Abstract
In eukaryotes, DNA N6-methyladenine (6mA) modification plays important roles in various cellular functions, such as chromatin dynamics, gene expression regulation, and DNA damage response. It remains largely unknown how eukaryotic DNA 6mA methyltransferases (MTases) recognize their substrates. Here, we reported the structures of DNA-bound eukaryotic 6mA MTase complexes. The MTA1 complex (MTA1c) in ciliates is composed of MTA1, MTA9 (or MTA9-B), p1 and p2 subunits. Cryo-electron microscopy structures of MTA1c–DNA complexes reveal that DNA lies on the surface of the MTA1–MTA9/9-B dimer and is clamped by the p1 N-terminal region. The target deoxyadenosine is flipped out of the DNA duplex and approaches the catalytic center. Unmethylated and hemi-methylated DNA substrates bind MTA1c with differential conformational dynamics. Our structural and biochemical studies shed light on the activation and substrate recognition of MTA1c and provide a framework for understanding the molecular mechanism of DNA 6mA modification in eukaryotes.
Similar content being viewed by others
Introduction
Epigenetic modifications of DNA regulate diverse cellular functions across all kingdoms of life, contributing to developmental processes and immune system regulation1. C5-methylcytosine is the most extensively studied and best-characterized DNA modification type in eukaryotes2. N6-methyladenine (6mA) modification of DNA was initially discovered in bacteria in the 1950s and is widespread in prokaryotes. Bacterial 6mA modification plays pivotal roles in the restriction-modification system, DNA replication, DNA damage response and transcriptional regulation3. Later, 6mA modification was found in ciliates and algae4,5,6, and more recently, it was widely detected in genomes of plants and metazoans, including mammals7,8,9,10,11,12,13,14,15. Despite its relatively lower abundance in multicellular organisms compared to prokaryotes and protists, 6mA has been implicated in the regulation of various biological processes, including gene transcription, chromatin structure, epigenetic inheritance, DNA damage repair and cell cycle1,16,17,18,19,20. Notably, mitochondrial DNA in mammalian cells exhibited a significantly higher level of 6mA compared to nuclear genomic DNA21,22. Elevated 6mA level at regions of stress-induced DNA double helix destabilization antagonizes SATB1 to regulate gene expression in early embryonic development23. In humans, changes of 6mA levels have been found in different types of cancer cell lines and the samples of tumor patients. For instance, gastric, liver, and triple-negative breast cancer samples have been reported to contain decreased 6mA levels, whereas glioblastoma stem cells and primary samples exhibit increased 6mA levels15,24,25. Targeting the 6mA regulator suppressed the growth of glioblastoma stem cells24. These findings underscore the importance of 6mA modification in tumorigenesis.
In prokaryotes, numerous methyltransferases (MTases) that mediate DNA methylation on N6-adenine have been identified. Biochemical and structural studies have revealed how prokaryotic MTases recognize DNA substrates and catalyze methyl transfer26. By contrast, eukaryotic 6mA MTases are less well characterized. Several potential DNA 6mA MTases have been reported, including TAMT-1 in Tetrahymena thermophila27, DAMT-1 and METL-9 in Caenorhabditis elegans8,28, and N6AMT1, METTL4 and METTL3–METTL14 complex in mammals15,22,29. TAMT-1, METL-9 and METTL4 preferentially mediate the methylation of single-stranded DNA (ssDNA)22,27,28. The METTL3–METTL14 complex, known for its MTase activity towards RNA, could modify N6-adenine in ssDNA and double-stranded (dsDNA) with unpaired regions around the target deoxyadenosine site in vitro29,30. Recently, Muir and Landweber labs identified and characterized a 6mA MTase complex, namely MTA1 complex (MTA1c), in ciliates Oxytricha trifallax and Tetrahymena thermophila, which exclusively acts on dsDNA31. 6mA was first detected in Tetrahymena in the 1970s4. 6mA occurs exclusively at ApT sites in linker DNA32,33,34,35 and possesses an essential function in positioning nucleosomes in Tetrahymena27. The characterized MTA1c is composed of MTA1 (also named AMT1), MTA9 (also named AMT7, or MTA9-B/AMT6), p1 and p2 subunits31,36, forming two distinct complexes—MTA1cMTA9 and MTA1cMTA9-B (Fig. 1a). MTA1, MTA9 and MTA9-B belong to the MT-A70-like MTase family, which also includes METTL3, METTL4, METTL14, DAMT-1 and TAMT-1 proteins. MTA1 contains key elements for catalysis, while MTA9 and MTA9-B are pseudo-enzymes. Both p1 and p2 possess a homeobox-like domain (HLD) with potential DNA-binding activity. MTA1c specifically methylates dsDNA at ApT dinucleotide sites, but not ssDNA or RNA31. 6mA reduced nucleosome occupancy on Oxytricha genomic DNAs in vitro, and a frameshift mutation in MTA1 resulted in 6mA reduction and sexual cycle failure31. MTA1 was independently characterized by another group as a key factor for 6mA deposition in Tetrahymena37. MTA1 loss influenced nucleosome distribution and gene expression and caused severe growth and developmental defects37. The structures of the MTA1c complex in the absence of DNA substrates have been recently reported38,39. However, the mechanism by which MTA1c interacts with and modifies DNA substrates remains unknown.
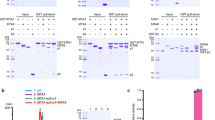
a Illustration of domain organization of Tetrahymena MTA1c components MTA1, MTA9/MTA9-B, p1 and p2. Abbreviations: CTD, C-terminal domain; CTR, C-terminal region; HLD, homeobox-like domain; HTH, helix-turn-helix; IR, insertion region; NH, N-terminal helices; NTD, N-terminal domain; NTR, N-terminal region. b Km and kcat values were calculated based on the MTase activities of MTA1cMTA9-B on two types of dsDNA in the presence of 20 μM SAM. For umDNA, the assay was performed in the presence of 1 μM MTA1cMTA9-B and 0–10 μM umDNA for 2 h at 30 °C. For hmDNA, the assay was performed in the presence of 0.125 μM MTA1c and 0–10 μM hmDNA for 0.5 h at 30 °C. Data are represented as mean ± SD from independent measurements (n = 3). c Kd values for MTA1cMTA9-B binding to DNA substrates were measured by fluorescence polarization experiments. 0–2 μM MTA1cMTA9-B was incubated with 1 nM 6-FAM-labeled umDNA or hmDNA. Data are represented as mean ± SD from independent measurements (n = 3). d Cryo-EM map of MTA1cMTA9-B in the apo state. e–g Cryo-EM structures of MTA1cMTA9-B in apo (e), SAH-bound (f), and SAM-bound (g) states. MTA1, MTA9-B, p1 and p2 are represented in a cartoon and colored green, cyan, slate and salmon, respectively. SAH and SAM are represented in magenta stick. h Cryo-EM structure of SAM-bound MTA1cMTA9-B with MTA1 D209A mutation. i Cryo-EM structure of SAM-bound MTA1cMTA9. MTA9 is colored dark cyan. j Structural comparison of MTA1c with MTA9 and MTA9-B. MTA9 and MTA9-B are aligned and zoomed in. Source data are provided as a Source Data file.
In this study, using cryogenic electron microscopy (cryo-EM), we have determined the structures of Tetrahymena MTA1c in complex with both unmethylated DNA (umDNA) and hemi-methylated (hmDNA) substrates, which are the first structures of substrate-bound eukaryotic DNA N6-adenine MTases and those of MT-A70-like family MTases. Both DNA substrates have extensive contacts with the substrate-binding loop of MTA1. They are clamped by the MTA1–MTA9/MTA9-B heterodimer and the p1 N-terminal region (NTR) that contains an HLD, while p2 HLD does not directly contact DNA. The target deoxyadenosine is flipped out of the DNA duplex and inserts into the catalytic site, poised for catalysis. We observed distinct local conformations of MTA1c upon binding to umDNA and hmDNA, together with biochemical analysis, offering insights into why MTA1c exhibits a clear preference for catalyzing hmDNA substrates. Our study thus establishes the mechanism by which MTA1c mediates DNA 6mA modification. Structural comparison with other MTases suggests a universal mechanism for the activation and substrate recognition of the MT-A70-like family DNA and RNA MTases.
Results
Biochemical characterization of Tetrahymena MTA1c
To study MTA1c, we expressed Tetrahymena MTA1c components in Escherichia coli. MTA1 and MTA9/MTA9-B were co-purified, and p1 and p2 were purified separately (Supplementary Fig. 1a). Because the yield of MTA9-B is better than that of MTA9 when coexpressed with MTA1, our biochemical analysis focused on MTA1c containing MTA9-B. The MTA1–MTA9-B heterodimer with p1 and p2 exhibited MTase activity on 27 bp dsDNA containing ApT dinucleotides. In contrast, MTA1–MTA9-B heterodimer alone or in combination with either p1 or p2 only had basal level activity (Supplementary Fig. 1b), consistent with previous findings31. Remarkably, the MTase activity of MTA1cMTA9-B dramatically increased in the presence of N6-hemi-methyladenine within the ApT motif (Supplementary Fig. 1b). Even the MTA1–MTA9-B–p1 subcomplex exhibited MTase activity on hmDNA at a level comparable to that of the intact MTA1c on umDNA (Supplementary Fig. 1b). Neither the subcomplex nor the holocomplex of MTA1cMTA9-B exhibited activity on DNA–RNA hybrid (Supplementary Fig. 1c).
