Abstract
Deciphering the pre-malignant cell of origin (COO) of different cancers is critical for understanding tumor development and improving diagnostic and therapeutic strategies in oncology. Prior work demonstrates that somatic mutations preferentially accumulate in closed chromatin regions of a cancer’s COO. Leveraging this information, we combine 3,669 whole genome sequencing patient samples, 559 single-cell chromatin accessibility cellular profiles, and machine learning to predict the COO of 37 cancer subtypes with high robustness and accuracy, confirming both the known anatomical and cellular origins of numerous cancers, often at cell subset resolution. Importantly, our data-driven approach predicts a basal COO for most small cell lung cancers and a neuroendocrine COO for rare atypical cases. Our study also highlights distinct cellular trajectories during cancer development of different histological subtypes and uncovers an intermediate metaplastic state during tumorigenesis for multiple gastrointestinal cancers, which have important implications for cancer prevention, early detection, and treatment stratification.
Similar content being viewed by others
Introduction
Cancer development is a complex, multi-step process driven by genetic and epigenetic alterations that accumulate over time. A fundamental question in oncology is understanding the cell of origin (COO), or the cellular progenitor that leads to malignant transformation1. Identifying the COO is critical not only for understanding tumorigenesis but also for improving cancer prevention, early detection, risk stratification, and targeted treatments1,2. For example, precursor lesions for esophageal adenocarcinoma arise from metaplastic changes in esophageal epithelial cells due to chronic acid reflux; understanding the COO in this context has led to targeted interventions, such as endoscopic surveillance and radiofrequency ablation, to prevent malignant progression3,4. Moreover, studies in prostate cancer have demonstrated that tumors originating from basal versus luminal epithelial cells exhibit distinct molecular profiles and clinical outcomes and respond differently to androgen deprivation therapy1,5,6.
Vast progress has been made in understanding the COO of different cancers using genetically-engineered mouse models1. However, it is also critical to directly study neoplastic processes using human samples that bypass limitations due to interspecies differences7. Recent advances in transcriptomic, genomic, and epigenomic profiling have emerged as powerful tools for tracing human cancer origins. Machine learning (ML) approaches have utilized extensive collections of normal and tumor bulk sequencing data to classify various cancer types according to their tissue of origin, with most methods relying on transcriptomic data8,9,10,11,12. While these approaches often achieve high prediction accuracy, the selected gene features have shown inconsistencies across different studies12. More recently, these and other methods have utilized single-cell transcriptomics data to infer the COO of several cancer types of interest13,14,15,16, but have not been scaled to predict COOs in a pan-cancer fashion. Additionally, approaches to detect the COO by modeling the relationship between normal and cancer transcriptomic data have limitations, as gene expression can be altered by the tumor microenvironment17, dedifferentiation18, and oncogenic reprogramming18,19, which can obscure the true cellular beginnings of a cancer.
Genetic data offers a more reliable means of tracing a human cancer’s COO, as the mutational landscape of cells is predominantly composed of passenger somatic mutations that accumulate over the lifespan of an individual before malignant transformation occurs. In addition, we and others have demonstrated that the underlying epigenome of the normal COO shapes the genomic distribution of somatic mutations20,21,22, which tend to accumulate in closed chromatin regions that are less accessible by DNA repair mechanisms23,24. Our team first exploited the inherent relationship between epigenomic features (e.g., histone modifications, chromatin accessibility, DNA replication timing) of normal cells and the mutational landscape, detected using whole genome sequencing (WGS), to predict the corresponding COO of different cancers using a Random Forest (RF) and a linear model21,25. Polak et al.21 also demonstrated that chromatin features from normal tissues are better predictors of the somatic mutational landscape than gene expression data. More recently, Yang et al. 26 used extreme gradient boosting27 (XGBoost) for COO prediction, which improved prediction speed and accuracy compared to RF, especially for tumor types with low mutation density. However, the aforementioned studies21,25,26 were based on bulk tissue epigenomic data that lacks the resolution to identify the specific cellular populations that give rise to different cancers.
Recent advances in high-throughput, single-cell Assay for Transposase-Accessible Chromatin (scATAC-seq) have been used to profile millions of human fetal and adult cells and map chromatin accessibility across hundreds of cell types28,29,30,31,32,33,34. We reasoned that combining this data with the plethora of publicly available WGS35,36,37 data can greatly enhance the resolution and scale in predicting the COO of different cancers.
Here, we assemble an extensive scATAC-seq dataset and leverage our ML framework dubbed SCOOP–Single-cell Cell Of Origin Predictor–to predict the COO of 37 cancer types. Unlike previous bulk transcriptomic and epigenomic approaches, single-cell chromatin accessibility data enables deconvolution of complex tissues and identification of cancer precursor cells at cell subset granularity. Our model demonstrates high accuracy and robustness, confirming known COOs of numerous tumor types while also generating unexpected hypotheses about several cancers. Most notably, SCOOP challenges the long-held theory that small cell lung cancer (SCLC) arises primarily from neuroendocrine cells, showing instead a predominantly basal COO, in agreement with a concurrent study employing cellular lineage tracing in SCLC genetically-engineered mouse models38. Interestingly, our data-driven approach also finds a role for neuroendocrine cells in the genesis of atypical SCLC and less aggressive carcinoid tumors. Moreover, SCOOP identifies a metaplastic-like stomach goblet cell as the COO for five different gastrointestinal cancers, indicating convergent cellular trajectories toward tumorigenesis, which has important implications for cancer prevention and early detection screenings. Taken together, our study establishes a cost-effective and scalable approach to infer a human cancer’s COO at cellular resolution by integrating normal tissue scATAC-seq and WGS data from tumor clinical samples.
Results
SCOOP improves cellular resolution and accuracy of COO predictions
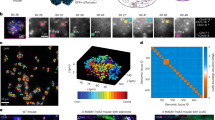
To predict the COO for 37 cancers of interest (Supplementary Data 1), SCOOP uses as input one megabase pair binned (Supplementary Data 2) single-nucleotide variant (SNV) count profiles aggregated across WGS patient samples, and similarly binned scATAC-seq aggregate profiles from a compendium of 559 normal cell subsets spanning 32 adult and 15 fetal tissue types (Fig. 1a; Supplementary Data 3; Methods). SCOOP leverages the binned scATAC-seq profiles and a ML model (XGBoost) to predict the mutation density of a given cancer (e.g., lung adenocarcinoma, LUAD). It then iteratively reduces the set of scATAC-seq cell features through backward feature selection to identify the most informative cell subset (e.g., alveolar type II (AT2) cells), which represents the predicted COO. The model is trained 100 times using different train/test splits and random seeds (100 SCOOP runs; Methods), resulting in 100 COO predictions.
a Left: Illustration of how SCOOP uses single-cell assay for transposase-accessible chromatin using sequencing (scATAC-seq) data to predict the cell-of-origin (COO) (e.g., alveolar type 2, or AT2, cells) associated with a given cancer’s mutation profile (e.g., lung adenocarcinoma, or LUAD). SCOOP takes as input a binned whole-genome sequencing (WGS) profile of cancer single-nucleotide variants (SNVs) and similarly binned scATAC-seq profiles from various normal cell subsets, where each cell subset is followed by a dataset indicator for that cell subset: D1 for28, D2 for29, D5 for34, D9 for45. The SNV and scATAC-seq profiles (features) are passed into a machine learning model, XGBoost, which predicts the COO through a process of backward feature selection (Methods). Right: Box plots of the feature importance distribution (100 SCOOP runs) of the top 5 COO predictions for LUAD (n = 37; predicted COO in red) amongst lung cell subsets (Methods). Also displayed is the number of times a cell subset appeared in the top 5 features across 100 runs (n). One-sided Mann-Whitney test p-values are displayed. Tumor and cell illustrations created in BioRender. Tsankov, A. (2025) https://BioRender.com/qu5wvua. b Test set variance explained (%) by the predicted COOs (red) for 8 cancer types studied in21 (Melanoma, n = 107; Hepatocellular carcinoma, n = 314; Colorectal adenocarcinoma, n = 52; Multiple myeloma, n = 23; Esophageal adenocarcinoma, n = 97; Glioblastoma, n = 39; Lung adenocarcinoma, n = 37; Lung squamous cell carcinoma, n = 47). Error bars show the standard error of the mean (SEM) across 100 SCOOP runs. One-sided Mann-Whitney test p-values are displayed. Each cell subset is followed by a dataset indicator for that cell subset: D1 for28, D2 for29, D3 for31, D4 for32, D5 for34. c Box plots of the feature importance distribution (100 SCOOP runs) of the top 5 COO predictions (predicted COO in red, similar cell subsets in pink) amongst lung cell subsets for epithelioid pleural mesothelioma (PM, n = 44), lung squamous cell carcinoma (LUSC, n = 47), and small cell lung carcinoma (SCLC, n = 107). Also displayed is the number of times a cell subset appeared in the top 5 features across 100 runs (n). One-sided Mann-Whitney test p-values are displayed, where Bonferroni correction for multiple hypothesis testing was used. d UMAP dimensionality reduction of individual lung cancer WGS samples binned mutation profiles (dots) colored by cancer type (adenocarcinoma, n = 37; epithelioid mesothelioma, n = 44; small cell lung cancer, n = 109; squamous cell carcinoma, n = 47). e UMAP dimensionality reduction of individual SCLC WGS sample binned mutation profiles (dots) from36,48 (aSCLC from48, n = 11; aSCLC from36, n = 2; SCLC-A, n = 37; SCLC-N, n = 4; SCLC-P, n = 6; SCLC-Y, n = 1; Undefined, n = 57). f Box plots of the feature importance distribution (100 SCOOP runs) of the top 5 COO predictions (predicted COO in red, similar cell subsets in pink) amongst lung cell subsets for atypical small cell lung cancer (aSCLC from48, n = 11; aSCLC from36, n = 2). Also displayed is the number of times a cell subset appeared in the top 5 features across 100 runs (n). One-sided Mann-Whitney test p-values are displayed, where Bonferroni correction for multiple hypothesis testing was used. g Box plots of the feature importance distribution (100 SCOOP runs) of the top 5 COO predictions amongst lung cell subsets for SCLC-A (n = 37; predicted COO in red, similar cell subsets in pink). Also displayed is the number of times a cell subset appeared in the top 5 features across 100 runs (n). One-sided Mann-Whitney test p-values are displayed, where Bonferroni correction for multiple hypothesis testing was used. h Percentage of cycling cells across lung epithelial cell types estimated using scRNA-seq data (Methods), where predicted COOs in our study are shown in red. i SCOOP’s predicted COO for different lung cancers: AT2, mesothelial, and neuroendocrine cells for LUAD, epithelioid PM, and aSCLC, respectively, and basal cells for both LUSC and SCLC. Lung model created in BioRender. Tsankov, A. (2025) https://BioRender.com/2vhmu6l. Cell type abbreviations are defined in Supplementary Data 3. Box plot vertical lines show 25th, 50th (median), and 75th percentiles, with horizontal whiskers extending to a maximum distance of 1.5 × interquartile range from the hinge. Data beyond the whisker ends are plotted individually.
Using SCOOP, we were able to recapitulate previous tissue-level COO predictions21 for eight well-established cancer types but often at higher cellular resolution and accuracy, given our use of single-cell rather than bulk epigenomic data (Fig. 1b; Supplementary Fig. 1). For instance, SCOOP predicted bone marrow B cells to give rise to multiple myeloma (MM) – which is supported by the literature39 – in contrast to a prior, more general hematopoietic COO prediction21,25. Also, melanoma was predicted to originate from melanocytes and glioblastoma (GBM) from fetal-like brain cell subsets, in agreement with previous bulk predictions21,25 and expected COOs40,41,42. Interestingly, our model suggested hepatoblasts as the COO for hepatocellular carcinoma (HCC), a type of hepatic progenitor cells (HPC) capable of differentiating into mature hepatocytes, with hepatocytes ranking second (Supplementary Fig. 1). Two competing theories implicate mature hepatocytes and HPCs as the source of HCC43, and our analysis adds weight to the conjecture that hepatoblast-like HPCs might be the primary COO of HCC. Finally, for LUAD and lung squamous cell carcinoma (LUSC), SCOOP pinpointed the specific cell type implicated in tumorigenesis44 – lung AT2 cells, and lung basal cells, respectively – again providing higher resolution and accuracy than previous tissue-level predictions21,25 of breast epithelial cells for both of these two cancers.
SCOOP uncovers a basal COO for most small cell lung cancers
We next conducted an in-depth analysis of lung cancers, including two subtypes which have not been considered in previous works: pleural mesothelioma (PM) and small cell lung cancer (SCLC). For this analysis, we also added a fetal lung scATAC-seq dataset45 and restricted the feature space to only include lung cell subsets (Methods). To visualize SCOOP’s reproducibility, we displayed the number of appearances (n) and the feature importance of the 5 most informative cell subsets after backward feature selection following 100 SCOOP runs (Fig. 1a, c; Methods). SCOOP accurately and robustly predicted AT2 and basal cells as the COO of LUAD and LUSC44, respectively, showing significantly higher feature importance than the next most predictive feature (Fig. 1a, c; Mann-Whitney test, \(p < 1{0}^{-19}\)). Additionally, SCOOP’s prediction of mesothelial cells as the COO of epithelioid PM is in line with current models of mesothelial oncogenesis46.
