Abstract
Bacteriophages (phages), viruses capable of infecting and lysing bacteria, are a promising alternative for treating infections from hypervirulent, antibiotic-resistant pathogens like Klebsiella pneumoniae, though narrow host range and phage resistance remain challenges. In this study, the hypervirulent K. pneumoniae NTUH-K2044 was used to purify phage ΦK2044, while two ΦK2044-resistant strains were used to purify two further phages: ΦKR1, and ΦKR8 from hospital sewage. A detailed characterization showed that ΦK2044 specifically killed KL1 capsule-type K. pneumoniae, while ΦKR1 and ΦKR8 targeted 13 different capsular serotypes. The phage cocktail (ΦK2044 + ΦKR1 + ΦKR8) effectively killed K. pneumoniae in biofilms, pre-treatment biofilm formation, and delayed phage-resistance. The phage cocktail improved 7-day survival in Galleria mellonella and mouse models and showed therapeutic potential in a catheter biofilm model. In summary, this proof-of-principle phage cocktail has a broad host range, including hypervirulent and highly drug-resistant K. pneumoniae, and serves as a promising starting point for optimizing phage therapy.
Similar content being viewed by others
Introduction
With the global burden of antimicrobial-resistance (AMR) taxing healthcare systems around the world, new therapies are being developed to treat antimicrobial-resistant infections1,2,3. One such approach is phage therapy4,5,6,7. Bacteriophages, as viruses that kill bacteria through cell lysis, possess advantages such as host specificity and self-amplification in infection sites, and side effects from phage treatments appear to be minimal8. Furthermore, numerous studies have reported that bacteria in infection sites, under the pressure of phages, can undergo adaptive trade-offs in evolution, including growth inhibition, reduced virulence and the restoration of sensitivity to antibiotics9,10. Therefore, phages may serve as a novel biological antimicrobial agent with dual antibacterial and antivirulence functions, offering significant potential for treating multidrug-resistant or invasive bacterial infections. Phages have been designated as urgent investigational new drugs (eINDs) by the United States Food and Drug Administration (FDA) for combating multidrug-resistant bacteria11.
In the list of pathogens requiring urgent attention, Klebsiella pneumoniae and particularly carbapenem-resistant K. pneumoniae is a significant threat in clinical settings12,13. A member of the Klebsiella pneumoniae species complex (Klebsiella spp.), these bacteria are commensal components of gut microbiomes because they live on plant materials and, as a result, are prevalent in invertebrate and vertebrate guts14,15,16,17. When K. pneumoniae is released from the gut lumen, as a result of disease or as an unintended consequence of surgery, it can enter other tissues and become a pathogen capable of causing liver abscesses, kidney and urinary tract infections and bloodstream infections. It has come to consistently rank among the top three pathogens causing urinary tract infections18 and has also increased in prevalence as a cause of meningitis, with a significant increase in regions such as South Korea, Southeastern China, and the West Indies19. In severe cases, K. pneumoniae initiates sepsis, posing significant challenges in treatment with high mortality rates. Mouse models of Klebsiella spp. infection mimic this scenario via intra-peritoneal injection that causes disseminated post-infection tissue pathology20.
A characteristic feature of Klebsiella spp. is the polysaccharide capsule secreted to surround each bacterial cell21. The carbohydrate composition of the capsular polysaccharide can vary, with more than 180 different capsule types recognized22, with KL1 and KL2 identified as the predominant capsule types holding epidemiological significance23. Indeed, several KL1 or KL2 hypervirulent K. pneumoniae strains which have hypermucinous and highly invasive characteristics have gained attention due to poor treatment outcomes following infection by these strains24,25. It is becoming evident that many phages that kill K. pneumoniae strains are specific to a single capsule type, which is a significant issue in the development of phage therapeutics.
The capsular polysaccharide also contributes significantly to the ability of Klebsiella spp. to form biofilms, a growth context in which they can develop both drug- and phage-resistance phenotypes26,27. Indeed, whether biofilms have formed or not, there is a concern that the use of single-phage treatments would induce the emergence of phage-resistant phenotypes in infection sites resulting in treatment failure28. To address the issues of phage resistance, limited host range “phage cocktails” comprised of multiple phages are being considered as a more effective biocontrol approach6,29.
This study utilized the hypervirulent strain of K. pneumoniae NTUH-K2044, which has capsule type KL1, to isolate phages with a view to creating a phage cocktail with therapeutic potential. Initial phage isolation experiments discovered a potent phage, ΦK2044, that killed K. pneumoniae NTUH-K2044 rapidly. However, phage ΦK2044 is specific for capsule type KL1 and prolonged treatment with phage ΦK2044 selected for phage-resistant variants of K. pneumoniae. These phage-resistant variants were then used as hosts to select for two further phages: ΦKR1 and ΦKR8, which had a broader host range. A cocktail of the three phages was mixed and found to be superior in killing planktonic and biofilm forms of K. pneumoniae and in rescuing infected animals from the consequences of K. pneumoniae-induced pathology and death. The results demonstrate the advantages of this phage cocktail, which combines stable lytic capabilities with a broad host range, offering novel perspectives and strategies for optimizing phage therapy.
Results
Isolation and characterization of phages ΦK2044, ΦKR1 and ΦKR8
While it is antibiotic-sensitive, Klebsiella pneumoniae NTUH-K2044 is a hypervirulent bacterium of epidemiological significance associated with septicemia, meningitis and liver abscesses and was the starting point for this study. Using K. pneumoniae NTUH-K2044 as an isolation host, phage ΦK2044 was purified from hospital sewage. Phage ΦK2044 generated large plaques with halos, characteristic of a phage that can hydrolyze the capsule of hypervirulent K. pneumoniae (Fig. 1A). Initial measurements suggest that the plaques formed by ΦK2044 have a central clear zone (diameter 1–3 mm) surrounded by a turbid halo (diameter 3–10 mm), which is consistent with the action of a polysaccharide depolymerase. High-titre stocks of the phage were prepared and transmission electron microscopy revealed a Podo-like morphotype, where ΦK2044 virions have a polyhedral capsid of 60-80 nm without an evident tail or protrusions (Supplementary Fig. 1A). In vitro testing showed the phage virions to be stable against a range of temperature and pH conditions and in the presence of human serum (Supplementary Fig. 2). Genome sequencing revealed the phage to belong to the Autographiviridae family, and that it is closely related to a previously isolated phage called NTUH-K2044-K1-130 (Fig. 1B, Supplementary Figs. 3A, 4A and Supplementary Table 1). Annotation of the genome suggested a depolymerase (OZ76_gp02) is encoded by the phage ΦK2044 (Supplementary Fig. 5; Supplementary Table 4), consistent with the halo morphology of the plaques formed by phage ΦK2044 (Fig. 1A). In the course of these experiments, it became apparent that phage-resistant mutants of hypervirulent K. pneumoniae NUTH-K2044 were readily recovered, at a frequency of ~3.5 × 10−6. Compared to NTUH-K2044, these phage-resistant mutant strains have become smaller in population and less mucoid (Supplementary Fig. 6A), with more compact cell pellets (Supplementary Fig. 6B). Quantitative measurements of the capsule also show that it is diminished in the phage-resistant K. pneumoniae strains R1 and R8 (Supplementary Fig. 6C). Under phage pressure, the host NTUH-K2044 undergoes fitness cost changes related to capsule-associated phenotypes, so we sought to use this phage-host system to address the prospect of developing a phage cocktail that could suppress the evolution of the phage-resistance phenotype.
A Plaque morphology of phage ΦK2044 on plates of K. pneumoniae NTUH-K2044. B Proteomic tree analysis of phages that infect generated using ViPTree. The branch lengths represent genomic similarity based on normalized tBLASTx scores plotted on a logarithmic scale. The bacterial host genus for each phage are detailed in the outer ring, and the viral subfamilies or genera are highlighted in the inner ring. Phage ΦK2044 is indicated with a red star. Plaque morphology of (C) phage ΦKR1 and (D) phage ΦKR8 on plates of K. pneumoniae NTUH-K2044. E Proteomic tree analysis, with the bacterial host genus for each phage in the outer ring, and the viral subfamilies or genera in the inner ring. Phages ΦKR1 and ΦKR8 indicated with red stars. F One-step growth curves for phages ΦK2044, ΦKR1 and ΦKR8. In each graph, the latent period (L) and burst size (B) is indicated.