To facilitate subsequent biochemical and structural investigations, MTA1cMTA9-B components were co-expressed and co-purified to obtain a homogenous sample (Supplementary Fig. 1d). Co-purified MTA1cMTA9-B displayed higher activity compared to the sample with mixed components, suggesting the properties of components are not optimal when expressed separately. We also measured the activity of MTA1cMTA9 on umDNA and hmDNA. Similar to MTA1cMTA9-B, MTA1cMTA9 prefers hmDNA as a substrate (Supplementary Fig. 1e, f). We further measured the Km and kcat of MTA1cMTA9-B for umDNA and hmDNA substrates. The Km value for MTA1cMTA9-B towards umDNA (1.49 µM) is slightly smaller than that of hmDNA (2.45 µM), but the kcat on hmDNA (1.11 min−1) was significantly higher than that for umDNA (0.0102 min−1) (Fig. 1b). We compared the DNA binding activities of MTA1cMTA9-B to two DNA molecules. Consistent with the enzymatic data, the DNA binding affinities, as shown by electrophoresis mobility shift assay (EMSA) and fluorescence polarization results, are highly similar (Fig. 1c and Supplementary Fig. 1g). The Kd values for MTA1cMTA9-B towards umDNA and hmDNA are 0.048 µM and 0.036 µM, respectively (Fig. 1c). Thus, factors other than DNA-binding affinity contribute to differences in catalytic activities of MTA1c on two types of DNA substrates.
We further applied multi-angle light scattering coupled with size exclusion chromatography (SEC–MALS) to analyze the interaction between MTA1cMTA9-B and two types of DNA substrates. The calculated molecular weight (Mw) of MTA1cMTA9-B is comparable to its theoretical Mw (Supplementary Fig. 1h and Supplementary Table 1). The addition of 27 bp umDNA or hmDNA to MTA1cMTA9-B caused an increase of 17 kDa (Supplementary Fig. 1h and Supplementary Table 1), which is nearly identical to the theoretical MW of the tested DNA, indicating MTA1cMTA9-B forms a 1:1 complex with umDNA/hmDNA in vitro.
Structures of Tetrahymena MTA1c
To obtain the structures of MTA1c in complex with DNA substrates, we systematically analyzed wild-type or catalytically inactive mutant D209A of MTA1cMTA9-B in the presence of dsDNA, with or without the methyl donor S-adenosyl methionine (SAM) or the product S-adenosyl homocysteine (SAH). We determined a series of MTA1cMTA9-B structures by single-particle cryo-EM analysis (Supplementary Figs. 2–6 and Supplementary Tables 2, 3). DNA densities were observed in cryo-EM maps only when wildtype MTA1cMTA9-B was briefly incubated with SAM in the presence of either umDNA or hmDNA. Based on these trials, we further determined the high-resolution structure of MTA1cMTA9 in the presence of hmDNA and SAM, as well as the structure of the complex in apo and umDNA-bound states (Supplementary Figs. 7, 8 and Supplementary Table 4). The cryo-EM maps of MTA1c alone in the absence or presence of the ligand SAM or SAH were resolved at overall resolutions ranging from 2.5 Å to 3.1 Å, and those of MTA1c in complexes with umDNA and hmDNA in the presence of SAM were refined to overall 2.9–3.3 Å resolutions.
To build the models of MTA1c in different states, the AlphaFold2 models of MTA1, MTA9, MTA9-B, p1 and p2 were docked into cryo-EM maps40, followed by manual building and real-space refinement (Fig. 1d–i, Supplementary Figs. 2–8 and Supplementary Tables 2–4). MTA1cMTA9 adopts a conformation closely resembling the previously reported structure39. The NTRs of MTA1 and MTA9/MTA9-B are largely disordered except for one helix in the former and two short helices in the latter (Fig. 1e, i). Both N-terminal helices (NH) are connected to their MTase domains by a loop region. We observed notable structural differences in the insertion regions (IRs) of MTA9 and MTA9-B, which directly contact their respective NHs (Fig. 1j). Compared to MTA9, MTA9-B possesses a shorter IR in the MTase domain, and the ordered region of MTA9-B IR in the structure displays a distinct fold from MTA9 IR (Fig. 1a, j). The second and third helices in MTA9 IR are replaced with one short helix in MTA9-B. A helix-turn-helix (HTH) motif in the middle region of p1 was modeled based on the electron density, but its NTR, which contains an HLD, and the long C-terminal region (CTR), are invisible. p2 can be divided into three parts: the N-terminal domain (NTD), the middle HLD and the C-terminal domain (CTD). In the absence of DNA substrates, the folded region of MTA1c adopts a seahorse-shaped conformation, encompassing a body formed by the MTA1–MTA9/MTA9-B heterodimer associated with p2 NTD and a head formed by MTA1 NH, p1 HTH domain and the remaining part of p2. Mutations in the interfaces of four subunits obviously decreased or even disrupted the MTase activity of MTA1cMTA9-B (Supplementary Fig. 9). Interestingly, mutations M21E/L25E and F42E of p2 NTD did not influence the complex formation but disrupted MTA1cMTA9-B activity, suggesting the hydrophobic interaction between p2 NTD and MTA1 is not crucial for the complex assembly but it can stabilize the active conformation of MTA1 catalytic site, which is consistent with previous finding38. The densities of SAH and SAM were observed in cryo-EM maps (Supplementary Figs. 3–8). SAM and SAH exclusively bind to MTA1 but not MTA9 or MTA9-B (Fig. 1f–i). SAM or SAH binding did not induce an overall conformational change in MTA1c, albeit slight local motions were observed.
hmDNA binding by MTA1cMTA9
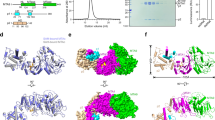
We obtained several cryo-EM maps of DNA-bound MTA1c, in which the local resolutions of DNA substrates vary. The DNA density in the cryo-EM map of MTA1cMTA9–umDNA–SAM complex is weak, suggesting the dynamic interaction between them (Supplementary Figs. 7 and 10a). However, we observed clear DNA density in the cryo-EM map of MTA1cMTA9–hmDNA–SAM complex (Fig. 2a and Supplementary Fig. 8), which indicates the more specific binding of hmDNA to MTA1cMTA9. We unequivocally built the model of the central region of hmDNA in this map (Fig. 2b). In this structure, the DNA molecule is positioned on the surface of the MTA1–MTA9 heterodimer and adjacent to their interface (Fig. 2c). p1 NTR, which includes a DNA-binding domain HLD, clamps the hmDNA together with the MTA1–MTA9 heterodimer. Although p2 HLD was suggested to have DNA-binding ability, no direct interaction was observed between p2 HLD and hmDNA. This is consistent with our EMSA results, which showed no binding between p2 and DNA (Supplementary Fig. 1i). The hmDNA is distorted and exhibits a ~ 30° bending in the central target region (Fig. 2b). The MTA1 gate loop 1 (β2/α2 loop, also named fence or active site loop 1) and the interface loop, which we renamed the substrate-binding loop due to its extensive contacts with the DNA substrate, locate on the two sides of the hmDNA duplex (Fig. 2d). The gate loop 1 of MTA1 binds to the major groove of hmDNA, while the substrate-binding loop contacts the minor groove of the distorted DNA double helix, which exhibits a much wider minor groove compared to a canonical B-form DNA duplex (Fig. 2d).
a Cryo-EM map of the MTA1cMTA9–hmDNA–SAM complex. Two strands of hmDNA are colored wheat and gray, respectively. b Cryo-EM density of hmDNA in the MTA1cMTA9–hmDNA–SAM complex is shown in transparent surface. The structure of hmDNA is shown in cartoon and a stick. The flipped residues dA0 and dT0’ are labeled. c Overall structure of MTA1c MTA9 in complex with hmDNA and SAM. d The substrate-binding loop and gate loop 1 of MTA1 contact and distort the hmDNA duplex. Their structures and densities, as well as those of hmDNA, are shown. e dA0 binds to the catalytic site in MTA1. The target dA0 is shown in wheat stick. The residues involved in interacting with dA0 are highlighted in yellow stick representation. f His291 in the interface loop of MTA1 and Gln380 in MTA9 insert into the hmDNA duplex. g Arg221 in the gate loop1 of MTA1 inserts into hmDNA and contacts mdA-1’ (6mA). h Phe293 in the substrate-binding loop of MTA1 and His379 in MTA9 contact hmDNA. i MTA1cMTA9–hmDNA interface. Residues in MTA1 and MTA9 involved in hmDNA binding are highlighted in yellow stick representation. j Relative MTase activity of MTA1c mutants on hmDNA and umDNA. For umDNA, the assay was performed in the presence of 1 μM MTA1cMTA9-B or its mutants for 2 h at 30 °C. For hmDNA, the assay was performed in the presence of 0.25 μM MTA1cMTA9-B or its mutants for 0.5 h at 30 °C. Data are represented as mean ± SD from independent measurements (n = 3). The numbering of corresponding residues in MTA9 is indicated in parentheses. Source data are provided as a Source Data file.
