Abstract
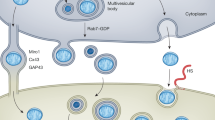
Mitochondria are essential intracellular organelles responsible for energy production. Over the past two decades, unconventional intercellular mitochondrial transfer has been reported, but the nature of the transport intermediates, the efficiency of the process, and the cellular mechanisms involved in their uptake and putative integration by acceptor cells remain poorly understood. This gap in knowledge is especially significant given the potential therapeutic applications of mitochondrial transplantation. In this study, we use quantifiable cell biology and biochemical approaches to assess intercellular mitochondria exchange. Our findings suggest that low amount of free mitochondria can be released into conditioned media and subsequently internalized by recipient cells, primarily via fluid-phase uptake, although alternative or concurrent endocytic pathways may also contribute. Notably, we show that a subset of internalized mitochondria escapes the endosomal compartment, reaches the cytosol, and may integrate into the host cell’s pre-existing mitochondrial network.
Similar content being viewed by others
Introduction
Intercellular transfer of mitochondria has been described across the eukarytic kingdom, from yeast1 to humans2, including plants3. Both natural and artificial mitochondrial transfer have been implicated in key cellular processes such as energy metabolism, tissue repair, and maintenance of cellular homeostasis4. Recently, horizontal mitochondrial transfer has been observed between cancer cells and neighboring immune cells, highlighting its potential role in cancer progression and T cell–dependent antitumor responses5,6. However, the precise mechanisms and broader physiological roles of mitochondrial transfer remain under investigation, both ex vivo and in vivo.
Mitochondrial transfer was thought to occur exclusively through tunneling nanotubes7. However, recent studies have revealed alternative mechanisms, including mitochondria encapsulated within extracellular vesicles8,9 or freely circulating in the extracellular space (including blood)10,11, which may also mediate intercellular mitochondrial transfer. As early as the 1980s, it was demonstrated that the uptake of isolated mitochondria could rescue the dysfunctional mitochondrial phenotype of recipient cancer cells12, supporting the hypothesis that free mitochondria may act as transport intermediates. This idea echoes the endosymbiotic theory of mitochondrial origin13, where ancient proteobacteria were engulfed and integrated into host cells through an endosymbiotic process, ultimately supporting cellular function. The fate of transferred mitochondria remains unclear as contradictory studies reported integration or not within the mitochondrial network of the host cell14,15.
Building on our previously developed and validated methodology and molecular tools for characterizing extracellular vesicle uptake at the cellular level16, we now apply a mitochondrial cargo–based approach in combination with well-established cell models to track the fate mitochondria. We quantitatively assess their presence and form in conditioned media, and analyze their uptake and intracellular localization in recipient cells. Our findings indicate that a substantial portion of mitochondria are present in free form within the conditioned media. These “putative” transport intermediates behave as isolated mitochondria and are taken up via a process resembling macropinocytosis. Notably, fewer than 10% of internalized mitochondria appear capable of escaping endosomal compartments and reaching the cytosol, where they may integrate into the host mitochondrial network.
Identification and characterization of the putative transport intermediate
To investigate this trafficking event, we engineered donor cell lines expressing NanoLuciferase (NLuc)-tagged versions of OMP2517, an outer mitochondrial membrane protein with the tag exposed on the cytosolic side, and COX8a18, an inner mitochondrial membrane protein with the NLuc tag localized within the intermembrane space (Fig. 1A). Both constructs were also HA-tagged to enable immunofluorescence and immunoblot detection. These methods confirmed the expected mitochondrial localization of both proteins and verified their expression as intact constructs (Fig. 1B, C). This validation establishes the two constructs as reliable probes for tracking mitochondria with high sensitivity, using NLuc detection16. Utilizing chimeric proteins helps to bypass artifacts previously encountered in studies involving labile and diffusible fluorescent dyes19.
A Topology of the endogenous protein tagged in this study (black), and the transiently expressed protein used in this study (orange). B Plasmid validation of Nluc-HA-OMP25 and COX8a-Nluc-HA. HeLa cells were transiently transfected with either plasmid, processed for immunofluorescence using an anti-HA antibody, and imaged by confocal microscopy. Images are representatives of three independent experiments. C Immunoblots of cell lysates (CL) from wild-type HeLa cells or HeLa cells transiently expressing Nluc-HA-OMP25 or COX8a-Nluc-HA. Equal amounts of protein were analyzed by immunoblotting for HA (upper gel) and Actin (lower gel). Blots are representative of two independent experiments.
We then used size exclusion chromatography to analyze the content of conditioned media emanating from NLuc-HA-OMP25 and COX8a-NLuc-HA expressing cells, and compared it to intact mitochondria isolated from the same donor cells. In conditioned media, we observed two main peaks: fraction 4 (F4), representing large particles consistent with extracellular organelles, contained less than 1% of the total NLuc activity (0.176% ± 0.004 for NLuc-HA-OMP25 and 0.132% ± 0.0026 for COX8a-NLuc-HA), while the majority was found in fraction 16 (F16), corresponding to smaller particles, likely free proteins or membrane debris (Fig. 2A). For the isolated mitochondria, fraction 4’ (F4’) accounted for 16.69% ± 0.954 for NLuc-HA-OMP25 and 20.89% ± 0.354 for COX8a-NLuc-HA, of the NLuc activity, with 15.11% ± 0.17 for NLuc-HA-OMP25 and 14.8% ± 0.108 for COX8a-NLuc-HA found in fraction 16’, corresponding to degradation products (Fig. 2B). This was confirmed with a third independent mitochondria marker (Sup Fig. 1A-C). We then measured the number and size of particles present in fractions F4, F16, and F4’ (Fig. 2C). No particles were detected in F16, consistent with the presence of free proteins or debris smaller than 30 nm. In contrast, F4 and F4’ exhibited similar particle profiles, with diameters ranging from 100 to 500 nm.
A, B SEC Fractionation profiles from conditioned media (CM) A or isolated mitochondria (IM) B derived from cells expressing either Nluc-HA-OMP25 (upper graph) or COX8a-Nlsuc-HA (lower graph). NLuc activity (blue) and protein concentration (red) were measured across SEC fractions (1 to 18). F4 contains the putative transport intermediate and F4’ contains isolated mitochondria. C Particle size distribution of fractions F4, F4′, and F16 as assessed by nanoparticle tracking analysis (NTA). D Immunoblots of cell lysates (CL), F4, F4′, and F16, probed for EV markers (CD63, CD9), ER marker (calreticulin), and mitochondrial marker (Nluc-HA-OMP25). Equal amounts of protein were loaded. Blots are representatives of two independents experiments. E Protease protection assay on Nluc-HA-OMP25 and COX8a-Nluc-HA in IM and CM samples, treated or not with proteinase K and/or β-OG. Each dot represents an independent biological replicate; each value is the mean of three technical replicates. Error bars indicate standard deviation.
