Abstract
The generation of neural organoids from human pluripotent stem cells holds great promise in modelling disease and screening drugs, but current approaches are difficult to scale due to undesired organoid fusion. Here we develop a scalable cerebral cortical organoid platform by screening biocompatible polymers that prevent the fusion of organoids cultured in suspension. We identify a cost-effective polysaccharide that increases the viscosity of the culture medium, significantly enhancing the yield of cortical organoids while preserving key features such as regional patterning, neuronal morphology and functional activity. We further demonstrate that this platform enables straightforward screening of 298 FDA-approved drugs and teratogens for growth defects using over 2,400 cortical organoids, uncovering agents that disrupt organoid growth and development. We anticipate this approach to provide a robust and scalable system for modelling human cortical development, and facilitate efficient compound screening for neuropsychiatric disorders-associated phenotypes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Gene expression data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE232581 (ref. 35). The data in this study are provided in the Source Data files and available on request from the corresponding author. Source data are provided with this paper.
Code availability
The custom code used for calcium imaging analysis has been deposited on Zenodo at https://doi.org/10.5281/zenodo.15092416 (ref. 36).
References
Qian, X., Song, H. & Ming, G. L. Brain organoids: advances, applications and challenges. Development 146, dev166074 (2019).
Chiaradia, I. & Lancaster, M. A. Brain organoids for the study of human neurobiology at the interface of in vitro and in vivo. Nat. Neurosci. 23, 1496–1508 (2020).
Kelley, K. W. & Pasca, S. P. Human brain organogenesis: toward a cellular understanding of development and disease. Cell 185, 42–61 (2022).
Hofer, M. & Lutolf, M. P. Engineering organoids. Nat. Rev. Mater. 6, 402–420 (2021).
Roth, J. G. et al. Advancing models of neural development with biomaterials. Nat. Rev. Neurosci. 22, 593–615 (2021).
Pasca, A. M. et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 12, 671–678 (2015).
Sloan, S. A., Andersen, J., Pasca, A. M., Birey, F. & Pasca, S. P. Generation and assembly of human brain region-specific three-dimensional cultures. Nat. Protoc. 13, 2062–2085 (2018).
Trevino, A. E. et al. Chromatin accessibility dynamics in a model of human forebrain development. Science 367, eaay1645 (2020).
Yoon, S. J. et al. Reliability of human cortical organoid generation. Nat. Methods 16, 75–78 (2019).
Blair, J. D., Hockemeyer, D. & Bateup, H. S. Genetically engineered human cortical spheroid models of tuberous sclerosis. Nat. Med. 24, 1568–1578 (2018).
Lopez-Tobon, A. et al. Human cortical organoids expose a differential function of GSK3 on cortical neurogenesis. Stem Cell Rep. 13, 847–861 (2019).
Pasca, A. M. et al. Human 3D cellular model of hypoxic brain injury of prematurity. Nat. Med. 25, 784–791 (2019).
Khan, T. A. et al. Neuronal defects in a human cellular model of 22q11.2 deletion syndrome. Nat. Med. 26, 1888–1898 (2020).
Bowles, K. R. et al. ELAVL4, splicing, and glutamatergic dysfunction precede neuron loss in MAPT mutation cerebral organoids. Cell 184, 4547–4563.e17 (2021).
Garcia-Ochoa, F., Santos, V. E., Casas, J. A. & Gomez, E. Xanthan gum: production, recovery, and properties. Biotechnol. Adv. 18, 549–579 (2000).
Tonnesen, J., Inavalli, V. & Nagerl, U. V. Super-resolution imaging of the extracellular space in living brain tissue. Cell 172, 1108–1121.e15 (2018).
Chambers, S. M. et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280 (2009).
Sloan, S. A. et al. Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron 95, 779–790.e6 (2017).
Yang, X. et al. Kirigami electronics for long-term electrophysiological recording of human neural organoids and assembloids. Nat. Biotechnol. 42, 1836–1843 (2024).
Vertesy, A. et al. Gruffi: an algorithm for computational removal of stressed cells from brain organoid transcriptomic datasets. EMBO J. 41, e111118 (2022).