Two base pairs around the target site of the hmDNA duplex are disrupted, with the deoxyadenosine for methylation (dA0) in the target strand flipped out and inserted into the catalytic pocket surrounded by the gate loop 1, the gate loop 2 (β6/α4 loop) and the β3-β5 strands (Fig. 2e). Within the catalytic pocket, dA0 engages with residues P210 and W212 in the DPPW motif of the gate loop 1 and contacts residues L214, V223, I225 and Y227 in the gate loop 1, I254 in the β3/α3 loop, and S332 and K334 from the gate loop 2. In this configuration, the N6 atom of dA0 forms a hydrogen bond with the catalytic residue D209 in the DPPW motif and establishes contact with the methyl group of SAM, positioning it for a nucleophilic attack during the methyl transfer reaction. Disruption of the dA0–dT0’ base pair is caused by the wedging of H291 in the substrate-binding loop of MTA1 and Q380 in the β5/β6 loop of MTA9 into the duplex (Fig. 2f). MTA1 H291 packs against the nucleotides at the − 1 and + 1 positions (dG-1 and dT1) in the target strand of the DNA duplex via π–π–π stacking and hydrogen bonds. This residue also directly contacts the methyl group of N6-methyl-deoxyadenosine (mdA-1’) in the complementary strand. This specific interaction likely stabilizes the flipped conformation and facilitates catalysis. Furthermore, MTA9 Q380 anchors mdA-1’ and dC1’ in the complementary strand. The base pair formed by mdA-1’ in the complementary strand and dT1 in the target strand is also disrupted. Residue R221 in the gate loop 1 of MTA1 inserts into the hmDNA duplex and separates mdA-1’ and dC-2’ (Fig. 2g). The guanidinium group of R221 is enwrapped by dT1, mdA-1’ and dC-2’, involving π-π stacking, hydrogen bond, and electrostatic interactions. The insertion of R221 and the repositioning of mdA-1’ extrude dT0’ from base stacking in the hmDNA duplex (Fig. 2h). The residue H379 in the β5/β6 loop of MTA9 stacks with dT0’, and together with the ribose of dG2, sandwiches the residue F293 in the substrate-binding loop of MTA1 via lone pair–π–π stacking.
The substrate-binding loop and the gate loops 1/2 of MTA1 and the β5/β6 loop of MTA9 have extensive interactions with hmDNA. Besides the residues mentioned above, the basic residues K286 and K289 from the substrate-binding loop of MTA1 and R381 in the β5/α5 loop of MTA9 contact the phosphate backbone of the DNA complementary strand via ion-pairing interactions (Fig. 2i). In addition, the basic residue K280 in the β4 strand of MTA1 anchors the phosphate of the methylation target dA0. Residue R328 in the gate loop 2 of MTA1 packs with the ribose of dG-1 in the target strand. Residues R366 in the β5 strand and K374 in the β5/β6 loop of MTA9 contact the target strand on the other side of hmDNA. Furthermore, residues T282, N284 and Q296 from the substrate-binding loop and Q331 from the gate loop 2 in MTA1 contribute to hydrogen bonds with the hmDNA substrate.
To assess the significance of residues in the DNA-binding interface of MTA1 and MTA9, we mutated these residues in MTA1 and the corresponding residues in MTA9-B. These mutations had varying effects on the DNA binding ability of MTA1c in EMSA (Supplementary Fig. 11). The mutations of positively charged residues, such as R221A, K280E, K286A/K289E in MTA1, showed the most severely reduced DNA binding ability. The vast majority of mutants either exhibited drastically reduced or completely lost MTase activity on hmDNA (Fig. 2j), underscoring the vital role of these residues in hmDNA binding and catalysis. These mutants also displayed a deficiency in the methylation of umDNA (Fig. 2j), indicating MTA1c uses the same sites for umDNA recognition and methylation. Mutation of H291 in MTA1 to phenylalanine (H291F) could still support π–π–π stacking but significantly eliminated SAH production (Fig. 2j), suggesting the hydrogen bonding interaction contributed by H291 plays an indispensable role in maintaining the optimal conformation for MTA1c activity on DNA substrates.
hmDNA recognition by p1 NTR in MTA1cMTA9
In the structure of hmDNA-bound MTA1c, we observed nearly complete NTR of p1 (Figs. 2a, 3a). p1 NTR contains a helical domain flanked by the N-terminal tail (N-tail) and a linker connected to its HTH domain. On the top side, the N-tail of p1 folds back to contact its HTH (Fig. 3b). It spans along the MTase domain of MTA1 and interacts with p2 NTD. Two serine residues, Ser13 and Ser15, contribute to hydrogen bonds to R358 and E371 in MTA1 (Fig. 3b). On the bottom side, the α5/α6 loop of p1 NTR forms interaction with the β5/β6 loop of MTA9, involving the residues D110 and N111 in p1 and E376, R378 and H379 in MTA9 (Fig. 3c). Deletion of the N-tail in p1 or mutation of corresponding interface residues in MTA9-B abolished the MTase activity of MTA1cMTA9-B on both hmDNA and umDNA (Fig. 3d), supporting the interaction of p1 NTR with MTA1 and MTA9/MTA9-B are crucial for MTA1c function.
a Structure of p1 NTR in MTA1cMTA9–hmDNA–SAM complex. p1 NTR clamps hmDNA together with MTA1 and MTA9-B. b p1 N-tail interacts with MTA1 and p2. MTA1 is shown in a cartoon and transparent surface. p1 and p2 are represented in a cartoon. The interacting residues in MTA1 and p1 are shown in stick. c p1 NTR interacts with MTA9. d Relative MTase activity of MTA1c mutants on hmDNA and umDNA. Data are represented as mean ± SD from independent measurements (n = 3). e p1 NTR interacts with hmDNA. p1 NTR and hmDNA are shown in a cartoon and surface, respectively. f, g The interface between p1 NTR and hmDNA. The residues in p1 NTR that are involved in hmDNA-binding are highlighted in yellow stick representation. h Kd values for p1 NTR binding to DNA substrates were measured by fluorescence polarization experiments. 0–2 μM p1 (1–150) was incubated with 1 nM 6-FAM-labeled umDNA or hmDNA. Data are represented as mean ± SD from independent measurements (n = 3). The numbering of corresponding residues in MTA9 is indicated in parentheses. Source data are provided as a Source Data file.
The helical domain in the p1 NTR, which is composed of seven helices and includes the HLD, directly recognizes hmDNA (Fig. 3e). Like the DNA binding mode of the canonical homeobox domain, the α3 helix of the p1 HLD, which is the DNA-recognition helix, embeds into the major groove of the hmDNA duplex (Fig. 3f). The residue M64 contacts the base of dA-3 in the target strand, and the residues Q61 and R68 interacts with the phosphate backbone of dA-3–dT-5 through hydrogen bonds and ion pairs. The residue Y63 makes contacts with mdA-1’ and dC-2’ in the complementary strand, involving both hydrogen bonds and hydrophobic interactions. In addition, the α2 helix of p1 HLD engages with the backbone of dC-2’–dA-4’ positions in the complementary strand (Fig. 3f). The residues K44, W45, and K46 in the α2 helix of the p1 HLD are involved in DNA binding. The region following the HLD in p1 helical domain covers the surface and anchors the major and minor grooves of central hmDNA duplex (Fig. 3g). In this region of p1 NTR, the residues Q73 and S74 in the short α3/α4 loop, K101, N103 and N107 in the long α5/α6 loop, and K114, R117 and K118 in the α6 helix contact the phosphate backbone of nucleotides dT-5, dA5 and dA6 in the target strand and mdA-1’–dT2’ in the complementary strand of hmDNA.
To confirm DNA binding by p1 NTR, we used EMSA and fluorescence polarization experiments to test its interaction with hmDNA and umDNA. p1 NTR directly binds both types of DNA with similar affinity in vitro (Fig. 3h and Supplementary Fig. 1j). The measured Kd values for p1 NTR towards umDNA and hmDNA are 0.037 µM and 0.035 µM, respectively, which are comparable to those for the MTA1cMTA9-B holocomplex (Figs. 1c and 3h), indicating p1 NTR plays a dominant role in DNA binding. Based on our structural information, we mutated residues K44 and K46 in the α2 helix, M64 and R68 in the α3 helix, and K114 and K117 in the α6 helix of p1 NTR. All three double mutations reduced DNA binding to MTA1cMTA9-B and drastically attenuated MTA1cMTA9-B activity on both hmDNA and umDNA (Fig. 3d and Supplementary Fig. 11).
