Abstract
Rumen microbiotas are known to influence the fat deposition (FD) in sheep, but controversy over causality remains unresolved. Here, we performed microbiome-wide association studies (MWAS), microbiome genome-wide association analysis (mbGWAS) and bidirectional mendelian randomization (MR) analyses on 1,150 sheep with genotype data from whole-genome resequencing, 16S rRNA sequencing and multilevel FD-traits data. We quantified the proportion of individual variation in FD-traits explained by host genetics, rumen microbiota, and their interaction effects. We identified 32 rumen microbiota biomarkers including Bifidobacterium that were associated with FD-traits (Padj <0.05). Further, utilizing five MR methods, we identified eight causal associations between marker genera and FD-traits (Padj <0.05), including Butyrivibrio, Olsenella, p-2534-18B5 gut group, Prevotellaceae UCG-003, and Pseudobutyrivibrio causing forward causal effects on FD, and changes in Flexilinea and Suttonella induced by FD. To our knowledge, this is the inaugural attempt to employ MR in sheep to investigate the causal relationships between gastrointestinal microbiota and complex phenotypes, underscoring the potential for developing interventions related to adipose deposition in sheep from the perspective of the rumen microbiome.
Similar content being viewed by others
Introduction
Fat is an essential nutrient in the human and animal bodies, playing a crucial role in maintaining vital functions and overall health1. However, excessive fat deposition (FD) in adipose tissue leads to obesity, a complex condition associated with various serious health issues. Obesity not only increases the risk of type-2 diabetes, cardiovascular diseases, and various cancers but also poses significant challenges to socio-economic aspects and public health2. In the livestock industry, sheep, known for their high FD capacity, cannot overlook the FD status within their bodies3. Sheep’s excessive fat deposition is predominantly evident in subcutaneous adipose tissues, particularly in the tail and backfat, and in visceral fat, especially in the perirenal and caul fat (greater omentum)3,4. The prevalent over-accumulation of fat in sheep within the current intensive farming systems has adverse implications for their health and welfare, predisposing them to metabolic disorders, inflammation, and a reduced capacity to combat diseases4. The energy storage in adipose tissues is also correlated with crucial economic traits related to survival and production, such as feed efficiency, immune response, reproductive performance, and meat quality5,6,7,8. Furthermore, excessive FD influences consumer preferences for mutton and its by-products. These issues are critical not only for the sustainable development of the sheep industry but also pose health risks to human consumers. Consequently, modulating fat deposition has become a key target for sheep breeders around the world.
With the successful application of rumen microbiota transplantation studies, modulation of the rumen microbiome has emerged as a significant regulatory option for a variety of complex diseases and phenotypes in sheep, including rumen acidosis9 and metabolic function10. As the most diverse digestive ecosystem described in the animal kingdom, rumen microbiota has functions such as cellulose and hemicellulose degradation11. Its fermentation end products, volatile fatty acids (VFA), provide more than 70% of the energy source for sheep12. These VFAs are mainly absorbed by the rumen epithelium and converted into acetyl-CoA and propionyl-CoA in the liver, which are then used as raw materials to participate in metabolic processes such as glycogen synthesis, gluconeogenesis, and cholesterol synthesis, ultimately affecting host FD13. In addition, the metabolites produced by rumen microbiota can promote rumen epithelial development by regulating host gene expression, which is closely related to host physiological functions14. Currently, several microbial markers associated with the FD-trait in sheep have been identified15,16,17,18,19. However, these studies have merely established correlations, and it remains unclear whether these microbial markers possess the potential for practical application in modulating fat deposition. Therefore, elucidating the causal relationship between the rumen microbiota and the FD-trait in sheep is an important and challenging problem in agriculture and animal production fields.
Mendelian Randomization (MR) is a genetic variable analysis method that follows Mendel’s laws of inheritance. It uses genetic variations as instrumental variables to infer causal relationships between exposure factors and outcomes20. In recent years, with the accumulation of data from genome-wide association analysis (GWAS), MR analysis has become an effective method for inferring causal relationships and has been widely applied in studying the relationship between gut microbiota and fat deposition, including childhood obesity21, abdominal fat22, and body fat percentage23. Furthermore, the application of MR methods has extended beyond human studies, particularly in the field of farm animal research, such as inferring causal relationships between gene expression and complex traits in pig and beef cattle24,25. Therefore, introducing MR studies to investigate the causal relationship between rumen microbial communities and FD in sheep is a novel and valuable approach.
In this study, our aims to systematically evaluate the rumen microbial effects on multilevel or hierarchical FD-traits through large-scale single homogenous sheep population (n = 1150), identify rumen microbiota associated with FD-traits and their host genetic effects using microbiome-wide association studies (MWAS), microbiome genome-wide association analysis (mbGWAS), and finally perform MR analysis to clarify the causal relationship between specific rumen microbiota and FD. This may offer valuable insights for regulating rumen microbiota transplantation and improving optimal microbiome selection to manipulate FD in sheep. This is also the first attempt to explore causal associations between gastrointestinal microbiome and complex traits using MR analysis in sheep, highlighting the significant importance of host genetic data in guiding research on rumen microbiome interventions.
Methods
Animals and sample collection
For this study, a total of 1150 healthy male Hu lambs (the most dominant breed in China’s intensive sheep farming system) were randomly selected and raised under standardized conditions (the study design flow: Fig. 1). The lambs were housed with their natural mothers from birth until weaning at 56 days of age, after which they were moved to the Minqin experimental farm of Lanzhou University and maintained in individual pens until slaughter at 180 days of age. All lambs received the same feeding regimen (Diet composition see Supplementary Table 1) and were not administered any antibiotics or other antimicrobial substances during the study period. At 180 days of age (standard commercial slaughter age), the animals were fasted for 12 h and their weight, length, and body mass index (BMI; Body Weight(kg)/[Body Length (m)]2) were measured. Whole blood was collected from the jugular vein using sodium heparin anticoagulation tubes and stored at –20 °C until further analysis.
Following the standard procedure practiced in commercial abattoirs, all animals in this experiment were then slaughtered. The greater omentum fat (Omentum-FW), the perirenal fat (Perirenal-FW) and tail fat (Tail-FW) were removed from each animal and weighed using electronic weigh scales with a precision of 0.01 kg. The detailed procedure for fat tissue collection is as follows: (1) Greater omentum: Following slaughter, the carcasses were suspended. An incision was made along the ventral midline of the abdominal wall, extending from the pubic symphysis to the xiphoid process of the sternum, to fully expose the viscera within the abdominal cavity. The entire gastrointestinal tract was meticulously isolated from the carcass and placed on a rectangular surgical tray. Surgical scissors were carefully employed to dissect along the edge of the greater omentum, sequentially separating it from the four-chambered stomach, ceasing at the second segment of the duodenum and the greater curvature of the abomasum. The weight of greater omentum was subsequently determined using an electronic scale. (2) Perirenal fat: Utilizing a scalpel, the entire renal system was isolated from the carcass along both sides of the lumbar vertebrae and placed on a rectangular surgical tray. Subsequently, the surgical blade was used to longitudinally section the perirenal adipose tissue along the outer margin of the kidneys, achieving complete separation of the kidneys from the adipose tissue. The weight of perirenal fat was then ascertained using an electronic scale. (3) Tail fat: Using a scalpel, the entire caudal vertebrae were carefully separated from the tail fat by making a longitudinal incision along the caudal vertebrae starting from the lumbar to caudal vertebral junction. The tail fat was subsequently separated from the carcass by incision at the lumbar-caudal vertebral junction. Weight of tail fat was measured using an electronic scale. The total fat weight (Total-FW) was obtained by adding up all the above fat tissues physical weight values. The relative weight of each fat tissue (RFW) was calculated by the percentage of total body weight occupied by the tissue weight. The carcasses were then stored overnight for 12 h maintained at 4 °C prior to recording backfat thickness (BF) and rib fat thickness (RFT) data. The BF was measured by cutting the carcass transversely at the posterior end of the 12th rib and measuring the thickness of the fat layer directly above the midpoint of the eye muscle between the 12th and 13th ribs using a vernier calliper. The RFT was assessed by measuring the thickness of adipose tissue at the 12th/13th rib intersection 11 cm away from the midline using a vernier calliper, and is used to represent the fat content of the carcass. All procedures of isolation of the fat tissue were performed by certified veterinarians in animal handling. Following slaughter, rumen fluid samples were promptly collected from the rumen of each animal. The rumen contents were processed through four layers of cheesecloth to separate the liquid component, which was subsequently transferred to sterilized centrifuge tube and cryopreserved at -80°C for 16S rRNA sequencing and VFA profiling analyses.
Volatile fatty acid analysis
The concentration of rumen VFAs in ruminal chyme was determined using gas chromatography, an adaptation of a previously published method26. For the VFA assay, ruminal fluid was centrifuged at 5400 rpm for 10 min, with a centrifugal radius of 14.5 cm and a relative centrifugal force of 4731 g. Thereafter, 1.0 ml of the supernatant was mixed with 0.2 ml of a 25% (w/v) metaphosphoric acid solution, which included 2-ethylbutyric acid as an internal standard at a concentration of 2 g/L. This mixture was incubated at 4 °C for 30 min, followed by a secondary centrifugation at 10,000 rpm, with a centrifugal radius of 5 cm and a relative centrifugal force of 5595 g, for an additional 10 min at 4 °C. The supernatant was then carefully collected and filtered through a 0.45 μm filter (0.45 μm Syringe Filters). The clarified supernatant was subsequently transferred to a vial for gas chromatographic analysis.
