Abstract
Host-pathogen interaction influences many non-infectious diseases, including metabolic diseases. Helicobacter hepaticus (H. hepaticus) has been found in some metabolic dysfunction-associated steatotic liver disease (MASLD) patients, however, the causal link and underlying mechanisms remain unclear. Here we report that H. hepaticus infection or overexpression of CdtB of H. hepaticus induces lipid deposition in hepatocytes, both in vivo and in vitro. Furthermore, we identify that CdtB translocates to mitochondria with the help of Hsp90, interacts with ATP5A1, reduces mitochondrial respiratory complex V activity, damages mitochondria, and disrupts lipid metabolism. Mechanistically, CdtB-induced lipogenesis depends on the CdtB-mitochondrial ROS-mTORC1-SREBP1 axis and CdtB-mediated NONO expression to enhance nuclear localization of SREBP1 that promote the de novo fatty acid synthesis in the hepatocytes. Neutralization of CdtB significantly alleviates hepatic lipidosis in mice upon H. hepaticus infection. Furthermore, the nucleic acid of H. hepaticus has been detected in the liver tissues of some patients with MASLD, which suggests a certain correlation between liver infection with H. hepaticus and the occurrence and progression of MASLD. Our findings highlight the critical role of CdtB in the pathogenesis of H. hepaticus infection-induced hepatic lipidosis and its potential as a therapeutic target.
Similar content being viewed by others
Introduction
The liver plays a pivotal role in lipid metabolism, serving as the primary site for lipid uptake, synthesis, oxidation, and distribution to peripheral tissues1,2. Excessive consumption of fat and carbohydrates is recognized as a significant risk for the development of MASLD3. The diagnosis of MASLD necessitates the presence of hepatic steatosis and at least one cardiometabolic risk factor, such as obesity, impaired glucose metabolism, hypertension, hypertriglyceridemia, or low high-density lipoprotein (HDL) cholesterol levels, in the absence of concurrent alcohol consumption3. Along with the sequencing technologies, the gut microbiota has been recognized as a vital factor in maintaining human and animal health and mediating a variety of diseases4. In the advanced stages of MASLD, there is typically a high prevalence of intestinal dysbiosis and the emergence of specific pathogens5. Patients with severe liver fibrosis often exhibit pronounced intestinal dysbiosis6, characterized by an overgrowth of pathobionts such as Escherichia coli among other pathogens7. Furthermore, the mounting evidence suggests a significant role of gut microbes in the progression of MASLD via the gut-liver axis8,9.
H. hepaticus is an elusive and emerging zoonotic enterohepatic Helicobacter species10 that primarily colonizes the liver and lower intestine and has been associated with colitis, colorectal cancer, and extraintestinal diseases in a number of mouse strains11. Recently, epidemiologic studies have shown that the serum H. hepaticus antibodies are positive in 50% of patients with primary liver hepatocellular carcinoma12. The percentage was significantly higher than the rates observed in individuals with benign liver tumors (7.7%) and normal liver tissue (6.3%)12. The prevalence rates of H. hepaticus specific antigen in patients with gallstones and pancreatic cancer were 18.2% and 10.5%, respectively13. Like other phylogenetically distant Proteobacteria, H. hepaticus possesses cytolethal distending toxin (CDT), which contributes to its pathogenicity. CDT is a heterotrimeric holotoxin composed of three subunits, CdtA, CdtB and CdtC, of which CdtB is most conserved among CDT-harboring bacteria and exerts its effect as a DNase I-like property. CdtA and CdtC, which serve as regulatory subunits, are thought to attach to the cholesterol-rich lipid rafts on cell membranes, which enhance CdtB to enter cells by endocytosis and translocate into the nucleus eventually14. A recent study demonstrated that Campylobacter jejuni (C. jejuni) CdtB induced mitochondrial damage15 through nuclear impairment16, leading to disrupted lipid metabolism characterized by impaired β-oxidation of fatty acids.
Lipid deposition in the liver is typically attributed to abnormal fat synthesis or dysfunction in fat metabolism17. Conditions such as nutrient stress, starvation, and thermogenesis trigger the translocation of fatty acids into mitochondria for β-oxidation. Mitochondrial dysfunction, reduced mitochondrial β-oxidation capacity, has been reported to be involved in the progression of MASLD2,18,19. Furthermore, the impairment of mitochondrial activity also elicits inflammatory and fibrotic signals, thereby establishing an unfavorable cellular microenvironment within the liver and promoting hepatic steatosis and damage2,20. In contrast to studies on fat catabolism21, not much is known about mitochondria in relation to hepatic de novo lipogenesis (DNL), i.e. the synthesis of new fat by the liver from carbohydrate or certain fructose. The regulation of DNL is primarily regulated by SREBP1, the key transcription factor involved in lipogenesis22,23. Recent evidences suggest that SREBP1 may be regulated by the mTOR signaling pathway24,25, while the retention of SREBP1 in the nucleus is dependent on the presence of NONO26.
Previous reports have shown that the deficiency of CdtB in H. hepaticus results in the loss of its intestinal and hepatic pathogenicity11,27. Here, we present a study aimed at explaining the mechanism underlying CdtB in liver injury induced by H. hepaticus infection. It was found that H. hepaticus infection can cause microvesicular steatosis in hepatocytes, which can then progress to macrovesicular steatosis. Mechanistically, CdtB could enter mitochondria through the assistance of Hsp90 and cause mitochondrial damage due to its interaction with ATP synthase subunit alpha (ATP5A1), and the decreased activity of ATP5A1 suppresses mitochondrial respiratory complex V, which further activates the mTORC1/P70S6K/SREBP1 signaling; meanwhile, CdtB mediated NONO expression strengthens the nuclear localization of SREBP1 which mediates de novo synthesis of fatty acids. Therefore, we believe that CdtB is a critical pathogenic factor in hepatic steatosis induced by H. hepaticus infection. Given its widespread carriage by various enteric pathogens, the relationship between CDT and liver/intestinal health deserves further attention.
Results
H. hepaticus-induced hepatic steatosis is dependent of CdtB
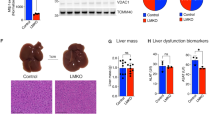
It has been reported that H. hepaticus infection is associated with MASLD28 and CdtB is the dominant virulence factor of H. hepaticus29. To investigate the role of CdtB in lipid accumulation, a CdtB-deficient strain (H. h ΔCdtB) was generated and infection experiments were performed. The colonization ability of the wild-type H. h strain (H. h WT) and H. h ΔCdtB was first examined and found to be comparable (Fig. S1a-b). It is noteworthy that the early infection with the H. h WT resulted in the formation and deposition of microvesicular lipid droplets (LD) in hepatocytes, which subsequently coexisted with both microvesicular and macrovesicular LD during the subsequent infection phase. In contrast, this phenomenon was significantly reduced in mice infected with the H. h ΔCdtB (Fig. 1a, b). Conversely, the excessive fat deposition induced by H. hepaticus infection can be alleviated by the absence of CdtB in a high-fat diet model (Fig. S1c). To further elucidate the function of the CdtB toxin in lipid deposition in hepatic cells, a liver-specific CdtB-expressing recombinant adenovirus (rAd-CdtB) was generated and administered via tail intravenous injections (i.v.). The results demonstrated that the overexpression of CdtB resulted in a significant increase in LD in the mouse liver compared to the control group at different time points after infection (Fig. 1c). Additionally, the elevation of triglyceride (TG) levels in serum (Fig. 1d) suggests a potential inclination for MASLD development in mice infected with the H. h WT. To further substantiate these findings, a human hepatocellular cell line Huh-7 was infected with the H. h WT or the H. h ΔCdtB, which revealed a diminished size of LD in the H. h ΔCdtB-infected cells compared with H. h WT-infected cells (Fig. 1e). Similarly, transfection with pCMV-CdtB in Huh-7 cells demonstrated a dose-dependent increase in cytoplasmic lipid deposition (Fig. 1f). Similar findings were observed in HepG2 and Hep3B cells (Fig. S2a). Furthermore, the mouse liver organoids also exhibited similar findings, with a more prominent LD observed following CDT treatment (Fig. 1g, h). These data collectively suggest that H. hepaticus induces hepatic steatosis in a CdtB-dependent manner.
a, b H&E (a) and Oil Red O (b) staining of liver sections from mice infected with Mock, wild type (WT) or CdtB knockout (ΔCdtB) H. hepaticus at 1, 3, 5 months post-infection (MPI) (n = 5 mice). c Oil Red O staining of liver sections from mice following tail vein injection with rAd-vector or rAd-CdtB at 17, 42, 60 days post-infection (DPI) (n = 5 mice). d Serum triglyceride (TG) levels in mice with Mock, WT or ΔCdtB H. hepaticus infection (n = 5 mice). e, f Oil Red O staining of Huh-7 cells infected with WT or ΔCdtB H. hepaticus (e) or transfected with pCMV or pCMV-CdtB (f) in a dose-dependent manner for 24 h. g, h Oil Red O staining of mouse liver organoids infected with WT or ΔCdtB H. hepaticus (g) or treated with CdtA/C or holotoxin CDT (h) for 48 h. Scale bars:50 µm (b, c, g, h) or 100 µm (a, e, f). Statistical analysis was performed by two-way ANOVA followed by Dunnett’s multiple comparisons test (b–d). Source data are provided as a Source Data file.
