Abstract
Short-read genome sequencing (GS) is a powerful technique for investigating the genetic etiologies of rare diseases, capturing diverse genetic variations that are challenging to approach with exome sequencing (ES). We performed GS on 260 families with intellectual disability/developmental delay. GS detected potentially disease-related variants in 55 of the 260 families, with structural resolution by long-read sequencing or optical genome mapping, and functional assessment by RNA sequencing. Excluding 31 theoretically ES-resolvable cases, GS yielded likely pathogenic variants in 17 of 229 as well as variants of unknown significance in 7 of 229, totaling 10.5%. These variants implicated several new etiological mechanisms: a microduplication syndrome involving ATP6V0C; disturbed interactions of TBL1XR1 and NR2F1 with putative cis-regulatory elements by chromosomal rearrangements; and a CCG repeat expansion near the CHD3 transcription start site. This study highlights the critical role of GS in clinical diagnostics and its potential to advance understanding of genetic disorders.
Similar content being viewed by others
Introduction
Genome sequencing using short-read technology (hereafter referred to as GS) is a powerful and scalable approach for scrutinizing the genetic underpinnings of rare diseases. Unlike exome sequencing (ES), which has been extensively used in this decade, GS captures the diverse spectrum of genetic variations, such as non-coding gene variants, deep-intronic variants, intergenic variants, and a variety of structural variants (SVs). Moreover, the recent decline in sequencing costs makes it feasible to apply GS to rare disease patients on a large scale. In this context, several studies have undertaken GS analysis on hundreds to thousands of rare disease cases and demonstrated its diagnostic superiority over ES1,2,3,4,5,6. However, these studies have not fully exploited the strengths of GS, which confined their analyses to specific variant categories, resulting in underestimations of the genuine diagnostic potential of GS and few new etiologies proposed.
In this study, we performed GS on hundreds of patients previously undiagnosed by ES and having a syndromic intellectual disability (ID)/developmental delay (DD), one of the most prevalent categories of rare diseases. We exhaustively investigated a wide range of genetic variations, including non-coding variants, deep intronic variants, and repeat expansions. Consequently, we achieved a relatively high diagnostic yield among the ES-negative patients and succeeded in proposing several new etiological mechanisms underlying ID/DD that are challenging to address with ES.
Results
GS detects small variants and SVs that are inaccessible by ES despite at exons
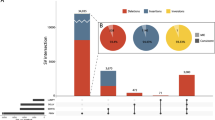
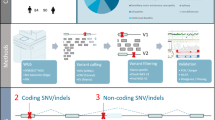
We performed GS on 1113 samples, including 260 ES-negative families having an ID/DD, consisting of 246 trios, 9 quads, 3 duos, and 2 singletons, along with 331 additional healthy control samples, totaling 844 healthy control samples (Table 1 and Fig. 1). After identifying potentially disease-related small variants at known ID/DD related protein-coding and non-coding genes, including RNU4-2, and new candidate genes, such as HIC1 (Table 2; Fig. 2a, Supplementary Figs. S1, S2; and Supplementary Data 1 and Material), we then examined SVs. Utilizing multiple SV callers, we uncovered a diverse array of SVs (Table 2; Supplementary Fig. S3; and Supplementary Data 2 and Material). These SVs that we detected involved single exon CNVs and an inversion, which are challenging to approach with ES, as well as large SVs and exonic mobile element insertions (MEIs) (Supplementary Fig. S4 and Supplementary Material). Besides protein-coding genes, we observed that Pt2190 carried a de novo deletion spanning the transcription start site of a non-coding gene, CHASERR (Fig. 2b). Three previously reported cases harbored similar deletions and exhibited upregulation of the flanking gene CHD2, which is presumed to underlie the pathogenesis7. We therefore sought to determine the CHD2 expression level in Pt2190’s LCL in two independent RNA-seq batches. After removing outlying samples in PCA biplots, Pt2190 had the highest CHD2 expression level among 22 samples in the first batch and 18 samples in the second batch while the CHASERR expression level was approximately half that of control samples (Supplementary Fig. S5). Furthermore, Pt2190 presented with a phenotype resembling that of the previous cases: short stature, low-set ears, small chin, bradycardia, atrial septal defect, severe global DD, seizures, hypotonia, myoclonus, and abnormal movements, along with enlarged ventricles and delayed myelination observed in brain MRI7. These suggest that Pt2190’s condition is attributable to CHASERR haploinsufficiency.
This figure was created in BioRender. Hamanaka, K. (2025) https://BioRender.com/f32v286.
a De novo small variants in the critical region of RNU4-2 in Pt0712 and Pt2092. Lollipop plot: Pathogenic/Likely pathogenic variants in ClinVar (https://ftp.ncbi.nlm.nih.gov/pub/clinvar/vcf_GRCh38/clinvar_20250608.vcf.gz). Green rectangle: RNA secondary structure. Heatmap plot: aggregated allele counts of single-nucleotide variants at each genomic position, derived from gnomAD v4.1.0 (https://gnomad.broadinstitute.org/data). b De novo deletion affecting CHASERR in Pt2190. Shown is an IGV view of read coverage and reads supporting the deletion. Read coloring follows the conventions of IGV for paired-end alignments (https://igv.org/doc/desktop/#UserGuide/tracks/alignments/paired_end_alignments).
