Abstract
Ribosomally synthesized and post-translationally modified peptides (RiPPs) are a diverse group of natural products. The lasso peptide class of RiPPs adopt a unique [1]rotaxane conformation formed by a lasso cyclase, conferring diverse bioactivities and remarkable stability. The prediction of lasso peptide properties, such as substrate compatibility with a particular lasso cyclase or desired biological activity, remains challenging due to limited experimental data and the complexity of substrate fitness landscapes. Here, we develop LassoESM, a tailored language model that improves lasso peptide property prediction. LassoESM embeddings enable accurate prediction of substrate compatibility, facilitate identification of novel non-cognate cyclase–substrate pairs, and enhance prediction of RNA polymerase inhibitory activity, a biological activity of several known lasso peptides. We anticipate that LassoESM and future iterations will be instrumental in the rational design and discovery of lasso peptides with tailored functions.
Similar content being viewed by others
Introduction
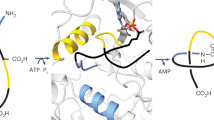
Ribosomally synthesized and post-translationally modified peptides (RiPPs) are structurally diverse natural products. RiPP precursor peptides are genetically encoded and many biosynthetic enzymes are tolerant to alternative substrates, making RiPPs attractive engineering targets for a variety of medical and other applications1. Lasso peptides are a particularly promising scaffold for RiPP engineering due to their modifiable and stabilized lariat knot-like structures1,2. During lasso peptide biosynthesis, the precursor peptide is bound by the RiPP recognition element, and a leader peptidase releases the core region of the substrate that undergoes conversion into the mature product. An ATP-dependent lasso cyclase then folds the core peptide into its characteristic threaded shape, which is kinetically trapped by macrolactam formation (Fig. 1A)3,4. The N-terminal amine (core position 1) and the side chain of Asp or Glu, typically located at core position 7, 8, or 9, form the macrolactam with large residues in the tail acting as steric locks to maintain threadedness. Lasso peptides are predominantly derived from bacteria and in some rare cases from archaea. In eukaryotic organisms, Niemyska et al. identified lasso-like motifs in proteins, stabilized by disulfide bridges or amide linkages5. These proteins, however, should not be confused with lasso peptides, which all share the same fold, right-handed chirality, and have molecular weights typically below 2 kDa.
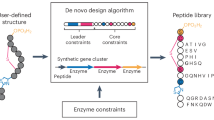
A Lasso peptide biosynthesis requires a leader peptidase, RiPP recognition element (RRE), and lasso cyclase to tie a linear core peptide into the lariat-like knot. B LassoESM was built upon the ESM-2 architecture and further pre-trained on lasso peptides using a domain-adaptive approach with masked language modeling. The resulting LassoESM embeddings were utilized for three downstream tasks: predicting lasso cyclase substrate tolerance, identifying substrate compatibility between non-cognate pairs of lasso cyclases and substrate peptides, and predicting RNAP inhibitory activity (numbers indicate enrichment values, estimating RNAP inhibitory activity).
Several lasso cyclases have shown high substrate tolerances, such as the fusilassin cyclase (FusC: WP_104612995.1), which was statistically shown to cyclize millions of variants with multi-site changes to the ring6, and the microcin J25 cyclase (McjC: WP_256498469.1), which can accommodate many diverse single-site and multi-site changes7,8. Additionally, lasso peptides exhibit antibacterial, antiviral, and anticancer activities, making them a compelling scaffold for engineering macrocyclic peptides for drug discovery purposes4,9. Despite their potential, chemical synthesis of lasso peptides remains challenging, with only one report that has yet to be replicated10. Therefore, designing novel lasso peptides with desired bioactivities will require sequences that are compatible with available lasso cyclases. However, profiling the substrate tolerance of lasso peptide biosynthetic enzymes remains challenging, hampering the generation of customized lasso peptides for biomedical and other industrial applications.
Different in silico approaches can be utilized to discover and design novel lasso peptides. For instance, RODEO predicts the native precursor peptides that are proximally encoded to the lasso peptide biosynthetic enzymes using linear combination scoring and a supervised machine learning approach11. RODEO matches substrates with enzymes but does not attempt to predict whether a biosynthetic enzyme would tolerate an alternative peptide substrate. Additionally, RODEO only predicts precursor peptides and their associated biosynthetic enzymes within a small local genomic context, which precludes the possibility of identifying potential hybrid peptide-enzyme pairs. Molecular dynamics (MD) simulation is a powerful computational approach for studying how lasso peptides or other RiPPs fold and how they interact with their biosynthetic enzymes during folding12,13,14. However, MD simulations are computationally expensive, limiting their applicability for the broadscale customization of lasso peptides with desired properties. In addition, a recent study presented a biophysical modeling framework to guide the design and biosynthesis of de novo RiPPs by systematically combining enzymes from diverse RiPP gene clusters15.
Deep learning is a type of artificial intelligence that can learn complex patterns and features from large datasets. It has emerged as a useful tool for RiPP engineering and design. For example, deep learning models have been trained on large-scale substrate specificity profiling data of RiPP-modifying enzymes from mRNA display technique. These trained models have facilitated the understanding of RiPP enzyme substrate tolerance and enabled the design of a diverse, highly modified variant library for selection against a target16,17,18. In addition to substrate tolerance prediction, deep learning has been applied to predict the bioactivity of RiPP variant libraries. For instance, DeepLasso predicts whether a variant of the lasso peptide ubonodin will retain RNA polymerase (RNAP) inhibitory activity19. The model demonstrates the utility of artificial intelligence beyond substrate compatibility prediction for lasso peptide biosynthesis. However, these methods required uncharacteristically large datasets for model training. In most cases, the experimental data that defines lasso peptide substrate tolerance and bioactivity is severely limited, necessitating methods that perform well in data scare scenarios.
Protein language models (PLMs) are a powerful framework for improving the performance of downstream tasks in situations with limited labeled data20,21,22,23,24,25. Most PLMs are trained using masked language modeling, in which the model is trained to predict the identity of masked amino acids given the local amino acid sequence. PLMs have been used to improve predictions for protein structure25, protein properties26,27,28, and mutational effects20,27,29,30. Effective PLMs require massive training datasets and large model size, which enable learning of general patterns present in natural proteins. However, PLMs are predominantly trained on protein sequences rather than peptides, and their ability to represent peptides is understudied. Since peptides differ from proteins in physical characteristics, such as shorter length, simpler three-dimensional structures, and conformational dynamics, we hypothesized that PLMs may have limited abilities to accurately represent peptide sequences, particularly lasso peptides, which exhibit a unique knot-like shape. As a result, the effectiveness of PLMs in performing predictive tasks related to lasso peptides appears to be a logical mismatch. Previous studies have demonstrated that domain-adaptive pretraining can further enhance the performance of large language models within a specific domain by learning the knowledge of the given domain31,32,33. For example, pretraining on DNA-binding protein sequences has improved model performance in four DNA-binding, protein-related downstream tasks32. Similarly, pretraining on a dataset focused on the substrate preference of one enzyme can enable accurate prediction of which substrates will be compatible with another enzyme in the same biosynthetic pathway33. Due to the diversity of RiPP classes and their unique sequence attributes, it is possible that class-specific RiPP language models could further improve downstream predictive tasks.
In this work, we used a domain adaptation method to further pre-train ESM-2 (Evolutionary Scale Modeling) on lasso peptide datasets, resulting in a lasso peptide-tailored language model named LassoESM. Compared to the more generic ESM-2, LassoESM showed superior performance in predicting the substrate tolerance of a known lasso cyclase, even with small quantities of training data. A human-in-the-loop approach, where a human expert iteratively validates new, unseen sequences and incorporates them into the training data, was used to further verify and improve the classification model34. To predict compatible lasso cyclase-substrate peptide pairs, LassoESM embeddings were combined with a cross-attention layer to effectively model the interactions between the lasso cyclase and peptide, leading to better model performance than when only the generic PLM was used. Finally, LassoESM embeddings were shown to predict lasso peptide RNAP inhibition activity more accurately than a generic PLM and DeepLasso19. With expanded lasso peptide property datasets, LassoESM could facilitate their biomedical application by guiding lasso cyclase selection for rational engineering campaigns.