To our surprise, SCOOP’s prediction for SCLC COO (Fig. 1c; Supplementary Fig. 2a when training with cell features across tissues) – basal cells – challenged the prevalent theory44 that SCLC arises primarily from pulmonary neuroendocrine cells (PNECs), also present in our feature set. Our findings are further bolstered by a previous study showing that inactivation of tumor suppressors Rb1, Pten, and Tp53 in Rbl1-null murine basal cells can give rise to SCLC47. At the time of revising our manuscript, a landmark study utilized multiple genetically-engineered mouse models to show that a tuft-like subtype of SCLC can originate from basal cells but not PNECs, and provided additional experimental evidence pointing to a basal COO for other SCLC subtypes states38. Moreover, we observed that most LUSC and SCLC patients’ mutational density profiles clustered together and separately from LUAD and PM patients’ subclusters (Fig. 1d; Methods). This suggests an intrinsic similarity in the somatic mutational landscapes of LUSC and SCLC and further supports a shared basal COO. Of note, the LUSC and SCLC WGS datasets35,36 used by SCOOP had distinctly different genetic drivers, including an expected high frequency of RB1 and TP53 mutations in SCLC and NFE2L2, CDKN2A, TP53, and PIK3CA mutations in LUSC cases, respectively (Supplementary Fig. 2b).
While SCOOP uncovered basal cells as the predominant COO of SCLC, we further investigated if this was the case for different SCLC subtypes. A recent study identified a rare subset of SCLC tumors that lacked RB1 and TP53 alterations and instead exhibited extensive chromothripsis48. These tumors were also associated with never- or light-smokers, not customary for most SCLCs, and were hence named “Atypical SCLC” (aSCLC) due to their unique pathogenesis characteristics48. The binned SNV profiles of aSCLC cases from two independent studies36,48 clustered separately from all other SCLC tumors, indicating a distinct WGS mutational density for aSCLC (Fig. 1e). Remarkably, SCOOP predicted a pulmonary neuroendocrine COO in both aSCLC patient cohorts36,48 (Fig. 1f, Supplementary Fig. 3a). In contrast, SCOOP still predicted a basal COO for the ASCL1+ neuroendocrine (SCLC-A; Fig. 1g) and three other previously defined SCLC molecular subtypes49,50,51,52,53–NEUROD1+ neuronal (SCLC-N), POU2F3+ tuft-like (SCLC-P), and YAP1+ (SCLC-Y)–although the smaller WGS sample sizes for the latter three subtypes warrants a lower degree of confidence in their predicted COO (Supplementary Fig. 3b). These four molecular subtypes did not co-cluster based on their mutational profile, unlike aSCLC samples (Fig. 1e). Supporting our neuroendocrine COO prediction for aSCLC, Rekhtman and colleagues demonstrated that aSCLC tumors exhibit histogenetic similarity to pulmonary carcinoids48–a class of low-grade neuroendocrine tumors. The study further noted that aSCLC tumors show higher overall survival compared to other SCLC subtypes48, which is consistent with the lower proliferation rate of neuroendocrine versus basal cells that we observed in lung homeostasis (Fig. 1h). Taken together, our data-driven approach agrees with the accepted COO for mesothelioma, LUAD, and LUSC and importantly provides strong genetic evidence for a basal COO in most human SCLCs and a neuroendocrine COO for atypical SCLC cases (Fig. 1i).
SCOOP achieves cell subset granularity in COO predictions
We next examined SCOOP’s ability to discern the COO within intestinal and hematopoietic regenerative lineages, focusing on microsatellite stable (MSS) colorectal cancer (CRC), chronic lymphocytic leukemia (CLL), and acute myeloid leukemia (AML). CRC tumors are typically classified into two major subtypes based on their microsatellite status: microsatellite stable (MSS) and microsatellite instable (MSI)54. Correlating aggregate mutational density of 51 MSS CRC35 with normal colon scATAC-seq data meta-cells31 (Fig. 2a; Methods), we observed the highest association with intestinal epithelial stem cells, the apex of the gut regenerative hierarchy. In agreement with this analysis and prior knowledge55, SCOOP also identified colon stem cells as the most informative feature and COO of MSS CRC when trained using only normal gut scATAC-seq data28,29,31 (Fig. 2b) as well as normal cell subsets across tissues (Supplementary Fig. 4a).
a Left: UMAP of normal colon single-cell assay for transposase-accessible chromatin using sequencing (scATAC-seq) data31 colored by cell annotations (n = 43,626 cells; 27 samples). Middle: Intestinal epithelial cell regenerative hierarchy. Regenerative hierarchy created in BioRender. Tsankov, A. (2025) https://BioRender.com/axbxmm9. Right: Visualization of the Pearson correlation coefficient (r) between aggregated microsatellite stable (MSS) colorectal cancer (CRC, n = 51) single-nucleotide variants (SNV) profile and colon scATAC-seq meta-cells (Methods). The strongest anti-correlations (r ≈ −0.79, red/bottom end of the scale) are concentrated in stem cells, while the weakest anti-correlations (r ≈ −0.675, blue/top end of the scale) occur in enterocytes. b Box plots of the feature importance distribution (100 SCOOP runs) of the top 5 cell-of-origin (COO) predictions amongst normal colon cell subsets28,29,31 for MSS CRC (n = 51; predicted COO in red). Each cell subset is followed by a dataset indicator for that cell subset: D1 for28, D2 for29, D3 for31. Also displayed is the number of times a cell subset appeared in the top 5 features across 100 runs (n). One-sided Mann-Whitney test p-value is displayed. c Top Left: UMAP of all peripheral blood mononuclear cells (PBMC) and bone marrow scATAC-seq cell subsets from32 (n = 33,513 cells; 10 samples). Bottom Left: Hematopoietic regenerative hierarchy. Regenerative hierarchy created in BioRender. Tsankov, A. (2025) https://BioRender.com/24n2gzz. Right: Visualization of the Pearson correlation coefficient (r) between aggregated chronic lymphocytic leukemia (CLL, n = 90) and acute myeloid leukemia (AML, n = 13) SNV profiles and scATAC-seq meta-cells. For CLL, the strongest anti-correlations (r ≈ −0.63, red/bottom end of the scale) are observed in B cells, while the weakest anti-correlations (r ≈ −0.51, blue/top end of the scale) occur in PBMC T cells. For AML, highest anti-correlations (r ≈ −0.38) are enriched in myeloid (e.g., granulocyte-monocyte progenitors, or GMP, GMP/Neutrophils, or GMP.Neut, common myeloid progenitor and lymphoid-primed multipotent progenitor, or CMP.LMPP) and erythroid progenitor populations, whereas lymphoid lineages (B, T, and NK cells) tend to have the lowest anti-correlations (r ≈ −0.30). d Box plots of the feature importance distribution (100 SCOOP runs) of the top 5 COO predictions amongst blood and bone marrow cell subsets from dataset D432 for CLL (n = 90) and AML (n = 13) aggregated SNV profiles (predicted COO in red, similar cell subsets in pink). Also displayed is the number of times a cell subset appeared in the top 5 features across 100 runs (n). One-sided Mann-Whitney test p-values are displayed, where Bonferroni correction for multiple hypothesis testing was used when more than one comparison was made. Cell type abbreviations are listed in Supplementary Data 3. Box plot vertical lines show 25th, 50th (median), and 75th percentiles, with horizontal whiskers extending to a maximum distance of 1.5 × interquartile range from the hinge. Data beyond the whisker ends are plotted individually.
We next leveraged scATAC-seq data from bone marrow and peripheral blood mononuclear cells (PBMCs)32 to investigate the COO of CLL and AML. Meta-cell correlation analysis showed that CLL was most anti-correlated with bone marrow B cells, but not PBMC B cells (Fig. 2c; top right). In agreement, SCOOP predicted bone marrow B cells as the COO (Fig. 2d; Supplementary Fig. 4a when training with cell features across tissues), supporting the prevailing hypothesis that B cells give rise to CLL56. In contrast, AML was highly anti-correlated with multiple myeloid progenitors, possibly due to the high interpatient heterogeneity observed in this leukemia57, most notably with bone marrow early erythrocyte (Early.Eryth) and granulocyte macrophage progenitors (GMP; Fig. 2c, bottom right). Prior work has shown leukemia stem cell (LSC) transcriptional activity resembles lymphoid-primed multipotent progenitors (LMPPs) and GMPs rather than hematopoietic stem cells (HSCs), suggesting that LSC transformation largely occurs at the progenitor stage, either directly from progenitors with abnormal self-renewal capabilities or from HSCs upon further differentiation58. Additionally, studies using murine leukemia models and various genetic modifications indicate that both HSCs and committed myeloid progenitor cells can evolve into LSCs, which phenotypically and molecularly resemble committed myeloid progenitor cells59,60. Curiously, SCOOP supported these studies, where the two most informative cell features were highly similar bone marrow cell subsets GMP and GMP/Neutrophils (GMP.Neut) – both multipotent myeloid progenitors (Fig. 2d). To evaluate the robustness of our AML prediction, we utilized another bone marrow scATAC-seq data61 (Supplementary Data 4) to independently train our model, and also found GMP.Neut myeloid progenitors as the most informative epigenetic feature (Supplementary Fig. 4b; Methods). Thus, our analyses bolster the hypothesis that myeloid lineage differentiation is a prerequisite for AML development62. Worth noting, our meta-cell correlation analysis highlights SCOOP’s capacity to exploit complex relationships that may not be easily captured by correlation analyses (see correlation-based predictions in Supplementary Data 1). In sum, SCOOP identified the COO for MSS CRC (colon stem cells) and CLL (B cells) at cell subset resolution while also highlighting the role of multipotent myeloid progenitors in AML development.
Distinct COOs for cancers with different histologies
Beyond displaying cell subset granularity, SCOOP also identified histological cancer subtypes with different COOs. We first examined three subtypes of kidney renal cell carcinoma (RCC): clear cell RCC (ccRCC), papillary RCC (pRCC), and chromophobe RCC (chRCC). Both ccRCC and pRCC are thought to originate from the proximal tubule in the kidney, while it is suspected that chRCC originates from the distal tubule63. SCOOP predicted proximal tubule progenitor-like (PTPL) cells as the ccRCC and pRCC COO and collecting duct, intercalated cell type A (ICA) from the distal tubule as the chRCC COO, confirming prior work while again demonstrating cell subset granularity in its prediction (Fig. 3a; Supplementary Fig. 5 when training with cell features across tissues). In agreement, individual patient somatic mutation profiles demonstrate higher similarity between ccRCC and pRCC in comparison to chRCC (Fig. 3b).
a Box plots of the feature importance distribution (100 SCOOP runs) of the top 5 cell-of-origin (COO) predictions (predicted COO is shown in red) amongst kidney-related cell subsets for papillary renal cell carcinoma (pRCC, n = 32), clear cell renal cell carcinoma (ccRCC, n = 111), and chromophobe renal cell carcinoma (chRCC, n = 43). Each cell subset is followed by a dataset indicator for that cell subset: D2 for29, D6 for33. Also displayed is the number of times the feature appeared in the top 5 features across the 100 runs (n). One-sided Mann-Whitney test p-values are displayed. Kidney model created in BioRender. Tsankov, A. (2025) https://BioRender.com/ht2q3vc. b UMAP of individual kidney cancer whole-genome sequencing (WGS) samples binned mutational profiles (dots) colored by cancer subtype (chRCC, n = 43; ccRCC, n = 111; pRCC, n = 32). c UMAP dimensionality reduction of individual pancreatic cancer WGS samples binned mutation profiles (dots) colored by cancer subtype (adenocarcinoma, n = 232; neuroendocrine, n = 47). d Left: UMAP of stomach and pancreas scATAC-seq data from29 (n = 58,175 cells; 12 samples). Thinner dashed line encompasses pancreatic cells, whereas thicker dashed line demarcates stomach cells. Right: UMAPs displaying the Pearson correlation coefficient (r) between aggregated pancreas adenocarcinoma (PDAC, n = 232) and pancreatic neuroendocrine tumor (PNET, n = 47) mutational profiles and single-cell assay for transposase-accessible chromatin using sequencing (scATAC-seq) data meta-cells. For PDAC, the strongest anti-correlations (r ≈ −0.60, red/bottom end of the scale) are observed in stomach goblet and parietal and chief cells, while the weakest anti-correlations (r ≈ −0.52, blue/top end of the scale) occur in stromal cells. For PNET, highest anti-correlations (r ≈ −0.52) are enriched in pancreas islet endocrine cells, whereas stromal and pancreas acinar cells tend to have the lowest anti-correlations (r ≈ −0.47). e Box plots of the feature importance distribution (100 SCOOP runs) of the top 5 COO predictions amongst pancreas- and stomach-related cell subsets28,29 for PDAC (n = 232) and PNET (n = 47; predicted COOs highlighted in red, similar cell subsets in pink). Each cell subset is followed by a dataset indicator for that cell subset: D1 for28, D2 for29. One-sided Mann-Whitney test p-values are displayed, with Bonferroni correction for multiple hypothesis testing. f Accepted model of colorectal cancer (CRC) COOs agrees with SCOOP’s predictions: colon goblet cells for microsatellite instable (MSI), and intestinal epithelial stem cells for MSS. Intestinal model created in BioRender. Tsankov, A. (2025) https://BioRender.com/001gcxg. g Box plot of the feature importance distribution (100 SCOOP runs) of the top 5 COO predictions amongst colon-related cell subsets28,29,31 for CRC, MSI (n = 7; predicted COO in red). Each cell subset is followed by a dataset indicator for that cell subset: D1 for28, D2 for29, D3 for31. One-sided Mann-Whitney test p-values are displayed. Cell type abbreviations are defined in Supplementary Data 3. Box plot vertical lines show 25th, 50th (median), and 75th percentiles, with horizontal whiskers extending to a maximum distance of 1.5 × interquartile range from the hinge. Data beyond the whisker ends are plotted individually.