He, Z. et al. An integrated transcriptomic cell atlas of human neural organoids. Nature 635, 690–698 (2024).
Zhang, Y. et al. Fast and sensitive GCaMP calcium indicators for imaging neural populations. Nature 615, 884–891 (2023).
Adam, M. P., Polifka, J. E. & Friedman, J. M. Evolving knowledge of the teratogenicity of medications in human pregnancy. Am. J. Med. Genet. C 157C, 175–182 (2011).
Xing, J. et al. In vitro micropatterned human pluripotent stem cell test (microP-hPST) for morphometric-based teratogen screening. Sci. Rep. 7, 8491 (2017).
Hande, K. R. Clinical applications of anticancer drugs targeted to topoisomerase II. Biochim. Biophys. Acta 1400, 173–184 (1998).
Meng, X. et al. Assembloid CRISPR screens reveal impact of disease genes in human neurodevelopment. Nature 622, 359–366 (2023).
Pasca, S. P. et al. A nomenclature consensus for nervous system organoids and assembloids. Nature 609, 907–910 (2022).
Negraes, P. D. et al. Altered network and rescue of human neurons derived from individuals with early-onset genetic epilepsy. Mol. Psychiatry 26, 7047–7068 (2021).
Boutin, M. E. et al. A multiparametric calcium signal screening platform using iPSC-derived cortical neural spheroids. SLAS Discov. 27, 209–218 (2022).
Miura, Y. et al. Engineering brain assembloids to interrogate human neural circuits. Nat. Protoc. 17, 15–35 (2022).
Miura, Y. et al. Generation of human striatal organoids and cortico-striatal assembloids from human pluripotent stem cells. Nat. Biotechnol. 38, 1421–1430 (2020).
Pantazis, C. B. et al. A reference human induced pluripotent stem cell line for large-scale collaborative studies. Cell Stem Cell 29, 1685–1702 e1622 (2022).
Fleck, J. S. et al. Resolving organoid brain region identities by mapping single-cell genomic data to reference atlases. Cell Stem Cell 28, 1177–1180 (2021).
Susaki, E. A. et al. Versatile whole-organ/body staining and imaging based on electrolyte-gel properties of biological tissues. Nat. Commun. 11, 1982 (2020).
Miura, Y. & Pașca, S.P. Scalable production of human cortical organoids using a biocompatible polymer. Datasets. NCBI GEO https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE232581 (2025).
Miura, Y. et al. Scalable production of human cortical organoids using a biocompatible polymer. Zenodo https://doi.org/10.5281/zenodo.15092416 (2025).
Acknowledgements
We thank members of the Pașca laboratory at Stanford University for scientific inputs, and N. Eckman for help with viscosity measurement. This work was supported by the Stanford Brain Organogenesis Big Idea Grant from the Wu Tsai Neurosciences Institute (to S.P.P. and S.C.H.), US National Institutes of Health (NIH) BRAINS Award MH107800 (to S.P.P.), R01 EB027171 and R01 MH137333 (to S.C.H.), the NYSCF Robertson Stem Cell Investigator Award (to S.P.P.), the Kwan Research Fund (to S.P.P.), the Coates Foundation (to S.P.P.), the Senkut Research Funds (to S.P.P.), The Ludwig Foundation (to S.P.P.), the Chan Zuckerberg Initiative Ben Barres Investigator Award (to S.P.P.), Stanford Medicine Dean’s Fellowship (to Y.M.), the US National Science Foundation (NSF) awards CBET 2033302, DMR 2103812 and DMR 2427971 (to S.C.H.), a TAA Young Investigator Award (to Y.M.), a Stanford Maternal and Child Health Research Institute (MCHRI) Postdoctoral Fellowship (to Y.M.), the Stanford Bio-X Undergraduate Summer Research Program (to S.D.P.), and SNSF Postdoc.Mobility Grant 222016 (to M.A.).
