Abstract
Next-generation vaccines are essential to address the evolving nature of SARS-CoV-2 and to protect against emerging pandemic threats from other coronaviruses. These vaccines should elicit broad protection, provide long-lasting immunity and ensure equitable access for all populations. In this study, we developed a panel of chimeric, full-length spike antigens incorporating mutations from previous, circulating and predicted SARS-CoV-2 variants. The lead candidate (CoVEXS5) was produced through a high-yield production process in stable CHO cells achieving >95% purity, demonstrated long-term stability and elicited broadly cross-reactive neutralising antibodies when delivered to mice in a squalene emulsion adjuvant (Sepivac SWE™). In both mice and hamsters, CoVEXS5 immunisation reduced clinical disease signs, lung inflammation and organ viral titres after SARS-CoV-2 infection, including following challenge with the highly immunoevasive Omicron XBB.1.5 subvariant. In mice previously primed with a licenced mRNA vaccine (Comirnaty XBB.1.5, termed mRNA-XBB), CoVEXS5 boosting significantly increased neutralising antibody (nAb) levels against viruses from three sarbecoviruses clades. Boosting with CoVEXS5 via systemic delivery elicited CD4+ lung-resident memory T cells, typically associated with mucosal immunisation strategies, which were not detected following mRNA-XBB boosting. Vaccination of hamsters with CoVEXS5 conferred significant protection against weight loss after SARS-CoV-1 challenge, compared to mRNA-XBB immunisation, that correlated with anti-SARS-CoV-1 nAbs in the sera of vaccinated animals. These findings highlight the potential of a chimeric spike antigen, formulated in an open-access adjuvant, as a next-generation vaccine candidate to enhance cross-protection against emerging sarbecoviruses in vaccinated populations globally.
Similar content being viewed by others
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has underscored the critical role of vaccines in controlling infectious diseases. Since the pandemic’s onset, multiple COVID-19 vaccines have been developed and deployed worldwide1. These vaccines, employing diverse technological platforms such as mRNA, viral vectors, protein in adjuvant and inactivated viruses, have significantly reduced infection rates, severe disease and mortality2. However, the efficacy of licensed vaccines has been constantly challenged by the emergence of SARS-CoV-2 variants, which can evade immune responses and outpace the introduction of updated vaccine formulations3. Moreover, the immunity conferred by these vaccines wanes over time, an effect most apparent with mRNA vaccines, necessitating multiple booster shots per year for those most at risk4. These challenges are compounded by the high costs and logistical hurdles associated with vaccine distribution, particularly in low- and middle-income countries, where resources are limited. Thus, there is a need for vaccines that not only offer broad and lasting immunity but are also affordable and easy to distribute globally5.
SARS-CoV-2 belongs to the Sarbecovirus subgenus of the genus Betacoronavirus, which also includes SARS-CoV-1, the cause of the 2002–2004 SARS outbreak. The COVID-19 pandemic intensified surveillance efforts to identify novel coronaviruses present in wildlife around the globe with the capacity to infect humans. In southeast Asia, many coronaviruses closely related to SARS-CoV-1 and -2 have been identified in bats and in other mammals such as pangolins, which may serve as reservoir or bridging hosts; some of these viruses can utilise human ACE2-dependent entry and replicate in human cells6,7. Such viruses pose a threat to human health in the event of a spillover, and the increasing frequency of pathogen emergence from animal reservoirs provides strong evidence that future pandemics similar to COVID-19 will occur8. To protect against these pandemic threats, a shift beyond the current reactionary vaccine development paradigm towards preparedness is required. Currently we lack effective tools to control another outbreak of a novel sarbecovirus.
Given its pivotal role in viral entry, the spike protein is the major antigenic component of most commercially available COVID-19 vaccines9,10. We have shown that levels of anti-spike neutralising antibodies (nAbs) strongly correlate with protection against symptomatic and severe COVID-1911,12. However, evolution of SARS-CoV-2 quickly resulted in variants with increased transmissibility and reduced nAb recognition and, consequently, vaccine efficacy13. Furthermore, nAbs elicited from current COVID-19 vaccines or prior SARS-CoV-2 infection do not effectively neutralise all novel sarbecoviruses found in nature, suggesting these viruses pose a risk of zoonotic spillover and adaption to humans14,15. In response to these challenges, research is now directed at developing vaccines that are broadly protective, either covering a cross-section of SARS-CoV-2 variants or spanning sarbecovirus clades16. Many of these approaches have combined immunogenic regions of the spike antigen; the receptor binding domain (RBD) is often selected due to the ease of manufacturing compared to the full-length protein16. However, full-length spike protein contains a broader selection of immunogenic epitopes; many dominant T-cell epitopes fall outside the RBD, and T-cell reactivity remains conserved across variants17. This suggests that specific T-cells may facilitate disease control in the absence of a strong neutralising antibody response.
We have previously demonstrated that full-length trimeric spike antigens produced in an optimised, scalable and chemically defined production process18, maintain long-term stability and are highly immunogenic in animal models19. We built on this technology to develop a panel of chimeric spike antigens (CSA) incorporating mutations precited to impact immunogenicity and stability. The lead candidate, CoVEXS5, showed high protein yield, purity, stability and elicited broadly reactive nAbs in mice and hamsters, reducing clinical disease signs and viral titres after both SARS-CoV-2 and SARS-CoV-1 infection. CoVEXS5 also boosted T-cell immunity and sarbecovirus nAbs in mice previously primed with an approved COVID-19 vaccine, highlighting its potential as a next-generation vaccine for broad sarbecovirus protection.
