Abstract
Bacteria often encounter physico-chemical stresses that disrupt division, leading to filamentation, where cells elongate without dividing. Although this adaptive response improves survival, it also exposes filaments to significant mechanical strain, raising questions about the mechanochemical feedback in bacterial systems. In this study, we investigate how mechanical strain modifies the geometry of bacterial filaments and influences the Min oscillatory system, a reaction-diffusion network central to division in Escherichia coli. Through a multidisciplinary approach combining quantitative fluorescence microscopy, biophysical modeling, microfluidics, and patterned growth substrates, we demonstrate that Escherichia coli filaments undergo a growth-induced buckling instability. This phenomenon modulates the spatiotemporal patterning of the Min system. Moreover, we show that synergistic mechanochemical effects determine the location of the division site after stress relief, effectively creating a mechanical “memory” for cytokinesis. Our findings underscore the critical role of mechanical forces and geometric effects in bacterial filamentation and provide new insights into the functional implications of mechanobiology in microbial systems.
Similar content being viewed by others
Introduction
Bacterial division has been extensively studied, typically under favorable growth conditions1,2,3. However, bacteria frequently face environmental stresses of diverse kinds, both physical and chemical, that require phenotypic adaptation4,5,6. Filamentation, a phenomenon in which bacteria stop division, but neither genetic replication nor elongation (that is, growth), is a common adaptive response to various stressors, including antibiotic exposure7,8,9,10, nutrient depletion11, oxygen deficiency12, and temperature changes13. The filamentation phenotype provides bacteria with advantages, such as colonizing heterogeneous adhesive surfaces14 or facilitating cell-to-cell dissemination during host invasion15; but it also poses challenges to bacterial cells. Thus, cells must cope with sizes up to hundreds of micrometers16,17 (in contrast to a size of a few microns when cells do not filament). Consequently, bacterial filaments withstand significant mechanical stresses18, compared to those in regular-sized cells, leading to increased susceptibility to bending and buckling forces19,20.
The interplay between mechanical forces and signaling pathways, i.e., the mechanosensing properties, of bacteria during filamentation remains an unresolved question21. Notably, in filaments of rod-shaped bacteria, such as E. coli and B. subtilis, the cell wall growth rate has been shown to depend on mechanical stress, and evidence suggests that MreB (an actin homolog) acts as a membrane curvature sensor18,22,23. However, how mechanical stimuli and curvature modulate other key signaling processes at the core of the filamentation phenomenon, such as the division machinery, is unclear.
The regulation of cell division has been extensively studied in E. coli24,25,26. In particular, the positioning of prospective septa, i.e., the so-called Z-rings27,28, is controlled by the Min oscillatory system (MinC, MinD, and MinE proteins), which follows a reaction-diffusion scheme29,30. Thus, when E. coli undergoes filamentation, the Min system exhibits a standing-wave, multinode pattern, whose potential division sites are located at the minima of the time-averaged Min signal27,31,32. Interestingly, experiments in deformed spheroplasts33, in E. coli with aberrant shapes34,35, and on patterned substrates36, suggest that MinD localizes in regions of high curvature. While the molecular mechanism underlying this phenomenon is, to the best of our knowledge, still under debate, a plausible reason is the reported positive feedback loop between the local membrane curvature and protein concentration37. This is supported by models in cocci-shaped bacteria, where slight deformations cause a micron-scale change in global membrane curvature during cell elongation, which may act as cues to select the plane of division38. These facts raise the intriguing question of whether or not, in bacterial filaments, prone to bend and buckle, Min patterning is influenced by mechanical stress and curvature (or the other way around). Moreover, given that Min patterning determines the prospective locations for cell cleavage, does mechanical stress and changes in the filament’s geometry condition the division sites in filaments when environmental stress ends and cytokinesis resumes?
Herein, we address these questions using a multidisciplinary approach that combines quantitative fluorescent time-lapse microscopy, micro-patterning of growth substrates, biophysical models, Fluorescence Recovery After Photobleaching (FRAP) analyses, and microfluidics. Our results reveal that bacterial filaments undergo growth-induced buckling instability that leads to a mechanochemical interplay between the Min oscillatory system and the curvature, preceded by the positioning of FtsZ rings. Further, we explore the post-stress consequences of such an interplay and conclude that it sets preferential division sites. Altogether, our study sheds light on the mechanobiology of bacterial filaments and their functional role.
Results
E. coli growing filaments undergo a buckling instability
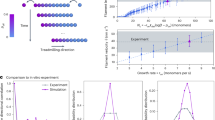
In our experiments, we used the antibiotic aztreonam to induce E. coli filamentation. Antibiotic-induced filamentation is widely used as a model to study bacterial adaptation under stress39,40. In clinically relevant settings, such as urinary tract infections (UTIs) caused by pathogenic E. coli, antibiotic treatments trigger filamentation via SOS response activation and division inhibition by SulA17. Furthermore, during infection, filamentation may occur through alternative pathways to that of SulA, such as the DamX-mediated pathway41. In this work, we chose aztreonam, which specifically inhibits FtsI - the cell division protein responsible for catalyzing peptidoglycan cross-linking at the septum42. At sub-lethal concentrations, aztreonam maintains membrane integrity and allows Z-ring assembly while preventing cell division, thereby inducing filament formation9,43. In our case, E. coli cultures were grown in M9 minimal medium to an OD600 of 0.3 before inoculation onto 2% agarose pads containing 10 μg/ml of aztreonam (Methods). Under these conditions, time-lapse microscopy revealed cell filamentation (Fig. 1A, Supplementary Movie 1). Growing filaments exhibited a buckling instability, which became more likely for filaments longer than ~10 μm (Supplementary Fig. 1). In our study, we utilized the DH5α strain, which exhibits a lower Young’s modulus compared to K1244, thereby potentially facilitating the buckling phenomenon. This instability is, at first, counterintuitive, since cell elongation could be thought as a tensile stress, whereas buckling develops due to compressive stresses37. In that regard, our estimation of the critical load required to induce filament buckling is on the order of \({{\mathcal{O}}}\left(1{0}^{1}\right)\) nN (Methods). Buckling led to high curvature values with radii smaller than the characteristic length of non-filamentous E. coli cells: ∣κ∣ > 0.5 μm−1 (Fig. 1A). In this context, we used ∣κ∣ as a proxy for mechanical strain.
A Phase contrast images of a representative E. coli filament growing on 10 μg/ml aztreonam agarose pads. Following cell segmentation and tracking (Methods), the curvature of the filaments is computed along the midline. At a given location (e.g., yellow point) the curvature, ∣κ∣, is computed as the inverse of the radius of the circumference that is tangent to that point (green circle): ∣κ∣ = 1/R. As time progresses and the filament grows, the snapshots reveal a buckling instability where large values of ∣κ∣ are reached by the central region of the filament (see E). B Cartoon of the mechanical toy model. Forces are exerted by a chain of connected springs (black). Nodes (grey circles) are subjected to elastic and bending forces. Due to cellular growth, the elastic energy relies on the time-dependent distance, L(t), between consecutive nodes (Methods). Bending energy depends on the angle θ as determined by neighboring segments. C Snapshots of a simulated growing filament (initial length: 50 μm; doubling time: 50 min). Color scales for each time point indicate the elastic (top) and bending (bottom) energies using pair-wise moving average coordinates (Methods). As time progresses, elastic energy accumulates by the central region. Buckling leads to a relaxation of the elastic energy at the expense of an increase of the bending energy (black triangles). D Ratio of the total energy between soft and stiff filaments (soft: \(\widetilde{\Omega }=0.9\); stiff: \(\widetilde{\Omega }=2\); sample size: n = 100 simulations). As time progresses, the panel shows that stiff filaments that are not able to buckle increase their energy. E Density histograms of the curvature of filaments as a function of the relative position as obtained in experiments (left) and simulations (pair-wise moving average coordinates; right). Sample size: n = 22 (experiments) and n = 100 (simulations). The color scale represents probability. Larger curvatures (buckling) accumulate in the central region of the filament.
To understand this phenomenon, we developed a mechanical toy model. The model consists of a 2D chain of massless nodes connected by linear springs. Segments connecting nodes are also subjected to bending forces (Methods, Fig. 1B). The springs were assumed to be identical (i.e., same elastic and bending constants). In the model, we neglected inertial effects and included energy dissipation. Thus, the model comprises the minimal components required to mimic the filament mechanics at low Reynolds numbers by disregarding the 3D structure of the cells, the adhesion forces to a substrate, and plastic deformation effects (see Discussion).
The theoretical analysis and our simulations confirmed that, as expected, if the equilibrium length of the springs, l0, is kept constant, then the simulated filaments reach a straight configuration regardless of the initial condition (Methods). However, if growth is included such that the equilibrium length depends on time, i.e., \({l}_{0}\left(t\right)\), then buckling develops (Supplementary Movie 2). Consequently, our model argues in favor of cellular growth as the driving force underlying buckling in agreement with recent studies on bacterial chains45. On the one hand, the growth of the springs located at the middle of the chain is dampened by the growth of the surrounding springs that push back. On the other hand, springs located by the poles of the chain find less opposition to growth. As a result, elastic energy accumulates at the central region of filaments (compressive stress), and energy relaxation can only be achieved by buckling. This buckling dynamics is revealed in our model by bending events at different length scales. Thus, local stresses at a scale of few nodes cause zig-zag configurations in the chain of springs. In addition, at a larger length scale, there is macroscopic buckling as revealed by the changes of the local curvature of the midline as experimentally shown (e.g., Fig. 1A). In order to distinguish between these different, yet complementary, phenomena, and properly compare simulations and experiments, we implemented a moving average approach (Methods).
As shown in Fig. 1C, our model reproduces that the accumulation of elastic energy (compressive stresses) due to growth decreases at the expense of an increase of the bending energy. The total energy decrease due to the buckling instability can be observed in Fig. 1D by comparing filaments with different (dimensionless) bending moduli: \(\widetilde{\Omega }=0.9\) (soft filament) vs. \(\widetilde{\Omega }=2\) (stiff filament). Thus, as time progresses, the simulations show that stiff filaments that are not able to buckle heap up energy with respect to soft filaments. The model also predicts that since compressive stresses accumulate at the middle of the filament, buckling is more likely to develop at that location. In that regard, Fig. 1E shows that in both, experiments and simulations, the region of the filament with the largest curvature is the central one. The experimental Fig. 1E also allows us to estimate the onset of buckling for ∣κ∣ > 0.5 μm−1, as these values are reached only at the central region of the filament.
In summary, the mechanical model explains the buckling phenomenon observed in growing E. coli filaments. Buckling events are more likely by the central region because the filament ends are able to elongate freely, whereas the middle section must exert larger forces in order to push and displace the rest of the filament. This mismatch between the growing dynamics of different regions of the filament leads to an accumulation of compressive stresses that ultimately cause the filaments to buckle.
