Abstract
Advancements in computational techniques have enhanced our understanding of human brain development, particularly through high-dimensional data from magnetic resonance imaging (MRI). One notable approach is the brain-age prediction framework, which predicts biological age from neuroimaging data and calculates the brain age gap (BAG), a marker of deviation from chronological age. Most commonly applied to adult samples, this approach is now increasingly used in children and adolescents. However, several considerations must be taken into account when applying brain-age prediction in youth. In this Perspective, we outline important challenges and provide recommendations for researchers as well as future directions for the field.
Similar content being viewed by others
Introduction
The human brain undergoes profound structural and functional changes throughout childhood and adolescence1. Neuroimaging advances over the last 25 years have established reproducible patterns of brain development2. Morphometric studies report reduced gray matter volume, monotonic cortical thinning, and surface area increases in childhood, followed by decreases in adolescence3. White-matter volume progressively increases throughout childhood and adolescence, mirrored by higher fractional anisotropy (FA) and lower mean diffusivity (MD), indicating microstructural changes measured by diffusion tensor imaging (DTI)4. Functional connectivity strength changes from childhood to adulthood, with an increase in network integration (stronger within-network connectivity) and segregation (weaker between-network connectivity)5. Overall, brain development in childhood and adolescence involves complex, dynamic changes reflecting reorganization and optimization6 that is shaped by the interplay of genetics and the environmental context.
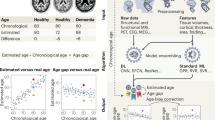
While MRI offers high spatial resolution, providing detailed information on the structure and function of the human brain, this information is multidimensional and complex, resulting in different brain characteristics being examined in isolation. Machine learning reduces this complexity by building statistical models of the brain based on MRI datasets. One example is brain-age prediction, which reduces brain MRI features into a summary score reflecting normative brain health and integrity7. To calculate brain age, researchers train models on large MRI datasets from individuals across different ages. The model learns patterns that predict age from brain characteristics and is then applied to new scans. By comparing predicted and chronological ages, researchers can assess deviations from typical age-related brain structure (see Fig. 1).
The difference between brain-predicted age and chronological age, termed brain age gap (BAG), indexes this deviation. A higher brain age than chronological age indicates an older-looking brain, often interpreted as accelerated maturation in childhood and adolescence8, and as decline in adulthood and senescence9. Inversely, a lower brain age may indicate delayed maturation in youth and potentially better brain health in adulthood and senescence10.
Numerous studies have linked inter-individual variation in BAG to multiple phenotypes, including cognitive functioning11, cardiometabolic health12,13,14,15, lifestyle factors16, mental disorders17,18, and neurodegenerative disorders (e.g., stroke, Alzheimer’s)19,20. However, most studies have focused on adult populations, rely on cross-sectional designs, and primarily use T1-weighted MRI data. Recent years have seen a slow but steady increase in brain-age prediction application in youth samples, but this expansion brings forth a new set of challenges that necessitate thorough discussion with its propagators and the neuroimaging field at large. The dynamic nature of age-related brain changes during youth poses significant challenges in interpreting BAG. Brain-age models may collapse complex, overlapping developmental patterns into one metric21. In a unidimensional model, delayed or accelerated development in different regions can average out. Further concerns include differences in imaging modalities, scanner acquisitions, and model features19,22, applying models outside their training ranges, limited longitudinal data, and ignoring key maturational processes (e.g., pubertal development16) or socio-environmental factors (e.g., early-life adversity23).
The objective of this Perspective is to discuss the advancement of brain-age prediction for child and adolescent research. We define childhood according to the pediatric staging in the AAP Bright Futures guidelines24 and the Centers for Disease Control and Prevention developmental milestone framework25, treating childhood as roughly 3–9 years (spanning early and middle-childhood stages). We define adolescence according to the World Health Organization26 as 10–19 years. We use youth as an umbrella term covering both periods. Reviewing all studies is not within the scope of this paper, but we will outline key similarities, limitations, and offer recommendations for the field. We also discuss key methodological challenges when applying brain-age models to child and adolescent cohorts, while acknowledging that some of these challenges overlap with adult and lifespan issues.
Overview of existing literature
Brain-age prediction has been investigated in relation to a number of domains in youth, including mental health, genetics, physical development, cognition, and environmental factors. Here, we briefly review key findings in the literature. For methodological details (e.g., samples, models, features), see Supplementary Table 1.
Mental health
A number of different mental health outcomes have been related to both positive and negative BAGs in youth. Negative BAGs have been related to generalized anxiety27, Autistic Spectrum Disorder symptom severity28, attention deficit hyperactivity disorder symptoms29, elevated Child Behavior Checklist scores30, and lower Children’s Global Assessment Scale scores indicating greater functional impairment31. Positive BAGs have been related to depression and functional impairment32, psychosis, obsessive-compulsive symptoms, general psychopathology33, and a schizophrenia diagnosis34. Additionally, a higher BAG has been associated with psychosis risk in a sample of clinically high-risk youth35. Longitudinally, a deceleration in BAG was found for high familial risk adolescents who developed a mood disorder36, while a greater increase in BAG was found for adolescent females (but not males) with internalizing problems37.
The highlighted studies suggest that BAG carries prognostic information, positioning it as a putative early-warning tool. Clinical translation, however, demands more research to be carried out in addition to several safeguards. For example, BAG values could be interpreted against age- and sex-specific reference curves derived from large, harmonized cohorts—akin to the morphometric brain charts now available38. Reliability should generalize across scanners and pipelines, and incremental validity over established clinical and demographic predictors must be demonstrated in longitudinal cohorts. Until such benchmarks are met, BAG remains a promising research marker—useful for group-level risk stratification—rather than an individual-level clinical biomarker. The risk of misclassification and stigmatizing young people underlines this cautious stance.
