Abstract
The removal of leaked radioactive iodine isotopes in humid air environments holds significant importance in nuclear waste management and nuclear accident mitigation. In this study, high-throughput computational screening and machine learning were combined to reveal the iodine capture performance of 1816 metal-organic framework (MOF) materials under humid air conditions. Initially, the relationship between the structural characteristics of MOF materials (including density, surface area and pore features) and their adsorption properties was explored, with the aim of identifying the optimal structural parameters for iodine capture. Subsequently, two machine learning regression algorithms—Random Forest and CatBoost, were employed to predict the iodine adsorption capabilities of MOF materials. In addition to 6 structural features, 25 molecular features (encompassing the types of metal and ligand atoms as well as bonding modes) and 8 chemical features (including heat of adsorption and Henry’s coefficient) were incorporated to enhance the prediction accuracy of the machine learning algorithms. Feature importance was assessed to determine the relative influence of various features on iodine adsorption performance, in which the Henry’s coefficient and heat of adsorption to iodine were found the two most crucial chemical factors. Furthermore, four types of molecular fingerprints were introduced for providing comprehensive and detailed structural information of MOF materials. The 20 most significant Molecular ACCess Systems (MACCS) bits were picked out, revealing that the presence of six-membered ring structures and nitrogen atoms in the MOF framework were the key structural factors that enhanced iodine adsorption, followed by the presence of oxygen atoms. This work combined high-throughput computation, machine learning, and molecular fingerprints to comprehensively and systematically elucidate the multifaceted factors governing the iodine adsorption performance of MOFs in humid environments, establishing a robust and profound guideline framework for accelerating the screening and targeted design of high-performance MOF materials.
Similar content being viewed by others
Introduction
As an efficient and low-carbon energy source, nuclear energy plays a significant role in the global energy landscape, particularly in the context of global climate change, where it provides significant support for achieving carbon neutrality goals1,2. However, the rapid expansion of nuclear energy industry is accompanied by potential environmental and safety risks3, especially in the handling of nuclear waste and during nuclear accidents, where the leakage of radioactive substances poses a severe threat to both the environment and human health. Among the radioactive isotopes involved in spent nuclear fuel reprocessing or nuclear accidents, iodine isotopes, particularly 131I and 129I, are of particular concern due to their volatility and strong bioaccumulation, which result in significant long-term impacts on the environment and human health4. The half-life of 131I is only 8 days; although its radiation is intense, the associated risks are generally short-term, primarily entering the human body via inhalation or the food chain, leading to acute health issues such as thyroid cancer. In contrast, 129I has an exceptionally long half-life (~ 1.57 × 107 years), enabling it to persist in the biosphere and cause sustained threats to ecosystems. Consequently, the efficient removal of radioactive iodine isotopes has become an urgent requirement for ensuring nuclear safety and reducing environmental contamination5,6,7,8.
Metal-organic frameworks (MOFs), as novel porous materials formed by metal clusters coordinated with organic ligands, have gained considerable attention as potential iodine adsorbents due to their highly tunable structures, large surface areas, and excellent porosity9,10. However, in the real nuclear industry and spent nuclear fuel reprocessing, high-humidity air environments are prevalent, thus demanding more robust iodine adsorption properties from MOF materials6,11,12. In recent years, many researchers have investigated the iodine adsorption behavior of various MOFs in humid environments: Nenoff and her co-workers explored the competitive I2 sorption by Cu-BTC from humid gas streams (about 3.5% relative humidity) at 75 °C and ambient pressure, revealing a remarkable iodine capacity of ~175 wt% with a derived I2/H2O adsorption selectivity of 1.513. Thallapally’s group reported I2 adsorption capacities and mechanisms in two microporous MOFs in the presence of 33% and 43% relative humidity (RH), in which SBMOF-1 and SBMOF-2 exhibited the 15 wt% and 35 wt% uptake, respectively14. Zhang et al. systematically studied the influence of H2O molecules on the iodine adsorption properties of different zeolitic imidazolate frameworks (ZIFs) using grand canonical Monte Carlo (GCMC) simulations, highlighting the competitive adsorption behavior between H2O and I2, particularly for hydrophilic materials15. Other MOF materials including MIL-101-Cr-TED, MIL-101-Cr-HMTA and ECUT-300, also were used to explore the iodine capture performance in a water-containing system16,17,18,19. In our previous work, grand canonical Monte Carlo (GCMC) and density functional theory methods were employed to investigate the iodine adsorption performance of 21 chemically stable MOF materials in high-humidity environments, and influence of different structural factors were revealed20. However, despite these advances, researches on the iodine adsorption behavior of MOFs under humid conditions remains limited, and a comprehensive insight of the key factors influencing iodine adsorption based on a larger number of MOF materials is still needed.