Next, we performed immunoblot analysis on each fraction to broadly assess their protein composition (Fig. 2D). F4, derived from conditioned media, was enriched in the EV markers CD63 and CD9, as expected, and also contained the NLuc-tagged mitochondrial cargo (NLuc-HA-OMP25). F4’, originating from isolated mitochondria, was enriched in NLuc-HA-OMP25 and showed only trace levels of EV markers. F16 showed no detectable EV markers and primarily contained degraded forms of OMP25-HA-NLuc, consistent with debris or degradation products. Importantly, ER markers were absent from all tested fractions, suggesting that the material present in the conditioned media is not the result of cell lysis.
Together, these results suggest that only a small fraction of mitochondria released into the conditioned media may act as transport intermediates, either as free mitochondria or enclosed within extracellular vesicles.
To distinguish between these two possibilities, we performed well-established protease protection assay20,21 based on the following rationale: if the transport intermediate is free mitochondria, NLuc-HA-OMP25, being exposed on the outer membrane, should be sensitive to protease, while COX8a-NLuc-HA, located within the intermembrane space, should remain resistant unless detergent is added. Conversely, if the mitochondria are enclosed within extracellular vesicles (EVs), both NLuc-HA-OMP25 and COX8a-NLuc-HA should be protected from protease in the absence of detergent, as we have previously shown for well-established EV cargo16,20.
We subjected fractions F4 and F16 (from conditioned media) as well as F4’ and F16’ (from isolated mitochondria) to proteinase K protection assay. Both F4 and F4’ exhibited similar behavior, with NLuc-HA-OMP25 being protease-sensitive and COX8a-NLuc-HA remaining resistant, consistent with free mitochondria. In contrast, both constructs in F16 and F16’ were sensitive to protease, suggesting these fractions contain degradation products (Fig. 2E). These findings indicate that F4 likely represents free mitochondria, ruling out the presence of mitochondria enclosed within EVs. Importantly, these results were confirmed using endogenous mitochondrial markers, which exhibited the same topology (Sup Fig. 2A-C).
Cellular characterization and quantification of free mitochondria uptake
We leveraged our highly sensitive luminescence-based assay16 to quantitatively measure the uptake of the different fractions derived from NLuc-HA-OMP25 and COX8a-NLuc-HA expressing donor cells. Equal amounts of NLuc activity from F4 (“free mitochondria” from conditioned media), F16 (small particles or debris from conditioned media), and F4’ (intact mitochondria isolated from donor cell) were loaded onto unlabeled acceptor cells and incubated for 24 hours to assess steady-state uptake. We found that a small but measurable fraction (1-2%) of both F4 and F4’ were taken up by the acceptor cells, whereas F16 uptake was virtually undetectable (Fig. 3A). Subsequent kinetic and dose-response studies revealed that luminescence activity in acceptor cells increased over time (Fig. 3B) and was proportional to the amount of F4 (“free mitochondria” in conditioned media) and F4’ (control isolated mitochondria) (Fig. 3C). Interestingly, uptake of F4’ was consistently—but not always statistically—slightly higher than that of F4, possibly reflecting some loss of integrity of free mitochondria in the conditioned media prior to harvest.
A Unlabeled HeLa acceptor cells were incubated for 24 hours with fractions F4, F4′, or F16 derived from donor cells expressing Nluc-HA-OMP25 or COX8a-Nluc-HA. NLuc activity was measured after 24 h. Input NLuc activity was set to 100%. Three independent experiments were performed, each including three technical replicates. Each dot represents a replicate. Error bars indicate standard deviation, center represent the means. *Statistical significance was assessed by unpaired, two-tailed t-test comparing F4 or F4′ to F16; p < 0.01. B, C Uptake kinetics and dose response. Acceptor cells were incubated with F4 or F4′ (from Nluc-HA-OMP25-expressing cells, 50,000 total NLuc activity units) for various durations B, or for 24 h with increasing amounts of F4 or F4′ input C. Three independent experiments were performed, each including four technical replicates. Each dot represents a replicate. D Temperature block experiment. Unlabeled acceptor cells were incubated with F4 and F4′ (from Nluc-HA-OMP25-expressing cells) for 4 h at either 37 °C or 4 °C. NLuc activity was then measured. Three independent experiments were performed, each including four technical replicates. Each dot represents a replicate. Statistical significance was assessed by unpaired, two-tailed t-test comparing F4 or F4′ at 37 °C vs. 4 °C; p < 0.01.
Since donor cells were initially transiently transfected, we considered the possibility that plasmid DNA encoding NLuc-HA-OMP25 could be released—either freely or within EVs—into the conditioned media, potentially confounding the interpretation of mitochondrial uptake. To rule out this possibility, we tested both F4 and F4’ fractions for the presence of the plasmid, which was not detected (Sup Fig. 3A). Importantly, both fractions were positive for mitochondrial DNA (Sup Fig. 3B), consistent with the presence of mitochondria in these preparations and in agreement with our earlier findings. To definitively rule out any bias introduced by transient transfection, we generated a HeLa cell clone stably expressing NLuc-HA-OMP25 and used it as a donor to produce the putative transport intermediate. The resulting F4 fraction was then applied to various adherent acceptor cell lines, including HeLa, A431, and SKOV3 cells (Sup Fig. 3C). In all tested cell types, F4 uptake was consistent with that observed using transiently transfected donor cells, thereby confirming that the mitochondrial transfer is not an artifact of plasmid contamination. Moreover, these results demonstrate that the transfer mechanism operates across multiple cell lines.
To investigate the mechanism of mitochondrial uptake, we applied a 4 °C temperature block—a classical approach to inhibit energy-dependent endocytosis while preserving high-affinity “ligand”–receptor interactions. This temperature-based assay has historically been pivotal in the discovery of bona fide receptors, such as the LDL receptor22. We hypothesized that if free mitochondria were recognized and captured by a high-affinity receptor on the surface of acceptor cells, NLuc activity would still be detectable at 4 °C. However, luciferase activity was virtually undetectable in cells incubated with F4 or F4’ at 4 °C for up to 4 h (Fig. 3D), suggesting minimal, if any, receptor-mediated binding under these conditions.
Previous studies have implicated heparan sulfate in mitochondrial uptake23, which is not incompatible with our findings. In such a model, heparan sulfate moieties may facilitate surface docking of mitochondria via electrostatic interactions, which could allow limited association without the affinity characteristic of canonical receptors. This would explain why mitochondrial uptake appears relatively inefficient—marked by slow kinetics and lack of saturation—when compared to ligand–receptor systems such as LDL uptake.
We next used confocal imaging to trace the intracellular fate of isolated mitochondria in acceptor cells. Colocalization analyses were performed using the OMP25-based mitochondrial probe alongside endogenous markers of early endosomes (EEA1) and lysosomes (Lamp1) (Fig. 4A–D). Mitochondria were found to colocalize with endosomal markers (39%) and lysosomal markers (30%), indicating that internalized mitochondria follow the canonical endo-lysosomal trafficking route. Additionally, co-incubation with red-labeled dextran—a fluid-phase uptake marker24, revealed strong colocalization with internalized mitochondria (Fig. 4E, F).
