Abstract
Cross-reactive antibodies targeting multiple epitopes have been identified in Sarbecoviruses, but the precise molecular mechanism(s) behind the crossreactivity remain poorly understood. Here, we isolate 3D1, a broadly neutralizing antibody (bnAb) derived from a human combinatorial antibody library targeting the conserved HR1 domain. 3D1 uniquely recognizes a β-turn fold comprising a 6-mer peptide (pepDVVNQN/Q) that forms during a pre-hairpin transition state, occurring exclusively before membrane fusion during viral infection. 3D1 effectively neutralizes a wide range of live SARS-CoV-2 wild-type strains except for Omicron, which evades neutralization due to a detrimental point mutation (Q954H). Notably, this cryptic epitope reveals a signature motif that extends throughout the core region of coronaviruses and is also present in various RNA viruses, including HIV and Marburgvirus. 3D1 functions as a natural or background antibody capable of binding to a diverse array of non-self antigens. 3D1’s cross-reactivity underscores the effectiveness of the library approach, which encompasses the entire antibody repertoire.
Similar content being viewed by others
Introduction
The development of potent therapeutic monoclonal antibodies (mAb) with broad neutralizing capacity has become a critical priority in combating RNA viruses, given their rapid antigenic evolution and persistent emergence of pathogenic variants1. Antibodies produced in the body as a natural immune response are triggered by infections or vaccinations. The identification of broad neutralizing antibodies (bnAb) that recognize cross-reactive epitopes (CRE) and tolerate diversity within core epitopes against HIV or Influenza has sparked increased interest in exploring common epitopes and vulnerabilities2,3,4,5,6. Several studies have demonstrated the presence of CREs in the natural immune response against coronaviruses from all genera7,8,9.
The synthetic immune system, utilizing combinatorial antibody library technology10,11, comprises 1011 diverse human antibody sequences and encompasses a comprehensive “fossil record” of adaptive and spontaneous humoral immune responses from the community12,13. The diversity of the synthetic immune system is at least three orders of magnitude larger than the natural B-cell repertoire of an individual11. The chemical nature and vast diversity of this synthetic system enable rapid screening (or evolution) within minutes to hours, in contrast to the days to years required for antibodies to adapt within a natural immune system. Additionally, the community “fossil records,” inherited by a synthetic immune system10,12, ensure comprehensive coverage of rare and specialized antibodies, including cross-reactive antibodies, and provide retrospective tracing of immunological imprinting14,15,16.
Pathogenic human coronaviruses (HCoV), including SARS-CoV-1, MERS-CoV, and SARS-CoV-2, along with HIV and Marburg virus, belong to a super-family of viruses that encode Class I enveloped glycoproteins17. These viruses pose serious threats to global public health18,19,20,21,22,23,24. One prominent feature of Class I viral fusion machinery is the trimeric glycoprotein displaying on virion surface, as observed in coronaviruses, retroviruses, filoviruses, orthomyxoviruses, and paramyxoviruses24,25. This trimeric structure is crucial for membrane fusion, which forms an intermediate structure that exposes three heptad repeat 1 domains (HR1) required for progression to the six-helix bundle (6-HB) after receptor-binding26,27,28. Subsequently, three α-helices of the heptad repeat 2 domain (HR2) pack antiparallel into the grooves of the coiled-coil of trimeric HR1s. The functionally and structurally conserved intermediate, often referred to as the “pre-hairpin intermediate,” is the target of peptidomimetic or small molecule inhibitors, as well as various antibodies29,30,31,32.
The HR1, within the S2 subunit of the spike protein from α, β, γ, and δ genera of coronaviruses, is highly conserved and serves as a fusion-promoting hexameric helical scaffold33. Several anti-SARS-CoV-2 antibodies (e.g., S2P6, B6, 28D9 and CC40.8) target the exterior of the S1 subunit, recognizing a relatively conserved stem helix region in the S2 subunit of the spike protein to inhibit membrane fusion34,35,36. However, the cross-immunoreactivities of these antibodies are restricted within the β-Coronavirus genera. In our search for other targeting epitopes for broad neutralizing antibodies, we analyzed viral sequences that are highly conserved and exposed during the process of viral entry. We hypothesized that these conserved epitopes, exposed only when needed, are the most vulnerable sequences for viral survival. The heptad repeat domain 1 (HR1), corresponding to NHR in HIV, within the S2 subunit of the spike protein from α, β, γ, and δ genera of coronaviruses is highly conserved and serves as a fusion-promoting hexameric helical scaffold, along with the HR2 domain (corresponding to CHR in HIV), that triggers membrane fusion33.
Here, we design a peptide antigen based on a conserved consecutive motif in HR1 to screen a naïve combinatorial library for high-affinity mAbs. One candidate, 3D1, potently neutralizes authentic SARS-CoV-2 and variants of concern (VOC), except Omicron, and exhibits pan-coronavirus inhibitory activity against pseudovirus entry. Structural analyses reveal that 3D1 targets a conserved receptor-binding epitope (CRE) that shares both a consensus amino acid sequence and a conserved secondary structure. By recognizing this characteristic fold, 3D1 demonstrates remarkable tolerance for epitope variation across viral families.
Results
Selection of monoclonal antibodies that neutralize a broad spectrum of coronaviruses
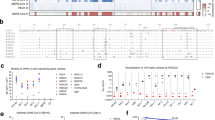
Since the emergency of coronaviruses in humans in 1965, mankind has experienced an ever-increasing threat of coronavirus mediated pandemics (Fig. 1a). The receptor-binding domain (RBD), which is less conserved and highly mutated, is not the optimal therapeutic target to prevent the spillover of zoonotic sarbecoviruses (Supplementary Fig. 1). Sequence alignment of the S2 HR1 regions from 19 sarbecoviruses revealed high homology among SARS-CoV-2, SARS-CoV-1, and its zoonotic orthologs (Fig. 1b). The HR1 fusion cores (HR1FC) can be further grouped into seven representative clusters based on sequence identity (HR1a – g, Fig. 1b).
a Emerging timeline for human coronaviruses, among which Disease X represents a threat of a potential future coronavirus-mediated pandemic. b Sequence alignment of HR1 domains of sarbecoviruses from four clades (SARS-CoV-2 numbering). NTD, RBD, CTD1 and 2, FP, HR1, HR2, TM and CT represent N-terminal domain, receptor binding domain, C-terminal domain 1 and 2, fusion peptide, heptad repeat 1 (HR1), heptad repeat 2 (HR2), transmembrane domain and cytoplasmic terminus, respectively.
Using the intact 32-mer peptidic HR1FC of SARS-CoV-2 (residues 924-955) in the S protein (HR1c, Fig. 1b) as the immunogen, we identified three mAbs in the scFv-IgG1 format--3A1, 3D1 and 3E2, after 3-rounds of panning, from a pre-Covid-19 näive human combinatorial antibody library37 (Fig. 2a, b). All three antibodies exhibited high somatic hypermutation (SHM) levels (14 to 27 mutations), suggesting antigen-driven maturation, potentially from viral replication (Fig. 2b). Germline origins were inferred using the IMGT database. Notably, 3D1 derived from IGHV7-4-1*02, a germline rarely or never observed in natural SARS-CoV-2 infections.
a Enrichement of combinatorial antibodies targeting HR1FCSC2. The input and output phage numbers in the three rounds of panning are summarized in the table (mean value) and histogram. Data (mean ± SD) are calculated from three plates with different dilution gradients. b Inferred germline genes encoding the variable regions of the enriched antibodies and their corresponding mutation frequencies. c Cross-interaction of 3D1 with the HR1FC peptides of SARS-CoV-1 and SARS-CoV-2, along with 3D1 epitope mapping against the full-length HR1 fusion core (HR1FC), the C-terminal of HR1FC (HR1C), the N-terminal of HR1FC (HR1N), and the minimal pepDVVNQN. d Cross-neutralization of 3D1 against the SARS-CoV-1 (green) and SARS-CoV-2 (blue) pseudoviruses. Each titration point in (c, d) represents the mean of independent replicates, with standard deviations shown as error bars (mean ± SD, n = 3).
3D1 demonstrated sub-nanomolar cross-reactivity with the HR1FC peptides from SARS-CoV-1 and SARS-CoV-2 and potently neutralized pseudoviruses of both (Fig. 2c, d). Epitope mapping localized 3D1 binding to the C-terminal HR1FC domain (HR1C; Fig. 2c). To define the minimal epitope, we assessed 3D1’s binding affinity to truncated HR1FC peptides via ELISA. A 6-mer peptide (950DVVNQN955), bound 3D1 with affinity comparable to longer HR1C and HR1FC peptides (Fig. 2c).