Physical and pubertal development
Studies consistently link BAG to pubertal development. Earlier pubertal timing, as measured via “puberty age,” is related to a higher BAG39. Additionally, higher parent-8,16 and youth-report8 pubertal development scale (PDS) scores have been related to an increased BAG, and annualized change in parent-report PDS has been related to annualized change in BAG16. Using a classifier trained to discriminate pre- versus post-menarche status, continuous menarche class probabilities have also been positively related to BAG40. Limited research examines BAG with other biological markers in youth. Preliminary evidence suggests BAG correlates with EpiAGE (an epigenetic aging measure)41, and both BAG and its change over time appear heritable42.
Cognition
The relationship between BAG and cognition remains ambiguous, with studies reporting positive31, negative43, or no relationship21 between BAG and cognition during childhood and adolescence. Additionally, work has reported conflicting findings within the same study, related to different age ranges or models8,44. Notably, cognition measures vary widely, ranging from composite batteries (NIH Toolbox8,21 and Penn Computerized Neurocognitive Battery45), to specific tasks (e.g., Flanker Task31, working memory and numerical ability46). Mixed results have been found for IQ43,47, possibly due to differences in model features and samples.
Environmental factors and life experiences
BAG has also been linked to a variety of environmental factors and life experiences, such as premature birth, socioeconomic status, and adversity. BAG appears to be higher in adolescents who are born very premature47. Longitudinally, neighborhood disadvantage in early adolescence is associated with a positive BAG, which decreases across adolescence48. In Cohen et al.30, a lower relative brain age (calculated using the residuals from regressing predicted age on chronological age) correlated with lower parental occupational prestige, lower public assistance enrollment, and more parent psychiatric diagnoses (but not parental education or income-to-needs ratio). An older BAG was likewise associated with environmental adversity; a composite score of multiple socioeconomic and adverse experience variables32. In an emotion-circuitry model, childhood abuse was linked to a lower BAG49. Dimensions of adversity have also been differentially related to BAG, such that a lower BAG is associated with factors related to emotional neglect, and an older BAG is associated with caregiver psychopathology, trauma exposure, family aggression, substance use, separation from biological parent, and socioeconomic disadvantage and neighborhood safety23.
With more brain-age prediction studies emerging in youth and inconsistent findings across various phenotypes, standard practices warrant scrutiny. Below, we highlight key challenges researchers should consider in order to ensure responsible, thorough neuroscience research.
Challenges
Over the past decade, brain-age prediction has been increasingly used in child and adolescent populations to assess brain developmental stages. However, interpreting these models—specifically the estimated age and the BAG—remains challenging in young individuals, where effects of genetic and early-life factors are already observable42,50, and may mask the subtle effects of the variable of interest being investigated. There are also a number of methodological considerations, including universal challenges that may be more pronounced in youth, including age bias, multi-site scanner corrections, sample size, and design limitations, as well as youth-specific challenges such as nonmonotonic trajectories of brain development. Below, we summarize key challenges researchers should consider when applying brain-age prediction to neurodevelopmental samples.
Issue 1: What does BAG represent in children and adolescents?
The BAG is defined as the difference between an individual’s predicted brain age and their chronological age. In this section, we use BAG as a generic shorthand for a brain-age deviation, whether expressed as the raw difference or as an age-corrected delta; the statistical distinctions and bias-correction procedures are treated in detail later, in Issue 6.
Existing research has not established what degree of variability in BAG is typical, and what may reflect substantially accelerated or decelerated development, meaning more work is needed to quantify the stability of BAG estimates over time and what factors underlie individual brain-age estimates21. Additionally, research has not yet determined whether BAGs persist across childhood and adolescence, or how common it is for someone to exhibit a BAG that narrows/converges with increasing age. Because this period is characterized by variable, nonlinear brain development51, and BAG condenses thousands of features into one global summary score, regional or mode-specific aging signals may overlook important regional nuances52. For instance, subcortical structures (e.g., amygdala, nucleus accumbens) often mature earlier than the prefrontal cortex53. This asynchrony can result in a developing brain that may appear “on time” globally yet harbor simultaneously delayed and accelerated tissue-specific trajectories. For example, multidimensional or tissue-specific clocks (e.g., mode-specific BAG, regional white-matter age) have revealed genetic associations invisible to a global score52,54. Furthermore, most models act as “black boxes,” obscuring which features contribute to model predictions55,56. These features may also vary over time and across individuals in terms of weight of contribution57.
In developing brains, deviations from the average may not signify pathology but could reflect normal variability, especially considering the high level of individual variance during childhood and adolescence51. For example, in adult samples, it is largely accepted that physical activity58, cardiometabolic risk factors12,13, and other environmental factors such as socioeconomic status and education46,59, influence BAG, with these influences accumulating over time. Because adult brain trajectories (e.g., increase in DTI FA, decrease in DTI MD, cortical thickness, and surface area38,60,61) are better established, interpreting BAG in adulthood is relatively more straightforward.
In contrast, youth studies are often plagued by narrow age ranges and nonmonotonic brain patterns (e.g., cortical surface area increases until ~10–11 years, then declines62,63), where it is likely that (1) negative lifestyle factors may not yet manifest as atypical brain development, and (2) quadratic/curvilinear effects may be hard for models to interpret. Moreover, some research has demonstrated the differential impact of factors related to emotional neglect being associated with delayed maturation, while other factors, such as parental psychopathology and disadvantageous SES to be associated with accelerated maturation23, meaning that in individuals experiencing co-occurring factors, this might reveal no deviation from typical development despite a larger net sum of a harsher environment.