Exploring the cellular origins of pancreatic ductal adenocarcinoma (PDAC) and pancreatic neuroendocrine tumor (PNET), we also observed intrinsic differences in tumors’ mutational profiles that cluster by histological subtypes (Fig. 3c). Lineage-tracing studies have demonstrated that acinar cells with oncogenic KRAS mutation (present in >90% of PDAC cases), and not ductal cells, can give rise to PDAC while undergoing a process known as acinar-to-ductal metaplasia (ADM)64,65. Meanwhile, PNETs are thought to arise from islet cells, which are part of the endocrine system of the pancreas66. In agreement, we observed that PDAC and PNET aggregated mutational profiles were most anti-correlated with stomach goblet cells and islet endocrine cells (Fig. 3d), respectively, and SCOOP further reinforced these results when trained using all pancreas and stomach cell features (Fig. 3e) and, in the case of PDAC, all cell features (Supplementary Fig. 5). Stomach goblet cells secrete mucus to protect the stomach lining, matching previous bulk-level prediction of stomach mucosa and suggesting metaplasia transformation in PDAC25, which we explore further in the next section. Our PNET COO prediction of pancreatic islet endocrine cells was also in agreement with PNET bulk-tissue prediction25, but came in second when training SCOOP across different tissues to the highly similar colon endocrine cells (Supplementary Fig. 5), highlighting the benefit of restricting SCOOP’s feature space to anatomically relevant cell subsets.
Motivated by previous studies arguing that the MSS CRC subtype arises from stem cells in the colon crypt, while the MSI subtype arises from gastric metaplasia67 (Fig. 3f), we leveraged SCOOP to investigate this hypothesis in more detail. To capture the metaplasia cell state relevant to MSI tumor COO, we added precancerous polyp scATAC-seq data31 to our feature space. SCOOP supported the hypothesis that MSI tumors likely arise from a metaplasia cell state distinct from MSS CRC tumorigenesis trajectory and further pointed specifically to polyp colon goblet cells as the COO (Fig. 3g; Supplementary Fig. 5 when training with cell features across tissues).
Multiple gastrointestinal cancers develop via a metaplastic intermediate
To further explore the hypothesis that PDAC and MSI CRC originate from intermediate metaplastic cell populations, we analyzed scRNA-seq datasets that induce metaplasia (pancreatic or intestinal) in response to injury or presence of oncogenic drivers. In particular, we investigated human chronic pancreatitis patient samples68, a pancreas injury mouse model69 that triggers ADM, PDAC development following induction of KrasG12D and p53 genetic alterations70, and precancerous cells from human colonoscopy samples67. We obtained highly specific human stomach goblet cell markers71, human pancreas acinar cell markers68 (the alternative COO for PDAC64,65), mouse pancreas acinar cell markers69, and mouse stomach goblet cell markers72 from several relevant scRNA-seq datasets (Methods; Supplementary Data 5; Supplementary Fig. 6a). Comparing acinar and stomach goblet cell signature scores across both acinar and metaplastic cells obtained from human chronic pancreatitis samples revealed a higher transcriptional similarity of metaplastic cells with stomach goblet cells than with acinar cells (Fig. 4a), supporting their role as the COO during injury-induced metaplasia in human patient samples. As in human chronic pancreatitis, we also found that metaplastic cells resemble stomach goblet cells rather than pancreatic acinar cells in scRNA-seq data from a mouse pancreas injury model69 that induces inflammation, pancreatic tissue reprogramming, and ADM69 (Fig. 4b).
a Left: UMAP of epithelial cells (n = 1161; 2 samples) from human chronic pancreatitis single-cell RNA sequencing (scRNA-seq data)68, colored by human stomach goblet cell module score. Warm colors (red, right end of the scale) indicate high module scores, whereas cold colors (blue, left end of the scale) indicate low module scores. Right: Violin plots comparing human pancreatic acinar and stomach goblet cell module scores in acinar (top) and metaplasia (bottom) cell clusters. Two-sided Mann-Whitney test p-values are displayed; *\(p < 0.0001.\) b Left: UMAP of epithelial cells (n = 13,362; 4 samples) from mouse pancreas injury model scRNA-seq data69, colored by mouse stomach goblet cell module score. Warm colors (red, right end of the scale) indicate high module scores, whereas cold colors (blue, left end of the scale) indicate low module scores. Right: Violin plots comparing mouse pancreatic acinar and stomach goblet cell module scores in acinar (top) and metaplasia (bottom) cell clusters. Two-sided Mann-Whitney test p-values are displayed; *\(p < 0.0001.\) c Violin plots comparing mouse pancreatic acinar and stomach goblet cell module scores in epithelial cells (21 samples) per experimental condition: normal (N1), regenerating (N2), pre-malignant (K1-K4), and malignant (K5, K6). Mouse models and treatment conditions are represented on the x-axis. Two-sided Mann-Whitney test p-values are displayed; *\(p < 0.0001.\) Mouse model illustration created in BioRender. Tsankov, A. (2025) https://BioRender.com/2uc0u4y. d Violin plots comparing human colon stem and goblet cell module scores in precancerous stem-like (left) and metaplastic (right) cell clusters (55 samples). Two-sided Mann-Whitney test p-values are displayed; *\(p < 0.0001.\) e Box plots of the feature importance distribution (100 SCOOP runs) of the top 5 cell-of-origin (COO) predictions for biliary (n = 34), esophageal (n = 97), and stomach cancer (n = 68; predicted COOs are highlighted in red, similar cell subsets in pink). Each cell subset is followed by a dataset indicator for that cell subset: D1 for28, D2 for29, D3 for31, D4 for32, D5 for34, D6 for33. One-sided Mann-Whitney test p-values are displayed, with Bonferroni correction for multiple hypothesis testing in the case of stomach and esophageal adenocarcinoma. f Left: Box plots of the feature importance distribution (100 SCOOP runs) for the most predictive scATAC-seq feature (highlighted in red, similar cell subsets in pink) for the binned mutational profile of intestinal metaplasia whole-genome sequencing (WGS) samples74 (n = 5). Also displayed is the number of times the feature appeared in the top 5 features across 100 SCOOP runs (n). Each cell subset is followed by a dataset indicator for that cell subset: D1 for28, D2 for29, D3 for31, D4 for32, D5 for34, D6 for33. One-sided Mann-Whitney test p-values are displayed, with Bonferroni correction for multiple hypothesis testing. Middle: Bar plots of the number of times cell subsets appeared as the top feature across 100 runs of SCOOP with the most frequently appearing feature highlighted in red. Exact binomial test p-values are shown, with Bonferroni correction for multiple hypothesis testing. Right: Box plots displaying the test set variance explained (test \({R}^{2}\)) by the model runs (n) for which goblet cells (title) were the top predicted feature when 10, 5, 2, and 1 features remained following backward feature selection. Box plot horizontal (or vertical) lines show 25th, 50th (median), and 75th percentiles, with vertical (or horizontal) whiskers extending to a maximum distance of 1.5 × interquartile range from the hinge. Data beyond the whisker ends are plotted individually.
To trace the cellular dynamics and role of metaplasia during tumorigenesis, we leveraged time-course scRNA-seq data collected from various stages of a state-of-the-art mouse model for PDAC development70, starting with normal pancreatic cells (stages N1-N2), during pre-neoplasia (stages K1-K2) and benign neoplasia (stages K3-K4) following induction of KrasG12D mutation, as well as during primary tumor formation (K5) and metastasis (K6) following induction of p53 genetic alteration (Fig. 4c). Scoring epithelial cells for acinar and stomach goblet signatures across different stages of PDAC formation, we observed a loss of acinar cell identity following induction of KrasG12D mutation (K1-K6) and a simultaneous increase of stomach goblet signature expression, especially during benign neoplasia (Fig. 4c). We additionally observed that gastric-like and ADM cell populations identified in70, primarily present in stages K1-K4 of PDAC development, had the highest similarity to our stomach goblet cell signature, which argues that these populations may represent the key transitional cell states during metaplasia-driven tumorigenesis (Supplementary Fig. 6b). To investigate tumor development in CRC, we obtained scRNA-seq data of metaplastic and stem-like precancerous cells from human colonoscopy samples67. We observed higher transcriptional similarity between precancerous stem-like cells and normal colon stem cells as well as between metaplastic cells and normal colon goblet cells (Fig. 4d). This aligns well with our COO predictions (Fig. 3f-g) and with the accepted models for CRC progression67, where MSS CRC is posited to originate from intestinal epithelial stem cells, while MSI CRC is likely derived from metaplastic cells.
Similar to PDAC and MSI CRC, three other gastrointestinal cancers – biliary, esophageal, and stomach adenocarcinoma – were predicted to arise from stomach goblet cells (Fig. 4e; Supplementary Fig. 6c), which matches previous bulk-level predictions of stomach mucosa and also suggests metaplasia transformation in these cancers25,73. To directly link the stomach goblet cell epigenome with metaplasia, we obtained WGS data from intestinal metaplasia samples collected during gastric cancer progression74. Stomach goblet cells were indeed the most predictive epigenomic feature for intestinal metaplasia mutational profiles when running SCOOP (Fig. 4f, Supplementary Fig. 6d), validating our claim that stomach goblet cell COO prediction represents an intermediate metaplastic state during tumorigenesis for multiple gastrointestinal cancers. In agreement with SCOOP’s predictions, esophageal adenocarcinoma formation is thought to undergo a transitional metaplasia phase, called Barrett’s esophagus75. Moreover, two biliary cancer studies investigating metaplastic lesions of extrahepatic bile ducts found that goblet cells were the predominant cell type within those lesions76,77. Taken together, our work argues that multiple gastrointestinal cancers likely undergo an intermediate metaplasia state resembling stomach goblet cells during tumorigenesis.