Differential binding of MTA1c to umDNA and hmDNA
We reconstituted the cryo-EM maps of MTA1cMTA9-B with SAM in the presence of either umDNA or hmDNA (Supplementary Figs. 5, 6). The local resolutions of DNA substrates in these maps are lower than that of the MTA1cMTA9–hmDNA complex. We docked the models of MTA1cMTA9-B subunits and DNA substrates into these two cryo-EM maps and manually adjusted the models based on the density, followed by real-space refinement. Superimposition of the structures of hmDNA-bound MTA1cMTA9 and MTA1cMTA9-B revealed that the hmDNA adopts a similar conformation to interact with two paralog complexes (Supplementary Fig. 10b, c). The overall binding modes of umDNA and hmDNA substrates to MTA1cMTA9-B exhibit remarkable similarity (Fig. 4a–d), indicating that MTA1c utilizes the same structural elements to bind umDNA and hmDNA and supporting that mutations in the hmDNA-binding interface also disturb the activity of MTA1c on umDNA. Although MTA1cMTA9-B adopts overall similar modes to contact hmDNA and umDNA substrates, conformational differences between the two complexes are observed. Notably, p1 NTR in the hmDNA-bound MTA1cMTA9-B has a rotation accompanying DNA reconfiguration compared to that of umDNA-bound MTA1cMTA9-B, featuring the closer contact of the α3 helix of p1 HLD with hmDNA (Fig. 4e, f). Compared to the umDNA, the hmDNA is positioned closer to MTA1cMTA9-B (Fig. 4f). In addition, the density for the p1 N-tail is not observed, and the gate loop 1 of MTA1 is disordered in umDNA-bound MTA1cMTA9-B.
a, b Cryo-EM map (a) and structural model (b) of MTA1cMTA9-B–umDNA–SAM complex. c, d Cryo-EM map (c) and structural model (d) of MTA1cMTA9-B–hmDNA–SAM complex. e Structures of MTA1cMTA9-B in complexes with umDNA and hmDNA were superimposed on the MTA1 subunit. The MTA1cMTA9-B–umDNA complex is colored dark gray, while the MTA1cMTA9-B–umDNA complex is colored slate. The pink dash represents the relative movement of p1 NTR. f Conformational difference of DNA molecules in the structural models of MTA1cMTA9-B–umDNA/hmDNA complexes. The pink dash represents the relative motion of two DNA molecules. g High-resolution melting curves of umDNA, hmDNA, and mDNA. Tm values are represented as mean ± SD from independent measurements (n = 3).
Besides the map mentioned above, 3D classification generated two additional maps of the MTA1cMTA9-B–umDNA complex, representing different conformations for umDNA binding (Supplementary Fig. 10d–f). These two structures of MTA1cMTA9-B–umDNA complex resemble that of MTA1cMTA9–umDNA complex, in which the NTR of p1 and the gate loop 1 of MTA1 are invisible, and the umDNA has poor density (Supplementary Fig. 10a, e, f). Among the maps of MTA1c–umDNA complexes, the location and orientation of umDNA relative to MTA1c exhibit differences (Supplementary Fig. 10a, d–f). Compared to the map of more complete MTA1c–DNA complexes discussed above, the three maps without p1 NTR density miss the DNA density on the left side of MTA1–MTA9/MTA9-B (Supplementary Fig. 10a, e, f). Together, these structural observations suggest that umDNA binding to MTA1c is highly dynamic, and therefore most binding events between umDNA and MTA1c are nonproductive, failing to efficiently initiate the downstream methyl transfer reaction.
Early studies suggested that 6mA destabilizes base pairing or stacking of dsDNA41,42. We applied a high-resolution melting experiment to determine the melting temperatures (Tm) of our umDNA and hmDNA, as well as DNA with 6mA on both strands (mDNA). The hmDNA and mDNA exhibit significantly lower Tm values than the umDNA (48.4 °C, 47.3 °C, and 54.3 °C for hmDNA, mDNA, and umDNA, respectively) (Fig. 4g), supporting that the presence of 6mA destabilizes DNA duplex structure. Except for the specific hmDNA recognition by MTA1c, 6mA-induced DNA destabilization further allows MTA1c to bind hmDNA more feasibly and thus efficiently approach the target deoxyadenosine for methylation.
Comparison of DNA recognition by MTA1c and bacterial 6mA DNA MTases
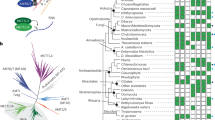
MTA1 and MTA9/MTA9-B are members of the MT-A70-like family of MTases, which evolved from DNA 6mA MTases of bacterial restriction-modification system43. In bacteria, 6mA is typically modified by a monomeric or homodimeric enzyme. Eukaryotes have evolved to adopt different forms of MTases, ranging from single proteins like METTL4 and binary METTL3–METTL14 complex to quaternary MTA1c with functionally diversified subunits. In comparison with bacterial 6mA MTases, the fold of MTA1 and its assembly with MTA9/MTA9-B mostly resemble those of Caulobacter crescentus cell cycle-regulated DNA methyltransferase (CcrM) and Escherichia coli EcoP15I modification (Mod) enzyme44,45 (Fig. 5a–c). Both CcrM and EcoP15I Mod function as a homodimer44,45. CcrM and EcoP15I Mod utilize one protomer to recognize and clamp DNA substrate, resembling a hybrid of MTA9 and p1 of MTA1c, while the other protomer engages in the methyl transfer reaction (Fig. 5a–c). CcrM, EcoP15I Mod and MTA1c apply their substrate-binding loops to contact DNA substrates and flip the target deoxyadenosine. However, DNA substrates adopt different conformations in the three complexes. DNA is nearly parallel to the MTA1–MTA9 interface but exhibits an obvious derivation angle when bound to CcrM and EcoP15I Mod dimers (Fig. 5a–c), which may facilitate DNA recognition by restriction enzyme as observed in the structure of EcoP15I holoenzyme44. In addition, the conformations of their substrate-binding loops and gate loop 1 also diversify to accommodate changes in DNA shape.
a Structure of MTA1cMTA9–hmDNA–SAM complex. b Structure of Caulobacter crescentus CcrM–DNA–sinefungin complex (PDB ID: 6PBD). c Structure of Escherichia coli EcoP15I Mod–DNA complex (PDB ID: 4ZCF). d–f The catalytic pockets in MTA1cMTA9 (d), CcrM (e) and EcoP15I Mod (f). The key residues involved in catalysis and the target deoxyadenosine, as well as ligand SAM and sinefungin, are shown in magenta sticks.
While the conserved [D/N/S/H]PP[Y/F/W] motif is essential for bacterial 6mA MTase activity43, this motif in MTA1, as well as in METTL3 and METTL4, is exclusively DPPW (Supplementary Fig. 12a). Unlike bacterial 6mA MTases that retain activity with NPP[Y/F/W] motif, the D209N mutation in the DPPW motif of MTA1c abolished its activity (Fig. 2j). In the structures of DNA-bound CcrM and EcoP15I Mod, the tyrosine from the [D/N]PPY motif is positioned at the bottom of the target deoxyadenosine, forming π–π stacking. However, the residue W212 in the DPPW motif of MTA1 orients outward to contact flanking helices, albeit the contact with the target deoxyadenosine is maintained (Fig. 5d–f). Instead, another hydrophobic residue, I254, together with W212, contributes to positioning the target deoxyadenosine in MTA1. Therefore, despite adopting an overall similar core fold, the eukaryotic 6mA MTase MTA1c exhibits distinct features from its prokaryotic ancestors, possibly reflecting their specific roles in eukaryotes that require more dynamic regulation in chromatin structure and gene expression.
Structural comparison of MT-A70-like MTases
MT-A70-like family of MTases includes METTL3, METTL4 and METTL14 (Fig. 6a and Supplementary Fig. 12a). METTL3 and METTL14 predominantly participate in N6-methyladenosine modification of RNA46,47, while METTL4 mediates the N6-methylation of adenosine or 2’-O-methyladenosine in U2 small nuclear RNA, besides its role in DNA 6mA modification as mentioned above22,48,49,50,51,52. Like MTA1 and MTA9/MTA9-B, METTL3 and METTL14 form an active heterodimeric MTase, in which METTL3 binds the methyl donor SAM and contains conserved elements for methyl transfer, while the pseudo-enzyme METTL14 serves as a regulatory subunit. They share a highly conserved core with an eight-stranded β sheet flanked by several helices (Fig. 6b–d). METTL3 and METTL4 possess a loop corresponding to the substrate-binding loop of MTA1, which should hold their substrates during methylation.
a Illustration of the domain organization of Tetrahymena thermophila (Tt) MTA1, MTA9 and MTA9-B, Homo sapiens (Hs) METTL3 and METTL14, and Arabidopsis thaliana (At) METTL4. The MTase domains are aligned at their C-termini. Abbreviations: CTE C-terminal extension; MID middle domain. b–d Structural comparison between MTA1cMTA9 (b), METTL3–METTL14 complex (PDB ID: 5IL1) (c) and METTL4 (PDB ID: 7CV7) (d) in two different views. The ligand SAM is shown in sticks. e Model for MTA1c-mediated DNA methylation. DNA substrate is initially captured by p1, which is connected to MTA1 through p2. p1 together with MTA1 and MTA9 (or MTA9-B) clamps DNA near the catalytic center on MTA1. Then MTA1, assisted by p2, transfers the methyl group from SAM to the DNA substrate. The cartoon model was drawn in Adobe Illustrator.