Results
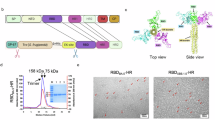
Selection of broadly immunogenic CSA
We sought to identify CSA that are highly immunogenic and produce a cross reactive response to diverse SARS-CoV-2 variants. Forty chimeric spike constructs were initially designed, which was downselected to 13 constructs after modelling for trimer formation (Supplementary Fig. 1A). All protein antigens developed are chimeric spike ectodomains with six proline mutations added to improve expression and thermostability9 and the furin cleavage site removed. Spike mutations were selected based on those known or predicted to impact infectivity and/or immunogenicity20, or identified through in vitro evolution studies21 (Supplementary Fig. 1C). Plasmids encoding each of the 13 constructs were transfected into CHOEXpress® cells to generate stable pools. Twelve of 13 (92%) constructs resulted in protein expression. Seven of these chimeric spike constructs were expressed as trimeric molecules with a range of initial productivity, from 50 to 400 mg/L, which increased upon subsequent optimisation (Supplementary Fig. 1B). The seven trimeric constructs were purified (one-step affinity resin) and all displayed >90% purity, as measured by HPLC-SEC. CSA05 and CSA07 were produced at the highest level (Supplementary Fig. 1B).
The seven trimeric CSA were formulated in Sepivac SWE™, an open-access oil-in-water emulsion and delivered to mice intramuscularly (i.m.) at days 1 and 21. At 42 days post the first immunisation, the nAb titres (IU/ml) in plasma were determined using a panel of spike-pseudotyped lentiviruses. No nAbs were observed in adjuvant alone groups (not shown). As represented by the heatmap of nAb responses (Fig. 1A), most CSAs demonstrated appreciable nAb levels against the majority of variants tested, although CSA04 and CSA12 displayed limited induction of cross-neutralising Abs. Both of these spike variants share the Omicron BA.1 RBD, which may account for their poor cross-reactivity. Notably, CSA05 and CSA07 displayed strong activity against early pandemic variants (Ancestral, Alpha, Beta, Delta) and most Omicron subvariants tested (Fig. 1B). Conversely, vaccination with Ancestral spike only showed good nAb activity against early pandemic variants (non-Omicron), while BA.1 spike immunisation resulted in neutralisation that was restricted to the early omicron subvariants (Fig. 1B). To select the CSA candidates for progression, we used a ranking system that examines the area under the curve (AUC) when nAb levels are plotted against the genetic distance between SARS-CoV-2 variants22. This allowed us to devise a score for each candidate based on their ability to neutralise a wide cross-section of variants (Fig. 1B with individual plots shown in Fig. 1C). The top candidate based on this ranking was CSA05, which also showed good manufacturability (Supplementary Fig. 1B), long-term stability (Supplementary Table 1) and existed in a trimeric conformation (Supplementary Fig. 2). We also observed that individuals infected with SARS-CoV-2 (prior to vaccination) had IgG antibodies that bound to CSA05, demonstrating the antigen is immunologically recognised during natural infection (Supplementary Fig. 3). CSA05 combined with the Sepivac SWETM adjuvant was termed CoVEXS5 and progressed to further analysis.
A Neutralisation of pseudoviruses by plasma from mice immunised with CSA candidates. Mice were vaccinated twice, 3 weeks apart with 5 μg of each spike antigen formulated in SWETM adjuvant. Twenty-one days after the last dose, nAb titres against pseudotyped viruses was determined. Grey bars indicate data points not determined. B Area under the curve (AUC) to demonstrate breadth of nAbs across different variants. Phylogenetic difference is based on variant RBD amino acid sequence. The results are expressed as the number of amino acid substitutions per 100 amino acids, relative to ancestral virus. C Individual plots for AUC analysis. Spk spike, Ancstrl Ancestral.
Protection against virulent SARS-CoV-2 in mice and hamsters
To determine if CoVEXS5-induced immunity was protective against SARS-CoV-2 infection, K18-hACE2 were vaccinated twice, 3 weeks apart with CoVEXS5, at three concentrations of CSA05 formulated 1:1 vol/vol with Sepivac SWETM. Twenty-eight days later, mice were challenged intranasally with SARS-CoV-2 (Delta variant; Fig. 2A). Mice sham-vaccinated with PBS succumbed to infection within 6 days displaying substantial deterioration in their condition (Fig. 2B) with ~15% weight loss (Fig. 2C). This was accompanied by extensive lung inflammation with substantial increases in inflammatory cells in the airways (Fig. 2D). Vaccination with all doses of CoVEXS5 completely protected against infection, with no observable change in clinical score (Fig. 2B) or weight loss (Fig. 2C). Vaccination with higher doses of CoVEXS5 reduced BALF inflammation, with partial reduction at the lower dose of 0.1 μg (Fig. 2D). However, all CoVEXS5 doses resulted in no detectable infectious virus in the airways, lungs and brain (Fig. 2E). Thus, CoVEXS5 is sufficient to completely protect mice from SARS-CoV-2 infection and prevent pathogenic inflammation in the lung.
A K18-hACE2 mice (n = 6) were immunised twice with sham (PBS) or CoVEXS5 at 5, 1 or 0.1 μg of CSA05 formulated 1:1 with SWETM and at 28 days challenged with 1 × 103 PFU SARS-CoV-2 Delta. B Clinical scores at day 6 post-infection. C Percentage of initial body weight loss in male K18-hACE2 mice (n = 6/group). D Total inflammatory cells in bronchoalveolar lavage fluid (BALF). E Viral titres in BALF, lung or brain homogenates were determined using plaque assay. Significant differences between placebo and vaccinated group were determined by one-way ANOVA with Tukey’s multiple comparison test; **p < 0.01, ***p < 0.001. A created with Biorender.
To determine whether efficacy extended across species and to more diverse SARS-CoV-2 variants, hamsters were immunised with two doses of CoVEXS5 or Novavax NVX-CoV2373 as a comparator (Fig. 3A). Both vaccines significantly protected animals from weight loss after challenge with the Delta variant (Fig. 3B) or the highly immune-evasive Omicron XBB.1.5 variant (Fig. 3C), compared to placebo controls. In vaccinated animals challenged with the Delta variant, there was complete clearance of virus in the majority of animals in both nasal turbinates and the lung, irrespective of the CoVEXS5 dose used (Fig. 3D). In animals challenged with Omicron XBB.1.5, all vaccines significantly reduced viral load compared to sham-vaccinated animals, although to a lesser extent than the protection observed with Delta variant challenge. A greater reduction in viral load was seen with the 20 μg compared to the 5 μg dose of CoVEXS5. These findings demonstrate that CoVEXS5 effectively induces protective immunity against diverse SARS-CoV-2 variants, with efficacy comparable to the NVX-CoV2373 vaccine and enhanced protection observed at higher dosing.