Results
LassoESM improves lasso peptide substrate tolerance prediction
Representative PLMs like ESM25 and ProtBERT21 have been trained on general protein sequences to learn sequence patterns across natural proteins in a self-supervised manner. Compared to general proteins, lasso peptides are much shorter (15–20 amino acids) and have a unique lariat knot-like structure1,4. The development of a lasso peptide-specific language model has the potential to enhance the lasso peptide representation capabilities and improve the performance of downstream predictive tasks. To address this, we adapted the ESM-2 architecture25, which contains 650 million parameters, as a starting point and further trained it on a dataset of 4485 unique, high-scoring lasso core peptides identified by RODEO. Using the masked language modeling approach, we developed LassoESM, a specialized language model tailored to lasso peptides. Pretrained PLMs can enhance protein property prediction from sequences through transfer learning, where their learned weights and representations are repurposed for downstream tasks35. In our study, we utilized LassoESM embeddings in various downstream lasso peptide-related predictive tasks, including predicting lasso cyclase substrate tolerance, identifying non-natural cyclase-substrate peptide pairs, and predicting RNAP inhibitory activity of lasso peptides (Fig. 1B).
Previous studies have shown that FusC exhibits a range of substrate tolerability, from promiscuous to highly intolerant, depending on the specific core position being varied6,14,36. However, the rules that govern substrate compatibility are poorly understood, making substrate tolerance prediction challenging. A total of 1121 fusilassin (FusA, also known as fuscanodin37) variants were confirmed as substrate or non-substrate sequences via cell-free biosynthesis (CFB) (Fig. 2A). This dataset was selected to evaluate if LassoESM could improve the substrate tolerance prediction for FusC. None of these fusilassin variant sequences were used for training LassoESM. Embeddings for all 1121 sequences generated by LassoESM were fed into various downstream classification models, including Random Forest (RF)38, Adaptive Boosting (AdaBoost)39, Support Vector Machine (SVM)40, and Multi-Layer Perceptron (MLP)41. Hyperparameter optimization was conducted using 10-fold cross-validation (Table S1), and the performance of the downstream classifiers was evaluated by AUROC score and balanced accuracy. Previously, we developed PeptideESM, a peptide-specific language model trained on 1.5 million unique peptide sequences33. We evaluated PeptideESM embeddings for lasso peptide substrate tolerance prediction. Among these models, SVM demonstrated the highest performance (Figs. 2B and S2). Across the various downstream classification models, PeptideESM and LassoESM provided superior embeddings for FusC substrate specificity prediction compared to that from the general PLM (VanillaESM) and the one-hot baseline representation method (Fig. 2B). To visualize the separation of substrate and non-substrate sequences captured by these embeddings, we applied t-SNE42 to project them into a two-dimensional space (Fig. S3). We also compared LassoESM embeddings with four additional representation methods, PeptideCLM (a SMILES-based Chemical Language Model)43, Extended-Connectivity Fingerprints (ECFP)44 and two biophysical descriptor sets45,46, to thoroughly assess their effectiveness in substrate tolerance prediction (Fig. S2).
A Core peptide sequence of fusilassin overlaid with substrate tolerance information. The n value is the number of sequences tested at the indicated position(s) by CFB, with the percentage of n sequences tolerated by the biosynthetic enzymes given. Bold indicates the macrolactam-forming residues. B AUROC score and balanced accuracy of the SVM model for the fusilassin variant dataset trained on embeddings from one-hot encoding (blue), VanillaESM (pink), PeptideESM (yellow), and LassoESM (green). Data are presented as mean \(\pm \,\)SD. n = 5 independent repeats of 10-fold cross-validation. VanillaESM vs. PeptideESM: AUROC, p = 0.0377; Balanced Accuracy, p = 0.0012. VanillaESM vs. LassoESM: AUROC, p = 0.0006; Balanced accuracy, p = 0.0119. p-value was calculated using a two-sided t-test. *p < 0.05, ** p < 0.01, *** p < 0.001. C Core peptide sequence of MccJ25 overlaid with substrate tolerance information. Each position was site-saturated with all other proteinogenic amino acids (n = 20) prior to heterologous expression7. The percentage of sequences that were cyclized and exported is given. Bold indicates the macrolactam-forming residues. D Same as (B), except for MccJ25. Data are presented as mean \(\pm \,\)SD. n = 5 independent repeats of 10-fold cross-validation. VanillaESM vs. PeptideESM: AUROC, p = 0.8759; Balanced accuracy, p = 0.7751. VanillaESM vs. LassoESM: AUROC, p = 0.0006; Balanced Accuracy, p = 0.0013. p-value was calculated using a two-sided t-test. E AUROC score and balanced accuracy of the SVM model trained on a fraction of the fusilassin variant training data using LassoESM embeddings and evaluated on the remaining data. Data are presented as mean ± SD. n = 10 random seeds, with a fraction of the dataset randomly selected for training in each seed. F Model accuracy at different predicted probabilities of the SVM model trained on the fusilassin variant dataset using LassoESM embeddings. Data are presented as mean ± SD. n = 10 random seeds. For each random seed, 80% of the fusilassin variant dataset was used for training and the remaining 20% for testing. The source data are available in the Source data file.
Microcin J25 (MccJ25) is a distinct lasso peptide whose biosynthetic enzymes are reported to be highly substrate-tolerant. A total of 552 MccJ25 variants have been experimentally evaluated as substrates or non-substrates in the literature (Fig. 2C)7,8,47,48,49,50,51. None of these Mccj25 variant sequences were used for training LassoESM. Analogous to fusilassin, LassoESM significantly outperformed VanillaESM and one-hot encoding to predict the substrate tolerance of McjC (Fig. 2D), and SVM displayed the highest performance (Figs. 2D and S4). These results show that LassoESM provides effective representations that enhance the accuracy of substrate tolerance predictions for two disparate lasso peptide cyclase, FusC and McjC, which have a sequence identity of only 22%.
An advantage of using language model embeddings for downstream classification tasks is their ability to enhance accuracy in data-scarce settings35. LassoESM, pretrained specifically on lasso peptide sequences, has learned sequence features unique to this peptide class, enabling it to generate informative and effective representations. This makes it particularly valuable for applications where labeled datasets are limited. In many biochemical tasks, including RiPP biosynthetic enzyme compatibility prediction, researchers often face a lack of large, well-annotated datasets due to the overwhelming size of the sequence space. Thus, we evaluated the accuracy of the best downstream classification model, SVM, trained on datasets of differing sizes using LassoESM embeddings. For each training dataset size, we conducted 10 random selections of samples at different random seeds and evaluated the AUROC score and balanced accuracy on the remaining data samples. Even with only 20% of the training dataset (224 samples), the average AUROC score was 0.78, and the average balanced accuracy was 0.71 (Fig. 2E). The results indicated that the classification model performs well even with a small dataset when using tailored language model embeddings. However, expanding the dataset with more diverse samples enhances model performance.
In classification tasks, the predicted probability is often interpreted as a measure of model uncertainty. However, the output from SVM is the distance from the decision hyperplane, not a probability. To address this, Platt scaling52 is commonly employed, where a logistic regression model is applied to the SVM decision values, effectively transforming distances into probability estimates ranging from 0 to 1. These probabilities represent the likelihood of a sequence being a substrate for the lasso cyclase. To assess whether these predicted probabilities effectively captured the uncertainty of the downstream classification model trained on the fusilassin dataset, we analyzed the distribution of the model accuracy across different probability ranges. Specifically, we plotted the accuracy distributions for the SVM models trained on LassoESM embeddings for fusilassin variant sequences (Fig. 2F). The results indicated that the model demonstrated high accuracy when the predicted probabilities were near 0 or 1. Conversely, predictions with probabilities between 0.4 and 0.6 exhibited a higher proportion of false classifications. This result suggested that the predicted probability was faithfully representing model uncertainty.
Human-in-the-loop feedback improves fusilassin substrate specificity prediction
Next, we assessed the accuracy of LassoEM in predicting the substrate tolerance of sequences not included in the training set. Human-in-the-Loop validation is a strategy where human experts actively participate in the decision-making process alongside artificial intelligence, providing oversight and iterative feedback to enhance model accuracy and reliability34. In our study, the workflow begins by classifying new sequences using the optimized SVM trained on LassoESM embeddings from an initial dataset of 1121 fusilassin variant sequences. The SVM model predicts the labels (substrate or non-substrate) of uncharacterized fusilassin variants. Selected sequences are then experimentally validated, and the newly labeled data are incorporated into the training set for model retraining. This iterative process continues until the model achieves the desired performance (Fig. 3A).