Narrow age ranges in youth samples also mean BAGs in youth typically reflect weeks or months. How viable it is to look at this error score in the context of one particular measure that is meant to account for much of this explained variance should involve a level of skepticism. Further, a positive or negative BAG should not be equated with a direct acceleration or delay of the underlying biological maturation curve. Rather, BAG should be best regarded as a summary deviation—a proxy that aggregates many influences (sampling error, technical variance, lifestyle, genetic liability) into a single score. While such deviations have shown to be informative for brain-health phenotypes, attributing them to altered maturational processes or a causal gauge for developmental tempo will require more longitudinal modeling55,64 (see Issue 3 for further discussion). Lastly, many adult studies differentiate between healthy and disorder-specific populations. Youth samples have the added layer of these studies potentially being carried out before the onset of clinical diagnoses for some of these individuals.
Recommendation
Interpretation of BAG should be done within the context of normative developmental variations, i.e., recognizing that small deviations may fall within the range of typical developmental variability65. Considering confidence intervals and effect sizes rather than relying solely on point estimates may be better suited to convey the practical significance of between-group differences in BAG. Researchers should also be cautious when attributing clinical significance to minor deviations and should consider longitudinal assessments to observe changes over time, including nonlinear change in BAG tempo.
Additionally, responsible and precise language should be used when describing and interpreting the results of BAG-focused analyses. Specifically, it may be prudent to avoid the use of language such as “accelerated” and “decelerated” maturation when it is not yet clear whether BAG reflects ongoing maturational processes during childhood and adolescence. Instead, more neutral and precise terms could be beneficial, such as “older/younger appearing brain” or “positive/negative brain age gap”.
With mode-specific analyses uncovering 34 genetically informed aging axes in adults52, adapting such multi-axis clocks to youth could expose tissue- or network-specific maturational lags that a single BAG obscures. Here, regional brain-age models could be a promising avenue. While a number of recent studies in adult populations have applied regional brain-age models, this approach has been infrequently used in youth populations49. If regional brain-age prediction is not feasible, researchers may consider specifying the features used in the model and providing each feature’s contribution. See Ball et al.66 for an example of region contribution extracted from the Manifold structure for tissue volume.
Interpreting feature contributions can be useful for understanding the model in the context of known developmental changes, or relationships to the variables of interest, despite weight maps being complicated67. Tools like vip (variable importance plots) and SHAP (Shapley additive explanations) can reveal feature importance even in complex models68. SHAP offers a model-agnostic framework for evaluating feature influence in linear, nonlinear, and deep-learning approaches69. Methodological recommendations for the nonlinearity of youth brain development are addressed in Issue 5.
Issue 2: Model training and sample choice
Model training and validation are critical for robust brain-age prediction. Models trained on data that do not represent the target population may lead to domain mismatch70, such as using adult-trained models on youth samples. Therefore, training on youth-specific data or including a substantial number of youth participants in the training set is important. The choice of modeling technique also matters71. Models may differ in key ways, such as their ability to extrapolate predictions beyond the sample observed in training. Tree-based algorithms such as random forests yield predictions confined to the observed training range71, whereas parametric or kernel-based methods can mathematically extrapolate—albeit often with high uncertainty—beyond that range.
Sample size is likewise critical55. Smaller datasets (and consequently lower power)72 are particularly problematic for neurodevelopmental studies, where inter-individual variability is high and thousands of participants may be required for robust brain-wide associations73. In comparison to traditional regression approaches, machine learning methods such as brain age approaches have two samples: a training set and a test set. Both sets must be sufficiently large, and different minimum sample sizes are potentially needed for each set, i.e., for model training versus model application and testing.
An additional consideration is whether models should be trained with sex-specificity in mind due to brain development variations in sex and pubertal development42. For instance, male youth exhibit more variability in brain structure than female youth74, and pubertal timing can influence brain development75, independently of chronological age. Brain-age models are able to robustly classify male and female brains66 despite small mean differences and neuroanatomical overlap. Research has reported 81% accuracy in sex prediction66, with higher BAG in female youth21, likely reflecting accelerated maturation in mid-to-late adolescence42. Research indicates about a 1-year difference at ages 14–16, with some convergence at 18 years of age, with males catching up to females42. This highlights the importance of accounting for sex and puberty during critical developmental periods.
Recommendations
First, ensure the training data reflects the target population to capture unique developmental patterns. The Brain Age Standardized Evaluation (BASE) provides a framework for evaluating model training and robust performance assessment76.
Second, use adequately large training and testing samples. Smaller datasets often fail to capture the high inter-individual variability in youth. Empirical learning curves show that Mean Absolute Error (MAE)—defined as the average of the absolute differences between each person’s predicted brain age and their actual chronological age—plateaus at roughly 20 high-quality, well-controlled scans per 6-month age bin. This equates to about 250–300 participants across a typical 6-year (11–17 years) window, with only marginal gains in accuracy after48,77. However, this is a practical minimum for studies using atlas-level features and classical regressors. We recommend (i) plotting a learning curve to confirm where your plateau lies and (ii) treating these numbers as starting points rather than hard cut-offs. Larger cohorts (>500) can still boost cross-scanner and cross-ethnic generalizability and provide the statistical power needed for smaller developmental differences. Multi-site harmonization, transfer-learning, and normative modeling all benefit from larger cohorts even when MAE has leveled off. Moreover, pooling multiple datasets and using cross-validation (e.g., k-fold or leave-one-out) can mitigate overfitting and yield reliable estimates. If a dataset is limited, applying a pre-trained model may be preferable to training a new one on insufficient data. Ideally, the training set should include data from a heterogeneous variety of scanners, as this helps generalization to external samples56.
Finally, consider stratifying models by sex or pubertal status to account for biological variability in development. Covering the entire span of puberty is especially helpful for capturing critical developmental trends.
Issue 3: Design
The current literature is limited by a reliance on cross-sectional designs and limited reproducibility. While cross-sectional studies can provide valuable snapshots of developmental differences, they are insufficient for testing hypotheses about the speed, timing, or trajectory of brain development78. This undermines claims of delayed or accelerated maturation during this highly variable and nonlinear period.