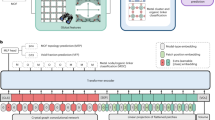
Nowadys, high-throughput computational screening based on molecular simulations offers a rapid approach to evaluating the iodine adsorption performance of MOFs under humid conditions21,22,23. Furthermore, the rapid development of artificial intelligence has ushered in a new research paradigm that combines data science with chemistry24,25. Machine learning has proven to be an efficient tool for analyzing computational data related to gas adsorption behaviors of MOFs, for revealing structure-property relationships, identifying promising MOF adsorbents and even guiding MOF structures design and modification26,27,28,29. In this work, we first selected 1816 I2-accessible MOF materials (with pore limiting diameter > 3.34 Å - the kinetic diameter of I2) from the well-established CoRE MOF 2014 database established by Chuang et al.30, and employed GCMC simulations via using RASPA software to study their I2 adsorption performance under humid air conditions31. Subsequently, three different types of descriptors (structural, molecular, and chemical features) were explored, and machine learning algorithms were utilized to predict iodine adsorption performance and reveal the relationships between various descriptors and iodine adsorption performance. Finally, molecular fingerprint technique was employed to comprehensively identify the influence of structural features on the iodine adsorption, providing valuable insights and guidelines for the future targeted design of MOF materials.
Results
Structure-performance relationships
To identify the optimal structural features, the relationships between the 1816 MOF structures and iodine adsorption performance were investigated in Fig. 1 and Fig. S1. Structural characteristics of MOFs included the pore limiting diameter (PLD), largest cavity diameter (LCD), void fraction (φ), pore volume, surface area and density. When LCD was less than 4 Å (Fig. 1a), the spatial steric hindrance between the I2 molecules and the pore walls resulted in negligible iodine adsorption. When 4 Å < LCD < 5.5 Å, an increase in LCD reduced the steric hindrance, thereby adsorption interaction between the framework materials and iodine molecules became the dominant factor, which led to an increase in both iodine adsorption capacity and selectivity. However, when LCD exceeded 5.5 Å, further enlargement of the channel size diminished the interaction between MOFs and iodine molecules, which intensified the desorption of I2 in the pores and resulted in a continuous decline in both adsorption capacity and selectivity. To identify MOF materials with optimal iodine adsorption properties, we could find the ideal value for LCD lay between 4 and 7.8 Å. As for porosity (with the optimal value for iodine adsorption in the range of 0 to 0.17) in Fig. 1b, iodine adsorption capacity and selectivity initially increased (φ < 0.09) and then decreased (0.09 < φ < 0.6). The relationships between density and iodine adsorption performance also followed a similar trend (Fig. 1c): at low densities (under 0.9 g/cm3), the increase in density promoted iodine adsorption due to the greater number of available adsorption sites; however, when the density exceeded 0.9 g/cm³, the steric hindrance effect gradually increased and excessively compact pore structures limited iodine adsorption; with the density further surpassing 2.2 g/cm3, iodine uptake amount would fall below 100 cm3/g. Furthermore, the optimal values of pore volume, PLD and surface area for iodine capture were also identified, which lay at the range of 0 ~ 0.18 cm3/g, 3.34 ~ 7 Å and 0 ~ 540 m2/g, respectively (Fig. 1d and Fig. S1). In order to facilitate the comparison of the adsorption behavior of different molecules, structure-performance relationships of MOF materials for H₂O adsorption have been delineated (Fig. S2), in which the optimal structural parameters exhibited a broader range (for instance, the optimal LCD could reach up to 11 Å and the optimal φ could attain 0.48); this is likely attributed to the larger kinetic diameter of H₂O molecules compared to I₂. The above results explained that relatively small pore sizes of MOF materials could confer the advantage during competitive iodine adsorption in humid conditions.
Machine learning
After the aforementioned analysis, we initially employed six structural descriptors - PLD, LCD, φ, pore volume, surface area and density, to train machine learning algorithms for predicting iodine gas adsorption in MOF materials under humid conditions (Fig. 2a, d). Two different machine learning algorithms (including random forest and CatBoost model) were trained and compared32,33. After training the model with only structural parameters as the simplest feature set, we gradually incorporated more comprehensive feature sets, including “structural + molecular descriptors” (Fig. 2b, e) and “structural + molecular + chemical descriptors” (Fig. 2c, f). For molecular descriptors, each molecular feature corresponded to specific elemental, hybridization, and bonding types. For carbon (C) and nitrogen (N) elements, the atomic types included C_1, C_2, C_3, and C_R (or N_1, N_2, N_3, and N_R), depending on the nature of single, double, triple, and ring bonds. Oxygen (O) atoms can form double bonds and ring bonds, defined as O_2 and O_R, respectively, as well as central tetrahedral oxygen (denoted as O_3_f) or central trigonal oxygen (denoted as O_2_z)34. For hydrogen (H), fluorine (F), chlorine (Cl), and bromine (Br), the atomic types are designated as H_, F_, Cl, and Br. Additionally, tetrahedral four-coordinate phosphorus for organo-metallic coordination is defined as P_3+q, along with sulfur atoms connected via cyclic bonds (denoted as S_R)35. Regarding metal atoms, only the predominant metal species within the MOF were considered: descriptors for these metals included metal ratio (the molar ratio relative to all atoms), atomic number, atomic weight, atomic radius, polarizability, electron affinity, and Mulliken electronegativity. In chemical descriptors, Henry’s coefficient and heat of adsorption of I2, H2O, N2 and O2 in MOF materials were considered and defined as I2_Henry (I2_heat), H2O_Henry (H2O_heat), N2_Henry (N2_heat) and O2_Henry (O2_heat), respectively.
