Abstract
In the leaves of most herbaceous plants, stomata exist in both the adaxial (upper) and abaxial (lower) surfaces1,2. Many previous studies have reported that stomata on the abaxial surface are more responsive to light than those on the adaxial surface3,4,5,6,7,8,9. However, the underlying molecular mechanism is still unclear. Here, by examining the model plants Arabidopsis and tobacco, we confirmed that the distinct feature occurred at the guard cell level. Next, with both single-cell RNA sequencing and transcriptome sequencing, we compared gene expression patterns of adaxial and abaxial guard cells and highlighted the possibility of different utilization of potassium ion (K+) channels. Via in silico OnGuard simulation and genetic modifications, we found that adaxial and abaxial guard cells rely on different K+in channels, which control K+ influx for stomatal opening. The present study provides insights into understanding the distinct stomatal light response of different leaf surfaces.
Similar content being viewed by others
Main
Stomata, central pore structures surrounded by a pair of guard cells (GCs), are typically present in the leaf epidermis and serve as an essential pathway for gas exchange between plants and the environment10. Plants have evolved complex signalling networks to regulate stomatal movement and development, consequently enhancing photosynthesis and water use efficiencies11,12. In most herbaceous plants, stomata are distributed on both the adaxial (upper) and abaxial (lower) surfaces of leaves1,2 (Fig. 1a). However, because of the differential micro-environment, especially light conditions6,13,14, adaxial and abaxial stomata exhibit different characteristics. The distinct features of adaxial and abaxial stomata have been reported in vitro and in vivo, using Vicia faba, Zea mays, Helianthus annuus and Xanthium strumarium, among others3,4,5,6,7,8,9. It has been shown that the light responses of adaxial stomata were significantly weaker than abaxial stomata, and the distinctions between both stomata were probably inherent7. Nevertheless, few studies have focused on the inherently different features of adaxial and abaxial stomata. Here, we systematically examined the different properties of adaxial and abaxial stomata in model plants Arabidopsis thaliana and Nicotiana tabacum, via single-cell RNA sequencing (scRNA-seq) and transcriptome sequencing (RNA-seq), we analysed gene expression patterns of the two types of GC and then investigated the underlying mechanism of distinct light responses between adaxial and abaxial stomata.
a, Arabidopsis leaf orientations of the adaxial surface (ad, left) and abaxial surface (ab, right). Scale bar, 1 cm. b, The stomatal size in the abaxial and adaxial surfaces of WT1 and WT2. The data are shown as mean ± s.d. (n = 3); each sample contains 15 stomata. Statistical analysis was carried out using a two-tailed Student’s t-test. c, The stomatal density in the abaxial and adaxial surfaces of WT1 and WT2. The data are shown as mean ± s.d. (n = 3); each sample contains ten microscopic fields. Statistical analysis was carried out using a two-tailed Student’s t-test. d, The stomatal index in the abaxial and adaxial surfaces of WT1 and WT2. The data are shown as mean ± s.d. (n = 3); each sample contained ten microscopic fields. Statistical analysis was carried out with a two-tailed Student’s t-test. e,f, The light response curves for the abaxial and adaxial surfaces in WT1 (e) and WT2 (f). Values are presented as the means of stomatal conductance ± s.d. (n = 8). g,h, The light response curves for the average per stoma of the abaxial and adaxial surfaces in WT1 (g) and WT2 (h). Values are presented as the means of stomatal conductance per stoma (\({g}_{\rm{s}}^{* }\)) ± s.d. (n = 8). Statistical analysis was carried out with a two-tailed Student’s t-test. The asterisks indicate statistically significant differences. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. i, Representative stomata of the abaxial and adaxial surfaces in WT1 and WT2 after exposure to 3 h of darkness or 200 µmol m−2 s−1 light treatment. Data shown are representative of three independent experiments with similar results. Scale bar, 10 μm. j, The stomatal apertures in the abaxial and adaxial epidermis of WT1 and WT2 after 3 h of dark or light treatment. The data are shown as mean ± s.d. (n = 3); each sample contains 25 stomata. Light conditions were consistent with those in i. Refer to the Methods for specific details on epidermal treatment. Statistical differences were assessed using a two-tailed Student’s t-test.
Using two common wild-type (WT) plants, Columbia-0 (named WT1) and gl1 (named WT2), we first characterized the stomatal properties on both surfaces of Arabidopsis plants. In both WT plants, the stomatal size on the adaxial surface was comparable to that on the abaxial surface (Fig. 1b), but stomatal density and stomatal index on the adaxial surface were significantly lower than those on the abaxial surface (Fig. 1c,d). Gas-exchange properties were evaluated using a LI-600 porometer. The stomatal conductance of both surfaces was increased with the increasing light intensities, but that on the abaxial surface were always significantly higher than on the adaxial surface (Fig. 1e,f). We also calculated the average stomatal conductance per stoma (\({g}_{\rm{s}}^{* }\)) by dividing the stomatal conductance by stomatal density to evaluate the difference between single stoma on both surfaces. The results were consistent with those of stomatal conductance (Fig. 1g,h), showing that the distinction of light response probably happened at the single stoma level. Then, to verify this possibility, we isolated the leaf epidermis and examined the single stomatal aperture directly. As shown in Fig. 1i,j, in both WT plants, the abaxial stomatal apertures were approximately 70% wider than the adaxial ones when exposed to light for 3 h, but no differences were detected in the closure of these stomata in the darkness. Similar results were also found in Arabidopsis under high-light growth conditions (Extended Data Fig. 1a–c) and in the other model plant, N. tabacum (Extended Data Fig. 2). These findings collectively suggest that the single stoma of the abaxial surface was more responsive to light than that of the adaxial surface, which implied that the GCs on both surfaces might be different.