A–D Confocal micrographs showing uptake of free mitochondria derived from Cherry-OMP25 expressing donor cells, internalized by acceptor cells for 4 h. After incubation, cells were fixed and processed for immunofluorescence against EEA1 A or LAMP1 C to label early endosomes and lysosomes, respectively, using green-labeled secondary antibodies. Red squares indicate regions of colocalization shown at higher magnification. B, D Quantification of colocalization. EEA1 or LAMP1 fluorescence was measured for each mitochondria-associated (red-positive) ROI, as well as for control EEA1-/LAMP1-negative ROIs. The dashed line indicates the maximum fluorescence intensity in EEA1-/LAMP1-negative ROIs. Values above this threshold were considered positive for colocalization. Three independent experiments were performed; 320 B and 324 ROIs D from at least 50 cells, from three independent samples were analyzed. Scale bar: 10 μm. E, F Confocal micrographs showing uptake of free mitochondria derived from Nluc-HA-OMP25–expressing donor cells after 4-h internalization. Acceptor cells were co-treated with Texas Red dextran and processed for immunofluorescence against HA using green-labeled secondary antibodies. Red squares highlight regions of colocalization at higher magnification. F Quantification of colocalization with dextran was performed as described above. Scale bar: 10 μm. 351 ROIs from 60 cells from three independent samples were analyzed.
Together, these results suggest that free mitochondria are internalized via a low-efficiency, non-receptor-mediated process. The observed colocalization with dextran supports macropinocytosis as the primary mode of uptake, followed by trafficking through the classical endo-lysosomal pathway.
Mitochondria delivery through endosomal escape
To assess and quantify the integration of internalized free mitochondria into the mitochondrial network of acceptor cells, we adapted our previous split-NLuc complementation assay utilizing HiBit/LgBit technology25. We engineered donor cells expressing HiBit-tagged OMP25 and acceptor cells expressing complementary LgBit-tagged OMP25. While neither HiBit nor LgBit emit luminescence on their own, their proximity restores NLuc activity, producing a detectable luminescent signal.
To validate the split-mitochondria proteins, we used size exclusion chromatography to measure luminescence in the various fractions by complementing HiBit-OMP25 with recombinant LgBit. The results (Fig. 5 A, B) mirrored those from Fig. 2, confirming the validity of the split-mitochondria system.
A, B SEC fractionation profiles of conditioned media A or isolated mitochondria B derived from cells expressing HiBiT-OMP25. NLuc activity was measured in fractions 1–18 following complementation with recombinant LgBiT and is shown in blue. C Quantification of cytosolic delivery of HiBiT-OMP25 from fractions F4, F4′, or F16 into acceptor HeLa cells expressing LgBiT. Three independent experiments were performed, each including three technical replicates. Each dot represents a replicate. Error bars indicate standard deviation, center represents the means. *Statistical significance was assessed by unpaired, two-tailed t-test comparing F4 or F4′ to F16; p < 0.01.
Next, we performed a protein complementation assay to track potential mitochondrial exchange between donor and acceptor cells. Equal amounts of HiBiT-OMP25 from F4 (free mitochondria in conditioned media), F16 (degradation products), and F4’ (intact mitochondria isolated from donor cells) were applied to LgBiT-OMP25–expressing acceptor cells and incubated for 24 h. Protein complementation was detected in cells treated with both F4 and F4’, but not with F16, consistent with previous uptake data (Fig. 5C).
Three hypotheses could explain the observed complementation of the outer membrane protein: (I) cytosolic delivery of donor mitochondria; (II) cytosolic delivery followed by clustering of donor and acceptor mitochondria; or (III) integration of donor mitochondria into the acceptor cell’s mitochondrial network (Fig. 6A). To distinguish among these possibilities, we generated different combinations of split donor and acceptor cell lines: donor cells expressing either HiBiT-OMP25 or HiBiT-COX8, and acceptor cells expressing LgBiT-OMP25, LgBiT-COX8, or cytosolic LgBiT. Each hypothesis if true would be revealed by a unique combinatorial complementation pattern that can be predicted (Fig. 6A, lower table).
A Schematic representation of three possible outcomes following cytosolic delivery of extracellular mitochondria: Hypothesis I: Individual donor mitochondria are delivered into the cytosol and remain isolated, with none too moderate complementation with outermembrane LgBit-protein or cytosolic LgBit. Hypothesis II: Donor mitochondria cluster with acceptor cell mitochondria. In this case, outer membrane (OM) protein complementation is expected to be positive, while inner membrane (IM) proteins should remain negative. Hypothesis III: Donor mitochondria fully integrate into the acceptor cell’s mitochondrial network. Both OM and IM proteins are expected to show positive complementation in during homotypic exchange. A summary table outlines the expected outcomes for each donor/acceptor combination under these three models. B extracellular mitochondria from donor cells expressing either HiBiT-OMP25 or HiBiT-COX8 were incubated for 24 hours with acceptor cells expressing either cytosolic LgBiT, LgBiT-OMP25, or LgBiT-COX8. NLuc activity was measured to assess complementation. Three independent experiments were performed, each including three technical replicates. Each dot represents a replicate. Error bars indicate standard deviation, center represents the means. *Statistical significance was assessed by unpaired, two-tailed t-test comparing homotypic combinations (OMP25/OMP25 and COX8/COX8) to the heterotypic OMP25/LgBiT control; p < 0.01.
Our results showed that complementation occurred exclusively in homotypic combinations (i.e., OMP25–OMP25 and COX8–COX8), with no signal observed in heterotypic or cytosolic combinations. This pattern supports hypothesis III, suggesting that functional complementation requires integration of donor mitochondria into the acceptor cell’s mitochondrial network. In particular, COX8-driven complementation implies content mixing between donor and host mitochondria, further suggesting mitochondrial fusion and network incorporation. Although our complementation results are fully compatible with fusion between delivered donor mitochondria and host mitochondria, it remains to be definitively demonstrated whether full integration into the endogenous mitochondrial network occurs, or if internalized mitochondria instead form localized aggregates with host mitochondria. Clarification of this point will likely be enabled by emerging super-resolution microscopy techniques.
To test the hypothesis that internalized mitochondria can escape endosomes, we performed confocal microscopy on cells expressing Galectin-3-GFP, a well-established reporter of endosomal membrane rupture26,27, Galectin 3 shows a diffuse, barely detectable GFP signal in the cytosol under normal conditions. However, upon endosomal rupture, Galectin 3 binds to glycans found on the luminal side of the endosomes, producing distinct GFP puncta. As a positive control, we used LLOME, a known endosomal membrane disruptor27, which generated clear GFP puncta (Fig. 7A).