Gliomas likely arise from fetal-like multipotent progenitor cells
Since SCOOP can accurately link distinct histological subtypes with their respective COO, we next examined the genesis of different brain cancers. In light of prior research indicating a significant role of fetal-like neural stem cells (NSCs) and oligodendrocyte progenitor cells (OPCs) in brain tumorigenesis41,42,78,79, we leveraged two additional scATAC-seq datasets that extensively characterized fetal and adult brain cell subsets’ chromatin accessibility30,80. Interestingly, SCOOP predicted that medulloblastoma (MB), GBM, pilocytic astrocytoma (PA), and oligodendroglioma (OG) all originate from fetal-like cell subsets (Fig. 5a; Supplementary Fig. 7 when training with cell features across tissues), in agreement with previous bulk epigenomic data modeling21,25. For MB, SCOOP predicted granule neurons from fetal cerebellum as the COO, closely matching the known anatomical and cellular origin (cerebellar granule neuron precursor cells) for one of the four major subtypes of MB, the Sonic Hedgehog (SHH) subtype79. The other major subtypes – WNT, Group 3, Group 4 – are thought to arise from cell types that were not present in our scATAC-seq dataset: rhombic lip cells for WNT81 and primitive progenitor cells for the latter two subtypes81,82.
a Box plots of the feature importance distribution (100 SCOOP runs) of the top 5 cell-of-origin (COO) predictions (COO highlighted in red, similar cell subsets in pink) amongst brain-related cell subsets28,29,30,80 for glioblastoma (GBM, n = 39), oligodendroglioma (OG, n = 18), pilocytic astrocytoma (PA, n = 89), and medulloblastoma (MB, n = 141). Each cell subset is followed by a dataset indicator for that cell subset: D1 for28, D2 for29, D7 for30, D8 for80. Also displayed is the number of times the feature appeared in the top 5 features across the 100 runs (n). One-sided Mann-Whitney test p-values are displayed, with Bonferroni correction for multiple hypothesis testing in the case of PA. Brain model created in BioRender. Tsankov, A. (2025) https://BioRender.com/f8uyajw. b Left: UMAP of fetal brain single-cell assay for transposase-accessible chromatin using sequencing (scATAC-seq) data cell subsets from30 (n = 31,074 cells; 13 samples). Right: UMAPs displaying the Pearson correlation coefficient (r) between aggregated PA (n = 89), GBM (n = 39), and OG (n = 18) mutational profiles and scATAC-seq meta-cells. For PA, the strongest anti-correlations (r ≈ −0.50, red/bottom end of the scale) are observed in oligodendrocyte progenitor cells (OPCs) and multipotent glial progenitor cells (mGPCs), while the weakest anti-correlations (r ≈ −0.33, blue/top end of the scale) occur in a subset of interneurons and glutamatergic neurons. For GBM, highest anti-correlations (r ≈ −0.48) are enriched in mGPCs, whereas lowest anti-correlations (r ≈ −0.32) occur in a subset of interneurons. For OG, the strongest anti-correlations (r ≈ −0.56, red/bottom end of the scale) are observed in OPCs and mGPCs, while the weakest anti-correlations (r ≈ −0.42, blue/top end of the scale) occur in a subset of glutamatergic neurons. c Test set variance explained (%) by the predicted COOs across 6 additional cancer types (B-cell lymphoma, n = 105; Myeloproliferative neoplasm, n = 23; Breast adenocarcinoma, n = 195; Leiomyosarcoma; n = 34; Thyroid adenocarcinoma, n = 48; Uterus adenocarcinoma, n = 44). The predicted COO for each cancer type is highlighted in red (also see Supplementary Fig. 8). Error bars show the standard error of the mean (SEM), after 100 SCOOP runs. One-sided Mann-Whitney test p-values are displayed. Each cell subset is followed by a dataset indicator for that cell subset: D1 for28, D2 for29, D4 for32. d Model performance as a function of WGS sample size for tumors with high (lung squamous cell carcinoma, or LUSC, melanoma), moderate (hepatocellular carcinoma, or HCC, GBM), and low (chromophobe renal cell carcinoma, or chRCC, Medulloblastoma, or MB) tumor mutation burden (TMB) tumors. Top: Distribution of variance explained across 100 SCOOP runs for different number of WGS samples, randomly subsampled. Bottom: Number of appearances of the expected COO as the top feature across 100 SCOOP runs for different whole-genome sequencing (WGS) sample sizes. Filled and hollow dots indicate that the expected COO was and wasn’t the most frequently appearing feature, respectively. Cell type abbreviations are listed in Supplementary Data 3. Box plot horizontal (or vertical) lines show 25th, 50th (median), and 75th percentiles, with vertical (or horizontal) whiskers extending to a maximum distance of 1.5 × interquartile range from the hinge. Data beyond the whisker ends are plotted individually.
For both pilocytic astrocytoma (PA) and oligodendroglioma (OG), SCOOP predicted the COO to be OPCs from the fetal cerebral cortex (Fig. 5a). Despite its namesake, recent studies have suggested that PA originates from the oligodendrocyte lineage83. Furthermore, PA cancer cells have been compared with various brain cell types using single cell transcriptomic atlases, and it was found that they exhibited a gene expression signature most similar to OPCs84. Finally, for GBM, while SCOOP originally predicted Oligo/Astrocytes from the fetal brain (Fig. 1b) as the COO, after adding the comprehensive fetal brain dataset, the COO was more specifically predicted as multipotent glial progenitor cells (mGPCs) from the fetal cerebral cortex. This is further supported by our recent fetal brain cell atlas, showing high transcriptional similarity of GBM malignant cells to multipotent progenitor fetal cell populations85. We observed highly similar cell subset specificity when conducting a meta-cell correlation analysis on scATAC-seq data from30 (Fig. 5b). One limitation of our glioma findings is that our feature set does not contain adult NSC epigenomes, which have been implicated in the genesis of adult GBM and OG42. However, our datasets contained fetal multipotent progenitor cells (mGPC, nIPC, radial glial cells) that can give rise to different glial and neuronal populations akin to NSCs78.
Pan-cancer COO predictions across 37 cancer types
We next analyzed WGS data from B-cell lymphoma, myeloproliferative neoplasm (MPN), breast adenocarcinoma, leiomyosarcoma, thyroid, and endometrial cancer, and found that SCOOP’s predictions matched one of the putative COOs for each of these cancers86,87,88,89 (Fig. 5c; Supplementary Fig. 8a). In the case of leiomyosarcoma, SCOOP predicted stromal cells as the COO, which show high chromatin accessibility at established smooth muscle (the conjectured COO87) cell marker loci (Supplementary Fig. 8b). For endometrial cancer, the predicted COO – PAEP/MECOM positive cells from the placenta – corresponds to endometrial epithelial cells29 that are believed to give rise to these tumors89. MPN most likely arises from HSCs90 but there is evidence suggesting that it may also originate from committed hematopoietic progenitors similar to HSCs91. SCOOP’s top three COO predictions for MPN were bone marrow CD34+ early basophils, GMP, and common myeloid progenitor (CMP)/LMPP (CMP.LMPP; Supplementary Fig. 8b, when using tissue-specific and all features), all of which represent multipotent hematopoietic progenitors with close proximity to HSCs in their epigenome and regenerative hierarchy (Fig. 2c). Worth noting, MPN had the second lowest average number of mutations per sample (µ = 805) among the cancer types we analyzed (Supplementary Data 1), which may explain the observed variance in its prediction. Finally, while SCOOP predicted basal epithelial cells from mammary tissue as the COO for breast adenocarcinoma – as opposed to luminal cells (present in our dataset) that are considered a more predominant COO25,92 – basal cells still constitute a possible COO25,92.
We also report COO predictions for other cancer types with available WGS data (Supplementary Fig. 9), which we categorized as either 1) matching a proxy cell type, 2) missing all expected COOs scATAC-seq data, or 3) demonstrating low variance explained (<10%). Category 1 included cervical as well as head and neck squamous cell carcinoma COO predictions of esophageal epithelial cells, which was the closest comparative cell profile in our dataset and in25 to the expected COO for these tumors (Supplementary Data 1). Osteosarcoma also fell into Category 1, matching stromal cells from the heart, as it is expected to arise primarily from mesenchymal cells93. Category 2 included bladder cancer, prostate cancer, and ovarian cancer (Supplementary Fig. 9; Supplementary Data 1). SCOOP identified stomach goblet cells as the top feature for bladder transitional cell carcinoma, in agreement with prior tissue-level stomach mucosa prediction25. Unlike other metaplasia-associated neoplasms in our study, our feature set did not contain the expected epithelial COO (transitional cells94) for bladder cancer, reducing our confidence in this prediction. Finally, Category 3 consisted of breast lobular carcinoma, for which the median variance explained of the top feature was lower than 10% (Supplementary Fig. 9).
To establish a simple baseline for SCOOP’s prediction accuracy, we compared its predictions across 31 cancer types for which a putative COO (Supplementary Data 1) was present in our scATAC-seq database (Supplementary Data 3), with those predicted by correlating (Spearman and Pearson) cancer mutational profiles with all scATAC-seq features and picking the most anti-correlated feature as the COO. Given the inherent difficulty in establishing “ground-truth” COOs for human cancers, for our benchmarking analysis we considered a given COO prediction to be correct if it was supported by at least one peer-reviewed publication (enumerated in Supplementary Data 1). We found that SCOOP outperformed both correlation approaches, achieving an accuracy of 30/31, compared to 26/31 and 13/31 for Spearman and Pearson correlation, respectively (Supplementary Fig. 10a; Supplementary Data 1). Additionally, we did not find a significant association between the heterogeneity in SCOOP’s predictions and the heterogeneity in WGS profiles (Supplementary Fig. 10b; Methods). To strengthen our confidence in SCOOP’s predictions, we acquired additional WGS data from the Clinical Proteomic Tumor Analysis Consortium95 (CPTAC; Methods) available for 5 cancer types with COO predictions in our study–LUAD, LUSC, GBM, RCC, and PDAC. Running SCOOP on WGS data from these independent cohorts reproduced our original predictions for all 5 cancers (Supplementary Fig. 10c). As additional validation, we split the original HCC data from PCAWG (Fig. 1b) by research study origin (France, Japan, United States) and found that WGS data from all three independent cohorts predicted hepatoblast as the COO (Supplementary Fig. 10d). Overall, SCOOP outperforms correlation analysis and achieves highly reproducible COO predictions across independent WGS datasets.
Furthermore, we quantified the relationship between WGS sample size and SCOOP’s prediction accuracy for 6 cancers with low, medium, and high average tumor mutational burden (TMB; Supplementary Data 1; Methods). Interestingly, for both medium and high TMB cancers examined, as few as 5 WGS samples were enough for SCOOP to identify a correct COO in at least 68% of runs (≥42.4% variance explained), while low TMB tumors (e.g., chRCC, MB) required 30 samples to achieve similar levels of performance (correct COO ≥ 69% of runs; Fig. 5d). Moreover, in order to glean some insights into the model’s predictions, we quantified SCOOP’s prediction accuracy for the same 6 cancer types at the bin level, analyzing the bins for which mutations were well predicted by the model (defined as “most-accurate”), and those for which mutations were not well predicted (defined as “under-” or “over-predicted”; Supplementary Fig. 11a; Methods). We first observed that the “most-accurate” and “under/over-predicted” bins did not significantly overlap across cancer types (Supplementary Fig. 11b). The “most-accurate” and “under/over-predicted” bins showed a higher and lower chromatin accessibility compared to other bins, respectively, when surveying the predicted COO epigenome for all 6 cancer types (Supplementary Fig. 11c; Methods). Moreover, the “under/over-predicted” and “most-accurate” regions contained genes with lower and higher expression, respectively, relative to all other genes when analyzing normal tissue-of-origin-matched bulk RNA-sequencing data from the Genotype-Tissue Expression project96 (GTEx; Supplementary Fig. 11d). In agreement, the bin categories also showed consistent enrichments of activating (Histone 3, Lysine 27 Acetylation, H3K27Ac) and repressive (Histone 3, Lysine 9 trimethylation; H3K9me3) chromatin marks across all cancers with available normal tissue-of-origin-matched ChIP-seq data from the Epigenomics Roadmap project97 (Supplementary Fig. 11e-f; Methods). Taken together, lower prediction accuracy bins reside in unique genomic regions containing inaccessible chromatin, lowly expressed genes, and heterochromatic histone marks (H3K9me) in the predicted normal COO.
Discussion
Our work combined machine learning, WGS, and scATAC-seq data to predict the cell of origin of 37 human cancer subtypes. Our approach employing single-cell epigenomic data marks a significant advancement, offering greatly improved resolution in the cellular subsets, developmental, and regenerative hierarchies underlying the genesis of cancer. For most cancers examined in our study, the predicted COOs and anatomical location were highly reproducible and aligned closely with those posited by prior research, serving as a validation of both the existing scientific consensus and the accuracy and reliability of our methodology.
Beyond validation, the true potential of our approach lies in its capacity for data-driven hypothesis generation, particularly for cancers with ambiguous or unknown cellular and anatomical origins. Our study’s surprising finding of a basal COO for most SCLCs, which has historically been thought to arise from neuroendocrine cells based on histological and transcriptomic similarities44, exemplified the potential of our ML approach to uncover unexpected cellular origins that will open future avenues for research. Substantiating our computational approach, a concurrent study employing cellular lineage tracing in genetically-engineered mouse models of SCLC supported the prediction that basal cells may constitute the predominant COO in SCLC, as they can give rise to all the SCLC subtypes upon transformation and in proportions matching human epidemiological data38. Moreover, genetic alterations enriched in tuft-like SCLC (i.e., Myc gain, Pten loss) gave rise to the appropriate subtypes when tumors were initiated from a basal cell, but not from a neuroendocrine cell38. These findings combined with our data-driven approach utilizing human tumor samples have important clinical implications for prevention, early detection, and treatment of SCLC. For example, early screening programs and smoking cessation strategies that target individuals with molecular changes in bronchial basal cells98 can be revised to account for not only LUSC but also SCLC incidence.
In contrast to most SCLCs, SCOOP predicted a neuroendocrine COO for rare aSCLC cases lacking TP53 and RB1 genetic alterations. In accordance, previous work showed that aSCLC shares histogenetic features with pulmonary carcinoids48, indolent neuroendocrine tumors that are also not associated with smoking and shown to occur in younger individuals99,100. Given that aSCLC is posited to arise from pulmonary carcinoids via chromotripsis48, our results by extension argue that pulmonary carcinoids also arise from neuroendocrine cells. While our work elucidates the different COOs across SCLC subtypes, it will be fascinating to investigate the cellular beginnings of other neuroendocrine tumors across tissues. For example, olfactory neuroblastomas101 display subtype heterogeneity resembling that of SCLC and were shown to arise from globose basal cells using genetically-engineered mouse models101.