CdtB induces mitochondrial dysfunction and lipid metabolism disorder in hepatocytes
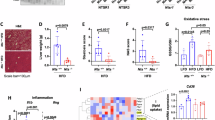
Microvesicular steatosis is a common feature of liver damage-associated diseases such as fatty acid β-oxidation deficiency, mitochondrial DNA depletion and deletion syndromes, alcoholic foamy degeneration, and inborn errors of metabolism, etc.30, suggesting that mitochondrial activity or function may be altered following H. hepaticus infection. Our findings demonstrated that CdtB was predominantly localized in the cytoplasm, accompanied by disrupted mitochondrial morphology, as evidenced by immunofluorescence staining of Hep3B cells 24 h post-transfection (Fig. 2a). Furthermore, the submicroscopic pathological injuries were also observed in the liver section post rAd-CdtB i.v. by transmission electron microscopy (TEM), including mitochondrial damage and LD (Fig. 2b). Moreover, CdtB-overexpressing cells demonstrated augmented MitoSOX staining compared with the control cells, suggesting that CdtB elevates mitochondrial ROS (mtROS) and oxidative damage (Fig. 2c). To determine whether the mitochondrial dysfunction is primarily regulated by the detrimental effects of CdtB in mitochondria rather than in the nucleus, a mitochondrial-localized CdtB plasmid (COX8A-CdtB) was constructed and transfected into Huh-7 cells. The findings demonstrated a comparable reduction in ATP generation in COX8A-CdtB-transfected cells relative to CdtB-transfected cells (Fig. 2d). A comparable impact of COX8A-CdtB on MMP and oxidative stress was also observed in Hep3B cells through TMRM and DHE staining (Fig. S2b, c). Mitochondrial oxidative damage frequently results in the release of biomolecules such as mitochondrial DNA (mtDNA)31. Consistent with this notion, significant increase of mtDNA and cell death were observed in cells transfected with CdtB, which can be reversed by treatment with Cyclosporin A (CsA), an inhibitor of the mitochondrial permeability transition pore (Fig. 2e, f and Fig. 2h). In addition, the overexpression of CdtB in the mouse liver resulted in an increase of mtDNA in the serum ((Fig. 2g), while nuclear DNA was difficult to detect in the normal mice (Fig. S2d), so as the H. h WT-infected mice compared with the H. h ΔCdtB-infected mice (Fig. S2e). Additionally, CdtB, but not cis-Platinum (CDDP), a positive control for nuclear DNA damage, resulted in a reduction in mitochondrial membrane potential (MMP) in hepatic cells (Fig. 2i), indicating that the mitochondrial dysfunction induced by CdtB is not dependent on nuclear DNA damage. Mitochondrial dysfunction is typically associated with impaired cellular metabolic processes and energy production. In accordance with this, the expression of CPT1A, a rate-limiting enzyme of β-oxidation, was markedly diminished in Hep3B cells in the presence of CdtB (Fig. 2j), thereby indicating an impairment in fatty acid catabolic activity. Similarly, a reduction in ATP generation in the liver was observed in rAd-CdtB-treated mice at different time points (Fig. S2f). Collectively, these findings suggest that CdtB induces hepatocytes lipidosis through regulation of cell-intrinsic mitochondria function and fatty acid catabolism.
a Immunofluorescence images of mitochondrial morphology (TOM20, green) in Hep3B cells transfected with pCMV-vector or pCMV-CdtB (CdtB mAb, red) for 24 h. b Transmission electron microscopy images of liver sections from mice with rAd-vector or rAd-CdtB tail vein injection (i.v.) at 7 days post-infection (DPI). Red triangle: Damaged mitochondria with loss of ridges and reduced inner and outer membrane integrity; Yellow triangle: Lipid droplet; Green triangle: Autophagosome; Blue triangle: Autophagosome containing damaged mitochondria; Purple triangle: Autolysosome. c MitoSOX Red staining of mitochondrial superoxides in HEK293A cells transfected with pCMV or pCMV-EGFP-CdtB. d ATP production in Huh-7 cells transfected with pCMV-vector, pCMV-CdtB or pCMV-COX8A-CdtB for 24 h (n = 3 independent experiments). e, f Analysis of mitochondrial DNA (mtDNA) in purified cytoplasmic fractions of Hep3B cells transfected with pCMV or pCMV-CdtB (e), followed by treatment with vehicle control (NC) or CsA (500 ng/mL) (f) for 24 h (n = 3 independent experiments). g Serum mtDNA level following intravenous administration of mice with rAd-vector or rAd-CdtB (n = 5 mice). h Flow cytometry analysis of Hep3B cell death labeled with propidium iodide (PI) following various treatments. i Flow cytometry analysis of mitochondrial membrane potential by tetramethylrhodamine methyl ester (TMRM) in Hep3B cells (n = 3 independent experiments) transfected with pCMV-EGFP or pCMV-EGFP-CdtB or treated with cis-Platinum (CDDP) (10 μM) for 24 h. j Western blot analysis of CPT1a expression of Hep3B cells infected with wild type (WT), or CdtB knockout (ΔCdtB) H. hepaticus. Scale bars represent 2 μm (b, original) or 1 μm (b, enlarged), 10 µm (a) or 50 µm (c). Statistical analysis was determined by one-way ANOVA (d, i), two-way ANOVA (f, g) followed by Dunnett’s multiple comparisons test, or unpaired two-tailed Student’s t test (e). Source data are provided as a Source Data file.
CdtB causes mitochondrial damage through ATP5A1 with the assistance of Hsp90
To gain insight into the mechanism of CdtB translocation to mitochondria, a luminescent split protein expression system was employed. CdtB was expressed using pBIFC-CdtB-VC155, while the other component of GFP (Venus) was expressed with the COX8A signal peptide using pBIFC-COX8A-VN173. This enabled the detection of functional fluorescence upon CdtB-VC155 translocation to mitochondria and interaction with COX8A-VN173. As illustrated in Fig. 3a and Fig. S2g, the presence of filamentous green fluorescent cells was observed in cells co-transfected with CdtB-VC155 and COX8A-VN173, indicating that CdtB was translocated to mitochondria and induced mitochondrial damage and fragmentation (Fig. 3a). Additionally, the expression of C. jejuni-CdtB using the luminescent split protein system yielded comparable results (Fig. S2g).
a Live cell images of Hep3B cells transfected with pBIFC-COX8A-VN173, pBIFC-CdtB-VC155 and pBIFC-C. j-CdtB-VC155 along with their corresponding reverse validation plasmids. b Western blot analysis of CdtB content in mitochondrial fractions. Hep3B cells were transfected with either pCMV-Myc-EGFP or pCMV-Myc-EGFP-CdtB for 24 h. Whole-cell lysates (cell), cytoplasmic (cyto), and mitochondrial components (mito) were probed with CTCF, β-actin, and TOM20 antibody, respectively. c Co-immunoprecipitation (Co-IP) assay of the interaction between CdtB and ATP5A1 in Hep3B cells transfected with pCMV-Myc-EGFP or pCMV-Myc-EGFP-CdtB. d Immunofluorescence images of the subcellular localization of CdtB (green) and ATP5A1 (red) in Hep3B cells transfected with pCMV-vector or pCMV-CdtB for 24 h. e The activity of mitochondrial respiratory complex V in Hep3B cells transfected with pCMV-vector or pCMV-CdtB for 24 h (n = 3 independent experiments). Scale bars: 10 µm (d) or 50 µm (a). Statistical analysis was performed using unpaired, two-tailed Student’s t test (e). Source data are provided as a Source Data file.
CdtB induces DNA damage depends on its DNase I activity32. To ascertain whether the translocation of CdtB requires its DNase I activity, a mutant plasmid encoding CdtB (H265L) without DNase I activity was constructed. The subsequent findings demonstrated that the H265L mutation did not affect the mitochondrial transport of CdtB (Fig. 3b and S2g). The occurrence of mutCdtB in mitochondria was found to be approximately 8%, while CdtB was identified in mitochondria at a rate of around 9% (calculated using the following formula: 150 μL mitochondrial lysate/1000 μL cytoplasmic lysate × the gray-scale value of mito-CdtB/ the gray-scale value of cyto-CdtB) (Fig. 3b). To identify the target of CdtB in mitochondria, immunoprecipitation-mass spectrometry (IP-MS) was performed using CdtB-overexpressing Hep3B cell lysis. The results demonstrated that ATP5A1 (ATPA), the alpha subunit of the mitochondrial ATP synthase F1 complex, achieved a higher score among all the candidates (Fig. S3a). The involvement of ATP5A1 in ATP production and mitochondrial function has been reported, while its reduced activity is also associated with the generation of ROS and oxidative damage to mitochondria33. The direct interaction between CdtB and ATP5A1 was confirmed by a co-immunoprecipitation (co-IP) assay (Fig. 3c). Meanwhile, immunofluorescence analysis also demonstrated the co-localization of CdtB with ATP5A1 (Fig. 3d) and its inhibitory effect on the activity of mitochondrial respiratory complex V (Fig. 3e).