A Confocal micrographs of acceptor HeLa cells expressing Galectin-3-GFP after 4 h of incubation with Nluc-HA-OMP25 mitochondria (middle panel) or Texas Red dextran (lower panel). As a posissstive control for endosomal damage, cells were treated with LLOMe for 30 minutes (upper panel). Cells were immunolabeled against HA using red-labeled secondary antibodies. Red squares highlight regions of Galectin-3 colocalization shown at higher magnification. Scale bar: 10 μm. B Quantification of Galectin-3-GFP colocalization with red-labeled mitochondria versus red dextran. Three independent experiments were performed. A total of 150 dextran-positive and 350 mitochondria-positive puncta were analyzed. Statistical significance was assessed by unpaired, two-tailed t-test; p < 0.01. C Proposed model: Extracellular mitochondria are internalized via fluid-phase uptake and trafficked through the endosomal pathway. Endosomal membrane rupture enables their escape into the cytosol, where they may integrate into the acceptor cell’s mitochondrial network.
Remarkably, around 9% of the internalized mitochondria colocalized with Galectin 3-GFP puncta (Fig. 7A, B), indicating that these mitochondria can damage endosomal membranes, facilitating their escape into the cytosol. Importantly Galectin3-GFP puncta were virtually absent from cell treated with the fluid phase marker (Fig. 7A, B) indicating some level of specificity. This suggests th at after internalization, mitochondria can induce endosomal membrane rupture, allowing their release into the cytosol and subsequent integration into the acceptor cell’s mitochondrial network.
Discussion
The simplest interpretation of our data supports a model in which “free mitochondria” released by donor cells can be internalized by recipient cells—at least in part through the fluid-phase—escape from endosomal compartments, to be delivered into the cytosol, where they may ultimately merge with the recipient cell’s existing mitochondrial network (Fig. 7C), Although measurable, this process seems poorly efficient, in our ex vivo experimental set-up. Importantly, we identified endosomal escape following internalization as a key cellular mechanism enabling mitochondrial transfer. This finding provides mechanistic clarity to earlier studies that qualitatively reported mitochondrial uptake and network integration using 3D super-resolution imaging, but did not capture the escape process itself15. Conversely, other studies failed to observe mitochondrial integration following artificial transplantation14. a discrepancy likely explained by their use of split-GFP systems, which lack the sensitivity and signal amplification provided by the bulk split-NLuc assay used in our study28.
As always, the working model proposed here will need to be refined and re-evaluated through continued efforts to address several key aspects of this process.
Core aspects of this process concern the nature of the transport intermediate:
Recent studies have revived interest in functional intercellular exchange of mitochondria2, including the possibility of free mitochondria as transport intermediates29, an hypothesis consistent with our findings. This mode of transport might be exclusive or concomitant to others, including the more established of tunneling nanotubes. When comparing both mode of transport through the split system with direct co-culture (where transfer can occur via both diffusible intermediates and nanotunneling) and media exchange through 1 μm or 200 nm cut-off filtration (where transfer can occur to either large of small intermediates, respectively) we observed greater transfer efficiency in the co-culture setup than with the 1 μm filtration condition) and no transfer was detected following 200 nm filtration (Sup Fig. 4). These results suggest that mitochondrial exchange may rely on both nanotunneling and a large diffusible transport intermediate.
Biochemical characterization using protease protection assays strongly supports the notion that this intermediate corresponds to free mitochondria. This interpretation is further validated by our systematic comparisons across uptake and delivery experiments, in which the behavior of material derived from conditioned media closely parallels that of isolated mitochondria. Importantly, the observed endosomal escape mechanism also supports the free mitochondria model, as it is incompatible with the expected fate of mitochondria encapsulated in extracellular vesicles (EVs), which would require additional steps such as membrane fusion to release their content into the cytosol16,30. Despite the rigor of our biochemical and cellular analyses, further investigation is required to fully characterize the “free mitochondria” population, highly diluted within other components of the secretome. Future priorities include high-resolution morphological analysis using electron microscopy and assessment of mitochondrial functionality through metabolic profiling. Although previous studies suggest that transplanted mitochondria can restore function in deficient recipient cells, we did not directly evaluate mitochondrial functionality here—an important limitation to be addressed.
Another critical aspect of our study is the low yield of recoverable transport intermediates from conditioned media, which raises questions about the underlying secretion mechanism. Nonetheless, this release appears to be biologically specific and regulated, as other organelles such as the ER were not detected in the extracellular space. While intact and respiratory competent mitochondria have previously been observed in blood10, our SEC isolation data—along with the high Nluc activity observed in the F16 fraction, which corresponds to free protein and small debris—suggest that the majority of mitochondria released into conditioned media are not stable in the extracellular environment. To resolve these questions, in vivo studies will be essential. Recent advances in live inter-organ imaging using physiologically relevant animal models31,32, offer a promising path to better understand the fate, function, and physiological significance of extracellular mitochondria in more native environments.
Another core aspect concerns the fate of the uptaken mitochondria within the acceptor cells:
Our data show a lack of efficient binding of mitochondria to the surface of acceptor cells at 4 °C, a temperature that only reveals high-affinity protein-protein interactions. These findings suggest that mitochondrial uptake is not mediated by a high-affinity receptor, unlike what has been observed for other physiological relevant entities such as lipoproteins22 or viruses33. We and others have previously reported similarly limited uptake efficiencies for extracellular vesicles, which follow macropinocytosis and alternative routes16,34. Earlier study has proposed a role for heparan sulfates in mitochondrial uptake23. One possibility is that the accumulation of multiple low-affinity interactions may suffice to capture a limited number of mitochondria. This raises the question of how such inefficient uptake can support the effective regulation of a related physiological function. Alternatively, these results may indicate that specific donor–acceptor tissue pairing is required for more efficient transfer, pointing toward a physiological role that remains to be fully elucidated.
Although we propose that mitochondrial delivery involves an initial step of endosomal escape, future studies are needed to identify the molecular machinery and mechanisms responsible for endosomal membrane destabilization or rupture. Reactive oxygen species (ROS) and putatively exposed cardiolipin, are appealing candidates among others. Investigating this process is particularly challenging as these molecules—typically confined to the reducing environment of the cytosol—would now be exposed to the extracellular milieu, which may lack the essential factors or cofactors required for their function. This unusual topological context presents a paradigm that requires further investigation and clarification.
Our results indirectly support mitochondria integration that may use the mitochondria fusion machinery, that is more complex that initially thought35. Several studies have suggested that the mitochondrial network is heterogeneous, and it remains unclear whether integrated mitochondria merge with the existing network or remain sequestered within specific subdomains. Highly sensitive and rapid super-resolution imaging technologies will be essential to address these questions.
Mitochondrial transfer has been proposed to play a role in a wide range of physiological functions36,37. However, how can such a low-efficiency delivery mechanism support functional transfer? At first glance, this may seem difficult to reconcile. However, when considering that1 mitochondria contain their own DNA and autonomous nucleoids, which control the inheritance of mitochondrial DNA38, and2 mitochondria form numerous contacts with other organelles, providing lipids necessary for membrane expansion and duplication39, the process may be efficient enough to colonize recipient cells with limited number of exogenous mitochondria, similarly to what is observed with bacteria and viruses, masters of cell’s invasion. This interpretation aligns with earlier studies that showed that a very limited number of injected-mitochondria is sufficient to completely replace endogenous mitochondrial DNA in mitochondria-deficient cells40,41. More recently, it has been demonstrated that the transplantation of a single mitochondria per cell equivalent significantly increases ATP levels within 24 hours, further supporting the merging and functional integration of mitochondria14.