Min patterning is modulated by the local curvature of filaments
In order to study how buckling-induced mechanical stress affects signaling proteins related to the filamentation phenomenon, we focused on the Min system. The MinCDE oscillatory system prevents FtsZ polymerization and, consequently, plays an important role in the filamentation and cell division processes46,47,48. In particular, MinD protein localizes either at the cell membrane or in the cytosol, depending on its ATP activity and it is a proxy for the activity and localization of MinC and MinE (see Supplementary Fig. 2). In our study, we used an E. coli strain that expresses the recombinant fluorescent protein YFP:MinD (Methods).
We tracked and measured, simultaneously, MinD and the curvature of the middle cell axis of E. coli filaments by quantitative time-lapse microscopy (Fig. 2, see also Supplementary Movie 3). In our analyses, we focused on growing filaments exhibiting two key characteristics: (1) constant MinD concentration (intensity per unit area) over time, and (2) persistent standing wave patterns. As for (1), since our analyses rely on fluorescent microscopy data quantification, we assume that all MinD molecules fold correctly and are fluorescent-active. We note that constant MinD concentration indicates a constant MinD production as the body mass increases in agreement with recent results49 (see Supplementary Fig. 3 and Methods). Furthermore, the maintained standing wave oscillations (see below) suggest a stable MinE/MinD ratio during the observation period50. This standing wave pattern allowed us to pinpoint the location of the non-functional FtsZ septa during filamentation (regions with a sustained low concentration of MinD). Interestingly, we observed that MinD concentration (i.e., the amplitude of the pattern) was dependent on the level of curvature, Fig. 2A. In particular, the average intensity of the MinD signal seemed to increase in buckled regions and, concurrently, the imaging data suggested that the nodes of the MinD pattern (i.e., local minima) tallied with large curvature peaks.
A Fluorescence channel (YFP, false color scale) of the illustrative filament shown in Fig. 1A (average Z-projection of the last 5 min). The zoomed region (circle) highlights the increase of MinD in buckled regions. In addition, the white arrows indicate the anti-correlation effect between MinD and locations with a high curvature. B Top: Kymographs of the filament shown in (A) representing curvature (top) and MinD intensity (middle) profiles along the filament middle axis as a function of time. Color scales account for the curvature (μm−1) and for MinD concentration (arbitrary units). The zoomed region in the MinD kymograph indicates the period, T, and the wavelength, λ, of the MinD standing-wave pattern. Bottom: Overlay of the time average of MinD and curvature profiles. Black arrows highlight the locations pinpointed in (A) by means of white arrows. Averages are computed within the region delimited by the white dashed lines. C Violin plots showing the distribution of values of the Pearson’s correlation coefficient between MinD and curvature (sample size: n = 19 filaments where high and low curvature regions were analyzed separately, Supplementary Fig. 4B, Methods). Black solid lines represent the mean (Low: 0.22; High: −0.14). D Normalized MinD intensity (mean and standard deviation) as a function of the curvature in non-patterned agarose pads (same dataset as in (C)) and in micro-patterned growth substrates (n = 20 filaments). Solid lines are, in both cases, a logarithmic fit: Eqn. (1), Methods. Inset: Illustrative phase contrast image of an E. coli filament growing in a micro-patterned agarose pad (scale bar: 10 μm).
To quantitatively assess the interplay between MinD patterning and buckling, we computed kymographs and performed temporal averages of MinD and curvature profiles (Fig. 2B). As expected, the number of nodes of the standing wave increased as the filament grew. However, other properties of the pattern, such as the wavelength (at any given time) and the period (at any given location) were not affected by buckling (Supplementary Fig. 4A). Further, these values were fairly conserved among filaments: \({{\uplambda}}=\left(7.6\pm 1.0\right)\,\mu {{\rm{m}}}\) and \(T=\left(55\pm 10\right)\) s (Supplementary Fig. 4A), in agreement with previous studies27. Our analysis confirmed that MinD peaks avoided regions with high curvature (∣κ∣ > 0.5 μm−1). To deepen into this effect, we computed the Pearson’s correlation coefficient between the averaged MinD and the curvature profiles (Methods) and differentiate between regions with high (∣κ∣ > 0.5 μm−1) and low (∣κ∣ < 0.5 μm−1) curvature values (Supplementary Fig. 4B). The results, Fig. 2C, revealed a significant anti-correlation in high-curvature regions and a positive correlation in low-curvature regions (Mann-Whitney U test: α = 5 ⋅ 10−2 significance level). As for the quantitative relationship between MinD concentration and buckling, we computed the value of the normalized intensity (per filament) of the wave envelope as a function of the curvature, Fig. 2D (Methods, Supplementary Fig. 4C). The analysis confirmed an increase in MinD as a function of the curvature, which reaches a plateau for curvature values above 0.5 μm−1 (Fig. 2D non-patterned agarose). In order to verify that the phenomenon of MinD correlation and accumulation as a function of the curvature is independent of MinD concentration, we performed experiments decreasing the plasmid induction 10 fold (Supplementary Fig. 5). Our results support that the phenomena observed for the previous plasmid induction are also valid at this MinD concentration. Likewise, we also tested whether the mechanical instability and its effects on MinD patterning are strain-dependent. For this, we replicated the experiment using a K12 strain (Supplementary Movie 4) that confirms the results obtained previously for Dh5α.
Given that our results indicate that buckling instability naturally develops in the central region of filaments (Fig. 1E), we further tested whether MinD accumulation preferentially occurs at regions of higher local curvature versus at locations by the middle (where growth-induced compressive stresses accumulate). To that end, we designed micro-patterned growth substrates (Methods, Supplementary Fig. 6), where we externally control the bending and curvature of the filaments along their body length (inset Fig. 2D and Supplementary Movie 5). Our analysis of MinD intensity as a function of the curvature revealed in this case a similar trend (Fig. 2D: patterned agarose). We notice that micro-patterned pads could not be used to perform a MinD-curvature correlation study (i.e., Fig. 2C). As mentioned above, that analysis requires to achieve local curvatures above 0.5 μm−1. However, we observed that E. coli filaments would escape the regions of the guiding growth patterns with curvatures larger than 0.4 μm−1. Therefore, our data support that MinD accumulation is driven by local curvature rather than by growth-induced mechanical strain, and, overall, the existence of a modulation of Min patterning due to curvature effects. These findings align with previous studies conducted on non-filamentous E. coli cells37 (Discussion).
MinD pattern modulation is driven by a diffusion drop in buckled regions
Previous studies have shown that membrane curvature and mechanical stress alter the assembly dynamics of proteins as well as their diffusive properties37,51,52,53,54,55. In that regard, since local changes in protein dynamics lead to local modifications in protein concentration, we wondered whether the modulation of the MinD pattern in buckled regions is driven by this physical phenomenon. To test this hypothesis, we performed Fluorescence Recovery After Photobleaching (FRAP) assays and numerical simulations. Given the oscillatory behavior of MinD, we devised a methodology of FRAP data that weights up the standing-wave patterning properties of the signal (Methods, Fig. 3A). Our analysis relies on the fact that a standing wave can be decomposed as the sum of two waves traveling in opposite directions with phase velocities c = λ/T. To characterize the effect of buckling in protein dynamics, we photobleached MinD and computed the dimensionless ratio between the fluorescence recovery speed, vr, and c. Here, vr is defined as vr = d/tr, where d is the distance from a MinD peak in the non-photobleached domain (emitter) to the location where the recovery (≥ 30%) of the MinD signal is observed (receiver), and tr is the recovery time following photobleaching (Methods, Fig. 3B). We note that the normalization of vr with respect to c accounts for possible differences (i.e., variability) among filaments with respect to their wave properties. Consequently, the larger vr/c, the faster the signal propagation due to the transport mechanism underlying patterning.
A Cartoon illustrating the FRAP experimental quantification (see text). B Top: Composite of bright-field and MinD fluorescence of a filament before photobleaching. The colored area highlights the photobleached region. The blue circle indicates the pulse for which the fluorescence recovery is analyzed (receiver). The orange circle highlights the non-photobleached pulse before the buckled region (emitter). Bottom: Kymograph of the FRAP experiment performed over the filament shown in the top. C Violin plots showing the dimensionless velocity recovery, vr/c, of the pulse’s recovery for straight (n = 17) and buckled (n = 8) filaments. Black lines account for the mean (Straight: 1.40; Buckled: 1.18). Buckled filaments reveal a larger recovery, thus indicating a slower diffusive transport.
To test this approach, we performed several control experiments (Supplementary Fig. 7). In the case of non-filamentous bacteria showing one pole-to-pole MinD pulse, photobleaching the pulse precludes recovery within the temporal observation window (~2.5 × T, Supplementary Fig. 7A). However, photobleaching one cell pole while there was a signal outside the photobleached region led to MinD recovery within the observation window (Supplementary Fig. 7B). This hints that signal recovery requires MinD traveling from a non-photobleached region within the filament. To further investigate this, we conducted the same experiment using short filaments. Consistent with the previous results, when we photobleached all of the MinD (Supplementary Fig. 7C), we observed no signal recovery. However, when we photobleached one of the pulses, while leaving at least one other MinD pulse intact, recovery was detected (Supplementary Fig. 7D). We applied the aforementioned methodology to buckled (∣κ∣max > 0.5 μm−1) and straight (∣κ∣max < 0.5 μm−1) filaments. Our results, Fig. 3B, indicate that buckling slows down the velocity recovery vr/c, therefore suggesting either a decrease in protein diffusivity or a change in the attachment/detachment kinetics in curved regions, as previously shown53,54,55,56,57,58,59.
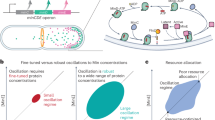
To further clarify our findings from the FRAP experiments, we conducted numerical simulations. In that regard, since the cross-sectional shape of the bacterial filaments remains unchanged due to bending/buckling, we do not expect that the transport properties of MinD in the cytoplasm are affected60. Consequently, we focus in our simulations on the effect of curvature on membrane diffusivity and on the binding kinetics of Min proteins. Our approach is based on a signaling model that accounts for the main molecular interactions of Min species, including oligomerization and the MinE-mediated hydrolysis of MinD-ATP (Fig. 4A and Methods)61.
A Cartoon of the Min oscillatory system model, adapted from61. B Spatio-temporal evolution of MinD obtained in simulations of a growing filament. Top: Kymograph of a filament with an initial length of 50 μm growing for its doubling time (50 min). For \(t\in \left(0,20\right)\) min (dashed purple line), no buckling is implemented. For \(t\in \left(20,50\right)\) min (solid purple line), the membrane diffusion coefficients of proteins are set to a 30% of their values within a 3 μm-wide band (Methods). Bottom: Temporal averages of MinD (arbitrary units) for a period of 20 minutes immediately before (dashed purple line) and a period of 30 minutes immediately after (solid purple line) the diffusion drop within the region delimited by the white dashed lines shown in the kymograph. The green line delimits the region where the simulated curvature reaches ∣κ∣ ~1μm−1 as represented in the inset.