Accordingly, two resistant variants (R1 and R8) were used as host bacterial strains in phage isolation trials. Two phages, ΦKR1 and ΦKR8, were identified and characterized. Phages ΦKR1 and ΦKR8 formed homogeneous clear plaques on the bacterial lawns of their hosts R1 and R8, respectively (Fig. 1C, D). Plaques generated by phages ΦKR1 and ΦKR8 had relatively small diameters (measuring approximately 0.5–1.5 mm and 1–2 mm, respectively) with no lytic halo. High-titre stocks of the phages were prepared and showed similar morphologies by electron microscopy, with ovoid-shaped capsids measuring 100–110 nm in length and tails approximately 250 nm in length (Supplementary Fig. 1B, 1C). Genome sequencing revealed that both phages have large genomes (ΦKR1 is 165,172 bp and ΦKR8 is 168,081 bp) and belong to the Straboviridae (Supplementary Fig. 7, 8). While both phages belong to the Drulisvirus genus within the Straboviridae, phage ΦKR1 is most closely related to a previously isolated phage called PKO11131 while phage ΦKR8 is more closely related to a phage called Mineola32 (Fig. 1E, Supplementary 3B, 4B; Supplementary Tables 2, 3).
An important consideration for determining the potency of phages is the optimal multiplicity of infection (MOI), defined as the ratio of phages to bacteria, that yields the maximum progeny. The optimal MOI for phage ΦK2044 was found to be 0.001, while for ΦKR1 and ΦKR8, it was 1 (Supplementary Table 5). Under their respective optimal MOIs, growth curves were generated to determine the latent period and burst size of the phages. The latent period, which is the infection-replication time taken before measurable phage progeny begin to be released, was approximately 10 min for phage ΦK2044, while ΦKR1 and ΦKR8 had latent periods of around 20 min (Fig. 1F). The burst size, representing the number of phages generated from each bacterial cell lysis, was approximately 300, 65, and 60 PFU/cell for phages ΦK2044, ΦKR1, and ΦKR8, respectively. In vitro tests revealed that all three phages were stable across a range of temperature and pH conditions and could withstand incubation in human serum (Supplementary Fig. 2).
Determination of phage host range
Having been isolated on K. pneumoniae NUTH-K2044, the three phages can infect this strain with a KL1 capsule type. Furthermore, six other strains in our clinical collection of KL1 capsule type were also susceptible to killing by each of the three phages (Table 1). To address whether any of the phages had a broader host range, we tested a total of 49 clinical isolates covering 12 other capsule types: KL2, KL5, KL9, KL10, KL19, KL47, KL53, KL54, KL57, KL63, KL64, KL74. The antibiotic resistance profiles of these strains are presented in Supplementary Table 6. After initial screening through spot assays, hosts that formed clear plaques were further tested for their efficiency of plating (EOP) values. The results of the EOP assay for the three phages on 49 clinical isolates of K. pneumoniae are presented in Table 1. Phage ΦK2044 exhibited high specificity, selectively targeting only the KL1 capsular serotype of K. pneumoniae. This is not uncommon for Klebsiella phages. In contrast, phages ΦKR1 and ΦKR8 showed varying degrees of lytic activity against another 10 capsule types, resulting in the lysis of approximately 50% of the tested strains. Notably, they show lysis rates of 83% (5/6) and 39% (7/18) against the highly prevalent multi-drug resistance (MDR) K. pneumoniae capsule types KL47 and KL64, respectively, demonstrating potential for treating MDR strains. The broad host range of phages ΦKR1 and ΦKR8 is an advantageous feature that could be useful in the development of broad-spectrum phage cocktails.
Frequency of phage-resistant Klebsiella and in vitro killing curve of phage cocktail
The measurable frequency of phage resistance phenotypes in K. pneumoniae NTUH-K2044 exposed to a single phage (i.e. ΦK2044) was 3.5 × 10−6. Ideally, the selection pressure exerted by a cocktail of phages should quell resistance phenotypes and the use of a three-phage cocktail significantly reduced the frequency of resistance mutations (P < 0.001) (Fig. 2A). We therefore sought to address the relative effects of this three-phage cocktail on killing of strains of K. pneumoniae. The cocktail was assayed against the entire set of strains in Table 1, and yielded a killing spectrum equivalent to the best individual phage. In most strains, the cocktail’s efficiency in killing bacteria exceeds that of any single component phage.
A The resistance mutation frequency calculated after treatment of K. pneumoniae NTUH-K2044 with either phage Φ2044 alone or the phage cocktail. The data were analyzed by a one-way ANOVA, followed by Tukey’s post hoc test (***P < 0.001). B In vitro infection curves (24 h) of individual phages and phage cocktail. Cells (107 CFU/mL) of the indicated K. pneumoniae strains of capsule type KL1 were transferred to a 96-well plate, mixed with the indicated phage (1 × 109 PFU/mL) or phage cocktail and optical density (OD600) measurements were plotted as a readout of bacterial cell growth. The measurements were taken at 60 min intervals for 12 h and then measured again at 24 h. C Equivalent measurements were undertaken with strains of the indicated capsular serotypes.
From this survey, we selected 10 strains of K. pneumoniae susceptible to the phage cocktail, and their growth was assessed by measuring optical density (OD600): (a) in the absence of phages, (b) in the presence of phage ΦK2044, (c) in the presence of phage ΦKR1, (d) in the presence of phage ΦKR8, or (d) in the presence of the cocktail of all three phages. Under the influence of a single phage, continuous growth of resistant bacteria was observed for up to 6–8 h. The three-phage cocktail generated more persistent lytic activity, from 12 to 24 h (Fig. 2B, C). Thus, compared to treatment with a single wild-type phage ΦK2044, the phage cocktail demonstrated advantages of relative broad-spectrum activity and delayed resistance development.
Confocal Laser Scanning Microscopy and quantitation reveal the anti-biofilm activity of phage cocktail
To extend the results on in vitro growth, we sought to address whether the phage cocktail could impact biofilm formation by K. pneumoniae NTUH-K2044. To this end, we employed SYTO®9 and propidium iodide fluorescent dyes to assess bacterial viability within biofilms. In this experimental system, the green fluorescence of SYTO®9 represents living bacterial cells, and the red fluorescence of propidium iodide designates killed bacterial cells. As biofilms constitute three-dimensional structures, we captured z-stack images with a thickness of 30 µm for comparative analyses (Fig. 3). In the untreated control group, the majority of bacterial cells were living and exhibited green fluorescence (Fig. 3A). Following treatment with the phage cocktail, the bacterial biofilms displayed predominantly red fluorescence in cells, indicating a notable cell death in the biofilm, more pronounced than that seen with treatment of phage ΦK2044 alone (Fig. 3B, C). The phage cocktail demonstrated the ability to penetrate the matrix components of mature biofilms, enhancing its efficacy in eradicating bacteria within the biofilm.
Confocal laser scanning microscopy images (Z-axis layer scan) showing fluorescence staining of mature biofilms of NTUH-K2044 after 24 h of growth. Each image is a merge of the layers of green (from SYTO®9 staining) and red fluorescence (from propidium iodide staining), where green fluorescence represents bacterial viability and red fluorescence represents bacterial death. A The mature biofilm formed in the absence of phage (that is, the LB broth control group). mB The mature biofilm after 24 h of treatment with the phage ΦK2044. C The mature biofilm after 24 h of treatment with the three-phage cocktail.
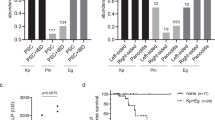
In order to quantify the ability of the phage cocktail to kill bacteria in a biofilm context, we used crystal violet staining to quantify the amount of biofilm and colony counting to enumerate the number of living bacteria within the biofilm. Four strains of K. pneumoniae with different capsular serotypes were selected as experimental strains, and bacterial biofilms were treated with phages at MOI = 10. In the first series of experiments in Fig. 4, individual phages or the phage cocktail were applied at the start of the experiment in order to measure their ability to inhibit the formation of the biofilms formed by each of the four strains. Phage ΦK2044 inhibited biofilm formation in NTUH-K2044 but not the other three strains of K. pneumoniae (Fig. 4A). Individually, the other two phages (ΦKR1 and ΦKR8) had limited impact in preventing biofilm formation, as was also reflected in their potency to kill bacteria within the biofilm (Fig. 4B). The phage cocktail exhibited significant biofilm inhibition across all experimental strains, inhibiting both the biofilm biomass and the number of viable bacteria within the biofilm (P < 0.001).