The quantification of VFA concentrations was executed using a TRACE-1300 series gas chromatograph (TRACE 1300, Thermo Scientific, Milan, Italy). The gas chromatographic conditions are delineated as follows: A capillary column DB-FFAP with dimensions of 15 meters in length, 0.32 millimeters in internal diameter, and 0.25 micrometers in film thickness was employed. The sample injection volume was precisely set to 1 microliter with a split ratio of 50:1. Both the injection port and the detector were kept at a constant temperature of 240 degrees Celsius. The flow rates for the carrier and auxiliary gases were established at 35 milliliters per minute for hydrogen flame gas, 20 milliliters per minute for nitrogen carrier gas, and 350 milliliters per minute for air. The temperature gradient for the analysis was programmed to commence with an isothermal hold at 50 degrees Celsius for a duration of 5 min, followed by a linear ramp to 190 degrees Celsius over a period of 2 min at a rate of 25 degrees Celsius per minute. Subsequently, the temperature was increased to 200 degrees Celsius over 5 min at a rate of 10 degrees Celsius per minute, concluding with a final ramp to 220 degrees Celsius, which was sustained for another 5 min at the same rate of 10 degrees Celsius per minute.
DNA extraction and quantification
Sheep genomic DNA was extracted from each animal blood sample using the EasyPure Blood Genomic DNA Kit (Tiangen Bio Company, Beijing, China) following the manufacturer’s instructions. For the extraction of rumen microbial DNA, approximately 200 mg of each rumen fluid sample was thawed on ice and homogenized. Subsequently, rumen microbial genomic DNA was extracted utilizing the EasyPure Stool Genomic DNA Kit (TransGen Biotech, catalog number EE301-01, Beijing, China), following the manufacturer’s prescribed protocol. The Nanodrop 2000 spectrophotometer (Thermo Scientific) was employed to evaluate the concentration and purity of the DNA extract for successful DNA isolation, and 1% agarose gel electrophoresis was used to assess the completeness of the DNA sample. At this stage, no DNA samples were removed.
Genotyping
The sheep genomic DNA samples (n = 1150) were randomly fragmented and sheared into fragments of ~350 bp in length using standard Covaris sonication. The fragmented DNAs were end-repaired, dA-tailed, ligated with Illumina adaptors, and amplified using PCR with 500 bp inserts for library construction. All qualified libraries were then sequenced on an Illumina HiSeq XTen platform with the PE150 mode. The quality assessment of raw sequence data and deletion of low-quality sequences was performed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The clean reads were mapped against the sheep reference genome (Oar_rambouillet_v1.0, https://mart.ensembl.org/Ovis_aries_rambouillet/Info/) by employing Burrows-Wheeler-Alignment Tool. Subsequently, potential PCR duplicate reads were marked and removed using SAMBAMBA (https://github.com/lomereiter/sambamba) and indexed in SAMtools (http://github.com/samtools/samtools). The variants were detected by Genome Analysis Toolkit (GATK; https://software.broadinstitute.org/gatk/) HaplotypeCaller and GenotypeGVCFs modules. To obtain high-quality Single Nucleotide Polymorphisms (SNP), we initially applied the GATK VariantFiltration module to rigorously filter the resulting VCF-dataset based on the robust criteria: FS > 60.0; QD < 10.0; MQ < 40.0; ReadPosRankSum < -8.0; MQRankSum < -12.5. Subsequently, we utilized VCFtools to exclude Insertion/Deletion mutations (InDels), retaining only bi-allelic genetic variations and removing SNPs with depths below 5. Following this, we conducted further SNP refinement using PLINK, setting the minor allele frequency threshold at 0.05 and a call rate of 70%. Finally, 15,252,471 SNPs were obtained through these procedures, spanning 27 chromosomes and encompassing 1150 animals, for subsequent analyses (reached a 7.05-fold depth; Supplementary Table 2).
Amplicon sequencing and data processing
For quality-assessed rumen microbial genomic DNA, dilution was performed to serve as a template for amplification. We selected the V3-V4 hypervariable regions of the bacterial 16S rRNA gene for amplification, utilizing specific primers 341 F/806 R (5ʹ-CCTAYGGGRBGCASCAG-3ʹ and 5ʹ-GGACTACNNGGGTATCTAAT-3ʹ). To differentiate among various samples within the same library, a 12 bp unique barcode sequence was incorporated at the 5’ end of the primers prior to amplification. After the purification and quantification, the qualified amplicons were sequenced on an Illumina NovaSeq PE250 platform according to the manufacturer’s standard protocol. After sequencing, paired-end reads of each sample were trimmed and assembled using FLASH software. After trimming control with FastQC, chimerical reads were removed by UCHIME algorithm (http://drive5.com/uchime). Following, the filtered sequences were processed using the DADA2 pipeline to generate amplicon sequence variants (ASVs) and taxonomic assignments were performed using the Naive Bayes pretrained SILVA taxonomic data set (version 138). To eliminate potential false or low-abundance ASVs resulting from sequencing errors or other sources of noise and to avoid sequence artifacts, we employed the “filter-features” function in the QIIME2 software to apply a threshold of presence in two or more samples and a minimum total frequency greater than 10 for filtering out rare features (--p-min-samples 2 and --p-min-frequency 10)27,28,29. Sequences affiliated with “mitochondria” or “chloroplasts” were meticulously excluded from the ASVs to ensure the accuracy of microbial community analysis. A rarefied ASV count table was generated using the QIIME2 feature-table rarefy commands with --p-sampling-depth 34,736 (the minimum library size method). The preliminary ASV tables of rumen microbes contained 1,150 individuals and 14,679 ASVs (assigned 872 genera; Supplementary Table 3). Alpha diversity indicators were calculated based on microbial ASVs using the microeco R-package, and principal coordinate analysis (PCoA) was performed using the vegan and ape packages.
Descriptive statistics, Spearman and Pearson correlation, genetic correlation and microbial correlation between FD-phenotypes
Descriptive statistics, Spearman and Pearson correlation were completed in psych R-package, and the corrplot R-package was used for data visualization purposes. The adjusted phenotypes were calculated by subtracting the estimates of the fixed effects (birthplace and rearing season), which were estimated using a linear least squares approach by linear fixed effects model. The principal component analysis (PCA) was performed using vegan R-package. We also estimated genetic correlations (rG) and microbial correlations (rM) of FD-traits using a multi-trait model in HIBLUP. The model is as follows:
Where y is a vector of phenotypic values, b is a vector of fixed effects of birthplace and rearing season, and W is the corresponding design matrix; g is a vector of additive genetic effects following a distribution of \({\bf{N}}({\bf{0}}, {\bf{G}}{{\boldsymbol{\sigma }}}_{{\boldsymbol{g}}}^{{\bf{2}}}{)}\), where G is GRM and \({{\boldsymbol{\sigma }}}_{{\boldsymbol{g}}}^{{\bf{2}}}\) is the additive genetic variance; m is a vector of rumen microbial effects following a multinomial distribution of \({\bf{N}}({\bf{0}},{\bf{M}}{\sigma }_{m}^{2})\), where M is the MRM and \({{\boldsymbol{\sigma }}}_{{\boldsymbol{m}}}^{{\bf{2}}}\) is the rumen microbial variance; K1 and K2 are the incidence matrices corresponding to the g and m, and e is the residuals following a distribution of \({\bf{N}}\left({\bf{0}},{\bf{I}}{{\boldsymbol{\sigma }}}_{{\boldsymbol{e}}}^{{\bf{2}}}\right)\), where I is an identity matrix and \({{\boldsymbol{\sigma }}}_{{\boldsymbol{e}}}^{{\bf{2}}}\) is the residual variance. Population structure was accounted for by incorporating the top five host genetic principal components (PC) as covariates in the model. The rG and rM were calculated using the following formula:
where rG is the genetic correlation between traits X and Y; \({{\boldsymbol{COV}}}_{{{\boldsymbol{G}}}_{{\boldsymbol{XY}}}}\) is the genetic covariance matrix of traits X and Y; \({{\boldsymbol{\sigma }}}_{{{\boldsymbol{G}}}_{{\boldsymbol{X}}}}\) and \({{\boldsymbol{\sigma }}}_{{{\boldsymbol{G}}}_{{\boldsymbol{Y}}}}\) are the genetic standard deviation of traits X and Y; rM is the microbial correlation between traits X and Y; \({{\boldsymbol{COV}}}_{{{\boldsymbol{M}}}_{{\boldsymbol{XY}}}}\) is the microbial covariance matrix of traits X and Y; \({{\boldsymbol{\sigma }}}_{{{\boldsymbol{G}}}_{{\boldsymbol{X}}}}\) and \({{\boldsymbol{\sigma }}}_{{{\boldsymbol{G}}}_{{\boldsymbol{Y}}}}\) are the microbial standard deviation of traits X and Y.
Assessment of the overall relationship between the rumen microbiome and host genetics
To assess the resemblance among sheep based on their rumen bacterial abundance, we adopted five microbial similarity/relationship matrix construction inference approaches, Euclidean, Bray-Curtis, Constrained Correspondence Analysis (CCA), Detrended Correspondence Analysis (DCA) and Microbial Relationship Matrix (MRM), using the ASV abundance data. Euclidean distance and Bray-Curtis dissimilarity were measured using the vegdist function in vegan R-package. CCA and DCA were calculated using the ordinate function with Bray-Curtis dissimilarity as the distance metric. The MRM was created following the procedure reported by Wen et al.30 and Tang et al.31. For genetic relationship matrix (GRM), we used HIBLUP software to create the matrix of all pairs of individuals within the animal cohort based on all SNPs. In order to investigate whether host genetics have an impact on rumen microbiota, Mantel test was done to evaluate similarities of GRM and all microbial matrices with 1000 permutations.