GS disentangles the architecture of complex SVs, including a high-copy-number amplification of ATP6V0C
GS could delineate complex SV architectures in several cases, with support from T-LRS or OGM (Fig. 3, Supplementary Figs. S6–S8). In Pt2074, we found a de novo 16p13.3 multiplication involving ATP6V0C. Despite the complex copy number profile, only two breakpoints were discovered, presumably due to the overlapping SegDup regions (Fig. 3a), and even long-read GS to about 30x coverage around the multiplication could not resolve the hidden breakpoints. Optical genome mapping assembled a hypothetical overall structure, part of which appeared twice, likely due to misassembly (Supplementary Fig. S6a). Two alternative structures, excluding either duplicated part, were well consistent with the GS-based copy number profile except in SegDup regions (Fig. 3b and Supplementary Fig. S6b). Gene expression levels increased proportionally to gene copy numbers in LCL RNA-seq (Fig. 3c). Pt2074 presented with intellectual disability, cerebellar ataxia, mild cerebral atrophy, and marked cerebellar atrophy as well as dysphagia, amblyopia, and hyperopia; two cases with overlapping duplication also had similar clinical features in the DECIPHER database (Fig. 3d). As no triplosensitive gene was recognized in this duplicated interval, these data together propose a new microduplication syndrome on 16p13.3, whose pathogenesis may be attributed to ATP6V0C (see Discussion).
A de novo 16p13.3 multiplication in Pt2074. a Upper, copy number profiles estimated using CNView. Lower, adjoined breakends identified through GS are illustrated as curved lines in the IGV snapshots. SegDup is displayed as in the UCSC genome browser (https://genome.ucsc.edu/cgi-bin/hgTrackUi?g=genomicSuperDups). The copy number profiles are presented in bins indicated on the top right corner, with gray intervals representing the mean ± two standard deviations of 1330 samples. Read colors follow IGV conventions. b The overall structure estimated from optical genome mapping data. Breakpoints, inferred from optical genome mapping data, are colored black. c Bar plot of gene expression levels in RNA-seq of Pt2074’s LCL (replicate n = 2) relative to control samples (n = 17). Individual replicates and control samples are indicated by black dots over the red and blue bars, respectively. d Two DECIPHER cases with similar 16p13.3 duplications to Pt2074. Blue bar: duplication; black bar: multiplication in Pt2074. c, d Gene models are taken from the gnomAD constraint metrics track of the UCSC genome browser (https://genome.ucsc.edu/cgi-bin/hgTrackUi?db=hg19&g=gnomadPLI) and color-coded according to LOEUF.
GS detects intergenic SVs disturbing interactions of TBL1XR1 and NR2F1 with cis-regulatory elements
Variants may exert their impact by disrupting enhancer-promoter or promoter-promoter interactions. We identified a de novo translocation, t(3;10)(q26.32;q11.23)dn, in Pt2286. This SV directly disrupted an autosomal recessive gene, OGDHL, at 10q11.23, but no additional deleterious variant was found in OGDHL. Meanwhile, this SV did not directly disrupt any gene on 3q26.32, but we found that the haploinsufficiency phenotype of TBL1XR1, close to the breakpoint, was compatible with clinical features of Pt2286; the patient showed short stature, low weight various dysmorphic features, such as long face, protruding lip, deep-set eyes, fleshy ears, short and broad nose, hearing impairment, global DD, a variety of seizures including West syndrome, atonic seizures, tonic seizures, hyperkinetic seizures, and myoclonic seizures, and cerebral atrophy. All of these features except the long face and hyperkinetic seizures were consistent with the phenotype of TBL1XR1 haploinsufficiency8,9,10,11,12,13,14,15. A previous HFFc6 micro-C experiment revealed that the TBL1XR1 TSS physically contacted to many sites within the relocated genomic region, including the LOC105374235 TSS (Fig. 4a). Some of these contact sites were located in open chromatin regions identified through assay for transposase-accessible chromatin sequencing (ATAC-seq) and demarcated with histone modifications such as histone H3 lysine 27 acetylation (H3K27ac) and histone H3 lysine 4 monomethylation (H3K4me1), but not histone H3 lysine 4 trimethylation (H3K4me3) in neuroblastoma cell lines such as BE(2)-C and SH-SY5Y, suggesting that they are putative enhancers (Fig. 4b). Contrary to our expectation, RNA-seq of Pt2286’s LCL showed no obvious decrease in TBL1XR1 expression, which may be explained by cell-type specificity in gene regulation.
Chromatin landscape around TBL1XR1 (a) and NR2F1 (c). From top to bottom, the figure displays the contact map and chromatin loop calls of an HFFc6 micro-C, coverage tracks of ATAC-seq and histone ChIP-seq including H3K27ac, H3K4me3, and H3K4me1 modifications in BE(2)-C and SH-SY5Y, and GENCODE V44 gene models colored according to UCSC conventions (https://genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid=2280748172_eutv8PURT0qZPmMVtttVc16EEnGI&c=chr12&g=wgEncodeGencodeV44). Dotted lines indicate SV breakpoints observed in Pt2286 (a) and Pt2800 as well as in a previous study (c)20. b, d Enlarged views of the (a) and (c) panels. The top tracks depict the regions interacting with the TBL1XR1 TSS (b) or NR2F1 TSS (d). To the left of the coverage tracks are the depth-normalized coverage intervals on the Y-axis.