A Human-in-the-loop workflow for optimizing the fusilassin cyclase substrate tolerance model. Embeddings for training data sequences were extracted from LassoESM. These embeddings were used to train a classifier, which then predicted the labels of unseen library sequences. Experimental validation was through CFB and MALDI-TOF-MS. The labeled data are then passed back into the workflow to improve model accuracy. B Experimental method to validate unseen sequences. The selected fusilassin variant sequences were produced using CFB and analyzed using MALDI-TOF-MS. C Balanced accuracy of the chosen samples across three rounds of experimental testing. The source data for (C) are provided in Tables S3–S10 and Figs. S6–S12.
A library containing all 18 randomized sites on fusilassin has 2018 (2.6 × 1023) library members, which is too large to analyze reasonably. Therefore, two smaller libraries were selected for investigation. Library 1 varied positions 2, 3, 4, 13, and 14 (n = 3.2 million sequences), while library 2 varied positions 7, 8, 10, and 11 (n = 160,000 sequences). These positions were selected because they comprise a continuous surface on fusilassin, which could potentially engage with a target in future lasso peptide engineering campaigns (Fig. S5). No sequences from either library were present in our initial training dataset.
The SVM model trained on LassoESM embeddings was used to classify all library sequences as substrates or non-substrates. A random selection of 118 fusilassin variant sequences (round 1) was experimentally validated (57 from library 1 and 61 from library 2) in CFB containing purified fusilassin biosynthetic enzymes. Lasso peptide cyclization was analyzed using MALDI-TOF-MS (Fig. 3B). The balanced accuracy of round 1 was 66% (Figs. 3C, S6–S9, Tables S3–S6). This result underscored the extrapolation capability of the classification model with LassoESM embeddings, as the model performed well despite not having seen sequences in libraries 1 and 2.
To improve the model’s performance, we introduced a Human-in-the-Loop approach. The results from round 1 were incorporated into the initial training dataset. Embeddings for the new data were created with LassoESM, and the SVM classification model was retrained on all available data (Fig. 3A). To verify the accuracy of the retrained model, 24 sequences were randomly selected from each library (round 2, n = 48) and evaluated using CFB. As anticipated, inclusion of round 1 data enhanced the model’s balanced accuracy to 75% (Figs. 3C, S10–S11, Tables S7–S8). In contrast, restricting the training data to the original dataset gave an accuracy of 69%.
The experimentally labeled data from rounds 1 and 2 were then incorporated into the training dataset, and the retrained SVM model was used to predict a more complex library (library 3), which varies positions 10–15 to encompass the entire fusilassin loop (library size of 64 million). A total of 30 fusilassin variant sequences were randomly selected from library 3 and evaluated using CFB. For these sequences, the model achieved a balanced accuracy of 84% (Figs. 3C, S12, and Table S9). The high accuracy further indicated that with the LassoESM embeddings, the trained SVM model demonstrated robust predictive accuracy even with sequences poorly represented in the initial training dataset. Table S10 summarizes the total number of sequences tested in CFB and the model’s balanced accuracy for each round of testing.
To further improve interpretability, we provide representative examples of model predictions and their experimental validation. In total, 196 fusilassin variants were tested across all rounds. Consistent with prior experimental observations, Met is well favored in the ring region, while Glu is not6,14. In the loop region, Leu is favored at position 14, whereas Asp is disfavored. These preferences are reflected in the experimentally validated sequences. For example, WLMMEWGLELIFHLPRFI was correctly predicted and confirmed as a substrate, whereas WIENEWGLELIFVDPRFI was predicted and validated as a non-substrate (bolded residues are different from the native fusilassin sequence). These examples demonstrate that the model not only provides accurate predictions but also captures known sequence–function relationships relevant to substrate tolerance.
A general model for predicting lasso cyclase-lasso peptide pairs
LassoESM embeddings can also effectively predict more general lasso cyclase specificity tasks. For instance, diverse lasso peptides could be screened in silico or rationally engineered to bind a target of interest. However, extensive engineering may obscure which lasso cyclase can produce a desired lasso peptide. Thus, we designed a general model to predict any lasso cyclase-lasso peptide pair. A set of 6599 unique lasso cyclase-lasso peptide pairs predicted via RODEO were used as training data11,14. To create the negative samples, an equal number of non-natural cyclase-lasso peptide pairs were created by randomly mismatching cyclase and peptide pairs. Because most characterized lasso cyclases are specific for only one acceptor residue4, the selected lasso peptides in the negative training set were verified to have a different acceptor residue from the native substrate (see “Methods”). In total, this dataset includes 6333 unique cyclases, with most cyclases having only one true peptide substrate and one mismatched peptide substrate. For example, the FusC appears twice in the dataset (once with its true substrate and once with a mismatched peptide), representing only 0.015% of the total dataset.
The general model uses VanillaESM to extract embeddings for the lasso cyclase sequences and LassoESM to extract embeddings for the lasso peptide sequences. Rather than treating these two embeddings separately and concatenating them, a cross-attention layer was used to effectively capture interactions between lasso cyclases and lasso peptides (Fig. 4A). The cross-attention layer emphasizes the amino acids in the lasso cyclase that likely interact with the lasso peptide substrate, thereby reducing noise and enhancing the model’s focus (see Methods). The model performance was evaluated under four different conditions: random, unseen cyclase, unseen peptide, and unseen cyclase & unseen peptide (Fig. 4B). In the random condition, the dataset was randomly split into 70% for training, 15% for validation, and 15% for testing, resulting in an accuracy of 0.8870. The other three conditions simulate out-of-distribution scenarios, where the training and test sets have no overlapping cyclase or peptide sequences. Specifically, in the unseen cyclase scenario, all cyclases in the test set are absent from the training set, while in the unseen peptide scenario, none of the peptides in the test set appear in the training set. In the most stringent case, both cyclases and peptides in the test set are completely absent from the training data, ensuring a rigorous evaluation of the model’s generalization performance. Under this condition, the model achieved an accuracy of 0.8281, demonstrating reliable and strong predictive performance for out-of-distribution data.
A The model architecture utilizes VanillaESM for cyclase embeddings and LassoESM for lasso peptide embeddings, incorporating a cross-attention layer to capture interactions between them. The attention-reweighted embeddings of lasso cyclases and peptides are averaged and then concatenated before being supplied to the MLP model to predict cyclase compatibility. The trained model is applied to predict the compatibility of other lasso peptides with FusC. B The model accuracy was evaluated on four dataset splits (random, unseen cyclase, unseen peptide, and unseen cyclase & peptide). Data are presented as mean ± SD. n = 3 random seeds for data splitting. C The model accuracy was assessed across different cyclase sequence similarities. The test set was divided into subsets based on maximum sequence similarity to those in the training set. Each bar represents the accuracy for cyclases within specific similarity ranges. D Ablation study evaluating the role of the cross-attention layer and LassoESM embeddings by accuracy scores of the general model. Data are presented as mean ± SD. n = 3 random seeds for data splitting. The source data are available in the Source data file.
Naturally occurring lasso cyclases predicted by RODEO exhibit varying degrees of sequence similarity, with an average pairwise sequence identity of 42% across the dataset. To determine if model accuracy correlates with cyclase sequence similarity, we calculated the maximum pairwise sequence identity for each test cyclase relative to all cyclases in the training set. The results showed that model accuracy improves as the sequence identity of test cyclases increases compared to those in the training set (Fig. 4C). For test cyclases with >50% sequence identity to any training cyclase, the accuracy exceeded 0.85. When sequence identity surpassed 70%, the model’s performance remained relatively stable.
Ablation analysis was conducted to evaluate the role of the important components in the model architecture. As shown in Fig. 4D, removing the cross-attention layer decreased accuracy from 0.8870 to 0.8673, indicating the benefits of the cross-attention layer in enhancing model performance. In contrast to the model employing average pooling across all amino acids, the cross-attention layer enhances the model’s ability to focus on the amino acids that determine enzyme substrate specificity, thereby reducing the bias introduced by non-essential amino acids in the cyclase. Using VanillaESM to represent lasso peptides instead of LassoESM also resulted in a notable reduction in model accuracy. This demonstrates the effectiveness of LassoESM in predicting cyclase specificity for lasso peptides. We also observed that when the same language model was used to generate embeddings for both lasso cyclase and lasso peptides, the cross-attention layer failed to learn useful information, resulting in model performance that was no better than simple concatenation of the embeddings.