Structural comparison highlights differences in the top regions of METTL3, METTL4 and METTL14 (Fig. 6b–d). METTL3 has an additional short β strand and an extra α helix at its C-terminus (Fig. 6c)53,54,55. METTL4 features a larger structured NTR followed by a middle domain (MID)48 (Fig. 6d). The core eight-stranded β sheet of both METTL3 and METTL4 forms a nine-stranded β sheet with an additional β strand either from METTL3 CTR or METTL4 MID. This β sheet in METTL3 collaborates with the C-terminal α helix to seal the hydrophobic pocket on its top region. The four-stranded β sheet of METTL4 MID, along with its CTR, occupies the other side of its core β sheet. These additional structural elements of METTL3 and METTL4 appear to play a similar role as p2 of MTA1c to stabilize the SAM-binding pocket allosterically. In addition, we identified a positively charged residue in the substrate-binding loops of METTL3 and METTL4 that occupies a position analogous to H291 in MTA1, suggesting a comparable role in base flipping (Supplementary Fig. 12a).
Comparing the structures of MTA9-B and METTL14, we found that their NTRs possess distinct folds, but both directly contact the substrate-binding loop in their partners MTA1 and METTL3, respectively (Fig. 6b, c). Except for their NTRs, MTA9-B and METTL14 adopt highly similar secondary structures to interact with their partners MTA1 and METTL3. However, MTA1–MTA9 and METTL3–METTL14 have distinct substrate preferences. The former exclusively adds a methyl group to dsDNA, while the latter prefers ssRNA. Recently, the METTL3–METTL14 complex was found to exhibit MTase activity on ssDNA and dsDNA with mismatched deoxyadenosine29. Superimposing the crystal structure of the METTL3–METTL14 complex onto our cryo-EM structure of DNA-bound MTA1–MTA9-B complex revealed that dsDNA binding to METTL3–METTL14 is hindered by a steric clash with two interacting helices from the NTR and CTR of METTL14 (Supplementary Fig. 13a–c). Residues in MTA9-B that contact DNA substrate are conserved in MTA9 but not in METTL14 (Supplementary Fig. 12b), suggesting this region might determine the types of preferred substrates, either dsDNA or ssRNA/ssDNA. Nevertheless, we cannot rule out the possibility of the dsDNA-binding ability of METTL3–METTL14 complex, as substantial conformational changes in these two helices may occur upon contact with dsDNA. METTL4 does not require a partner for catalysis, while a long, bent α helix from its NTR located under the bottom of the MTase domain contacts the substrate-binding loop, indicating this helix plays a role akin to MTA9 and METTL14 in stabilizing the loop for substrate binding as proposed previously48 (Fig. 6d).
Both MTA1c and the METTL3–METTL14 complex possess additional substrate-recognition modules: the p1 in the former and two CCCH zinc finger motifs of METTL3 and C-terminal RGG motifs of METTL14 in the latter. The CCCH and RGG motifs are indispensable for the activity of METTL3–METTL14 complex54,55,56. While METTL4 currently lacks an extra identified substrate-binding region, AlphaFold2-predicted METTL4 structures of full-length human and Arabidopsis display a loop in its NTR containing a positively charged fragment, suggesting a potential role in initial substrate recognition40 (Supplementary Fig. 13d, e).
Therefore, despite variations in the component number and structural features, MTA1c, the METTL3–METTL14 complex, and METTL4 share three structural characteristics with functional similarity contributing to stabilizing the MTase domain for SAM binding, enhancing substrate recruitment by the target-recognition module and maintaining active conformation of the substrate-binding loop for DNA or RNA methylation.
Discussion
6mA DNA modification recently appeals to broad interests due to its emerging roles in the regulation of chromatin structure, gene expression, and DNA damage repair, and its links to different pathological conditions in eukaryotes. In this study, we biochemically and structurally characterized the eukaryotic 6mA writer MTA1c. MTA1c exhibits inherent MTase activity towards both umDNA and hmDNA substrates, with a strong preference for the latter. MTA1c demonstrates comparable affinities for both types of DNA substrates but distinct turnover rates. The optimal MTase activity of MTA1c relies on the catalytic subunit MTA1, the catalytically inactive subunit MTA9/MTA9-B, and two additional subunits, p1 and p2. Within this complex, MTA1 and MTA9/MTA9-B provide a basic platform for DNA methylation, where MTA9/MTA9-B positions MTA1 substrate-binding loop, while p2 stabilizes the methyl donor pocket and tethers the DNA-recognition module p1 to the catalytic module MTA1–MTA9/9-B (Fig. 6e). Our structures of MTA1c in complexes with DNA substrates manifest that the four subunits of MTA1c corporately encompass DNA substrate and position the target deoxyadenosine proximal to the catalytic center in MTA1. Our mutagenesis experiments validated the importance of the interactions among these four subunits and those with DNA substrates for effective DNA methylation. MTA1c utilizes a base flipping mechanism for target recognition, which is universally employed by nucleic acid processing enzymes participating in DNA/RNA modification, DNA replication, repair and recombination, and RNA transcription57,58. Upon DNA binding, a conserved histidine residue in the substrate-binding loop of MTA1 wedges into the DNA duplex and enforces the target deoxyadenosine to flip out and insert into the catalytic site. p1 NTR clamps DNA substrates in coordination with MTA1–MTA9/MTA9-B heterodimer. Without p1 NTR, the DNA binding is dynamic and unstable, as observed in three of four umDNA-bound MTA1c structures. Moreover, the affinities of p1 NTR alone binding to umDNA and hmDNA are comparable to those of MTA1c, suggesting that the measured similar affinity of MTA1c to umDNA and hmDNA is mainly contributed by p1 NTR. Therefore, p1 likely participates in recognizing DNA substrates during initial recruitment, positioning substrates on the MTA1–MTA9/MTA9-B surface, and facilitating substrate approaching the catalytic pocket during methyl transfer reaction (Fig. 6e).
The cryo-EM analysis on umDNA- and hmDNA-bound MTA1c revealed profound differences in local resolution and conformational dynamics. Specifically, the MTA1c–hmDNA complex exhibits a well-defined DNA-binding interface and a stable conformation. In contrast, the MTA1c–umDNA complex shows lower local resolution and conformational heterogeneity. We observed specific interactions between MTA1 residues and N6-methyladenine in the high-resolution cryo-EM structure of MTA1cMTA9–hmDNA complex. These differences suggest that although both substrates are similarly recruited by the p1 subunit, the presence of the methyl group in hmDNA stabilizes the catalytically active conformation of MTA1c. In addition, the hmDNA substrate is more easily melted compared to umDNA in vitro, potentially decreasing the energy barrier for the separation of two DNA strands and the base flipping of the target deoxyadenosine, and further contributing to the significantly higher activity of MTA1c to the former. The substantially higher activity on hmDNA suggests MTA1c mainly functions as a maintenance MTase for 6mA31,37. Recently, two MT-A70 family proteins, AMT2 and AMT5, were identified as de novo 6mA MTases in Tetrahymena59. Further structural studies on substrate-bound AMT2 and AMT5 will uncover the mechanism of substrate specificity of different eukaryotic 6mA MTases.
The NTRs of MTA1 and MTA9/MTA9-B and the CTR of p1 are unstructured regions based on 3D structure prediction by AlphaFold. MTA1 NTR is highly enriched in charged residues, with each of aspartic acid, glutamic acid, and lysine comprising more than 10% of its composition. The theoretical pI of this region is 5.21, indicating it is acidic and may negatively regulate the DNA-binding affinity and enzymatic activity of MTA1c. A similar feature is present in p1 CTR, which has an even lower pI of 3.45 for residues 261–360. The MTA1/MTA9/MTA9-B NTRs and p1 CTR are unresolved in the previously reported structures and ours. The potential regulatory roles of these disordered regions within MTA1c subunits remain to be elucidated. Recently, a study reported a functional discrepancy between MTA9 and MTA9-B36. MTA1cMTA9 is coupled with transcription-associated epigenetic marks, including H2A.Z and H3K4me3, whereas MTA1cMTA9-B associates with DNA replication to initiate maintenance methylation36. The NTRs and IRs of MTA9 and MTA9-B are highly diverse (Supplementary Fig. 12b). The difference between MTA9 and MTA9-B likely underlies distinct interaction potentials with binding partners and contributes to functional divergence between the paralogs. Indeed, a PCNA-binding motif was identified only in MTA9-B36. In contrast, the bromodomain-containing protein IBD2 associates with MTA9, but not with MTA9-B36.
MTA1 and MTA9 belong to the MT-A70-like family of MTases, which are responsible for methylating adenine in DNA or RNA substrates in eukaryotes. This family evolved from bacterial DNA 6mA MTases43. MTA1c and bacterial 6mA DNA MTases adopt a differential local conformation in the catalytic pocket to contact the target deoxyadenosine (Fig. 5). The π–π stacking used by the [D/N/S/H]PP[Y/F/W] motif of bacterial 6mA DNA MTases to support the target deoxyadenosine is not present in MTA1c, suggesting a divergent mechanism for DNA recognition and catalysis. Beyond MTA1c, structural information of eukaryotic MT-A70-like MTases is currently limited to the mammalian METTL3–METTL14 complex and plant METTL4 in the absence of nucleic acid substrate48,53,54,55, except the structures of METTL4 bound to a single 2’-O-methyladenosine nucleotide48 and its bound to a dsDNA that does not locate at the putative substrate binding site (PDB ID: 7DPE). Compared to MTA1c, the METTL3–METTL14 complex has fewer subunits, and METTL4 does not require an additional partner. Despite their different substrate preferences, sequence and structural analysis strongly suggest that the three MTases share several functionally similar modules to recruit nucleic acid substrates and stabilize the SAM-binding pocket and substrate-binding loop (Fig. 6). These modules are either intrinsic to the active enzyme itself or contributed by its binding partner(s). Our structural insights and biochemical characterization support that nucleic acid recognition and substrate-binding loop stabilization modules of MTA1c are essential for substrate access to the catalytic center. These modules would be similarly required for the METTL3–METTL14 complex and METTL4, albeit further structural studies are necessary to validate this hypothesis and explore the functions of diverse nucleic acid recognition modules.