In our simulations we implemented an exponential growth of the filaments (i.e., elongation) and the aforementioned changes in protein dynamics in a 3 μm region —approximately the width of a MinD peak— where ∣κ∣ ~1 μm−1 (Methods). As for the effect this has on membrane diffusivity on Min proteins, we estimated that such curvature implies a ~30% decrease of its original value54. Under those conditions, the MinD accumulation pattern in buckled regions observed in our simulations resembled the experimental results (cf. Fig. 2B middle and Fig. 4B).
As for the effect on patterning caused by changes in the binding rate of Min proteins to the cell membrane, previous experiments in septins have shown that going from a flat filament to the simulated curvature implies an approximate 100% increase in the binding rate (see Fig. 2B in ref. 59). Under these conditions, our simulations indicate that Min patterning is disrupted (Supplementary Fig. 8A), which contrasts with our experimental observations (Fig. 2B, middle). In fact, the same experimental figure suggests that the binding rate remains constant for ∣κ∣ ranging from 0.1 to 0.4 μm−1, for which we already see MinD accumulation in our experiments (Fig. 2D). To investigate whether a specific range of binding rates could lead to accumulation, we tested values down to a 10% increase, finding this to be the minimum increase that does not disrupt MinD patterning and resembles the scenario we observe experimentally (Supplementary Fig. 8B).
In summary, FRAP experiments and numerical simulations support the idea that buckling modifies the membrane diffusivity of Min proteins, which in turn causes protein accumulation.
Post-stress filament division sites are favored by high-curvature regions preceded by FtsZ and absence of DNA
Herein we have shown that curvature peaks positively correlate with sustained MinD signal minima (i.e., as shown in Fig. 2C, high curvature —indicating large mechanical strain— and MinD maxima are anti-correlated). This observation prompted us to investigate whether Min patterning and mechanical effects might synergistically determine preferred division sites in post-stress conditions.
To verify this, we conducted microfluidic experiments adapting the protocol established by Wehrens et al.32 (Methods). Cells were initially cultured in M9 medium under normal growth conditions, then switched to stressor medium (M9 + aztreonam) for 3 − 5 hours to induce filamentation. During this period, we observed progressive cell elongation followed by buckling. Division was restored by returning cells to pure M9 medium, at which point the filamented bacteria resumed cytokinesis16,32,62 (Fig. 5A and Supplementary Movie 6).
A Time-lapse of bacteria growing in a microfluidic chamber. After 1 h, aztreonam was added to the culture medium to induce filamentation. 5 h later, aztreonam was removed such that filaments resumed cytokinesis, and the curvature at the division locations was calculated. B 5 min average of phase contrast and fluorescent channels showing the location of MinD pulses just before the first division. The white arrows point at the 3 first divisions of the filament. C Probability density functions of the curvatures for filaments at the division points (in blue) and for the whole filaments just before the corresponding divisions (in yellow). The means of the distributions are 0.6 ± 0.4 and 0.3 ± 0.3 μm−1, respectively (sample size n = 125). D (left) Kymograph representing FtsZ fluorescence from a single cell to a filament. The y-axis is the position of the ring in relative value, and the x-axis is the time. (right) A kymograph representing the curvature of the cell. The axes are the same as in the left. The red triangles mark the point when the cell reaches a length of 8 μm. The dashed lines show the regions where the first 3 FtsZ rings are formed. E Probability density distribution of lag times between FtsZ ring appearance and a buckling event. On average, FtsZ precedes bending by 22 min, and the most probable value is 63 min. Each bin corresponds to 9 min (sample size n = 19). F 5 min average of fluorescent channel showing DNA distribution, avoiding curvature peaks (highlighted with white arrows) in a filament in the microfluidic device. G Violin plots showing the Pearson’s correlation coefficients between the DNA signal and curvature. The correlation was calculated separating the buckled region (high) and the non-buckled regions (low) as done with MinD in Fig. 2C (sample size n = 47). Solid lines represent the means of the distributions (Low: 0.02; High: −0.20).
Under these conditions, we selected buckled filaments with a maximum curvature exceeding 0.5 μm−1 and characterized the curvature at the division sites, ∣κ∣div. (Fig. 5B). For the analysis, we included divisions in filaments longer than 8 μm (either the originally selected ones or those resulting from divisions of longer filaments), ensuring that at least two division rings (or 3 sustained MinD pulses) were assembled in the cell (Fig. 5B). We note that the morphological differences observed in filaments growing in microfluidic traps compared to those on agarose pads are primarily attributable to the substrate. Filaments within microfluidic chips appear wider due to slight compression between the two rigid surfaces of the trap. Furthermore, these filaments experience greater frictional forces with the substrate (the two “sandwiching” stiff surfaces) than those growing on softer agarose pads, which favors the development of curvature, resulting in a more wiggly appearance63. Fig. 5C shows the probability density function (PDF) of the curvature at the division points (blue), revealing that filament division is more likely to occur at sites with ∣κ∣div.≥ 0.5 μm−1 (mean division curvature: \(\langle | \kappa {| }_{div.}\rangle=\left(0.6\pm 0.4\right)\,\mu {{{\rm{m}}}}^{-1}\)). In contrast, the PDF of the curvature of filaments, ∣κ∣fil., just before cytokinesis (Fig. 5C, orange) shows a distribution shifted towards lower curvatures (\(\langle | \kappa {| }_{fil.}\rangle=\left(0.3\pm 0.3\right)\,\mu {{{\rm{m}}}}^{-1}\)). Given that the threshold curvature value to observe a negative correlation between curvature maxima and MinD sustained minima is 0.5 μm−1, we calculated the fraction of divisions occurring above this threshold. Our results, revealed that 60% of divisions were above the threshold. If we normalize the curvature at the division site by the mean curvature of the filament, the divisions above the mean curvature would account for 82% of the total.
In this context, we also analyzed Z-ring dynamics (Supplementary Movie 7). To this end, we employed the MG1655 strain expressing gfp::ftsZ (Methods). We then quantified the correlation between the FtsZ signal and the curvature profile, as depicted in Fig. 5D. Given that FtsZ forms a dynamic Z-ring that disassembles (FtsZ depolymerization) before the onset of buckling, we computed the lag time to account for a potential temporal delay, as shown in Fig. 5E. Our results indicate that regions of high curvature are preceded by Z-rings by an average of 22 minutes. Importantly, the findings from these experiments suggest that divisome dynamics contribute to an increase in curvature (see Discussion). The role of the Min oscillatory system in influencing chromosome positioning and segregation has been a topic of debate64,65. In that regard, we additionally checked whether DNA segregation within the filaments followed the pattern dictated by MinD pulses, i.e., whether the DNA avoided the curvature peaks in filaments. To that end, we labeled and tracked the DNA in living filaments using SYTOX Orange (Methods). We found that DNA was distributed in a discrete manner along the filament avoiding curvature peaks, Fig. 5D. This avoidance became more pronounced as curvature increased (see Supplementary Fig. 9). These locations putatively mark the positioning of division septa when cytokinesis resumes32,62. To quantitatively assess the relationship between DNA distribution and the curvature of filaments, we computed the correlation of their profiles (Fig. 5E). Our results revealed a significant anti-correlation (Mann-Whitney U test: α = 5 ⋅ 10−2 significance level). This demonstrates that either the absence of DNA facilitates bending or that curvature can influence chromosomal positioning, in line with previous studies showing that DNA can “sense” the curvature of the cell walls and adjust accordingly38,64. Consequently, these findings endorse the view that regions of high curvature, and thus mechanical strain, play a crucial, functional role in determining preferential division sites in post-stress filaments.
Discussion
Previous investigations have studied E. coli filamentation and have established that Z-ring positioning is reallocated in response to the filament size32. Our study builds on this understanding by revealing how mechanical forces develop during E. coli filamentation resulting in an increase in curvature, which affects the Min system —the protein complex responsible for positioning the division ring. In particular, our findings provide a mechanistic explanation for filament buckling, linking three key factors: (1) FtsZ ring positioning, (2) local DNA depletion, and (3) mechanical strain accumulation. This spatiotemporal relationship emerges naturally from energy minimization principles: as filaments grow, elastic energy accumulates, which is subsequently reduced through bending events. In this context, previous studies have shown that E. coli filaments exhibit elastic behavior when subjected to transient external forces. However, sustained external forces lead to significant cell wall synthesis, resulting in plastic deformations in which the localized activity of MreB plays a key role19,20,66. Additionally, hydrodynamic and adhesion forces have also been shown to influence filament shape43. A point warranting discussion is the role of the 2D constrained growth employed in our experiments in the observed buckling phenomenon. Confinement undoubtedly exacerbates buckling, as a 3D environment typically offers more space for stress redistribution, potentially leading to a more gradual dissipation of instabilities63. However, buckling has also been observed during 3D bacterial growth, as evidenced, for instance, in UTIs caused by pathogenic E. coli17. We also emphasize that 2D confined growth of bacteria occurs naturally in various settings, such as the early stages of biofilm development63. This evidence suggests that a comprehensive quantitative description of the mechanical properties of filaments would require incorporating mechanisms and processes that were excluded from our model, as we deemed them unnecessary given the convincing results achieved using only a linear elastic theory.
Regarding the impact of the buckling instability and curvature on Min signaling, we demonstrated that MinD can adjust in response to both naturally occurring curvatures and controlled curvatures induced by patterned growth substrates. Specifically, MinD accumulates in regions of increasing curvature, and for curvatures exceeding 0.5 μm−1, our data indicated that MinD maxima avoid curvature peaks. This phenomenon may be attributed to the increased surface-to-bulk ratio characteristic of high-curvature regions, which can induce geometry-dependent behavior in mass-conserving reaction-diffusion systems27,67,68,69. In fact, previous studies suggested that increased protein binding leads to increased membrane occupancy, thereby reducing the diffusion of proteins across the membrane70 and that curvature changes the binding affinity of membrane proteins affecting their diffusion71. This, together with our simulations, points to a decrease in diffusion as the primarily responsible of the accumulation of MinD in regions of high curvature. Our FRAP experiments are consistent with this hypothesis. However, the observed differences in the FRAP results are not statistically significant (Mann-Whitney U test: α = 5 ⋅ 10−2 significance level), probably because despite having attempted to mitigate variations between filaments by normalizing the data considering each filament’s wavelength and period, this approach required these parameters to remain consistent before and after photobleaching, which was not always the case. In that regard, it is worth mentioning that some studies have reported that cumulative light excitation slowed down GFP:MinD oscillations by about 10 s, potentially influencing FRAP results72. Due to these challenges, we got a low number of bona fide replicates among all the samples taken. Thus, given the limited statistics, the FRAP results should be considered as an indication, rather than as clear evidence, to support the modification of the membrane diffusion parameter in the signalling model. We also note that Min patterning modulation may be influenced by mechanisms beyond slower diffusion in buckled regions. In that regard, some studies have shown changes in the membrane association and dissociation kinetics of some proteins due to membrane curvature effects58,59. However, to the best of our knowledge, in the case of Min proteins the only documented changes of the binding affinity are related to membrane composition in the context of synthetic vesicles73. To further assess the role played by changes in the binding rate, we performed numerical simulations (Supplementary Fig. 8). We concluded that mild changes in the binding rate constant disrupts MinD patterning, further supporting the idea that the main contribution to MinD accumulation is caused by a diffusion drop due to an increase in curvature.