A similar pathological basis was suspected for the chromothripsis of Pt2800 mentioned above. Pt2800 showed DD, spastic paraplegia, dystonia, dysphagia, and myopia. With the exception of paraplegia and dystonia, these findings were observed in the haploinsufficiency of NR2F1, which was proximal to one of the breakpoints of the chromothripsis15,16,17. Furthermore, a previous study reported a translocation with a similar breakpoint in a case of DD, speech delay, expressive language delay, congenital infantile left eye esotropia, and bilateral fifth finger clinodactyly, all of which were observed in the NR2F1-related neurodevelopmental disorder, although the original report attributed the condition to the direct disruption of lncNR2F1 by the translocation (Fig. 4c)16,18,19,20. The upstream region displaced from NR2F1 by the chromothripsis contained putative regulatory elements interacting with the NR2F1 promoter (Fig. 4d). The expression level of NR2F1 in Pt2808 could not be evaluated due to its scarce expression in LCL samples (https://www.gtexportal.org/home/).
GS detects intronic small variants and an intronic DLX4 retrotransposition that may impact splicing
We explored splicing-altering variants, including deep-intron ones, utilizing SpliceAI, a deep learning model predicting whether each base within a given sequence functions as a donor site, an acceptor site, or neither. Leveraging this tool, we discovered variants possibly altering splicing (Supplementary Figs. S9, S10, and Supplementary Material), such as DLX4 retrotransposition into a deep-intronic region of STXBP1 in Pt2178 (Fig. 5). In this case, a deep-intronic region of STXBP1 was adjacent to a DLX4 transcript start and end through a poly(A) sequence. Notably, we observed elevated coverage at DLX4 exons and split reads spanning DLX4 exon-exon junctions, together indicating a DLX4 transcript insertion. SpliceAI suggested that the retrotransposed transcript possessed an acceptor site in the exon 1 and a donor site at the exon-exon junction, although the very low expression of STXBP1 in LCL precluded experimental validation.
A de novo retrotransposition of a DLX4 transcript into STXBP1 intron 10 in Pt2178. Depicted are IGV snapshots of STXBP1 (top) and DLX4 (middle) alongside GENCODE transcript models, and SpliceAI score distribution along the mutant STXBP1 intron 10 sequence in Pt2178 (bottom). For the SpliceAI score, the upper portion illustrates donor site scores while the lower portion displays acceptor site scores. Blue bars: wild-type sequence scores; red bars: mutant sequence scores; blue rectangle: potential pseudo-exonization based on SpliceAI predictions.
GS detects repeat expansions outside coding regions
Our focus shifted towards tandem repeat expansions. To measure the length of tandem repeats, we utilized ExpansionHunter Denovo (EHdn), which counts two types of reads: anchored in-repeat reads (IRRs), which originate within tandem repeats and their mates originate in the surrounding unique sequence; paired IRRs, read pairs where both mates stem from within the same tandem repeat. These categories of reads inform long repeat size well beyond the GS read length. Applying EHdn to our GS data, we investigated repeat expansions rarely observed among 843 healthy control individuals genome-wide, with one control individual excluded due to its outlying number of expanded repeats. This analysis led to definite diagnoses of dentatorubral–pallidoluysian atrophy and congenital myotonic dystrophy in two cases (Supplementary Fig. S11 and Supplementary Material). Furthermore, we observed a rare increase in in-CCG-repeat read counts around a CHD3 transcription start site (TSS) in Pt481 and his mother (Fig. 6a). These anchored read counts were equivalent between Pt481 and his mother, and no paired in-CCG-repeat-reads were observed, but T-LRS apparently exhibited a de novo expansion of the Chr17:7,885,308-7,885,345 CCG repeat, with 244 units in Pt481 and 159 in his mother (Fig. 6b, c). T-LRS reads with CCG expansion were highly methylated, and Pt481 had a higher DNA methylation level than his mother, parallel to their repeat size (Fig. 6d). The TSS near the CCG repeat was the most predominant in aggregated CAGE data from various cells and tissues, although the Matched Annotation from NCBI and EMBL-EBI (MANE) considered a downstream TSS as canonical (Fig. 6d). Their CHD3 expression levels could not be examined due to the unavailability of LCL samples. Previously, Fazal et al. also applied EHdn to 40× GS data of 1115 individuals and 30 × 1000 Genomes GS data of 2504 individuals, both of which were generated using a 150 bp paired-end PCR-free protocol like our GS data21. In their data, five individuals had at least one in-CCG-repeat read tethered to the CHD3 TSS region, with ~10 or fewer depth-normalized counts, far lower than our patients (Fig. 6a): NA18615, 6.4; NA19085, 2.4; NA18525, 1.1; HG01440, 10.5; and HG02304, 3.2. Although direct comparisons should be made with caution, Pt481 and his mother likely have very rare CGG repeat sizes. CHD3 is a haploinsufficient gene responsible for global DD with lower penetrance in females; consistently, Pt481 presented with global DD, along with infantile spasms22. Overall, the rare CCG repeat expansion may cause CHD3 haploinsufficiency in Pt481 through DNA hypermethylation and consequent transcriptional silencing.
a Counts of in-CCG-repeat reads anchored to Chr17:7884501-7886140 in CHD3 among Pt481, his parents, and healthy control individuals. b–d T-LRS of the Chr17:7,885,308-7,885,345 CCG repeat at CHD3 in Pt481 and his mother. b Frequency of CCG repeat size in T-LRS reads. Red, forward read; blue, reverse read. c Waterfall plot showing the tri- or di-nucleotide composition of the CCG repeat expansion and flanking sequences in T-LRS reads. Tri- or di-nucleotides were colored as shown in the upper right corner. d DNA methylation status around the predominant CHD3 TSS. Top, GENCODE transcript models; middle, total CAGE counts aggregated from various cells and tissues (https://genome.ucsc.edu/cgi-bin/hgTrackUi?db=hg38&g=fantom5); bottom, T-LRS reads with (“exp.”) or without (“no-exp.”) CCG repeat expansion. Horizontal line: T-LRS read; black circle: methylated CpG; white circle: unmethylated CpG.