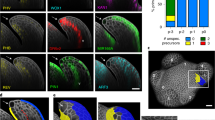
To analyse the potential types of GC, using Arabidopsis, GC protoplasts (GCPs) were isolated from rosette leaves through enzymatic digestion. Subsequently, the 10x Genomics Chromium platform was utilized to obtain 24,840 scRNA-seq, forming a scRNA-seq atlas (Fig. 2a). Both WT1 and WT2 were used for protoplast isolation to ensure reliable and repeatable results. Using t-distributed stochastic neighbourhood embedding (t-SNE) for dimensionality reduction and visualization, we observed highly similar cell distributions pattern from WT1 and WT2 (Fig. 2b). Combining the samples of WT1 and WT2 for cell clustering, 20 distinct clusters were identified through unsupervised clustering using Seurat (Extended Data Fig. 3a). According to previous studies and databases15,16, cell types were discerned by analysing the expression patterns of multiple marker genes in each cluster. On the basis of these patterns, all clusters were categorized into four groups: clusters 15 and 18 as epidermal cells; clusters 1–6, 8–9, 13–14, 16 and 19 as GCs; clusters 10–11, 17 and 20 as mesophyll cells; and clusters 7 and 12 as vascular cells (Extended Data Fig. 3a,c). Among them, GCs represented the highest proportion (Extended Data Fig. 3b). Moreover, it is interesting that the GC cluster could be further subdivided into two distinct subclusters, namely GC1 and GC2 (Fig. 2c), in which some genes related to stomatal light response were expressed quite differently (Extended Data Fig. 3d). However, whether these two subclusters represented the abaxial and adaxial GCs is unknown.
a, A flowchart of scRNA-seq of GCPs in Arabidopsis. b, A visualization of the distribution and numbers of protoplasts for two samples (WT1 and WT2) using t-SNE. c, Isolation of two cell clusters of GCPs for two samples (WT1 and WT2) using t-SNE. Each coloured dot represents individual cell types, namely GC1 and GC2. d, PCA of the scRNA-seq and RNA-seq for the GCPs. Among them, WT1-ab, WT2-ab, WT1-ad and WT2-ad represent the mean results of GC RNA-seq obtained through the tape–peel method in both WT plants. WT1-GC1, WT2-GC1, WT1-GC2 and WT2-GC2 represent the mean results of GCPs scRNA-seq in both WT plants. Percentages represent variance captured by principal components 1 and 2 in each analysis. e, A heat map showing the DEGs in the GCPs from the adaxial and abaxial surfaces of two WT plants by RNA-seq (n = 3). Data were Z-score normalized within a given parameter, and organized by hierarchical clustering. Biological processes are listed, such as response to high light intensity, RNA modification, response to heat, oxidation–reduction processes, transmembrane transport, stomatal movement and photosynthesis. f, qPCR analysis of K+in channels genes in GCPs of the abaxial and adaxial epidermis of WT1 and WT2. The columns and error bars in f represent the mean ± s.d. (n = 6). The letters indicate groups with statistically significant differences (P < 0.05). Data were analysed through one-way ANOVA and Fisher’s multiple comparison with two-tailed tests.
To further understand the two subclusters of GCs, we extracted the total RNA from isolated GCPs of the adaxial or abaxial surfaces for the RNA-seq analysis and compared the results with scRNA-seq. We developed the tape–peel method with enzymatic digestion to isolate GCPs from the adaxial and abaxial surfaces, respectively. To confirm that the tape–peel method was as efficient as the traditional blender method for epidermal isolation, stomatal aperture and fluorescein diacetate staining were performed to evaluate the activation of GCs. As shown in Extended Data Fig. 4, both stomatal aperture and fluorescent intensity were comparable between the tape–peel method and the blender method.
Then, three independent biological experiments using the tape–peel method for GCP isolation were performed in both WT plants. The mean results of RNA-seq analysis were labelled as WT1-ad and WT2-ad for adaxial GCs and WT1-ab and WT2-ab for abaxial GCs. By principal component analysis (PCA), as shown in Fig. 2d, we found that the results of transcriptomes could be clearly catalogued into two groups, in which WT1-GC1, WT2-GC1, WT1-ab and WT2-ab were in one group, while WT1-GC2, WT2-GC2, WT1-ad and WT2-ad were in the other group. This implies that the transcriptome patterns of adaxial and abaxial GCs are significantly different, and the two subclusters, GC1 and GC2, refer to these two kinds of GC, respectively. Although the WT-ab and GC1 (or WT-ad and GC2) were grouped together, they were not identical. Because the cluster analysis of scRNA-seq was based on calculating the mean expression and dispersion of all genes, whereas the RNA-seq data were explicitly derived from either adaxial or abaxial GCs, the RNA-seq results and the subsequent quantitative polymerase chain reaction (qPCR) validation more accurately represent the true distinction between adaxial and abaxial GCs.
We then compared the results of RNA-seq analysis of GCPs from the adaxial and abaxial GCs in detail. In two WT plants, over 70% of genes showed overlapped expression (Extended Data Fig. 5a), and the transcriptome patterns were quite similar to the correlation analysis (Extended Data Fig. 5b). Subsequently, we conducted Gene Ontology term enrichment analysis on the differentially expressed genes (DEGs) in the GCPs from the adaxial and abaxial surfaces of two WT plants to investigate the molecular mechanisms underlying biological processes in distinct GCs (Fig. 2e). The results indicated that processes such as response to high light intensity, RNA modification, and response to heat and oxidation–reduction processes are significantly enriched in the adaxial GCs. Meanwhile, processes significantly enriched in the abaxial GCs include transmembrane transport, stomatal movement and photosynthesis (Fig. 2e). Partially consistent with the results in single-cell sequencing, different GCs exhibit the selectivity on the expression levels of genes related to stomatal light response (Extended Data Fig. 5c). To further evaluate the expression differences of potential genes between adaxial and abaxial GCs, we performed qPCR analysis (Fig. 2f, Extended Data Fig. 6 and Supplementary Table 3). Among these, it is worth noting that the expression levels of inward-rectifying K+ (K+in) channel genes, KAT1, KAT2, AKT1 and AKT2, were significantly distinct in adaxial and abaxial GCs.
K+ influx, activated by plasma membrane hyperpolarization, is recognized as an important driving force for stomatal opening17,18. In Arabidopsis, suppressing the K+ currents could limit the process of stomatal opening19,20. Using qPCR assays, we found that KAT1 and AKT1 were relatively dominant in GCs compared with the other two K+in channel genes. KAT1 expression was approximately 2.2-fold higher in abaxial GCs, whereas AKT1 was approximately twofold higher in adaxial GCs (Fig. 2f and Supplementary Table 3). Western blotting analyses also demonstrated that the levels of KAT1 expressed in the abaxial side of WT plants were ~1.5-fold higher than in adaxial side, whereas those of AKT1 were ~1.8-fold higher in the adaxial than in abaxial side (Extended Data Fig. 7a,b). These findings implied the possibility that the differential proportions of KAT1 and AKT1 might contribute to the distinct stomatal light response.