Autoreactivity vs. polyreactivity of 3D1
To further assess the potential origin and self-reactivity risk of 3D1, we conducted an HEp-2 autoreactivity assay. 3D1 exhibited a measurable autoreactivity, similar to its putative germlinecounterpart (3D1-Germline) (Fig. 3a). The germline antibody was generated by reverting all 14 SHMs in the 3D1 heavy chain to the näive IGHV7-4-1*02 sequence and 5 SHMs in the light chain to IGLV2-8*01 sequence (Supplementary Fig. 2a, b). Notably, 3D1-Germline retained binding affinity for SARS-CoV-2 HR1FC (Supplementary Fig. 2c), suggesting that 3D1 may exist as a natural or background antibody without antigen-driven affinity maturation. The weak HEp-2 signal likely reflects non-specific binding or low-affinity interactions with the untested autoantigen panel (Fig. 3a, b).
a Autoreactivity measurement of 3D1 and 3D1-Germline in Hep-2 epithelial cells. Representative immunofluorescence microscopy photos demonstrate that the green fluorescence indicates positive binding of the tested antibodies to Hep-2 cells. Positive control (PC) and negative control (NC) samples were obtained from the serum of patients with and without autoimmune diseases. Scale bars represent 200 μm. b ELISA binding experiment between 3D1 and phospholipids, DNA, and other known auto-antigens. c Sequence alignment of HR1C sequences from all genera of coronaviruses. The area shaded in light green represents the minimal epitope sequence (P-4 to P+1 residues) of 3D1. The essential Gln (Q) at P0 position is shaded in orange, along with residues S937-955 of SARS-CoV-2. Residues involved in interactions with 3D1 are conserved across different genera of CoVs.
The nine human coronaviruses shown in Fig. 1a span the α, β, and δ genera (Fig. 3c). To evaluate the pan-coronaviral polyreactivity of 3D1, we evaluated its binding to HR1C peptides from these viruses: SARS-CoV-1, SARS-CoV-2, MERS-CoV, HCoV-229E, HCoV-NL63, Hu-PDCoV, CCoV-HuPn-2018, HCoV-OC43, and HCoV-HKU1. 3D1 bound strongly to HR1Cs from five coronaviruses (SARS-CoV-1, SARS-CoV-2, HCoV-229E, HCoV-NL63, and Hu-PDCoV) but showed no interaction with MERS-CoV, HCoV-OC43, HCoV-HKU1 and CCoV-HuPn-2018 (Supplementary Table 1). Sequence alignment indicated that the Q954 in the minimal epitope 950DVVNQN955 is critical for 3D1 binding (Fig. 2c). Substituting Q954 with Ala, Asn, and Thr abolished 3D1’s binding as confirmed by Bio-Layer Interferometry (BLI). Based on an array of 19-mer peptides with the conserved DVVNQN epitope from 18 extra viral isolates representing α, β, γ, and δ coronavirus genera, 3D1 dispalyed broad but variable affinities for HR1C peptides derived from diverse zoonotic hosts, including civets, bats, and pangolins (Fig. 3c, Supplementary Table 1).
Crystal structure of 3D1-scFv complex with the cognate epitope HR1C of SARS-CoV-2
To elucidate the structural basis of broad cross-reactivity of 3D1, we determined the crystal structures of 3D1-scFv bound to SARS-CoV-2 HR1C (HR1CSC2) and 3D1-Fab bound to HCoV-229E HR1C (HR1C229E). Initially, the binding affinity of 3D1-scFv to HR1C and the neutralizing activity of 3D1-scFv against SARS-CoV-2 pseudovirus were confirmed (Supplementary Fig. 3a, b). Size-Exclusion Chromatography (SEC) and Iso-Thermal Calorimetry (ITC) revealed that, unlike gp41-taregting antibodies (e.g., D5-5-HB, HK20-5-HB or 8066-gp41 trimer32,38,39,40) which form 3: 3 complexes, 3D1 bound to HR1CSC2 in a 1: 1 stoichiometry, suggesting a distinct binding mode (Supplementary Fig. 4).
The 3D1-scFv/HR1CSC2 co-crystal structure was resolved at 1.08 Å resolution (Fig. 4a, Supplementary Table 2). The co-crystal structure of 3D1-Fab complex with HR1C229E exhibited a diffraction resolution of 2.28 Å (Fig. 4b, Supplementary Table 2). In both structures, only the electron densities of the C-terminal residues and the C-terminal amide group were well defined and assigned to 950DVVNQN955-NH2 (pepDVVNQN) for SARS-CoV-2 and 835DVVNQQ840-NH2 (pepDVVNQQ) for HCoV-229E (Fig. 4c, d). The CDR residues of 3D1 involved in the interactions with either pepDVVNQN or pepDVVNQQ were similar (Supplementary Tables 3, 4).
Overall view of the 3D1-pepDVVNQN (a) and 3D1-pepDVVNQQ (b) complex structures. The heavy and light chains of 3D1 are shown as cyan and orange semi-transparent surfaces, respectively, with all six CDRs depicted as cartoons. The 3D1-bound peptides are shown in green and presented in cartoon and stick representations, with the main chain hidden. The underlying CDR loops are also depicted in cartoon representation. Refined 2Fo−Fc electron density maps of the peptides (contoured at 1σ and within 2 Å of selected atoms) are located between residues D950 and N955 of HR1SC2 (c) and residues D835 and Q840 of HR1229E (d), bound to 3D1. The peptides are shown in stick representation (green carbon). The N-terminus and C-terminus of each peptide are indicated (Nterm/Cterm). Interfaces between the heavy and light chains with pepDVVNQN (e) and pepDVVNQQ (f) are also shown. Type I β-turns are highlighted with transparent grey circles for peptides from HR1SC2 (g) and HR1229E (h) bound to 3D1. Intrapeptide hydrogen bonds that emulate a pseudo 310 turn between the side chain of the first Asn and the amide backbone of the third residue in the turn are shown as black dashed lines. Ramachandran plots for the dihedral angles of 3D1-bound pepDVVNQN (g, right panel) and pepDVVNQQ (h, right panel) are provided. Residues with dihedral angles indicative of canonical type I β-turns are colored green; otherwise, they are colored blue.
Both epitope peptides pepDVVNQN and pepDVVNQQ perch across the complementarity-determining region (CDR) junction between the heavy and light chains of 3D1 in an extended conformation, rather than adopting an amphipathic α-helix secondary structure as demonstrated by pre-fusion state or the 6-HB post-fusion structure (Fig. 4e, f). From the N-terminus, each peptide forms weak interactions with CDRL1 and CDRL3 before looping around CDRL3 and CDRH3, with C-terminal residues nestled in a pocket framed by CDRH1 and CDRH2. At the paratope-epitope interface, the buried surface area reached approximately 502.3 Å2 and 507.8 Å2 with the heavy chain contributing 345.4 Å2 and 360.9 Å2, and the light chain contributing 156.9 Å2 and 146.9 Å2, respectively, by PDBePISA (Fig. 4e, f). Notably, in both complex structures, the heavy chain of 3D1 accounted for over two-thirds of the buried surfaces at the antigen interfaces.
Analysis of dihedral angles revealed the presence of a type I β-turn (residue i to i + 2), consistent with β-type Ramachandran plots (Fig. 4g, h). The turn is stabilized by hydrogen bonds between the side chain of the first Asn (N) side chain (residue i) and the backbone amide of the C-terminus Asn (or Gln for HR1229E; residue i + 2). Moreover, the β-turns were further reinforced by intrapeptide hydrogen bonds involving N955SC2 or N838229E, facilitated by water molecules (Supplementary Fig. 5a–d). It is worth noting that three SHM sites, Y32, A33, and N52, located within the paratope of the 3D1 heavy chain, are also in close proximity to the epitope-binding pocket and contribute to hydrogen-bonding network through backbone interactions. Consistently, mutations of the side-chains in 3D1-germline did not affect the binding affinity of 3D1 (Supplementary Fig. 6).