Clinical impact of GS on diagnosis and treatment
We evaluated GS diagnostic value for cases negative for ES. We here assumed that ES could theoretically detect small variants with a median depth of eight or more in the gnomAD ES data1, as well as CNVs of three or more exons and exonic MEIs. Based on these assumptions, 31 cases were considered resolvable by ES (i.e., all variants listed in Table 2 were theoretically detectable for respective cases). In practical terms, however, five of the variants in the 31 cases were difficult to detect at the time of ES for various reasons: RBMX was not yet associated with disease; the SCN8A variant was located in an exon that was not part of the canonical transcript at the time; there were few supporting reads for the exonic MEI in SMC1A; and accurate splicing prediction tools were not yet available for CEP290 and WDR62. The variants in the remaining 26 cases were likely detectable at the time of ES and were presumably missed due to insufficiently thorough analysis. We classified pathogenicity of the variants in the other 24 theoretically ES-unresolvable cases using the guidelines of the American College of Medical Genetics and Genomics (ACMG), the Association for Molecular Pathology (AMP), and the Clinical Genome Resource (ClinGen), as detailed in Supplementary Information. According to these criteria, GS yielded Pathogenic/Likely Pathogenic variants (P/LP) in 7.4% (17/229) as well as variants of unknown significance (VUS) in 3.1% of cases (7/229), totaling 10.5%. A common alternative for detecting CNVs is chromosomal microarray analysis (CMA). As CMA detects CNVs larger than 20 kb (https://www.agilent.com/cs/library/usermanuals/public/K1201-90001.pdf), GS seemed to yield P/LP variants in 6.6% of CMA/ES-unresolvable cases (15/227) and VUS in 3.1% (7/227). Note that even variants categorized as VUS were also considered likely related to the diseases given the strong phenotypic match with the affected genes.
These diagnoses made by GS could impact clinical management in certain cases (Fig. 1). For instance, Pt2878 was identified with Niemann-Pick disease type C, for which two approved therapeutic drugs are available23. Pt7183 was diagnosed with generalized squalene synthase deficiency, a disorder characterized by the accumulation of intermediates in the mevalonate pathway mediated by 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) and a deficiency in cholesterol, the primary product. A localized manifestation of the disease in the skin due to somatic variants was mitigated using a topical ointment of cholesterol and statin, an HMGCR inhibitor24. Therefore, systemic administration of these agents may be effective for the generalized deficiency in Pt7183.
Discussion
By comprehensively analyzing GS data, we achieved a high diagnostic yield of ~7% for ES-negative patients with ID/DD, identified potentially disease-associated VUSs in ~3%, and proposed several new etiologies, including a microduplication syndrome of 16p13.3, disrupted interactions of TBL1XR1 and NR2F1 with cis-regulatory elements, and a CCG repeat expansion in the CHD3 intron 1.
Our semi-automated validation approach using IGV allowed us to detect a wide array of SVs. As the actual overall architecture of SVs cannot be deduced from SV calls alone (Supplementary Data 2), just looking at tables of SV calls would mislead us in interpreting their pathogenicity. Indeed, many SV calls indicating pathogenic SVs did not accurately represent their true structures (Supplementary Data 2). Therefore, high-throughput validation methods like ours or sophisticated computational tools that predict true overall structures are essential.
To our knowledge, this is the first report of a de novo NUMT insertion tethering a telomere sequence, which we observed in Pt448, and the mutation mechanism is unclear. NUMTs act as a sticking-plaster during the repair of double-strand breaks via the non-homologous end joining pathway25. As the reversal near the 2q terminus would excessively elongate the q arm in Pt448 (Supplementary Fig. S7a), it is plausible that the q arm was truncated and then repaired with a NUMT. It will be intriguing to investigate whether chromosomes with similar CNVs in other cases also have a NUMT preceding a telomere sequence.
Although we propose a new microduplication syndrome associated with a 400 kb region on 16p13.3, the specific gene responsible for this syndrome remains uncertain. Since haploinsufficient genes are supposed to be more likely triplosensitive, ATP6V0C is a good candidate, being the only gene currently recognized as haploinsufficient within this interval. Indeed, the clinical manifestations of Pt2074 resembled the phenotype of ATP6V0C haploinsufficiency. ATP6V0C encodes the c subunit of vacuolar ATPase, and nine c subunits and one c” subunit, encoded by ATP6V0B, make up the proton pump complex of the vacuolar ATPase26. Excess c subunit may disrupt this stoichiometry and in turn impair vacuolar ATPase functions. Thus, ATP6V0C triplosensitivity may underlie the microduplication syndrome.