Heritability, Microbiability and Holobiability
We estimated variance components and assessed the proportion of phenotypic variation explained by host genetics (G) and rumen microbiome (M), i.e., heritability (h2), microbiability (m2), and genome-by-microbiome interaction (G×M2) and holobiability (ho2)32 for all FD-traits (y), using a model GMP (y = G + M + G ×M) in HIBLUP (https://www.hiblup.com/). In general, the contributions of the G and M to the phenotype are defined in terms of h2 and m2, respectively. However, if both G and M are included in a model, then h2 and m2 can only reflect the contribution of direct genetic effects and direct microbial effects, and the effect of the interaction between G and M on the phenotype is quantified as G×M2. Concurrently, the holobiont effect, which encapsulates the combined influence of G and M on the phenotype, is quantified as ho2. The details of the GMP model are as follow:
Where y is a vector of phenotypic values, b is a vector of fixed effects including birthplace and rearing season, and W is the corresponding design matrix; g is a vector of additive genetic effects following a distribution of \({\bf{N}}({\bf{0}}, {\bf{G}}{{\rm{\sigma }}}_{g}^{2}{)}\), where G is GRM and \({{\boldsymbol{\sigma }}}_{{\boldsymbol{g}}}^{{\boldsymbol{2}}}\) is the additive genetic variance; m is a vector of rumen microbial effects following a distribution of \({\bf{N}}({\bf{0}}, {\bf{M}}{{\boldsymbol{\sigma }}}_{{\boldsymbol{m}}}^{{\boldsymbol{2}}}{)}\), where M is the MRM and \({{\boldsymbol{\sigma }}}_{{\boldsymbol{m}}}^{{\boldsymbol{2}}}\) is the rumen microbial variance; g × m is a vector of effects of host genetic and rumen microbiome interactions, K1, K2, and K3 are the incidence matrices corresponding to the g, m, and g × m in the model GMP, and e is the residuals following a distribution of \({\bf{N}}\left({\bf{0}}, {\bf{I}}{{\boldsymbol{\sigma }}}_{{\boldsymbol{e}}}^{{\boldsymbol{2}}}\right)\), where I is an identity matrix and \({{\boldsymbol{\sigma }}}_{{\boldsymbol{e}}}^{{\boldsymbol{2}}}\) is the residual variance. The phenotypic variance (\({{\boldsymbol{\sigma }}}_{{\boldsymbol{p}}}^{{\boldsymbol{2}}}\)) was the sum of all variance components: \({{\boldsymbol{\sigma }}}_{{\boldsymbol{p}}}^{{\boldsymbol{2}}}{=}{{\boldsymbol{\sigma }}}_{{\boldsymbol{g}}}^{{\boldsymbol{2}}}{+}{{\boldsymbol{\sigma }}}_{{\boldsymbol{m}}}^{{\boldsymbol{2}}}{+}{{\boldsymbol{\sigma }}}_{{\boldsymbol{g}}{\boldsymbol{\times }}{\boldsymbol{m}}}^{{\boldsymbol{2}}}{+}{{\boldsymbol{\sigma }}}_{{\boldsymbol{e}}}^{{\boldsymbol{2}}}\). The \({{\boldsymbol{\sigma }}}_{{\boldsymbol{g}}}^{{\boldsymbol{2}}}{/}{{\boldsymbol{\sigma }}}_{{\boldsymbol{p}}}^{{\boldsymbol{2}}}\), \({{\boldsymbol{\sigma }}}_{{\boldsymbol{m}}}^{{\boldsymbol{2}}}{/}{{\boldsymbol{\sigma }}}_{{\boldsymbol{p}}}^{{\boldsymbol{2}}}\), \({{\boldsymbol{\sigma }}}_{{\boldsymbol{g}}{\boldsymbol{\times }}{\boldsymbol{m}}}^{{\boldsymbol{2}}}{/}{{\boldsymbol{\sigma }}}_{{\boldsymbol{p}}}^{{\boldsymbol{2}}}\) and (\({{\boldsymbol{\sigma }}}_{{\boldsymbol{g}}}^{{\boldsymbol{2}}}{+}{{\boldsymbol{\sigma }}}_{{\boldsymbol{m}}}^{{\boldsymbol{2}}}{+}{{\boldsymbol{\sigma }}}_{{\boldsymbol{g}}{\boldsymbol{\times }}{\boldsymbol{m}}}^{{\boldsymbol{2}}}{)}{/}{{\boldsymbol{\sigma }}}_{{\boldsymbol{p}}}^{{\boldsymbol{2}}}\) were defined as h2, m2, G×M2 and ho2, respectively. Population structure was accounted for by incorporating the first five host genetic PC as covariates in the model.
Microbiome-wide association studies (MWAS)
The associations between 7 diversity metrics (including Shannon, Simpson indices, and the top 5 PCoA Axis) and FD-traits were first fitted and tested using a linear model and permutation tests (5000 permutations) implemented in the lmPerm R-package (https://CRAN.R-project.org/package=lmerPerm). Due to the low detection rate and taxonomic ambiguity of certain taxa, which provided limited information for association analysis, we excluded genera present in below 20% and unclassified taxa. In the end, 117 genera were utilized for further analysis, representing an average of 90.95% of the total abundance per sample. We then employed a two-part MWAS model described by Fu et al.33 and Wen et al.30 to ascertain associations between sheep FD-traits and the quantitative features (abundance) and binary features (presence/absence) of each bacterial genus. The zero-inflated nature of microbial abundance data presents a challenge in meeting the normal distribution assumption required by the quantitative model. To address this, we normalized the relative abundances using the centered log-ratio (CLR) method. Details of the two-part model are given below:
let y denote the Total-FW and Total-RFW value after adjustment for birthplace and rearing season; b be a binary feature of a specific genera, which is recoded as 0 for absent or 1 for present for each individual; q be the CLR transformed relative abundance of a specific genera. β1 and β2 represent the regression coefficients of the two models respectively, and e is the intercept. P values were derived from sequential permutation tests comprising 5000 permutations conducted using the lmPerm package. In each permutation, the rumen microbial feature was randomized among individuals, and the 2-part analysis was carried out on the permuted data. Significance was maintained at Bonferroni-adjusted P values < 0.05.
This approach considers the intricate features of microbial data and maximizes research efficiency. The association of each FD-trait with all bacterial genera was examined using both binary (B) and quantitative (Q) models, and ultimately each trait will yield 2 result sets, a binary analysis set and a quantitative analysis set for that trait. If only the association P value in the B model is below the cutoff threshold, it indicates that the influence is solely due to the presence or absence of the taxon, making the abundance of the taxon in the sample less significant. Conversely, if only the association P value in the Q model is below the cutoff threshold, it suggests a relationship between the abundance level of the taxon and the FD-trait, with no impact in the absence of the taxon. The biological explanation is that there exists another taxon with a similar function to it that has replaced it33. When the association P value is below the cutoff threshold in both the B and Q models, it signifies that both the presence/absence and abundance of the taxon affect the phenotype.
Venn diagram was used to show each significant MWAS result set’s shared and unique marker microbial features. The Pearson’s and Spearman’s correlation analyses was used to explore the relationship between marker genera. After merging the two correlation coefficient matrices, hierarchal clustering was conducted to identify potential taxa modules by using pheatmap R-package. Spearman’s correlation analyses were also conducted to reveal the correlations between marker genera and VFAs.
Microbiome genome-wide association study (mbGWAS)
To investigate the impact of host genetics on the FD-related rumen microbiota, we performed a mbGWAS analysis using all significant marker microbial features that were identified by the MWAS. The input data used for the mbGWAS were consistent with the MWAS, i.e., B-features were coded as 1 (relative abundance > 0) or 0 (relative abundance = 0) based on presence/absence; Q-features were normalized using the CLR method for raw relative abundance to achieve a normal distribution. All genera were categorized into the above microbial features for performing mbGWAS.
The mbGWAS for B-features used the Generalized Linear Mixed Model (GLMM) in GCTA (https://yanglab.westlake.edu.cn), which is a GWAS method specifically for binary traits that better addresses the inflated test statistic due to unbalanced case control. Nevertheless, considering that the extremely small sample sizes in the case-controls would lead to unstable values in the calculations, which would result in a negatively-defined Variance-Covariance matrix (https://gcta.freeforums.net/thread/42/error-1-matrix-invertible), we deleted the B-features of the marker genera with a detection rate of over 90%. Genera with low detection rates have been excluded from the upstream analysis (see Methods: MWAS). The mbGWAS for Q-features used the mixed linear model (MLM) in GCTA. For each mbGWAS, the individual’s birthplace and rearing season, as well as the first 5 eigenvectors from PCA were used as covariates. Finally, we set the genome-wide significance threshold at P < 3.28E–09 (0.05/NSNPs, N = 15,252,471) and suggestive significance threshold at P < 6.56E–08 (1/NSNPs) based on Bonferroni correction.
Bi-directional one-sample mendelian randomization (MR)
We evaluated the potential causal relationship between specific rumen microbiota and sheep FD using MR analysis in the TwoSampleMR R-package (https://mrcieu.github.io/TwoSampleMR) using 38 marker rumen microbial features identified by MWAS as exposures, selecting SNPs with significant associations with them as instrumental variables (IV), and two FD-traits as outcome variables. Further, we conducted backward MR analysis to investigate for reverse causation (FD-traits as exposure and rumen microbial features as outcome). In MR, three hypotheses were required to be satisfied for the genetic variant to be used as an IV: (1) the hypothesis of relevance: the IV is associated with the exposure factor; (2) the hypothesis of independence: the IV is unassociated with confounders; and (3) a hypothesis of exclusivity: the IV has no direct association with the outcome and only affects the outcome through the exposure factor. All the scripts of the MR analysis were performed with slight modifications based on https://github.com/Zukunft-Zhang/MendelRookie_English.
Data sources
Exposure and outcome data for MR analyses were obtained from present large homogenous populations. The GWAS summary statistics sets of FD-trait were obtained from our unpublished study. The phenotypic GWAS was performed in GCTA using the same 1150 animals as in the present study, the same genetic variants, fixed effect and the same mixed linear model as in the mbGWAS for quantitative features.