Throughout the above process, the prediction performance of both the random forest and CatBoost algorithms progressively improved. The accuracy of the machine learning algorithm was evaluated using R2 2, mean absolute error (MAE), and mean squared error (MSE). When only structural descriptors were used, the random forest algorithm exhibited a relatively low prediction accuracy (R2 = 0.438). After adding molecular descriptors to the feature set, the prediction accuracy of the random forest model improved, with R2 increasing to 0.592. Further incorporation of chemical descriptors led to the highest prediction accuracy (R2 = 0.900). Simultaneously, the MAE and MSE values of random forest model demonstrated a steady decrease: MAE reduced from 75.588 to 61.673, and finally to 23.378; MSE decreased from 14293.744 to 10387.059, and ultimately to 2547.433. For the CatBoost algorithm, the trends were similar to those observed with the random forest model, but overall, it exhibited better prediction performance. The hyperparameter tuning space and optimal hyperparameters are listed in Table S1. When using the “structural descriptors + molecular descriptors + chemical descriptors” feature set, the CatBoost algorithm achieved the highest R2 of 0.941, with MAE and MSE dropping to 18.276 and 1512.681, respectively. Additionally, by following the above process, the accuracy of the CatBoost algorithm in predicting the H₂O adsorption performance of MOF materials also gradually improved (Fig. S3), and the value of R2, MAE and MSE respectively reached 0.911, 5.672 and 166.554 based on “structural descriptors + molecular descriptors + chemical descriptors” feature set. The above results further confirmed that molecular and chemical descriptors significantly complemented the structural descriptors, playing a crucial role in accurately predicting gas molecules capture in MOF materials.
To investigate the contribution of different features in predicting iodine adsorption performance, the SHAP (SHapley Additive exPlanations) method was used to rank and explain the significance of various features in the CatBoost model (Fig. 3). Among the structural descriptors (Fig. 3a), LCD was identified as the most important descriptor, followed by void fraction and surface area (both of which exhibit a certain negative correlation with I2 adsorption performance). PLD and density of the MOF material ranked next in importance, while pore volume had the least significance among the six structural descriptors. In the “structural descriptors + molecular descriptors” set (considering only the top 20 descriptors in terms of importance), all structural descriptors, except for pore volume, ranked within the top 10, with LCD maintaining the highest importance. Among the molecular descriptors, the most significant was C_R (positively correlated with adsorption capacity), followed by the proportion of metal atoms (negatively correlated with adsorption capacity), and then H_R, N_R, and O_2 (all positively correlated with adsorption capacity). We speculated that iodine adsorption in MOF materials primarily relied on the organic ligands, with carbon rings, nitrogen, and oxygen atoms serving as key adsorption sites. In the “structural descriptors + molecular descriptors + chemical descriptors” set (considering only the top 20 descriptors), the most influential features were I2_Henry, followed by I2_Heat. Both features were positively correlated to iodine capture amounts. Notably, in contrast to N₂ and O₂, the adsorption heat of H₂O (H2O_heat) was strongly negatively correlated with iodine adsorption capacity (with the damping coefficient positively correlated), likely because under humid conditions, H₂O molecules are the main competitive species for iodine gas adsorption. In terms of H2O molecules adsorption (Fig. S4), H2O_heat possessed the highest relative importance, and metal atoms also exhibited relatively high significance with polarizability, metal ratio, Mulliken electronegativity and atomic radius positioning within the top six rankings. In contrast to the adsorption of I2 molecules, the atomic radius of the metal atoms exhibited a positive correlation with the adsorption of H2O molecules (Fig. S4b); furthermore, O_R replaced N_R as the most favorable ligand structure due to the strong hydrogen bonding interactions (Fig. S4c).
The correlation coefficients between different features were calculated and visualized as heatmaps. Among the pure structural descriptors (Fig. S5), a strong positive correlation (correlation coefficient > 0.6) was evident between PLD, LCD, void fraction, surface area, and pore volume, while density showed a strong negative correlation (correlation coefficient > -0.4) with the other five structural parameters. In the “structural descriptors + molecular descriptors” set (Fig. S6), metal ratio, metal atomic number, metal atomic weight, and metal atomic radius exhibited positive correlations with the density of the MOF material (with correlation coefficients of 0.32, 0.38, 0.39, and 0.28, respectively), due to the heavier nature of metal atoms compared to ligand atoms. Additionally, the metal atoms ratio was positively correlated with void fraction (with correlation coefficient of 0.23), whereas the correlations between porosity and the number of metal atoms, metal atomic weight, and metal atomic radius were relatively weak. This was because metal clusters, formed by a greater number of metal atoms as connecting nodes, would lead to the increased porosity, irrespective of the type of metal atoms. The metal ratio was negatively correlated with the number of most organic ligand atoms (including H_R, C_R, N_R, O_R, C_3, and N_3) (with correlation coefficient of -0.43, -0.42, -0.11, -0.10, -0.15, and -0.13), but was positively correlated with O_2 (with correlation coefficient of 0.31), which was likely due to the fact that a certain amount of O existed to form metal clusters in MOF materials by bonding with metals atoms. In the “structural + molecular + chemical descriptors” set (Fig. 4), I2_Henry showed a low correlation with both structural and molecular descriptors. N2_Henry, O2_Henry, and H2O_Henry exhibited significant positive correlations with most structural descriptors (including PLD, LCD, porosity, specific surface area, and pore volume), while N2_Heat, O2_Heat, and H2O_Heat were negatively correlated with the aforementioned descriptors. This was attributed to the fact that the larger size of cavity or pore channel in MOFs facilitated the diffusion of gas molecules within MOFs, but the reduced density of adsorption sites weakened the adsorption strength, leading to the lower adsorption heats. The very strong correlations between N2_Henry and O2_Henry (correlation coefficient = 0.98) and between N2_Heat and O2_Heat (correlation coefficient = 0.89) arose from the similar molecular structures of N₂ and O₂. I2_Heat was negatively correlated with the proportion of metal atoms (correlation coefficient of -0.27) and positively correlated with C_R (correlation coefficient of 0.21), further indicating that metal sites were not effective adsorption sites for I₂ molecules, which tended to be adsorbed near organic ligands (such as benzene rings).
