Abstract
Emerging evidence indicates that oral microbes are closely related to gastric microbes and gastric lesions, including gastric atrophy, intestinal metaplasia and gastric cancer (GC). Helicobacter pylori is a key pathogen involved in GC. However, the increasing prevalence of H. pylori-negative GC and gastric dysbiosis in GC patients emphasize the potential role of other microbial factors. In this review, we discussed the current evidence about the relationship between the oral–gastric microbial axis and oral and gastric health. Epidemiologic evidence indicates that poor oral hygiene is related to greater GC risk. Multiple oral-associated microbes are enriched in the stomach of GC patients. Once colonizing the stomach, oral-associated microbes Streptococcus anginosus and Prevotella melaninogenica, are involved in gastric inflammation or carcinogenesis. Microbial metabolites such as lactate, nitrite, and acetaldehyde promote malignant transformation. The stomach, as a checkpoint of microbial transmission in the digestive tract, is of great importance since the link between oral microbes and intestinal diseases has been emphasized. Still, new technologies and standardized metrics are necessary to identify potential pathogenetic microbes for GC and the core microbiota, interactions, richness, colonization, location and effect (CIRCLE). In the future, oral microbes could be candidates for noninvasive indicators to predict gastric diseases.
Similar content being viewed by others
Oral microbiota and gastric microbiota
The oral cavity has the second most complex microbial community in the human body1. More than 700 oral bacteria have been identified in the Human Oral Microbiome Database at present (HOMD :: Human Oral Microbiome Database, https://www.homd.org, eHOMD V3.1, updated on April 10th, 2023). High biodiversity is present across six representative niches: subgingival plaque, supragingival plaque, tongue and saliva, hard palate, mucosa, and keratinized gingiva. Streptococcus, Neisseria, Prevotella, Haemophilus, and Rothia are highly prevalent at most sites. Other primary genera of the oral microbiota in healthy individuals include Gemella, Porphyromonas, Alloprevotella, Pseudomonas, Treponema and Solobacterium2,3. Specifically, Streptococcus is nearly the most abundant genus in mucosal tissues, occupying 44–66% in the microbiota of hard palate, oral mucosa, and keratinized gingiva. Simonsiella was specifically detected in the hard palate2. In subgingival microbiota, Halomonas, Streptococcus and the anaerobes Actinomyces, Veillonella, Fusobacterium, are enriched in subgingival plaque2,4. Streptococcus and Neisseria have the highest relative abundance in supragingival microbiota5. Salivary microbiota is primarily derived from the tongue mucosa membrane, with the predominance of Streptococcus, Prevotella, Veillonella and Neisseria6.
The oral microbiota interacts extensively with external factors such as oral hygiene, diet, smoking, drinking alcohol, betel nut chewing, etc.7,8. Oral hygiene impacts oral microbiota greatly, maintaining the dental plaque in an immature state with high proportions of Streptococcus7. As for diet, although there is debate about whether it can affect the oral microbiota, some human studies indicated that diet affects the composition and diversity of the oral microbiota8. Exposure to nitrates, mainly from vegetables, increased oral health-associated Neisseria and Rothia and suppressed oral disease-associated Prevotella, Veillonella and Streptococcus9. Fermented foods can also transfer lactic acid bacteria to the oral cavity transiently in rodent models10. Lifestyles including drinking and smoking are also important factors. Drinking and smoking lead to lower species richness and altered composition of the oral mucosal microbiota, such as decreased Neisseria abundance11. Smoking is associated with α and β diversity in the upper gastrointestinal tract microbiome12. Several studies found the relative abundance of Lactobacillus is positively associated with smoking13. Bacteroides, Fusobacterium spp., Dialister invisus and Megasphaera micronuciformis are enriched in smokers’ oral microbiota12,14. Overall, the oral microbial changes affected by tobacco and alcohol are highly variable, probably due to differences in sampling sites, consumption type and frequency of tobacco and alcohol. The influence of betel nut chewing has also been initially revealed. The relative abundance of Streptococcus infantis was nearly four-fold higher in current betel nut chewers compared to past/never chewers, and that of Streptococcus anginosus was 16-fold higher in chewers with oral premalignant lesions15.
In healthy individuals, there are similar gastric compositions between the antrum and corpus, with a similar global microbiota community structure16. A systematic review of reports from the past half-century revealed that the most consistently detected genera of the gastric microbiota include Fusobacterium, Streptococcus, Haemophilus, Neisseria, Prevotella and Veillonella17, which are also highly prevalent in the oral cavity. Whether Helicobacter pylori is part of the healthy gastric microbiota remains controversial since this gastric carcinogen is also widely detected in healthy populations. It has been found in about half of the world’s population, but its prevalence and enrichment vary depending on demographic features, location and sanitation standards16,18. The prevalence of H. pylori in children has fallen below 10% in some developed areas, such as Germany, Japan, Korea, Taiwan and Hong Kong19. However, the incidence is still higher in rural areas and less economically developed regions. Different testing methods and participant criteria also cause heterogeneity in the results. Individuals infected with H. pylori exhibit distinct gastric ecosystems, which usually have lower α diversity than those without H. pylori20.
Among the oral, nasal, gastric and pulmonary microbiota within the same individual, the oral and gastric microbiota are most similar in bacterial composition and diversity, illustrating the microbial continuity from the mouth to stomach in healthy adults21. Notably, in the established oral–gut microbial axis, oral microbes migrate mainly through two pathways: the enteral route and the hematogenous route22,23. In this regard, it is possible that gastric microbiota could be affected by oral microbes since the stomach is a necessary stop of the enteral route. Oral microbes are the source of the microbiota in downstream organs and continuously seed the gastrointestinal tract during eating and swallowing. Vomiting and gastroesophageal reflux allow bacteria to travel retrogradely from the gastrointestinal tract to the oral cavity. This finding is the foundation of the correlation between oral and gastric microbiota under both physiological and pathological conditions. Since we know very little about the role of viruses and fungi in mouth-stomach relations, this review mainly focuses on bacteria.
Oral microbial involvement in altered gastric conditions during gastric carcinogenesis
Oral microbes and precancerous gastric lesions
Gastric microbiota shows a decrease in bacterial diversity from non-atrophic gastritis to GC in most cases, although some studies have reported contrasting findings24. In particular, the relative abundance of oral-associated microbes significantly altered in gastric microbiota of several precancerous stages, including atrophic gastritis (AG) and intestinal metaplasia (IM). Decreased diversity and distinct microbial compositions already exist in atrophy stages, and more co-occurrences of oral bacteria in the stomach occur as the composition of samples shifts away from the normal network25. Gastric juice analysis has shown that oral microbes Porphyromonas gingivalis, Campylobacter gracilis, and Granulicatella elegans were enriched in AG samples compared with non-AG samples, suggesting that oral pathogens may be associated with AG26. In H. pylori-eradicated individuals, the duration and development of gastric atrophy and IM are associated with oral microbes Peptostreptococcus, Streptococcus, Parvimonas, Prevotella, Rothia and Granulicatella27. However, some oral-associated microbes, including Streptococcus mutans, Streptococcus parasanguinis, and Streptococcus sanguinis, were depleted in both the oral and gastric microbiota of the IM28.
Human epidemiological studies connecting oral diseases to GC
Gastric cancer (GC) is a highly prevalent and lethal cancer that ranks fifth in incidence and fourth in mortality globally, accounting for one in thirteen cancer-related deaths. Notably, increased GC incidence among young adults has been observed in both low-risk and high-risk countries, which may be related to gastric microbiota dysbiosis29. Chronic H. pylori infection is the principal cause of noncardia GC and a contributing factor for cardia GC. Nevertheless, accumulating evidence suggests that microbial carcinogenic effects cannot be entirely attributed to H. pylori and that other microbes likely play a role in GC. In the USA, the prevalence of H. pylori-negative GC increased from 50% in 2007–2010 to 70% in 2015–201830,31. In other developed countries, however, the proportion is less impressive (24.7% in Germany, 14% in Italy, 14.2% in Japan, and 4% in South Korea)31,32,33,34,35,36. Less than 3% of H. pylori-infected people develop GC, and H. pylori eradication does not eliminate the long-term risk for GC development27. Toothbrushing once or less per day, tooth loss and denture-associated lesions are risk factors for GC, and irregular flossing is an effective GC risk predictor, suggesting that poor oral health may indicate higher GC risk37,38,39. A study involving 238 patients revealed that self-reported periodontitis was linked to a 52% greater risk of gastric adenocarcinoma (HR 1.52, 95% CI 1.13–2.04)40. Periodontitis was also closely related to a greater risk of GC mortality (HR 4.288, 95% CI 3.969–4.632)41. However, oral microbes and their correlation with GC have not been conclusively established. The positive relationship may be a consequence of shared risk factors, upstream drivers or the nonspecific systemic inflammatory response caused by periodontal inflammation42,43.
