Abstract
Triple-negative breast cancer (TNBC) is the most aggressive subtype, typically requiring neoadjuvant chemotherapy (NAC) as an obligatory component of the treatment regimen. Achieving a pathological complete response to NAC is associated with improved long-term outcomes for patients with TNBC. The functional status of the immune system plays a critical role in NAC efficacy. Herein, we presented the investigation of systemic and local immune landscape during the initial course of NAC treatment and identify factors that contribute to chemotherapy resistance of TNBC. Using single-cell RNA sequencing, we demonstrated that the transcriptional profile remained stable in a patient who responded to NAC, while a non-responder exhibited significant dysregulation in the expression of genes involved in stress response, apoptosis, immune cell proliferation, and differentiation within lymphocyte and monocyte populations. During the first course of NAC, circulating cytotoxic CD8 T cells in the non-responder patient overexpressed granzymes B and H, granulysin, and perforin. In contrast, expression of these factors decreased in CD8 T cells within the tumor. Finally, we identified for a first time a signature of myeloid-derived suppressor cells (MDSC) within the S100АhighMHClow monocyte population and calculated an MDSC score for both the responder and the non-responder TNBC patients. An elevated MDSC score in the non-responder was validated using data from an independent cohort of patients with poor NAC response. Our data underscores the importance of immune system functionality in determining chemotherapy efficacy in TNBC.
Similar content being viewed by others
Introduction
Breast cancer is a highly prevalent disease affecting women worldwide1,2. There are four intrinsic molecular subtypes of breast cancer: luminal A, luminal B, human epidermal growth factor receptor 2 (HER2) enriched, and triple-negative. Each subtype is characterized by unique gene signature, prognosis, clinical behavior, and treatment response3,4,5. Moreover, specific molecular subtypes are also distinguished according to the immune microenvironment profiles6. Triple-negative breast cancer (TNBC) has the poorest prognosis among all four types and is the most difficult to treat7. TNBC, defined by the lack of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) expression, accounts for up to 15% of all breast cancer cases4,7. Unlike ER or PR positive and HER2 subtypes that present specific targets for therapy, TNBC has no distinctive molecular markers, and no targeted drugs are currently available. While recently checkpoint inhibition has been approved for early-stage TNBC, NAC remains the backbone of the systemic treatment in early-stage TNBC.
It is well known that local and systemic immunity controls cancer development, therapy response, and progression8,9,10,11,12. However, conventional cytotoxic chemotherapy has a mixed impact on the antitumor immune response. On the one hand, it always induces leucopenia12. On the other, chemotherapy leads to immunogenic cell death of malignant cells, elimination of immunosuppressive populations such as T regulatory lymphocytes (Tregs) or myeloid-derived suppressor cells (MDSCs), and enhancement of the antitumor immune response12,13,14. Notably, a functionally active immune system is required to mediate the antitumor effect of chemotherapy15,16,17,18. However, the specific immune parameters that associated with the effectiveness of cytotoxic treatment have yet to be determined.
Preoperative neoadjuvant chemotherapy (NAC) is a widely used treatment strategy for non-metastatic TNBC. NAC favors breast-conserving surgery by reducing tumor size and down-staging lymph node status, eradicates micro-metastases19,20. In addition, pathological complete response (pCR) in TNBC is associated with improved long-term outcomes compared to patients with a residual tumor19,20. Nonetheless, not all TNBC patients benefit from NAC, which highlights the importance of differentiating between responders and non-responders. To date, a higher number of tumor-infiltrating lymphocytes (TILs) is considered a favorable factor for achieving pCR of tumors after NAC and increasing the duration of the relapse-free period21,22. In young TNBC patients without NAC, high level of TILs ( > 75%) also a positive predictive factor for long-term prognosis23. Therefore, patients with favorable and unfavorable outcomes in TNBC exhibit distinct immune compositions. However, the association of other immune cell populations in the tumor microenvironment and circulating blood to chemotherapy sensitivity is poorly characterized.
In this study, we hypothesized that NAC-induced changes in systemic and local immune responses differ between favorable and unfavorable outcomes in TNBC. Using single-cell RNA sequencing (scRNA-seq) of peripheral blood mononuclear cells (PBMCs) and tumor tissue from TNBC patients, one responder and one non-responder to NAC, we examined the dynamics in systemic and local immune parameters during the first course of NAC.
Results
Study design and analysis of blood single immune cell profiling
To resolve the landscape of the systemic immune response induced by NAC in early-stage TNBC patients, we collected peripheral blood before NAC and on days 3 and 21 after first course of NAC (Figure 1A, Supplementary Data 1). Then targeted scRNA-seq was performed using BD Rhapsody Immune Panel. Data from both patients were integrated using the Harmony tool (Fig. 1C). After standard data processing and quality control procedures, we obtained transcriptomic profiles for 6473 cells. The cells were divided into several major types according to their canonical lineage markers and cluster-specific marker genes (Fig. 1B, D). CD4+ T lymphocytes, CD8+ T lymphocytes, B cells, monocytes, and dendritic lineage cells were re-clustered to separate immune cell subtypes. Using the most significantly upregulated genes in each cluster, we identified 21 cell subtypes in total (Supplementary Data 2). CD4+ T cells were subdivided into five classes: TRBC2+ naive CD4+ T cells (CD4 Naive TRBC2), CCR7+ naive CD4+ T cells (CD4 Naive), effector memory CD4+ T cells (CD4 EM), terminal effector memory CD4+ T cells (CD4 TEM), and FOXP3+ regulatory T cells (Treg) (Fig. 1B, D; Supplementary Data 2). CD8+ T cells were subdivided into four classes: LEF+CCR7+ naive CD8+ T cells (CD8 Naive), GZM+ effector memory CD8+ T cells (CD8 TEM), MAIT cells (MAIT), and innate NK-like cytotoxic CD8+ T cells (iNKT). In addition, we identified three major B cell subsets: IGHM+ and IGHD+ naive B cells (B Naive), POU2AF1+ IGHA1+ memory B cells (B Memory), and plasma cells, or so-called antibody-secreting cells expressing high levels of immunoglobulin genes, such as IGHA1, IGHG1 and MZB1 (Plasma cells). A population of plasmacytoid DCs (pDCs) was also identified. Two populations were annotated as γδT cells, including γδT cells of the Vd2 and non-Vd2 subsets. Possibly due to the fact that we analyzed PBMCs of patients undergoing NAC, we found a small population of proliferative lymphocytes. As for myeloid cells (including monocytes and dendritic cells), we identified four transcriptionally distinct subsets: CD14highCD16−S100A+ classical monocytes (CD14MnS100A) exhibiting migratory properties (Supplementary Data 3), CD14highCD16−MHC+ classical monocytes (CD14MnMHC) exhibiting inflammatory properties (Supplementary Data 3), CD14−CD16high nonclassical monocytes (CD16 Mn), and CD1c+ conventional myeloid DCs (mDCs). These subtypes of cells were then used for downstream analyses.