Hamsters were vaccinated with CoVEXS5 (5 or 20 μg) or NVX-CoV2373 (5 μg) as shown in (A), and infected with either SARS-CoV-2 Delta or XBB.1.5 at 21 days post-final vaccination. Change in body weight after challenge is shown for Delta (B) or XBB.1.5 (C) challenge. Also shown are viral loads in nasal turbinates and the lung at Day 3 post challenge (Delta, D; XBB.1.5, E). Significant differences between group were determined by two-way (B, C) or one-way (D, E) ANOVA with Tukey’s multiple comparison test; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. A created with biorender.
CoVEXS5 boosts pre-existing immunity to provide broad sarbecovirus neutralisation
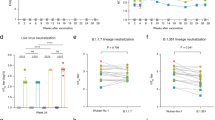
Given that the majority of the population has now been vaccinated and/or infected with SARS-CoV-2, we sought to determine whether the lead candidate, CoVEXS5, could boost pre-existing immunity. Mice were primed with a low dose (0.05 μg) of the Comirnaty Original/Omicron BA.4/5 (mRNA-BV) to mimic prior exposure with SARS-CoV-2 variants (Fig. 4A). This resulted in waning/low nAb (Fig. 4B) and T cell levels (Fig. 4C) over a 9 week time period, compared to a standard dose of 5 μg.
C57BL/6 mice (n = 6) were vaccinated i.m. with two doses of Comirnaty Original/Omicron BA.4/5 (mRNA-BV) at 2 weeks apart (0.05 or 5 μg per dose) (A) and kinetics of neutralising antibody (nAb) responses (B) or spike-specific T-cells secreting IFN-γ (C) were determined. Mice that received the 0.05 μg dose of mRNA-BV were boosted 7 weeks later with two doses of CoVEXS5 (1 μg) or Comirnaty XBB.1.5 (mRNA-XBB; 5 μg). Pre-and post-nAb levels were determined using a panel of spike-pseudotyped viruses representing SARS-CoV-2 variants (D) or non-SARS-CoV-2 sarbecoviruses (E). Numbers represent the fold change between pre- and post-boost nAb levels. A created with Biorender.
To define the impact of boosting with CoVEXS5, mRNA-BV-vaccinated mice were boosted with 1 μg of CoVEXS5 or with 5 μg Comirnaty Omicron XBB.1.5 (mRNA-XBB) as a comparator (Fig. 4A). Boosting with CoVEXS5 increased nAb levels against early pandemic variants (Ancestral, Delta) and omicron subvariants (Fig. 4D). This boosting effect ranged from ~6-fold to 100-fold, depending on the variant examined. Encouragingly, the impact of boosting with CoVEXS5 was apparent for the highly immune-evasive XBB.1.5 and JN.1 subvariants. mRNA-XBB was most effective at boosting XBB.1.5 and BA.4/5 nAb titres but was less effective against early SARS-CoV-2 variants (Fig. 4D). NAb responses against a panel of six non-SARS-CoV-2 sarbecoviruses from Clade 1a (SARS-CoV-1, WIV-1, LyRA11), Clade 1b (BANAL-52, pCoV-GX) and Clade 3 (BtKY72) were also assessed. These viruses were selected to determine the broad utility of the vaccine as they represent a cross-section of sarbecoviruses, and their spike sequences were not used for the design of the CSAs described in Fig. 1. CoVEXS5 immunisation resulted in increased nAbs against all viruses, notably BANAL-52 (34-fold), BtKY72 (20-fold) and WIV-1 (11-fold) (Fig. 4E). Thus, CoVEXS5 is able to stimulate nAb responses following priming with approved COVID-19 vaccines, resulting in augmented, broad-spectrum immunity against diverse SARS-CoV-2 variants and other sarbecoviruses.
CoVEXS5 boosting enhances lung-resident memory CD4 + T-cell responses
To assess the impact of CoVEXS5 immunisation on T-cell immunity, we analysed antigen-specific cytokine expression and cell phenotype both pre- and post-boosting. The low dose ‘priming’’ model allowed us to assess the impact of boosting when T-cell responses had waned to background levels (Fig. 4A). CoVEXS5 boosting resulted in expansion of blood CD4+ and CD8 + T-cells secreting IFN-γ (Fig. 5A). While the frequency of cytokine-secreting CD4 + T-cells in the circulation was low after boosting with either CoVEXS5 or mRNA-XBB (Fig. 5B), both vaccines led to high frequencies of functional CD8 + T-cells, with mRNA-XBB boosting leading to the highest responses (Fig. 5C). In the lung however, CoVEXS5 boosting resulted in cytokine-secreting CD4 + T-cell frequencies above background, which was not observed with mRNA-XBB boosting (Fig. 5B). Vaccination with either CoVEXS5 or mRNA-XBB led to similar levels of activated CD8 + T-cells in the lung, with frequencies markedly lower than that seen in circulation (Fig. 5C).