To evaluate LassoESM performance on experimentally verified cyclase-lasso peptide pairs, we used the trained model to predict the compatibility of FusC with other predicted naturally occurring lasso peptides. These sequences were tested using a chimeric engineering strategy, where the fusilassin leader peptide was fused to the selected core peptides. This enables the proper function of the RRE and leader peptidase, which are required for full cyclase activity. The set includes some chimeric peptides previously examined6, which were selected based on cyclase similarity to FusC. It also contains Glu8- and Glu9-mer core peptides selected to maximize sequence diversity while maintaining the loop hydrophobicity previously shown to be preferred by FusC14. Despite FusC’s low representation in the training set, the model successfully predicted 24 out of 35 tested peptide pairs (69% accuracy), including correctly identifying all three FusC-compatible peptides (Fig. S13 and Table S11). Notably, the model predicted the FusC-compatible peptides with probabilities exceeding 0.7, indicating high confidence. Due to competition between the cyclase and endogenous proteases in the CFB, some of the 11 incorrectly predicted peptides may be poor substrates that would be detected in a pure setting6. However, testing sequences by cell-extract based CFB is fast, significantly cheaper than pure cell-free systems, and more accurately reflects the heterologous expression conditions required for large scale lasso peptide production.
Prediction of RNA polymerase inhibition by ubonodin and klebsidin variants
In addition to applying LassoESM embeddings to cyclase specificity prediction, we evaluated performance in an RNAP inhibition prediction task. Previous high-throughput screening studies explored the RNAP inhibitory activity of ubonodin variants and developed a deep learning model, DeepLasso, to predict RNAP inhibitory activity19. VanillaESM, PeptideESM, and LassoESM were employed to extract embeddings for 8885 literature-reported ubonodin variant sequences, and an MLP regressor was trained to predict the enrichment values, which act as estimates for RNAP inhibitory activity. The dataset was randomly partitioned into training (70%), validation (15%), and test (15%) sets across 10 random seeds. The LassoESM embeddings model achieved the best performance (Fig. 5B–D), with Pearson and Spearman correlation coefficients of 0.83 and 0.78, respectively. LassoESM obtained a mean absolute error (MAE) of 1.31, lower than that reported for DeepLasso (MAE of 2.20). DeepLasso uses lasso peptide sequence and topology information as input to train a model consisting of three convolutional neural network layers, two bidirectional long short-term memory layers, and one attention layer19. These results show that LassoESM embeddings yield a scenario where a simple two-layer MLP architecture resulted in superior model performance. To further evaluate the model’s generalization and mitigate potential data leakage, we implemented a position-based data splitting strategy, ensuring that mutation positions in the training set did not appear in the test set. This approach imposes a more stringent evaluation by eliminating sequence context overlap between train and test sets. Notably, even under this more rigorous setup, the LassoESM model continued to outperform VanillaESM and PeptideESM (Fig. S14).
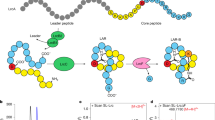
A Bubble diagram of ubonodin. B–D Model performance of the MLP regressor for prediction of RNAP inhibition activity for the ubonodin variant dataset using embeddings from VanillaESM (pink), PeptideESM (yellow), and LassoESM (green). Blue bars indicate the performance of DeepLasso19. Data are presented as mean ± SD. n = 10 random seeds for data splitting. VanillaESM vs. PeptideESM: Pearson correlation, p = 0.2425; Spearman correlation, p = 0.0368; Mean absolute error, p = 0.1616. VanillaESM vs. LassoESM: Pearson correlation, p = 6.1804 \(\times {10}^{-9}\); Spearman Correlation, p = 5.3240 \(\times {10}^{-6}\); Mean absolute error, p = 4.4822 \(\times {10}^{-8}\). E Bubble diagram of klebsidin. F–H Model performance of the AdaBoost regressor’s prediction of RNAP inhibition activity for the klebsidin variant dataset, using embeddings from VanillaESM (pink), PeptideESM (yellow), and LassoESM (green). Data are presented as mean \(\pm \,\)SD. n = 10 independent repeats of 10-fold cross-validation. VanillaESM vs. PeptideESM: Pearson correlation, p = 4.4438 \(\times {10}^{-7}\); Spearman correlation, p = 0.3622; Mean absolute error, p = 1.9884 \(\times {10}^{-9}\). VanillaESM vs. LassoESM: Pearson correlation, p = 1.1840 \(\times {10}^{-12}\); Spearman correlation, p = 4.3752 \(\times {10}^{-12}\); Mean absolute error, p = 2.0563 \(\times {10}^{-16}\). p-value was calculated using a two-sided t-test. *p < 0.05, ** p < 0.01, *** p < 0.001. N.S. non-significant. The source data are available in the Source data file.
Similarly, we utilized LassoESM embeddings for another lasso peptide, klebsidin, to predict the RNAP inhibitory activity of its variants. We collected 340 klebsidin single-site variant sequences with enrichment values from a previous report53. By searching for the best regression model, we trained an AdaBoost regressor on the klebsidin dataset53. Even with the limited dataset, the LassoESM embeddings lead to Pearson and Spearman correlation coefficients of ~0.80 and a MAE of ~0.74. These results show significant improvement over those from VanillaESM embeddings, namely a 9% increase in Pearson correlation, 7% increase in Spearman correlation, and 14% decrease in MAE (Fig. 5F–H).
Discussion
Lasso peptides are touted as promising scaffolds for drug development due to their remarkable stability and diverse biological activities. Many lasso peptide biosynthetic enzymes exhibit impressive substrate tolerance, allowing for the de novo design or rational engineering of novel lasso peptides with desired properties. However, predicting substrate tolerance and exploring sequence-activity relationships remains challenging due to the scarcity of experimentally labeled data. In this work, we utilized language models to improve the prediction of lasso peptide-related properties. To better capture lasso peptide features, we developed a lasso peptide-specific language model, LassoESM, which outperformed a generic PLM in a variety of downstream tasks, including predicting lasso cyclase substrate tolerance, identifying non-natural but compatible cyclase-substrate pairs, and predicting RNAP inhibition activity. Our results also show that embeddings from LassoESM enable accurate predictions even with small training sets, highlighting the potential of language models to address data scarcity and improve prediction accuracy. The pre-trained LassoESM offers optimal representations for lasso peptides and will be particularly useful for various lasso peptide-related tasks as high-throughput methods are developed for dataset generation.
Previous research has demonstrated that ESM-2 can recognize and store sequence motifs, which are specific patterns of amino acids that often appear together in proteins54. Since LassoESM was pre-trained on lasso peptide sequences, it is plausible that LassoESM learned to recognize the lasso fold pattern. Conversely, VanillaESM did not learn these specific sequence patterns because it was not supplied with the necessary sequences during training. Therefore, the underlying reason LassoESM outperforms VanillaESM in lasso peptide-related predictive tasks could be the tailored language model’s ability to learn the lasso fold.
While our computational workflow successfully characterized the substrate preferences of lasso biosynthetic enzymes, the molecular mechanisms that distinguish substrates from non-substrates remain unclear. MD simulations offer a promising avenue to uncover these mechanisms by pinpointing key amino acids that govern substrate selectivity from both structural and dynamic perspectives. However, the effectiveness of MD simulations is contingent upon having accurate structural information for lasso peptides and their biosynthetic enzymes. Recent advances in lasso peptide structure prediction have significantly enhanced our ability to model these systems. For instance, Juarez et al. developed LassoHTP—a software tool that constructs and models lasso peptide structures from input sequences and annotations of the ring, loop, and tail regions55. Building on this work, the same group introduced LassoPred, a web-based tool featuring an annotator-constructor architecture. In this tool, the annotator predicts up to three sets of sequence annotations, and the constructor converts each predicted set into a 3D lariat-like structure56. In addition to achieving accurate structural models, defining optimal reaction coordinates is essential for applying enhanced sampling methods to effectively capture the lasso peptide folding process57. Collectively, these new insights and technological advancements pave the way for future research that leverages MD simulations to deepen our understanding of substrate selectivity in lasso biosynthetic enzymes.