Clinical evidence showing connections of the oral microbiome and oral health to GC
The search strategy for the studies in Supplementary Table 1 is as follows. We searched the PubMed database for studies investigating the gastric microbiota of GC patients or patients with precancerous lesions from January 2017 to September 2024. The MESH terms search strategy is ((“Bacteria”[Mesh]) OR “Microbiota”[Mesh]) AND (“Stomach Neoplasms”[Mesh]), and supplemental textwords searching (microbiome OR microbe) AND (“Stomach Neoplasms”[Mesh]). The inclusion criteria are as follows: 1) gastric samples from GC patients for sequencing, 2) original studies of 16S rRNA sequencing or metagenome analysis, 3) studies published in English. There is no restriction on sample size or region. Duplicate studies based on the same data were excluded (Supplementary Fig. 1).
The gastric microbiota differs in terms of diversity, composition, species relative abundance, interactions, and metabolism based on disease status and temporal and spatial distribution (Supplementary Table 1). Twelve of the 32 studies reported decreased α diversity, eight studies reported increased α diversity, and four studies reported no significant difference. Another four studies could not be categorized into any of the above groups due to complex grouping. The following six genera were most frequently reported to be enriched in the GC microbiota: Streptococcus, Prevotella, Lactobacillus, Fusobacterium, Veillonella, and Helicobacter (Table 1). Most of these genera are oral-associated. Despite the limited amount of species-level research, we have identified several oral-associated species that make up the GC microbiota, which are Streptococcus anginosus, Streptococcus mitis, Streptococcus oralis, Prevotella melaninogenica, Prevotella oris, Prevotella intermedia, Fusobacterium nucleatum, and Veillonella parvula. At strain level, Aggregatibacter_segnis.t_GCF_000185305 and Porphyromonas_endodontalis.t_GCF_000174815 are enriched in GC, the latter being an oral commensal or opportunistic pathogens44. Strain level information is very limited as it relies on high-resolution sequencing methods, while the current study is dominated by short-reading 16S rRNA sequencing, especially V3-V4 region amplification. Besides metagenomics, full-length 16S sequencing is recommended because it probably provides confidence and taxonomic resolution at species and strain levels45. Other options such as DNA microarray may also be applied for high-throughput detection at the strain level in the future, as it was reported in cyanobacteria46. Defining the core microbiota often requires filtering the raw data based on taxon prevalence and relative abundance47. Here, these GC-enriched microbes identified through the literature review may be involved in the disease-associated core microbiota.
A meta-analysis of nine studies reported that gastric carcinogenesis is accompanied by shifts in the gastric microbiome accompanied by decreased microbial diversity and enrichment of oral microbes. Oral-originating bacteria have greater diversity and relative abundance in GC than in gastritis and IM after excluding H. pylori sequences48. Another meta-analysis revealed that a single genera is less effective as an universal diagnostic marker, but five genera assembles, Streptococcus, Peptostreptococcus, Selenomonas, Pseudomonas, and Prevotella, can effectively distinguish GC patients from non-GC patients in oral, gastric and fecal samples49. The oral-associated microbes Peptostreptococcus stomatis, S. anginosus, Parvimonas micra, Slackia exigua and Dialister pneumosintes have significant centralities identified by weighted node connectivity scores in the GC interaction network and have been confirmed to distinguish GC from superficial gastritis (SG)50. Microbial diversity is significantly lower in GC biopsies than in nonmalignant tissue, with Helicobacter, Lactobacillus, Streptococcus, and Prevotella showing significant enrichment even after adjusting for age, race and sex51. Lactobacillus is significantly more abundant in the GC mucosa and gastric fluid than in the mucosa and gastric fluid of SG patients, indicating its valuable diagnostic potential52.
Currently, most evidence remains at the relevance level rather than revealing causal relationships. The evidence from sequencing indicates that oral-associated microbes could be a marker of altered gastric conditions. However, whether oral-associated microbes have an impact on gastric carcinogenesis or development needs further evidence from mechanistic studies. Another issue is that the sequencing results are quite inconsistent. There are both positive and negative links between oral microbes and GC. The discrepancy of the studies is probably attributed to the populations, samples, study types, materials and methods, and data analysis used53. For example, a study in Portugal revealed that Neisseria, Streptococcus and Prevotella were inversely related to GC54, while these microbes were enriched in GC in many Asian studies (Supplementary Table 1).
Different classifications and staging seem to have corresponding gastric microbiota features. Lauren’s classification divides gastric adenocarcinoma into two histological subtypes: intestinal type and diffuse type55. The intestinal type is more prevalent, mainly influenced by environmental factors, and has better prognosis. The diffuse type is affected by genetic factors and has a worse prognosis. Regarding Lauren’s classification, lower diversity and species richness were found in the diffuse type compared with the intestinal type, but no significant differences at the genus level were observed56. Similarly, no significant difference in the composition of the gastric microbiota was found in the same gastric microhabitat among different Lauren’s classifications57. In addition, Fusobacterium nucleatum detected in GC tissue is associated with significantly worse overall survival in patients with diffuse-type rather than intestinal-type58. In gastric cardia adenocarcinoma patients, the α diversity is different in stage T2-T4, and the relative abundance of Helicobacter decreased and Prevotella increased with the more advanced tumor stage59. Tumor N stage (lymph node status) also affects gastric microbial diversity. Higher microbial diversity probably connects with a lower risk of lymph node metastasis in GC patients60. In gastric juice samples, Helicobacter is more correlated with early GC (stage I-II), and Streptococcus is more correlated with advanced GC (stage III-IV)61.
Oral health status, oral hygiene and periodontal pathogen burdens are correlated with GC. The combined results of three cohorts containing Asian people, African Americans, and European Americans showed that enrichment of Neisseria mucosa and Prevotella pleuritidis in mouth rinse samples was related to GC. The typical periodontal pathogen P. gingivalis is associated with increased GC risk in Asian people62. Other oral microbes, Treponema denticola, Tannerella forsythia and Actinobacillus actinomycetemcomitans, colonizing dental plaque are predictors of gastric precancerous lesions, including AG, IM and dysplasia38.
The tongue is another oral niche with high species richness. From normal to precancerous, early-stage GC and late-stage GC, the abundance of Alloprevotella increased gradually, while the abundance of Veillonella decreased gradually63. Thus, the altered tongue coating microbiota may be implied the gastric conditions. The tongue-coating microbiome-based and tongue image-based models are stable and effective, and promising for GC diagnosis. In addition, core shared oral bacteria between the tongue coating and gastric mucosa are associated with gastric disease and H. pylori infection status, which indicates that microbial dysbiosis in H. pylori-positive GC patients may be attributed to ectopic oral microbe colonization64.
Rodent models exploring the role of oral microbes in GC
Although H. pylori infection accelerated gastritis and gastrointestinal intraepithelial neoplasia, the pathogenic effect was diminished in the absence of commensal flora. This indicates the potential role of non-H. pylori microbes in promoting neoplasia in achlorhydric stomachs65. In germ-free insulin/gastrin transgenic mice, Streptococcus salivarius coinfection with H. pylori induced a higher gastric pathology score and more proliferating epithelial cells than H. pylori monoinfection66. Beyond coinfection with H. pylori, the role of other microbes in gastric pro-inflammation and premalignant changes has also gained some support. An in vivo study revealed that gastric mucosal microbiota transplantation from GC and IM patients promoted premalignant changes in the germ-free mouse stomach. This was mainly attributed to non-H. pylori microbes, as Helicobacter rarely colonize the gastric mucosa of recipients67. In specific pathogen-free mice, the oral pathogen P. melaninogenica promotes inflammatory cell infiltration in gastric tissues and activation of the STAT3 signaling pathway68.
