Abstract
Gut microbes play a vital role in host physiology, but whether specific bacterial functions contribute to the exceptional athletic performance of racehorses needs to be better understood. Here, we identify an association of gut butyrate-producing bacteria with athletic performance in racehorses (Thoroughbred horse). Butyrate-producing bacteria and microbial butyrate synthesis genes were significantly enriched in the racehorse gut, and the GC-MS results confirmed this conclusion. Using a mouse model, we demonstrated that sodium butyrate is sufficient to increase treadmill run time performance. We also show that butyrate improves the host response to exercise, significantly altering muscle fibre type in skeletal muscle, and increasing muscle mitochondrial function and activity. In addition, in-depth analysis of the published data showed that the gene for the synthesis of butyrate was also significantly enriched in the gut microbes of human athletes. Overall, our study indicates that gut microbial butyrate improves run time via the gut-muscle axis, providing novel insights into gut microbial functions and paving the way for improving athletic performance by targeted gut microbiome manipulation.
Similar content being viewed by others
Introduction
Because of their power and athletic ability, horses (Equus caballus) provide people with enjoyable sports and recreation, such as racing, hacking, hunting, show jumping, and eventing1,2. Horse racing is popular worldwide as a national pastime and to bet on and celebrate festivals. People want to see their horses run fast for prizes and glory. There are an estimated 59 million horses worldwide3, with an annual economic impact of approximately $300 billion4,5, and further understanding the factors affecting horse athleticism, therefore, would have a notable economic impact on the cost of caring for horses and the income from horse racing6. In addition, racehorses, which are routinely submitted to very intensive exercise during training and races, provide a good model for studying the factors that influence the athletic ability of human athletes7,8.
The gut microbiota of horses comprises bacteria, fungi, archaea, protozoa and viruses, which play important roles in host physiology, including the potential to improve athletic performance9,10,11. Growing evidence suggests that the gut microbiome profoundly affects the host’s athletic ability12,13. The athletic performance of germ-free mice was significantly worse than that of standard specific pathogen-free (SPF) mice, and transplanting the gut microbiota from SPF mice into germ-free mice improved performance by inducing changes in the antioxidant enzyme system and regulating skeletal muscle mass and function14. Scheiman et al. reported that Veillonella atypica in the gut of human athletes metabolises lactate into propionate, which can improve athletic performance15. Some studies have shown that exercise training results in significant changes in microbial compositions in the horse gut, and higher abundances of Firmicutes and Bacteroidetes were found in the racehorse gut at 48 h after exercise16,17. Mach et al. reported that gut microbiomes enriched in Lachnospiraceae taxa were negatively associated with cardiovascular fitness in elite endurance horses9. However, if and how the gut microbiome functions are responsible for better athletic performance in horses remains to be characterised.
Thoroughbred horses are the fastest gallopers of all horse breeds and have gained built-in biological mechanisms for racing prowess through years of selective breeding and tailored training programmes18,19. Horse racing represents a typical high-intensity short-duration effort20. During horse racing, muscle is activated close to the upper limit or maximal contraction duration, exerting tremendous physiological stress on the whole body21 and forcing the gut microbiome and the host to adapt to this adversity22,23,24. Many physiological changes occur in response to the intensity of physical activity, such as the recruitment of fast glycolytic pathways, increased mitochondrial biogenesis, acute inflammatory reactions, and an unbalanced redox system. Recent evidence shows the gut microbiota may change mitochondrial biogenesis by regulating key transcription factors or enzymes, such as ATP synthase 6 (ATPase6), the transcriptional regulator of mitochondrial DNA (Tfam), and uncoupling protein 1 (UCP1)25,26,27. Moreover, compelling new evidence suggests that the gut microbiota and its metabolites, including short-chain fatty acids (SCFAs), also contribute to intestinal anti-inflammation and intestinal barrier protection by inhibiting inflammasome responses28,29.
In this study, we aimed to investigate the functions and potential mechanisms of gut microbes contributing to the athletic performance of racehorses. We recruited gut samples of the racehorse group (n = 58) and non-racehorse group (n = 65), and conducted combined gut metagenomic, metatranscriptomics, and metabolomics approaches to identify gut microbial structures and functions associated with athletic performance. Furthermore, we investigate the beneficial effects of gut microbial butyrate on athletic performance via the gut-muscle axis. Our results indicate that gut microbial butyrate improves run time, thereby providing further rationale for targeted gut microbiome manipulation that could improve athletic performance.
Methods
Horse recruitment
A total of 58 racehorses (Thoroughbred horse) were used in this study. These racehorses were bred in the same racecourse and trained under the same conditions for more than 4 months. They all followed the same daily training programme, with a mean duration of the training, excluding hacking or round pen work, of 4 h/day, 2 h in the morning and 2 h in the afternoon, with resting on Saturday and Sunday. Training typically involves a warm-up on a horse walker, followed by ridden walking and trotting (usually on the way to the gallop), then two pieces of canter or speed work (2 km each, separated by a walk) on the racecourse professional track followed by recovery at a walk on the return to the stables. All horses were reared under similar rearing and management conditions. Commercially formulated fodder (concentrate diet A: 10% COOL ‘N’ COOKED MIX, Connolly’s RED MILLS, Goresbridge Co., Kilkenny, Ireland; or concentrate diet B: MDN, Beijing Redrunm Technology Company, Ltd, Beijing, China) and alfalfa hay were available ad libitum for each horse. All the horses were kept in separate stalls. At the time of sampling, these racehorses had been kept for 3 months according to the above breeding and training procedures. Racehorses that have achieved at least one top-three finish in races within their age-matched categories are considered to have good athletic performance, and these racehorses are included in this study. We evaluated the fitness of racehorses based on their mental state, veterinary evaluation and athletic performance. All horses had not received any antimicrobial treatment (antibiotic, anthelmintic or anti-inflammatory nonsteroidal treatments) in the 2 months before sampling.
In order to minimise the influence of various factors (diet, genetics, rearing and management conditions, etc.) on the identification of exercise-associated microbiota, we selected four breeds of horses from different breeding environments and conditions as non-racehorse controls, including 35 Yili horses, 10 Tibetan horses, 10 Yunnan pony horses and 10 Akhal-Teke horses. These non-racehorse controls (Yili, Tibetan, Yunnan pony and Akhal-Teke horses) have not received any special training and have not competed in the Horse Racing Series. They are not used for racing and training, but for visitors to visit. Yili and Akhal-Teke horses were fed a mix of pasture grasses supplemented with 5 kg/day concentrate feedstuffs (concentrate diet C: Maliang, Xinjiang Tiankang Feed Co., Ltd, Xinjiang, China, and alfalfa hay). Tibetan and Yunnan pony horses were grazed on natural pastures. All the horses had free access to water. In addition, all horses had not received any antimicrobial treatment (antibiotic, anthelmintic or anti-inflammatory nonsteroidal treatments) in the 2 months before sampling.
Faecal sampling
Faeces samples were collected from the rectum of Thoroughbred horse after a race in the Chinese National Horse Racing Series in 2020. These racehorses competed in races ranging from 1.1 to 2.3 km. All the races were concentrated in July and August. Within an hour of the game ending, one faecal sample from each animal was collected in sterile 50 ml cryovials (DNA and RNA-free) and immediately snap-frozen in liquid nitrogen. For the non-racehorses, fresh faecal samples were also obtained from the rectum. All samples were transferred to a laboratory and kept in a −80 °C freezer until the extraction of nucleic acid (DNA and RNA) and SCFA analysis. All details about the experimental horses and samples are listed in Supplementary Table 1.