Correction of the winner’s curse in GWAS
To avoid the winner’s curse of the GWAS on the causal relationships estimated using one-sample MR in the present study, we corrected the effect sizes of the GWAS summary data with a Bootstrap method from the winnerscurse R-package (https://amandaforde.github.io/winnerscurse/index.html), which is specially designed for one-sample estimation, to produce more accurate estimates of true associations. This method has demonstrated competitive performance in a wide variety of simulations and realistic tests conducted with the authors34.
The Selection of Instrumental Variables (IV)
To ensure the accuracy and validity of the causal relationship between the rumen microbiota and fat deposition, our study employed the following criteria for IV selection: (1) For forward MR, we employed three thresholds, strict and lenient, to select IVs associated with the exposure. We first selected SNPs associated with rumen microbial features using the more suggestive threshold P < 1E–06, aiming for these SNPs to be representative of microbial features. Furthermore, we validated these results by comparing the consistency of direction between the compromised threshold and other two strict thresholds [Genome-wide significant (6.56E–08) and significantly suggestive (3.28E–09)]. Our choice of 1E–06 as the primary cutoff was supported by two main reasons. The current study represents the first application of MR to infer causal relationships between gastrointestinal microbiota and complex phenotypes in sheep. Human microbiome studies have suggested that a P value threshold below 1E–06 is more suitable for MR analysis35,36,37, providing robust support for our choice of critical value. As illustrated in Supplementary Table sets, we experimented with various thresholds for IVs selection. However, we observed that mbGWAS yielded a limited number of ‘hits’ (SNPs associated with microbial features at P < 6.56E–08 or 3.28E–09). For several rumen microbiota, no variants showed significant association with the microbial features. Thus, we concluded that adopting a more lenient threshold such as 1E–06 is a pragmatic strategy for the selection of MR IVs for rumen microbiota. For the backward MR, for consistency, we also used the same three thresholds to select SNPs associated with two FD-traits. (2) To ensure the independence of each IV and eliminate the influence of linkage disequilibrium, we set the LD coefficient (R2) to 0.001 and the genetic distance to 10,000 kb. (3) To ensure the strength of the association between IVs and the exposure factor, weak IVs were excluded by calculating the F-statistic [F = R2(n-k-1)/k(1-R2), R2 represents the variance of exposure explained by the IVs, n is the sample size of the GWAS, and k is the number of IMs] and removing SNPs with F-values < 10. (4) To avoid violating the third assumption of MR analysis, the “exclusion restriction”, SNPs directly related to the outcome were removed. Furthermore, the MR-PRESSO method was used to detect SNPs acting as outliers, and causal relationship estimates were conducted after removing these outliers.
Mendelian Randomization and sensitivity analysis
We first employed five distinct MR methodologies to investigate whether the significant associations identified by MWAS were causal: inverse variance weighting (IVW)38, weighted mode (WM)39, MR-Egger regression40, weighted median estimator (WME)41, and simple mode method (SM)39. The IVW, proposed by Burgess et al. in 201338, is a weighted linear regression model that combines the Wald estimates of each SNP to obtain an overall estimate. The IVW method assumes that all SNPs are valid instrumental variables with strong causal inference capabilities, but the results may be influenced by horizontal pleiotropy. Studies have shown that when there is an association between IVs and exposure and no significant genetic heterogeneity, the IVW test is more effective than other methods41,42. Hence, in cases with two or more IVs, we prioritized the IVW results. In cases with only a single IV, we employed the Wald estimate method for testing43. In MR analysis, a Bonferroni-corrected P value of <0.05 [0.05/N, where N corresponds to the number of tests below the SNP selection threshold (N1E–06 = 38, N6.56E–08 = 21, and N3.28E–09 = 8 in the forward MR; all N = 38 in the backward MR)] indicates statistical significance of the results. Sensitivity analyses include tests for heterogeneity and horizontal pleiotropy. Cochran’s Q test was employed to analyse heterogeneity among IVs, with P < 0.05 indicating heterogeneity. Additionally, to determine if significant results are driven by a single SNP, a leave-one-out analysis was conducted, systematically removing each SNP, calculating the effect of the remaining SNPs, and observing if the results change significantly after each SNP removal. MR-Egger regression method was used to test for horizontal pleiotropy, with P < 0.05 indicating the presence of horizontal pleiotropy. In cases of heterogeneity and horizontal pleiotropy, the multiplicative random-effects IVW model (MRE-IVW) was utilized for analysis, allowing for overdispersion in the weighted linear regression, a statistical model commonly used in meta-analysis and statistics to address heterogeneity.
Results
Characteristics of fat deposition phenotypes in the large-scale sheep cohort
By meticulous dissection, we measured 11 FD-traits of 1150 sheep. All FD-traits had coefficients of variation (CV) above 20%, with BMI being the exception at 12%. Moreover, the absolute values of skewness and kurtosis coefficients for all traits were less than 1, indicating a normal distribution (Table 1 and Supplementary Table 4). Additionally, this Hu sheep population, as a Chinese dominant local breed, has not been strongly selected for any traits. These findings showed that the genetic background and gut microbiota of this animal population were in a relatively unaltered state, free from human intervention, and thus provide a suitable model for investigating the impact of host genetics and gut microbiota on complex traits.
To comprehensively investigate the relationship between the FD-traits, Pearson’s and Spearman’s correlation analysis was performed (Fig. 2a). All FD-traits were positively correlated and the average correlation coefficients observed over all pairs of analyses were 0.51. Total-FW and Total-RFW demonstrated moderate to strong positive correlation (coefficients: 0.28–0.91) with all other traits, which was statistically significant, suggesting that they may be used as representative indexes to evaluate FD levels in this study. This was further supported by PCA, which revealed respectively clear separations and gradual transition colors in the score plot based on the categorical (Low-FD: trait <0.5 SD below the mean; Medium-FD: trait ±0.5 SD above and below the mean; and High-FD: trait >0.5 SD above the mean) and continuous data (Fig. 2b–e and Supplementary Fig. 1).
a The correlation between the phenotype of sheep fat deposition. All sheep fat deposition traits were classified into two types: overall level and local level, based on their sources. The Spearman and Pearson correlation methods were used above and below the diagonal, respectively. The size and color of the squares, as well as the area and color of the pie chart, represent the magnitude of the correlation coefficient. “*” represents P value < 0.05, “**” represents P value < 0.01, and “***” represents P value < 0.001. b–e The principal component analysis (PCA) analysis was performed on all adjusted data of sheep fat deposition phenotypes to identify representative indicators. The horizontal axis represents the first principal component (PC), the vertical axis represents the second PC, and the percentage represents the contribution of the PC to the sample differences. Each point represents a sample. Figure (b) show the PCA results visualization based on the Total-FW grouping (categorical data). Samples in the same group are represented by the same color. H_Total-FW represents individuals with Total-FW values greater than “mean + 3sd” (n = 178); M_Total-FW represents individuals with Total-FW values between “mean + 3 SD” and “mean –3 SD” (n = 770); L_Total-FW represents individuals with Total-FW values less than “mean –3 SD” (n = 190). Figure (c) shows the PCA visualization based on the Total-RFW grouping, using the same grouping method as Total-FW (H_Total-RFW: n = 174, M_Total-RFW: n = 775, L_Total-RFW: n = 187). The figure (d, e) shows the PCA visualization based on the continuous values of Total-FW and Total-RFW, where the color changes represent the magnitude of Total-FW and Total-RFW values. f Above and below the diagonal lines are the “microbial correlation” and “genetic correlation” of sheep fat deposition phenotypes, respectively. All correlations were positively correlated. The size and color variation of the circles represent the high and low values. BMI body mass index, BF backfat thickness, GR Rib thickness, FW Absolute weight of fat (Measured using an electronic scale), RFW relative weight of fat (FW/body weight).
To investigate whether FD-traits are influenced by common genetic factors and rumen microbial communities, we conducted genetic and microbial correlation estimations (Fig. 2f). We observed that a substantial proportion (80%) of the genetic correlation estimates between FD-traits exhibited strong positive correlations (coefficients >0.5), while weak positive genetic correlations were observed in only 7% of cases (coefficients <0.2; mean: 0.70, range: 0.06–0.97). Additionally, microbial correlation estimates also indicated a strong positive microbial relationship between FD-traits (mean: 0.96, range: 0.36–0.99). Moreover, both Total-FW and Total-RFW showed high positive genetic correlations (mean:0.83, range: 0.65–0.99) and microbial correlations (mean: 0.99, range: 0.98–0.99) with other FD traits. Therefore, FD-traits were influenced by some shared genetic factors and rumen microbial taxa in sheep.