A In order to measure inhibition of biofilm formation, the indicated K. pneumoniae strains (capsular types KL1, KL2, KL47 or KL64) were grown as biofilms for 24 h of co-incubation with each of the phages or the phage cocktail, as indicated. At the end of the experiment, biofilms were stained with crystal violet and the stain was measured by spectrophotometry at 595 nm wavelength. B The inhibition of biofilm formation experiment was also evaluated by plating for colony forming units (CFU) of bacteria. The live bacterial counts after 24 h of co-incubation with each of the single phages or with the phage cocktail are presented. C In order to measure phage killing in mature biofilms, the indicated K. pneumoniae strains (capsular types KL1, KL2, KL47 or KL64) were grown for 24 h to establish mature biofilms and, after introducing phages the incubation was continued for a further 24 h. At the end of the experiment, biofilms were stained with crystal violet and the stain was measured by spectrophotometry at 595 nm wavelength. D The biofilm killing experiment was also evaluated by plating for CFU of bacteria. The live bacterial counts from mature biofilms after treatment with each of the single phages or with the phage cocktail. In all experiments, data were analyzed by a one-way ANOVA, followed by Tukey’s post hoc test (ns, not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001).
In the second series of experiments, individual phages or the phage cocktail were applied after mature biofilms had already been established by each of the four strains. Degradation of the biofilms was observed after 24 h in the presence of phage. For example, phage ΦK2044 reduced both biofilm mass and had some impact on cell viability in NTUH-K2044 but not the other three strains of K. pneumoniae (Fig. 4C), with phages ΦKR1 and ΦKR8 exhibiting broader strain impacts, particularly on MDR strains FK4724 and FK7014. The phage cocktail demonstrated the highest degree of killing the mature biofilms, as measured by reduced biofilm biomass (Fig. 4C) and by the number of viable bacteria within the biofilm (Fig. 4D) (P < 0.001). In summary, the phage cocktail effectively suppressed the formation of early-stage bacterial biofilms and displayed some capacity to eliminate bacteria from the deeper layers of mature biofilms. On the other hand, regardless of when the phage cocktail treatment is administered, the overall resistance rate of surviving bacteria within the biofilm is approximately 10% (Supplementary Fig. 9). This indicates that the cocktail exerts a sustained and effective bactericidal effect within the biofilm, with no significant induction of phage resistance observed.
In vivo efficacy of phage cocktail treatment in infected mouse
A mouse intra-peritoneal infection model and a subcutaneous catheter biofilm model were established to verify the in vivo efficacy of the phage cocktail in treating disseminated infections and locally targeting biofilms on non-biological surfaces. The specific protocol of the mouse infection model is depicted in Fig. 5A. Mice were injected intra-peritoneally with K. pneumoniae and died within 96 h after infection (Fig. 5B). In terms of phage rescue, the 7-day survival rate of mouse treated with individual phages increased to 40-70%. The phage cocktail demonstrated superior therapeutic effects, significantly improving the survival rate (approaching nearly 100%) and maintained the health condition of the mouse post-infection as monitored by body weight (Fig. 5C).
A Experimental procedure of the mouse intra-abdominal infection model. In this time-line, the 0 h time-point is defined as the time at which the mouse was infected with bacteria. B The 7-day survival curve of an infected mouse treated with the phage cocktail. The capsule type of each of the four K. pneumoniae strains (capsule types KL1, KL2, KL47, or KL64) is indicated. Data were analyzed by the Gehan-Breslow-Wilcoxon method in the Kaplan-Meier survival analysis (ns, not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001). C Health monitoring of the mouse was measured through body weight changes after the intra-abdominal infection. Each treatment group included 5 mice, and each experiment was repeated three times.
To determine whether the phage rescue had reduced the bacterial load in the tissues and blood of infected mouse, pathology samples were assessed. In the case of mouse infected with K. pneumoniae NTUH-K2044 the three individual phages each reduced the bacterial load in all tissues significantly. Except for the kidneys, the cocktail treatment demonstrated better in vivo clearance compared to single phage treatment, with the most significant effects observed in the blood. These differences were statistically significant after post hoc pairwise comparisons (P < 0.05) (Fig. 6A). In the case of mouse infected with the other three strains of K. pneumoniae, phage ΦK2044 had no impact on reducing the bacterial load in any tissues. The other two phages, ΦKR1 and ΦKR8, showed significant impact on clearing bacterial loads from all tissues tested, and in some cases that phage cocktail was observed to exert a better clearance than the individual phage treatments (P < 0.05). Overall, the bacterial load in tissues was shown to have decreased by over 5 log10 values, highlighting the rapid and substantial therapeutic effects of the phage cocktail in the treatment phase (P < 0.001).
Six tissues: heart, liver, spleen, lung, kidney and blood were sampled and the bacterial load from each was measured by colony formation. Each treatment group included 5 mice, and each experiment was repeated three times. Data were analyzed by a one-way ANOVA, followed by Tukey’s post hoc test (ns, not statistically significant; **P < 0.01; ***P < 0.001).
In order to be certain that the results from enumerated bacterial loads from tissues were reflective of tissue damage caused by the bacterial infections, tissue pathology slides were prepared from the heart, liver, spleen, lungs, and kidneys of mouse infected with K. pneumoniae NTUH-K2044 with or without treatment with the phage cocktail. The tissue pathology slides were assessed by standard hematoxylin and eosin (H&E) staining to evaluate the inflammatory infiltration into tissues (Fig. 7A). Consistent with it being hypervirulent, NTUH-K2044 was seen to have induced pronounced inflammation or tissue damage in all of the assessed organs. Diffuse neutrophil infiltration was observed in the lung tissues of infected mouse, with eosinophilic mucus secretion in some airways. Liver tissues showed localized hepatocyte ballooning with infiltration by granulocytes and lymphocytes. The spleen exhibited scattered lymphocyte infiltration and necrosis in numerous splenic nodules, accompanied by an abundance of megakaryocytes present. Perivascular congestion and infiltration of lymphocytes and granulocytes were evident around the renal pelvis of the kidneys.
A Histopathological examination of lung, liver, spleen and kidneys 24 h after treatment with the phage cocktail, stained with hematoxylin and eosin. B Experimental procedure of the mouse subcutaneous catheter biofilm model. In this time-line, the 0 h time-point is defined as the time at which the mouse was infected with biofilm-related catheter. C After surgical removal of the catheters from different treatment groups, they were subjected to ultrasonication and their viable bacterial load was measured. Data were analyzed by a one-way ANOVA, followed by Tukey’s post hoc test (***P < 0.001). D Representative schematic of the mouse subcutaneous catheter biofilm implantation model. Each treatment group included 4 mice, and each experiment was repeated three times.
However, 24 h of phage cocktail treatment significantly alleviated the inflammatory damage that was observed in each of the tissues. This demonstrates that the phage cocktail has a favorable in vivo anti-infection therapeutic effect. Since the mouse model is a complicated means to screen for phages and phage combinations, we also assessed a facile invertebrate model used for infection biology using the same four K. pneumoniae strains (NTUH-K2044, FK6048, FK4724 and FK7014) and found a similar impact from the phage cocktail using the experimentally tractable Galleria model that could provide means for future screening and/or optimization of phage cocktails (Supplementary Fig. 10).
The efficacy of the phage cocktail in combating biofilms in vivo was investigated using a mouse subcutaneous catheter implantation. The specific protocol for the mouse subcutaneous catheter biofilm model is illustrated in Fig. 7B. The creation of subcutaneous pouches on both sides of the mouse back simulated the microenvironment of in vivo catheter-related biofilm infections (Fig. 7D). We observed that timely administration of the phage cocktail significantly inhibited NTUH-K2044 biofilm formation on non-biological surfaces, reducing the number of viable bacteria within the biofilm on the catheter surface by over 4 log10 units (P < 0.001) (Fig. 7C). Consistent with the results from the intra-peritoneal infection model, bacterial load on the catheter surface decreased most significantly after treatment with the phage cocktail compared to any single phage (P < 0.05), highlighting the rapid and effective advantage of the phage cocktail in the local treatment of catheter-related biofilm infections.