Furthermore, Hsp90 was identified as another candidate associated with mitochondria dysfunction, and the interaction between CdtB and Hsp90 was confirmed by co-IP assay (Fig. 4a). The blocking Hsp90 with 17-AAG in Hep3B cells led to a significant reduction in the mitochondrial translocation of CdtB (Fig. S3b), as well as an attenuation in the collapse of MMP (Fig. 4b), the release of mtDNA into the cytoplasm (Fig. 4c), and a decrease in ATP production (Fig. 4d). Moreover, the treatment of 17-AAG was found to alleviate cell death and mitochondrial oxidative damage induced by CdtB, as demonstrated by PI or DHE staining, respectively (Fig. 4e, f). In addition to mitigating mitochondrial damage, the fatty acid β-oxidation activity was restored by 17-AAG relative to the CdtB transfected alone (Fig. 4g). Collectively, these results suggest that the functional significance of CdtB in cells requires the existence of Hsp90.
a The interaction between CdtB and Hsp90 in Hep3B cells. The cells were transfected with pCMV-Myc-EGFP or pCMV-Myc-EGFP-CdtB for 24 h prior to co-immunoprecipitation analysis (co-IP) assay. b–f 17-AAG inhibits CdtB-induced mitochondrial damage, as reflected by preservation of mitochondrial membrane potential (b), reduced cytosolic mtDNA levels (c), restored ATP production (d), decreased cell death (e) and suppression of mitochondrial ROS accumulation (f). Hep3B (b–e) or HEK293A (f) cells were treated with a vehicle control (NC) or 17-AAG (500 nM) for 12 h, followed by transfection with pCMV-EGFP or pCMV-EGFP-CdtB for an additional 24 h (n = 3 independent experiments). TMRM tetramethylrhodamine methyl ester, PI propidium iodide, DHE dihydroethidium. g 17-AAG restores CdtB-impaired mitochondrial β-oxidation capacity. Hep3B cells were treated as described above. Rotenone and cis-Platinum (CDDP) served as positive controls for mitochondrial and nuclear DNA damage, respectively (n = 3 independent experiments). Scale bar: 50 μm (f). Statistical significance was determined using the two-way ANOVA followed by Dunnett’s multiple comparisons test (b–d, g). Source data are provided as a Source Data file.
The CdtB-mtROS-mTORC1-SREBP1 axis promotes lipogenesis in the hepatocytes
Our subsequent objective was to ascertain the mechanism by which CdtB induces lipogenesis and subsequent lipid deposition in the liver. SREBP1 is a key transcription factor that regulates de novo fatty acid synthesis34. It was observed that the persistent nuclear localization of SREBP1 in hepatocytes was induced by rAd-CdtB treatment (Fig. 5a), indicating that CdtB is sufficient to promote de novo fatty acid synthesis. Similarly, the transcription and protein level of SREBP1 were upregulated in Hep3B cells following infection with the H. h WT in comparison to the H. h ΔCdtB, as well as its downstream genes (Fig. 5b, c and Fig. S4a). Furthermore, the upregulation of transcription of SREBP1-a, SREBP1-c, FASN, SCD1 and LPIN1 was found in Hep3B cells overexpressing CdtB in comparison to the control cells (Fig. S4b). Given the pivotal role of the mTORC1 signaling pathway in regulating the activation of SREBP124, an investigation was conducted to ascertain the role of mTORC1 signaling in CdtB-induced lipogenesis. The results demonstrated that the phosphorylation of mTOR (Ser2448) and its downstream target P70-S6 kinase were robustly increased in Hep3B cells infected with the H. h WT, which was abrogated by the deletion of CdtB (Fig. 5f). Furthermore, it was observed that the nuclear translocation of SREBP1, which was induced by CdtB, could be blocked by the mTORC1 inhibitor Rapamycin (Fig. 5d, e). These findings suggest that the nuclear translocation of SREBP1 induced by CdtB is facilitated by the activation of mTORC1.
a Immunohistochemistry analysis of SREBP1 distribution in the liver tissues of mice infected with rAd-vector or rAd-CdtB at 17, 42, 60 days post-infection (DPI). b, c Quantitative analysis of SREBP1 mRNA (b) and protein (c) levels of Hep3B cells infected with wild type (WT) or CdtB knockout (ΔCdtB) H. hepaticus (n = 3 independent experiments). d, e Immunofluorescence analysis (d) or Western blot (e) of SREBP1 or nSREBP1 (nuclear SREBP1) in Hep3B cells. The cells were transfected with pCMV or pCMV-CdtB, and subsequently treated with 100 nM rapamycin or 100 μM MitoTEMPO for 24 h. f Western blot analysis of mTOR and p70S6K phosphorylation in Hep3B cells infected with WT or ΔCdtB H. hepaticus. Scale bars: 50 µm (d) or 100 µm (a). Statistical significance was determined using the two-way ANOVA followed by Dunnett’s multiple comparisons test (b). Source data are provided as a Source Data file.
mtROS has been demonstrated to activate PI3K/AKT/mTOR signaling35. Our findings demonstrated that CdtB caused mitochondrial dysfunction with increased mtROS production (Fig. 2d), prompting us to investigate whether CdtB regulates mTORC1 activity and subsequent SREBP1 activation through mtROS. To test this hypothesis, MitoTEMPO, a mitochondrial superoxide scavenger, was employed to neutralize mtROS. The results demonstrated a comparable level of nuclear SREBP1 between control and CdtB-expressing cells (Fig. 5d, e). Meanwhile, the activation of mTOR signaling, which was induced by CdtB, was markedly diminished with the administration of MitoTEMPO (Fig. S4c). To confirm whether CdtB in mitochondria can regulate the activation of SREBP1, the mitochondrial-localized CdtB, Cox8A-CdtB, was transfected into Hep3B cells, resulting in a significant nuclear translocation of SREBP1 (Fig. S5a, b). Conversely, it was observed that nuclear DNA damage induced by CDDP did not promote the nuclear localization of SREBP1, whereas oxidative mitochondrial damage caused by rotenone did result in this effect (Fig. S5a). Moreover, the nuclear localization of SREBP1 in liver organoids was also observed, exhibiting a more pronounced presence in the CDT-treated group (Fig. 6a, b and S5c). Collectively, these findings suggest that H. hepaticus-induced lipogenesis is mainly dependent on CdtB-induced mtROS, which activates mTORC1 signaling and subsequent SREBP1 activation in hepatocytes.
a, b Immunofluorescence images of SREBP1 in mouse liver organoids infected with wild type (WT) or CdtB knockout (ΔCdtB) H. hepaticus (a) or treated with CdtA/C or holotoxin CDT (b) for 48 h. c Co-immunoprecipitation assay demonstrating the interaction between CdtB and NONO in Hep3B cells. Cells were transfected with either pCMV-Myc-EGFP or pCMV-Myc-EGFP-CdtB for 24 h prior to immunoprecipitation. d Immunohistochemical detection of γH2AX in liver sections from mice administered rAd-vector or rAd-CdtB at 17, 42, 60 days post-infection (DPI). e Western blot analysis of NONO expression in Hep3B cells following infection with WT or ΔCdtB H. hepaticus. f Immunofluorescence images of SREBP1 in Hep3B cells with either NONO overexpression or NONO knockout, transfected with pCMV or pCMV-CdtB. g Western blot analysis of SREBP1 in nuclear fractions of Hep3B cells and Hep3B NONO-knockout cells transfected with pCMV-Myc-EGFP or pCMV-Myc-EGFP-CdtB for 24 h. Cytoplasmic (cyto) and nuclear components (nuc) were probed with β-actin and CTCF antibodies, respectively. Scale bars: 20 µm (a, b), 50 µm (f) or 100 µm (d). Source data are provided as a Source Data file.
SREBP1-mediated lipid synthesis is stabilized by CdtB-NONO interaction
\({{\rm{P}}54}^{{\rm{nrb}}}\)/NONO is a novel regulator of SREBP1-a through maintaining SREBP1-a stability in the nucleus and subsequently promoting SREBP1-mediated lipid gene transcription and lipid production26. It is noteworthy that the IP-MS dataset (Fig. S3a) showed an interaction between CdtB and NONO, which was subsequently validated in vitro (Fig. 6c). It has been previously reported that the nuclear DNA damage can induce the activation of NONO36 and the DNA damage caused by CdtB was also confirmed in the liver by an increase in γH2AX (Fig. 6d). Next, the impact of H. hepaticus infection on NONO expression was investigated. The results demonstrated a significant elevation in NONO protein expression in Hep3B cells following infection with the H. h WT, which was dependent on the CdtB (Fig. 6e). This suggests that the CdtB may facilitate the nuclear expression of NONO via direct interaction, while COX8A-CdtB does not (Fig. S5d). To gain further insight into the role of the NONO pathway in the regulation of SREBP1 by CdtB, the NONO was knocked out in Hep3B cells (Fig. S5e), and the results revealed a significant decrease in nuclear localization of SREBP1 compared to the NONO-intact cells transfected with CtdB (Fig. 6f, g). In light of these findings, it can be posited that the presence of NONO is necessary for the CdtB-mitochondrial ROS-mTORC1-SREBP1 axis.
Neutralization of CdtB alleviates hepatic lipidosis caused by H. hepaticus infection
To investigate the therapeutic potential of CdtB in the context of H. hepaticus-induced hepatic lipidosis, a mouse monoclonal antibody (mAb) against CdtB was developed and administered to H. hepaticus-infected mice. Notably, the administration of CdtB mAb resulted in a reduction in the number of lipid vacuoles and Oil Red O staining areas observed in the livers of mice infected with H. hepaticus (Fig. 7a, b). In an in vitro study, exposure of the liver cells to CdtB resulted in cell cycle arrest at the G2/M phase, which was suppressed by the anti-CdtB mAb (Fig. 7c). In addition, the expression of both NONO and SREBP1 induced by H. hepaticus was suppressed by the neutralizing mAb (Fig. 7d). These results indicate that targeting CdtB may be a potential therapeutic strategy for H. hepaticus-induced hepatotoxicity.
a, b H&E-stained (a) or Oil Red O-stained (b) liver sections from mock- or H. hepaticus (H. h)-infected mice, with or without CdtB mAb treatment, at 1, 3, 5 months post-infection (MPI). c Cell cycle assay of Hep3B cells treated with CdtB in the presence or absence of CdtB mAb (n = 3 independent experiments). d Protein detection and semi-quantitative analysis of NONO, SREBP1 and nucleus SREBP1 of mice treated with H. hepaticus, H. hepaticus + CdtB mAb or Mock + CdtB mAb at 5 MPI (n = 3 mice). Scale bars: 50 µm (b) or 100 µm (a). Statistical significance was determined using the one-way ANOVA followed by Dunnett’s multiple comparisons test (c, d). Source data are provided as a Source Data file.