Author information
Authors and Affiliations
Contributions
G.N., Y.M. and S.P.P. conceived the project and designed experiments. G.N. performed the screening experiments for biocompatible polymers and drugs. Y.M. carried out differentiation experiments, 3D clearing and staining, immunocytochemistry, single-cell transcriptomics experiments and data analyses. S.D.P. conducted the differentiation experiments, 3D clearing, imaging and image analysis. M.V.T. performed the differentiation experiments, RNA extractions and qPCRs, LDH assay and characterization of organoids. J.G.R., S.S. and S.C.H. selected and prepared biocompatible polymers, and characterized properties of XG-supplemented media. M.A. performed differentiation experiments, immunocytochemistry, 3D clearing, characterization of XG-hCO cultures, and prepared scRNA-seq libraries. J.K. and Z.H. analysed the calcium imaging data. Y.M. and S.P.P. wrote the paper with input from all authors.
Corresponding author
Ethics declarations
Competing interests
G.N. was an employee of Daiichi-Sankyo Co., Ltd, during the duration of this study, but the company did not have any input on the design of experiments and interpretation of the data. Stanford University holds a patent that covers the generation of cortical organoids (US patent 62/477,858), which has been commercially licensed to STEMCELL Technologies. S.P.P. is listed as an inventor on this patent. The other authors declare no competing interests.
Peer review
Peer review information
Nature Biomedical Engineering thanks Woong Sun and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Scalable organoid culture using biocompatible polymers.
(a) Quantification of remaining organoids at day 25 of culture following supplementation with 0%, 0.001%, 0.01%, and 0.1% XG; n = 12 wells for each condition. One-way ANOVA result: ***P = 0.0003, F = 0.8485(5, 66), P = 0.99 Dunn’s multiple comparisons test for 0% versus 0.001%, *P = 0.03 for 0% versus 0.01%, **P = 0.003 for 0% versus 0.1%. Data show mean ± s.e.m. (b) Quantification of remaining organoids at day 25 following supplementation with 0.1% XG; n = 15 wells conditions starting with 5 organoids, 10 organoids or 15 organoids (3 hiPS cell lines). Kruskal–Wallis test, ANOVA result; ***P = 0.0002, ***P = 0.0003 with Dunn’s multiple comparisons test for 5 organoids versus 10 organoids, **P = 0.0046 for 5 organoids vs. 15 organoids, P > 0.9999 for 10 organoids vs. 15 organoids. Data show mean ± s.e.m. (c) Representative images of 10 cm dishes including hCO and hCO +XG at day 0 (top) and day 6 (bottom). (d) Number of organoids at days 0, 2, 4 and 6. n = 24 plates for control and n = 24 (days 0 and 4), 23 (day 2), and 22 (day 4) for hCO + XG. Two-tailed unpaired t-test, ****P < 0.0001. Data show mean ± s.e.m. (e) qPCR of hCO and hCO + XG (from day 0) at day 25; n = 16–22 organoids for untreated hCO, n = 10–11 organoids for hCO + XG from 3 differentiation experiments including 4 hiPS cell lines. Two-tailed Mann-Whitney test, P = 0.07 for FOXG1, P = 0.34 for EN1, P = 0.09 for EMX1, P = 0.07 for NKX2.1, P = 0.72 for FOXA2, P = 0.93 for RAX, P = 0.30 for HOXB4. Data show mean.
Extended Data Fig. 2 Reduced fusion of organoids following XG treatment.