Molecular basis for broad antigen-recognition of 3D1
Comparison of apo-3D1 structure with the 3D1-pepDVVNQN and 3D1-pepDVVNQQ, reveales a notable conformational change upon epitope binding (Fig. 5a). This transition creats an open “reclining chair”-shaped binding pocket with deep binding grooves in 3D1. Notably, the side-chain of K54 in the 3D1 heavy chain rotates from a 9 o’clock to 12 o’clock orientation (Fig. 5a). This movement enables formation of an additional K54-water-amide hydrogen bond, facilitated by the longer side-chain of Gln in pepDVVNQQ (Supplementary Fig. 5e, f). This observation supports the earlier observation that pepDVVNQQ has a larger buried surface-area and a higher percentage of heavy chain contribution in the complex structure (Fig. 4c, d), as well as its ~10-fold higher binding affinity (Supplementary Table 1).
a Crystal structures of apo 3D1 (left panel), 3D1-pepDVVNQN (middle panel), and 3D1-pepDVVNQQ (right panel) are shown as electrostatic potential surfaces. The quality of electron density at K54 of 3D1 (middle and right panels) is presented in a close-up view. Peptide epitopes are colored green and depicted in cartoon and stick representations, with the main chain hidden. b Overall view of interactions between 3D1 and pepDVVNQN. Residues in the paratope and epitope are shown as sticks. Detailed interactions between 3D1 and HR1SC2 are highlighted using black dashed lines. 3D1 is represented as a cartoon, with residues from pepDVVNQN shown in either stick or cartoon format. The light chain is depicted in orange, while the heavy chain is depicted in cyan. Residues engaging in interactions are shown as sticks and labeled. Residues involved in side chain-to-main chain hydrophilic interactions are underlined. Peptide residues that mediate both side chain-to-side chain and side chain-to-main chain interactions are boxed. c, d Featured binding properties of Q954SC2 with 3D1 mAb. The terminal acylamino group of the side chain of Q954SC2 forms a pair of hydrogen bonds with N35HFW2 (c), and Trp-Asn interactions are presented in (d). The center of the aromatic ring is indicated with red pseudo dots. e Sensorgram of 3D1 mutants binding to HR1C by BLI. The 3D1 mutants that significantly affect 3D1 mAb binding are shown in color, while the wild-type 3D1 is shown in black. f Alanine scan mutagenesis of the linear epitope of pepDVVNQN. The binding constants between 3D1 and each mutant peptide were measured by BLI. NA indicates no binding detected. The bar graph displays interaction contributions between corresponding epitopic residues and heavy (cyan color) or light chain (orange color) of 3D1 caculated by PISA. g Neutralization of SARS-CoV-2 pseudovirus wild-type and mutant strains. The wild-type virus is shown in blue, while the mutant viruses are depicted in assorted colors. Results are presented as mean ± SD, derived from two replicate experiments.
It illustrates the intricate molecular interactions between 3D1 and pepDVVNQN, highlighting the extensive hydrogen bond network formed between the antibody and the antigen (Fig. 5b). This network includes a far-reaching paratope-water-epitope hydrogen bond interaction that enhances recognition (Supplementary Fig. 5). A key aspect of the epitope is the Q954SC2 residue (or Q839 residue in HCoV-229E spike, referred to as Q839229E), which inserts its bulky and polar side chains into a binding cavity perpendicularly to the plane of the main chains. This interaction is precisely recognized by the side chain of the N35 residue on the CDRH2 framework region of 3D1 (N35HFW2) (Fig. 5c). Furthermore, the aromatic benzopyrrole groups of W105 in CDRH3 (W105H3) and W50HFW2 provide stabilization to the Q954SC2 side chain via CH–π, NH–π, and π–π interactions (Fig. 5d). BLI affinity studies demonstrated that a single mutation of either N35AHFW1 or W105AH3 in the heavy chain completely disrupted binding between a subset of 3D1 mutants and HR1CSC2 (Fig. 5e), underscoring the crucial role of these residues in antigen recognition. Conversely, a single mutation of W50AHFW2 resulted in approximately a 5-fold reduction in binding affinity compared to wild-type 3D1, with the affinity decreasing from 2.7 nM for the wild type to 14 nM for the mutant (Fig. 5e).
In addition, the hydroxyl side chains of Y93 and Y32 in the light chain region of 3D1 (Y93L3 and Y32L1) appear to form strong hydrogen bond interactions with the backbone carbonyl group of D950 and V952 in HR1CSC2, as well as with the carboxyl side chain of D950, respectively (Fig. 5b). The mutation of Y93AL3 abolished the 3D1-antigen interaction, while the mutation of Y32AL1 led to a two orders of magnitude reduction in binding affinity, decreasing from 2.7 nM for the wild type to 120 nM for the mutant (Fig. 5e).
Alanine scanning of the 6-mer pepDVVNQN, shows that Q954A completely abolished the binding affinity towards 3D1 and subsequent neutralization of Q954A-harbouring pseudoviruses (Fig. 5f, g). N955A caused milder effects (10-fold lower affinity, 100-fold reduced neutralization), while other substitutions had minimal impact (Fig. 5f).
Mode-of-action of 3D1 in coronavirus neutralization
The assembly of the 6-HB fusion core is a key step in spike-mediated membrane fusion of SARS-CoV-2 and host cells41,42. As illustrated in Fig. 7, the membrane fusion process of coronavirus infection comprises four steps including 1) interaction of S1 with hACE2 leading to cleavage of S1 from spike-ECD; 2) conformational change of S2-HR1; 3) insertion of the N-terminal fusion peptide (FP) of HR1 into the host cell membrane and 4) re-folding of HR1 and HR2 to form the 6-HB fusion core.
To demonstrate that the antiviral effect of 3D1 is due to its blockade of the membrane interaction between S1 and hACE2, a cell-cell fusion assay was established. HEK293T cell lines that overexpress either Spike (293T-S) or hACE2 (293T-hACE2) were generated. As shown in Fig. 6a, unlike the negative control of an isotypic antibody, 3D1 exhibited a dose-dependent inhibition of membrane fusion between 293T-S and 293T-hACE2 cells. Similarly, the formation of the 6-HB complex through a 1: 1 mixing of HR1FC and HR2 peptides was also inhibited by 3D1 in a dose-dependent manner, as evidenced by the native polyacrylamide gel electrophoresis (N-PAGE) results (Fig. 6b).
a Spike-hACE2-mediated cell-cell membrane fusion assay. Representative images of SARS-CoV-2 spike (expressed in HEK293T cells with EGFP) mediated syncytium formation with hACE2-expressing cells from three independent replicates. Scale bars represent 200 µm. b Dose-dependent inhibition of 3D1 on the 6-HB conformation between HR1FC and HR2 peptides of SARS-CoV-2, representative of 3 independent trials. c 3D1 interferes with the hACE2 induced 6-HB formation, which is resistant to proteinase K, as visualized by Western-blot, as shown as a representative image from three independent experiments. d Dose-dependent binding of 3D1 to HR1FC but not to other conformations of the ectodomain of spike. Each titration point represents the mean of indenpendt triplicates. Standard deviations shown as error bars (n = 3). e Comparison of the HR1 conformation of 3D1-bound with those of HR1 in different fusion states. The signature-fold of 950DVVNQN955 is displayed as sticks, while other residues in the helical structures of HR1 in the prefusion and post-fusion states are shown as lines.
To demonstrate whether subverted accessibility of HR1 on the fusogenic S2 would preclude spontaneous binding of 3D1 to the intact Spike protein, the full-length Spike of SARS-CoV-2 was utilized in pseudovirus assembly. Previous studies have shown that soluble hACE2 protein facilitates the conformational change of the membrane Spike by interacting with its S1 domain, leading to the rearrangement of the fusogenic S2 domain and the formation of the 6-HB fusion core, which can withstand proteolytic digestion by trypsin and proteinase K43,44. n the absence of the soluble hACE2 protein, the pseudoviral particles of SARS-CoV-2 containing the full-length Spike were completely digested, regardless of the presence of 3D1As shown in Fig. 6c, i. However, in the presence of soluble hACE2 protein, proteolytic digestion by trypsin and proteinase K resulted in a 60 kDa band corresponding to the 6-HB fusion core (Fig. 6c). 3D1 appeared to interfere with the formation of the 6-HB fusion core by interacting with the HR1 domain of the fusogenic S2 subunit that is exposed during the conformational change (Transition State). Consistantly, in the affinity binding experiment with Spike ECD, HR1FC, 3-HB and 6-HB, only HR1FC showed potent interaction with 3D1 (Fig. 6d).