Discussion
The rise of antibiotic-resistant (AMR) K. pneumoniae strains has severely limited clinical treatment options, and there are issues that limit the development of new drugs that could come to treat current AMR infections3. Phage therapy holds great potential and has advantages in treating bacterial infections, especially those caused by AMR bacteria. Having a history spanning over a century, this long-standing approach is heralded and yet in need of optimization and revival33. The current view is that cocktails of distinct phages could be the preferred therapeutic choice, but knowledge of how well phage cocktails might work remains at an early stage7.
Recent studies have reported phage cocktails targeting K. pneumoniae. For instance, a phage cocktail targeting K. pneumoniae of ST16 (capsule type KL51)34, a cocktail containing three lytic phages targeting ST11 (capsule type KL64) K. pneumoniae35, a cocktail of three phages targeting carbapenem-resistant (capsule type KL54) K. pneumoniae36. These studies show promise, with wound healing taking 10 days instead of 14 days in one study35, and an eight-phage cocktail having synergy with meropenem treatment to reduce biofilms34. Not every combination of phages will yield a good cocktail: in some studies, there is no significant increase in cocktail effects compared to individual phage treatments36, and other studies have suggested that increasing the number of phages may lead to antagonistic interactions between them37.
Our study focused on a hypervirulent (capsule type KL1) K. pneumoniae to derive an effective phage (ΦK2044) used that phage-host system to isolate phage-resistant variants, and then used those phage-resistant variant K. pneumoniae strains to derive additional effective phages (ΦKR1 and ΦKR8). In addition to its effectiveness against the antibiotic-sensitive hypervirulent isolation host, the cocktail was shown to kill AMR strains. All in all, the cocktail was shown to have excellent properties in that (i) it can lyse clinical K. pneumoniae strains with over 10 different capsular types, (ii) it can delay the development of phage-resistance phenotypes in K. pneumoniae, (iii) it can prevent the formation of K. pneumoniae biofilms of different capsule types, (iv) it can kill the cells within established K. pneumoniae biofilms of different capsule types, and (v) it can protect the animals from tissue damage by preventing the spread of K. pneumoniae in the peritoneal cavity while also reducing the formation of catheter-associated biofilms in vivo. Importantly, none of the component phages carry recognizable virulence genes or antimicrobial resistance genes, suggesting their suitability as safe options for phage therapy.
First, a cocktail that can lyse clinical K. pneumoniae strains with over 10 different capsular types. The initial aim of the cocktail was as a treatment for diverse K1 strains given that their hypermucinous and highly invasive characteristics often lead to poor drug-based treatment outcomes. The first phage isolated, ΦK2044, is highly specific with a narrow host-range to KL1 capsule type strains. Several previous studies have reported on narrow-spectrum host-range phages specific to KL2 capsule types, and these phages use the KL2 capsule as their primary receptor for host recognition38,39. On these same grounds, we suggest that phage ΦK2044 might use the KL1 capsular polysaccharide as its receptor. In addition to its capsule type specific host-range, analysis of the genome suggested a typical capsule depolymerase is encoded in the genome of phage ΦK2044. Also, the halo around the plaques from phage ΦK2044 is consistent with capsule degradation.
Phages ΦKR1 and ΦKR8 have a much broader host range, infecting K. pneumoniae strains of ten different capsule types (Table 1) including various multidrug-resistant strains (Supplementary Table 6). Some phages have multiple different capsule depolymerases, as is the case for phage PhiK64-1 that carries nine capsule depolymerases to infect seven capsule types of K. pneumoniae40. However, more commonly phages are agnostic to capsule type where they use proteins or lipids in the outer membrane (i.e. situated below the capsule layer) as their primary receptors4. Being Myoviridae phages ΦKR1 and ΦKR8 have a rich array of structural components that could serve as receptor-binding proteins, particularly tail fibers, tail spikes and central tail spikes (Supplementary Figs. 5, 7 and 8; Supplementary Table 4), and genome sequence analysis revealed that phages ΦKR1 and ΦKR8 encode many structural proteins given their genome sizes close to those of jumbo phages41. We have not yet identified the receptors on their host cell surfaces, but the diverse host range (Table 1) suggests that these are largely conserved proteins and or lipids in the outer membrane rather than capsular polysaccharides.
Furthermore, a cocktail that delays the development of phage-resistance phenotypes in K. pneumoniae. The phage cocktail (ΦK2044 + ΦKR1 + ΦKR8) was shown to delay the development of phage-resistance in K. pneumoniae NTUH-K2044. Torres-Barceló et al. outlined two main approaches for addressing phage resistance in phage therapy: by minimizing the evolution of phage resistance, or by harnessing the evolution of phage resistance42. The phage cocktail in our study effectively implements the former strategy. While we did not address how this was achieved, we speculate that under the pressure of phage ΦK2044 that NTUH-K2044 accumulated mutations that combined to reduce capsule production. This speculation is based on our initial observation of colony morphological phenotypes (Supplementary Fig. 6). Meanwhile, the genomic alignment data indicated that the phage-resistant mutants R1 and R8 may have experienced a deletion mutation in the capsule synthesis-related gene wzc (Supplementary Table 7). This would be consistent with our previous findings regarding the resistance mechanisms of KL2 capsular type K. pneumoniae and their fitness costs38, which may explain why the ΦK2044-resistant variant strains of K. pneumoniae enabled the purification of phages ΦKR1 and ΦKR8 that are not KL1-type specific in their host tropism.
Moreover, a cocktail that can prevent the formation of K. pneumoniae biofilms, and kill the cells within established K. pneumoniae biofilms. A biofilm is a specialized form of bacterial growth adapted to the environment for self-protection, characterized by the presence of an extracellular matrix and dense cell growth, forming microcolonies on living or non-living surfaces43. In K. pneumoniae, the genetic control of a well-characterized “biofilm switch” is understood and involves a decrease in capsule production44. According to the data from the National Institutes of Health, biofilms are present in over 80% of bacterial infections, and the formation of bacterial biofilms is a key factor in developing antibiotic resistance45. We showed here that treatment with the phage cocktail significantly inhibited the initial formation of K. pneumoniae biofilms. In addition, effective mature biofilm clearance was observed after 24 h of cocktail therapy due to cell death, as imaged by confocal scanning fluorescence microscopy. This finding is consistent with previous research that has indicated that persister cells in biofilms, which can become resistant to the effects of antibiotic drugs, can still be susceptible to attack by some phages46. We also found that among the colonies remaining after phage cocktail treatment of the biofilm, only a few survived due to the development of resistance, while most survived due to insufficient dosage or treatment duration (Supplementary Fig. 9). Therefore, there is potential for improvement through optimization of the dosage and administration frequency. The long-term efficacy and patterns of this phage cocktail against biofilms appear to be both notable and intriguing.
Finally, a cocktail that can exert antibacterial and anti-biofilm effects in vivo while protecting against tissue damage in mouse models of K. pneumoniae infection. In vivo, animal immune systems play a critical role synergistically with phages: phage therapy can reduce the bacterial load in an infection site to a point where immune surveillance can eliminate the remaining bacterial subpopulations47. In the larvae and mouse infection models, the safety of phages ΦK2044, ΦKR1, and ΦKR8 was demonstrated, as no deaths were observed in animals treated with the phages. The rescue of animals by the phage treatment was demonstrated, with the 7-day survival rate of larvae and mouse treated with the phage cocktail exceeding 80%, with a significant reduction in bacterial load in the organs of the mouse that were subject to pathology testing. We speculate that ΦK2044 will reduce sensitive bacteria armed with capsules to minimal levels, while any remaining phage-resistant bacterial cells are collectively cleared by ΦKR1, ΦKR8, and host immunity. This study did not explore the optimal treatment dosage, a limitation of our research and a prospect for future studies to carefully consider appropriate phage dosages and treatment durations.
The formation of biofilms on medical device catheters is a key factor in diffuse and recurrent infections48. Phage cocktails show promising potential in inhibiting local biofilms49. Thus, in addition to its efficacy in preventing bacterial dissemination within the mouse peritoneal cavity, we also validated the anti-biofilm activity of this phage cocktail on in vivo catheters. Consistent with the in vitro biofilm inhibition results, the phage cocktail effectively prevented biofilm formation on in vivo non-biological surfaces, with significant and stable therapeutic effects observed within the subcutaneous micro-environment in mice, without interference from the host immune system. However, the role of this phage cocktail in the biofilm-implant-host interaction warrants further in-depth investigation50.