The nucleic acid of H. hepaticus is present in the liver of MASLD patients
The RNA FISH detection reveals the presence of H. hepaticus in the liver tissue of 20% (2/10) of metabolic dysfunction-associated steatotic liver (MASL) patients and 16.67% (3/18) of MASH patients, but not in non-MAFLD liver (n = 4) (Fig. 8 and S6, Supplementary Data 1). It is noteworthy that the levels of ALT and AST in those H. hepaticus-positive MASL patients, who did not appear to be obviously overweight, are significantly higher than those in other H. hepaticus-negative MASL patients (Supplementary Data 1). This observation may be indicative of a potential association with H. hepaticus-induced hepatotoxicity.
RNA fluorescence in situ hybridization (FISH) targeting H. hepaticus 16S rRNA in liver tissues form one transplant donor and one MASLD patient. H. hepaticus (H.h) specific probe, labeled with Cy3 at 5’ end, was used to detect bacterial 16S rRNA, while DAPI was employed for nuclear counterstaining, scale bar: 25 μm.
Discussion
The MASLD progression includes metabolic dysfunction-associated steatotic liver (MASL), metabolic dysfunction-associated steatohepatitis (MASH), fibrosis, cirrhosis, and MASH-related hepatocellular carcinoma37. MASLD, defined as steatosis liver disease, is diagnosed through imaging or biopsy in patients with any of the following five cardiovascular risk factors: increased BMI or waist circumference, elevated fasting blood glucose levels, arterial hypertension, hypertriglyceridemia, and low plasma HDL-cholesterol. Other potential causes of chronic liver disease such as viral hepatitis or excessive alcohol consumption were not observed38,39. A recent study also reveals that H. hepaticus can colonize the human gut and may promote the progression of alcoholic steatohepatitis40. Here, we demonstrate the presence of H. hepaticus in the liver of a subset of patients with MASLD. The detection rate of H. hepaticus is 17.86% (5/28) (Supplementary Data 1), which is marginally lower than the rate (27.49%) detected by Alvarez et al.28 in the MASLD population of Guatemala. However, due to the limited number of liver biopsy samples from MASLD patients and the sensitivity of RNA FISH detection, the important role of H. hepaticus in the pathogenesis of human MASLD may still be underestimated.
H. hepaticus, a potential zoonotic bacterium colonizing in the intestine, whose antigen or DNA has been identified in individuals with liver and bile diseases. Our previous study demonstrated that H. hepaticus can induce colitis, hepatitis, and liver fibrosis in mice41. Several pathogenic factors of H. hepaticus such as CDT, hydrogenase and urease, have been identified in the past decades27,42. In this study, a mouse infection model was employed to elucidate the underlying mechanisms by which H. hepaticus contributes to hepatic lipidosis. CdtB could translocate to mitochondria while causing its dysfunction and blocking β-oxidation of fatty acids. Meanwhile, it activates SREBP1 to elicit lipid synthesis via the mTORC1/P70S6K signaling pathway, independently of which CdtB interacts with NONO to enhance the nuclear localization of SREBP1 in hepatic cells. Considering the concurrent elevation in serum TG levels, it is plausible to speculate that CdtB, a virulence gene of H. hepaticus, may contribute to the progression towards MASLD.
As supported by the overexpression of CdtB, which promotes hepatocyte steatosis, and attenuation of steatosis by CdtB neutralizing antibody in vivo (Fig. 7a, b), it is strongly suggested that liver steatosis caused by H. hepaticus is dependent on the virulence factor CdtB secreted by H. hepaticus, rather than direct bacterial infection of hepatocytes. CDTs have been extensively studied for their toxicity and hazards. Their DNase I activity causes DNA damage and cell cycle blockade, leading to cell senescence or death when located in the nucleus42,43,44. However, mitochondrial dysfunction induced by these toxins is rarely reported. The amino acid sequence analysis showed that there is no clear subcellular localization signal peptide in CdtB. Therefore, the intracellular distribution of CdtB requires the assistance of other proteins such as Hsp90, an important molecular chaperone protein that can be distributed to multiple organelles45,46,47 such as the nucleus and mitochondria. Under certain stress conditions, such as ischemia-reperfusion48 or hypoxia49, Hsp90 can be transported to the mitochondria and protect organelles from various damages. Although Hsp90 is essential for cell survival and resistance to stimuli, it can also be hijacked by various pathogenic microorganisms50,51. In this work, we found that the interaction of CdtB with Hsp90 confers mitochondrial access in the absence of a mito-localizing peptide. Furthermore, mitochondrially localized CdtB also induced changes in mitochondrial morphology through ATP5A1 and decreased the activity of mitochondrial respiratory complex V (Fig. 3e). ATP5A1 is an essential enzyme in ATP synthesis and plays an important role in cellular energy conversion52. The impaired mitochondrial respiratory complex V increased mitochondrial superoxide33, decreased the membrane potential, and caused mtDNA release. Mitochondrial damage or dysfunction is present in a variety of liver diseases, such as acute fatty liver of pregnancy53, mtDNA deletion and deficiency syndrome54, alcoholic foamy liver steatosis55, and MASLD. Furthermore, the mtDNA released by hepatocytes can be identified by STING in macrophages, which subsequently enhances the liver inflammation56. Notably, microvesicular steatosis was observed in the liver at the early infection stage, and then the macrovesicular LD in some hepatocytes was gradually obvious during the subsequent infection phase (Fig. 1a, b). Microvesicular steatosis usually occurs as a result of impaired mitochondrial β-oxidation, which in turn leads to the accumulation of fatty acids in the form of triglycerides and formation of small lipid droplets in the cytosol of hepatocytes57. Furthermore, the presence of microvesicular steatosis in liver biopsies may serve as a severity degree marker of MASLD30,58. Therefore, we proposed that mitochondrial injury may be involved in CdtB-induced steatosis as evidenced by damaged activity of mitochondrial respiratory complex V, mtDNA release and impairment of cellular fatty acid β-oxidation.
CdtB is a classical genetic toxin commonly found in several pathogenic microorganisms, including Escherichia coli, Shigella, C. jejuni, Actinobacillus companionis, Haemophilus dubliniensis59. CdtB has been shown to be associated with C. jejuni infection leading to diarrhea-predominant irritable bowel syndrome (IBS-D)60, acute enteritis, and colorectal cancer61. In addition, C. jejuni CdtB also shows mitochondrial localization (Fig. 3a). This suggests that CdtB-induced mitochondrial damage may be common to other CdtB-positive bacterial infections. It is noteworthy that CdtB induced direct mitochondrial damage via ATP5A1, and the injury was alleviated by a Hsp90 inhibitor. This suggests that Hsp90 could be an effective target for the treatment of diseases induced by CdtB-positive bacteria.
Fatty acid synthesis is another crucial factor influencing fat deposition in hepatocytes. The liver can activate the de novo fatty acid synthesis pathway through mTORC1/P70S6K/SREBP1 signaling when sufficient glucose is present23,62. mTORC1, a target of rapamycin, exerts control over cellular metabolism by regulating the transcription and translation of metabolic genes such as PGC-1α, SREBP1/2, and HIF-1α63. Extensive research has highlighted the pivotal regulatory role of mTORC1 in mitochondrial metabolism, thereby establishing it as a key determinant of mitochondrial function64. As mTOR directly perceives mitochondrial dysfunction65 or sense ROS through retrograde signals35,66, thereby regulating both the quantity and functionality of mitochondria. Consistently, our results show that mito-localized CdtB can activate the nuclear translocation of SREBP1 (Fig. S5a, b), while the removal of mtROS by MitoTEMPO can strongly inhibit its activation (Fig. 5d, e and Fig. S4c). These results indicate that CdtB can directly damage mitochondria and activate the mTORC1/P70S6K/SREBP1 pathway-mediated fatty acid de novo synthesis.
The cytotoxicity of CdtB is primarily characterized by its ability to induce nuclear DNA damage via its DNase I activity67. IP-MS experiments revealed that CdtB interacts with NONO, a nuclear scaffolding protein that acts as a bridge and protector for various transcription factors68, as well as a DNA damage-inducing protein36. NONO interacts with nuclear SREBP-1a, resulting in increased stability of the nuclear SREBP-1a protein. This subsequently facilitates SREBP-1-mediated transcription of lipid genes and promotes lipid production26. The results clearly demonstrate that CdtB promotes the activation and translocation of SREBP1 into the nucleus, meanwhile the upregulation of NONO (Fig. 6e) significantly enhances the nuclear localization of SREBP1, thereby further enhancing associated lipogenesis.