Longitudinal designs are essential for distinguishing between the speed and timing of maturation, clarifying to what extent variations in BAG reflect true deviations (e.g., accelerated or decelerated development). Cross-sectional estimates risk conflating group differences with developmental differences, as they cannot account for individual variability in brain development over time. This is particularly problematic in childhood and adolescence, when rapid, heterogeneous changes occur2,63. Longitudinal designs are uniquely positioned to identify sensitive periods or turning points in brain age trajectories, shedding light on whether deviations in BAG are transient or stable indicators of risk.
Recent work has quantified the extensive nature of individual variation in brain development during childhood and adolescence, illustrating the difficulty of differentiating altered developmental trajectories from normative variation51. Longitudinal data is also an avenue to explore the impact of not only single time point estimates, but also how changes in BAG may relate to different outcomes, and how these relationships can shift across development. For example, Rakesh et al.48 linked neighborhood disadvantage to a positive BAG in early adolescence, and a deceleration in BAG in later adolescence, suggesting timing-dependent effects. Though a cross-sectional BAG might indicate persistent risk, more longitudinal work is needed to confirm when BAG truly reflects accelerated or delayed maturation and how it relates to health concerns.
Recommendations
Longitudinal data is essential to address challenges in design and developmental variability79. By conducting longitudinal brain age studies, we can better differentiate between normative variation and altered developmental pathways, resulting in a clearer understanding of BAG and true maturational speed. This is particularly important when brain age metrics are coupled with youth clinical or behavioral assessments, where claims of atypical brain development may arise. Tracking the same individuals over time may reveal whether BAG deviations signify genuine acceleration or delay in maturation.
Issue 4: Model performance metrics
Model performance metrics, such as MAE and root mean square error (RMSE), are central to evaluating brain-age models but can be difficult to interpret across different studies and age ranges80, never mind developmental stages. In youth samples, MAEs are typically much lower (e.g., 0.5–1.5 years)19,55 than in adult populations, where values of 3–6 years are considered good performance55,56. However, with MAE being scale-dependent81, these raw metrics can be misleading without context. For example, an MAE of 0.35 years in a youth sample may appear to outperform models with MAEs of 3.5 years in adult samples. However, both represent an approximate deviation of 7% of their total age ranges (9–14 and 40–87, respectively). It remains a task of future research to determine how we compare these error and performance metrics across youth versus adult samples.
Relatedly, MAE and similar metrics are inherently influenced by the age range of the training and test samples80. Wider age ranges tend to increase prediction errors because they introduce more variability in brain structure and function. Conversely, narrower ranges, especially during periods of rapid anatomical change, can yield artificially low MAEs and r values that may not generalize to other contexts66,80. These findings underscore the importance of interpreting performance metrics in the context of age range, developmental stage, and variability.
Recommendations
To improve the interpretability and comparability of performance metrics across studies, researchers should consider reporting the MAE together with the chronological-age range of the test set and, where cross-study comparison is a goal, optionally add a normalized figure. This supplementary value contextualizes performance while providing a context-sensitive comparison. For example, providing MAE/RMSE for absolute error and the cross-validated predictive R2—the proportion of age variance explained in each held-out fold—listed fold-by-fold rather than as a single mean, in line with BASE76 and BabyPy82 guidelines. Developing shared reference datasets and benchmarking frameworks would further standardize practice and harmonize reporting—an especially important goal given the scarcity of distinct youth cohorts, which currently restricts opportunities for truly independent model evaluation.
Issue 5: Nonlinearity
Nonmonotonic and nonlinear brain patterns are especially pertinent during childhood and adolescence. While there is a growing expectation that nonlinear and ensemble algorithms (e.g., kernel methods, deep learning) will better capture these complexities, evidence shows that such methods do not necessarily outperform simpler linear models in practice83. In fact, research shows that regularized linear algorithms are as effective as nonlinear and ensemble algorithms, while significantly reducing computational costs84. A key factor is that neurodevelopmental datasets—often constrained by modest sample sizes and measurement noise—may not have sufficiently robust nonlinear signals for complex models to exploit, leaving linear approaches performing comparably well.
Moreover, deep convolutional architectures assume translation invariance and compositional structure, assumptions that may not readily apply to the fixed anatomical organization of the human brain. Schulz and colleagues83 demonstrate that when you artificially inject high levels of noise into a dataset, kernel and deep models eventually perform no better than linear models due to the noise washing out the higher-order patterns. Even when genuine nonlinear trajectories exist, the interpretability of black-box algorithms remains challenging. As we assume the existence of BAG in the age prediction model, a good predictive model for brain age estimates should not overfit the data and yield a perfect prediction for chronological age85, as that yields no meaningful variance in the BAG measure. Simpler methods grounded in known developmental principles can capture a large portion of the variance without risking overfitting, especially when samples are small or noisy.
Recommendations
Making recommendations for the challenge of nonlinearity is difficult. Ideally, we should consider nonlinear modeling techniques to better capture complex developmental trajectories and asynchronicities. Such models may help account for the dynamic and regionally specific growth spurts or regressive processes (e.g., pruning) that define childhood and adolescence6. Machine learning techniques capable of handling nonlinear effects, such as Gaussian Process Regression, XGBoost, and Support Vector Regression that use nonlinear mapping functions (i.e., kernels) to discover boundaries in the data by creating an implicit feature space86, or neural networks with appropriate regularization, may be better suited for predicting brain age in youth samples. Where sample size allows, researchers may also deploy multi-axis or modality-specific brain-age clocks that partition nonlinear maturation into distinct aging trajectories (see “Conclusion” for more). However, at current data scales and quality, researchers may find that linear or simpler nonlinear strategies provide a more transparent and practical starting point. Here, it may be more important to avoid overfitting by, e.g., imposing a higher level regularization87 and using lower-dimensional linear models. If you suspect strong nonlinear effects (e.g., quadratic or cubic age trends) and have enough participants spanning the age range of interest, you may consider kernel or neural-network approaches only after simpler spline or polynomial approaches (or well-powered linear models) have been tested. As sample sizes grow and noise reduction techniques improve, however, advanced nonlinear models may eventually prove valuable for elucidating subtle developmental irregularities that simpler approaches might overlook.