In summary, the phage cocktail used in this study demonstrated a broad host range and prolonged capacity for bacterial killing, presenting promising therapeutic outcomes both in vitro and in vivo (Fig. 8). We did not observe any cross-resistance among the phages, with their effects being additive and, in some respect, synergistic. This phage cocktail design strategy, based on elements of co-evolution of phages and phage-resistant bacterial variants, has the potential to serve as a framework for optimizing the treatment of recalcitrant K. pneumoniae bacterial infections.
Methods
Bacterial strains and bioinformatics analysis
In this study, 49 strains of K. pneumoniae isolated from clinical samples at the First Affiliated Hospital of Wenzhou Medical University were used in addition to the hypervirulent K. pneumoniae NTUH-K2044 purchased from ATCC51. These bacterial strains were characterized using Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF/MS) based on the manufacturer’s instructions (BioMérieux, Lyon, France). The strains were cultured in Luria-Bertani (LB) broth to which was added 30% glycerol before storage at −80 °C. Phage-resistant strains R1 and R8 were isolated after exposing NTUH-K2044 to phage ΦK2044 for 48 h in a double-layer agar plate. After five rounds of passaging and purification, R1 and R8 were confirmed as stable phage-resistant clones.
Whole genome sequencing (WGS) was performed on 49 clinical K. pneumoniae strains. Genomic DNA was extracted with an AxyPrep Bacterial Genomic DNA Miniprep kit (Axygen Scientific, Union City, CA, USA). The library with an average insert size of 400 bp was constructed using the NEBNext Ultra II DNA library preparation kit and then subjected to high-throughput sequencing using the Illumina Novaseq platform (paired-end run; 2 × 150 bp). Quality control of the raw sequenced reads was carried out using FastQC, and trimming was performed with fastp under default settings (phred quality ≥ Q15 and minimum length > 0)52. Trimmed reads were assembled de novo with SPAdes53. Annotations of the assemblies were done using Prokka54. The sequence types (ST) were assigned via mlst (https://github.com/tseemann/mlst)55. The capsular (K-locus) genotypes were identified using Kaptive56. Whole-genome alignments of the strains were performed using breseq version 0.38.357.
Isolation and purification of the phages
Phages were isolated from wastewater samples collected from the First Affiliated Hospital of Wenzhou Medical University, a hospital in the south-eastern coastal region of China. A mixture of wastewater and NTUH-K2044 bacterial culture (grown overnight with shaking) was incubated at 37 °C with shaking for 24 h to allow for phage proliferation. As described previously, a double-layer agar method was employed for phage isolation and purification58,59. Specifically, the logarithmic-phase bacterial culture and the phage-enriched solution were mixed and poured onto a semi-solid LB agar medium as the top layer. Independent plaques were selected and transferred to SM buffer, repeating the process 4-5 times until consistent plaque morphology was observed. The method for isolating phages from phage-resistant mutant strains followed the same protocol. The phage lysates were stored at 4 °C for subsequent experiments.
Phage biological characterization
To determine the multiplicity of infection (MOI), phage stocks of known titer were incubated with host bacteria at ratios of 0.0001, 0.001, 0.01, 0.1, 1, and 10. Overnight cultured bacterial suspension was adjusted to 1 × 108 CFU/mL for these experiments and the mixture was then incubated at 37 °C with shaking (160 rpm) for 5 h. After that time, the cultures were centrifuged at 10,000 rpm for 6 min to collect the supernatant, which was then sterilized by filtering through a 0.22 μm membrane filter. Subsequently, the double-layer agar plate method is used to determine the titer of the progeny phages at different MOI ratios.
The analysis of the one-step growth curve was conducted according to previously published methods60. Log-phase bacterial culture was mixed with phage at an optimal MOI. After incubation at 37 °C for 20 min, the mixture was centrifuged at 10,000 rpm for 6 min at 4 °C, and the supernatant was discarded. Subsequently, 20 mL of LB medium was added, and the culture was shaken at 37 °C and 160 rpm. Samples were collected continuously within 2 h, centrifuged at 10,000 rpm for 6 min, and the supernatant was filtered through a 0.22 μm membrane to remove bacteria. At various infection times, the phage titer was determined using a double-layer agar plate method. The one-step growth curve was plotted with the infection time as the x-axis and the phage titer as the y-axis (GraphPad Prism 8.0 software).
The basic morphology and structure of the phage were documented by the negative staining technique and transmission electron microscopy (TEM) as previously described in ref. 61. Phage suspension was pipetted onto a copper grid with a carbon-coated ultra-thin film (SolelyBio, China) and allowed to settle for 1 min. The excess liquid was removed using filter paper, and then 10 μL of 2% uranyl acetate (Sigma, Sydney, Australia) was added to the copper grid, followed by settling for 1 min. The grid was then air-dried at room temperature for 20 min. Transmission electron microscopy (HITACHI H-7650) was used for imaging at 80-120 kV to observe the morphology of the phages.
Spot test and efficiency of plating
Initial phage host range screening was conducted using 49 characterized strains of K. pneumoniae using spot testing as previously described in ref. 62. Bacteria were cultured overnight in LB medium. A mixture of 100 µL of this culture with 5 mL of 0.4% agar and LB medium was used to prepare a double-layered agar plate. Then, 5 µL of phage lysate (1×109 PFU/mL) was spotted onto the double-layered agar plate and incubated at 37 °C for 12 h. The sensitivity of the bacteria to the phages was determined by measuring the clearance zones where lysis occurred. Based on these clearance zones, bacteria were initially categorized as “lysing” or “non-lysing”. Subsequently, the efficiency of plating (EOP) for lysing bacteria was determined using double-layered agar plates. The EOP was calculated as the average plaque-forming units (PFU) of the target bacteria divided by the average PFU of the host bacteria determined in three independent measurements.
Determination of phage stability
The activity of phages was assessed under different temperatures, pH levels, and human serum conditions63. To conduct a heat stability test, phages were prepared in sterile saline-magnesium (SM) buffer (1 × 1010 PFU/mL) and incubated for 60 min at temperatures in the range of 4 °C to 80 °C. To evaluate pH stability, 100 μL of phages (1 × 1010 PFU/mL) were added to 900 μL of sterile SM buffer. The pH was adjusted to a range of 2–12 using either sodium hydroxide or hydrochloric acid, followed by incubation at 37°C for 60 min. To assess stability in human serum, 100 μL of phages (1 × 1010 PFU/mL) were incubated in 100% human serum for 2, 6, 12, and 24 h. Subsequently, the titer of the phages in the sterile SM buffer dilution series was determined using the double-layer agar method, reflecting their viability.
Whole genome sequencing and analysis
As described in detail previously, 5 mL of phage lysate (> 109 PFU/mL) was treated with MgCl2 (final concentration of 5 mM), DNase I and RNase A (final concentration of 10 μg/mL) at room temperature for 30 min. Phage virions were recovered by polyethylene glycol (PEG) precipitation using PEG 8000 and sodium chloride (final concentrations of 10% [wt/vol], and 1 M, respectively) at 4 °C overnight. The precipitate was centrifuged (17,000 × g for 15 min) and the pellet was resuspended in nuclease-free water (Promega, Sydney, Australia). Phages were exposed to Proteinase K (final concentration of 50 μg/mL), EDTA (final concentration of 20 mM) and sodium dodecyl sulphate (final concentration of 0.5% [vol/vol]) at 55°C for 1 h. An equal volume of phenol-chloroform-isoamyl alcohol (29:28:1) was added and thoroughly mixed before being centrifuged at 12,000 × g for 3 min. The aqueous phase was carefully removed, and an equal volume of isopropanol and 1/10 volume of sodium acetate (pH 5) was added. The mixture was left at −20 °C overnight to allow DNA precipitation. After centrifugation (12,000 × g for 10 min), the supernatant was removed, the DNA pellet was washed with 70% ethanol, air dried and resuspended in 30 μL of nuclease free water (Promega, Sydney, Australia)64.