A Schematic overview of the experimental design. B ScRNA-seq data integration via Harmony tool. Cells were colored according to their samples. C UMAP visualization of 6473 PBMCs grouped into 21 immune populations. D DotPlot of expression patterns across cell populations. The Y axis display the populations. The expression-related color depicts the average expression level, and the dot size depicts the percentage of cells expressing the genes across the population. E Histogram indicating the proportion of PBMCs of responder and non-responder patients. F Histogram indicating the proportion of PBMCs in both TNBC patients at three time points: before NAC and on days 3 and 21 after the 1st NAC course. TNBC triple negative breast cancer, PBMCs peripheral blood mononuclear cells, NAC neoadjuvant chemotherapy, NR_1 non-responder before NAC, NR_2 non-responder on days 3 after NAC NR_3, non-responder on days 21 after NAC, R_1 responder before NAC, R_2 responder on days 3 after NAC, R_3 responder on days 21 after NAC, Prolif proliferative T cells, Plasma plasma cells, Tregs regulatory T cells, MAIT mucosal-associated invariant T cells.
Responder and non-responder patients exhibit different immune profiles during chemotherapy
We compared the PBMC transcriptomic profiles of the non-responder with those of the responder to NAC. We calculated the proportions of various cell subsets in the blood samples of both patients normalized to the total number of adaptive and innate immune cells (Fig. 1E). The innate immune response differed between patients, with a higher proportion of iNKT cells in the PBMCs of the non-responder and a more prevalent NK cluster in the responder’s PBMCs. We assessed changes in the proportions of various immune cells associated with chemotherapy. Eight cell populations, including CD4+ and CD8+ lymphocytes, Tregs, NK cells, and two populations of classical monocytes differed in their proportion dynamics between patients with poor and good response to NAC (Fig. 1E; Supplementary Data 4).
Dynamic changes in the transcriptomic profile of lymphocyte populations during NAC
None of the naive CD4+ populations showed markedly differences between the responder and the non-responder during the NAC (Fig. 1E). However, an increase in CD4 TEM cells was observed only in the non-responder by day 21. CD8 TEM cells decreased in the responder by day 21 only but increased in the non-responder. B memory cells decreased in the responder by days 3 and 21, while in the non-responder, they increased by day 3 and decreased by day 21. Meanwhile, the percentage of naive B lymphocytes remained unchanged in the non-responder but decreased in the responder.
Regarding the innate immune response, the iNKT cell percentage decreased in the non-responder, while iNKT cells were almost absent in the responder. By klday 21, NK cell counts decreased in the responder and increased in the non-responder. Tregs exhibited an increase by day 3 and a subsequent decrease by day 21 in the responder, whereas in the non-responder, they decreased by day 3 and recovered by day 21 day.
To explore the dynamics of the transcriptome profile of immune cell populations induced by NAC, we applied the Limma package (Supplementary Data 5)24. This approach allowed us to identify statistically significant transcriptome dynamics over three research points (before NAC and 3 and 21 days after NAC) in responder and non-responder patients. For all lymphocyte populations, the top DEGs were factors related to stress response (FOSB, JUN, JUNB, DUSP1, and DUSP2), apoptosis (BAX), and immune cell proliferation, differentiation, migration and cytokine production (EGR1, TNFa, RGS1, and CXCR4) (Fig. 2A, B; Supplementary Fig. S2, Supplementary Data 6). This appears biologically relevant since the patients were receiving chemotherapy. Key transcription factors of proliferation and differentiation EGR1 and RGS1 were significantly decreased in the responder up to day 21 but increased in lymphocyte populations of the non-responder (Fig. 2B). It is noteworthy that the lymphocytes of the responder expressed stress response factors and transcription factors of proliferation and differentiation throughout the entire course of NAC. In contrast, cells from the non-responder produced these factors at a very low level.
Violin plots showcase the significant variations in gene expression identified by Limma analysis (adjusted p-value < 0.0001). A UMAP of T/NK populations from all samples. B The variations in gene expression dynamics of lymphocytes in responder and non-responder patients. Common genes for T and NK cells are shown. C Violin plots reflect the variations in gene expression dynamics of lymphocyte populations in responder and non-responder patients. DotPlot demonstrates the transcriptional dynamics in responder and non-responder up to day 3 and 21 after NAC and transcriptional changes between the patients. D Correlation between the CD27, СD52, CD69, and IKZF1 gene expression in tumor and overall survival in independent cohort of TNBC patients. E UMAP of monocyte populations from all samples. F The gene expression dynamics in responder and non-responder patients. Common genes for CD14-positive monocytes are shown. G Violin plots demonstrate the gene expression dynamics of monocyte populations in responder and non-responder patients. DotPlot shows the transcriptional dynamics in responder and non-responder up to day 3 and 21 after NAC and transcriptional changes between the patients. R responder, NR non-responder.
Next, we evaluated the involvement of lymphocytes in the anti-tumor immune response (Fig. 2B). The expression of the key antitumor immune response cytokine gene TNFa by lymphocyte populations was consistently high in the responding patient at all time points, while it was absent in the non-responder (Fig. 2B). Interestingly, INFg expression increased only in the non-responder by day 21. Among the factors mediating the migration of lymphocyte populations, the dynamics of CXCR4 expression was most prominent (Fig. 2B). In the responding patient, CXCR4 expression increased from day 0 to day 3 and decreased by day 21, while the opposite was true for the non-responding patient.
While populations of lymphocytes shared similarities in the expression patterns of genes involved in stress response, proliferation, and differentiation, individual variations were also noted for each population. As shown by the Limma statistical analysis (Fig. 2C), naive TRBC2, naive CD4, and Treg cells differed in the NAC-associated expression dynamics of genes responsible for T lymphocyte proliferation, activation, and differentiation, such as CD44, CD27, CD52, and CD6925,26,27,28. However, the differences between the responder and the non-responder were not very distinctive, although statistically significant. In the CD4 TEM population, transcription factors IKZF1 and BCL11B, which determine the differentiation of CD4+ cells towards Th2 polarization, decreased in the responder but increased in the non-responder during the follow-up29,30,31,32.
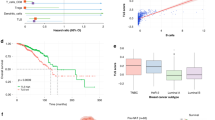
Notably, the genes differentially expressed in blood lymphocytes during NAC between responder and non-responder patients are also linked to cancer outcome in an independent cohort of TNBC patients. We used the Kaplan-Meier plotter tool to correlate overall survival with DEGs from lymphocyte clusters that were found significant according to Limma33. This analysis showed that overexpression of CD27, CD52, CD69, and IKZF1 markers in TNBC was observed in the patients with the better prognosis (Fig. 2D).