Faecal DNA extraction and metagenome sequencing
As reported in previous studies, a modified hexadecyltrimethylammonium bromide (CTAB) method was used for DNA extraction30. Briefly, 1000 µl of CTAB lysis buffer (0.1 M Tris-HCl [pH 8.0], 1.4 M NaCl, 0.02 M EDTA, and 2% CTAB; DNA- and RNA-free) and 20 µl of lysozyme (DNA- and RNA-free) was added to a 100 mg sample, and the lysis solution was incubated for 2–3 h at 65 °C after mixing well. After brief centrifugation, 950 µl of the supernatant was mixed with 950 µl of phenol–chloroform–isoamyl alcohol (25:24:1, pH 8.0, DNA- and RNA-free) solution. After centrifugation at 12,000 × g for 10 min, the supernatant was mixed with an equal volume of chloroform–isoamyl alcohol (24:1, DNA- and RNA-free). After centrifugation at 12,000 × g for 10 min, the supernatant was mixed with a ¾ volume of isopropanol, and the tube was placed at −20 °C for 20 min after mixing. Following another centrifugation at 12,000 × g for 10 min, the supernatant was discarded, and the precipitate was washed twice with 1 ml of 75% ethanol. After drying, 50 µl of ddH2O was added to dissolve the DNA sample. Next, 1 µl of RNase A was added, and the solution was incubated at 37 °C for 15 min to digest the RNA. Agarose gel electrophoresis, a NanoDrop Kit (Implen, CA, USA) and a Qubit® 2.0 fluorometer (Life Technologies, CA, USA) were used to evaluate the purity and precisely quantify the DNA. According to the manufacturer’s protocol, a metagenome library was constructed from 1 μg of DNA using a NEBNext® Ultra™ DNA Library Preparation Kit (New England Biolabs, USA). All metagenome libraries were purified with the AMPure XP system (Beckman Coulter, Brea, CA, USA). The quantity of each library was evaluated using a Qubit 2.0 fluorimeter (Invitrogen, Carlsbad, CA, USA). These metagenome libraries were subjected to paired-end sequencing using the Illumina NovaSeq 6000 platform with a read length of 2 × 150 base pairs (PE150).
Faecal RNA extraction and metatranscriptome sequencing
Total RNA was extracted from 0.5 g of centrifuged faecal content using TRIzol (Invitrogen, CA, USA) according to the manufacturer’s protocol. Briefly, samples were mixed with 200 mg of 0.1 mm zirconia beads (BioSpec Products, Bartlesville, OK, USA) and 1.2 ml of TRIzol using a Fast Prep-24 instrument (MP Biomedicals, Irvine, CA, USA). RNA precipitation was performed using a mix of isopropanol and a saline solution (1.2 M NaCl, 0.8 M disodium citrate)31. Next, 1 µl of DNase (NEB, Ipswich, MA, USA) was added, and the solution was incubated at 37 °C for 15 min to digest the DNA. The integrity and quantity of total RNA were assessed with an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). Subsequently, ~100 ng of purified RNA was used to construct a metatranscriptome library using a TruSeq RNA Library Prep Kit v2 (Illumina, San Diego, CA, USA) according to the manufacturer’s protocol. A high-sensitivity bioanalyzer chip (Agilent, Santa Clara, CA, USA) was used to assess the quality of the metatranscriptome library. These metatranscriptome libraries were subjected to paired-end sequencing using the Illumina NovaSeq 6000 platform with a read length of 2 × 150 base pairs (PE150).
Analysis of metagenome-assembled genomes (MAGs) and metatranscriptomes
Quality control of each dataset obtained from the Illumina sequencing platform was performed using Trimmomatic (ver. 0.35) to filter out low-quality bases (quality score < 20), low-quality reads, and adaptor contaminants32. These reads were aligned to the horse genome (available at NCBI sequence archive with the accession number EquCab3.0: GCA_002863925.1; Equus caballus. EquCab3.0) using Bowtie 2 software (ver. 2.2.4) to remove potentially contaminating reads with the parameters ‘end-to-end, sensitive, -I 200, -X 400’33. The remaining high-quality reads were further analysed.
Assembly and binning were carried out as previously described34. Cleaned data were assembled into scaffolds using SOAPdenovo (ver. 2.04) software with the parameters ‘-d 1, -M 3, -R, -u, -F, -K 55’35. MetaWRAP ver. 1.2.1 software (https://github.com/bxlab/metaWRAP) was subsequently used for metagenomic binning36. Duplicate bins were removed using dRep1.1.2 software with the parameters ‘-p 16, -comp 80, -con 10, -str 100, -strW 0’, and secondary clustering was performed at a threshold of 99% average nucleotide identity and at least 25% overlap between MAGs34. Assembled high-quality bins were assessed using CheckM (ver. 1.0.7) based on integrity criteria ≥80% and contamination ≤10%34,37. The metawrap quant_bins module was used with the options ‘metawrap quant_bins -b genomes/-o QUANT_BINS/’ to calculate the abundance of MAGs in each sample36. The Genome Taxonomy Database Toolkit (GTDB-Tk; ver. 1.3.0, release 95) was used to assign the taxonomy of the MAGs38. The parameters used were set to default. Finally, PhyloPhlAn ver. 3.0.51 software39 was used to construct a maximum-likelihood phylogenetic tree of high-quality MAGs with the following parameters: ‘--diversity low --fast --min_num_marker 50’. MetaGeneMark (prokaryotic GeneMark.hmm, ver. 2.10) was used to predict open reading frames40. DIAMOND (ver. 0.9.9.110) aligned the predicted genes with the integrated NR database (25 October 2019; https://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/). Genes and reads were mapped to the database to determine abundance using Bowtie ver. 2.2.4 with the parameters ‘--end-to-end, --sensitive, -I 200, -X 400’. The abundance of genes was calculated by counting the number of reads and normalising the result according to gene length41. To assess the contribution of the constructed MAGs to the functional potential of the gut microbiome, the predicted genes and proteins were compared with the Encyclopedia of Genes and Genomes (KEGG) database (2018-01-01, http://www.kegg.jp/kegg/) using DIAMOND software to obtain KEGG orthologues to determine the functional pathways enriched in the microbiota.
All clean reads were subjected to de novo metatranscriptome assembly by using Trinity (ver. 2.2.0) with default settings42,43, the sequences of all samples were integrated, and repeats (sequence identity threshold: 0.95) were deleted using CD-HIT-EST to obtain a unigene dataset44. Transdecoder software (ver. 5.5.0) (http://transdecoder.github.io) was used to predict potential coding regions within these transcripts with default settings43. The microbial source of each transcript was identified by comparing unigenes with extracted bacterial, fungal, archaeal, and viral sequences from the NCBI nr_euk reference database (25 October 2019; https://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/) using DIAMOND software (blastp, evalue ≤ 1e-5), which was also used to map unigenes to the Gene Ontology (GO) and Kyoto KEGG databases to obtain gene function information. The clean reads of each sample were mapped to a reference sequence spliced by Trinity. Transcripts and genes were normalised using fragments per kilobase of transcript sequence per million base pairs (FPKM)45. Differential expression analysis was performed using DEGseq (version 1.20.0)46. The resulting p values were adjusted using the Benjamini–Hochberg method47. GO and KEGG enrichment analyses of differentially expressed RNAs were implemented by the clusterProfiler R package (version 3.14.3), in which gene length bias was corrected. Enrichment was considered significant when the corrected p-value was less than 0.05.
Quantification of faecal SCFAs via GC–MS detection in horses
Ether was used to prepare ten mixed standard-concentration gradients of acetic acid (Sigma–Aldrich, St. Louis, MO, USA), propionic acid (Sigma–Aldrich), and butyric acid (Sigma–Aldrich) to generate a standard curve of SCFAs; the concentrations used were 0.02, 0.1, 0.5, 2, 10, 25, 50, 100, 250, and 500 μg/ml. Twenty faeces samples (10 from each group) were randomly selected. A total of 100 mg of each sample was water extracted. A volume of 675 μl supernatant fraction was added to an Eppendorf (EP) tube containing 50 μl of 15% phosphoric acid, 100 μl of a 125 μg/ml internal standard (isohexanoic acid) solution, and 400 μl of ether; the mixture was homogenised for 1 min and then centrifuged at 12,000 rpm for 10 min at 4 °C. The supernatant was analysed using a Thermo TRACE 1310-ISQ LT Gas Chromatography-Mass Spectrometer (GC–MS) (Thermo Scientific, Madison, WI, USA) equipped with a 7693 autosampler (Agilent Technologies, Santa Clara, CA, USA), a flame-ionisation detector and a capillary column of HP-5MS (30 m × 250 µm; AgilentJ&W Scientific, Folsom, CA, USA). SCFAs were quantified in conventional 70 eV electron ionisation mode using Mass Hunter software (Agilent Technologies, Santa Clara, CA, USA)48.