Our study also explored the implications of our findings in post-stress conditions that lead to the resumption of cytokinesis in filaments. Given MinD’s role in inhibiting the divisome assembly in E. coli, and the observed tendency of Min to avoid curvature peaks, we hypothesized that Z-rings that assemble at these peaks would operate preferentially. Indeed, in our microfluidic experiments, when the stressor medium was removed —once the filaments reached high curvature values— the majority of division events occurred at curvatures exceeding the mean filament curvature. Additionally, the cross-correlation analysis between the FtsZ signal and the curvature profiles indicates that Z-ring formation preferentially precedes regions of high curvature, suggesting a contribution of the division machinery to the enhancement of curvature. Thus, mechanical buckling conditions Min patterning such that the standing wave nodes avoid regions of high curvature and favors Z-ring assembly, which in turn eventually contributes to increased curvature (arguably because the subsequent depolymerization of FtsZ and peptidoglycan remodeling softens the cell wall). Furthermore, this observation aligned with our characterization of DNA distribution within the filaments, which also avoided curvature peaks. Thus, in agreement with the current knowledge about the causality relationship among all the components mentioned in our work, the Min system acts first, the nucleoid occlusion then refines the permissive region from MinCDE by ensuring Z-ring formation happens only after chromosome segregation begins74. The absence of DNA in turn facilitates bending driven by compressive forces because the cell is “emptier” in those regions (i.e., softer). Ultimately, these results suggest the existence of a mechanical “memory” for cytokinesis, where the interplay between mechanical strain and signaling mechanisms defines the preferential division sites.
In a broader context, Min’s sensitivity to the shape of its “container” is not unique to E. coli56,71. For example, in B. subtilis, an analogous complex to Fts, known as DivIVA, localizes to regions of highest curvature due to MinD’s preference for flatter membrane regions67. In Staphylococcus aureus, a cocci-shaped bacterium, some models suggest that a modification in membrane curvature during elongation is used by DivIVA to recognize the next division plane38. Mechanical deformations in minimal systems, such as liposomes containing Min proteins, have also been shown to alter Min patterning68. Specifically, MinE has been observed to distribute asymmetrically in response to liposome deformation. Interestingly, MinE accumulation and liposome deformation appear to mutually promote one another, suggesting a positive mechanochemical feedback loop that may play a role in our system.
Finally, while our study focuses on Min, other components of the divisome could be affected by changes in cell geometry and mechanical stress in the membrane. In fact, a previous study has reported that MreB is correlated with the nucleoid position in E. coli unperturbed cells, but this correlation decreases when the cell geometry changes75. Thus, studying other components involved in the elongation, division, and regulation of the mechanical properties of cells is crucial for the complete understanding of the relationship between signaling, filamentation, and cytokinesis, and its implications for bacterial adaptation and antibiotic resistance76. Also, in this study, we have focused on antibiotic-induced filamentation mediated by activation of the SOS response, leading to division inhibition via SulA. However, during UTIs, other pathways, such as those involving DamX41, have been shown to regulate bacterial cell shape. This raises the intriguing question of how filamentation in certain clinical contexts, which do not necessarily involve alterations in membrane composition, differs from the phenomenon described here. Notably, our results demonstrate that the underlying physical mechanism driving buckling still applies when using bacterial strains with varying membrane stiffness. Nonetheless, in UTIs where filamentation is actively regulated by DamX or alternative mechanisms, the buckling process may also be subject to active control. Future studies could shed light on how these regulatory pathways influence buckling dynamics, potentially guiding the development of new therapeutic strategies to combat UTIs.
Method
Strain and growth conditions
Experiments were conducted using the Escherichia coli strain DH5α (NZYTech), transformed with the plasmid pYLS68 (Plac-yfp::minD minE::cfp), generously provided by Dr. Shih77. Overnight cultures were initiated from a single colony and grown in LB medium supplemented with 100 μg/ml ampicillin at 37 °C with shaking at 220 rpm.
After overnight growth, cultures were diluted to a final OD600 of ~0.01 in minimal M9 medium containing 0.4% glucose, 1 mM thiamine hydrochloride, 0.2% casamino acids, 100 μg/ml ampicillin, and 100 μM IPTG for plasmid induction (except for Supplementary Fig. 5 where 10 μM IPTG was used). These cultures were incubated under the same conditions as the overnight cultures until they reached an OD600 of 0.2−0.3. Subsequently, they were further diluted to OD600 ~0.005 to prepare the working cell culture.
Agarose pads and patterned growth substrates
Low-melting-point agarose pads were prepared following the protocol described in ref. 78 using M9-supplemented medium and 10 μg/ml aztreonam to induce bacterial filamentation as previously described43. Subsequently, 2 μl of the working culture were inoculated onto the agarose pads. The pads were incubated at 37 °C for 15 min and then dried in a desiccator for 20 min. Once placed in the microscopy dish (IBIDI μ-Dish 35 mm, low), they were incubated at 37 °C for 2 h to allow the cells to develop the filamentation phenotype.
To pattern the agarose growth substrates, master mold structures were fabricated on standard silicon samples. The fabrication process employed electron-beam direct writing on a Poly(methyl methacrylate) (PMMA) resist film. The electron-beam exposure, performed with a Jeol JBX-8100FS tool, was optimized to achieve the desired dimensions using an acceleration voltage of 100 keV and a beam current of 20 nA. After developing the PMMA resist using a conventional 1: 3 MIBK/IPA developer, the resist patterns were transferred onto the silicon samples using an optimized Inductively Coupled Plasma-Reactive Ion Etching process with fluoride gases (SF6, C4F8). The remaining resist was removed with oxygen plasma.
The etched silicon patterns were used as molds for fabricating complementary patterns on PDMS. The silicon structures were silanized in gas with 1H, 1H, 2H, 2H-perfluorodecyltrichlorosilane (97%, stabilized with copper) for approximately 27 h by placing a droplet of the silanizer near the sample in a vacuum desiccator. A mixture of silicone elastomer base and curing agent (in a mass ratio of 1: 10) was poured onto the silanized silicon pattern and cured in an oven at 90 °C for 1 h. The cured PDMS stamp was cooled for 20 min at room temperature before being carefully peeled off from the silicon mold. This PDMS stamp contained a pattern complementary to that of the silicon mold.
To prepare the patterned agarose substrates, 2.5% low melting point agarose in culture medium with 1 μg/ml aztreonam was poured onto the PDMS stamp and covered with a 2 × 2 cm coverglass. After refrigeration for 15 min, the coverglass was removed, and the PDMS was separated from the agarose. Finally, 2 μl of cell culture with an OD600 of 0.2–0.3 was inoculated on the agar pad. The agar pads were dried in a laminar flow hood for 15 min before being transferred to a glass-bottom dish (IBIDI μ-Dish 35 mm, low). The same PDMS mold could be reused after proper cleaning: it was immersed in 100% ethanol, sonicated for 15 min, rinsed thoroughly with DI water, and dried in an oven at 60 °C for 1 h.
The patterns were designed using the open-source mask design tool Nazca Design79. Four designs were created to control the curvature of the filaments (Supplementary Fig. 6): three based on curved channels and one based on a lattice of pillars. The “wave” pattern featured a sequence of “s” curves formed by connecting three half-circular bends, each with a different radius of curvature (Supplementary Fig. 6A). The “snake” pattern consisted of a sequence of “s” curves, where each “s” was formed by two half-circular bends, with the radius of curvature alternating every half rotation (Supplementary Fig. 6B). For both patterns, the radius of curvature ranged from 0.35 μm to 12 μm. The “spiral” pattern was composed of half-circular bends with a radius of curvature increasing from 0.8 μm to 10.4 μm in increments of 0.8 μm per half rotation (Supplementary Fig. 6C). The width of the channels for these three patterns ranged from 0.7 μm to 1.1 μm. The fourth design was a triangular lattice of circles, each equidistant from its neighbors, with connected spaces in between. The diameters of the circles ranged from 2 μm to 10 μm, in 1 μm increments, and the lattice constant ranged from 2.7 μm to 10.7 μm (Supplementary Fig. 6D).
Microscope settings
Images were acquired using a Leica Thunder inverted microscope, equipped with a HC PL-APO CS2 63 × /1.40 oil objective and a CMOS camera (Leica DFC9000 GTC). Köhler illumination was applied to optimize image contrast in transmitted light mode (phase contrast). Fluorophore excitation was performed using the Spectra-X Light Engine LED lamp, which features 8 solid-state LEDs. For MinD observation, excitation was performed at 510 nm and YFP filter settings (Ex: 495/517 nm, Em: 527/551 nm) combined with an external filter wheel at 535 ± 35 nm; for FtsZ excitation at 475 nm and GFP filter settings (Ex: 450/490 nm, Em: 500/540 nm) and the external filter at 535 ± 35 nm; and for dyed DNA at 555 nm and SYTOX filter settings (Ex: 542/566 nm, Em: 578/610 nm) and the external filter at 590 ± 25 nm. The Leica Application Suite X (LAS X) software was used for microscope control and image acquisition. All experimental procedures were conducted at 37 °C with temperature regulation provided by the IBIDI heating system.
Unless otherwise specified, time-lapses were acquired with an interframe time of 10 s over a 30-min duration, using 20% light source power (Spectra-X nominal output power 500 mW), 30 ms exposure time, and 2 × 2 binning. While our plasmid produces upon induction MinE:CFP and YFP:MinD protein fusions, images were only taken in the YFP channel (MinD) due to the high photobleaching and phototoxicity when trying to get a signal in the CFP channel (MinE) at the desired image acquisition rate during our long-term experiments.
For MinD membrane localization, a Z-stack was acquired with a HC PL-APO 100 ×/1.40 oil objective. The Z-stack resolution was 0.27 μm for a total range of 2.43 μm. To remove out-of-plane light effects, a Leica Thunder Small-Volume Computational Clearing filter was applied to the stack. To characterize the fluorescence profile, a line was drawn with Fiji80, and the function “Plot profile” was used.
Image and data analysis
Using Fiji80, the background was removed from all images with a rolling ball radius of 50 pixels in the fluorescent channel. The brightness and contrast of the phase-contrast images were adjusted to maximize contrast based on the characteristics of each image. However, the levels of the fluorescence images were consistently adjusted between 200 and 5000 to ensure comparability. Segmentation was performed on the phase-contrast images using the WeKa plugin81. The resulting segmentation masks were then used to analyze both phase-contrast and fluorescence images with the MicrobeJ plugin82, allowing us to extract quantitative data.