According to the Limma analysis, responder and non-responder patients differed in the expression patterns of PRF1, IL2RB, and CD38 genes in NK lymphocytes. However, PRF1 and IL2RB were expressed at high levels both in responder and non-responder NK cells, with a slight increase on day 21. Only CD38, involved in the implementation of cytotoxic function and secretion of perforins and granzymes by NK cells34,35, demonstrated differences and was significantly upregulated up to day 21 in the non-responder.
Dynamic changes in the transcriptomic profile of monocytes populations during NAC
The population of classical monocytes, including CD14MnMHC and CD14MnS100A, markedly decreased by day 3 and then dramatically increased by day 21 in both patients (Fig. 1E). However, for the CD14MnS100A population, this increase was more pronounced in the responder than in the non-responder.
Using Limma, we detected a trend toward overexpression of genes FOSB, JUN, JUNB, DUSP1, DUSP2, RGS1, ARL4C, NAMPT, PIK3AP1 in the CD14MnS100A and CD14MnMHC monocyte populations of the non-responder up to day 21, compared to the responder (Fig. 2E, F). This finding was similar to that described above for lymphocyte populations. In addition, CD14MnMHC and CD14MnS100A monocytes showed altered expression of genes associated with inflammation and migration, respectively (Fig. 2G; Supplementary Data 6). On the 3rd day, differences were observed in the expression of adhesion and migration factors such as S100A9, S100A12, ICAM1, and ITGAX between the non-responder and the responder (Fig. 2E, F). However, their levels had returned to baseline by day 21. The level of FCER1G, which encodes the Fc fragment of IgE receptor Ig and is mainly expressed by monocytes/macrophages in the tumor microenvironment, decreased from day 3 in CD14MnS100A monocytes of the responder patient compared to the non-responder. In the CD14MnMHC population, gene expression of proinflammatory factors, such as IL1B and interferon-induced transmembrane proteins IFITM2 and IFITM3, remained steady in the responder, but decreased by day 3 and increased by day 21 in the non-responder (Supplementary Data 6). Notably, TNFa expression was almost absent in this monocyte population in the non-responder, while it was preserved in the responder on days 0 and 3 of NAC. In CD14MnMHC cells, the dynamics of the hemoglobin scavenger receptor gene CD163 also differed between responder and non-responder. After 3 days of treatment, CD163 expression was undetectable in the responder, whereas it remained stable in the non-responder.
Granzyme expression in CD8 cells differs between tumor and blood immune cells in the non-responder
CD8 cytotoxic lymphocytes that produce proteases, granzymes, and small protein granulysins, and perforins are key effectors of antitumor adaptive immunity36. Low tumor levels of CD8 T cell infiltration, granzyme B, and perforin-1 are associated with poor prognosis in breast cancer37,38,39,40,41. Given the prognostic significance of granzyme and perforin-1 expression in breast cancer, we were interested in exploring how their expression in tumor-infiltrating CD8 T cells correlates with their expression in circulating CD8 T cells during NAC.
Tumor biopsies from the non-responder were available for analysis only on days 3 and 21 days after NAC (Fig. 1A). We evaluated a total of 1542 cells from biopsy specimens and identified 10 immune cell populations (Fig. 3A). Cell types were identified by taking the top canonical cell markers, marker count coupled with average expression, and the greatest mean expression of the marker genes alone (Fig. 3B; Supplementary Data 2). Thus, populations of memory B, naive B, macrophages, T regs, naive CD4, CD4 TEM, CD4 follicular helper (CD4 fh), NK, NKT, and CD8 cells were identified. For further analyses, we focused only on the CD8 TEM population in the blood and tumor as major producers of granzymes and perforins (Supplementary Data 7)36. Transcription levels of granzymes GZMB, GZMY, and GZMH and perforin PRF1 differed between blood and tumor cells during NAC (Fig. 3C). While their expression increased in cytotoxic CD8 T blood cells from day 3 to 21. The opposite dynamics was observed in the tumor: the expression of GZMB, GNLY, GZMH, and PRF1 was decreased in CD8 T cells (Fig. 3C). Thus, the transcription patterns of granzymes and perforin during NAC were opposite in the blood and tumor of the non-responder patient (Fig. 3C).
A UMAP visualization of 1542 cells from TNBC tissues grouped into 10 immune populations. B DotPlot of expression patterns across cell populations. The Y axis display the populations. The expression-related color depicts the average expression level, and the dot size depicts the percentage of cells expressing the genes across the population. С Dynamics of granzymes and perforin expression in blood and tumor tissue in the responder and non-responder up to day 3 and 21 after NAC. D Correlation between the GZMB, GNLY, GZMH, and PRF1 gene expression in tumor and overall survival in independent cohort of TNBC patients. GZMB granzyme B, GNLY granulysin, GZMH granzyme H, PRF1 perforin 1.
Co-expression network analysis of monocytes reveals NAC response-associated gene modules
Next, we used HdWGCNA to identify the key molecular features of PBMCs in the responder and non-responder42. HdWGCNA is a comprehensive tool for analyzing co-expression networks in high-dimensional transcriptomics data. Following analysis of differential gene expression in PBMCs, we revealed five co-expression modules and calculated relative monocyte populations (Fig. 4A; Supplementary Fig. S3; Supplementary Data 8). Of the five modules, Mn-M3 showed the highest expression of genes: S100A12, S100A9, TREM1, F5, FCN1, GAPDH, CD14, CLEC4E, CD36, MGST1, VNN2, ITGAM, and CD163 (Fig. 4B, C; Supplementary Data 6) overlapping with the signature of monocytic myeloid-derived suppressor cells (moMDSC) according CellMarker 2.0 annotation43. Based on the moMDSCs signature, we calculated an MDSC score. We evaluated the temporal changes in the MDSC score of the Mn-M3 module in the responder and the non-responder throughout NAC. In the responder, the MDSC score dramatically decreased on day 3 after NAC, whereas the non-responder’s score increased from the point before NAC to the 21st day after treatment (Supplementary Fig. S4). When comparing the MDSC score between the responder and the non-responder patient, it was found that the MDSC score was significantly higher in the non-responder on day 3 and day 21st after NAC (p < 0.001) (Fig. 4D).
A UMAP plots as in Fig. 1C colored by co-expression modules. B Mn-M3 module regulatory network. C Overlap of monocytic MDSC signature with the signature of monocytes co-expression modules. D The MDCS score in the responder and non-responder up to day 3 and 21 after NAC. E Schematic overview of public dataset processing. F Histogram indicating the cell proportion of PBMCs of pCR and RD cohorts from public dataset. G The MDSC score after NAC in patients with RD and pCR calculated from public dataset. Mn co-expression module, MDSCs myeloid-derived suppressor cell, R responder, NR non-responder, NS no significant, NAC neoadjuvant chemotherapy, TNBC triple negative breast cancer, PBMCs peripheral blood mononuclear cells, pCR pathological complete response, RD residual disease. ***–p < 0.001.