In summary (Fig. 9), the hepatic lipid deposition caused by H. hepaticus infection is primarily due to the action of its virulence factor CdtB, which directly induces mitochondrial damage through ATP5A1, reduces cellular β-oxidation capacity with the assistance of Hsp90, and activates the mTORC1/P70S6K/SREBP1 signaling pathway leading to gene expression related to lipid synthesis. In addition, CdtB upregulates the expression of NONO, which further enhances the transcriptional activity of SREBP1. Importantly, CDT is present in several pathogenic bacteria, and it is crucial to evaluate its potential role in promoting lipid deposition in the liver and other organs, particularly in the context of metabolic syndrome-related diseases.
H. hepaticus is transported from the portal vein to the liver, releasing CDT around hepatocytes. Upon binding to cell surface receptors, the subunit CdtB is internalized into hepatocytes. In a process facilitated by Hsp90, CdtB is trafficked to mitochondria, where it interacts with ATP5A1, a core component of mitochondrial respiratory complex V, thereby inducing mitochondrial dysfunction, characterized by mitochondrial DNA release, elevated oxidative stress, diminished ATP production, and suppression of β-oxidation. This process can be impeded by the Hsp90 inhibitor, 17-AAG. The resultant increase in mitochondrial reactive oxygen species (mtROS) activates the mTORC1/P70S6K/SREBP1 signaling pathway, driving SREBP1 maturation and nucleus translocation. This activation promotes de novo lipogenesis, a process susceptible to inhibition by rapamycin. Additionally, a fraction of CdtB enter the nucleus, causing DNA damage and upregulating the expression of NONO, which stabilizes nuclear SREBP1 and sustains the transcriptional upregulation of lipogenic genes (Created in BioRender. Xia, L. (2025) https://BioRender.com/qwsavjs).
Methods
Animal experiments
The experimental animals were 144 four-week-old male BALB/c (BALB/cAnSlac) mice purchased from SLAC LABORATORY ANIMAL in Shanghai, China. Prior to infection, the mice were housed in a specific pathogen-free facility, and the infected control animals were also housed in the same facility. At five weeks of age, mice were infected intragastrically with 108 CFU/mL of H. hepaticus (wild-type or CdtB-deficient strain) at a volume of 100 μL per mouse, while the blank group was treated with PBS. To evaluate the MASLD model in the context of H. hepaticus infection, mice were inoculated with H. hepaticus via intraperitoneal injection and then subjected to a high-fat diet (HFD, 60% calcium from fat, Research Diet # XTHF60, Xietong Pharmaceutical, Jiangsu, China) for a period of 12 weeks. CdtB neutralizing antibody was injected into the tail vein at a dose of 0.5 mg per mouse every 14 days. Administration of rAd-vector or rAd-CdtB was performed at a dose of 1010 PFU per mouse via i.v. The mice were maintained under controlled conditions with a temperature of 24 °C ± 2 °C and humidity of 50% ± 5%, following a 12-h light /12-h dark cycle (lights on from 7 AM to 7 PM), and had ad libitum access to food and water. The animal experiments complied with the China Laboratory Regulation Act (2022) under Project License (SYXK(SU)2022-0044) authorized by Jiangsu Provincial Science and Technology Department and approved by the Institutional Animal Care and Use Committee (IACUC) of Yangzhou University. Experimental animals were handled according to the guiding principles outlined in China’s Animal Welfare Law as well as being approved by the Experimental Animal Ethics Committee of Yangzhou University (approval no. 202203042).
Human samples
MASLD patients were enrolled in this study according to the results of pathological examinations and the inclusion criteria reported by Ding, J. et al.69, and written informed consent was signed by each patient. The control biopsies were from 2 liver focal nodular hyperplasia patients and 2 liver transplant donors.
To detect H. hepaticus in liver sections, MASL patients (n = 10, 6 male/ 4 female) and MASH patients (n = 18, 12 male/ 6 female) were enrolled in this study. Liver tissue from the liver focal nodular hyperplasia and liver transplant donors (n = 4, 3 male/ 1 female) was used as the non-MASLD control. Baseline features of these subjects are shown in Supplementary Data 1. The liver tissues were prepared into paraffin sections for RNA FISH analysis. The protocol was approved by the Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University (approval no. 2016-246).
Recombinant adenovirus (rAd) production and delivery
In order to mitigate the toxicity of CdtB to other organs and cells, the replication-deficient type 5 adenovirus (rAd), which has been demonstrated to have an evident affinity for the liver70, was selected as the vehicle in vivo with i.v. administration. This approach can result in the majority of rAd being sequestered by the liver71. Moreover, the expression of CdtB was derived from a hepatocyte-specific promoter, the thyroxine-binding globulin (TBG) promoter, which reduces the impact on other organs. The adenovirus shuttle plasmid pShuttle-mcherry-TBG-EGFP-P2A-CdtB and its control adenovirus vector pShuttle-mcherry required for this study were generated through PCR amplification and connected by ligation PCR. Subsequently, the shuttle plasmid was linearized using Pme I and then transformed into BJ5183 competent cells harboring the adenovirus backbone plasmid (pAdEasy-1) to construct the full-length recombinant adenovirus plasmid. Following Pac I digestion, the linearized rAd-CdtB or rAd-vector plasmid was transfected into HEK293A cells for virus rescue. The recombinant adenovirus was amplified by HEK293A and subsequently purified using iodixanol gradient centrifugation. Finally, HEK293A cells were infected with the recombinant virus at various dilutions to determine its titer. Intravenous administration via the tail vein of rAd-vector or rAd-CdtB was performed at a dosage of 1010 PFU per mouse at the age of 5 weeks. These mice were sacrificed at 17, 42, and 60 days post-infection (DPI).
Cell culture and treatment
The human hepatocellular carcinoma cell lines Hep3B and Huh-7, as well as the human embryonic kidney cell line HEK293A, were cultured in DMEM (41966-029, Thermo Fisher) supplemented with 10% fetal bovine serum (3023A, Umedium). All cells were incubated at 37 °C in a 5% CO2 environment. Rapamycin (100 nM, HY-10219, MedChemExpress (MCE)), CsA (500 ng/mL, HY-B0579, MCE), 17-AAG (500 nM, HY-10219, MCE), MitoTEMPO (100 μM, HY-112879, MCE) and Rotenone (800 nM, HY-B1756, MCE) were administered for 24 h. Nuclear DNA damage was induced by treating with CDDP (10 μM, HY-17394, MCE) for 24 h. H. h WT and H. h ΔCdtB were used to infect the cells at an MOI of 1.
The cellular fatty acid oxidation activity was measured using Fatty Acid Oxidation Assay Kit (ab118183, Abcam); the intracellular ATP concentration was measured using Enhanced ATP Assay Kit (S0027, Beyotime); the total triglyceride content in mouse serum was measured using Amplex Red TG Assay Kit (S0219M, Beyotime); the cellular mitochondrial oxidative stress level was measured using DHE (S0063, Beyotime) and MitoSOX (M36008, Thermo Fisher); The activity of mitochondrial respiratory chain complex V in cells was measured using Micro Mitochondrial complex Ⅴ Activity Assay Kit (KTB1890, Abbkine).
Bacterial culture and collection
The CdtB mutant strain (H. h ΔCdtB strain) was generated and preserved in our laboratory. H. hepaticus strains ATCC 51449 (Hh3B1) (H. h WT strain) and H. h ΔCdtB strain were cultured on Brucella Agar (211086, BD) with 5% sheep blood (1001339-1, Qingdao Hope Bio-technology), under the conditions of microaerophilic atmosphere (80% N2 and 10% CO2, with 5% O2 and 5% H2) at 37 °C. The culture medium was supplemented with Helicobacter pylori Selective Supplement (Dent) antibiotics (SR0147E, Oxoid), followed by incubation for a period of 2-3 days. The bacterial cells were harvested in PBS solution at an optical density reading of OD600 = 1 prior to mice infection.
CDT expression and purification
E. coli BL21(DE3) carrying a pCold I plasmid encoding CdtA, CdtB, and CdtC genes of H. hepaticus were induced with 0.1 mM IPTG (I8070, Solarbio) at 16 °C for 20 h, respectively. The bacterial cells were pelleted and were resuspended in 20 mM Tris (pH 7.5) with 0.5 M NaCl, 0.1% Triton X-100 and 5% glycerol. Then, the bacterial cells were sonicated to release the soluble proteins, which were then purified using Ni-NTA agarose resin (L00250, GenScript). Following purification, CdtA, CdtB, and CdtC were mixed at a final concentration of 1 nM to prepare the holotoxin CDT (CdtA + CdtB + CdtC) or control toxin CdtA/C (CdtA + CdtC), respectively.