The coordinates of the medial axis obtained from MicrobeJ were analyzed to calculate the local curvature of the filaments using Mathematica83 (Fig. 1A). The Mathematica code employs finite differences to estimate the first and second derivatives at each point of the medial axis of the filament. The output is a list of point pairs representing curvature and normalized length. To verify the curvature results, the known curvature of the agarose-patterned substrates was used as a control. To reduce noise, curvature data were smoothed using 5-min time-averages. Additionally, the curvature profile was further smoothed when the maximum filament curvature was < 0.3 μm−1 to decrease noise. For filaments displaying buckling, this step was unnecessary as their curvatures typically reached ~1 μm−1, with a noise level of ~0.1 μm−1.
To estimate MinD’s concentration as a function of time, we quantified its cumulative fluorescent intensity in filaments (i.e., the total fluorescent signal of MinD in a filament) normalized by the area of the filament (Supplementary Fig. 3). The value of MinD concentration, ρMinD, results from a balance between different processes: the (unknown) MinD’s production, \(f\left({\rho }_{MinD}\right)\), the degradation (exponential), μ ⋅ ρMinD, and the protein dilution due to the increasing body mass, (1/V)(dV/dt)ρMinD (where V is the filament volume). That is, in general terms, the rate of change in MinD concentration as a function of time reads, \(d{\rho }_{MinD}/dt=f\left({\rho }_{MinD}\right)-\mu \cdot {\rho }_{MinD}-\left(\log \left(2\right)/\tau \right){\rho }_{MinD}=f\left({\rho }_{MinD}\right)-\alpha {\rho }_{MinD}\) where α is a constant (τ being the replication time) and where we used the fact that filaments increase their body mass exponentially32. Consequently, since our data indicates that dρMinD/dt ~0, i.e, \({\rho }_{MinD} \sim {{\rm{const.}}}\) (see Supplementary Fig. 3), it results that \(f\left({\rho }_{MinD}\right) \sim \alpha {\rho }_{MinD} \sim {{\rm{const.}}}\): the production remains constant with the increasing body mass. We note that this methodological approach of using the fluorescent signal to measure protein concentration has been used in the past and confirmed with Western Blot analyses49.
MinD intensity profiles obtained from MicrobeJ were analyzed to perform Fourier analysis of MinD oscillations in both space and time. These profiles were also used to compute the Pearson correlation between MinD intensity and curvature profiles using Mathematica. High- and low-curvature regions were defined using a curvature threshold of 0.5 μm−1. High-curvature regions were defined from the left valley of the first peak, where curvature exceeded 0.5 μm−1 to the right valley of the last peak, exceeding this threshold (Supplementary Fig. 4B). Low-curvature regions were defined from the filament poles to these valleys. The correlation function was applied separately to each of these three regions (i.e., low curvature–high curvature–low curvature) for every filament.
To analyze the relationship between MinD intensity and filament curvature, MinD intensity profiles were processed to determine the MinD envelope (Supplementary Fig. 4C). This approach avoided the influence of MinD pulse valleys. The maxima of MinD pulses were identified using the ‘FindPeaks’ function in Mathematica, followed by interpolation of these points and discretization to recover the original number of points. This process yielded a new MinD profile, representing the envelope of the original MinD signal. At each point along the filament’s arc length, s, the MinD intensity was normalized as:
Curvature profiles were processed similarly, but without normalization. Curvature values were then grouped into the following bins:[0, 0.05), [0.05, 0.15), [0.15, 0.25), [0.25, 0.35), [0.35, 0.45), [0.45, 1), and [1, ∞). These bins, along with their corresponding MinDnorm. values, were fitted to the function:
where κ is the curvature. The last bin was excluded from the fit as MinDnorm. saturates and does not follow Eqn (1). From the fit, we obtained a = 0.69 ± 0.07 and b = 0.08 ± 0.04 with R2 = 0.9.
For agarose patterns, the last two bins were absent, as filaments did not reach those curvature values. Fitting the data to the same equation yielded apat. = 1.00 ± 0.98 and bpat. = 0.15 ± 0.13 with R2 = 0.99.
FRAP experiments
The bacterial strain was grown, and agarose pads were prepared as described in “Strain and growth conditions” and “Agarose pads and patterned growth substrates”, respectively. Images were acquired using a ZEISS LSM 980 super-resolution confocal microscope equipped with a PLAN-Apochromat 63x/1.4 DIC M27-Oil lens, an Airyscan detector, and ZEN software. All experiments were performed at 37 °C using the built-in incubation system.
Images were captured every 3 s for a total duration of 2.5 min. A 488 nm laser was used at 0.5% power (nominal output power 500 mW) with a 57 μm pinhole. A master gain of 738 V and 304 V and digital offsets of −24116 and 0 were applied for the fluorescent and bright-field channels, respectively. Photobleaching was initiated after the fifth frame (15 s) by increasing the 488 nm laser power to 50% for 10 ms. The photobleached area was manually drawn using the polygonal selection tool to ensure it encompassed an entire MinD pulse. Recovery of fluorescence was monitored for a duration of 2.5 min, as photobleaching from repeated imaging precluded longer observations.
Photobleaching was performed on regular-length cells and filaments under various conditions. For regular-length cells, two scenarios were considered (Supplementary Fig. 7): when MinD was fully concentrated at one bacterial pole, with no fluorescence at the opposite pole (Supplementary Fig. 7A), and when fluorescence was transitioning between the poles, with fluorescence visible at both poles (Supplementary Fig. 7B). For filaments showing three MinD pulses, photobleaching targeted the central pulse when MinD fluorescence was fully concentrated there, with no fluorescence at the poles (Supplementary Fig. 7C). For filaments showing more than three pulses, photobleaching was performed on a pulse located away from the poles (Supplementary Fig. 7D).
To assess MinD diffusion in filaments, photobleaching areas were drawn to cover approximately half of the filament. Filaments were categorized as either straight (maximum curvature < 0.5 μm−1) or buckled (maximum curvature > 0.5 μm−1). The coordinate origin was set at the edge of the photobleached area closest to the filament’s midpoint. In straight filaments, the analyzed pulse was the second pulse from this origin, whereas in buckled filaments, it was the first pulse located after the buckling region. Typically, this pulse was the second photobleached pulse, but depending on the filament’s geometry, it could also be the first or third pulse (Fig. 3 A & B, orange and blue dots).
The analysis of the selected pulse was conducted in Fiji by defining a circular region of interest (ROI) and plotting the Z-axis profile. Recovery time was defined as the first frame in which the fluorescence intensity of the photobleached pulse recovered to at least 30% of its pre-photobleaching intensity. The MinD oscillation period was measured by analyzing pulses in the non-photobleached regions of the filament and performing a Fourier transform to determine the oscillation period.
Recovery time (tr) was normalized by dividing it by the characteristic MinD oscillation period (T) for each filament, yielding a dimensionless recovery time. The distance (d) between the analyzed pulse and the first non-photobleached pulse (Fig. 3A, blue and orange dots) was measured using the segmented line tool in Fiji and normalized by the characteristic wavelength of each filament (λ). Recovery velocity was computed as the ratio of the dimensionless distance to the dimensionless recovery time, yielding a dimensionless velocity (vr/c). This methodology allowed for the comparison of MinD dynamics between straight and buckled filaments, highlighting how mechanical deformation influences protein diffusion and recovery behavior.
Microfluidic experiments
A microfluidic CellASIC ONIX2 BA04A03 system was used to study filament division after removal of the filamentation stressor. For each experiment, a single row of the device was utilized, pre-emptying wells 1, 2, 6, 7, and 8. E. coli cells were grown as described in “Strain and growth conditions”, and 50 μl of culture was transferred to well 8 of the microfluidic device. Well 2 was filled with 200 μl of M9-supplemented media, while well 1 contained the same media supplemented with aztreonam at concentrations ranging from 1 to 10 μg/ml.
To operate the device, a custom sequence was used: 1. Media from well 2 was flowed at 5 psi for 5 min to pre-clean the device. 2. Cells were loaded by flowing E. coli (well 8) at 2 psi. 3. After loading, media from well 2 was flowed at 1 psi for 30 min to 1 h to stabilize the cells. 4. Media containing aztreonam (well 1) was then flowed at 1 psi for 3 to 5 h. 5. Finally, the flow was switched back to aztreonam-free media from well 2 for 3 h to observe filament division points under the microscopy system described in “Microscope settings”.
Division points were manually identified using the phase-contrast channel of microscopy time-lapses. Curvature at each division point was calculated for the frame immediately preceding division using the code described in ‘Image and data analysis’. The curvature value was averaged over the three nearest points along the filament to the division point. The probability density function (PDF) of curvatures at division points was obtained using MATLAB84 (‘histogram’ function using the ‘pdf’ normalization option). To compare this with the overall filament curvature, a second PDF was calculated for all curvatures of a filament. In this case, curvatures were smoothed using the MATLAB ‘smooth’ function. For this analysis, only filaments with a maximum curvature > 0.5 μm−1 and a minimum length of 8 μm were considered.
To characterize DNA distribution within the filaments, the same microfluidic setup and loading protocol were followed. After cell loading, just media was flowed at 1 psi for 30 min. Subsequently: 1. Media containing aztreonam was flowed at 1 psi for 3 h. 2. Aztreonam media supplemented with SYTOX Orange (Invitrogen) was flowed for 30 min to 1 h to stain DNA. 3. Same media without SYTOX was then flowed at 5 psi for 5 min to remove excess background fluorescence. 4. Time-lapses were acquired for 5 min (10 s interframe time) using phase-contrast and fluorescence channels with the microscopy set-up described in “Microscope settings”. Correlation between SYTOX intensity and filament curvature profiles was calculated using a customized MATLAB script, following the same protocol applied for MinD (see “Microscope settings”). The poles of filaments (1 μm from each end) were excluded from the analysis due to chromosome-free regions at the cell poles, consistent with previous findings62.
To characterize FtsZ distribution within the filaments, the same microfluidic setup and loading protocol were followed using E. coli MG1655 (Ptrc99A-mut2gfp::ftsZ). After cell loading: 1. Media was flowed at 1 psi for 30 min. 2. Aztreonam media was flowed for 3 h to promote filamentation. 3. Media without aztreonam was then flowed at 1 psi for 5 h. Time-lapses were acquired with 3 min interframe time using phase-contrast and fluorescence channels with the microscopy set-up described in “Microscope settings”. To characterize the temporal relationship between FtsZ and curvature, we calculated the cross-correlation between these two magnitudes for each point along the filament using the ‘xcorr’ function in Matlab with the ‘coef’ normalization. Prior to cross-correlation analysis, both the FtsZ and curvature signals were interpolated to have a common axis across the filament’s body. Then, both signals were binarized. The FtsZ signal was binarized using the ‘findpeaks’ function in Matlab. Peaks were identified by setting the ‘MinPeakHeight’ parameter to the mean fluorescent signal intensity of the entire filament at a given time point and the ‘MinPeakProminence’ parameter to 200. The values corresponding to these identified peaks were set to 1, while all other fluorescent values were set to 0. For the curvature signal, values greater than or equal to 0.5 were set to 1, and values below 0.5 were set to 0. Following binarization, the ‘xcorr’ function with ‘coef’ normalization was applied to determine the lag time between the appearance of an FtsZ ring (represented by a value of 1 in the binarized signal) and the occurrence of a curvature value ≥ 0.5 (also represented by a value of 1 in the binarized signal) for every spatial point along each filament. Since the delay difficults the direct correlation between Z-ring location and curvature value due to filament growth during that period, the position along the filament was normalized by its length at a given time (Fig. 5D).