We then tested whether the module signature follows the same expression pattern in an independent sample of patients. We used publicly available data from a study by Axelrod and colleagues that described the association of blood monocytes with the efficacy of NAC44. For this cohort of breast cancer patients, we selected bulk RNA-seq PBMC data from nine patients with TNBC who received NAC with anthracyclines/cyclophosphamide or taxanes alone (Fig. 4E). Within the bulk RNA-seq cohort, four patients had a complete pathologic response to NAC and five demonstrated residual disease. As bulk RNA-seq was used in this study, we applied deconvolution to convert data to a single-cell framework (Fig. 4E). Following deconvolution, 9 immune cell clusters were isolated, which had canonical markers corresponding to the markers of our single-cell RNA-seq clusters: B cells, CD4 T cells (CD4 T), CD8 cells (CD8 T), T regulatory cells (Treg), gamma delta T cells (gdT), NK cells (NK), non-classical СD16 positive cells (CD16 Mn), classical CD14 positive CD14 MHChigh, and CD14 S100Ahigh cells (CD14 Mn MHC and CD14 Mn S100A) (Fig. 4F). The Mn-M3 co-expression module was detected in the CD14S100 cluster of monocytes from the public data. The MDSC score of the Mn-M3 module was significantly higher in patients with residual disease compared to those in the pCR group (Fig. 4G).
Discussion
TNBC patients who do not respond to NAC have the poorest prognosis compare to responders. Previously, immune-related factors were identified to be associated with NAC response in TNBC. Specifically, high neutrophil-to-lymphocyte and platelet-to-lymphocyte ratios, along with low hemoglobin, albumin, lymphocyte, and platelet levels, were considered risk factors for poor NAC efficacy in TNBC45. However, to our knowledge, limited data are currently available on the transcriptional profiles of individual immune cell populations or the molecular pathways underlying the immune system’s role in NAC response. In this study, we investigated the temporal dynamics of immune cells in early-stage TNBC patients undergoing NAC and identified features of the immune landscape associated with NAC efficacy. Our findings indicate that the immune profile is linked to both NAC response and disease prognosis.
This study highlights the differences in the immune landscape between the NAC responder and non-responder. Initial differences in the composition of immune cells were detectable even before therapy. This primarily applies to lymphocytes mediating the innate immune response, natural killers, and iNKT cells. NK cell function is known to be impaired in breast cancer patients due to tumor-produced TGFb and PGE246,47. However, in our study, the responding patient showed an increased percentage of NK cells in the blood during NAC, whereas the non-responding patient appeared to lack a resource of NK cells to facilitate the effects of chemotherapy. Interestingly, the iNKT population showed the opposite trend, predominating in the non-responding patient during chemotherapy. iNKT cells are T lymphocytes with characteristics of both T cells and NK cells, and they have antitumor activity through the recognition of lipids and glycolipids presented by the monomorphic MHC-like molecule CD1d and through cytotoxic activity48. Nevertheless, this mechanism seems less effective in the non-responding patient. It is likely that despite the high proportion of iNKT cells, the lack of other interacting elements, such as adequate presentation of tumor antigens on antigen-presenting cells, which is often limited in tumors, may reduce their efficacy49.
Monocytes showed similar changes during NAC in both the responder and the non-responder. There was a sharp decrease in the monocyte population by day 3, followed by recovery by day 21. This pattern aligns with observations in other oncological contexts, where monocyte levels typically reach a minimum around days 5–7 days50. We suggest that this trend can be attributed to the biology of monocytes, as these cells have a short life span and are rapidly replenished in the bloodstream.
The TNBC patients demonstrated distinct gene expression signatures in immune cells depending on their response to NAC. In the responder, lymphocyte and monocyte populations consistently produced stress response factors such as FOSB, JUN, and JUNB51. In monocyte clusters, dual-specificity phosphatases DUSP1 and DUSP2 showed similar expression patterns in both patients. In T/NK cells, however, DUSP1 and DUSP2 remained stable in the responder patient but increased by day 21 in the non-responder. Notably, the level of EGR1, a transcription factor involved in differentiation and cytokine production, was stable throughout NAC treatment in the responder but increased dramatically in the non-responder. EGR1 expression is known to be triggered by mitogen-activated kinase (MAPK) signaling pathways, including the dual-specificity MAPK-regulatory phosphatase DUSP152,53. The regulatory role of EGR1 in lymphocyte response to chemotherapy and its relationship with MAPK signaling should be further explored. In the responder, RGS1 and CXCR4 that enable cell migration were consistently expressed in T/NK cells and upregulated at day 3 in monocytes54,55. In the non-responder, RGS1 expression was only detected in monocytes on day 21. Additionally, T/NK cells in the non-responder demonstrated downregulation of an anti-tumor cytokine TNFa. Our findings suggest that immune cells in the NAC responder are in a more favorable functional state associated with a better prognosis.
Markers of naïve CD4 cell activation, such as CD52, CD69, CD27, and CD44, showed high expression in the responder and the non-responder, indicating that both patients have the potential for an antitumor immune response25,56,57,58. However, according to Limma statistics, their expression was more pronounced in the responder. Interestingly, CD4 TEM cells in the non-responder significantly upregulated factors linked to Th2 polarization, such as IKZF1 and BCL11B, during NAC, whereas their expression remained stable in the responder. IKZF1 inhibits the interferon pathway in T cells, and its co-expression with BCL11B directs T cells toward Th2 differentiation31,59,60. It is also noteworthy that markers, which differed in expression during NAC between the responder and the non-responder, also showed prognostic significance in an independent cohort of patients undergoing a similar chemotherapy regimen. Improved overall survival correlated with elevated levels of CD27, CD52, and CD69, which are linked to the activation of T lymphocytes, along with IKZF1, known for its role in anti-inflammatory response (Fig. 2D). Furthermore, we observed increased expression of granzymes GZMB, GZMH, granylisin GNLY, and perforin PRF1 in circulating CD8 T cells of non-responder, whereas in the tumor tissue of the same patient granzymes and perforin-1 were downregulated. This aligns with the findings of a similar study, which reported that high expression of cytotoxic signature in blood correlates with residual disease after NAC in TNBC61. Thus, increased levels of cytotoxic T cell granzymes and perforin-1 in peripheral blood are associated with TNBC persistence, while increased levels of granzymes in tumor correlate with active antitumor defense and improved overall survival (Fig. 3D). The association of GZMB expression in tumor tissue with overall survival was demonstrated recently40. In summary, the involvement of lymphocytes in both blood and tumor plays an important role in determining disease outcome.