Potential mechanisms of oral–gastric microbial correlation
In this section, we describe the in vivo and in vitro studies on potential mechanisms of the oral–gastric microbial correlation. These factors include microbial survival and colonization of the stomach, microbial interactions, host–microbe interactions, and their oncogenic effects.
Survival and colonization strategies of oral microbes in stomach
The mouth is a reservoir of microorganisms that constantly complement the gastric flora. An individual swallows approximately 1500 billion oral microbes per day. Once the oral microbes reach the stomach, they will select and adapt to certain niches in the stomach. However, unfavorable factors such as persistent extremely low pH in the stomach reduce the live bacterial load to millions69. This filtering effect may explain why the upper gastrointestinal tract has greater richness and heterogeneity than the lower gastrointestinal tract16. There are several possible survival mechanisms for oral microbes. First, alkali-producing oral microbes neutralize gastric acid to shape a suitable microenvironment for survival70. Second, H. pylori changes the gastric microenvironment and helps them survive. H. pylori has a selective advantage in the stomach, mainly through the release of extracellular urease, which breaks down urea and neutralizes gastric acid. H. pylori can penetrate the mucus layer to reach epithelial cells and colonize adjacent glands, while other microbes are mainly distributed above the mucus layer in the healthy stomach66. Favorable circumstances, such as mucosal layer impairment and gastric acid neutralization, are thought to contribute to ectopic oral bacterium colonization in the stomach, resulting in potential pathogenic effects52. Some diseases may cause a decrease in stomach acid production, such as autoimmune AG and H. pylori-induced AG. As expected, the gastric microbiota of autoimmune AG has high diversity and enriches Streptococcus than the normal stomach. While H. pylori-induced AG showed lower bacterial abundances and diversity, with only increased proportions of Helicobacter71. This may be due to a combined factor of H. pylori infection and reduced gastric acid. Another gastric acid inhibitor is proton pump inhibitor (PPI) use, a first-line therapy in NSAID-related ulcers and gastroesophageal reflux disease. Zhou et al concluded that PPI users have altered gastric microbiota with higher α diversity and increased abundance of Streptococcus72. Although the origin of Streptococcus enriched in the stomach of PPI users is unknown, a recent randomized controlled trial revealed that PPI promotes oral-originated Streptococcus, especially Streptococcus anginosus, translocate to gut microbiota73. Accordingly, PPI may also impact the translocation of the oral microbiota to the stomach and then to the guts. However, whether gastric acid reduction has any effect is controversial because some diseases leading to achlorhydria, such as autoimmune gastritis, are not related to GC74. Also, it is uncertain whether PPI-induced changes in gastric flora contribute to higher GC risk, since new ideas suggest that there may be reverse causality between PPI use and GC, with short-term PPI intake (6 months to 3 years) being associated with higher GC risk, but long-term use of PPI (longer than 3 years) presenting no significant association with GC75. Third, under certain circumstances, the host presents an oral microbe-affinity mucin phenotype. Mucin mediates bacterial adhesion to epithelial cells. The intestinal mucin phenotype is prevalent in advanced GC and favors proinflammatory oral microbes forming strong co-occurrence networks. The oral microbes Neisseria, Prevotella and Veillonella present a high affinity for MUC13-overexpressing tumors and are involved in shaping the community structure76. In conclusion, the stomach is an important checkpoint for the gastrointestinal migration of oral microbes. It filters out many bacteria, while certain species harbor the gastric environment and thrive.
Gastric co-occurrence and co-exclusion microbial interactions
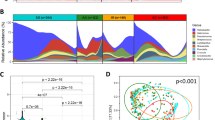
An increased intensity of co-occurrence and co-exclusion microbial interactions was detected among GC-enriched and GC-depleted bacteria during GC progression. The strongest microbial interactions were detected in the GC stage compared with all the other precancerous stages. The probiotics Lactobacillus salivarius and Lactobacillus fermentum exhibited co-occurrence interactions in SG, AG and GC50. An increasing co-occurrence of oral-associated bacteria was observed from normal to early precancerous states in the stomach, while co-exclusion interactions were found between oral and other bacteria25 (Fig. 1).
As gastric lesions develop from inflammation to cancer, the abundance of the dominant colonizer H. pylori decreases, while that of oral-associated microbes increases. Oral-associated microbes co-occur with each other and have complex interactions with other microbes. The direct effects and indirect effects are shown by solid and dashed arrows, respectively. Created with BioRender.com.
It is widely recognized that H. pylori is a gastric carcinogen, but whether non-H. pylori microbes play a role in GC is under debate74. There is evidence supporting that non-H. pylori microbes may promote gastric lesions by coinfecting with H. pylori. S. salivarius seems to have synergistic effects with H. pylori in vivo without changing the expression of host proinflammatory genes, including IL-1β, IL-17A and IFN-γ. In contrast, these genes were suppressed in mice infected with both H. pylori and Staphylococcus epidermidis, which indicates that S. epidermidis probably exerts a protective immunomodulatory effect66. P. gingivalis coincubated with H. pylori produces more gingipains, which enhance the migration of oral keratinocytes mediated by Toll-like receptor 477. The interaction between the gastric microbiota and H. pylori expands the understanding of H. pylori pathogenesis in GC. However, the role of microbes other than H. pylori in GC is unclear74. It is also important to determine the pattern of bacterial symbiosis and to what extent it affects GC78. Cross-species communication in the GC microbiota may be mediated by cross-feeding, quorum sensing, extracellular vesicles, diffusible signaling factors, etc., and these phenomena warrant further investigation.
Host responses to microbial invasion
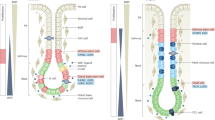
Host–microbe interactions are key drivers of homeostasis–dysbiosis transitions in the oral cavity and stomach. For example, the gastric microbiota possibly helps modulate the gastric immune microenvironment. An animal model suggested that gastric commensal microbes can promote the clearance of H. pylori by triggering the host immune response. The stomach commensal bacteria Bacteroidales Family S24-7 are involved in the expansion of Group 2 innate lymphoid cells (ILC2s) via IL-7 and IL-33. These microbes induce the secretion of IL-5 by ILC2s, which protect the stomach by eliminating IgA-coated pathogenic H. pylori79 (Fig. 2). Moreover, the oral pathogen P. melaninogenica and bile acids (BAs) have synergistic carcinogenic effects. The lipopolysaccharide (LPS)-producing bacterium P. melaninogenica has a strong positive correlation with taurodeoxycholic acid (TDCA), a secondary BA that is significantly enriched in the gastric juice in bile reflux gastritis and GC. TDCA and LPS promote gastric epithelial cell proliferation by activating the IL-6/JAK1/STAT3 pathway68.
Persistent H. pylori infection induces gastric gland atrophy and subsequently suppresses endocrine activity. The Bacteroidales family S24-7 activates ILC-2 to produce IL-5, contributing to diminished H. pylori. S. anginosus stimulates the recruitment of neutrophils and monocytes to mediate epithelial dysplasia. Oral-associated microbes produce carcinogens such as lactate, LPS, nitrite, and acetaldehyde. L-Lactate assists H. pylori in resisting complement C4b. P. melaninogenica LPS synergistic with TCDA promotes gastric epithelial cell proliferation by activating the IL-6/JAK1/STAT3 pathway. Acetaldehyde causes DNA damage and mutation in epithelial cells. Suppressed bacterial arginine degradation provides arginine availability for tumor cell growth. The direct effects and indirect effects are shown by solid and dashed arrows, respectively. Created with BioRender.com.
In addition to direct pathogenicity in the stomach, oral microbes can also affect distant sites through various indirect mechanisms80. According to the mucosal immune theory, the oral mucosa and gastric mucosa are system-wide immune organs, and stimulation of one part can lead to distal area changes81. Periodontal bacteria can migrate to distal organs through various modes, including bacteremia, the oro-pharyngeal route, the oro-digestive route and crossing the blood‒brain barrier. These pathogens also aggravate distal inflammation by activating innate and adaptive immunity82. For example, trained immunity is a form of innate immune memory that helps the host respond quickly to microbial stimuli but sometimes exacerbates chronic inflammation in a harmful way. Maladaptive innate immune training for myelopoiesis underlies the inflammatory comorbidities of periodontitis83. Moreover, these inflammatory comorbidities disturb the oral microbiota and alter its pathogenicity84.