Variance of sheep fat deposition explained by host genetics and rumen microbiota composition
In order to quantify the relative contribution of host genetics, rumen microbiota, host genome-by-microbiome interaction and holobiont effects to the FD traits, we calculated h2, m2, G×M2 and ho2 of the 11 FD-traits. The ho2 ranged from 17.62 to 68.14% and consisted mainly of contributions from host genetics and rumen microbiome, while interactions accounted for a very low proportion of the total. The mean h2 was 35.92%, ranging from 12.94% for RFT to 53.61% for Perirenal-FW (Fig. 3a and Supplementary Table 5). Most of the FD-traits (9/11) with a h2 greater than 30% (including 32.59% for Total-FW and 38.33% for Total-RFW). The FD-traits had low to moderate m2 with an average of 18.35% (Fig. 3a and Supplementary Table 5). BMI had the highest estimated m2 (32.62%) among the FD-traits measured, followed by the m2 of BF (26.08%), Tail-FW (21.11%), Total-FW (19.75%), Omentum-RFW (18.29%), Perirenal-RFW (16.87%), Omentum-FW (16.61%), Total-FW (15.91%), Tail-RFW (15.73%), Perirenal-FW (14.24%) and RFT (4.68%). However, in our observations, the mean G×M2 value was only 0.028%, ranging from 0 to 0.30%, implying that the interaction effect has a limited effect on traits. To further investigate the extent to which host genetics interact with the rumen microbiota, we assessed the correlation between the host genetic kinship matrix and the rumen microbial heterogeneity/relationship matrices using the mantel test. This process was performed 1000 times and the average correlations were 0.0034 with MRM, 0.056 with DCA, 0045 with CCA, 0.0089 with Euclidean and -0.0094 with Bray Curtis (Fig. 3b). This provides further evidence of a limited interaction between the sheep genome and the rumen microbiome.
a The individual differences in sheep fat deposition phenotypes explained by host genetics, rumen microbiome, and their interaction were estimated using the variance component method. Ho2 holobiability, BMI body mass index, BF backfat thickness, GR Rib fat thickness, FW absolute weight of fat (Measured using an electronic scale), RFW relative weight of fat (FW/body weight). b Mantel-test (based on Spearman’s rank correlation) was performed between the rumen microbial relationship/similarity matrices (based on ASV data) and the genome relationship matrix (GRM). Each square represents the correlation coefficient between two matrices, and the darker color indicates a higher correlation. GRM genomic relationships matrix, Euclidean: Euclidean distance, Bray-Curtis: Bray-Curtis dissimilarity, Jaccard: Jaccard index, CCA constrained correspondence analysis, DCA Detrended Correspondence Analysis, MRM Microbial Relationship Matrix.
Association of rumen microbiota with sheep fat deposition
To investigate the relationship between the rumen microbiome and fat deposition, we initially examined the association of the first five axis of a principal coordinates analysis (PCoA) of Bray-Curtis dissimilarities, representing global summary of variation in the sheep rumen microbiome (explaining 29.42%, 20.75%, 14.24%, 11.38% and 5.76% of the variation, respectively), with two FD representative indicators (Total-FW and Total-RFW). We observed that the first four PCo axes (P < 0.001) were all associated with Total-FW, while the first PCo axis (P = 0.046) was correlated with Total-RFW. Furthermore, we also observed associations between the Firmicutes: Bacteroidota ratio and Total-FW (P = 0.027), Simpson index and Total-RFW (P = 0.034).
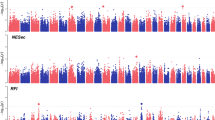
We next tested for association between the individual bacterial genera and two FD representative indicators. We identified 32 bacterial genera associated with two representative indicators at Bonferroni-adjusted P values less than 0.05, 32 genera were associated with Total-FW, and 5 genera were associated with Total-RFW (Fig. 4a–d and Supplementary Table 6). The majority of marker genera belonged to Firmicutes (62.50%), followed by Bacteroidota (21.88%), Actinobacteriota (6.25%), Proteobacteria (3.13%) and Chloroflexi (3.13%), as well as Verrucomicrobiota (3.13%). We found 17 significant associations (36%) of 14 marker genera with FD-traits detected by the binary model, and a total of 30 significant associations (64%) of 28 marker genera with two FD-traits detected by the quantitative model; a total of 10 bacterial genera were jointly detected in both the binary and quantitative models (implying that both presence/absence and abundance affect phenotype). While most marker genera were associated with Total-FW, five marker taxa were shared between all two FD-traits (Fig. 4e). In detail, the binary (B) and quantitative (Q) features of Bifidobacterium, the B-features of Defluviitaleaceae UCG-011 and Suttonella, and the Q-feature of Prevotellaceae UCG-003 were associated with all two FD-traits, and the B and Q features of Alloprevotella were associated with Total-RFW and Total-FW, respectively.
a–d Microbiome-wide association studies (MWAS). Identification of rumen microbial features associated with fat deposition phenotypes using MWAS. The results were visualized using Manhattan plots. Each circle represented a genus, and the X-axis indicated that all the genera were sequenced at the phylum level, and the Y-axis indicated the relevant statistical significance. Solid red lines and dashed red lines indicate Bonferroni corrected P value = 0.01 and 0.05, respectively. e Venn diagram depicting unique and shared marker rumen microbial features among four MWAS outcome datasets (2 Traits × 2 models). The box plots on the left show details of the distribution of marker microbial features among the four MWAS outcome datasets. f The bar graphs illustrate the detection rate and average relative abundance of marker microbiota. g Pearson (lower diagonal) and Spearman (upper diagonal) correlations between the rumen microbial genera associated with fat deposition in sheep. Blue and red indicated positive and negative correlations, respectively; The gray background labels represent the modules divided by clustering. h The Sperman correlation between marker genera related to fat deposition and rumen VFA profile. The size and color of the circle indicated the size of the correlation coefficient, while red and blue indicated positive and negative correlation, respectively. The square color block in the background indicated the P value of correlation test.
The interrelationships among marker microbiota and their correlation with VFAs
We further investigated the average relative abundance of marker genera, their detection rates, and potential interactions between them. The cumulative abundance of marker genera was as high as 20.27% and the average detection rate was 75%. The highest taxa (Rikenellaceae RC9 gut group) abundance totaled 6.32% and was present in all animals, while the lowest taxa (Suttonella) abundance was 0.01% and was observed in 25.04% of individuals (Fig. 4f and Supplementary Table 6). We observed that the higher cumulative abundance was mainly contributed by the 17 marker bacterial genera with a detection rate of >90% (occurring in at least 1035 or more individuals), which amounted to 19.17%, especially the four bacteria Rikenellaceae RC9 gut group, F082 (5.76%), Erysipelatoclostridiaceae UCG-004 (1.45%), and Lachnospiraceae ND3007 group (1.14%), which appeared in all animals. It is worth noting the potential false positives that can result from extremely imbalanced quantitative proportions in binary analysis models. Hence, caution about those B-features of marker taxa (Moryella, Defluviitaleaceae UCG-011, Veillonellaceae UCG-001 and Anaerovibrio) with detection rates greater than 90% is required, although they may be interesting candidates for further research.
We conducted Pearson’s and Spearman’s correlation analyses and identified 468 (45.70%) and 714 (69.73%) pairs of relationships between marker bacterial genera, respectively (excluding self-pairings). The average correlation coefficients were 0.12 and 0.19, respectively (Fig. 4g). In the significant Pearson’s relationships, the highest correlation coefficient was observed between F082 and Rikenellaceae RC9 gut group at 0.45, while in the significant Spearman’s relationships, it was between Pseudobutyrivibrio and Butyrivibrio at 0.68. We further divided the marker bacterial genus into six modules by hierarchical clustering, and taxa within the modules were highly positively correlated. Modules 1–3 were predominantly positively correlated with FD (83%), whereas modules 4–6 were predominantly negatively correlated (86%). Interestingly, we observed a higher detection rate (87.22% vs. 63.73%) and average abundance (17.6% vs. 2.64%) for modules 4–6 compared to modules 1–3; in particular, module 4 was present in all animals and had a cumulative abundance of 17.6%.
The VFAs, produced through the fermentation process by rumen microbiome, serve as a significant source of energy acquisition for sheep. Consequently, we have also investigated the relationship between the identified marker genera and the VFAs (Fig. 4h and Supplementary Table 7). Among the 224 microbiota-VFA relationship pairs, a total of 117 significant relationship pairs were observed (62 positive and 55 negative correlations; P < 0.05), accounting for 52.23% of the total. Except for Flexilinea, the remaining marker genera were associated with at least one type of VFA. The Ruminococcaceae UCG-001 and Erysipelatoclostridiaceae UCG-004 were associated with all VFA. The VFA with the highest number of significant correlations with the marker genera was Isobutyric acid proportion (26), followed by Isovaleric acid proportion (22), Acetic acid proportion (18), and Propionic acid proportion (16); while the least number of taxa were significantly associated with Total-VFA content, with only 11 relationship pairs. Interestingly, we observed that the direction of correlation of modules 1–3 with VFA is reversed from the direction of correlation of modules 4–6 with VFA. For example, the significant correlation between Acetic acid proportion and marker taxa is positive in Modules 1–3 and negative in Modules 4 to 6, and the significant correlation between Isobutyric acid proportion, Isovaleric acid proportion and marker taxa is negative in Modules 1–3, and positive correlation in Modules 4 to 6. In addition, FD phenotypes were most associated with Valeric acid proportion and Isovaleric acid proportion in the present animal population, with the presence of specific marker genera associated with these two VFAs in each module at the 0.1% significant level.
The effect of host genetics on fat deposition-associated microbiota
To study the effect of host genetics on marker microbiota, we performed mbGWAS on 38 microbial features associated with two FD representative indicators, using 15,252,471 SNPs from 1150 individuals (Supplementary Table 8). These 38 microbial features were identified by the two-part model MWAS after 5000 permutation tests, including 10 features with less than 90% detection rate out of 14 B-features and 28 Q-features. At the genome-wide significance level (P < 3.28E–09), we observed 77 associations of 76 genomic variants with 15 microbial features (including only one B-feature: Lachnospiraceae FD2005). Interestingly, among the above SNPs, we observed one SNP showing pleiotropy (a T-substitution to C variant, named here as Chr15:43988256 since no Reference SNP ID was assigned), associated with two microbial features. These two microbial features were all from the marker genus Lachnospiraceae FD2005, meaning that both B and Q-features of this taxon are associated with Chr15:43988256. Moreover, Chr15:43988256 was also the lead SNP in mbGWAS for these two bacterial features. At a more lenient suggestive significance threshold (P < 6.56E–08), we investigated a total of 401 associations from 397 SNPs and 33 microbial features, of which 391 associations were from 26 Q-features and a further 10 associations were from 7 B-features (Supplementary Table 9). As well, we also observed pleiotropy for four SNPs including Chr15:43988256, particularly Chr13:8302677 (within MACROD2 gene; C/T) and Chr16:19845722 (T/A), which were associated with all two features (B and Q-features) of genus Ruminococcaceae CAG-352 and Alloprevotella, respectively. Also, Chr21:52930312 was associated with all of the Q-features of Butyrivibrio and RF39.