Our study also identified goblet cells as the most predictive epigenomic feature for the somatic mutational landscape of intestinal metaplasia74 and several cancer types–PDAC, MSI CRC, biliary, esophageal, and stomach adenocarcinoma. These results are compatible with an intermediate metaplastic state contributing to tumorigenesis across diverse tissues and organs, which is further corroborated by prior studies64,65,67,75,102 and bulk epigenetic predictions25. Thus, our work highlights the widespread contribution of metaplasia to gastrointestinal cancers and underscores that the biological principles and epigenome of metaplastic transitions may be highly similar across tissues. These findings can inform the development of future metaplasia biomarkers for improving early detection across multiple cancers as well as the repurposing of successful prevention103, risk assessment, and treatment strategies.
Despite the greatly enhanced resolution, accuracy, and scale of SCOOP’s COO predictions, our ML approach has several limitations that will require future research. A fundamental impediment arises from previous observations21,25 that robust model prediction requires aggregation of WGS profiles from different tumor samples that may have different COOs. This presents challenges for obtaining personalized predictions, which can be remedied in the future by acquiring additional WGS data that can enable grouping of patient tumors with similar COOs. Another limitation pertains to the comprehensiveness of our single-cell atlas and data quality across tissues, potentially omitting relevant cell subsets that could serve as the COO for specific cancers. Finally, SCOOP identifies the pre-malignant cellular ancestor whose chromatin accessibility best explains a cancer’s cumulative somatic mutational landscape. In some cases, this cell type can differ from the normal cell that acquires the initial oncogenic hit, as appears to be the case for PDAC, other metaplasia-related cancers in our study, AML, and as shown previously in gliomas41,42 and other cancers; future modeling and experimental studies tracing the transition from precancerous to malignant cell states will be necessary to uncover the exact trajectory of cellular transitions involved in tumorigenesis.
SCOOP contrasts with previous approaches that necessitated individual experiments for each sample—for instance, conducting 100 separate experiments to generate 100 ATAC-seq profiles from bulk tissue. Instead, using publicly available scATAC-seq data, our approach successfully derived 559 distinct cell subset profiles from 42 tissue experiments. This enhanced efficiency not only facilitates the identification of a wider variety of cell types, but also paves the way for a cost-effective framework for COO identification by substantially decreasing the need for additional sequencing experiments and streamlining laboratory procedures. Future studies will also benefit from the increasing number and quality of WGS and scATAC-seq data being generated, and employ SCOOP to investigate rare cancers, such as aSCLC, and more refined histological and molecular subtypes not examined here. Moreover, expanded scATAC-seq sampling across the human body can further enhance our method’s ability to identify the anatomical location for a cancer’s COO, as demonstrated for MB (cerebellum). Our easy-to-use computational platform is accessible to all cancer biologists, requiring only the availability of WGS data for a cancer of interest and, if necessary, scATAC-seq data from the corresponding normal tissue to complement our 559 normal cell subsets already assembled.
Methods
Ethics statement
This research was conducted entirely with previously published data that has been deidentified by each study and, hence, does not require human subjects study protocol and complies with all relevant ethical regulations.
Whole genome sequencing (WGS) data acquisition and processing
All cancer WGS data besides that for mesothelioma37, SCLC36,48, multiple myeloma21, MSI-high CRC104, intestinal metaplasia74, and CPTAC95 cancers (Supplementary Fig. 10c) were obtained from the Pan-Cancer Analysis of Whole Genomes (PCAWG) study35. PCAWG samples were obtained both from the publicly available, International Cancer Genome Consortium (ICGC) portion of PCAWG via the ICGC data portal35, and from the restricted access TCGA portion of PCAWG via Bionimbus105. Only samples on the tumor whitelist from PCAWG were processed for analysis. For pleural mesothelioma, the somatic mutation genome locations in37 were provided directly by the authors. We restricted our analysis to epithelioid pleural mesothelioma, since the number of samples for the other two histological subtypes (sarcomatoid and biphasic) was very low (2 and 3 respectively). We also excluded samples that were classified as “Not Otherwise Specified.” For SCLC, the SNVs from36 were provided directly by the authors through the European Genome-Phenome Archive (EGA). The SCLC molecular subtypes of different WGS datasets were obtained from49. The aSCLC somatic WGS mutations from48 were provided directly by the authors. Multiple myeloma data was obtained and processed as in21. MSI-high data was obtained from TCGA. Finally, we acquired SNVs from WGS of 5 intestinal metaplastic samples directly from the authors of74. WGS VCF files and available clinical information for CPTAC-3 datasets (lung, brain, pancreas, and kidney cancers) were downloaded from the TCGA data portal (https://portal.gdc.cancer.gov/). Only variants marked as PASS were kept and their coordinates were converted to hg19 using the R package liftOver106 (v1.26.0).
We obtained SNVs from WGS data for each cancer type examined in our study and aggregated the variant counts across samples into 1 megabase pair bins, excluding sex chromosomes21. In brief, all somatic single nucleotide mutation data per cohort were converted to BED format, and intersected using BEDtools with the 1MB bins. The number of mutations per bin were then aggregated by cancer type/subtype. These bins exclude regions that overlap centromeres and telomeres, and regions where the fraction of mappable base pairs is lower than 0.92. Since one aSCLC WGS sample (“A07”) had an outlier number of mutations (189,396) compared to the rest of the WGS samples (average of 13,618 per sample), we corrected for the effect of the outlier by 1) excluding the outlier sample and 2) first scaling the mutation counts for each aSCLC WGS sample across bins to have a zero mean and unit variance before running SCOOP, and both approaches robustly predicted a neuroendocrine COO.
Somatic variant calling for MSI CRC WGS samples
For the microsatellite instability (MSI) samples, both tumor and matched normal BAM files containing mapped reads onto the human genome version hg38 were downloaded. We applied an ensemble consensus variant calling approach utilizing Mutect2107 (GATK v4.3.0.0), Strelka2108 (v2.9.10), and VarScan109 (v2.4.6), retaining only SNPs identified by at least two callers. Mutect2 analysis included the use of The Panel of Normals (PoN) and germline resources following GATK Best Practices. Subsequently, the coordinates of the filtered SNVs were converted to hg19 using the R package liftOver106 (v1.26.0).
scATAC-seq data acquisition and processing
scATAC-seq data for all cell subsets used in this study were obtained from multiple previously published scATAC-seq datasets. 222 fetal and adult cell subsets from 30 adult and 15 fetal tissues were obtained from scATAC-seq atlases in refs. 28,29. More specialized scATAC-seq data from adult brain80, blood and bone marrow32, colon31, lung34, and kidney33, as well as fetal lung45 and brain30, were also included in the final dataset (Supplementary Data 3). We also acquired an additional bone marrow validation scATAC-seq dataset from61 to validate our COO prediction for AML (Supplementary Data 4). We note that data from separate datasets were kept separate and were not merged. For all datasets except kidney33, fragment files were available and were migrated to hg19 if they were not already aligned to hg19 using the R package liftOver106 (v1.26.0). For kidney scATAC-seq data, we aligned FASTQ files to hg19 and obtained fragment files using Cell Ranger ATAC pipeline110 (v1.1.0). scATAC-seq data fragment counts were binned as described above for SNV counts from WGS data; in brief, scATAC-seq fragment counts across cells were aggregated into bins to obtain chromatin accessibility profiles for each cell subset. Each cell subset (feature) is followed by a dataset identifier: D1 for28, D2 for29, D3 for31, D4 for32, D5 for34, D6 for33, D7 for30, D8 for80, and D9 for45.
scATAC-seq data curation and annotation
scATAC-seq cell annotations for each individual dataset were obtained directly from the corresponding paper’s published materials, except for the lung dataset34 for which the annotations were not available at the time of writing, and for which we generated de novo annotations. After reviewing the publicly available annotations, we further refined them as follows: for the adult atlas28, brain80, blood and bone marrow32, kidney33, all cell types that had a number index at the end indicating cell-type sub-clusters were collapsed into a single combined cell type. For example, “Colon Epithelial Cell 1”, “Colon Epithelial Cell 2”, and “Colon Epithelial Cell 3” from Transverse Colon were annotated as “Colon Epithelial Cell”. For the blood and bone marrow dataset32, we additionally removed cell types annotated as unknown, and combined all T-cell subsets (i.e., all CD8 and CD4 subsets) into T cells. Except for the five blood cancers (MM, CLL, AML, BNHL, and MPN), where it was important to distinguish between CD34+ and CD34- bone marrow, we collapsed the annotation into a single bone marrow category (Supplementary Data 3 contains the uncollapsed numbers). For the lung dataset34, we used an analogous approach to that used in the original publication. In particular, the cell type annotation was informed by labels from scRNA-seq data and was implemented in ArchR111 (v1.0). These labels were transferred to the scATAC-seq data using the addGeneIntegrationMatrix function. Marker discovery was conducted de novo using the getMarkerFeatures function with the GeneScoreMatrix assay. The final annotation of scATAC-seq cells was carried out by mapping the newly discovered clusters to predicted RNA cell types, or, when no corresponding cell type was found in the RNA data, by linking them to the chromatin accessibility at the locus of known gene markers that were most prevalent in each cluster. For the fetal atlas29, we excluded all cell types that were annotated as unknown. For the fetal brain dataset30, we used the same annotations as displayed in the scATAC-seq UMAP of the original paper (see Fig. 1f in the latter paper), except we removed cell types annotated as unknown. We did not use cancer tissue scATAC-seq data from31; specifically, our compiled dataset included only normal and unaffected samples, both of which were considered as “normal” tissue. For MSI CRC modeling that is expected to undergo metaplasia, our modeling also included polyp samples that are expected to contain this stage in tumor development. For the bone marrow dataset61, we used the function addGeneIntegrationMatrix from ArchR111 (v1.0) to label transfer the annotations using the transcriptomics profiles in the multiome data from32, since the data from61 did not contain annotations for GMP.Neut (our predicted COO using the data from32). To remove low-quality cells, we filtered cells with transcription start sites (TSS) score less than 8 and number of fragments (nFrags) less than 3000. No annotation curation was done for the fetal lung dataset45.
After finalizing our annotations, we applied a filter of a minimum 100 cells per feature to exclude cell subsets with insufficient number of fragments to produce an accurate pseudo-bulk epigenetic profile, as quantified in112. If a given cell type did not meet this threshold, it was excluded from further analysis, except in the following cases:
-
adult pulmonary neuroendocrine cells (PNECs) from34 when conducting lung-specific analyses (Fig. 1c, f, g; Supplementary Figs. 2, 3, 10c), since they were conjectured to be the COO of SCLC. While our scATAC-seq data base contained only 41 adult cells from34, it also included 356 GHRL+ and 231 fetal PNECs from45, which is comparable to the number of mesothelial cells (n = 401 and n = 223 for adult and fetal, respectively).
-
GMP from61 when conducting the AML validation analysis (Supplementary Fig. 4), since it was of particular interest and did not meet the threshold (n = 77; Supplementary Data 4).
SCOOP feature space selection for modeling
We trained SCOOP on a variety of different feature spaces. Except when indicated otherwise, we trained our models using all tissues from 6 different datasets: blood/bone marrow32, colon31, lung34, kidney33, and two cross-tissue28,29 scATAC-seq atlases. Other scATAC-seq datasets30,45,61,80 were included in the analysis only when diving deeper into a select cancer type and interrogating the relevant tissue of its COO more comprehensively. Specifically, a fetal lung dataset45 was included for lung cancers (Fig. 1a,c; Supplementary Fig. 2-3, 10c), fetal30 and adult80 brain datasets for brain cancers (Fig. 5a; Supplementary Figs. 7, 10c), and a bone marrow dataset61 for AML validation results (Supplementary Fig. 4b). Moreover, as indicated in the main text, certain models were trained on specific tissue scATAC-seq features.
Cancer subtype classification
We used the metadata in cBioPortal113 to obtain MSI high vs MSS classification for CRC. In particular, if the metadata column “subtype” had “MSI” as a suffix, we considered the sample to be MSI high. Otherwise, it was considered to be MSS. We note that all samples for ColoRect-AdenoCA from PCAWG are MSS samples, except for one, which is MSI high. We used the metadata in ICGC35 to obtain pRCC and ccRCC classification for kidney cancers.