In summary, our structures of DNA-bound MTA1c offer insights into how eukaryotic DNA 6mA MTase differentially recognizes its substrates and how the catalysis is facilitated by various functional modules. This work also contributes to a broader understanding of the mechanisms of other MT-A70-like MTases, such as the METTL3–METTL14 complex, which is known for mediating N6-adenosine methylation of RNA and plays vital roles in chromatin dynamics and gene regulation in eukaryotes47,60,61,62.
Methods
Protein expression and purification
Tetrahymena MTA1, MTA9-B, p1 and p2 cDNAs were codon optimized and cloned into pET-derived vectors with an N-terminal 6×His-SUMO dual tag followed by a human rhinovirus 3 C or TEV protease cleavage site. MTA1 and MTA9-B were cloned into the pETDuet-1 vector with a C-terminal StrepII tag linked to MTA1 or a C-terminal TwinStrepII tag fused to MTA9-B, while p1 and p2 were cloned into the pET28a vector separately or into the pRSFDuet-1 vector for coexpression with MTA1–MTA9-B. p1 NTR (1–150) was cloned into the pET28a vector. Tetrahymena MTA9 was codon optimized and cloned into the pET-28a vector with an N-terminal 6 × His-SUMO dual tag followed by a human rhinovirus 3 C protease cleavage site and a C-terminal TwinStrepII tag. We screened and chose MTA9-B for our biochemical studies due to its higher yield when coexpressed with MTA1 in Escherichia coli. All mutants were constructed by overlapping PCR followed by DpnI digestion. Each protein or protein complex was expressed in Lemo21(DE3) E. coli cells (New England BioLabs) in Luria-Bertani broth with appropriate antibiotics and IPTG. Cells were resuspended in Lysis Buffer TN500 (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10% glycerol, 5 mM 2-mercaptoethanol) supplemented with 1 mM PMSF and 10 mM imidazole and then disrupted by a sonicator or a high-pressure homogenizer (ATS AH-1500). The cell lysate was cleared by centrifugation at 22,554 × g for 40 min at 4 °C. The supernatant was mixed with Ni Sepharose 6 Fast Flow (Cytiva) for 1 h at 4 °C and then applied to a gravity column. The resin was washed with Lysis Buffer TN500 supplemented with 30 mM imidazole, and the bound protein samples were eluted with Buffer TN150 (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10% glycerol, 5 mM 2-mercaptoethanol) supplemented with 400 mM imidazole. The His-SUMO dual tag was removed by home-made human rhinovirus 3 C protease digestion at 4 °C. The samples were then loaded onto a Resource Q (Cytiva). For the StrepII-tagged samples, the samples eluted from Resource Q were mixed with Strep-Tactin XT 4Flow (IBA) for 1 h at 4 °C, and the bound protein samples were eluted with Buffer TN150 with 50 mM D-biotin. The samples were further purified on a Superdex 200 Increase 10/300 GL column (Cytiva) pre-equilibrated with GF Buffer HN150 (20 mM HEPES pH 7.5, 150 mM NaCl, 5% glycerol and 1 mM DTT) for biochemical studies or GF Buffer HKGlu100 (20 mM HEPES pH 7.5, 100 mM KGlu, 0.5 mM TCEP) for structural studies. Samples were detected by SDS-polyacrylamide gel electrophoresis (SDS–PAGE) and Coomassie blue staining. The fractions containing pure samples were collected and concentrated with an Amicon Centrifugal Filter Unit (Merck Millipore). For biochemical experiments, samples were snap frozen in liquid nitrogen and stored at − 80 °C. Freshly purified samples were used for cryo-EM study.
DNA substrates
The following 27-nt DNA oligos were synthesized in Tsingke Biotechnology or Genscript Biotech for biochemical and structural experiments:
27ATrich_s: AACTTTCTTAACATCTTAACTTTAACT (numbered as dA-13–dT13 with the target site dA0 in the MTA1cMTA9–hmDNA complex structure)
27ATrich_6mA_s: AACTTTCTTAAC(m6dA)TCTTAACTTTAACT
27ATrich_a: AGTTAAAGTTAAGATGTTAAGAAAGTT
27ATrich_6mA_a: AGTTAAAGTTAAG(m6dA)TGTTAAGAAAGTT (numbered as dA-13’–dT13’ in the MTA1cMTA9–hmDNA complex structure)
27ATrich_Cy5.5_a: Cy5.5-AGTTAAAGTTAAGATGTTAAGAAAGTT
27ATrich_6FAM_a: 6-FAM-AGTTAAAGTTAAGATGTTAAGAAAGTT
27_RNA_a: AGUUAAAGUUAAGAUGUUAAGAAAGUU
These oligos were dissolved in 30 mM HEPES, pH 7.5, 100 mM KCl. Complementary oligos were mixed with an equal molar ratio, heated to 95 °C for 5 min, and then slowly cooled down to room temperature. 27ATrich_s and 27ATrich_a/27ATrich_Cy5.5_a/27ATrich_6FAM_a were annealed to produce unlabeled and fluorescently labeled umDNA substrates. 27ATrich_6mA_s and 27ATrich_a/27ATrich_Cy5.5_a/27ATrich_6FAM_a were annealed to produce unlabeled and fluorescently labeled hmDNA substrates. 27ATrich_6mA_s and 27ATrich_6mA_a were annealed to produce mDNA. 27ATrich_s and 27_RNA_a were annealed to produce a DNA–RNA hybrid. The annealed DNA duplexes and DNA–RNA hybrid were aliquoted and frozen at − 20 °C.
MTase activity assay
MTase-Glo Methyltransferase Assay kit (Promega) was used to measure the MTase activity of MTA1c. Typically, samples were incubated with 10 μM SAM and 10 μM DNA or DNA–RNA hybrid substrate in 8 μL of MTase Buffer (20 mM HEPES pH 7.5, 50 mM KCl, 3 mM MgCl2, 1 mM DTT, 1 mM EDTA, 0.1 mg/mL BSA, 5% glycerol) at 30 °C. 2 μL of 5 × MTase-GloTM were added and incubated at room temperature for 30 min. Then 10 μL of MTase-GloTM Detection Solution were added to the reaction mixture and incubated at room temperature for an additional 30 min. 18 μL reaction mixture was transferred to a White Opaque 384-well Microplate (PerkinElmer). Luminescence was measured with a Spark multimode microplate reader (TECAN). The concentration of SAH was calculated with a standard curve. Three replicates were performed in each experiment, and each experiment was performed twice independently and obtained similar results. Data analysis was carried out in Prism 10 (GraphPad).
Gel electrophoresis mobility shift assay (EMSA)
5 nM Cy5.5-labeled DNA duplexes were incubated with 2-fold serial dilutions of MTA1cMTA9-B (0–2 μM for wildtype protein or 0–0.25 μM for wildtype protein and mutants), p2 or p1 NTR (0–2 μM) on ice for 30 min in EMSA Buffer (20 mM HEPES pH 7.5, 50 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.01% IGEPAL® CA-630, 5% glycerol). Samples were separated on 5% native PAGE in 0.5 × TBE Buffer and scanned with Odyssey DLx Imaging System (LI-COR). The experiments were repeated twice with similar results.
Fluorescence polarization
1 nM 6-FAM-labeled DNA duplexes were incubated with 2-fold serial dilutions of MTA1cMTA9-B or p1 NTR (0–2 μM) on ice for 30 min in EMSA Buffer (20 mM HEPES pH 7.5, 75 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.01% IGEPAL® CA-630, 5% glycerol). Fluorescence polarization values were measured with a Spark multimode microplate reader (TECAN). Data was analyzed in Prism 10 (GraphPad) with the nonlinear regression model. Kd was calculated by the equation: FP = (FPmax – FPmin)[protein]/(Kd + [protein])) + FPfree, where FP is the polarization measured at a given protein concentration, FPmin is the polarization of 6-FAM-labeled DNA, FPmax is the maximum polarization, and [protein] is the protein concentration.
Multi-angle light scattering coupled with size exclusion chromatography (SEC-MALS)
30 μM of MTA1cMTA9-B and DNA were incubated for 30 min on ice in SEC-MALS Buffer (20 mM HEPES pH 7.5, 75 mM NaCl, 1 mM DTT). 50 μL of MTA1cMTA9-B or MTA1cMTA9-B–DNA mixture (∼ 4 mg/mL) was loaded into the TSKgel G3000SWXL column (Tosoh Bioscience) pre-equilibrated in SEC–MALS Buffer at a flow rate of 0.5 mL/min at 25 °C. The sample eluted from the TSKgel G3000SWXL column was loaded into an in-line DAWN® system with 18 detector angles and a 658 nm laser (Wyatt Technology). Refractive index was measured with an Optilab® refractometer (Wyatt Technology). Peak alignment and band broadening correction between the UV, MALS, and RI detectors were performed, and the weight average molecular weight (Mw), the number average molecular weight (Mn) and the polydispersity index (PDI) were calculated using ASTRA® 6.1 software (Wyatt Technology).
