a new Fusarium head blight resistance gene FhbRc2 from Roegneria ciliaris 3ScL chromosome arm into common wheat. Crop J 12, 1718–1726 (2024).
Cainong, J. C. et al. Chromosome engineering, mapping, and transferring of resistance to Fusarium head blight disease from Elymus tsukushiensis into wheat. Theor. Appl. Genet. 128, 1019–1027 (2015).
Yang, J. L., Baum, B. & Yen, C. A revision of the genus Roegneria K. Koch (Triticeae: Poaceae). J. Sichuan Agric. Univ. 26, 311–381 (2008).
Koch. Beitrage zueiner Florades Orientes. Linnaea 21, 289–443 (1848).
Yang, C., Baum, B. R., Johnson, D. A., Zhang, H. & Zhou, Y. Molecular diversity of the 5S nuclear ribosomal DNA in Campeiostachys with StHY haplome constitution. J. Syst. Evol. 58, 69–76 (2020).
Yen, C., Yang, J. & Baum, B. R. Biosystematics of Triticeae Vol. 4, 337–582 (China Agricultural Press, 2011).
Mason-Gamer, R. J. Origin of North American Elymus (Poaceae: Triticeae) allotetraploids based on granule-bound starch synthase gene sequences. Syst. Bot. 26, 757–768 (2001).
Lei, Y. et al. Phylogenetic relationships and the maternal donor of Roegneria (Triticeae: Poaceae) based on three nuclear DNA sequences (ITS, Acc1, and Pgk1) and one chloroplast region (trnL-F). J. Syst. Evol. 60, 305–318 (2022).
Hu, Q., Sun, D. & Sun, G. Molecular phylogeny revealed distinct origin of the Y and St genome in Elymus longearistatus (Triticeae: Poaceae). Mol. Phylogenet. Evol. 85, 141–149 (2015).
Fan, X. et al. Phylogenetic relationships and Y genome origin in Elymus L. sensu lato (Triticeae: Poaceae) based on single-copy nuclear Acc1 and Pgk1 gene sequences. Mol. Phylogenet. Evol. 69, 919–928 (2013).
Chen, C. et al. Chromosome-specific painting reveals the Y genome origin and chromosome rearrangements of the St genome in Triticeae. Plant Physiol 196, 870–882 (2024).
Zhang, C. et al. Phylogenetic analysis of questionable tetraploid species in Roegneria and Pseudoroegneria (Poaceae: Triticeae) inferred from a gene encoding plastid acety1-CoA carboxylase. Biochem. Syst. Ecol. 37, 412–420 (2009).
Lei, Y. et al. Phylogeny and molecular evolution of the DMC1 gene in the polyploid genus Roegneria and its affinitive genera (Poaceae: Triticeae). Bot. J. Linn. Soc. 186, 129–142 (2018).
Lei, Y. et al. Phylogenetic analysis of the species with awnless lemma in Roegneria (Poaceae, Triticeae) based on single copy of nuclear gene. DMC1. Biochem. Syst. Ecol. 65, 185–191 (2016).
Wang, H. et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 368, eaba5435 (2020).
Liu, S. et al. A telomere-to-telomere genome assembly coupled with multi-omic data provides insights into the evolution of hexaploid bread wheat. Nat. Genet. 57, 1008–1020 (2025).
Manni, M., Berkeley, M. R., Seppey, M., Simão, F. A. & Zdobnov, E. M. BUSCO Update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 38, 4647–4654 (2021).
Ou, S., Chen, J. & Jiang, N. Assessing genome assembly quality using the LTR Assembly Index (LAI). Nucleic Acids Res 46, e126 (2018).
Jia, K. et al. SubPhaser: a robust allopolyploid subgenome phasing method based on subgenome-specific k-mers. New Phytol 235, 801–809 (2022).
Luo, M. et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551, 498–502 (2017).
Zhai, X. et al. A chromosome level genome assembly of Pseudoroegneria Libanotica reveals a key Kcs gene involves in the cuticular wax elongation for drought resistance. BMC Genom 25, 253 (2024).
Zhang, X. et al. A chromosome-scale genome assembly of Dasypyrum villosum provides insights into its application as a broad-spectrum disease resistance resource for wheat improvement. Mol. Plant 16, 432–451 (2023).
Kuang, L. et al. The genome and gene editing system of sea barleygrass provide a novel platform for cereal domestication and stress tolerance studies. Plant Commun 3, 100333 (2022).
Yu, Z. et al. Characterization of chromosomal rearrangement in new wheat-Thinopyrum intermedium addition lines carrying Thinopyrum-Specific grain hardness genes. Agronomy 9, 18 (2019).
Jiang, C., Liu, X., Yang, Z. & Li, G. Chromosome rearrangement in Elymus dahuricus revealed by ND-FISH and Oligo-FISH painting. Plants 12, 3268 (2023).
Tang, Z., Yang, Z. & Fu, S. Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 55, 313–318 (2014).
Ma, Z. et al. Germplasms, genetics and genomics for better control of disastrous wheat Fusarium head blight. Theor. Appl. Genet. 133, 1541–1568 (2020).
Luo, Y. et al. Evaluation of Fusarium head blight resistance and identification of resistant germplasms in different populations of Roegneria kamoji. J. Plant Genet. Resour. 22, 725–733 (2021).
Feldman, M. & Levy, A. A. Origin and evolution of wheat and related Triticeae species. in Alien Introgression in Wheat: Cytogenetics, Molecular Biology, and Genomics 21–76 (Springer International Publishing, 2015).
Allen, G. C., Flores-Vergara, M. A., Krasynanski, S., Kumar, S. & Thompson, W. F. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325 (2006).
Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770 (2011).
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18, 170–175 (2021).