Genomic DNA was prepared from phage samples and sequenced using an Illumina Hiseq 2500 platform (~1 Gbp/sample)65. Fastp was used for adapter trimming and quality filtering when demultiplexing the raw reads. To annotate the genomes, open-reading frames were predicted using the Rapid Annotations function in Pharokka (https://github.com/gbouras13/pharokka)66. The genome sequence information was visualized and mapped using GView (https://server.gview.ca/)67. ViPTree, a web server provided through GenomeNet generates viral proteomic trees for the classification of viruses based on genome-wide similarities (http://www.genome.jp/viptree) and was used as previously described68. Genome alignment viewer DiGAlign was used to extend the alignment visualization capabilities implemented in ViPTree (https://www.genome.jp/digalign)69. STEP3 provided a robust computational approach to distinguish specific and universal features in phages to improve the quality of phage cocktails (http://step3.erc.monash.edu/)65.
Phage cocktail preparation and in vitro killing experiments
Each of the three phages, ΦK2044, ΦKR1 and ΦKR8, was diluted to 1 × 109 PFU/mL, and then mixed in equal volumes to create a cocktail, which was stored at 4 °C for future use. To evaluate the in vitro lytic capability of this phage cocktail, 180 μL of K. pneumoniae cells (107 CFU/mL) were transferred to a 96-well plate. They were then mixed with 20 μL of the phage cocktail (1 × 109 PFU/mL), resulting in an MOI of 10. The 96-well plate was shaken at 180 rpm, and optical density (OD600) was measured at specific time intervals (0, 4, 8, 12, 16, 20, 24 h) using a spectrophotometer (BioTek, Synergy, USA)34.
Detection of resistance mutation frequencies
The resistance mutation frequency of K. pneumoniae NTUH-K2044 under selection of the phage cocktail was determined using a turbidity-based method70. Exponential growth phase suspensions of bacteria were serially diluted (10-fold) until a dilution of 10−8 was achieved. Three aliquots (each 5 mL) of the NTUH-K2044 bacterial suspension from each dilution gradient were prepared. To each dilution, 100 μL of a single phage or the phage cocktail with the same titer was added, followed by incubation at 37 °C and 180 rpm for 18 h. The dilution with the lowest turbidity was selected for viable cell counting. The mutation frequency was calculated as the number of viable bacteria in the test tube divided by the initial bacterial count in the same tube, reflecting the frequency of resistant bacteria emerging after phage treatment.
Biofilm inhibition and clearance assay
The biofilm inhibition and mature biofilm eradication assays were both conducted in flat-bottomed 96-well microtiter plates71. Four strains of K. pneumoniae with different capsular types (KL1, KL2, KL47, KL64) were assayed. Initially, the bacterial strains were inoculated into the 96-well plates at a concentration of 1.5 × 106 CFU/mL. Phage alone or in a cocktail was co-cultured with bacteria at an MOI = 10. For the mature biofilm assay, pre-culturing of the bacterial suspension for 24 h was essential to initiate the formation of biofilms before introducing phages. After incubating the 96-well plates at 37 °C for 24 h, the planktonic cells were discarded, and the wells were washed twice with 1×PBS. Subsequently, the plates were air-dried to facilitate direct fixation of the biofilm. Staining using a 1% crystal violet (CV) solution (Solarbio, Beijing, China) was followed by destaining using ethanol. Between these steps, wells were rinsed with 1×PBS and left to air dry. Finally, absorbance for each well was measured at 595 nm using a microplate reader (BioTek, Synergy, USA). For viable cell counting within the biofilm and their phage resistance assessment, post-culture 24-well plates were sonicated for 10 min in an ultrasonic bath to disrupt the biofilm structure and release bacteria. After appropriate dilutions, bacterial aliquots from the biofilm were spotted on LB agar plates. The number of viable cells was calculated to determine the antibacterial efficacy against the biofilm. Additionally, a spot test method is used to assess the resistance of these colonies to phage treatment, and the overall resistance rate of the residual colonies after treatment is calculated70.
Confocal laser scanning microscopy
To validate the effect of the phage cocktail on mature bacterial biofilms, a live/dead cell staining assay was conducted using confocal laser scanning46. Briefly, NTUH-K2044 cells in the exponential growth phase were cultured for 24 h in confocal dishes to form mature biofilms. These biofilms were then exposed to phage ΦK2044 alone or to the phage cocktail (1 × 109 PFU/mL) and continuously treated for 24 h. After discarding the culture medium, cells were stained with SYTO®9 (100 µg/mL) and propidium iodide (PI) (50 µg/mL) at 37 °C. Using a confocal laser scanning microscope (Leica®, Japan), assessments of bacterial death were directly observed.
In vivo infection model of Galleria mellonella larvae
Phage killing of four strains of K. pneumoniae with different capsular types (KL1, KL2, KL47, KL64) was evaluated using a Galleria mellonella larvae infection model72,73. Larvae weighing between 250 and 300 mg and exhibiting comparable motility were selected. The larvae were divided into five groups: (i) PBS control; (ii) individual phage ΦK2044 treatment; (iii) individual phage ΦKR1 treatment; (iv) individual phage ΦKR8 treatment; and (v) phage cocktail treatment group. Each treatment group consisted of 10 larvae. Using a microinjector, bacterial suspensions (10 µL each) of varying concentrations were injected into the penultimate proleg. The infective dose for different strains was adjusted to 75% lethal dose (LD75) based on their virulence: NTUH-K2044 at 1.5 × 105 CFU/mL; FK6048 at 1.5 × 106 CFU/mL; FK4724 at 7.5 × 106 CFU/mL and FK7014 at 1.5 × 107 CFU/mL. After a 2 h infection period, phages were injected into the alternate proleg (1 × 109 PFU/mL, 10 µL each). Subsequently, the larvae were maintained at 37 °C and observed continuously for 7 days to evaluate the therapeutic efficacy of the combined treatment. Larvae showing no response to physical stimuli were considered dead.
In vivo infection model of mouse
The mouse were Specific Pathogen-Free (SPF) male ICR (Charles River Laboratories, Massachusetts, USA) with each weighing 25 g. The mouse intra-peritoneal infection model has been described, and the mouse were housed according to the Chinese national standards for laboratory animals36,74. The animal study received ethical approval from the Zhejiang Association for Science and Technology (ID: SYXK[Zhejiang]2021-0017), and experiments were conducted following the guidelines for the welfare and ethics of experimental animals75.
The mouse were divided into five groups: (i) PBS control; (ii) individual phage ΦK2044 treatment; (iii) individual phage ΦKR1 treatment; (iv) individual phage ΦKR8 treatment; and (v) phage cocktail treatment group. Each treatment group consisted of 5 mice. Initially, after a 3-day acclimatization period, bacterial suspensions of K. pneumoniae were injected into the mouse’s peritoneal cavities (100 µL per injection). The infective doses for different strains were adjusted to LD75 based on their virulence: NTUH-K2044 at 7.5 × 105 CFU/mL; FK6048 at 1.5 × 105 CFU/mL; FK4724 at 1.5 × 108 CFU/mL and FK7014 at 1.5 × 108 CFU/mL. Phage treatment (100 µL at 1 × 109 PFU/mL) was administered intra-peritoneally 2 h after bacterial inoculation. Untreated control mouse were also included. After 24 h of treatment, mouse were euthanized, and organs including the heart, liver, spleen, lungs, and kidneys, along with blood samples, were collected post-mortem. Organ tissues were weighed, homogenized, and assessed by bacterial colony counting.
Based on the same animal grouping, a subcutaneous catheter biofilm model in mice was established according to reference76. First, medical catheters (12FR, PVC catheter, UYun) were pre-cut into 0.5 cm fragments and soaked in bovine serum overnight. The catheters were then incubated in NTUH-K2044 (1.5 × 107 CFU/mL) bacterial suspension at 37 °C for 3 h to allow bacterial adhesion to the catheter surface. After a 3-day acclimatization period, the mice were anesthetized with an intra-peritoneal injection of 500 µL of 1.25% tribromoethyl alcohol (M2910, AibeiBio), the hair on their backs was shaved, and the skin was locally disinfected with povidone-iodine. A sterile pair of scissors was used to make a 5 mm skin incision, and the bacteria-inoculated catheter was implanted into a subcutaneous tunnel, followed by sealing the incision with 3 M Vetbond tissue glue. Each animal had two catheters implanted, one on each side. 4 h after catheter implantation, 100 µL of phage (1 × 109 PFU/mL) was injected into the infected subcutaneous tissue. After 24 h, the mice were euthanized, and the catheters were carefully retrieved via surgery. The catheters were treated in 1 mL PBS by ultrasonication at 40,000 Hz for 10 min, followed by serial dilution and bacterial load quantification.