Future research should focus on identifying the molecular and physicochemical triggers that facilitate mitochondria-mediated endosomal disruption and the key players involved in this process on both the mitochondrial and endosomal sides. A deeper understanding of the molecular machinery governing this process could enhance the efficiency of mitochondrial transplantation, paving the way for new therapeutic approaches. It may also provide valuable insights into the intercellular transfer of mitochondria in both physiological and pathological contexts.
Methods
Cell Culture and Transfection
HeLa, A-431 and SKOV-3 cells (from ATCC, Virginia, USA) were grown in DMEM (Gibco, Illinois, USA) supplemented with 10% fetal bovine serum (Gibco, Illinois, USA or Biosera, France) at 37 °C with 5% CO2. Cells were transfected at 60% to 70% confluency using Lipofectamine 2000 (Invitrogen, Massachusetts, USA) according to the manufacturer’s protocol. HeLA clone stably expressing NLuc-HA-OMP25 was selected with geneticin 10 μg/mL (Gibco, Illinois, USA).
Transport intermediate isolation
24 hours after transfection, donor cells were cultured in serum-free DMEM for 24 hours. Conditioned media was harvested and subjected to a 600 g centrifugation for 10 minutes at 4 °C, followed by a 2000g centrifugation for 20 minutes at 4 °C to remove cell debris. Conditioned media was then concentrated using Amicon Ultra-15, MWCO = 100 kDa (UFC9100, Millipore, MA). First, the filters were washed with 15 ml PBS to remove any trace of glycerol. Then, 15 ml of media was added and centrifuged at 4500 g for 15 minutes at 4 °C. The filtrate was discarded, and the concentrated media was saved. This step was repeated to filter the entire volume of conditioned media. All the concentrated media was recovered and pooled together. 500 μl of concentrated media was added to SEC column (qEVoriginal/70 nm Gen 2, Izon, Christchurch, New Zealand), and the separation was performed according to the manufacturer’s protocol. Fraction 1 was eluted in 2 ml, fraction 2 in 0.5 ml, and fraction 3 in 0.4 ml of PBS; these fractions were considered as buffer volume. Fractions 4 to 15 were eluted in 0.4 ml of PBS, and fraction 16 in 1 ml, with fractions 17 and 18 in 2 ml. These fractions were used for subsequent Nluc activity measurement, protease protection assay, uptake assay, and cytosolic delivery assay.
Intracellular mitochondria isolation
Mitochondria from cultured cells were isolated to serve as a positive control. Cells were scraped from culture dishes, washed with PBS, and centrifuged at 600 × g for 5 min at room temperature. Mitochondria were then isolated using the Abcam Mitochondria Isolation Kit for Cultured Cells (ab110170, Abcam, Cambridge, U.K.), following the manufacturer’s instructions with minor adjustments. Briefly, cells were subjected to five cycles of freeze–thaw to promote membrane disruption, resuspended in Reagent A, and incubated for 10 minutes. The suspension was homogenized using a Dounce homogenizer and centrifuged at 1000 × g for 10 min at 4 °C. The supernatant was retained, while the pellet was resuspended in Reagent B and subjected to the same process. The combined supernatants were centrifuged again at 1000 × g for 10 min at 4 °C to eliminate remaining cellular debris. The final supernatant was then centrifuged at 10,000 × g for 15 min, and the resulting pellet—containing crude mitochondria—was resuspended in Reagent C supplemented with protease inhibitors. These isolated mitochondria further purified by size exclusion chromatography using Izon qEVoriginal/70 nm Gen 2 SEC columns, as described above.
Immunoblot
Cells,isolated mitochondria (IM), Nluc-HA-OMP25 F4, F16 and F’4 were resuspended in lysis buffer (50 mM Tris, 150 mM NaCl, 1% Triton-X-100, protease/phosphatase inhibitor (PPI) cocktail (Roche, Switzerland), pH8) and incubated on ice for 30 min. Samples were then centrifuged at 15,000 g for 20 min. Supernatants (cell lysates) were collected. Protein concentrations of cell lysate (CL) and IM samples were measured using the Micro BCA™ Protein Assay kit (Thermo Scientific, IL). Samples were mixed with 4X Laemmli buffer (Bio-Rad, CA) and 10% β-mercaptoethanol (BME), then loaded onto 4–20% precast polyacrylamide gel (Bio-Rad, CA) in tris/glycine/SDS buffer (Bio-Rad, CA). After electrophoresis, proteins were transferred onto PVDF membranes (0.2 μm, Bio-Rad, CA) using the Trans-Blot Turbo system (Bio-Rad, CA). The Precision Plus Protein Standards (Bio-Rad, CA) was used as a protein weight ladder. Membranes were blocked in PBS with 0.05% Tween 20 and 5% milk at room temperature, then incubated overnight at 4 °C with 1/1000 primary antibody (α-Actin (MAB1501, Sigma-Aldrich, MA), α-HA (C29F4, cell signaling, MA), α-MFN2 (56889, Abcam, UK), α-PDHA1 (110334, Abcam, UK), α-CD63 (Cat # 556,019, BD Bioscience, New Jersey, U.S.A.), α-CD9 (Cat # cbl162, Millipore, Germany), calreticulin (22683, Abcam,UK) in PBS 0.05% Tween 20, 5% milk, and 0.02% sodium azide. Membranes were washed 3 times for 20 mins each, and finally incubated with 1/5000 HRP-coupled secondary antibody (α-mouse or α-rabbit, 115-035-003, Jackson ImmunoResearch, UK) in PBS 0.05% Tween 20. Membranes were revealed using the Clarity Western ECL Substrate (Bio-Rad, CA) and the ImageQuant LAS 400 imager (GE Healthcare Life Sciences, Chicago). Image analysis and quantification were performed using the ImageJ software.
Protease Protection Assay
Nluc-HA-OMP25 or Cox8a-Nluc-HA Fractions F4, F4’, F16 and F16’ were incubated in PBS with or without 0.1% βOG and 0.2 mg/mL Proteinase K (AM2542, Ambion, Texas, USA) for 3 hours at 4 °C. The same activity of nanoluciferase was used in each condition. Samples were diluted in 200 μL PBS, and 20 μL of Nano-Glo™ Live Cell assay reagent (20 μL buffer + 0.5 μL substrate) (N2012, Promega, Wisconsin, USA) was added to immediately measure the remaining NLuc activity in each well using an iD3 SpectraMax microplate reader (Molecular Devices, California, USA). In parallel, this assay was confirmed by conducting a protease protection assay assessment via immunoblot. F4’ and F16’ were incubated in PBS with or without 3.2 μg/mL proteinase K and 0.1% TritonX100 (Sigma-Aldrich, Missouri, USA) for 1 h on ice. The reaction was neutralized by adding 1 μL of boiled 0.2 M PMSF, immediately followed by loading buffer (100 °C, 15 min).