Mouse liver organoids culture and differentiation
The protocol for the isolation, culture, and differentiation of mouse liver organoids was referenced from Huch et al.72. Briefly, 3 healthy male BALB/c mice aged 6–8 weeks were selected and injected with 1 mg/kg mouse CCL4 5 days prior to mouse sacrifice. Subsequently, the liver samples were collected in aseptic conditions and the individual liver lobes were cut into small pieces. The liver pieces were then incubated at 37 °C with collagenase XI (C7657, Sigma-Aldrich) and dispase II (17105041, Gibco) for a period of 2–3 h. Subsequently, the digested mixture underwent several low-speed centrifugations (50 g) to remove suspended, fragmented hepatocytes. The precipitated cells and ductal tissue were resuspended in organoid expansion medium, containing 1 × N2 (17502048, Gibco), 1 × B27 Supplement Minus Vitamin A (12587010, Gibco), 10 mM nicotinamide (47865-U, Sigma-Aldrich), 1.25 mM NAC (A8199, Sigma-Aldrich), 15% L-WRN culture medium, 50 ng/mL EGF (315-09-500UG, PeproTech), 10 nM gastrin (304329, Novoprotein Scientific), 50 ng/mL HGF (315-23-20UG, PeproTech) and 100 ng/mL FGF10 (450-61-25UG, PeproTech) in Advance DMEM/F12 (12634010, Gibco), and mixed 1:1 with matrix gel (C0392, Corning). This suspension was then inoculated into 24-well plates. The liver organoids were cultivated in expansion medium for 5 days, after which they were cultured in differentiation medium, containing 1 × N2, 1 × B27, 10 mM nicotinamide, 1.25 mM NAC, 50 ng/mL HGF and 100 ng/mL FGF10, 3 µM CHIR-99021 (HY-10182, MCE), 1 µM dexamethasone (HY-14648, MCE), 1 µM A83-01 (HY-10432, MCE) in Advance DMEM/F12, for 9 days. The L-WRN cell line (CRL-3276, ATCC) was cultured in DMEM with 10% FBS, and the culture medium was harvested in accordance with the protocol established by Miyoshi et al.73.
Treatment of organoids with H. hepaticus or CDT
Liver organoids were treated with 1 × 107 CFU/mL of H. hepaticus or 1 nM CDT for 48 h, respectively. Then, the organoids were collected and fixed with 4% paraformaldehyde for Oil red O staining or indirect immunofluorescence assay.
RNA FISH analysis
The RNA FISH test was performed using the Fluorescence In Situ Hybridization detection kit (G3017, Servicebio, Wuhan, China). Briefly, the liver tissue sections were deparaffinized and rehydrated via xylene and ethanol. Subsequently, antigen repair was executed with an EDTA-citrate solution (pH 9.0), followed by permeabilization with proteinase K. The sections were then incubated with the Hep642 probe (5’ Cy3-labeled, 200 nM) in a solution consisting of 15% formamide, overnight at 40 °C. Then, the sections were stained with DAPI and observed under a fluorescence microscope (Leica, Wetzlar, Germany). The sequence of Hep642 probe was 5’-CCCACACTCCAGATACAGT-3’74. The cross-reactivity assay between the Hep642 probe and H. pylori was excluded by the detection of stomachs from Mongolian gerbils infected with H. pylori for four months (Fig. S7). The total bacterial 16S rRNA probe, EUB338 (5’ Cy3-labeled, 200 nM, 5’-GCCTCCCGTAGGAGT-3’), was utilized as a positive reference for the RNA FISH assay. The Hep642 and EUB338 probes were synthesized by General Biol (Anhui, China).
DNA extraction and PCR analysis
Mice were euthanized at 1, 3, and 5 months post-infection (MPI). Liver samples from the same location were collected, followed by extraction of tissue and bacterial DNA using the TIANamp Genomic DNA Kit (DP304-03, TIANGEN) according to the manufacturer’s instructions. Quantification of H. hepaticus in the liver was performed using the HH1450 gene primer (RTF2: ATGAAAGAGACTTTATTGCTTCA; RTR2: AGCCTGTGCATACCCTCATA)75. The Applied Biosystems StepOne Real-Time PCR System (ABI) was utilized for qPCR analysis of H. hepaticus levels in the liver with ChamQ SYBR qPCR Master Mix (Q311-02, Vazyme). The colonization level of H. hepaticus was estimated by normalization against mouse chromosomal DNA. The extraction and quantification of mtDNA in blood samples was performed according to Zhang et al.76.
Quantitative real-time PCR analysis
Total RNA was extracted from cells using RNAiso Plus (9108, Takara). RNA was then reverse transcribed into cDNA using HiScript III RT SuperMix for qPCR (+gDNA wiper) kit (R323, Vazyme) following the manufacturer’s instructions. qPCR analysis was performed to assess the mRNA levels of SREBP1-a, SREBP1-c, FASN, SCD1, and LPIN1, along with the housekeeping gene β-actin, on a StepOne real-time PCR system using ChamQ SYBR qPCR Master Mix. The relative expression of the target mRNA was determined by calculating 2−ΔΔCt method. Primers used in this study were as follows: SREBP1-a (5′-TCAGCGAGGCGGCTTTGGAGCAG-3′, 5′-CATGTCTTCGATGTCGGTCAG-3′)77; SREBP1-c (5′-GGAGGGGTAGGGCCAACGGCCT-3′, 5′-CATGTCTTCGAAAGTGCAATCC-3′)77; FASN (5′-AAGGACCTGTCTAGGTTTGATGC-3′, 5′-TGGCTTCATAGGTGACTTCCA-3′); SCD1 (5′-TCTAGCTCCTATACCACCACCA-3′, 5′-TCGTCTCCAACTTATCTCCTCC-3′); LPIN1 (5′-AGCCTCATACCCTAATTCGGAT-3′, 5′-CCTTTCCGTGGACTTGCTGA-3′). The primer sequences for FASN, SCD1, and LPIN1 were obtained from Primerbank (https://pga.mgh.harvard.edu/primerbank/). To quantify cytoplasmic mtDNA, mitochondrial and cytoplasmic fractions were separated by the Cell Mitochondria Isolation Kit (C3601, Beyotime). qPCR was used to detect mtDNA and nDNA, respectively. Primers used in this experiment were as follows: hmito (5′-CACTTTCCACACAGACATCA-3′, 5′-TGGTTAGGCTGGTGTTAGGG-3′); GAPDH (5′-AGGGCCCTGACAACTCTTTT-3′, 5′-TTACTCCTTGGAGGCCATGT-3′)78.
Flow cytometry analysis
After transfection with pCMV-vector or pCMV-CdtB, cells were harvested and stained with 5 μM Propidium Iodide (PI) (C0080, Solarbio) or 100 nM Tetramethylrhodamine, methyl ester (TMRM) (T4058, US Everbright, Suzhou, China) according to the manufacturer’s instructions. The percentage of dead cells, cell cycle or the mitochondrial membrane potential was then determined by flow cytometry (BD Biosciences, Franklin Lakes, USA).
Histopathology evaluation
Liver tissues from each mouse were fixed in 4% paraformaldehyde for 24 h at necropsy and embedded in paraffin for subsequent microscopic examination. Tissues were then sectioned at 5 μm slices and stained with hematoxylin and eosin (H&E). Additionally, Oil Red O staining was performed to assess the extent of oil in frozen sections. The percentage of positive area within the liver sections was quantified using ImageJ software.
Immunohistochemistry and indirect immunofluorescence analysis
Prior to immunohistochemistry, paraffin-embedded sections were subjected to deparaffinization and rehydration through a graded alcohol. Subsequently, the sections underwent treatment with 3% hydrogen peroxide followed by antigen retrieval using EDTA-citrate solution before being incubated with bovine serum albumin. The diluted primary antibodies, including anti-H. hepaticus (H.h) pAb (laboratory-made), anti-NONO (1:500, ab133574, Abcam) and anti-SREBP1 (1:500, 14088-1-AP, Proteintech) antibody, were then applied overnight at 4 °C. Finally, the sections were developed using 3,3N-diaminobenzidine tetrahydrochloride (DAB) (AR1000, Boster), and counterstained with hematoxylin for 1 min before observation under a light microscope.
Prior to indirect immunofluorescence, cells or liver organoids were fixed with 4% paraformaldehyde, broken with 0.25% Triton X-100, blocked with 5% BSA, and incubated with diluted primary antibodies. The following antibodies were used: anti-CdtB, anti-TOM20 (1:500, ab186735, Abcam), anti-ATP5a1 (1:500, A11217, ABclonal), anti-Albumin (1:500, 66051-1-Ig, Proteintech) and anti-SREBP1 (1:500). Subsequently, the cells or liver organoids were incubated with CoraLite488-conjugated goat anti-mouse antibody (1:1000, SA00013-1, Proteintech) or CoraLite594-conjugated donkey anti-rabbit antibody (1:500, SA00013-8, Proteintech), and then observed under a fluorescence microscope (Leica).
Transmission electron microscopy assay
Mice injected with rAd-vector or rAd-CdtB were sacrificed at 7 DPI and liver tissues of 1-3 mm³ were collected and immediately put into 2.5% glutaraldehyde, then stored in a refrigerator at 4 °C overnight and sent to Hangzhou Yanqu Information Technology Co., Ltd for TEM. The HITACHI HT7800 were used for observation.
Western blot analysis
Cell proteins were obtained using RIPA Lysis Buffer. Cell mitochondria and cytoplasm were isolated using the Cell Mitochondria Isolation Kit. The cytoplasmic and nuclear components were isolated using the Nuclear and Cytoplasmic Protein Extraction Kit (P0027, Beyotime). 40 µg protein from each sample was loaded onto a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), separated by electrophoresis, and transferred to polyvinylidene fluoride (PVDF) membranes. Then, the membranes were incubated with primary antibodies for 16 h at 4 °C, respectively. including ant-CPT1a (1:1000, A5307, ABclonal), anti-TOM20 (1:5000, A19403, ABclonal), anti-Myc-tag (1:10000, AE026, ABclonal), anti-Hsp90 (1:5000, ab203085, Abcam), anti-mouse IgG (1:1000, 58802S, light chain specific, CST), anti-ATP5a1 (1:10000), anti-SREBP1(1:1000), anti-NONO (1:5000, ab133574, Abcam), anti-p-mTOR (1:1000, 2971S, CST), anti-mTOR (1:1000, 2972S, CST), anti-p-P70S6K (1:1000, 9205S, CST), anti-P70S6 (1:1000, 9202S, CST), anti-β-actin (1:5000, 4967S, CST) and anti-CTCF (1:1000, 2899S, CST). The membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies (anti-Mouse IgG (H + L) AS003; anti-Rabbit IgG (H + L) AS014, ABclonal) at a dilution of 1:5000 for 1 h. Proteins were visualized using NcmECL High (P2300, NCM Biotech). The presentation of full scan blots was provided in the Source Data file.