In summary, we developed a lasso peptide-tailored language model that provides effective representations for lasso peptides and enhances lasso peptide-related predictive tasks. Our efficient pipeline for characterizing the substrate preferences of lasso peptide biosynthetic enzymes and identifying lasso cyclase-lasso peptide pairs is a powerful tool for chemists aiming to rationally design functional and biocompatible lasso peptides for myriad biomedical and industrial applications. In the future, our pipeline could be extended to predict compatible sequences for additional enzymes in biosynthetic gene clusters (BGCs), including those responsible for secondary modifications such as glycosylation, aspartimide formation, and biaryl crosslinking in lasso peptide core sequences.
Methods
Dataset construction for LassoESM
A bioinformatics tool, RODEO, was developed to automate the identification of RiPP BGCs, including lasso peptide BGCs, from genomes available in GenBank11. RODEO utilizes profile Hidden Markov Models and a SVM classifier to locate RiPP BGCs and predict the most likely precursor peptide, respectively. A previous study identified 7701 non-redundant, high-scoring lasso precursor peptides using RODEO6. After removing identical entries (if two distinct organisms encode the same core sequence, only one was retained), 4485 unique core sequences were retained for pre-training LassoESM.
Model architecture and Pre-training
ESM-2 is a PLM trained on ~65 million non-redundant protein sequences using a masked language modeling approach25. Masked language modeling has been shown as a powerful pretraining technique for language models. In our work, we adapted the ESM-2 (with 33 layers and 650 million parameters) architecture as the starting point for pretraining on lasso peptide datasets. During pre-training, 15% of the amino acids in the sequences were randomly masked. The model was trained to predict the identity of masked amino acids based on the surrounding sequence context, as shown in Eq. (1):
where M represents a set of indices of the randomly masked amino acids. The model was optimized to minimize the negative likelihood of true amino acid \({x}_{i}\) given the surrounding context \({x}_{{context}}\) in the lasso peptide sequences. By updating the full 650 million parameters of the ESM-2 model during pretraining, we enabled the model to learn specialized sequence patterns unique to lasso peptides, named LassoESM.
LassoESM was trained for 20 epochs with a learning rate of 5 × 10−5 and a batch size of 8 on a single NVIDIA GeForce RTX 3090 GPU with 12 GB memory. To reduce the memory required, GaLore (Gradient Low-Rank Projection) was integrated into the training process58. GaLore is a training strategy that allows full-parameter learning but can significantly reduce the memory requirements traditionally associated with optimizer states and gradients during the training process. During training, the loss to be minimized is the mean cross-entropy loss between the predicted and the ground truth masked amino acid. The epoch with the least validation loss was saved as the final model. LassoESM is publicly available on Hugging Face (https://huggingface.co/ShuklaGroupIllinois/LassoESM).
Fusilassin labeled dataset construction
The fusilassin substrate tolerance dataset contained substrate and non-substrate sequences confirmed via CFB and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) from the following previously published libraries: 5 × NNK ring library6, 5 × NNK loop library6,14, and three 2 × NNK loop panels (Note: NNK encodes for all 20 common proteinogenic amino acids and a stop codon)14. The dataset also contained a variety of published and unpublished variants of fusilassin. All sequences were verified as substrates or non-substrates using CFB with purified enzymes: 10 µM each of maltose-binding protein (MBP)-FusB, MBP-FusE, and MBP-FusC. The reactions were analyzed for lasso cyclization with MALDI-TOF-MS.
Training of downstream models
Lasso peptide substrate tolerance prediction
Three language models were used in this study: VanillaESM (650 million parameters ESM-2)25, PeptideESM33, and LassoESM. The last layer of these language models was utilized to generate information-rich representations, also known as embeddings. For each amino acid in each labeled lasso peptide sequence, the last layer of the language model generated a 1280-dimensional vector. The final embeddings for each peptide sequence were obtained by calculating the average of all the amino acid vectors, which were then fed into downstream models.
A total of 1121 fusilassin variant sequences were collected and experimentally labeled as substrates (656 sequences) or non-substrates (465 sequences). Embeddings were generated using one-hot encoding, VanillaESM, PeptideESM, and LassoESM to represent these sequences. The embeddings were then used to train various downstream classification models, including RF, AdaBoost, SVM, and MLP. Stratified 10-fold cross-validation was employed to optimize the hyperparameters of supervised classification models. The optimized hyperparameters for all downstream models are in Table S1.
To ensure a fair evaluation, we employed stratified 10-fold cross-validation on the fusilassin dataset. In this approach, the dataset was divided into 10 folds, with each fold containing 112 variant sequences. Each fold maintained the same ratio of substrate to non-substrate sequences as the entire dataset. The model was trained 10 times, with each iteration using 9 folds for training and 1 fold for testing. Model performance was evaluated using AUROC and balanced accuracy. The average results across the 10 iterations were reported as the model’s performance. We repeated the stratified 10-fold cross-validation process 5 times, with a different split of the dataset in each repetition. This approach minimizes any potential bias introduced by specific splits. All downstream classification models were implemented using Scikit-learn59.
We incorporated four additional peptide representation approaches: PeptideCLM (a SMILES-based Chemical Language Model)43, ECFP44 and two sets of biophysical descriptors45,46. We applied pretrained PeptideCLM (full model with 23 million parameters) to encode each lasso peptide sequence into a 768-dimensional vector43. For ECFP representations, we used 2048-bit ECFP6 fingerprints to encode the lasso peptide sequences44. In addition, we evaluated two previously developed biophysical descriptor sets. In the first set, each amino acid in the lasso peptide sequence was represented by five multidimensional patterns that summarize a large portion of the known variability among amino acids45. In the second set, each amino acid was represented by 19 interpretable descriptors derived from AAindex via PCA and varimax rotation46.
The same approach was applied to MccJ25 variant sequences to predict the substrate tolerance for the microcin J25 cyclase. In this case, 552 MccJ25 variants were collected from the literature, with 413 sequences labeled as substrates and 139 as non-substrates7,8,47,48,49,50,51. The optimized hyperparameters for all downstream models are in Table S2.
Prediction of lasso cyclase-substrate peptide pairs
A total of 6599 unique lasso cyclase-substrate peptide pairs were identified using RODEO11. The dataset includes some class I, class III, and class IV lasso peptides; however, the vast majority (>90%) of the pairs are class II lasso peptides. To generate synthetic, non-natural, cyclase-peptide pairs as negative samples, we first generated all possible non-natural pairs for each cyclase. From these, we retained only those pairs in which the acceptor residue of the peptide differed from the original acceptor residue, as the acceptor residue is the most restrictive and is exclusively either Asp or Glu (only two examples are known where a lasso cyclase tolerates Asp and Glu60,61). Finally, one lasso peptide was randomly selected from this pool for each cyclase, generating 6599 synthetic non-natural cyclase-peptide pairs as negative samples.
LassoESM was used to generate embeddings for lasso peptides, while VanillaESM was employed to generate embeddings for lasso cyclases. For each amino acid in the lasso peptide sequence, the last layer of LassoESM produced a 1280-dimensional vector, resulting in a matrix of dimensions \({d}_{1280}*m\), where \(m\) is the length of the lasso peptide. For each amino acid in the lasso cyclase sequence, the last layer of VanillaESM generated a 1280-dimensional vector, resulting in a matrix with \({d}_{1280}*n\), where \(n\) is the length of the lasso cyclase. Padding was added at the end of the substrate peptide and lasso cyclase sequences to ensure that all substrate peptide embeddings had the same dimensions and that all lasso cyclase embeddings had the same dimensions.
These two matrices were then supplied into a cross-attention layer, where the lasso peptide embedding matrix served as the query (Q) and the lasso cyclase embedding matrix as the key (K) and value (V). An attention mask was applied to every padding position, as well as the BOS (Beginning of Sentence) and EOS (End of Sentence) tokens, to ensure the model ignored these elements. The attention mechanism was computed as shown in Eq. (2):
where \({d}_{k}\) is the dimensionality of K. The dot product \(Q{K}^{T}\) generates an attention matrix, where each element represents the relevance of a specific cyclase residue to a given peptide residue. Residues in the cyclase that are more relevant for peptide interaction receive higher attention scores. The attention weight matrix was used to reweight the cyclase embedding matrix. The reweighted cyclase matrix was then concatenated with the lasso peptide matrix and fed into an MLP model. The MLP model consisted of two hidden layers with 512 and 64 neurons. The full model, including the cross-attention layer, was trained using the Adam optimizer, with a batch size of 32 and a learning rate of 0.001. The training was conducted for 25 epochs using PyTorch62.