To further investigate the impact of KAT1 and AKT1 on stomatal behaviour, we took advantage of the OnGuard model, to explore the in silico changes for stomatal behaviours. The OnGuard platform is an effective tool that considers membrane ion transport, osmotically active solute metabolism and their dynamic interactions to accurately describe and predict stomatal physiology21. In our modelling, we first differentiated inward-rectifier channels into KAT1-like (KAT) and AKT1-like (AKT) channels. The key distinction lies in the activation of AKT1 by cytosolicCa2+ (refs. 22,23). By manipulating the ratio of KAT/AKT and the expression levels of total K+in channels, we simulated the diurnal changes in stomatal behaviour. These simulations encompassed different ratios of KAT/AKT following standard 8-h/16-h day/night cycles (Supplementary Table 1). The other parameter settings remained consistent with previous descriptions24,25,26. Notably, changes in the total number of K+in channels had a significant impact on stomatal aperture, with a decrease in the total amount of K+in channels resulting in a decrease in stomatal conductance (Fig. 3a). The maximum stomatal conductance during the day showed a clear linear correlation with the ratio of KAT/AKT, indicating that an increase in the KAT ratio leads to an increase in stomatal conductance. When the total amount of K+in channels decreases, increasing the ratio of KAT/AKT can partially compensate for the loss of stomatal conductance (Fig. 3b).
a, The OnGuard model output for stomatal conductance involves plants with varying AKT/KAT ratio and K+ channel numbers. The specific AKT/KAT ratio and K+ number details are provided in Supplementary Table 1. The light cycle is represented by white and grey, with white denoting the light phase and grey representing the dark phase. b, The steady state of stomatal conductance and its dependence on the AKT/KAT ratio and K+ number from the model (surface plot), including the data in a.
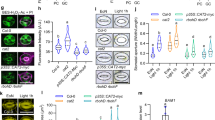
To verify the simulation using the OnGuard model, and examine our prediction that the adaxial and abaxial GCs use different K+in channels, we investigated transgenic plants overexpressing KAT1 and AKT1 (KAT1ox and AKT1ox) using the strong GC promoter GC1 (Fig. 4a), and mutant plants (kat1 and akt1) with T-DNA insertion lines (Fig. 4b). The expression levels of KAT1 and AKT1 in the epidermis of abaxial and adaxial surfaces in WT1, WT2, KAT1ox#1, KAT1ox#2, AKT1ox#1, AKT1ox#2, kat1 and akt1 are shown in Extended Data Fig. 7c. Western blotting analysis demonstrated that the KAT1 levels in the levels of KAT1 expressed in abaxial of KAT1ox#1 and KAT1ox#2 were ~2.1-fold and 1.6-fold higher than those in the WT2, respectively, while the AKT1 levels in the adaxial of AKT1ox#1 and AKT1ox#2 were ~1.7-fold and 1.6-fold higher than those in the WT2 (Extended Data Fig. 7d). Although the same promoter (GC1 promoter) was used in the construction of both KAT1ox and AKT1ox, it is noteworthy that the expression level of gene KAT1 differed significantly between adaxial and abaxial GCs, whereas no such difference was observed for gene AKT1. This suggests possible distinct transcriptional regulation mechanisms for genes KAT1 and AKT1 in adaxial and abaxial GCs, respectively. Further studies will be required to elucidate the underlying mechanisms. Subsequently, we investigated the stomatal light responses in these plants. For the abaxial stomata, overexpression of KAT1 or AKT1 did not alter their stomatal aperture, conductance and K+ influx rate (Fig. 4d,f,j), while kat1 mutants showed decreased stomatal aperture (Fig. 4c), stomatal conductance (Fig. 4e) and K+ influx rate (Fig. 4i). Similar results are also seen in \({g}_{\rm{s}}^{* }\) after eliminating stomatal density effects (Fig. 4g,h). However, the stomatal aperture (Fig. 4c), stomatal conductance or \({g}_{\rm{s}}^{* }\) (Fig. 4e,g) and K+ influx rate (Fig. 4i) in the akt1 mutant were similar to WT, indicating that KAT1 may play a predominant role in the abaxial surface. In the case of the adaxial stomata, AKT1ox plants demonstrated elevated stomatal aperture, K+ influx rate and conductance or \({g}_{\rm{s}}^{* }\) compared with the WT, while the opposite was observed in akt1 (Fig. 4c–j). According to the measurement of K+ flux analysis, a significant decrease in K+ flux was detected in the GCs on the adaxial surface of akt1, while no impact on the abaxial surface was observed (Fig. 4i). The above results indicate that AKT1 may serve as the primary K+in channel in the GCs on the adaxial surface. Interestingly, overexpression of either KAT1 or AKT1 leads to a rescue in stomatal aperture and conductance on the adaxial surface to a level comparable to that in the abaxial surface, suggesting that the K+ transport capacity limited the adaxial stomatal response to light. We also examined the expression level of PIFs because it was reported that the PIFs directly regulate the expression of KAT127. As predicted, the expression levels of PIFs were obviously higher in abaxial GCs than in adaxial GCs (Extended Data Fig. 8), which is consistent with previous findings27.
a, A schematic illustration showing that the overexpression of KAT1 and AKT1 using the strong GC promoter GC1 forms transgenic plants KAT1ox#1, KAT1ox#2, AKT1ox#1 and AKT1ox#2. b, A schematic illustration showing the gene structure of AT5G46240 and AT2G26650 and the approximate location of the corresponding T-DNA insertion. The AT5G46240 and AT2G26650 genes consist of 9 and 11 exons, respectively (shown as black boxes). c,d, The increment of stomatal apertures in the abaxial and adaxial epidermis of WT1, kat1 and akt1 (c) and WT2, KAT1ox#1, KAT1ox#2, AKT1ox#1 and AKT1ox#2 (d) after 3 h of darkness or 200 µmol m−2 s−1 light treatment. Values are presented as mean ±s.d. (n = 3). The letters indicate groups with statistically significant differences (P < 0.05). Data were analysed through ANOVA and Fisher’s multiple comparison. e,f, The increment of stomatal conductance in the abaxial and adaxial epidermis of WT1, kat1 and akt1 (e) and WT2, KAT1ox#1, KAT1ox#2, AKT1ox#1 and AKT1ox#2 (f) after 1 hour 200 µmol m−2 s−1 light treatment. Values are presented as mean ± s.d. (n = 9 biologically independent samples). The letters indicate groups with statistically significant differences (P < 0.05). Data were analysed through ANOVA and Fisher’s multiple comparison. g,h, The \({g}_{\rm{s}}^{* }\) in the abaxial and adaxial epidermis of WT1, kat1 and akt1 (g) and WT2, KAT1ox#1, KAT1ox#2, AKT1ox#1 and AKT1ox#2 (h) after 1 hour 200 µmol m−2 s−1 light treatment. Values are presented as mean ± s.d. (n = 9 biologically independent samples). The letters indicate groups with statistically significant differences (P < 0.05). Data were analysed through ANOVA and Fisher’s multiple comparison. i,j, Quantification of K+ influx rates from WT and transgenic plants. Difference in net K+ influx rates across the GC plasma membrane of the abaxial epidermis and adaxial epidermis in WT1, kat1 and akt1 (i) and WT2, KAT1ox#1, KAT1ox#2, AKT1ox#1, AKT1ox#2 (j). The negative values represent K+ net influx. Values in i and j are presented as mean ± s.d. (n = 6 biologically independent samples). The letters indicate groups with statistically significant differences (P < 0.05). Data were analysed through ANOVA and the Fisher’s multiple-comparison test. k, The difference in the conformation of constitutively active KAT1 and AKT1 between the cytoplasmic domains with four-fold symmetry predicted by AlphaFold3. Among them, R177C and D379A represent the constitutively active mutant of KAT1 and AKT1, respectively, which exhibit larger internal diameter and currents. l, A simplified model for the selectivity of K+ channels in GCs on the abaxial and adaxial surfaces.