Butyrate treatment experiment in mice
Thirty C57BL/6 mice (male), about 8 weeks old (±1 week), were purchased from the Experimental Animal Center of Huiji District, Zhengzhou City (Zhengzhou, Henan, China). All mice were housed in the Animal Genetic Engineering Laboratory at Shihezi University. The mice were randomly divided into two groups, namely, the sodium chloride group (Control, n = 15) and the butyrate group (Butyrate, n = 15). Mice were housed in SPF individually ventilated cages (2 mice per cage) under a temperature-controlled room (23 °C ± 3 °C) and relative humidity (60 ± 10%) conditions. The room was maintained on a 12:12-h dark-light cycle, and the light intensity was not less than 300 lux. The diet (Experimental Mice Maintenance Feed (AIN-93), XIETONG SHENGWU, Nanjing, China, 3530 kcal/kg) was standardised and identical for all groups, it included corn, wheat, fish meal, chicken meal, soybean meal, fat, soybean oil, sunflower oil, amino acids, vitamins, minerals. After 7 days of acclimatisation, animals were gavaged with 20 ml of sodium chloride and 20 ml of butyrate, respectively, for 42 consecutive days. The mice in the butyrate group were given water containing 0.1 M sodium butyrate (Solarbio, Beijing, China), and those in the control group received water containing 0.1 M sodium chloride (Solarbio, Beijing, China) to balance osmotic pressure. The processed water was stored in a −20 °C freezer, and the water of all mice was changed daily at 10:00 a.m. All mice had free access to water and food. Next, all mice underwent treadmill run experiments on the 42nd day of butyrate or sodium chloride treatment (see below for further details). Next, we lifted the tail of the mice and gently massaged the lower abdomen of the mice with our fingers. Fresh faeces were collected in a sterile EP tube and stored in a −80 °C refrigerator.
Treadmill run experiment in mice
Three days prior to starting the experiment, mice were acclimated to the treadmill (No. XR-PT-10B; Shanghai XinRuan Information Technology Co., Ltd, Shanghai, China) by walking at 10 m/min for 10 min every day for three consecutive days. Athletic performance was based on exhaustion time. For exhaustion measurements, mice fasted for 6 h before exercise. Mice were run on the treadmill with a runway with 11° of inclination. The exercise regimen started with the shock grid ON at 10 m/min for 30 s, and the speed was increased by 1 m/3 min up to 20 m/min and then held at 20 m/min until exhaustion. The electric shock intensity was 2 mA, and the electric shock tolerance time was 10 s at a frequency of 50 Hz. According to the stimulation of the experimental animal, the system automatically identifies and records the exhaustion time and stops the stimulation of the experimental animal simultaneously.
Mice grip strength testing and tissue sample collection
Three days after the treadmill test, grip strength was assessed in mice in both the control and butyrate groups using a force metre (ELECALL, Zhejiang, China). In brief, mice were lifted and held by the tail, causing the forelimbs to grasp the pull-bar assembly connected to the force metre. The mouse was drawn along a straight line leading away from the sensor until the grip was broken, and the peak amount of force in grams was recorded. Measurement of forelimb grip strength of each mouse was repeated 5 times. Then, the mean maximum strength was used for data analysis. After completing the measures, the mice were euthanised.
Quantification of serum SCFAs via GC–MS in mice
Serum samples from 12 randomly selected mice (6 from each group) in the butyrate-treated and control groups were analysed following the same method as for horse faeces.
Mice euthanasia and sampling
Mice were euthanised by cervical dislocation. Immediately after, blood was collected via cardiac puncture. The small intestine and skeletal muscle (hindleg muscle) were quickly harvested for further processing. Portions of the skeletal muscle (hindleg muscle) and small intestine were fixed in 4% paraformaldehyde (Biosharp, Shanghai, China) for histological examination (see below). The remaining samples were immediately frozen in liquid nitrogen and stored at −80 °C for further analysis.
Mice tissue qRT-PCR analysis
Total RNA was extracted from mice’s skeletal muscles (hindleg muscle) using TRIzol (Invitrogen, CA, USA). From purified RNA, cDNA was synthesised using an RT-PCR Kit (Takara, Dalian, China). As reported in previous studies13, the expression levels of marker genes related to muscle fibre types and mitochondrial function were analysed by qRT-PCR in all samples. All primers were designed with Primer 5.0 software, and the primer sequences for these genes are listed in Supplementary Table 2. qRT-PCR was performed using SYBR Green (TaKaRa Biotech, Dalian) according to the manufacturer’s protocol. β-Actin was used as an internal reference gene. Each sample was run in triplicate. Real-time RT-PCR was conducted using the following reaction system: 10 μl of SYBR Premix DimerEraser, 2 μl of cDNA, 0.6 μl each of the upstream and downstream primers, and 6.8 μl of RNase-free ddH2O water. qRT-PCR was performed with the following thermocycling conditions: initial denaturation at 95 °C for 30 s, followed by 55 cycles of 95 °C for 5 s, 58 °C for 10 s, and 72 °C for 10 s. Relative gene expression was analysed by the ΔΔCt method49.
Mitochondrial DNA copy number quantification
Three mice were randomly selected from the butyrate treatment and control groups for mitochondrial DNA analysis. Total DNA was extracted from mice’s skeletal muscles (hindleg muscle) using the Rapid Animal Genomic DNA Isolation Kit (Sangon Biotech, Shanghai, China). The ratio of mitochondrial DNA copy number to nuclear DNA was determined by qPCR13. Mitochondrial DNA was quantified using the NADH dehydrogenase subunit 1 (ND1) gene. Nuclear DNA was quantified using the β-like globin (β-globin) gene.
Histological analyses
At the end of the experiment, the skeletal muscle (hindleg muscle) tissues of three mice were randomly selected from the butyrate treatment group and the control group for histological analyses at room temperature. These samples were fixed in 4% paraformaldehyde for 24 h. Then, the samples were removed and immersed in 100% ethanol for 24 h for dehydration and processed for paraffin embedding. The samples were cut into ~6 µm-thick sections using a rotary microtome (RM2255, Leica Biosystems, Germany) and stained with haematoxylin-eosin (Solarbio, Beijing, China) to quantify total fibres and cross-sectional area. A total of 100 fibres from each mouse were measured. Images were obtained using a microscope (OLYMPUS-BX 50, Olympus Optical, Tokyo, Japan) at 20× magnification50.
Public populations of human athletes and non-athletes
Raw sequencing data of human populations from ten athletes (Professional international rugby union players) and ten non-athletes (People with a sedentary lifestyle) were downloaded from the ENA (http://www.ebi.ac.uk/ena/data/, accession number PRJEB15388), mainly from a recently published athlete’s paper51. These data were reanalysed following the analytical process for horse gut metagenomic data described in this article.
Statistical analysis
The results of all qRT-PCRs were analysed by one-way analysis of variance (ANOVA), and all results are expressed as the mean ± SD. The relative abundance of microbial taxa was arcsine square root transformed, and ANOVA was used to compare the differences between racehorses and non-racehorses42. Based on the Shannon and Simpson indices, the alpha diversity was compared among groups by the Kruskal–Wallis test. Principal coordinates analysis (PCoA) was used to visualise racehorse and non-racehorse gut microbial communities at the bacterial genus level based on the Bray‒Curtis difference matrix using PERMANOVA32. As previously reported, transcripts with relative abundances greater than 0.01% were selected for further analysis52. Read counts were normalised in FPKM to account for differences in gene length53. Differential abundances of functional classes and transcripts were compared between racehorses and non-racehorses. All other statistical analyses were conducted in R ver. 3.6.2. The GraPhlAn ver. 1.1.3 and the ggtree packages in R ver. 3.6.2 were used to draw phylogenetic trees54,55. Box plots and scatter plots were drawn with the ggplot2 package56.