Wingett, S. et al. HiCUP: pipeline for mapping and processing Hi-C data. F1000Research 4, 1310 (2015).
Zhang, X., Zhang, S., Zhao, Q., Ming, R. & Tang, H. Assembly of allele-aware, chromosomal-scale autopolyploid genomes based on Hi-C data. Nat. Plants 5, 833–845 (2019).
Durand, N. C. et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst 3, 99–101 (2016).
Wu, T. D. & Watanabe, C. K. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21, 1859–1875 (2005).
Tarailo-Graovac, M. & Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Current Protocols in Bioinformatics 25, 4.10.1–4.10.14 (2009).
Ou, S. & Jiang, N. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol 176, 1410–1422 (2018).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797 (2004).
Rice, P., Longden, I. & Bleasby, A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16, 276–277 (2000).
Ma, J. & Bennetzen, J. L. Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA 101, 12404–12410 (2004).
Ling, H. et al. Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature 557, 424–428 (2018).
Maccaferri, M. et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 51, 885–895 (2019).
International Wheat Genome Sequencing Consortium (IWGSC). et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, r7191 (2018).
Mascher, M. et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433 (2017).
The International Brachypodium Initiative. et al. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768 (2010).
Goff, S. A. et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100 (2002).
Jiao, Y. et al. Improved maize reference genome with single-molecule technologies. Nature 546, 524–527 (2017).
Gish, W. & States, D. J. Identification of protein coding regions by database similarity search. Nat. Genet. 3, 266–272 (1993).
Birney, E., Clamp, M. E. & Durbin, R. GeneWise and Genomewise. Genome Res 14, 988–995 (2004).
Keller, O., Kollmar, M., Stanke, M. & Waack, S. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 27, 757–763 (2011).
Korf, I. Gene finding in novel genomes. BMC Bioinform 5, 59 (2004).
Burge, C. & Karlin, S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94 (1997).
Blanco, E., Parra, G. & Guigó, R. Using geneid to identify genes. Current Protocols in Bioinformatics 18, 4.3.1–4.3.28 (2002).
Majoros, W. H., Pertea, M. & Salzberg, S. L. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20, 2878–2879 (2004).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Kent, W. J. BLAT-the BLAST-like alignment tool. Genome Res 12, 656–664 (2002).
Haas, B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol 9, R7 (2008).
Boeckmann, B. et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 31, 365–370 (2003).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28, 27–30 (2000).
Pruitt, K. D., Tatusova, T. & Maglott, D. R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35, D61–D65 (2007).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402 (1997).
Hunter, S. et al. InterPro: the integrative protein signature database. Nucleic Acids Res 37, D211–D215 (2009).
Quevillon, E. et al. InterProScan: protein domains identifier. Nucleic Acids Res 33, W116–W120 (2005).
Emms, D. M. & Kelly, S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol 16, 157 (2015).
Li, L. et al. Genome sequences of five Sitopsis species of Aegilops and the origin of polyploid wheat B subgenome. Mol. Plant 15, 488–503 (2022).
Li, G. et al. A high-quality genome assembly highlights rye genomic characteristics and agronomically important genes. Nat. Genet. 53, 574–584 (2021).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at arXiv https://doi.org/10.48550/arXiv.1303.3997 (2013).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303 (2010).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Han, F., Lamb, J. C. & Birchler, J. A. High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc. Natl. Acad. Sci. USA 103, 3238–3243 (2006).
Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27, 573–580 (1999).
Zhou, C. et al. Oatk: a de novo assembly tool for complex plant organelle genomes. Genome Biol 26, 235 (2025).
Kuraku, S., Zmasek, C. M., Nishimura, O. & Katoh, K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res 41, W22–W28 (2013).
Hindson, B. J. et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 83, 8604–8610 (2011).
Hou, B. et al. A novel strategy for detoxification of deoxynivalenol via modification of both toxic groups. Food Chem 456, 139886 (2024).
Acknowledgements
We thank Professor Haiqin Zhang (Sichuan Agricultural University) for providing the R. kamoji accessions and insightful guidance. We thank the Chinese Crop Germplasm Resources Information System for providing the germplasm resources of Th. intermedium. We thank Professor Ruifen Li (Beijing Academy of Agriculture and Forestry Sciences) for providing the H. brevisubulatum accession. We thank the Supercomputing Center at Shandong Agricultural University for technical support. This work was supported by the National Key Research and Development Program of China (2022YFF1001504) to H.W.W.; the Key R&D Program of Shandong Province, China (2023LZGC022) to S.L.S.; the Key R&D Program of Shandong Province, China (2022LZGC002) to L.R.K.; the National Natural Science Foundation of China (U23A20181) to H.W.W.; and the Shandong Provincial Natural Science Foundation (ZR2021YQ19) to S.L.S.
Author information
Authors and Affiliations
Contributions
S.L.S., H.W.W., and L.R.K. designed the project. S.L.S., L.Y.C., Y.C.H., Q.Y.C., and F.F.Y. performed bioinformatics analysis. N.X.C., Y.C.X., W.Y.G., S.S.Q., W.X.Z., C.M.D., J.Q.L., Y.X.L., L.H., and C.Z.J. conducted experiments. S.L.S., N.X.C., and Q.Y.C. wrote the paper with input from all authors. Z.W.L., L.Z., Z.J.Y., H.W.W., and L.R.K. revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Simon Krattinger, Haiqin Zhang, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sun, S., Che, N., Chen, L. et al. Analysis wheat wild relatives Thinopyrum intermedium and Roegneria kamoji genomes reveal different polyploid evolution paths. Nat Commun 16, 7693 (2025). https://doi.org/10.1038/s41467-025-63007-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-63007-y