Mitochondrial uptake and delivery assay
Nluc activity in each fraction was measured by adding 5 μL of the fraction to 95 μL of PBS and 20 μL of Nano-Glo™ live reagent or 50 μL of the Nano-Glo reagent (N2012/ N1120 Promega, Wisconsin, USA) in each well of a white 96-well plate (ThermoFischer, MA, USA). Luminescence activity was read using the iD3 SpectraMax microplate reader. Wild-type HeLa acceptor cells were seeded 24 h before the uptake experiment to reach 80% confluency in white/clear bottom 96-well plates (Thermofisher, MA, USA). The same Nluc activity in each fraction was added in serum-free DMEM. For temperature dependent experiment, fractions were added in HBSS (ThermoFisher, MA, USA). Cells were incubated with mitochondria at either 4 °C or 37 °C for 4 h, or for the specifically indicated time. After incubation, cells were washed three times with PBS. Finally, 50 μL of PBS and 20 μL of Nano-Glo™ live reagent were added to each well, and luminescence activity was read.
Mitochondrial cytosolic delivery assay
Nluc activity in each fraction was measured by adding 5 μL of the fraction, 2.5 μL of PBS, 2.5 μL of 0.1% Triton, and 90 μL of recombinant LgBiT (Promega, Wisconsin, USA) diluted at 1/10,000 to each well in a white 96-well plate, along with 20 μL of Nano-Glo™ live reagent (Promega, Wisconsin, USA). Luminescence activity was read using an iD3 SpectraMax microplate reader. HeLa acceptor cells were seeded in a 24-well plate. 24 hours before the experiment, they were transfected with Lgbit OMP25 plasmid as recommended. Six hours later, they were trypsinized and transferred to a white/clear bottom 96-well plate to reach 80% confluency on the day of experiment. The same Nluc activity in each fraction was added in serum-free DMEM. Cells were incubated at 37 °C for 24 h. After incubation, cells were washed three times with PBS. Finally, 50 μL of PBS and 20 μL of Nano-Glo™ live reagent were added to each well, and luminescence activity was read.
Confocal microscopy
Acceptor cells were seeded 24 h before the experiment in a 24-well plate on top of coverslips to reach 50–60% confluency. For early endosomes colocalization: Cells were incubated with mCherry OMP25 IM for 1 and 4 h at 37 °C. For lysosomes colocalization: Cells were incubated with Nluc HA OMP25 IM for 1 to 4 h at 37 °C. For macropinocytosis: Cells were incubated with red dextran (ThermoFisher, MA, USA) at 20 mg/ml and with Nluc HA OMP25 IM for 30 min and 4 hr. For Galectin 3 colocalization: Cells transfected with Gal3 GFP plasmid 24 hours before experiment, were incubated with Nluc HA OMP25 IM for 4 and 24 h. In addition, control cells were treated for 30 min with 500 μM LLOME (L7393-500MG, Sigma-Aldrich, Missouri, USA). After incubation, cells were washed with PBS, fixed for 15 min at room temperature with 4% PFA, washed 3 times with PBS and permeabilized for 10 min in 0.1% Triton-X-100. Cells were then blocked in 1% BSA for 1 h. Primary antibodies (EEA1 (2411S, Cell signaling, MA), Lamp1 (15665S, Cell signaling, MA) HA (C29F4, cell signaling, MA), were incubated for 2 h at room temperature at dilutions of 1/200,1/100, and 1/1000, respectively, in PBS. After three washes in PBS, samples were incubated for 1 hour at room temperature with secondary antibodies (α-mouse or α-rabbit coupled to AlexaFluor 488 or 546 (ThermoFisher, MA, USA) at a dilution of 1/2000 in PBS. Following additional washes, samples were mounted with a ProLong Diamond Antifade Mountant (Invitrogen) and Images were acquired using an LSM 880 confocal microscope with a 63× oil immersion objective (ZEISS, Baden-Württemberg, Germany). Image analysis and colocalization quantification were performed using ImageJ software (NIH, Maryland, USA). Briefly, to assess compartment colocalization, the fluorescent signal coming from acceptor cell protein in region of interest (ROI) corresponding to each IM positive dot or negative ROI (approximatively the same size) for EEA1, LAMP1, and DEXTRAN was measured.
Nanoparticle Tracking Analysis
NTA was performed using the ZetaView x20 (Particle Metrix, Ammersee, Germany) with the following parameters: laser 488 nm, scatter, 11 positions, 1 cycle, sensitivity 80, shutter 100, pH = 7 entered, T °C sensed. All samples were diluted into filtered PBS.
Statistical Analysis
Unpaired, two-tailed t-test were performed using the Prism software (GraphPad, San Diego, California U.S.A.) on biological replicates.
Amplification of the mitochondrial genome
DNA was extracted from F4, F16 and from isolated mitochondria using Plasmid mini prep Kit (Macherey Nagel, France) and then amplified by long range PCR performed in a 50 μL total volume using Phusion™ Plus DNA Polymerase kit (F630S, ThermoFisher, MA, USA), using PCR thermocycler (Bio-Rad, California, USA). Thermal cycling conditions were as follows: 95 °C (2:00) + [95 °C (0:15) + 68 °C (10:00)] × 30 cycles + 68 °C (20:00) + 4 °C ( ∞ ). PCR amplifications were performed with five pairs of overlapping primers (Mito1/Mito2/Mito3/hmt1/hmt2). All PCR amplified products were loaded on a 0.8% agarose gel to check the size of amplicons with reference to the 1Kb plus DNA ladder (ThermoFisher, MA, USA).
Plasmids
mCherry-OMP25 construct was a gift from David Tareste, who replace GFP by mCherry in pCB7-GFPOM (Malka et al., EMBO reports VOL 6 | NO 9 | 2005). Gal3-EGFP was a gift from Ludger Johannes. Lgbit construct was obtained by removing Nluc-CD63 sequence from Nluc-CD63 construct (Emeline bonsergent et al.). PCR amplified Lgbit sequence from Lgbit plasmid (Promega, Wisconsin, USA), was then inserted. Lgbit-CD63 construct was obtained by removing RFP sequence from RFP-CD63 (a gift from Walther Mothes), using Age1 and Xho1 restriction enzyme (NEB, Massachusetts, USA). PCR amplified Lgbit sequence from Lgbit construct, with the following primers: (forward) GCCACCGGTCTCACCATGGTCTTCACACTCGA, (Reverse) GCCCTCGAGAACTGTTGATGGTTACTCGGA, was then inserted using the same enzymes.