(a) Representative images of untreated hCO (top), xanthan gum-supplemented (hCO + XG, middle) and STEMdiffTM Neural Organoid Basal Medium 2 cultured hCO (hCO + STEMdiffTM NOBM, bottom). Scale bar: 5 mm. (b) Graph showing remaining organoids after 5 days of culture; n = 24 wells tested for untreated hCO, n = 24 wells tested for hCO + XG, n = 24 wells tested for hCO + ST from 2 differentiation experiments of 2 hiPS cell lines. Kruskal–Wallis test, ANOVA result: ****P < 0.0001, ****P < 0.0001 with multiple comparisons for untreated hCO versus hCO + XG, **P = 0.002 for hCO + XG versus hCO + ST. Data show mean ± s.e.m. (c) Immunostaining for SOX2 (yellow), PAX6 (magenta), and Hoechst (cyan) in hCO, hCO+XG, and hCO + ST at day 25. Scale bar: 200 µm, and 50 µm for an inset. (d) Quantified data of SOX2+ / Hoechst+ cells and PAX6+ / Hoechst+ cells in untreated hCO, hCO + XG and hCO + ST at day 25. n = 6 organoids for untreated hCO, n = 4 organoids for hCO + XG and n = 7 organoids for hCO + ST from 2 differentiation experiments of 1 hiPS cell line. Ordinary one-way ANOVA, P = 0.25 for SOX2+ / Hoechst+ cells and P = 0.70 for PAX6+ / Hoechst+ cells. Data show mean ± s.e.m. (e) Density of progenitor ventricular-like zones. n = 6 for hCO, n = 4 for hCO + XG, and n = 7 for hCO + ST. Ordinary one-way ANOVA, P = 0.37. Data show mean ± s.e.m.
Extended Data Fig. 3 Characterization of XG-hCO.
(a) Quantification of remaining organoids at day 25 of culture following supplementation with 0.1% XG or 0.1% HA; n = 18 wells for each condition (3 hiPS cell lines). Kruskal–Wallis test, ANOVA result: ****P < 0.0001, **P = 0.0013 with Dunn’s multiple comparisons test for control versus 0.1% XG, *P = 0.047 for control versus 0.1% HA, ****P < 0.0001 for 0.1% XG versus 0.1% HA. Data show mean ± s.e.m. (b) Area of hCO at day 3, 10 and 15 cultured with XG or regular culture medium; n = 409 organoids for untreated hCO at day 3, n = 516 organoids for hCO +XG at day 3, n = 208 organoids for untreated hCO at day 10, n = 479 organoids for hCO + XG at day 10, n = 143 organoids for untreated hCO at day 15, n = 308 organoids for hCO + XG at day 15, from 4 differentiation experiments including 4 hiPS cell lines. Two-way ANOVA ****P < 0.0001, P = 0.99 for day 3, ****P < 0.0001 for day 10, ****P < 0.0001 for day 15 with Šidák multiple comparison tests. Data show mean ± s.d. (c) Area of hCO at day 7, and 15 cultured with XG or regular culture medium in individual well culture experiments; n = 81 organoids for untreated hCO at day 7, n = 80 organoids for hCO +XG at day 7, n = 74 organoids for untreated hCO at day 15, n = 64 organoids for hCO +XG at day 15 from 3 differentiation experiments including 3 hiPS cell lines. Two-way ANOVA ***P = 0.0004, P = 0.99 for day 7, and ****P < 0.0001 for day 15 with Šidák multiple comparison test. Data show mean ± s.d.
Extended Data Fig. 4 Cortical patterning and cytotoxicity analysis of XG-hCO.
(a) 3D immunostaining of CUBIC-cleared XG-hCO. SOX2: Cyan, RedDot2: gray. Scale bar: 200 µm. (b) Immunostaining for SOX2 (yellow), PAX6 (magenta), and Hoechst (cyan), and quantification of SOX2+ / Hoechst+ cells and PAX6+ / Hoechst+ cells in untreated hCO and hCO + XG at day 25. n = 33 organoids for untreated hCO, n = 28 organoids for hCO + XG from 3 differentiation experiments of 3 hiPS cell lines for SOX2, and n = 22 organoids for untreated hCO, n = 22 organoids for hCO + XG from 2 differentiation experiments of 3 hiPS cell lines for PAX6. Two-tailed Mann-Whitney test, P = 0.17 for SOX2 and P = 0.71 for PAX6. Scale bar: 200 µm. Data show mean ± s.e.m. (c) LDH cytotoxicity assay for day 25 hCO or hCO + XG; n = 39 wells for untreated hCO, n = 39 wells for hCO + XG from 3 differentiation experiments including 4 hiPS cell lines. Two-tailed Mann-Whitney test, P = 0.64. Data show mean ± s.e.m. (d) Immunostaining of cleaved Caspase-3 (c-Cas3) on day 25 untreated hCO and hCO + XG. Scale bar: 200 µm. (e) qPCR of hCO and XG-hCO cultured as single organoids; n = 5 organoids for hCO, n = 5 organoids for hCO + XG from 4 differentiation experiments of 2 hiPS cell lines. Two-tailed Mann-Whitney test, P = 0.54 for FOXG1, P > 0.9999 for EN1, P = 0.84 for EMX1, P = 0.69 for NKX2.1, P = 0.69 for FOXA2, P = 0.16 for RAX, P = 0.55 for HOXB4. Data show mean ± s.e.m.