Results
Enrichment of microbes involved in butyrate synthesis in racehorses
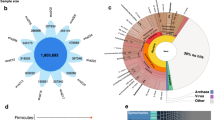
To reveal the gut microbes associated with the athletic performance of horses, we collected faeces samples from 58 racehorses (Thoroughbred horse) and 65 non-racehorse controls. The non-racehorse controls included four breeds of horses (35 Yili, 10 Tibetan, 10 Yunnan pony and 10 Akhal-Teke horses) (Supplementary Table 1). These faecal samples were subjected to metagenomic and metatranscriptomic next-generation sequencing. After quality filtering and host sequence decontamination, nearly 938 GB of high-quality metagenomic data was available for further analyses (Supplementary Table 3), with an average sequencing depth per sample of 50.9 million paired reads (Supplementary Table 3). A total of 98.9% of the taxonomic reads belonged to bacteria, and the remaining 1.1% belonged to viruses, archaea, and eukaryotes. A total of 900 high-quality (defined as >80% completeness and <10% contamination) metagenome-assembled genomes (MAGs) were created using gene compartmentation (Fig. 1A, Supplementary Table 4)57, which made it possible to study differences in gut microbial communities between racehorses and non-racehorses at the species level. According to the GTDB-Tk, this MAG repertoire was assigned to 897 bacteria and 3 archaeon, involving bacteria from the Bacteroidota and Firmicutes, followed by Spirochaetota, Proteobacteria, Fibrobacterota, Verrucomicrobiota, Elusimicrobiota, Actinobacteriota, Myxococcota, Campylobacterota, and Abditibacteriota (Supplementary Table 4). However, only 774 and 53 MAGs were classified at the genus and species level, respectively. Given the abundance and variety of carbohydrates in the horse diet, most MAGs encoded enzymes that degrade polysaccharides (Supplementary Table 5).
A Basic information of the assembled MAGs. Phylogenetic tree of MAGs from racehorses and non-racehorses’ gut, generated by PhyloPhlAn. The coloured circles represent MAGs. B The principal coordinate analysis (PCoA) of taxonomical composition was performed on all samples from racehorses (n = 58) and non-racehorses, including Yili horse (n = 35), Tibetan horse (n = 10), Yunnan pony horse (n = 10) and Akhal-Teke horse (n = 10). C Top 30 differentially abundant MAGs in the racehorse and non-racehorse horse gut. Colours from blue to red represent an increasing abundance of MAGs. D The differences in the abundance of genes encoding key enzymes involved in butyrate synthesis between the racehorse and non-racehorse. The enzymes, which show a significant difference in abundance between racehorses and at least one of four non-racehorse breeds, were labelled in red in the butyrate synthesis pathway.
To identify the differences in the gut microbiome composition between racehorses and the four breeds of non-racehorses (Yili, Tibetan, Yunnan pony and Akhal-Teke horses), we first performed principal coordinate analysis (PCoA, Bray‒Curtis distance) of the 900 MAGs abundance. We found differences in the gut microbial communities between racehorses and the four breeds of non-racehorses (Fig. 1B), and that the separation of microbial communities was statistically significant between racehorses and each of the four breeds of non-racehorses (Supplementary Fig. 1). Furthermore, when the four breeds of non-racehorses were combined into a whole control group, the microbial communities of racehorses were still significantly different from that of the combined non-racehorses controls (Supplementary Fig. 1, Adonis PERMANOVA, p < 0.001). Compared with the combined non-racehorse controls, the Simpson index of the racehorses decreased significantly (Wilcoxon test, p < 0.05), but the difference in Shannon index was not statistically significant (Supplementary Fig. 2).
We then investigated which microbial taxa were underpinned in each group at the genus level. Olivibacter and Alicyclobacillus genera were most abundant in the racehorse gut (Supplementary Fig. 3). A total of 190 MAGs were differentially abundant between groups (Supplementary Fig. 4, Supplementary Tables S6 and S7). MAGs ascribed to Olivibacter (HFM56.bin.4, HFM51.bin.9, HFM92.bin.23 and HFM46.bin.11), Prevotella (HFM73.bin.4 and HFM51.bin.6), Paludibacteraceae (HFM96.bin.1), Saccharofermentans (HFM88.bin.17), Ruminococcus (HFM53.bin.6) and Colidextribacter (HFM80.bin.13) taxa were among the top significantly enriched in the racehorse gut (Fig. 1C, Supplementary Tables S6 and S7). The most abundant microbial genomes in racehorses were HFM56.bin.4 (Olivibacter).
Having identified genus- and species-level differences in the microbiota of racehorses and non-racehorses, we next aimed to predict the potential functions of individual microbiota constituents in these animals. Metagenomic analysis of KEGG pathways revealed significant differences in gut microbiota function between racehorses and non-racehorses (Fig. 1D). A total of 35 KEGG pathways and 21 of them are metabolism, including butanoate metabolism, fatty acid degradation, Metabolism of cofactors and vitamins and Metabolism of terpenoids and polyketides, were significantly differentially abundant (adjusted P < 0.05) between racehorses and non-racehorses (Supplementary Table 8). Interestingly, out of all carbohydrate metabolic pathways, only the butanoate metabolism pathway was enriched in the racehorse gut, indicating a potentially critical role for butanoate in the gut.
To validate the significance of the association between butyrate and racehorses, we next focused on the butanoate metabolism pathway to reveal the key steps linking the athletic performance of the racehorse due to butyrate as a potential ergogenic aid for exercise and training58. Analysis of the metagenomic enzymes revealed that some critical genes involved in butyrate synthesis were significantly enriched in the butanoate metabolism pathway. At least one enzyme in all steps involved in the gut synthesis of butyrate in racehorses was significantly enriched when compared with those in four non-racehorse breeds (Fig. 1D, Supplementary Fig. 5, Supplementary Table 9). Especially, butyrate kinase (EC 2.7.2.7), which was a key enzyme for butyrate synthesis in the final step in the butyrate pathway, was significantly more abundant in racehorses than in all four control breeds.
Differences in intestinal butyrate synthesis between racehorses and non-racehorses
After identifying the enrichment of butyrate-producing bacteria in racehorses, we next aimed to evaluate whether there was a significant difference in gut microbial butyrate synthesis between racehorses and non-racehorses. Gut microbes produce butyrate through two main pathways. We used metatranscriptomics data to analyse the differences in the expression of the enzymatic machinery related to the butyric acid synthesis pathway between racehorses and non-racehorses, including the butyryl-CoA: acetate CoA-transferase pathway (but) and the butyrate kinase (buk) pathways. We found a significant difference in transcription of butyrate pathway genes in the gut microbiota of racehorses compared to their control counterparts (Supplementary Fig. 6, Supplementary Table 10). The expression levels of the enzymes trans-2-enoyl-CoA reductase (NAD+) (EC 1.3.1.44) and butyrate kinase (EC 2.7.2.7) were significantly higher (40-fold and 3-fold, respectively) in the racehorse gut microbiota than in the non-racehorse gut microbiota (Fig. 2A, Supplementary Table 11).
A Bar graph showing the levels of transcripts encoding critical enzymes for butyrate synthesis, according to the metatranscriptomic analysis of horse faeces. The enzymes here are the same as those shown in Fig. 1E. B Pod plots of SCFA concentrations in the faeces of racehorses and non-racehorses analysed by targeted metabolomics. Numbers in parentheses indicate the fold change in the content of the corresponding SCFA in the intestines of racehorses relative to non-racehorses.