Nluc–OMP25 construct was obtained by removing CD63 sequence from Nluc-CD63 (Emeline bonsergent et al.), using Xho1 and Mfe1 restriction enzyme (NEB, Massachusetts, USA). PCR amplified OMP25 sequence from mCherry-OMP25 construct with the following primers: (forward) GCCCTCGAGGGCGACGGAGAGCCGAGT,(Reverse) GCCCAATTGTCAGGTTCAGGGGGAGGTGT was then inserted using the same enzymes. Nluc-HA-OMP25 construct was obtained by removing Nluc sequence from Nluc-OMP25 plasmid using Age1 and Xho1 restriction enzyme (NEB, Massachusetts, USA). PCR amplified Nluc -HA sequence from the Nluc–OMP25 construct with the following primers: (forward) CGGGAATTCACCGGTATGGTCTTCACACTC, (Reverse) CGG CTC GAG TGCGTAGTCTGGTACGTCGTACGGATAACTAGTCGCCAGAATGCGTTCGCA, was then inserted using the same enzymes.
Lgbit-OMP25 construct was obtained by removing CD63 sequence from Lgbit-CD63 plasmid, using Xho1 and Mfe1 restriction enzyme (NEB, MA). PCR amplified OMP25 sequence from mCherry-OMP25 construct with the following primers: (forward) GCCCTCGAGGCGACGGAGAGCCGAGT, (Reverse) GCCCAATTGTCAGGTTCAGGGGGAGGTGT, was then inserted using the same enzymes. Hibit-OMP25 construct was obtained by removing GFP-Hsp70 sequence from GFP-hsp70 plasmid (15215, Addgene, MA), using Age1 and Mfe1 restriction enzyme (NEB, MA). PCR amplified Hibit-OMP25 sequence from mCherry-OMP25 construct with the following primers: (forward) GCCACCGGTACCATGACTAGTGTGAGCGGCTGGCGGCTGTTCAAGAAGATTAGCGGATCCTCCGGTGGATCGAGCGGTGGGAATTCTGGTGGAGGATCCGCTAGCGGCGACGGAGAGCCG, (Reverse) GCCCAATTGTCAGGTTCAGGGGGAGGTGT, was then inserted using the same enzymes. Cox8a-Nluc-HA construct was obtained by removing Snap tag sequence from pSNAPf-Cox8A plasmid (101129, Addgene, MA), using EcoRI and Xho1 restriction enzyme (NEB, Massachusetts, USA). PCR amplified Nluc HA sequence from Nluc OMP25 construct with the following primers: (forward) CGGGAATTCACCGGT ATGGTCTTCACACTC, (Reverse) CGGCTCGAGTTATGCGTAGTCTGGTAC GTCGTACGGATAACTAGTCGCAGAATGCGTTCGCA, was then inserted using the same enzymes. Cox8a-Hibit construct was obtained by removing GFP-Hsp70 from from GFP-hsp70 plasmid (15215, Addgene, MA), using Mfe1 and Nhe1 restriction enzyme (NEB, MA). PCR amplified Cox8a-Hibit sequence from pSNAPf-Cox8A plasmid (101129, Addgene, MA) with the following primers: (Forward): GCCGCTAGCTACCATGTCCGTCCTGA. (Reverse): GCCCAATTGTTAGCTAATCTTCTTGAACAGCCGCCAGCCGCTCACACTAGTCATGCTAGCGGATCCTCCACCAGAATTCCCACCGCTCGATCCACCGGAGGATCCGGTGAATTCACCGGT was then inserted using the same enzymes. Cox8a-Lgbit construct was obtained by removing snap tag from pSNAPf-Cox8A plasmid (101129, Addgene, MA) using EcoRi and XhoI restriction enzyme (NEB, MA). PCR amplified Lgbit sequence from Lgbit construct with the following primers: (Forward) GCCGAATTCACCATGGTCTTCACACTCGA (Reverse) GCCCTCGAGTTAACTGTTGATGGTTACTCGGA was then inserted using the same enzymes. Tom20-Nluc construct was obtained by removing sfCherry+11 sequence from Tom20 plasmid (83033, Addgene, MA), using Age1 and Not1 restriction enzyme (NEB, MA). PCR amplified Nluc sequence from Nluc-CD63 construct with the following primers: (forward) GCCACCGGTTATGGTCTTCACACTCGAAGAT, (Reverse) CGGGCG GCCGCTTACGCCAGAATGCGTTCGCA, was then inserted using the same enzymes. Tom20-Nluc HA construct was obtained by removing Tom20-sfcherry+11 sequence from Tom20 plasmid (83033, Addgene, MA), using Nhe1 and Not1 restriction enzyme (NEB, MA). PCR amplified Nluc Tom20 HA sequence from Nluc Tom20 construct with the following primers: (forward) GCCGAACCGTCAGATCCGCTA, (Reverse) CGGGCGGCCGCTTATGCGTAGTCTGGTACGTCGTACGGATACGCCAGAATGCGTTCGCA, was then inserted using the same enzymes.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data presented in this study are available and have been uploaded as supplemental material. Source data are provided with this paper.
References
Fukuda, H. & Kimura, A. Transfer of mitochondria into protoplasts of Saccharomyces cerevisiae by mini-protoplast fusion. FEBS Lett. 113, 58–60 (1980).
Spees, J. L., Olson, S. D., Whitney, M. J. & Prockop, D. J. Mitochondrial transfer between cells can rescue aerobic respiration. Proc. Natl. Acad. Sci. USA 103, 1283–1288 (2006).
Bergthorsson, U., Adams, K. L., Thomason, B. & Palmer, J. D. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 424, 197–201 (2003).
Im, G. B. & Melero-Martin, J. M. Mitochondrial transfer in endothelial cells and vascular health. Trends Cell Biol. https://doi.org/10.1016/J.TCB.2025.04.004 (2025).
Baldwin, J. G. et al. Intercellular nanotube-mediated mitochondrial transfer enhances T cell metabolic fitness and antitumor efficacy. Cell 187 (2024).
Ikeda, H. et al. Immune evasion through mitochondrial transfer in the tumour microenvironment. Nature https://doi.org/10.1038/s41586-024-08439-0 (2025).
Rustom, A., Saffrich, R., Markovic, I., Walther, P. & Gerdes, H. H. Nanotubular highways for intercellular organelle transport. Science 303, 1007–1010 (2004).
Nicolás-Ávila, J. A. et al. A network of macrophages supports mitochondrial homeostasis in the heart. Cell 183, 94–109.e23 (2020).
Crewe, C. et al. Extracellular vesicle-based interorgan transport of mitochondria from energetically stressed adipocytes. Cell Metab. 33, 1853–1868.e11 (2021).
Al Amir Dache, Z. et al. Blood contains circulating cell-free respiratory competent mitochondria. FASEB J. 34, 3616–3630 (2020).
Boudreau, L. H. et al. Platelets release mitochondria serving as substrate for bactericidal group IIA-secreted phospholipase A2 to promote inflammation. Blood 124, 2173–2183 (2014).
Clark, M. A. & Shay, J. W. Mitochondrial transformation of mammalian cells. Nature 295, 605–607 (1982).