In summary, combining the different conformational states of HR1 (Fig. 6e), it’s hypothesized that 3D1 opportunistically hijacks the unraveled motif 950DVVNQN955 in the transition state of a helix-turn-helix structure, originally positioned in the α13 helix adjacent to the inflection point of the transition from loop to helix between the α12 and α13 helices (Supplementary Fig. 7), thereby preventing formation of a long extended helix bundle intermediate (3-HB) (Fig. 7).
The process occurs through four distinct stages: (1) pre-fusion, (2) transition, (3) intermediate, and (4) post-fusion states. The antibody 3D1 (red) binds specifically to the hinge region connecting the central helix and fusion peptide (FP), which adopts a transition-state conformation during the pre-fusion to intermediate stage conversion.
Broad neutralization of 3D1 against all coronavirus genera
To investigate whether the molecular interaction between 3D1 and epitopes in the HR1C domain could lead to neutralization against the corresponding coronaviruses, we conducted a pseudoviral infection assay. Pseudoviral particles were constructed using Spike proteins from nine human-infecting coronaviruses, along with zoonotic sarbecoviral orthologs that are known to target the hACE2 receptor45,46,47. The zoonotic viruses studied included BANAL-20-52 and PCoV-GX/P2V/2017 from the SARS-CoV-2 clade, as well as WIV1 and Rs33671 from the SARS-CoV-1 clade. Remarkably, 3D1 exhibited potent pseudoviruse neutralizing activity, with IC50 values in the picomolar to nanomolar range, against 7/9 HCoVs and tested SARS-CoV-1/2 ortholog zoonotic viruses (Fig. 8a, b). While 3D1 effectively neutralized the pseudoviruses for MERS-CoV and CCoV-HuPn-2018 at picomolar concentrations, it did not show any binding interactions with the corresponding HR1C peptides. However, when we used the full-length HR1FC peptides from MERS-CoV and CCoV-HuPn-2018, 3D1 revealed comparable binding affinities in the nanomolar range (Supplementary Table 1), suggesting that the corresponding epitope sequences are situated in the N-terminal region of HR1 (Supplementary Fig. 8a, b).
a Cross-neutralization of 3D1 against pseudovirions harboring the spike proteins of human coronaviruses HCoV-229E, HCoV-NL63, SARS-CoV-2, SARS-CoV-1, MERS-CoV, CCoV-HuPn-2018, and Hu-PDCoV. b Cross-neutralization of 3D1 against pseudovirions harboring the spike proteins of BANAL-20-52, WIV1, Rs3367, and PCoV-GX/P2V/2017. c Cross-neutralization of 3D1 against pseudovirions harboring the spike proteins of wild-type (WT) and variants Beta, Lambda, and Delta of SARS-CoV-2. d Plaque reduction neutralization test for authentic SARS-CoV-2 viruses, both wild-type and VOCs, based on Vero E6 cells using 3D1. All above data (shown as mean ± SD) are from three independent replicates.
The SARS-CoV-2 6-mer epitope pepDVVNQN is conserved in VOCs (B.1.1.7/Alpha, B.1.351/Beta and P.1/Gamma) and VOIs (B.1.617.1/Kappa and C.37/ Lambda). In contrast, the VOC Delta (B.1.617.2) and Omicron (B.1.1.529) feature D950N and N954H substitutions, respectively. Pseudovirus particles from the Alpha, Beta, Lamda, Delta and Omicron variants were constructed and tested against 3D1 in the pseudoviral infection assay. As anticipated, while 3D1 displayed strong neutralizing activity against the Alpha, Beta, Lambda, and Delta variants, it showed no neutralizing effect against the Omicron variant (Fig. 8c). The emergence and spread of Omicron appear to be associated with its capability to evade most of the host immune surveillance.
To validate the neutralization effects of 3D1 observed in the above pseudoviral infection assay, we performed a Plaque Reduction Neutralization Test (PRNT) on live SARS-CoV2 virus and its variants by applying gradient 3D1 to prevent virus plaque formation on Vero cells in a BSL-3 laboratory. Consistent with the results from pseudoviral assays, 3D1 exhibited significant neutralization against authentic wild type (Eng 2/20) and the VOCs of SARS-CoV-2 from Alpha to Delta except for Omicron (B.1.1.529) (Fig. 8d). The 3D1 antibody, thus, has the potential to confer protection against future sarbecoviral infections resulting from spillover events across multiple host species. However, the actual epitope exposure during natural infection/vaccination and 3D1 protective capacity remain to be confirmed in vivo.
Cross-reactivity towards other RNA viruses by 3D1
The specific binding motif of 3D1 appears to be present in both the N- and C-terminal regions of the coronavirus HR1 domain, as evidenced by 3D1 strong binding of MERS 983VGITQQ988 and CCoV 1117TVQLQN1122 and effective neutralization of MERS-CoV and CCoV-HuPn-2018 pseudoviruses. It is highly likely that this signature motif is common among RNA viruses beyond just coronaviruses. Indeed, sequence alignment of the GenBank database for RNA viruses reveals several sequences that exhibit features of the 3D1 epitope. They are the “LIKNQN” motif in the Filovirus family shared by the Musoke and Angola strains of Marburg virus (MARV) (Supplementary Fig. 9a), and the “GIVQQQ” motif in the NHR from gp41 domain (analogous to the HR1 region) found in HXB2 (HIV-1) and ROD (HIV-2) strains (Supplementary Fig. 9b). The “GIVQQQ” motif is conserved in 7,936 out of 8070 strains in the HIV sequence database as of 2019. Additionally, the “A/EVVLQN” motif in the HR1 region of human endogenous retroviruses (HERV) is conserved among β-, γ-, and δ-retroviruses48. Encouragingly, similar to coronaviruses, 3D1 demonstrated nanomolar binding affinity for all three identified epitope sequences (Supplementary Fig. 9c–e). Thus, the presence of the 3D1 epitopic motif in the viral glycoprotein may indicate broader applicability for 3D1 beyond coronavirus family, pending further functional validation in the future research.
Discussion
The symbiotic relationship between viral infections and the host immune system during evolution is a complex interplay that has shaped both viral pathogens and their hosts. Viral infections exert selective pressure, driving the evolution of host immune defenses, while hosts develop adaptive mechanisms to combat and tolerate viral infections. To break the symbiotic cycle, altenative approaches are necessary to eradicate viruses that strive to dominant and cause widespread transmission. There is a pressing need to identify the vulnerabilities of viruses and uncover hidden targets for neutralizing antibody responses.
The conserved heptad-repeat (HR) region, which is shared among a wide range of coronaviruses, is essential for their survival. Specifically, HR1 plays a crucial role in the membrane-fusion process, a decisive step for viral entry into the host. Clearly, the HR region is a major target for neutralizing antibody response. Literature reports indicate that 97.9% of COVID-19 patients developed high titers of IgG antibodies responding to the HR domain49. On the other hand, HR1 is the most highly protected region in coronaviruses, being only transiently exposed during membrane fusion to evade host immune attack. Moreover, b against SARS-CoV-1Antibodies isolated from convalescent patients against the conserved HR domain of the SARS-CoV-2 Spike protein are generally broad-spectrum neutralizing50. Furthermore, a subunit vaccine candidate consisting of HR1 and HR2 has demonstrated significant efficacy against most SARS-CoV-2 variants in animal models51.
Using the highly conserved C-terminal HR1 sequence (HR1C), we discovered a monoclonal antibody named 3D1 from a pre-pandemic combinatorial antibody library. Through epitope mapping and structural analyses, we identified a antibody recognition sequence that is present across all four genera of coronaviruses. Similar to how short peptides bind to class I MHC52,53 molecules during T cell responses, 3D1 binds to peptides composed of 5-6 amino acids with two anchor positions (P0 and P+1) located near the C-terminal end. This epitope is characterized by the consensus sequence “XΨΨXQQ(N)”, featuring a distinctive β-turn scaffold. In this sequence, X (at positions of P−4 and P−1) can be any amino acid, Ψ (at positions of P−3 and P−2) typically includes hydrophobic residues, Q (at positions of P0 or P+1) represents the single-letter amino acid code for glutamine, and N (at position of P+1) signifies asparagine. The glutamine at P0 is strictly conserved and crucial for recognition by 3D1. The P+1 position of the epitope, which curves into the binding pocket, can accommodate only glutamine or asparagine. Substituting glutamine at this position typically enhances 3D1 binding affinity tenfold, consistent with the stronger hydrogen bonding associated with the longer propionamido group.