Critical loading force
The same filaments used to analyze the DNA distribution in the microfluidic device were also employed to determine the critical buckling length. Time-lapse movies were acquired at 5 min intervals with an interframe time of 10 s. These time-lapses were projected into a single image that represents the average intensity of the signal of all frames using the ‘z project function’ in Fiji, using the average intensity option. Then, this projected image was thresholded to isolate the filaments, resulting in a binary mask. The MicrobeJ plugin was used to process this mask and obtain the medial axis, as described in “Image and data analysis”.
Using the medial axis data, custom-written Mathematica code was employed to calculate the maximum curvature of each filament profile. These curvature values were then compared to the corresponding filament lengths, which were also obtained using MicrobeJ. The critical buckling load was calculated using the Euler formula:
where k = 0.25 is the effective length factor for the filament85. The length of the filaments at which buckling started, \(\hat{L}\), was chosen to be 10 μm (see Supplementary Fig. 1). EI represents the flexural rigidity of the filament, where E is the Young’s modulus and I is the second moment of inertia. The Young’s modulus, E, depends on the bacterial strain used. For the Dh5α strain used in our experiments, E is reported to range between 2 and 3 MPa44. For a thin hollow cylinder with radius r and wall thickness b, the second moment of inertia, I, can be expressed as I = πr3b86. Previous studies have reported a wall thickness of b = 4 nm for E. coli19,87. Using a radius of r = 0.5 μm88, we calculate a flexural rigidity of EI = 4.7 ⋅ 10−21 N ⋅ m2. Substituting these values into Eqn. (2), we obtain a critical loading force of F = 7.4 nN.
Mechanical model
E. coli filaments were modeled as a 2D chain of initial length L0 = 50 μm, defined by N = 161 nodes. Each node, i, was represented by Cartesian coordinates ri = (xi, yi) using a custom code written in R89. The characteristic distance between consecutive nodes was \({l}_{c}=\frac{{L}_{0}}{N-1}=0.31\) μm. This resolution was chosen to ensure approximately six nodes per 0.3 μm radius, corresponding to a maximum curvature of 3 μm−1. The chain length L was selected to match the filament lengths observed in the experimental data. The initial coordinates for the chain were set as \({{{\bf{r}}}}_{i}=\left(i-1,i-1\right)\), with i ranging from 0 to N − 1. A random perturbation was applied to the yi coordinates, such that \({{{\bf{r}}}}_{i}=\left(i-1,i-1+{{\mathcal{N}}}\left(0,0.5\right)\right)\), where \({{\mathcal{N}}}\left(0,0.5\right)\) denotes a normal distribution with 0 mean and 0.5 variance.
Two active forces were incorporated into the model: elastic and bending. For the elastic term, nodes were connected by linear springs with an elastic constant K and an equilibrium length \({l}_{0}\left(t\right)\) that depends on time to simulate filament growth. The elastic energy for a node i, \({E}_{{{{\rm{elastic}}}}_{i}}\), is given by:
where Ri−1 = ri−1 − ri and Ri+1 = ri+1 − ri.
The bending energy, which promotes filament flattening, depends on the angle formed by three consecutive nodes. Normalized vectors \({\hat{{{\bf{R}}}}}_{j}={{{\bf{R}}}}_{j}/| {{{\bf{R}}}}_{j}|\) were used to ensure that the bending energy remains independent of the distances between nodes:
Thus, the bending energy reaches a minimum when the angle formed by three consecutive nodes is 180° and its maximum when the angle is 0°. To perform the simulations we implemented a dimensionless form of the energy, \(\tilde{{E}_{i}}={E}_{i}/{E}_{c}\), defined as a function of the characteristic energy \({E}_{c}=K{l}_{c}^{2}\):
where \(\tilde{\Omega }=\frac{\Omega }{K{l}_{c}^{2}}\).
The equation for the dynamics of a node reads \(m{\ddot{{{\bf{r}}}}}_{i}=-\nabla {E}_{i}-\gamma {\dot{{{\bf{r}}}}}_{i}\), m being the “mass” of a node and γ a friction coefficient (energy dissipation). Disregarding the inertia term (low Reynolds number) we finally obtain that \({\dot{{{\bf{r}}}}}_{i}=-\frac{1}{\gamma }\nabla {E}_{i}\) or, as a function of dimensionless units: \({\dot{\tilde{{{\bf{r}}}}}}_{i}=-\tilde{\nabla }\tilde{{E}_{i}}\); where a characteristic time tc = γ/K, linked to the dissipation of the elastic energy, was defined. Filament growth was implemented by increasing the preferred length between nodes, \({l}_{0}\left(t\right)\), as a function of time exponentially: \({l}_{0}\left(t\right)={l}_{0}\cdot {2}^{t/\tau }\), τ being the doubling time. In our simulations, the parameters shown in Table 1 were used.
The curvature at node i was defined as the inverse of the radius of the circle passing through nodes i −1, i, and i + 1, resulting in N − 2 curvature points. Statistics were generated by conducting 100 simulations with random seeds to initialize the perturbation of the y-coordinates. Simulation results were registered every 10 s to align with the experimental time-lapse interval. In simulations, to properly compare with the experiments, we implemented a pair-wise moving average that connects the middle points of adjacent springs connecting a node. That is, given a set of M coordinates of a filament, \({{\mathcal{C}}}=\left({{{\bf{r}}}}_{1},{{{\bf{r}}}}_{2},{{{\bf{r}}}}_{3},\ldots ,,{{{\bf{r}}}}_{M-1},{{{\bf{r}}}}_{M}\right)\) we generate the set of M coordinates \(\hat{{{\mathcal{C}}}}=\left(\frac{{{{\bf{r}}}}_{1}+{{{\bf{r}}}}_{2}}{2},\frac{{{{\bf{r}}}}_{2}+{{{\bf{r}}}}_{3}}{2}, \ldots ,\frac{{{{\bf{r}}}}_{M-1}+{{{\bf{r}}}}_{M}}{2}\right)\).
Growth-induced elastic energy accumulation
Let us consider a chain of N (even) nodes as described in the previous section. For simplicity, the nodes are constrained to move along the x-axis, and only elastic energy is considered. Initially, the nodes are symmetrically positioned with respect to x = 0, such that: \({x}_{1}\left(t=0\right)=-{l}_{0}\left(0\right)\frac{\left(N-1\right)}{2}\), \({x}_{2}\left(t=0\right)=-{l}_{0}\left(0\right)\frac{\left(N-3\right)}{2}\), \({x}_{3}\left(t=0\right)=-{l}_{0}\left(0\right)\frac{\left(N-5\right)}{2}\), …, \({x}_{N}\left(t=0\right)=-{l}_{0}\left(0\right)\frac{\left(1-N\right)}{2}\). Consequently, \({\left\vert {x}_{i+1}-{x}_{i}\right\vert }_{t=0}={l}_{0}\left(0\right),\) indicating that the system is initially in mechanical equilibrium. The dynamics of the nodes is then described by \(\frac{d{{\bf{x}}}}{dt}=\frac{K}{\gamma }\left[{\mathbb{A}}\cdot {{\bf{x}}}+{\mathbb{B}}\right]\) with \({{\bf{x}}}={\left({x}_{1},{x}_{2}, \ldots ,{x}_{N-1},{x}_{N}\right)}^{T}\) and \({\mathbb{A}}\) and \({\mathbb{B}}\) read,
Due to the symmetry of the problem, the dimensionality of the system can be reduced to N/2, as \({x}_{1}\left(t\right)=-{x}_{N}\left(t\right)\), \({x}_{2}\left(t\right)=-{x}_{N-1}\left(t\right)\), and so on. This linear system can be solved, and it can be shown that \({\left\vert {x}_{i+1}-{x}_{i}\right\vert }_{t} > 0\,\forall \,t\). As expected, the exponential growth of the equilibrium length, \({l}_{0}\left(t\right)={l}_{0}{2}^{\frac{t}{\tau }}\), leads to increasing separation of the nodes over time.
Furthermore, elastic energy accumulates in the central springs compared to the springs at the ends of the chain. For instance, in the simplest case of N = 4 (one central spring surrounded by two springs at the ends), the ratio of the lengths between the end spring and the central spring over short times is given by:
This implies that, regardless of the spring constant K, the dissipation coefficient γ, or the doubling time τ, elastic energy preferentially accumulates in the central spring due to growth.
Min signaling model
A signaling model based on that of Walsh et al.61 was used for the simulations presented in Figs. 4 and Supplementary Fig. 8. The model captures the most fundamental processes about the biochemistry of MinD and MinE species (including olygomerization of species and the MinE-mediated hydrolysis of MinD-ATP) and their transport properties. The model considers that proteins can be in different cellular interacting compartments (cytoplasm bulk, cytoplasm vicinity of the membrane, and the membrane) of exponentially growing filaments. The equations describing the model read:
In these equations Dz and \({E}_{{2}_{z}}\) represent MinD and MinE species either in the bulk of the cytoplasm (z = b) or in the vicinity of the cell membrane (z = m), whereas d, d2, e2, and d2e2 represent species in the membrane (see61 for details). This model differs from that of Walsh and coworkers by explicitly defining different regions in the cell cytoplasm. The model equations are integrated in a one-dimensional exponentially growing spatial domain (doubling time td) using a custom-made Mathematica83 code. The parameter values used in our simulations are (see61):
The parameter ωbm is defined by the diffusivity properties of species in the cytoplasm and determines the rate to travel among different cytoplasmic regions as follows (see Table 2). The characteristic time related to a diffusive transport process is given by \(\tau={l}_{c}^{2}/{{\mathcal{D}}}\), where lc is a characteristic length scale and \({{\mathcal{D}}}\) is the diffusion constant. In our case, \({l}_{c} \sim (1{0}^{-1})\mu m\) (i.e., E. coli cell radius) and \({{\mathcal{D}}} \sim {{\mathcal{O}}}(1{0}^{1})\mu {m}^{2}\cdot {s}^{-1}\) (i.e., the cytoplasmic diffusivities of MinD and MinE). Thus, \(\tau \sim {{\mathcal{O}}}(1{0}^{-3})s\) and, consequently, we estimated the rate as ωbm ~1/τ ~103 s−1.