Mice were vaccinated as shown in Fig. 4a. The proportions of CD44 + CD4+ or CD44 + CD8 + T-cells expressing cytokine in the blood before and after CoVEXS5 boosting (A), or in the blood or lung after mRNA-XBB or CoVEXS5 boosting (B, C), was determined by flow cytometry. Representative flow cytometry plots (D) and proportion (E) of CD4 + CD45:IV− T-cells expressing CD49a, CD69 or CD103 after boosting. CD4 + CD45:IV- T-cells expressing both CD69 and CD49a, or CD69 and CD103 in the lung post-boost is also shown (F). Cytokine release of CD4 + CD45:IV- T-cells after restimulation with Spike peptide pool is shown in (G). Significant differences between groups were determined by one-way ANOVA with Tukey’s multiple comparison test; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
The finding that CoVEXS5 boosting led to preferential expansion of CD4 + T-cells in the lung, compared to mRNA-XBB, led us to further investigate the phenotype of these cells. CoVEXS5-vaccinated mice displayed enrichment of lung parenchymal-residing (intravascular negative, IV−) CD4 + T-cells, which expressed CD69, CD49a and/or CD103, key markers of tissue residency (Fig. 5D)23. CD4 + CD49 + IV− cells were the most prominent population identified (Fig. 5E), however cells expressing multiple tissue-resident memory T-cells (TRM) were also observed (i.e., CD4 + CD69 + CD49 + IV−) (Fig. 5F). Notably, the proportion of CD4 + T-cells expressing TRM markers after mRNA-XBB boosting did not differ to responses observed in control mice (Fig. 5D, E). After antigen restimulation, only CD4 + IV− T-cells from CoVEXS5 boosted-mice produced cytokine (Fig. 5G). In the CD8 + T-cell compartment, comparably lower frequencies of TRM cells were observed, and these did not differ appreciably between vaccine groups (Supplementary Fig. 4). Thus, CoVEXS5 boosting enhances lung-resident memory CD4 + T-cell responses, promoting tissue-resident phenotypes and cytokine production, distinguishing it from mRNA-XBB, which primarily drives circulating CD8 + T-cell expansion.
Protection against SARS-CoV-1 following CoVEXS5 vaccination
As CoVEXS5 was able to boost nAb responses to a broad cross-section of sarbecoviruses (Fig. 4), we assessed protection against SARS-CoV-1 in hamsters (Fig. 6A), a virus distinct from SARS-CoV-2 within the sarbecovirus subgenus. Both CoVEXS5 and mRNA-XBB significantly protected animals from weight loss following SARS-CoV-1 infection, with CoVEXS5 showing a clear dose-response effect (Fig. 6B). Notably, CoVEXS5 provided significantly greater protection against weight loss than mRNA-XBB (Fig. 6B). CoVEXS5 vaccination (high dose) led to near-complete recovery of body weight by day 10 post-infection, whereas mRNA-XBB-vaccinated animals remained ~7.5% below their starting weight. Quantitative assessment of viral load revealed only modest reductions in all vaccinated groups compared to placebo controls, with a significant reduction only observed in caudal lung tissue across the three vaccine groups (Fig. 6C). Only animals vaccinated with CoVEXS5 showed anti-SARS-CoV-1 nAb in the sera of vaccinated animals at early timepoints post-challenge, before the impact of the challenge virus on nAb titres was observed (Fig. 6D). Thus, CoVEXS5 induced stronger cross-protection against SARS-CoV-1 infection in hamsters compared to mRNA-XBB, significantly reducing weight loss, eliciting early nAb responses and demonstrating a clear dose-dependent protective effect.
Hamsters were vaccinated with CoVEXS5 (5 or 20 μg) or mRNA-XBB (5 μg) as shown in (A), and infected with SARS-CoV-1 Tor-2 variant at 21 days post-final vaccination. Change in body weight after challenge (B), viral loads in nasal turbinates or lung at Day 3 post challenge (C) and neutralisation titres to SARS-CoV-1 live virus (D) is also shown. Significant differences between group were determined by two-way (B) or one-way (C) ANOVA with Tukey’s multiple comparison test; *p < 0.05, **p < 0.01, ****p < 0.0001. A created with Biorender.
Discussion
The ongoing evolution of SARS-CoV-2 results in the emergence of new viral variants capable of evading pre-existing immunity, necessitating the continual updating of vaccines. A vast pool of antigenically distinct sarbecoviruses also circulate in nature, posing a further risk of zoonotic spillovers to humans. Thus, the development of next-generation vaccines that can elicit broad sarbecovirus protection and can be easily distributed is essential.
The vaccine candidate described in this report, CoVEXS5, combines a novel highly stable chimeric spike antigen with the open-access adjuvant Sepivac SWETM. The spike antigen in CoVEXS5 has been engineered not only for stability and optimal expression but also to incorporate key mutations from other SARS-CoV-2 variants, enhancing the breadth of protection against evolving strains. Importantly, CoVEXS5 elicits robust and cross-reactive immune responses against a range of SARS-CoV-2 variants, including the highly immune-evasive Omicron XBB.1.5 and JN.1 subvariants, and against a broad range of diverse sarbecoviruses. The protective efficacy of CoVEXS5 was demonstrated in both mice (Fig. 2) and hamsters (Figs. 3, 6) against diverse SARS-CoV-2 variants, and SARS-CoV-1. These findings are promising for the potential use of CoVEXS5 in humans, where cross-species efficacy is a valuable indicator of broad protective capabilities.
Approved COVID-19 vaccines are based on single or bivalent spike antigens and are unable to maintain efficacy over time and as new variants emerge13. They require updated formulations and frequent booster doses to sustain protection. We combined mutations into full-length spike proteins to identify stable and cross-reactive antigens. Some chimeric spikes contained variant-based RBD sequences, such as CSA04 and CSA12, which had a BA.1-based RBD. However, both CSA04 and CSA12 performed poorly against non-Omicron variants; in fact, these chimeric spikes ranked lower than the wild-type BA.1 spike antigen, showing that certain combination of mutations are detrimental to immunogenicity (Fig. 1). The two best ranked chimeric spikes, CSA05 and CSA07, contained identical RBD mutations, derived from various circulating variants, or mutations that at the time of antigen design (around the time of BA.1/BA.2 emergence) were predicted to arise. Encouragingly, the RBDs of CSA05 and CSA07 contain a mix of mutations that either have been preserved in the majority of Omicron sublineages (T478K24), have independently arisen in later omicron lineages (L452R in BA.4/525) or that are being increasingly observed in new omicron sublineages such as JN.1 (A475V26).