Current data indicate that the state of monocytes is associated with therapeutic outcome in breast cancer44,62,63,64. At the same time, our understanding of monocyte populations is evolving with advances in single-cell analysis. In this study, we identified two clusters within the CD14+ classical monocytes based on enrichment of either S100А or MHC transcripts: CD14MnS100 and CD14MnMHC (Fig. 1C). Within the CD14MnS100 monocyte cluster, we identified a phenotypic subset with a signature of monocytic MDSC (moMDSC; Fig. 4). However, differences between monocytes, enriched by S100A transcripts (S100АhighMHClow) and the traditional MDSC monocytic lineage are still not fully understood. Given the distinct MDSC gene expression signature in CD14MnS100 monocytes, we hypothesize that CD14S100АhighMHClow cells may belong to the moMDSC population. Previously, moMDSC cells or monocytes with low in MHC expression (CD14S100АhighMHClow) detected by single-cell sequencing or flow cytometry have been described as exhibiting an immunosuppression65,66,67. In our study, the non-responder TNBC patient showed an elevated MDSC score, which we validated using data from an independent cohort of patients with poor response to NAC (Fig. 4E, H). Probably, CD14S100АhighMHClow cells belongs to immunosuppression population and may be as a marker of low NAC efficacy.
Although this study provides some evidence on the association of the immune cell parameters with NAC efficacy in TNBC patients, it has some limitations. The inherent limitations of the 1 vs 1 comparison responder to non-responder, which makes statistics challenging. In addition, analysis of cryopreserved samples, which means that granulocytes and granulocyte MDSCs were lysed during sample processing and are therefore absent; hence only blood mononuclear cell populations were analyzed. Finelly, protein validation is not included in this study. Hence these limitations indicate further directions for the research progression.
In conclusion, we demonstrate transcriptomic immune changes during the first course of NAC in two TNBC patients: one who achieved pCR with chemotherapy and another with non-pCR and rapid progression. We observed that NAC responder displayed anti-tumor immune activity through high expression of TNFα and IL1β in monocytes, as well as TNFα, IKZF1, EGR1, and RGS1 in lymphocytes. The strategy of the non-responder’s immune system characterized by deficiency in immune activation, manifested as impaired stress-response genes expression, dysregulation of cytotoxic anti-tumor activity and increased presence immunosuppressive cells with CD14S100АhighMHClow phenotype. Additional research involving a larger patient cohort is necessary to validate the findings for potential application in clinical practice.
Materials and Methods
Patients
The study enrolled two women who had been diagnosed with TNBC and underwent NAC (Supplementary Data 1). Their ER, PR, and HER2 status was determined in accordance with the American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) guidelines68. Both patients received 2–4 courses of adriamycin with cyclophosphamide followed by 4 NAC courses of taxanes before surgery. The first patient received a partial mastectomy and showed no evidence of tumors cells in the breast tissue or lymph nodes (regardless of the presence of an in-situ component) during postoperative pathological evaluation, which was defined as pCR. This patient remained disease-free for more than 36 months. In contrast, the second patient demonstrated progression of TNBC during the fourth cycle of NAC. Therefore, the NAC was interrupted and a radical mastectomy was performed. Two months after the surgery, metastases developed in her pelvic bones, leading to the patient’s death two months later. In the context below, the patient with a poor response to therapy and unfavorable outcome will be referred to as ‘Non-responder,’ while the patient with a good response and favorable outcome will be termed ‘Responder’. The sample collection timelines are described in the Results section (Fig. 1A). The study was approved by the Ethics Committee of the Саnсеr Rеsеаrсh Institute of Tomsk National Research Medical Center of the Russian Academy of Sciences (Study ID: No. 16 of 29 August 2022). All subjects gave written informed consent in accordance with the Declaration of Helsinki.
Sample collection and processing
Peripheral blood samples were collected from both TNBC patients at three time points: before NAC and on days 3 and 21 after the administration of the 1st NAC course. The chemotherapy process took place in one day, beginning with an intravenous injection of Adriamycin and subsequently administering cyclophosphamide. PBMCs were separated using Ficoll density gradient centrifugation within 1 h of blood collection and preserved in a freezing medium consisting of 90% fetal bovine serum (FBS) and 10% DMSO for future use.
For non-responder patient were available biopsy samples of tumor tissue on days 3 and 21 after the first course of NAC. The sample taken before NAC largely consisted of adipose tissue and could not be included into experiment. Each sample was collected using two 16-gauge needles. Samples were placed in the RPMI-1640 medium (Gibco, USA) and enzymatically digested no more than 20 min after collection. Tissues were washed extensively with 1× PBS with 200 U/mL penicillin. For collagenase/hyaluronidase treatment, tissue specimens were mechanically dissociated using a scalpel to remove vascular material, transferred to a solution of 300 U/ml collagenase and 100 U/ml hyaluronidase (STEMCELL Technologies, USA) in DMEM media, and then incubated at 37 °C and 600 × g for 30 min in a shaking incubator (BioSan TS-100, Poland). Once mucin clots formed, 5 U/ml of DNA hydrolyzing enzyme DNAase I (Sigma Aldrich, USA) was added to the suspension, followed by incubation under the same conditions for 10 min. To remove cell aggregates and debris, the cell suspension was filtered using a sterile cell strainer with a 70 μm pore diameter (Becton Dickinson, USA). Immediately after dissociation, the cell suspension was incubated with ROCK inhibitor (10 μM/ml) for 1 h, followed by cryopreservation in a solution of 95% FBS with 5% DMSO on ice.
After isolation, cryovials with PBMCs or tumor cell suspensions were transferred in Mr. Frosty Freezing Containers (Thermo Fisher Scientific, USA) containing isopropyl alcohol for gradient cooling and placed in a freezer, where they were stored for 24 h at −80 °C. Then the tubes were transferred to liquid nitrogen for long-term storage for up to 6 months at −196 °C. For scRNA-seq, cells were removed from storage and rapidly thawed by immersion in a 37 °C water bath with gentle agitation. Each cryovial was thawed individually until only a small amount of ice crystals remained. Samples were immediately transferred to round-bottom tissue culture tubes. Then, 10 ml of RPMI 1640 containing 10% FBS was added over 4 min with agitation, and the tubes were centrifuged immediately at 300 × g for 5 min. The cells were then suspended in PBS containing 2% bovine serum albumin, followed by cell counting and viability assessment.
Single-cell RNA sequencing
The cellular viability of all PBMCs and tissue samples exceeded 75%, as evaluated by Calcein (BD Bioscience, USA) vs DRAQ7 (Thermo Fisher Scientific, USA) via flow cytometry. According to the manufacturer’s protocol, 8 samples (6 PBMCs and 2 tumor cells suspensions) were labeled by Sample Tag and pooled for single-cell capture with the BD Rhapsody Single-Cell Analysis system (BD Bioscience, USA). 1,700 cells from each sample were included in the pool. Single-cell suspensions with PBS were loaded into microfluidic devices using the BD Rhapsody Single-Cell Analysis System (BD Bioscience, USA). Subsequently, the scRNA-seq libraries were constructed according to the protocol of the Immune Response Targeted Panel (Human) Single Cell RNA Library Kit (BD Bioscience, USA). Individual libraries were diluted and pooled according to the manufacturer’s recommendations. Finally, Illumina NextSeq 2000 sequencing was performed using NextSeq 2000 P2 reagents (100 cycles).
