Abstract
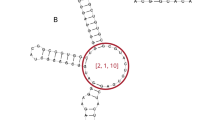
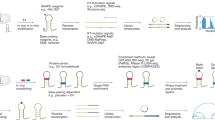
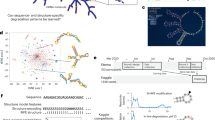
RNA design aims to find a sequence that can fold into a target secondary structure. It can create artificial RNA molecules for specific functions, with wide applications in medicine. It is computationally challenging due to two levels of combinatorial explosion: the exponentially large design space and the exponentially many competing structures per design. Popular methods such as local search cannot keep up with these combinatorial explosions. We instead employ two techniques from machine learning, continuous optimization and Monte-Carlo sampling. We start from a distribution over all valid sequences, and use gradient descent to improve the expectation of an arbitrary objective function. We define novel coupled-variable distributions to model the correlation between nucleotides. We then use sampling to approximate the objective, estimate the gradient, and select the final candidate. Our work consistently outperforms state-of-the-art methods in key metrics including Boltzmann probability and ensemble defect, especially on long and hard-to-design structures.
Similar content being viewed by others
Data availability
The designed sequences generated in this study are provided in the Supplementary Informatio/Source Data file. Source data are provided with this paper.
Code availability
The SamplingDesign source code is available at https://github.com/weiyutang1010/SamplingDesign, under Apache 2.0 license. The specific version of the code associated with this publication is archived in Zenodo and is accessible via https://doi.org/10.5281/zenodo.1767402148.
References
Eddy, S. R. Non-coding RNA genes and the modern RNA world. Nature Reviews Genetics 2, 919–929 (2001).
Doudna, J. A. & Cech, T. R. The chemical repertoire of natural ribozymes. Nature 418, 222–228 (2002).
Bachellerie, J. P., Cavaillé, J. & Hüttenhofer, A. The expanding snoRNA world. Biochimie 84, 775–790 (2002).
Zhou, T. et al. RNA design via structure-aware multifrontier ensemble optimization. Bioinformatics 39, i563–i571 (2023).
Portela, F. An unexpectedly effective Monte Carlo technique for the RNA inverse folding problem. BioRxiv (2018).
Hofacker, I. L. et al. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 125, 167–167 (1994).
Lorenz, R. et al. ViennaRNA Package 2.0. Algorithms Mol. Biol. 6, 26 (2011).
Andronescu, M., Fejes, A. P., Hutter, F., Hoos, H. H. & Condon, A. A New Algorithm for RNA Secondary Structure Design. Journal of Molecular Biology 336, 607–624 (2004).
Busch, A. & Backofen, R. INFO-RNA – a fast approach to inverse RNA folding. Bioinformatics 22, 1823–1831 (2006).
Bellaousov, S., Kayedkhordeh, M., Peterson, R. J. & Mathews, D. H. Accelerated RNA secondary structure design using preselected sequences for helices and loops. RNA 24, 1555–1567 (2018).
Garcia-Martin, J. A., Clote, P. & Dotu, I. RNAiFOLD: a constraint programming algorithm for RNA inverse folding and molecular design. J. Bioinfo. Comp. Bio.11 (2013).
Zadeh, J. N. et al. NUPACK: Analysis and design of nucleic acid systems. J. Comp. Chem. 32, 170–173 (2011).
Dotu, I. et al. Complete RNA inverse folding: computational design of functional hammerhead ribozymes. Nucleic Acids Research 42, 11752–11762 (2014).
Yamagami, R., Kayedkhordeh, M., Mathews, D. H. & Bevilacqua, P. C. Design of highly active double-pseudoknotted ribozymes: a combined computational and experimental study. Nucleic Acids Research 47, 29–42 (2018).
Schwab, R., Ossowski, S., Riester, M., Warthmann, N. & Weigel, D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. The Plant Cell 18, 1121–1133 (2006).
Hamada, M. In silico approaches to RNA aptamer design. Biochimie 145, 8–14 (2018).
Bauer, G. & Suess, B. Engineered riboswitches as novel tools in molecular biology. Journal of biotechnology 124, 4–11 (2006).
Findeiß, S., Etzel, M., Will, S., Mörl, M. & Stadler, P. F. Design of artificial riboswitches as biosensors. Sensors 17, 1990 (2017).
Norn, C. et al. Protein sequence design by conformational landscape optimization. Proceedings of the National Academy of Sciences 118, e2017228118 (2021).
Bonnet, É, Rzazewski, P. & Sikora, F. Designing RNA secondary structures is hard. J. Comp. Bio 27, 302–316 (2020).
Matthies, M., Krueger, R., Torda, A. & Ward, M. Differentiable Partition Function Calculation for RNA. Nucleic Acids Research (2023).
Dai, N., Zhou, T., Tang, W. Y., Mathews, D. H. & Huang, L. EnsembleDesign: messenger RNA design minimizing ensemble free energy via probabilistic lattice parsing. Proceedings of ISMB (2025).
Yang, X., Yoshizoe, K., Taneda, A. & Tsuda, K. RNA inverse folding using Monte Carlo tree search. BMC bioinformatics 18, 468 (2017).