Molecular Fingerprint
In order to comprehensively reveal the factors that positively or negatively influenced iodine adsorption performance, thereby paving the way for future molecular design, four types of molecular fingerprint (including Molecular ACCess Systems (MACCS), PubChem, AtomPairs2D and Estate fingerprint) were employed in place of the previously used molecular descriptors. MACCS and PubChem represented two of the most widely used fingerprints derived from substructure key information: the MACCS fingerprint consisted of a set of 166 structural keys, constructed using SMART patterns; while the PubChem fingerprint originated from the PubChem database, encompassing 881 types of structural keys represented as binary substructure encodings. The APFP fingerprint encoded a total of 780 atomic pairs based on their topological distances. Estate summarized the microscopic structure of materials through a 79-byte representation. Based on the presence or absence of bits, molecular fingerprint digitized the molecular features of MOF materials, thereby providing microscopic insights into the structure of exceptional materials. These molecular fingerprints, in conjunction with previous structural and chemical descriptors, were applied to train machine learning models for the prediction of iodine adsorption performance under humid conditions. Although the prediction performance showed a slight decrease based on “structure + molecular fingerprint + chemical descriptors” set compared to the prior “structure + molecular + chemical descriptors” set, because the encoding of molecular fingerprint solely indicated the presence or absence of specific features and other information such as the quantity or proportion in a single MOF unit cell were missed, which limited their ability in prediction of uptake amount; however, their comprehensive inclusion of various structural categories enabled them to serve as excellent interpretative tools.
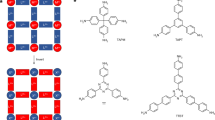
After comparing the prediction accuracies of the four molecular fingerprints (Fig. 5a, Fig. S7 and Table S2), MACCS molecular fingerprint exhibited its superiority in machine learning (R2 = 0.927, MAE = 20.057, and MSE = 1651.391). The 20 most significant MACCS bits were ranked and accompanied by detailed interpretations (Fig. 5b and Table S3). Additionally, the autocorrelation coefficients of the molecular fingerprints were presented in the heatmap to reveal the interrelationships among the fingerprints (Fig. S8). The top two, Bit_158 and Bit_75, demonstrated the significant positive impact of nitrogen (N) atoms in MOF materials on iodine adsorption performance. The strong correlations between Bit_158 and Bit_156 (with correlation coefficient of 0.9), and between Bit_158 and Bit_45 (with correlation coefficient of 0.73) further highlighted that the presence of N atoms, whether in rings or directly bonded to carbon (C) atoms, promoted the iodine adsorption in humid air conditions. Bit_163 and Bit_162, which had a correlation coefficient of 0.6, represented that six-membered aromatic ring was another important structural feature. Six-membered aromatic ring likely provided electron-rich adsorption sites due to their large π-bonds, thereby enhancing the iodine adsorption. Bit_6 and Bit_12, which were associated with the type of metal, represented lanthanide metal and group IB (or IIB) metal elements, respectively. The negative correlation (-0.38) between these two molecular fingerprints arose from the relatively homogeneous nature of the metal clusters in these materials; however, the former demonstrated a negative effect on I2 adsorption, while the latter had a positive impact (consistent with previous findings, where a negative correlation was observed between the metal atomic radius and iodine adsorption performance). The relationships between MACCS molecular fingerprint and other structural (or chemical) features were also illustrated (Fig. S8). It could be found that although Bit_6 (lanthanide metals) and Bit_12 (group IB/IIB metals) showed little correlation with I2_Henry and I2_Heat, Bit_6 enhanced the adsorption of H2O in MOF materials (correlation coefficient with H2O_Heat of 0.65), while Bit_12 weakened the adsorption of H2O (correlation coefficient with H2O_Heat of -0.41); thus, it could be concluded that group IB/IIB metals played the significant role in promoting competitive adsorption of I2 under humid conditions compared to lanthanide metals.