Single-cell RNA-seq data analysis
FASTQ files were processed via the standard BD Rhapsody analysis pipeline with default parameters (BD Biosciences) on Seven Bridges (https://www.sevenbridges.com) per the manufacturer’s instructions. Initial processing included quality control, read alignment, cell identification, UMI deduplication, and count gene-barcode matrix. Resulting gene-barcode matrices with distribution-based error correction (DBEC) UMI counts were imported to Seurat in the R environment for further analysis69. Gene-barcode matrices were additionally filtered to retain cells with at least 25 detected genes and 100 UMI counts. SCTransform with default parameters was applied to normalize gene expression and select variable genes for the following dimension reduction and clustering70. Linear dimension reduction was performed with PCA on 150 features chosen with the SelectIntegrationFeature function in Seurat. Harmony batch correction was applied on the first 15 principal components (PCs) in order to remove the batch effect from individual samples71. The first 15 harmony-corrected PCs were used in Louvain clustering under resolution = 0.5 and UMAP dimension reduction with default parameters in Seurat. Clustering resulted in 21 cell clusters, which were annotated using SingleR automatic annotation with default parameters using Monaco et al. as a reference and manually by cluster markers using the Human Protein Atlas database with the Human Blood Atlas (Supplementary Fig. S1)72. Marker detection was performed using the Wilcoxon test in the FindAllMarkers function on the RNA assay after log normalization in the NormalizeData function of Seurat. Marker genes having log2FC > 0.58 and FDR < 0.01 (Benjamini-Hochberg correction) were considered significant. High-dimensional weighted gene co-expression network analysis (HdWGCNA) was performed to identify key gene modules associated with NAC response using default settings and minModuleSize = 10, mergeCutHeight = 0.15 for modules detection42. Module scores were calculated using top 25 hub-genes via UCell method73. Coexpression network plots were generated using the HubGeneNetworkPlot function from the hdWGCNA package, visualizing the top 20 hub genes within each correlation module. Module significance was evaluated by the Wilcoxon test. PBMC populations were selected from scRNA-seq data to create a gene expression correlation matrix, construct weighted gene co-expression networks, and perform module detection. Module-trait relationship analysis identified modules significantly associated with the NAC efficacy, and hub genes within significant modules were identified based on their intra-module connectivity. Time-point and group-wise analyses of differential gene expression were performed using the Limma package with the limma-trend mode, which was shown to have robust performance in identifying differentially expressed genes (DEGs) in scRNAseq data24,74. Gene set enrichment analyses was performed for cell cluster functional annotation and NAC-induced differential pathway annotation using the enricher function in clusterProfiler with default settings75. Visualizations were created with Seurat, ggplot2, and pheatmap packages76,77. Complete information about all packages producing the results of the publication is available in the supplementary materials as the output from session info in R environment (Supplementary Note 1).
Public data collection and analysis
The PBMC data from a bulk RNA-seq of nine TNBC patients after NAC were obtained from the NCBI GEO dataset under the accession number GSE201085 (Supplementary Data 1). Cell type deconvolution tools, namely DWLS and Granulator with NNLS, were employed to estimate the cell type proportions78. Our PBMC scRNA-seq was used as reference in the DWLS and the bulk Monaco et al. PBMC RNA-seq reference in the NNLS72,79,80. The bulk deconvolution process was implemented in R.
The KM-plotter tool which includes clinical cohorts from Gene Expression Omnibus (GEO), European Genome-phenome Archive (EGA), and The Cancer Genome Atlas (TCGA) was used to evaluate the correlation between gene expression and survival rates in 333 TNBC patients (https://kmplot.com/analysis/)81. All patients had a basal subtype of breast cancer defined according to the St Gallen and PAM50 recommendations. All patients had NAC treatment. The log-rank test was used to estimate the difference in Kaplan-Meier survival analysis with a p-value < 0.05 considered statistically significant.
Data availability
Single cell RNA-Seq data were deposited into the Gene Expression Omnibus database under accession number GSE289825 and are available at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE289825.
References
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer J. clinicians 74, 229–263 (2024).
Wilkinson, L. & Gathani, T. Understanding breast cancer as a global health concern. Br. J. Radiol. 95, 20211033 (2022).
Sørlie, T. et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA 98, 10869–10874 (2001).
Pereira, A., Siegrist, J., Lizarraga, S. & Pérez-Medina, T. Clustering Molecular Subtypes in Breast Cancer, Immunohistochemical Parameters and Risk of Axillary Nodal Involvement. J. Personalized Med. 12, https://doi.org/10.3390/jpm12091404 (2022).
Perou, C. M. et al. Molecular portraits of human breast tumours. Nature 406, 747–752 (2000).
Klopfenstein, Q. et al. Evaluation of tumor immune contexture among intrinsic molecular subtypes helps to predict outcome in early breast cancer. J. Immunother. Cancer 9, https://doi.org/10.1136/jitc-2020-002036 (2021).
Zagami, P. & Carey, L. A. Triple negative breast cancer: Pitfalls and progress. NPJ breast cancer 8, 95 (2022).
Gerashchenko, T., et al. Breast Cancer Immune Landscape: Interplay Between Systemic and Local Immunity. Adv. Biol. 8, e2400140 (2024).
Michaud, D. S. et al. Understanding the Role of the Immune System in the Development of Cancer: New Opportunities for Population-Based Research. Cancer Epidemiol., Biomark. Prev. : a Publ. Am. Assoc. Cancer Res., cosponsored Am. Soc. Preventive Oncol. 24, 1811–1819 (2015).
Correia, A. L. Locally sourced: site-specific immune barriers to metastasis. Nat. Rev. Immunol. 23, 522–538 (2023).
Wang, L., et al. Breast cancer induces systemic immune changes on cytokine signaling in peripheral blood monocytes and lymphocytes. EBioMedicine 52, 102631 (2020).
Hiam-Galvez, K. J., Allen, B. M. & Spitzer, M. H. Systemic immunity in cancer. Nat. Rev. Cancer 21, 345–359 (2021).
Fumet, J. D., Limagne, E., Thibaudin, M. & Ghiringhelli, F. Immunogenic Cell Death and Elimination of Immunosuppressive Cells: A Double-Edged Sword of Chemotherapy. Cancers 12, https://doi.org/10.3390/cancers12092637 (2020).
Sprooten, J., et al. Trial watch: chemotherapy-induced immunogenic cell death in oncology. Oncoimmunology 12, 2219591 (2023).
Casares, N. et al. Caspase-dependent immunogenicity of doxorubicin-induced tumor cell death. J. Exp. Med. 202, 1691–1701 (2005).
Obeid, M. et al. Calreticulin exposure dictates the immunogenicity of cancer cell death. Nat. Med. 13, 54–61 (2007).