The generalizability of the GS diagnostic yield observed in this study is subject to several limitations. The 260 cases analyzed here were randomly selected from those previously analyzed in our laboratory. However, there is potential bias in the cohort, as many referrals came from epilepsy clinics or from physicians suspecting known syndromes. Furthermore, most patients likely had not undergone prior microarray testing, as such testing was not widely implemented as a routine clinical practice in Japan until recently (2021). If performed, the additional yield from GS might have been slightly reduced, as microarray can sometimes detect large CNVs affecting only a single exon. In addition, our GS analysis was conducted as part of translational research and included labor-intensive steps, such as manual inspection of BND calls using IGV. This may have contributed to a higher diagnostic yield than that typically achievable in standard clinical settings. Therefore, in more diverse real-world contexts, the diagnostic yield of GS may be somewhat lower than reported here.
This study demonstrates the potential of GS to uncover previously unrecognized disease etiologies. The next step will be to establish the pathogenicity of these novel candidates. To achieve this, it will be essential to apply GS at scale, accumulate cases with similar genetic findings, and validate promising candidates using functional approaches such as genome editing.
Thus, this study demonstrates that GS could be highly beneficial for making genetic diagnoses and uncovering novel etiologies beyond the reach of ES, inspiring nation-wide programs for GS diagnosis of rare disease patients ongoing in various countries.
Methods
Participant enrollment
The study was approved by the institutional review board of Yokohama City University School of Medicine (Yokohama, Japan) and National Center for Global Health and Medicine (Tokyo, Japan). Written informed consent was obtained from the patients or their legal guardians. This study conformed to the Declaration of Helsinki. The 260 patients were suspected of having rare syndromes with varying degrees of ID and/or DD by attending physicians (Table 1). These individuals were referred to the Department of Human Genetics, Yokohama City University for genetic diagnosis, both domestically and through international collaborations with countries such as Brazil and Israel. However previous ES testing revealed no diagnostic variants. Cases were referred from hospitals located in Japan (n = 195), Brazil (n = 37), Israel (n = 25), and other countries (n = 3). Additional healthy participants were referred by hospitals in Japan (n = 287), Brazil (n = 26), Israel (n = 12), and other countries (n = 6).
GS
We extracted genomic DNA from peripheral blood leukocytes using QuickGene-610L (Fujifilm, Tokyo, Japan) according to the manufacturer’s protocol and performed GS as previously described in detail27. Variant calls were annotated with their functional consequences, deleteriousness, and allele frequencies, including Sorting Intolerant From Tolerant (SIFT), Polymorphism Phenotyping v2 (Polyphen-2), and CADD, using ANNOVAR v2019May (https://annovar.openbioinformatics.org/en/latest/) and UTRannotator (https://github.com/ImperialCardioGenetics/UTRannotator). Additionally, we manually annotated calls with precomputed SpliceAI scores (spliceai_scores.masked.indel.hg38.vcf.gz, https://github.com/Illumina/SpliceAI?tab=readme-ov-file); allele frequency from several large-scale GS projects, such as the Trans-Omics for Precision Medicine (TOPMed) program (https://legacy.bravo.sph.umich.edu/freeze8/hg38/downloads), the Tohoku University Tohoku Medical Megabank Organization (ToMMo) (https://grch38.togovar.org/downloads/gem_j_wga/grch38/liftover/), and National Center Biobank Network (NCBN) (https://humandbs.dbcls.jp/en/hum0331-v1); and poly(A) signal sites (https://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/release_35/gencode.v35.polyAs.gtf.gz).
SV analysis
We utilized a total of five SV callers: Manta v1.6.0 (https://github.com/Illumina/manta), Delly v0.8.3 (https://github.com/dellytools/delly), Smoove v0.2.6 (https://github.com/brentp/smoove), Canvas v1.40.0 (https://github.com/Illumina/canvas), CNVnator v0.3.3, implemented in SVE (https://github.com/timothyjamesbecker/SVE), and MELT v2.2.0 (https://melt.igs.umaryland.edu/)28,29,30,31,32,33,34,35. For Delly, Smoove, and MELT, we generated multi-sample VCFs encompassing all samples using their joint genotyping functions according to the developers’ protocols (https://github.com/dellytools/delly, https://github.com/brentp/smoove, and https://melt.igs.umaryland.edu/manual.php). Manta and Canvas, while also capable of joint genotyping, are limited to a small number of samples; thus, we performed joint genotyping for each family, split each family-level VCF into individual-level VCFs, and merged all VCFs using Jasmine v1.1.4 (https://github.com/mkirsche/Jasmine), which consolidates SVs with the same class (e.g., deletion) and proximate breakpoints across individuals. Meanwhile, CNVnator lacks a built-in joint genotyping function, therefore we generated VCFs for each sample and merged them using Jasmine. Based on these multi-sample VCFs, allele frequency was calculated for each SV. For Delly, we omitted the last “delly filter” step to enhance its sensitivity. The SV calls were annotated using AnnotSV v3.0.1 (https://github.com/lgmgeo/AnnotSV). To validate SV calls on a large scale, we employed IGV batch scripts to automatically capture IGV snapshots in given settings (https://igv.org/doc/desktop/#UserGuide/tools/batch/). IGV images of the start, end, and/or entire region of SV calls were captured and manually inspected. Allele frequencies were referenced from the gnomAD-SV v4.1.0 (https://gnomad.broadinstitute.org/ and https://storage.googleapis.com/gcp-public-data--gnomad/release/4.1/genome_sv/gnomad.v4.1.sv.sites.vcf.gz).