RNA polymerase inhibition prediction
A total of 11,363 published ubonodin variant sequences with a measurable enrichment value were collected19. After removing sequences containing stop codons, 8885 ubonodin variants remained in the dataset. The last layers of VanillaESM, PeptideESM, and LassoESM were used to extract embeddings for each amino acid in the ubonodin sequences. The final embedding for each sequence was obtained by averaging the embeddings of all amino acids in the sequence. These averaged embeddings were then utilized to train an MLP regressor. The MLP model consisted of two hidden layers with 256 and 32 neurons. MLP was trained using the Adam optimizer, with a batch size of 16 and a learning rate of 0.001. The model was trained for 100 epochs using PyTorch62, and early stopping was applied to monitor the validation loss.
A similar approach was applied to another lasso peptide, klebsidin. A total of 340 klebsidin variant sequences were obtained from a previous publication53. The embeddings of these sequences were supplied to various downstream regression models, including RF, AdaBoost, SVM, and MLP. Among these, the AdaBoost model achieved the best performance on the klebsidin dataset, as determined by the average R² score from 10-fold cross-validation. The optimal hyperparameters for AdaBoost were n_estimators = 200 and learning_rate = 1.
Experimental materials and methods
Synthetic DNA primers and ultramers were ordered from Integrated DNA Technologies. New England Biolabs supplied the Q5 DNA polymerase and the Gibson assembly mix. Chemical reagents were from Sigma-Aldrich unless otherwise noted. Sequencing was performed at the Core DNA Sequencing Facility at the University of Illinois at Urbana-Champaign.
Molecular biology techniques
The fusilassin biosynthetic genes were previously cloned into a modified pET28 vector that provides an N-terminal MBP tag for solubility and stability14,36. This vector was used for the expression of the fusilassin leader peptidase (FusB, WP_011291590.1), RiPP Recognition Element (RRE, FusE, WP_011291591.1), and an E. coli codon-optimized cyclase (FusCop, WP_104612995.1). A previously reported pET28-mbp-fusA (precursor peptide) construct was used as a template to prepare all the fusilassin variants needed for this study36.
Protein purification
The MBP-tagged FusB, FusE, and FusCop proteins were expressed using E. coli BL21(DE3) cells. MBP-FusB and MBP-FusE were expressed in LB media containing 50 µg/mL kanamycin and induced at OD600 0.6 with a final concentration of 0.5 M IPTG for 3 h at 37 °C. MBP-FusCop was expressed in LB media containing 50 µg/mL kanamycin and 3% ethanol. It was induced at OD600 0.8 with a final concentration of 0.5 M IPTG for 3 h at 37 °C14,36. The proteins were purified via amylose resin column chromatography. 1 L of induced cells were harvested and then resuspended in lysis buffer [50 mM Tris-HCl pH 7.4, 500 mM NaCl, 2.5% glycerol (v/v), and 0.1% Triton X-100], 100 mg lysozyme, and 500 µL of protease inhibitor cocktail (6 mM PMSF, 0.1 mM leupeptin, 0.1 mM E64)]. Cells were lysed with sonication and the lysate was clarified via centrifugation. The clarified supernatant was applied to a pre-equilibrated column containing 5 mL of amylose resin at 4 °C. After washing with 10 column volumes of wash buffer [50 mM Tris-HCl pH 7.4, 500 mM NaCl, and 2.5% glycerol (v/v)], the proteins were eluted with 30 mL of elution buffer [50 mM Tris-HCl pH 7.4, 300 mM NaCl, 2.5% glycerol (v/v), and 10 mM maltose] at 4 °C. Protein concentration and buffer exchange was done using a 30,000 kDa Amicon Ultra centrifugal filter (EMD Millipore) and protein storage buffer [50 mM HEPES pH 7.5, 300 mM NaCl, 0.5 mM tris-(2-carboxyethyl)-phosphine (TCEP), 2.5% glycerol (v/v)]. Protein concentrations were determined with a Nanodrop OneC instrument measuring the absorbance at 280 nm and by Bradford assay. Extinction coefficients were calculated using the Expasy ProtParam tool63. Protein purity was determined by visual inspection of a Coomassie-stained gel after separation via SDS-PAGE (Fig. S1).
Overlap extension PCR to construct fusilassin library members tested in CFB
DNA templates encoding the FusA variants were prepared by overlap extension PCR64. In the first PCR round, two pieces of DNA, each containing a complementary region, were made using the wild-type pET28-mbp-fusA vector as a template and primers from Supplementary Data Table 1. The two DNA pieces were then purified via a Qiagen spin column PCR cleanup kit. In the second PCR round, the two DNA pieces were allowed to anneal together and primers CFB_F and CFB_R were used to amplify the completed linear DNA product (Supplementary Data Table 1). The PCR product was purified using the Qiagen spin column PCR cleanup kit. DNA concentrations were determined by absorbance at 260 nm using a Nanodrop OneC instrument.
Generation of linear DNA for chimeric lasso peptide sequences tested in CFB
Ultramer oligos encoding the fusilassin leader peptide and the desired core peptide were amplified by PCR and cloned into the pET28-mbp vector using Gibson assembly (Supplementary Data Table 1). After sequence confirmation by Sanger sequencing, the linear DNA encoding the MBP-tagged chimeric peptide was amplified via PCR using primers CFB_F and CFB_R. The DNA concentration was determined on a Nanodrop OneC instrument at an absorbance of 260 nm.
Cell-free biosynthesis reagent preparation
E. coli cellular lysates were prepared by growing E. coli BL21(DE3) in 1 L of 2X YTPG (10 g/L yeast extract, 16 g/L tryptone, 3 g/L KH2PO4, 7 g/L K2HPO4, 5 g/L NaCl, 18 g/L glucose) at 37 °C while shaking. The cells were induced at OD600 0.5 with 0.1 M IPTG (final). At OD600 ~ 3.0, the cells were harvested and washed twice with 400 mL ice-cold S30A buffer [10 mM Tris, pH = 8.2, 14 mM magnesium acetate, 60 mM potassium glutamate, 2 mM 1,4-dithiothreitol (DTT, freshly added)]. The pellet was then resuspended in 40 mL of ice-cold S30A and transferred to a pre-weighed 50 mL tube. The cells were spun down, the supernatant removed, and the pellet flash frozen in liquid nitrogen. The next day, the cells were thawed on ice and resuspended in cold S30A buffer at 1 mg/g cell pellet. The French Press (single pass, 10,000 psi) was used to lyse the cells and then the membrane debris was spun down at 12,000 × g for 45 min at 4 °C. The supernatant was removed into a fresh tube, aliquoted into 100 µL aliquots, and stored at −80 °C. The total protein concentration was determined via Bradford assay.
Cell-free biosynthesis energy mix was prepared similarly to Sun et al65. Briefly, 240 mM stock concentrations were made for all amino acids except for Leucine, which 200 mM. 2.5 mL of water was combined with 125 µL of each amino acid stock to make a 4X amino acid solution. 700 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) at pH 8.0, 21 mM ATP, 21 mM GTP, 12.6 mM CTP, 12.6 mM UTP, 2.8 mg/mL tRNA (Sigma-Aldrich), 3.64 mM coenzyme A (CoA), 4.62 mM nicotinamide adenine dinucleotide (NAD), 10.5 mM cAMP, 0.95 mM folinic acid, 14 mM spermidine, and 420 mM 3-phosphoglyceric acid were combined to make a 14X energy solution stock. Stock solutions of 1 M maltodextrin, 4 M potassium glutamate, and 500 mM magnesium glutamate were also prepared. 10 mL of the energy buffer contained 5.6 mL of the 4× amino acid solution, 1.6 mL of the 14X energy solution, 780 µL of 1 M maltodextrin, 780 µL of the 4 M potassium glutamate, 400 µL of the 500 mM magnesium glutamate and 840 µL of water. The energy buffer was vortexed well and then aliquoted and stored at −80 °C.