Furthermore, the MACCS fingerprints Bit_138, Bit_69, Bit_100 and Bit_131, representing different forms of hydrogen (H) (whether connected to C or non-C elements), and Bit_143 and Bit_139, representing different forms of oxygen (O) (either as part of a ring structure or as hydroxyl groups), all demonstrated the positive role of H and O atoms in enhancing iodine adsorption in MOF structures. Nevertheless, modifications involving N atoms appeared to offer the greater advantages over O atoms (Overall, Bit_158 and Bit_75 had the higher importance ranking compared to Bit_143 and Bit_139). This was because the presence of N atoms, compared to O atoms, exerted the evident suppressive effect on both pore size and the adsorption of H2O molecules, both of which were more favorable for iodine adsorption in humid environments; specifically, the correlation coefficients between N-related molecular fingerprints (Bit_158, Bit_75, Bit_156, and Bit_45) and pore volume were -0.27, -0.28, -0.26, and -0.26, respectively, and their correlations with H2O_Heat were -0.12, -0.18, -0.13, and -0.17; in contrast, O-related molecular fingerprints (Bit_143 and Bit_139) had the positive correlation coefficients with pore volume of 0.2 and 0.046, respectively, and correlation coefficients with H2O_Heat were 0.24 and 0.029. The role of H atoms lay between that of the N and O atoms: the correlation coefficients between H-related molecular fingerprints (Bit_138, Bit_69, Bit_100 and Bit_131) and pore volume were at the range of -0.1 ~ -0.2, and the correlation with H2O_Heat were also weak (with negative correlation coefficients less than -0.1).
Finally, the top six MOF materials with the best iodine adsorption performance were picked out, and microscopic insights into the common fingerprints present shared by most of these MOFs were provided (Fig. 6 and Table S4). These MOF structures shared a common set of molecular fingerprints: Bit_45, Bit_75, Bit_156, Bit_158, and Bit_163, all of which indicated the presence of six-membered rings and N atoms, and N atoms were part of the six-membered rings or directly coordinated with metal atoms. Furthermore, the metal atoms in these structures were all transition metals from the fourth period (including Zn, Co, Ni, and Mn), which were characterized by the smaller atomic radii and atomic numbers compared to lanthanide elements. Four of these MOF materials had Bit_97 molecular fingerprint, representing the presence of O atoms. To derive more generalizable conclusions, all MOF materials were categorized into different groups based on their adsorption performance, and the occurrence frequencies of the aforementioned MACCS bits were analyzed (Table S5). The proportions of Bit_45, Bit_75, Bit_156, Bit_158, Bit_163 and Bit_97 in the entire MOF material database were 35.8%, 37.2%, 51.7%, 50.9%, 72.8%, and 30.7%, respectively. Among the top 30 high-performance MOF materials, their frequencies increased to 76.7%, 70.0%, 73.3%, 76.7%, 86.7% and 56.7%, respectively; whereas in the 300 lowest-performing MOFs, their frequencies dropped to 22.0%, 27.7%, 47.3%, 47.7%, 45.3% and 26.3%, respectively. Additionally, Bit_6 (representing lanthanide elements) was more prevalent in MOFs with poorer adsorption performance, appearing in 23.0% of the bottom 300 MOFs but entirely absent in the top 30 MOFs. These findings suggested that the presence of six-membered rings, nitrogen atoms, oxygen atoms, small metal atomic radii, and low metal atomic numbers favored iodine adsorption in MOFs. This further validated the previous analysis and provided valuable insights for the future design of high-performance MOF materials.
Discussion
In summary, large-scale GCMC simulations were employed to investigate iodine adsorption performance (including adsorption capacity and selectivity) for 1816 MOF materials from CoRE MOF database under humid conditions. Two machine learning algorithms, Random Forest and CatBoost, were utilized to predict the iodine adsorption performance of MOF materials, gradually incorporating three different types of descriptors to enhance the prediction accuracy of the models: 6 structural features (including pore characteristics, density, and surface area), 25 molecular features (including the types of metal and ligand atoms as well as their bonding modes), and 8 chemical features (including adsorption heat and Henry’s coefficient). SHAP method was used to rank the importance of these descriptors, and correlation coefficients were employed to reveal the relationships among features. Four types of molecular fingerprints were also generated in place of molecular features and combined with the CatBoost algorithm to predict iodine adsorption performance. The top 20 MACCS bits were extracted, demonstrating the most significant role of six-membered rings and N atoms in MOF materials, followed by O atoms. Among the metal sites, the lighter transition metal elements were found to be more favorable for iodine adsorption compared to lanthanide elements. This comprehensive and systematic study shed light on the iodine adsorption performance of MOF materials under humid conditions, providing valuable insights for the future screening and design of high-performance MOF materials.