We visualized copy number profiles along specified genomic regions using CNView (https://github.com/RCollins13/CNView). Reads were counted in 10 kb bins across the genome in each of 1330 samples using the “counts” option of the bedtools v2.19.0 coverage function and CRAM files. The 1330 samples included all the samples in this study as well as other samples (n = 217) sequenced in the same manner. These 1330 resulting bedgraph files were merged using the bedtools unionbedg function and then processed with CNView. CNView extracts the coverage of a given genomic region and its flanking 5 Mb and normalizes each sample by dividing the coverage of each bin by the median bin-wise coverage of the sample. We then computed the mean and standard deviation of the normalized coverage of each bin in all samples as a reference. In Pt448, Pt2361, and Pt2539, who had subchromosomal CNVs, read counts were normalized based on the entire genome, instead of their flanking 5 Mb. In male Pt7147 with Xp11.22 multiplication, this whole CNView analysis was restricted to 665 male samples.
Targeted long-read sequencing
Genomic DNA was extracted from blood or lymphoblastoid cell lines (LCLs) and was sheared to 20 kb or 40 kb using the Megaruptor 2 (Diagenode, Seraing, Belgium), or DNA fragments less than 20 kb were removed with the Short Read Eliminator Kit (Pacific Biosciences, Menlo Park, CA, US). Sequencing libraries were constructed by the Ligation Sequencing Kit (SQK-LSK114, Oxford Nanopore Technologies, Oxford, UK) as previously described36. Approximately 25–50 fmol of the library was loaded onto a flow cell (FLO-MIN114, R10.4.1) for sequencing on the GridION Mk1 (Oxford Nanopore Technologies) with the adaptive sampling option to enrich 0.25% or 0.5% of the whole genome that contains regions of interest. Basecalling was performed with Guppy v.6.5.7 (https://community.nanoporetech.com/docs/prepare/library_prep_protocols/Guppy-protocol/v/gpb_2003_v1_revax_14dec2018). For Pt481 and his mother, who harbored an expanded CCG repeat in the CHD3 intron 1, modified base calling for 5-methylcytosine was performed in super accuracy mode, and the reads were aligned to the reference genome using LAST v1389 (https://gitlab.com/mcfrith/last) for repeat sizing or minimap2 v.2.14 (https://github.com/lh3/minimap2) for DNA methylation analysis. Repeat size was measured in each individual read using tandem-genotypes v.1.9.0 (https://github.com/mcfrith/tandem-genotypes). Reads with or without repeat expansion were extracted and stored separately in BAM files using SAMtools (https://github.com/samtools/). Based on these allele-specific BAM files, DNA methylation status along the genome was plotted using methylartist (https://github.com/adamewing/methylartist). Waterfall plots of the repeat sequence composition were generated using RepeatAnalysisTools (https://github.com/PacificBiosciences/apps-scripts/tree/master/RepeatAnalysisTools).
Epigenetic landscape
To depict the epigenetic landscapes of genomic regions, we downloaded depth-normalized coverage bigWig files of ATAC-seq and histone ChIP-seq including H3K27ac, H3K4me3, and H3K4me1 modifications in BE(2)-C and SH-SY5Y cell lines from the ChIP-Atlas (https://github.com/inutano/chip-atlas/wiki) (Supplementary Data 3). We computed the average coverage of each experiment type of each cell line using the “mean” function of wiggletools (https://github.com/Ensembl/WiggleTools), generating a wiggle-formatted file. This file was in turn converted to a bigWig file using wig2bed (https://github.com/bedops/bedops/releases/download/v2.4.41/bedops_linux_x86_64-v2.4.41.tar.bz2) and bedGraphToBigWig (http://hgdownload.soe.ucsc.edu/admin/exe/linux.x86_64/bedGraphToBigWig) and then visualized in IGV. In addition, we downloaded the hic-format file of a micro-C experiment of human foreskin fibroblasts (HFFc6) (https://data.4dnucleome.org/files-processed/4DNFIPC7P27B/@@download/4DNFIPC7P27B.hic) and visualized the chromatin contact map utilizing the Juicebox web application (https://aidenlab.org/juicebox/). To identify chromatin loops, we obtained a pairs-format file from the experiment (https://data.4dnucleome.org/files-processed/4DNFINYO612N/@@download/4DNFINYO612N.pairs.gz) and converted it to a hic-format file using the “pre” function of juicer_tools_1.22.01.jar (https://github.com/aidenlab/juicer/wiki/Download). Subsequently, loops were called at 5 or 10 kb resolution with a default 20% false discovery rate using Mustache ver. 1.3.2 (https://github.com/ay-lab/mustache/). Loops arising from the TBL1XR1 TSS (Chr3:177,190,00-177,205,000) or NR2F1 TSS (Chr5:93,575,000-93,590,000) were then visualized using CoolBox (https://gangcaolab.github.io/CoolBox/).
Detection of repeat expansion using Expansion Hunter Denovo (EHdn)
We searched repeat expansion utilizing EHdn ver. 0.9.0 following developers’ instructions (https://github.com/Illumina/ExpansionHunterDenovo/tree/master/documentation). Briefly, we generated a JavaScript Object Notation (JSON) file encompassing anchored and paired IRR count profiles for each sample employing the “profile” function. The JSON files were integrated into a single JSON file using the “merge” function and then transformed into two tables depicting counts of IRR anchored at each locus and paired IRR of each motif using outlier.py, an accompanying auxiliary script. The IRR counts were normalized to a depth of 30×. Repeat loci in the former file were annotated with RefSeq genes using annotate_ehdn.sh, another script.