Carcinogenicity of the oral–gastric microbiome
H. pylori is currently recognized as a human GC carcinogen85. It causes gastric mucosa atrophy and precancerous lesions through various virulence factors, including CagA and VacA. CagA leads to nuclear β-catenin accumulation and procarcinogenic gene transcription. VacA induces the vacuolation of epithelial cells, eventually causing cell apoptosis and autophagy86.
The potential carcinogenicity of non-H. pylori microbes also warrants attention (Fig. 2). Microbiota dysbiosis characterized by decreased diversity and significant enrichment of oral-associated microbes is acknowledged in GC. In addition, GC risk factors such as cigarette smoking and alcohol use may also interact with the oral and gastrointestinal microbiota. Alcohol is associated with GC by multiple mechanisms of carcinogenesis including the by-product acetaldehyde, alcohol-induced inflammation, immune surveillance impairment, DNA methylation changes, etc,30,87. Most importantly, acetaldehyde is the group 1 carcinogen recognized by the International Agency for Research on Cancer88. Microbes with high alcohol dehydrogenase activity and can utilize alcohol to produce acetaldehyde. For example, the oral microbes Streptococcus mitis, S. salivarius, Neisseria mucosa, Neisseria sicca and Streptococcus mutans can synthesize acetaldehyde, which contributes to DNA damage and point mutations89. S. anginosus is also carcinogenic via its production of acetaldehyde and stimulation of neutrophil and monocyte recruitment, which mediate epithelial dysplasia90. Besides, α diversity of oral microbiota and gut microbiota was significantly lower in patients with alcohol dependence, and oral-gut microbiota overlap was higher (9 genera) than in healthy controls (3 genera), which suggests shifted oral and gut microbiota in alcohol-dependent patients91. Smoking is a risk factor for intestinal-type GC rather than diffuse-type30. The GC risk increases in correlation with cigarette consumption per day and duration of regular smoking30. Currently, we know little about the crosstalk of smoking and the gastric microbiota. The effects of smoking on the oral microbiota were discussed in Part I, but the involvement of oral microbiota in smoking-related carcinogenesis or development also remains to be uncovered. Note that gut microbiota dysbiosis and increased TDCA in the colon were found in mice exposed to cigarettes, which may activate oncogenic MAPK/ERK signaling in the colonic epithelium92. Also, smoking and drinking may play an indirect role by suppressing mucosal immunity. For example, cigarette smoking suppresses NOD-like receptor family pyrin domain containing 3 inflammasome and the secretion of IL-1β and IL-18, therefore inhibiting oral mucosal defense against Candida albicans in rat models93. IL-10 inhibition induced CD8+ cell dysfunction accelerated GC development induced by H.pylori infection and chronic alcohol use94.
In addition, the molecular mechanisms by which oral microbes participate in GC are being explored. S. anginosus is another species enriched in the gastric mucosa of GC patients. Recent evidence suggests that long-term S. anginosus infection induces gastritis-atrophy-metaplasia-dysplasia serial lesions in conventional mice and promotes GC progression in tumor allograft mice. It promoted cell proliferation and inhibited apoptosis through direct interactions with gastric epithelial cells via the TMPC-ANXA2-MAPK axis95.P. gingivalis is a vital periodontal pathogen. P. gingivalis lipopolysaccharide (LPS) exposure may lead to gastric barrier destruction, macrophage activation and TNFα release. LPS and TNFα might activate TLR2-β-catenin signaling in GC96.
Lactobacillus plays a complex role in gastric dysbiosis. It is not only more enriched in the human GC microbiome compared to SG, but is also highly correlated with all the differentially abundant metabolites in the bile secretion, amino acid biosynthesis, and tryptophan metabolism pathways between GC tumor and nontumor tissues52,97. However, Lactobacillus is also thought to be a probiotic that maintains a lower environmental pH by producing lactic acid, thus inhibiting pathogenic bacteria. Additionally, Lactobacillus provides colonization resistance, degrades nitrosamines and produces anti-inflammatory and anticancer substances such as short-chain fatty acids (SCFAs) and exopolysaccharides98.
Other microbial products are also involved in carcinogenesis, including nitrosation metabolism, lactate synthesis, arginine degradation, and LPS. The carcinogenic nitrosating microbial community is enriched in the GC microbiota. Elevated nitrite levels were detected in GC compared with SG and IM, with the nitrite and N-nitroso compound producers Neisseria, Veillonella, Fusobacterium and Lactobacillus enriched in GC mucosa samples52. Bacterial functional analysis revealed greater richness of the nitrate reductase gene nrfA in the GC group than in the noncancer group99. According to a recent meta-analysis, dietary nitrite intake increased GC (RR 1.33, 95% CI 1.0-1.73). Nitrates and nitrites are precursors of N-nitroso compounds, which participate in carcinogenesis by inducing DNA-damaging metabolites100.
Lactate, an important substance in the acidic tumor microenvironment, has a complex role in mediating host–microbiota interactions. Lactate is a nutrient for cancer cells. It helps hp evade host complement immunity, which may promote malignant transformation of the gastric epithelium. H. pylori utilizes L-lactate to prevent C4b accumulation on its surface and subsequently resists complement activation via the classical pathway, especially in the antrum. The remarkable lactate depletion observed in the antrum of H. pylori-infected mice supports this conclusion101. Although the lactate source has not been fully elucidated, the oral bacteria Abiotrophia, Leptotrichia, Lactobacillus and Streptococcus are important L-lactate producers102.
Arginine is a nutrient that microbes compete with hosts for. It directly affects tumor growth and is a substrate for arginine deiminase-positive bacteria such as Streptococcus sanguis, which produce ammonia to resist acid stress98. Suppressed arginine degradation was detected in early gastric neoplasia patients, which indicates increased arginine availability for tumor cell growth103. Although the role of arginine in cancer is controversial104, microbial metabolism may affect the tumor microenvironment by regulating arginine levels.
In response to direct or indirect challenges by the above microbes, the host mainly relies on the gastric mucosal barrier for defense. For example, H. pylori infection first exacerbates the production of Th17 cytokines, followed by increased IgA levels in the lumen and reduced production of Muc5ac in the corpus. This mechanism allows the host to maintain barrier integrity and activate immunopathogenic responses to H. pylori infection105. After persistent H. pylori infection, gastric mucosal lesions may progress to the IM, low-grade intraepithelial neoplasia, and early GC stages. Beyond the “point of no return”, other factors may play a role106. The oral–gastric microbial axis may be a potential driving factor.
Oral–gastric microbial axis and its systemic effect
The emerging oral–gastric microbial axis
As the oral–intestinal microbial association is increasingly recognized, the stomach, as a checkpoint to microbial transmission in the digestive tract, has attracted increasing attention. We reviewed the correlation of oral-associated microbes in the mouth and stomach with GC and the evidence regarding oral-associated microbe migration, interactions with host/other microbes, and carcinogenicity. Here, we present the concept of the oral–gastric microbial axis, which refers to the dynamic interconnection of oral and gastric health status and local microecology. It is facilitated through microbial exchange between the oral cavity and stomach and promoted by microbial interspecies as well as microbial-host interactions by signaling, function, and metabolites (Fig. 3).
The representative oral microbiota in healthy individuals contains the major gastric microbiota in healthy individuals, although most oral microbes are blocked by the mucus barrier secreted by the healthy gastric mucosa. However, under certain circumstances, such as persistent H. pylori infection, the weakened gastric mucus barrier allows oral-associated microbes to invade. Ectopically colonized microbes promote gastric carcinogenesis through direct contact or products. Oral and gastric dysbiosis have potential effects on the brain, probably triggering feedback on lifestyle and self-maintenance. The direct effects and indirect effects are shown by solid and dashed arrows, respectively. Created with BioRender.com.