To simulate a specific degree of curvature in our in silico filaments within an experimentally observed arc-length of s = 3 μm, we used the fact that s = θ ⋅ R = θ/∣κ∣, where θ and R are the angle and the radius of curvature, respectively. By imposing that ∣κ∣ = 1 μm−1, it follows that the simulated bending angle is θ ≃ 172° (see inset of Fig. 4B). To determine how the membrane diffusivity of species changes due to this simulated bending, we used a logarithmic dependency. This relationship dates back to the foundational work by Saffman and Delbrück and has been experimentally and numerically validated by posterior studies that also linked curvature with protein accumulation in agreement with our data54. The theory states that in the low Reynolds number regime, the membrane diffusivity \({{{\mathcal{D}}}}_{m}\propto \log \left(R/a\right)\), where a is the characteristic protein size (\(a \sim {{\mathcal{O}}}(1{0}^{-3})\mu m\) in the case of Min proteins90,91). Consequently, the diffusivity if ∣κ∣ = 1 μm−1 compared to the diffusivity at ∣κ∣ = 10−1 μm−1 (a value experimentally determined to be residual and indistinguishable from zero) reads \({{{\mathcal{D}}}}_{m}{| }_{| \kappa |=1}\simeq 0.7\times {{{\mathcal{D}}}}_{m}{| }_{| \kappa |=0.1}\) (i.e., ~30% decrease).
As for how the MinD membrane binding rate, ωbd, changes as the curvature increases (∣κ∣ = 10−1 μm−1 ⟶ ∣κ∣ = 1 μm−1) we use values obtained from septins (membrane-binding nanoscopic proteins), see Fig. 2B of59. Following that study, in our case we implemented a two-fold change (i.e., 100% increase): ωbd∣∣κ∣=1 ≃ 2 × ωbd∣∣κ∣=0.1 (Supplementary Fig. 8A). We then tested different values of ωbd down to 10%, the minimum increase which did not disrupt MinD patterning (Supplementary Fig. 8B).
In our simulations we integrated the equations for 10 min in a non-growing filament until a steady standing-wave formed. From this point, we integrated the equations for td = 50 min in a growing filament. Membrane diffusion drops or MinD binding increases due to curvature (i.e., filament bending) were implemented after 20 min of growth. In the in silico kymographs shown in this study, MinD accounts for the total concentration of MinD regardless of its cellular localization (i.e., both cytoplasmic and membrane-bound) or the olygomerization state (i.e., monomeric, multimeric, or MinE bound)54,55,92,93.
Statistics and Reproducibility
Statistical analyses were performed using Mathematica and no statistical method was used to predetermine sample size. Filaments in contact with each other were discarded for the analysis of Fig. 1E (left) to analyze only those whose curvature appears naturally. For Figs. 2 and 5, only filaments that grow and present a standing wave MinD pattern were selected. For Fig. 3, those filaments whose MinD pattern was altered (or disrupted) after photobleaching were discarded.
Representative images of filaments (Fig. 1A, Fig. 2A, Fig. 2D insert, Fig. 3B, Fig. 5A, B, and Fig. 5F) are examples of the sample size indicated in their corresponding analyzed plots.
Low-curvature and high-curvature correlation data were compared using the Mann-Whitney U test in Fig. 2 (p = 0.002) and Fig. 5 (p = 0.001) with statistically significant results. Straight and buckled filaments in Fig. 3 (p = 0.5) were compared using the Mann-Whitney U test with no statistically significant results. p < 0.05 was considered to indicate a statistically significant difference. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw data generated in this study have been deposited in the OSF database under accession code https://osf.io/8FHXR/94.
Code availability
The simulation codes generated in this study have been deposited in the OSF database under accession code https://osf.io/8FHXR/94.
References
Bi, E. & Lutkenhaus, J. FtsZ ring structure associated with division in Escherichia coli. Nature 354, 161–164 (1991).
Hale, C. A. & De Boer, P. A. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88, 175–185 (1997).
Ghigo, J., Weiss, D. S., Chen, J. C., Yarrow, J. C. & Beckwith, J. Localization of FtsL to the Escherichia coli septal ring. Mol. Microbiol. 31, 725–737 (1999).
Justice, S. S., Hunstad, D. A., Cegelski, L. & Hultgren, S. J. Morphological plasticity as a bacterial survival strategy. Nat. Rev. Microbiol. 6, 162–168 (2008).
Matuła, K. et al. Phenotypic plasticity of Escherichia coli upon exposure to physical stress induced by ZnO nanorods. Sci. Rep. 9, 8575 (2019).
O’Boyle, N., Connolly, J. P. R., Tucker, N. P. & Roe, A. J. Genomic plasticity of pathogenic Escherichia coli mediates D-serine tolerance via multiple adaptive mechanisms. Proc. Natl Acad. Sci. USA 117, 22484–22493 (2020).
Rolinson, G. N. Effect of -lactam antibiotics on bacterial cell growth rate. Microbiology 120, 317–323 (1980).
Domadia, P., Swarup, S., Bhunia, A., Sivaraman, J. & Dasgupta, D. Inhibition of bacterial cell division protein FtsZ by cinnamaldehyde. Biochem. Pharmacol. 74, 831–840 (2007).
Guru, A. et al. The cell division protein ZapE is targeted by the antibiotic aztreonam to induce cell filamentation in Escherichia coli. FEBS Lett. 597, 2931–2945 (2023).
Kjeldsen, T. S., Sommer, M. O. & Olsen, J. E. Extended spectrum β-lactamase-producing Escherichia coli forms filaments as an initial response to cefotaxime treatment. BMC Microbiol. 15, 63 (2015).
Wainwright, M., Canham, L. T., Al-Wajeeh, K. & Reeves, C. L. Morphological changes (including filamentation) in Escherichia coli grown under starvation conditions on silicon wafers and other surfaces. Lett. Appl. Microbiol. 29, 224–227 (1999).
Strahl, H. & Hamoen, L. W. Membrane potential is important for bacterial cell division. Proc. Natl Acad. Sci. 107, 12281–12286 (2010).
Jones, T., Gill, C. & McMullen, L. The behaviour of log phase Escherichia coli at temperatures that fluctuate about the minimum for growth. Lett. Appl. Microbiol. 39, 296–300 (2004).
Möller, J., Emge, P., Vizcarra, I. A., Kollmannsberger, P. & Vogel, V. Bacterial filamentation accelerates colonization of adhesive spots embedded in biopassive surfaces. N. J. Phys. 15, 125016 (2013).
Tran, T. D., Ali, M. A., Lee, D., Félix, M.-A. & Luallen, R. J. Bacterial filamentation as a mechanism for cell-to-cell spread within an animal host. Nat. Commun. 13, 693 (2022).
Söderström, B., Pittorino, M. J., Daley, D. O. & Duggin, I. G. Assembly dynamics of FtsZ and DamX during infection-related filamentation and division in uropathogenic e. coli. Nat. Commun. 13, 3648 (2022).
Justice, S. S., Hunstad, D. A., Seed, P. C. & Hultgren, S. J. Filamentation by Escherichia coli subverts innate defenses during urinary tract infection. Proc. Natl Acad. Sci. 103, 19884–19889 (2006).
Wong, F. et al. Mechanical strain sensing implicated in cell shape recovery in Escherichia coli. Nat. Microbiol. 2, 17115 (2017).
Amir, A., Babaeipour, F., McIntosh, D. B., Nelson, D. R. & Jun, S. Bending forces plastically deform growing bacterial cell walls. Proc. Natl Acad. Sci. 111, 5778–5783 (2014).
Caspi, Y. Deformation of filamentous Escherichia coli cells in a microfluidic device: A new technique to study cell mechanics. PLos One 9, e83775 (2014).
Gordon, V. D. & Wang, L. Bacterial mechanosensing: the force will be with you, always. J. Cell Sci. 132, jcs227694 (2019).
Shi, H., Bratton, B. P., Gitai, Z. & Huang, K. C. How to build a bacterial cell: MreB as the foreman of E. coli construction. Cell 172, 1294–1305 (2018).
Hussain, S. et al. MreB filaments align along greatest principal membrane curvature to orient cell wall synthesis. eLife 7, e32471 (2018).
Bramkamp, M. & Van Baarle, S. Division site selection in rod-shaped bacteria. Curr. Opin. Microbiol. 12, 683–688 (2009).
Taheri-Araghi, S. et al. Cell-size control and homeostasis in bacteria. Curr. Biol. 25, 385–391 (2015).
Karasz, D. C., Weaver, A. I., Buckley, D. H. & Wilhelm, R. C. Conditional filamentation as an adaptive trait of bacteria and its ecological significance in soils. Environ. Microbiol. 24, 1–17 (2022).
Ramm, B., Heermann, T. & Schwille, P. The E. coli MinCDE system in the regulation of protein patterns and gradients. Cell. Mol. Life Sci. 76, 4245–4273 (2019).
Rowlett, V. W. & Margolin, W. The Min system and other nucleoid-independent regulators of Z ring positioning. Front. Microbiol. 6, 478 (2015).
Lutkenhaus, J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu. Rev. Biochem. 76, 539–562 (2007).
Barrows, J. M. & Goley, E. D. FtsZ dynamics in bacterial division: what, how, and why? Curr. Opin. Cell Biol. 68, 163–172 (2021).
MacCready, J. S. & Vecchiarelli, A. G. In long bacterial cells, the Min system can act off-center. Mol. Microbiol. 109, 268–272 (2018).
Wehrens, M. et al. Size laws and division ring dynamics in filamentous Escherichia coli cells. Curr. Biol. 28, 972–979.e5 (2018).
Renner, L. D. & Weibel, D. B. Cardiolipin microdomains localize to negatively curved regions of Escherichia coli membranes. Proc. Natl Acad. Sci. 108, 6264–6269 (2011).
Varma, A., Huang, K. C. & Young, K. D. The Min System as a general cell geometry detection mechanism: Branch lengths in Y-Shaped Escherichia coli cells affect min oscillation patterns and division dynamics. J. Bacteriol. 190, 2106–2117 (2008).
Männik, J. et al. Robustness and accuracy of cell division in Escherichia coli in diverse cell shapes. Proc. Natl Acad. Sci. 109, 6957–6962 (2012).
Zieske, K., Schweizer, J. & Schwille, P. Surface topology assisted alignment of Min protein waves. FEBS Lett. 588, 2545–2549 (2014).
Rombouts, J., Elliott, J. & Erzberger, A. Forceful patterning: theoretical principles of mechanochemical pattern formation. EMBO Rep. 24, e57739 (2023).
Ramos-León, F. & Ramamurthi, K. S. How do spherical bacteria regulate cell division? Biochem. Soc. Trans. 53, 447–460 (2025).
Ultee, E., Ramijan, K., Dame, R. T., Briegel, A. & Claessen, D. Stress-induced adaptive morphogenesis in bacteria. In Advances in Microbial Physiology, vol. 74, 97–141 (Elsevier).
Pourahmad Jaktaji, R. & Pasand, S. Overexpression of SOS genes in ciprofloxacin-resistant Escherichia coli mutants. Gene 576, 115–118 (2016).
Khandige, S. et al. DamX Controls Reversible Cell Morphology Switching in Uropathogenic Escherichia coli. mBio 7, 10–1128 (2016).