The causal relationship between rumen microbiota and sheep fat deposition
To investigate the potential causal relationship between rumen microbiota and fat deposition in sheep, we attempted to further assess potential causal relationship using bi-directional MR. In our study, a total of 38 microbial features were included in the MR analysis (Supplementary Table 8). To mitigate potential effects of the winner’s curse, all effect sizes were corrected (Supplementary Table 9). Following rigorous IV screening, we retained 1031 (all 38 features), 249 (only 21 features), and 56 (only 8 features) non-redundant SNPs associated with microbial features for forward MR analysis using three threshold lines of 1E–06, 6.56E–08, and 3.28E–09, respectively. Consistently, we also used the above 3 thresholds to identify 44 (NTotal-FW = 20; NTotal-RFW = 26), 10 (NTotal-FW = 5; NTotal-RFW = 6), and 1 (only from Total-RFW) non-redundant SNPs associated with the two FD-traits for reverse causal inference, respectively. All SNPs had F-statistic values (Fval) above 10, ranging from 10 to 1,723, indicating a low risk of weak instrumental bias.
In the forward MR analysis, we identified eight potential causal relationships (Bonferroni-corrected P < 0.05) between five microbial features and two FD-traits (Fig. 5). Notably, the Q-features of Butyrivibrio and p-2534-18B5 gut group exhibited negative causal effects on all FD-traits (Fig. 6a, b, e, f). Additionally, the Q-features of Prevotellaceae UCG-003 and Pseudobutyrivibrio also showed negative causal associations with Total-FW (Fig. 6g, h). Specifically, we found that the Q-features of Olsenella were positively causally associated with all FD-traits (Fig. 6c, d). An increase of one standard deviation in Olsenella abundance led to approximately a 1.13-fold increase in Total-FW (OR = 1.13, 95% CI: 1.08–1.18) and a 1.17-fold increase in Total-RFW (OR = 1.17, 95% CI: 1.09–1.26). The estimated effects of the causal associations between microbial features and FD-phenotypes were consistent in direction and magnitude across different methods, indicating the reliability of the results. In the backward MR analysis (with FD-phenotype as exposure and microbial features as outcomes), we did not detect potential causal effects of FD-phenotype on the five microbial features (Bonferroni-corrected P > 0.05) similar to those identified in the forward MR analysis. Interestingly, we found some evidence for a causal association of sheep FD leading to changes in the rumen microbiota in the backward MR (Bonferroni-corrected P < 0.05; Fig. 5). Total-RFW was negatively causally associated to both B and Q features of Flexilinea, which means that an increase in Total-RFW causes a decrease in abundance and absence of Flexilinea (Fig. 6i, j). A positive causal association between Total-RFW and B-features of Suttonella was demonstrated (Fig. 6k). Here, we also found no evidence of a potential causal effect of these three microbial features on the FD-phenotype. In the leave-one-out analysis, no single SNP strongly drove the overall estimates (Supplementary Tables 34–43). Among the identified causal relationships, we observed heterogeneity and horizontal pleiotropy only in the causal association between Butyrivibrio and two FD-traits (Supplementary Tables 11–12, 15–16, 19–20, 24–25, 28–29, 32). Therefore, we selected the results of the MRE-IVW model. Furthermore, we also compared the MR results which were obtained with IVs at different association thresholds (including 6.56E–08, and 3.28E–09) and found that the relationship between Butyrivibrio and Total-FW had a consistent direction (Bonferroni-corrected P < 0.05). The full MR result sets for both instrumental variable selection and MR analysis were reported in the Supplementary Tables 10–43.
Discussion
Emerging evidence suggests that rumen microbiota plays a crucial role in sheep fat deposition. The metabolites generated by the rumen microbiome serve as signals or substrates that modulate the host’s absorption, storage, and energy acquisition, thereby participating in fat metabolism. However, the causal relationships of rumen microbial taxa and FD remain elusive. Here, we characterized 11 fat deposition traits and rigorously determined a series of rumen microbiota associated with two FD representative indicators (Total-FW and Total-RFW) in sheep. We identified genomic variations associated to marker microbes and employed one-sample bi-directional MR to unveil 11 causal relationships between rumen microbial features and two FD indicators, indicating the potential utility of microbial interventions in fat deposition. While mechanistic studies with rumen fluid transplantation and direct-fed microbials have been popular, our data-driven analysis underscored the relevance of rumen microbes that have not yet been widely cultured and characterized for reducing sheep FD, which may be particularly relevant to East Asia undergoing a transition from grazing to large-scale intensive sheep production (house feeding) system.
Fat deposition in sheep is not limited to a specific site, but rather exists in multiple sites such as the visceral fat44. The amount and distribution of FD in different sites vary, making it difficult to evaluate the overall level of FD45. Therefore, it is necessary to consider the FD in multiple sites comprehensively. The BMI was initially designed as an indicator to assess obesity status and health risks in humans. Due to its low cost, simplicity, and non-invasive nature, it has gradually been introduced into the comprehensive evaluation of whole-body FD in sheep46,47. However, due to significant physiological and biological differences between sheep and humans, the limitations of BMI in assessing sheep FD have become apparent. Sheep exhibit substantial differences in body size, skeletal structure, muscle mass, and FD compared to humans questioning the stability and accuracy of BMI as an evaluation tool. Similar shortcomings in assessing obesity using BMI have been observed in human studies as it fails to accurately distinguish between lean muscle and fat48. Therefore, direct measurements of total fat mass and its proportion in total body mass are considered a more authentic assessment of whole-body fat composition and a more accurate standard for measuring FD48. In recent years, researchers have begun using new technologies and methods to evaluate animal’s body-fat levels such as dual-energy X-ray absorptiometry technology, bioelectrical impedance analysis, 3D body scanning, magnetic resonance imaging, and computed tomography scanning. These novel technologies can more precisely measure the levels of FD in various animal tissues and provide a more comprehensive assessment of whole-body FD levels.
m2 estimation is a quantitative genetic concept introduced in microbiome analysis, which is a tool used to quantify the impact of gut microbiota abundance on host phenotypes49. In humans, Fu et al.33 used cross-validation and additive model regression to evaluate the contribution of gut microbiota. In addition, Difford et al. constructed an MRM based on the structural features of metagenomics and 16S data and used variance component analysis to estimate the proportion of phenotype variation inferred from gut microbiota, drawing on the estimation method of host SNP heritability50,51. Currently, this method has been widely applied in farm animal research, including complex traits in pigs, chickens, and cattle30,31,52. Our m2 estimates suggest that the rumen microbial communities have an impact on various FD-phenotypes, but the extent of their contributions shows distinct tissue specificity.
Although the association between rumen microbial communities and FD-traits in sheep has been extensively studied in recent years, MR analysis, as a valuable method for assessing causal relationships, is being attempted for the first time in sheep. In human studies, obesity is defined as the excessive accumulation of body fat. A substantial number of MR studies have already identified causal relationships between human obesity and gut microbiota, particularly using measures such as fat mass and body fat percentage (reflecting total fat accumulation in adipose tissue) as phenotypic assessment indicators for obesity22,23. This insight from human studies has inspired our current research, suggesting that similar tasks in the field of animal science could be a significant direction for MR studies. Conducting MR analysis on gut microbiota and productivity phenotypes may stimulate further mechanistic and intervention studies.
The microbial genera Butyrivibrio, Pseudobutyrivibrio, Prevotellaceae UCG-003, and p-2534-18B5 gut group have been shown to have a protective effect against excessive FD, while Olsenella may be a potential risk factor for FD. Butyrivibrio and Pseudobutyrivibrio, which evolved from a common ancestor53, have been previously reported to be associated with intramuscular fat phenotype54. As dominant genera in the rumen ecosystem, they form an important group of butyrate-producing bacteria, playing a crucial role in acquiring energy from food in the rumen55. The carbohydrate-active enzymes of these taxa mainly belong to the glycoside hydrolase families 2, 3, 5, 13, and 43, with many intra-family isoforms, contributing to metabolic plasticity and resilience in response to dietary changes55. Prevotellaceae UCG-003 is important in regulating host metabolic functions, actively participating in hemicellulose degradation and playing an important role in protein and starch degradation56,57. Similarly originating from the phylum Bacteroidota, the p-2534-18B5 gut group may also be involved in protein degradation. Its relationship with FD is a new finding, and its function in the gastrointestinal tract of ruminants is not yet clear. Olsenella is also a dominant rumen bacterial genus, producing lactic acid and succinic acid, which are related to fatty acids and feed efficiency in sheep58,59. Studies in chickens60, pigs61, mice62, and humans23 support the positive correlation of this taxa with host FD. Particularly in human gut microbiota MR studies investigating the relationship between Olsenella and body fat percentage23, a positive forward causal relationship with body fat percentage was observed, consistent with the MR results of the current study. The interaction between Olsenella and host fat metabolism may have similar biological mechanisms across different species, indicating evolutionary conservation. Although a causal relationship between Bifidobacterium and two FD-traits was not observed at the current corrected MR P value. (PQ feature→Total-FW: 0.02, PB feature→Total-FW: 0.18, PQ feature→Total-RFW: 0.007, PB feature→Total-RFW: 0.22), the association of all features of this taxon with all traits is encouraging. This outcome may highlight the efficacy of the present study and the generalizability of our findings. Bifidobacterium is ubiquitously present in the rumen of ruminant animals, and probiotic strains of Bifidobacterium have been developed for use in humans to control FD63. These applications have begun to extend to farm animals, including sheep64,65.