We next measured faeces’ SCFA levels in faeces (as a proxy of the colon production) using targeted gas chromatography-mass spectrometry (GC‒MS) to identify further the difference in gut microbial butyrate synthesis between racehorses and non-racehorses. Acetate, propionate and butyrate comprised >95% of the total SCFA pool and were present at a molar rate of 72.3%:14.6%:8.1% in faeces. We found that butyrate (P < 0.001), propionate (P < 0.001), caproate (P < 0.01), valerate (P < 0.01), and branched-chain fatty acids such as iso-butyrate (P < 0.001) and iso-valerate (P < 0.001) levels were significantly higher in racehorse faeces than in non-racehorses’ faeces (Fig. 2B). Among the SCFAs present at similar concentrations (acetate, propionate and butyrate), butyrate showed the most significant difference between groups (~8.6-fold, P < 0.001), implying a critical role for this SCFA in the racehorse gut.
Sodium butyrate enhances the athletic performance of mice
To test whether gut microbes’ enhancement of host athletic performance is at least partly attributable to butyrate, we compared the performance of butyrate-treated mice and control mice (saline vehicles) on a motorised, speed-controlled, modular treadmill system spanning 6 weeks. The butyrate-treated group (butyrate) had a significantly prolonged exercise time on the treadmill compared with the sodium chloride-treated control group (Control) (Fig. 3A, P < 0.05). In addition, to assess changes in muscle physiology, we performed an involuntary physical exercise test of muscle strength in the same treated and control mice to determine the effect of butyrate on muscle strength. Butyrate-treated mice had greater grip strength (P < 0.05, Fig. 3B) than their control counterparts. Together, these results suggest that butyrate may improve the athletic performance of the host.
Butyrate treatment can significantly improve the athletic performance (A) and muscle grip (B) of mice. C The results of the targeted metabolomic study showed that butyrate gavage could significantly increase the concentration of butyrate in mouse serum without changing the concentrations of other SCFAs. * significant, ** highly significant.
While butyrate is traditionally considered the principal energy source for intestinal epithelial cells59 and up to 95% of microbial-produced butyrate has been estimated to be consumed by the colonocytes, it is worth considering whether butyrate is distributed beyond the gut to the central nervous system and peripheral tissues such as liver, and brown and white adipose tissues, and used as a substrate for mitochondrial oxidation, lipid production, and gluconeogenesis, regulating the whole body energy homoeostasis. Therefore, we used targeted GC‒MS to measure SCFA levels in butyrate-treated and sodium chloride-treated mice serum. The results showed that the serum butyrate content in butyrate-treated mice was significantly higher than that in nontreated mice (P < 0.01, Fig. 3C). There were no differences between the two groups in the levels of other SCFAs, including acetate, propionate, valerate, caproate, iso-butyrate, and iso-valerate (Fig. 3C). Taken together, these results indicate that microbially synthesised butyrate in the gut is a critical metabolite that may enhance host athletic performance.
Butyrate acts on skeletal muscle to promote athletic performance in the mice treadmill model
Direct evidence for butyrate’s role in regulating muscle function is emerging60. To elucidate the mechanism by which butyrate mediates enhanced muscle strength and athletic performance in mice, we analysed the expression of several exercise-related genes in the skeletal muscle and the small intestine, including those involved in redox reactions, energy metabolism, mitochondrial function, muscle fibre type composition, inflammation, and motor neuron signalling, following the treadmill running. Considering that skeletal muscle composition and functions directly affect athletic performance, we first investigated the effect of oral butyrate on skeletal muscle. Out of 14 genes (Myog, Tnnt1, Myh2, Lrp4, Rapsn, Ppargc1α, Lrp4, Tnnt3, Myh4, Pax7, Myod, Foxo3, Myh1, Mb and Myosin) analysed, qRT-PCR results showed that the expression levels of skeletal muscle differentiation genes such as myogenin (Myog), myosin heavy chain IIa (Myh2) and peroxisome proliferator-activated receptor γ coactivator-1 (Ppargc1α) were increased (P = 0.05) in the skeletal muscles of the butyrate-treated mice compared with those in the skeletal muscles of the control mice. Furthermore, the expression levels of LDL receptor-related protein 4 (Lrp4), which is important for neuromuscular junction assembly and function, were also higher in the skeletal muscles of the butyrate-treated mice than those of the control mice. In addition, we observed increased expression of critical genes for myofiber contractility, namely troponin T Type 1 (Tnnt1) and Troponin T Type 3 (Tnnt3) (P < 0.05) (Fig. 4A). Conversely, the expression level of the muscle atrophy marker muscle RING-finger was lower than that in control mice. Still, the difference was insignificant (Fig. 4A). Histological analysis showed that the diameter of muscle fibres increased significantly in the skeletal muscles of the butyrate-treated mice than in those of the control mice (Fig. 4B, C). These results indicate that the fibre type of skeletal muscles in mice treated with butyrate changes profoundly. The coupled histological analysis supported these molecular transcriptional results. The increase in the proportion of ganglion head, fast twitch (type II, glycolytic), and slow twitch (type I, oxidative) muscles, which are rich in mitochondria and stimulate FAO for ATP production, may be the most direct cause of the increased motor performance and strength in mice.
qRT-PCR analysis shows the relative expression of genes related to muscle fibre type (A), mitochondrial function (D) and energy metabolism (E) in butyrate-treated mice (Butyrate) and control mice (Control). β-actin levels are used as a control. Representative images of haematoxylin–eosin-stained muscle (B) pictures. The results shown are representative of n = 3 independent experiments. C Mean myofiber diameter for Control and Butyrate group. F Quantitative analysis of the mitochondrial DNA (mtDNA) ratio to nuclear DNA in gastrocnemius muscles from Control and Butyrate mice. *significant, ** highly significant.
Muscle mitochondria are energy supply centres of cells, and their quantity and function affect athletic performance by regulating muscle fibre contraction60. The gene expression of mitochondrial biogenesis markers61,62, including ATPase6, mitochondrial cytochrome b (Cytb), cytochrome c oxidase subunit 2 (Cox2), a transcriptional regulator of mitochondrial DNA (Tfam), and UCP1, was measured in hindleg skeletal muscle tissues to investigate differences in mitochondrial function between butyrate-treated mice and control mice. The expression levels of these five marker genes in muscle were higher in mice treated with butyrate than in control mice, with significant differences for ATPase6, Cytb, Cox2 and Tfam (P < 0.05) (Fig. 4D). Mitochondrial oxidative phosphorylation (OXPHOS), which combines electron transport with cell respiration and ATP synthesis, and fatty acid β oxidation are the two metabolic pathways that are central for energy production. Mitochondria can also use the enzymatic pathways of the tricarboxylic acid (TCA) cycle to generate ATP (about 20% of ATP). Therefore, we investigated key enzymes of mitochondrial energy metabolism, including succinate dehydrogenase (SDH), muscle-type phosphofructokinase (Pfkm), pyruvate kinase, lactate dehydrogenase (LDH), and pyruvate dehydrogenase (PDH), to evaluate the oxidative metabolic capacity of skeletal muscle. qRT-PCR results showed that the expression levels of SDH, Pfkm, LDH and PDH were significantly higher in butyrate-treated mice than in control mice (Fig. 4E), evoking increased capacity for OXPHOS and -oxidation of fatty acids or ketone bodies and thus, energy production. In addition, mitochondrial DNA content tended to be higher in the butyrate-treated group related to the control, suggesting increased biogenesis. Still, the difference was insignificant (P = 0.1, Fig. 4F).
Potential conservation of gut microbial butyrate synthesis in a human athlete cohort
To test whether gut microbial butyrate synthesis was positively associated with the athletic performance of human athletes, we reanalysed the dataset available by Barton et al. 51, and tested differences in gut microbiota between elite professional athletes and nonathlete healthy controls Consistent with findings from a previous study51, we found a significant separation of the gut microbiota between the athlete group and control group (Fig. 5A). Relative to the control group, the athlete group had a higher abundance of butyrate-producing bacteria, including Bacteroides63, Veillonella64, Prevotella65, and Romboutsia66,67,68 (Fig. 5B, Supplementary Table 12). Relative abundance analysis of butyrate synthesis genes encoded by the microbiota revealed that key enzymes in the butyrate synthesis pathway were significantly enriched in the athlete’s gut (Fig. 5B, Supplementary Table 13).