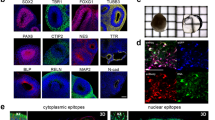
Extended Data Fig. 5 Single-cell RNAseq analysis of XG-hCO.
(a) UMAP projection of cell type makers of hCO, and XG-supplemented hCO. (b) Heatmap showing differentially expressed genes per each cluster. (c) Plot showing the Pearson’s correlation of the normalized average gene expression between untreated hCO and hCO + XG. (d) Violin plots showing gene expressions of SOX2, PAX6, BCL11B, and SATB2 in hCO and hCO + XG.
Extended Data Fig. 6 Cortical layer marker expression and neuronal morphology in XG-hCO.
(a) Immunostaining for CTIP2 (magenta) and Hoechst (cyan) at day 62 (left panels, n = 3 for hCO and n = 4 for XG-hXO), GFAP (green), MAP2 (magenta) and Hoechst (cyan) at day 100 (middle panels, n = 8 for hCO and n = 6 for XG-hXO), SATB2 (magenta) and Hoechst (cyan) at day 100 (right panels, n = 6 for hCO and n = 6 for XG-hXO) in hCO and XG-hCO. Scale bars: 100 µm for left and middle panels, and 200 µm for right panels. (b) Confocal live Imaging of virally labeled eYFP+ neurons in hCO and XG-hCO. Representative images of neurons expressing eYFP in hCO and hCO + XG. Quantification of soma area, soma circularity and the number of neurites. n = 95 neurons from 5 organoids for hCO, n = 100 neurons from 6 organoids for hCO + XG from 2 differentiation experiments of 2 hiPS cell lines. Two-tailed Mann-Whitney test, P = 0.86 for soma area, P = 0.52 for soma circularity, and P = 0.15 for the number of neurites. Data show mean ± s.e.m.
Extended Data Fig. 7 Effects of doxorubicin on hCO.
(a) Organoid growth ratio of hCO treated with Doxo for 8 days or DMSO or H2O treated hCO; n = 12 organoids for control (DMSO), n = 12 organoids for control (H2O), n = 14 organoids for Doxo treated hCO from 3 hiPS cell lines. One-way ANOVA: F = 2.310(2, 35), P = 0.69 with Tukey’s multiple comparisons test for control (DMSO) versus control (H2O), **P = 0.0013 for control (DMSO) versus Doxo, ***P = 0.0001 for control (H2O) versus Doxo. Data show mean ± s.e.m. (b) Organoid growth ratio of hCO treated with Doxo per hiPS cell lines from data in (e); n = 4 organoids for each condition (KOLF2.1 J and 1205-4), n = 4 organoids for control and n = 6 organoids for Doxo-treated hCO (1208-2); two-tailed unpaired t-test, **P = 0.0058 for KOLF2.1 J, ****P < 0.0001 for 1208-2, and ***P = 0.0003 for 1205-4. Data show mean ± s.e.m. (c) Organoid growth ratio of hCO treated with Doxo added to control medium or XG-supplemented medium; n = 12 organoids for Doxo (Control), n = 14 organoids for Doxo (XG) including 3 hiPS cell lines. Two-tailed unpaired t-test, P = 0.08. Data show mean ± s.e.m.
Supplementary information
Supplementary Tables 1–6
Supplementary Tables 1–6.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Narazaki, G., Miura, Y., Pavlov, S.D. et al. Scalable production of human cortical organoids using a biocompatible polymer. Nat. Biomed. Eng (2025). https://doi.org/10.1038/s41551-025-01427-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41551-025-01427-3