Issue 6: Corrections and other biases
Brain-age models face biases that can impact their accuracy, interpretability, and generalizability. These include age dependence, which can be addressed through bias correction80 as well as batch effects from multi-site MRI datasets, which can be mitigated through harmonization techniques88,89.
The most widely reported index—BAG—is a raw difference between an individual’s predicted and chronological age. Because this is algebraically proportional to the out-of-sample prediction error, it is necessarily correlated with age, leading to systematic overestimation in younger participants and underestimation in older ones. Smith and colleagues90 showed that, in the extreme case where imaging features carry no true age signal, BAG collapses to a simple linear function of chronological age, so any downstream association with cognition, psychopathology, or environmental risks being a proxy for residual age effects. To mitigate this “regression-to-the-mean” bias, several corrected variants are now common, such as regressing out age effects from model predictions, including chronological age as a covariate in analyses, and correcting predictions using slope and intercept adjustments91,92. While these methods can reduce bias, they are not without trade-offs. Certain correction techniques, particularly those based on regression adjustments, can artificially inflate model performance metrics such as R2 and reduce error measures93.
Multi-site datasets, such as the Adolescent Brain and Cognitive Development Study94 and IMAGEN95, are invaluable for training and testing brain-age models. However, these datasets are often subject to systematic differences introduced by varying imaging sites and scanner protocols96. Without correction, scanner/site effects inflate apparent inter-individual variance and can bias BAG estimates if not addressed. Several methods have been proposed to address site and scanner effects, for example, including site/scanner as a covariate in statistical analyses. Alternatively, one can utilize a suite of harmonization tools such as NeuroHarmonize, CovBat, RAVEL, cross-sectional- and longitudinal ComBat, which have been shown to reduce scanner-induced variability effectively88,97. These approaches reduce feature-level variance attributable to technical artifacts, although recent work98 shows that such corrections do not invariably improve brain-age prediction accuracy. Despite their utility, harmonization techniques must be applied cautiously to avoid data leakage99.
Recommendations
With no consensus on the best correction method, it is advisable for researchers to assess the degree of bias in raw predictions before applying corrections and report both corrected and uncorrected metrics, as recommended by de Lange et al.80, and visualize residuals across the age span to provide transparency and enable meaningful comparisons.
When addressing multi-site effects, harmonization techniques can be particularly useful for reducing variability in brain measures due to technical artifacts. Harmonization parameters should be learned only on the training data within each cross-validation fold and then applied unchanged to the held-out test set. Estimating them on the full dataset before the split leaks information from test to train and can inflate performance, whereas re-estimating them separately on the test set avoids leakage but puts train and test features on different scales, undermining comparability. Well-designed pipelines, therefore, fit the correction in the training partition and apply that fixed transformation to the test partition, ensuring bias removal without overfitting. Segmentation routines, such as using standard adult reference data100 can also introduce systemic bias. Franke and colleagues101 avoided this issue by using the Template-O-Matic toolbox102, which generates a sample-specific template where tissue segmentation does not rely on prior information maps but rather solely on voxel intensity. Deep learning methods are also a promising avenue here, using data-driven representations of various global and local data features and removing the reliance on data preprocessing to extract meaningful features55 (Table 1).
Conclusion
Our work highlights the potential for refining the use of the brain age framework in developmental samples. As an exciting frontier in child and adolescent neurodevelopmental research, brain-age prediction offers a powerful way to capture the unique and dynamic changes occurring during these critical periods. Yet, challenges such as how to interpret BAG given the nonmonotonic and nonlinear brain patterns in youth, model training and sample size, lack of longitudinal datasets, insufficient reporting of multiple model performance metrics, and other biases such as site and scanner variability are emblematic of broader methodological issues in developmental neuroscience. Addressing these challenges, alongside others discussed in this Perspective paper, is crucial not only for improving methodologies but also for ensuring that these models yield meaningful insights about the developing brain.
Moving forward, the field would benefit from establishing standard best practices for applying brain-age prediction in youth populations and improving efforts that foster reproducibility and cross-study integration in brain age research. Progress may also come from expanding brain-age prediction beyond a single chronological clock. Data-driven analyses in adults reveal multiple orthogonal aging axes52 and tissue-specific or multimodal models such as BrainAgeNeXt103 detect white-matter and cross-modal signals that a global score misses. Tailoring these multi-axis frameworks to youth cohorts may offer a more nuanced, biologically specific picture of neurodevelopmental timing and tempo. Lastly, we also encourage open science practices, including pre-registering studies, sharing model code and weight maps, and providing detailed methodology. This would facilitate replication, cross-sample validation, and continued innovation in youth brain-age prediction.
References
Blakemore, S.-J. Imaging brain development: the adolescent brain. NeuroImage 61, 397–406 (2012).
Mills, K. L. et al. Structural brain development between childhood and adulthood: convergence across four longitudinal samples. NeuroImage 141, 273–281 (2016).
Mills, K. L. & Tamnes, C. K. Longitudinal structural and functional brain development in adolescence. in The Oxford Handbook of Developmental Cognitive Neuroscience 75–98 (Oxford University Press, 2022).
Lebel, C. & Deoni, S. The development of brain white matter microstructure. NeuroImage 182, 207–218 (2018).
Rosenberg, B. M., Mennigen, E., Monti, M. M. & Kaiser, R. H. Functional segregation of human brain networks across the lifespan: an exploratory analysis of static and dynamic resting-state functional connectivity. Front. Neurosci. 14, 561594 (2020).