A PCoA of the taxonomy of the gut microbiota of human athletes (n = 10) and controls (n = 10). B Genes encoding enzymes for butyrate synthesis were enriched in human athlete gut microbes. The bar chart shows the differences in the abundance of genes encoding key enzymes in butyrate synthesis between human athletes and control individuals. *significant, ** highly significant.
Discussion
Horses are raised worldwide as a source of mechanical energy for sports, leisure, and transportation. The gut microbiota is critical in horses’ health, athletic performance, and behaviour6,7,8. However, the need for more functional data on gut microbiota on athletic performance has dramatically hindered our understanding of microbial roles and our ability to regulate the athletic performance of hosts via the gut microbiota. More importantly, insights into the effects of the gut microbiome on athletic performance in racehorses, as an exercise training model, may provide information to optimise gut microbiota functionality for both human athletes and fitness people. Here, we performed a metagenomic and metatranscriptomics analysis of the gut microbiome of racehorses and non-racehorses and identified an association between gut microbial butyrate and athletic performance.
The activity of fibre degraders like Olivibacter, Prevotella, Paludibacteraceae, Saccharofermentans, Ruminococcus and Colidextribacter members likely contributed to the creation of butyrate resources, leading to its local and systemic availability. Notably, elevated abundances of Olivibacter, Prevotella and Ruminococcus were also observed in human athlete microbiomes or postexercise69,70. The abundance of butyrate-producing species matched increased gut microbiome diversity and coincided with increased faecal butyrate concentration. Reconstruction of metabolic pathways for butyrate production based on community gene expression also indicated that butyrate synthesis was more active in racehorses. These butyrate-producing bacteria in racehorses may produce higher relevant goods for host energy requirements, better gut immune function and inflammation regulation, indirectly impacting physical performance and recovery. The relationship between physical activity, the gut microbiome, and butyrate, coupled with the considerable malleability of the microbiota relative to host genomes, opens the possibility of influencing gut health and athletic performance via microbiota manipulation. Additionally, this knowledge may be developed into bacteria-based indicators for the early selection of future racehorses with different athletic abilities. However, establishing specific performance-enhancing marker microbes requires further experimental research71.
A mouse model was used to tie together the butyrate production and athletic performance. Butyrate treatment significantly improved limb grip and athletic performance in mice. Although previous reports indicate that butyrate can be utilised as an energy source in intestinal epithelial cells52, we demonstrated using targeted metabolomics that butyrate can act locally, and be carried into the bloodstream to skeletal muscle while improving athletic performance. Having observed that butyrate directly enhances mice’s athletic performance and muscle strength, we next found that butyrate treatment in mice can significantly increase the function of mitochondria and energy metabolism in skeletal muscle, directly improving the availability of ATP, which is critical for skeletal muscle contractile activity in explosive-power or spring events lasting from seconds to minutes. Because the intramuscular stores of ATP are relatively small, activating other mitochondrial pathways is essential. This finding is consistent with studies showing that butyrate increases mitochondrial biogenesis, function, and energy expenditure72,73. Upon further assessment of changes in skeletal muscle, we found that butyrate treatment significantly increased neuromuscular junction assembly and function and fast twitch and slow twitch muscles, which are all beneficial for athletic performance and strength13,74. Building upon previous reports indicating that butyrate can significantly increase muscle mass in aged mice75, our study revealed another effect of butyrate on muscle. Mitochondrial and muscle changes provide the physiological basis for butyrate-mediated enhancement of host athletic performance14. Overall, the results of this study are consistent with previous reports that butyrate benefits host skeletal muscle. However, the models used in this study differ from those in previous studies based on germ-free mice, aged mice, or cells76,77,78,79. This study demonstrated that the benefits of butyrate synthesised by gut microbes are robust, especially regarding athletic performance, and paves the way to use pre-, pro- or postbiotics as therapeutic options to target gut athletes’ health and improve athletic performance.
In addition, our analysis using an independent cohort of human athletes showed that butyrate-producing bacteria are significantly enriched in the athletes’ gut microbiota compared to non-athletes. The butyrate synthesis genes were also improved in the human athlete gut, which addressed concerns about the conclusions regarding butyrate raised in the study from which the metagenomic data were procured51. Moreover, considering the increased faecal butyrate concentrations in human athletes’ gut, as Barton et al.51 reported, we believe that the butyrate content in athletes positively correlates with their athletic performance. Therefore, butyrate has the same critical role in promoting athletic performance in different species, such as humans, mice, and horses. Although the microbiota constituents that synthesise butyrate in the gut of different species are not identical, our study indicates that at least the pattern of microbial butyrate synthesis in the gut of other species under systemic stress induced by resistance exercise is relatively conserved80,81,82. During the co-evolution of the gut microbiota with its host83, butyrate-synthesising microbes may have been selected by the host due to their production of this critical substance that improves bodily functions.
Notably, diet is an important factor affecting the functional composition of the gut microbial system. Some studies highlight that dietary has extreme impact on gut microbial diversity of professional athletes51,84. The differences in microbial butyrate synthesis observed in this study were likely due to the different diet and genetic backgrounds between racehorses and non-racehorses, although we collected horses of various breeds in different dietary states as controls in an attempt to eliminate the influence of diet and genetic background on the gut microbiota85. Moreover, other factors influencing exercise metabolism and performance, including training status, sex, age, emotional and physical stress, and environmental conditions, should be noted in future work86,87. It is interesting to explore the difference in butyrate-producing bacteria in horses kept under similar conditions but with differing athletic performance or training intensity in future research88.
To meet the increased energy needs of exercise, we propose a model for microbe-mediated enhancement of performance in which the butyrate produced by gut bacteria may enter circulation as an energy supply and target metabolism for an ergogenic benefit during athletic events (Fig. 6). The model includes the following points: (1) Butyrate enters circulation and may serve as an important energy source for some tissues and organs (e.g. skeletal muscle), which promotes athletic performance. (2) Butyrate may regulate skeletal muscle fibre type and promote skeletal muscle fibre remodelling to improve host athletic performance. (3) Butyrate increases the number and energy metabolism efficiency of mitochondria, which provide energy to skeletal muscle for high-intensity exercise, thereby improving host athletic performance. (4) The butyrate metabolic pathway yields increased amounts of ATP, which may then be effectively utilised for bacterial propagation and thus further increase the abundance and gut colonisation of performance-enhancing microbes. Although the above hypothesis requires further experimental verification, the model contributes to our understanding of the excellent athletic performance of racehorses.
In conclusion, we demonstrated that butyrate-producing bacteria and/or butyrate contribute to athletic performance across species. The butyrate-producing bacteria may be developed into novel probiotics for improving performance by regulating the gut microbiota. Several butyrate-producing bacteria in the gut or butyrate concentration in blood could be used as a potential marker for optimising the sports training of elite athlete candidates. Therefore, we propose a regulatory model for synergy in the microbe-gut-skeletal muscle-mitochondria axis to enhance host athletic performance. Our findings provide new insights and a paradigm for improving host athletic performance by studying gut microbial ecosystems.
Data availability
All original sequences obtained in this work have been deposited in the National Center for Biotechnology Information (NCBI) under project numbers PRJNA1089803 and PRJNA973826. The data can also be obtained from the corresponding author upon reasonable request or from the included supplementary information files.
References
Wade, C. M. et al. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326, 865–867 (2009).
Plancade, S. et al. Unraveling the effects of the gut microbiota composition and function on horse endurance physiology. Sci. Rep. 9, 9620 (2019).
Jastrzębska, E., Daszkiewicz, T., Górecka-Bruzda, Górecka-Bruzda, A. & Feliś, D. Current situation and prospects for the horse meat market in Poland and the world. Med. Weter. 75, 196–202 (2019).