Relative quantification of capsular polysaccharides
The content of capsular polysaccharide (CPS) in K. pneumoniae strains was tested using the phenol-sulfuric acid method77. The main component of capsular polysaccharide (CPS), glucuronic acid, was hydrolyzed in a sulfuric acid solution containing sodium tetraborate. The resulting hydrolysis products further reacted with phenol to form a pink-colored derivative, with absorbance measured at 520 nm showing a linear relationship with CPS. Briefly, bacterial cultures were treated with 1% Zwittergent 3–14 and 100 mM citric acid, incubated at 50 °C for 30 min. After centrifugation, the supernatant (300 µL) was mixed with 1.2 mL of anhydrous ethanol and incubated at 4 °C for 20 min. After centrifugation at 14,000 rpm for 5 min, the pellet was dried and resuspended in 200 µL of distilled water. Then, 1.2 mL of 12.5 mM sodium borate was added, and the mixture was incubated at 100 °C for 5 min. Finally, 40 µL of 0.5% NaOH and 0.15% phenol solution were added, and absorbance at 520 nm was measured after 5 min at room temperature.
Statistical analysis
Except for the in vitro lysis curves, which were repeated twice, all other experiments were conducted with three repetitions, with each experiment including three biological or technical replicates. The survival curves were tested using the Gehan-Breslow-Wilcoxon method in the Kaplan-Meier survival analysis. The remaining statistical significance was performed using one-way analysis of variance (ANOVA), with post hoc testing conducted using Tukey’s multiple comparisons. The results are expressed as mean ± standard deviation (SD), and P-values < 0.05 were considered statistically significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001 and ns, P > 0.05 for all analyses. Data were analyzed by GraphPad Prism 8.0 statistical software.
Data availability
The original contributions presented in the study are included in the article; further inquiries can be directed to the corresponding authors. The complete genomic sequence of ΦK2044, ΦKR1, and ΦKR8 has been deposited in GenBank under the accession numbers PP442062 (Phi_K2044), PP442063 (Phi_KR1), and PP442064 (Phi_KR8). The genome data of 49 K. pneumoniae strains have been submitted to NCBI under the BioProject accession number PRJNA844536.
References
Aggarwal, R. et al. Antibiotic resistance: a global crisis, problems and solutions. Crit Rev Microbiol. 50, 896–921 (2024).
Abel, K. et al. System-wide approaches to antimicrobial therapy and antimicrobial resistance in the UK: the AMR-X framework. Lancet Microbe. 5, e500–e507 (2024).
Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 400, 2221–2248 (2022).
Evans, S. R. et al. The Future Ain’t What It Used to Be…Out With the Old…In With the Better: Antibacterial Resistance Leadership Group Innovations. Clin. Infect. Dis. 77, S321–s330 (2023).
Gelman, D. et al. Clinical Phage Microbiology: a suggested framework and recommendations for the in-vitro matching steps of phage therapy. Lancet Microbe 2, e555–e563 (2021).
Hatfull, G. F., Dedrick, R. M. & Schooley, R. T. Phage Therapy for Antibiotic-Resistant Bacterial Infections. Annu Rev. Med 73, 197–211 (2022).
Strathdee, S. A. et al. Phage therapy: From biological mechanisms to future directions. Cell 186, 17–31 (2023).
Green, S. I. et al. A Retrospective, Observational Study of 12 Cases of Expanded-Access Customized Phage Therapy: Production, Characteristics, and Clinical Outcomes. Clin. Infect. Dis. 77, 1079–1091 (2023).
Fujiki, J. et al. Fitness Trade-Offs between Phage and Antibiotic Sensitivity in Phage-Resistant Variants: Molecular Action and Insights into Clinical Applications for Phage Therapy. Int J. Mol. Sci. 24, 15628 (2023).
Gordillo Altamirano, F. et al. Bacteriophage-resistant Acinetobacter baumannii are resensitized to antimicrobials. Nat. Microbiol 6, 157–161 (2021).
Fauconnier, A. Phage Therapy Regulation: From Night to Dawn. Viruses 11, 352 (2019).
Kingston, R. et al. Excess resource use and cost of drug-resistant infections for six key pathogens in Europe: a systematic review and Bayesian meta-analysis. Clin. Microbiol Infect. 30, S26–s36 (2024).
Wyres, K. L. & Holt, K. E. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr. Opin. Microbiol 45, 131–139 (2018).
Jurkevitch, E. Riding the Trojan horse: combating pest insects with their own symbionts. Micro. Biotechnol. 4, 620–627 (2011).
Hadapad, A. B. et al. Diversity of bacterial communities in the midgut of Bactrocera cucurbitae (Diptera: Tephritidae) populations and their potential use as attractants. Pest Manag Sci. 72, 1222–1230 (2016).
Schukken, Y. et al. The “other” gram-negative bacteria in mastitis: Klebsiella, serratia, and more. Vet. Clin. North Am. Food Anim. Pr. 28, 239–256 (2012).
Aylward, F. O., Currie, C. R. & Suen, G. The Evolutionary Innovation of Nutritional Symbioses in Leaf-Cutter Ants. Insects 3, 41–61 (2012).
Flores-Mireles, A. L. et al. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol 13, 269–284 (2015).
Piednoir, P. et al. Spontaneous community-acquired bacterial meningitis in adults admitted to the intensive care units in the Caribbean French West Indies: Unusual prevalence of Klebsiella pneumonia. Int J. Infect. Dis. 100, 473–475 (2020).
Walker, K. A. & Miller, V. L. The intersection of capsule gene expression, hypermucoviscosity and hypervirulence in Klebsiella pneumoniae. Curr. Opin. Microbiol 54, 95–102 (2020).
Wang, H. et al. Nanomechanics measurements of live bacteria reveal a mechanism for bacterial cell protection: the polysaccharide capsule in Klebsiella is a responsive polymer hydrogel that adapts to osmotic stress. Soft Matter 9, 7560–7567 (2013).
Holt, K. E. et al. Correction to: Diversity and evolution of surface polysaccharide synthesis loci in Enterobacteriales. Isme J. 16, 1478 (2022).
Kochan, T. J. et al. Klebsiella pneumoniae clinical isolates with features of both multidrug-resistance and hypervirulence have unexpectedly low virulence. Nat. Commun. 14, 7962 (2023).
Choby, J. E., Howard-Anderson, J. & Weiss, D. S. Hypervirulent Klebsiella pneumoniae - clinical and molecular perspectives. J. Intern Med 287, 283–300 (2020).
Ali, S. et al. Potential therapeutic targets of Klebsiella pneumoniae: a multi-omics review perspective. Brief. Funct. Genomics 21, 63–77 (2022).
Łusiak-Szelachowska, M., Weber-Dąbrowska, B. & Górski, A. Bacteriophages and Lysins in Biofilm Control. Virol. Sin. 35, 125–133 (2020).
Li, Y. & Ni, M. Regulation of biofilm formation in Klebsiella pneumoniae. Front Microbiol 14, 1238482 (2023).
Nikolich, M. P. & Filippov, A. A. Bacteriophage Therapy: Developments and Directions. Antibiotics (Basel) 9, 135 (2020).
Bleriot, I. et al. Improving phage therapy by evasion of phage resistance mechanisms. JAC Antimicrob. Resist 6, dlae017 (2024).
Eriksson, H. et al. A suggested new bacteriophage genus, “Kp34likevirus”, within the Autographivirinae subfamily of Podoviridae. Viruses 7, 1804–1822 (2015).
Park, E. A. et al. Characterization and genome analysis of novel bacteriophages infecting the opportunistic human pathogens Klebsiella oxytoca and K. pneumoniae. Arch. Virol. 162, 1129–1139 (2017).
Boeckman, J. X. et al. Complete Genome Sequence of Klebsiella pneumoniae Myophage Mineola. Microbiol Resour. Announc 8, e00257–19 (2019).
Uyttebroek, S. et al. Safety and efficacy of phage therapy in difficult-to-treat infections: a systematic review. Lancet Infect. Dis. 22, e208–e220 (2022).
Martins, W. et al. Effective phage cocktail to combat the rising incidence of extensively drug-resistant Klebsiella pneumoniae sequence type 16. Emerg. Microbes Infect. 11, 1015–1023 (2022).