Methods
Simulation method
GCMC simulations were employed using RASPA software to investigate the adsorption behavior of I₂ within the MOFs at an environment of 423 K and 1 bar. The temperature of 423 K was relevant to operational conditions in the nuclear industry6,36,37. To replicate the high humidity environment encountered during the post-treatment phase of spent nuclear fuel, the mixed gas system was composed of 300 ppm I₂, 68.5% N₂, 18.4% O₂, and 12.2% H₂O, achieving a relative humidity of 100%6. Throughout the simulation, the MOFs were treated as fixed rigid structures with periodic boundary conditions. Supercells were utilized as necessary to ensure that the system dimensions exceeded twice the cutoff distance (12 Å). In addition, the selectivity of I2 during adsorption was calculated as the following equation8,37:
where \({X}_{{I}_{2}}\) and \({Y}_{{I}_{2}}\) denoted the uptake amounts and gas phase concentration of I2; \({X}_{{others}}\) and \({Y}_{{others}}\) were the uptake amounts and gas phase concentration of other gas components (N2, O2 and H2O).
All GCMC simulations comprised an equilibration phase of 50,000 cycles, followed by a production phase of 50,000 cycles. Each cycle involved the movement of all adsorbed molecules, encompassing the insertion, deletion, translation, rotation, reinsertion, identity change, and swap processes. Iodine molecules were modeled as spherical entities, with van der Waals parameters derived from the viscosity of pure iodine38. Water molecules were represented using the transferable intermolecular potential (TIP3P) model (rOH = 0.9527 Å and θ∠HOH = 104.52°), which was a model empirically validated to accurately describe hydrogen bonding interactions39,40. N₂ and O₂ molecules were modeled as three-site representation41,42. The relevant molecular model parameters were referenced from previous published work20. The Universal Force Field (UFF) was employed to establish the Lennard-Jones (LJ) parameters for the MOF structures43. Interatomic interactions were described using LJ and electrostatic potential energy functions as below:
where \(U({r}_{{ij}})\) denoted the non-bonded interaction energy between atoms i and j; the first term represented the van der Waals non-bonded potential energy, and the second term accounted for the Coulombic electrostatic interaction energy. \({r}_{{ij}}\) signified the interatomic distance, \({\sigma }_{{ij}}\) was the depth of the Lennard-Jones (LJ) potential well, \({q}_{i}\) and \({q}_{j}\) represented the partial charges of atoms \(i\) and \(j\), respectively, and \({\varepsilon }_{0}\) was the vacuum dielectric constant.
Material descriptors
For structural descriptors, PLD and LCD were computed using the Zeo + + software package, based on Voronoi tessellation44. The void fraction was calculated using the RASPA software package with helium atoms (kinetic diameter = 2.58 Å) as probes. Meanwhile, the surface area, pore volume, and density were determined by RASPA using nitrogen molecules (kinetic diameter = 3.64 Å) as probes. To identify the molecular descriptors within the MOF structure, the Python program lammps_interface was utilized based on the UFF4MOF force field34,45,46. As for the chemical descriptors, adsorption heat and Henry’s coefficient were calculated under infinite dilution conditions using RASPA based on an NVT-MC system (MC referred to the Monte Carlo method), with simulations conducted for 10,000 cycles.
The OpenBabel and PaDEL-Descriptor software were performed to compute four types of molecular fingerprint47,48: MACCS, PubChem, AtomPairs2D and Estate49,50,51,52. OpenBabel was an open-source chemical toolbox designed to convert CIF files into SDF files compatible with PaDEL-Descriptor. And the PaDEL-Descriptor software processed the SDF format structural files, ultimately yielding the required four types of molecular fingerprint.
Machine learning
Two machine learning regression algorithms - Random Forest and CatBoost, were implemented in Python 3.9 using the scikit-learn package32,33,53. Both of these algorithms offered advantages in terms of low computational costs and good interpretability compared to other machine learning model such as neural network algorithm54. For each ML algorithm, the dataset was randomly divided into training sets and test sets, of which 80% were used for the training of the model and 20% for testing. Additionally, for optimizing and validating the models, we tuned the related hyperparameters using cross-validation. The accuracy of the models was evaluated using R2, MAE, and MSE, whose relevant equations were as follows:
where \(n\) represented the number of instances in the training or testing set, \(Y\) denoted the predicted values from the machine learning algorithms, \(\bar{Y}\) signified the mean of the model’s predictions, and \(\hat{Y}\) represented the computed values for the MOF materials.
Data availability
The datasets generated and/or analyzed during the current study are available at the following GitHub repository: https://github.com/gcshan82/ML4MOF.git. The data and codes employed in the current work can also be obtained from the corresponding author upon request.
Code availability
Code is publicly available at the following GitHub repository: https://github.com/gcshan82/ML4MOF.git.
References
Chu, S. & Majumdar, A. Opportunities and challenges for a sustainable energy future. Nature 488, 294–303 (2012).
Adamantiades, A. & Kessides, I. Nuclear power for sustainable development: current status and future prospects. Energ. Policy 37, 5149–5166 (2009).
Bowyer, T. W. et al. Elevated radioxenon detected remotely following the Fukushima nuclear accident. J. Environ. Radioact. 102, 681–687 (2011).
Frohlich, E. & Wahl, R. The current role of targeted therapies to induce radioiodine uptake in thyroid cancer. Cancer Treat. Rev. 40, 665–674 (2014).
Soelberg, N. R. et al. Radioactive iodine and krypton control for nuclear fuel reprocessing facilities. Sci. Technol. Nucl. Ins. 2013, 1–12 (2013).