Orpin, C. G. Isolation of cellulolytic phycomycete fungi from the caecum of the horse. Microbiology 123, 287–296 (1981).
Cross, P. Global Horse Statistics Internal 02 2019. (HiPoint Agro Bedding Corp, 2019).
Musser, W. N. et al. Economic impact of horse racing in Maryland. Policy Anal. Rep. 99, 1 (1999).
Harkins, J. D., Beadle, R. E. & Kamerling, S. G. The correlation of running ability and physiological variables in Thoroughbred racehorses. Equine Vet. J. 25, 53–60 (1993).
Hinchcliff, K. W. & Geor, R. J. The horse as an athlete: a physiological overview. in iEquine Exercise Physiology: The Science of Exercise in the Athletic H/iiorse./i (ed. Hinchcliff, K. W. & Geor, R. J.) 454–454 (CABI Digital Library, 2008).
Mach, N. et al. Mining the equine gut metagenome: poorly-characterized taxa associated with cardiovascular fitness in endurance athletes. Commun. Biol. 5, 1032 (2022).
Edwards, J. E. et al. Multi-kingdom characterization of the core equine fecal microbiota based on multiple equine (sub) species. Anim. Microbiome 2, 1–16 (2020).
Kauter, A. et al. The gut microbiome of horses: current research on equine enteral microbiota and future perspectives. Anim. Microbiome 1, 1–15 (2019).
Le Roy, T. et al. Dysosmobacter welbionis is a newly isolated human commensal bacterium preventing diet-induced obesity and metabolic disorders in mice. Gut 71, 534–543 (2022).
Yoon, H. S. et al. Akkermansia muciniphila secretes a glucagon-like peptide-1-inducing protein that improves glucose homeostasis and ameliorates metabolic disease in mice. Nat. Microbiol. 6, 563–573 (2021).
Lahiri, S. et al. The gut microbiota influences skeletal muscle mass and function in mice. Sci. Transl. Med. 11, eaan5662 (2019).
Scheiman, J. et al. Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat. Med. 25, 1104–1109 (2019).
Górniak, W. et al. Effect of intense exercise on the level of bacteroidetes and Firmicutes Phyla in the digestive system of Thoroughbred racehorses. Animals 11, 290 (2021).
Janabi, A. H. D., Biddle, A. S., Klein, D. & McKeever, K. H. Exercise training-induced changes in the gut microbiota of Standardbred racehorses. Comp. Exerc. Physiol. 12, 119–130 (2016).
Thiruvenkadan, A. K., Kandasamy, N. & Panneerselvam, S. Inheritance of racing performance of Thoroughbred horses. Livest. Sci. 121, 308–326 (2009).
Bower, M. A. et al. The genetic origin and history of speed in the Thoroughbred racehorse. Nat. Commun. 3, 643 (2012).
McGowan, C. Clinical pathology in the racing horse: the role of clinical pathology in assessing fitness and performance in the racehorse. Vet. Clin. North Am. Equine Pract. 24, 405–421 (2008).
Allaam, M. et al. Physiological and hemato-chemical evaluation of thoroughbred race horse after exercise. IJAVMS 8, 81–93 (2014).
Mach, N. & Fuster-Botella, D. Endurance exercise and gut microbiota: a review. J. Sport Health Sci. 6, 179–197 (2017).
Clark, A. & Mach, N. Exercise-induced stress behavior, gut-microbiota-brain axis and diet: a systematic review for athletes. J. Int. Soc. Sports Nutr. 13, 43 (2016).
Crowson, M. M. & McClave, S. A. Does the intestinal microbiome impact athletic performance?. Curr. Gastroenterol. Rep. 22, 53 (2020).
Rhodes, M. A. et al. Carbon monoxide, skeletal muscle oxidative stress, and mitochondrial biogenesis in humans. Am. J. Physiol. Heart Circ. Physiol. 297, H392–H399 (2009).
Viña, J. et al. Mitochondrial biogenesis in exercise and in ageing. Adv. Drug Deliv. Rev. 61, 1369–1374 (2009).
McKie, G. L. & Wright, D. C. Biochemical adaptations in white adipose tissue following aerobic exercise: from mitochondrial biogenesis to browning. Biochem. J. 477, 1061–1081 (2020).
Xu, T. et al. The regulatory roles of dietary fibers on host health via gut microbiota-derived short chain fatty acids. Curr. Opin. Pharmacol. 62, 36–42 (2022).
Chen, R. et al. Transplantation of fecal microbiota rich in short chain fatty acids and butyric acid treat cerebral ischemic stroke by regulating gut microbiota. Pharm. Res. 148, 104403 (2019).
Griffith, G. W., Ozkose, E., Theodorou, M. K. & Davies, D. R. Diversity of anaerobic fungal populations in cattle revealed by selective enrichment culture using different carbon sources. Fungal Ecol. 2, 87–97 (2009).
Béra-Maillet, C. et al. Development of a RT-qPCR method for the quantification of Fibrobacter succinogenes S85 glycoside hydrolase transcripts in the rumen content of gnotobiotic and conventional sheep. J. Microbiol. Methods 77, 8–16 (2009).
Li, F., Hitch, T. C. A., Chen, Y., Creevey, C. J. & Guan, L. L. Comparative metagenomic and metatranscriptomic analyses reveal the breed effect on the rumen microbiome and its associations with feed efficiency in beef cattle. Microbiome 7, 6 (2019).
Qin, N. et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 513, 59–64 (2014).
Stewart, R. D. et al. Compendium of 4941 rumen metagenome-assembled genomes for rumen microbiome biology and enzyme discovery. Nat. Biotechnol. 37, 953–961 (2019).
Luo, R. et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1, 18 (2012).
Uritskiy, G. V., DiRuggiero, J. & Taylor, J. MetaWRAP-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6, 158 (2018).
Glendinning, L., Stewart, R. D., Pallen, M. J., Watson, K. A. & Watson, M. Assembly of hundreds of novel bacterial genomes from the chicken caecum. Genome Biol. 21, 34 (2020).
Chaumeil, P. A., Mussig, A. J., Hugenholtz, P. & Parks, D. H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36, 1925–1927 (2019).
Asnicar, F. et al. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat. Commun. 11, 2500 (2020).
Zeller, G. et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 10, 766 (2014).
Li, J. et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 32, 834–841 (2014).
Xiao, T., Yan, A., Huang, J. D., Jorgensen, E. M. & Shah, N. P. Comparative PEptidomic And Metatranscriptomic Analyses Reveal Improved Gamma-amino Butyric Acid Production Machinery in Levilactobacillus brevis strain NPS-QW 145 cocultured with Streptococcus thermophilus Strain ASCC1275 during milk fermentation. Appl. Environ. Microbiol. 87, e01985–20 (2020).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013).
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
Huang, Y. et al. Metatranscriptomics reveals the functions and enzyme profiles of the microbial community in Chinese Nong-Flavor Liquor Starter. Front. Microbiol. 8, 1747 (2017).
Edgcomb, V. P. et al. Gene expression profiling of microbial activities and interactions in sediments under haloclines of E. Mediterranean deep hypersaline anoxic basins. ISME J. 10, 2643–2657 (2016).
Gifford, S. M., Sharma, S., Rinta-Kanto, J. M. & Moran, M. A. Quantitative analysis of a deeply sequenced marine microbial metatranscriptome. ISME J. 5, 461–472 (2011).
Furuhashi, T., Sugitate, K., Nakai, T., Jikumaru, Y. & Ishihara, G. Rapid profiling method for mammalian feces short chain fatty acids by GC-MS. Anal. Biochem. 543, 51–54 (2018).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001).
Ning, F. L. et al. Activating BK channels ameliorates vascular smooth muscle calcification through Akt signaling. Acta Pharm. Sin. 43, 624–633 (2022).
Barton, W. et al. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut 67, 625–633 (2018).
Wang, W. L. et al. Application of metagenomics in the human gut microbiome. World J. Gastroenterol. 21, 803–814 (2015).