Mode-of-action analysis suggests that 3D1 exerts its broad antiviral activity by binding to the transition state in the membrane fusion process, thereby blocking viral infection of host cells. Unlike antibodies reported to target the conserved HR1 and HR2 domains isolated from convalescent patients, such as 76E1, EK1, and EK1C4, which recognize the pre-fusion and intermediate HR bundle configurations9,33,54, may 3D1 specifically binds to the transiently exposed β-turn configuration of a 6-mer epitope peptide during membrane fusion. Interestingly, the HR1-targeting EK1 antibody, known for its broad neutralizing activity against coronaviruses, has also been reported to exhibit cross-inhibitory activity against HIV55.
The combinatorial antibody library, functioning as a synthetic immune system, encompasses not only the complete antibody repertoire of an individual or a population of individuals but, more importantly, preserves the “fossil record” of their immunological history. This “fossil record” enable for the recovery of genetic information pertaining to an individual’s immunological history, regardless of whether the specific antibody is currently being produced. The 3D1 antibody identified in this study exemplifies a unique and rare antibody that targets signature motifs evolved during the interaction between evolving viruses and antibodies. 3D1 exhibits a high degree of somatic mutation from rare germline genes of IGHV7-4-1*02 and IGLV2-8*01, indicating a long history of evolution and adaptation to coronaviruses. However, the antibody showed no affinity maturation over the course of natural selection. 3D1’s highly specific binding energy reveals a signature sequence motif that appears to play a role in the conformational change of the tandemly repeated core region of coronaviruses. Identifying such signature motifs, which are essential for viral infection, could greatly aid in the design of super-antibodies that may one day exert broad neutralizing activity before an infection is established. Collectively, this work may inspire future development of antiviral agents.
Methods
Cell lines
The cell lines Vero (CRL-1586, E6 being a subclone), and HEK293T (CRL-3216) used in this study were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). We obtained the Huh-7 cell line (SCSP-526) from the National Collection of Authenticated Cell Cultures (Shanghai, China), where technicians had immortalized the parental cells originally sourced from ATCC. All cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; # SH30243.FS, Hyclone) supplemented with 10% fetal bovine serum (FBS; # FB-1058, biosera). Stable cell lines of 293T-hACE2 were previously generated via lentiviral transduction12. Expi293 cells were purchased from Thermo Fisher Scientific and cultured in 293 F medium (#UP1000, Uninon-Biotech).
Plasmid constructs
The spike protein genes of SARS-CoV-2 strains (Wuhan-Hu-1, Beta /B.1.351, Gamma /P.1, Delta /B.1.617.2, Lambda /C.37, and Omicron /B.1.1.529), as well as those from SARS-CoV-1-S, WIV1-S, Rs3367-S, MERS-CoV-S, Hu-PDCoV-S and among others, were synthesized and constructed into a pcDNA3.1(+) vector by Azenta for subsequent use.
Sequence alignment and phylogenetic analysis of coronaviruses
Sequence alignment was completed using the online tools Clustal Omega, ESPript, and WebLogo. Phylogenetic trees were constructed using MEGAX, with sequences primarily downloaded from UniProt. The accession numbers used for phylogenetic analysis are as follows: SARS-CoV-2 (P0DTC2), SARS-CoV-1 (P59594), HCoV-229E (P15423), HCoV-NL63 (Q6Q1S2), HCoV-HKU1 (U3NAI2), HCoV-OC43 (U3NAI2), MERS-CoV (R9UQ53), Bat SARS-CoV Rf1/2004 (Q0QDZ0), BtCoV BM48-31/BGR/2008 (E0XIZ3), SADS-CoV (A0A2P1G7F5), BtCoV-HKU2 (A8JNZ2), Feline CoV/FCoV (C5IGD1), Feline Infectious Peritonitis Virus (FIPV) (Q66951), Porcine delta coronavirus (W8Q9Y7), Munia coronavirus HKU13-3514 (B6VDY7), BCoV (P25191), Mouse Hepatitis Virus/MHV (P11224), BtCoV-HKU4 (A3EX94), BtCoV-HKU5 (A3EXD0), SL-CoV WIV1 (U5WI05), SL-CoV Rs3367 (U5WHZ7), BtCoV-HKU9 (A3EXG6), Avian infectious bronchitis virus/IBV (P11223), Beluga whale coronavirus SW1 (B2BW33), Bottlenose dolphin HKU22 (V5TFD8), RaTG13 (A0A6B9WHD3), BtCoV-HKU3 (Q3LZX1), Civet SARS-CoV 007/2004 (Q3ZTF3), TGEV (P07946), Canine coronavirus/CCoV (D2WXL7), Sparrow deltacoronavirus/SpDCoV (A0A2Z4EVU6_9NIDO). Additionally, the following sequences were downloaded from NCBI: Pangolin coronavirus GX/P2V/2017 (QVT76606.1), Pangolin coronavirus GD/1/2019 (QLR06867.1), RpYN06 (MZ081381), RmYN02 (MW201982), Hu-PDCoV (MW685622), BANAL-20-247/Laos/2020 (MZ937004), BANAL-20-52/Laos/2020 (MZ937000), Montifringilla taczanowskii coronavirus/MtCoV (MT215336) and CCoV-HuPn-2018 (MW591993). Lastly, RshSTT182 (EPI_ISL_852604) is available in the GISAID database.
Synthesis of SARS-CoV-2 spike HR1FC peptide and others
N-terminal biotinylated peptides and N-terminal 6xHis-tagged peptides corresponding to the HR1FC sequence of SARS-CoV-2 (residue number range: 924-955) were chemically synthesized at Sangon Biotech. Peptides corresponding to the HR1FC sequences of six other human coronaviruses SARS-CoV-1, MERS-CoV, HCoV-HKU1, HCoV-OC43, HCoV-229E, and HCoV-NL63, were synthesized at Genscript ProBio. HR1C peptides, individually fused to the C-terminus of a His6 tag, were synthesized from the spikes of the following coronaviruses: SARS-CoV-2 (residues 937–955), SARS-CoV-1 (residues 938–956), RpYN06 (residues 910–928), BANAL-20-247/Laos/2020 (residues 891–909), BM48−31 (residues 923–941), Rf1 (residues 938–956), HCoV-229E (residues 822–840), HCoV-NL63 (residues 1003–1021), and SADS-CoV (residues 799–817), all synthesized by Genscript ProBio. Additionally, for crystallization studies, HR1CSC2 without a His tag was synthesized by Genscript ProBio. Sequences of different peptides are provided in Supplementary Table 5.
ELISA assay
Different peptides were immobilized on high protein-binding microtiter plates (#4420404, ThermoFisher Scientific) at a concentration of 2 μg/mL in phosphate-buffered saline (PBS; #41403ES76, Yeasen) overnight at 4 °C. The plates were washed the next morning with PBS containing 0.05% Tween 20 (PBS-T) and then blocked with PBS containing 3% BSA (#A23088, ABCONE) for 1 hour at 37 °C. Following this, the plates were incubated with serial dilutions of mAbs for 60 min at room temperature. After four washing steps with PBS containing PBS-T, the antibodies were serially diluted again and incubated for 1.5 h at 37 °C. Horseradish peroxidase (HRP)-conjugated goat anti-human IgG antibody, diluted 1:1000 (#109-006-098, Jackson Immuno Research), was used to detect the binding of mAbs, and the plates were subsequently developed with ABTS (#002024, ThermoFisher Scientific). After a 45 min incubation, absorbance was measured at 405 nm using a microplate spectrophotometer (Envision, Perkin Elmer). All ELISA experiments were performed in duplicate, and affinity measurements, represented as KD in nanomolar concentrations, were calculated using Graphpad Prism 10 by performing a non-linear regression. All experiments were performed in triplicate.
Protein expression and purification
Antibodies in the forms of 3D1-scFv, 3D1scFv-Fc, and IgG were produced in Expi293 cells, which were grown in suspension using 293 F medium at 37 °C in a humidified 8% CO2 incubator with rotation at 130 rpm. Cells were grown to a density of 2 million cells per mL, transfected using PEI (#40816ES03, Yeasen), and cultivated for 3 to 5 days. Proteins with a human Fc tag were purified from clarified supernatants using a HiTrap Protein A affinity column (Cytiva) operated with an ÄKTA FPLC purifier (Cytiva) and UNICORN software version 5.11 (Build 407) with PBS as the mobile phase. Proteins were buffer-exchanged into PBS from the elution buffer of 0.1 M acetate acid (pH 3.0) and concentrated using centrifugal filters (#UFC901096PK, Amicon Ultra) before being flash frozen. Pouch-packed 3D1scFv-Fc and IgG solutions were frozen at −80 °C until use.