It is known that members of the Shaker family, such as KAT1 and AKT1, possess six transmembrane domains and can form either homologous or heterologous tetramers to fulfil their functions28,29. Utilizing AlphaFold330, an advanced AI model for protein structure and interaction prediction, we predicted the continuously active tetrameric structures of KAT1 and AKT1, achieving a pTM score, which measures the global folding reliability of the complex with scores >0.5 indicating structural similarity to native conformations, exceeding 0.6 (Extended Data Fig. 9a,b). The structures reveal that the internal diameter of the constriction of KAT1 is 1.19 Å larger than that of AKT1 (Fig. 4k). Owing to the established positive correlation between the ion channel internal diameter and its functional activity31, the K+ flux through the KAT1-formed ion channel significantly exceeds that facilitated by the AKT1-formed channel. This observation is consistent with the cryo-electron microscopy analysis of the two types of complex32,33,34.
In summary, our study highlights the molecular distinction between adaxial and abaxial stomata by the preference of K+in channels. Compared with abaxial stomatal GCs, adaxial ones rely on AKT1, which possesses a lower K+ transport capacity than KAT1, resulting in a lower K+ influx and smaller stomatal aperture under light conditions (Fig. 4l). It is known that the adaxial leaf surface is always more exposed in higher-light conditions than the abaxial leaf surface; thus, downregulating the adaxial stomatal light response via AKT1 is probably beneficial for preventing excessive water loss. As most previous studies on stomata are based on abaxial GCs, the present study on the distinction of adaxial and abaxial GCs provides a further understanding of how leaves balance the carbon uptake and water loss. It will be interesting to explore the unique regulation of the adaxial GCs, which may contribute to improving plant water use efficiency for the advancement of sustainable agriculture.
Methods
Plant materials and growth conditions
Arabidopsis and N. tabacum were used in this study. Arabidopsis Col-0, used as the WT1, was the background ecotype of kat1 (SALK_127506C) and akt1 (SALK_061400C) transgenic plants. Arabidopsis Col-5 (Columbia (Col), carrying the homozygous recessive gl1) (https://www.arabidopsis.org/ecotype?key=94), used as the WT2, was the background ecotype of transgenic plants, KAT1ox and AKT1ox. All experimental plants were cultured in a mixture with soil and vermiculite (1:1) and planted in a growth cabinet. The growth conditions are 16 h light (80 µmol m−2 s−1)/8 h dark cycle at 24 °C in 50–60% (Pa/Pa) humidity. All the plants in the growth cabinet were watered twice a week.
Vector construction and plant transformation
The plasmid vectors used for transformation were constructed according to a previous method35. In brief, a transformation vector was constructed by replacing the corresponding region of the CaMV35S in pPZP211 (pPZP211-GC1). cDNA fragments of KAT1 (At5g46240) and AKT1 (At2g26650) were amplified using the primers listed in Supplementary Table 2. The amplified DNA fragments were inserted into pPZP211-GC1 using XbaI. Subsequently, all plant transformation plasmids were introduced into Agrobacterium tumefaciens strain GV3101, followed by their use for plant transformation using standard protocols35. Homozygous F3 plants were used in the experiments.
Measurement of stomatal traits
Stomatal aperture and size were measured according to a previous method with some modifications35. Epidermal fragments were harvested using a blender from overnight dark-adapted mature rosette leaves and then treated with 200 µmol m−2 s−1 light or dark for 3 h at room temperature. Stomatal apertures in the abaxial and adaxial epidermis were evaluated microscopically (BA210 Digital LED, MOTIC CHINA GROUP) using Motic Images Plus 2.0 and Image Analysis System 11. Compared with the adaxial surface, the abaxial surface exhibits a higher stomatal density and the epidermal cells are more wrinkled (Supplementary Fig. 1.). Following the distinction between the adaxial and abaxial epidermis, the stomatal aperture was measured (ImageJ 1.53). We performed three independent experiments (n = 3); in each independent experiment, two to three leaves from three plants were used and 20–30 stomatal apertures were randomly measured. Stomatal density and index were measured as described previously7. We performed three independent experiments (n = 3); in each independent experiment, three leaves from three plants were used and ten fields of view were measured randomly.
Measurement of stomatal conductance
Stomatal conductance was measured using the LI-600 Porometer (LI-COR) in the abaxial surface or adaxial surface of leaves. Light response curves were measured with leaves after the overnight dark adaptation. The intensity of white light (0, 20, 50, 100, 150, 200 and 500 µmol m−2 s−1) was increased step by step for about 120 min. In other experiments, stomatal conductance was measured after a treatment duration of 60 min with 200 µmol m−2 s−1 light. Fully expanded leaves of N. tabacum or rosette leaves of Arabidopsis from 4-week-old plants were used in these experiments. In these experiments, we used three to nine biologically independent samples (n = 3–9).