Norbom, L. B. et al. New insights into the dynamic development of the cerebral cortex in childhood and adolescence: integrating macro- and microstructural MRI findings. Prog. Neurobiol. 204, 102109 (2021).
Franke, K., Ziegler, G., Klöppel, S. & Gaser, C. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: exploring the influence of various parameters. NeuroImage 50, 883–892 (2010).
Whitmore, L. B., Weston, S. J. & Mills, K. L. BrainAGE as a measure of maturation during early adolescence. Imaging Neurosci. 1, 1–21 (2023).
Elliott, M. L. et al. Brain-age in midlife is associated with accelerated biological aging and cognitive decline in a longitudinal birth cohort. Mol. Psychiatry 26, 3829–3838 (2021).
Seitz-Holland, J., Haas, S. S., Penzel, N., Reichenberg, A. & Pasternak, O. BrainAGE, brain health, and mental disorders: a systematic review. Neurosci. Biobehav. Rev. 159, 105581 (2024).
Anatürk, M. et al. Prediction of brain age and cognitive age: quantifying brain and cognitive maintenance in aging. Hum. Brain Mapp. 42, 1626–1640 (2021).
Beck, D. et al. Adipose tissue distribution from body MRI is associated with cross-sectional and longitudinal brain age in adults. NeuroImage Clin. 33, 102949 (2022).
Beck, D. et al. Cardiometabolic risk factors associated with brain age and accelerate brain ageing. Hum. Brain Mapp. 43, 700–720 (2022).
Beck, D. et al. Dissecting unique and common variance across body and brain health indicators using age prediction. Hum. Brain Mapp. 45, e26685 (2024).
de Lange, A.-M. G. et al. Multimodal brain-age prediction and cardiovascular risk: The Whitehall II MRI sub-study. NeuroImage 222, 117292 (2020).
Holm, M. C. et al. Linking brain maturation and puberty during early adolescence using longitudinal brain age prediction in the ABCD cohort. Dev. Cogn. Neurosci. 60, 101220 (2023).
Kaufmann, T. et al. Common brain disorders are associated with heritable patterns of apparent aging of the brain. Nat. Neurosci. 22, 1617–1623 (2019).
Tønnesen, S. et al. Brain age prediction reveals aberrant brain white matter in schizophrenia and bipolar disorder: a multisample diffusion tensor imaging study. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 5, 1095–1103 (2020).
Franke, K. & Gaser, C. Longitudinal changes in individual BrainAGE in healthy aging, mild cognitive impairment, and Alzheimer’s disease. GeroPsych J. Gerontopsychol. Geriatr. Psychiatry 25, 235–245 (2012).
Subramaniapillai, S. et al. Sex differences in brain aging among adults with family history of Alzheimer’s disease and APOE4 genetic risk. NeuroImage Clin. 30, 102620 (2021).
Ball, G., Kelly, C. E., Beare, R. & Seal, M. L. Individual variation underlying brain age estimates in typical development. NeuroImage 235, 118036 (2021).
Han, L. K. M. et al. Brain aging in major depressive disorder: results from the ENIGMA major depressive disorder working group. Mol. Psychiatry 26, 5124–5139 (2020).
Beck, D. et al. Dimensions of early-life adversity are differentially associated with patterns of delayed and accelerated brain maturation. Biol. Psychiatry 97, 64–72 (2024).
American Academy of Pediatrics. Bright Futures: Guidelines for Health Supervision of Infants, Children, and Adolescents 4th edn. https://www.scribd.com/document/410609912/Bright-Futures-Guidelines-for-Health-Supervision-of-Infants-Children-and-Adolescents-4TH-pdf (American Academy of Pediatrics, 2017).
Centers for Disease Control and Prevention (CDC). Positive parenting tips: preschoolers (3–5 years). https://www.cdc.gov/child-development/positive-parenting-tips/preschooler-3-5-years.html (CDC, 2024).
World Health Organization (WHO). Adolescent health. https://www.who.int/health-topics/adolescent-health (WHO, 2025).
Zhou, Z. et al. Differential effects of generalized anxiety and separation anxiety on brain structural development during adolescence. J. Affect. Disord. 339, 478–485 (2023).
Tunç, B. et al. Deviation from normative brain development is associated with symptom severity in autism spectrum disorder. Mol. Autism 10, 46 (2019).
Kurth, F. et al. Preliminary evidence for a lower brain age in children with attention-deficit/hyperactivity disorder. Front. Psychiatry 13, 1019546 (2022).
Cohen, J. W. et al. Relative brain age is associated with socioeconomic status and anxiety/depression problems in youth. Dev. Psychol. 60, 199–209 (2024).
Luna, A. et al. Maturity of gray matter structures and white matter connectomes, and their relationship with psychiatric symptoms in youth. Hum. Brain Mapp. 42, 4568–4579 (2021).
Drobinin, V. et al. The developmental brain age is associated with adversity, depression, and functional outcomes among adolescents. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 7, 406–414 (2022).
Cropley, V. L. et al. Brain-predicted age associates with psychopathology dimensions in youths. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 6, 410–419 (2021).
Truelove-Hill, M. et al. A multidimensional neural maturation index reveals reproducible developmental patterns in children and adolescents. J. Neurosci. 40, 1265–1275 (2020).
Chung, Y. et al. Use of machine learning to determine deviance in neuroanatomical maturity associated with future psychosis in youths at clinically high risk. JAMA Psychiatry 75, 960–968 (2018).
de Nooij, L. et al. Longitudinal trajectories of brain age in young individuals at familial risk of mood disorder from the Scottish Bipolar Family Study. Wellcome Open Res. 4, 206 (2020).
MacSweeney, N. et al. Multimodal brain age indicators of internalizing problems in early adolescence: a longitudinal investigation. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 10, 475–484 (2024).