Under physiological conditions, oral commensals, such as streptococci, can regulate the structure and function of the oral microbiome through physical mechanisms, antibacterial products and host immune response modulation107,108. In pathological cases, oral microbes may be involved in GC and other gastric diseases, as mentioned above. However, the impact of H. pylori on oral health is relatively limited. A clinical trial showed that H. pylori-positive patients have worse periodontal disease stages and grades, but their saliva samples were H. pylori negative109. In addition, the high prevalence of oral H. pylori in subgingival plaque has no correlation with a greater risk of oral cancer110. Thus, there may be an association between H. pylori gastric infection and periodontitis, but no clear evidence connects H. pylori and oral cancer.
From an ecological viewpoint, H. pylori infection essentially represents a state of gastric dysbiosis. H. pylori infection induces decreased diversity, species evenness, richness, and a less connected microbial network in the gastritis and GC microbiota25,57. Gastric dysbiosis may persist long after H. pylori eradication, which may account for primary and metachronous GC111. Oral microbes present centrality and shape the structure of the disordered community50,76. Oral microbe-dominated communities may contribute to diseases through direct contact and indirect interactions through metabolites (e.g., lactate, BAs, SCFAs) and immune regulation. Mucosal immunity also responds differently to microbes during inflammation and homeostasis112. Understanding the development of mucosal immunity will aid in understanding the role of the local microbiota and its indirect impact on distal disease. Such concepts have also been raised in the field of the oral–gut axis. Although the role of innate and adaptive immunity in the oral cavity and stomach homeostasis–dysbiosis remains to be revealed, this linkage provides a new understanding of the potential oral–gastric axis.
Therapeutic applications of the oral–gastric microbial axis
Periodontal therapy is an effective method for controlling dental plaque, and it appears to have synergistic effects with H. pylori eradication treatment. H. pylori-positive patients have additional periodontal improvement when receiving periodontal therapy combined with eradication treatment109. Also, periodontal therapy improves gastric H. pylori eradication and nonrecurrence rates in parallel with conventional systemic eradication therapy113. A longitudinal study analyzed the saliva and fecal microbiota of periodontitis patients before and after periodontal therapy. Periodontal treatment not only mitigated oral dysbiosis, but also altered the gut microbiota, indicating its impact on gastrointestinal microecology114. In addition, a study identified dental caries as a risk factor for H. pylori eradication failure. This may be attributed to the protective effect of persistent oral H. pylori from oral biofilms and poor blood circulation in infected teeth115.
Oral and gastric applications of probiotics have also been conducted separately. In animal models, oral probiotics improve preclinical, microbiological and immunological outcomes of periodontitis. These probiotics usually contain lactobacilli, bifidobacteria, and streptococci116. In caries research, probiotics and prebiotics (arginine, fluoride, etc,.) have been shown to have good anticaries effects117. Novel bacterial replacement therapy reduces the oral pathogenicity of S. mutans without destroying the oral ecology118. The European guidelines state that specific probiotics, such as Lactobacillus and Saccharomyces boulardii, are worth considering as complementary therapies to improve H. pylori eradication rates and reduce adverse events119,120,121. However, there is no therapeutic research based on the oral–gastric microbial axis.
Potential influence of the oral–gastric-brain functional axis on GC
The brain receives signals from the oral cavity and stomach and then adjusts its diet and hygiene practices, which is integral to the overall feedback loop. Communication between the brain and the upper gastrointestinal tract is mediated primarily through the nervous and endocrine systems. Nutritional intake also has an impact on oral and gastric health.
Ghrelin, known as a hunger hormone, is broadly secreted by many sites, including the gastric mucosa, oral mucosa and gingival fibroblasts, and predominantly stimulates food intake through receptors in the brain122. In the stomach, low baseline serum ghrelin concentrations are associated with an elevated risk of cardia and noncardia gastric adenocarcinoma123. In the oral cavity, ghrelin inhibits the proinflammatory factor IL-1β, which acts as a strong inflammatory mediator in periodontitis122,124.
Oral health influences the stomach by affecting masticatory and digestive functions. Insufficient toothbrushing is related to increased GC risk, which may be attributed to the accumulation of dental plaque and carcinogenic metabolites39. Deficient mastication modifies the host’s nutrient bioaccessibility and results in higher contents of harmful nondigested nitrogen at the end of gastrointestinal digestion, which is possibly fermented by microbes125. Although the oral–gastric-brain axis has a potentially intriguing functional link, additional convincing evidence and careful debates are needed, and this topic warrants further research.
Concluding remarks and future directions
The GC microbiota is characterized by progressive dysbiosis with enrichment of oral-associated microbes. Stomach disease is often accompanied by alterations in the oral microbiome. Oral diseases are also associated with the GC pathogen H. pylori. The correlation of oral-associated microbes in the stomach with gastric carcinogenesis and disease development has been supported by several high-quality sequencing studies, guiding us to elucidate a refined oral and gastric microbial landscape. However, most GC-associated microbiologic analyses are performed at the genus level, and the conclusions are inconsistent, making it difficult to identify specific species and their carcinogenetic patterns. In addition, the gastric microbiota corresponding to clinical features, such as GC classification and staging, has not been well studied. In the future, new technology and standardized metrics are needed to identify potential pathogenetic microbes and the core microbiota, location, interaction, effect, richness and colonization (CIRCLE) (Fig. 4). Further detection of the absolute abundance of species is needed to determine the actual oral–gastric microbial biomass and the true relationship between the microbiota and diseases. Changes in other microorganisms, such as fungi and viruses, also require further investigation. As microbial single-cell DNA/RNA/spatially resolved transcriptomic (SRT) sequencing has been achieved126, advanced technologies in the field of eukaryotes, such as single-cell multimodal omics being applied to the microbiome, will provide further insights within the CIRCLE framework. Another question, whether the relationship between microbial dysbiosis and GC is causal or correlative, is still unresolved. High-quality longitudinal evidence on this topic is urgently needed to overcome the existing cognitive bottleneck. Computerized methods can also be of some help in causal-inference analytics. For instance, Transkingdom Network Analysis has been applied to characterize the role of the microbiome in cervical cancer, lymphoma and melanoma127. Artificial intelligence is promising in handling huge amounts of data and building dynamic models, but its ethical issue and academic integrity need attention. Additionally, the mechanisms by which the oral–gastric microbial axis regulates the shift in host homeostasis–dysbiosis remain to be explored. First, the patterns of oral microbe survival and colonization in the stomach need to be investigated. The impact of oral-associated microbes on the gastric environment, including how oral-associated microbes reprogram host metabolism, remodel oral and gastric ecology and mucosal immunity, and initiate epithelial malignancy, warrants further investigation. Furthermore, how the brain and systemic immunity are affected by oral-associated microbes remains unclear. Clinically, GC is mainly diagnosed through gastroscopy and pathological biopsy, and many patients have already developed advanced GC when they feel sick. The development of noninvasive indicators such as microbial features of the tongue coating and saliva is highly important for early GC screening. Finally, the impact of dental plaque management on GC prevention needs to be evaluated, as limited evidence suggests potential benefits for gastric health.
Six aspects to explore the unknown universe of oral–gastric microbes. New technology and standardized metrics are called on to identify potential pathogenetic microbes and their core microbiota, location, interaction, effect, richness and colonization (CIRCLE). Prospective solutions to these issues are listed on the left. ISH, in situ hybridization; SRT, spatially resolved transcriptomics; SAHMI, single-cell analysis of host–microbiome interactions; WGS, whole-genome sequencing; WES, whole-exome sequencing; RNA-seq, whole-transcriptome sequencing.
Data availability
Details of the systematically reviewed studies in section 2.3 are available in the supplementary files. For further information, please contact the corresponding authors.
References
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 486, 207–214 (2012).
Caselli, E. et al. Defining the oral microbiome by whole-genome sequencing and resistome analysis: the complexity of the healthy picture. BMC Microbiol. 20, 120 (2020).
Chen, X. et al. The interactions between oral-gut axis microbiota and Helicobacter pylori. Front. Cell. Infect. Microbiol. 12, 914418 (2022).
Park, O. J. et al. Pyrosequencing Analysis of Subgingival Microbiota in Distinct Periodontal Conditions. J. Dent. Res. 94, 921–927 (2015).