Isolation of GCPs
GCPs of Arabidopsis were isolated according to a previous method with some modifications36,37. GCPs for scRNA-seq were isolated using the traditional enzymolysis approach with cellulase R-10 or RS (Yakult Pharmaceutical Industry) and macerozyme R10 (Yakult Pharmaceutical Industry). GCPs of adaxial or abaxial stomata were isolated from different epidermises. Using time tape (Performance Flatback Tape 2525, 3M) to adhere closely to the adaxial surface of the rosette leaves, and then using magic tape (Scotch Magic Greener Tape Dispensered Rolls, 3M) to peel off the abaxial surface38,39 (Supplementary Fig. 2), we separated the epidermal strips of the adaxial and abaxial surfaces of 4-week-old Arabidopsis fully expanded rosette leaves, and then the mesophyll tissues and tapes were removed by a blender with cold distilled water. More than 300 leaf pieces were used for each experiment. These experiments were repeated at least three times using different plants.
scRNA-seq and data processing
The extracts of the protoplasts of GCs in WT1 and WT2 rosette leaves were used for scRNA-seq. Cell capture and cDNA synthesis Using Single Cell 3′ Library and Gel Bead Kit V3.1 (10x Genomics, 1000075) according to the manufacturer’s protocol. The libraries were sequenced using an IlluminaNovaseq6000 sequencer with a sequencing depth of at least 100,000 reads per cell with a paired-end 150-bp (PE150) reading strategy (performed by CapitalBio Technology). The Loupe Browser 6 and Cell Ranger (6.0.1) obtained from the 10x Genomics website was utilized for demultiplexing cellular barcodes and mapping reads to the TAIR10 reference genome. We processed the unique molecular identifier count matrix using Seurat 3.0 (R package). Quality control was conducted on the samples according to a previous method40, resulting in the retention of 24,840 single cells for downstream analysis. PCA was used for dimensionality reduction, and visualization was realized through t-SNE. The mean expression and dispersion were computed for all cells41, and cell types were identified through DEGs.
High-throughput RNA-seq analysis
Total RNA was isolated from the GCPs of abaxial epidermis and adaxial epidermis of WT1 and WT2 using the E.Z.N.A. Plant RNA Kit (Omega) according to the manufacturer’s instructions. We constructed the cDNA library for the GCPs of the abaxial epidermis and adaxial epidermis of WT1 and WT2 using VAHTS Universal V6 RNA seq Library Prep Kit for Illumina (Vazyme) and sequenced the data using a NextSeq 500 system (Illumina) according to a previous method42. Experiments were repeated three times with different GCPs. Gene expression values were reported in counts units. Normalization of read counts and statistical analyses were performed using the RStudio (R v4.3.2) software package.
Quantitative reverse-transcription PCR analysis
Owing to the low concentration of RNA in the GCPs, we used the Single Cell Sequence Specific Amplification Kit (Vazyme, P621) to perform reverse transcription and amplification reactions on trace amounts of total RNA43. We mix the tested gene primers to prepare the Assay pool, with a final concentration of 0.1 μM for each primer. According to the instructions, we prepared a reaction system and then performed reverse transcription and amplification reactions in a PCR instrument to obtain cDNA. The gene expression levels in GCPs of the abaxial epidermis and the adaxial epidermis were analysed using LightCycler 480 SYBR Green I Master (Roche) on a LightCycler 96 Instrument (Roche). The UBQ10 (At4g05320) genes were used as internal controls and expression levels were analysed using the 2−ΔΔCT method. We performed three independent experiments (n = 3). The specific sets of qPCR primers for gene amplification are listed in Supplementary Table 2.
Western blot
Western blot analysis was conducted following a previously established protocol with several modifications44,45. Total protein was extracted by grinding the abaxial epidermis fragments and adaxial epidermis fragments of WT (WT1 and WT2 mix), and homogenate was solubilized by adding a sodium dodecyl sulfate sample buffer. The solubilized sample was centrifuged at 12,000g for 10 min, and the resulting supernatant was subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred electrophoretically to nitrocellulose membranes (Bio-Rad). Nitrocellulose membranes were blocked and incubated at room temperature for 1 h, then with polyclonal rabbit anti-AKT1 or anti-KAT1 primary antibody (diluted 2,000-fold, PHYTOAB) at 4 °C overnight. A goat anti-rabbit immunoglobulin G (IgG, diluted 5,000-fold, EASYBIO) was used as the secondary antibody, and actin protein was detected using anti-actin antibody (diluted 2,000-fold, TDYBIO) as a control. The relative amount of KAT1 and AKT1 was quantified as the ratio to actin (ImageJ 1.53).
Measurement of K+ flux in GCs
The net K+ fluxes at the GCs in isolated epidermal strips were determined by a non-invasive micro-test technique (NMT 100 Series, Xuyue (Beijing) Sci. & Tech). The preparations and calibrations of K+-selective electrodes were performed according to a previous method46. The K+ backfilling buffer contains 100 mM of KCl, and the selective cocktail of K+ is K-LIX (XY-SJ-K-10). After each test, the same selective electrodes was recalibrated according to the same procedures and standards. Adaxial and abaxial surface epidermis strips were peeled with tweezers from 4-week-old Arabidopsis leaves and stuck to the middle of a Petri dish47. After equilibrating epidermal strips in buffer solution for 10 min, measurements were taken in the dark for 5 min, followed by exposure to light at 200 µmol m−2 s−1 for 10 min. K+-selective electrodes were moved perpendicularly to the GC, and the K+ fluxes were determined between two positions separated by 10 μm. The buffer solution contained 1 mM of KCl and 0.1 mM of CaCl2, with pH 6.0. In these experiments, we used six biologically independent samples (n = 6). Net ion fluxes were calculated using imFluxes V3.0 software.
OnGuard
OnGuard3 48, available for download at https://plantscienceglasgow.org/, is an advanced software built on the HoTSig platform49,50. The modelling process involves driving OnGuard3 through a diurnal light–dark cycle, similar to previous studies24,25,26. Additional steps in pCO2 and light are imposed on this cycle as required, and all model outputs are derived from this comprehensive cycle. Apoplastic solute contents are kept constant, while primary, energy-dependent transport, sucrose and malic acid synthesis within the GCs are coupled to light, following established relationships. It is worth noting that all other model parameters remain fixed, meaning the properties of individual transporters, metabolism and buffering reactions respond solely to changes in the model variables influenced by the encoded parameters (Supplementary Appendix 1).
Statistical analysis
Significance was determined according to the Student’s t-test using Microsoft Excel (2019). Unless otherwise indicated, statistical significance was concluded when the P value was less than 0.05. Analyses of variance (ANOVAs) were performed using SPSS 25.0 Statistics. Datasets were first examined for normality and then compared using one-way ANOVA followed by least significant difference (LSD) tests.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data analysed during this study are provided in the Letter and its Supplementary Information. The scRNA-seq and RNA-seq data have been deposited in a public database (BioStudies) under accession numbers S-BSST1965 and S-BSST1966. Source data are provided with this paper.
References
Drake, P. L., de Boer, H. J., Schymanski, S. J. & Veneklaas, E. J. Two sides to every leaf: water and CO2 transport in hypostomatous and amphistomatous leaves. N. Phytol. 222, 1179–1187 (2019).
Muir, C. D. Light and growth form interact to shape stomatal ratio among British angiosperms. N. Phytol. 218, 242–252 (2018).