XGBoost regression model
We trained XGBoost27 (Extreme Gradient Boosting) regression models using XGBRegressor from the Python XGBoost package (v1.7.4). XGBoost is an advanced implementation of the gradient boosting algorithm, designed for speed and performance. It builds an ensemble of decision trees in a sequential manner, where each tree corrects the errors of its predecessor. XGBoost employs a regularized model formalization to control over-fitting, making it robust to noisy data. The algorithm is parallelizable across both cores in a CPU and machines in a distributed setting, resulting in significantly faster training times compared to traditional gradient boosting. This enabled us to perform robustness analysis by performing 100 runs for each model.
Our choice of XGBoost was motivated by recent analyses showing that XGBoost outperforms RF models in efficiency and accuracy for bulk tissue COO prediction26. Based on our own experimention, we observed a five-fold decrease in run time when using XGBoost compared to RF when using all normal cell features.
We frame our task as a machine learning regression problem as follows: each genomic bin corresponds to a training example (i.e., data point) where the input features (i.e., “predictors” or “independent variables”) are the aggregated scATAC counts from the various cell subsets for that bin, and the label (i.e., “response variable” or “target variable” or “dependent variable”) is the mutation count for that bin. Put differently, each genomic bin is characterized by two sets of numbers; one set (input) is composed of multiple “features” where a particular “feature” constitutes an aggregated scATAC count for a particular cell subset, while the other set (output) is comprised of a single number: the aggregated number of mutations within that bin.
Robustness analysis
After partitioning the genome into bins, we grouped contiguous bins into 10 train/test folds. We used each fold as a test fold 10 times and used the remaining 9 folds for training and validation using cross-validation with a 90/10% split of training and validation respectively. For each of the 10 runs of cross validation and model testing per fold, 10 different seeds were used for seeding the XGBoost model building process, and, when training models with more than one feature, the feature importance calculation. Since we used each of the 10 folds as a test set 10 times (with a different seed each time), we in effect obtained 100 estimates of model performance on an unbiased test set.
Feature importance calculation
Feature importance was calculated using the permutation_importance function from scikit-learn114 (v1.3.0), setting the “n_repeats” parameter to 10, after picking the best model according to cross-validation. This function implements a permutation importance mechanism: for each feature in the feature space, it randomly permutes its values and keeps all other features constant, then measuring the effect of this permutation on a model’s chosen performance metric by comparing the change in performance to the baseline performance when no permutation is performed. The larger the negative effect of this process on the chosen performance metric, the more important a feature is deemed to be. Since this permutation is a random process, it is useful to perform this process multiple times, and measure the mean effect of permutation on performance. We chose to repeat the process 100 times. We note that since we used cross-validation, feature importance was calculated on each validation fold and averaged across folds; since we used a 90/10% train/validation split, which implies 10 train/validation folds, and for each fold we computed 10 permutation scores (n_repeats = 10), this in effect means that we performed 100 permutations and averaged these for any given feature.
Feature selection
If the initial feature space had more than 20 features, we next selected the top 20 features according to our feature importance score. Otherwise, the entire unmodified feature space was used to proceed. Using this reduced (or unmodified) feature space, we then performed iterative backward feature selection until only a single feature remained, which in most cases we would expect to correspond to the COO for the cancer type under consideration (see Robustness box plots and COO prediction sections for more details). To be more specific, after reducing the feature space to 20 features, we trained a new model using this reduced feature space, picked the best model according to mean performance across validation folds when performing cross-validation, ranked the 20 features based on the chosen model, and eliminated the bottom feature. This was then repeated iteratively.
We note that performing this process may aid in alleviating the potential bias that can be induced by having correlated features. As an example, suppose our dataset contains cell types A, B, and C, where B and C are highly correlated. When computing feature importance, cell types B and C may be ranked lower than they would have been ranked otherwise if the other cell type were absent. Thus, if the model ranks B above C and we remove cell type C, cell type B can now more fairly compete against cell type A for the top spot. There is the further issue that B and C may be arbitrarily ranked above one another by the model, which motivates training the model multiple times using different random seeds.
Model evaluation metric
We assessed model performance by computing the \({R}^{2}\) score (i.e., variance explained) of our model. This is computed as
where RSS is the residual sum of squares, and TSS is the total sum of squares. We emphasize here that it is possible to obtain a negative value for \({R}^{2}\), if the model performs worse than a simple mean model. This situation occurs when the RSS is greater than TSS, which means that the model’s predictions are on average further from the actual values than the simple mean of the data.
Hyperparameter optimization
Hyperparameter optimization was performed using the automated hyperparameter search framework Optuna115 (v3.3.0). We used the default hyperparameter optimization strategy, Tree-structured Parzen Estimator (TPE), which is a Bayesian optimization strategy. Briefly, we note that, in contrast to the naive but commonly employed strategy of randomly choosing and testing model hyper-parameters setting (e.g., grid search or random search, which is an inefficient and suboptimal strategy), Bayesian optimization strategies like TPE take advantage of the history of model performance under different hyperparameter settings to cleverly explore the hyperparameter search space, and exploit settings that performed well to narrow down the search space for optimal hyperparameters.
Each training run in Optuna is called a “study.” Each study consists of multiple “trials,” each corresponding to a specific model hyperparameter setting. At the end of a study, the best model hyperparameters are chosen based on the trial that performed best according to some pre-specified metric. In our case, this is the mean variance explained across validation folds. In other words, we fix a hyperparameter setting, compute its mean performance across validation folds during cross validation, report this number as the performance for the hyperparameter setting in question, and repeat this process for different hyperparameter settings. We note that the number of trials per study must be specified, and we set it to 50 (i.e., the “n_trials” parameter of the study.optimize function is set to 50). In practice, this means that 50 hyperparameter settings are tested per training run of the model. We emphasize that during backward feature selection, this process is repeated from scratch (i.e., for each new feature space, 50 different hyperparameter settings are tested) and the best models chosen for different feature spaces very likely differ in their hyperparameter settings.
Table 1 lists the XGBoost model hyperparameters and the corresponding ranges of values we searched over. The full description of each hyperparameter can be found at https://xgboost.readthedocs.io/en/release_1.7.0/python/python_api.html, under xgboost.XGBRegressor.
Robustness box plots
For the robustness box plots (e.g., Fig. 1c), we display, across 100 runs, the feature importance of different features when the model was trained on x features (i.e., after conducting backward feature selection and being left with x features). We further restricted the display to only show the top 5 features, where we define “top 5 features” as the features that appeared most frequently in the top 5 across 100 runs of the model. Also displayed is the number of times the feature appeared in the top x features across the 100 runs (n). The y-axis is ordered first by n, and ties are broken by the median feature importance, with the top-ranking feature appearing at the top of the y-axis. This feature is highlighted in red, and if the second top-ranking feature represented a highly similar cell subset, it was highlighted in pink. We set x = 5 for main figures and trained SCOOP with cancer tissue specific cell features, while in Supplementary Figures we displayed box plots with x = 10, 5, and 2 and trained SCOOP with cell features across tissues.
COO prediction
For each cancer type, the most frequent cell subset appearing as the top feature across 100 SCOOP runs was predicted as the COO. The COO prediction is consistently highlighted in red throughout the manuscript. The second most frequently appearing cell subset, if representing a highly similar cell subset to the COO, was highlighted in pink. Except for MPN (Supplementary Fig. 8a; see discussion in manuscript), this prediction also corresponded to the most informative feature in the robustness box plots when 5 features remained for all “high confidence” COO predictions (all predictions except those in Supplementary Fig. 9). For the benchmarking analysis (Supplementary Fig. 10a) we considered a COO prediction to be correct when this prediction was supported by evidence in the research literature (supporting literature is listed for each prediction in Supplementary Data 1).
For MPN, when using blood and bone marrow features, CD34+ CLPs – as opposed to CD34+ early basophils – appeared most frequently when only 5 features remain in the backward selection (Supplementary Fig. 8a). While they do not represent the exact same cell type, they both represent multipotent hematopoietic progenitors and support the hypothesis that MPN originates from a committed hematopoietic progenitor similar to HSCs91 (see main text).
Statistical testing for XGBoost model results
For Figs. 1b and 5c, to assess if the variance explained distribution of the top feature (based on mean variance explained) across 100 runs of the model had a significantly higher median compared to the next most important feature, we used a one-sided Mann-Whitney test.
Similarly, to assess if the feature importance distribution of the top robustness box plot feature across 100 runs of the model had a significantly higher median compared to the next most important feature, we used a one-sided Mann-Whitney test.
To check if the most frequently appearing top feature showed up as the top feature across 100 runs of the model, significantly more so than the second most frequently appearing top feature, we used a one-sided, exact binomial test. Under the null hypothesis, we would expect each feature to appear ½(x+y) times on average, where \(x\) and \(y\) are the actual number of times the first and second most frequently appearing top feature actually appeared as the top feature, respectively. In one case, the first and second top features corresponded to essentially the same feature and came from the same dataset (e.g., GMP and GMP.Neut from bone marrow mononuclear cells; Fig. 2d, bottom). In this case, we compared the combined appearances of the top 2 features to the third top feature. In other cases, where the top 2 features were similar but not coming from the same dataset (e.g., Fig. 1c), we compared the appearances of the top feature to the third feature. The exact comparison done is indicated on the bar plots by square brackets.
WGS sample SNV profile UMAPs
To visualize WGS mutational density profiles across samples we used Uniform Manifold Approximation and Projection (UMAP) dimensionality reduction as implemented in Seurat116 (v4) except we considered our feature space to be genomic bins rather than genes. Briefly, we normalized the mutational data using the NormalizeData function with the parameter normalization.method set to “LogNormalize.” Since we have a total of 2,128 bins only, we used all bins in the projection and did not run FindVariableFeatures. We then ran RunPCA with npcs set to “30.” Finally, we ran RunUMAP with dims set to “1:15.” In the case of lung, we set min.dist to “0.2” instead of “0.3”.
scATAC-seq data analysis via ArchR
Cell type UMAPs
To plot the UMAPs of the scATAC-seq data from various cell types, we used ArchR111 (v1.0). Specifically, we ran addIterativeLSI with resolution “0.2”. Then, for Figs. 2a and 5b (data from31 and30, respectively), we further ran addHarmony due to the presence of batch effects (not present in Fig. 2c). Finally, we ran addUMAP with nNeighbors set to “30” and minDist set to “0.5.”
Marker gene score computation and visualization
Expression of cell type marker genes was inferred from chromatin accessibility at a gene’s locus using ArchR’s gene score method111. Gene scores were visualized using plotEmbedding with colorBy set to “GeneScoreMatrix” and quantCut set to have a range of “0.01” to “0.95” prior to imputation using addImputeWeights (Supplementary Figs. 6a, 8b)111.
Meta-cell correlation analysis
We began by first sampling meta-cells (i.e., groups of similar cells) in scATAC-seq data using a K-nearest-neighbor (KNN) approach. Meta-cells were generated by randomly identifying seed cells for which 500 nearest neighbor cells were then selected based on a KNN graph generated in the latent semantic indexing (LSI) space. Meta-cells were allowed to overlap up to 80%. The function used to conduct this analysis is a customized version of the ArchR111 (v1.0) function addCoAccessibility.
After obtaining meta-cells, we summed the scATAC-seq fragment counts across cells for that meta-cell, representing an aggregated fragment profile. We then correlated all meta-cell profiles with the cancer mutation profile of interest using Pearson’s correlation coefficient. Finally, for each individual cell (i.e., not meta-cell), we assigned it a correlation score corresponding to the mean correlation of the meta-cells it was assigned to. We then plotted the correlation on the scATAC-seq data UMAP per cell. We note that for Fig. 2a, we filtered out cells with a fragment count lower than 10,000 before running any of the previous steps, since we noticed that the meta-cell correlation for that particular dataset correlated with the number of fragments when the fragment count was too low.
Metaplasia-related single-cell transcriptomics data acquisition and analysis
To quantify the transcriptional similarity between goblet cells (our COO predictions) and metaplastic cells, we analyzed multiple single-cell transcriptomics datasets related to metaplasia including human chronic pancreatitis68, ADM in response to pancreatic injury69, KrasG12D-induced neoplasia70, precancerous colorectal states67, human stomach71, and human colon polyps31. These datasets included scRNA-seq data from human and mouse models with the exception of human chronic pancreatitis and colon polyps data being single-nucleus RNA sequencing (snRNA-seq).
Data preprocessing prior to marker discovery and module score computation
We directly utilized the processed Seurat objects shared by the authors for all the aforementioned datasets, except for the pancreas injury model study69. For the latter, we created the Seurat object using the available count matrices and associated metadata, which had already been filtered for high quality cells69. All clustering and cell type annotations were retained from the original publications or accompanying supplementary files. For the human adult pancreas68 dataset, we pooled all acinar cell subpopulations from the provided Seurat object to discover generalizable acinar cell markers. For deriving colon goblet and stem cell markers from the colon polyps dataset31, we only used intestinal epithelial cells. We also converted HUGO Gene Nomenclature Committee (HGNC) gene symbols to Ensembl gene identifiers (Ensembl IDs) using biomaRt117 v2.56.1 (useMart, getBM) to facilitate downstream analyses. Since the precancerous colon dataset67 used in Fig. 4d used Ensembl IDs as features, this conversion was needed to calculate module scores consistently across different scRNA-seq datasets.