Tongu, M., et al. Metronomic chemotherapy with low-dose cyclophosphamide plus gemcitabine can induce anti-tumor T cell immunity in vivo. Cancer Immunol., Immunother.: CII 62, 383–391 (2013).
Lu, Y. et al. Complement Signals Determine Opposite Effects of B Cells in Chemotherapy-Induced Immunity. Cell 180, 1081–1097.e1024 (2020).
Gass, P. et al. Prediction of pathological complete response and prognosis in patients with neoadjuvant treatment for triple-negative breast cancer. BMC cancer 18, 1051 (2018).
Zhang, J. et al. Pathologic response as predictor of recurrence, metastasis, and survival in breast cancer patients receiving neoadjuvant chemotherapy and total mastectomy. Am. J. cancer Res. 10, 3415–3427 (2020).
Luen, S. J. et al. Prognostic implications of residual disease tumor-infiltrating lymphocytes and residual cancer burden in triple-negative breast cancer patients after neoadjuvant chemotherapy. Ann. Oncol. : Off. J. Eur. Soc. Med. Oncol. 30, 236–242 (2019).
de Moraes, F. C. A., Souza, M. E. C., Sano, V. K. T., Moraes, R. A. & Melo, A. C. Association of tumor-infiltrating lymphocytes with clinical outcomes in patients with triple-negative breast cancer receiving neoadjuvant chemotherapy: a systematic review and meta-analysis. Clin. Transl. Oncol.: Off. Publ. Federation Span. Oncol. Societies Natl Cancer Inst. Mex. 27, 974–987 (2025).
de Jong, V. M. T. et al. Prognostic Value of Stromal Tumor-Infiltrating Lymphocytes in Young, Node-Negative, Triple-Negative Breast Cancer Patients Who Did Not Receive (neo)Adjuvant Systemic Therapy. J. Clin. Oncol. : Off. J. Am. Soc. Clin. Oncol. 40, 2361–2374 (2022).
Soneson, C. & Robinson, M. D. Bias, robustness and scalability in single-cell differential expression analysis. Nat. methods 15, 255–261 (2018).
Borst, J., Hendriks, J. & Xiao, Y. CD27 and CD70 in T cell and B cell activation. Curr. Opin. Immunol. 17, 275–281 (2005).
Lo, J. W. et al. A population of naive-like CD4(+) T cells stably polarized to the T(H) 1 lineage. Eur. J. Immunol. 52, 566–581 (2022).
Wang, J., et al. CD52 Is a Prognostic Biomarker and Associated With Tumor Microenvironment in Breast Cancer. Front. Genet. 11, 578002 (2020).
Radulovic, K. et al. The early activation marker CD69 regulates the expression of chemokines and CD4 T cell accumulation in intestine. PloS one 8, e65413 (2013).
Kuehn, H. S., Boast, B. & Rosenzweig, S. D. Inborn errors of human IKAROS: LOF and GOF variants associated with primary immunodeficiency. Clin. Exp. Immunol. 212, 129–136 (2023).
Kuehn, H. S., Nunes-Santos, C. J. & Rosenzweig, S. D. IKAROS-Associated Diseases in 2020: Genotypes, Phenotypes, and Outcomes in Primary Immune Deficiency/Inborn Errors of Immunity. J. Clin. Immunol. 41, 1–10 (2021).
Lorentsen, K. J. et al. Bcl11b is essential for licensing Th2 differentiation during helminth infection and allergic asthma. Nat. Commun. 9, 1679 (2018).
Butcher, M. J. & Zhu, J. Recent advances in understanding the Th1/Th2 effector choice. Fac. Rev. 10, 30 (2021).
Karlsson, M. et al. A single-cell type transcriptomics map of human tissues. Sci. Adv. 7, https://doi.org/10.1126/sciadv.abh2169 (2021).
Zambello, R., Barilà, G., Manni, S., Piazza, F. & Semenzato, G. NK cells and CD38: Implication for (Immuno)Therapy in Plasma Cell Dyscrasias. Cells 9, https://doi.org/10.3390/cells9030768 (2020).
Morra, M., Zubiaur, M., Terhorst, C., Sancho, J. & Malavasi, F. CD38 is functionally dependent on the TCR/CD3 complex in human T cells. FASEB J.: Off. Publ. Federation Am. Societies Exp. Biol. 12, 581–592 (1998).
Cassioli, C. & Baldari, C. T. The Expanding Arsenal of Cytotoxic T Cells. Front. Immunol. 13, 883010 (2022).
Sun, Y. P., Ke, Y. L. & Li, X. Prognostic value of CD8(+) tumor-infiltrating T cells in patients with breast cancer: A systematic review and meta-analysis. Oncol. Lett. 25, 39 (2023).
Oshi, M. et al. CD8 T Cell Score as a Prognostic Biomarker for Triple Negative Breast Cancer. Int. J. Mol. Sci. 21, https://doi.org/10.3390/ijms21186968 (2020).
Vihervuori, H. et al. Tumor-infiltrating lymphocytes and CD8(+) T cells predict survival of triple-negative breast cancer. J. cancer Res. Clin. Oncol. 145, 3105–3114 (2019).
Mizoguchi, K. et al. Granzyme B Expression in the Tumor Microenvironment as a Prognostic Biomarker for Patients with Triple-Negative Breast Cancer. Cancers 15, https://doi.org/10.3390/cancers15184456 (2023).
Guan, X. et al. Perforin 1 in Cancer: Mechanisms, Therapy, and Outlook. Biomolecules 14, https://doi.org/10.3390/biom14080910 (2024).
Morabito, S., Reese, F., Rahimzadeh, N., Miyoshi, E. & Swarup, V. hdWGCNA identifies co-expression networks in high-dimensional transcriptomics data. Cell Rep. Methods 3, 100498 (2023).
Hu, C. et al. CellMarker 2.0: an updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic acids Res. 51, D870–d876 (2023).
Axelrod, M. L. et al. Peripheral Blood Monocyte Abundance Predicts Outcomes in Patients with Breast Cancer. Cancer Res. Commun. 2, 286–292 (2022).
Lou, C., Jin, F., Zhao, Q. & Qi, H. Correlation of serum NLR, PLR and HALP with efficacy of neoadjuvant chemotherapy and prognosis of triple-negative breast cancer. Am. J. Transl. Res. 14, 3240–3246 (2022).
Mamessier, E. et al. Human breast cancer cells enhance self tolerance by promoting evasion from NK cell antitumor immunity. J. Clin. Investig. 121, 3609–3622 (2011).
Rahimian, L., et al. Reduced Expression of Natural Killer Cell-Related Activating Receptors by Peripheral Blood Mononuclear Cells from Patients with Breast Cancer and Their Improvement by Zoledronic Acid. Asian Pac. J. cancer Prev.: APJCP 23, 1661–1669 (2022).