Goh, C.-H., Oku, T. & Shimazaki, K. Properties of proton pumping in response to blue light and fusicoccin in GCPs isolated from adaxial epidermis of Vicia leaves. Plant Physiol. 109, 187–194 (1995).
Wang, X. Q., Wu, W. H. & Assmann, S. M. Differential responses of abaxial and adaxial guard cells of broad bean to abscisic acid and calcium. Plant Physiol. 118, 1421–1429 (1998).
Driscoll, S. P., Prins, A., Olmos, E., Kunert, K. J. & Foyer, C. H. Specification of adaxial and abaxial stomata, epidermal structure and photosynthesis to CO2 enrichment in maize leaves. J. Exp. Bot. 57, 381–390 (2006).
Wang, Y., Noguchi, K. & Terashima, I. Distinct light responses of the adaxial and abaxial stomata in intact leaves of Helianthus annuus L. Plant Cell Environ. 31, 1307–1316 (2008).
Wang, Y., Noguchi, K. & Terashima, I. Photosynthesis-dependent and -independent responses of stomata to blue, red and green monochromatic light: differences between the normally oriented and inverted leaves of sunflower. Plant Cell Physiol. 52, 479–489 (2011).
Mott, K. A. & Peak, D. Effects of the mesophyll on stomatal responses in amphistomatous leaves. Plant Cell Environ. 41, 2835–2843 (2018).
Goh, C. H., Oku, T. & Shimazaki, K. Photosynthetic properties of adaxial guard cells from Vicia leaves. Plant Sci. 127, 149–159 (1997).
Chen, Z. H. et al. Molecular evolution of grass stomata. Trends Plant Sci. 22, 124–139 (2017).
Clark, J. W. et al. The origin and evolution of stomata. Curr. Biol. 32, R539–R553 (2022).
Wang, Y., Wang, Y., Tang, Y. & Zhu, X. G. Stomata conductance as a goalkeeper for increased photosynthetic efficiency. Curr. Opin. Plant Biol. 70, 102310 (2022).
Mott, K. A., Gibson, A. C. & O’Leary, J. W. The adaptive significance of amphistomatic leaves. Plant Cell Environ. 5, 455–460 (2006).
Lawson, T., Terashima, I., Fujita, T. & Wang, Y. in The Leaf: A Platform for Performing Photosynthesis (eds Adams III, W. W. & Terashima, I.) 141–161 (Springer, 2018).
Liu, Z. et al. Global dynamic molecular profiling of stomatal lineage cell development by single-cell RNA sequencing. Mol. Plant 13, 1178–1193 (2020).
Jin, J. et al. PCMDB: a curated and comprehensive resource of plant cell markers. Nucleic Acids Res. 50, D1448–D1455 (2022).
Schroeder, J. I., Raschke, K. & Neher, E. Voltage dependence of K+ channels in guard- cell protoplasts. Proc. Natl Acad. Sci. USA. 84, 4108–4112 (1987).
Riedelsberger, J. et al. Distributed structures underlie gating differences between the Kin channel KAT1 and the Kout channel SKOR. Mol. Plant 3, 236–245 (2010).
Kwak, J. M. et al. Dominant negative guard cell K+ channel mutants reduce inward-rectifying K+ currents and light-induced stomatal opening in Arabidopsis. Plant Physiol. 127, 473–485 (2001).
Szyroki, A. et al. KAT1 is not essential for stomatal opening. Proc. Natl Acad. Sci. USA. 98, 2917–2921 (2001).
Blatt, M. R., Jezek, M., Lew, V. L. & Hills, A. What can mechanistic models tell us about guard cells, photosynthesis, and water use efficiency? Trends Plant Sci. 27, 166–179 (2022).
Li, L., Kim, B.-G., Cheong, Y. H., Pandey, G. K. & Luan, S. A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc. Natl Acad. Sci. USA. 103, 12625–12630 (2006).
Li, K. L., Tang, R. J., Wang, C. & Luan, S. Potassium nutrient status drives posttranslational regulation of a low-K response network in Arabidopsis. Nat. Commun. 14, 360 (2023).
Wang, Y. Z. et al. Systems dynamic modeling of a guard cell Cl− channel mutant uncovers an emergent homeostatic network regulating stomatal transpiration. Plant Physiol. 160, 1956–1967 (2012).
Wang, Y. Z., Hills, A. & Blatt, M. R. Systems analysis of guard cell membrane transport for enhanced stomatal dynamics and water use efficiency. Plant Physiol. 164, 1593–1599 (2014).
Wang, Y. Z. et al. Unexpected connections between humidity and ion transport discovered using a model to bridge guard cell-to-leaf scales. Plant Cell 29, 2921–2939 (2017).
Rovira, A. et al. PIF transcriptional regulators are required for rhythmic stomatal movements. Nat. Commun. 15, 4540 (2024).
Lefoulon, C. The bare necessities of plant K+ channel regulation. Plant Physiol. 187, 2092–2109 (2021).
Véry, A. A. & Sentenac, H. Molecular mechanisms and regulation of K+ transport in higher plants. Ann. Rev. Plant Biol. 54, 575–603 (2003).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Geng, Y., Niu, X. & Magleby, K. L. Low resistance, large dimension entrance to the inner cavity of BK channels determined by changing side-chain volume. J. Gen. Physiol. 137, 533–548 (2011).
Li, S. et al. Cryo-EM structure of the hyperpolarization-activated inwardly rectifying potassium channel KAT1 from Arabidopsis. Cell Res. 30, 1049–1052 (2020).
Lu, Y. et al. Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis. Nat. Commun. 13, 5682 (2022).
Latorre, R. et al. Molecular coupling between voltage sensor and pore opening in the Arabidopsis inward rectifier K+ channel KAT1. J. Gen. Physiol. 122, 459–469 (2003).
Wang, Y. et al. Overexpression of plasma membrane H+-ATPase in guard cells promotes light-induced stomatal opening and enhances plant growth. Proc. Natl Acad. Sci. USA. 111, 533–538 (2014).
Ueno, K., Kinoshita, T., Inoue, S., Emi, T. & Shimazaki, K. Biochemical characterization of plasma membrane H+-ATPase activation in guard cell protoplasts of Arabidopsis thaliana in response to blue light. Plant Cell Physiol. 46, 955–963 (2005).
Hayashi, M., Inoue, S. I., Ueno, Y. & Kinoshita, T. A Raf-like protein kinase BHP mediates blue light-dependent stomatal opening. Sci. Rep. 7, 45586 (2017).
Shi, W. et al. Hydrogen peroxide is required for light-induced stomatal opening across different plant species. Nat. Commun. 15, 5081 (2024).