Comtet-Marre, S. et al. Metatranscriptomics reveals the active bacterial and eukaryotic fibrolytic communities in the rumen of dairy cow fed a mixed diet. Front. Microbiol. 8, 67 (2017).
Asnicar, F., Weingart, G., Tickle, T. L., Huttenhower, C. & Segata, N. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ 3, e1029 (2015).
Yu, G., Smith, D. K., Zhu, H., Guan, Y. & Lam, T. T. Y. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36 (2017).
Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 3, 180–185 (2011).
Bowers, R. M. et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 35, 725–731 (2017).
Bongiovanni, T., Yin, M. O. L., Heaney, L. M. & Liam, M. The athlete and gut microbiome: Short-chain fatty acids as potential ergogenic aids for exercise and training. Int. J. Sports Med. 42, 1143–1158 (2021).
Hamer, H. M. et al. The role of butyrate on colonic function. Aliment. Pharmacol. Ther. 27, 104–119 (2008).
Barger, K., Langsetmo, L., Orwoll, E. S. & Lustgarten, M. S. Investigation of the diet-gut-muscle axis in the osteoporotic fractures in men study. J. Nutr. Health Aging 24, 445–452 (2020).
Perry, C. G. et al. Inhibiting myosin-ATPase reveals a dynamic range of mitochondrial respiratory control in skeletal muscle. Biochem. J. 437, 215–222 (2011).
Glancy, B. et al. Mitochondrial reticulum for cellular energy distribution in muscle. Nature 523, 617–620 (2015).
Rodriguez-Castaño, G. P. et al. Bacteroides thetaiotaomicron starch utilization promotes quercetin degradation and butyrate production by Eubacterium ramulus. Front. Microbiol. 10, 1145 (2019).
Cha, K. H. et al. Effects of fermented milk treatment on microbial population and metabolomic outcomes in a three-stage semi-continuous culture system. Food Chem. 263, 216–224 (2018).
Balakrishnan, B. et al. Prevotella histicola protects from arthritis by expansion of allobaculum and augmenting butyrate production in humanized mice. Front. Immunol. 12, 609644 (2021).
Wu, H. et al. Neuroprotective effects of an engineered Escherichia coli Nissle 1917 on Parkinson’s disease in mice by delivering GLP-1 and modulating gut microbiota. Bioeng. Transl. Med. 8, e10351 (2022).
Tabone, M. et al. The effect of acute moderate-intensity exercise on the serum and fecal metabolomes and the gut microbiota of cross-country endurance athletes. Sci. Rep. 11, 3558 (2021).
Xiao, M. et al. Anti-fatigue property of the oyster polypeptide fraction and its effect on gut microbiota in mice. Food Funct. 11, 8659–8669 (2020).
Petersen, L. M. et al. Community characteristics of the gut microbiomes of competitive cyclists. Microbiome 5, 1–13 (2017).
Clarke, S. F. et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut 63, 1913–1920 (2014).
Nichols, A. W. Probiotics and athletic performance: a systematic review. Curr. Sports Med. Rep. 6, 269–273 (2007).
Gao, Z. et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 58, 1509–1517 (2009).
Rose, S. et al. Butyrate enhances mitochondrial function during oxidative stress in cell lines from boys with autism. Transl. Psychiatry 8, 1–17 (2018).
Huang, Y., Gao, S., Jun, G., Zhao, R. & Yang, X. Supplementing the maternal diet of rats with butyrate enhances mitochondrial biogenesis in the skeletal muscles of weaned offspring. Br. J. Nutr. 117, 12–20 (2017).
Zhang, Y. et al. Butyrate promotes slow-twitch myofiber formation and mitochondrial biogenesis in finishing pigs via inducing specific microRNAs and PGC-1α expression. J. Anim. Sci. 97, 3180–3192 (2019).
Hill, C., James, R. S., Cox, V. M., Seebacher, F. & Tallis, J. Age-related changes in isolated mouse skeletal muscle function are dependent on sex, muscle, and contractility mode. Am. J. Physiol. Regul. Integr. Comp. Physiol. 319, R296–R314 (2020).
Walsh, M. E. et al. The histone deacetylase inhibitor butyrate improves metabolism and reduces muscle atrophy during aging. Aging Cell14, 957–970 (2015).
Kundu, P. et al. Neurogenesis and prolongevity signaling in young germ-free mice transplanted with the gut microbiota of old mice. Sci. Transl. Med. 11, eaau4760 (2019).
Chang, J. G. et al. Treatment of spinal muscular atrophy by sodium butyrate. Proc. Natl. Acad. Sci. USA 98, 9808–9813 (2001).
Yu, C. et al. Effect of exercise and butyrate supplementation on microbiota composition and lipid metabolism. J. Endocrinol. 243, 125–135 (2019).
Matsumoto, M. et al. Voluntary running exercise alters microbiota composition and increases n-butyrate concentration in the rat cecum. Biosci. Biotechnol. Biochem. 72, 572–576 (2008).
Campbell, S. C. & Wisniewski, P. J. Exercise is a novel promoter of intestinal health and microbial diversity. Exerc. Sport Sci. Rev. 45, 41–47 (2017).
Groussin, M., Mazel, F. & Alm, E. J. Co-evolution and co-speciation of host-gut bacteria systems. Cell Host Microbe 28, 12–22 (2020).
Han, M. et al. Stratification of athlete’ gut microbiota: the multifaceted hubs associated with dietary factors, physical characteristics and performance. Gut Microbes 12, 1842991 (2020).
Park, T., Yoon, J., Yun, Y. & Unno, T. Comparison of the fecal microbiota with high- and low performance race horses. J. Anim. Sci. Technol. 66, 425–437 (2024).
Hargreaves, M. & Spriet, L. L. Skeletal muscle energy metabolism during exercise. Nat. Metab. 2, 817–828 (2020).
Kruk, J., Aboul-Enein, B. H., Duchnik, E. & Marchlewicz, M. Antioxidative properties of phenolic compounds and their effect on oxidative stress induced by severe physical exercise. J. Physiol. Sci. 72, 1–24 (2022).
Singh, V. et al. Butyrate producers, “The Sentinel of Gut”: their intestinal significance with and beyond butyrate, and prospective use as microbial therapeutics. Front. Microbiol. 13, 1103836 (2023).
Acknowledgements
This work was supported financially by the Tianshan Talent Project [to S.W.H. and W.N.], the Tianchi Talent Project [to C.Y.L. and to X.Y.L.] and the National Natural Science Foundation of China [32360016, 32400045 to C.Y.L.], the Third Xinjiang Scientific Expedition Program [2021xjkk0605 to S.W.H., and 2022xjkk0804 to W.N.], the Research Foundation for Advanced Talents [RCZK202363, RCZK202361 and KX6222], the Interdisciplinary project [KX6157] and the Foundation of State Key Laboratories for Sheep Genetic Improvement and Healthy Production [2021ZD08].
Author information
Authors and Affiliations
Contributions
C.L., X.L., N.M., W.N. and S.H. wrote the paper. C.L., X.L., K.L. and J.X. performed the multi-omics data analyses. C.L., X.L. and K.L. performed the metagenomics, metatranscriptomics, and metabolomics experiments. C.L., X.L. and K.L. performed the mice experiments and qPCR analysis. C.L., Z.L., P.Z., L.W., C.L. and J.Y. performed the collection of samples. W.N. and S.H. supervised the study. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
The Animal Care Committee of Shihezi University approved all of the procedures involving animals. The study was performed in accordance with the ethical standards established in the 1964 Declaration of Helsinki and subsequent amendments. Winners and riders provided their informed consent before the start of sampling procedures with the animals.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, C., Li, X., Liu, K. et al. Multiomic analysis of different horse breeds reveals that gut microbial butyrate enhances racehorse athletic performance. npj Biofilms Microbiomes 11, 87 (2025). https://doi.org/10.1038/s41522-025-00730-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41522-025-00730-w