Espinoza, J. L. et al. Supragingival Plaque Microbiome Ecology and Functional Potential in the Context of Health and Disease. mBio 9, https://doi.org/10.1128/mBio.01631-18 (2018).
Santacroce, L. et al. Oral microbiota in human health and disease: A perspective. Exp. Biol. Med. (Maywood, N.J.) 248, 1288–1301 (2023).
Wade, W. G. Resilience of the oral microbiome. Periodontology. 86, 113–122 (2021).
Weyrich, L. S. The evolutionary history of the human oral microbiota and its implications for modern health. Periodontology 85, 90–100 (2021).
Moran, S. P., Rosier, B. T., Henriquez, F. L. & Burleigh, M. C. The effects of nitrate on the oral microbiome: a systematic review investigating prebiotic potential. J. oral. Microbiol. 16, 2322228 (2024).
Ibarlucea-Jerez, M. et al. Fermented food consumption modulates the oral microbiota. NPJ Sci. food 8, 55 (2024).
Thomas, A. M. et al. Alcohol and tobacco consumption affects bacterial richness in oral cavity mucosa biofilms. BMC Microbiol. 14, 250 (2014).
Vogtmann, E. et al. Association between tobacco use and the upper gastrointestinal microbiome among Chinese men. Cancer Causes Control: CCC 26, 581–588 (2015).
Rothman, J. A. et al. Oral microbial communities in children, caregivers, and associations with salivary biomeasures and environmental tobacco smoke exposure. mSystems 8, e0003623 (2023).
Tan, J., Lamont, G. J. & Scott, D. A. Tobacco-enhanced biofilm formation by Porphyromonas gingivalis and other oral microbes. Mol. oral. Microbiol. 39, 270–290 (2024).
Hernandez, B. Y. et al. Betel nut chewing, oral premalignant lesions, and the oral microbiome. PloS one 12, e0172196 (2017).
Vasapolli, R. et al. Analysis of Transcriptionally Active Bacteria Throughout the Gastrointestinal Tract of Healthy Individuals. Gastroenterology 157, 1081–1092.e1083 (2019).
Rajilic-Stojanovic, M. et al. Systematic review: gastric microbiota in health and disease. Alimentary Pharmacol. therapeutics 51, 582–602 (2020).
Malfertheiner, P. et al. Helicobacter pylori infection. Nat. Rev. Dis. Prim. 9, 19 (2023).
Liou, J. M. et al. Screening and eradication of Helicobacter pylori for gastric cancer prevention: the Taipei global consensus. Gut 69, 2093–2112 (2020).
Ruan, W., Engevik, M. A., Spinler, J. K. & Versalovic, J. Healthy Human Gastrointestinal Microbiome: Composition and Function After a Decade of Exploration. Digestive Dis. Sci. 65, 695–705 (2020).
Bassis, C. M. et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio 6, e00037 (2015).
Kitamoto, S., Nagao-Kitamoto, H., Hein, R., Schmidt, T. M. & Kamada, N. The Bacterial Connection between the Oral Cavity and the Gut Diseases. J. Dent. Res. 99, 1021–1029 (2020).
Pacheco-Yanes, J., Reynolds, E., Li, J. & Mariño, E. Microbiome-targeted interventions for the control of oral-gut dysbiosis and chronic systemic inflammation. Trends Mol. Med. 29, 912–925 (2023).
Chen, C. C. et al. The interplay between Helicobacter pylori and gastrointestinal microbiota. Gut microbes 13, 1–22 (2021).
Ndegwa, N. et al. Gastric Microbiota in a Low-Helicobacter pylori Prevalence General Population and Their Associations With Gastric Lesions. Clin. Transl. Gastroenterol. 11, e00191 (2020).
Dong, T. et al. Gastric bacteria as potential biomarkers for the diagnosis of atrophic gastritis. Mol. Biol. Rep. 50, 655–664 (2023).
Sung, J. J. Y. et al. Gastric microbes associated with gastric inflammation, atrophy and intestinal metaplasia 1 year after Helicobacter pylori eradication. Gut 69, 1572–1580, (2020).
Wu, F. et al. Oral and gastric microbiome in relation to gastric intestinal metaplasia. Int. J. cancer 150, 928–940 (2022).
Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: a cancer J. clinicians 71, 209–249 (2021).
Thrift, A. P., Wenker, T. N. & El-Serag, H. B. Global burden of gastric cancer: epidemiological trends, risk factors, screening and prevention. Nat. Rev. Clin. Oncol. 20, 338–349 (2023).
Nguyen, T. H. et al. Prevalence of Helicobacter pylori Positive Non-cardia Gastric Adenocarcinoma Is Low and Decreasing in a US Population. Digestive Dis. Sci. 65, 2403–2411 (2020).
Meimarakis, G. et al. Helicobacter pylori as a prognostic indicator after curative resection of gastric carcinoma: a prospective study. Lancet Oncol. 7, 211–222 (2006).
Marrelli, D. et al. Negative Helicobacter pylori status is associated with poor prognosis in patients with gastric cancer. Cancer 115, 2071–2080 (2009).
Matsuo, T. et al. Low prevalence of Helicobacter pylori-negative gastric cancer among Japanese. Helicobacter 16, 415–419 (2011).
Ono, S. et al. Frequency of Helicobacter pylori -negative gastric cancer and gastric mucosal atrophy in a Japanese endoscopic submucosal dissection series including histological, endoscopic and serological atrophy. Digestion 86, 59–65 (2012).
Kim, H. J. et al. Comparison between Resectable Helicobacter pylori-Negative and -Positive Gastric Cancers. Gut liver 10, 212–219 (2016).
Ndegwa, N. et al. Association between poor oral health and gastric cancer: A prospective cohort study. Int. J. cancer 143, 2281–2288 (2018).
Sun, J. et al. Chronic Periodontal Disease, Periodontal Pathogen Colonization, and Increased Risk of Precancerous Gastric Lesions. J. Periodontol. 88, 1124–1134 (2017).
Zhang, T. et al. Poor oral hygiene behavior is associated with an increased risk of gastric cancer: A population-based case-control study in China. J. Periodontol. 93, 988–1002 (2022).
Lo, C. H. et al. Periodontal disease, tooth loss, and risk of oesophageal and gastric adenocarcinoma: a prospective study. Gut 70, 620–621 (2021).
Sung, C. E. et al. Periodontitis, Helicobacter pylori infection, and gastrointestinal tract cancer mortality. J. Clin. Periodontol. 49, 210–220 (2022).
Acharya, C., Sahingur, S. E. & Bajaj, J. S. Microbiota, cirrhosis, and the emerging oral-gut-liver axis. JCI insight 2, https://doi.org/10.1172/jci.insight.94416 (2017).
Baima, G. et al. Periodontitis and risk of cancer: Mechanistic evidence. Periodontology, https://doi.org/10.1111/prd.12540 (2023).
Hu, Y. L., Pang, W., Huang, Y., Zhang, Y. & Zhang, C. J. The Gastric Microbiome Is Perturbed in Advanced Gastric Adenocarcinoma Identified Through Shotgun Metagenomics. Front. Cell. Infect. Microbiol. 8, 433 (2018).
Johnson, J. S. et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 10, 5029 (2019).
Shu, H. Y. et al. CyanoStrainChip: A Novel DNA Microarray Tool for High-Throughput Detection of Environmental Cyanobacteria at the Strain Level. Environ. Sci. Technol. 58, 5024–5034 (2024).
Neu, A. T., Allen, E. E. & Roy, K. Defining and quantifying the core microbiome: Challenges and prospects. Proceedings of the National Academy of Sciences of the United States of America 118, https://doi.org/10.1073/pnas.2104429118 (2021).
Li, Y. et al. Meta-analysis reveals Helicobacter pylori mutual exclusivity and reproducible gastric microbiome alterations during gastric carcinoma progression. Gut microbes 15, 2197835 (2023).
Chen, J. et al. Leveraging existing 16S rRNA microbial data to identify diagnostic biomarker in Chinese patients with gastric cancer: a systematic meta-analysis. mSystems, e0074723, https://doi.org/10.1128/msystems.00747-23 (2023).
Coker, O. O. et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 67, 1024–1032 (2018).
Abate, M. et al. A Novel Microbiome Signature in Gastric Cancer: A Two Independent Cohort Retrospective Analysis. Ann. Surg. 276, 605–615 (2022).