Churkin, A. et al. Design of RNAs: comparing programs for inverse RNA folding. Briefings in bioinformatics 19, 350–358 (2018).
Mittal, A., Turner, D. H. & Mathews, D. H. NNDB: An Expanded Database of Nearest Neighbor Parameters for Predicting Stability of Nucleic Acid Secondary Structures. Journal of Molecular Biology436 (2024).
Zhou, T., Tang, W. Y., Mathews, D. H. & Huang, L. Undesignable RNA Structure Identification via Rival Structure Generation and Structure Decomposition. Proceedings of RECOMB (2024).
Zhou, T., Tang, W. Y., Mathews, D. H. & Huang, L. Scalable Identification of Minimum Undesignable RNA Motifs on Loop-Pair Graphs. Proceedings of RECOMB (2024).
Ward, M., Courtney, E. & Rivas, E. Fitness functions for RNA structure design. Nucleic Acids Research 51, e40–e40 (2023).
Lafferty, J., McCallum, A. & Pereira, F. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. Proceedings of ICML (2001).
Vaswani, A. et al. Attention is all you need. Proceedings of NIPS (2017).
Kahn, H. & Harris, T. E. Estimation of particle transmission by random sampling. NBS. applied math series 12, 27–30 (1951).
Zhang, H. et al. Algorithm for optimized mRNA design improves stability and immunogenicity. Nature 621, 396–403 (2023).
Wayment-Steele, H. K. et al. Theoretical basis for stabilizing messenger rna through secondary structure design. Nucleic acids research 49, 10604–10617 (2021).
Runge, F., Franke, J., Fertmann, D., Backofen, R. & Hutter, F. Partial RNA design. Bioinformatics 40, i437–i445 (2024).
Deigan, K. E., Li, T. W., Mathews, D. H. & Weeks, K. M. Accurate SHAPE-directed RNA structure determination. Proceedings of the National Academy of Sciences 106, 97–102 (2009).
Zadeh, J. N., Wolfe, B. R. & Pierce, N. A. Nucleic acid sequence design via efficient ensemble defect optimization. Journal of computational chemistry 32, 439–452 (2011).
Zhang, H., Zhang, L., Mathews, D. H. & Huang, L. LinearPartition: linear-time approximation of RNA folding partition function and base-pairing probabilities. Bioinformatics36 (2020).
Kingma, D. P. & Ba, J. Adam: A method for stochastic optimization. arXiv:1412.6980 (2014).
Nesterov, Y. A method for unconstrained convex minimization problem with the rate of convergence O(1/k2). Dokl. Akad. Nauk. SSSR 269, 543 (1983).
Huang, L. et al. LinearFold: linear-time approximate RNA folding by 5’-to-3’ dynamic programming and beam search. Bioinformatics 35, i295–i304 (2019).
Taneda, A. MODENA: a multi-objective RNA inverse folding. Adv. Appl. Bioinform. Chem. 4, 1 (2011).
Eastman, P., Shi, J., Ramsundar, B. & Pande, V. S. Solving the RNA design problem with reinforcement learning. PLoS Comp. Bio. 14, e1006176 (2018).
Runge, F., Stoll, D., Falkner, S. & Hutter, F. Learning to design RNA. arXiv:1812.11951 (2018).
Anderson-Lee, J. et al. Principles for predicting RNA secondary structure design difficulty. J. Mol. Bio. 428, 748–757 (2016).
Koodli, R. V. et al. Redesigning the EteRNA100 for the Vienna 2 folding engine. BioRxiv 2021–08 (2021).
Adamczyk, B., Antczak, M. & Szachniuk, M. RNAsolo: a repository of cleaned PDB-derived RNA 3D structures. Bioinformatics 38, 3668–3670 (2022).
Badura, J., Rybarczyk, A. & Zok, T. Comprehensive datasets for RNA design, machine learning, and beyond. Scientific Reports 15, 21417 (2025).
Tang, W. Y., Dai, N., Zhou, T., Mathews, D. H. & Huang, L. SamplingDesign: RNA Design via Continuous Optimization with Coupled Variables and Monte-Carlo Sampling (2025). https://doi.org/10.5281/zenodo.17674022.
Acknowledgements
This work was supported in part by NSF grants 2009071 (L.H.) and 2330737 (L.H. and D.H.M.).
Author information
Authors and Affiliations
Contributions
L.H. conceived and directed the project, and developed the sampling framework. W.Y.T. implemented the whole system, and analyzed and visualized the results. N.D. suggested the coupled variable for pairs, guided the softmax parameterization, and ran with the baseline21. T.Z. implemented the projected gradient descent and contributed to data analysis, esp. for the SAMFEO baseline. D.H.M. suggested the coupled variable for mismatches, and guided the data analysis and visualizations. All authors wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tang, W.Y., Dai, N., Zhou, T. et al. SamplingDesign: RNA design via continuous optimization with coupled variables and Monte-Carlo sampling. Nat Commun (2026). https://doi.org/10.1038/s41467-025-67901-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-67901-3