Immunoprecipitation, immunoblot and mass spectrometry identification
Whole cells were lysed using a cell lysis buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 1% NP40, and 1 mM EDTA with protease inhibitors. The lysate was then centrifuged at 12000 rpm for 15 min at 4 °C, and the resulting supernatants were collected as input for immunoprecipitation (IP) in a volume of 5%. Following that, the supernatants were incubated with 15 μL Anti-c-Myc beads (88842, Thermo Fisher) for 4 h at 4 °C and washed 4 times with cell lysis buffer. The immune complexes bound to the beads were then solubilized overnight at 4 °C in a sample buffer prior to separation by SDS-PAGE gel electrophoresis. After gel separation, the proteins were transferred to a PVDF membrane for immunoblotting (IB). The left immune complexes were mixed with 4 × protein loading buffer and subjected to boiling at a temperature of 100 °C for a duration of 10 min. After cooling, it was transported to GENE CREATE on dry ice for mass spectrometry identification.
Statistics and reproducibility
No data was excluded from the analyses. The statistical analysis and data visualization were performed using GraphPad 8.0 software. Sample sizes (n) are indicated in the figure legends and P-values are indicated in the individual figures. Significance was determined by unpaired two-tailed Student’s t test, one-way or two-way ANOVA followed by Dunnett’s multiple comparisons test. Data are expressed as the means ± SD. Animal experiments were performed once due to ethical considerations. All non-animal experiments were repeated independently three times with similar results. At least 3 independent samples were included in each experiment. Source data are provided as a Source Data file.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The mass spectrum data generated in this study have been deposited in the ProteomeXchange via the PRIDE database under accession code PXD062676. All constructs generated in this study are available from the corresponding author upon request. All data supporting the article are included in the main article, Supplementary Information and the Source Data file. Source data are provided with this paper.
References
Blouin, A., Bolender, R. P. & Weibel, E. R. Distribution of organelles and membranes between hepatocytes and nonhepatocytes in the rat liver parenchyma. A stereological study. J. Cell Biol. 72, 441–455 (1977).
Di Ciaula, A. et al. Nonalcoholic Fatty Liver Disease (NAFLD). Mitochondria as players and targets of therapies?. Int J. Mol. Sci. 22, 5375 (2021).
Huttasch, M., Roden, M. & Kahl, S. Obesity and MASLD: Is weight loss the (only) key to treat metabolic liver disease?. Metabolism 157, 155937 (2024).
Ji, Y., Yin, Y., Sun, L. & Zhang, W. The molecular and mechanistic insights based on gut-liver axis: nutritional target for Non-Alcoholic Fatty Liver Disease (NAFLD) Improvement. Int J. Mol. Sci. 21, 3066 (2020).
Sharpton, S. R., Schnabl, B., Knight, R. & Loomba, R. Current concepts, opportunities, and challenges of gut microbiome-based personalized medicine in Nonalcoholic Fatty Liver Disease. Cell Metab. 33, 21–32 (2021).
Feng, G. et al. Recompensation in cirrhosis: unravelling the evolving natural history of nonalcoholic fatty liver disease. Nat. Rev. Gastroenterol. Hepatol. 21, 46–56 (2024).
Loomba, R. et al. Gut microbiome-based metagenomic signature for non-invasive detection of advanced fibrosis in human Nonalcoholic Fatty Liver Disease. Cell Metab. 25, 1054–1062.e1055 (2017).
Vallianou, N. G. et al. NAFLD/MASLD and the Gut-Liver Axis: From Pathogenesis to Treatment Options. Metabolites 14, 366 (2024).
Ha, S., Wong, V. W., Zhang, X. & Yu, J. Interplay between gut microbiome, host genetic and epigenetic modifications in MASLD and MASLD-related hepatocellular carcinoma. Gut 74, 141–152 (2024).
Akhlaghi, H., Javan, A. J. & Chashmi, S. H. E. Helicobacter pullorum and Helicobacter canadensis: Etiology, pathogenicity, epidemiology, identification, and antibiotic resistance implicating food and public health. Int. J. Food Microbiol. 413, 110573 (2024).
Fox, J. G., Ge, Z., Whary, M. T., Erdman, S. E. & Horwitz, B. H. Helicobacter hepaticus infection in mice: models for understanding lower bowel inflammation and cancer. Mucosal Immunol. 4, 22–30 (2011).
Yang, J., Ji, S., Zhang, Y. & Wang, J. Helicobacter hepaticus infection in primary hepatocellular carcinoma tissue. Singap. Med. J. 54, 451–457 (2013).
Shimoyama, T. et al. Serological analysis of Helicobacter hepaticus infection in patients with biliary and pancreatic diseases. J. Gastroenterol. Hepatol. 25, S86–S89 (2010).
Boesze-Battaglia, K. et al. Cholesterol-rich membrane microdomains mediate cell cycle arrest induced by Actinobacillus actinomycetemcomitans cytolethal-distending toxin. Cell. Microbiol. 8, 823–836 (2006).
Canonico, B. et al. Monocyte response to different Campylobacter jejuni lysates involves endoplasmic reticulum stress and the lysosomal(-)mitochondrial axis: when cell death is better than cell survival. Toxins (Basel) 10, 239 (2018).
Gu, J. et al. Campylobacter jejuni cytolethal distending toxin induces GSDME-dependent pyroptosis in colonic epithelial cells. Front. Cell Infect. Microbiol. 12, 853204 (2022).
Pessayre, D., Mansouri, A. & Fromenty, B. Nonalcoholic steatosis and steatohepatitis. V. Mitochondrial dysfunction in steatohepatitis. Am. J. Physiol. Gastrointest. Liver Physiol. 282, G193–G199 (2002).
Annesley, S. J. & Fisher, P. R. Mitochondria in Health and Disease. Cells 8, 680 (2019).
Al Amir Dache, Z. & Thierry, A. R. Mitochondria-derived cell-to-cell communication. Cell Rep. 42, 112728 (2023).
Nassir, F. & Ibdah, J. A. Role of mitochondria in nonalcoholic fatty liver disease. Int. J. Mol. Sci. 15, 8713–8742 (2014).
Dong, J. et al. Mic19 depletion impairs endoplasmic reticulum-mitochondrial contacts and mitochondrial lipid metabolism and triggers liver disease. Nat. Commun. 15, 168 (2024).
Engelking, L. J., Cantoria, M. J., Xu, Y. & Liang, G. Developmental and extrahepatic physiological functions of SREBP pathway genes in mice. Semin. Cell Dev. Biol. 81, 98–109 (2018).
Ma, N. et al. PPDPF alleviates hepatic steatosis through inhibition of mTOR signaling. Nat. Commun. 12, 3059 (2021).
Han, J. & Wang, Y. mTORC1 signaling in hepatic lipid metabolism. Protein Cell 9, 145–151 (2018).
Kim, J. & Guan, K. L. mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Biol. 21, 63–71 (2019).
Zhu, Z. et al. p54(nrb)/NONO regulates lipid metabolism and breast cancer growth through SREBP-1A. Oncogene 35, 1399–1410 (2016).
Cao, S. et al. Helicobacter hepaticus promotes liver fibrosis through oxidative stress induced by hydrogenase in BALB/c mice. Helicobacter 28, e13001 (2023).
Alvarez, C. S. et al. Associations between Helicobacter pylori with nonalcoholic fatty liver disease and other metabolic conditions in Guatemala. Helicobacter 25, e12756 (2020).
Pere-Vedrenne, C. et al. The Cytolethal Distending Toxin Subunit CdtB of Helicobacter hepaticus Promotes Senescence and Endoreplication in Xenograft Mouse Models of Hepatic and Intestinal Cell Lines. Front. Cell Infect. Microbiol. 7, 268 (2017).
Tandra, S. et al. Presence and significance of microvesicular steatosis in nonalcoholic fatty liver disease. J. Hepatol. 55, 654–659 (2011).
Kowalczyk, P. et al. Mitochondrial oxidative stress—a causative factor and therapeutic target in many diseases. Int J. Mol. Sci. 22, 13384 (2021).
Elwell, C. A. & Dreyfus, L. A. DNase I homologous residues in CdtB are critical for cytolethal distending toxin-mediated cell cycle arrest. Mol. Microbiol. 37, 952–963 (2000).
Ni, R. et al. Mitochondrial Calpain-1 Disrupts ATP Synthase and Induces Superoxide Generation in Type 1 Diabetic Hearts: A Novel Mechanism Contributing to Diabetic Cardiomyopathy. Diabetes 65, 255–268 (2016).
Shimano, H. & Sato, R. SREBP-regulated lipid metabolism: convergent physiology - divergent pathophysiology. Nat. Rev. Endocrinol. 13, 710–730 (2017).
Lin, X. et al. ROS/mtROS promotes TNTs formation via the PI3K/AKT/mTOR pathway to protect against mitochondrial damages in glial cells induced by engineered nanomaterials. Part Fibre Toxicol. 21, 1 (2024).
Trifault, B. et al. Nucleolar detention of NONO shields DNA double-strand breaks from aberrant transcripts. Nucleic Acids Res. 52, gkae022 (2024).