Lawrence, S. 2nd, Pang, Q., Kong, W. & Chen, S. Stomata tape-peel: an improved method for guard cell sample preparation. J. Vis. Exp. 137, e57422 (2018).
Han, X. et al. Time series single-cell transcriptional atlases reveal cell fate differentiation driven by light in Arabidopsis seedlings. Nat. Plants 9, 2095–2109 (2023).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Yang, T. et al. Chromatin remodeling complexes regulate genome architecture in Arabidopsis. Plant Cell 34, 2638–2651 (2022).
Sheng, X. et al. The mitochondrial protease LONP1 maintains oocyte development and survival by suppressing nuclear translocation of AIFM1 in mammals. EBioMedicine 75, 103790 (2022).
Sutter, J. U., Campanoni, P., Tyrrell, M. & Blatt, M. R. Selective mobility and sensitivity to SNAREs is exhibited by the Arabidopsis KAT1 K+ channel at the plasma membrane. Plant Cell 18, 935–954 (2006).
Reintanz, B. et al. AtKC1, a silent Arabidopsis potassium channel α-subunit modulates root hair K+ influx. Proc. Natl Acad. Sci. USA. 99, 4079–4084 (2002).
Liu, B. et al. The anion channel SLAH3 interacts with potassium channels to regulate nitrogen-potassium homeostasis and the membrane potential in Arabidopsis. Plant Cell 35, 1259–1280 (2023).
Wei, J. et al. Phytomelatonin receptor PMTR1-mediated signaling regulates stomatal closure in Arabidopsis thaliana. J. Pineal. Res. 65, e12500 (2018).
Jezek, M. et al. Guard cell endomembrane Ca2+-ATPases underpin a ‘carbon memory’ of photosynthetic assimilation that impacts on water-use efficiency. Nat. Plants 7, 1301–1313 (2021).
Chen, Z. H. et al. Systems dynamic modeling of the stomatal guard cell predicts emergent behaviors in transport, signaling, and volume control. Plant Physiol. 159, 1235–1251 (2012).
Hills, A., Chen, Z. H., Amtmann, A., Blatt, M. R. & Lew, V. L. OnGuard, a computational platform for quantitative kinetic modeling of guard cell physiology. Plant Physiol. 159, 1026–1042 (2012).
Acknowledgements
We are grateful to J. I. Schroeder (University of California, San Diego) and C. Wang (Northwest A&F University) for advice on the potassium ion channel study. We express our sincere gratitude to Y. Gao (Peking University) for his insightful advice on the applications of AlphaFold3. We thank X. Liu (Beijing LI-COR), Z. Ren, B. Suolang and L. Luo (Peking University) for their technical support. This work was supported by National Science Foundation of China (grant no. 31972937 to Yin W.).
Author information
Authors and Affiliations
Contributions
Yin W. and Yizhou W. conceptualized the project and designed the majority of the experiments. J.W. participated in designing the experiments and performed most of the experiments, including the data analysis. K.H., M.L., Y.L. and Yizhou W. contributed to the experiments. Yin W. acquired the funding. J.W. wrote the original draft of the manuscript. Yin W., Yizhou W., W.T., Y.Z. and L.-M.F. revised and edited the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Lynne Yenush and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Stomatal light response of the abaxial and adaxial surfaces in high-light-grown Arabidopsis.
a. Increment of stomatal apertures in the abaxial and adaxial epidermis of WT1, WT2 under high light (200 µmol·m−2·s−1) growth after 3 h of darkness or 200 µmol·m−2·s−1 light treatment. The data are shown as means ± SD (n = 3). Statistical analysis was done with two-tailed Student’s t-test. b-c. Increment of stomatal conductance (b) and \({g}_{s}^{* }\) (c) in the abaxial and adaxial epidermis of WT1, WT2 under high light (200 µmol·m−2·s−1) growth after 1 h 200 µmol·m−2·s−1 light treatment. The data are shown as means ± SD (n = 3), Statistical analysis was done with two-tailed Student’s t-test.
Extended Data Fig. 2 Stomatal properties of the abaxial and adaxial surfaces in N. tabacum.
a. Stomatal density in the abaxial and adaxial surfaces of N. tabacum. The data are shown as means ± SD (n = 3), each sample contained 10 microscopic fields. Statistical analysis was done with two-tailed Student’s t-test. b. Stomatal size in the abaxial and adaxial surfaces of N. tabacum. The data are shown as means ± SD (n = 3), each sample contained 15 stomata. Statistical analysis was done with two-tailed Student’s t-test. c. Stomatal index in the abaxial and adaxial surfaces of N. tabacum. The data are shown as means ± SD (n = 3), each sample contained 10 microscopic fields. Statistical analysis was done with two-tailed Student’s t-test. d. The increment of stomatal conductance in response after white light (200 µmol·m−2·s−1) on the abaxial and adaxial surfaces of N. tabacum. Values are presented as the means of increment of stomatal conductance ± SD (n = 3). e. The stomatal conductance of average per stoma in response after white light (200 µmol·m−2·s−;1) on the abaxial and adaxial surfaces of N. tabacum. Values are presented as the means of \({g}_{s}^{* }\) ± SD (n = 3). Statistical analysis was done with two-tailed Student’s t-test. f. Representative stomata of the abaxial and adaxial surfaces in N. tabacum after exposure to 3 h of darkness or 200 µmol·m−2·s−1 light treatment. Scale bar = 10 μm. g. Stomatal apertures in the abaxial and adaxial epidermis of N. tabacum after 3 h of darkness or light treatment. The data are shown as means ± SD (n = 3), each sample contained 20 stomata. Light conditions were consistent with those in (f). Refer to the Materials and Methods for specific details on epidermal treatment. Statistical differences were assessed using the two-tailed Student’s t-test.
Extended Data Fig. 3 Generation of cell atlas and identification of cell types with representative marker genes.
a. According to the expression patterns of marker genes in each cell cluster, the twenty cell clusters are divided into four types: EC (epidermal cell), GC (guard cell), MC (mesophyll cell), VC (vascular cell). b. Relative abundance of each cell type expressed as a percentage of the whole population of cells. c. Expression patterns of cell-type marker genes for the twenty cell clusters. The dot size indicates the proportion of cluster cells expressing the marker gene, and the colour represents the average expression level in that cluster. d. Differences in the average expression levels of stomatal-light-response related genes in GC1 and GC2 clusters for WT1, WT2 and mixed samples. The columns and error bars in (d) represent the means ± SD (n = 3). Statistical differences were assessed using the two-tailed Student’s t-test.