The CHOEXpress® platform we use allowed high-yield production of chimeric antigens, with the lead CSA05 protein approaching yields of 1 g/L (Supplementary Fig. 1) and demonstrating long-term stability at 4 °C (Supplementary Table 1). This is particularly crucial for global vaccination efforts, especially in resource-limited settings where cold chain logistics pose significant challenges. When combined with the open-access Sepivac SWE™ adjuvant, this represents a significant step towards facilitating vaccine accessibility. The SWETM adjuvant is a squalene oil-in-water emulsion, similar in composition to MF-59®, a class of adjuvants with a strong safety record in humans27. Importantly, we observed cross-reactive immunity with a single chimeric antigen, including against divergent sarbecoviruses. This differs to other approaches to develop pan-sarbecovirus vaccines, that require multiple protein components to achieve broad immune responses28,29. Single chimeric antigens will have reduced complexity and cost of vaccine development, making them suitable for broad global distribution.
Globally, the majority of people have been vaccinated with at least a primary series of COVID-19 vaccines encoding ancestral spike antigen. We approximated this scenario in mice by priming with a low dose of the Comirnaty Original/Omicron BA.4/5 vaccine and showed that boosting with CoVEXS5 significantly increased nAb levels against SARS-CoV-2 variants and antigenically distinct sarbecoviruses from different lineages (Fig. 4). The Sepivac SWETM adjuvant may contribute to these cross-reactive responses, an effect seen when this adjuvant was combined with bivalent and trivalent spike antigen combinations30. The observation that CoVEXS5 boosting preferentially enhances lung- TRM CD4 + T-cell responses highlight a key mechanistic distinction compared to mRNA-XBB. TRM cells are critical for rapid mucosal immune responses at pathogen entry sites23. The increased frequency of lung-resident CD4 + TRM cells following CoVEXS5 immunisation, characterised by expression of residency markers such as CD69 and CD49a, and their capacity for rapid cytokine production, contrasts with the predominantly circulating CD8 + T-cell responses induced by mRNA-XBB (Fig. 5). Lack of CD4 + TRM after mRNA vaccination has been observed previously in humans31, while the presence of mucosal TRM correlate strongly with improved clinical outcome after SARS-CoV-2 infection32. Notably, we were able to induce lung-resident memory following systemic (intramuscular) vaccine delivery, which contrasts with most approaches that rely primarily on mucosal routes to achieve effective local immunity33.
The robust CD4 + TRM response induced by CoVEXS5 immunisation likely contributes to the enhanced protection against clinical disease (weight loss) following SARS-CoV-1 infection compared to mRNA-XBB (Fig. 6). Previous studies highlight that both the presence and quality of T-cell responses during SARS-CoV-2 infection are associated with disease severity32,34, supporting the superior protective effect observed with CoVEXS5. Additionally, CoVEXS5 uniquely induced detectable anti-SARS-CoV-1 nAbs post-vaccination, distinguishing it from mRNA-XBB (Fig. 6). Neutralising antibody titres strongly predict protection against severe COVID-1913, and even modest vaccine-induced nAb titres can significantly mitigate disease severity in humans35. Therefore, the absence of significant lung-resident TRM induction and delayed nAb responses following mRNA-XBB vaccination likely contributed to its reduced protective efficacy against SARS-CoV-1.
In conclusion, CoVEXS5 represents a significant step forward in the development of an accessible pan-sarbecovirus vaccine. These findings also emphasise the importance of vaccine strategies that effectively generate lung-resident TRM cells to enhance mucosal immunity and highlight the potential advantages of protein-based vaccines for inducing durable, localised immune memory responses, critical for protection against respiratory viral infections. The robust immune responses elicited by CoVEXS5, its ability to boost pre-existing immunity, and its efficacy in diverse animal models underscore its potential as a next-generation vaccine. Future research should focus on clinical trials to confirm these findings in humans and explore the full potential of CoVEXS5 in pandemic preparedness. Additionally, our chimeric antigen design may be applied to other families of viruses to provide cross-protection against antigenically diverse members.
Methods
Chimeric spike design and expression
Sequences of spike proteins of available variants were collected from public database (GISAID). Selected mutations from variants and predicted potential mutations were incorporated into a Hexapro spike sequence ‘backbone’’ and sequences were run through ROBETTA36 and SWISS-MODEL37 for homology structure prediction for trimer formation. DNA constructs were synthetised by ATUM (Menlo Park, USA) and provided with CMC relevant documentation. The furin cleavage-site RRAR was mutated to be non-functional. The transmembrane domain and the C-terminal intracellular tail were removed and replaced by a T4 foldon sequence in trimer designs38. A four amino acid C-terminal ‘C-tag’’ was added to purify chimeras that could not bind commercially available resins39.
Transfection and culturing of CHOExpress® cells (ExcellGene SA, Monthey, Switzerland) was performed as previously described18. After transfections, suspension cells were selected with puromycin, stable pools selected with high secretion and clonal cell lines were obtained by image-assisted cell distribution (f-sight, Cytena GmbH, Freiburg, Germany). The lead clonal cell lines were used for scale-up in an optimised fed-batch process at the 0.2, 10 and 50 L bioreactor scale of operation. The production medium utilised was EX-CELL® Advanced™ CHO Fed-batch medium (Merck, St. Louis, Missouri, United States). Bioreactors were seeded at a density of 5 x 105 cells/mL and maintained at 37 °C for 4 days. Production culture fluids were harvested, clarified using Harvest RC (3MTM), and proteins purified via affinity chromatography. The loading, washing, and elution steps were performed on an NGL COVID-19 resin (Repligen, Waltham, United States) as per the manufacturer’s recommendations. The eluted product was further purified using Capto adhere anion exchange resin (Cytiva, Marlborough, United Sates), followed by tangential flow filtration with a 100 kD cut-off to isolate trimeric spike proteins.
Mouse immunisation
Female C57BL/6 mice (6–8 weeks of age) were purchased from Australian BioResources (Moss Vale, Australia) or Walter and Eliza Hall Institute of Medical Research (Parkville, Australia); K18-hACE2 mice were bread as hemizygous at Centenary Institute, Newtown, Australia. All mice were housed at the Centenary Institute in specific pathogen-free conditions. All mouse experiments were performed according to the National Health and Medical Research Council Australian code for the care and use of animals for scientific purposes, and were approved by the Sydney Local Health District (SLHD) Animal Ethics and Welfare Committee (ethics approval number 2020-009).