The reverse MR revealing the impact of Flexilinea is an intriguing discovery. An increase in individual adiposity leads to a decrease in the abundance of this taxon, as well as leading to disappearance. A randomized controlled trial (RCT) in sheep confirmed a reduction in Flexilinea abundance in response to higher dietary metabolic loads66. Another reverse causality involves Suttonella. A RCT study indicates that severe feed restriction leads to a decrease in sheep body weight (with the accompanying loss of body fat). This decrease in body weight is also accompanied by a reduction in ruminal Suttonella abundance67. Furthermore, the feed restriction increases the relative abundance of gene families involved in lipid metabolism67. These RCT provide indirect support for our observed potential causal relationships, demonstrating that there may be underlying biological facts behind inferred causal relationships.
This study has several limitations. (1) The restriction in the number of SNPs may have led to IV bias. A stringent threshold was applied during the SNP selection process, resulting in the exclusion of certain taxa from the MR analysis. To encompass a broader range of SNPs, we moderately relaxed the significance threshold for the association between instrumental variables and exposure factors. Nevertheless, the F-statistics values for the SNPs included in this study were all greater than 10, which effectively mitigates the potential weak instrument bias in MR analysis. (2) Following the findings by Sanna and colleagues in human studies68, current sample size is not sufficient to fully capture the genetic effects of the majority of the rumen microbiota in sheep. (3) Certain findings from our present research on Hu sheep, the predominant breed in intensive sheep farming in China, may not be generalizable to other breeds. Additionally, the housing of all animals in individual pens hinders the transfer of gut microbiota between individuals, complicating a comprehensive understanding of sheep microbial diversity. (4) The study’s exclusive use of male samples introduces a gender bias, and the universality of these results in the female population requires further investigation. Future research will employ a variety of methods, including multi-omics analysis, confounding factor studies, and even wet-lab experiments, to more comprehensively elucidate the interplay between the rumen microbiota and FD.
In conclusion, through estimation of microbiability in a large-scale homogeneous population, as well as MWAS, mbGWAS, and MR analyses, we observed that specific rumen microbial taxa are potential causal agents for sheep FD. Further investigation is needed in the future to delineate the biological mechanisms underlying these presumed causal relationships. Our data-driven methodologies highlight the immense potential of mbGWAS and MR in attaining a comprehensive understanding of the microbiome in agricultural animals. These approaches are adept at elucidating the intrinsic mechanisms and at informing interventions from the perspective of gastrointestinal microbiota, thereby enhancing the health status and growth performance of animals.
Data availability
All results data analysed during this study are included in this published article and its supplementary information files. Individual-level raw data including host genetics and 16S rRNA sequencing data have been uploaded to the Genome Sequence Archive (GSA) database (https://ngdc.cncb.ac.cn/gsa/). Whole genome resequencing data numbers are CRA019576 (https://ngdc.cncb.ac.cn/gsa/s/4Ae3cx2L) and CRA019589 (https://ngdc.cncb.ac.cn/gsa/s/ueg474Z4), and 16S rRNA sequencing data accession number is CRA019574 (https://ngdc.cncb.ac.cn/gsa/s/5H8V3Xu2). Any other questions can be directed to the corresponding author (wangweimin@lzu.edu.cn).
Code availability
The underlying code for this study is available in GitHub and can be accessed via this link https://github.com/Zukunft-Zhang/MendelRookie.v2. Additional visualization code can be obtained from the corresponding author (wangweimin@lzu.edu.cn) upon reasonable request.
Change history
14 December 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41522-024-00620-7
References
Sakers, A., De Siqueira, M. K., Seale, P. & Villanueva, C. J. Adipose-tissue plasticity in health and disease. Cell 185, 419–446 (2022).
Malik, V. S., Willett, W. C. & Hu, F. B. Global obesity: trends, risk factors and policy implications. Nat. Rev. Endocrinol. 9, 13–27 (2013).
Li, X. et al. Whole-genome resequencing of wild and domestic sheep identifies genes associated with morphological and agronomic traits. Nat. Commun. 11, 2815 (2020).
Xu, Y. X. et al. Whole-body adipose tissue multi-omic analyses in sheep reveal molecular mechanisms underlying local adaptation to extreme environments. Commun. Biol. 6, 159 (2023).
Wood, J. et al. Fat deposition, fatty acid composition and meat quality: A review. Meat Sci. 78, 343–358 (2008).
Murphy, T., Loerch, S., McClure, K. & Solomon, M. Effects of restricted feeding on growth performance and carcass composition of lambs. J. Anim. Sci. 72, 3131–3137 (1994).
Zhang, X. et al. Association of residual feed intake with growth and slaughtering performance, blood metabolism, and body composition in growing lambs. Sci. Rep. 7, 12681 (2017).
Zhang, X. et al. Effect of feed efficiency on growth performance, body composition, and fat deposition in growing Hu lambs. Anim. Biotechnol. 34, 183–198 (2023).
Liu, J., Li, H., Zhu, W. & Mao, S. Dynamic changes in rumen fermentation and bacterial community following rumen fluid transplantation in a sheep model of rumen acidosis: implications for rumen health in ruminants. FASEB J. : Off. Publ. Federation Am. Societies Exp. Biol. 33, 8453–8467 (2019).
Fu, L. et al. Inoculation of newborn lambs with ruminal solids derived from adult goats reprograms the development of gut microbiota and serum metabolome and favors growth performance. J. Agric. Food Chem. 72, 983–998 (2024).
Mizrahi, I., Wallace, R. J. & Moraïs, S. The rumen microbiome: Balancing food security and environmental impacts. Nat. Rev. Microbiol. 19, 553–566 (2021).
Zhang, Y. et al. Characterization of the rumen microbiota and its relationship with residual feed intake in sheep. Anim.: Int. J. Anim. Biosci. 15, 100161 (2021).
Alexander, C., Swanson, K. S., Fahey, G. C. & Garleb, K. A. Perspective: Physiologic importance of short-chain fatty acids from nondigestible carbohydrate fermentation. Adv. Nutr. (Bethesda, Md.) 10, 576–589 (2019).
Newbold, C. J. & Ramos-Morales, E. Review: Ruminal microbiome and microbial metabolome: Effects of diet and ruminant host. Anim.: Int. J. Anim. Biosci. 14, s78–s86 (2020).
Krause, T. R. et al. The relationship between the rumen microbiome and carcass merit in Angus steers. J. Anim. Sci. 98, https://doi.org/10.1093/jas/skaa287 (2020).
Kim, M., Park, T., Jeong, J. Y., Baek, Y. & Lee, H. J. Association between Rumen microbiota and marbling score in Korean Native Beef Cattle. Animals: An Open Access Journal from MDPI 10, https://doi.org/10.3390/ani10040712 (2020).
Zhang, Y. et al. Exploring the ruminal microbial community associated with fat deposition in lambs. Animals : An Open Access Journal from MDPI 11, https://doi.org/10.3390/ani11123584 (2021).
Xiang, J. et al. A comparative analysis of carcass and meat traits, and rumen bacteria between Chinese Mongolian sheep and Dorper × Chinese Mongolian crossbred sheep. Anim.: Int. J. Anim. Biosci. 16, 100503 (2022).
Zhang, Y. M. et al. Relationships between rumen microbes, short-chain fatty acids, and markers of white adipose tissue browning during the cold season in grazing Mongolian sheep (Ovisaries). J. Therm. Biol. 110, 103386 (2022).
Lawlor, D. A., Harbord, R. M., Sterne, J. A., Timpson, N. & Davey Smith, G. J. S. I. M. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat. Med. 27, 1133–1163 (2008).
Li, Q., Gao, J., Luo, J., Lin, D. & Wu, X. J. F. I. P. Mendelian randomization analyses support causal relationship between gut microbiota and childhood obesity. Front Pediatr. 11, 1229236 (2023).
Xu, Q. et al. Mendelian randomization analysis reveals causal effects of the human gut microbiota on abdominal obesity. J. Nutr. 151, 1401–1406 (2021).
Wang, X. et al. Exploring causal effects of gut microbiota and metabolites on body fat percentage using two‐sample Mendelian randomization. Diab. Obes. Metab. 26, 3541–3551 (2024).
Teng, J. et al. A compendium of genetic regulatory effects across pig tissues, 1-12 (2024).
Cai, W. et al. The eQTL colocalization and transcriptome-wide association study identify potentially causal genes responsible for economic traits in Simmental beef cattle. J. Anim. Sci. Biotechnol. 14, 78 (2023).
Erwin, S., Marco, G. & Emery, A. E. Volatile fatty acid analysis of blood and rumen fluid by gas chromatography. (1961).
Xia, Y. & Sun, J. in Bioinformatic and Statistical Analysis of Microbiome Data: From Raw Sequences to Advanced Modeling with QIIME 2 and R 65-94 (Springer, 2023).
Endo, H., Umezawa, Y., Takeda, S. & Suzuki, K. J. M. E. Haptophyte communities along the Kuroshio current reveal their geographical sources and ecological traits. Mol. Ecol. 32, 110–123 (2023).
Rajeev, M., Jung, I., Song, J., Kang, I. & Cho, J.-C. J. A. Comparative microbiota characterization unveiled a contrasting pattern of floc-associated versus free-living bacterial communities in biofloc aquaculture. 577, 739946 (2023).
Wen, C. et al. Joint contributions of the gut microbiota and host genetics to feed efficiency in chickens. Microbiome 9, 126 (2021).
Tang, S. et al. Screening of microbes associated with swine growth and fat deposition traits across the intestinal tract. Front Microbiol 11, 586776 (2020).
Boggio, G. M. et al. Host and rumen microbiome contributions to feed efficiency traits in Holstein cows. J. Dairy Sci. 107, 3090–3103 (2024).
Fu, J. et al. The gut microbiome contributes to a substantial proportion of the variation in blood lipids. Circulation Res. 117, 817–824 (2015).