Extended Data Fig. 4 Comparison of epidermal isolation between tape-peel method and blender method.
a. Representative stomata of the abaxial and adaxial surfaces in WT1 and WT2 through the method of blender and tape-peel after exposure to darkness or light for 3 h. Light conditions were consistent with those in (b). Data shown are representative of 3 independent experiments with similar results. Scale bar = 10 μm. b. Stomatal apertures in the abaxial and adaxial epidermis of WT1 and WT2 after 3 h of darkness or 200 µmol·m−2·s−1 light treatment. In this figure, the box represents the epidermis obtained using a blender, while the shaded portion indicates the epidermis obtained through the method of separation using tape-peel. Refer to the Materials and Methods for specific details on epidermal treatment. The data are shown as means ± SD (n = 3). Statistical analysis was done with two-tailed Student’s t-test. c. Fluorescein diacetate staining showing viability of stomata of the abaxial and adaxial surfaces in WT1 and WT2 using the method of blender and tape-peel. Data shown are representative of 9 independent experiments with similar results. Scale bar = 50 μm. d. The percentage of active stomata was determined from the abaxial and adaxial surfaces of WT1 and WT2 using the blender and tape-peel method. The columns and error bars in (d) represent the means ± SD (n = 9). Statistical differences were assessed using the two-tailed Student’s t-test.
Extended Data Fig. 5 DEGs analysis of RNA-seq from GCPs on the adaxial and abaxial surfaces of WT1 and WT2.
a. Venn diagram representing the overlapping expressed genes in GCPs on the adaxial and abaxial surfaces of WT1 and WT2 (FPKM > 1), where FPKM represents fragments per kilobase of transcript per million mapped reads. b. Correlation analysis of RNA-seq from the GCPs of adaxial and abaxial surfaces of WT1 and WT2 (Spearman correlation coefficient, fit line by LM). c. Differences in the average expression levels of stomatal-light-response related genes in GCPs of abaxial and adaxial surface for two samples (WT1 and WT2) used RNA-seq analysis. The columns and error bars in (c) represent the means ± SD (n = 6). Statistical differences were assessed using the two-tailed Student’s t-test.
Extended Data Fig. 6 Key genes in light response signalling pathway expression levels in GCPs of the abaxial and adaxial.
qPCR analysis of expression levels of key genes in light response signalling pathway in GCPs of the abaxial and adaxial of leaves (WT1 and WT2 mix). The columns and error bars represent the means ± SD (n = 3). Statistical analysis was done with two-tailed Student’s t-test.
Extended Data Fig. 7 KAT1 and AKT1 expression levels in transgenic plants.
a. Western blot of the epidermis of abaxial and adaxial surfaces in leaves (WT1 and WT2 mix), separated by SDS-PAGE and detected with a KAT1 and AKT1-specific antibody. Refer to the Materials and Methods for specific details. b. Actin protein was detected using anti-actin antibody as a control. The relative amount of KAT1 and AKT1 was quantified as the ratio of KAT1 and AKT1 to actin signal intensity. The columns and error bars in (b) represent the means ± SD (n = 4). Statistical differences were assessed using the two-tailed Student’s t-test. c. qPCR analysis of K+in channels genes in abaxial and adaxial epidermis of WT1, WT2, KAT1ox#1, KAT1ox#2, AKT1ox#1, AKT1ox#2, kat1 and akt1. The columns and error bars represent the means ± SD (n = 3). The letters indicate groups with statistically significant differences (P < 0.05. Data were analysed through ANOVA and the Fisher’s multiple comparison). d. Western blot analysis on total protein extracts from the epidermis of abaxial and adaxial surfaces in WT2, KAT1ox#1, KAT1ox#2, AKT1ox#1, AKT1ox#2. Refer to the Materials and Methods for specific details. Actin protein was detected using anti-actin antibody as a control. The relative amount of KAT1 and AKT1 was quantified as the ratio of KAT1 and AKT1 to actin signal intensity. The columns and error bars in (d) represent the means ± SD (n = 3). The letters indicate groups with statistically significant differences (P < 0.05. Data were analysed through ANOVA and the Fisher’s multiple comparison).
Extended Data Fig. 8 The expression levels of PIFs in the epidermis of the adaxial and abaxial surfaces.
a. Differences in the average expression levels of PIFs related genes in GC1 and GC2 clusters for WT1, WT2 and mixed samples in single-cell transcriptome analysis. The columns and error bars in (a) represent the means ± SD. Statistical differences were assessed using the two-tailed Student’s t-test. b. Differences in the average expression levels of PIFs related genes in GCPs of abaxial and adaxial surface for two samples (WT1 and WT2) used RNA-seq analysis. The columns and error bars in (b) represent the means ± SD (n = 6). Statistical differences were assessed using the two-tailed Student’s t-test. c. qPCR analysis of PIFs of the abaxial and adaxial epidermis of WT1. The columns and error bars represent the means ± SD (n = 3). Statistical differences were assessed using the two-tailed Student’s t-test. Among them, PIFs represent the combined effect of PIF1, PIF3, and PIF4.
Extended Data Fig. 9 AlphaFold3 accurately predicted the structures of KAT1 and AKT1.
Cartoon model of the AlphaFold3 output (https://www.alphafoldserver.com) for a representative KAT1 (a) or AKT1 (b). AlphaFold3 produces a per-residue confidence score (pLDDT) between from 0 to 100. Regions with low pLDDT maybe unstructured in isolation. Predicting the confidence level of the relative positions of two residues in a structure using Predicted Aligned Error (PAE). Structure shows dark green patches for the domains, indicating that they are well defined.
Supplementary information
Supplementary Information
Supplementary Tables 1–3, Figs. 1 and 2 and Appendix 1.
Source data
Source data for Fig. 1
Statistical source data.
Source data for Fig. 2
Statistical source data.
Source data for Fig. 4
Statistical source data.
Source data for Extended Data Fig. 1
Statistical source data.
Source data for Extended Data Fig. 2
Statistical source data.
Source data for Extended Data Fig. 3
Statistical source data.
Source data for Extended Data Fig. 4
Statistical source data.
Source data for Extended Data Fig. 5
Statistical source data.
Source data for Extended Data Fig. 6
Statistical source data.
Source data for Extended Data Fig. 7
Statistical source data.
Source data for Extended Data Fig. 7
Unprocessed western blots.
Source data for Extended Data Fig. 8
Statistical source data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wei, J., Hu, K., Liu, M. et al. Guard cells on the adaxial and abaxial leaf surfaces use different compositions of potassium ion channels to drive light-induced stomatal opening. Nat. Plants 11, 1260–1269 (2025). https://doi.org/10.1038/s41477-025-02026-5
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41477-025-02026-5