Haefner, D. R. & Tranter, T. J. Methods of Gas Phase, Capture of Iodine from Fuel Reprocessing Off-Gas: A Literature Survey; INL/EXT-07-12299, Idaho National Laboratory, Idaho Falls (2007).
Zhou, J. B., Hao, S., Gao, L. P. & Zhang, Y. C. Study on adsorption performance of coal based activated carbon to radioactive iodine and stable iodine. Ann. Nucl. Energy 72, 237–241 (2014).
Nandanwar, S. U., Coldsnow, K., Utgikar, V., Sabharwall, P. & Eric Aston, D. Capture of harmful radioactive contaminants from off-gas stream using porous solid sorbents for clean environment - A review. Chem. Eng. J. 306, 369–381 (2016).
Li, J. R., Kuppler, R. J. & Zhou, H. C. Selective gas adsorption and separation in metal-organic frameworks. Chem. Soc. Rev. 38, 1477–1504 (2009).
Rosen, A. S. et al. Machine learning the quantum-chemical properties of metal-organic frameworks for accelerated materials discovery. Matter 4, 1578–1597 (2021).
Ge, Y. X. et al. Understanding water adsorption and the impact on CO2 capture in chemically stable covalent organic frameworks. J. Phys. Chem. C. 122, 27495–27506 (2018).
Duan, J. G., Jin, W. Q. & Kitagawa, S. Water-resistant porous coordination polymers for gas separation. Coord. Chem. Rev. 332, 48–74 (2017).
Sava, D. F. et al. Competitive I2 sorption by Cu-BTC from humid gas streams. Chem. Mater. 25, 2591–2596 (2013).
Banerjee, D. et al. Iodine adsorption in metal organic frameworks in the presence of humidity. ACS Appl. Mater. Interfaces 10, 10622–10626 (2018).
Yuan, Y., Dong, X., Chen, Y. & Zhang, M. Computational screening of iodine uptake in zeolitic imidazolate frameworks in a water-containing system. Phys. Chem. Chem. Phys. 18, 23246–23256 (2016).
Taghipour, F. & Evans, G. J. Radiolytic organic iodide formation under nuclear reactor accident conditions. Environ. Sci. Technol. 34, 3012–3017 (2000).
Riley, B. J., Vienna, J. D., Strachan, D. M., McCloy, J. S. & Jerden, J. L. Materials and processes for the effective capture and immobilization of radioiodine: A review. J. Nucl. Mater. 470, 307–326 (2016).
Li, B. et al. Capture of organic iodides from nuclear waste by metal-organic framework-based molecular traps. Nat. Commun. 8, 485 (2017).
Zhang, H. P. et al. Efficient organic iodide capture by a mesoporous bimetallic-organic framework. Cell Rep. Phys. Sci. 3, 100830 (2022).
Tan, H. & Shan, G. Computational screening and functional tuning of chemically stable metal organic frameworks for I2/CH3I capture in humid environments. iScience 27, 109096 (2024).
Daglar, H. & Keskin, S. Recent advances, opportunities, and challenges in high-throughput computational screening of MOFs for gas separations. Coord. Chem. Rev. 422, 213470 (2020).
Wilmer, C. E. et al. Large-scale screening of hypothetical metal-organic frameworks. Nat. Chem. 4, 83–89 (2011).
Gantzler, N., Deshwal, A., Doppa, J. R. & Simon, C. M. Multi-fidelity Bayesian optimization of covalent organic frameworks for xenon/krypton separations. Digit. Discov. 2, 1937–1956 (2023).
Yan, Y. et al. Machine learning and in-silico screening of metal–organic frameworks for O2/N2 dynamic adsorption and separation. Chem. Eng. J. 427, 131604 (2022).
Teng, Y. & Shan, G. Interpretable machine learning for materials discovery: predicting CO2 adsorption properties of metal–organic frameworks. APL Mater. 12, 081115 (2024).
Wu, X. et al. Mapping the porous and chemical structure-function relationships of trace CH3I capture by metal-organic frameworks using machine learning. ACS Appl. Mater. Interfaces 14, 47209–47221 (2022).
Yao, Z. et al. Inverse design of nanoporous crystalline reticular materials with deep generative models. Nat. Mach. Intell. 3, 76–86 (2021).
Yang, P. et al. Analyzing acetylene adsorption of metal–organic frameworks based on machine learning. Green. Energy Environ. 7, 1062–1070 (2022).
Kim, B., Lee, S. & Kim, J. Inverse design of porous materials using artificial neural networks. Sci. Adv. 6, eaax9324 (2020).
Chung, Y. G. et al. Computation-ready, experimental metal–organic frameworks: a tool to enable high-throughput screening of nanoporous crystals. Chem. Mater. 26, 6185–6192 (2014).
Dubbeldam, D., Calero, S., Ellis, D. E. & Snurr, R. Q. RASPA: molecular simulation software for adsorption and diffusion in flexible nanoporous materials. Mol. Simula. 42, 81–101 (2015).
Breiman, L. Random forests. Mach. learn. 45, 5–32 (2001).