Bethlehem, R. A. I. et al. Brain charts for the human lifespan. Nature 604, 525–533 (2022).
Dehestani, N., Whittle, S., Vijayakumar, N. & Silk, T. J. Developmental brain changes during puberty and associations with mental health problems. Dev. Cogn. Neurosci. 60, 101227 (2023).
Gottschewsky, N., Kraft, D. & Kaufmann, T. Developmental brain changes during puberty and associations with mental health problems. Biol. Sex Differ. 15, 25 (2024).
Mareckova, K. et al. Longitudinal study of epigenetic aging and its relationship with brain aging and cognitive skills in young adulthood. Front. Aging Neurosci. 15, 1215957 (2023).
Brouwer, R. M. et al. The speed of development of adolescent brain age depends on sex and is genetically determined. Cereb. Cortex 31, 1296–1306 (2021).
Lewis, J. D., Evans, A. C. & Tohka, J. T1 white/gray contrast as a predictor of chronological age, and an index of cognitive performance. NeuroImage 173, 341–350 (2018).
Ullman, H. & Klingberg, T. Timing of white matter development determines cognitive abilities at school entry but not in late adolescence. Cereb. Cortex 27, 4516–4522 (2017).
Erus, G. et al. Imaging patterns of brain development and their relationship to cognition. Cereb. Cortex 25, 1676–1684 (2015).
Steffener, J. et al. Differences between chronological and brain age are related to education and self-reported physical activity. Neurobiol. Aging 40, 138 (2016).
Hedderich, D. M. et al. Increased brain age gap estimate (BrainAGE) in young adults after premature birth. Front. Aging Neurosci. 13, 653365 (2021).
Rakesh, D. et al. Neighborhood disadvantage and longitudinal brain-predicted-age trajectory during adolescence. Dev. Cogn. Neurosci. 51, 101002 (2021).
Keding, T. J. et al. Differential patterns of delayed emotion circuit maturation in abused girls with and without internalizing psychopathology. Am. J. Psychiatry 178, 1026–1036 (2021).
Vidal-Pineiro, D. et al. Individual variations in ‘brain age’ relate to early-life factors more than to longitudinal brain change. eLife 10, e69995 (2021).
Mills, K. L. et al. Inter-individual variability in structural brain development from late childhood to young adulthood. NeuroImage 242, 118450 (2021).
Smith, S. M. et al. Brain aging comprises many modes of structural and functional change with distinct genetic and biophysical associations. eLife 9, e52677 (2020).
Mills, K. L., Goddings, A.-L., Clasen, L. S., Giedd, J. N. & Blakemore, S.-J. The developmental mismatch in structural brain maturation during adolescence. Dev. Neurosci. 36, 147–160 (2014).
Lee, P.-L. et al. Regional rather than global brain age mediates cognitive function in cerebral small vessel disease. Brain Commun. 4, fcac233 (2022).
Cole, J. & Franke, K. Predicting age using neuroimaging: innovative brain ageing biomarkers. Trends Neurosci. 40, 681–690 (2017).
Liem, F. et al. Predicting brain-age from multimodal imaging data captures cognitive impairment. NeuroImage 148, 179–188 (2017).
Brown, T. T. et al. Neuroanatomical assessment of biological maturity. Curr. Biol. 22, 1693–1698 (2012).
Sanders, A.-M. et al. Linking objective measures of physical activity and capability with brain structure in healthy community dwelling older adults. NeuroImage Clin. 31, 102767 (2021).
Busby, N. et al. Lower socioeconomic status is associated with premature brain aging. Neurobiol. Aging 130, 135–140 (2023).
Beck, D. et al. White matter microstructure across the adult lifespan: a mixed longitudinal and cross-sectional study using advanced diffusion models and brain-age prediction. NeuroImage 224, 117441 (2021).
Westlye, L. T. et al. Life-span changes of the human brain white matter: diffusion tensor imaging (DTI) and volumetry. Cereb. Cortex 20, 2055–2068 (2010).
Raznahan, A. et al. How does your cortex grow? J. Neurosci. 31, 7174–7177 (2011).
Wierenga, L. M., Langen, M., Oranje, B. & Durston, S. Unique developmental trajectories of cortical thickness and surface area. NeuroImage 87, 120–126 (2014).
Liang, H., Zhang, F. & Niu, X. Investigating systematic bias in brain age estimation with application to post-traumatic stress disorders. Hum. Brain Mapp. 40, 3143–3152 (2019).
Lenroot, R. K. & Giedd, J. N. Brain development in children and adolescents: insights from anatomical magnetic resonance imaging. Neurosci. Biobehav. Rev. 30, 718–729 (2006).
Ball, G., Adamson, C., Beare, R. & Seal, M. L. Modelling neuroanatomical variation during childhood and adolescence with neighbourhood-preserving embedding. Sci. Rep. 7, 17796 (2017).
Haufe, S. et al. On the interpretation of weight vectors of linear models in multivariate neuroimaging. NeuroImage 87, 96–110 (2014).
Greenwell, B. M. & Boehmke, B. C. Variable importance plots—an introduction to the vip Package. R. J. 12, 343 (2020).
Lundberg, S. M. et al. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2, 56–67 (2020).
Wood, D. A. et al. Optimising brain age estimation through transfer learning: a suite of pre-trained foundation models for improved performance and generalisability in a clinical setting. Hum. Brain Mapp. 45, e26625 (2024).
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001).
Button, K. S. et al. Power failure: Why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci. 14, 365–376 (2013).
Marek, S. et al. Reproducible brain-wide association studies require thousands of individuals. Nature 603, 654–660 (2022).
Wierenga, L. M. et al. A key characteristic of sex differences in the developing brain: greater variability in brain structure of boys than girls. Cereb. Cortex 28, 2741–2751 (2018).