To generate Fab, 3D1 IgG was digested with Papain (#P5306-25MG, SigmaAldrich) at 4 °C overnight, and then incubated with Protein-A beads at 4 °C for an additional 2 h to remove the Fc fragments. The 3D1-scFv with a hexahistidine tag was purified via an Ni-Excel column (Cytiva). Finally, 3D1-scFv and 3D1 Fab were concentrated and further purified by size-exclusion chromatography using a Superdex 75 10/300 GL column (Cytiva). The selected fractions were pooled and concentrated again for further use.
Affinity determination by Biolayer Interferometry
The determination of monoclonal antibody binding with selected peptides was performed using biolayer interferometry (BLI) on an Octet RED96 (FortéBio/Sartorius). Activated Ni-NTA biosensors were used to capture N-terminal 6xHis-conjugated peptides, which were diluted in Octet buffer (PBS plus 0.1% Tween 20) to a final concentration of 0.05 mg/mL. The biosensors were first immersed in the peptide solution for 5–10 s and then transferred to Octet buffer for 60 sec to remove unbound peptides and establish a baseline. After establishing the baseline, the sensors were immersed in antibody (scFv-Fc) solutions at various concentrations to record the progressive binding signals for 300 s, followed by transfer to Octet buffer for dissociation for 300 s. The generated data were analyzed using FortéBio/Sartorius analysis software (version 12.0) for correction, curve fitting, and calculation of KD values, and the results were plotted using Graphpad Prism 10.
Crystallization
Apo 3D1scFv (10 mg/mL) and a mixture of 3D1scFv (15 mg/mL) with the HR1CSC2 at a twofold molar ratio were screened for crystallization using 576 conditions from Hampton Research and Qiagen on a Mosquito (TTP LabTech) liquid handler. The same procedure was applied to 3D1 Fab in complex with the HR1C229E for the crystal screen.
Crystallization trials were set up using the vapor diffusion method in sitting drops containing 0.1 μL of protein and 0.1 μL of reservoir solution. Optimized crystals of apo 3D1scFv, 3D1scFv-HR1CSC2, and 3D1Fab- HR1C229E were grown in drops containing the following conditions: [0.1 M BIS-TRIS buffer at pH 5.5 and 25% (w/v) polyethylene glycol 3350], [0.1 M HEPES buffer at pH 7.5 and 25% (w/v) polyethylene glycol 8000], [1 M lithium chloride, 0.1 M trisodium citrate buffer at pH 4.0 and 20% (w/v) polyethylene glycol 6000], and [0.2 M ammonium sulfate, 0.1 M BIS-TRIS buffer at pH 5.5 and 25% (w/v) polyethylene glycol 3350], respectively, at 16 °C.
Structural determination
Crystals were harvested by soaking in reservoir solution supplemented with 20% (v/v) glycerol or methanol, and then flash-cooled and stored in liquid nitrogen until data collection. Diffraction data were collected at cryogenic temperatures (100 K) at the Shanghai Synchrotron Radiation Facility (SSRF) on beamline BL19U1, with a beam wavelength of 0.97853 Å. Datasets were indexed, integrated, and scaled using the HKL-2000 package52. Data collection and processing statistics are outlined in Supplementary Table 2. The structures were solved by molecular replacement using PHASER in PHENIX53, with a homology model generated by SWISS-MODEL and PIGSPro for either apo 3D1scFv or 3D1Fab-pepDVVNQN as the search model. Iterative model building and refinement were carried out in COOT54 and PHENIX53, respectively, combined with additional manual building cycles. Clear Fo-Fc density was observed in the 3D1 combining site for the peptide after several rounds of refinement. The peptide was manually built into the difference density Fo-Fc map, followed by further rounds of refinement of the complex in PHENIX.refine and manual building cycles in COOT. Buried surface area (BSA) and paratope residues, as well as their interactions, were identified by accessing the PISA program at the European Bioinformatics Institute website55. The φ and ψ angle distributions were generated in Ramachandran plots embedded in COOT. The electrostatic potential was calculated using APBS and PDB2PQR56.
Isothermal titration calorimetry assay
Isothermal titration calorimetry (ITC) experiments were performed using a PEAQ-ITC instrument (Malvern) at 25 °C. All protein samples were prepared in assay buffer containing 25 mM Tris-HCl (pH 8.0; # T97644-1L, ABCONE) and 50 mM NaCl (#B548121-0100, Sangon Biotech). The assay was conducted in multi-injection mode by titrating 40 μL of titrant from the syringe into 200 μL of titrate in the cell. To characterize the interaction between the antibody and HR1C, titrations were performed with 20 injections of 500 μM peptide into 33 μM 3D1(scFv) at intervals of 150 s. The thermodynamic parameters were obtained by nonlinear least-squares fitting of the data using the PEAQ-ITC analysis software (Malvern). Heat changes in the reactions were corrected for dilution effects by buffer subtraction, and the final figures were prepared using Origin software (OriginLab).
Inhibition of SARS-CoV-2 spike-mediated cell-cell fusion
293T-hACE2 cells were used as target cells and HEK293T cells that were transiently transfected with SARS-CoV-2 spike-P2A-EGFP (HEK293T/S/EGFP) were set as effector cells. Effector cells and target cells were co-cultured in the presence or absence of 3D1 antibody at the indicated concentrations for fusion. After cultivating at 37 °C for 6 h, the rates of cell-cell fusion were evaluated using a fluorescence microscope (EVOS M5000, Life Technologies).
Preparation of pseudovirions
Briefly, NL4-3 psudoviruses were produced using the following constructs: pcDNA3.1 plasmids encoding spikes from SARS-CoV-1, Rs3367, WIV-1, RaTG13, MERS-CoV and HCoV-229E, SARS-CoV-2 with C-terminal 19 amino acid deletion also including the VOCs (Delta and Beta) and the VOI Lambda, as well as HCoV-NL63 (containing a C-terminal 18–amino acid deletion), respectively. Plasmids encoding the S gene and the co-package vector pNL4-3.luc.RE were co-transfected into HEK293T cells in 10 cm dishes using Lipofectamine 3000 reageats (#L3000015, ThermoFisher Scientific). The supernatants containing pseudovirus were harvested at 48 h post-transfection, filtered (0.45 μm pore size, Sartorius) and mixed with the Lenti-X Concentrator (#631231, Clontech) overnight at 4 °C. The mixture was then centrifuged at 1500 g for 45 min at 4 °C. The cell pellets were collected and re-suspended in a DMEM medium and stored at -80 °C until use.
Pseudovirus neutralization assay
Briefly, effector cells (293T-hACE2 cells for SARS-CoV-2 including variants, SARS-CoV-1, HCoV-NL63, Rs3367, WIV1, BANAL-52 and GX/P2V/2017, HEK293T/hAPN cells for HCoV-229E, Hu-PDCoV, Huh-7 cells for MERS and HEK293T/fAPN cells for CCoV-HuPn-2018) were seeded 96-well, white-bottom plates at a density of 1 × 104/well and cultivated overnight. The virus was then diluted in DMEM with 10% FBS mixed with an equal volume of serially diluted antibodies and incubated at 37 °C. One hour later, the mixtures were transferred to effector cells. The cells were incubated at 37 °C for 48 h. PsV infection efficacy was evaluated by luciferase activity using Bright-Lumi Firefly Luciferase Reporter Gene Assay Kit (#11404ES80, Yeasen). Fifty microliter of luciferase substrate was added to each well, and the relative luminescence unit (RLU) values were measured on an Envision plate reader (PerkinElmer, Ensight). The concentration of antibody required for a 50% decrease in foci number (IC50) was calculated using a nonlinear curve fitting algorithm in Graphpad Prism 10.