Dorogush, A. V., Ershov, V. & Gulin, A. CatBoost: gradient boosting with categorical features support. Adv. Neural Inf. Process. Syst. 31, 1–7 (2018).
Coupry, D. E., Addicoat, M. A. & Heine, T. Extension of the universal force field for metal-organic frameworks. J. Chem. Theory Comput. 12, 5215–5225 (2016).
Rappé, A. K., Casewit, C. J., Colwell, K. S., Goddard, W. A. & Skiff, W. M. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc. 25, 10024–10035 (1992).
Lan, Y. S., Tong, M. M., Yang, Q. Y. & Zhong, C. L. Computational screening of covalent organic frameworks for the capture of radioactive iodine and methyl iodide. CrystEngComm 19, 4920–4926 (2017).
Wu, X. et al. In Silico Tuning of the Pore Surface Functionality in Al-MOFs for Trace CH3I Capture. ACS Omega 6, 18169–18177 (2021).
Hirschfelder, J. O. H., Curtiss, C. F., Bird, R. B. & Mayer, M. G. Molecular theory of gases and liquids. Am. Sci. 43, 60–64 (1955).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Zhang, K., Nalaparaju, A., Chen, Y. & Jiang, J. Biofuel purification in zeolitic imidazolate frameworks: the significant role of functional groups. Phys. Chem. Chem. Phys. 16, 9643–9655 (2014).
Potoff, J. J. & Siepmann, J. I. Vapor-liquid equilibria of mixtures containing alkanes, carbon dioxide, and nitrogen. Aiche J. 47, 1676–1682 (2001).
Zhang, L. & Siepmann, J. I. Direct calculation of Henry’s law constants from Gibbs ensemble Monte Carlo simulations: nitrogen, oxygen, carbon dioxide and methane in ethanol. Theor. Chem. Acc. 115, 391–397 (2006).
Rappe, A. K., Casewit, C. J., Colwell, K. S., Goddard, W. A. & Skiff, W. M. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc. 114, 10024–10035 (2002).
Willems, T. F., Rycroft, C. H., Kazi, M., Meza, J. C. & Haranczyk, M. Algorithms and tools for high-throughput geometry-based analysis of crystalline porous materials. Micropor. Mesopor. Mat. 149, 134–141 (2012).
Boyd, P. G., Moosavi, S. M., Witman, M. & Smit, B. Force-field prediction of materials properties in metal-organic frameworks. J. Phys. Chem. Lett. 8, 357–363 (2017).
Addicoat, M. A., Vankova, N., Akter, I. F. & Heine, T. Extension of the universal force field to metal organic frameworks. J. Chem. Theory Comput. 10, 880–891 (2014).
O’Boyle, N. M. et al. Open Babel: An open chemical toolbox. J. Cheminformatics 3, 1–14 (2011).
Yap, C. W. PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 32, 1466–1474 (2011).
Carhart, R. E., Smith, D. H. & Venkataraghavan, R. Atom pairs as molecular features in structure-activity studies: definition and applications. J. Chem. Inf. Comp. Sci. 25, 64–73 (1985).
Ruiz, I. L. & Nieto, M. Á. G. A new data representation based on relative measurements and fingerprint patterns for the development of QSAR regression models. Chemom. Intell. Lab. 176, 53–65 (2018).
Durant, J. L., Leland, B. A., Henry, D. R. & Nourse, J. G. Reoptimization of MDL keys for use in drug discovery. J. Chem. Inf. Comput. Sci. 42, 1273–1280 (2002).
Seo, M., Shin, H. K., Myung, Y., Hwang, S. & No, K. T. Development of natural compound molecular fingerprint (NC-MFP) with the dictionary of natural products (DNP) for natural product-based drug development. J. Cheminformatics 12, 1–17 (2020).
Pedregosa, F. et al. Scikit-learn: machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Gorishniy, Y., Rubachev, I., Khrulkov, V. & Babenko, A. Revisiting deep learning models for tabular data. Adv. Neural Inf. Process. Syst. 34, 18932–18943 (2021).
Acknowledgements
The work was carried out at National Supercomputer Center in Tianjin, and the calculations were performed on Tianhe new generation supercomputer. This work was financially supported by the National Key R&D Program of China (No.2022YFB4703403 and No. 2016YFC1402504). Dr. G.C. Shan particularly acknowledges additional support from the College of Science Distinguished Alumni Award granted by the College of Science at the City University of Hong Kong (CityUHK). Finally, this groundbreaking study is dedicated to commemorating the 120th anniversary of Fudan University to be celebrated in May 2025.
Author information
Authors and Affiliations
Contributions
H.Y.T. designed and implemented the workflow, performed the computation, generated the data, and prepared the manuscript. Y.K.T. provided importance advice for machine learning and revised the paper. G.C.S. gave scientific and technical advice throughout the work, revised and reviewed the manuscript, and supervised the project. All authors proofread and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tan, H., Teng, Y. & Shan, G. High throughput computational screening and interpretable machine learning for iodine capture of metal-organic frameworks. npj Comput Mater 11, 115 (2025). https://doi.org/10.1038/s41524-025-01617-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41524-025-01617-2