Mice were immunised twice, three weeks apart i.m. Each vaccine contained 5, 1 or 0.1 μg of different stabilised full length spike antigen (chimeric, Ancestral, Delta or BA.1) in endotoxin free PBS with 25 μl of Sepivac SWE™ adjuvant (1:1 volume ratio; Seppic, France). For boosting experiments mice received a prime with 0.05 μg of Comirnaty Omicron XBB.1.5 vaccine twice, 2 weeks apart i.m., rested seven weeks before being boosted with chimeric spike in SWETM (1 μg of antigen) or Comirnaty Omicron XBB.1.5 (5 μg; twice, 2 weeks apart.) In some experiments, mice were injected with 3 μg of anti-CD45 biotin (Becton Dickinson, New Jersey, USA) intravenously prior to organ collection.
Pseudovirus neutralisation assays
Replication-deficient SARS-CoV-2 Spike pseudotyped lentivirus particles were generated by co-transfecting a GFP-luciferase, mTAG-BFP2 or LSSmOrange encoding lentivirus vector and a spike expression construct together with lentivirus packaging and helper plasmids into 293 T cells using Fugene HD (Promega, Wisconsin, USA) as previously described40. To determine nAb titres (NT50), pseudovirus particles were incubated with serially diluted plasma samples at 37 °C for 1 h prior to spinoculation (800 x g) of ACE2 over-expressing 293 T-cells. Seventy-two hours post-transduction, cells were fixed and stained with SYTO™ 60 Red Fluorescent Nucleic Acid Stain (Invitrogen) as per the manufacturers instructions, imaged used an Opera Phenix high content screening system (Revvity, Massachusetts, USA) and the percentage of GFP, mTAG-BFP2 or LSSmOrange positive cells was enumerated (Harmony® high-content analysis software, Revvity). NT50 titres were converted to international units (IU)/ml using WHO working standard 21/234. For each pseudovirus variant the fold change in NT50 from ancestral was calculated and used to transform IU/ml for each serum sample. To determine the breath of neutralisation across variants, neutralisation titres for each were plotted on the y-axis, while the x-axis displayed the genetic distance of the RBD for each pseudotyped spike virus relative to ancestral RBD. This generated a unique curve for each mouse plasma sample. AUC was measured for each curve generated using GraphPad Prism. For testing of human sera, samples were collected from patients 5–32 days (Ancestral) or 14–-28 days (Delta) post positive PCR swab in Sydney, Australia during March 2020 or June 2021, respectively, as described41. Ethics approval was granted by the RPA ethics committee, human ethics number X20-0117 and 2020/ETH00770, or the Ethics Committees of the Northern SLHD and the University of New South Wales, NSW Australia (ETH00520). Written or verbal consent was obtained from all patients.
Flow cytometry
Blood was collected from the lateral tail vein and peripheral blood mononuclear cells (PBMCs) were isolated via Histopaque 1083 (Merck) stratification. Spleen and lung samples were processed as described previously42. To assess cytokine secretion by Spike-specific T cells, murine PBMCs, lung, or spleen, cells were stimulated for 4 h with 5 μg/mL of full length Ancestral spike with 1 μg/mL of Spike peptide (amino acid positions aa 538–54643), or for 30 min with 1 μg/mL of Peptivator Spike peptide pool (Miltenyi biotec) and then supplemented with Protein Transport Inhibitor Cocktail (Life Technologies, Carlsbad, USA) for a further 12 or 4 h, respectively. Cells were surface stained with fixable blue dead cell stain (Life Technologies) and marker-specific fluorochrome-labelled antibodies (Supplementary Table 2). Cells were then fixed and permeabilised using the BD Cytofix/CytopermTM kit (Becton Dickinson) according to the manufacturer’s protocol and intracellular staining was performed to detect cytokines. All samples were acquired on an Aurora five-laser spectral cytometer (Cytek Biosciences, Fremont, United States) and assessed using FlowJoTM analysis software v10.10 (Treestar, Oregon, USA).
Mouse SARS-Cov-2 challenge
Male hemizygous K18-hACE2 mice were challenged as described previously40. Briefly, mice were anaesthetised with isoflurane followed by intranasal challenge with 1 × 103 PFU SARS-CoV-2 (Delta strain). Mice were weighed and monitored daily. Mice were euthansed by carbon dioxide (CO2) inhalation. Clinical scores were defined as follows: No clinical score refers to mice that have no visible signs of disease, they may have experienced body weight loss but do not show any phenotypic alterations to body condition or behaviour; Category 1 mice are typically lethargic, with some slight hunched posture and ruffling; Category 2 mice are lethargic and slow to move, with clear hunching, ruffling, signs of laboured breathing, closed or partially closed eyes; Category 3 mice are moribund; requiring immediate euthanasia, typically cannot hold themselves up, cold to the touch, shallow and laboured breathing.
At day 6 post-infection, mice were euthanised and BALF was collected and enumerated using a haemocytometer. Tissue was homogenised using a gentle MACS tissue homogeniser, after which homogenates were centrifuged (300 × g, 7 min) to pellet cells, followed by collection of supernatants for viral quantification by plaque assay. VeroE6 cells (CellBank Australia, Australia) were grown in Dulbecco’s Modified Eagles Medium (Thermo Fisher, Waltham, United States) supplemented with 10% heat-inactivated foetal bovine serum (Sigma-Aldrich, Saint Louis, United States) at 37 °C/5% CO2. Cells were placed into a 24-well plate at 1.5 × 105 cells/well and allowed to adhere overnight. The following day, virus-containing samples (BALF, Lung and brain homogenates) were serially diluted in modified eagles medium (MEM), cell culture supernatants removed from the VeroE6 cells and 250 μL of virus-containing samples was added to cell monolayers. After 1 h, 250 μL of 0.6% agar/MEM solution was gently overlaid onto samples and placed back into the incubator. At 72 h post-infection, each well was fixed with an equal volume of 8% paraformaldehyde solution (4% final solution) for 30 min at RT, followed by several washes with PBS and incubation with 0.025% crystal violet solution for 5 min at RT to reveal viral plaques.