GamS was prepared using E. coli BL21(DE3) pBAD-gamS similar to previously described protocols6,66. Briefly, a 1 L culture was grown in LB with 50 µg/mL ampicillin at 37 °C to OD600 0.45. The cells were induced with 0.25% w/v arabinose at 37 °C for 4 h. After harvesting, the pelleted cells were resuspended in lysis buffer [50 mM Tris pH 8.0, 500 mM NaCl, 5 mM imidazole, 0.1% Triton X-100, 3 mg/mL lysozyme, 2 μM leupeptin, 2 μM benzamidine HCl, 2 μM E64, and 30 mM phenylmethylsulfonyl fluoride]. After sonication, the lysate was clarified via centrifugation and the proteins were purified using 5 mL nickel-nitrilotriacetic acid (Ni-NTA) resin. The resin was washed with 10 column volumes of wash buffer (50 mM Tris pH 8.0, 500 mM NaCl, 25 mM imidazole) and eluted in 15 mL of elution buffer (50 mM Tris pH 8.0, 500 mM NaCl, 250 mM imidazole). The eluted proteins were concentrated using a 30 kDa Amicon Ultra centrifugal filter (EDM Millipore) and buffer exchanged into protein storage buffer [50 mM HEPES pH 7.5, 300 mM NaCl, 0.5 mM tris-(2-carboxyethyl)-phosphine (TCEP), 2.5% glycerol (v/v)].
Cell-free biosynthesis reactions
The cell-free biosynthesis reactions were carried out at 5 µL volume. Each reaction contained 50 ng of linear DNA, 1.75 µL of E. coli cell lysate, 2.25 µL of energy buffer, 0.053 µL of IPTG (50 mM stock), 0.16 µL DTT (100 mM stock), 0.25 µL GamS (125 µM stock), and a final concentration of 10 µM each MBP-FusB, MBP-FusE, and MBP-FusCop. The reactions were allowed to sit at room temperature for 16-18 h (overnight) and then were quenched with 60% (v/v) acetonitrile. The samples were spun down and then spotted onto a MALDI-TOF-MS plate using sinapic acid (80% acetonitrile, 0.1% formic acid) for MALDI-TOF-MS analysis. MALDI-TOF-MS data were acquired at the University of Illinois Urbana-Champaign in the School of Chemical Sciences on a Bruker UltraXtreme instrument in reflector positive mode. The MALDI-TOF-MS data was analyzed using Bruker FlexAnalysis version 3.4 software. Each reaction containing a unique template was repeated three times. Data in the Supplementary Information shows one representative spectrum from the replicates.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All datasets (including protein accessions) used in this study are available on GitHub at https://github.com/ShuklaGroup/LassoESM/tree/main/data and on Zenodo at https://doi.org/10.5281/zenodo.16429863. Source data are provided with this paper as a Source Data file. The sequences for the proteins used in the experimental portion can be found using the following NCBI accessions: FusB: WP_011291590.1, FusE: WP_011291591.1, FusC: WP_104612995.1. Source data are provided with this paper.
Code availability
The code used to train LassoESM and to perform downstream analyses in the study are available on GitHub at: https://github.com/ShuklaGroup/LassoESM and on Zenodo at https://doi.org/10.5281/zenodo.16429863. The pre-trained LassoESM model is available for download on Hugging Face at: https://huggingface.co/ShuklaGroupIllinois/LassoESM.
References
Montalbán-López, M. et al. New developments in RiPP discovery, enzymology and engineering. Nat. Prod. Rep. 38, 130–239 (2021).
Hegemann, J. D. Factors governing the thermal stability of lasso peptides. ChemBioChem 21, 7–18 (2020).
Hegemann, J. D., Zimmermann, M., Xie, X. & Marahiel, M. A. Lasso peptides: an intriguing class of bacterial natural products. Acc. Chem. Res. 48, 1909–1919 (2015).
Barrett, S. E. & Mitchell, D. A. Advances in lasso peptide discovery, biosynthesis, and function. Trends Genet. 40, 950–968 (2024).
Niemyska, W. et al. Complex lasso: new entangled motifs in proteins. Sci. Rep. 6, 36895 (2016).
Si, Y., Kretsch, A. M., Daigh, L. M., Burk, M. J. & Mitchell, D. A. Cell-free biosynthesis to evaluate lasso peptide formation and enzyme–substrate tolerance. J. Am. Chem. Soc. 143, 5917–5927 (2021).
Pavlova, O., Mukhopadhyay, J., Sineva, E., Ebright, R. H. & Severinov, K. Systematic structure-activity analysis of microcin J25*. J. Biol. Chem. 283, 25589–25595 (2008).
Pan, S. J. & Link, A. J. Sequence diversity in the lasso peptide framework: discovery of functional microcin J25 variants with multiple amino acid substitutions. J. Am. Chem. Soc. 133, 5016–5023 (2011).
Ongpipattanakul, C. et al. Mechanism of action of ribosomally synthesized and post-translationally modified peptides. Chem. Rev. 122, 14722–14814 (2022).
Chen, M., Wang, S. & Yu, X. Cryptand-imidazolium supported total synthesis of the lasso peptide BI-32169 and its D-enantiomer. Chem. Commun. 55, 3323–3326 (2019).
Tietz, J. I. et al. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat. Chem. Biol. 13, 470–478 (2017).
da Hora, G. C. A. et al. Lasso peptides: exploring the folding landscape of nature’s smallest interlocked motifs. J. Am. Chem. Soc. 146, 4444–4454 (2024).
Mi, X. et al. Sequence controlled secondary structure is important for the site-selectivity of lanthipeptide cyclization. Chem. Sci. 14, 6904–6914 (2023).
Barrett, S. E. et al. Substrate interactions guide cyclase engineering and lasso peptide diversification. Nat. Chem. Biol. 21, 412–419 (2025).
Glassey, E., Zhang, Z., King, A. M., Niquille, D. L. & Voigt, C. A. De novo design of ribosomally synthesized and post-translationally modified peptides. Nat. Chem. 17, 233–245 (2025).
Vinogradov, A. A., Chang, J. S., Onaka, H., Goto, Y. & Suga, H. Accurate models of substrate preferences of post-translational modification enzymes from a combination of mRNA display and deep learning. ACS Cent. Sci. 8, 814–824 (2022).
Chang, J. S., Vinogradov, A. A., Zhang, Y., Goto, Y. & Suga, H. Deep learning-driven library design for the de novo discovery of bioactive thiopeptides. ACS Cent. Sci. 9, 2150–2160 (2023).
Vinogradov, A. A., Bashiri, G. & Suga, H. Illuminating substrate preferences of promiscuous F420H2-dependent dehydroamino acid reductases with 4-track mRNA display. J. Am. Chem. Soc. 146, 31124–31136 (2024).
Thokkadam, A. et al. High-throughput screen reveals the structure–activity relationship of the antimicrobial lasso peptide ubonodin. ACS Cent. Sci. 9, 540–550 (2023).
Rives, A. et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc. Natl. Acad. Sci. USA 118, e2016239118 (2021).
Elnaggar, A. et al. ProtTrans: toward understanding the language of life through self-supervised learning. IEEE Trans. Pattern Anal. Mach. Intell. 44, 7112–7127 (2022).
Brandes, N., Ofer, D., Peleg, Y., Rappoport, N. & Linial, M. ProteinBERT: a universal deep-learning model of protein sequence and function. Bioinformatics 38, 2102–2110 (2022).
Alley, E. C., Khimulya, G., Biswas, S., AlQuraishi, M. & Church, G. M. Unified rational protein engineering with sequence-based deep representation learning. Nat. Methods 16, 1315–1322 (2019).
Elnaggar, A. et al. Ankh ☥: optimized protein language model unlocks general-purpose modelling. Preprint at https://doi.org/10.1101/2023.01.16.524265 (2023).
Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023).
Rao, R. et al. Evaluating protein transfer learning with TAPE. Adv. Neural Inf. Process. Syst. 32, 9689–9701 (2019).
Hie, B. L. et al. Efficient evolution of human antibodies from general protein language models. Nat. Biotechnol. 42, 275–283 (2024).
Thumuluri, V. et al. NetSolP: predicting protein solubility in Escherichia coli using language models. Bioinformatics 38, 941–946 (2022).