Data availability
There are restrictions to the availability of the GS data presented here due to the protection of personal information. The genetic variants reported in this study are scheduled to be publicly available through the Medical Genomics Japan Variant Database (MGeND) (https://mgend.ncgm.go.jp/).
Code availability
The codes generated during this study are available at https://github.com/hamanakakohei/NPJGenomicMed2025.
References
Smedley, D. et al. 100,000 genomes pilot on rare-disease diagnosis in health care - preliminary report. N. Engl. J. Med. 385, 1868–1880 (2021).
van der Sanden, B. et al. The performance of genome sequencing as a first-tier test for neurodevelopmental disorders. Eur. J. Hum. Genet. 31, 81–88 (2023).
Lindstrand, A. et al. Genome sequencing is a sensitive first-line test to diagnose individuals with intellectual disability. Genet. Med. 24, 2296–2307 (2022).
Bertoli-Avella, A. M. et al. Successful application of genome sequencing in a diagnostic setting: 1007 index cases from a clinically heterogeneous cohort. Eur. J. Hum. Genet. 29, 141–153 (2021).
Stranneheim, H. et al. Integration of whole genome sequencing into a healthcare setting: high diagnostic rates across multiple clinical entities in 3219 rare disease patients. Genome Med. 13, 40 (2021).
Turro, E. et al. Whole-genome sequencing of patients with rare diseases in a national health system. Nature 583, 96–102 (2020).
Ganesh, V. S. et al. Neurodevelopmental disorder caused by deletion of CHASERR, a lncRNA Gene. N. Engl. J. Med. 391, 1511–1518 (2024).
Slavotinek, A. et al. Pierpont syndrome associated with the p.Tyr446Cys missense mutation in TBL1XR1. Eur. J. Med. Genet. 60, 504–508 (2017).
Vaqueiro, A. C. et al. Expanding the spectrum of TBL1XR1 deletion: report of a patient with brain and cardiac malformations. Eur. J. Med. Genet. 61, 29–33 (2018).
Lemattre, C. et al. TBL1XR1 mutations in Pierpont syndrome are not restricted to the recurrent p.Tyr446Cys mutation. Am. J. Med. Genet. A 176, 2813–2818 (2018).
Tamma, P. L., Streff, H. & Murali, C. N. A novel de novo pathogenic variant in TBL1XR1 as a new proposed cause of Pierpont syndrome. Am. J. Med. Genet. A 191, 1576–1580 (2023).
Zaghlula, M. et al. Current clinical evidence does not support a link between TBL1XR1 and Rett syndrome: description of one patient with Rett features and a novel mutation in TBL1XR1, and a review of TBL1XR1 phenotypes. Am. J. Med. Genet. A 176, 1683–1687 (2018).
Saitsu, H. et al. A girl with West syndrome and autistic features harboring a de novo TBL1XR1 mutation. J. Hum. Genet. 59, 581–583 (2014).
Ren, M., Zheng, H., Lu, X., Lian, W. & Feng, B. Expanding the genotypic and phenotypic spectrum associated with TBL1XR1 de novo variants. Gene 886, 147777 (2023).
Jurkute, N. et al. Pathogenic NR2F1 variants cause a developmental ocular phenotype recapitulated in a mutant mouse model. Brain Commun. 3, fcab162 (2021).
Chen, C. A. et al. The expanding clinical phenotype of Bosch-Boonstra-Schaaf optic atrophy syndrome: 20 new cases and possible genotype-phenotype correlations. Genet. Med. 18, 1143–1150 (2016).
Kaiwar, C. et al. Novel NR2F1 variants likely disrupt DNA binding: molecular modeling in two cases, review of published cases, genotype-phenotype correlation, and phenotypic expansion of the Bosch-Boonstra-Schaaf optic atrophy syndrome. Cold Spring Harb. Mol. Case Stud. 3, https://doi.org/10.1101/mcs.a002162 (2017).
Mio, C. et al. Missense NR2F1 variant in monozygotic twins affected with the Bosch-Boonstra-Schaaf optic atrophy syndrome. Mol. Genet. Genom. Med. 8, e1278 (2020).
Starosta, R. T. et al. Bosch-Boonstra-Schaaf optic atrophy syndrome (BBSOAS) initially diagnosed as ALG6-CDG: Functional evidence for benignity of the ALG6 c.391T>C (p.Tyr131His) variant and further expanding the BBSOAS phenotype. Eur. J. Med. Genet. 63, 103941 (2020).
Ang, C. E. et al. The novel lncRNA lnc-NR2F1 is pro-neurogenic and mutated in human neurodevelopmental disorders. Elife 8, https://doi.org/10.7554/eLife.41770 (2019).
Fazal, S. et al. Large scale in silico characterization of repeat expansion variation in human genomes. Sci. Data 7, 294. https://doi.org/10.1038/s41597-020-00633-9 (2020).
van der Spek, J. et al. Inherited variants in CHD3 show variable expressivity in Snijders Blok-Campeau syndrome. Genet. Med. 24, 1283–1296 (2022).
Niemann-Pick double win. Nat. Biotechnol. 42, 1479 (2024).
Saito, S. et al. Gene-specific somatic epigenetic mosaicism of FDFT1 underlies a non-hereditary localized form of porokeratosis. Am. J. Hum. Genet. 111, 896–912 (2024).