EASL-EASD-EASO Clinical practice guidelines on the management of metabolic dysfunction-associated steatotic liver disease (MASLD). J. Hepatol. 81, 492–542 (2024).
Lazarus, J. V. et al. Reply: a multi-society Delphi consensus statement on new fatty liver disease nomenclature. Hepatology 79, E93–E94 (2024).
Rinella, M. E. et al. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. Hepatology 78, 1966–1986 (2023).
Qian, M. et al. Aryl hydrocarbon receptor deficiency in intestinal epithelial cells aggravates alcohol-related liver disease. Cell Mol. Gastroenterol. Hepatol. 13, 233–256 (2022).
Cao, S. et al. Helicobacter hepaticus infection-induced IL-33 promotes hepatic inflammation and fibrosis through ST2 signaling pathways in BALB/c mice. Biochem Biophys. Res. Commun. 525, 654–661 (2020).
Qian, M., Cao, S., Wang, T., Xu, X. & Zhang, Q. Apoptosis triggered by cytolethal distending toxin B subunit of Helicobacter hepaticus is aggravated by autophagy inhibition in mouse hepatocytes. Biochem. Biophys. Res. Commun. 598, 40–46 (2022).
Pons, B. J., Vignard, J. & Mirey, G. Cytolethal distending toxin subunit B: a review of structure-function relationship. Toxins (Basel) 11, 595 (2019).
Shenker, B. J., Walker, L. P., Zekavat, A., Korostoff, J. & Boesze-Battaglia, K. Aggregatibacter actinomycetemcomitans cytolethal distending toxin-induces cell cycle arrest in a Glycogen Synthase Kinase (GSK)−3-dependent manner in oral keratinocytes. Int J. Mol. Sci. 23, 11831 (2022).
Mahony, D., Parry, D. A. & Lees, E. Active cdk6 complexes are predominantly nuclear and represent only a minority of the cdk6 in T cells. Oncogene 16, 603–611 (1998).
Didelot, C. et al. Interaction of heat-shock protein 90 beta isoform (HSP90 beta) with cellular inhibitor of apoptosis 1 (c-IAP1) is required for cell differentiation. Cell Death Differ. 15, 859–866 (2008).
Islam, S., Jayaram, D. T., Biswas, P. & Stuehr, D. J. Functional maturation of cytochromes P450 3A4 and 2D6 relies on GAPDH- and Hsp90-dependent heme allocation. J. Biol. Chem. 300, 105633 (2024).
Liu, M. et al. Pinacidil ameliorates cardiac microvascular ischemia-reperfusion injury by inhibiting chaperone-mediated autophagy of calreticulin. Basic Res. Cardiol. 119, 113–131 (2024).
Sharma, P., Ma, J. X. & Karamichos, D. Effects of hypoxia in the diabetic corneal stroma microenvironment. Exp. Eye Res. 240, 109790 (2024).
Dalidowska, I., Orlowska, A., Smreczak, M. & Bieganowski, P. Hsp90 activity is necessary for the maturation of rabies virus polymerase. Int J. Mol. Sci. 23, 6946 (2022).
Hou, G. et al. An aquatic virus exploits the IL6-STAT3-HSP90 signaling axis to promote viral entry. PLoS Pathog. 19, e1011320 (2023).
Song Ba, Y., Wang Ma, F., Wei Ma, Y., Chen Ba, D. & Deng Ba, G. ATP5A1 participates in transcriptional and posttranscriptional regulation of cancer-associated genes by modulating their expression and alternative splicing profiles in Hela cells. Technol. Cancer Res. Treat. 20, 15330338211039126 (2021).
Ramanathan, R. & Ibdah, J. A. Mitochondrial dysfunction and acute fatty liver of pregnancy. Int J. Mol. Sci. 23, 3595 (2022).
Zou, R. et al. DNA-PKcs promotes sepsis-induced multiple organ failure by triggering mitochondrial dysfunction. J. Adv. Res. 41, 39–48 (2022).
Uchida, T., Kao, H., Quispe-Sjogren, M. & Peters, R. L. Alcoholic foamy degeneration–a pattern of acute alcoholic injury of the liver. Gastroenterology 84, 683–692 (1983).
Yu, Y. et al. STING-mediated inflammation in Kupffer cells contributes to progression of nonalcoholic steatohepatitis. J. Clin. Invest 129, 546–555 (2019).
Natarajan, S. K., Eapen, C. E., Pullimood, A. B. & Balasubramanian, K. A. Oxidative stress in experimental liver microvesicular steatosis: role of mitochondria and peroxisomes. J. Gastroenterol. Hepatol. 21, 1240–1249 (2006).
Germano, C. W. et al. Microvesicular steatosis in individuals with obesity: a histological marker of non-alcoholic fatty liver disease severity. Obes. Surg. 33, 813–820 (2023).
Lai, Y. R. et al. From DNA damage to cancer progression: potential effects of cytolethal distending toxin. Front. Immunol. 12, 760451 (2021).
Leite, G. et al. Cytolethal distending toxin B inoculation leads to distinct gut microtypes and IBS-D-like microRNA-mediated gene expression changes in a rodent model. Gut Microbes 16, 2293170 (2024).
He, Z. et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut 68, 289–300 (2019).
Shen, S. et al. SIRT1/SREBPs-mediated regulation of lipid metabolism. Pharm. Res. 199, 107037 (2024).
de la Cruz Lopez, K. G., Toledo Guzman, M. E., Sanchez, E. O. & Garcia Carranca, A. mTORC1 as a regulator of mitochondrial functions and a therapeutic target in cancer. Front. Oncol. 9, 1373 (2019).
Anjum, S. et al. TORC1 mediated regulation of mitochondrial integrity and calcium ion homeostasis by Wat1/mLst8 in S. pombe. Int J. Biol. Macromol. 253, 126907 (2023).
Desai, B. N., Myers, B. R. & Schreiber, S. L. FKBP12-rapamycin-associated protein associates with mitochondria and senses osmotic stress via mitochondrial dysfunction. Proc. Natl Acad. Sci. USA 99, 4319–4324 (2002).
Schieke, S. M. & Finkel, T. Mitochondrial signaling, TOR, and life span. Biol. Chem. 387, 1357–1361 (2006).
Guerra, L., Cortes-Bratti, X., Guidi, R. & Frisan, T. The biology of the cytolethal distending toxins. Toxins (Basel) 3, 172–190 (2011).
Feng, P. et al. NONO and tumorigenesis: more than splicing. J. Cell Mol. Med. 24, 4368–4376 (2020).
Ding, J. et al. Integrative multiomic analysis identifies distinct molecular subtypes of NAFLD in a Chinese population. Sci. Transl. Med. 16, eadh9940 (2024).
Waddington, S. N. et al. Adenovirus serotype 5 hexon mediates liver gene transfer. Cell 132, 397–409 (2008).
Shayakhmetov, D. M., Li, Z. Y., Ni, S. & Lieber, A. Analysis of adenovirus sequestration in the liver, transduction of hepatic cells, and innate toxicity after injection of fiber-modified vectors. J. Virol. 78, 5368–5381 (2004).
Huch, M. et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494, 247–250 (2013).
Miyoshi, H. & Stappenbeck, T. S. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc. 8, 2471–2482 (2013).
Chan, V. et al. Visualization of Helicobacter species within the murine cecal mucosa using specific fluorescence in situ hybridization. Helicobacter 10, 114–124 (2005).
Ge, Z. et al. Cytolethal distending toxin is essential for Helicobacter hepaticus colonization in outbred Swiss Webster mice. Infect. Immun. 73, 3559–3567 (2005).
Zhang, Q., Itagaki, K. & Hauser, C. J. Mitochondrial DNA is released by shock and activates neutrophils via p38 map kinase. Shock 34, 55–59 (2010).
Zhou, R. H. et al. Vascular endothelial growth factor activation of sterol regulatory element binding protein: a potential role in angiogenesis. Circulation Res. 95, 471–478 (2004).
Jansen, M. P. B. et al. Urinary mitochondrial DNA associates with delayed graft function following renal transplantation. Nephrol. Dialysis Transplant.35, 1320–1327 (2020).
Acknowledgements
The authors thank Professor Xiaoyan Du (Capital Medical University, Beijing, China) for providing the gastric tissue of Mongolian gerbils infected with H. pylori. This work was supported by the National Natural Science Foundation of China (32273004 to Q.Z.), Key R&D plan of Jiangsu Province (BE2020674 to Q.Z.), Qing Lan project of Jiangsu province (to Q.Z.), Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD to Q.Z.), Postgraduate Research and Practice Innovation Program of Jiangsu Province (KYCX21_3209 to S.J.).
Author information
Authors and Affiliations
Contributions
Q.Z. proposed the research; Q.Z., M.Z., and L.Z. designed the research; S.J. performed most experiments; R.B., L.Y., L.L., and T.Z. performed additional experiments; Q.Z, S.J., and L.Z. analyzed data; S.Z., L.Z., J.Y., T.W., and J.Z. discussed part of the experimental design; S.J, L.Z, T.W., L.Z., and Q.Z. contributed to writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Kaori Hayashi, Abraham S Meijnikman, Erina Nishimura, and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jin, S., Zhu, L., Bao, R. et al. Helicobacter hepaticus promotes hepatic steatosis through CdtB-induced mitochondrial stress and lipid metabolism reprogramming. Nat Commun 16, 7954 (2025). https://doi.org/10.1038/s41467-025-63351-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-63351-z