Hamster SARS-CoV-2 challenge
The hamster challenge studies were performed at the vaccine and infectious disease organisation (VIDO, University of Saskatchewan, Canada). The University of Saskatchewan’s University Animal Care Committee and Animal Research Ethics Board approved the animal work as per guidelines of the Canadian Council of Animal Care’s criteria. Male Golden Syrian hamsters (7–8 weeks old) were purchased from Charles River Laboratories (Charles River, Kingston, U.S.A.). Hamsters were immunised i.m. twice three weeks apart in the quadriceps once with a total volume of 100 μl. Each vaccine contained five or 20 μg of chimeric spike antigen in endotoxin free PBS with SWETM, 100 μl of Novavax NVX-CoV2373 (1 μg of the ancestral spike antigen formulated in Matrix-M adjuvant) or Comirnaty Omicron XBB.1.5 (5 μg). Three weeks after the second immunisation, animals were challenged intranasally with 1 × 105 TCID50 of SARS-CoV-2 variant Omicron (XBB.1.5), 1 × 105 TCID50 of SARS-CoV-2 variant Delta (B.1.617.2) or SARS-CoV-1 (Tor-2). Administration of the challenge virus was in both nares with 50 μL/nare. Body weight of each animal was measured daily. Animals were euthanised by CO2 inhalation either at 3 days post-challenge or at 10 days post-challenge. At necropsy, blood, lung tissues and nasal turbinate were collected for assessment of lesions, infectious virus quantification and histopathological examination. Infectious virus was determined by TCID50 analysis. Assays were conducted in 96-well plates using Vero 76 cells (ATCC CRL-1587). TCID50 was determined by microscopic observation of the cytopathic effect on cells.
Statistical analysis
The significance of differences between experimental groups was evaluated by either one-way or ANOVA, with pairwise comparison of multi-grouped data sets achieved using Tukey’s post-hoc test, as indicated in the relevant figure legend. Differences were considered statistically significant when p ≤ 0.05. Statistical analysis was performed using GraphPad Prism Version 10.2.1.
Data availability
CSA sequences are available at GenBank with the following accession numbers: PV539588 (CSA01), PV539589 (CSA02), PV539590 (CSA03), PV539591 (CSA04), PV539592 (CSA05), PV539593 (CSA07), PV539594 (CSA12). The data used and/or analysed during the current study is obtained within the manuscript and supplementary information, or available from the corresponding author on reasonable request.
References
Brice, Y., Morgan, L., Kirmani, M., Kirmani, M. & Udeh, M. C. COVID-19 vaccine evolution and beyond. Neurosci. Insights 18, 26331055231180543 (2023).
Mahrokhian, S. H., Tostanoski, L. H., Vidal, S. J. & Barouch, D. H. COVID-19 vaccines: Immune correlates and clinical outcomes. Hum. Vaccin Immunother. 20, 2324549 (2024).
Triccas, J. A. & Steain, M. C. Australia’s COVID-19 vaccine journey: progress and future perspectives. Microbiol. Aust. 45, 27–31 (2024).
Menegale, F. et al. Evaluation of waning of SARS-CoV-2 vaccine–induced immunity: a systematic review and meta-analysis. JAMA Netw. Open 6, e2310650 (2023).
Triccas, J. A., Kint, J. & Wurm, F. M. Affordable SARS-CoV-2 protein vaccines for the pandemic endgame. npj Vaccines 7, 89 (2022).
Temmam, S. et al. Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature 604, 330–336 (2022).
He, W. T. et al. Virome characterization of game animals in China reveals a spectrum of emerging pathogens. Cell 185, 1117–1129.e1118 (2022).
Marani, M., Katul, G. G., Pan, W. K. & Parolari, A. J. Intensity and frequency of extreme novel epidemics. Proc. Natl. Acad. Sci. 118, e2105482118 (2021).
Hsieh, C. L. et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369, 1501–1505 (2020).
Calvaresi, V. et al. Structural dynamics in the evolution of SARS-CoV-2 spike glycoprotein. Nat. Commun. 14, 1421 (2023).
Khoury, D. S. et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 27, 1205–1211 (2021).
Cromer, D. et al. Predicting vaccine effectiveness against severe COVID-19 over time and against variants: a meta-analysis. Nat. Commun. 14, 1633 (2023).
Cromer, D. et al. Neutralising antibody titres as predictors of protection against SARS-CoV-2 variants and the impact of boosting: a meta-analysis. Lancet Microbe 3, e52–e61 (2022).
Seifert, S. N. et al. An ACE2-dependent sarbecovirus in Russian bats is resistant to SARS-CoV-2 vaccines. PLoS Pathog. 18, e1010828 (2022).
Nie, J. et al. Functional comparison of SARS-CoV-2 with closely related pangolin and bat coronaviruses. Cell Discov. 7, 21 (2021).
Cankat, S., Demael, M. U. & Swadling, L. In search of a pan-coronavirus vaccine: next-generation vaccine design and immune mechanisms. Cell Mol. Immunol. 21, 103–118 (2024).
Sette, A., Sidney, J. & Crotty, S. T-cell responses to SARS-CoV-2. Annu. Rev. Immunol. 41, 343–373 (2023).
Pino, P. et al. Trimeric SARS-CoV-2 Spike proteins produced from CHO cells in bioreactors are high-quality antigens. Processes 8, 1539 (2020).
Counoupas, C. et al. High-titer neutralizing antibodies against the sars-cov-2 delta variant induced by alhydroxyquim-II-adjuvanted trimeric spike antigens. Microbiol. Spectr. 10, e0169521 (2022).
Yao, Z., Zhang, L., Duan, Y., Tang, X. & Lu, J. Molecular insights into the adaptive evolution of SARS-CoV-2 spike protein. J. Infect. 88, 106121 (2024).
Zahradnik, J. et al. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat. Microbiol. 6, 1188–1198 (2021).
Aguilar-Bretones, M., Fouchier, R. A., Koopmans, M. P. & van Nierop, G. P. Impact of antigenic evolution and original antigenic sin on SARS-CoV-2 immunity. J. Clin. Invest 133 e162192 (2023).