Look, A., Burns, D., Tews, I., Roghanian, A. & Mansour, S. Towards a better understanding of human iNKT cell subpopulations for improved clinical outcomes. Front. Immunol. 14, 1176724 (2023).
Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 12, 31–46 (2022).
Patysheva, M., et al. Monocyte programming by cancer therapy. Front. Immunol. 13, 994319 (2022).
Ren, F. J., Cai, X. Y., Yao, Y. & Fang, G. Y. JunB: a paradigm for Jun family in immune response and cancer. Front. Cell. Infect. Microbiol. 13, 1222265 (2023).
Woodson, C. M. & Kehn-Hall, K. Examining the role of EGR1 during viral infections. Front. Microbiol. 13, 1020220 (2022).
Patysheva, M. R., Prostakishina, E. A., Budnitskaya, A. A., Bragina, O. D. & Kzhyshkowska, J. G. Dual-Specificity Phosphatases in Regulation of Tumor-Associated Macrophage Activity. Int. J. Mol. Sci. 24, https://doi.org/10.3390/ijms242417542 (2023).
Wu, M. et al. Regulator of G protein signaling 1 is a potential target in gastric cancer and impacts tumor-associated macrophages. Cancer Sci. 115, 1085–1101 (2024).
Mezzapelle, R. et al. CXCR4/CXCL12 Activities in the Tumor Microenvironment and Implications for Tumor Immunotherapy. Cancers 14, https://doi.org/10.3390/cancers14092314 (2022).
Cibrián, D. & Sánchez-Madrid, F. CD69: from activation marker to metabolic gatekeeper. Eur. J. Immunol. 47, 946–953 (2017).
Barbour, M., Wood, R., Harte, T., Bushell, T. J. & Jiang, H. R. Anti-CD52 antibody treatment in murine experimental autoimmune encephalomyelitis induces dynamic and differential modulation of innate immune cells in peripheral immune and central nervous systems. Immunology 165, 312–327 (2022).
Estess, P., DeGrendele, H. C., Pascual, V. & Siegelman, M. H. Functional activation of lymphocyte CD44 in peripheral blood is a marker of autoimmune disease activity. J. Clin. Investig. 102, 1173–1182 (1998).
Powell, M. D., Read, K. A., Sreekumar, B. K. & Oestreich, K. J. Ikaros Zinc Finger Transcription Factors: Regulators of Cytokine Signaling Pathways and CD4(+) T Helper Cell Differentiation. Front. Immunol. 10, 1299 (2019).
Lyon de Ana, C., Arakcheeva, K., Agnihotri, P., Derosia, N. & Winandy, S. Lack of Ikaros Deregulates Inflammatory Gene Programs in T Cells. J. Immunol. (Baltim., Md.: 1950) 202, 1112–1123 (2019).
Axelrod, M. L. et al. Changes in Peripheral and Local Tumor Immunity after Neoadjuvant Chemotherapy Reshape Clinical Outcomes in Patients with Breast Cancer. Clin. Cancer Res. : Off. J. Am. Assoc. Cancer Res. 26, 5668–5681 (2020).
Valdés-Ferrada, J. et al. Peripheral Blood Classical Monocytes and Plasma Interleukin 10 Are Associated to Neoadjuvant Chemotherapy Response in Breast Cancer Patients. Front. Immunol. 11, 1413 (2020).
Patysheva, M., et al. Effect of Early-Stage Human Breast Carcinoma on Monocyte Programming. Front. Oncol. 11, 800235 (2021).
Knöbl, V., et al. Monocyte subsets in breast cancer patients under treatment with aromatase inhibitor and mucin-1 cancer vaccine. J. Transl. Med. 22, 913 (2024).
Bao, Y., Mo, J., Ruan, L. & Li, G. Increased monocytic CD14+HLADRlow/- myeloid-derived suppressor cells in obesity. Mol. Med. Rep. 11, 2322–2328 (2015).
Yao, R. Q., et al. Single-cell transcriptome profiling of sepsis identifies HLA-DR(low)S100A(high) monocytes with immunosuppressive function. Mil. Med. Res. 10, 27 (2023).
Mengos, A. E., Gastineau, D. A. & Gustafson, M. P. The CD14(+)HLA-DR(lo/neg) Monocyte: An Immunosuppressive Phenotype That Restrains Responses to Cancer Immunotherapy. Front. Immunol. 10, 1147 (2019).
Hammond, M. E. et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch. Pathol. Lab. Med. 134, e48–e72 (2010).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. methods 16, 1289–1296 (2019).
Monaco, G. et al. RNA-Seq Signatures Normalized by mRNA Abundance Allow Absolute Deconvolution of Human Immune Cell Types. Cell Rep. 26, 1627–1640.e1627 (2019).
Andreatta, M. & Carmona, S. J. UCell: Robust and scalable single-cell gene signature scoring. Computational Struct. Biotechnol. J. 19, 3796–3798 (2021).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids Res. 43, e47 (2015).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics : a J. Integr. Biol. 16, 284–287 (2012).
Hadley Wickham, D. N., and Thomas Lin Pedersen. (Springer, 2016).
Kolde, R. Pretty Heatmaps. Version 1.0.12. (2018-12-26).
Pfister, S, K. V. & Ferrero, E. granulator: Rapid benchmarking of methods for *in silico* deconvolution of bulk RNA-seq data. https://doi.org/10.18129/B9.bioc.granulator (2025).
Tsoucas, D. et al. Accurate estimation of cell-type composition from gene expression data. Nat. Commun. 10, 2975 (2019).
Abbas, A. R., Wolslegel, K., Seshasayee, D., Modrusan, Z. & Clark, H. F. Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PloS one 4, e6098 (2009).
Győrffy, B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Computational Struct. Biotechnol. J. 19, 4101–4109 (2021).
Acknowledgements
The authors kindly acknowledge Andrey Evteev for his support in the processing of blood and tumor tissue samples. The work was carried out on the equipment provided by the Tomsk Regional Common Use Center and the Core Facility of Medical Genomics at Tomsk NRMC. The research was funded by the Russian Science Foundation (grant #22-75-10128).
Author information
Authors and Affiliations
Contributions
M.R.P. – investigation, methodology, formal analysis, data curation, original draft, visualization; P.S.I. – software, formal analysis, visualization; A.A.F. - investigation, methodology, visualization; O.D.B., M.A.V. – resources; E.Y.G. – resources, conceptualization; N.V.C.– conceptualization; E.V.D. - review & editing; T.S.G. – project administration, funding acquisition, conceptualization, investigation, formal analysis, review & editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Patysheva, M.R., Iamshchikov, P.S., Fedorenko, A.A. et al. Single-cell transcriptomic profiling of immune landscape in triple-negative breast cancer during neoadjuvant chemotherapy. npj Syst Biol Appl 11, 72 (2025). https://doi.org/10.1038/s41540-025-00549-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41540-025-00549-3