Luo, Y. et al. ECNet is an evolutionary context-integrated deep learning framework for protein engineering. Nat. Commun. 12, 5743 (2021).
Zhao, J., Zhang, C. & Luo, Y. Contrastive fitness learning: reprogramming protein language models for low-N learning of protein fitness landscape. International Conference on Research in Computational Molecular Biology (Springer, 2024), pp. 470–474.
Gururangan, S. et al A. Don’t stop pretraining: adapt language models to domains and tasks. Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics. 8342–8360 (2020). https://doi.org/10.18653/v1/2020.acl-main.740.
Zeng, W., Dou, Y., Pan, L., Xu, L. & Peng, S. Improving prediction performance of general protein language model by domain-adaptive pretraining on DNA-binding protein. Nat. Commun. 15, 7838 (2024).
Clark, J. D., Mi, X., Mitchell, D. A. & Shukla, D. Substrate prediction for RiPP biosynthetic enzymes via masked language modeling and transfer learning. Digital Discovery. 4, 343–354 (2025).
Mosqueira-Rey, E., Hernández-Pereira, E., Alonso-Ríos, D., Bobes-Bascarán, J. & Fernández-Leal, Á. Human-in-the-Loop Machine Learning: A State of the Art. Artif. Intell. Rev. 56, 3005–3054 (2023).
Li, F.-Z.; Amini, A. P.; Yue, Y.; Yang, K. K.; Lu, A. X. Feature reuse and scaling: understanding transfer learning with protein language models. Preprint at https://doi.org/10.1101/2024.02.05.578959 (2024).
DiCaprio, A. J., Firouzbakht, A., Hudson, G. A. & Mitchell, D. A. Enzymatic reconstitution and biosynthetic investigation of the lasso peptide fusilassin. J. Am. Chem. Soc. 141, 290–297 (2019).
Koos, J. D. & Link, A. J. Heterologous and in vitro reconstitution of fuscanodin, a lasso peptide from thermobifida fusca. J. Am. Chem. Soc. 141, 928–935 (2019).
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001).
Schapire, R. E. Explaining AdaBoost. In Empirical Inference: Festschrift in Honor of Vladimir N. Vapnik (eds Schölkopf, B., Luo, Z. & Vovk, V.) 37–52 (Springer, 2013).
Hearst, M. A., Dumais, S. T., Osuna, E., Platt, J. & Scholkopf, B. Support vector machines. IEEE Intell. Syst. Appl. 13, 18–28 (1998).
Popescu, M.-C., Balas, V. E., Perescu-Popescu, L. & Mastorakis, N. Multilayer perceptron and neural networks. WSEAS Trans. Circuits Syst. 8, 579–588 (2009).
Maaten, L. & van der Hinton, G. Visualizing data using T-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Feller, A. L. & Wilke, C. O. Peptide-aware chemical language model successfully predicts membrane diffusion of cyclic peptides. J. Chem. Inf. Model. 65, 571–579 (2025).
Schissel, C. K. et al. Deep learning to design nuclear-targeting abiotic miniproteins. Nat. Chem. 13, 992–1000 (2021).
Atchley, W. R., Zhao, J., Fernandes, A. D. & Drüke, T. Solving the protein sequence metric problem. Proc. Natl. Acad. Sci. USA 102, 6395–6400 (2005).
Georgiev, A. G. Interpretable numerical descriptors of amino acid space. J. Comput. Biol. 16, 703–723 (2009).
Pan, S. J., Cheung, W. L., Fung, H. K., Floudas, C. A. & Link, A. J. Computational design of the lasso peptide antibiotic microcin J25. Protein Eng. Des. Sel. 24, 275–282 (2011).
Knappe, T. A. et al. Introducing lasso peptides as molecular scaffolds for drug design: engineering of an integrin antagonist. Angew. Chem. Int. Ed.50, 8714–8717 (2011).
Ducasse, R. et al. Sequence determinants governing the topology and biological activity of a lasso peptide, microcin J25. ChemBioChem 13, 371–380 (2012).
Hegemann, J. D. et al. Rational improvement of the affinity and selectivity of integrin binding of grafted lasso peptides. J. Med. Chem. 57, 5829–5834 (2014).
Ritter, S. C., Yang, M. L., Kaznessis, Y. N. & Hackel, B. J. Multi-species activity screening of microcin J25 mutants yields antimicrobials with increased specificity towards pathogenic salmonella species relative to human commensal Escherichia coli. Biotechnol. Bioeng. 115, 2394–2404 (2018).
Platt, J. Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. Adv. Large Margin Classif. 10, 61–74 (2000).
Hills, E., Woodward, T. J., Fields, S. & Brandsen, B. M. Comprehensive mutational analysis of the lasso peptide klebsidin. ACS Chem. Biol. 17, 998–1010 (2022).
Zhang, Z. et al. Protein language models learn evolutionary statistics of interacting sequence motifs. Proc. Natl. Acad. Sci. USA. 121, e2406285121 (2024).
Juarez, R. J. et al. LassoHTP: a high-throughput computational tool for lasso peptide structure construction and modeling. J. Chem. Inf. Model. 63, 522–530 (2023).
Ouyang, X. et al. LassoPred: a tool to predict the 3D structure of lasso peptides. Nat. Commun. 16, 5497 (2025).
da Hora, G. C. A., Oh, M., Nguyen, J. D. M. & Swanson, J. M. J. One descriptor to fold them all: harnessing intuition and machine learning to identify transferable lasso peptide reaction coordinates. J. Phys. Chem. B 128, 4063–4075 (2024).
Zhao, J. et al. GaLore: memory-efficient LLM training by gradient low-rank projection. Preprint at https://doi.org/10.48550/arXiv.2403.03507 (2024).
Pedregosa, F. et al. Scikit-learn: machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Li, Y. et al. Characterization of sviceucin from streptomyces provides insight into enzyme exchangeability and disulfide bond formation in lasso peptides. ACS Chem. Biol. 10, 2641–2649 (2015).
Reyna-Campos, A. O. et al. Heterologous expression of lasso peptides with apparent participation in the morphological development in streptomyces. AMB Express 14, 97 (2024).
Paszke, A. et al. PyTorch: an imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 32, 8024–8035 (2019).
Gasteiger, E. et al. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook (ed. Walker, J. M.) 571–607 (Humana Press, 2005).
Higuchi, R., Krummel, B. & Saiki, R. K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16, 7351–7367 (1988).
Sun, Z. Z. et al. Protocols for implementing an Escherichia coli based TX-TL cell-free expression system for synthetic biology. J. Vis. Exp. 79, e50762 (2013).
Yim, S. S., Johns, N. I., Noireaux, V. & Wang, H. H. Protecting linear DNA templates in cell-free expression systems from diverse bacteria. ACS Synth. Biol. https://doi.org/10.1021/acssynbio.0c00277 (2020).
Acknowledgements
This work was supported by National Institutes of Health grants (R35GM142745 and R21AI167693 to D.S.; R35GM158411 and R01AI144967 to D.A.M.). S.E.B. was supported by a National Science Foundation Graduate Research Fellowship (DGE 21-46756) and the University of Illinois Urbana-Champaign Illinois Distinguished Fellowship. The authors thank Joseph D. Clark for discussing LassoESM pretraining and Tanner J. Dean for discussing the cross-attention layer implementation. The authors also thank Soumajit Dutta and Song Yin for their valuable input during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
X.M., S.E.B., D.A.M. and D.S. conceived and designed the study. X.M. designed and implemented the computational framework. S.E.B. conducted experiments and analyzed experimental data. All authors discussed the results and contributed to the final manuscript. D.A.M. and D.S. supervised the research and acquired funding for the study.
Corresponding authors
Ethics declarations
Competing interests
S.E.B. and D.A.M. are inventors on a provisional patent application filed by the University of Illinois at Urbana-Champaign covering lasso cyclase engineering (US Provisional Application Ser. No. 63/673,853). D.A.M. is also a co-founder and owns stock in Lassogen, Inc. The other authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Ivan Koludarov and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mi, X., Barrett, S.E., Mitchell, D.A. et al. LassoESM a tailored language model for enhanced lasso peptide property prediction. Nat Commun 16, 8545 (2025). https://doi.org/10.1038/s41467-025-63412-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-63412-3