Sasson, S. C., Gordon, C. L., Christo, S. N., Klenerman, P. & Mackay, L. K. Local heroes or villains: tissue-resident memory T-cells in human health and disease. Cell Mol. Immunol. 17, 113–122 (2020).
Carabelli, A. M. et al. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat. Rev. Microbiol. 21, 162–177 (2023).
Focosi, D., Quiroga, R., Mcconnell, S., Johnson, M. C. & Casadevall, A. Convergent evolution in SARS-CoV-2 spike creates a variant soup from which new COVID-19 waves emerge. Int. J. Mol. Sci. 24, 2264 (2023).
Yang, S. et al. Fast evolution of SARS-CoV-2 BA.2.86 to JN.1 under heavy immune pressure. Lancet Infect. Dis. 24, e70–e72 (2024).
O’Hagan, D. T., van der Most, R., Lodaya, R. N., Coccia, M. & Lofano, G. World in motion” - emulsion adjuvants rising to meet the pandemic challenges. npj Vaccines 6, 158 (2021).
Halfmann, P. J. et al. Broad protection against clade 1 sarbecoviruses after a single immunization with cocktail spike-protein-nanoparticle vaccine. Nat. Commun. 15, 1284 (2024).
Cohen, A. A. et al. Mosaic RBD nanoparticles protect against challenge by diverse sarbecoviruses in animal models. Science 377, eabq0839 (2022).
Garg, R. et al. Efficacy of a stable broadly protective subunit vaccine platform against SARS-CoV-2 variants of concern. Vaccine 42, 125980 (2024).
Pieren, D. K. J. et al. Limited induction of polyfunctional lung-resident memory T cells against SARS-CoV-2 by mRNA vaccination compared to infection. Nat. Commun. 14, 1887 (2023).
Zhu, A. et al. Robust mucosal SARS-CoV-2-specific T-cells effectively combat COVID-19 and establish polyfunctional resident memory in patient lungs. Nat. Immunol. 26, 459–472 (2025).
Park, S. C., Wiest, M. J., Yan, V., Wong, P. T. & Schotsaert, M. Induction of protective immune responses at respiratory mucosal sites. Hum. Vaccin Immunother. 20, 2368288 (2024).
Bacher, P. et al. Low-avidity CD4(+) T-cell responses to SARS-CoV-2 in unexposed individuals and humans with severe COVID-19. Immunity 53, 1258–1271.e1255 (2020).
Gilbert, P. B. et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science 375, 43–50 (2022).
Kim, D. E., Chivian, D. & Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 32, W526–W531 (2004).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Efimov, V. P. et al. Fibritin encoded by bacteriophage T4 gene wac has a parallel triple-stranded alpha-helical coiled-coil structure. J. Mol. Biol. 242, 470–486 (1994).
Jin, J. et al. Accelerating the clinical development of protein-based vaccines for malaria by efficient purification using a four amino acid C-terminal ``C-tag”. Int J. Parasitol. 47, 435–446 (2017).
Ashley, C. L. et al. Optimisation of a multiplexed, high throughput assay to measure neutralising antibodies against SARS-CoV-2 variants. J Virol Methods 332, 115073 (2025).
Baird, S. et al. A unique cytotoxic CD4(+) T cell-signature defines critical COVID-19. Clin. Transl. Immunol. 12, e1463 (2023).
Counoupas, C. et al. Mycobacterium tuberculosis components expressed during chronic infection of the lung contribute to long-term control of pulmonary tuberculosis in mice. npj Vaccines 1, 16012 (2016).
Zhuang, Z. et al. Mapping and role of T cell response in SARS-CoV-2-infected mice. J. Exp. Med. 218, e20202187 (2021).
Acknowledgements
This work was supported by the Coalition for Epidemic Preparedness Innovations (CEPI) as part of the Broadly Protective Beta-Coronavirus Programme. We are grateful to the University of Sydney Drug Discovery Initiative and the Sydney Infectious Diseases Institute for support through their seed funding programmes. We thank the Vaccine and Infectious Disease Organisation for undertaking hamster challenge studies and helpful discussions; Charles Baily, Centenary Institute, Sydney, Australia for provision of lentivirus packaging and helper plasmids; Stuart Turville, Kirby Institute UNSW, Sydney, Australia for providing SARS-CoV-2 spike plasmid (Ancestral) and ACE2 293T cells; Nathaniel Landau, NYU Grossman School of Medicine, NY, USA for providing SARS-CoV-2 Delta spike plasmid Rowena Bull, Kirby Institute UNSW, Sydney, Australia for sera from SARS-CoV-2-infected individuals. We thank Céline Lemoine, Falko Apel and Morgane Joessel from the Vaccine Formulation Institute, Switzerland for helpful discussions. We acknowledge the support of the University of Sydney Advanced Cytometry Facility and the animal facility at the Centenary Institute. The authors thank the study participants for their contribution to this research and the clinical staff who collected the samples.
Author information
Authors and Affiliations
Contributions
C.C., P.P., V.K.M., M.J.W., F.M.W. and J.A.T., conceived the study. C.C., E.C., M.D.J., C.A., E.E., J.T., S.A., S.M. and N.G.H. performed data acquisition. All authors including J.A., L.J.S., G.S. and W.J.B. undertook data analysis and interpretation of data. C.C., P.P., V.K.M., M.J.W., M.S., T.C., P.M.D., N.C., P.M.H., F.M.W. and J.A.T. contributed to resources and funding acquisition. The initial manuscript draft was prepared by J.A.T. and M.S. with input from all other authors. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
C.C., P.P., M.J.W., F.M.W. and J.A.T. are listed as inventors on international patent application WO2023214385A1, which describes the methodology used in this manuscript for generating CSA. All other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Counoupas, C., Chan, E., Pino, P. et al. An adjuvanted chimeric spike antigen boosts lung-resident memory T-cells and induces pan-sarbecovirus protective immunity. npj Vaccines 10, 89 (2025). https://doi.org/10.1038/s41541-025-01144-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41541-025-01144-7