Roger, A. J., Muñoz-Gómez, S. A. & Kamikawa, R. The origin and diversification of mitochondria. Curr. Biol. 27, R1177–R1192 (2017).
Lin, R. Z. et al. Mitochondrial transfer mediates endothelial cell engraftment through mitophagy. Nature 629, 660–668 (2024).
Cowan, D. B. et al. Transit and integration of extracellular mitochondria in human heart cells. Sci. Rep. 7 (2017).
Bonsergent, E. et al. Quantitative characterization of extracellular vesicle uptake and content delivery within mammalian cells. Nat. Commun. 12 (2021).
Nemoto, Y. & De Camilli, P. Recruitment of an alternatively spliced form of synaptojanin 2 to mitochondria by the interaction with the PDZ domain of a mitochondrial outer membrane protein. EMBO J. 18, 2991–3006 (1999).
Patterson, T. E. & Poyton, R. O. COX8, the structural gene for yeast cytochrome c oxidase subunit VIII. DNA sequence and gene disruption indicate that subunit VIII is required for maximal levels of cellular respiration and is derived from a precursor which is extended at both its NH2 and COOH termini. J. Biol. Chem. 261, 17192–17197 (1986).
Chen, C., Li, H., Zhang, J., Cheng, S. C. Exploring the limitations of mitochondrial dye as a genuine horizontal mitochondrial transfer surrogate. Commun. Biol. 7 (2024).
Bonsergent, E. & Lavieu, G. Content release of extracellular vesicles in a cell-free extract. FEBS Lett. 593, 1983–1992 (2019).
Sabatini, D. D. & Blobel, G. Controlled proteolysis of nascent polypeptides in rat liver cell fractions: II. Location of the polypeptides in rough microsomes. J. Cell Biol. 45, 146–157 (1970).
Goldstein, J. L. & Brown, M. S. Binding and degradation of low density lipoproteins by cultured human fibroblasts. Comparison of cells from a normal subject and from a patient with homozygous familial hypercholesterolemia. J. Biol. Chem. 249, 5153–5162 (1974).
Brestoff, J. R. et al. Intercellular mitochondria transfer to macrophages regulates white adipose tissue homeostasis and is impaired in obesity. Cell Metab. 33, 270–282.e8 (2021).
Vogel, G., Thilo, L., Schwarz, H. & Steinhart, R. Mechanism of phagocytosis in Dictyostelium discoideum: phagocytosis is mediated by different recognition sites as disclosed by mutants with altered phagocytotic properties. J. Cell Biol. 86, 456–465 (1980).
Sahr, T. et al. Translocated Legionella pneumophila small RNAs mimic eukaryotic microRNAs targeting the host immune response. Nat. Commun. 13 (2022).
Radulovic, M. et al. ESCRT-mediated lysosome repair precedes lysophagy and promotes cell survival. EMBO J 37 (2018).
Skowyra, M. L., Schlesinger, P. H., Naismith, T. V., Hanson, P. I. Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 360 (2018).
Teo, S. L. Y. et al. Unravelling cytosolic delivery of cell penetrating peptides with a quantitative endosomal escape assay. Nat. Commun. 12, 3721 (2021).
Hayakawa, K. et al. Transfer of mitochondria from astrocytes to neurons after stroke. Nature 535, 551–555 (2016).
Joshi, B. S., de Beer, M. A., Giepmans, B. N. G. & Zuhorn, I. S. Endocytosis of extracellular vesicles and release of their cargo from endosomes. ACS Nano 14, 4444–4455 (2020).
Hyenne, V. et al. Studying the fate of tumor extracellular vesicles at high spatiotemporal resolution using the zebrafish embryo. Dev. Cell 48, 554–572.e7 (2019).
Verweij, F. J. et al. Live tracking of inter-organ communication by endogenous exosomes in vivo. Dev. Cell 48, 573–589.e4 (2019).
White, J., Kartenbeck, J. & Helenius, A. Fusion of Semliki forest virus with the plasma membrane can be induced by low pH. J. Cell Biol. 87, 264–272 (1980).
de Jong, O. G. et al. A CRISPR-Cas9-based reporter system for single-cell detection of extracellular vesicle-mediated functional transfer of RNA. Nat. Commun. 11 (2020).
Tábara, L. C. et al. MTFP1 controls mitochondrial fusion to regulate inner membrane quality control and maintain mtDNA levels. Cell 187, 3619–3637.e27 (2024).
Z. Al Amir Dache, A.R. Thierry, Mitochondria-derived cell-to-cell communication. Cell Rep. 42 (2023).
Borcherding, N. & Brestoff, J. R. The power and potential of mitochondria transfer. Nature 623, 283–291 (2023).
Meeusen, S. & Nunnari, J. Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome. J. Cell Biol. 163, 503–510 (2003).
J.C. Casler, C.S. Harper, A.J. White, H.L. Anderson, L.L. Lackner, Mitochondria-ER-PM contacts regulate mitochondrial division and PI(4)P distribution. J. Cell Biol. 223 (2024).
King, M. P. & Attardi, G. Injection of mitochondria into human cells leads to a rapid replacement of the endogenous mitochondrial DNA. Cell 52, 811–819 (1988).
King, M. P. & Attardi, G. Human cells lacking mtDNA: Repopulation with exogenous mitochondria by complementation. Science (1979) 246, 500–503 (1989).
Acknowledgements
Work in G.L laboratory is supported by INSERM, “Agence Nationale de la Recherche” (ANR-19-CE18-0020 Excelldisc, ANR-20-CE15-0021 BIOEV, ANR-21-CE11-0009-EVfusion, ANR-24-CE13-4736 EVEM, ANR-24-CE11-5626 FUMESPIME) and Chaire d’Excellence Idex (ANR-18-IDEX-0001, IdEx Université Paris Cité). Z.A.A.D. received a postdoctoral fellowship from “Fondation pour la Recherche Medicale” (FRM: SPF202110013921). We acknowledge the Imaging core facility of BioMedTech (INSERM US36/CNRS UAR2009, Université Paris Cité) and Everzom for NTA access. We thank Christian Wunder and Ludger Johannes from Institut Curie, Paris, France for the gift of Galectin3-GFP plasmid, David Tareste from the institute of psychiatry and neuroscience of Paris for the gift of mCherry-OMP25 plasmid, and Walther Mothes from Yale University, New Haven, USA for the gift of RFP-CD63 plasmid.
Author information
Authors and Affiliations
Contributions
Z.A.D. designed and performed the experiments, and analyzed the data. J.D. and M.C. performed experiments and analyzed data. G.L. supervised the study. All authors wrote or revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Peer review
Peer review information
Nature Communications thanks Jonathan Brestoff, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Al Amir Dache, Z., Chevé, M., Dancourt, J. et al. Quantitative cellular characterization of extracellular mitochondria uptake and delivery. Nat Commun 16, 9053 (2025). https://doi.org/10.1038/s41467-025-64147-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-64147-x