Neutralization assay against authentic SARS-CoV-2 viruses
The study was performed in the CL3 Facility of the University of Oxford operating under license from the HSE, on the basis of an agreed Code of Practice, Risk Assessments (under ACDP) and Standard Operating Procedures. The microneutralization protocol is similar to that described before56. 1.5% carboxymethyl cellulose-containing overlay was used to prevent satellite focus formation. Twenty hours post-infection, the monolayers were fixed with 4% paraformaldehyde, permeabilized with 2% Triton X-100 (#20107ES76, Yeasen) and stained for N protein using mAb EY 2 A. After development with a peroxidase conjugated antibody and TrueBlue peroxidase substrate, infectious foci were enumerated by ELISPOT reader. Data were analyzed using four parameter logistic regression (Hill equation) in Graphpad Prism 10. Vero-TMPRSS2 cells were seeded into black-walled, clear-bottom 96-well plates at 2 × 104 cells/well and cultured overnight at 37 °C. The next day, 9-point 5-fold serial dilutions of mAbs were prepared in infection media (DMEM supplement with 10% FBS). The different SARS-CoV-2 strains were diluted in infection media at a final MOI of 0.01 PFU/cell, added to the mAb dilutions and incubated for 30 min at 37 °C. Media was removed from the cells, mAb-virus complexes were added and incubated at 37 °C for 18 h. Cells were fixed with 4% PFA (#P69575-500ML, ABCONE), permeabilized with Triton X-100 and stained with an antibody against the viral nucleocapsid protein (#40143-R001, SinoBiologicals) followed by a staining with the nuclear dye Hoechst 33342 (#H21492, ThermoFisher Scientific) and a goat anti-rabbit Alexa Fluor 647 antibody (#A-21245, ThermoFisher Scientific). Cells were imaged with an automated multimode plate reader (Biotek, Cytation 5).
Native polyacrylamide gel electrophoresis
HR1FC and HR2 peptides were dissolved in PBS (pH 7.4) at a final concentration of 25 μM. HR1P with or without indicated-concentration 3D1 were incubated at 25 °C for 30 min, followed by the addition of HR2 peptide. The mixture was incubated for another 30 min and segregated by 18% Tris-glycine gel with constant 120 V at room temperature for 2 h. The gel was stained with Coomassie blue staining (#P0017, Beyotime Biotechnology) and imaged with a Tanon 2500-B scanner (Tanon Science & Technology).
Autoreactivity assay with HEp-2 cell
According to the manufacturer’s instructions, the autoreactivity assay was performed using a HEp-2 anti-nuclear antibodies kit (#4220-12CN, Medical and Biological Laboratories). Briefly, 35 μL of 0.1 mg/mL antibodies were loaded to the wells in a slide pre-seeded with fixed and permeabilized HEp-2 cells and incubated for 20 min at room temperature. Positive serum from autoimmune patients and negative serum from healthy donors provided by the kit were used as controls. After washing twice (5 min each), the FITC-conjugated secondary anti-human antibody was incubated with the cells for 20 min at room temperature. The slide was then washed and mounted with a coverslip before observation on a fluorescent microscope (ZEISS, LSM710) with a 20× objective.
Human Antiphospholipid IgG ELISA
The Human Antiphospholipid IgG ELISA Kit (#ml038148, mlbio) employs a solid-phase sandwich enzyme-linked immunosorbent assay (ELISA) methodology. Standards of known analyte concentration and test samples were loaded into microplate wells. Following incubation with a biotin-conjugated detection antibody, wells were washed, and horseradish peroxidase (HRP)-labeled avidin was added. After further incubation and washing to remove unbound conjugates, substrates A and B were introduced simultaneously. Color development was catalyzed by the HRP-avidin-biotin complex, with optical density (450 nm) proportional to antiphospholipid IgG concentration in samples.
Assessment of autoantigen reactivity
Reactivity of the 3D1 antibody against lupus-associated autoantigens was evaluated using the Sai’er Biotechnology Anti-Nuclear Antibody (ANA) microarray chip (12/14-antigen panel). 3D1 was tested at an initial concentration of 6.6 mg/mL, serially diluted in assay buffer to final ratios of 1:41, 1:81, 1:161, and 1:321. Following manufacturer’s protocol, diluted samples were applied to antigen-coated chips and incubated at 25 °C for 60 min. After washing, bound antibodies were detected using fluorescently labeled anti-human IgG (Cy3-conjugated, 1:2000). Signal quantification was performed using a GenePix 4400 A microarray scanner. No specific binding to any of the 14 systemic lupus erythematosus (SLE)-associated autoantigens was observed across all tested dilutions. This work was completed by CRO service from Shenzhen Sciarray Biotechn Co., Ltd.
Western-blot analyses of soluble hACE2-induced conformational change of Spike
Pseudovirions consisting of the spike protein of SARS-CoV-2 were thawed and incubated with either an equal volume of buffer, 10 μM of the hACE2 ectodomain (#PHA002, Sanyou Bio), or 1 μM of 3D1 for 30 min or 2 h on ice. Refolding into the postfusion conformation was detected by the appearance of a proteinase K (Pro-K; # HY-108717, MCE) resistant band migrating at approximately 55 kDa. Trypsin (5 μg/mL; # HY-129047-10 mg, MCE) was then added to these samples and incubated for 30 min at 37 °C. Subsequently, the samples were supplemented with 10 μg/mL Pro-K and incubated for 30 min at 4 °C. A 5X SDS-PAGE loading buffer (#P0015L, Beyotime Biotechnology) was then added to all samples prior to boiling at 95 °C for 5 min. The samples were run on a 4–12% Bis-Tris PAGE gel (#M00653, GenScript) and transferred to PVDF membranes (#FFP71, Beyotime Biotechnology). An anti-S2 SARS-CoV-2 polyclonal primary antibody (1:2,000 dilution; # 40590-T62, SinoBiologicals) and an HRP-conjugated goat anti-rabbit secondary antibody (1:3,000 dilution, #ab205718, Abcam) were used for Western blotting. Images were developed using a ChemiDoc (Bio-Rad) imager.
Statistics of data
All statistical tests were performed with Graphpad Prism 10 software. Values of measurements were expressed as mean with error bar of standard deviation (SD) in figures. Numbers to get the mean (n value) were chosen according to assay features and indicated in each figure legend.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data needed to evaluate the conclusions of the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. The coordinates and structure factors of the crystal structures have been deposited in the Protein Data Bank (PDB) database, under accession codes 7YD3 (apo 3D1 scFv; https://www.rcsb.org/structure/7YD3), 7Y8J (3D1 scFv in complex with pepDVVNQN; https://www.rcsb.org/structure/7Y8J), and 7YI6 (3D1 Fab in complex with pepDVVNQQ; https://www.rcsb.org/structure/7YI6). Source data are provided with this paper.
References
Walker, L. M. & Burton, D. R. Passive immunotherapy of viral infections: ‘super-antibodies’ enter the fray. Nat. Rev. Immunol. 18, 297–308 (2018).
Xu, R. et al. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 328, 357–360 (2010).
Burton, D. R. & Hangartner, L. Broadly Neutralizing Antibodies to HIV and Their Role in Vaccine Design. Annu Rev. Immunol. 34, 635–659 (2016).
Morgand, M. et al. V1/V2 Neutralizing Epitope is Conserved in Divergent Non-M Groups of HIV-1. J. Acquir Immune Defic. Syndr. 71, 237–245 (2016).
Sok, D. & Burton, D. R. Recent progress in broadly neutralizing antibodies to HIV. Nat. Immunol. 19, 1179–1188 (2018).
Stadlbauer, D. et al. Broadly protective human antibodies that target the active site of influenza virus neuraminidase. Science 366, 499–504 (2019).
Dacon, C. et al. Broadly neutralizing antibodies target the coronavirus fusion peptide. Science 377, 728–735 (2022).
Low, J. S. et al. ACE2-binding exposes the SARS-CoV-2 fusion peptide to broadly neutralizing coronavirus antibodies. Science 377, 735–742 (2022).
Sun, X. et al. Neutralization mechanism of a human antibody with pan-coronavirus reactivity including SARS-CoV-2. Nat. Microbiol 7, 1063–1074 (2022).
Lerner, R. A. Combinatorial antibody libraries: new advances, new immunological insights. Nat. Rev. Immunol. 16, 498–508 (2016).
Lerner, R. A. Manufacturing immunity to disease in a test tube: the magic bullet realized. Angew. Chem. Int Ed. Engl. 45, 8106–8125 (2006).
Qiang, M. et al. Neutralizing Antibodies to SARS-CoV-2 Selected from a Human Antibody Library Constructed Decades Ago. Adv. Sci. (Weinh.) 9, e2102181 (2022).
Fan, Y. et al. Encoding and display technologies for combinatorial libraries in drug discovery: The coming of age from biology to therapy. Acta Pharm. Sin. B 14, 3362–3384 (2024).
Meng, B. et al. Inhibitory antibodies identify unique sites of therapeutic vulnerability in rhinovirus and other enteroviruses. Proc. Natl Acad. Sci. USA 117, 13499–13508 (2020).
Xu, R. et al. Preferential recognition of avian-like receptors in human influenza A H7N9 viruses. Science 342, 1230–1235 (2013).