Puertas, M. J. & González-Sánchez, M. Insertions of mitochondrial DNA into the nucleus-effects and role in cell evolution. Genome 63, 365–374 (2020).
Wang, L., Wu, D., Robinson, C. V., Wu, H. & Fu, T. M. Structures of a complete human V-ATPase reveal mechanisms of its assembly. Mol. Cell 80, 501–511 (2020).
Kawai, Y. et al. Exploring the genetic diversity of the Japanese population: insights from a large-scale whole genome sequencing analysis. PLoS Genet. 19, e1010625 (2023).
Chen, X. et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32, 1220–1222 (2016).
Rausch, T. et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28, i333–i339 (2012).
Pedersen, B. S., Ryan, L. & Quinlan, A.R. smoove: structural-variant calling and genotyping with existing tools. https://github.com/brentp/smoove (2020).
Roller, E., Ivakhno, S., Lee, S., Royce, T. & Tanner, S. Canvas: versatile and scalable detection of copy number variants. Bioinformatics 32, 2375–2377 (2016).
Ivakhno, S., Roller, E., Colombo, C., Tedder, P. & Cox, A. J. Canvas SPW: calling de novo copy number variants in pedigrees. Bioinformatics 34, 516–518 (2018).
Abyzov, A., Urban, A. E., Snyder, M. & Gerstein, M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res.21, 974–984 (2011).
Gardner, E. J. et al. The Mobile Element Locator Tool (MELT): population-scale mobile element discovery and biology. Genome Res. 27, 1916–1929 (2017).
Becker, T. et al. FusorSV: an algorithm for optimally combining data from multiple structural variation detection methods. Genome Biol. 19, 38 (2018).
Miyatake, S. et al. Rapid and comprehensive diagnostic method for repeat expansion diseases using nanopore sequencing. NPJ Genom. Med. 7, 62 (2022).
Acknowledgements
We thank all of the participants for their cooperation in this research. This work was supported by AMED under grant numbers JP24ek0109674, JP24ek0109760, JP24ek0109617, JP24ek0109648 and JP24ek0109677 (N. Matsumoto); JSPS KAKENHI under grant numbers JP23K27520 (S. Miyatake), JP23K27568 and JP23K18278 (T. Mizuguchi), JP22K15901 (A. Fujita), JP22K15646 (K. Hamanaka) and JP24K02230 (N. Matsumoto); the Takeda Science Foundation (T. Mizuguchi, N. Matsumoto); and The Ichiro Kanehara Foundation for the Promotion of Medical Science & Medical Care (S. Miyatake). The funding source had no role in the conduct of the study.
Author information
Authors and Affiliations
Contributions
Conceptualization: K.Ha, N.M.; Methodology: K.Ha, A.F., S.M., K.M., E.K., Y.Uc.; Formal analysis: K.Ha, A.F., K.M.; Investigation: K.Ha. A.F., S.M., K.M., E.K., Y.Uc., N.Ts., M.Sak., K.Iw., N.N., Y.Ut., L.F., M.T., Q.L., R.S., T.It., K.Sa., S.O., S.K., H.F., Y.In., Y.K., Y.O., K.T., T.Mi., T.G., K.Ic., I.K., M.F., K.K., T.S., K.Shim., K.O., Y.Ue., K.C., K.Yu., N.Ta., M.Y., A.D., K.Hi., T.Yan., T.Yam, K.Shir., T.F.M., A.F.V., D.L., H.Y., E.I., Y.S., M.M., K.Su., A.I., M.Sas., Y.W., J.I.T., C.A.K., K.Yo., J.T., T.Mo., Y.Iz., Y.H., N.O., T.Ik., H.O., M.K.; Resources: T.G., K.I., I.K., M.F., K.K., T.S., K.Shim., K.O., Y.Ue., K.C., K.Yu., N.Ta., M.Y., A.D., K.Hi., T.Yan., T.Yam, K.Shir., T.F.M., A.F.V., D.L., H.Y., E.I., Y.S., M.M., K.Su., A.I., M.Sas., Y.W., J.I.T., C.A.K., K.Yo., J.T., T.Mo., Y.Iz., Y.H., N.O., T.Ik., H.O., M.K.; Writing - Original Draft: K.Ha. A.F., N.M.; Writing - Review & Editing: S.M., K.M., E.K., Y.Uc., N.Ts., M.Sak., K.Iw., N.N., Y.Ut., L.F., M.T., Q.L., R.S., T.It., K.Sa., S.O., S.K., H.F., Y.In., Y.K., Y.O., K.T., T.Mi., T.G., K.I., I.K., M.F., K.K., T.S., K.Shim., K.O., Y.Ue., K.C., K.Yu., N.Ta., M.Y., A.D., K.Hi., T.Yan., T.Yam., K.Shir., T.F.M., A.F.V., D.L., H.Y., E.I., Y.S., M.M., K.Su., A.I., M.Sas., Y.W., J.I.T., C.A.K., K.Yo., J.T., T.Mo., Y.Iz., Y.H., N.O., T.Ik., H.O., M.K.; Supervision: N.M.; Project administration: N.M.; Funding acquisition: K.Ha. A.F., S.M., T.Mi., N.M. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hamanaka, K., Fujita, A., Miyatake, S. et al. Genome sequencing provides high diagnostic yield and new etiological insights for intellectual disability and developmental delay. npj Genom. Med. 10, 60 (2025). https://doi.org/10.1038/s41525-025-00521-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41525-025-00521-4