High-resolution melting experiment
The reaction mixture for the high-resolution melting analysis contains 10 µL of 2 × EvaGreen dye (Biotium), 5 µL of 50 µM sense strand ssDNA, and 5 µL of 50 µM antisense strand ssDNA in 30 mM HEPES pH 7.5, 100 mM KCl. Samples were subjected to the following melting program on CFX384 Touch Real-Time PCR Detection System (Bio-Rad): 1) denaturation at 95 °C for 5 min, 2) slow cooling to 25 °C for 5 min with 0.1 °C decrements every 1 s, and 3) continuous heating at 0.1 °C increments every 10 s with fluorescence acquisition from 25 to 95 °C. Three replicates were performed in each experiment. Data were analyzed with CFX Maestro Software and Prism 10 (GraphPad).
Strep pulldown experiment
StrepII-tagged MTA1 and untagged MTA9-B, p1 and p2 proteins were mixed with 10 μL Strep-Tactin 2 NUPharose Fast Flow (NUPTEC) in Binding Buffer HN150 (20 mM HEPES pH 7.5, 150 mM NaCl, 1 mM DTT, 0.01% IGEPAL® CA-630, 5% glycerol) at 4 °C for 1 h. The resin was washed four times with 1 mL of Binding Buffer HN150, and then the bound proteins were eluted with 50 μL of Binding Buffer HN150 supplemented with 50 mM D-biotin. Elution samples were analyzed by 4–20% SDS-PAGE and Coomassie blue staining. The experiments were repeated twice with similar results.
Cryo-EM specimen preparation and data processing
To prepare DNA-bound MTA1c samples, purified MTA1c with full-length proteins was incubated with unmodified or hmDNA substrates. 1 mM SAM was added to MTA1c after the incubation of MTA1c with DNA, and the mixture was incubated briefly. In the case of MTA1cMTA9-B (D209A), DNA and 0.1 mM SAM were added to MTA1cMTA9-B simultaneously. To prepare MTA1cMTA9-B–SAH sample, purified MTA1cMTA9-B was incubated with 1 mM SAH. After the incubation, the samples were applied onto glow-discharged holey carbon grids (Quantifoil Au 400 mesh, R1.2/1.3). Different detergents were used as an additive to optimize particle distribution on cryo-EM grids. After screening, 0.025–0.05% n-octyl-β-D-glucoside was added to the samples before applying the samples to the grids. The grids were blotted for 4.5 s at 100% humidity using a Vitrobot Mark IV System (Thermo Fisher Scientific) before plunging into liquid ethane. The grids were screened on a Glacios Cryo-TEM (Thermo Fisher Scientific).
A total of 4105, 2870, 12,678 and 4910 movie stacks were collected for the samples of MTA1cMTA9-B–hmDNA, MTA1cMTA9-B (D209A)–umDNA–SAM, MTA1cMTA9-B–umDNA–SAM and MTA1cMTA9-B–hmDNA–SAM, respectively, on Titan Krios G3i Cryo-TEM (Thermo Fisher Scientific) operating at 300 kV equipped with a K3 Camera (Gatan) and BioQuantum K3 Imaging Filter (Gantan Model 1967) at a nominal magnification of 81,000 × (corresponding to 1.0773 Å/pixel) and an accumulated dose of 50 e–/Å2. Motion correction was performed with the MotionCorr2 program63. A total of 3634, 11,231 and 6718 movie stacks for the samples of MTA1cMTA9-B–SAH, MTA1cMTA9–umDNA–SAM and MTA1cMTA9–hmDNA–SAM were collected on Titan Krios G4 Cryo-TEM (Thermo Fisher Scientific) operating at 300 kV equipped with a Falcon 4i Direct Electron Detector and Selectris X Imaging Filter at a nominal magnification of 130,000 × (corresponding to 0.92 Å/pixel) and an accumulated dose of 50 e–/Å2. Patch motion correction was performed in CryoSPARC64. Patch CTF estimation and the following processing steps were performed in CryoSPARC64. Micrographs with better than 6 Å resolution and relative ice thickness ranging from 1 to 1.1 were selected for particle picking. Generally, particles were initially picked up with Topaz using a pretrained model65. Class averages representing projections in different orientations selected from the second round of 2D classification were used as templates for Topaz model training. Then the generated model was used to pick particles for the whole dataset. Particles were sorted by 2D classification, and particles with clear structural features were chosen for generating initial maps. After several cycles of ab-initio reconstruction and heterogeneous refinement, particles from the best class of heterogeneous refinement were applied to non-uniform refinement, followed by local refinement66. For MTA1cMTA9-B–umDNA/hmDNA–SAM samples, particles from the best class of the first round of heterogeneous refinement were imported into RELION-467. 3D classification and focused 3D classification on the DNA part were performed to separate DNA-bound MTA1c particles68. These particles were transferred back to CryoSPARC for homogenous reconstruction and non-uniform refinement. For MTA1cMTA9–umDNA/hmDNA–SAM samples, particles from the best class of the first round of heterogeneous refinement were applied to focused 3D classification on the DNA part in CryoSPARC. The particles within the 3D class that displayed clear and complete DNA density were further applied to non-uniform refinement. DeepEMhancer was used to sharpen maps and improve map quality69.
Model building and refinement
AlphaFold2 predicted models were docked into the cryo-EM map of MTA1cMTA9-B–SAH and MTA1cMTA9–SAM complexes in UCSF Chimera40,70. The initial models were iteratively manually built in Coot71 and refined with real-space refinement in Phenix72,73. The refined models were docked to other cryo-EM maps of MTA1c complexes, followed by manually building in Coot and real-space refinement in Phenix. Secondary structure, base pairing, and stacking restraints were applied during refinement. Model validation was performed in Molprobity74,75. Representative cryo-EM images, 2D class averages and 3D maps of each complex are shown in Supplementary Figs. 2–8. Data collection and processing, model refinement and statistics are shown in Supplementary Tables 2–4. Structural images were generated in UCSF Chimera and ChimeraX70,76 and PyMOL (Schrodinger).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The cryo-EM maps generated in this study have been deposited in the Electron Microscopy Data Bank (EMDB) under the accession codes EMD-38777, EMD-38780, EMD-38781, EMD-38782, EMD-38786, EMD-38787, EMD-63956, EMD-63961, EMD-63963, EMD-63967, and EMD-63970. The atomic coordinates have been deposited in the Worldwide Protein Data Bank (wwPDB) under the accession codes 8XYL, 8XYP, 8XYQ, 8XYX, 9U92, 9U9E, 9U9K, and 9VU6. The wwPDB datasets used for analysis in this study include 4ZCF, 5IL1, 6PBD, 7CV7, and 7DPE. Source data are provided in this paper.
References
Boulias, K. & Greer, E. L. Means, mechanisms and consequences of adenine methylation in DNA. Nat. Rev. Genet. 23, 411–428 (2022).
Dunn, D. B. & Smith, J. D. Occurrence of a new base in the deoxyribonucleic acid of a strain of Bacterium coli. Nature 175, 336–337 (1955).
Wion, D. & Casadesus, J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 4, 183–192 (2006).
Gorovsky, M. A., Hattman, S. & Pleger, G. L. 6 N)methyl adenine in the nuclear DNA of a eucaryote, Tetrahymena pyriformis. J. Cell Biol. 56, 697–701 (1973).
Cummings, D. J., Tait, A. & Goddard, J. M. Methylated bases in DNA from Paramecium aurelia. Biochim. Biophys. Acta 374, 1–11 (1974).
Hattman, S., Kenny, C., Berger, L. & Pratt, K. Comparative study of DNA methylation in three unicellular eucaryotes. J. Bacteriol. 135, 1156–1157 (1978).
Fu, Y. et al. N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell 161, 879–892 (2015).
Greer, E. L. et al. DNA Methylation on N6-Adenine in C. elegans. Cell 161, 868–878 (2015).
Zhang, G. et al. N6-methyladenine DNA modification in Drosophila. Cell 161, 893–906 (2015).
Koziol, M. J. et al. Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat. Struct. Mol. Biol. 23, 24–30 (2016).
Liu, J. et al. Abundant DNA 6mA methylation during early embryogenesis of zebrafish and pig. Nat. Commun. 7, 13052 (2016).
Wu, T. P. et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature 532, 329–333 (2016).
Liang, Z. et al. DNA N(6)-Adenine Methylation in Arabidopsis thaliana. Dev. Cell 45, 406–416 (2018).
Zhou, C. et al. Identification and analysis of adenine N(6)-methylation sites in the rice genome. Nat. Plants 4, 554–563 (2018).
Xiao, C. L. et al. N(6)-Methyladenine DNA Modification in the Human Genome. Mol. Cell 71, 306–318 (2018).
Li, X. et al. The exploration of N6-deoxyadenosine methylation in mammalian genomes. Protein Cell 12, 756–768 (2021).
Shen, C., Wang, K., Deng, X. & Chen, J. DNA N(6)-methyldeoxyadenosine in mammals and human disease. Trends Genet. 38, 454–467 (2022).