Lang, S. et al. Antibody 27F3 Broadly Targets Influenza A Group 1 and 2 Hemagglutinins through a Further Variation in V(H)1-69 Antibody Orientation on the HA Stem. Cell Rep. 20, 2935–2943 (2017).
Schibli, D. J. & Weissenhorn, W. Class I and class II viral fusion protein structures reveal similar principles in membrane fusion. Mol. Membr. Biol. 21, 361–371 (2004).
Drosten, C. et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med 348, 1967–1976 (2003).
Zaki, A. M., van Boheemen, S., Bestebroer, T. M., Osterhaus, A. D. & Fouchier, R. A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med 367, 1814–1820 (2012).
Layne, S. P., Hyman, J. M., Morens, D. M. & Taubenberger, J. K. New coronavirus outbreak: Framing questions for pandemic prevention. Sci. Transl. Med. 12, https://doi.org/10.1126/scitranslmed.abb1469 (2020).
Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020).
Simon, V., Ho, D. D. & Abdool Karim, Q. HIV/AIDS epidemiology, pathogenesis, prevention, and treatment. Lancet 368, 489–504 (2006).
Kortepeter, M. G., Dierberg, K., Shenoy, E. S. & Cieslak, T. J. Marburg virus disease: A summary for clinicians. Int J. Infect. Dis. 99, 233–242 (2020).
Kielian, M. & Rey, F. A. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol 4, 67–76 (2006).
White, J. M., Delos, S. E., Brecher, M. & Schornberg, K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem Mol. Biol. 43, 189–219 (2008).
Pu, J., Wang, Q. & Jiang, S. Peptide-Based HIV Entry Inhibitors. Adv. Exp. Med Biol. 1366, 15–26 (2022).
Xu, C. et al. Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. Sci. Adv. 7, https://doi.org/10.1126/sciadv.abe5575 (2021).
Dey, S., Pahari, P., Mukherjee, S., Munro, J. B. & Das, D. K. Conformational dynamics of SARS-CoV-2 Omicron spike trimers during fusion activation at single molecule resolution. Structure 32, 1910–1925.e1916 (2024).
Frey, G. et al. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc. Natl Acad. Sci. USA 105, 3739–3744 (2008).
Xue, J. et al. Efficient treatment and pre-exposure prophylaxis in rhesus macaques by an HIV fusion-inhibitory lipopeptide. Cell 185, 131–144.e118 (2022).
Roymans, D. et al. Binding of a potent small-molecule inhibitor of six-helix bundle formation requires interactions with both heptad-repeats of the RSV fusion protein. Proc. Natl Acad. Sci. USA 107, 308–313 (2010).
Luftig, M. A. et al. Structural basis for HIV-1 neutralization by a gp41 fusion intermediate-directed antibody. Nat. Struct. Mol. Biol. 13, 740–747 (2006).
Xia, S. et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 5, eaav4580 (2019).
Pinto, D. et al. Broad betacoronavirus neutralization by a stem helix-specific human antibody. Science 373, 1109–1116 (2021).
Wang, C. et al. A conserved immunogenic and vulnerable site on the coronavirus spike protein delineated by cross-reactive monoclonal antibodies. Nat. Commun. 12, 1715 (2021).
Zhou, P. et al. A human antibody reveals a conserved site on beta-coronavirus spike proteins and confers protection against SARS-CoV-2 infection. Sci. Transl. Med. 14, eabi9215 (2022).
Gao, C., Brümmer, O., Mao, S. & Janda, K. D. Selection of Human Metalloantibodies from a Combinatorial Phage Single-Chain Antibody Library. J. Am. Chem. Soc. 121, 6517–6518 (1999).
Sabin, C. et al. Crystal structure and size-dependent neutralization properties of HK20, a human monoclonal antibody binding to the highly conserved heptad repeat 1 of gp41. PLoS Pathog. 6, e1001195 (2010).
Gustchina, E. et al. Complexes of neutralizing and non-neutralizing affinity matured Fabs with a mimetic of the internal trimeric coiled-coil of HIV-1 gp41. PLoS One 8, e78187 (2013).
Louis, J. M. et al. Binding of HIV-1 gp41-directed neutralizing and non-neutralizing fragment antibody binding domain (Fab) and single chain variable fragment (ScFv) antibodies to the ectodomain of gp41 in the pre-hairpin and six-helix bundle conformations. PLoS One 9, e104683 (2014).
Matsuyama, S. & Taguchi, F. Two-step conformational changes in a coronavirus envelope glycoprotein mediated by receptor binding and proteolysis. J. Virol. 83, 11133–11141 (2009).
Jackson, C. B., Farzan, M., Chen, B. & Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 23, 3–20 (2022).
Walls, A. C. et al. Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion. Cell 176, 1026–1039.e1015 (2019).
Park, Y. J. et al. Antibody-mediated broad sarbecovirus neutralization through ACE2 molecular mimicry. Science 375, 449–454 (2022).
Temmam, S. et al. Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature 604, 330–336 (2022).
Xia, L. Y. et al. Characterization of a pangolin SARS-CoV-2-related virus isolate that uses the human ACE2 receptor. Sci. China Life Sci. 67, 1502–1513 (2024).
Ge, X. Y. et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503, 535–538 (2013).
Ruigrok, K. et al. X-ray Structures of the Post-fusion 6-Helix Bundle of the Human Syncytins and their Functional Implications. J. Mol. Biol. 431, 4922–4940 (2019).
Liu, B. et al. Recovered COVID-19 patients with recurrent viral RNA exhibit lower levels of anti-RBD antibodies. Cell Mol. Immunol. 17, 1098–1100 (2020).
Elshabrawy, H. A., Coughlin, M. M., Baker, S. C. & Prabhakar, B. S. Human monoclonal antibodies against highly conserved HR1 and HR2 domains of the SARS-CoV spike protein are more broadly neutralizing. PLoS One 7, e50366 (2012).
Pang, W. et al. A variant-proof SARS-CoV-2 vaccine targeting HR1 domain in S2 subunit of spike protein. Cell Res 32, 1068–1085 (2022).
Shin, S. et al. Antigen Recognition Determinants of γδ T Cell Receptors. Science 308, 252–255 (2005).
Adams, J. J. et al. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity 35, 681–693 (2011).
Xia, S. et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res 30, 343–355 (2020).
Yu, D. et al. Pan-coronavirus fusion inhibitors possess potent inhibitory activity against HIV-1, HIV-2, and simian immunodeficiency virus. Emerg. Microbes Infect. 10, 810–821 (2021).
Skelly, D. T. et al. Two doses of SARS-CoV-2 vaccination induce robust immune responses to emerging SARS-CoV-2 variants of concern. Nat. Commun. 12, 5061 (2021).
Acknowledgements
This work was supported by the Ministry of Science and Technology of China fund (Grant No. 2021YFA1100800), the Jiangsu Basic Research Center for Synthetic Biology (Grant No. BK20233003), Shanghai Frontiers Science Center for Biomacromolecules and Precision Medicine and the Oxford Glycobiology Endowment, and the University of Oxford’s COVID-19 Research Response Fund. The authors also thank the technical support from BL19U1 beamline (https://cstr.cn/31129.02.NFPS.BL19U1) of National Facility for Protein Science in Shanghai.
Author information
Authors and Affiliations
Contributions
G.Y., L.Y. and A.L. conceived and designed the study. L.Y., F.W., L.Z., X.S., Q.H., Z.L., H.L., Y.Z. and L.L. performed biochemical and molecular biological experiments, including antibody selection, characterization, crystallization, as well as cell functional studies. M.H. and J.B. performed the authentic virus neutralization experiments and analyzed the data. L.Y., F.W and X.D drew the figures. L.Y., F.W. and G.Y. analyzed all the data and wrote the manuscript. A.L., R.A.D. and N.Z. provided manuscript edits and suggestions. All authors reviewed and edited the paper.
Corresponding authors
Ethics declarations
Competing interests
G.Y. and L.Y. are inventors on a patent application describing the cross-reactive antibody 3D1 (Case No. P240115226CN, Application No. 2025105093316). The other authors declare that they have no competing interests.
Peer review
Peer review information
Nature Communications thanks Juan C. Almagro and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yan, L., Wang, F., Hill, M. et al. A broadly neutralizing antibody recognizes a unique epitope with a signature motif common across coronaviruses. Nat Commun 16, 7580 (2025). https://doi.org/10.1038/s41467-025-63101-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-63101-1