Feng, Y. et al. Safety and efficacy of a phage cocktail on murine wound infections caused by carbapenem-resistant Klebsiella pneumoniae. Int J. Antimicrob. Agents 63, 107088 (2024).
Fang, C. et al. Isolation and characterization of three novel lytic phages against K54 serotype carbapenem-resistant hypervirulent Klebsiella pneumoniae. Front Cell Infect. Microbiol 13, 1265011 (2023).
Haines, M. E. K. et al. Analysis of Selection Methods to Develop Novel Phage Therapy Cocktails Against Antimicrobial Resistant Clinical Isolates of Bacteria. Front Microbiol 12, 613529 (2021).
Tang, M. et al. Phage resistance formation and fitness costs of hypervirulent Klebsiella pneumoniae mediated by K2 capsule-specific phage and the corresponding mechanisms. Front Microbiol 14, 1156292 (2023).
Dunstan, R. A. et al. Epitopes in the capsular polysaccharide and the porin OmpK36 receptors are required for bacteriophage infection of Klebsiella pneumoniae. Cell Rep. 42, 112551 (2023).
Pan, Y. J. et al. Klebsiella Phage ΦK64-1 Encodes Multiple Depolymerases for Multiple Host Capsular Types. J. Virol. 91, e02457–16 (2017).
Yuan, Y. & Gao, M. Jumbo Bacteriophages: An Overview. Front Microbiol 8, 403 (2017).
Torres-Barceló, C., Turner, P. E. & Buckling, A. Mitigation of evolved bacterial resistance to phage therapy. Curr. Opin. Virol. 53, 101201 (2022).
Grooters, K. E. et al. Strategies for combating antibiotic resistance in bacterial biofilms. Front Cell Infect. Microbiol 14, 1352273 (2024).
Wilksch, J. J. et al. MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 7, e1002204 (2011).
Luo, Y. et al. Mechanisms and Control Strategies of Antibiotic Resistance in Pathological Biofilm. J. Microbiol Biotechnol. 31, 1–7 (2021).
Asghar, S. et al. Genomic characterization of lytic bacteriophages A¥L and A¥M infecting ESBL K. pneumoniae and its therapeutic potential on biofilm dispersal and in-vivo bacterial clearance[J]. Microbiol Res 262, 127104 (2022).
Roach, D. R. et al. Synergy between the Host Immune System and Bacteriophage Is Essential for Successful Phage Therapy against an Acute Respiratory Pathogen. Cell Host Microbe 22, 38–47.e34 (2017).
Kadirvelu, L. et al. A review on antimicrobial strategies in mitigating biofilm-associated infections on medical implants. Curr. Res Micro. Sci. 6, 100231 (2024).
Gordon, M. & Ramirez, P. Efficacy and Experience of Bacteriophages in Biofilm-Related Infections. Antibiotics (Basel) 13, 125 (2024).
Cometta, S., Hutmacher, D. W. & Chai, L. In vitro models for studying implant-associated biofilms - A review from the perspective of bioengineering 3D microenvironments. Biomaterials 309, 122578 (2024).
Lin, M. H. et al. Phosphoproteomics of Klebsiella pneumoniae NTUH-K2044 reveals a tight link between tyrosine phosphorylation and virulence. Mol. Cell Proteom. 8, 2613–2623 (2009).
Chen, S. et al. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Prjibelski, A. et al. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinforma. 70, e102 (2020).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Jolley, K. A. & Maiden, M. C. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinforma. 11, 595 (2010).
Wyres, K. L. et al. Identification of Klebsiella capsule synthesis loci from whole genome data. Micro. Genom. 2, e000102 (2016).
Deatherage, D. E. & Barrick, J. E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151, 165–188 (2014).
Kropinski, A. M. et al. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol. Biol. 501, 69–76 (2009).
Bonilla, N. et al. Phage on tap-a quick and efficient protocol for the preparation of bacteriophage laboratory stocks. PeerJ 4, e2261 (2016).
Dunstan, R. A. et al. Mechanistic Insights into the Capsule-Targeting Depolymerase from a Klebsiella pneumoniae Bacteriophage. Microbiol Spectr. 9, e0102321 (2021).
Brown, T. L. et al. Characterization and formulation into solid dosage forms of a novel bacteriophage lytic against Klebsiella oxytoca. PLoS One 12, e0183510 (2017).
Khan Mirzaei, M. & Nilsson, A. S. Isolation of phages for phage therapy: a comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS One 10, e0118557 (2015).
Liu, S. et al. APTC-C-SA01: A Novel Bacteriophage Cocktail Targeting Staphylococcus aureus and MRSA Biofilms. Int J. Mol. Sci. 23, 6116 (2022).
Yoo, S. et al. Designing phage cocktails to combat the emergence of bacteriophage-resistant mutants in multidrug-resistant Klebsiella pneumoniae. Microbiol Spectr. 12, e0125823 (2024).
Thung, T. Y. et al. Component Parts of Bacteriophage Virions Accurately Defined by a Machine-Learning Approach Built on Evolutionary Features. mSystems 6, e0024221 (2021).
Bouras, G. et al. Pharokka: a fast scalable bacteriophage annotation tool. Bioinformatics 39, btac776 (2023).
Petkau, A. et al. Interactive microbial genome visualization with GView. Bioinformatics 26, 3125–3126 (2010).
Nishimura, Y. et al. ViPTree: the viral proteomic tree server. Bioinformatics 33, 2379–2380 (2017).
Nishimura, Y. et al. DiGAlign: Versatile and Interactive Visualization of Sequence Alignment for Comparative Genomics. Microbes Environ. 39, ME23061 (2024).
Yu, L. et al. A guard-killer phage cocktail effectively lyses the host and inhibits the development of phage-resistant strains of Escherichia coli. Appl Microbiol Biotechnol. 102, 971–983 (2018).
Sanchez, B. C. et al. Development of Phage Cocktails to Treat E. coli Catheter-Associated Urinary Tract Infection and Associated Biofilms. J. Front Microbiol 13, 796132 (2022).
Feng, J. et al. Characterization and genome analysis of phage vB_KpnS_SXFY507 against Klebsiella pneumoniae and efficacy assessment in Galleria mellonella larvae. Front Microbiol 14, 1081715 (2023).
Forti, F. et al. Design of a Broad-Range Bacteriophage Cocktail That Reduces Pseudomonas aeruginosa Biofilms and Treats Acute Infections in Two Animal Models. Antimicrob. Agents Chemother. 62, e02573–17 (2018).
Thanki, A. M. et al. Development of a Phage Cocktail to Target Salmonella Strains Associated with Swine. Pharm. (Basel) 15, 58 (2022).
Macarthur Clark, J. A. & Sun, D. Guidelines for the ethical review of laboratory animal welfare People’s Republic of China National Standard GB/T 35892-2018 [Issued 6 February 2018 Effective from 1 September 2018]. Anim. Model Exp. Med 3, 103–113 (2020).
Gong, Y. et al. Gold nanoclusters cure implant infections by targeting biofilm. J. Colloid Interface Sci. 674, 490–499 (2024).
Choi, M. J. & Ko, K. S. Loss of hypermucoviscosity and increased fitness cost in colistin-resistant Klebsiella pneumoniae sequence type 23 strains. Antimicrob. Agents Chemother. 59, 6763–6773 (2015).
Acknowledgements
This work was supported by research grants from the National Natural Science Foundation of China (no.82472338), the Health Department of Zhejiang Province of the People’s Republic of China (no.2024KY1260), Key Laboratory of Clinical Laboratory Diagnosis and Translational Research of Zhejiang Province (2022E10022), and the Australian National Health & Medical Research Council (NHMRC) Investigator Award (2016330).
Author information
Authors and Affiliations
Contributions
H.C., H.L. and M.T. conceived and designed the experiments. H.C., D.Z., and Y.G. performed the experiments. C.Z. and Z.M. analyzed the data. H.C. wrote the paper with input from all co-authors. T.Z., T.L. and R.D. Reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, H., Liu, H., Gong, Y. et al. A Klebsiella-phage cocktail to broaden the host range and delay bacteriophage resistance both in vitro and in vivo. npj Biofilms Microbiomes 10, 127 (2024). https://doi.org/10.1038/s41522-024-00603-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41522-024-00603-8