Zhang, X., Blumenthal, R. M. & Cheng, X. A Role for N6-Methyladenine in DNA Damage Repair. Trends Biochem. Sci. 46, 175–183 (2021).
Feng, X. & He, C. Mammalian DNA N(6)-methyladenosine: Challenges and new insights. Mol. Cell 83, 343–351 (2023).
Kong, Y. et al. Critical assessment of DNA adenine methylation in eukaryotes using quantitative deconvolution. Science 375, 515–522 (2022).
Koh, C. W. Q. et al. Single-nucleotide-resolution sequencing of human N6-methyldeoxyadenosine reveals strand-asymmetric clusters associated with SSBP1 on the mitochondrial genome. Nucleic Acids Res. 46, 11659–11670 (2018).
Hao, Z. et al. N(6)-Deoxyadenosine methylation in mammalian mitochondrial DNA. Mol. Cell 78, 382–395 (2020).
Li, Z. et al. N(6)-methyladenine in DNA antagonizes SATB1 in early development. Nature 583, 625–630 (2020).
Xie, Q. et al. N(6)-methyladenine DNA Modification in Glioblastoma. Cell 175, 1228–1243 (2018).
Sheng, X., Wang, J., Guo, Y., Zhang, J. & Luo, J. DNA N6-Methyladenine (6mA) Modification Regulates Drug Resistance in Triple Negative Breast Cancer. Front. Oncol. 10, 616098 (2020).
Cheng, X. & Roberts, R. J. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 29, 3784–3795 (2001).
Luo, G. Z. et al. N(6)-methyldeoxyadenosine directs nucleosome positioning in Tetrahymena DNA. Genome Biol. 19, 200 (2018).
Ma, C. et al. A novel N(6)-Deoxyadenine methyltransferase METL-9 modulates C. elegans immunity via dichotomous mechanisms. Cell Res. 33, 628–639 (2023).
Woodcock, C. B. et al. Human MettL3-MettL14 complex is a sequence-specific DNA adenine methyltransferase active on single-strand and unpaired DNA in vitro. Cell Discov. 5, 63 (2019).
Yu, D. et al. Human MettL3-MettL14 RNA adenine methyltransferase complex is active on double-stranded DNA containing lesions. Nucleic Acids Res. 49, 11629–11642 (2021).
Beh, L. Y. et al. Identification of a DNA N6-adenine methyltransferase complex and its impact on chromatin organization. Cell 177, 1781–1796 (2019).
Bromberg, S., Pratt, K. & Hattman, S. Sequence specificity of DNA adenine methylase in the protozoan Tetrahymena thermophila. J. Bacteriol. 150, 993–996 (1982).
Pratt, K. & Hattman, S. Nucleosome phasing in Tetrahymena macronuclei. J. Protozool. 30, 592–598 (1983).
Wang, Y., Chen, X., Sheng, Y., Liu, Y. & Gao, S. N6-adenine DNA methylation is associated with the linker DNA of H2A.Z-containing well-positioned nucleosomes in Pol II-transcribed genes in Tetrahymena. Nucleic Acids Res. 45, 11594–11606 (2017).
Karrer, K. M. & VanNuland, T. A. Methylation of adenine in the nuclear DNA of Tetrahymena is internucleosomal and independent of histone H1. Nucleic Acids Res. 30, 1364–1370 (2002).
Wang, Y. et al. Dual modes of DNA N(6)-methyladenine maintenance by distinct methyltransferase complexes. Proc. Natl. Acad. Sci. USA 122, e2413037121 (2025).
Wang, Y. et al. A distinct class of eukaryotic MT-A70 methyltransferases maintain symmetric DNA N6-adenine methylation at the ApT dinucleotides as an epigenetic mark associated with transcription. Nucleic Acids Res. 47, 11771–11789 (2019).
Chen, J. et al. Structural basis for MTA1c-mediated DNA N6-adenine methylation. Nat. Commun. 13, 3257 (2022).
Yan, J. et al. Structural insights into DNA N(6)-adenine methylation by the MTA1 complex. Cell Discov. 9, 8 (2023).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Engel, J. D. & von Hippel, P. H. Effects of methylation on the stability of nucleic acid conformations. Studies at the polymer level. J. Biol. Chem. 253, 927–934 (1978).
Peng, S., Padva, A. & LeBreton, P. R. Ultraviolet photoelectron studies of biological purines: the valence electronic structure of adenine. Proc. Natl. Acad. Sci. USA 73, 2966–2968 (1976).
Iyer, L. M., Zhang, D. & Aravind, L. Adenine methylation in eukaryotes: Apprehending the complex evolutionary history and functional potential of an epigenetic modification. Bioessays 38, 27–40 (2016).
Gupta, Y. K., Chan, S. H., Xu, S. Y. & Aggarwal, A. K. Structural basis of asymmetric DNA methylation and ATP-triggered long-range diffusion by EcoP15I. Nat. Commun. 6, 7363 (2015).
Horton, J. R. et al. The cell cycle-regulated DNA adenine methyltransferase CcrM opens a bubble at its DNA recognition site. Nat. Commun. 10, 4600 (2019).
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G. & Rottman, F. M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247 (1997).
Liu, J. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014).
Luo, Q. et al. Structural insights into molecular mechanism for N(6)-adenosine methylation by MT-A70 family methyltransferase METTL4. Nat. Commun. 13, 5636 (2022).
Goh, Y. T., Koh, C. W. Q., Sim, D. Y., Roca, X. & Goh, W. S. S. METTL4 catalyzes m6Am methylation in U2 snRNA to regulate pre-mRNA splicing. Nucleic Acids Res. 48, 9250–9261 (2020).
Chen, H. et al. METTL4 is an snRNA m(6)Am methyltransferase that regulates RNA splicing. Cell Res. 30, 544–547 (2020).
Gu, L. et al. CG14906 (mettl4) mediates m(6)A methylation of U2 snRNA in Drosophila. Cell Discov. 6, 44 (2020).
Kweon, S. M. et al. An adversarial DNA N(6)-methyladenine-sensor network preserves polycomb silencing. Mol. Cell 74, 1138–1147 (2019).
Wang, X. et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534, 575–578 (2016).
Wang, P., Doxtader, K. A. & Nam, Y. Structural basis for cooperative function of mettl3 and mettl14 methyltransferases. Mol. Cell 63, 306–317 (2016).
Sledz, P. & Jinek, M. Structural insights into the molecular mechanism of the m(6)A writer complex. Elife 5, e18434 (2016).
Scholler, E. et al. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. RNA 24, 499–512 (2018).
Hong, S. & Cheng, X. DNA Base flipping: A general mechanism for writing, reading, and erasing DNA modifications. Adv. Exp. Med. Biol. 945, 321–341 (2016).
Roberts, R. J. & Cheng, X. Base flipping. Annu. Rev. Biochem. 67, 181–198 (1998).
Cheng, T. et al. Identification and characterization of the de novo methyltransferases for eukaryotic N(6)-methyladenine (6mA). Sci. Adv. 11, eadq4623 (2025).
Sendinc, E. & Shi, Y. RNA m6A methylation across the transcriptome. Mol. Cell 83, 428–441 (2023).
Murakami, S. & Jaffrey, S. R. Hidden codes in mRNA: Control of gene expression by m(6)A. Mol. Cell 82, 2236–2251 (2022).
Shi, H., Wei, J. & He, C. Where, when, and how: Context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74, 640–650 (2019).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods14, 290–296 (2017).
Bepler, T. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods16, 1153–1160 (2019).
Rubinstein, J. L. & Brubaker, M. A. Alignment of cryo-EM movies of individual particles by optimization of image translations. J. Struct. Biol. 192, 188–195 (2015).
Kimanius, D., Dong, L., Sharov, G., Nakane, T. & Scheres, S. H. W. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem. J.478, 4169–4185 (2021).
Bai, X. C., Rajendra, E., Yang, G., Shi, Y. & Scheres, S. H. Sampling the conformational space of the catalytic subunit of human gamma-secretase. Elife 4, https://doi.org/10.7554/elife.11182 (2015).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018).
Afonine, P. V. et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Struct. Biol. 74, 814–840 (2018).
Williams, C. J. et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Acknowledgements
We would like to thank H. Yu for manuscript review and insightful suggestions. We thank the staff at the Cryo-EM Facility of Westlake University for technical assistance. This work was supported by the High-level Talents Special Support Program of Zhejiang Province (2023R5245 to Z.S.), the “Pioneer” and “Leading Goose” R&D Program of Zhejiang (2024SSYS0033 to Z.S.), the National Natural Science Foundation of China (32271264 to Z.S.), and the Westlake Education Foundation (to Z.S.).
Author information
Authors and Affiliations
Contributions
Z.S. initiated and supervised the study. Q.X. and Y.X. contributed to protein purification. Q.X. contributed to biochemical experiments, cryo-EM sample preparation, data collection, and model building. Z.S. contributed to cryo-EM data processing, model building, and data analysis. Z.S. wrote the manuscript with the input from all co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Yogesh Gupta and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xu, Q., Xie, Y. & Shi, Z. Mechanism for the substrate recognition by a eukaryotic DNA N6-adenine methyltransferase complex. Nat Commun 16, 8690 (2025). https://doi.org/10.1038/s41467-025-63738-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-63738-y