Normalized and scaled gene expression data were already available in the Seurat objects from67,68. Since normalized data was not available for70, and normalized and scaled data was not available for31,69,71, we proceeded as follows. For70, we performed log2-transformation after adding a pseudocount of 0.1, consistent with the methods described in the original manuscript. For the other three datasets31,69,71, we applied Seurat’s standard analysis pipeline using the functions NormalizeData, FindVariableFeatures, and ScaleData in succession as done in118.
Marker discovery, dimensionality reduction, and module score computation
After preprocessing all objects as described above, we used Seurat’s FindMarkers function to derive human stomach goblet cell markers71, human pancreas acinar cell markers68, mouse pancreas acinar cell markers69, human colon goblet and stem cell markers31. Due to unavailability of normal mouse stomach scRNA-seq data with well-annotated goblet cells, we obtained mouse stomach goblet cell markers from the Mouse Cell Atlas 3.072. After gathering all aforementioned sets of markers, we computed their module scores in normal and metaplastic scRNA-seq and snRNA-seq datasets using Seurat’s AddModuleScore function. Generation of UMAPs and visualization of module scores (Fig. 4a, b) was performed using Seurat’s RunPCA and RunUMAP functions after keeping the top 20 principal components.
Proliferation rate quantification using scRNA-seq data
To identify the percentage of lung epithelial cells that are proliferating/cycling at homeostasis, we used scRNA-seq data from healthy lung donors34. Seurat’s CellCycleScoring function was used to compute the module score for the expression of genes linked to either G1/S or G2/M phase of the cell cycle119. Cells with either a G1/S or G2/M phase score greater than 0.1 were classified as cycling and all other cells were considered non-cycling as done in120. The percentage of cycling cells for each lung epithelial cell was displayed in Fig. 1h, where cell types were ordered from most to least proliferative.
Variance explained as a function of sample size
To classify cancers into low, medium, and high TMB, we computed the average number of mutations per WGS sample for each cancer type (Supplementary Data 1). Following this, we segmented the cancer types into these three classifications by employing quantile division. We then picked 2 cancers with high confidence COO predictions from each category. Per cancer, for each specific number of WGS samples n, we randomly sampled n patients 100 times, aggregated the sampled patient data SNV profiles, and ran SCOOP on the sampled aggregated mutation profile, using a different random seed for each run.
Association between model prediction heterogeneity and cancer mutation heterogeneity
We investigated the relationship between model prediction and cancer mutation heterogeneity via a linear regression (Supplementary Fig. 10b). To quantify SCOOP’s prediction heterogeneity, we computed the Gini impurity for each COO prediction, which is calculated as
where n is the number of cell types predicted as the COO, and \({p}_{i}\) is the proportion of predictions (out of the 100 runs of the model) that corresponded to cell type i. As n increases and the proportion of predictions becomes more equally distributed across the n categories, Gini impurity increases (i.e., higher Gini impurity corresponds to higher heterogeneity in prediction). To quantify mutational heterogeneity, we computed the average pairwise correlation between any two binned (1 Mbp) WGS samples for each cancer type and defined a cancer type’s mutational similarity as the mean pairwise WGS sample correlation. We note that mutational similarity is inversely related to mutational heterogeneity.
Analysis of bin prediction accuracy
Bin categorization
For each of the 6 cancer types in Fig. 5d, we classified bins into four categories based on standard residuals. Specifically, we computed
where \({e}_{i}={y}_{i}-\hat{{y}_{i}}\) is the residual – the difference between the observed number of mutations \({y}_{i}\) and the average predicted mutation \(\hat{{y}_{i}}\) across 10 seeds – and \(\hat{\sigma }({e}_{i})\) is the estimated standard deviation of the residuals. The standard residual quantifies the model’s accuracy in predicting mutations. Higher absolute values of the standardized residual indicate lower prediction accuracy. We defined bins corresponding to lowest standardized residuals (within the top 5th percentile) as the “most-accurate” regions. If the standard residual was greater than 2, we classified the bin as “under-predicted”, indicating that the average predicted number of mutations is lower than the observed mutations. When the standard residual was less than −2, indicating that the average prediction exceeds the number of observed mutations, we defined the bin to be “over-predicted.” All other bins were classified as “rest”.
Gene expression and epigenomic data acquisition and processing
To compare gene expression across different categories of bins, we downloaded tissue gene expression data from GTEx (RNA-Seq, normalized using TPM)96 that matched the normal tissue of origin for each cancer’s COO prediction. To quantify the histone occupancy across different bin categories, we downloaded H3K27Ac (activation marker) and H3K9me3 (repressive marker) ChIP-seq data from the NIH Roadmap Epigenomics Project97 that matched the normal tissue of origin for each cancer’s COO prediction.
Comparing expression and epigenetic data between bin categories
For GTEx RNA-seq data, since multiple samples are available for each tissue, we calculated the average TPM per gene across all samples within each tissue. For the ChIP-seq data, we quantified the number of reads in each bin and converted them to Reads Per Kilobase per Million (RPKM), or
For tissues with multiple samples, we calculated the average RPKM across samples per bin. We applied the same normalization method to our assembled scATAC-seq dataset. We quantified the statistical significance of differences between bin categories for the various assays (RNA-seq, ChIP-seq, scATAC-seq) across bin categories using the Mann-Whitney test.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The publicly available scATAC-seq data used in this study are available under the following accession codes in GEO (GSE18446228, GSE14968329, GSE24461880, GSE13936932, GSE20134831, GSE24016834, GSE16654733, GSE16217030, GSE21646461) and ArrayExpress (E-MTAB-1126645) databases. The publicly available scRNA-seq data used in this study are available under the following accession codes or project IDs in GEO (GSE17238069, GSE20793870), HCA (cfece4d2-f18d-44ad-a46a-42bbcb5cb3b771), EGA (EGAS0000100465368), HTAN (HTA11_202167, HTA10_202231), and dbGaP (phs000424.v10.p296) databases. The publicly available WGS datasets used in this study are available under the following accession codes or project IDs in dbGaP (phs001287.v21.p695, phs000178.v11.p8121, phs003676.v1.p148) and EGA (EGAD0000100844737, EGAS0000100092536, EGAS0000100706774) databases. All data supporting the findings of this study are available within the paper and its supplementary information files. Source data are provided with this paper.
Code availability
All code associated with this project can be found at our Github page at https://doi.org/10.5281/ZENODO.16753796122 and https://github.com/TsankovLab/SCOOP, which contains instructions on how to reproduce our results.
References
Visvader, J. E. Cells of origin in cancer. Nature 469, 314–322 (2011).
Pandya, R. et al. The cell of cancer origin provides the most reliable roadmap to its diagnosis, prognosis (biology) and therapy. Med. Hypotheses 157, 110704 (2021).
Shaheen, N. J. et al. Upper endoscopy for gastroesophageal reflux disease: best practice advice from the clinical guidelines committee of the American College of Physicians. Ann. Intern. Med. 157, 808–816 (2012).
Shaheen, N. J. et al. Radiofrequency ablation in Barrett’s esophagus with dysplasia. N. Engl. J. Med. 360, 2277–2288 (2009).
Lawson, D. A. et al. Basal epithelial stem cells are efficient targets for prostate cancer initiation. Proc. Natl Acad. Sci. USA 107, 2610–2615 (2010).
Wang, X. et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature 461, 495–500 (2009).
Kersten, K., de Visser, K. E., van Miltenburg, M. H. & Jonkers, J. Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol. Med. 9, 137–153 (2017).
Vibert, J. et al. Identification of tissue of origin and guided therapeutic applications in cancers of unknown primary using deep learning and RNA sequencing (TransCUPtomics). J. Mol. Diagn. 23, 1380–1392 (2021).
Moiso, E. et al. Developmental deconvolution for classification of cancer origin. Cancer Discov. 12, 2566–2585 (2022).
Divate, M. et al. Deep learning-based pan-cancer classification model reveals tissue-of-origin specific gene expression signatures. Cancers (Basel) 14, 1185 (2022).
Hong, J., Hachem, L. D. & Fehlings, M. G. A deep learning model to classify neoplastic state and tissue origin from transcriptomic data. Sci. Rep. 12, 9669 (2022).
Štancl, P. & Karlić, R. Machine learning for pan-cancer classification based on RNA sequencing data. Front. Mol. Biosci. 10, 1285795 (2023).
Zhang, Y. et al. Single-cell analyses of renal cell cancers reveal insights into tumor microenvironment, cell of origin, and therapy response. Proc. Natl Acad. Sci. USA 118, e2103240118 (2021).
Graf, M. et al. Single-cell transcriptomics identifies potential cells of origin of MYC rhabdoid tumors. Nat. Commun. 13, 1544 (2022).
Hu, L. et al. Single-cell RNA sequencing reveals the cellular origin and evolution of breast cancer in BRCA1 mutation carriers. Cancer Res. 81, 2600–2611 (2021).
De Bie, J. et al. Single-cell sequencing reveals the origin and the order of mutation acquisition in T-cell acute lymphoblastic leukemia. Leukemia 32, 1358–1369 (2018).
Cooper, L. A. D. et al. The tumor microenvironment strongly impacts master transcriptional regulators and gene expression class of glioblastoma. Am. J. Pathol. 180, 2108–2119 (2012).
Friedmann-Morvinski, D. & Verma, I. M. Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO Rep. 15, 244–253 (2014).
Suvà, M. L., Riggi, N. & Bernstein, B. E. Epigenetic reprogramming in cancer. Science 339, 1567–1570 (2013).
Polak, P. et al. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat. Genet. 49, 1476–1486 (2017).
Polak, P. et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature 518, 360–364 (2015).
Haradhvala, N. J. et al. Mutational Strand Asymmetries in Cancer Genomes Reveal Mechanisms of DNA Damage and Repair. Cell 164, 538–549 (2016).
Polak, P. et al. Reduced local mutation density in regulatory DNA of cancer genomes is linked to DNA repair. Nat. Biotechnol. 32, 71–75 (2014).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Kübler, K. et al. Tumor mutational landscape is a record of the pre-malignant state. bioRxiv 517565. https://doi.org/10.1101/517565 (2019)
Yang, S. et al. COOBoostR: An Extreme Gradient Boosting-Based Tool for Robust Tissue or Cell-of-Origin Prediction of Tumors. Life 13, 71 (2023).
Chen, T. & Guestrin, C. XGBoost: A Scalable Tree Boosting System. in Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (ACM, New York, NY, USA). https://doi.org/10.1145/2939672.2939785. (2016).
Zhang, K. et al. A single-cell atlas of chromatin accessibility in the human genome. Cell 184, 5985–6001.e19 (2021).
Domcke, S. et al. A human cell atlas of fetal chromatin accessibility. Science 370, eaba7612 (2020).
Trevino, A. E. et al. Chromatin and gene-regulatory dynamics of the developing human cerebral cortex at single-cell resolution. Cell 184, 5053–5069.e23 (2021).
Becker, W. R. et al. Single-cell analyses define a continuum of cell state and composition changes in the malignant transformation of polyps to colorectal cancer. Nat. Genet. 54, 985–995 (2022).
Granja, J. M. et al. Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat. Biotechnol. 37, 1458–1465 (2019).
Wang, Q. et al. Single-cell chromatin accessibility landscape in kidney identifies additional cell-of-origin in heterogenous papillary renal cell carcinoma. Nat. Commun. 13, 31 (2022).
Shah, V. S. et al. Single cell profiling of human airway identifies tuft-ionocyte progenitor cells displaying cytokine-dependent differentiation bias in vitro. Nat. Commun. 16, 5180 (2025).
ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020).
George, J. et al. Comprehensive genomic profiles of small cell lung cancer. Nature 524, 47–53 (2015).
Creaney, J. et al. Comprehensive genomic and tumour immune profiling reveals potential therapeutic targets in malignant pleural mesothelioma. Genome Med. 14, 58 (2022).
Ireland, A. S. et al. Nature https://doi.org/10.1038/s41586-025-09503-z (2025).
Johnsen, H. E. et al. The myeloma stem cell concept, revisited: from phenomenology to operational terms. Haematologica 101, 1451–1459 (2016).
Castro-Pérez, E., Singh, M., Sadangi, S., Mela-Sánchez, C. & Setaluri, V. Connecting the dots: Melanoma cell of origin, tumor cell plasticity, trans-differentiation, and drug resistance. Pigment Cell Melanoma Res. 36, 330–347 (2023).
Liu, C. et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell 146, 209–221 (2011).
Zong, H., Parada, L. F. & Baker, S. J. Cell of origin for malignant gliomas and its implication in therapeutic development. Cold Spring Harb. Perspect. Biol. 7, a020610 (2015).