Herting, M. M. & Sowell, E. R. Puberty and structural brain development in humans. Front. Neuroendocrinol. 44, 122–137 (2017).
Dular, L. & Špiclin, Ž.Alzheimer’s Disease Neuroimaging Initiative BASE: Brain Age Standardized Evaluation. NeuroImage 285, 120469 (2024).
Griffiths-King, D., Wood, A. G. & Novak, J. Predicting ‘Brainage’ in late childhood to adolescence (6-17yrs) using structural MRI, morphometric similarity, and machine learning. Sci. Rep. 13, 15591 (2023).
Kraemer, H. C., Yesavage, J. A., Taylor, J. L. & Kupfer, D. How can we learn about developmental processes from cross-sectional studies, or can we? Am. J. Psychiatry 157, 163–171 (2000).
Foulkes, L. & Blakemore, S.-J. Studying individual differences in human adolescent brain development. Nat. Neurosci. 21, 315–323 (2018).
de Lange, A.-M. G. et al. Mind the gap: performance metric evaluation in brain-age prediction. Hum. Brain Mapp. 43, 3113–3129 (2022).
Hyndman, R. J. & Koehler, A. B. Another look at measures of forecast accuracy. Int. J. Forecast. 22, 679–688 (2006).
Biondo, F. et al. BabyPy: a brain-age model for infancy, childhood, and adolescence. Preprint at https://www.biorxiv.org/content/10.1101/2025.02.05.636598v2 (2025).
Schulz, M.-A. et al. Different scaling of linear models and deep learning in UKBiobank brain images versus machine-learning datasets. Nat. Commun. 11, 4238 (2020).
Han, J., Kim, S. Y., Lee, J. & Lee, W. H. Brain age prediction: a comparison between machine learning models using brain morphometric data. Sensors 22, 8077 (2022).
Niu, X., Taylor, A., Shinohara, R. T., Kounios, J. & Zhang, F. Multidimensional brain-age prediction reveals altered brain developmental trajectory in psychiatric disorders. Cereb. Cortex 32, 5036–5049 (2022).
Hofmann, T., Schölkopf, B. & Smola, A. J. Kernel methods in machine learning. Ann. Stat. 36, 1171–1220 (2008).
Bashyam, V. M. et al. MRI signatures of brain age and disease over the lifespan based on a deep brain network and 14 468 individuals worldwide. Brain 143, 2312–2324 (2020).
Beer, J. C. et al. Longitudinal ComBat: a method for harmonizing longitudinal multi-scanner imaging data. NeuroImage 220, 117129 (2020).
Lombardi, A. et al. Extensive evaluation of morphological statistical harmonization for brain age prediction. Brain Sci. 10, 364 (2020).
Smith, S. M., Vidaurre, D., Alfaro-Almagro, F., Nichols, T. E. & Miller, K. L. Estimation of brain age delta from brain imaging. NeuroImage 200, 528–539 (2019).
de Lange, A.-M. G. & Cole, J. Commentary: Correction procedures in brain-age prediction. NeuroImage Clin. 26, 102229 (2020).
Beheshti, I., Nugent, S., Potvin, O. & Duchesne, S. Bias-adjustment in neuroimaging-based brain age frameworks: a robust scheme. NeuroImage Clin. 24, 102063 (2019).
Butler, E. R. et al. Pitfalls in brain age analyses. Hum. Brain Mapp. 42, 4092–4101 (2021).
Casey, B. J. et al. The Adolescent Brain Cognitive Development (ABCD) study: imaging acquisition across 21 sites. Dev. Cogn. Neurosci. 32, 43–54 (2018).
Mascarell-Maričić, L. et al. The IMAGEN study: a decade of imaging genetics in adolescents. Mol. Psychiatry 25, 2648–2671 (2020).
Han, X. et al. Reliability of MRI-derived measurements of human cerebral cortical thickness: the effects of field strength, scanner upgrade and manufacturer. NeuroImage 32, 180–194 (2006).
Fortin, J.-P. et al. Harmonization of cortical thickness measurements across scanners and sites. NeuroImage 167, 104–120 (2018).
Yu, Y. et al. Brain-age prediction: systematic evaluation of site effects, and sample age range and size. Hum. Brain Mapp. 45, e26768 (2024).
Marzi, C. et al. Efficacy of MRI data harmonization in the age of machine learning: a multicenter study across 36 datasets. Sci. Data 11, 115 (2024).
Wilke, M., Schmithorst, V. J. & Holland, S. K. Normative pediatric brain data for spatial normalization and segmentation differs from standard adult data. Magn. Reson. Med. 50, 749–757 (2003).
Franke, K., Luders, E., May, A., Wilke, M. & Gaser, C. Brain maturation: predicting individual BrainAGE in children and adolescents using structural MRI. NeuroImage 63, 1305–1312 (2012).
Wilke, M., Holland, S. K., Altaye, M. & Gaser, C. Template-O-Matic: a toolbox for creating customized pediatric templates. NeuroImage 41, 903–913 (2008).
La Rosa, F. et al. BrainAgeNeXt: advancing brain age modeling for individuals with multiple sclerosis. Imaging Neurosci. 3, imag_a_00487 (2025).
Acknowledgements
This work was supported by the Research Council of Norway (#288083, #323951), the South-Eastern Norway Regional Health Authority (#2021070), and the University of Oregon College of Arts and Sciences.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to every part of the Perspectives. Both authors read and approved the final manuscript and agree to be accountable for all aspects of the work. A specific overview of tasks is listed as follows. Dani Beck and Lucy Whitmore: conceptualization, project administration, investigation, software, visualization, writing—original draft, writing—reviewing and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Laura Han and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Whitmore, L., Beck, D. Current challenges and future directions for brain age prediction in children and adolescents. Nat Commun 16, 7771 (2025). https://doi.org/10.1038/s41467-025-63222-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-63222-7