He, C. et al. Convergent dysbiosis of gastric mucosa and fluid microbiome during stomach carcinogenesis. Gastric Cancer: Off. J. Int. Gastric Cancer Assoc. Jpn. Gastric Cancer Assoc. 25, 837–849 (2022).
Bakhti, S. Z. & Latifi-Navid, S. Oral microbiota and Helicobacter pylori in gastric carcinogenesis: what do we know and where next? BMC Microbiol. 21, 71 (2021).
Ferreira, R. M. et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 67, 226–236 (2018).
Assumpção, P. P. et al. The diffuse-type gastric cancer epidemiology enigma. BMC Gastroenterol. 20, 223 (2020).
Lehr, K. et al. Microbial composition of tumorous and adjacent gastric tissue is associated with prognosis of gastric cancer. Sci. Rep. 13, 4640 (2023).
Liu, X. et al. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine 40, 336–348 (2019).
Boehm, E. T. et al. Fusobacterium nucleatum is associated with worse prognosis in Lauren’s diffuse type gastric cancer patients. Sci. Rep. 10, 16240 (2020).
Shao, D. et al. Microbial characterization of esophageal squamous cell carcinoma and gastric cardia adenocarcinoma from a high-risk region of China. Cancer 125, 3993–4002 (2019).
Lei, L. et al. Distinct oral-associated gastric microbiota and Helicobacter pylori communities for spatial microbial heterogeneity in gastric cancer. mSystems 9, e0008924 (2024).
Wei, Q. et al. Analysis of bacterial diversity and community structure in gastric juice of patients with advanced gastric cancer. Discov. Oncol. 14, 7 (2023).
Yang, Y. et al. Prospective study of oral microbiome and gastric cancer risk among Asian, African American and European American populations. Int. J. cancer 150, 916–927 (2022).
Yuan, L. et al. Development of a tongue image-based machine learning tool for the diagnosis of gastric cancer: a prospective multicentre clinical cohort study. EClin. Med. 57, https://doi.org/10.1016/j.eclinm.2023.101834 (2023).
Wu, Z. F. et al. Helicobacter pylori infection is associated with the co-occurrence of bacteria in the oral cavity and the gastric mucosa. Helicobacter 26, e12786 (2021).
Lofgren, J. L. et al. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology 140, 210–220, (2011).
Shen, Z. et al. Gastric Non-Helicobacter pylori Urease-Positive Staphylococcus epidermidis and Streptococcus salivarius Isolated from Humans Have Contrasting Effects on H. pylori-Associated Gastric Pathology and Host Immune Responses in a Murine Model of Gastric Cancer. mSphere 7, e0077221 (2022).
Kwon, S.-K. et al. Human gastric microbiota transplantation recapitulates premalignant lesions in germ-free mice. Gut 71, 1266 (2022).
Wang, S. et al. Bile Acid-Microbiome Interaction Promotes Gastric Carcinogenesis. Adv. Sci. (Weinh., Baden.-Wurtt., Ger.) 9, e2200263 (2022).
Schmidt, T. S. et al. Extensive transmission of microbes along the gastrointestinal tract. eLife 8, https://doi.org/10.7554/eLife.42693 (2019).
Barbour, A., Elebyary, O., Fine, N., Oveisi, M. & Glogauer, M. Metabolites of the oral microbiome: important mediators of multikingdom interactions. FEMS Microbiol. Rev. 46, https://doi.org/10.1093/femsre/fuab039 (2022).
Parsons, B. N. et al. Comparison of the human gastric microbiota in hypochlorhydric states arising as a result of Helicobacter pylori-induced atrophic gastritis, autoimmune atrophic gastritis and proton pump inhibitor use. PLoS Pathog. 13, e1006653 (2017).
Zhou, C., Bisseling, T. M., van der Post, R. S. & Boleij, A. The influence of Helicobacter pylori, proton pump inhibitor, and obesity on the gastric microbiome in relation to gastric cancer development. Comput. Struct. Biotechnol. J. 23, 186–198 (2024).
Xiao, X. et al. Proton pump inhibitors alter gut microbiota by promoting oral microbiota translocation: a prospective interventional study. Gut 73, 1098–1109 (2024).
Engstrand, L. & Graham, D. Y. Microbiome and Gastric Cancer. Digestive Dis. Sci. 65, 865–873 (2020).
Sassano, M. et al. Intake of Proton-Pump Inhibitors and Gastric Cancer within the Stomach Cancer Pooling (StoP) Project. Cancer Epidemiol., Biomark. Prev. : a Publ. Am. Assoc. Cancer Res., cosponsored Am. Soc. Preventive Oncol. 32, 1174–1181 (2023).
Oosterlinck, B. et al. Mucin-microbiome signatures shape the tumor microenvironment in gastric cancer. Microbiome 11, 86 (2023).
Soto, C. et al. Porphyromonas gingivalis-Helicobacter pylori co-incubation enhances Porphyromonas gingivalis virulence and increases migration of infected human oral keratinocytes. J. oral. Microbiol. 14, 2107691 (2022).
Schorr, L., Mathies, M., Elinav, E. & Puschhof, J. Intracellular bacteria in cancer-prospects and debates. NPJ Biofilms Microbiomes 9, 76 (2023).
Satoh-Takayama, N. et al. Bacteria-Induced Group 2 Innate Lymphoid Cells in the Stomach Provide Immune Protection through Induction of IgA. Immunity 52, 635–649.e634 (2020).
Baker, J. L., Mark Welch, J. L., Kauffman, K. M., McLean, J. S. & He, X. The oral microbiome: diversity, biogeography and human health. Nat. Rev. Microbiol. https://doi.org/10.1038/s41579-023-00963-6 (2023).
Gill, N., Wlodarska, M. & Finlay, B. B. The future of mucosal immunology: studying an integrated system-wide organ. Nat. Immunol. 11, 558–560 (2010).
Hajishengallis, G. & Chavakis, T. Local and systemic mechanisms linking periodontal disease and inflammatory comorbidities. Nat. Rev. Immunol. 21, 426–440 (2021).
Li, X. et al. Maladaptive innate immune training of myelopoiesis links inflammatory comorbidities. Cell 185, 1709–1727.e1718 (2022).
Graves, D. T., Corrêa, J. D. & Silva, T. A. The Oral Microbiota Is Modified by Systemic Diseases. J. Dent. Res. 98, 148–156 (2019).
DiMaio, D., Emu, B., Goodman, A. L., Mothes, W. & Justice, A. Cancer Microbiology. J. Natl. Cancer Inst. 114, 651–663 (2022).
Bessède, E. & Mégraud, F. Microbiota and gastric cancer. Semin. cancer Biol. 86, 11–17 (2022).
Hatta, W. et al. The Impact of Tobacco Smoking and Alcohol Consumption on the Development of Gastric Cancers. Int. J. Mol. Sci. 25, https://doi.org/10.3390/ijms25147854 (2024).
IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Personal habits and indoor combustions. IARC monographs on the evaluation of carcinogenic risks to humans 100, 1–538 (2012).
Tagaino, R. et al. Metabolic property of acetaldehyde production from ethanol and glucose by oral Streptococcus and Neisseria. Sci. Rep. 9, 10446 (2019).
Stasiewicz, M. & Karpiński, T. M. The oral microbiota and its role in carcinogenesis. Semin. cancer Biol. 86, 633–642 (2022).
Hu, L. et al. The association between oral and gut microbiota in male patients with alcohol dependence. Front. Microbiol. 14, 1203678 (2023).
Bai, X. et al. Cigarette smoke promotes colorectal cancer through modulation of gut microbiota and related metabolites. Gut 71, 2439–2450 (2022).
Ye, P. et al. Long-term cigarette smoking suppresses NLRP3 inflammasome activation in oral mucosal epithelium and attenuates host defense against Candida albicans in a rat model. Biomedicine Pharmacother. = Biomedecine pharmacotherapie 113, 108597 (2019).
Aziz, F. et al. Gastric tumorigenesis induced by combining Helicobacter pylori infection and chronic alcohol through IL-10 inhibition. Carcinogenesis 43, 126–139 (2022).