Forde, A., Hemani, G. & Ferguson, J. Review and further developments in statistical corrections for Winner’s Curse in genetic association studies. PLoS Genet 19, e1010546 (2023).
Li, C., Pang, D., Lin, J., Yang, T. & Shang, H. Shared genetic links between frontotemporal dementia and psychiatric disorders. BMC Med. 20, 131 (2022).
Fan, J. et al. Cross-talks between gut microbiota and tobacco smoking: a two-sample Mendelian randomization study. BMC Med. 21, 163 (2023).
Li, P. et al. Causal effects of sedentary behaviours on the risk of migraine: A univariable and multivariable Mendelian randomization study. European journal of pain (London, England), https://doi.org/10.1002/ejp.2296 (2024).
Burgess, S., Butterworth, A. & Thompson, S. G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665 (2013).
Hartwig, F. P., Davey Smith, G. & Bowden, J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 46, 1985–1998 (2017).
Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525 (2015).
Bowden, J., Davey Smith, G., Haycock, P. C. & Burgess, S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314 (2016).
Lawlor, D. A., Harbord, R. M., Sterne, J. A., Timpson, N. & Davey Smith, G. Mendelian randomization: Using genes as instruments for making causal inferences in epidemiology. Stat. Med. 27, 1133–1163 (2008).
Wong, S. H. et al. Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology 153, 1621–1633.e1626 (2017).
Fang, S., Xiong, X., Su, Y., Huang, L. & Chen, C. 16S rRNA gene-based association study identified microbial taxa associated with pork intramuscular fat content in feces and cecum lumen. BMC Microbiol 17, 162 (2017).
Bakhtiarizadeh, M. R., Salehi, A., Alamouti, A. A., Abdollahi-Arpanahi, R. & Salami, S. A. Deep transcriptome analysis using RNA-Seq suggests novel insights into molecular aspects of fat-tail metabolism in sheep. Sci. Rep. 9, 9203 (2019).
Chavarría-Aguilar, L. M. et al. Relationship between body fat depots and body mass index in Pelibuey ewes. 124-126 (2016).
Lourencon, R. V. et al. Effects of nutritional plane at breeding on feed intake, body weight, condition score, mass indexes, and chemical composition, and reproductive performance of hair sheep. Anim. (Basel). 13, 735 (2023).
Xiong, Y. et al. Positive association between body fat percentage and hyperuricemia in patients with hypertension: The China H-type hypertension registry study. Nutr. Metab. Cardiovasc Dis. 31, 3076–3084 (2021).
Wen, C. L., Sun, C. J. & Yang, N. [The concepts and research progress: from heritability to microbiability]. Yi chuan = Hereditas 41, 1023–1040 (2019).
Difford, G. F. et al. Host genetics and the rumen microbiome jointly associate with methane emissions in dairy cows. PLoS Genet 14, e1007580 (2018).
Ross, E. M., Moate, P. J., Marett, L. C., Cocks, B. G. & Hayes, B. J. Metagenomic predictions: from microbiome to complex health and environmental phenotypes in humans and cattle. PLoS One 8, e73056 (2013).
Li, F. et al. Host genetics influence the rumen microbiota and heritable rumen microbial features associate with feed efficiency in cattle. Microbiome 7, 92 (2019).
Pidcock, S. E. et al. Phylogenetic systematics of Butyrivibrio and Pseudobutyrivibrio genomes illustrate vast taxonomic diversity, open genomes and an abundance of carbohydrate-active enzyme family isoforms. Microbial Genomics 7, https://doi.org/10.1099/mgen.0.000638 (2021).
Wang, J. et al. Association between rumen microbiota and marbling grade in Hu sheep. Front Microbiol 13, 978263 (2022).
Shen, J., Liu, Z., Yu, Z. & Zhu, W. Monensin and Nisin affect rumen fermentation and microbiota differently in vitro. Front Microbiol 8, 1111 (2017).
Cui, Y. et al. Whole-plant corn silage improves rumen fermentation and growth performance of beef cattle by altering rumen microbiota. Appl Microbiol Biotechnol. 106, 4187–4198 (2022).
Ramšak, A. et al. Unravelling the genetic diversity of ruminal bacteria belonging to the CFB phylum. FEMS Microbiol. Ecol. 33, 69–79 (2000).
Daghio, M. et al. A diet supplemented with hazelnut skin changes the microbial community composition and the biohydrogenation pattern of linoleic acid in the rumen of growing lambs %J Italian Journal of Animal Science. Ital. J. Anim. Sci. 20, 1256–1263 (2021).
Spillane, C., Waters, S. M., Mcloughlin, S., Claffey, N. & Diskin, M. G. J. F. I. M. Rumen Microbiome Composition Is Altered in Sheep Divergent in Feed Efficiency. Front. Microbiol. 11, https://doi.org/10.3389/fmicb.2020.01981 (2020).
Xiang, H., Gan, J., Zeng, D., Li, J. & Li, H. J. F. I. M. Specific microbial taxa and functional capacity contribute to chicken abdominal fat deposition. Front. Microbiol. 12, 643025 (2021).
Mejicanos, G. A., González-Ortiz, G. & Nyachoti, C. M. J. J. O. A. S. Effect of dietary supplementation of xylanase in a wheat-based diet containing canola meal on growth performance, nutrient digestibility, organ weight, and short-chain fatty acid concentration in digesta when fed to weaned pigs. Journal of ANIMAL SCIENCE, https://doi.org/10.1093/jas/skaa064 (2020).
Wei, B. et al. Modulation of the microbiome-fat-liver axis by lactic acid bacteria: A potential alleviated role in high-fat-diet-induced obese mice. J. Agric. food Chem. 71, 10361–10374 (2023).
Sung, H. K., Youn, S. J., Choi, Y., Eun, S. W. & Shin, S. M. Body fat reduction effect of bifidobacterium breve B-3: A randomized, double-blind, placebo comparative clinical trial. Nutrients 15, https://doi.org/10.3390/nu15010028 (2022).
Krehbiel, C., Rust, S., Zhang, G. & Gilliland, S. Bacterial direct-fed microbials in ruminant diets: Performance response and mode of action. J. Anim. Sci. 81, E120–E132 (2003).
Adjei-Fremah, S. et al. Effect of probiotic supplementation on growth and global gene expression in dairy cows. J. Appl. Anim. Res. 46, 257–263 (2018).
Wang, Q. et al. Effects of dietary energy levels on rumen fermentation, microbiota, and gastrointestinal morphology in growing ewes. Food Sci. Nutr. 8, 6621–6632 (2020).
Hu, F., Xue, Y., Guo, C., Liu, J. & Mao, S. The response of ruminal fermentation, epithelium-associated microbiota, and epithelial barrier function to severe feed restriction in pregnant ewes. J. Anim. Sci. 96, 4293–4305 (2018).
Sanna, S., Kurilshikov, A., van der Graaf, A., Fu, J. & Zhernakova, A. Challenges and future directions for studying effects of host genetics on the gut microbiome. Nat. Genet 54, 100–106 (2022).
Acknowledgements
This study was funded by the National Natural Science Foundation of China (32260818 and 31960653), National Key R&D Program of China (2021YFD1300901), West Light Foundation of the Chinese Academy of Sciences, and Major Science and Technology Special Project of Gansu Province, P.R.China (22ZD6NC069). The funder played no role in study design, data collection, analysis and interpretation of data, or the writing of this manuscript. We gratefully acknowledge all participants of the Dual Ten Thousand Sheep Genome Project for their contributions to this work. All the computation works were supported by the Supercomputing Center of Lanzhou University. The author would like to thank Prof. Congjiao Sun and Prof. Xiaolei Liu for their great support for this study. Chaoliang Wen and Shi Tang provided invaluable help with the coding of the microbiability. The first author, Dr. Yukun Zhang, wants to thank his parents and girlfriend for their continuous support. Finally, the project is indebted to the open-source community and all the contributors to the open-source code libraries that the algorithm implementation has been built on.
Author information
Authors and Affiliations
Contributions
Y.K.Z., W.M.W. and F.D.L conceived and designed the study. W.M.W., Y.K.Z., X.X.Z., C.L., H.B.T., X.X.W., C.C.L., D.Y.Z., Y.Z., X.L.L., J.B.C., L.M.Z., D.X., X.B.Y. and F.D.L. were responsible for sample collection. D.X., C.C.L., D.Y.Z., Y.Z., X.L.L., J.B.C., L.M.Z., D.X., X.B.Y. and Y.K.Z. was responsible for the volatile fatty acid determinations. Y.K.Z. integrated host Genome, Microbiome and Phenome data for full bioinformatics analysis and visualization. Y.K.Z. wrote the first draft of the manuscript and responded to review comments. W.M.W., F.D.L, Z.H.J. and Y.K.Z. revised and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All animal experiments and procedures were conducted under the approval and guidance of the Animal Ethics Committee of Lanzhou University (Nos.: 2020-01 and 2021-02).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, Y., Zhang, X., Li, C. et al. Rumen microbiome and fat deposition in sheep: insights from a bidirectional mendelian randomization study. npj Biofilms Microbiomes 10, 129 (2024). https://doi.org/10.1038/s41522-024-00606-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41522-024-00606-5
This article is cited by
-
Altered rumen bacterial flora is associated with increased lipogenesis of adipose tissue in obese dairy cows before calving
Microbiome (2026)
-
A guide to network analysis, multi-omics integration, and applications in livestock microbiome research
World Journal of Microbiology and Biotechnology (2026)
-
Prevotella stercorea increases fat deposition in Jinhua pigs fed alfalfa grass-based diets
Journal of Animal Science and Biotechnology (2025)
-
Expression of LGALS3 and XPNPEP2 genes and their association with growth traits in sheep
Tropical Animal Health and Production (2025)