Schneller, D. & Angel, P. Cellular Origin of Hepatocellular Carcinoma. in Hepatocellular Carcinoma (ed. Tirnitz-Parker, J. E. E.) (Codon Publications, Brisbane (AU)).
Ferone, G., Lee, M. C., Sage, J. & Berns, A. Cells of origin of lung cancers: lessons from mouse studies. Genes Dev. 34, 1017–1032 (2020).
He, P. et al. A human fetal lung cell atlas uncovers proximal-distal gradients of differentiation and key regulators of epithelial fates. Cell 185, 4841–4860.e25 (2022).
Carbone, M. et al. Malignant mesothelioma: facts, myths, and hypotheses. J. Cell. Physiol. 227, 44–58 (2012).
Lázaro, S. et al. Differential development of large-cell neuroendocrine or small-cell lung carcinoma upon inactivation of 4 tumor suppressor genes. Proc. Natl Acad. Sci. USA 116, 22300–22306 (2019).
Rekhtman, N. et al. Chromothripsis-mediated small cell lung carcinoma. Cancer Discov. 15, 83–104 (2025).
Rudin, C. M. et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat. Rev. Cancer 19, 289–297 (2019).
Megyesfalvi, Z. et al. Clinical insights into small cell lung cancer: Tumor heterogeneity, diagnosis, therapy, and future directions. CA Cancer J. Clin. 73, 620–652 (2023).
Sutherland, K. D., Ireland, A. S. & Oliver, T. G. Killing SCLC: insights into how to target a shapeshifting tumor. Genes Dev. 36, 241–258 (2022).
Poirier, J. T. et al. New approaches to SCLC therapy: From the laboratory to the clinic. J. Thorac. Oncol. 15, 520–540 (2020).
Owonikoko, T. K. et al. YAP1 expression in SCLC defines a distinct subtype with T-cell-inflamed phenotype. J. Thorac. Oncol. 16, 464–476 (2021).
Li, K., Luo, H., Huang, L., Luo, H. & Zhu, X. Microsatellite instability: a review of what the oncologist should know. Cancer Cell Int 20, 16 (2020).
Huels, D. J. & Sansom, O. J. Stem vs non-stem cell origin of colorectal cancer. Br. J. Cancer 113, 1–5 (2015).
Delgado, J., Nadeu, F., Colomer, D. & Campo, E. Chronic lymphocytic leukemia: from molecular pathogenesis to novel therapeutic strategies. Haematologica 105, 2205–2217 (2020).
Zeng, A. G. X. et al. Precise single-cell transcriptomic mapping of normal and leukemic cell states reveals unconventional lineage priming in acute myeloid leukemia. bioRxiv https://doi.org/10.1101/2023.12.26.573390 (2023).
Goardon, N. et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell 19, 138–152 (2011).
Krivtsov, A. V. et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL–AF9. Nature 442, 818–822 (2006).
Shlush, L. I. et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 506, 328–333 (2014).
Hu, Y. et al. Multiscale footprints reveal the organization of cis-regulatory elements. Nature https://doi.org/10.1038/s41586-024-08443-4 (2025).
Ye, M. et al. Hematopoietic Differentiation Is Required for Initiation of Acute Myeloid Leukemia. Cell Stem Cell 17, 611–623 (2015).
Cairns, P. Renal cell carcinoma. Cancer Biomark. 9, 461–473 (2010).
Rooman, I. & Real, F. X. Pancreatic ductal adenocarcinoma and acinar cells: a matter of differentiation and development? Gut 61, 449–458 (2012).
Kopp, J. L. et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 22, 737–750 (2012).
Di Domenico, A. et al. Epigenetic landscape of pancreatic neuroendocrine tumours reveals distinct cells of origin and means of tumour progression. Commun. Biol. 3, 740 (2020).
Chen, B. et al. Differential pre-malignant programs and microenvironment chart distinct paths to malignancy in human colorectal polyps. Cell 184, 6262–6280.e26 (2021).
Tosti, L. et al. Single-nucleus and in situ RNA-sequencing reveal cell topographies in the human pancreas. Gastroenterology 160, 1330–1344.e11 (2021).
Ma, Z. et al. Single-cell transcriptomics reveals a conserved metaplasia program in pancreatic injury. Gastroenterology 162, 604–620.e20 (2022).
Burdziak, C. et al. Epigenetic plasticity cooperates with cell-cell interactions to direct pancreatic tumorigenesis. Science 380, eadd5327 (2023).
Tsubosaka, A. et al. Stomach encyclopedia: Combined single-cell and spatial transcriptomics reveal cell diversity and homeostatic regulation of human stomach. Cell Rep. 42, 113236 (2023).
Wang, R. et al. Construction of a cross-species cell landscape at single-cell level. Nucleic Acids Res 51, 501–516 (2023).
Wardell, C. P. et al. Genomic characterization of biliary tract cancers identifies driver genes and predisposing mutations. J. Hepatol. 68, 959–969 (2018).
Huang, K. K. et al. Spatiotemporal genomic profiling of intestinal metaplasia reveals clonal dynamics of gastric cancer progression. Cancer Cell 41, 2019–2037.e8 (2023).
McDonald, S. A. C., Lavery, D., Wright, N. A. & Jansen, M. Barrett oesophagus: lessons on its origins from the lesion itself. Nat. Rev. Gastroenterol. Hepatol. 12, 50–60 (2015).
Hoang, M. P., Murakata, L. A., Padilla-Rodriguez, A. L. & Albores-Saavedra, J. Metaplastic lesions of the extrahepatic bile ducts: a morphologic and immunohistochemical study. Mod. Pathol. 14, 1119–1125 (2001).
Van Patten, K. & Jain, D. Benign tumors and tumor-like lesions of the gallbladder and extrahepatic biliary tract. Diagn. Histopathol. (Oxf.) 16, 371–379 (2010).
Alcantara Llaguno, S. R. & Parada, L. F. Cell of origin of glioma: biological and clinical implications. Br. J. Cancer 115, 1445–1450 (2016).
Yang, Z.-J. et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell 14, 135–145 (2008).
Li, Y. E. et al. A comparative atlas of single-cell chromatin accessibility in the human brain. Science 382, eadf7044 (2023).
Wang, J., Garancher, A., Ramaswamy, V. & Wechsler-Reya, R. J. Medulloblastoma: From molecular subgroups to molecular targeted therapies. Annu. Rev. Neurosci. 41, 207–232 (2018).
Hovestadt, V. et al. Resolving medulloblastoma cellular architecture by single-cell genomics. Nature 572, 74–79 (2019).
Okonechnikov, K. et al. Mapping pediatric brain tumors to their origins in the developing cerebellum. Neuro. Oncol. 25, 1895–1909 (2023).
Reitman, Z. J. et al. Mitogenic and progenitor gene programmes in single pilocytic astrocytoma cells. Nat. Commun. 10, 3731 (2019).
Ramos, S. I. et al. An atlas of late prenatal human neurodevelopment resolved by single-nucleus transcriptomics. Nat. Commun. 13, 7671 (2022).
Shingleton, J. et al. Non-Hodgkin Lymphomas: Malignancies Arising from Mature B Cells. Cold Spring Harb. Perspect. Med. 11, a034843 (2021).
Lee, C.-H. et al. A panel of antibodies to determine site of origin and malignancy in smooth muscle tumors. Mod. Pathol. 22, 1519–1531 (2009).
Pstrąg, N., Ziemnicka, K., Bluyssen, H. & Wesoły, J. Thyroid cancers of follicular origin in a genomic light: in-depth overview of common and unique molecular marker candidates. Mol. Cancer 17, 116 (2018).
Ren, X. et al. Single-cell transcriptomic analysis highlights origin and pathological process of human endometrioid endometrial carcinoma. Nat. Commun. 13, 6300 (2022).
Mead, A. J. & Mullally, A. Myeloproliferative neoplasm stem cells. Blood 129, 1607–1616 (2017).
Haeno, H., Levine, R. L., Gilliland, D. G. & Michor, F. A progenitor cell origin of myeloid malignancies. Proc. Natl Acad. Sci. USA 106, 16616–16621 (2009).
Keller, P. J. et al. Defining the cellular precursors to human breast cancer. Proc. Natl Acad. Sci. USA 109, 2772–2777 (2012).
Abarrategi, A. et al. Osteosarcoma: Cells-of-Origin, Cancer Stem Cells, and Targeted Therapies. Stem Cells Int 2016, 3631764 (2016).
Shin, K. et al. Cellular origin of bladder neoplasia and tissue dynamics of its progression to invasive carcinoma. Nat. Cell Biol. 16, 469–478 (2014).
Li, Y. et al. Proteogenomic data and resources for pan-cancer analysis. Cancer Cell 41, 1397–1406 (2023).
GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues Science 369, 1318–1330 (2020).
Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Spira, A. et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat. Med. 13, 361–366 (2007).
Caplin, M. E. et al. Pulmonary neuroendocrine (carcinoid) tumors: European Neuroendocrine Tumor Society expert consensus and recommendations for best practice for typical and atypical pulmonary carcinoids. Ann. Oncol. 26, 1604–1620 (2015).
Rekhtman, N. Lung neuroendocrine neoplasms: recent progress and persistent challenges. Mod. Pathol. 35, 36–50 (2022).
Finlay, J. B. et al. Olfactory neuroblastoma mimics molecular heterogeneity and lineage trajectories of small-cell lung cancer. Cancer Cell 42, 1086–1105.e13 (2024).
Sugano, K., Moss, S. F. & Kuipers, E. J. Gastric intestinal metaplasia: Real culprit or innocent bystander as a precancerous condition for gastric cancer? Gastroenterology 165, 1352–1366.e1 (2023).
Maisonneuve, P. & Lowenfels, A. B. Epidemiology of pancreatic cancer: an update. Dig. Dis. 28, 645–656 (2010).
Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Heath, A. P. et al. Bionimbus: a cloud for managing, analyzing and sharing large genomics datasets. J. Am. Med. Inform. Assoc. 21, 969–975 (2014).
Bioconductor Package Maintainer. liftOver: Changing genomic coordinate systems with rtracklayer::liftOver. Bioconductor (2024). R package version 1.26.0. https://bioconductor.org/packages/liftOver. https://doi.org/10.18129/B9.bioc.liftOver.
Benjamin, D. et al. Calling Somatic SNVs and Indels with Mutect2. bioRxiv 861054 https://doi.org/10.1101/861054 (2019).
Kim, S. et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods 15, 591–594 (2018).
Koboldt, D. C. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
Satpathy, A. T. et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 37, 925–936 (2019).
Granja, J. M. et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet. 53, 403–411 (2021).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, l1 (2013).
Pedregosa, F. et al. Scikit-learn: Machine Learning in Python. arXiv [cs.LG] (2012).
Akiba, T., Sano, S., Yanase, T., Ohta, T. & Koyama, M. Optuna: A Next-generation Hyperparameter Optimization Framework. in Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining 2623–2631 (Association for Computing Machinery, New York, NY, USA, 2019).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Durinck, S., Spellman, P. T., Birney, E. & Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191 (2009).
Giotti, B. et al. Single cell view of tumor microenvironment gradients in pleural mesothelioma. Cancer Discov. 14, 2262–2278 (2024).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016).
Soni, N. et al. Single-cell dissection of the genotype-immunophenotype relationship in glioblastoma. Brain. https://doi.org/10.1093/brain/awaf129 (2025).
Cancer Genome Atlas Research Network et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 (2013).
Bairakdar, M., Lee, W., Giotti, B., Kumar, A. & Tsankov, A. TsankovLab/SCOOP: SCOOP. (Zenodo,). https://doi.org/10.5281/ZENODO.16753796. (2025).
Acknowledgements
We thank all members of the Tsankov lab for their feedback. This project was funded by the Chan Zuckerberg Initiative (CZI) Data Insights Grant 2022-249299 to P.P., R.K., and A.M.T. This work was also supported in part by the Croatian National Science Foundation Project PREDI-COO (Project number: HRZZ IP-2019-04-9308) to P.S. and R.K and through the computational and data resources and staff expertise provided by Scientific Computing and Data at the Icahn School of Medicine at Mount Sinai and supported by the Clinical and Translational Science Awards (CTSA) grant UL1TR004419 from the National Center for Advancing Translational Sciences.
Author information
Authors and Affiliations
Contributions
P.P. and A.M.T. conceptualized the project. M.D.B., W.L., P.S., B.G., A.K., and R.K. assembled and processed the data. M.D.B., W.L., B.G., A.K., P.S., R.K, and A.M.T. performed formal analyses. M.D.B., W.L., B.G., A.K., and A.M.T. wrote the manuscript with valuable feedback from D.H., E.W., B.G., P.S., P.P., R.K., and approval from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bairakdar, M.D., Lee, W., Giotti, B. et al. Learning the cellular origins across cancers using single-cell chromatin landscapes. Nat Commun 16, 8301 (2025). https://doi.org/10.1038/s41467-025-63957-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-63957-3