Fu, K. et al. Streptococcus anginosus promotes gastric inflammation, atrophy, and tumorigenesis in mice. Cell 187, 882–896.e817 (2024).
Oriuchi, M. et al. Porphyromonas gingivalis Lipopolysaccharide Damages Mucosal Barrier to Promote Gastritis-Associated Carcinogenesis. Digestive Dis. Sci. 69, 95–111 (2024).
Dai, D. et al. Interactions between gastric microbiota and metabolites in gastric cancer. Cell death Dis. 12, 1104 (2021).
Garbacz, K. Anticancer activity of lactic acid bacteria. Semin. cancer Biol. 86, 356–366 (2022).
Gantuya, B. et al. Gastric mucosal microbiota in a Mongolian population with gastric cancer and precursor conditions. Alimentary Pharmacol. therapeutics 51, 770–780 (2020).
Seyyedsalehi, M. S. et al. Association of Dietary Nitrate, Nitrite, and N-Nitroso Compounds Intake and Gastrointestinal Cancers: A Systematic Review and Meta-Analysis. Toxics 11, https://doi.org/10.3390/toxics11020190 (2023).
Hu, S. & Ottemann, K. M. Helicobacter pylori initiates successful gastric colonization by utilizing L-lactate to promote complement resistance. Nat. Commun. 14, 1695 (2023).
Velusamy, S. K., Sampathkumar, V., Ramasubbu, N., Paster, B. J. & Fine, D. H. Aggregatibacter actinomycetemcomitans colonization and persistence in a primate model. Proc. Natl. Acad. Sci. USA 116, 22307–22313 (2019).
Png, C. W. et al. Mucosal microbiome associates with progression to gastric cancer. Theranostics 12, 48–58 (2022).
Pirzadeh, M., Khalili, N. & Rezaei, N. The interplay between aryl hydrocarbon receptor, H. pylori, tryptophan, and arginine in the pathogenesis of gastric cancer. Int. Rev. Immunol. 41, 299–312 (2022).
Brackman, L. C. et al. IL-17 receptor A functions to help maintain barrier integrity and limit activation of immunopathogenic response to H. pylori infection. Infect. Immun. 92, e0029223 (2024).
Wang, S. T., Yang, H. W., Zhang, W. L., Li, Z. & Ji, R. Disruption of the gastric epithelial barrier in Correa’s cascade: Clinical evidence via confocal endomicroscopy. Helicobacter 29, e13065 (2024).
Baty, J. J., Stoner, S. N. & Scoffield, J. A. Oral Commensal Streptococci: Gatekeepers of the Oral Cavity. J. Bacteriol. 204, e0025722 (2022).
Lei, L. et al. Carbohydrate Metabolism Regulated by Antisense vicR RNA in Cariogenicity. J. Dent. Res. 99, 204–213 (2020).
Tsimpiris, A. et al. Periodontitis and Helicobacter pylori Infection: Eradication and Periodontal Therapy Combination. Eur. J. Dent. 16, 145–152 (2022).
Sekar, R., Murali, P. & Junaid, M. Quantification of Helicobacter pylori and its oncoproteins in the oral cavity: A cross-sectional study. Oral diseases, https://doi.org/10.1111/odi.14141 (2022).
Watanabe, T. et al. Long-term persistence of gastric dysbiosis after eradication of Helicobacter pylori in patients who underwent endoscopic submucosal dissection for early gastric cancer. Gastric Cancer: Off. J. Int. Gastric Cancer Assoc. Jpn. Gastric Cancer Assoc. 24, 710–720 (2021).
Spindler, M. P., Mogno, I., Suri, P., Britton, G. J. & Faith, J. J. Species-specific CD4(+) T cells enable prediction of mucosal immune phenotypes from microbiota composition. Proc. Natl. Acad. Sci. USA 120, e2215914120 (2023).
Ozturk, A. Periodontal Treatment Is Associated With Improvement in Gastric Helicobacter pylori Eradication: An Updated Meta-analysis of Clinical Trials. Int. Dent. J. 71, 188–196 (2021).
Baima, G. et al. Effect of Periodontitis and Periodontal Therapy on Oral and Gut Microbiota. J. Dent. Res. 103, 359–368 (2024).
Iwai, K. et al. Association between failed eradication of 7-day triple therapy for Helicobacter pylori and untreated dental caries in Japanese adults. Sci. Rep. 14, 4043 (2024).
Nguyen, T., Brody, H., Radaic, A. & Kapila, Y. Probiotics for periodontal health-Current molecular findings. Periodontology 87, 254–267 (2021).
Yu, X., Devine, D. A. & Vernon, J. J. Manipulating the diseased oral microbiome: the power of probiotics and prebiotics. J. oral. Microbiol. 16, 2307416 (2024).
Pan, W. et al. A new gcrR-deficient Streptococcus mutans mutant for replacement therapy of dental caries. TheScientificWorldJournal 2013, 460202 (2013).
Malfertheiner, P. et al. Management of Helicobacter pylori infection-the Maastricht V/Florence Consensus Report. Gut 66, 6–30 (2017).
Szajewska, H., Horvath, A. & Kołodziej, M. Systematic review with meta-analysis: Saccharomyces boulardii supplementation and eradication of Helicobacter pylori infection. Alimentary Pharmacol. therapeutics 41, 1237–1245 (2015).
Lv, Z. et al. Efficacy and safety of probiotics as adjuvant agents for Helicobacter pylori infection: A meta-analysis. Exp. therapeutic Med. 9, 707–716 (2015).
Słotwińska, S. M. Ghrelin and oral diseases. Cent.-Eur. J. Immunol. 45, 433–438 (2020).
Pritchett, N. R. et al. Serum ghrelin and esophageal and gastric cancer in two cohorts in China. Int. J. cancer 146, 2728–2735 (2020).
Cheng, R., Wu, Z., Li, M., Shao, M. & Hu, T. Interleukin-1β is a potential therapeutic target for periodontitis: a narrative review. Int. J. oral. Sci. 12, 2 (2020).
Ribes, S. et al. Oral impairments decrease the nutrient bioaccessibility of bread in the elderly. Food Hydrocoll. 135, 108202 (2023).
Wang, Q. et al. Computational methods and challenges in analyzing intratumoral microbiome data. Trends Microbiol. https://doi.org/10.1016/j.tim.2023.01.011 (2023).
Newman, N. K. et al. Transkingdom Network Analysis (TkNA): a systems framework for inferring causal factors underlying host-microbiota and other multi-omic interactions. Nature protocols, https://doi.org/10.1038/s41596-024-00960-w (2024).
Acknowledgements
We appreciate Professor Xuesong He from ADA Forsyth Institute for providing contributing suggestions and language guidance. We thank Postgraduate Yiting Cheng and Han Jiang for their assistance with the figure design. This work was supported by the National Natural Science Foundation of China (NO. 82170948, 82270972, 82401091, 81970948) and Sichuan Science and Technology Program (2024YFFK0069, 2024NSFSC0543). The funder played no role in the research. We thank American Journal Expert for providing language editing after each draft.
Author information
Authors and Affiliations
Contributions
MY Xia and L Lei contributed to the study conception, study design, data acquisition and interpretation, figure drawing, drafted and critically revised the manuscript. MM Li and HY Zhang contributed to the data acquisition and interpretation, figure drawing, and critical revision of the manuscript. JK Hu, LY Zhao and WQ Xu contributed to the data acquisition and interpretation and critically revised the manuscript. R Cheng and T Hu contributed to the study conception, design and critically revised the manuscript. All authors gave final approval and agreed to be accountable for all aspects of the work.
Corresponding authors
Ethics declarations
Competing interests
All authors declare no financial or nonfinancial competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xia, M., Lei, L., Zhao, L. et al. The dynamic oral–gastric microbial axis connects oral and gastric health: current evidence and disputes. npj Biofilms Microbiomes 11, 1 (2025). https://doi.org/10.1038/s41522-024-00623-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41522-024-00623-4
This article is cited by
-
The interleukin gene landscape: understanding its influence on inflammatory mechanisms in apical periodontitis
Molecular Biology Reports (2025)
-
Streptococcus anginosus: the potential role in the progression of gastric cancer
Journal of Cancer Research and Clinical Oncology (2025)