Arends, S. J. R. & Weiss, D. S. Inhibiting cell division in Escherichia coli has little if any effect on gene expression. J. Bacteriol. 186, 880–884 (2004).
Jahnke, J., Terrell, J., Smith, A., Cheng, X. & Stratis-Cullum, D. Influences of adhesion variability on the “living” dynamics of filamentous bacteria in microfluidic channels. Molecules 21, 985 (2016).
Tuson, H. H. et al. Measuring the stiffness of bacterial cells from growth rates in hydrogels of tunable elasticity. Mol. Microbiol. 84, 874–891 (2012).
McMahon, S. G., Neu, J. C. & Chen, J. Kinking and buckling instability in growing bacterial chains. bioRxiv, https://doi.org/10.1101/2025.01.12.632655 (2025).
Bernhardt, T. G. & De Boer, P. A. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell 18, 555–564 (2005).
Tonthat, N. K. et al. Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check: Mechanism of SlmA-mediated nucleoid occlusion. EMBO J. 30, 154–164 (2011).
Tsang, M. & Bernhardt, T. G. A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Mol. Microbiol. 95, 925–944 (2015).
Parada, C. M. et al. Growth-dependent concentration gradient of the oscillating Min system in Escherichia coli. J. Cell Biol. 224, e202406107 (2025).
Ren, Z. et al. Robust and resource-optimal dynamic pattern formation of min proteins in vivo. Nat. Phys. 21, 1160–1169 (2025).
Genova, L. A. et al. Mechanical stress compromises multicomponent efflux complexes in bacteria. Proc. Natl Acad. Sci. USA 116, 25462–25467 (2019).
Ivanov, V. & Mizuuchi, K. Multiple modes of interconverting dynamic pattern formation by bacterial cell division proteins. Proc. Natl Acad. Sci. 107, 8071–8078 (2010).
Gov, N. S. Diffusion in curved fluid membranes. Phys. Rev. E 73, 041918 (2006).
Daniels, D. R. & Turner, M. S. Diffusion on membrane tubes: A highly discriminatory test of the Saffman-Delbrück theory. Langmuir 23, 6667–6670 (2007).
Domanov, Y. A. et al. Mobility in geometrically confined membranes. Proc. Natl Acad. Sci. 108, 12605–12610 (2011).
Klaus, C. J. S., Raghunathan, K., DiBenedetto, E. & Kenworthy, A. K. Analysis of diffusion in curved surfaces and its application to tubular membranes. Mol. Biol. Cell 27, 3937–3946 (2016).
Kusters, R., Paquay, S. & Storm, C. Confinement without boundaries: anisotropic diffusion on the surface of a cylinder. Soft Matter 11, 1054–1057 (2015).
Jin, R., Cao, R. & Baumgart, T. Curvature dependence of BAR protein membrane association and dissociation kinetics. Sci. Rep. 12, 7676, https://www.nature.com/articles/s41598-022-11221-9.
Shi, W. et al. Curvature sensing as an emergent property of multiscale assembly of septins. Proc. Natl Acad. Sci. 120, e2208253120.
Bauer, M., Godec, A. & Metzler, R. Diffusion of finite-size particles in two-dimensional channels with random wall configurations. Phys. Chem. Chem. Phys. 16, 6118–6128 (2014).
Walsh, J. C., Angstmann, C. N., Duggin, I. G. & Curmi, P. M. G. Molecular interactions of the min protein system reproduce spatiotemporal patterning in growing and dividing Escherichia coli cells. PLOS ONE 10, e0128148.
Raghunathan, S., Chimthanawala, A., Krishna, S., Vecchiarelli, A. G. & Badrinarayanan, A. Asymmetric chromosome segregation and cell division in DNA damage-induced bacterial filaments. Mol. Biol. Cell 31, 2920–2931 (2020).
Yaman, Y. I., Demir, E., Vetter, R. & Kocabas, A. Emergence of active nematics in chaining bacterial biofilms. Nat. Commun. 10, 2285 (2019).
Wu, F. et al. Cell boundary confinement sets the size and position of the E. coli chromosome. Curr. Biol. 29, 2131–2144.e4 (2019).
Di Ventura, B. et al. Chromosome segregation by the Escherichia coli Min system. Mol. Syst. Biol. 9, 686 (2013).
Wong, F., Garner, E. C. & Amir, A. Mechanics and dynamics of translocating MreB filaments on curved membranes. eLife 8, e40472 (2019).
Feddersen, H., Würthner, L., Frey, E. & Bramkamp, M. Dynamics of the Bacillus subtilis Min system. mBio 12, e00296–21 (2021).
Fu, M. et al. Mechanochemical feedback loop drives persistent motion of liposomes. Nat. Phys. 19, 1211–1218 (2023).
Burkart, T., Wigbers, M. C., Würthner, L. & Frey, E. Control of protein-based pattern formation via guiding cues. Nat. Rev. Phys. 4, 511–527 (2022).
Horton, M. R., Höfling, F., Rädler, J. O. & Franosch, T. Development of anomalous diffusion among crowding proteins. Soft Matter 6, 2648 (2010).
Tamemoto, N. & Noguchi, H. Reaction-diffusion waves coupled with membrane curvature. Soft Matter 17, 6589–6596 (2021).
Downing, B. P. B., Rutenberg, A. D., Touhami, A. & Jericho, M. Subcellular Min oscillations as a single-cell reporter of the action of polycations, protamine, and gentamicin on Escherichia coli. PLoS ONE 4, e7285 (2009).
Kanwa, N., Kohyama, S., Fröhlich, L., Desai, A. & Schwille, P. Mutual dependence between membrane phase separation and bacterial division protein dynamics in synthetic cell models. Angew. Chem. Int. Ed. 64, e202417800 (2025).
Margolin, W. Ftsz and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 6, 862–871 (2005).
Spahn, C. et al. Transertion and cell geometry organize the Escherichia coli nucleoid during rapid growth, bioRxiv, https://doi.org/10.1101/2023.10.16.562172 (2023).
Delaby, M. et al. Phenotypic plasticity in bacterial elongation among closely related species, bioRxiv, https://doi.org/10.1101/2024.11.07.622495 (2024).
Shih, Y.-L., Fu, X., King, G. F., Le, T. & Rothfield, L. Division site placement in E.coli: mutations that prevent formation of the MinE ring lead to loss of the normal midcell arrest of growth of polar MinD membrane domains. EMBO J. 21, 3347–3357 (2002).
Deter, H. S., Dies, M., Cameron, C. C., Butzin, N. C. & Buceta, J. A cell segmentation/tracking tool based on machine learning. In Rebollo, E. & Bosch, M. (eds.) Computer Optimized Microscopy: Methods and Protocols, Methods in Molecular Biology, 399–422 https://doi.org/10.1007/978-1-4939-9686-5_19 (Springer New York).
Design, N. Nazca design: An open-source framework for photonic IC design, https://nazca-design.org/.
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Arganda-Carreras, I. et al. Trainable Weka Segmentation: A machine learning tool for microscopy pixel classification. Bioinformatics 33, 2424–2426 (2017).
Ducret, A., Quardokus, E. M. & Brun, Y. V. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 1, 1–7 (2016).
Inc., W. R. Mathematica, Version 12.2. https://www.wolfram.com/mathematica, Champaign, IL, 2024.
Inc., T. M. Matlab version: 9.13.0 (r2022b) (2022). https://www.mathworks.com.
Jahnke, J. P. et al. “Living” dynamics of filamentous bacteria on an adherent surface under hydrodynamic exposure. Biointerphases 12, 02C410 (2017).
Wang, S., Arellano-Santoyo, H., Combs, P. A. & Shaevitz, J. W. Actin-like cytoskeleton filaments contribute to cell mechanics in bacteria. Proc. Natl Acad. Sci. 107, 9182–9185 (2010).
Gan, L., Chen, S. & Jensen, G. J. Molecular organization of gram-negative peptidoglycan. Proc. Natl Acad. Sci. 105, 18953–18957 (2008).
Riley, M. Correlates of smallest sizes for microorganisms. In Size Limits of Very Small Microorganisms: Proceedings of a Workshop, 21–25 (National Academies Press (US)) https://www.ncbi.nlm.nih.gov/books/NBK224751/.
R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria https://www.R-project.org/ (2021).
Erickson, H. P. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol. Procedures Online 11, 32–51.
Hayashi, I. Structural and functional studies of MinD ATPase: implications for the molecular recognition of the bacterial cell division apparatus. EMBO J. 20, 1819–1828.
Saffman, P. G. & Delbrück, M. Brownian motion in biological membranes. Proc. Natl Acad. Sci. USA 72, 3111–3113 (1975).
Aimon, S. et al. Membrane shape modulates transmembrane protein distribution. Dev. Cell 28, 212–218 (2014).
Nadal, M., Grobas, I. & Buceta, J. E. coli filament buckling modulates min patterning and cell division. https://doi.org/10.17605/OSF.IO/8FHXR (2025).
Mathelié-Guinlet, M., Asmar, A. T., Collet, J.-F. & Dufrêne, Y. F. Lipoprotein lpp regulates the mechanical properties of the E. coli cell envelope. Nat. Commun. 11, 1789 (2020).
Acknowledgements
We thank Dr. Shih for providing us with the pYLS68 plasmid and Prof. Ulrich Schwarz for his valuable comments during the preparation of this manuscript. We are also grateful to Enrique Navarro Raga from SCSIE at UV for his assistance with the FRAP assays and to Amadeu Griol from NTC for his support with the fabrication methodology. I.G. acknowledges funding from the Margarita Salas Fellowship (Ref. MS21-128) and the HORIZON-MSCA-2022-PF-01-01 program (grant no. 101107228). I.D., J.H., and A.M. acknowledge support from the European Commission through the CHIRALFORCE Pathfinder project (grant no. 101046961) and from the Conselleria de Educación, Universidades y Empleo under the NIRVANA grant (PROMETEO Program, CIPROM/2022/14). J.B. acknowledges support from grant PID2022-137436NB-I00 and the research network RED2022-134573-T funded by ‘Ministerio de Ciencia e Innovación’ (MCIN/AEI/10.13039/501100011033) and by ERDF ‘A way of making Europe’ by the E.U. Additional support to J.B. was provided by the E.U. COST action CA22153 ‘European Curvature and Biology Network’ (EuroCurvoBioNet).
Author information
Authors and Affiliations
Contributions
M.N.: experiments, data analysis, writing, reviewing and editing. L.G.: numerical simulations (mechanical model). I.D.: micro-fabrication and writing (methods). J.H. and A.M.: micro-fabrication. I.G.: experiments, data analysis, writing, reviewing, editing, conceptualization and supervision. J.B.: writing, reviewing, editing, data analysis, numerical simulations (signaling model), conceptualization, funding acquisition, and project supervision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Nadal, M., Guitou, L., Diez, I. et al. E. coli filament buckling modulates Min patterning and cell division. Nat Commun 16, 8193 (2025). https://doi.org/10.1038/s41467-025-63509-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-63509-9